Graphical abstract
Method name: Effect of airborne particulate matter on DNA methylation
Keywords: PM10 (PM with aerodynamic diameter ≤10 μm), Epigenetic, 5-Methylscytosine (5-mC), 5-Hydroxymethylscytosine (5-hmC), In vitro DNA methylation, Air pollution
Abstract
The International Agency for Research on Cancer (IARC) has defined outdoor air pollution and PM as the human carcinogen (Group 1), which mainly cause an increased risk of lung cancer. Scientists have considered epigenetic modifications as a possible mechanism to deal with adverse effects of air pollution. This study aimed to compare the effect of airborne PM10 (PM with aerodynamic diameter ≤10 μm) on in vitro global methylation in human peripheral blood mononuclear cells (PBMCs). PM10 was sampled in metropolitan Tehran, the capital of Iran. PBMCs were extracted from whole blood of healthy males and treated with PM10 suspension at concentrations of 50–300 μg/mL for 4 h. Untreated cells were used as the negative control. Genomic DNA was extracted from each sample using the DNA blood mini kit according to the manufacturer’s instruction. Moreover, 5-methylsytosine (%5-mC) and 5-hydroxymethylcytosine (%5-hmC) were measured by the enzyme-linked immunosorbent assay (ELISA) method. %5-mC and %5-hmC in each sample was compared with negative control and reported as difference %5-mC and %5-hmC.
Specifications Table
| Subject Area | Select one of the following subject areas:
|
| More specific subject area: | Epigenetic in environmental science |
| Method name: | Effect of airborne particulate matter on DNA methylation |
| Name and reference of original method |
For DNA extraction see: https://www.qiagen.com/us/shop//sample-technologies/dna/qiaamp-dna-mini-kit/#resources For analysis of 5-mC see: https://www.zymoresearch.eu/5-mc-dna-elisa-kits For analysis of 5-hmC see: https://www.zymoresearch.eu/quest-5-hmc-dna-elisa-kits |
| Resource availability | NA |
Method
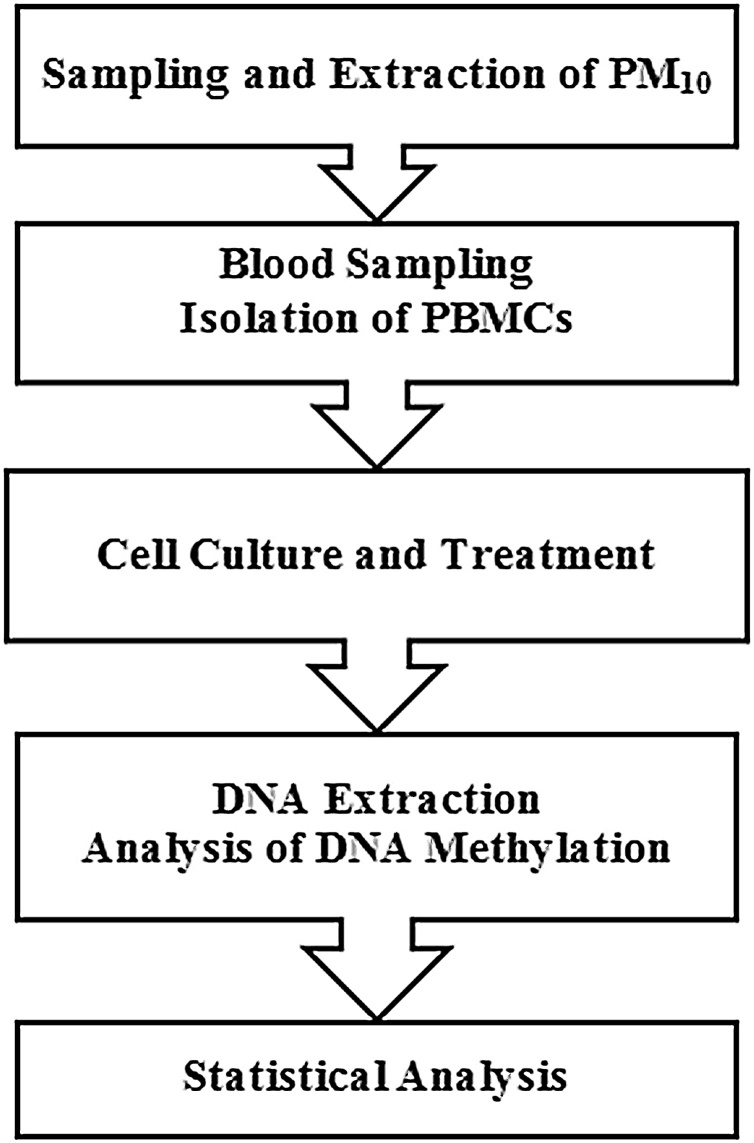
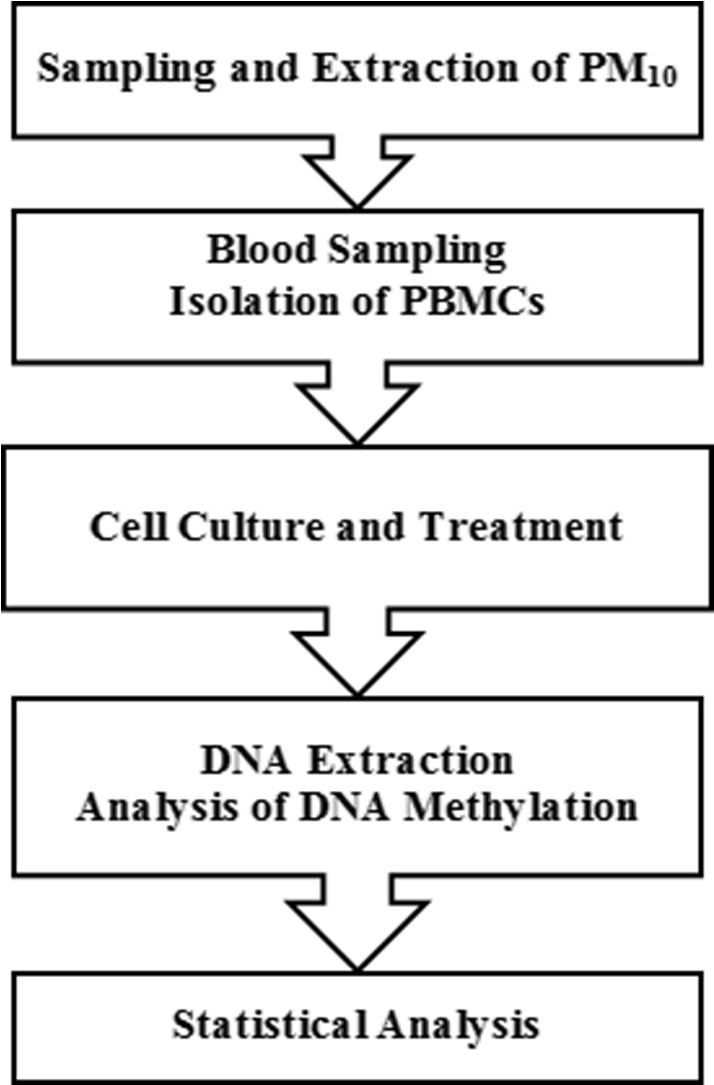
The study stages were as follows: PM10 sampling; preparation of PM10 suspension; blood sampling and isolation of human peripheral blood mononuclear cells (PBMCs); cell culture and treatment; DNA extraction and analysis of DNA methylation and statistical analysis. All the stages are exhibited in Fig. 1.
Fig. 1.
Flow diagram of the study stages.
PM10 sampling
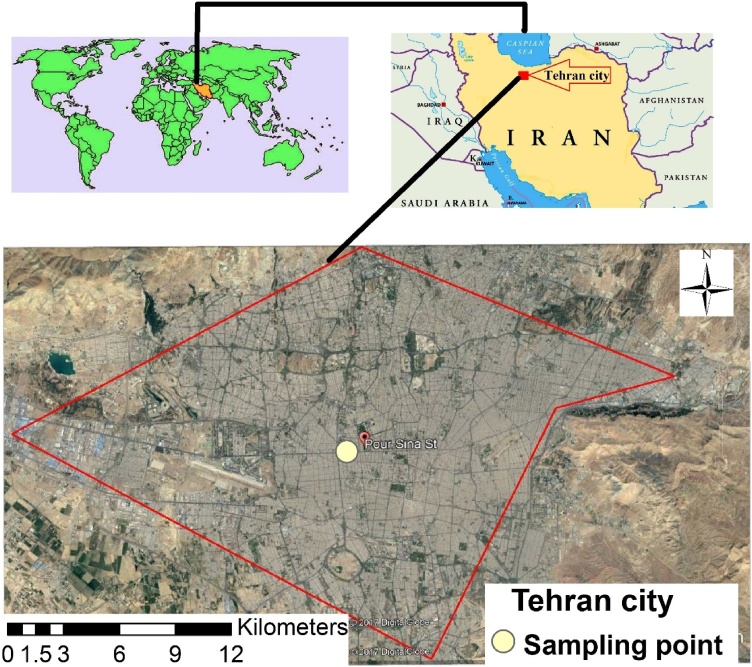
Sampling was described in detail in our previous work [1]. Briefly, PM10 was sampled in Tehran, Iran (35°70′66.00″N, 51°39′38.55″E) (Fig. 2). Sampling was done as 24 h using a high-volume sampler (1.3–1.7 m3/min) (Grasebey, USA) included fiberglass filter (8 × 10 inch, grade G 653 Whatman, USA). Firstly, filters were heated for 2 h at 180 °C and maintained in a dark and dry place. Then, they were weighed with an analytical balance (±10 mg) before and after sampling to calculate the sampled PM mass. Weather information was obtained from a local meteorological monitoring station (Mehrabad).
Fig. 2.
Map of the study area and PM10 sampling point.
Preparation of PM10 suspension
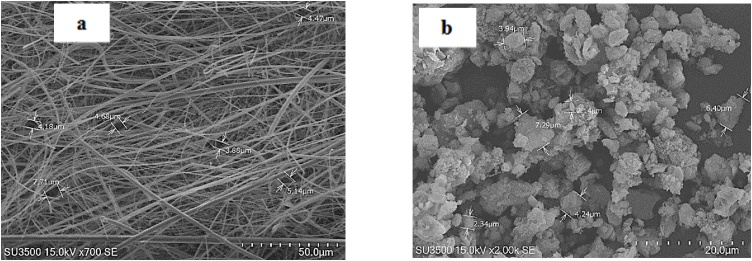
Particle extraction for biological assay was done as the dry ultrasonic method [2]. Firstly, filters were cut into the small pieces and placed in 50 ml centrifuge tubes. Tubes were placed in an ultrasonic bath (Elma-ultrasonic, Germany) for 45 min followed by smooth sweeping with a brush. PM10 was collected into endotoxin-free vials, weighted and then stored at −18 °C until their use [[3], [4], [5]]. In the dry extraction method, presence of fiberglass fibers in PM samples is negligible and cannot cause noticeable biologic impacts on cells [5]. Morphological structure of blank filter and extracted samples were investigated by using a scanning electron microscope (SEM) (HITACHI, SU3500, Japan) (Fig. 3). Fiberglass fibers in the blank filter (Fig. 3a) were not observed in the extracted particles (Fig. 3b). Therefore, it can be concluded that dry extraction method used in this study prevented contamination of the PM10 samples with fiberglass fibers. The presence of fibers in the extracted samples is serious for biological assay because these minerals have their own inherent toxicity [6]. Immediately before use, extracted particles were autoclaved and suspended in RPMI-1640 culture medium to prepare PM10 suspension at different concentrations.
Fig. 3.
SEM images of the blank filter (a) and extracted PM10 (b).
Blood sampling and isolation of peripheral blood mononuclear cells (PBMCs)
This study was approved by the ethics committee of the Tehran University of Medical Sciences (IR.TUMS.SPH.REC.1395.841). Written informed consent was obtained from volunteers and their parents before initiating the study.
PBMCs were isolated from 20 mL whole blood sample from five males (16–18 years old), that enrolled in this study according to defined inclusion criteria, by density centrifugation on Ficoll-Hypaque gradient. Briefly, whole blood was diluted using 40 mL Ca2+/Mg2+ -free PBS (Biosera, France) in a laminar flux hood and gently mixed. Then, 30 mL Ficoll-Hypaque solution (Biosera, France) was added into the diluted blood and isolated PBMCs by density centrifugation (22 min, 2000 rpm, no acceleration, no brake). Layer of PBMCs was collected and washed by lysis buffer and isolation buffer and again centrifuged (400 g, 14 min, acceleration 6, brake 4). Next, PBMCs were suspended in 1 mL complete RPMI-1640 culture medium (Gibco BRL, San Diego, CA) and counted by trypan blue exclusion [7]. The viability of the cells was over 95%. Subsequently, the cells were maintained in a humidified incubator at 37 °C with 5% (v/v) CO2 in air.
Cell cultures and treatment
PBMCs were cultured at a density of 500,000 cells/mL in RPMI-1640 culture medium for 24 h before treatment with PM suspension in an incubator at 37 °C with 5% (v/v) CO2. Further, cells from each donor were separately treated during 4 h with PM10 suspension at six concentrations of 50, 100, 150, 200, 250, and 300 μg/mL. The untreated cells were used as negative control in all the experiments. All the experiments at six concentrations were conducted in triplicate to confirm repeatability of results. After 4 h, the culture medium was collected and centrifuged at 5000 rpm for 8 min to separate cells from the supernatant. Finally, the cells were stored at −20 °C until DNA extraction [1].
DNA extraction and analysis of DNA methylation
Genomic DNA was extracted from each sample using the QIAamp DNA blood mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instruction. DNA concentration was measured based on absorbance at 260 nm using Nanodrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Global %5-mC and %5-hmC in the extracted DNA samples were measured by the enzyme-linked immunosorbent assay (ELISA) method with 5-mC and Quest 5-hmC™ DNA ELISA kits (both from Zymo Research, Orange, CA, USA) using 100 ng extracted genomic DNA according to the manufacturers’ protocols. All samples were assayed in duplicate. The detection limit per 100 ng single-stranded DNA was 0.5% for 5-mC and 0.02% for 5-hmC [1].
Statistical analysis
PM10 concentrations in the methylation test at six levels (50, 100, 150, 200, 250 and 300 μg/mL) were considered as independent variable. The effect of PM10 concentration on the difference of %5-mC or %5-hmC (change of %5-mC and %5-hmC in the treated cells in respect to the negative control) as the response variables was investigated using R software version 3.4.3 [8,9]. Significant level was defined as p-value lower than 0.05. According to the Shapiro–Wilk test for the 5-mC difference (p = 0.01), the effect of PM10 concentration on the value of 5-mC was investigated using the non-parametric test of Scheirer Ray Hare in Fisheries Stock Analysis (FSA) package [10] and the results were reported as median ± interquartile range (IQR). Normality and homogeneity of the 5-hmC difference were approved considering the p-value of concentration 0.11 in the Shapiro–Wilk test and 0.89 in the Bartlett test. Thus, analysis of variance (ANOVA) test was applied to compare the 5-hmC average. Moreover, Dunn test and TukeyHSD were respectively selected as post-hoc tests to assess the 5-mC and 5-hmC average between different PM10 concentrations.
Conflict of interest
The authors declare that they have no competing financial interests.
Acknowledgements
This work was part of a PhD thesis of Maryam Faraji (first author), a student of the Tehran University of Medical Sciences (TUMS). The authors acknowledged the following support: the Institute for Environmental Research (IER) of Tehran University of Medical Sciences (TUMS) (grant number 95-03-46-32844) and Iran National Science Foundation (INSF) (grant number 95831261). The authors would like to thank the IER and INSF for financially supports. Also, they are grateful to the Immunology, Asthma and Allergy Research Institute (IAARI) for technically support and staff collaboration.
References
- 1.Faraji M., Pourpak Z., Naddafi K., Nodehi R.N., Nicknam M.H., Shamsipour M. Effects of airborne particulate matter (PM10) from dust storm and thermal inversion on global DNA methylation in human peripheral blood mononuclear cells (PBMCs) in vitro. Atmos. Environ. 2018;195:170–178. [Google Scholar]
- 2.Faraji M., Nodehi R.N., Naddafi K., Pourpak Z., Alizadeh Z., Rezaei S. Cytotoxicity of airborne particulate matter (PM10) from dust storm and inversion conditions assessed by MTT assay. J. Air Pollut. Health. 2018;3(3):135–142. [Google Scholar]
- 3.Bonner J.C., Rice A.B., Lindroos P.M., O’brien P.O., Dreher K.L., Rosas I. Induction of the lung myofibroblast PDGF receptor system by urban ambient particles from Mexico City. Am. J. Respir. Cell Mol. Biol. 1998;19(4):672–680. doi: 10.1165/ajrcmb.19.4.3176. [DOI] [PubMed] [Google Scholar]
- 4.Alfaro-Moreno E., Martínez L., García-Cuellar C., Bonner J.C., Murray J.C., Rosas I. Biologic effects induced in vitro by PM10 from three different zones of Mexico City. Environ. Health Perspect. 2002;110(7):715. doi: 10.1289/ehp.02110715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Osornio-Vargas ÁR., Bonner J.C., Alfaro-Moreno E., Martínez L., García-Cuellar C., Rosales SP-d-L. Proinflammatory and cytotoxic effects of Mexico City air pollution particulate matter in vitro are dependent on particle size and composition. Environ. Health Perspect. 2003;111(10):1289. doi: 10.1289/ehp.5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alfaro-Moreno E., Torres V., Miranda J., Martínez L., García-Cuellar C., Nawrot T.S. Induction of IL-6 and inhibition of IL-8 secretion in the human airway cell line Calu-3 by urban particulate matter collected with a modified method of PM sampling. Environ. Res. 2009;109(5):528–535. doi: 10.1016/j.envres.2009.02.010. [DOI] [PubMed] [Google Scholar]
- 7.Nazarpour R., Zabihi E., Alijanpour E., Abedian Z., Mehdizadeh H., Rahimi F. Optimization of human peripheral blood mononuclear cells (PBMCs) cryopreservation. Int. J. Mol. Cell. Med. 2012;1(2):88. [PMC free article] [PubMed] [Google Scholar]
- 8.Sarkar D. Springer Science & Business Media; 2008. Lattice: Multivariate Data Visualization with R. [Google Scholar]
- 9.Venables W.N., Ripley B.D. 2003. Modern Applied Statistics with S (statistics and Computing) [Google Scholar]
- 10.Ogle D.H. 2017. FSA: Fisheries Stock Analysis. [Google Scholar]