Summary
BCL-2-related ovarian killer (BOK) is a pro-apoptotic BAX-like member of the BCL-2 family with suggested tumor suppressor activity. The molecular mechanisms regulating BOK expression are poorly understood and fail to explain a frequent lack of concordance between protein and transcript levels. Here, we describe a potent post-transcriptional mechanism that negatively regulates BOK expression mediated by conserved (AU/U)-rich elements within its 3’ UTR. Using proteomics approaches we identified TRIM28 as a key component associating with U-rich elements in the human BOK 3’ UTR, resulting in a dramatic reduction of BOK expression. TRIM28 is overexpressed in several cancers, correlating with poor patient outcome, whereas the BOK locus is frequently deleted or its expression downregulated in human cancers. Data mining indicated that, for certain cancers, high TRIM28 and low BOK expression are significantly correlated in the stratum of patients with the worst survival, suggesting that this mechanism might be of potential therapeutic value.
Subject Areas: Molecular Mechanism of Gene Regulation, Cancer
Graphical Abstract
Highlights
-
•
BOK mRNA is destabilized by AU-(mouse) or U-rich (human) elements within its 3’ UTR
-
•
Mutation of these ARE/URE sequences results in increased BOK RNA and protein levels
-
•
TRIM28 represses BOK expression by associating with the UREs of human BOK mRNA
-
•
Inverse correlation of TRIM28 and BOK levels predicts survival in selected cancers
Molecular Mechanism of Gene Regulation; Cancer
Introduction
The abundance of a protein results from the interplay of epigenetic, (post-)transcriptional, and (post-)translational events. The post-transcriptional control of gene expression is based on the recruitment of RNA-binding proteins (RBP) (Glisovic et al., 2008) to homing sequences on the target messenger RNA (mRNA) via RNA-recognition motifs (Lunde et al., 2007). Prominent mechanisms in that respect include micro RNA (miRNA) (Biggar and Storey, 2015) and AU-, U- or GU-rich elements mediating RNA decay (Vlasova-St Louis and Bohjanen, 2014).
Contrary to miRNA-based regulation, AU-rich element (ARE)-based mechanisms do not require annealing of complementary RNA strands but a direct recruitment of RBP to the target transcript. These elements are preferentially localized in the 3′ untranslated region (3’ UTR) of the mRNA. Initially, this mechanism was described only for the pentamer AUUUA, whereas later experimental evidence revealed that extended variants reaching up to 13-mers, as well as GU- and U-rich sequence patterns, were likewise able to modulate the half-life of transcripts (Fallmann et al., 2016). Moreover, the impact of these elements on mRNA stability is determined by the nature of the recruited RBP, which can be grouped into mRNA stabilizers, such as ELAVL1 (Wigington et al., 2015), NCL (Sengupta et al., 2004), and ZMAT3 (Vilborg et al., 2009), or mRNA destabilizing members, such as HNRNPD (Ishimaru et al., 2010) and ZFP36 (Murata et al., 2005).
Although ARE-mediated mRNA modulation is known to influence the landscape of gene expression (Bakheet et al., 2018), its impact on members of the BCL-2 family, which are key regulators of the intrinsic (mitochondrial) apoptotic pathway, is poorly understood, with the notable exception of BCL-2 itself. The expression of BCL-2 has been shown to be influenced by the balance of HNRNPD and NCL recruited to the 3’ UTR of its transcript, with implications for the integrity of the mouse B cell compartment upon deletion of critical AREs therein (Diaz-Munoz et al., 2015, Schiavone et al., 2000).
BCL-2-related ovarian killer (BOK) is a member of the BCL-2 family closely related to the pro-apoptotic multi-BH-domain containing members BAX and BAK (Fernandez-Marrero et al., 2017, Ke et al., 2018, Zheng et al., 2018). Although BOK has a particularly high affinity for the ER and associated membranes, its enforced accumulation or overexpression leads to intrinsic apoptosis, suggesting that cellular BOK levels need to be tightly regulated in healthy cells (Echeverry et al., 2013, Llambi et al., 2016, Rabachini et al., 2017). Interestingly, there are indications of selective loss of BOK in cancer, as the genomic locus containing the human BOK gene is deleted in a substantial percentage (average 15%) of human cancers (Beroukhim et al., 2010). We recently showed that BOK is downregulated in patients with late stage (lymph node positive) versus early stage non-small cell lung cancer (NSCLC), with high BOK protein levels being predictive of extended patient survival (Moravcikova et al., 2017). Likewise, BOK was shown to be downregulated and of prognostic value in colorectal carcinoma (Carberry et al., 2018). Current understanding of the mechanism(s) regulating these changes in BOK abundance is meagre, restricted to epigenetic regulation by promoter methylation (Moravcikova et al., 2017) or selective proteasomal turnover, which seems to be linked to the localization of BOK at the ER and regulated by the ERAD pathway and the interaction of BOK with inositol triphosphate receptors (Llambi et al., 2016, Schulman et al., 2013, Schulman et al., 2016).
Tripartite Motif Containing 28 (TRIM28/TIF1-β/KAP1) is a co-repressor multidomain protein acting together with Krüppel-Associated Box Zinc Finger Protein (KRAB-ZNF) transcription factors (Friedman et al., 1996, Moosmann et al., 1996), nucleosome remodeling deacetylase (NuRD), and heterochromatin-associated protein 1 (HP1) (Cheng et al., 2014) to silence multiple genes at the chromatin level. These interactions sustain the role of TRIM28 as a regulatory hub in multiple processes such as the maintenance of cell stemness (Hu et al., 2009), inhibition of p53 activity (Okamoto et al., 2006), transcriptional repression (Groner et al., 2010), and epithelial-mesenchymal transition (EMT) (Venkov et al., 2007). Interestingly, despite its key role in regulating chromatin activity (Alexander et al., 2015) and direct gene transcription (Bunch et al., 2014), TRIM28 lacks obvious DNA-binding domains and thus depends on adaptor proteins to localize to the DNA (Iyengar and Farnham, 2011). The functional pleiotropism of TRIM28 has been linked to tumorigenesis, and studies with human cancer cell lines or xenografts highlight the contribution of TRIM28 to sustained mTOR activation and cell cycle progression (Li et al., 2018), sustained Warburg effect (Jin et al., 2017), and maintenance of the cancer stem cell niche (Czerwinska et al., 2017). Moreover, from a clinical perspective, TRIM28 has been reported to be overexpressed in cervical (Li et al., 2018), hepatocellular (Jin et al., 2017), breast (Czerwinska et al., 2017), and ovarian (Cui et al., 2014) cancers, in all of which it is associated with a poor clinical prognosis.
Here, we describe a regulatory circuit controlling BOK expression at the transcript level, involving conserved (AU/U)-rich elements (ARE/URE) present in the 3’ UTRs of both human and mouse BOK mRNA. Remarkably, negative regulation of human BOK expression through mRNA destabilization depends on the unexpected association of TRIM28 with these URE sites. Furthermore, TRIM28-dependent repression specifically targets BOK, whereas it does not affect the pro-apoptotic multidomain BCL-2 family members BAX and BAK. This TRIM28-mediated negative regulation of BOK might have important clinical implications since TRIM28highBOKlow levels were associated with poor patient survival in certain cancer (sub-)types.
Results
The 3′ Untranslated Region of Mouse Bok mRNA Contains a Strong Negative-Regulating AU-Rich Element
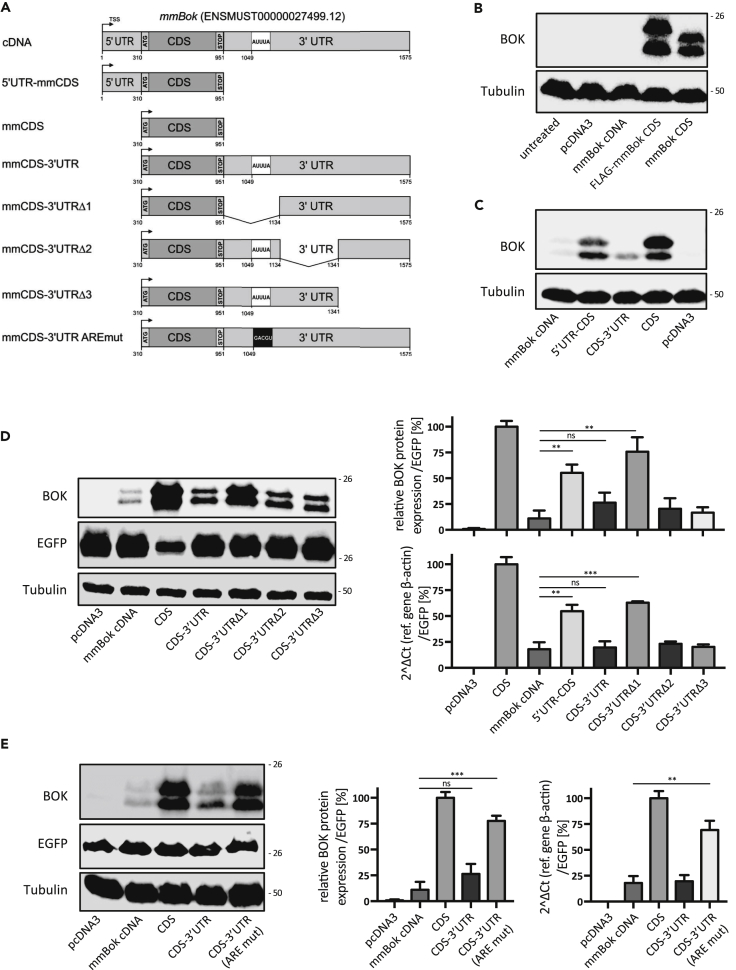
We noticed that the architecture of the mouse Bok mRNA dramatically influences protein expression levels upon transient transfection into the human embryonic kidney cell line HEK293T, which expresses barely detectable levels of endogenous BOK. To further study this phenomenon, we generated a series of expression constructs consisting of the Bok coding sequence (CDS) harboring permutations of the 5′ and 3′ UTR or truncated versions of the latter (Figure 1A). Although transfection of Bok CDS (or FLAG-tagged Bok CDS) resulted in strong protein expression, it was barely detectable when Bok cDNA was used (Figure 1B). This was an indication for the presence of one or more negative regulatory elements in either the 5’ and/or the 3’ UTR of Bok mRNA. As depicted in Figure 1C, the expression of Bok CDS containing either the 3’ UTR or the 5’ UTR demonstrated that it is the 3’ UTR that is responsible for the observed repression.
Figure 1.
Identification of a Negative Regulatory Element in the 3′ UTR of Mouse Bok
(A–E) (A) List of constructs generated to narrow down the region containing destabilizing elements in the mouse Bok transcript sequence. Western blot analysis of BOK levels in total protein extracts prepared from HEK293T cells transiently transfected with (B) Bok cDNA or Bok CDS (with or without N-terminal FLAG tag), (C) Bok CDS with or without its 5’ UTR and/or 3’ UTR, (D) Bok CDS combined with the full length or truncated version of its 3’ UTR, or (E) Bok CDS combined with its wild-type 3’ UTR or harboring discrete point mutations in a predicted AU-rich element (ARE) therein. Each immunoblot is representative of at least three independent experiments. Tubulin and/or EGFP were used to normalize the transfection efficiency and gel loading. The double band detected for BOK corresponds to the full-length protein and a shorter version translated from a cryptic start codon at methionine 15 (Schulman et al., 2016). Bar graphs to the right of panels D and E show the quantification of BOK levels from quantitative immunoblots (using near-infrared fluorochromes) and real-time quantitative PCR. The data are presented as mean ± SD, N = 3. Statistical analysis was performed using a one-way ANOVA followed by a Tukey post-hoc test in GraphPad Prism. Significance levels, p > 0.05ns, p < 0.01**, p < 0.001***.
To map the position of the regulatory sequence(s) present in the 3’ UTR, we generated three deletion mutants of the mouse Bok CDS-3’UTR construct, in which similarly sized parts within the 3’ UTR were removed (3’ UTRΔ1, 183 bp; 3’ UTRΔ2, 207 bp; and 3’ UTRΔ3, 234 bp) (Figure 1A). Co-transfection of these mutants (together with pcDNA3/EGFP at a ratio of 1:10 for normalization of transfection efficiencies) into HEK293T cells and subsequent analyses by qPCR and western blotting demonstrated that region 1 (most proximal to the stop codon) contained all of the negative regulatory element(s) (Figure 1D). Furthermore, the concordance observed in Figure 1D (right panel) between transcript and protein levels indicated that the negative regulatory element likely affects Bok mRNA stability, resulting in its degradation.
We next made use of the AREsite2 prediction tool (http://nibiru.tbi.univie.ac.at/AREsite2/) (Fallmann et al., 2016) to identify regulatory elements in the mouse Bok 3’ UTR and found one sequence corresponding to an ARE located within the 3’ UTR region 1 (position 1,049–1,053, sequence AUUUA) (Figure 1A). As shown in Figure 1E, site-directed mutagenesis of this ARE site (AUUUA -> GACGU) restored both Bok mRNA and protein expression to levels comparable with expression of the CDS alone. Taken together, these data demonstrate that a single ARE site within the proximal region of the 3’ UTR strongly compromises mouse Bok mRNA stability, resulting in decreased protein expression.
The Mouse Bok ARE Sequence Suppresses the Expression of Unrelated Coding Sequences
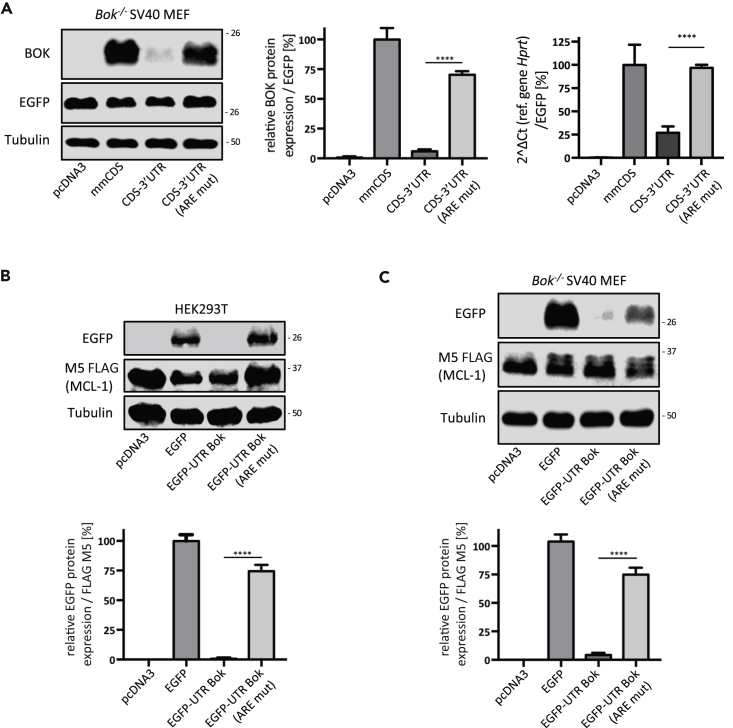
Considering that the initial experiments were performed with mouse Bok expression constructs in a human cell line we next wanted to confirm our findings in an autologous cell model, namely, SV40 immortalized Bok-/- mouse embryonic fibroblasts (MEF) (Echeverry et al., 2013). Similar to HEK293T cells, transfection of the mouse Bok CDS into Bok-/- MEFs resulted in a strong protein signal by western blotting, whereas it was barely detectable when the entire cDNA was transfected (Figure 2A). Mutation of the ARE site restored BOK protein levels to approximately 70% of the positive control, demonstrating that the ARE-mediated regulatory mechanism is functional in both human and mouse cells (Figure 2A). qPCR analysis confirmed that BOK protein levels correlated with mRNA levels (Figure 2A).
Figure 2.
Validation of Mouse BOK 3’ UTR Regulatory Function in Autologous Cells and an Unrelated Coding Sequence
(A) The functionality of the ARE-mediated regulation mechanism in autologous cells was evaluated at the protein and mRNA levels by transiently transfecting mouse Bok CDS, CDS-3’UTR, and CDS-3’UTR(AREmut) expression constructs into Bok−/− SV40 MEF. Transfection efficiency was normalized to a co-transfected pcDNA3/EGFP vector.
(B and C) (B) Expression of chimeric EGFP constructs flanked by mouse Bok's 5′ and 3′-UTR, with or without ARE-specific mutation, transiently transfected into HEK293T cells or (C) Bok−/− SV40 MEF. The transfection efficiency was normalized to a co-transfected pcDNA3/FLAG-MCL-1 vector. The data of quantitative immunoblotting are presented as mean ± SD, N = 3.
Statistical analysis was performed using a one-way ANOVA followed by a Tukey post-hoc test in GraphPad Prism. Significance levels, p < 0.0001****.
The previous results supported the existence of a conserved regulation of Bok expression through its 3’ UTR. We next tested whether the 3’ UTR could also modulate unrelated coding sequences. To this purpose, we evaluated the expression of enhanced green fluorescent protein (EGFP) flanked by the 5′ and 3′ UTRs of mouse Bok, with or without mutation of the ARE site. In line with our previous results, the inclusion of the Bok 3’ UTR massively reduced EGFP expression in both HEK293T and Bok−/− SV40 MEF, whereas the ARE mutation led to a significant rescue of EGFP levels (Figures 2B and 2C). These data indicate that the ARE sequence in the mouse Bok 3’ UTR is a genuine repressor of gene expression irrespective of the upstream coding sequence.
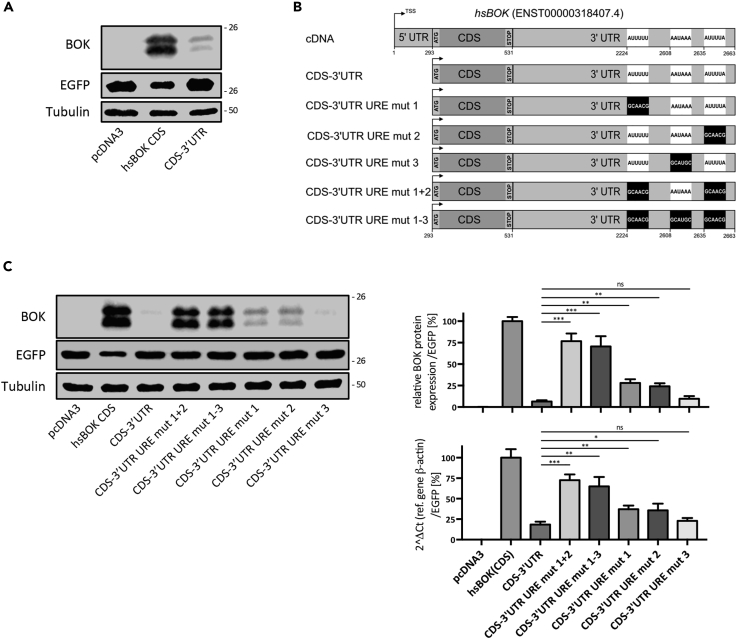
The Human BOK 3’ UTR Contains U-Rich Regulatory Elements
Despite similar exon-intron organization of the mouse and human BOK genes, a remarkable difference resides in the lengths of their respective 3’ UTRs, with the human 3’ UTR being three times longer than its mouse counterpart; nevertheless, human BOK expression was also found to be strongly repressed by its 3’ UTR (Figure 3A). Using in silicio tools described earlier, we screened for the existence of ARE sites in the human BOK 3’ UTR, also including related GU/CU- and U-rich elements in the analysis. Contrary to the mouse transcript, the human 3’ UTR lacks classical ARE sites (core sequence AUUUA) but contains six stretches of non-canonical U-rich elements (five URE, motifs W1−2UUUW1−2, and one AWTAAA motif, W=U/A) (Figure S1). These six sequences overlapped to some degree and could be reduced to three potential U-rich elements we termed URE1, 2 and 3, respectively (Figure 3B). Site-directed mutagenesis of these sites showed that individual destruction of either URE1 or 2 partially recovered BOK mRNA and protein expression, whereas mutation of URE3 did not show any effect (Figure 3C). However, simultaneous mutation of URE1 and 2, or of all three URE sites (URE1-3), significantly recovered BOK expression to levels approaching those seen for the transfection control with the CDS (Figure 3C). These results indicate that the 3’ UTR-dependent negative regulation of BOK is conserved between mice and humans. Furthermore, similar to the single destabilizing ARE site in the mouse Bok transcript, there are two U-rich elements (URE1 and 2) present in the human BOK 3’ UTR exerting a comparable regulatory role.
Figure 3.
Discovery of Analogous Negative Regulatory Elements in the 3’ UTR of Human BOK Transcript
(A) Transient transfection into HEK293T cells of human BOK CDS or its coding sequence fused to its 3’ UTR, highlighting the occurrence of negative regulators in the 3’ UTR.
(B) List of constructs generated used to validate the functionality of the predicted U-rich destabilizing elements (URE) within the human BOK transcript sequence (see also Figure S1).
(C) Representative immunoblot showing BOK levels in HEK293T cells transiently transfected with the indicated human BOK expression constructs. The right panel shows the levels of BOK obtained from quantitative immunoblotting using near-infrared fluorochromes and real-time quantitative PCR quantification from three independent experiments. The data are presented as mean ± SD, N = 3.
Statistical analysis was performed using a one-way ANOVA followed by a Tukey post-hoc test in GraphPad Prism. Significance levels, p > 0.05ns, p < 0.05*, p < 0.01**, p < 0.001***.
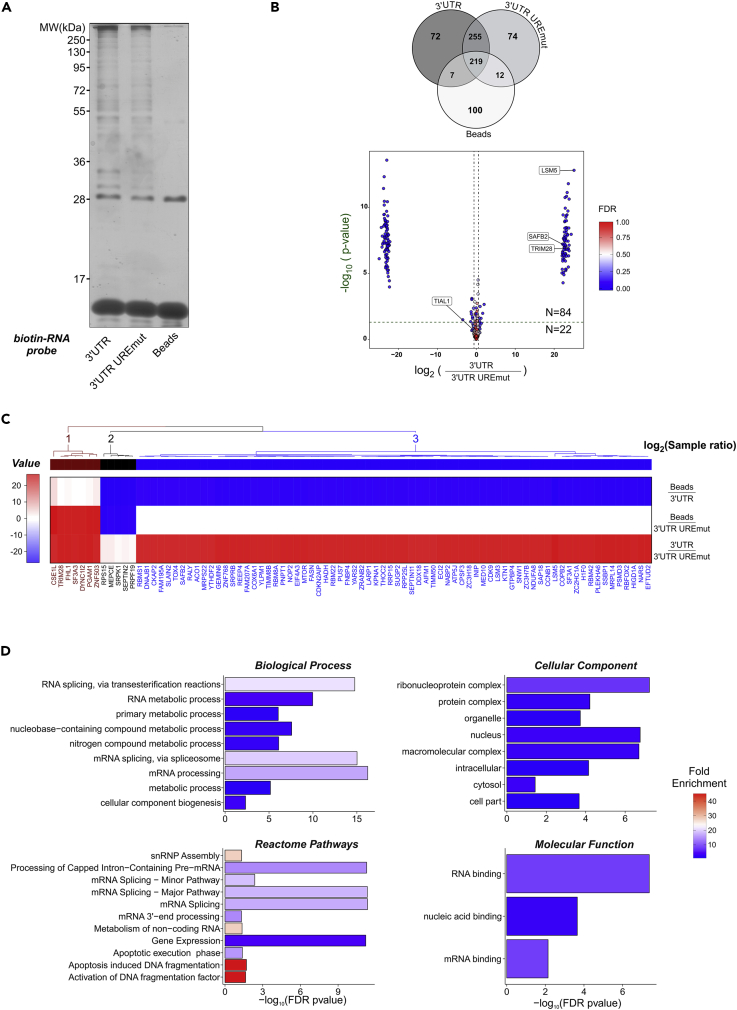
Identification of URE-Binding Proteins in the 3’ UTR of Human BOK by Proteomics Approaches
To identify proteins binding to the URE elements within the human BOK 3’ UTR and being critical for its repression, we performed a protein pull-down from HEK293T-derived cell lysate using synthetic biotinylated RNA probes encoding human BOK CDS bearing the wild-type or the URE mutated (UREmut1-3) version of the 3’ UTR, followed by identification by mass spectrometry. Initial analysis using silver staining on SDS-PAGE-separated samples already indicated differences in the protein binding to each probe compared with the beads-only negative control (Figure 4A). Overall, 749 hits were identified, of which 246 (33%) were unique to at least one of the three studied samples (Figure 4B and Table S1). A total of 101 hits were overrepresented more than 1.5 times in the samples pulled down with the wild-type probe compared with the UREmut1-3 probe (Figure 4B). Additional filtering criteria using adjusted p values and false discovery rate led to a final list of 84 candidates that were clustered in three major categories according to the differential abundance among the probes and the background (Figure 4C). Of particular interest were the resulting hits in clusters 1 and 3, as they displayed the most dramatic differences between wild-type and UREmut1-3 probes; thus, candidates from these clusters were selected for the validation round. Furthermore, we performed an overrepresentation gene ontology test with the selected 106 candidates (Figure 4D) and found a significant enrichment in proteins involved in biological processes related to RNA metabolism, processing, and splicing. This group contained several proteins known to assemble into (macro)molecular complexes, which, surprisingly, are not only restricted to the above-mentioned splicing and mRNA 3’-end processing pathways, but also involved in DNA fragmentation during apoptosis, according to the annotation of the Reactome Pathway database (Fabregat et al., 2018).
Figure 4.
Identification of Proteins Binding to the UREs within the Human BOK 3’ UTR
(A and B) HEK293T lysates were incubated with biotinylated RNA probes encoding human BOK CDS-3’UTR or CDS-3’UTR(UREmut1-3), a version with the three predicted URE sites mutated. Pull-down fractions were analyzed for the presence of RNA-binding proteins by SDS-PAGE and silver staining (A) and shotgun tandem mass spectrometry (MS/MS). The proteins detected were represented with a Venn diagram reflecting the count of detected proteins per sample type (B upper panel) and a volcano plot with the ratio and statistical significances shown between the indicated probes (B lower panel). Vertical (magenta) and horizontal (green) dashed lines mark the thresholds for absolute fold change (≥±1.5) and p values (p ≤ 0.05). A continuous color scale is applied to each point in accordance to the estimated false discovery rate (FDR <1%). Values corresponded to averages from three independent experiments.
(C) Clustering of the paired sample ratio abundances for the identified hits.
(D) A total of 106 candidates with a positive fold change (≥1.5) were analyzed in the database Panther (http://pantherdb.org/) for overrepresentation within four gene ontology categories performing a Fischer test with FDR correction. Bars represent the -log10-transformed corrected p value and are filled according to the calculated fold enrichment against the corresponding human genome background.
TRIM28 Negatively Regulates Human BOK Expression through Association with 3’ UTR Contained U-Rich Elements
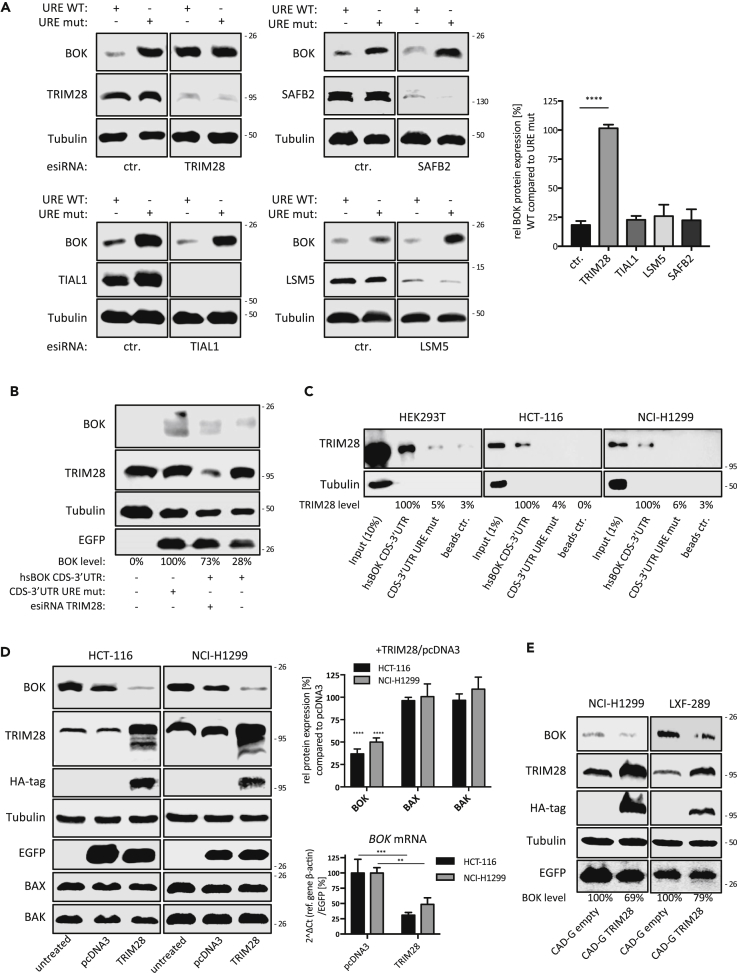
To validate URE-binding candidates for their role in destabilizing BOK mRNA in living cells, we performed transfections of pooled endoribonuclease-prepared short interfering RNAs (esiRNA) in HEK293T cells stably expressing either the human BOK CDS-3’UTR or its URE mutated version (UREmut1-3). We evaluated several candidates belonging to the clusters defined in Figure 4C (TRIM28, LSM5, SAFB2) as well as one hit (TIAL1) that fell below the selection threshold (p value ≤ 0.05, fold change 3’ UTR/3’ UTR UREmut1-3 ≥ 1.5) but is a known regulator of mRNA stability, which served as a negative control (Wigington et al., 2015, Zhao et al., 2014).
From this validation round, only knockdown of TRIM28 produced a robust and statistically significant recovery of BOK protein levels in cells expressing exogenous BOK fused to the wild-type 3’ UTR but not the URE1-3 mutated version (Figure 5A). TRIM28, also known as KAP1/TIF1-β, is a known co-repressor of gene expression at the chromatin level (Hosoya et al., 2013). To strengthen our finding of TRIM28 acting at the mRNA level rather than at the genomic BOK locus, we transiently co-transfected TRIM28-specific esiRNA with BOK-3’UTR or BOK-3’UTR(UREmut1-3) expression plasmids into HEK293T cells and confirmed that the knockdown of TRIM28 only rescued BOK expression in those cells receiving the BOK-3’UTR construct, up to 70% compared with the control BOK-3’UTR(UREmut1-3) (Figure 5B). Furthermore, we corroborated the presence of endogenous TRIM28 protein in pull-down fractions from lysates prepared from the human cell lines HEK293T, HCT-116, and NCI-H1299 using biotinylated CDS-3’UTR RNA probe. However, the signal was reduced to non-specific binding (comparable with beads-only control) when using the CDS-3’UTR(UREmut1-3) probe, strengthening the evidence that TRIM28 indeed binds to the UREs present in the 3’ UTR of BOK (see Figures 5C and S2 for predicted RNA-binding motifs in TRIM28).
Figure 5.
Validation of Selected BOK Negative Regulator Candidates
(A) HEK293T cells stably expressing human BOK constructs CDS-3’UTR or CDS-3’UTR(UREmut1-3) were transfected with validated esiRNAs targeting the candidates LSM5, TRIM28, SAFB2, and TIAL1. The recovery of BOK protein levels was used to determine involvement of a candidate in the URE-mediated repression of BOK. Immunoblots shown are representative of three independent experiments, and the quantification (right panel) of the signal is presented as mean ± SD. Statistical differences were determined using a one-way ANOVA followed by a Tukey post-hoc test.
(B) BOK recovery in HEK293T cells transiently co-transfected with human BOK CDS-3’UTR and esiRNA targeting TRIM28. The 3’UTR(UREmut1-3) construct was used as a 100% reference.
(C) Demonstration of the specific recruitment of endogenous TRIM28 to the 3’ UTR of human BOK in three independent cell lines using the indicated biotinylated synthetic probes (see Figure S2 for prediction of RNA binding motifs in TRIM28).
(D) Transient overexpression of HA-tagged TRIM28 in the lung cancer cell line NCI-H1299 or the colon carcinoma cell line HCT-116 leads to downregulation of endogenous BOK without impacting on the levels of BAX or BAK. Quantitative immunoblots are representative of three independent experiments; data are presented as mean ± SD. Statistical differences were detected using an unpaired t test, comparing with the BOK levels obtained on the sample transfected with the empty vector. Downregulation of BOK by overexpressed HA-TRIM28 correlates with reduced BOK mRNA levels (means ±SD, N = 3).
(E) Stable expression of HA-tagged TRIM28 in the lung cancer cell lines NCI-H1299 and LFX-289 leads to a reduction of endogenous BOK levels. See Figure S3B for quantification.
Significance levels, p < 0.01**, p < 0.001***, p < 0.0001****.
This functional link between BOK repression and TRIM28 could further be demonstrated by performing the opposite approach, in which we transiently overexpressed TRIM28 in the BOK-proficient colorectal cancer cell line HCT-116 or the NSCLC cell line NCI-H1299. As shown in Figure 5D, overexpression of HA-tagged TRIM28 resulted in a significant reduction of >50% of endogenous BOK protein levels compared with the control vector. This modulation of endogenous BOK expression was independently confirmed in NCI-H1299 and LXF-289 lung cancer cell lines stably transduced with HA-TRIM28 (Figures 5E, S3B, and S3C). Of note, this functional association with TRIM28 was specific for BOK within the pro-apoptotic multi-domain family members, since the expression levels of BAX and BAK remained unchanged under the same experimental conditions (Figure 5D). Given the high level of homology between human and mouse TRIM28, we transfected human TRIM28 into MEF and likewise found reduction in endogenous (mouse) BOK protein levels, supporting a similar mode of action of TRIM28-mediated BOK repression in mouse cells (Figure S3A). Taken together, these data demonstrate that TRIM28 recruitment to URE sites in the 3’ UTR of human BOK mRNA results in a potent repression of BOK cellular abundance.
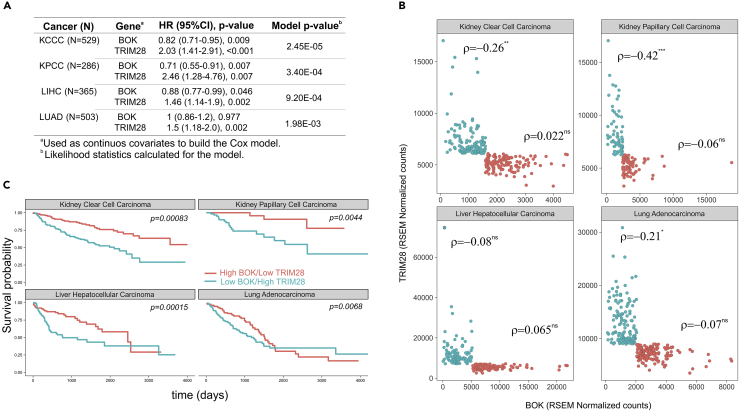
BOK and TRIM28 are Negatively Correlated in Selected Types of Cancer
Our previous results demonstrated a negative impact of TRIM28 levels on BOK expression. We next sought to address whether this phenomenon could be detected in the publicly available Cancer Genome Atlas (TCGA) database. As previously reported, we found that elevated transcript levels of TRIM28 constitute a risk factor in several cancer types, whereas BOK transcript levels mostly show an opposite trend (i.e., decreased in cancer), with the notable exception of adrenocortical cancer and skin cutaneous melanoma (Figures S4 and S5). Simultaneous modeling of BOK and TRIM28 levels using multiparametric Cox hazard ratio models in primary tumors derived from patients with kidney clear cell or papillary cell carcinoma as well as hepatocellular carcinoma indicated a significant reduction in the risk of death for patients with elevated BOK levels, which contrasts with the increased risk of death associated with high TRIM28 expression (Figure 6A). Supporting these models, patients who simultaneously had BOK levels above and TRIM28 levels below their respective population medians exhibited the best survival rate, in stark contrast to those patients belonging to the opposite strata (Figure 6B see Table S2). Interestingly, in kidney malignancies, TRIM28 and BOK expressions are negatively correlated in the patient stratum with the highest mortality rate, whereas it is less clear for liver and lung carcinomas (Figure 6C see Table S2). Overall, these data strengthen the interrelationship between TRIM28 and BOK expression indicating that, in these selected cancers, the negative correlation between TRIM28 and BOK may potentially be of prognostic clinical value.
Figure 6.
Potential Clinical Implications of the TRIM28 and BOK Interrelationship
(A–C) (A) Multiparametric Cox hazard ratio modeling of BOK and TRIM28 mRNA levels in several cancer types. Number of patients (N), 95% confidence interval of the hazard ratio function (HR 95% CI), significance (p value), and overall significance of the model (Model p value) are tabulated. Patients were classified according to their BOK and TRIM28 levels with respect to each gene population median expression into two strata (“high”/“low”) and further analyzed for overall survival (B) and correlation of the two genes within each strata (C). Differences in survival were detected with the log rank test, and correlations were determined using the Spearman's estimate. Significance levels, p > 0.05ns, p < 0.05*, p < 0.01**, p < 0.001***. See Table S2 for original and full data.
Discussion
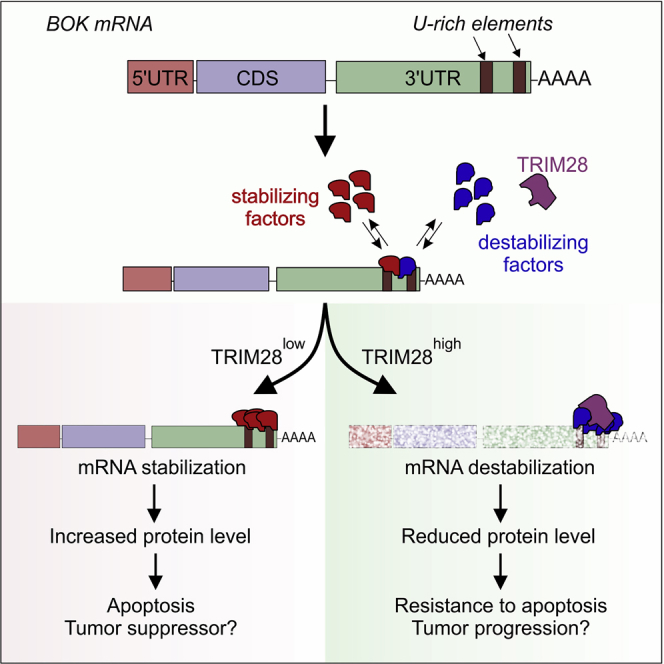
BOK is emerging as a BCL-2 family member of increasing interest, both during development as well as in its role as a potential tumor suppressor or modulator of cancer behavior (Carberry et al., 2018, Ke et al., 2018, Moravcikova et al., 2017, Rabachini et al., 2017). Owing to the pro-apoptotic response resulting from its accumulation, BOK expression seems to be tightly regulated at various levels. Here we expand this complexity by describing the existence of AU- and U-rich elements (ARE/URE) within the 3’ UTR of mouse or human BOK mRNAs, respectively, both of which contribute to the negative regulation of BOK expression.
Our model provides an alternative yet non-excluding explanation to the previously described epigenetic and proteasomal regulation of BOK levels (Llambi et al., 2016, Moravcikova et al., 2017). The existence of (A/U)RE could uncouple the transcription rate of the BOK genomic locus from its total protein concentration, explaining previously observed inconsistent correlations between BOK protein and mRNA levels in some mouse tissues and NSCLC biopsies (Echeverry et al., 2013, Moravcikova et al., 2017).
In our experimental setting, the (A/U)RE elements in the mouse and human BOK 3’ UTRs exclusively acted as a negative regulators, which even worked for unrelated coding sequences, contrasting with the dual role of ARE sequences in both stabilization and destabilization of the mRNA of other genes, such as BCL-2 (Diaz-Munoz et al., 2015, Schiavone et al., 2000). This dominance of the BOK 3’ UTR to suppress its transcript abundance and protein expression also suggests that the post-transcriptional control of BOK may dominate over its translational regulation, e.g. ribosome scanning or cap-(in)dependent translation, which is traditionally exerted by the 5’ UTR region (Leppek et al., 2018).
In human cells, we showed that 3’ UTR-dependent repression of BOK involves the recruitment of TRIM28 to the URE elements. This is a striking observation since all known repressing activities of TRIM28 occur at the chromatin level (Bunch et al., 2014, Li et al., 2017), and more importantly, TRIM28 lacks classical nucleic-acid-binding motifs (Iyengar and Farnham, 2011). Considering this, TRIM28 is likely recruited as part of a multiprotein complex to the 3’ UTR-URE sites rather than directly binding to the RNA. However, the latter possibility warrants further investigation since analysis of TRIM28 amino acid sequence with the Web tool catRAPIDsignature (Livi et al., 2016) predicts multiple modules with potential RNA-binding activity (Figure S2). Furthermore, experimental evidence from the Landthaler group, characterizing the transcript-bound proteome in HEK293T cells, detected TRIM28 in two of three mass spectrometry runs (Baltz et al., 2012), supporting the possibility of true mRNA binding of TRIM28. Regarding the underlying mechanism of mRNA degradation, it will be interesting to investigate whether TRIM28 requires its SUMO E3 ligase activity or not. Involvement of E3 ligase activity in such a context has been described for other RNA-binding E3 ligases, such as MEX-3C, which mediates a RING-dependent destabilization of MHC-I transcripts (Cano et al., 2012).
The repression of BOK in human cells was TRIM28 dependent as demonstrated both by RNA interference and overexpression of TRIM28 in several independent cell lines. This mechanism seems peculiar for BOK, since our mass spectrometry data showed absence of ELAVL1 and tristetraprolin (TTP), two of the most common mediators of ARE-based RNA decay in the human transcriptome (Bakheet et al., 2018). Another feature of TRIM28-mediated regulation seems to be the specificity for BOK over its pro-apoptotic multidomain relatives BAX and BAK. This might result from the type, abundance, and position of the predicted URE sites in the respective 3’ UTRs of these particular genes, which at the same time greatly diverges from the best studied ARE-regulated protein of the whole family, the BCL-2 gene (Figure S1).
Our proteomic approach combined with independent protein pull-downs using biotinylated RNA probes undoubtedly placed TRIM28 at the mRNA level; separating its known role at the chromatin from our discovered function at the mRNA level was challenging. Therefore, we performed our assays using transient transfections and collecting the samples within a short temporal window, as well as included some assays using stable transfections. These assays indicated that relatively high levels of TRIM28 may be required to repress BOK mRNA. It is further possible that TRIM28 may affect BOK protein stability directly (e.g., in LFX-289 cells), and further investigation in that direction is needed. The transient approach allowed us to minimize the likelihood of chromosomal integration of the transfected BOK expression constructs, increasing the chance of a transcript-based regulatory mechanism.
Our results provide a potential value of the TRIM28-dependent, URE-mediated decay of BOK mRNA in certain cancers, particularly in subtypes of kidney and lung carcinomas, considering their inverse correlation in the stratum of patients with the worst overall survival and/or risk of event. It should be noted, however, that, as a limitation of these results, such an inverse correlation was only shown at the mRNA level but not yet interrogated at the protein level. TRIM28 is increased in several cancerous malignancies, and increased TRIM28 is a poor prognosis factor in early-stage NSCLC (Liu et al., 2013), glioma (Qi et al., 2016), and colorectal cancer (Fitzgerald et al., 2013). Furthermore, TRIM28 increases the expression of the ATP-binding cassette genes ABCG2 and ABCB1, thus contributing to chemoresistance to paclitaxel and cisplatin of epithelial ovarian cancer cells (Hu et al., 2015). On the contrary, pro-apoptotic BOK is currently discussed as a potential tumor suppressor gene, since its genomic locus is deleted across various cancers (Beroukhim et al., 2010) and elevated levels of this protein are associated with enhanced overall survival of patients with lymph-node-positive lung cancer (Moravcikova et al., 2017). It is currently not clear whether TRIM28-mediated repression of BOK levels primarily affects apoptotic responses or whether recently described “non-apoptotic” roles of BOK need to be considered as well (D'Orsi et al., 2016, Kalkat et al., 2013, Moravcikova et al., 2017, Rabachini et al., 2017). Along that line, a molecular process potentially linking TRIM28 and BOK could be EMT. Knockdown of TRIM28 was shown to impair EMT in ovarian cancer cells by upregulation of E-cadherin (CHD1) and reduction of vimentin (VIM) and N-cadherin (CDH2) (Deng et al., 2017). Along the same line, Chen and colleagues reported that, in NSCLCs, TGFβ-induced EMT is dependent on induction of TRIM28, strongly suggesting that TRIM28 contributes to EMT in NSCLC (Chen et al., 2014). TRIM28 was further shown to promote EMT and metastasis in breast cancer (Wei et al., 2016). Strikingly these phenotypes reported for downregulation or overexpression of TRIM28 with regards to EMT are inversely mimicked by BOK, as we have recently shown for NSCLC cells that overexpression of BOK antagonizes TGFβ-induced EMT (Moravcikova et al., 2017), thereby further underlining the connection and opposing functions of TRIM28 and BOK in a cancer context.
In summary, we have identified a post-transcriptional negative regulation of BOK expression through association of TRIM28 with distinct URE present in the 3’ UTR of the BOK transcript, resulting in destabilization and decay of the latter. This mechanism seems to rather specifically affect BOK, while sparing other pro-apoptotic multi-domain BCL-2 family members. Future efforts should focus on integrating this regulatory mechanism into the various (patho)physiological functions that have been described for BOK, in particular corroborating its relevance to cancer biology and its potential use by cancer cells to evade current chemotherapeutic regimens and/or to promote malignant transformation, including metastasis.
Limitations of the Study
Recent advances in elucidating the role of BOK in health and disease suggest that its abundance could be a critical factor to the cellular fate. Nevertheless, we lack comprehensive understanding of how BOK levels are regulated. Our work provides another layer of BOK regulation, involving the description of destabilizing (AU/U)-rich elements in the 3’ UTR of its mRNA. We currently do not know if and how strongly these regulatory elements contribute to physiology in development or in adult life. One of the destabilizing factors identified to associate with the U-rich elements of human BOK 3’ UTR is TRIM28. It is not clear whether TRIM28 directly binds to mRNA or indirectly as part of a macromolecular protein complex and through which mechanism TRIM28 induces mRNA destabilization.
Methods
All methods can be found in the accompanying Transparent Methods supplemental file.
Acknowledgments
We thank Drs. X. Wang and P. Jost (Munich, Germany) for scientific discussion and the Proteomics and Mass Spectrometry Core Facility at the Department for BioMedical Research (DBMR), University of Bern, for running the mass spectrometry, interpreting the data, and evaluating the primary results. This work was supported by the Swiss National Science Foundation (project grant 31003A_173006, to T.K.). Y.F.-M. was a PhD student of the Graduate School of Cellular and Biomedical Sciences of the University of Bern.
Author Contributions
Conceptualization, Y.F.-M. and T.K.; Investigation, D.B., E.L., Y.F.-M.; Writing-Original Draft, Y.F.-M., D.B., and T.K.; Writing-Review & Editing, Y.F.-M. and T.K.; Visualization, Y.F.-M. and D.B.; Formal Analysis, Y.F.-M.; Funding Acquisition, T.K.
Declaration of Interests
The authors declare no competing interests.
Published: November 30, 2018
Footnotes
Supplemental Information includes Transparent Methods, five figures, and two tables, and can be found with this article online at https://doi.org/10.1016/j.isci.2018.11.005.
Supplemental Information
Details the identified proteins, peptide numbers, principal-component analysis, clustered analysis, t test, and FDR correction for each independent replicate pulled down with the BOK-3’ UTR wild-type or URE-mutated RNA probes.
Dataset containing TRIM28 and BOK gene expression matched to clinical parameters in 33 cancer types by patient ID following the TCGA nomenclature. Includes detailed description of the original data source as well as the manipulations performed in this work to stratified patients according to the selected gene expression.
References
- Alexander K.A., Wang X., Shibata M., Clark A.G., Garcia-Garcia M.J. TRIM28 controls genomic imprinting through distinct mechanisms during and after early genome-wide reprogramming. Cell Rep. 2015;13:1194–1205. doi: 10.1016/j.celrep.2015.09.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakheet T., Hitti E., Al-Saif M., Moghrabi W.N., Khabar K.S.A. The AU-rich element landscape across human transcriptome reveals a large proportion in introns and regulation by ELAVL1/HuR. Biochim. Biophys. Acta. 2018;1861:167–177. doi: 10.1016/j.bbagrm.2017.12.006. [DOI] [PubMed] [Google Scholar]
- Baltz A.G., Munschauer M., Schwanhausser B., Vasile A., Murakawa Y., Schueler M., Youngs N., Penfold-Brown D., Drew K., Milek M. The mRNA-bound proteome and its global occupancy profile on protein-coding transcripts. Mol. Cell. 2012;46:674–690. doi: 10.1016/j.molcel.2012.05.021. [DOI] [PubMed] [Google Scholar]
- Beroukhim R., Mermel C.H., Porter D., Wei G., Raychaudhuri S., Donovan J., Barretina J., Boehm J.S., Dobson J., Urashima M. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463:899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggar K.K., Storey K.B. Insight into post-transcriptional gene regulation: stress-responsive microRNAs and their role in the environmental stress survival of tolerant animals. J. Exp. Biol. 2015;218:1281–1289. doi: 10.1242/jeb.104828. [DOI] [PubMed] [Google Scholar]
- Bunch H., Zheng X., Burkholder A., Dillon S.T., Motola S., Birrane G., Ebmeier C.C., Levine S., Fargo D., Hu G. TRIM28 regulates RNA polymerase II promoter-proximal pausing and pause release. Nat. Struct. Mol. Biol. 2014;21:876–883. doi: 10.1038/nsmb.2878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano F., Bye H., Duncan L.M., Buchet-Poyau K., Billaud M., Wills M.R., Lehner P.J. The RNA-binding E3 ubiquitin ligase MEX-3C links ubiquitination with MHC-I mRNA degradation. EMBO J. 2012;31:3596–3606. doi: 10.1038/emboj.2012.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carberry S., D'Orsi B., Monsefi N., Salvucci M., Bacon O., Fay J., Rehm M., McNamara D., Kay E.W., Prehn J.M.H. The BAX/BAK-like protein BOK is a prognostic marker in colorectal cancer. Cell Death Dis. 2018;9:125. doi: 10.1038/s41419-017-0140-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L., Munoz-Antonia T., Cress W.D. Trim28 contributes to EMT via regulation of E-cadherin and N-cadherin in lung cancer cell lines. PLoS One. 2014;9:e101040. doi: 10.1371/journal.pone.0101040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C.T., Kuo C.Y., Ann D.K. KAPtain in charge of multiple missions: emerging roles of KAP1. World J. Biol. Chem. 2014;5:308–320. doi: 10.4331/wjbc.v5.i3.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Yang S., Fu X., Feng J., Xu S., Ying G. High levels of KAP1 expression are associated with aggressive clinical features in ovarian cancer. Int. J. Mol. Sci. 2014;16:363–377. doi: 10.3390/ijms16010363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czerwinska P., Shah P.K., Tomczak K., Klimczak M., Mazurek S., Sozanska B., Biecek P., Korski K., Filas V., Mackiewicz A. TRIM28 multi-domain protein regulates cancer stem cell population in breast tumor development. Oncotarget. 2017;8:863–882. doi: 10.18632/oncotarget.13273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Orsi B., Engel T., Pfeiffer S., Nandi S., Kaufmann T., Henshall D.C., Prehn J.H. Bok is not pro-apoptotic but suppresses poly ADP-ribose polymerase-dependent cell death pathways and protects against excitotoxic and seizure-induced neuronal injury. J. Neurosci. 2016;36:4564–4578. doi: 10.1523/JNEUROSCI.3780-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng B., Zhang S., Zhang Y., Miao Y., Meng X., Guo K. Knockdown of Tripartite Motif Containing 28 suppresses the migration, invasion and epithelial-mesenchymal transition in ovarian carcinoma cells through down-regulation of Wnt/beta-catenin signaling pathway. Neoplasma. 2017;64:893–900. doi: 10.4149/neo_2017_611. [DOI] [PubMed] [Google Scholar]
- Diaz-Munoz M.D., Bell S.E., Turner M. Deletion of AU-rich elements within the Bcl2 3'UTR reduces protein expression and B cell survival in vivo. PLoS One. 2015;10:e0116899. doi: 10.1371/journal.pone.0116899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echeverry N., Bachmann D., Ke F., Strasser A., Simon H.U., Kaufmann T. Intracellular localization of the BCL-2 family member BOK and functional implications. Cell Death Differ. 2013;20:785–799. doi: 10.1038/cdd.2013.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabregat A., Jupe S., Matthews L., Sidiropoulos K., Gillespie M., Garapati P., Haw R., Jassal B., Korninger F., May B. The reactome pathway knowledgebase. Nucleic Acids Res. 2018;46:D649–D655. doi: 10.1093/nar/gkx1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fallmann J., Sedlyarov V., Tanzer A., Kovarik P., Hofacker I.L. AREsite2: an enhanced database for the comprehensive investigation of AU/GU/U-rich elements. Nucleic Acids Res. 2016;44:D90–D95. doi: 10.1093/nar/gkv1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Marrero Y., Bleicken S., Das K.K., Bachmann D., Kaufmann T., Garcia-Saez A.J. The membrane activity of BOK involves formation of large, stable toroidal pores and is promoted by cBID. FEBS J. 2017;284:711–724. doi: 10.1111/febs.14008. [DOI] [PubMed] [Google Scholar]
- Fitzgerald S., Sheehan K.M., O'Grady A., Kenny D., O'Kennedy R., Kay E.W., Kijanka G.S. Relationship between epithelial and stromal TRIM28 expression predicts survival in colorectal cancer patients. J. Gastroenterol. Hepatol. 2013;28:967–974. doi: 10.1111/jgh.12157. [DOI] [PubMed] [Google Scholar]
- Friedman J.R., Fredericks W.J., Jensen D.E., Speicher D.W., Huang X.P., Neilson E.G., Rauscher F.J., 3rd KAP-1, a novel corepressor for the highly conserved KRAB repression domain. Genes Dev. 1996;10:2067–2078. doi: 10.1101/gad.10.16.2067. [DOI] [PubMed] [Google Scholar]
- Glisovic T., Bachorik J.L., Yong J., Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582:1977–1986. doi: 10.1016/j.febslet.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner A.C., Meylan S., Ciuffi A., Zangger N., Ambrosini G., Denervaud N., Bucher P., Trono D. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet. 2010;6:e1000869. doi: 10.1371/journal.pgen.1000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosoya T., Clifford M., Losson R., Tanabe O., Engel J.D. TRIM28 is essential for erythroblast differentiation in the mouse. Blood. 2013;122:3798–3807. doi: 10.1182/blood-2013-04-496166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Kim J., Xu Q., Leng Y., Orkin S.H., Elledge S.J. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. doi: 10.1101/gad.1769609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu M., Fu X., Cui Y., Xu S., Xu Y., Dong Q., Sun L. Expression of KAP1 in epithelial ovarian cancer and its correlation with drug-resistance. Int. J. Clin. Exp. Med. 2015;8:17308–17320. [PMC free article] [PubMed] [Google Scholar]
- Ishimaru D., Zuraw L., Ramalingam S., Sengupta T.K., Bandyopadhyay S., Reuben A., Fernandes D.J., Spicer E.K. Mechanism of regulation of bcl-2 mRNA by nucleolin and A+U-rich element-binding factor 1 (AUF1) J. Biol. Chem. 2010;285:27182–27191. doi: 10.1074/jbc.M109.098830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S., Farnham P.J. KAP1 protein: an enigmatic master regulator of the genome. J. Biol. Chem. 2011;286:26267–26276. doi: 10.1074/jbc.R111.252569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Pan Y., Wang L., Zhang L., Ravichandran R., Potts P.R., Jiang J., Wu H., Huang H. MAGE-TRIM28 complex promotes the Warburg effect and hepatocellular carcinoma progression by targeting FBP1 for degradation. Oncogenesis. 2017;6:e312. doi: 10.1038/oncsis.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkat M., Garcia J., Ebrahimi J., Melland-Smith M., Todros T., Post M., Caniggia I. Placental autophagy regulation by the BOK-MCL1 rheostat. Autophagy. 2013;9:2140–2153. doi: 10.4161/auto.26452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke F.F.S., Vanyai H.K., Cowan A.D., Delbridge A.R.D., Whitehead L., Grabow S., Czabotar P.E., Voss A.K., Strasser A. Embryogenesis and adult life in the absence of intrinsic apoptosis effectors BAX, BAK, and BOK. Cell. 2018;173:1217–1230.e17. doi: 10.1016/j.cell.2018.04.036. [DOI] [PubMed] [Google Scholar]
- Leppek K., Das R., Barna M. Functional 5' UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 2018;19:158–174. doi: 10.1038/nrm.2017.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Wang Z., Lu G. TRIM28 promotes cervical cancer growth through the mTOR signaling pathway. Oncol. Rep. 2018;39:1860–1866. doi: 10.3892/or.2018.6235. [DOI] [PubMed] [Google Scholar]
- Li J., Xi Y., Li W., McCarthy R.L., Stratton S.A., Zou W., Li W., Dent S.Y., Jain A.K., Barton M.C. TRIM28 interacts with EZH2 and SWI/SNF to activate genes that promote mammosphere formation. Oncogene. 2017;36:2991–3001. doi: 10.1038/onc.2016.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L., Zhao E., Li C., Huang L., Xiao L., Cheng L., Huang X., Song Y., Xu D. TRIM28, a new molecular marker predicting metastasis and survival in early-stage non-small cell lung cancer. Cancer Epidemiol. 2013;37:71–78. doi: 10.1016/j.canep.2012.08.005. [DOI] [PubMed] [Google Scholar]
- Livi C.M., Klus P., Delli Ponti R., Tartaglia G.G. catRAPID signature: identification of ribonucleoproteins and RNA-binding regions. Bioinformatics. 2016;32:773–775. doi: 10.1093/bioinformatics/btv629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F., Wang Y.M., Victor B., Yang M., Schneider D.M., Gingras S., Parsons M.J., Zheng J.H., Brown S.A., Pelletier S. BOK is a non-canonical BCL-2 family effector of apoptosis regulated by ER-associated degradation. Cell. 2016;165:421–433. doi: 10.1016/j.cell.2016.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde B.M., Moore C., Varani G. RNA-binding proteins: modular design for efficient function. Nat. Rev. Mol. Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann P., Georgiev O., Le Douarin B., Bourquin J.P., Schaffner W. Transcriptional repression by RING finger protein TIF1 beta that interacts with the KRAB repressor domain of KOX1. Nucleic Acids Res. 1996;24:4859–4867. doi: 10.1093/nar/24.24.4859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moravcikova E., Krepela E., Donnenberg V.S., Donnenberg A.D., Benkova K., Rabachini T., Fernandez-Marrero Y., Bachmann D., Kaufmann T. BOK displays cell death-independent tumor suppressor activity in non-small-cell lung carcinoma. Int. J. Cancer. 2017;141:2050–2061. doi: 10.1002/ijc.30906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Morita N., Hikita K., Kiuchi K., Kiuchi K., Kaneda N. Recruitment of mRNA-destabilizing protein TIS11 to stress granules is mediated by its zinc finger domain. Exp. Cell Res. 2005;303:287–299. doi: 10.1016/j.yexcr.2004.09.031. [DOI] [PubMed] [Google Scholar]
- Okamoto K., Kitabayashi I., Taya Y. KAP1 dictates p53 response induced by chemotherapeutic agents via Mdm2 interaction. Biochem. Biophys. Res. Commun. 2006;351:216–222. doi: 10.1016/j.bbrc.2006.10.022. [DOI] [PubMed] [Google Scholar]
- Qi Z.X., Cai J.J., Chen L.C., Yue Q., Gong Y., Yao Y., Mao Y. TRIM28 as an independent prognostic marker plays critical roles in glioma progression. J. Neurooncol. 2016;126:19–26. doi: 10.1007/s11060-015-1897-8. [DOI] [PubMed] [Google Scholar]
- Rabachini T., Fernandez-Marrero Y., Montani M., Loforese G., Sladky V., He Z., Bachmann D., Wicki S., Villunger A., Stroka D. BOK promotes chemical-induced hepatocarcinogenesis in mice. Cell Death Differ. 2017;25:706–718. doi: 10.1038/s41418-017-0008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiavone N., Rosini P., Quattrone A., Donnini M., Lapucci A., Citti L., Bevilacqua A., Nicolin A., Capaccioli S. A conserved AU-rich element in the 3' untranslated region of bcl-2 mRNA is endowed with a destabilizing function that is involved in bcl-2 down-regulation during apoptosis. FASEB J. 2000;14:174–184. doi: 10.1096/fasebj.14.1.174. [DOI] [PubMed] [Google Scholar]
- Schulman J.J., Wright F.A., Han X., Zluhan E.J., Szczesniak L.M., Wojcikiewicz R.J. The stability and expression level of Bok are governed by binding to inositol 1,4,5-trisphosphate receptors. J. Biol. Chem. 2016;291:11820–11828. doi: 10.1074/jbc.M115.711242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman J.J., Wright F.A., Kaufmann T., Wojcikiewicz R.J. The Bcl-2 protein family member Bok binds to the coupling domain of inositol 1,4,5-trisphosphate receptors and protects them from proteolytic cleavage. J. Biol. Chem. 2013;288:25340–25349. doi: 10.1074/jbc.M113.496570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta T.K., Bandyopadhyay S., Fernandes D.J., Spicer E.K. Identification of nucleolin as an AU-rich element binding protein involved in bcl-2 mRNA stabilization. J. Biol. Chem. 2004;279:10855–10863. doi: 10.1074/jbc.M309111200. [DOI] [PubMed] [Google Scholar]
- Venkov C.D., Link A.J., Jennings J.L., Plieth D., Inoue T., Nagai K., Xu C., Dimitrova Y.N., Rauscher F.J., Neilson E.G. A proximal activator of transcription in epithelial-mesenchymal transition. J. Clin. Invest. 2007;117:482–491. doi: 10.1172/JCI29544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilborg A., Glahder J.A., Wilhelm M.T., Bersani C., Corcoran M., Mahmoudi S., Rosenstierne M., Grander D., Farnebo M., Norrild B. The p53 target Wig-1 regulates p53 mRNA stability through an AU-rich element. Proc. Natl. Acad. Sci. U S A. 2009;106:15756–15761. doi: 10.1073/pnas.0900862106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova-St Louis I., Bohjanen P.R. Post-transcriptional regulation of cytokine signaling by AU-rich and GU-rich elements. J. Interferon Cytokine Res. 2014;34:233–241. doi: 10.1089/jir.2013.0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei C., Cheng J., Zhou B., Zhu L., Khan M.A., He T., Zhou S., He J., Lu X., Chen H. Tripartite motif containing 28 (TRIM28) promotes breast cancer metastasis by stabilizing TWIST1 protein. Sci. Rep. 2016;6:29822. doi: 10.1038/srep29822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigington C.P., Jung J., Rye E.A., Belauret S.L., Philpot A.M., Feng Y., Santangelo P.J., Corbett A.H. Post-transcriptional regulation of programmed cell death 4 (PDCD4) mRNA by the RNA-binding proteins human antigen R (HuR) and T-cell intracellular antigen 1 (TIA1) J. Biol. Chem. 2015;290:3468–3487. doi: 10.1074/jbc.M114.631937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Zhao J., Hou M., Wang Y., Zhang Y., Zhao X., Zhang C., Guo D. HuR and TIA1/TIAL1 are involved in regulation of alternative splicing of SIRT1 pre-mRNA. Int. J. Mol. Sci. 2014;15:2946–2958. doi: 10.3390/ijms15022946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J.H., Grace C.R., Guibao C.D., McNamara D.E., Llambi F., Wang Y.M., Chen T., Moldoveanu T. Intrinsic instability of BOK enables membrane permeabilization in apoptosis. Cell Rep. 2018;23:2083–2094.e6. doi: 10.1016/j.celrep.2018.04.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Details the identified proteins, peptide numbers, principal-component analysis, clustered analysis, t test, and FDR correction for each independent replicate pulled down with the BOK-3’ UTR wild-type or URE-mutated RNA probes.
Dataset containing TRIM28 and BOK gene expression matched to clinical parameters in 33 cancer types by patient ID following the TCGA nomenclature. Includes detailed description of the original data source as well as the manipulations performed in this work to stratified patients according to the selected gene expression.