Abstract
Small heat shock protein (sHSP)-B7 (HSPB7) is a muscle-specific member of the non-ATP-dependent sHSPs. The precise role of HSPB7 is enigmatic. Here, we disclose that zebrafish Hspb7 is a kinetically privileged sensor that is able to react rapidly with native reactive electrophilic species (RES), when only substoichiometric amounts of RES are available in proximity to Hspb7 expressed in living cells. Among the two Hspb7-cysteines, this RES sensing is fulfilled by a single cysteine (C117). Purification and characterizations in vitro reveal that the rate for RES adduction is among the most efficient reported for protein-cysteines with native carbonyl-based RES. Covalent-ligand binding is accompanied by structural changes (increase in β-sheet-content), based on circular dichroism analysis. Among the two cysteines, only C117 is conserved across vertebrates; we show that the human ortholog is also capable of RES sensing in cells. Furthermore, a cancer-relevant missense mutation reduces this RES-sensing property. This evolutionarily conserved cysteine-biosensor may play a redox-regulatory role in cardioprotection.
Graphical Abstract
The intrinsic nucleophilicity of the amino acid cysteine has been exploited by chemical biologists and biologists alike. In the 1930s, the susceptibility of the activity of the cysteine protease, papain, to alkylating agents was reported.1 Many believed—as was later proven to be the case—that this was an indication of a catalytic cysteine. Work of early chemical biologists such as Lowe and Kaiser in the 1970s2 and 1980s3 not only established the functionality of protein-bound cysteines, but also that these nucleophilic residues could be harnessed for traditional chemical transformations on enzymes. Since those early days, a large number of mainly in vitro cysteine transformations have been delineated.4
Over the recent years, appreciation for the role of intrinsic protein reactivity at cysteine in living systems has expanded; it is now known that some proteins and enzymes contain biologically relevant off-active-site nucleophilic cysteines. Many such cysteines engage in a noncanonical, nonenzymatic signaling mode mediated by reactive electrophilic species (RES).5−8 Electrophile signaling is an umbrella term to describe endogenous processes in which an apparently nontargeted RES modifies a specific RES-sensor protein and triggers a specific downstream response. It remains unknown how many of these nonactive-site sensor-cysteines are encoded in the genome.9 Furthermore, mechanisms explaining both how the RES-reaction enhancement comes about and how phenotypic outputs are engendered are unclear.
Thus, there are few predictive tools that can help identify these functional sensor-cysteines.10 Computational modeling data indicate that charge in the vicinity of cysteine can improve nucleophilicity.11,12 Such an arrangement likely promotes RES adduction in some cases; however, in other cases, this does not appear to be a significant factor.13 Nevertheless, accumulating data show that low-occupancy RES modifications are sufficient modulators of specific pathways at single-target-protein resolution. For instance, 4-hydroxynonenal (HNE) and several other native enals/enones—only available in limited amount and only within subcellular microenvironments—can alkylate the key sensor protein Keap1,14−19 one of many redox-sensitive upstream antagonists of cytoprotective transcription factor Nrf2.20 Low-occupancy RES-modifications of Keap1 alone are sufficient to elicit gain-of-function Nrf2 activation. Furthermore, a recent example21 uncovered an evolutionarily conserved cysteine in a variable-loop region within a specific isoform of kinase Akt (Akt3) that enables it to react specifically with HNE in vivo when only a substoichiometric amount of HNE is made available in proximity.22 Under identical settings, other Akt isoforms are not as HNE-sensitive. HNEylation of Akt3 selectively inhibits Akt3-kinase-activity likely through a dominant-negative mechanism. On the other hand, an E2-conjugation modifier Ube2V2, upon low-occupancy RES-binding, modulates ubiquitin signaling to protect against genome injury in cells and zebrafish.23 Thus, RES-sensor proteins have a tendency to be kinetically privileged in engagement with a specific RES and function through dominant signaling mechanisms.7,9 These properties are likely necessary for signaling to occur, because native RES signaling-mediators are ephemeral and are likely promiscuous. Thus, to ensure that the correct cellular messages are triggered and relayed, signal amplification mechanisms must be in place and sensor proteins should react rapidly with (a) specific electro-phile(s).
We recently demonstrated a proof-of-concept medium-throughput screen to discover such privileged sensors.21 The screen is built using our in-house-developed T-REX precision RES-targeting toolset,16 in combination with the commercially available Halo ORF clone library that enables any human/mouse gene to be pilot-tested for their RES-sensing ability in living cells. T-REX enables a specific protein of interest (POI) in live cells16 (or fish21,23/worms14) to be given first refusal to a substoichiometric amount of native RES at a precise space and time. This POI-specific targeted RES perturbation in vivo is accomplished by introducing a bioinert, cell/organism-permeable HaloTag-targetable small-molecule photocaged precursor to a specific RES such as HNE (“Ht-PreHNE”15−18 hereafter), to living systems expressing the functional Halo-POI fusion protein, followed by photouncaging (see Figure 1A, as well as Figure S1 in the Supporting Information). With this screening platform, we discovered Hspb7 from zebrafish is a kinetically privileged sensor of HNE.
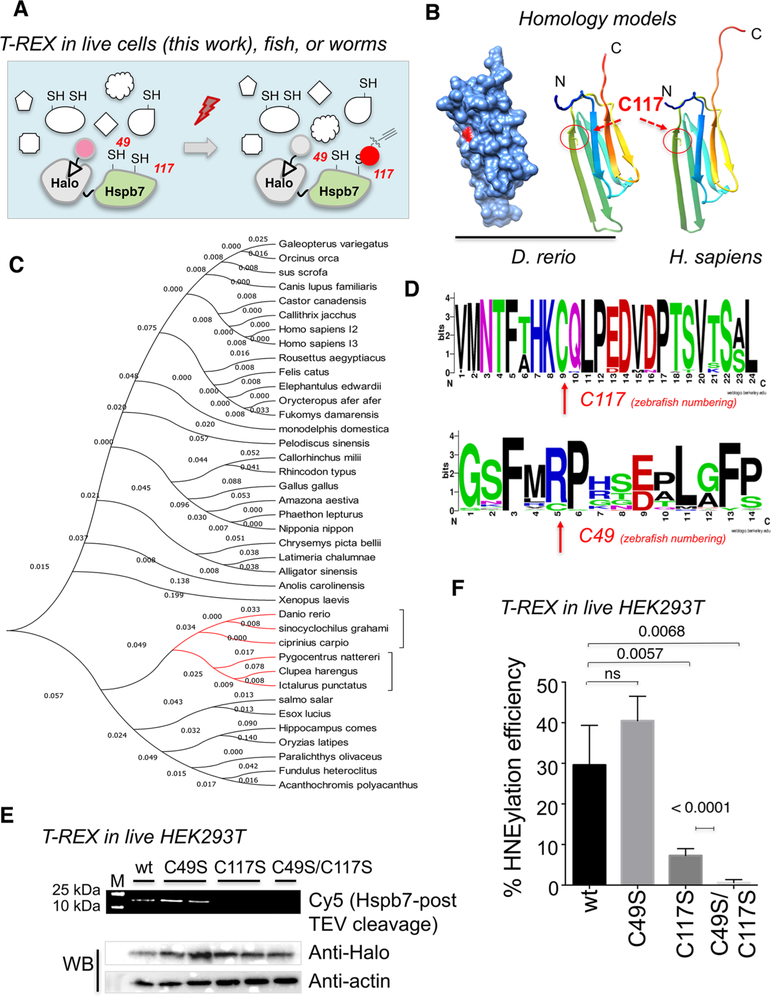
Figure 1.
C117S is a conserved sensor of HNE. (A) Schematic for T-REX in living systems. Also see Figure S1 in the Supporting Information. (B) Homology models of (left) zebrafish Hspb7 (surface-accessible and ribbon) and (right) human HSPB7 (ribbon). Note that, for the surface-accessible model, sulfur within C117 is colored red. (C) Phylogenetic analysis of HSPB7s. Clade colored red contains C49. Also see Figures S2 and S3 in the Supporting Information. (D) Sequence logos for C117 and C49. (E) HEK293T cells were transfected with respective plasmids encoding Halo-HSPB7(wt/mutants), grown for 24 h, then treated with Ht-PreHNE (20 μM, 2 h), washed and exposed to light. Cells were lysed and HNEylated-proteins were labeled with Cy5-azide; see Figure S1 for workflow. Lysates were resolved by SDS-PAGE and imaged by Cy5, and blotted for the stated proteins. (F) Quantitation of data in panel (E).
HSPB7—also known as cardiovascular HSP—is a member of the family of small heat shock proteins (sHSPs). These proteins appear to be non-heat-shock-regulated, energy-independent molecular chaperones.24−26 However, aside from its expression being highly enriched in striated muscle (such as the heart),27−29 little is known about HSPB7. Since sHSPs are mostly not regulated like canonical HSPs, novel functional cues that modulate sHSPs are of active pursuit. Thus, our discovery of native lipid-derived RES-signal-sensing capability by HSPB7 presents a new potential mode of regulating these pathophysio-logically relevant biological chaperones. Importantly, the only other sHSP currently implicated in redox regulation is HSPB1.30
Additional nuanced aspects of HSPB7 further provide compelling reasons to characterize its sensing capability: (1) HSPB7 contains only two cysteines (see Figure 1B, as well as Figure S2 in the Supporting Information) (cf. human Keap1 contains 27 cysteines; Akt3 contains 8 cysteines), suggesting that privileged sensing is not correlated with cysteine-richness of a protein; (2) HSPB7 is a small (18 kDa) protein (vs >60 kDa for Keap1 and Akt3). Indeed, HSPB7 presents one of the simplest HNE-sensors that we have identified thus far via TREX. Thus, HSPB7 is also an ideal model on which to evaluate our T-REX data in vitro, and to establish how transposable our live-cell-based target-identification of bona fide HNE sensors is to biochemical experiments on purified proteins in isolation. With this aim, we here report the kinetically privileged cysteine within HSPB7 that supports its HNE sensitivity in both isolated systems and in live cells.
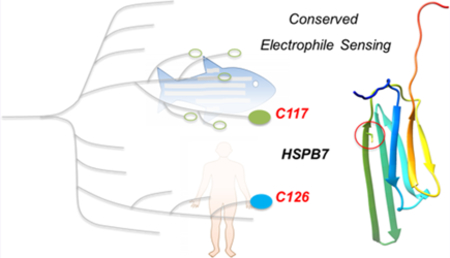
Successful purification of recombinant wild-type (wt) zebrafish Hspb7 allowed us to further characterize HNE-induced secondary structural changes using circular dichroism. Importantly, simplicity of the protein with few (only two) cysteines (see Figure 1B, as well as Figure S2) enabled assessment of time-dependent covalent-ligand binding on this cysteine. On the basis of sequence conservation across the vertebrate taxa (Figures 1C and 1D, as well as Figures S2 and S3 in the Supporting Information), we finally documented that the human ortholog senses HNE in living cells.
To begin investigating RES sensing by HSPB7, we used TREX (vide infra, Figure 1A and Figure S1). We chose to study zebrafish (Danio rerio) Hspb7 because, as detailed above, we have proven that T-REX functions effectively in zebrafish—a model system in which initial developmental genetic studies of Hspb7 have been successfully carried out.29 Among the two cysteines that zebrafish hspb7 encodes (C49 and C117; zebrafish numbering) (Figure 1D, as well as Figure S2), C117 is conserved in humans, and indeed across the entire HSPB7 family from all species we surveyed carrying this gene (Figure S3). Since neither X-ray nor nuclear magnetic resonance (NMR) structures have been obtained for HSPB7, we used homology modeling to model both zebrafish Hspb7 and human HSPB7 (sharing 64% sequence identity), using the4.5 Å crystal structure of human HSPB6 as the template (PDB ID: 5LTW) (Figure 1B). 31% identity is shared between human HSPB6 and either zebrafish Hspb7 or human HSPB7 (Figure S2). The model indicates that the conserved cysteine (C117) is surface-accessible in both zebrafish and human proteins, and lies on the edge of the folded core of the protein (Figure 1B). The second cysteine (C49) lies in a mobile loop. This cysteine is not present in humans (Figure S2). Indeed, C49 is only present in a small number of fish related to zebrafish (a clade colored red in the phylogenetic tree, see Figure 1C). Assuming that one of the two cysteines within Hspb7 senses HNE, the sensor residue is either highly conserved, or specific to a small number of fish related to D. rerio. Either way, we were interested in understanding the role of electrophile sensing in this small protein.
Since there are only two cysteine residues within zebrafish Hspb7, we identified the sensor by sequential mutagenesis. Halo-Hspb7 analogues were engineered with the two cysteines separately mutated to serine, C49S and C117S. The double cysteine to serine (C49S/C117S) mutant was also created. TREX (Figure 1A and Figure S1) is an ideal tool to evaluate precision HNE-sensing capability among mutants in living systems, because only substoichiometric amounts of HNE are liberated (thus leading to little cellular perturbation or off-target effects such as background ROS production, as confirmed by our previous studies9,14−16,18,19,21−23). In addition, because TREX builds on a quasi-intramolecular setup, similar expression levels are not necessary for a comparison of RES-modification efficiency (a value transposable to covalent ligand occupancy on POI) across wild-type (wt)/mutant-proteins. Nevertheless, all mutants were expressed similarly in cells and showed similar cellular localization by immunofluorescence (see Figure 2A). C49S sensed HNE equally as well as wt in live 293T cells: under T-REX (~40% ± 7% POI RES-labeling efficiency, vs ~30% ± 10% for wt (see Figures 1E and 1F). However, the alternative point mutant, C117S, showed significantly reduced sensing (~5% POI RES-labeling efficiency). The double mutant showed virtually no sensing. The sensing capacity of C117 is on the order of the most potent sensing residues we have discovered so far,16,21,23 including Keap1, which is a quintessential RES-sensor protein.16−18 Therefore, we conclude that the primary sensing residue is C117 (Figure 1B), although C49 may have some weak sensing capacity [similar to the less-potent sensors that we have characterized (such as Ube2 V1)23]. Interestingly, C117 is not present evenly in closely related HSPBs (Figure S3), such as HSPB3 or other sHSPs implicated in heart health.31 Thus, HSPB7 has a unique HNE-sensing mode that occurs on a residue (C117 in zebrafish) that is common to human and numerous higher eukaryotes (Figures 1B and 1C and Figure S2).
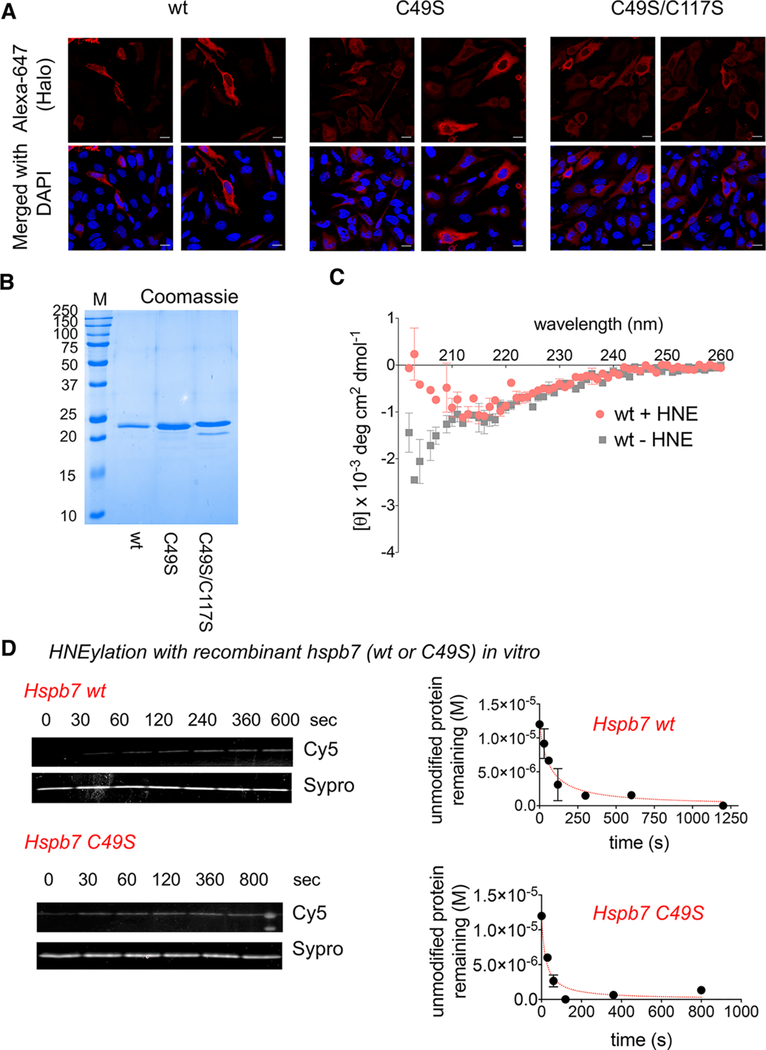
Figure 2.
Zebrafish Hspb7 mutants behave similarly to wt both in cells and in vitro. (A) HeLa cells were transfected with the Halo-TEV-Hspb7 construct and after 2 days, fixed with MeOH, then blocked, stained, with rabbit anti-Halo antibodies, followed by antirabbit IgG conjugated to Alexa 647. Cells were analyzed by confocal imaging. (B) The stated proteins (without Halo tag) were cloned into pET28 and purified using nickel affinity resin. Purified proteins were analyzed by SDS-PAGE. (C) Hspb7 wt (2.8 μM) was treated with HNE (1 equiv) or dimethylsulfoxide (DMSO) for 30 min at 37 °C. After this time, samples were analyzed by CD. (D) (Left) The indicated recombinant protein (12 μM) was treated with HNE-alkyne (12 μM) at room temperature and aliquots were removed as a function of time and diluted into chilled buffer. Samples were derivatized using Cy5-azide Click coupling (reporting on HNEylation) and Sypro-Ruby (total protein). (Right) Quantitation of total unlabeled protein remaining as a function of time and fit to a homodimerization equation.
Based on this newly discovered sensing cysteine, we next hypothesized that, if HNEylation of C117 were of functional significance, modification would likely cause structural perturbation. To this end, we first successfully established an optimized procedure to express and isolate recombinant zebrafish Hspb7 from E. coli BL21 (Figure 2B). The protein is prone to aggregation, especially in the absence of salt and glycerol. Gel filtration analysis shows the monomeric globular state. Circular dichroism (CD) analysis at RT showed significant β sheet structure (33%) with little α helix (<6%) (Figure 2C), which broadly agrees with the available CD spectra of the rat HSPB7, as well as our structure homology model (Figure 1B). Thus, zebrafish Hspb7 adopts a similar structure to the rat ortholog. Remarkably, pretreatment with HNE (1 equiv, for 30 min at 37 °C) followed by CD-data analysis at room temperature resulted in a significant change in the CD trace, compared to the spectra obtained from the same protein subjected to identical preincubation period in the absence of HNE (Figure 2C). Modeling this spectrum of HNEylated protein with K2D2 program indicated that ~50% of the protein adopted a β-sheet. These findings further supported that the RES-sensing ability of HSPB7 is likely functionally relevant. To ensure that ligand binding was saturating under the preincubation conditions and to further support that changes observed above were not a result of nonspecific protein aggregation/HNE-association to the protein independent of RES sensing, we undertook similar analyses on the three functional mutants following pretreatment with 6 equiv of HNE to protein (see Figure S4 in the Supporting Information).
The T-REX setup is uniquely designed to identify kinetically privileged sensors in living systems. Not only are limited amounts of a specific RES (such as HNE) presented to the POI in its native microenvironment, the setup also allows for competition between reactivity of the POI-cysteine and the innate diffusibility of the liberated RES (see Figure S1, inset). In other words, RES-chemotype to POI-cysteine-target functional engagement is tested under RES-limited conditions mimicking physiologic signaling. Therefore, we have proposed—and, in a few instances (Keap1 and Ube2 V2), validated—that functional cysteines identified in such a manner should manifest efficient kinetics of RES-adduction. Recombinant zebrafish Hspb7 is an ideal model to further test this hypothesis.
Therefore, we have examined the time-dependent labeling efficiency of the protein in vitro. Treatment of 12 μM recombinant Hspb7 with 12 μM alkyne-functionalized HNE, followed by rapid dilution into chilled buffer at various time points and subsequent Cy-azide dye-conjugation by Click coupling, showed that the protein was labeled by HNE. Reaction with HNE was rapid and occurred both at 37 °C and RT (below the typical growth temperature of fish). A similar efficiency of labeling was obtained with 36 μM protein and 36 μM alkyne-functionalized HNE. Consistent with cell-based TREX-data that showed C49S was a marginally better HNE sensor (Figure 1E–F), recombinant C49S also reacted faster with HNE in vitro (Figure 1D). These data provide compelling evidence that Hspb7(C117) is the HNE sensor.
Although practical limitations of the above assay posed challenges to quantify an accurate rate of HNE labeling on nonenzymatic proteins, wherein labeling is not correlated with changes in activity, we were intrigued by this rapid covalent adduction. At concentrations of 12 μM HspB7 and 12 μM HNE, labeling saturated within ~5 min (Figure 2D). This HNEylation rate compares favorably with several known HNE-sensing proteins: bovine glutathione-S-reductase (GSR) is inhibited in a two-step processes by HNE with an apparent second-order rate constant of 500 M−1 s−1 (derived by dividing kinact by Ki, i.e., assuming a sequential two-step inhibition process);32 human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is inhibited by racemic HNE with an apparent rate constant of 3 M−1 s−1 (half-life expected to be hours under these conditions);33 and human HSP70 is modified by HNE with an apparent IC50 of ~50−100 μM after 16 h of treatment.34 Because inhibition and modification by HNE are not necessarily correlated, we evaluated GSR, GAPDH, and HSP70 (all from humans) in our gel-based labeling assay (see Figure 2D, as well as Figure S5 in the Supporting Information).
We found that GSR was not labeled over the time scale that HSPB7 was labeled under identical conditions. Preincubation with 3 mM TCEP prior to the start of labeling reaction did not alter the outcome. Increasing the protein and HNE concentrations in the initial reaction mixture (prior to dilution for the subsequent Click coupling step), and prolonging the assay incubation time showed some labeling, consistent with GSR being HNE-sensitive, on the order of minutes to hours at 36 μM protein and 36 μM HNE [estimated apparent second-order rate constant, >10 M−1 s−1, a value ~10-fold faster than reaction of HNE with GSH (1 M−1 s−1)]35 (see Figure S5A). Differences between expected association rates and those in the literature could, in part, reflect that the GSR used is from humans, the enzymes used may not be fully functional/active, or neither enzyme inhibition step may occur through enzyme HNEylation. Alternatively, the “slow step” measured in the previous report32 could represent an inhibition pathway that occurs through a second-order reaction between GSR and HNE that is not linked to the fast inhibition step. Viewed through this lens (and acknowledging the other sources of error we discussed), our data, which showed significant labeling of GSR by 36 μM HNE (see Figure S5A, left panel) is similar to the inhibition data reported in the literature (reported 50% activity inhibition after 1 h with treatment with 10 μM HNE). In our assay, increasing the HNE concentration to 360 μM resulted in a similar extent of labeling being achieved within a much shorter incubation time (<1 min) (see Figure S5A, right panel).
GAPDH was also HNE-sensitive, but rapid labeling was observed only in excess HNE (360 μM) (see Figure S5B), validating its HNE sensitivity in our assay conditions. HSP70 (10 μM, which is the maximum concentration that we were able to achieve for this protein in vitro) failed to be labeled by HNE, following prolonged incubation with excess HNE (either 36 or 360 μM) over the course of several hours. This profile (GSR > GAPDH ≫ HSP70) agrees with literature data on overall HNE sensitivity. Furthermore, we can also infer that there is no nonspecific labeling of thiols under our reaction conditions, since labeling does not match the number of cysteines in each protein of human origin: HSPB7 (2 cysteines) > GSR (10 cysteines) > GAPDH (3 cysteines) ≫ HSP70 (5 cysteines).
Based on these data, we predicted that the human ortholog is also sensitive to HNE. Accordingly, we set about investigating whether human HSPB7 is a sensor of HNE. Despite sharing 64% identity, a closer inspection revealed that several residues around C117 differed between the two proteins [see Figure 1D (top panel) vs Figure 3A]. Importantly, the residue adjacent to the sensor cysteine in human HSPB7 is a glutamine, whereas it is an arginine (118) in zebrafish Hspb7. There is also a threonine (114) to alanine mutation in humans. It is likely that local changes in side-chain physicochemical properties—and, hence, local structure/stereoelectronic properties—may influence sensing capability; it could then be argued that a loss of positive charge adjacent to the sensor cysteine could prevent human HSPB7 from sensing HNE. However, it is also possible that both local and remote changes in the environment around the sensor cysteine can also influence HNE sensing. Therefore, we have investigated to what extent the human protein senses HNE in living HEK293T cells under bulk HNE exposure (under the conditions where cell viability is not perturbed) and found that HSPB7 is efficiently HNEylated under these settings (Figure 3B).
Figure 3.
Human HSPB7 also senses HNE, and the conserved disease-relevant neighbor residue (H115, zebrafish numbering) functionally perturbs C117-sensing capability. (A) Homology models of zebrafish Hspb7 depicting the proximal H115 (zebrafish numbering), with respect to C117. (B) HEK293T cells were transfected with HSPB7-Myc-FLAG. After 24 h, cells were treated with HNE (20 μM) for 2 h, then cells were harvested and lysed. Lysates were labeled with biotin azide via Click coupling and then enriched by streptavidin. Protein was eluted by heating in Laemeli buffer, and inputs and elutions were resolved by SDS-PAGE and analyzed by anti-FLAG Western blot. Inset shows a schematic for pulldown. Elution blot (top) probing for HNEylated-FLAG-tagged protein shows a major band of slightly shifted molecular weight (MW). This MW shift is likely because eluted protein has biotin attached to it. The shift could also be due to intramolecular cross-linking; however, a faint band of similar MW to the band in input blot (lower) is also observable. (C) HEK293T cells were transfected with the plasmid encoding Halo-Hspb7, grown for 24 h, then treated with Ht-PreHNE (20 μM, 2 h), washed, and exposed to light [or no light (controls)]. Cells were lysed and light-exposed samples were treated with TEV and all samples were subjected to Click coupling with Cy5-azide. Lysates were resolved by SDS-PAGE and imaged by Cy5, and analyzed by Sypro-Ruby. (Inset shows the quantitation.)
Interestingly, our homology model indicates that C117 lies on a β-sheet (Figure 1B). In this structural arrangement, alternating side chains point outward in opposite directions, meaning that adjacent residues are not spatially close to each other. Closer inspection showed that a histidine (115; residue 124 in human) points toward C117 (Figure 3A), raising the possibility that this residue is more important for sensing than the flanking charged residues. H115 is mutated to aspartate in the Catalogue of Somatic Mutations in Cancer (COSMIC)-database (cancer.sanger.ac.uk ). We accordingly investigated whether the H115D mutation affected sensing of zebrafish Hspb7 using T-REX. This protein was expressed similarly to the wt protein and showed similar localization by IF (Figure S6A in the Supporting Information). However, the protein was unable to sense HNE by T-REX (Figure 3C and Figure S6B in the Supporting Information). Thus, residues remote from the sensor cysteine can be critical for HNE sensing.
In summary, the remarkable sensing ability of HSPB7 can be ascribed almost exclusively to a single conserved privileged sensor cysteine, whereas, in quintessential RES-sensor proteins such as Keap1, our data16−18 and those of others20,36 implicate numerous cysteines in sensing. In the broader picture, this work constitutes the first opportunity to undertake an in vitro biochemical analysis of a sensor-protein identified by T-REX. The result gives high confidence that T-REX is indeed able to tease out proteins with unique ability to react with a specific RES chemotype in controlled physiological settings. The HNEylation of zebrafish Hspb7 is highly efficient and is superior to other HNE-sensitive proteins previously reported by independent laboratories. Such efficiency designates Hspb7-(C117) residue as being among the most efficient RES-sensing protein-cysteines. The fact that Hspb7 is not an enzyme (and C117 not being an enzymatically active residue) further underscores the significance of this finding, in relation to expanding our knowledge about and mining of nonenzymatic functional cysteine that might ultimately be druggable by covalent small molecules.
Interestingly, the sensing ability is not especially dependent on adjacent residues. Zebrafish Hspb7 has two flanking positive charges surrounding the sensor residue, whereas the human protein has only one; both proteins are sensors (see Figure 1D and Figure S2). Furthermore, mutation of residues not adjacent to the sensor residue (e.g., H115) can exert a much more significant effect on sensing capacity (see Figures 3B and 3C, as well as Figure S6 in the Supporting Information), highlighting the need to appreciate structural and spatial context of the sensor cysteine. Importantly, the H115D mutation is documented in cancer, and while such a result does not prove that H115D is a cancer-causing mutation, it does raise the tantalizing possibility that hampered redox-sensing ability could be linked to disease states; such a finding warrants further investigation. On a more fundamental level, our finding that Hspb7(H115D) does not sense HNE implicates some form of misregulation in a protein that expresses and localizes similarly to wt. Thus, redox sensing may be a good method to investigate “activity” of proteins for which there is no clearly defined assay.
The efficient HNEylation of C117 represent “rate enhancement” of reaction of cysteine nucleophiles with HNE in the absence of an “active site”. Furthermore, it is many-fold faster than the kinetics of association of cysteine thiolates with HNE (~1 M−1 s−1).12,22 This comparison gives good credence to the fact that HSPB7—similar to other privileged sensors of ROS—probably has mechanisms other than lowering pKa to promote rapid association. HNE-binding sites for some HNE sensors have been delineated,32 and it is possible that such a site may exist in HSPB7. However, other “tricks” employed by enzymes to elevate association kinetics could also be at play. Further experimentation, such as NMR studies, may elucidate how privileged sensing comes about in HSPB7. Given the rapid rate of adduction, HSPB7 may not need any assistance to compete effectively for HNE in the cellular milieu, especially if there are phenotypically dominant mechanisms that trigger a sensing function. While many of the nuts and bolts of electrophile sensing have yet to be understood, and the kinetic requirements for what would constitute a sensor are also unknown and difficult to rigorously assess in vitro, in relation to cellular context, it is worth noting that sensing by hitherto unknown sensors that do not appear to show up in any of the traditional electrophile-sensing screens can nevertheless be efficient and significant.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge A. Van Hall-Beauvais and P. Huang (Aye Lab) for TEV-protease preparation; the Crane Lab (Department of Chemistry and Chemical Biology, Cornell University) for the use of the CD spectrometer.
Funding
Research instrumentation, supplies, and personnel in this work are partly or fully supported by NSF CAREER (No. CHE-1351400), Beckman Young Investigator, NIH New Innovator (No. 1DP2GM114850), ONR Young Investigator (N00014–17-1–252), and Sloan Fellowship (FG-2016–637) award programs (to Y.A.); Burroughs Wellcome Fund (BWF) CRTG [to Y.A. and T.E. (host)]; No. R35 HL135778 (T.E.); Cornell University Graduate School Fellowship and NSF Graduate Research Fellowship (No. DGE-1650441, to D.A.U.); Hill Undergraduate Summer Fellowship from Cornell Chemistry & Chemical Biology Department (to S.L.S.); Tanner Dean’s Scholarship from Cornell College of Arts & Sciences (to I.M.E); NSF MRI (No. CHE-1531632, PI: Y.A.) for Cornell University core-facility NMR instrumentation; No. NIH 1S10RR025502 (PI: R. M. Williams) for Cornell Imaging Center.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acschembio.7b00925.
Methods, materials, and supporting tables and figures (PDF)
The authors declare no competing financial interest.
In accordance with the nomenclature adopted by the ZFIN (The Zebrafish Information Network) database, the zebrafish protein and gene are designated as Hspb7 and hspb7, respectively. The human orthologs are designated as HSPB7 and HSPB7, respectively.
■REFERENCES
- (1).Balls AK, and Lineweaver H (1939) Isolation and properties of crystalline papain. J. Biol. Chem 130, 669–686. [Google Scholar]
- (2).Clark PI, and Lowe G (1978) Conversion of the Active-Site Cysteine Residue of Papain into a Dehydro-serine, a Serine and a Glycine Residue. Eur. J. Biochem 84, 293–299. [DOI] [PubMed] [Google Scholar]
- (3).Kaiser E, and Lawrence D (1984) Chemical mutation of enzyme active sites. Science 226, 505–511. [DOI] [PubMed] [Google Scholar]
- (4).Chalker JM, Bernardes GJ, Lin YA, and Davis BG (2009) Chemical modification of proteins at cysteine: opportunities in chemistry and biology. Chem.—Asian J 4, 630–640. [DOI] [PubMed] [Google Scholar]
- (5).Schopfer FJ, Cipollina C, and Freeman BA (2011) Formation and signaling actions of electrophilic lipids. Chem. Rev 111, 5997–6021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Jacobs AT, and Marnett LJ (2010) Systems Analysis of Protein Modification and Cellular Responses Induced by Electrophile Stress. Acc. Chem. Res 43, 673–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Long MJ, and Aye Y (2016) The Die Is Cast: Precision Electrophilic Modifications Contribute to Cellular Decision Making. Chem. Res. Toxicol 29, 1575–1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Wang C, Weerapana E, Blewett MM, and Cravatt BF (2014) A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles,. Nat. Methods 11, 79–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Long MJC, and Aye Y (2017) Privileged Electrophile Sensors: A Resource for Covalent Drug Development. Cell Chem. Biol 24, 787–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Fomenko DE, Xing W, Adair BM, Thomas DJ, and Gladyshev VN (2007) High-Throughput Identification of Catalytic Redox-Active Cysteine Residues. Science 315, 387–389. [DOI] [PubMed] [Google Scholar]
- (11).Van Laer K, Oliveira M, Wahni K, and Messens J (2014) The concerted action of a positive charge and hydrogen bonds dynamically regulates the pKa of the nucleophilic cysteine in the NrdH-redoxin family. Protein Sci. 23, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Doorn JA, and Petersen DR (2003) Covalent adduction of nucleophilic amino acids by 4-hydroxynonenal and 4-oxononenal. Chem.-Biol. Interact 143–144, 93–100. [DOI] [PubMed] [Google Scholar]
- (13).Jao S-C, English Ospina SM, Berdis AJ, Starke DW, Post CB, and Mieyal JJ (2006) Computational and Mutational Analysis of Human Glutaredoxin (Thioltransferase): Probing the Molecular Basis of the Low pKa of Cysteine 22 and Its Role in Catalysis. Biochemistry 45, 4785–4796. [DOI] [PubMed] [Google Scholar]
- (14).Long MJC, Urul DA, Chawla S, Lin HY, Zhao Y, Haegele JA, Wang Y, and Aye Y (2018) Precision Electrophile Tagging in Caenorhabditis elegans. Biochemistry 57, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Long MJ, Lin HY, Parvez S, Zhao Y, Poganik JR, Huang P, and Aye Y (2017) β-TrCP1 Is a Vacillatory Regulator of Wnt Signaling. Cell Chem. Biol 24, 944–957.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Parvez S, Long MJ, Lin HY, Zhao Y, Haegele JA, Pham VN, Lee DK, and Aye Y (2016) T-REX on-demand redox targeting in live cells. Nat. Protoc 11, 2328–2356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Lin HY, Haegele JA, Disare MT, Lin Q, and Aye Y (2015) A generalizable platform for interrogating target- and signal-specific consequences of electrophilic modifications in redox-dependent cell signaling. J. Am. Chem. Soc 137, 6232–6244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Parvez S, Fu Y, Li J, Long MJ, Lin HY, Lee DK, Hu GS, and Aye Y (2015) Substoichiometric hydroxynonenylation of a single protein recapitulates whole-cell-stimulated antioxidant response.J. Am. Chem. Soc 137, 10–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Fang X, Fu Y, Long MJ, Haegele JA, Ge EJ, Parvez S, and Aye Y (2013) Temporally controlled targeting of 4-hydroxynonenal to specific proteins in living cells. J. Am. Chem. Soc 135, 14496–14499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hayes JD, and Dinkova-Kostova AT (2014) The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci 39, 199–218. [DOI] [PubMed] [Google Scholar]
- (21).Long MJC, Parvez S, Zhao Y, Surya SL, Wang Y, Zhang S, and Aye Y (2017) Akt3 is a privileged first responder in isozyme-specific electrophile response. Nat. Chem. Biol 13, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Long MJ, Poganik JR, and Aye Y (2016) On-Demand Targeting: Investigating Biology with Proximity-Directed Chemistry. J. Am. Chem. Soc 138, 3610–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Zhao YL, Long MJC, Wang Y, Zhang S, and Aye Y (2018) Ube2V2 Is a Rosetta Stone Bridging Redox and Ubiquitin Codes, Coordinating DNA Damage Responses. ACS Cent. Sci, DOI: 10.1021/acscentsci.7b00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Hilton GR, Lioe H, Stengel F, Baldwin AJ, and Benesch JLP (2013) Small Heat-Shock Proteins: Paramedics of the Cell In Molecular Chaperones (Jackson S, Ed.); pp 69–98 Springer: Berlin, Heidelberg. [DOI] [PubMed] [Google Scholar]
- (25).Eyles SJ, and Gierasch LM (2010) Nature’s molecular sponges: small heat shock proteins grow into their chaperone roles. Proc. Natl. Acad. Sci. U. S. A 107, 2727–2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Mercer EJ, Lin Y, Cohen-Gould L, and Evans T (2018) HSPB7 is a cardioprotective chaperone facilitating sarcomeric proteostasis. Dev. Biol, DOI: 10.1016/j.ydbio.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Elicker KS, and Hutson LD (2007) Genome-wide analysis and expression profiling of the small heat shock proteins in zebrafish. Gene 403, 60–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Krief S, Faivre J-F, Robert P, Le Douarin B, Brument-Larignon N, Lefrére I, Bouzyk MM, Anderson KM, Greller LD, Tobin FL, Souchet M, and Bril A (1999) Identification and Characterization of cvHsp: a novel human small stress protein selectively expressed in cardiovascular and insulin-sensitive tissues. J. Biol. Chem 274, 36592–36600. [DOI] [PubMed] [Google Scholar]
- (29).Rosenfeld GE, Mercer EJ, Mason CE, and Evans T (2013) Small heat shock proteins Hspb7 and Hspb12 regulate early steps of cardiac morphogenesis. Dev. Biol 381, 389–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Christians ES, Ishiwata T, and Benjamin IJ (2012) Small heat shock proteins in redox metabolism: implications for cardiovascular diseases. Int. J. Biochem. Cell Biol 44, 1632–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Marunouchi T, Abe Y, Murata M, Inomata S, Sanbe A, Takagi N, and Tanonaka K (2013) Changes in Small Heat Shock Proteins HSPB1, HSPB5 and HSPB8 in Mitochondria of the Failing Heart Following Myocardial Infarction in Rats. Biol. Pharm. Bull 36, 529–539. [DOI] [PubMed] [Google Scholar]
- (32).Vander Jagt DL, Hunsaker LA, Vander Jagt TJ, Gomez MS, Gonzales DM, Deck LM, and Royer RE (1997) Inactivation of glutathione reductase by 4-hydroxynonenal and other endogenous aldehydes. Biochem. Pharmacol 53, 1133–1140. [DOI] [PubMed] [Google Scholar]
- (33).Hiratsuka A, Hirose K, Saito H, and Watabe T (2000) 4-Hydroxy-2(E)-nonenal enantiomers: (S)-selective inactivation of glyceraldehyde-3-phosphate dehydrogenase and detoxification by rat glutathione S-transferase A4−4. Biochem. J 349, 729–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Carbone DL, Doorn JA, Kiebler Z, Sampey BP, and Petersen DR (2004) Inhibition of Hsp72-Mediated Protein Refolding by 4-Hydroxy-2-nonenal. Chem. Res. Toxicol 17, 1459–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Witz G (1989) Biological interactions of α,β-unsaturated aldehydes. Free Radical Biol. Med 7, 333–349. [DOI] [PubMed] [Google Scholar]
- (36).Bryan HK, Olayanju A, Goldring CE, and Park BK (2013) The Nrf2 cell defence pathway: Keap1-dependent and -independent mechanisms of regulation. Biochem. Pharmacol 85, 705–717. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.