Abstract
Identifying chemicals that provide a specific function within a product, yet have minimal impact on the human body or environment, is the goal of most formulation chemists and engineers practicing green chemistry. We present a methodology to identify potential chemical functional substitutes from large libraries of chemicals using machine learning based models. We collect and analyze publicly available information on the function of chemicals in consumer products or industrial processes to identify a suite of harmonized function categories suitable for modeling. We use structural and physicochemical descriptors for these chemicals to build 41 quantitative structure–use relationship (QSUR) models for harmonized function categories using random forest classification. We apply these models to screen a library of nearly 6400 chemicals with available structure information for potential functional substitutes. Using our Functional Use database (FUse), we could identify uses for 3121 chemicals; 4412 predicted functional uses had a probability of 80% or greater. We demonstrate the potential application of the models to high-throughput (HT) screening for “candidate alternatives” by merging the valid functional substitute classifications with hazard metrics developed from HT screening assays for bioactivity. A descriptor set could be obtained for 6356 Tox21 chemicals that have undergone a battery of HT in vitro bioactivity screening assays. By applying QSURs, we were able to identify over 1600 candidate chemical alternatives. These QSURs can be rapidly applied to thousands of additional chemicals to generate HT functional use information for combination with complementary HT toxicity information for screening for greener chemical alternatives.
Introduction
At the heart of green chemistry is the concept that throughout the development and production of chemicals, risk should be minimized by minimizing hazard.1 The most straightforward process for achieving this is by the design and selection of chemicals with reduced toxicity. This idea is emphasized in the fourth principle of green chemistry which states that “Chemical products should be designed to preserve efficacy of function while reducing toxicity”.1 While this principle focuses on reducing toxicity as a hallmark of green chemistry, it also states that it is vital that the function of the chemical constituent is maintained while addressing toxicity concerns. In order to uphold this principle, not only is there a need to classify chemicals that may be used in the manufacture of consumer products by toxicity, but also by function. Approaches that evaluate both function and toxicity in a high-throughput manner would allow formulation chemists and process engineers to rapidly identify potential functional substitutes meeting some toxicity criteria for further evaluation.
A hindrance to the joint evaluation of toxicity and function is the lack of toxicity information for many chemicals in commerce: this makes it difficult to identify chemical alternatives on the basis of hazard. In the United States, there are 85 000 chemicals that are available for public use that are also controlled under the Toxic Substance Control Act (TSCA).2 When combined with chemicals that are not covered under TSCA (food additives, cosmetic ingredients, pharmaceuticals, and active ingredients in pesticides) there were, as of 2006, more than 100 000 unique chemicals in the U.S. market.3 In addition, there is no high-quality in vivo toxicity data for roughly three quarters of these chemicals and there is not even limited toxicity data for one third of these chemicals.4–7 However, as described by Judson8 there are multiple approaches for characterizing the likely differential toxicities of green alternatives, including quantitative structure–activity relationships (QSARs) and high-throughput screening (HTS). The cross-agency Tox21 consortium9 and the Toxicity Forecasting (ToxCast) program10 of the United States Environmental Protection Agency (EPA) to date have evaluated over 8000 chemicals using HTS to identify potential hazard as quantified by in vitro bioactivity. This library of tested chemicals includes consumer product ingredients, food additives, industrial process chemicals, and human and veterinary pharmaceuticals.
While the Tox21 and ToxCast HTS programs have focused efforts on prioritizing and testing chemicals for bioactivity, other programs both internal and external to the EPA have focused on the next logical step – how to replace chemicals that are considered hazards either to human health or to the environment with safer chemicals. Two such programs within the EPA are the Safer Choice Program (formerly the Design for the Environment Program)11,12 and the Program for Assisting and Replacing Industrial Solvents (PARIS III).13 Safer Choice is a program within the EPA that aims to provide US consumers with a resource to find household products with formulations that are safer for the consumer and the environment. Products displaying the Safer Choice Label contain chemicals (all of which have known functional uses) that have met general and function-specific hazard criteria, (e.g., carcinogenicity, genetic toxicity, neurotoxicity). PARIS III uses chemical similarity with health and environment impact criteria to identify alternatives from a library of solvents.14,15 While both of these tools are useful in aiding chemical alternatives assessments, they are currently applied in a relatively low-throughput fashion (i.e., a single chemical is evaluated and multiple alternatives are returned). A useful complementary approach would be the ability to screen large libraries of chemical structures for potential functional substitutes, analogous to how HTS of potential drugs is performed in the pharmaceutical industry. Such a screening process would identify sets of chemicals for further targeted evaluation.
Tickner et al. recently described a framework for alternatives assessment formulated around the idea of “functional substitution” and proposed that grouping chemicals by their function enables comparative evaluation with regards to hazards and potential exposures.55
Here we develop classification models for identifying in a HT manner chemical functional substitutes from large libraries of chemicals that could then be screened for differential toxicity using HT approaches. To do this, we have used random forest classification on publicly available data and chemical descriptors to develop quantitative structure–use relationship (QSUR) models that are capable of predicting the function of a chemical, specifically, in consumer products. Just as QSARs have aided in high-throughput screening of in vivo potency for large libraries of molecules by correlating molecular activity with its structure and physicochemical properties,16 our QSURs should elucidate which features and properties of a chemical give rise to its function and identify chemicals with similar properties and features that could also fulfill that role. As an example case study, we apply our validated QSUR classifier models to chemicals in the Tox21 chemical library which contains structural and bioactivity information for non-pharmaceutical chemicals suspected of being prevalent in areas of human exposure.
Methods
Functional use data
We recently described the development of a chemical Functional Use (FUse) database for use in HT prediction of chemical functional use and weight fraction in consumer products by using physicochemical properties and broad use categories.17 These data were collected from publicly available chemical functional use lists; the largest source of information was the European Chemical Agency’s (ECHA) Cosmetic Ingredient (CosIng) database.18 Here we expanded the database to include additional functional use categories and chemical use sectors by collecting additional information from online government, industry, and manufacturer sources (Table 1). These sources were identified by internet searches of the words “chemical”, “function”, “role”, and “use”. Further, various consumer product websites were examined to determine if product ingredients were given functional uses by the manufacturer. While more information was available on functional use, some websites did not provide open access. As the intent was to make the data used in this work available to the public (ESI Table 1†), data from proprietary websites were not included in this database. Each source had a unique set of functional use categories; during an initial screening of the data, similar functional uses were combined to eliminate redundancy. For example, a “viscosity controller” and “viscosity controlling agent” were both considered to be a “viscosity controlling agent”. After this step, the data contained 32 263 records of chemical-functional use pairs, 14 272 unique chemicals, i.e., unique chemical abstract service registration numbers (CASRNs), and 224 unique functional use category descriptions. The functional use categories were then further harmonized as described below.
Table 1.
Number of unique pairings of a chemical with a reported functional use from each of the internet sources used to build the FUse dataset
| Source | Unique Chemical-functional use pairs |
|---|---|
| CosIng18 | 18 351 |
| SpecialChem19 | 5994 |
| Adhesives | 2192 |
| Coatings | 538 |
| Polymer additives | 3264 |
| International Fragrance Association20 | 2993 |
| Food Flavorings Database21 | 2685 |
| ACToR UseDB22 | 1203 |
| Safer Choice Ingredient List23 | 1065 |
| America Cleaning Institute24 | 713 |
| SC Johnson25 | 101 |
| The Clorox Company26 | 98 |
| method27 | 40 |
Harmonization of functional use
For the library of chemicals under study, 224 unique reported functions were identified in FUse. Many reported functions were considered redundant. For example, one source would list a chemical as a surfactant, while another would list the same chemical as a cleaner. To prevent building many models for functional uses that are very similar, we first harmonized the functional uses of chemicals within FUse in order to obtain a one-to-one mapping of a chemical to a single functional use. In other words, after harmonization, each chemical had a unique harmonized function. We applied a hierarchical clustering analysis28 (HCA) to identify commonalities in reported functions for groups of chemicals, and reduce the dimensionality (i.e., redundancy) of the data for quantitative structure–use relationship (QSUR) modeling. HCA is a widely-used approach that has been used for grouping chemicals by properties or other descriptors for various chemometric analyses.29 A binary array – one value for each unique reported function – was assigned to each chemical. A value of one was assigned to an array element if a chemical had a particular reported function in any data source and a value of zero if not; this array can be thought of as a “fingerprint” of the chemical in terms of its likely function(s). Casting the dataset in this way allowed the pairwise distance between two chemicals’ reported function fingerprints to be computed; this was done via the Jaccard distance metric.30 The distances were then clustered using the centroid linkage method.28 An optimal number of clusters that described a significant proportion of the variance in distance (in essence, a significant amount of the variation among chemicals with respect to reported function) was determined from a scree plot.31 Each cluster was given a consensus label – a harmonized function (HF) – based upon the original reported functions represented in the cluster. The steps in the data curation (including reduction of the overall available records due to missing information) are shown in Table 2.
Table 2.
Details of the number of chemicals, reported functions for each step in the curation of the QSUR training set
| Data curation step | Unique chemicals | Unique functions |
|---|---|---|
| FUse database | 14 272 | 224 |
| Function harmonization via cluster analysis | 14 272 | 137 |
| QSUR Set 1: Merged QSAR-ready structures with structural descriptors | 5806 | 98 |
| Filter QSUR Set 1 to functions containing at least 10 chemicals | 5666 | 49 |
| QSUR Set 2: Merged QSAR-ready structures with structural and physicochemical descriptors | 4791 | 84 |
| Filter QSUR Set 2 to functions containing at least 10 chemicals | 4667 | 43 |
Chemical descriptors
The EPA’s Distributed Structure-Searchable Toxicity (DSSTox) Database Network contains highly curated mappings of CASRNs to two-dimensional, QSAR-ready structures of chemicals.32,33 DSSTox was used to match structures to each unique CASRN found in FUse. Structures in the form of simplified molecular-input line-entry system (SMILES) codes were found for 5806 of 14 272 CASRNs. Verified SMILES strings were used as input into the ChemoTyper34 application to obtain ToxPrint35 descriptors (ESI Table 2†). These publicly-available descriptors, annotating 729 chemical substructures, were created with the intent of providing better fragment coverage of chemical substructures contained within toxicity databases. In addition, predicted physicochemical descriptors (properties) for 4791 of the 5806 chemicals were obtained using the U.S. EPA’s Estimation Program Interface (EPI) Suite.36 The physicochemical properties used were molecular weight, vapor pressure, water solubility, Henry’s Law constant, the log of the octanol–air partition coefficient – log(Koa), the log of the octanol–water coefficient – log(Kow), half-life of a chemical in soil, sediment, water, and air, and the persistence of a chemical in the environment (ESI Table 3†). Thus, a full set of descriptors for a model using only structural descriptors had 729 descriptors, while a full set of descriptors for a model using both structural and physicochemical properties had 740 descriptors (729 structural descriptors + 11 physicochemical properties). Descriptors were removed if they had constant values across the entire dataset. Only HFs that contained at least ten chemicals were used in the model training sets; the number of chemicals and functional uses in the training sets for both sets of descriptors are shown in Table 2.
QSUR models and validation
A suite of QSUR classification models for functional use were built using the random forest classification method.37 Random forests are grown by building multiple individual decision trees using randomly sampled subsets of training data descriptors. The classification returned by the final random forest model is the ensemble average of all decisions trees grown within this “forest” of trees; the fraction of the trees returning a positive result quantifies the probability, or confidence in the classification.
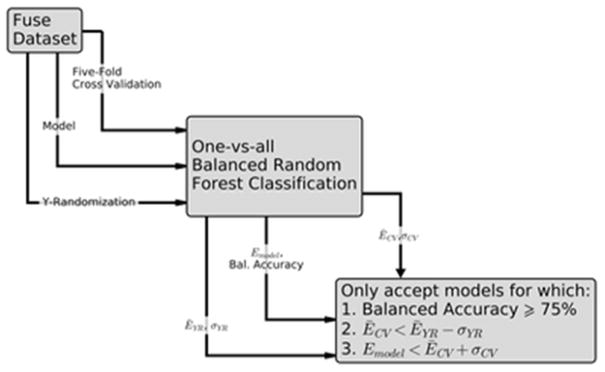
One-vs.-all balanced random forest models were constructed for each HF via the randomForest package38 of the R statistical software program.39 Two sets of QSUR models were constructed: one using only the structural descriptors, another using both structural and physicochemical descriptors. The workflow shown in Fig. 1 was used to validate both sets of models. Due to the large number of descriptors, 10 000 decision trees were grown for each model. As a one-vs.-all model implies, each HF was transformed into a binary variable (equal to 1 if the chemical had a given function and 0 if it did not have that functional use) and a model built for this binary variable. Most HFs had a small number of chemicals relative to the size of the overall dataset; to avoid bias in classification toward the negative class, balanced random forest was used (which samples the same number of positive and negative samples for each tree).40 An added benefit of using the random forest classification method to build our QSUR models is that we obtain information about the important features for classifying each HF, via the mean decrease in the Gini index41 which essentially measures the impurity of each parent node compared to the two children nodes. The purer the children nodes are, the lower the Gini index, and the higher the importance of a descriptor.
Fig. 1.
QSUR workflow used to create functional use predictions based on structural and physicochemical descriptors.
The performance of the HF models were evaluated with five-fold cross-validation (CV).42 The data were split so that approximately the same number of chemicals in each functional use category were present in each fold. The model classification error (the number of incorrect predictions made by a model divided by the total number of predictions made by a model), sensitivity, specificity, and balanced accuracy43 were obtained for each fold, and summary statistics across folds were calculated.
The models were further validated using the method of Y-randomization.44 In this process, models for each function are built using randomly permuted descriptors to ensure that the success of the classification model was not due to a chance correlation between one or more descriptors and function. Models were built using 100 random permutations of the descriptors and the mean classification error was computed for comparison with the mean classification error calculated using 5-fold CV.
Validation of all QSURs began with an evaluation of the balanced accuracy of the resulting model; if this value was less than 75%, the models were immediately considered to be invalid. After this, two criteria dependent on each QSUR’s model, 5-fold CV, and Y-randomization classification errors, were used to further validate the models: (1) the mean 5-fold CV error (ĒCV) must be less than the lower bound of the Y-randomization error (ĒYR – σYR) and (2) the model error Emodel must be less than the upper bound of the 5-fold CV error (ĒCV + σCV). If a model failed to meet any of these criteria, it was considered an invalid model. As there was no other set of data which allowed external validation of our QSURs, the average 5-fold CV error was used to evaluate the external predictability of these models rather than being used to optimize the parameters of the models.
In cases where both sets of models (i.e., those constructed using only structural descriptors and those constructed using both structural descriptors and physicochemical properties) yielded a valid model, the better of the two models was determined to be the model with the highest balanced accuracy. Balanced accuracy (the average of the true positive rate and the true negative rate) was selected as the deciding metric in order to optimize the ability of each model to make both true positive (chemical has a function) and true negative (chemical does not have a function) predictions.
Screening for functional substitutes with QSUR models and HTS bioactivity data
We demonstrate the application of our QSUR models for function in high-throughput alternative identification by applying them to a known library of chemical structures for which high-throughput bioactivity data are available. The Tox21 library of chemicals has been screened using a battery of in vitro assays for bioactivity in a concentration response format, with those chemicals causing reproducible, concentration-dependent bioactivity determined to be “hits”.45 All data are publically available from the Integrated Chemical Safety for Sustainability ToxCast dashboard.46 A full descriptor set (containing both structural and physicochemical descriptors) could be obtained for 6672 of the Tox21 chemicals; of these, 2182 chemicals had an HF in FUse. Function classifications were made for all chemicals, even those present in FUse, since they may be functional substitutes for other HFs.
In order to ensure that predictions made on the structure library are within the same chemical structure space as the training set, and are thus valid predictions, we computed the domain of applicability of each model by using the method described by Golbraikh, Tropsha, and others.47,48 Using this method, the Tanimoto distance49 matrix between all chemicals with a given functional use in the training set is calculated. A cutoff distance (Dcutoff) for the domain of applicability is determined via the relationship Dcutoff = D̄ + Zσ, where D̄ and σ are, respectively, the average and standard deviation of the distance between chemicals of a functional use in the training set and Z is a similarity threshold, set at 0.5. We next computed the distance between each chemical in the Tox21 case study predicted to have a functional use and its nearest function neighbor in the training set. If the distance between these chemicals is less than the cutoff distance, then the model for that HF is considered valid for a given chemical.
Chemicals identified as functional substitutes using the QSURs were then assessed using a bioactivity index (BAI) previously developed for prioritization of chemicals identified in non-targeted analyses of environmental media.50 The BAI was calculated from assay results obtained from the EPA’s Tox21 repository (version 20141022).46 The index incorporated results from sixteen assays covering five pathways known to be altered upon exposure to environmental contaminants (aryl hydrocarbon receptor, androgen receptor, estrogen receptor alpha, nuclear factor of kappa light polypeptide gene enhancer in B cells 1, and the peroxisome proliferator-activated receptor gamma).51 In order to achieve consistency across assay results for all five pathways, the number of hit calls (or positive assay results) for each chemical was averaged and normalized by the total number of hit calls for the assays in which that chemical was tested. This resulted in a BAI from 0% (no observed bioactivity) to 100% (all assay tests indicate bioactivity).50
Results
Harmonization of functional use
Rather than building predictive models for redundant reported functions, we aimed to develop a smaller number of representative uses for modeling, which increased the number of example chemicals per functional use and led to some chemicals being identified as serving many different functional uses. After manually eliminating obvious redundancy, there were 244 reported functions and single chemicals still shared multiple functional uses; in one instance, a single chemical possessed 28 reported functions. We therefore applied hierarchical clustering analysis (HCA) to develop harmonized functions (HF). 269 HF clusters – i.e., clusters of similar reported functions – were identified by HCA. These particular clusters capture a significant amount of the variance in pairwise distance between chemicals as described in the Methods section. The most frequently occurring functional uses within each cluster were used to assign a unique HF category label to each cluster. There were several clusters that contained fewer than ten chemicals, and exhibited many reported functions for each chemical. These clusters were combined together to form a “ubiquitous” HF – that is, the chemicals had functional uses that were found in multiple places in functional use space. The creation of the ubiquitous function category was done to provide an opportunity consolidate chemicals that would otherwise be discarded due to small cluster size into a single cluster. In addition, a ubiquitous function allows investigation of chemical features that would contribute to a chemical being able to serve many functional roles. After this reduction there were 137 HF categories.
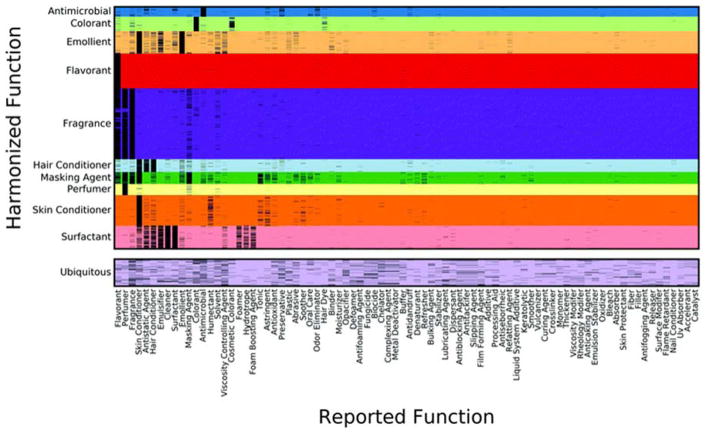
A subset of the resulting harmonized functions (HFs) using HCA are illustrated in Fig. 2. Reported functions (i.e., the function “fingerprints” used to group chemicals into harmonized categories) are shown for the ten largest HFs and the ubiquitous category. In some cases, an HF captured multiple reported functions, indicating that chemicals in that use category could serve multiple functional uses. For example, the “surfactant” HF has such commonly occurring reported functions as ‘surfactant’, ‘cleaner’, ‘hydrotrope’, and ‘emulsifier’. In contrast, the ‘ubiquitous’ HF had no clear grouping of reported function, rather chemicals in this cluster have multiple uses across most of the functional uses listed from the sources. We also find that there are stark HFs such as ‘flavorant’, in which each chemical is known to be only a flavorant in contrast to the fragrance HF, which contains chemicals that are classified as flavorants in addition to fragrances and perfumers.
Fig. 2.
Overlap in reported functions and harmonized functions (HF). The function “fingerprints” of chemicals in the 10 largest HF clusters and the ubiquitous HF cluster in terms of the most frequently occurring original reported functions. Reported functions are on the horizontal axis; harmonized functions are on the vertical axis.
QSUR models and validation
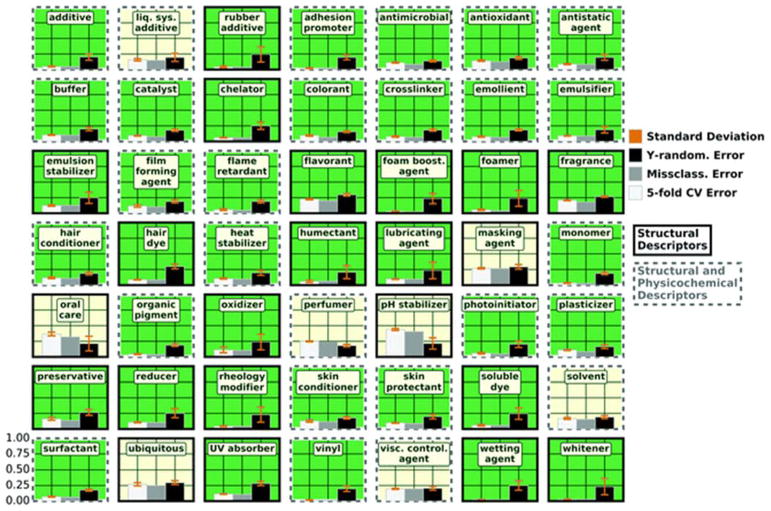
After matching all chemicals in the 137 HF categories to chemicals with available descriptors and eliminating HFs with fewer than 10 chemicals, quantitative structure–use relationship (QSUR) models could be constructed for 49 HFs. Model validation results for these 49 HFs using both structural and physicochemical descriptors are shown in Fig. 3. The bar graph for each model compares the mean 5-fold CV classification error, the mean Y-randomization classification error, and the model classification error. As the balanced accuracies of either QSUR set was typically higher than balanced accuracies of models constructed with only physicochemical properties (see Isaacs et al.17), models using only physicochemical properties were not re-constructed for this analysis. Analyses were performed for each HF in each of the two QSUR sets (i.e., the set of QSURs using structural descriptors, and the set of QSURs using both structural and physicochemical property descriptors). When using only structural descriptors, our workflow yielded valid models for 39 out of 49 HFs. By adding physicochemical descriptors to the QSURs we were able to obtain valid models for two additional HFs for a total of 41 valid models; these results improved upon our previous models that used solely physicochemical descriptors.17 As mentioned in the Methods, when both sets of QSURs gave valid models for an HF, the QSUR model with the highest balanced accuracy was chosen for application purposes.
Fig. 3.
Mean 5-fold cross validation error (white), mean Y-randomization error (black), and model classification error (gray) for each of the 49 harmonized function models constructed. Standard deviations of the 5-fold CV and Y-randomization are shown in orange. Harmonized function QSURs constructed with structural descriptors have a black outline and QSURs constructed with both structural and physicochemical descriptors have a dashed gray outline. A green background indicates a valid model; a pale yellow background indicates an invalid model. The model with the highest balanced accuracy is shown.
There were 8 HFs for which no valid classifier model could be built using either set of descriptors: liquid system additive, masking agent, oral care, perfumer, pH stabilizer, solvent, ubiquitous, and viscosity controlling agent. In general, these categories are much broader than categories that had better models. For example, a viscosity controlling agent could be used to either thicken or thin a solution, which would require two different properties (a thickening or thinning property), however a chelating agent only requires one property (forming multiple single bonds to a single metal ion). It can be assumed that were there an ability, in some cases, to define more specific HFs (e.g., viscosity increasing agent and viscosity decreasing agent rather than viscosity controlling agent) these models would show improvement. It also draws attention to the need for improved, canonical classification of functional uses, either in product reporting or via harmonization methods. For example, the most commonly occurring reported functions within the HF viscosity controlling agent are “viscosity controlling”, “bulking”, and “skin conditioning”. In contrast, the HF labeled rheology modifier, a category that should be just as vague as viscosity controlling agent, has “viscosity modifier”, “thickener”, and “rheology modifier” as the most frequently occurring reported function. Having a large number of thickeners in the HF (as opposed to thickeners and thinners), is likely the reason for this QSUR’s validity. Indeed, property-based methods similar to those used in the field of solvent classification52–54 could be applied in tandem with our method to further refine our HF categorizations. These methods usually classify solvents into sub-categories (e.g., polar vs. non-polar, protic vs. aprotic, etc.). This sub-classification of solvents could result in improved models for the prediction of solvents. As most of our solvents were only labeled as “solvent” in the data sources, further sub-classification was not possible. This is the most probable cause for a poor QSUR for solvents. In the future, further refining of our HFs into subcategories based on properties or structure-based classification could result in better models.
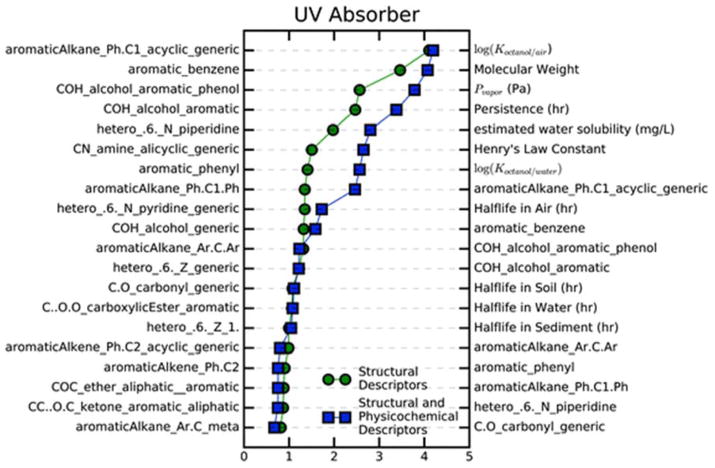
Aside from the power of validated QSURs to predict functions associated with large libraries of chemical structures, these models can aid in understanding the relationship between individual chemical descriptors and use (similarly to how QSARs are used to link biological activity to descriptors). This is readily done via the Gini impurity index, which in addition to aiding in the ensemble learning process of the random forest algorithm, also provides a measure of the relative importance of each descriptor used in a model. Using this model, we are able to identify key features of HFs. The most important descriptors for each of the valid HF models is given in ESI Fig. 1–11.† As an example, mean decrease of the Gini impurity index for the most important descriptors in the UV absorber QSUR are shown in Fig. 4. Here, it can be seen that using only structural descriptors, our models identify aromatic and nitrogen containing substructures as the most important, which are functional group requirements that respectively, make up antioxidants and hindered amines light stabilizers (HALS): two typical categories of industrial UV absorbers. Using this information, one could quickly identify key features of a HF and use these features to identify functional substitutes from other data sources for which full descriptor sets were not available.
Fig. 4.
Ranking of the mean decrease in the Gini index of descriptors for the UV absorber QSURs by importance. The rankings of structural descriptors are shown on the left axis (green circles), and the rankings using both structural and physicochemical descriptors is shown on the right axis (blue squares).
Screening for functional substitutes using QSUR models and HTS bioactivity data
To demonstrate how the QSUR models could be used to screen large libraries of structures, we apply the 41 valid models to predict function of chemicals in the Tox21 library, and assess the associated bioactivity of chemicals within each HF using available high-throughput screening (HTS) data. We then compare the bioactivity of each classified chemical (each “functional substitute”) in each HF with a threshold bioactivity index (BAI),50 calculated as a fixed percentile of the bioactivity index (BAI) of chemicals with the same HF in the functional use database – FUse (i.e., known chemicals with a given HF). By doing so, we can identify in a high-throughput manner a suite of “candidate alternatives” associated with each HF and a given threshold BAI. Chemicals were said to be functional substitutes for an HF if (1) the classification probability returned by the HF QSUR was greater than or equal to a threshold probability (Pr) of 80%, (2) the chemical was not a true positive prediction, and (3) the chemical was within the domain of applicability for the HF model. A chemical was defined to be a candidate alternative for a given HF if it met the following two criteria: (1) the chemical met the criteria of a functional substitute and (2) the BAI for the chemical was below the threshold BAI. Here we selected a threshold BAI for each HF equal to the 75th percentile of the chemicals in the HF in FUse. The threshold BAI for each HF are given in ESI Table 4.† The benefit of using a threshold Pr (80%) and BAI (75th percentile) is that these values can be adjusted depending on the desired stringency of the screening process.
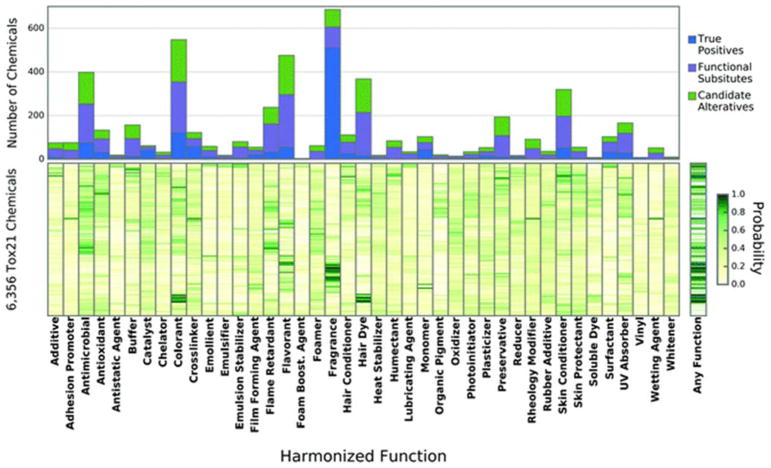
We applied these screening methods to the 6356 chemicals in the Tox21 library which were tested in our selected Tox21 assays (i.e., had a BAI value) and for which structural and physicochemical descriptors could be obtained (i.e., could obtain an HF prediction). Details of how many chemicals from the Tox21 library were unavailable for candidate alternative selection is provided in Table 3. Functional use predictions for each of the chemicals were made using the 41 valid HF classifier models, resulting in the 41-by-6356 matrix of Pr values depicted in the heat map in Fig. 5 (values of predictions with probabilities of at least 80% are available in ESI Table 5†). Predictions were made for all chemicals, even the 2142 chemicals present in FUse, since they may be an alternative for other HFs. Approximately 88% of the predictions yielded a probability less than or equal to 50%, which is consistent of the low false positive rates of the models (minimum false positive rate = 0, mean = 0.15, max = 0.46). The confusion matrices of each model is provided in ESI Table 4.† Of the 260 596 predictions of Pr made, 4412 (roughly 50% of chemicals) returned a probability of ≥80%. The chemicals with multiple HF classifications at a Pr ≥ 80% typically were assigned HFs that were very similar to one another such as emulsifier and surfactant, colorant and hair dye, or skin conditioner and skin protectant, which shows good consistency between models of similar HF. Of the 4412 predictions that classified a chemical to a HF at the threshold Pr, 1326 predictions were for chemicals that were known to have that function in FUse; that is, there were 1326 true positive predictions of HF out of 1544 chemicals in both FUse and Tox21 that had a valid HF model. As these chemicals were in the model training set, they were excluded as alternatives for further BAI screening. However, if that chemical was predicted to have another function aside from its known HF, it was still screened as a candidate alternative for that HF. The remaining 3086 predictions (2227 chemicals) were then screened to determine if they were functional substitutes. There were 2198 functional substitutes (1686 unique chemicals) identified by our screening process (ESI Table 5†).
Table 3.
Number of chemicals available for candidate alternative screening using the developed BAI and harmonized function predictions
| Data curation step | Unique chemicals |
|---|---|
| Tox21 chemical library | 8599 |
| Full QSAR descriptor set available | 6672 |
| BAI available | 6356 |
Fig. 5.
Prediction of harmonized function for 6356 chemicals from the Tox21 library. The horizontal axis represents each of the 41 harmonized functions that were deemed to be valid QSURs. The vertical axis of the lower heat map represents the unique 6356 chemicals for which function predictions were made. Pale yellow indicates a low probability for a chemical to have a functional use, while dark green indicates a high probability of a chemical having a function. The histogram above indicates how many true positive, functional substitutes, or candidate alternatives were predicted for each harmonized function.
The bioactivity indices (BAI) of the chemicals that passed the function screening for each HF were compared to the threshold BAI for that HF. As noted above, the threshold BAI was selected to be the 75th percentile of chemicals known to have each HF. For example, if a chemical predicted to be a colorant had a lower BAI than the 25% of chemicals reported to be a colorant with the highest BAI then that chemical was considered a candidate alternative for colorant. By applying the screening criteria, we found that we were able to identify 1674 candidate alternatives spanning 39 of the 41 HF categories (ESI Table 5†). Table 4 shows how many chemicals were true positive HF predictions, functional substitutes, and candidate alternatives for each HF. The HF categories for which there were no identified functional substitutes were foam boosting agents or vinyls. As classification of a functional substitute is requirement for a chemical to be considered as a candidate alternative, there were no candidate alternatives identified for foam boosting agents or vinyls.
Table 4.
Summary of the number of chemicals from the Tox21 Library that were true positive predictions of chemicals in FUse, functional substitute predictions, and candidate alternatives for that functional use
| Harmonized function | True positive predictions of FUse | Functional substitutes | Candidate alternatives |
|---|---|---|---|
| Additive | 11 | 37 | 27 |
| Adhesion promoter | 4 | 37 | 35 |
| Antimicrobial | 78 | 175 | 145 |
| Antioxidant | 33 | 60 | 40 |
| Antistatic agent | 6 | 8 | 5 |
| Buffer | 15 | 79 | 63 |
| Catalyst | 44 | 12 | 6 |
| Chelator | 5 | 14 | 13 |
| Colorant | 124 | 230 | 194 |
| Crosslinker | 61 | 33 | 28 |
| Emollient | 13 | 27 | 19 |
| Emulsifier | 3 | 10 | 5 |
| Emulsion stabilizer | 8 | 48 | 24 |
| Film forming agent | 23 | 18 | 14 |
| Flame retardant | 35 | 126 | 77 |
| Flavorant | 57 | 239 | 180 |
| Foam boosting agent | 3 | 0 | 0 |
| Foamer | 1 | 35 | 26 |
| Fragrance | 513 | 93 | 80 |
| Hair conditioner | 28 | 50 | 33 |
| Hair dye | 18 | 196 | 154 |
| Heat stabilizer | 1 | 11 | 6 |
| Humectant | 2 | 52 | 30 |
| Lubricating agent | 1 | 24 | 9 |
| Monomer | 46 | 30 | 27 |
| Organic pigment | 3 | 11 | 6 |
| Oxidizer | 4 | 6 | 4 |
| Photoinitiator | 6 | 16 | 12 |
| Plasticizer | 16 | 19 | 18 |
| Preservative | 13 | 95 | 86 |
| Reducer | 8 | 5 | 4 |
| Rheology modifier | 2 | 46 | 43 |
| Rubber additive | 3 | 17 | 16 |
| Skin conditioner | 54 | 143 | 123 |
| Skin protectant | 8 | 27 | 20 |
| Soluble dye | 2 | 4 | 2 |
| Surfactant | 35 | 44 | 24 |
| UV absorber | 30 | 89 | 47 |
| Vinyl | 8 | 0 | 0 |
| Wetting agent | 1 | 27 | 24 |
| Whitener | 0 | 5 | 5 |
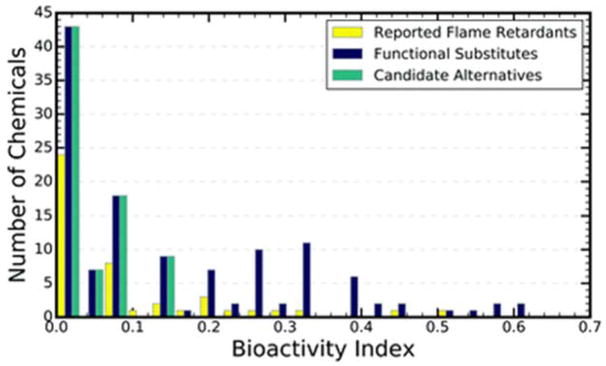
We demonstrate the utility of using this approach by examining the BAI distribution for the HF of flame retardants (shown in Fig. 6). There were 45 chemicals in the Tox21 Library that were identified in FUse to have an HF of flame retardant. The 75thpercentile BAI for these 45 chemicals was 0.125. Table 4 shows that our models predicted 126 chemicals in that same library to be functional substitutes for flame retardants, and that from those substitutes, we were able to identify 77 candidate alternatives. Looking only at known and predicted flame retardants with a BAI less than 0.04 (that is, the lowest histogram bin in Fig. 6), we were able to expand the number of flame retardants in this bin from 24 to 43 (24 known + 43 functional substitutes). Because the BAI in this bin is lower than the 75th percentile, we found that all 43 functional substitutes in this bin were also candidate alternatives.
Fig. 6.
Histogram of the bioactivity index for chemicals with either a known or predicted flame retardant function. Yellow indicates that a chemical was reported to be a flame retardant, violet represents chemicals that were predicted to be flame retardant functional substitutes, while teal indicates that chemicals were candidate alternatives to known flame retardants.
Discussion
Arguments have been made that the method of “drop-in” replacements – replacing a chemical with a structurally similar chemical – can lead to regrettable substitution (that is, replacement chemicals with similar toxicity due to similar structural features).55 Indeed this is the idea behind the International Chemical Secretariat’s (ChemSec) SINimilarity Tool, which identifies if a potential alternative’s structure is too similar to any chemicals on ChemSec’s Substitute It Now (SIN) List.56 Our approach to identifying alternatives based on structural and toxicological information, allows screening of any chemical likely to provide a functional use that is less bioactive than the chemical being replaced. In the case of UV absorbers, this would allow formulators to compare chemicals containing aromatic rings as well as those with sterically hindered amines, thus broadening the search for alternatives to other corners of chemical space.
Admittedly, our implementation of hierarchical clustering analysis (HCA) for functional use harmonization is not the only way to reduce the dimensionality of our FUse data set. The method was chosen because it is a cursory approach to high-throughput categorization, which can be easily automated, and thus allow thousands of chemicals to be incorporated into the FUse dataset. Indeed, other approaches based on grouping physicochemical properties by cluster analysis or principal components analysis (PCA) have been used to categorize solvents for evaluation or selection.52–54 However these methods were less desirable for our purpose, as they are not easily translatable to high-throughput classification.
The bioactivity index used here is intended merely as an illustrative example of potential bioactivity of interest.50 Although high-throughput screening (HTS) data are now available for over 1000 assay endpoints in the ToxCast project57 and greater than 50 assays in Tox21, we focused on five transcription factors known to play important roles in disease pathogenesis, plus a set of cytotoxicity/viability assays to account for general cell-stress and toxicity. The bioactivity index used when screening for alternatives could be broadened to include additional assay endpoints, or narrowed to answer focused questions (e.g., endocrine disruption).
This work demonstrated our ability to rapidly screen a chemical library of roughly 6400 chemicals in order to identify 1674 chemicals that could undergo additional high-tiered screening for functional substitution and toxicity. The set of identified candidate alternatives could further be evaluated with additional in vivo toxicity data or high-level exposure data, or compared with refined information on chemicals previously excluded from a particular functional use (for toxicity, cost, or other reasons). The workflow for our screening methods is highly flexible, allowing one to not only build new models for new HFs, but to also tune the prediction probability and bioactivity index threshold to be more stringent or lax, depending on the user’s need. In addition, we demonstrated the applicability of the QSUR models in HTS by using one high-throughput (HT) metric of toxicity – bioactivity as measured by HTS assays. The functional substitutes identified here could also be screened for potential toxicity using bioactivity metrics developed using structure alerts from other HT tools (e.g., OECD’s QSAR Toolbox,58 and Lhasa’s Derek59). We have demonstrated the application of these approaches to a library of over 6400 chemicals. However, the real power of these methods is that they can be applied rapidly to any number of chemicals for which curated structures are available. The US EPA has recently made great progress in collecting and providing information on large numbers of chemicals, including curated structures, via the Computational Toxicology (CompTox) dashboard.46 The DSSTox database underlying this dashboard currently contains data for over 720 000 chemicals, plans are in place to include the HF model predictions for all chemicals with curated structures as part of the CompTox dashboard in the near future, similar to how the results for QSAR models for physicochemical properties are already publicly available. In addition, raw FUse data (reported/harmonized functions and data sources) will be available on the CompTox dashboard in the near future.
In addition to screening for functional substitutes, the QSUR models developed here have several other promising applications. The models can be used to parameterize HT empirical and mechanistic models that predict the exposure of humans to thousands of chemicals for screening and prioritization, such as those being developed by EPA’s ExpoCast project. Wambaugh et al. developed a linear regression model from easily-obtained general chemical use heuristics that explained more than half the variability in parent chemical exposures inferred from CDC NHANES biomonitoring data.60 The QSUR models could be used to develop additional functional heuristics that could improve the characterization of exposure variability. Furthermore, Isaacs et al. recently developed property-based QSUR models for function and weight fractions for chemicals known to be in specific types of personal care products.17 The structure-based QSURs developed here will expand these methods to address additional function categories and consumer product types. The QSUR models are also being used to interpret the results of new non-targeted analyses of chemicals in various media in support of HT screening and prioritization of chemicals on the basis of exposure potential. For example, the EPA’s ExpoCast project is performing analyses of environmental media (e.g., house dust), biological media, and consumer products using high resolution mass spectrometry. These analyses return large numbers of tentatively identified chemical structures. Applying the QSURs to these structures can provide evidence of sources of chemicals found in the media via these HT analyses. Bioactivity indices created from structure alerts and these models can be applied to theorized chemicals (e.g., a database of molecules that could be synthesized using metabolic engineering). In addition, the important descriptors of each model provide insight into initial functional groups that should be included in the hypothetical molecules. In this way newer, greener functional substitutes could be prioritized for synthesis rather than chemicals that are more likely to be bioactive.
Conclusion
In keeping with the principles of green chemistry, we have developed methods for identifying lists of predicted functional substitutes that can be further screened using measures of differential toxicity (such as HTS or metrics predicted using QSAR or read-across methods). The QSUR models expand the HT aspects of green design beyond these hazard criteria to include chemical use. We were able to build valid classifier models for 41 harmonized function categories; the number of valid models will increase as we expand our function, property descriptor, and structural descriptor databases and further refine our definition categories. These models have promising potential for application in alternatives assessment and exposure-based chemical prioritization.
Supplementary Material
Acknowledgments
The United States Environmental Protection Agency, through its Office of Research and Development’s Chemical Safety for Sustainability research program, provided funding and managed the research described here. This research was supported in part by an appointment to the Postdoctoral Research Program at the National Exposure Research Laboratory, administered by the Oak Ridge Institute for Science and Education through Interagency Agreement No. DW-89-92298301-0 between the U.S. Department of Energy and the U.S. Environmental Protection Agency. The views expressed in this article are those of the authors and do not necessarily represent the views or policies of the U.S. Environmental Protection Agency. Reference to commercial products or services does not constitute endorsement. The authors would like to thank Drs Brandall Ingle and Antony Williams for their helpful review of the manuscript.
References
- 1.Poliakoff M, Fitzpatrick JM, Farren TR, Anastas PT. Science. 2002;297:807–810. doi: 10.1126/science.297.5582.807. [DOI] [PubMed] [Google Scholar]
- 2.U. S. Code, Toxic Substances Control Act, Title 15, Sections 2601–2629.
- 3.Muir DC, Howard PH. Environ Sci Technol. 2006;40:7157–7166. doi: 10.1021/es061677a. [DOI] [PubMed] [Google Scholar]
- 4.Anastas P, Teichman K, Hubal EC. J Exposure Sci Environ Epidemiol. 2010;20:395–396. doi: 10.1038/jes.2010.28. [DOI] [PubMed] [Google Scholar]
- 5.Judson R, Richard A, Dix DJ, Houck K, Martin M, Kavlock R, Dellarco V, Henry T, Holderman T, Sayre P. Environ Health Perspect. 2009;117:685–695. doi: 10.1289/ehp.0800168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wambaugh JF, Setzer RW, Reif DM, Gangwal S, Mitchell-Blackwood J, Arnot JA, Joliet O, Frame A, Rabinowitz J, Knudsen TB. Environ Sci Technol. 2013;47:8479–8488. doi: 10.1021/es400482g. [DOI] [PubMed] [Google Scholar]
- 7.Egeghy PP, Vallero DA, Hubal EAC. Environ Sci Policy. 2011;14:950–964. [Google Scholar]
- 8.Judson R. Handbook of Green Chemistry. Wiley-VCH Verlag GmbH & Co; KGaA: 2010. [Google Scholar]
- 9.Tice RR, Austin CP, Kavlock RJ, Bucher JR. Environ Health Perspect. 2013;121:756–765. doi: 10.1289/ehp.1205784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dix DJ, Houck KA, Martin MT, Richard AM, Setzer RW, Kavlock RJ. Toxicol Sci. 2007;95:5–12. doi: 10.1093/toxsci/kfl103. [DOI] [PubMed] [Google Scholar]
- 11.Lavoie ET, Heine LG, Holder H, Rossi MS, Lee RE, Connor EA, Vrabel MA, DiFiore DM, Davies CL. Environ Sci Technol. 2010;44:9244–9249. doi: 10.1021/es1015789. [DOI] [PubMed] [Google Scholar]
- 12.U. S. Environmental Protection Agency. [accessed April 2016];Safer Choice. https://www.epa.gov/saferchoice.
- 13.U. S. Environmental Protection Agency. Program for Assisting the Replacement of Industrial Solvents. Apr, 2016. [Google Scholar]
- 14.Harten PF. presented in part at the 18th Annual Green Chemistry & Engineering Conference; North Bethesda, MD, USA. 2014. [Google Scholar]
- 15.U. S. Environmental Protection Agency. Program for Assisting the Replacement of Industrial Solvents (PARIS III) User’s Guide. http://nepis.epa.gov/Adobe/PDF/P100HVTD.pdf.
- 16.Leach AR. Molecular Modelling: Principles and Applications. 2. Prentice Hall; 2001. [Google Scholar]
- 17.Isaacs KK, Goldsmith MR, Egeghy P, Phillips KA, Brooks R, Hong T, Wambaugh JF. Toxicol Rep. 2016;3:723–732. doi: 10.1016/j.toxrep.2016.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.European Commission. Cosmetic Ingredient Database. http://ec.europa.eu/growth/tools-databases/cosing/index.cfm?fuseaction=search.simple.
- 19.SpecialChem. [accessed December 2014]; http://www.specialchem.com/
- 20.International Fragrance Association. [accessed March 2015];Ingredients List. http://www.ifraorg.org/en-us/ingredients.
- 21.European Commission. [accessed May 2015];Fl@voring Substances. http://ec.europa.eu/food/food/chemicalsafety/flavouring/database/index.cfm.
- 22.U. S. Environmental Protection Agency. [accessed January 2015];Aggregated Computational Toxicology Resource. http://actor.epa.gov/actor/faces/ACToRHome.jsp;jsessionid=195C8616B5B394EC92C3356AFD9D6F4A.
- 23.U. S. Environmental Protection Agency. [accessed April 2015];Safer Chemical Ingredients List. http://www.epa.gov/saferchoice/safer-ingredients.
- 24.American Cleaning Institute. [accessed March 2015];Cleaning Product Ingredient Inventory. http://www.cleaninginstitute.org/Ingredient_Inventory/
- 25.Johnson SC. [accessed March 2015];What’s Inside SC Johnson? http://www.whatsinsidescjohnson.com/us/en.
- 26.The Clorox Company. [accessed March 2015];Ingredient List. https://www.thecloroxcompany.com/products/ingredients-inside/
- 27. [accessed March 2015];method, Ingredient List. http://methodhome.com/beyond-the-bottle/ingredients/
- 28.Miyamoto S. In: Torra V, Narukawa Y, López B, Villaret M, editors. Modeling Decisions for Artificial Intelligence: 9th International Conference, MDAI 2012; Girona, Catalonia, Spain. November 21–23, 2012; Berlin Heidelberg, Berlin, Heidelberg: Springer; 2012. pp. 1–10. Proceedings. [Google Scholar]
- 29.Leach AR, Gillet VJ. An Introduction to Chemoinformatics. Springer Science & Business Media; 2007. [Google Scholar]
- 30.Toldo R, Fusiello A. In: Forsyth D, Torr P, Zisserman A, editors. Computer Vision – ECCV 2008: 10th European Conference on Computer Vision; Marseille, France. October 12–18, 2008; Berlin Heidelberg, Berlin, Heidelberg: Springer; 2008. pp. 537–547. Proceedings, Part I. [Google Scholar]
- 31.Zhu M, Ghodsi A. Comput Stat Data Anal. 2006;51:918–930. [Google Scholar]
- 32.U. S. Environmental Protection Agency. [accessed October 2015];DSSTox. http://www.epa.gov/ncct/dsstox/index.html.
- 33.Mansouri K, Abdelaziz A, Rybacka A, Roncaglioni A, Tropsha A, Varnek A, Zakharov A, Worth A, Richard AM, Grulke CM. Environ Health Perspect. 2016;124(7):1023–1033. doi: 10.1289/ehp.1510267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Molecular Networks GmbH. The ChemoTyper. 2013 https://chemotyper.org/
- 35.Yang C, Tarkhov A, Marusczyk J, Bienfait B, Gasteiger J, Kleinoeder T, Magdziarz T, Sacher O, Schwab CH, Schwoebel J, Terfloth L, Arvidson K, Richard A, Worth A, Rathman J. J Chem Inf Model. 2015;55:510–528. doi: 10.1021/ci500667v. [DOI] [PubMed] [Google Scholar]
- 36.U. S. Environmental Protection Agency. Estimation Programs Interface Suite for Microsoft Windows v 4.11. https://www.epa.gov/tsca-screening-tools/epi-suitetm-estimation-program-interface.
- 37.Breiman L. Mach Learn. 2001;45:5–32. [Google Scholar]
- 38.Liaw A, Wiener M. R News. 2002;2:18–22. [Google Scholar]
- 39.The R Project for Statistical Computing, Version 3.1.2 (Pumpkin Helmet) 2014. [Google Scholar]
- 40.Chen C, Liaw A, Breiman L. D. o. Statistics Report 666. University of California; Berkeley: 2004. Using random forest to learn imbalanced data. [Google Scholar]
- 41.Louppe G, Wehenkel L, Sutera A, Geurts P. Advances in Neural Information Processing Systems. 2013:431–439. [Google Scholar]
- 42.Gramatica P. QSAR Comb Sci. 2007;26:694–701. [Google Scholar]
- 43.Fawcett T. Pattern Recognit Lett. 2006;27:861–874. [Google Scholar]
- 44.Rücker C, Rücker G, Meringer M. J Chem Inf Model. 2007;47:2345–2357. doi: 10.1021/ci700157b. [DOI] [PubMed] [Google Scholar]
- 45.Judson R, Houck K, Martin M, Richard AM, Knudsen TB, Shah I, Little S, Wambaugh J, Setzer RW, Kothya P, Phuong J, Filer D, Smith D, Reif D, Rotroff D, Kleinstreuer N, Sipes N, Xia M, Huang R, Crofton K, Thomas RS. Toxicol Sci. 2016 doi: 10.1093/toxsci/kfw148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.U. S. Environmental Protection Agency. iCSS ToxCast Dashboard. http://actor.epa.gov/dashboard/
- 47.Golbraikh A, Shen M, Xiao ZY, Xiao YD, Lee KH, Tropsha A. J Comput-Aided Mol Des. 2003;17:241–253. doi: 10.1023/a:1025386326946. [DOI] [PubMed] [Google Scholar]
- 48.Tropsha A, Golbraikh A. Curr Pharm Des. 2007;13:3494–3504. doi: 10.2174/138161207782794257. [DOI] [PubMed] [Google Scholar]
- 49.Willett P, Winterman V, Bawden D. J Chem Inf Comput Sci. 1986;26:36–41. [Google Scholar]
- 50.Rager JE, Strynar MJ, Liang S, McMahen RL, Richard AM, Grulke CM, Wambaugh JF, Isaacs KK, Judson R, Williams AJ, Sobus JR. Environ Int. 2016;88:269–280. doi: 10.1016/j.envint.2015.12.008. [DOI] [PubMed] [Google Scholar]
- 51.Rager JE, Fry RC. In: Network Biology. Zhang WJ, editor. Nova Science Publishers; 2013. pp. 81–130. [Google Scholar]
- 52.Chastrette M, Rajzmann M, Chanon M, Purcell KF. J Am Chem Soc. 1985;107:1–11. [Google Scholar]
- 53.Gu CH, Li H, Gandhi RB, Raghavan K. Int J Pharm. 2004;283:117–125. doi: 10.1016/j.ijpharm.2004.06.021. [DOI] [PubMed] [Google Scholar]
- 54.Tobiszewski M, Tsakovski S, Simeonov V, Namieśnik J, Pena-Pereira F. Green Chem. 2015;17:4773–4785. [Google Scholar]
- 55.(a) Tickner JA, Geiser K, Rudisill C, Schifano J. Chemical Alternatives Assessments, Royal Society of Chemistry. 2013:256–295. [Google Scholar]; (b) Tickner JA, Schifano JN, Blake A, Rudisill C, Mulvihill MJ. Environ Sci Technol. 2015;49:742–749. doi: 10.1021/es503328m. [DOI] [PubMed] [Google Scholar]
- 56.International Chemical Secretariat. SINimilarity Tool. 2015 Nov; http://sinimilarity.chemsec.org/
- 57.Richard AM, Judson RS, Houck KA, Grulke CM, Volarath P, Thillainadarajah I, Yang C, Rathman JF, Martin MT, Wambaugh JF. Chem Res Toxicol. 2016;29:1225–1251. doi: 10.1021/acs.chemrestox.6b00135. [DOI] [PubMed] [Google Scholar]
- 58.The Organisation for Economic Co-operation and Development. QSAR Toolbox. http://www.oecd.org/chemicalsafety/risk-assessment/theoecdqsartoolbox.htm.
- 59.Lhasa Limited. Derek Nexus. 2015 http://www.lhasalimited.org/products/derek-nexus.htm.
- 60.Wambaugh JF, Wang A, Dionisio KL, Frame A, Egeghy P, Judson R, Setzer RW. Environ Sci Technol. 2014;48:12760–12767. doi: 10.1021/es503583j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.