Abstract
The olive moth -Prays oleae Bern.- remains a significant pest of olive trees showing situation dependent changes in population densities and in severity of damages. The genetic variability of olive moth was assessed on three main olive orchards regions in Portugal by three different markers (COI, nad5 and RpS5), suggesting high species diversity albeit with no obvious relation with a regional pattern nor to an identified ecological niche. Selected COI sequences obtained in this study were combined with those available in the databases for Prays genus to generate a global dataset. The reconstruction of the Prays phylogeny based on this marker revealed the need to revise Prays oleae to confirm its status of single species: COI data suggests the co-existence of two sympatric evolutionary lineages of morphologically cryptic olive moth. We show, however, that the distinct mitochondrial subdivision observed in the partial COI gene fragment is not corroborated by the other DNA sequences. There is the need of understanding this paradigm and the extent of Prays variability, as the disclosure of lineage-specific differences in biological traits between the identified lineages is fundamental for the development of appropriate pest management practices.
Introduction
Olive is an ancient ubiquitous crop of considerable socioeconomic importance, being a major agro-ecosystem in the Mediterranean basin. For the Mediterranean region, three main olive pests have been recognized: the olive fruit fly, Bactrocera oleae Gmelin, the olive moth, Prays oleae Bern. and the black scale, Saissetia oleae Bern. [1–3]. The importance of the last two has decreased as a whole due to advances in olive pest management [3], but regional relevance persists.
The olive moth, Prays oleae (Lepidoptera, Yponomeutidae) remains an abundant pest of olive trees throughout the Mediterranean and the Black Sea, the Middle East and Canary Islands [4]. Undertaking three generations per year, and with the larval stages attacking different organs of the tree, its’ action can increase fruit fall and damage leaves, flowers and fruits. The olive moth is thus being held responsible for high losses in the olive yield [5], lowering tree growth, fruit set and fruit/oil quality. In north Portugal this moth competes in importance with the olive fruit fly, being considered the most important olive tree pests due to the large production losses [6]. There is empirical indication that the seriousness of the losses due to Prays oleae are highly variable, depending both on time (crop seasons) and space (regions).
The degree of synchrony between adult emergence and the olive fruit suitability for oviposition by egg-laying moth females varies greatly from year to year [7]. This synchrony can account for part of the seasonal variability of the losses caused by the olive moth. Predators (like ants, chrysopids, anthocorids and spiders; e.g.[8]) and parasitoids (mainly egg parasitoid Trichogramma species, but others have also been referenced, e.g. [9–11]) are also likely responsible for the observed variability, both seasonal and regional. The state and composition of the functional diversity associated to the olive grove is highly affected by crop management practices (including tillage and the use of pesticides) impacting also at a landscape/regional scale (e.g. [12,13]).
Agricultural systems, including olive groves, form a mosaic at a landscape scale shifting both in time and space, that might induce differentiation and determine the population structure of the Prays oleae (as it depends on olive trees for survival). Because genetic variation is essential for the adaptability of a population, the selection of fitness‐related traits might be driving changes in population densities and severity of the pest.
In this study we look into the genetic variability of Prays oleae on three main olive grove regions in Portugal by means of sequencing two selected mitochondrial DNA amplicons and a nuclear DNA amplicon. To the best of our knowledge, this is the first systematic study looking into the population(s) genetic variability of Prays oleae. The reconstruction of the Prays phylogeny based on COI revealed the need to revise Prays oleae to confirm its status of single species and assess its relation with other Prays species, in particular with Prays fraxinella. Furthermore, the phylogenetic and the network analyses of the variability here performed suggest cryptic species diversity of the olive moth, albeit not clearly linked to regional patterns.
Methodology
Taxon sampling and data collection
No specific permissions are required to sample olive moth. All the samples were obtained from the monitoring services of the Portuguese Ministry of Agriculture or from private land with the permission of the owners and did not involve endangered or protected species. Biological material was collected from 28 sites (Fig 1) using commercial sticky traps with specific pheromones (Biosani) during the summer of 2017. The installation and collection of the traps was partially performed by local associations and/or by the Regional Services for Agriculture (S1 Table). The traps stayed for at least a week at the designated olive locations, most of them used to monitoring the pest, and were then transported to the laboratory in individual plastic bags. The trapped adults, putatively belonging to Prays oleae, were collected from the traps and stored at -20°C in 70% ethanol until DNA extraction. Individuals were allowed to dry on filter paper prior to DNA extraction. DNA from whole body tissue was extracted following extraction protocols using CTAB extraction buffer [14] after being ground up with a plastic pestle. Proteins were removed with 24 : 1 isoamylalcohol : chloroform, and DNA precipitated with isopropanol. DNA extracts were eluted in 50 μL of sterile water. All extraction products were stored at -20°C and later used directly in the PCR.
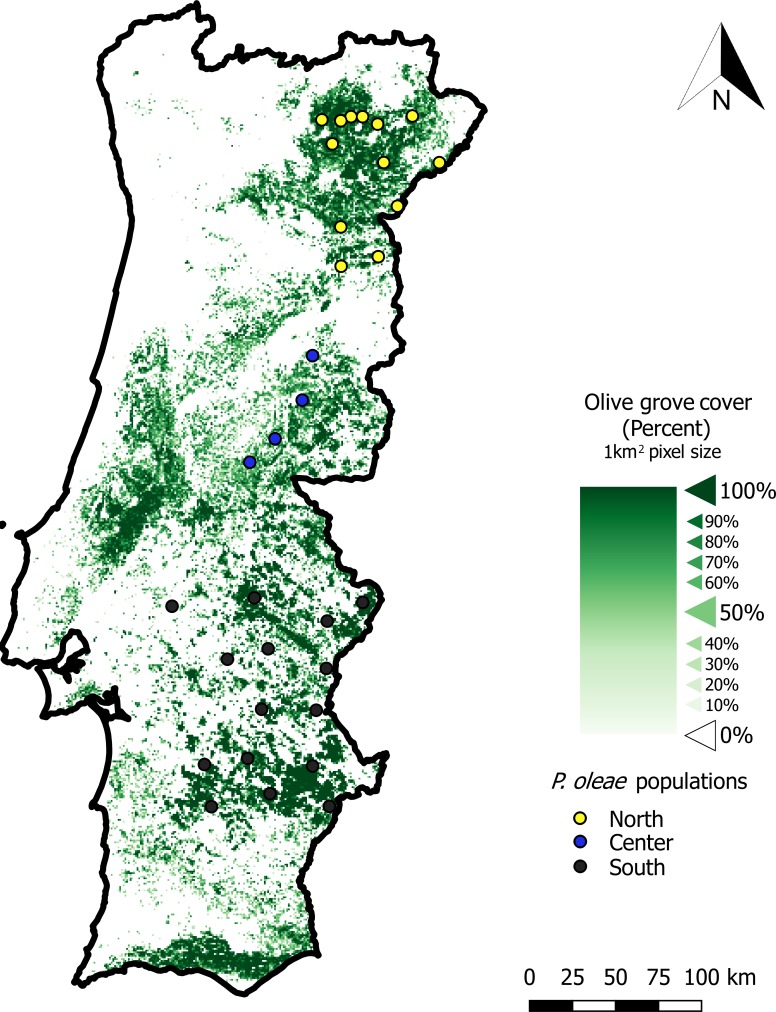
Fig 1. Prays sampling sites (colored dots), discriminated by the three regions (North, Center and South).
Green color on background represents the density of olive groves per square kilometer, ranging from low density (light green) to high density (dark green).
Prays sequences
Two mitochondrial genes (COI and nad5) and one protein-coding nuclear gene region (RpS5) were amplified using the following primer pairs: 1) LCO1490 (5’- GGT CAA CAA ATC ATA AAG ATA TTG G -3' and HCO2198 (5’- TAA ACT TCA GGG TGA CCA AAA AAT CA -3’) for a fragment of the cytochrome c oxidase subunit I (COI) gene [15]; 2) nad5_fw (5’- TTA TAT CCT TAG AAT AAA ATC C -3’) and nad5_rev (5’- TTA GGT TGA GAT GGT TTA GG -3’) for a fragment of the NADH dehydrogenase subunit 5 (nad5) gene [16] and 3) RpS5_f (5’- ATG GCN GAR GAR AAY TGG AAY GA -3’) and RpS5_r (5’- CGG TTR GAY TTR GCA ACA CG -3’) for a fragment of the ribosomal protein S5 (RpS5) gene [17]. PCR reactions were conducted using 1 μl of the extracted DNA in a standard 25 μl reaction, with 0.5 pmol/μl of each primer, 1.5 mM MgCl2, 0.5 mM dNTPs and 0.04 U/ml Taq DNA polymerase. The cycle protocol involved initial denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, gene-specific annealing temperatures (55°C for COI and nad5, and 53°C for RpS5) for 30 s and 72°C for 1 min, and an extension cycle of 72°C for 7 min; the PCR product was purified using the NZYGelpure kit (from NZYTech, Lda) and sequencing was done commercially (Macrogen Inc.). The sequences were assembled, edited and aligned using the CLC Main Workbench version 7.5.1 (Qiagen Aarhus A/S, Denmark) (all sequences generated in this study and their GenBank accession numbers are in S2 Table). The COI sequences obtained in this study were organized as haplotypes, and a subset of representative COI haplotypes were selected based on high level of differentiation (no. of differences) and on their representativeness in the sample. These were combined with those available for the genus Prays from GenBank to generate a global dataset.
Phylogenetic analysis
The phylogenetic reconstruction analysis based on the COI sequences was performed in BEAST v.4.2.8 [18]. We selected the Gamma Site Model with 4 gamma categories, and rate frequencies were estimated. All other settings were left as default, including the chain length of 10 000 000 generations. The output of BEAST was analysed in the software Tracer v.1.6 to determine chain convergence and burnin. The majority rule consensus tree was obtained from the trees sampled in the analysis using the program TreeAnnotator v.2.4.8, considering a burn-in of 10% (first 1000 trees were removed). Reconstructions with Maximum Likelihood and Neighbor-Joining methods as implemented in Mega 7.0 [19] were performed for testing for congruence between methods (S1 and S2 Figs). To test the monophyly of Prays oleae, we inferred the phylogeny once without constraints and once with all accessions of P. oleae constrained to be monophyletic, to evaluate the likelihood of this alternative phylogenetic relationship. Bayes factors were used to test if the topological constrained topology was significantly different than the unconstrained topology, and was measured using twice the difference of −ln likelihood (2lnBF) with 2lnBF = 0–2 meaning not worth a mention, 2lnBF = 2–6 meaning positive support, 2lnBF = 6–10 meaning strong support, and 2lnBF > 10 meaning decisive support [20].
Variability and population structure
Sequence variability analyses of the three DNA fragments analysed were performed in DnaSP v. 4.0 [21]. For haplotype and nucleotide diversity estimates (Hd and Pi), we chose to analyze synonymous and non-synonymous sites jointly because if analyzed separately, the number of sites would have been too low to yield reliable results [22]. Tajima’s D statistics compares the average number of pairwise differences with the number of segregating sites [23]. Over the all sequenced fragments, linkage disequilibrium was measured using the ZnS statistic (the squared allele frequency correlation r2 [24]) on the basis of the parsimony informative sites. Statistical significance for ZnS and Tajima’s D was assessed by coalescent simulations with 10 000 replicates as implemented in DnaSP v. 4.0 [21], conducted considering all segregating sites and an intermediate level of recombination. These analyses were performed for the sequence data of the three amplicons independently and concatenated. A haplotype network approach was chosen for a concise representation of the dataset obtained in this work, both concatenated and all three regions separately. The haplotype networks were constructed in PopART [25] using TCS network (95% connection limit).
Ecological niche modelling
Georeferenced Prays oleae capture sites were used to obtain Ecological Niche Models (ENM’s) employing the maximum entropy algorithm in Maxent 3.4.0 (Maximum entropy modeling of species geographic distributions). Predictions were based on a set of 16 environmental data maps including bioclimatic [26] and land cover derived maps (S3 Table). This information was prepared for the area where Prays captures were registered (NUTS3 European administrative limits were considered) and exported to ASCII grid format with 1 km2 resolution using QGIS [27]. Among highly correlated covariables (Pearson correlation coefficient R > 0.75, ENMTools 1.4.4 [28,29]) only the ones presenting the highest percentage of importance to the model in a preliminary run was retained for further procedures [30] (S3 Table). Prediction models were run for 10 interactions using 50% random records to test each run, and the model quality was accessed by area under the curve (AUC scores above 0,7 are acceptable for a good model performance, [31]).
For the main Prays groups (defined in the phylogenetic analyses), the ENM’s similarity was accessed by calculating Schoener’s D [32] and Hellinger’s I [29] indices and then preforming an identity test (run with 300 pseudoreplicates, ENMTools 1.4.4 [28]).
Results
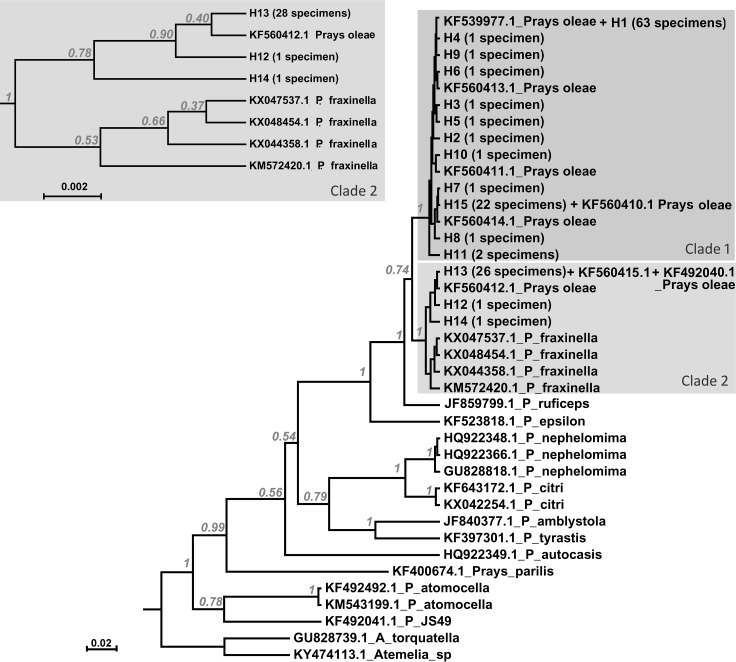
The reconstructed unconstrained phylogeny of the genus Prays based on COI (Fig 2) resolved Prays oleae as paraphyletic. This model is significantly better than when monophyly of P. oleae samples is constrained (2·lnBF = 33.17; with BI ln [unconstrained model] = -2830.90 and BI ln [alternative model] = -2847.49).
Fig 2. Phylogenetic relationship between Prays oleae and other Prays species with data available on GenBank (accession code given on tree), based on the COI amplicon.
The phylogeny corresponds to the majority rule consensus tree of trees sampled in a Bayesian analysis, and the posterior probability values are shown for main nodes (xml input files available in S1 File). Two Atemelia species were used as outgroups. Similar topologies were obtained using Maximum Likelihood and Neighbor-Joining methods as implemented in Mega 7.0 [19] (S1 and S2 Figs) and will further not be discussed. The top left grey window highlights a clade showing Prays oleae specimens nesting in the Prays fraxinella clade, an unexpected result. Prays oleae samples were collected from Portugal with exception of KF560413.1, KF560414.1, and KF560415.1 which were collected in Tunisia and KF492040.1 from Spain. The tip labels represent either the haplotypes and number of specimens with that same haplotype (details in S2 Table) or the GenBank accession number.
The three amplicons–COI, nad5 and RpS5– were sequenced for 128 specimens spanning relevant olive groves areas in Portugal and their variability (number of haplotypes, polymorphic sites and diversity estimates) is presented in Table 1. Because of the observed non-monophyly of Prays oleae (Fig 2), we present the same estimates separately for the samples nested in clade 1 or in clade 2 (Table 1). Levels of nucleotide diversity for the mtDNA amplicons are equivalent between sequenced regions, with the numbers of non-synonymous changes being half of the synonymous ones. An exception to this is in clade 2, where the two mitochondrial regions behave differently (Table 1). Contributing to this might be the low number of samples analysed. The fragment of the nuclear gene encoding for the ribosomal protein S5a (RpS5) shows almost equivalent numbers of non-synonymous and synonymous sites (except for clade 2; again low number of samples needs to be kept in mind). Considering the full dataset together, no linkage disequilibrium was detected and the Tajima’s D statistics was non-significant for all markers suggesting that these DNA sequences have evolved randomly (‘neutrality’) (Table 2). When analyzing clade 1 and clade 2 separately, the Tajima’s D statistics for the mitochondrial COI marker is in both cases significant (Table 2).
Table 1. Sequence variability analyses of the three DNA fragments analysed, considering the complete dataset and partitioned by clade as identified in Fig 2.
| COI | nad5 | RpS5 | all markers | |
|---|---|---|---|---|
| Number of sequences | 128 | 128 | 128 | 128 |
| Number of sites (bp) | 620 | 676 | 517 | 1813 |
| Number of haplotypes | 26 | 31 | 19 | 85 |
| Polymorphic sites (S) | 43 | 41 | 22 | 106 |
| Parsimony informative | 21 | 25 | 8 | 54 |
| Total number of mutations | 46 | 41 | 22 | 109 |
| Synonymous changes | 32 | 27 | 12 | 71 |
| Non-Synonymous | 14 | 14 | 10 | 38 |
| Haplotype diversity (Hd) | 0.754 | 0.891 | 0.733 | 0.984 |
| Aver. nucleotide diff. (k) | 6.243 | 7.435 | 1.857 | 15.536 |
| Nucleotide diversity (Pi) | 0.010 | 0.011 | 0.004 | 0.008 |
| Prays oleae Clade 1 | ||||
| Number of sequences | 101 | 101 | 101 | 101 |
| Number of haplotypes | 23 | 27 | 17 | 68 |
| Polymorphic sites (S) | 23 | 39 | 16 | 78 |
| Parsimony informative | 7 | 21 | 7 | 35 |
| Total number of mutations | 23 | 39 | 16 | 78 |
| Synonymous changes | 16 | 27 | 9 | 52 |
| Non-Synonymous | 7 | 12 | 7 | 26 |
| Haplotype diversity (Hd) | 0.663 | 0.870 | 0.752 | 0.983 |
| Aver. nucleotide diff. (k) | 1.698 | 4.176 | 1.827 | 7.701 |
| Nucleotide diversity (Pi) | 0.003 | 0.006 | 0.003 | 0.004 |
| Prays oleae Clade 2 | ||||
| Number of sequences | 27 | 27 | 27 | 27 |
| Number of haplotypes | 3 | 7 | 9 | 17 |
| Polymorphic sites (S) | 12 | 23 | 12 | 47 |
| Parsimony informative | 0 | 20 | 3 | 23 |
| Total number of mutations | 12 | 22 | 12 | 46 |
| Synonymous changes | 5 | 20 | 8 | 33 |
| Non-Synonymous | 7 | 2 | 4 | 13 |
| Haplotype diversity (Hd) | 0.145 | 0.601 | 0.561 | 0.869 |
| Aver. nucleotide diff. (k) | 0.889 | 5.322 | 1.692 | 7.903 |
| Nucleotide diversity (Pi) | 0.002 | 0.008 | 0.003 | 0.004 |
Table 2. Population genetics inferences based on Tajima's D, the site frequency spectrum (SFS) of mutations; and ZnS, the statistical association among those (linkage disequilibrium).
The same statistics are presented for the dataset portioned by clade as identified in Fig 2.
| Tajima´s D | Significance* | ZnS | Significance* | |
|---|---|---|---|---|
| All Prays oleae sequences (n = 128) | ||||
| COI | -0.650 | p = 0.26; [-1.54, 1.94] | 0.123 | p = 0.61; [-0.03, 0.34] |
| nad5 | -0.049 | p = 0.56; [-1.53, 1.88] | 0.118 | p = 0.59; [0.04, 0.31] |
| RpS5 | -1.544 | p = 0.01; [-1.32, 1.57] | 0.076 | p = 0.592; [0.03, 0.16] |
| All markers | -0.660 | p = 0.19; [-1.19, 1.14] | 0.062 | p = 0.297; [0.04, 0.14] |
| Prays oleae Clade 1 (n = 101) | ||||
| COI | -1.818 | p = 0.00*; [-1.36, 1.44] | 0.018 | p = 0.00*; [-0.03, 0.16] |
| nad5 | -1.385 | p = 0.02; [-1.34, 1.42] | 0.095 | p = 0.74; [0.04, 0.15] |
| RpS5 | -1.139 | p = 0.07; [-1.37, 1.66] | 0.063 | p = 0.38; [0.03, 0.20] |
| All markers | -1.589 | p = 0.00*; [-1.37, 1.66] | 0.033 |
p = 0.00*; [0.04, 0.15] |
| Prays oleae Clade 2 (n = 27) | ||||
| COI | -2.386 | p = 0.00*; [-1.45, 1.49] | 0.834 | p = 1.00; [0.06, 0.34] |
| nad5 | -0.389 | p = 0.31; [-1.41, 1.37] | 0.631 | p = 1.00; [0.07, 0.27] |
| RpS5 | -1.524 | p = 0.02; [-1.43, 1.50] | 0.212 | p = 0.85; [0.06, 0.33] |
| All markers | -1.332 | p = 0.01; [-1.27, 1.35] | 0.222 | p = 0.94; [0.08, 0.26] |
*p-value; 99% confidence interval
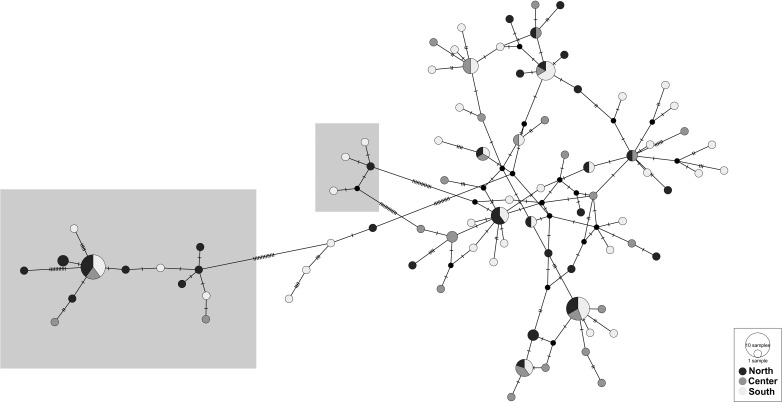
The haplotype network showed a complex and diversified topology, consisting of one main star-like arrangement with satellites and a slightly more complex adjacent network (Fig 3, haplotype networks partitioned per gene are presented in S3 Fig). No obvious relation with sampling location (North, Center or South) is observed, being the clearest pattern coming from the two P. oleae clades previously identified in Fig 2.
Fig 3. TCS haplotype networks based on the three markers (COI, nad5 and RpS5).
Each circle represents a sequence; the size of the circle is proportional to number of individuals with a particular haplotype sequence. The connections are mutational steps between individuals. The grey windows highlight the Prays oleae clade 2 (Fig 2).
Maxent ENM’s for both clades showed a good predictive ability (clade 1, test AUC ± SD = 0.80 ± 0.07; clade 2, test AUC ± SD = 0.86 ± 0.07). The percentage contribution of each predictor showed, as obviously expected, that “distance to homogeneous olive groves (> 1 ha)” had the greatest contribution to clade 1 (79.5%) and clade 2 (87.7%) models, being followed by continentally (in one or another index form; Table 3). The overlap indices obtained for clade 1 and clade 2 models (Schoener’s D = 0.72, Hellinger’s I = 0.94) were not inferior to the null distribution of 300 pseudoreplicates (Schoener’s D ± SD = 0.71 ± 0,04,Hellinger’s I ± SD = 0.92 ± 0,02), indicating identical niche models.
Table 3. Relative importance to ENM-variables for both identified Prays oleae clades.
The absence of contribution score (-) means the variable was not included in the respective clade final model.
| Variables description | Contribution (%) | |
|---|---|---|
| clade 1 | clade 2 | |
| Distance to homogeneous olive groves (> 1 ha) | 79.5 | 87.7 |
| Simple continentality (Rivas-Martínez 2007, 2008, 2011) | 11.6 | - |
| Distance to riparian gallery | 5.5 | - |
| Mean temperature of the warmest month of the year | 1.8 | - |
| Density of olive groves per km2 | 1.5 | 3.2 |
| Simple continentality index | - | 4.8 |
Discussion
The unconstrained topology (Fig 2) resolves Prays oleae as non-monophyletic, questioning its’ species status. Constraining the olive moth to monophyly resulted in a topology significantly worse than the unconstrained one. From the samples obtained in the present work, about one third forms a well-supported clade with Prays fraxinella (clade 2), while the rest is sister to that clade (clade 1) (Fig 2). Only eight records of Prays oleae were available for the cytochrome c oxidase subunit I (COI) gene, comprising samples from Portugal (4), Tunisia (3) and Spain (1), but specimens belonging to both Prays oleae putative clades were found in all three countries suggesting the existence of two sympatric evolutionary lineages of cryptic olive moths with no described phenotypical differences.
However, we need to acknowledge the possibility that the distinct mitochondrial subdivision observed in the partial COI gene fragment of P. oleae might not be well corroborated by other DNA sequences (particularly nuclear), genital morphology, mating behaviour or ecological niche. Actually, and contrary to our hypothesis, the observed non-monophyly of Prays oleae does not seems to be reflected in a different use of the habitat by the specimens belonging to the identified clades (the niche modelling showed no niche differentiation for the variables used). If we are indeed dealing with two differentiated lineages, they either have the same ecological niche or we were unable to identify the divergent axis of their niche.
Also population statistics were congruent regardless of grouping the samples by P. oleae clade. Tajima’s D statistics provided the main exception, with the mitochondrial marker COI being significant only when analyzing the two clades separately. The significant negative value of Tajima’s D suggests either a recent selective sweep (or linkage to it) or a recent population expansion following a bottleneck, as these values are negative when there is an excess of rare variants. The fact that this was only observed for the COI marker and that the impact of difference in sample size on estimates of Tajima’s D is difficult to track analytically (because both sampling strategy and species demographic history can have impact on the estimate; see, for example [33]), we cannot conclude on population dynamics.
The selection of markers for a given analyses is critical for results interpretation. For phylogenetic studies, the advantages of mitochondrial DNA are well known: 1) strict maternal transmission; 2) high mutation rate and 3) conserved simple structure, allowing the design of “universal” primers. Protein coding genes seem to be the most useful when dealing with taxonomic levels such as families, genera and species, and amongst these, the COI is found to be the best and most widely used molecular marker for DNA barcoding, species identification and evolutionary studies [34]. In the case of the present study, and based on the reported reliability of the COI gene, we could conclude that P. oleae variability and its non-monophyly suggest cryptic species diversity questioning the phylogenetic relation between P. oleae and other Prays species, in particular with P. fraxinella. However, and considering the results and constraints presented, any conclusions require caution. A taxonomic revision is doubtlessly needed, as a correct identification is the basis for the management of species in the ecosystem and hence crucial for risk assessment and pest control in the agricultural context. Taxon sampling influences phylogenetic inferences, and it should be broadened to include more representatives of the species within the genus Prays and from a wider geographical area. While no complete genomes and only the Prays oleae mitogenome are available [35], the phylogeny within this genus needs to be tackled through the analyses of more molecular markers than the COI gene, towards an understanding of speciation and eventual hybridizations.
Molecular genetic methods have been uncovering cryptic lineages [22,36–38] but the extent to which this genetic variation affects phenotypic traits is unclear. The genetic divergence observed within cryptic lineage complexes may result in differences other than morphological ones: traits related to intraspecific competition and predator avoidance, for instance, can vary, making generalizations regarding their response to environmental variables inappropriate if made assuming one single lineage [39]. In the case of the olive moth, a recognized pest of olives, the foretold existence of two cryptic lineages with potential deviation in traits might have a high impact in the agro-ecosystem management. It urges thus to confirm the findings of the present work, by expanding the sampling throughout the species distribution. Furthermore, the disclosure of lineage-specific differences in biological traits between the identified lineages is fundamental for the development of appropriate pest management practices.
Supporting information
(PDF)
(PDF)
TCS haplotype networks of sampled specimens, considering a) the mitochondrial marker COI, b) the mitochondrial marker nad5, and c) the nuclear marker RpS5. Each circle represents a sequence; the size of the circle is proportional to number of individuals with a particular haplotype sequence.
(PDF)
Locations with alphabetic code only were sampled by local associations and/or by the Regional Directorates for Agriculture.
(PDF)
Code corresponds to the location code as in Table 1 followed by specimen number.
(PDF)
(PDF)
(ZIP)
Acknowledgments
The authors wish to thank Nuno Gracinhas Guiomar for the facilitation of the maps and support in the analyses and the valuable suggestions of the reviewers.
Data Availability
All sequences are available from the NCBI database; accession numbers are included in S2 Table. Alignments, scripts and tree files were submitted as supplemental material.
Funding Statement
This work, including TN and LG research fellowships, was supported by the project ‘Integrated protection of the Alentejo olive grove. Contributions to its innovation and improvement against its key enemies’ with the reference ALT20-03-0145-FEDER-000029, co-financed by the European Union through the European Regional Development Fund, under the ALENTEJO 2020 (Regional Operational Program of the Alentejo). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Haber G, Mifsud D. Pests and Diseases Associated With Olive Trees in the Maltese Islands (Central Mediterranean). Cent Mediterr Nat. 2007;4: 143–161. [Google Scholar]
- 2.Delrio G. Biological control of olive pests in the Mediterranean region. Integrated Protection of Olive Crops wprs Bull 53 2010. pp. 85–92. [Google Scholar]
- 3.Haniotakis GE. Olive pest control: Present status and prospects. Integrated Protection of Olive Crops wprd Bull. 2005. pp. 1–9. [Google Scholar]
- 4.Tzanakakis ME. Seasonal development and dormancy of insects and mites feeding on olive: a review. Netherlands J Zool. 2003;52: 87–224. [Google Scholar]
- 5.Ramos P, Campos M, Ramos JM. Long-term study on the evaluation of yield and economic losses caused by Prays oleae Bern. in the olive crop of Granada (southern Spain). 1998;17: 645–647. [Google Scholar]
- 6.Bento A, Torres L, Lopes J. Avaliação de prejuízos causados pela traça da oliveira, Prays oleae (Bern.) em Trás-os-Montes. Rev Ciencias Agrar. 2001;24: 89–96. [Google Scholar]
- 7.Ramos P, Rosales R, Sabouni I, Garrido D, Ramos JM. Crop losses due to olive moth mediated by ethylene. Pest Manag Sci. 2008;724: 720–724. 10.1002/ps [DOI] [PubMed] [Google Scholar]
- 8.Morris TI, Campos M, Kidd NAC, Jervis MA, Symondson WOC. Dynamics of the predatory arthropod community in Spanish olive groves. 1999;
- 9.Pelekassis CD. A contribution to the study or nomenclature, taxonomy, biology, ecology and the natural parasitisation of the olive kernel borer (Prays oleae (Bernard) Lesne). 1962.
- 10.Hegazi E, Herz A, Hassan SA, Khafagi WE, Agamy E, Zaitun A, et al. Field efficiency of indigenous egg parasitoids (Hymenoptera, Trichogrammatidae) to control the olive moth (Prays oleae, Lepidoptera, Yponomeutidae) and the jasmine moth (Palpita unionalis, Lepidoptera, Pyralidae) in an olive plantation in Egypt. Biol Control. 2007;43: 171–187. 10.1016/j.biocontrol.2007.07.009 [Google Scholar]
- 11.Nave A, Gonçalves F, Teixeira R, Costa CA, Campos M, Torres L. Hymenoptera parasitoid complex of Prays oleae (Bernard) (Lepidoptera: Praydidae) in Portugal. 2017; 502–512. 10.3906/zoo-1603-50 [Google Scholar]
- 12.Altieri MA. The ecological role of biodiversity in agroecosystems In: Paoletti MG, editor. Invertebrate Biodiversity as Bioindicators of Sustainable Landscapes. Amsterdam: Elsevier; 1999. pp. 19–31. 10.1016/B978-0-444-50019-9.50005-4 [DOI] [Google Scholar]
- 13.Tscharntke T, Klein AM, Kruess A, Steffan-Dewenter I, Thies C. Landscape perspectives on agricultural intensification and biodiversity–ecosystem service management. Ecol Lett. 2005;8: 857–874. 10.1111/j.1461-0248.2005.00782.x [Google Scholar]
- 14.Hunt GJ. Insect DNA Extraction Protocol In: Micheli MR, Bova R, editors. Fingerprinting Methods Based on Arbitrarily Primed PCR. Berlin, Heidelberg: Springer Berlin Heidelberg; 1997. pp. 21–24. 10.1007/978-3-642-60441-6_3 [Google Scholar]
- 15.Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol Mar Biol Biotechnol. 1994;3: 294–299. 10.1371/journal.pone.0013102 [PubMed] [Google Scholar]
- 16.Wei SJ, Shi BC, Gong YJ, Jin GH, Chen XX, Meng XF. Genetic Structure and Demographic History Reveal Migration of the Diamondback Moth Plutella xylostella (Lepidoptera: Plutellidae) from the Southern to Northern Regions of China. PLoS One. 2013;8 10.1371/journal.pone.0059654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wahlberg N, Wheat CW. Designing Novel Nuclear Markers for Genomic DNA Extractions of Lepidoptera. Syst Biol. 2008;57: 231–242. 10.1080/10635150802033006 [DOI] [PubMed] [Google Scholar]
- 18.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7: 214 10.1186/1471-2148-7-214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol Biol Evol. 2016;33: 1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grummer JA, Bryson RW Jr., Reeder TW. Species Delimitation Using Bayes Factors: Simulations and Application to the Sceloporus scalaris Species Group (Squamata: Phrynosomatidae). Syst Biol. 2014;63: 119–133. 10.1093/sysbio/syt069 [DOI] [PubMed] [Google Scholar]
- 21.Rozas J, Sanchez-DelBarrio JC, Messeguer X, Rozas R. DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics. 2003;19: 2496–2497. 10.1093/bioinformatics/btg359 [DOI] [PubMed] [Google Scholar]
- 22.Larsson H, Gyllenstrand N, Lascoux M. Distribution of long-range linkage disequilibrium and Tajima ‘ s D values in scandinavian populations of Norway spruce (Picea abies). Genes|Genomes| Genet. 2013;3: 795–806. 10.1534/g3.112.005462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123: 585–595. doi: PMC1203831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelly JK. A test of neutrality based on interlocus associations. Genetics. 1997;146: 1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leigh JW, Bryant D. POPART: full-feature software for haplotype network construction. Methods inEcology andEvolution. 2015;6: 1110–1116. 10.1111/2041-210X.12410 [Google Scholar]
- 26.Monteiro-Henriques T, Martins MJ, Cerdeira JO, Silva P, Arsénio P, Silva, et al. Bioclimatological mapping tackling uncertainty propagation: Application to mainland Portugal. Int J Climatol. 2016;36: 400–411. 10.1002/joc.4357 [Google Scholar]
- 27.QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation Project. [Internet]. 2018. Available: http://qgis.osgeo.org
- 28.Warren DL, Glor RE, Turelli M. ENMTools: A toolbox for comparative studies of environmental niche models. Ecography (Cop). 2010;33: 607–611. 10.1111/j.1600-0587.2009.06142.x [Google Scholar]
- 29.Warren DL, Glor RE, Turelli M. Environmental niche equivalency versus conservatism: Quantitative approaches to niche evolution. Evolution (N Y). 2008;62: 2868–2883. 10.1111/j.1558-5646.2008.00482.x [DOI] [PubMed] [Google Scholar]
- 30.Pearson RG, Raxworthy CJ, Nakamura M, Townsend Peterson A. Predicting species distributions from small numbers of occurrence records: A test case using cryptic geckos in Madagascar. J Biogeogr. 2007;34: 102–117. 10.1111/j.1365-2699.2006.01594.x [Google Scholar]
- 31.Pearce J, Ferrier S. Evaluating the predictive performance of habitat models developed using logistic regression. Ecol Modell. 2000;133: 225–245. 10.1016/S0304-3800(00)00322-7 [Google Scholar]
- 32.Schoener TW. The anolis lizards of Bimini: Resource partitioning in a complex fauna. Ecology. 1968;49: 704–726. [Google Scholar]
- 33.Städler T, Haubold B, Merino C, Stephan W, Pfaffelhuber P. The impact of sampling schemes on the site frequency spectrum in nonequilibrium subdivided populations. Genetics. 2009;182: 205–216. 10.1534/genetics.108.094904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mandal S De, Chhakchhuak L, Gurusubramanian G, Kumar NS. Mitochondrial markers for identification and phylogenetic studies in insects–A Review. DNA Barcodes. 2014;2: 1–9. 10.2478/dna-2014-0001 [Google Scholar]
- 35.van Asch B, Blibech I, Pereira-Castro I, Rei FT, da Costa LT. The mitochondrial genome of Prays oleae (Insecta: Lepidoptera: Praydidae). Mitochondrial DNA. 2014;1394: 1–2. 10.3109/19401736.2014.982579 [DOI] [PubMed] [Google Scholar]
- 36.Roy V, Demanche C, Livet A, Harry M. Genetic differentiation in the soil-feeding termite Cubitermes sp. affinis subarquatus: occurrence of cryptic species revealed by nuclear and mitochondrial markers. BMC Evol Biol. 2006;6: 102 10.1186/1471-2148-6-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matute DR, McEwen JG, Puccia R, Montes B a, San-Blas G, Bagagli E, et al. Cryptic speciation and recombination in the fungus Paracoccidioides brasiliensis as revealed by gene genealogies. Mol Biol Evol. 2006;23: 65–73. 10.1093/molbev/msj008 [DOI] [PubMed] [Google Scholar]
- 38.Schmidt ÆBC, Derhousoff J, Mclean ÆJA, Humble ÆLM. In the dark in a large urban park: DNA barcodes illuminate cryptic and introduced moth species. 2009; 3825–3839. 10.1007/s10531-009-9682-7 [Google Scholar]
- 39.Feckler A, Zubrod JP, Thielsch A, Schwenk K, Schulz R, Bundschuh M. Cryptic species diversity: an overlooked factor in environmental management? 2014; 958–967. 10.1111/1365-2664.12246 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF)
(PDF)
TCS haplotype networks of sampled specimens, considering a) the mitochondrial marker COI, b) the mitochondrial marker nad5, and c) the nuclear marker RpS5. Each circle represents a sequence; the size of the circle is proportional to number of individuals with a particular haplotype sequence.
(PDF)
Locations with alphabetic code only were sampled by local associations and/or by the Regional Directorates for Agriculture.
(PDF)
Code corresponds to the location code as in Table 1 followed by specimen number.
(PDF)
(PDF)
(ZIP)
Data Availability Statement
All sequences are available from the NCBI database; accession numbers are included in S2 Table. Alignments, scripts and tree files were submitted as supplemental material.