Abstract
Efficiently producing transparent analyses may be difficult for beginners or tedious for the experienced. This implies a need for computing systems and environments that can efficiently satisfy reproducibility and accountability standards. To this end, we have developed a system, R package, and R Shiny application called adapr (Accountable Data Analysis Process in R) that is built on the principle of accountable units. An accountable unit is a data file (statistic, table or graphic) that can be associated with a provenance, meaning how it was created, when it was created and who created it, and this is similar to the ‘verifiable computational results’ (VCR) concept proposed by Gavish and Donoho. Both accountable units and VCRs are version controlled, sharable, and can be incorporated into a collaborative project. However, accountable units use file hashes and do not involve watermarking or public repositories like VCRs. Reproducing collaborative work may be highly complex, requiring repeating computations on multiple systems from multiple authors; however, determining the provenance of each unit is simpler, requiring only a search using file hashes and version control systems.
Introduction
Reproducibility, accountability and transparency are increasingly accepted as core principles of science and statistics. While some of the impetus for the interest in reproducibility has come from high-profile incidents that reached the sphere of the general public (Carroll, 2017), there is a consensus that reproducibility is fundamental to scientific communication and to the acceleration of the scientific process (Wilkinson et al., 2016). Furthermore, transparency and reproducibility are deemed necessary by the American Statistical Association’s Ethical Guidelines for Statistical Practice (American Statistical Association, 2016). There have been significant strides in improving the set of available tools for constructing reproducible analyses such as the concept and implementation of literate computing, version control systems, and mandates by journals and sponsors that create standards for submitting data, code, and systems for reproducibility. Also, interest in data science has expanded the number of researchers and students who are undertaking data analyses, which underlies the need for tools manageable for a broad range of user expertise. While there is commercial software such as SAS (SAS, Cary, NC) and Stata (StataCrop, College Station, TX, USA) and open-source tools that have systems for reproducible computations, these may have significant challenges. Both open-source and commercial tools could encounter a number of barriers such as 1) the tool is focused on one dimension of the reproducibility problem rather than offering a complete system, 2) the tool may be hard to identify, 3) they may have subtle differences in capabilities, making it hard for users to select among them, 4) documentation or support are in short supply, and 5) they require degrees of sophistication that beginners lack.
Before the introduction of R, Becker and Chambers (1988) proposed a framework for auditing data analyses that could replicate and verify calculations called the AUDIT S-plus (Becker et al., 1988) system. Their auditing system would not only extract the computing steps required to produce the analysis, but also would identify the patterns of the creation and use of particular objects within the directed acyclic graph of computations. Currently, many R packages facilitate reproducible analyses with literate programing or by tracking computational results. Some of these are listed within the CRAN Task View: Reproducible Research (Zeileis, 2005). The knitr (Xie, 2015) and rmarkdown (Allaire et al., 2017; Baumer et al., 2014) packages are widely used and enable literate programing by weaving R code and results into the same document in many formats. Tracking computational results with the cacher packgage (Peng, 2008) can be useful for efficient storage and reproducibility, and the rctrack package (Liu and Pounds, 2014) tracks computations including random seeds in a read-only format without modifications of existing R code. The package RDataTracker (Lerner and Boose) provides a very granular level of tracking R objects within a script with minimal overhead on the user, and the recordr (Slaughter et al., 2015) package tracks dependencies within an R script and does not require the modification of the code, but this facility is limited to specific read/write methods (e.g. read.csv,write.csv,ggsave,readLines,writeLines, etc.). The remake (Fitzjohn et al., 2015) package proposes a different model for managing the complexity of analyses. Rather than instrumenting arbitrary or minimally modified R code for tracking, remake provides a structured approach for projects in which dependencies are documented explicitly. An interesting addition is the archivist package (Biecek and Kosinski, 2017) that can store R code and R objects, handle multiple versions thereof, and has many features that enhance sharing and browsing results.
Besides R, other open source programming languages and computing environments are developing reproducibility tools such as Python’s Sumatra (Davison et al., 2014) which stores computations for simulation or analysis as an electronic lab notebook. These notebooks represent a type of dynamic document (Becker, 2014) that combines data, code, and results within the same object. Dynamic documents enable a very high level of transparency, and while technically challenging to implement, are becoming an increasingly prevalent means of producing and sharing results. The tool Jupyter (Ragan-Kelley et al., 2014), originally developed to be polyglot including languages such as Python, is a web-based electronic notebook built for literate programming which is extendable to multiple languages, recently including R. The Jupyter system has many features such as interactivity, ability to share notebooks, graphical user interfaces, and the potential for high performance computing. Other systems such as Traverna (Wolstencroft et al., 2013) and Galaxy (Afgan et al., 2016) are geared toward workflow development and allow multiple programing languages, web-based sharing and integrate high performance computing. The development of compendia is also important to reproducibility, Gentleman and Temple Lang (2007) introduced the concept of a compendium that is comprised of data, code, and the computational results that also allows the distribution of the software tools. This goes beyond reproducibility by distributing a large set of general tools thereby enabling other researchers to use and assess these methodologies in different contexts.
We present the adapr (Accountable Data Analysis Process in R) package to facilitate reproducible and accountable computations. The objective is to use a structured, transparent, and self-contained system that would allow users to quickly and easily generate reports and manage the complexity of analyses while minimizing overhead. Unlike other tools such as Jupyter, Traverna and Galaxy, adapr is built in R for R and is geared toward simplicity for beginning R users with some minimal alterations to standard R coding. adapr is an R package that uses a R shiny (Chang et al., 2017) graphical user interface to interact with the directed acyclic graph (DAG) that represents the data analysis. This structured system would facilitate multiple data analysts in interpreting or collaborating on projects. Although adapr is capable of literate programming using R Markdown, adapr is designed for producing files (graphics, tables, etc.) that are distinct from the code that produced them and can be shared within a scientific collaboration. We call these files accountable units because the system can provide their providence, meaning the data, code dependencies, date, and authorship. This functionality is sustained by computing the cryptographic hashes (Rabin et al., 1981) of the result files and storing these alongside the R code within the version control system Git (Hamano and Torvalds, 2005). It is important to note that the primary data are not version tracked, but only the file hashes, this leads to a check of the identity of the data, but does not allow for reproducing data if modified so that primary data should be stored in another archive. The reasons for this are that 1) the analysis should not modify the primary data, 2) the primary datasets should be stored in a secured version system or database, and 3) tracking versions of the primary data is most efficiently done using a database rather that system such as Git.
The idea of accountable units is similar to that of verifiable computational results (VCRs) proposed by Gavish and Donoho (2011). These authors recognized the difficulties and of ensuring long-term re-execution of source code, and argued the only way of ensuring this is to virtualize and store the full computing platform. They further argued that the source code, the inputs, and output are most important to evaluating the reasoning behind published results rather than strict computational reproduction, which can be time-consuming and technically challenging. There is a bright distinction between collecting a history of computations compared to retaining and sharing the ability to reproduce the final results. We have emphasized elsewhere (Gelfond et al., 2014) that, beyond reproducibility, the analytical history is essential to accountable data analyses based upon formal theories of accountability (Jagadeesan et al., 2009). These theories imply that an evaluator (e.g. peer reviewer) cannot make a valid assessment of a communication (e.g. scientific analysis) unless the evaluator is aware of a verifiable context and history. Furthermore, the computational time required to identify a history is that of text search, which most often can be accomplished much more simply and quickly than attempting to recompute the analysis. We see adapr as fulfilling and assisting in creating some of the necessary components of VCRs. While adapr is not intended as a tool for instrumenting arbitrarily structured R code to track computation input/output, this package is aimed at providing a transparent organizational structure while establishing provenance and reproducibility assuming the stability of the computing platform.
Setting up adapr and defining a project
The adapr package is available from CRAN (Comprehensive R Archive network, cran.r-project.org) with the latest package version available on GitHub (github.com) that can be installed with adaprUpdate(). In order to initially set up adapr, there is a helper function defaultAdaprSetup() that will ask a short sequence of questions (default project directories, preferred username, version control preference, etc.) and will create the first project ‘ adaprHome’ within the project directory (e.g. by default the computer’s ‘ Document’ directory). This setup function must be executed in ordered for adapr to function properly. If RStudio (RStudio Team, 2016) is used, this setup function can utilize the RStudio environment to locate system resources such as pandoc automatically. The setup function creates a directory at the top level of the user directories called ‘ ProjectPaths’ with two files ‘ projectid_2_directory_adapr.csv’ and ‘ adapr_options.csv’ that are required to locate projects and configure adapr, respectively. The location of the ProjectPaths options directory can be changed from the default by setting an option within the R Profile using the statement options(adaprHomeDir='myPathToOptions').
An adapr project is a set of analyses based on a given collection of data that is structured as a directed acyclic graph. The project is contained within a folder (a file directory with the same name as the project) with all data, R scripts, and results. A new project can be created with the function initProject that can be invoked as initProject('myProject'). When a new project is initialized, the project is created as a single folder within a file system (OS X, Linux or Windows) with the following structure:
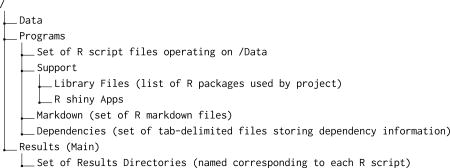
The fundamental unit of the project is an R script that is linked to three corresponding elements. The first element is a ‘ Results’ folder with the same name as the R Script that resides within the project’s main ‘ Results’ directory. The second is an R markdown file with the same name (with file extension ‘ .Rmd’) within the Markdown directory. The third element is a dependency file with the same name as the script (with the file extension .txt) within the ‘ Dependency’ directory. The dependency file contains the read/write history of the R script. When a project is initialized by default, a ‘ read_data.R’ file is created and executed automatically. This, in turn, creates a results directory ‘ Results/read_data.R’ and an R markdown file ‘ Markdown/read_data.Rmd’ and a corresponding dependency file ‘ Dependency/read_data.txt’.
In addition to the main project directory, each project is associated with a ‘ Publish’ directory that can be used to share selected results with collaborators. The Publish directory could be a Dropbox, file server or web server directory.
Initializing projects and adapr R scripts
The function call projectInit('myProject') in the R console will create a new project that by default contains a single R script ‘ read_data.R’. A side effect of this function is to automatically set current project to the new project. Lastly, the project’s settings can also be changed with the function relocateProject by specifying the project and the new settings. It is also possible to import projects that were already built by a collaborator. For example,
relocateProject('importedProjectID',project.path='parentDirectory')
imports a new project called ‘ importedProjectID’ that is within a subdirectory of ‘ parentDirectory’.
The adapr package contains a sample project called ‘ adaprTest’ that can be loaded into the users file system with the command loadAdaprTest(). To create another R script within that project, one can execute the following console commands:
>loadAdaprTest()
>setProject('adaprTest')
>makeScript(r='postProcess.R',description='mixed model analysis',
project.id=getProject())
The setProject function specifies the project and the getProject function retrieves the project ID in other functions by default so that the user does not have to respecify the project. The makeScript function will, by default, automatically open the corresponding script with the default R file browser. Each script (e.g. ‘ postProcess.R’) is given a text string description (e.g. "mixed model analysis"), and also, every file that is read or written within a project has an associated text string description. Timely descriptions are an important part of tracking because they facilitate the gradual accumulation of metadata that promotes efficient transparency rather than an automated code instrumentation, which may not easily generate precise, correct, or human-interpretable metadata tags. When a program is created with makeScript, the program is executed, creating a script-specific directory within the parent ‘ Results’ directory, R markdown file, and the dependency file. The adapr R scripts have a structured header that identifies the project and the script, and a footer that writes the read/write dependencies into a file corresponding to the R script within the ‘ Program/Dependency’ directory. The header and footer that are automatically created by makeScript and are shown below.
rm(list=ls()) #Clears R workspace
set.seed(2011) # Sets random seed
library('adapr')
source.file <-'read_data.R' #Name of file matching filename /Programs directory name
project.id <- 'project_name' # Name of project
source_info <- create_source_file_dir(source.description='reads data')
# Source description will be linked to R script
# Library statements here (e.g.\, Library('ggplot2'))
# Begin program body here
# End Program Body
dependency.out <- finalize_dependency()
Within the adapr script, the key step within the header is the create_source_file_dir function that creates the source_info object of type ‘ list’. The source_info object is stored within the R options and can be retrieved using getSourceInfo(). This is used to collect the read/write information within the body of the R script by wrapping the read/write commands within three R commands. The first is the Read function:
Read(file.name = 'data.csv', description = 'Data file',
read.fcn = guess.read.fcn(file.name), …)
This command returns the file object read by the function read.fcn and tracks the description and the file hash with "…" passed as additional arguments to the read function read.fcn. Note that in the usage example below, it is transparent and parsimonious, especially given that the Read function defaults to reading from the project’s data directory:
Dataset <- Read('datafile.csv','my first dataset')
The argument read.fcn could be any function that given a file, returns an R object, and so, Read is not limited to flat files. To facilitate the identification of a project’s data files, the listDatafiles() function call produces a list of files within the data directory.
The second command is the Write function:
Write(obj = NULL, file.name = 'data.csv', description = 'Result file',
write.fcn = guess.write.fcn(file.name), date = FALSE, …)
This example writes object ’cars’ into the Results directory as a comma separated value (CSV) file
Write(cars,'cars.csv','speed and distance')
and this example writes object ’cars’ into the Results directory as a R data object.
Write(cars,'cars.rda','speed and distance')
This command writes the R object obj using the function write.fcn and tracks the description and the file hash with "…" passed as additional arguments to write.fcn. If the file.name argument has a .rda extension then the R object is written to the results directory as an R data object. The third function used by adapr creates graphics files:
Graph(file.name = 'ouput.png', description = 'Result file',
write.fcn = guess.write.fcn(file.name), date = FALSE, …)
This command opens a graphics device for plotting using the function write.fcn and tracks the description and the file hash with "…" passed as additional arguments to write.fcn. The function guess.write.fcn uses the file suffix to determine the type of file to write (e.g. pdf, png, etc). The date variable is a logical that determines whether a date is added to the filename. This function is followed by graphics command set and closed by a dev.off() or graphics.off() statement. Below is an example.
Graph('histogram.png','hist of random normals')
hist(rnorm(100))
graphics.off()
This code snippet creates a file in the results directory of the R script called ‘ histogram.png’ with a histogram of 100 standard normal variables.
Another key function is loadFlex invoked as the following:
LoadedObject <- loadFlex('process_data.R/intermediate_result.rda')
The loadFlex function reads and returns an R data object with the .rda file extension written by another R script within the project. The loadFlex function could also read any intermediate result file type if the argument read.fcn is specified. This function is critical because it allows the R scripts within a project to build on the results of other R scripts in a transparent and structured manner. This function searches through all results for a file name that matches the argument string to identify the R object to load, and returns the corresponding R object (assigned to LoadedObject in the example). This function must follow the call Write(intermediate,'intermediate_result.rda','normalized data') within another R script ( process_data.R in the example) to pass preprocessed data from one R script to another or to collect key results from multiple R scripts. It is important to note that loadFlex requires prior use of a Write function because adapr does not, by default, collect the entire workspace of each R script. The convenience function call listBranches() returns a data frame of all intermediate results available for loading along with their descriptions.
By transferring and tracking objects from one script to the next, adapr promotes the idea of modularity. In this manner, the scripts can have a specific function rather than one script being burdened with all analytical tasks. Modularity promotes transparency by allowing a collaborator or reviewer to quickly identify which scripts are responsible for which analytic tasks, and allowing a collaborator to isolate their contribution to a particular task within a script that is linked to scripts created by others. Lastly, modularizing scripts to perform specific tasks facilitates transforming scripts into functions themselves. Modularization makes clear what the inputs (arguments to loadFlex) and outputs (arguments to Write) of a particular script are. R functions that are specific to a project are stored within the ‘ Programs/support_functions’ directory. Any R script within this directory will be sourced and loaded within the create_source_file_dir function within the adapr script header. A function can be created within this directory by using the function call makeFunction('mySpecialPlot') in the R console that will create a file ‘ Programs/support_funcionts/mySpecialPlot.R’ and by default will open the function’s file for editing.
The adapr footer contains the critical function call finalize_dependency() that transforms the R script’s file input/output (I/O) stored within the object getSourceInfo() and writes this data to the project’s ‘ Dependency’ directory in the form of a tab-delimited file with the same prefix as the R script. The dependency data within the file includes the R script name, R script description, R script hash, R script file modification time, R script run time, the project ID, the target file that is read/written, the direction of dependency (read or write), the target file description, target file hash, and the target file modification time. The script header function create_source_file_dir and footer function finalize_dependency execute many side effects that are necessary for tracking and these are listed in Table 1.
Table 1.
Side effects of header and footer functions
| Function | Side effects |
|---|---|
| create_source_file_dir | Create directories for Project and Results |
| Set project id for R session | |
| Initialize the file tracking object source_info | |
| Gather file information on all project files | |
| Initialize Git for project (if using Git) | |
| Add dependencies files to Git | |
| Initialize archivist repo for script | |
| Run all R scripts in ‘ support_function’ directory | |
| Run all R scripts in script specific ‘ support_function/myRscript.R’ directory | |
| Create R markdown file with same file prefix (if not already done) | |
| Create publication file (if not already done) | |
| finalize_dependency | Pass workspace to R markdown file and render |
| Store file modification time and hash for all dependencies | |
| Write dependency information to ‘ Dependency’ directory | |
| Add dependency file and R session information to Git |
For some critical analyses or data objects, it may be useful to store a version history. This means storing more than one version of the object itself, not just the code to reproduce the object. This can be done using the archivist package that has been incorporated into adapr by creating an archivist archive for each adapr script in the R script’s ‘ Result’ directory. Using the function call arcWrite(myRobj,"my object description") in the R script file stores the object within an archivist archive. If the R script that wrote the object was ‘ myRscript.R’, this may be loaded into another R Script by using the statement myObject <-arcRead('myRscript.R','my object description') that assigns the value of that object to myObject.
Literate programing using R Markdown
An R Markdown file is created automatically within the ‘ Programs/Markdown’ directory when an adapr R script is first created and executed. This file has the ‘ .Rmd’ suffix and is rendered when the R script is executed as a side-effect of the footer finalize_dependency() function within the corresponding R script. Additional R markdown files can be created using the createMarkdown function and rendered within the ‘ Results’ directory with the ‘ renderRmd’ function. Executing the R script will pass its workspace to the markdown file R commands and render these markdown files. By pairing each R script with a markdown file, adapr allows the R script to perform transformations and analytical tasks that can be more easily summarized, formatted, and displayed within the R markdown file.
The ‘ .Rmd’ file can also be executed by using the "knit" button in RStudio if the file header has the header line scriptLoader(project.id,R Script) executed (this is commented out by default). Additionally, file input/output can be tracked using the line if(checkRmdMode())){dependency.out <-finalize_dependency()} as the final line in the markdown file. These lines are automatically included in the R markdown files, but are commented out. The RStudio "knit" button can facilitate exploring and debugging; however, it does not direct the rendered markdown into the results directory, and the rendered markdown file would go to the ‘ Markdown’ directory.
Managing R packages
By default, projects share a common R library that is specified at setup. Sometimes users may prefer that project has a project-specific library, instead of the default R library. This may be useful for improving consistency in computations, although the larger libraries can use substantial drive storage. The user may specify library preferences at initialization by setting the following argument within the projectInit function: initProject('myProject',project.libraryTF=libOption). The project.libraryTF parameter libOption may be set to one of 3 values ( "TRUE", "FALSE", or "packrat"). The default value FALSE implies the use of the default library. The value TRUE will use the project specific library located in the Programs/Support/Library directory, and the value "packrat" will imply the packrat (Ushey et al., 2016) to manage R packages. The packrat package allows the isolation of the package dependencies for R projects and has many features including isolation of the computing environment, cross-platform compatibility and cleaning out unused packages. adapr does set up or operate packrat, and this responsibility is left to the user. We have found that the RStudio IDE is helpful for the initialization of packrat rather than R console commands.
adapr facilitates loading and tracking R packages using the Library function and by tracking the loaded libraries within the R session information. R packages needed for each R script can be loaded with the Library function within the R script header. This function will install and/or load the package if it is not installed or loaded. A use case would be Library('ggplot2','cran') or Library('limma','bioc'). At the end of the program body, the finalize_dependency function writes the R session information to a file. The getLibrary() statement will read all of these session information files for a project and collect all of the packages and their versions. This information can be useful for reproducibility, and these packages can be installed using the installLibrary() function call. The installLibrary function has an argument to install the specific versions, and the packages are installed from CRAN, MRAN (Microsoft R Application Network, https://mran.microsoft.com), Bioconductor (Huber et al., 2015), or GitHub. Currently, only the latest compatible version of Biocondunctor packages will be installed. If the packrat package is used for a project, then it is recommended to use packrat functions and the RStudio IDE for managing package versions. An example of how to use these functions is shown below: The following series of console commands loads the example project stored within the adapr R package.
>loadAdaprTest()
This sets the project.
>setProject('adaprTest')
This returns the directory containing the project library.
>getProjectLibrary()
This returns the project library’s package information.
>getLibrary()
This installs project library.
>installLibrary()
There are many technical intricacies of tracking R package use within a project, and there has been significant effort build stable solutions for package reproducibility such as packrat and switchr (Becker et al., 2017). These difficulties are not currently completely handled by adapr and is an area of active development.
Summarizing and visualizing projects
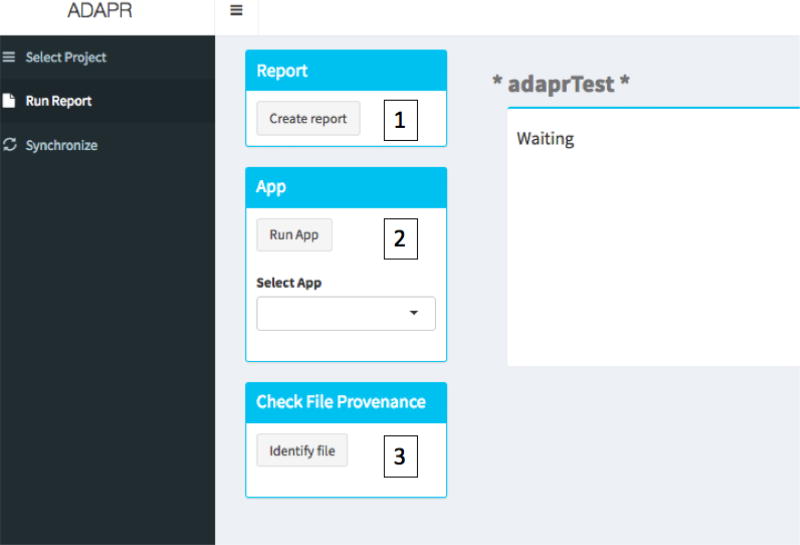
The reportProject function and the app can summarize all programs and results within a single HTML document with descriptions, dependency information, and links to programs and results. The app can be launched with within the R console with the function adaprApp(). The report information can be obtained within the Report tab of the app in Figure 1. After clicking the "Report" button, the HTML project summary is written to the ‘ Results/Tree_controller’.R/ directory. The Report tab shown in figure also is capable of launching project R Shiny apps that access project data or Results.
Figure 1.
Run report tab. 1. The Create Report button will create an html report listing all R script input/outputs. 2. RunApp selects and launches App within the project App directory. 3. The Identify file button initiates a file select dialogue, and the version control history based upon the file hash is given.
Synchronizing and version control
Two critical parts of accountability and reproducibility are addressed by the syncProject and commitProject functions as well as within the Synchronize tab of the app. The syncProject function can be called in the R console with syncProject() after a setProject('myProject') call. It will synchronize data, scripts, and results for a project. The project is synchronized if two criteria are met: 1) the file modification times stored within the R scripts dependency files are the same as the modification times within the file system, and 2) if the cryptographic hash (data fingerprint) of the file hash of each file within the project is consistent with all of the dependency files. The syncProject function called by syncProject() detects lack of synchrony and can execute the corresponding R scripts that are needed to achieve synchrony. It is possible that the DAG has structures ("forks") that allow the R scripts to be run in parallel because they are not dependent on one another. We have added experimental functions parallelSync and monitorParallelSync that allow parallel execution of the DAG by different cores. Note that monitorParallelSync should be executed in a separate R session and will indicate which cores are working on which part of the project DAG. For larger projects, an alternative to synchronizing the whole project is to only synchronize the results of a particular R script. This involves only synchronizing the parts of the DAG that that R script depends upon. This can be done with the syncTrunk('myscript.R','myProject') function call. This is illustrated in the code snippet below, and Figure 2 shows the results of the last graphProject() statement.
>loadAdaprTest() # Loads the project stored within the adapr R package
>setProject('adaprTest') # Sets the project
>syncProject() # Synchronizes the project
>openScript('read_data.R') # Open R script
We could make some minor change in ‘ read_data.R’ and then execute the following to synchronize the project.
>graphProject() # Shows project DAG with synchronization status
>runScript('read_data.R') # Executes the script in a clean R environment
>syncTrunk('graph_cardata.R') # Executes scripts needed to synchronize
results from 'graph_cardata.R'
>graphProject() # Shows project DAG with synchronization status
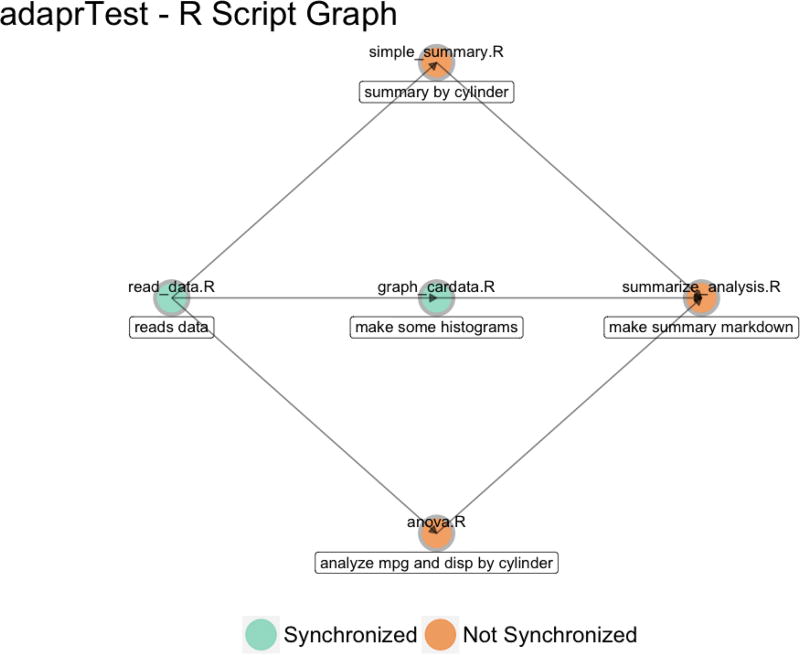
Figure 2.
Resulting graph from graphProject() in code snippet. The R Script Graph shows dependencies of R scripts in the "adaprTest" project with their synchronization status after syncTrunk(’graph_cardata.R’), which synchronizes results from the R script ‘ graph_cardata.R’.
adapr optionally uses the Git version control system, which can be operated with the commitProject() function and the Synchonize tab. The Synchronize tab is shown in Figure 3. Git must be installed for this feature to work. There are platform independent and free Git clients available for download (https://git-scm.com/downloads/guis). While adapr handles Git tracking and snapshots, it does not currently revert the current version of the project back to a previous snapshot. We recommend that reversions be performed using Git directly or through a Git client, which have more features and visualization capabilities. The function commitProject() and button within the app takes a project snapshot of all R scripts and adds the synchronization status (synchronized or not) to the commit message. Because the file names and file hashes/fingerprints are stored within dependency files that are tracked by the version control system, any data, R script, or result file can be matched by its hash/fingerprint within the version control history to determine its provenance. The adapr function Digest is a wrapper for functions in the digest (Eddelbuettel et al., 2017) package and is used the compute the cryptographic hash (not serialized) of a file, and this file hash can be identified by a Git history search to determine its provenance within a project. adapr uses Git operating functions that were adapted from those within the devtools (Wickham and Chang, 2015) and git2r (Widgren and others, 2017). If Git is used, adapr will automatically perform a commit after syncProject. This allows the user to capture all result file hashes and to identify which results were obtained in a synchronized state. It is important to note that adapr automatically adds all R script files into the project’s Git repository, but it does not automatically use formal version control for data or results files because these files may be too large for the version control system to efficiently track the changes. If the user would like to store multiple versions of a result, then we recommend using the arcWrite function. However, by default, the file hash is computed and stored within the ‘ Dependency’ directory for all files that are read or written within the project so that all relevant files and their provenances can be identified if a commit was made when the file was read/written. File histories can be identified within the Run Report tab that executes a file hash search and returns the Git commit associated with the file including the date, author, and file description. The Git history of a file is also obtained with the gitProvenance function with arguments project.id and filepath that opens a file selection window and runs a Git search for the hash of the file within the project.
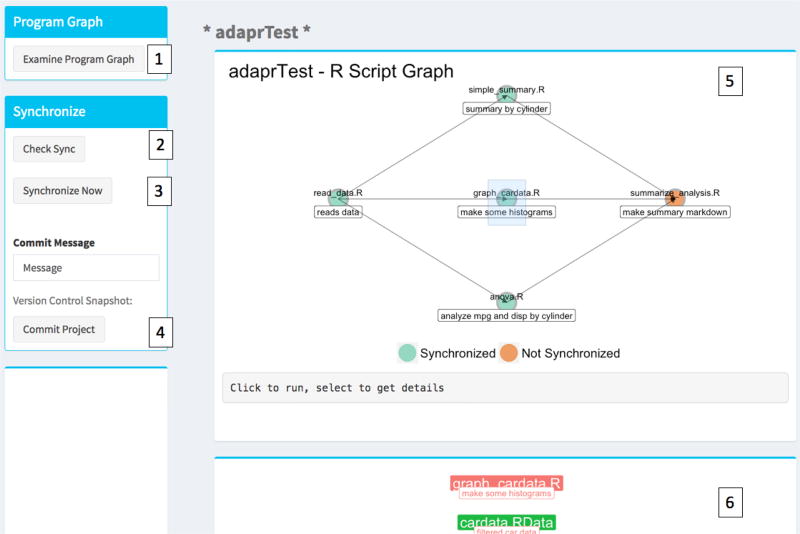
Figure 3.
Synchronize Tab. 1. Examine program graph creates or updates two interactive plots. 2. Check the synchronization status and synchronize project by running necessary R scripts. 3. Syncronize project. A pop-up progress bar will open. 4. Commit message with Git version control snapshot of project. Commit Project button operates Git. 5. R Script Graph shows dependencies of R scripts. Clicking on button runs the R scripts. Selecting or brushing button opens panel 6. 6. Displays the inputs and output of the selected R script. Clicking on the file label opens the file or selecting opens the file folder.
There is an interactive project map feature within the "Synchronize" tab. The "Examine Program Graph" button displays the DAG of all the project’s R scripts colored by their synchronization states. These scripts can be executed by clicking on the colored point, and their file I/O can be examined by selecting or "brushing" over the colored point. This reveals the file I/O graphic in an adjacent panel. These files are labeled by their descriptions and can be opened by clicking on the file or their file directories may be browsed by selecting or "brushing" over the colored file labels. The graph may alternatively be displayed using the graphProject() function. A limitation is that very large DAGs or very large input/output lists for an R script can overwhelm the visual space of these graphics. Future versions will address this issue.
Sharing results
Specified result files from the project can be copied to a shared ‘ Publish’ directory using the publishProject('myProject') function. This allows users to share or update a list a selected results with one command. The publishProject function reads a file within the ‘ Programs/Support_functions’ directory that contains a list of files that are selected for publication (i.e. copied to the project publication directory, which may be a shared server folder, a web server, or cloud server such as Dropbox). Publishing can be automated by including the publishProject function within an adapr R script. The file that contains paths to the results that will be published can be opened for editing with the browsePubFiles('myProject') function.
Configuration
The adapr package can be updated and configured within the setAdaprOptions and getAdaprOptions functions. An option can be set with the syntax setAdaprOptions("optionName", "optionValue"). The key options are summarized in Table 2. For example, the use of Git can be activated by setAdaprOptions('git','TRUE'). Git itself can be configured using Git commands or within R using the adapr function call gitConfigure(username, email).
Table 2.
List of adapr options
| Purpose | Option Name | Value |
|---|---|---|
| Path to system resources | PATH | Paths separated by system delimiter |
| Specify use of Git | git | TRUE or FALSE |
| Default project path | project.path | File path |
| Default publication path | publish.path | File path |
| Default R library | library | File path to default library |
| Specify use of adapr beta version | adaprBeta | TRUE or FALSE |
Convenience functions
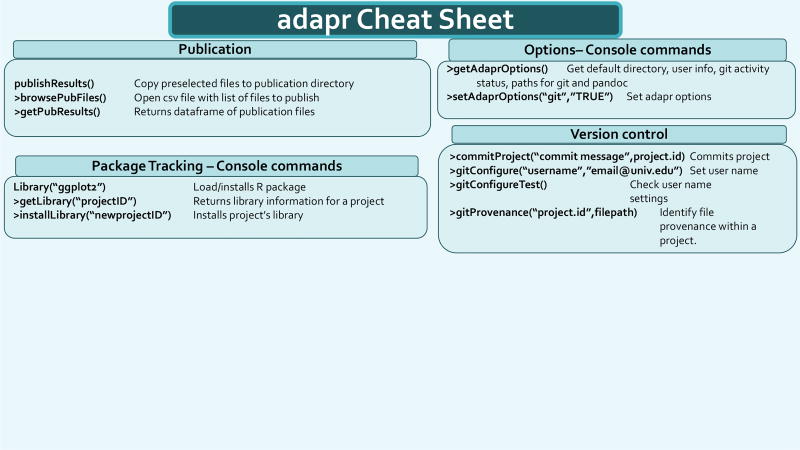
We have found that certain project oriented functionality can save time and effort, and we have compiled a list of functions that we have found useful in Table 3. These can be used without specifying the project argument after the setProject('myProject') function. The two pages of the function summary "cheat sheet" that is opened with the adaprSheet() function call is shown in Figures 4 and 5.
Table 3.
Convenience functions
| Purpose | Function example |
|---|---|
| Browse main function cheat sheet for adapr | adaprSheet() |
| Show file usage information for all projects | fileInfoProjects() |
| Open project directory in file browser | showProject() |
| Open project or source results directory in file browser | showResults() |
| Open project’s R script or R markdown file lists R scripts if not specified | openScript() |
| List intermediate result objects | listBranches() |
| Plot synchronization status of project scripts | graphProject() |
| Execute R script within a project optionally create a markdown log | runScript('read_data.R', 'adaprTest',logRmd=TRUE) |
| Search R scripts for string | searchScripts('lmer') |
| Copy data file to project data directory | importData() |
| Return results directory within R script | resultsDir() |
| Return data directory within R script | dataDir() |
| Track file reads without using Read | ReadTrack('arraydata.idat', 'read by limma package') |
| Track file writes without using Write | WriteTrack('myFile.docx', 'From ReporteRs package') |
Figure 4.
Cheat sheet snapshot. This figure shows first out of two pages of the function summary cheat sheet. This is opened from the R console using the function call adaprSheet().
Figure 5.
Cheat sheet snapshot. This figure shows second out of two pages of the function summary cheat sheet. This is opened from the R console using the function call adaprSheet().
Conclusion
Becker and Chambers (1988) have written that "the complexity of the structure that represents a realistically large analysis makes a thorough interactive investigation very challenging. New statistical, computational, and graphical techniques will be needed." Since then, there have been many strides towards that goal. The adapr package is a demonstration of how scientific reproducibility and accountability can be accelerated by automatically structuring, tracking, and implementing analyses. Beginning R programmers may benefit from this approach by not having to create their own system, and while more experienced analysts have likely created their own methods for structuring analyses, it is possible that there are certain features of adapr that they could integrate into their systems. These features include adding a standard structure to project directories, checking the synchronization status of the project, selectively synchronizing part of the DAG, linking short text descriptions to data and results files, creating a HTML summary of the full project, interactive project maps, and version control integration with file hash search capability for results and programs. This integrated version control feature tracks file hashes of data, programs, and results and can capture the evolution of the analysis, not just the final results. This points to an important challenge: the analyses in many scientific articles are extremely intricate and necessarily become more complex as the data are inevitably explored. This makes version control system indispensable for unraveling the history of analysis.
Limitations
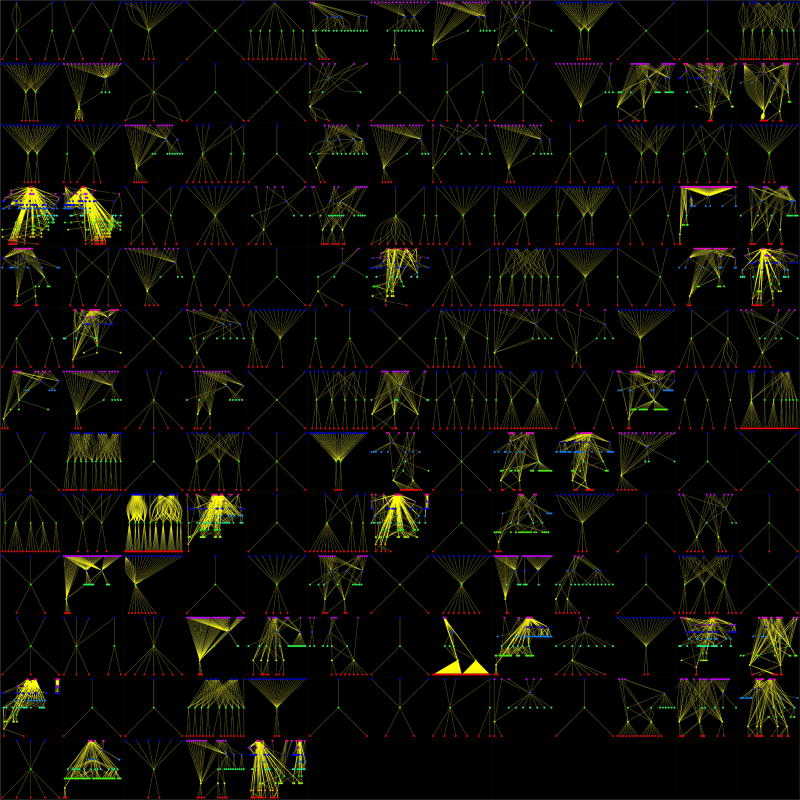
The limitations of adapr include the inability to directly track changes in external databases, file hash computation can be slow for files that are multiple gigabytes, the requirements for the user to use adapr read/write functions for tracking, and the package management system is less capable than packages such as packrat and switchr. Currently, adapr can be used with external database input functions, but these would not be tracked as inputs with the Read function because they are not within the file system. We are considering options for tracking external database input for future adapr versions. One workaround for this is to have an R script within the project that downloads the relevant database queries and writes these objects to the file system with a Write function, and these query results can be read into another R script and tracked as a dependency using the loadFlex function. The adapr package is also limited by its dependency on other R packages and by the fact that running the Shiny app while programming would require two R sessions. We have found that using project oriented functions ( makeScript,syncProject,commitProject,initProject,setProject, etc.) within the console makes for a more fluid experience, but this requires that the user have an idea of which functions to utilize, which is why we created the adaprSheet() function to give a quick overview. Also, beginning R users may be just learning the art of programming, which could make using a structured programming system like adapr difficult if that structure is violated. There are some safeguards built into adapr, but violating the header/footer structure can result in errors. We have used adapr in a class with first-time programmers, and this has greatly informed our development. For example, the function makeScript originally only created the script within the file system, but now, it also opens the script with the default R program so that the students do not have to search for it. The students use a single function Library function instead of two functions install.packages and library. Also, we have found that the runScript function is very useful for teaching reproducibility as it lets the students run the script in a clean environment. Students can also be introduced to R through the R scripts, and after a few classes, can be introduced to R markdown. This allows time to get familiar with R, before adding the additional complexity of R markdown syntax. Because each adapr script is paired with and passes its environment to an R markdown file, the students can see how R markdown can nicely format the objects that they have previously created. While a limitation of this work is that the adapr user base is small, one of the key aspects of the system is that it has been tested on a variety of projects. The adapr package and earlier versions have been tested in over 160 projects. These projects were conducted by five statisticians in an academic health center (AHC). The projects included clinical trials, genomic analyses, experimental data from basic sciences, as well as projects created specifically to test adapr. Figure 6 represents the directed acyclic graphs with structures related to the complexity of the projects.
Figure 6.
The directed acyclic graphs of 160 projects that used the adapr package. Each panel is a project, and points are files (data files, R scripts, and output files) organized by their dependencies (flowing from top to bottom) within that project.
Other potential benefits
Additional goals of adapr include efficiency and transparency. The AHC environment creates a high demand for statistical analyses as well as significant pressure to reduce the time-to-completion. We have studied process of statistical consulting in some detail (Schmidt et al., 2016), and the use of the system has not led to any significant increase in lead times, but it has subjectively improved the ease of collaboration among analysts. The use of any structured system like adapr can be highly beneficial when a long dormant project becomes active again or a project is passed from one analyst to another. Another advantage of the system is that reports and interactive maps allow users who are not familiar with R the opportunity to browse the analysis, which promotes transparency. These maps also help the users, regardless of experience level, to orient themselves to large or complex analyses. Structuring projects in directories that are systematically scanned for activity (e.g. every 30 minutes) allows automated tracking of the work patterns within projects (e.g. file modification times of R scripts or markdown files within the projects directory). This facilitates the objective estimation of work-hours that are useful for reporting, billing, or resource allocation.
Lastly, given that accountability has been recently added as a requirement for authorship by the International Committee of Medical Journal Editors (Resnik et al., 2015; International Committee of Medical Journal Editors, 2016), scientific reviewers or readers may benefit from knowing that an adapr data analysis had a high degree of accountability or reproducibility. Hence, we suggest adding the statement "The data analysis was conducted using an accountable process." to the ’Statistical methods’ or ’Data analysis’ section of an article that used adapr. This designates that: 1) a documented system was used for the version-controlled analytic history, 2) the provenance of results can be checked, and 3) that the analyses and results have descriptions, a documented structure, and a means of reproduction. Note that version control systems such as Git store the author’s name and email associated with commits so that these naturally document contributions.
Acknowledgments
The authors thank Emily Burnett for her contribution to early versions of the user interface. The authors would like to thank the editor and two anonymous reviewers for substantial improvement of this work. The project described was supported by the National Cancer Institute and the National Center for Advancing Translational Sciences, National Institutes of Health, through the grants GM070335, NIA Shock Center P30AG013319, NIA Pepper Center P30AG044271, CTRC P30 Cancer Center Support Grant CA054174 and the Clinical and Translational Science Award (CTSA) UL1 TR001120. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Contributor Information
Jonathan Gelfond, Department of Epidemiology and Biostatistics, UT Health San Antonio, TX, USA, gelfondjal@uthscsa.edu.
Martin Goros, Department of Epidemiology and Biostatistics, UT Health San Antonio, TX, USA, goros@uthscsa.edu.
Brian Hernandez, Department of Epidemiology and Biostatistics, UT Health San Antonio, TX, USA, hernandezbs@uthscsa.edu.
Alex Bokov, Department of Epidemiology and Biostatistics, UT Health San Antonio, TX, USA, bokov@uthscsa.edu.
Bibliography
- Afgan E, Baker D, van den Beek M, Blankenberg D, Bouvier D, Čech M, Chilton J, Clements D, Coraor N, Eberhard C, et al. The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2016 update. Nucleic Acids Research. 2016:1. doi: 10.1093/nar/gkw343. https://doi.org/10.1093/nar/gkw343 URL . [DOI] [PMC free article] [PubMed]
- Allaire J, Cheng J, Xie Y, McPherson J, Chang W, Allen J, Wickham H, Atkins A, Hyndman R, Arslan R. rmarkdown: Dynamic Documents for R. R package version 1.6 2017 URL https://CRAN.R-project.org/package=rmarkdown.
- A. American Statistical Association. [Accessed: 2017-11-20];ASA Ethical Guidelines ethical guidelines for statistical practice. 2016 http://www.amstat.org/asa/files/pdfs/EthicalGuidelines.pdf.
- Baumer B, Cetinkaya-Rundel M, Bray A, Loi L, Horton NJ. R markdown: Integrating a reproducible analysis tool into introductory statistics. arXiv preprint arXiv:1402.1894. 2014 https://doi.org/10.1063/pt.5.028530 URL .
- Becker G. Dynamic Documents for Data Analytic Science. University of California, Davis: 2014. [Google Scholar]
- Becker G, Barr C, Gentleman R, Lawrence M. Enhancing reproducibility and collaboration via management of R package cohorts. Journal of Statistical Software. 2017;82(1):1–17. doi: 10.18637/jss.v082.i01. URL https://doi.org/10.18637/jss.v082.i01. [DOI] [Google Scholar]
- Becker RA, Chambers JM. Auditing of data analyses. SIAM Journal on Scientific and Statistical Computing. 1988;9(4):747–760. https://doi.org/10.1137/0909049 URL . [Google Scholar]
- Becker RA, Chambers JM, Wilks AR. The new s language. Pacific Grove, Ca.: Wadsworth & Brooks; 1988. https://doi.org/10.2307/2531523 1988 . URL . [Google Scholar]
- Biecek P, Kosinski M. archivist: An R package for managing, recording and restoring data analysis results. Journal of Statistical Software. 2017;82(11):1–28. doi: 10.18637/jss.v082.i11. [DOI] [Google Scholar]
- Carroll A. Science needs a solution for the temptation of positive results. the upshot. New York Times: May 29, 2017. https://doi.org/10.3886/icpsr09104 URL . [Google Scholar]
- Chang W, Cheng J, Allaire J, Xie Y, McPherson J. shiny: Web Application Framework for R. R package version 1.0.5 2017 URL https://CRAN.R-project.org/package=shiny.
- Davison A, Mattioni M, Samarkanov D, Teleczuk B. Sumatra: a toolkit for reproducible research. Implementing reproducible research. 2014:57–79. https://doi.org/10.1201/b16868 URL .
- Eddelbuettel D, Lucas A, Tuszynski J, Bengtsson H, Urbanek S, Frasca M, Lewis B, Stokely M, Muehleisen H, Murdoch D, Hester J, Wu W, Kou Q, Onkelinx T, Lang M, Simko V. digest: Create Compact Hash Digests of R Objects. R package version 0.6.12 2017 URL https://CRAN.R-project.org/package=digest.
- Fitzjohn R, Falster D, Cornwell remake: Make-like declarative workflows in r. 2015 URL https://github.com/richfitz/remake.
- Gavish M, Donoho D. A universal identifier for computational results. Procedia Computer Science. 2011;4:637–647. https://doi.org/10.1016/j.procs.2011.04.067 URL . [Google Scholar]
- Gelfond JA, Klugman CM, Welty LJ, Heitman E, Louden C, Pollock BH. How to tell the truth with statistics: The case for accountable data analyses in team-based science. Journal of translational medicine & epidemiology. 2014;2(2) https://doi.org/10.1002/sim.4282 URL . [PMC free article] [PubMed] [Google Scholar]
- Gentleman R, Temple Lang D. Statistical analyses and reproducible research. Journal of Computational and Graphical Statistics. 2007;16(1):1–23. https://doi.org/10.1198/106186007x178663 URL . [Google Scholar]
- Hamano J, Torvalds L. Git-fast version control system. 2005 [Google Scholar]
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, Gottardo R, Hahne F, Hansen KD, Irizarry RA, Lawrence M, Love MI, MacDonald J, Obenchain V, Ole’s AK, Pag‘es H, Reyes A, Shannon P, Smyth GK, Tenenbaum D, Waldron L, Morgan M. Orchestrating high-throughput genomic analysis with Bioconductor. Nature Methods. 2015;12(2):115–121. doi: 10.1038/nmeth.3252. https://doi.org/10.1038/nmeth.3252 URL . [DOI] [PMC free article] [PubMed] [Google Scholar]
- I. International Committee of Medical Journal Editors. Defining the role of authors and contributors. [Accessed: 2017-12-30];2016 http://www.icmje.org/recommendations/browse/roles-and-responsibilities/defining-the-role-of-authors-and-contributors.html.
- Jagadeesan R, Jeffrey A, Pitcher C, Riely J. Computer Security–ESORICS 2009. Springer; 2009. Towards a theory of accountability and audit; pp. 152–167. [Google Scholar]
- Lerner BS, Boose ER. Collecting provenance in an interactive scripting environment [Google Scholar]
- Liu Z, Pounds S. An r package that automatically collects and archives details for reproducible computing. BMC bioinformatics. 2014;15(1):1. doi: 10.1186/1471-2105-15-138. https://doi.org/10.1186/1471-2105-15-138 URL . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng RD. Caching and distributing statistical analyses in r. Journal of Statistical Software. 2008 URL https://doi.org/10.18637/jss.v026.i07.
- Rabin MO, et al. Fingerprinting by random polynomials. Center for Research in Computing Techn., Aiken Computation Laboratory, Univ.; 1981. [Google Scholar]
- Ragan-Kelley M, Perez F, Granger B, Kluyver T, Ivanov P, Frederic J, Bussonier M. The jupyter/ipython architecture: a unified view of computational research, from interactive exploration to communication and publication. AGU Fall Meeting Abstracts. 2014;1:07. [Google Scholar]
- Resnik DB, Tyler AM, Black JR, Kissling G. Authorship policies of scientific journals. Journal of medical ethics, pages medethics–2015. 2015 doi: 10.1136/medethics-2015-103171. https://doi.org/10.1136/medethics-2015-103171 URL . [DOI] [PMC free article] [PubMed]
- RStudio Team. RStudio: Integrated Development Environment for R. RStudio, Inc.; Boston, MA: 2016. URL http://www.rstudio.com/ [Google Scholar]
- Schmidt S, Goros M, Parsons HM, Saygin C, Wan H-D, Shireman PK, Gelfond JA. Improving initiation and tracking of research projects at an academic health center a case study. Evaluation & the Health Professions. 2016 doi: 10.1177/0163278716669793. https://doi.org/10.1177/0163278716669793 0163278716669793. URL . [DOI] [PMC free article] [PubMed]
- Slaughter P, Jones C, Jones M, Palmer L. recordr: Provenance tracking for r. 2015 URL https://github.com/NCEAS/recordr.
- Ushey K, McPherson J, Cheng J, Atkins A, Allaire J. packrat: A Dependency Management System for Projects and their R Package Dependencies. R package version 0.4.8-1 2016 URL https://CRAN.R-project.org/package=packrat.
- Wickham H, Chang W. devtools: Tools to Make Developing R Packages Easier. R package version 1.8.0 2015 URL http://CRAN.R-project.org/package=devtools.
- Widgren S, et al. git2r: Provides Access to Git Repositories, 2017. R package version 0.19.0 URL https://CRAN.R-project.org/package=git2r.
- Wilkinson MD, Dumontier M, Aalbersberg IJ, Appleton G, Axton M, Baak A, Blomberg N, Boiten J-W, da Silva Santos LB, Bourne PE, et al. The fair guiding principles for scientific data management and stewardship. Scientific data. 2016;3:160018. doi: 10.1038/sdata.2016.18. https://doi.org/10.1038/sdata.2016.18 URL . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolstencroft K, Haines R, Fellows D, Williams A, Withers D, Owen S, Soiland-Reyes S, Dunlop I, Nenadic A, Fisher P, et al. The taverna workflow suite: designing and executing workflows of web services on the desktop, web or in the cloud. Nucleic acids research. 2013:gkt328. doi: 10.1093/nar/gkt328. https://doi.org/10.1093/nar/gkt328 URL . [DOI] [PMC free article] [PubMed]
- Xie Y. Dynamic Documents with R and knitr. Vol. 29. CRC Press; 2015. [Google Scholar]
- Zeileis A. Cran task views. R News. 2005;5(1):39–40. [Google Scholar]