Abstract
In order to profile the lipidome for untargeted lipidomics applications, analysis by ultra-high performance liquid chromatography – high resolution mass spectrometry (UHPLC-HRMS) typically requires the extraction of lipid content from sample matrices using matrix-specific conditions. The Folch, Bligh-Dyer, and Matyash extraction methods, while promising approaches, were originally tailored to specific matrices (brain tissue, fish muscle, and E. coli, respectively). Each of these methods have specific solvent ratios that must be adhered to achieve optimal extraction. Thus, the sample-to-solvent ratios for these methods should be optimized for the sample matrix of interest prior to employment. This study evaluated the appropriate sample-to-extraction solvent ratios for human plasma-based lipidomics studies. An advantage of employing biphasic lipid extractions is the ability to investigate both the aqueous and organic layers for increased analyte coverage in untargeted studies. Therefore, this work also evaluated the multi-omic capability of each lipid extraction method for plasma in an effort to provide a workflow capable of increasing analyte coverage in a single extraction, thus providing a more complete understanding of complex biological systems. In plasma, a decrease in sample-to-solvent ratios from 1:4, 1:10, 1:20, to 1:100 (v/v) resulted in a gradual increase in the peak area of a diverse range of metabolite (aqueous layer) and lipid (organic layer) species for each extraction method. The Bligh-Dyer and Folch methods yielded the highest peak areas at every plasma sample-to-solvent ratios for both metabolite and lipid species. Depending on the lipid class of interest, the Folch or Bligh-Dyer method is best suited for analysis of human plasma at a 1:20 (v/v) sample to total solvent ratio.
Keywords: lipidomics, metabolomics, multi-omics, liquid-liquid extraction, sample preparation, LC-MS, plasma, NIST SRM 1950
INTRODUCTION
Lipids are diverse in structure and serve a variety of critical roles in several metabolic activities including cell signaling, transport, protein sorting, and biosynthetic pathways.[1, 2] Lipidomics is the comprehensive study of lipids present within a cell, tissue, organism, and/or biological system.[1–3] Literature has reported that fluctuations in the lipidome due to an external perturbation, diet, and/or disease development/progression appear well before the appearance of protein biomarkers.[2, 4] Recent advancements in mass spectrometry and bioinformatics have led to an increased detection and annotation of lipid species within the community.[1, 5–8] More specifically, ultra-high performance liquid chromatography – high resolution mass spectrometry (UHPLC-HRMS) has provided a promising means for lipid analysis with improved sensitivity and specificity by reducing complications associated with the presence of co-eluting and isobaric compounds in lipid-rich regions of chromatograms. These advancements have expanded the cellular profiling of lipids and, as a result, have enhanced the understanding of lipid contributions to human health.
In order to investigate changes in the lipidome, analysis by UHPLC-HRMS requires the extraction of lipid content from cells and other sample matrices (e.g., liquid-liquid extraction) using precise solvent ratios that include organic solvents.[6, 9] The solvent system should extract the lipid content in an unbiased manner without the introduction of non-lipid material (e.g., sugars, peptides, amino acids, and other water-soluble compounds), which can be trapped within lipid micelles and carried into the organic layer.[6, 9, 10] The chosen solvent extraction system should also fully dissociate and deactivate any lipolytic enzymes. Polarity is an important component in the selection of an ideal solvent extraction system. Polar solvents are necessary for the dissociation of amphipathic complex lipids from cellular membranes. These lipid types do not normally dissolve in non-polar solvents. However, caution should be advised, as the solvent system should not be too polar to cause a chemical reaction with other lipid classes such as triacylglycerols or disrupt the extraction of the non-polar lipids.[9, 11, 12] The recovery of the lipidome using a lipid liquid-liquid extraction system is generally dependent on the sample matrix (e.g., cells, plasma, tissue, plant material) and the effective partitioning of lipid classes into the organic layer.[6, 9]
While many solvent extraction systems have been proposed for lipidomic studies, there are three extractions that are commonly employed: Folch [13], Bligh-Dyer [14], and Matyash [10]. The Folch is a widely cited and employed extraction protocol for untargeted lipid analysis. Although the Folch extraction was originally established for lipid isolation in brain using 20 volumes of chloroform:methanol:water (CHCl3:MeOH:H2O, 8:4:3, v/v/v) per volume of tissue (e.g., assumption 1 g of tissue equates to 1 mL tissue volume), this extraction method has been applied to other sample types (e.g., plasma, urine, tissue, plants, and cells) [6, 15–18]. The Bligh-Dyer extraction method (CHCl3:MeOH:H2O 2:2:1.8 (v/v/v)) was developed as a rapid method for isolating lipids in fish muscle (i.e., 1 part sample for 3 parts solvent) that reduced the amount of chloroform used in the Folch extractions. One reason for the popularity of these two methods is that a combination of methanol and chloroform is used to non-selectively and reproducibly extract a broad range of lipid classes from a wide variety of sample matrices. Methanol serves an important role in the biphasic lipid extraction as literature has shown solvent mixtures containing acetonitrile and chloroform, as opposed to methanol and chloroform, to not be as effective in extracting a broad range of lipid species. In each biphasic system, ice-cold methanol is added to disrupt hydrogen bonding and/or electrostatic networks between the lipid-lipid and/or lipid-protein biomolecules.[6, 19] Literature has shown short-chain alcohols, such as methanol and ethanol, increase the critical micelle concentration and reduce hydrophobic interactions, causing destabilization of micelles by perturbing surface tension.[11] The non-polar organic solvents in each case are used to extract hydrophobically bound lipids, and water is used for phase separation as well as to increase the insolubility of lipid species in the aqueous phase.[19]
More recently, the Matyash or methyl tert-butyl ether (MTBE) extraction was reported as an alternative solvent extraction system for lipidomics, providing comparable results to the Folch and Bligh-Dyer methods for plasma and E. coli, but without the use of chloroform.[10, 20, 21] The ratios of MTBE/MeOH/H2O for the Matyash method are 10:3:2.5 (v/v/v), respectively. These three liquid-liquid extraction methods incorporate a binary mixture of methanol with a non-polar organic solvent (chloroform or MTBE) to extract diverse lipid classes. With a range of solvent extraction systems available in literature, along with the ability of each method to be modified using buffers, salts, and/or antioxidants, matrix-dependent optimization is necessary to ensure the optimal extraction of lipid content in a reproducible manner.
The Folch, Bligh-Dyer, and Matyash methods, while promising approaches, were tailored to specific matrices. The Folch method suggests a 1:20 sample-to-solvent ratio [22] and the Bligh-Dyer method originally implemented a 1:3 sample-to-solvent ratio (i.e., the reported Bligh-Dyer ratio does not account for the volume of water in tissues). Thus, these respective ratios have only been optimized for the original matrices reported in literature and should be vetted for use with other sample matrices of interest such as plasma and cell lines. More specifically, mammalian cell lipidomics studies incorporate total cell count as opposed to weight or volume measurements. Therefore, it is challenging to adopt preexisting sample-to-solvent ratios to applications involving cells. In addition, the sample-to-solvent ratios should be optimized for specific cell lines as the lipid composition varies.
Optimization of the sample-to-solvent ratio is vital for complete lipid extraction. An increase in the solvent volume will potentially result in the increase of recovered lipid content as an environment is established that is more suitable for lipids. Therefore, this study evaluated the sample-to-solvent ratios for human plasma-based lipidomics studies using the Folch, Matyash, and Bligh-Dyer extraction protocols, in an effort to optimize methods for lipidomics profiling workflows. An additional advantage of these extraction methods is the possibility of performing multi-omic analyses, where both phases of a single biphasic extraction are analyzed (i.e., aqueous phase for metabolomics analysis and organic phase for lipidomics analysis). The aqueous phase was compared to the traditional 80% methanol metabolite extraction for plasma [23–25]. Analysis of both phases in a multi-omics approach increases the analyte coverage per sample extraction, reduces sample preparation time, increases efficiency, and thus provides a more complete and efficient understanding of complex biological systems.[15]
MATERIALS AND METHODS
Materials
Exogenous triacylglycerol (TAG) lipid standards were purchased from Sigma-Aldrich (St. Louis, MO). Exogenous lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidylglycerol (PG) lipid standards were purchased from Avanti Polar Lipids (Alabaster, AL). Analytical grade formic acid, chloroform, MTBE, and MeOH were purchased from Fisher-Scientific (Hampton, NH). All mobile phase solvents were Fisher Optima LC/MS grade (acetonitrile, isopropanol, and water (H2O)). Six lipid classes (LPC, PC, PS, PG, PE, and TG) were chosen to represent the most abundant lipid classes, including a balance of hydrophobic-hydrophilic neutral and charged lipids. Metabolite internal standards were purchased from Cambridge Isotopes (L-lysine-4,4,5,5-d4) and C/D/N Isotopes (creatine-methyl-d3, D-leucine-d10, L-tryptophan-2,3,3-d3, caffeine-methyl-d3, L-phenylalanine-3,3-d2, and D/L-carnitine-trimethyl-d9) (Tewksbury, MA and Quebec, Canada, respectively).
Lipid Extraction
For plasma, the National Institute of Standards and Technology (NIST) Standard Reference Material (SRM) 1950 - Metabolites in Frozen Human Plasma (50 μL) was extracted according to the Folch, Bligh-Dyer, and Matyash protocols outlined below with the internal standards spiked before the extraction. The addition of internal standards during the lipid extraction can be used to assess the effects of an incomplete recovery on a lipid class. Therefore, there are minor secondary effects associated with the recovery of individual lipid species within a lipid class compared to the internal standard. The plasma-to-total solvent volume ratios (v/v) before re-extraction were as follows: 1:4, 1:10, 1:20, and 1:100. Total solvent volumes were 200 μL, 500 μL, 1000 μL, and 5000 μL as shown in Figure S1. An outline of the applied extraction volumes can be found in Table S1. Solvent extraction blanks containing no plasma matrix were extracted alongside each biological sample. Plasma lipid extracts were reconstituted in 200 μL of isopropanol.
[Folch method [13]]:
Chloroform:methanol (2:1, v/v) was added to each sample according to the original Folch procedure allowing for a CHCl3:MeOH:H2O ratio of 8:4:3 (v/v/v). Briefly, ice-cold methanol and chloroform in the amounts previously outlined (Table S1) were added directly to the sample. The suspension was vortexed occasionally to bring about physical mixing and the sample was incubated on ice for 30 min. After the addition of water, which was used to separate the aqueous and organic layers, the suspension was incubated on ice for an additional 10 min. Samples were centrifuged at 2000 rpm for 5 min at 4 °C. The lower phase (organic) layer was transferred to a new tube. The aqueous layer was re-extracted with 1 mL of 2:1, v/v chloroform/methanol. The chloroform layers were combined for analysis. The aqueous layer was centrifuged at 2000 rpm for 5 min at 4 °C and collected for metabolite analysis. Extracts were then dried under nitrogen.
[Bligh-Dyer method [14]]:
Chloroform:methanol (1:1, v/v) was added to each sample according to the original Bligh-Dyer procedure allowing for a CHCl3:MeOH:H2O ratio of 2:2:1.8 (v/v/v). Briefly, ice-cold methanol and chloroform in the amounts previously outlined (Table S1) were added directly to the sample. The sample was vortexed occasionally to bring about physical mixing and incubated on ice for 30 min. After the addition of water, the suspension was incubated on ice for an additional 10 min. Samples were centrifuged at 2000 rpm for 5 min at 4 °C. The lower phase (organic) layer was transferred to a new tube. The aqueous layer was re-extracted with 1 mL of 1:1, v/v chloroform/methanol. The chloroform layers were combined for analysis. The aqueous layer was centrifuged at 2000 rpm for 5 min at 4 °C and collected for metabolite analysis. Extracts were then dried under nitrogen.
[Matyash method [10]]:
All samples were extracted according to the procedure found in literature.[4,5] To each sample, methanol and MTBE was added according to the amounts outlined in Table S1. The sample was incubated on ice and subjected to an orbital shaker for 1 hr. After the addition of water, the mixture was incubated on ice for 10 min and centrifuged at 2000 rpm for 10 min. The organic (upper) layer was collected, and the aqueous layer was re-extracted with 1 mL of 10:3:2.5, v/v/v MTBE/MeOH/water. The MTBE layers were combined for analysis. The aqueous layer was centrifuged at 2000 rpm for 5 min at 4 °C, dried under nitrogen and collected for metabolite analysis.
Metabolite Extraction
Metabolites were isolated from SRM 1950 (50 μL) using ice-cold 80% MeOH (200 μL, 500 μL, 1000 μL, and 5000 μL), while accounting for the water fraction in plasma, to represent the total volume in a 1:4, 1:10, 1:20, and 1:100 (v/v) respective sample-to-total solvent ratio. All plasma samples were thawed on ice for 10 min, 80% methanol was added, and the resulting mixture was vortexed and centrifuged at 2000 rpm for 10 min to pellet the protein and cellular content. The supernatant was transferred to a new tube, and the contents dried under nitrogen. Plasma metabolite extracts were reconstituted in 100 μL of 0.1 % formic acid in water. A flow diagram of the experimental design can be found in the Supplemental Information (Figure S1).
Analysis of Human Plasma Metabolite and Lipid Extracts by UHPLC-HRMS
Mass spectra were acquired on a Thermo Scientific Orbitrap Fusion Lumos Tribrid mass spectrometer equipped with a heated electrospray ionization (HESI II) probe in positive and negative ion mode. HESI and mass spectrometric parameters for metabolite extracts were as follows in positive/negative ion mode, respectively: spray voltage: 3.5/2.5 kV, sheath gas: 35/40 arbitrary units, auxiliary nitrogen pressure: 8/10 arbitrary units, sweep gas: 1/2 arbitrary units, ion transfer tube and vaporizer temperatures: 275 °C and 300 °C, and S-lens level: 30. HESI and mass spectrometric parameters for lipid extracts were as follows in positive/negative ion mode, respectively: spray voltage: 3.5/2.5 kV, sheath gas: 40/35 arbitrary units, auxiliary nitrogen pressure: 15 arbitrary units, sweep gas: 1/0 arbitrary units, ion transfer tube and vaporizer temperatures: 325 °C and 300/275 °C, and RF lens level: 30. Full scan, data-dependent MS/MS (top10-ddMS2), and data-independent acquisition mode data were collected at m/z 150 to m/z 2000 (lipidomics), corresponding to the mass range of expected lipids, and m/z 50 to m/z 750 (metabolomics) for metabolites.
A Thermo Scientific Vanquish UHPLC system (Thermo Scientific, San Jose, CA) was coupled to the Orbitrap Fusion Lumos Tribrid for the chromatographic separation of metabolites and lipids. The autosampler temperature was maintained at 4 °C for all experiments. Solvent extraction blanks and reconstitution blanks were jointly analyzed over the course of a batch (10 – 15 samples) to evaluate the mass accuracy (< 0.35 ppm error) and instrument variability (<2.9 %CV retention time deviation and <7.4 %CV peak area deviation) across the runs. A Waters Acquity C18 BEH column (2.1 × 100 mm, 1.7 μm particle size, Waters, Milford, MA) was maintained at 60 °C and used for all lipidomic studies. The injection volume was 5 μL in positive and 10 μL in negative ion mode with a mobile phase flow rate of 450 μL min−1. The gradient program consisted of mobile phase C [60:40 acetonitrile/water] and mobile phase D [90:8:2 isopropanol/acetonitrile/water], each containing 10 mM ammonium formate and 0.1% formic acid.[26] The total run time was 22 min, including a 5-min equilibration. An ACE Excel 2 C18-PFP column (2.1 × 100 mm, 1.7 μm particle size, Advanced Chromatography Technologies Ltd, Aberdeen, Scotland) was maintained at 30 °C for the metabolomic studies. The injection volume was 15 μL in positive ion mode and 20 μL in negative ion mode with a mobile phase flow rate of 350 μL min−1. The gradient program consisted of mobile phase A [0.1% formic acid in water] and mobile phase B [acetonitrile modified with 0.1% formic acid]. The total run time was 28 min, including a 3-min equilibration.
Data Processing
All UHPLC-HRMS data were collected and initially processed by Thermo Xcalibur Workstation software version 2.2. Raw data files were centroided and converted to a useable format (mzXML) using MSConvert. Metabolite data processing and analysis was performed with XCMS R Script, Thermo Compound Discoverer, and MetaboAnalyst 3.0, respectively. Lipid data processing was performed using MZmine [27] to generate a feature list (m/z, retention time, and peak area). Peak detection was performed by mass detection, chromatogram smoothing, peak deconvolution, deisotoping, feature alignment, and gap-filling. Lipid annotation was performed on the top10-ddMS2 files using Thermo LipidSearch (Score A & B) and LipidMatch, the latter of which can be accessed at <http://secim.ufl.edu/secim-tools/> [7].
RESULTS AND DISCUSSION
Accessibility of Organic Layer for Lipid Extractions
To determine which lipid extraction method and solvent extraction volume provided the highest peak areas from lipid isolation in human plasma, the most commonly employed lipid extraction methodologies were compared (e.g., Folch, Bligh-Dyer, and Matyash).
Access to the organic layer is an important consideration in the selection of a lipid extraction method. Due to the fact that chloroform has a higher density compared to the methanol/water aqueous layer, the organic layer remains on the bottom of the Folch and Bligh-Dyer biphasic solvent extraction systems. Therefore, the user must pipette through the aqueous and protein layer to obtain the organic layer. There is difficulty in collecting the organic layer as non-lipid contaminants and salts can be transferred, lipid content can get lost in the process, and there is more room for analyst variability based on skill. To increase recovery of the lipid content, the aqueous layer in this work was re-extracted with CHCl3:MeOH (2:1, v/v) in the Folch extraction and CHCl3:MeOH (1:1, v/v) in the Bligh-Dyer extraction, which added to the total time spent during sample preparation and the overall extraction volume, thus decreasing the high-throughput aspect of lipidomic profiling. The density of MTBE allows for the desired organic layer to remain on the top, aiding in the collection of the lipid content. Based on visual observation, the Matyash method produced a more intact protein pellet at the bottom of the aqueous layer, which aided in the collection of the organic and aqueous layers. However, the Matyash method required more time than the other methods as the protocol recommends an additional 1 hour incubation step on an orbital shaker. As reported, this hour incubation step can be reduced using low-power sonication or a microwave oven.
Analysis of Sample-to-Total Solvent Volume Ratios for Human Plasma
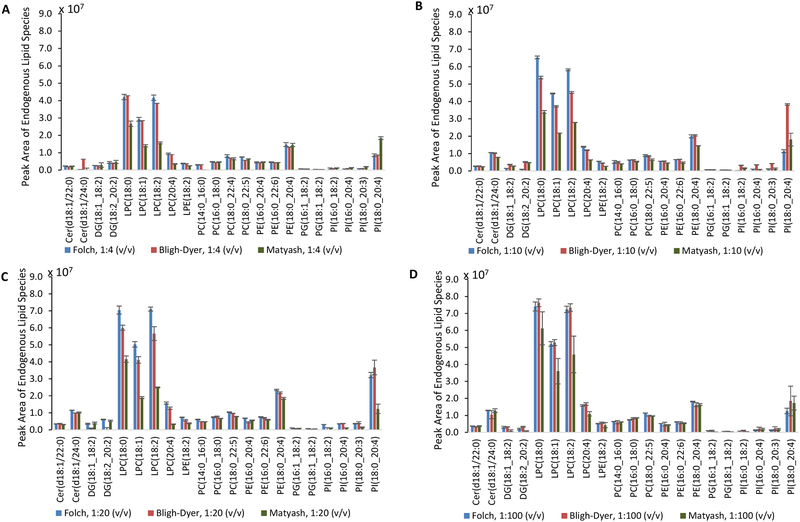
For the Folch method, the 1:10, 1:20, and 1:100 (v/v) sample-to-solvent ratios provided higher peak areas for Cer(d18:1/22:0) and many LPC/PC lipid classes compared to the 1:4 (v/v) sample-to-solvent ratio (Figure 1). As the solvent extraction volume increased starting with the 1:4 (v/v) sample-to solvent ratio, the variability increased for DG and PI lipid species. However, for the PCs, the variability decreased as the solvent extraction volume increased. For the Bligh-Dyer method, the 1:10, 1:20, and 1:100 (v/v) sample-to-solvent ratios provided higher peak areas for Cer(d18:1/24:0), all LPCs, and many PC/PE/PG lipid classes compared to the 1:4 (v/v) sample-to-solvent ratio (Figure 1). As the solvent extraction volume increased for the Bligh-Dyer method, smaller differences were observed in the variability of higher-abundant lipid species. For the Matyash method, the 1:10, 1:20, and 1:100 (v/v) sample-to-solvent ratios only provided higher peak areas for Cer(d18:1/24:0) and PC(14:0_16:0). The Matyash method exhibit low repeatability for the PI lipid class with %CVs ranging from 45.9 – 56.4 % for the 1:20 (v/v) sample-to-solvent extraction volume.
Fig. 1:
Comparison of endogenous lipid species extracted from SRM 1950 plasma with a 1:4 [A], 1:10 [B], 1:20 [C] and 1:100 [D] (v/v) sample-to-total solvent ratio. The (*) denotes a significant difference between the Bligh-Dyer and Matyash methods, (#) denotes a significant difference between the Folch and Matyash method, and (^) denotes a significant difference between the Bligh-Dyer and Folch method according to ANOVA with the Bonferroni correction, p-value < 0.05. Error bars represent the standard deviation of the mean.
As the plasma sample-to-solvent ratio decreased from 1:4, 1:10, 1:20, and 1:100 (v/v), many of the Cer, LPC, PC, and PE lipid species increased in peak area for the Bligh-Dyer, Folch, and Matyash methods as shown in Figure 1. Anova comparisons across sample-to-solvent ratios for each lipid extraction method can be found in Table S2. This finding correlates to studies where the sample-to-solvent ratio was decreased from 1:20 (v/v) to 1:30 (v/v) for homogenized fish tissues to yield higher intensities of endogenous lipid species.[9] Literature has also suggested that the total solvent volume of 2:1 CHCl3:MeOH (v/v) be 17 times higher than the tissue volume to effectively extract lipids as well as to associate weak hydrogen bonds between chloroform and water.[9]
The Bligh-Dyer and Folch methods provided higher peak areas for LPC lipid species at every sample-to-solvent ratio (Figure 1). The Bligh-Dyer and Folch methods provided comparable results at the 1:4 (v/v) and 1:100 (v/v) sample-to-solvent ratios. However, at the 1:10 (v/v) sample-to-solvent ratio, the Folch method provided higher peak areas for LPCs, the most polar of the lipid species investigated in this work, compared to the Bligh-Dyer and Matyash methods. At the 1:20 (v/v) sample-to-solvent ratio, the Folch method provided higher peak areas for LPCs compared to the Matyash method. As the sample-to-total solvent ratio decreased from 1:20 (v/v) to 1:100 (v/v), the variability amongst replicates decreased for PCs, suggesting that the use of a higher volume of extraction solvent was needed for this lipid class (Table S3). Compared to the Bligh-Dyer method, the Folch lipid extraction provided higher variability for the DG lipid class, a fairly low-abundant lipid class, at the 1:10, 1:20, and 1:100 (v/v) sample-to-solvent ratios. While taking into account peak area and repeatability for the Cer, LPC, LPE, PC, PG, and PI lipid classes, the 1:20 (v/v) sample-to-solvent ratio provided the best results for the Folch and Matyash method.
The Matyash extraction method provided lower extracted peak areas for polar lipid species such as LPCs compared to the Folch and Bligh-Dyer method. Literature has shown mixtures of methanol with more nonpolar solvents that do not have an acidic proton (e.g., tetrachloride and tetrachlorethylene) to solubilize reduced amounts of water, resulting in a lower lipid extraction efficiency.[9] The complete list of extracted peak areas for each lipid species and for each extraction method, annotated using LipidMatch in positive and negative ionization mode, can be found in the Supplemental Information (NegIDed.csv and PosIDed.csv).
Applicability to Multi-Omics Applications
The metabolome encompasses a large polarity range which spans from hydrophilic to hydrophobic compounds.[34] A single analytical platform will not encompass the entire metabolome. However, the coupling of a metabolite and lipid extraction from the same sample will increase the coverage of the metabolome, overall efficiency, and the throughput for sample preparation. A single sample can be used to provide a wider snapshot of the metabolism associated with the sample/individual. Therefore, the aqueous and organic layers of the plasma biphasic lipid extraction were collected and analyzed to demonstrate potential multi-omic capabilities.
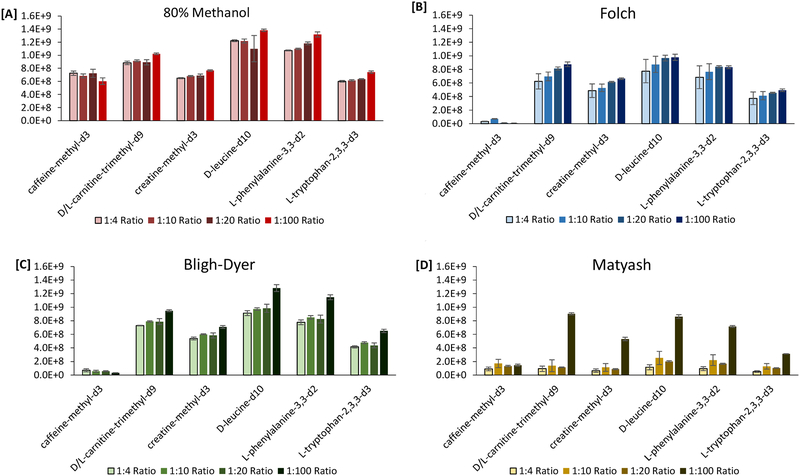
For human plasma studies, the aqueous layer of the Folch, Bligh-Dyer, and Matyash extractions was isolated and compared to a commonly used metabolite isolation protocol, 80% methanol. At every sample-to-solvent ratio, the 80% methanol protocol yielded a higher peak area for the metabolite internal standards and endogenous species compared to the aqueous phase of the biphasic methods (Figure 2 and Figure S2). This lower peak area for the Folch, Bligh-Dyer, and Matyash methods suggests that metabolites are lost in a biphasic extraction. A decrease in the plasma sample-to-solvent ratio (v/v) from 1:4, 1:10, 1:20, to 1:100 resulted in a gradual increase in the peak area of the metabolite ISTD from the aqueous layer of each extraction method (Table S4–S5). The Bligh-Dyer extraction method produced comparable results to the Folch extraction method for exogenous metabolite peak areas at every plasma sample-to-solvent ratios except for the 1:100 (v/v) ratio where the Bligh-Dyer method generated higher peak areas for L-tryptophan-2,3,3-d3. (Figure S3). Compared to the 80% methanol extraction method, the Folch and Bligh-Dyer method yielded the closest results at a 1:20 (v/v) sample-to-solvent ratio and the Bligh-Dyer method was closest at a 1:100 (v/v) sample-to-solvent ratio (Table S4–S5). With increasing solvent volume up to the 1:20 (v/v) sample-to-solvent ratio, the variability in the aqueous extract decreased for the Folch method, except for caffeine (Table S6). This finding may indicate that with the Folch extraction, metabolites partition better into the aqueous phase. Caffeine resulted in the highest %CVs for each extraction method, suggesting that a biphasic extraction may be non-optimal for this metabolite. Decreasing the sample-to-solvent extraction ratio from 1:20 (v/v) to 1:100 (v/v) resulted in a significant increase in the metabolite peak area for the Folch and Bligh-Dyer method (Table S7). However, an increase in the variability was observed for the 1:100 (v/v) sample-to-solvent ratio compared to the 1:20 (v/v) sample-to-solvent ratio for all metabolites investigate using the Bligh-Dyer method and two amino acid metabolites with the Folch method (Table S6). The Matyash method greatly benefitted from the 1:100 sample-to-solvent ratio for analysis of the aqueous layer (Figure 2). These same findings were observed in endogenous metabolite peaks (Figure S2). However, even with the increase in peak area for the Matyash method with the 1:100 sample-to-solvent ratio, higher peak areas were obtained from the Folch and Bligh-Dyer methods at the 1:20 (v/v) sample-to-solvent ratio. Therefore, a plasma sample-to-solvent ratio of 1:20 (v/v) is most applicable for multi-omics analysis of the metabolite and lipid content from a Folch and Bligh-Dyer extraction. The Matyash method requires a plasma sample-to-solvent ratio of at least 1:100 (v/v) to have comparable results to Bligh-Dyer and Folch at 1:20 ratio. The Matyash has a larger ratio of non-polar solvent (MTBE) to methanol and water (10:3:2.5, v/v/v), resulting in its inability to effectively partition metabolites into the aqueous layer. The additional solvent extraction volume for the 1:20 (v/v) and 1:100 (v/v) sample-to-solvent ratios reduced the variability in the peak areas for exogenous metabolite species compared to the 1:4 (v/v) and 1:10 (v/v) sample-to-solvent ratios (Table S6). However, this additional solvent volume results in a higher aqueous volume that is to be dried down and thus limits its applicability for implementation in a high-throughput protocol.
Fig. 2:
Comparison of plasma ISTD peak areas for the 80% MeOH [A], Folch [B], Bligh-Dyer [C], and Matyash [D] extraction methods for each human plasma sample-to-solvent ratio (1:4, 1:10, 1:20, and 1:100). Error bars represent the standard deviation of the mean.
CONCLUSIONS
The solvents or steps used for extraction bias the absence and/or concentration of biomolecules. Extraction methods are sample-dependent and thus should be optimized for each matrix in an effort to increase qualitative and quantitative coverage and recovery of a wide range of compounds in untargeted studies. For human plasma studies, the Bligh-Dyer and Folch extraction methods, regardless of the solvent extraction volume, yielded higher peak areas. The 1:20 (v/v) plasma to total solvent ratio provided higher peak areas for low abundant lipid species such as Cer and PI. Therefore, the Bligh-Dyer and/or Folch method should be employed using a 1:20 (v/v) sample-to-solvent ratio for untargeted lipidomics studies. This work highlights the necessity to optimize lipid extraction protocols for targeted studies as each lipid class responded differently to the lipid extraction method in total peak area and repeatability measurements. Analysis of the aqueous layer for multi-omics applications showed that the 1:20 (v/v) sample-to-solvent ratio yielded higher metabolite peak areas and reduced variability. The Matyash method requires a larger solvent extraction volume compared to the Bligh-Dyer and Folch methods to yield comparable results. Future work includes the analysis of multiple volumes of plasma, trace metabolite and lipid species, and additional analyte classes to ensure that the suggested sample-to-solvent ratios remain optimal.
Supplementary Material
ACKNOWLEDGEMENTS
This research was done in collaboration between Core 1 and Core 3 of the Southeast Center for Metabolomics (SECIM) <http://secim.ufl.edu/> [NIH grant #U24 DK097209]. The authors would like to thank the Matthews Laboratory in the College of Medicine Department of Pathology, Immunology and Laboratory Medicine at the University of Florida for the donation of samples and resources.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLAIMER
Certain commercial equipment, instruments, or materials are identified in this paper to specify adequately the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology; nor does it imply that the materials or equipment identified are necessarily the best for the purpose.
REFERENCES
- [1].van Meer G, de Kroon AI, Lipid map of the mammalian cell, J Cell Sci, 124 (2011) 5–8. [DOI] [PubMed] [Google Scholar]
- [2].Petkovic M, Vocks A, Muller M, Schiller J, Arnhold J, Comparison of different procedures for the lipid extraction from HL-60 cells: a MALDI-TOF mass spectrometric study, Z Naturforsch C, 60 (2005) 143–151. [DOI] [PubMed] [Google Scholar]
- [3].Rupasinghe TWT, Lipidomics: Extraction Protocols for Biological Matrices, in: Roessner U, Dias DA (Eds.) Metabolomics Tools for Natural Product Discovery: Methods and Protocols, Humana Press, Totowa, NJ, 2013, pp. 71–80. [DOI] [PubMed] [Google Scholar]
- [4].Berman ES, Fortson SL, Checchi KD, Wu L, Felton JS, Wu KJ, Kulp KS, Preparation of single cells for imaging/profiling mass spectrometry, J Am Soc Mass Spectrom, 19 (2008) 1230–1236. [DOI] [PubMed] [Google Scholar]
- [5].Ivanova PT, Milne SB, Myers DS, Brown HA, Lipidomics: a mass spectrometry based systems level analysis of cellular lipids, Curr Opin Chem Biol, 13 (2009) 526–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Reis A, Rudnitskaya A, Blackburn GJ, Mohd Fauzi N, Pitt AR, Spickett CM, A comparison of five lipid extraction solvent systems for lipidomic studies of human LDL, Journal of Lipid Research, 54 (2013) 1812–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Koelmel JP, Kroeger NM, Ulmer CZ, Bowden JA, Patterson RE, Cochran JA, Beecher CWW, Garrett TJ, Yost RA, LipidMatch: an automated workflow for rule-based lipid identification using untargeted high-resolution tandem mass spectrometry data, BMC Bioinformatics, 18 (2017) 331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Ma X, Chong L, Tian R, Shi R, Hu TY, Ouyang Z, Xia Y, Identification and quantitation of lipid C=C location isomers: A shotgun lipidomics approach enabled by photochemical reaction, Proc Natl Acad Sci U S A, 113 (2016) 2573–2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Christie WW, Preparation of lipid extracts from tissues, in: Christie WW (Ed.) Advances in Lipid Methodology - Two, Oily Press, Dundee, Bridgwater, U.K., 1993, pp. 193–213. [Google Scholar]
- [10].Matyash V, Liebisch G, Kurzchalia TV, Shevchenko A, Schwudke D, Lipid extraction by methyltert-butyl ether for high-throughput lipidomics, Journal of lipid research, 49 (2008) 1137–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Zahler P, Niggli V, The Use of Organic Solvents in Membrane Research, in: Korn E (Ed.) Methods in Membrane Biology, Springer; US: 1977, pp. 1–50. [Google Scholar]
- [12].Iverson SJ, Lang SL, Cooper MH, Comparison of the Bligh and Dyer and Folch methods for total lipid determination in a broad range of marine tissue, Lipids, 36 (2001) 1283–1287. [DOI] [PubMed] [Google Scholar]
- [13].Folch J, Lees M, Stanley GHS, A simple method for the isolation and purification of total lipides from animal tissues, The Journal of Biological Chemistry, 226 (1957) 497–509. [PubMed] [Google Scholar]
- [14].Bligh EG, Dyer WJ, A rapid method of total lipid extraction and purification, Can. J. Biochem. Physiol, 37 (1959) 911–917. [DOI] [PubMed] [Google Scholar]
- [15].Nakayasu ES, Nicora CD, Sims AC, Burnum-Johnson KE, Kim YM, Kyle JE, Matzke MM, Shukla AK, Chu RK, Schepmoes AA, Jacobs JM, Baric RS, Webb-Robertson BJ, Smith RD, Metz TO, MPLEx: a Robust and Universal Protocol for Single-Sample Integrative Proteomic, Metabolomic, and Lipidomic Analyses, mSystems, 1 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Knittelfelder OL, Weberhofer BP, Eichmann TO, Kohlwein SD, Rechberger GN, A versatile ultra-high performance LC-MS method for lipid profiling, J Chromatogr B Analyt Technol Biomed Life Sci, 951–952 (2014) 119–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Dunn SR, Thomas MC, Nette GW, Dove SG, A lipidomic approach to understanding free fatty acid lipogenesis derived from dissolved inorganic carbon within cnidarian-dinoflagellate symbiosis, PLoS One, 7 (2012) e46801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Fraher D, Sanigorski A, Mellett NA, Meikle PJ, Sinclair AJ, Gibert Y, Zebrafish Embryonic Lipidomic Analysis Reveals that the Yolk Cell Is Metabolically Active in Processing Lipid, Cell Rep, 14 (2016) 1317–1329. [DOI] [PubMed] [Google Scholar]
- [19].Patterson RE, Ducrocq AJ, McDougall DJ, Garrett TJ, Yost RA, Comparison of blood plasma sample preparation methods for combined LC-MS lipidomics and metabolomics, J Chromatogr B Analyt Technol Biomed Life Sci, 1002 (2015) 260–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abbott SK, Jenner AM, Mitchell TW, Brown SH, Halliday GM, Garner B, An improved high-throughput lipid extraction method for the analysis of human brain lipids, Lipids, 48 (2013) 307–318. [DOI] [PubMed] [Google Scholar]
- [21].Dietmair S, Timmins NE, Gray PP, Nielsen LK, Krömer JO, Towards quantitative metabolomics of mammalian cells: Development of a metabolite extraction protocol, Anal. Biochem, 2010, pp. 155–164. [DOI] [PubMed] [Google Scholar]
- [22].Seppanen-Laakso T, Oresic M, How to study lipidomes, Journal of Molecular Endocrinology, 42 (2009) 185–190. [DOI] [PubMed] [Google Scholar]
- [23].Galanos DS, Kapoulas VM, Isolation of polar lipids from triglyceride mixtures, Journal of Lipid Research, 3 (1962) 134–136. [Google Scholar]
- [24].Nelson GJ, Isolation and Purification of Lipids from Animal Tissues, in: Perkins EG (Ed.) Analysis of Lipids and Lipoproteins, American Oil Chemists Society, Champaign, IL, pp. 1–22. [Google Scholar]
- [25].Boyd EM, The extraction of blood lipids, Journal of Biological Chemistry, 114 (1936) 223–234. [Google Scholar]
- [26].Strober W, Trypan blue exclusion test of cell viability, Current Protocols in Immunology, Appendix 3 (2001) Appendix 3B. [DOI] [PubMed] [Google Scholar]
- [27].Ulmer CZ, Yost RA, Chen J, Mathews CE, Garrett TJ, Liquid Chromatography-Mass Spectrometry Metabolic and Lipidomic Sample Preparation Workflow for Suspension-Cultured Mammalian Cells using Jurkat T lymphocyte Cells, Journal of Proteomics & Bioinformatics, 8 (2015) 126–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Ulmer CZ, Koelmel JP, Ragland JM, Garrett TJ, Bowden JA, LipidPioneer: A Comprehensive User-Generated Exact Mass Template for Lipidomics, J. Am. Soc. Mass Spectrom, 28 (2017) 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Issac G, McDonald S, Astarita G, Lipid Separating using UPLC with Charged Surface Hybrid Technology, 2011, pp. 1–8. [Google Scholar]
- [30].Ulmer CZ, Patterson RE, Koelmel JP, Garrett TJ, Yost RA, A Robust Lipidomics Workflow for Mammalian Cells, Plasma, and Tissue Using Liquid-Chromatography High-Resolution Tandem Mass Spectrometry, in: Bhattacharya SK (Ed.) Lipidomics: Methods and Protocols, Springer; New York, New York, NY, 2017, pp. 91–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Patti GJ, Tautenhahn R, Siuzdak G, Meta-analysis of untargeted metabolomic data from multiple profiling experiments, Nat Protoc, 7 (2012) 508–516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS, MetaboAnalyst 2.0--a comprehensive server for metabolomic data analysis, Nucleic Acids Res, 40 (2012) W127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Pluskal T, Castillo S, Villar-Briones A, Oresic M, MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data, BMC Bioinformatics, 11 (2010) 395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dettmer K, Nurnberger N, Kaspar H, Gruber MA, Almstetter MF, Oefner PJ, Metabolite extraction from adherently growing mammalian cells for metabolomics studies: optimization of harvesting and extraction protocols, Analytical and bioanalytical chemistry, 399 (2011) 1127–1139. [DOI] [PubMed] [Google Scholar]
- [35].Ulmer CZ, Yost RA, Garrett TJ, Global UHPLC/HRMS Lipidomics Workflow for the Analysis of Lymphocyte Suspension Cultures, in: Wood P (Ed.) Lipidomics, Springer; New York, New York, NY, 2017, pp. 175–185. [Google Scholar]
- [36].Leon Z, Garcia-Canaveras JC, Donato MT, Lahoz A, Mammalian cell metabolomics: experimental design and sample preparation, Electrophoresis, 34 (2013) 2762–2775. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.