Abstract
The origin of species would not have been possible without high fidelity DNA replication and complex genomes evolved with mechanisms that control the initiation of DNA replication at multiple origins on multiple chromosomes such that the genome is duplicated once and only once. The mechanisms that control the assembly and activation of the replicative helicase and the initiation of DNA replication in yeast and Xenopus egg extract systems have been identified and reviewed [1,2]. The goal of this review is to organize currently available data on the mechanisms that control the initiation of DNA replication in human cells.
Preface for a general audience.
DNA replication is targeted by chemotherapeutic drugs as cancer cells generally proliferate faster than most normal cells, and most of them have acquired mutations that inactivate the mechanisms that ensure genome stability. A precise understanding of the mechanisms that initiate DNA replication will allow the rational design of clinical trials of new agents and combinations that target DNA replication. At this time our understanding of the mechanisms that initiate DNA replication is largely derived from experiments performed in highly tractable yeast and Xenopus egg extract systems. The objective of this review is to summarize what is currently known about the initiation of DNA replication in human cells.
Complexity and timing of the initiation of DNA replication.
The human genome evolved with mechanisms that assemble and activate the replicative helicase to initiate DNA unwinding and replication at ~104 origins (indirect estimations showed ~22,000 in HeLa [3], ~50,000 in HeLa Kyoto [4]). The MCM2-7 hexamer is loaded onto DNA to license origins in G1 phase and Cdc7 (working in complex with Dbf4 and also known and DDK for Dbf4-dependent kinase) and Cdk2 kinase activities in S phase initiate the assembly of CDC45. MCM2-7 and GINS and activation of the replicative helicase CMG [5]. The spatiotemporal pattern of replication foci identified by replisome components and pulses of labelled nucleotides is consistent from one cell cycle to the next and identifies regions of the genome that replicate in early, middle and late S phase [3, 6].
Origins of replication in human cells have been mapped only partially [7-9], and it appears that sets of active origins may be different for different human cell lines [8], The ~50,000 origins that replicate the human genome are selected from a large excess of “licensed” origins (origins that have loaded MCM helicases). Quantitative studies identified ~ 10 fold excess of chromatin-loaded MCM complexes over active origins in Xenopus egg extract [10] and ~20-50 fold in budding yeast [11]. However, there are few estimations for human cells. Bukhari et al. [12] in their classic widely cited study demonstrated that about 10-20% of 106 MCM complexes in HeLa cells were tightly bound to DNA, making it 2-4 fold excess over active origins. It is worth noting that this study was performed on asynchronous cells by quantifying western blots after cell fractionation, and therefore may not be very accurate. Another study by Ibarra et al. [13] showed that knocking down MCM subunits to 5-10% of their normal level was enough for cell survival, which indirectly implies a 10-20 fold excess. Further investigation is required to determine the exact excess of loaded MCM over the number of active origins in human cells. This excess is different in different human cell types and is expected to define their sensitivity to replication stress.
Activation of additional replicative helicases at origins that would otherwise be passively replicated is observed after stress [14]. This plasticity in origin use is a simple mechanism to recover DNA replication between stalled and collapsed replication forks. Origin use is also affected by oncogene expression [15].
The mechanisms that determine the spatiotemporal sequence of origin firing and identify which origins fire in any given region of the genome are not clear. Several models attempt to describe the observed replication dynamics.
A stochastic model of origin firing is supported by computational modeling [16-18]. The most recent model assumes that origins in euchromatin and facultative heterochromatin regions fire stochastically with a domino-like progression that places later firing origins in close proximity to recently fired origins. This model requires an unidentified mechanism that inhibits origin firing at a distance of 7 to 120 kbp around the replication fork, a distance that corresponds to the chromatin loops [16]. This model was able to accurately predict the observed spatiotemporal pattern of replication foci, leaving the underlying mechanism(s) to be elucidated.
An MCM loading efficiency-dependent model of origin firing is supported by observations made in yeast [19], where a higher density of MCM loading on chromatin correlates with earlier origin firing [20]. It is proposed that the number of MCM complexes loaded per origin defines its probability of firing [21]. This model doesn’t contradict the stochastic model, but rather defines the starting point for the domino-like progression, suggested by computational modelling. To date, there is no evidence that a higher density of MCM loading on chromatin correlates with earlier origin firing in human systems.
Chromatin structure has been shown to be a major factor regulating the initiation of DNA replication. Rif1 (Rap 1-interacting-factor-1) has been identified as a critical component of replication timing regulation in human cells [22]. Rif1 has been shown to regulate chromatin loop structures and to be co-localized with mid-S-phase replication foci [22]. Rif1 depletion causes increased Cdc7 activity and increased DNA synthesis in early S-phase as well as major changes in replication timing domains [22]. Since Rif1 defines chromatin loop structures, it is likely a component of the mechanism that inhibits origin firing across a loop, that is predicted by the stochastic computational model, possibly through the regulation of Cdc7 activity.
Topologically associating domains (TADs) are conserved 3D domains within the genome that were identified by studying 3D chromatin organization using Hi-C approach [23]. Recently TADs have been shown to correlate with replication domains [24]. Depending on the cell line, a TAD may be an early or late replicating unit, but the replication timing tends to be the same for all the parts of the domain: it depends on domain’s association with lamina and transcriptional repression/chromatin state. TADs and their association with replication timing were demonstrated in yeast as well [25].
Epigenetic regulation is an important factor determining the initiation of DNA replication. While some histone modification marks are common for all active origins, others are specific for a certain subset of replication initiation sites. For example, it has been shown that the activation of origins near transcription start sites require H3K14 acetylation by BRPF3-HBO1 [26]. Recently, H4K20 tri-methylation in heterochromatin has been shown to be critical for the replication timing program [27]. Another histone modification that controls the replication timing program is acetylation. One of the critical origin firing factors – treslin/TICRR – has been shown to directly interact with the acetyl-histone binding proteins BRD2 and BRD4. Disruption of treslin recruitment to acetylated chromatin results in abnormal or mixed replication patterns [28].
Some proteins that regulate origin firing by unknown mechanisms have been identified. RepID was shown to bind a subset of replication origins in human cells and be crucial for their activation [29]. The authors suggest that RepID may be a member of a family of proteins that regulate different subsets of origins in mammalian cells. SIRT1 phosphorylated at T530 was shown to prevent excessive origin firing [30]. SIRT1 is a deacetylase that may prevent the initiation of DNA replication by removing the acetylation marks from the surrounding chromatin. ATR kinase activity is also implicated in the regulation of DNA replication in the absence of stress by recent observations by our group and others that ATR kinase inhibition induces origin firing [31-33].
Replication complex assembly
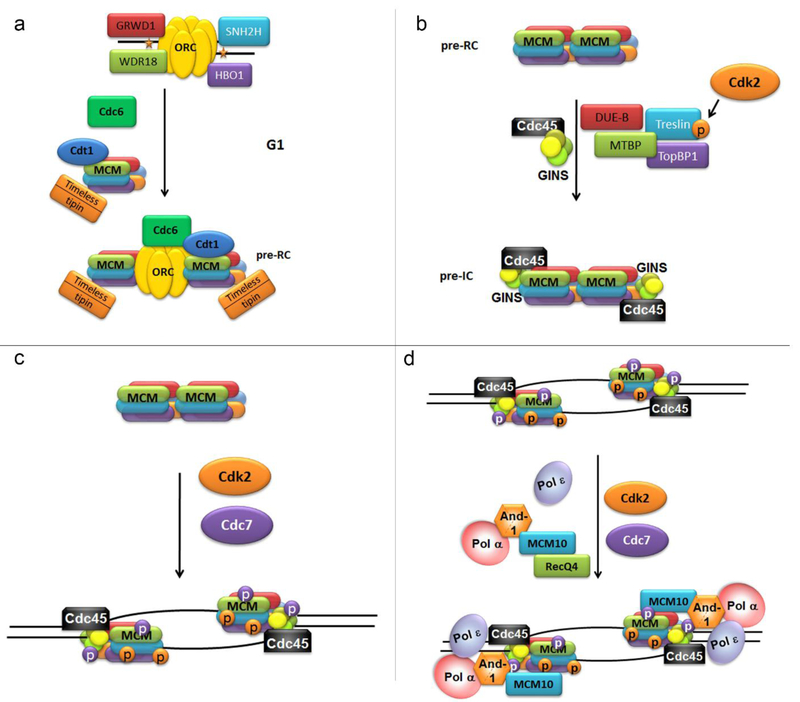
The assembly of the replisome in human cells and all eukaryotic systems begins with origin licensing in the G1 phase of the cell cycle (Figure 1a).
Figure 1.
Replication initiation in human cells. 1a. MCM loading; 1b.CMG assembly; 1c. CMG activation. 1d. Full replication complex
MCM loading on chromatin in G1 phase human cells, just like yeast, requires ORC (Origin Recognition Complex)[34, 35], Cdt1 [36], and Cdc6 [37] and these proteins need to be tightly regulated to prevent re-loading of the replicative helicase and re-replication in S phase [38]. However, the mechanisms preventing re-replication are quite different between species. While yeast ORC complex remains bound to replication origins throughout the cell cycle, in mammalian cells ORC1 subunit is selectively degraded starting at the onset of S-phase[39], and Cdk1/Cyclin A-dependent phosphorylation prevents its binding to chromatin until mitosis is complete[40]. While yeast Cdc6 is rapidly degraded in S-phase, its human counterpart is very stable and exported from the nucleus in a Cdk2 phosphorylation-dependent manner [41-43]. Cdt1 is quickly degraded at the onset of S-phase in human cells [44], partial conservation of this mechanism is reviewed in [45]. In human cells [46] and Xenopus egg extracts [47] Cdt1 can also be inhibited by binding to an inhibitor protein - geminin. Geminin is absent in yeast.
MCM loading on chromatin requires additional proteins in human cells:
-
-
HBO1 (human acetylase binding to ORC1; also known as KAT7 and MYST2) has been shown to interact with ORC, Cdt1 and MCM and possess specific H4-acetyltransferase activity that is critical for MCM chromatin loading [48].
-
-
SNF2H (sucrose nonfermenting 2 homolog) is a non-essential chromatin remodeler that functions in MCM loading downstream of Cdt1 [49].
-
-
GRWD1 (glutamate-rich WD40 repeat containing 1) protein was identified as a Cdt1 interacting partner and was subsequently shown to function in MCM loading, presumably through histone binding and chromatin remodeling [50].
-
-
WDR18 (WD repeat domain 18) or hIPI3 (human homolog of yeast Ipi3p) was shown to interact with ORC and MCM subunits and to function in MCM loading [51].
Three of these additional proteins are chromatin remodelers, suggesting that histone modification is important for MCM loading in humans.
While ORC marks replication origins and directly participates in MCM loading on DNA, it appears that MCM and ORC do not co-localize [52]. MCM helicases seem to not localize to replication foci in the live cell imaging experiments either [53]. These observations together with the excessive loading of MCM on chromatin, discussed above, constitute the MCM paradox reviewed in [54].
Origin licensing in G1 phase is followed by the assembly of the replicative helicase, CMG, through the recruitment of CDC45 and GINS to the loaded MCM (Figure 1b).
CDC45 is an essential component of the CMG helicase that facilitates RPA loading on ssDNA in human cells [55]. CDC45 appears to be rate-limiting for the initiation of DNA replication in human cells: overexpression of CDC45 causes increased origin firing and ssDNA accumulation [56]. However, there are complex mechanisms regulating CDC45 recruitment to MCM.
It has been clearly established that in yeast Cdc45 loading requires Sld3, Sld7 proteins and DDK kinase activity [57]. Treslin (main candidate for the role of Sld3 homolog in human cells) and TopBP1 were demonstrated to interact with Cdc45 [58, 59] and be required for its recruitment to chromatin in human cells [59, 60]. Treslin was also shown to be phosphorylated by cyclin A/Cdk2 and this promoted the treslin-TopBP1 interaction and CDC45 loading [58, 61]. It has been demonstrated that MTBP (MDM2 binding protein) forms a complex with treslin and TopBP1 and that MTBP functions in CMG assembly in human cells [62].
In contrast, bimolecular fluorescence complementation (BiFC) analyses revealed that TopBP1 is dispensable for CMG formation in human cells [63]. This observation contradicts both the data described above and the finding that TopBP1-deficient cells had lower levels of Cdk2 activity, impaired loading of replication components on chromatin, and failed to enter S-phase [64], so further investigation is required to resolve this contradiction.
More recently two more players in Cdc45 loading were identified in Xenopus: DUE-B (DNA unwinding element-binding protein) [65] and GEMC1 [66]. DUE-B is another candidate for the functional role of yeast Sld3 in human cells, and its C-terminal phosphorylation by Cdc7 (that can be reversed by PP2A) was shown to be important for Cdc45 chromatin association in human cells [67]. While a human homolog of Xenopus GEMC1 has been identified [68], there’s no evidence that it plays a role in the initiation of DNA replication in human cells.
Thus, the proteins that are likely required for CDC45 loading in humans are Cdk2-phosphorylated treslin, Cdc7-phosphorylated DUE-B, MTBP and TopBP1.
It has been shown that CDC45 loading at origins near transcription start sites in human cells requires H3K14 acetylation by BRPF3-HBO1 [26]. A recent study [69] demonstrated that demethylation of histone H3K9 by Kdm4d was critical for CDC45 recruitment while it does not affect origin licensing step. This observation suggests that one of the players in the “CDC45 recruitment team” described above is regulated by this chromatin modification. Another study showed that treslin interacts with acetylated chromatin [28]. This may suggest that CMG assembly is regulated by histone acetylation through the recruitment of treslin to facilitate CDC45 incorporation.
The GINS complex is an essential component of CMG helicase [70], but little is known about the mechanism(s) underlying the recruitment of GINS to MCM/CDC45 in human cells. DNA polymerase epsilon (pol ε) is required to recruit GINS to MCM helicase in yeast [71], but there is no evidence that this is the case in human cells. It has also been shown that the binding of GINS and CDC45 to CMG may be facilitative, that is the binding of GINS promotes the binding of CDC45, and vice versa [63]. The recruitment of GINS to CMG is one of the least studied aspects of the initiation of DNA replication in human cells.
Additional factors demonstrated to be important for origin firing in human cells are MCM10, RECQ4 and And-1 (Ctf4 in yeast) (Figure 1d). MCM10, RecQ4, and And-1 form a complex in a Cdc7- and Cdk2-dependent manner, and the recruitment of this complex to CMG helicase is an essential step in origin firing in human cells [72]. MCM10 also interacts with ssDNA [73], MCM2-7 [74] and CDC45 (in a DNA-dependent manner) [75], while And-1 has been shown to form a stable complex with DNA polymerase alpha [76] and interact with GINS in a manner dependent on Sld5 phosphorylation by Cdc7, but independent of DNA [33]. These data indicate that RECQ4-MCM10-And-1-polymerase alpha complex is recruited to the CMG helicase through the Cdc7-dependent interaction between And-1 and GINS, and that this is facilitated by MCM10 binding to the MCM hexamer. A DNA-dependent interaction between MCM10 and CDC45 stabilizes the complex once the helicase is activated and ssDNA is available.
The recruitment of the main replicative DNA polymerases - delta and epsilon - to origins is not well studied in human cells. While in yeast polymerase epsilon is recruited as a part of a well-characterized complex with GINS and other proteins [71], no such complex has been described in human cells. Interactions between the human GINS complex and three DNA polymerases – alpha, delta and epsilon – has been demonstrated using purified proteins [77] and in human cells [74], as in Xenopus [78], polymerase epsilon was shown to be loaded after Cdc45, but before polymerase alpha. The molecular mechanism underlying the recruitment is not known.
Timeless (TIM) is a homolog of the fruit fly gene responsible for the circadian clock system [79]. TIM and its interacting partner TIPIN have been shown to be essential for DNA replication in humans [80] and their yeast homologs Tof1 and Csm3 are required for maximum processivity in yeast [81]. While both yeast [82] and human [83] TIM-TIPIN complexes have been shown to travel with the replication fork, no molecular mechanism for their recruitment has been identified. A recent study [84] indicates that TIM directly interacts with the MCM helicase before it is loaded on chromatin, and that disruption of TIM leads to MCM loading on chromatin and CMG assembly outside of S-phase as well as delayed initiation of DNA replication. These data suggest that TIM-TIPIN are recruited to the replication complex together with the MCM complex at the time of origin licensing in G1 phase and while they are not essential for MCM loading, they are required for timely and efficient replication initiation.
The signaling required for the initiation of DNA replication
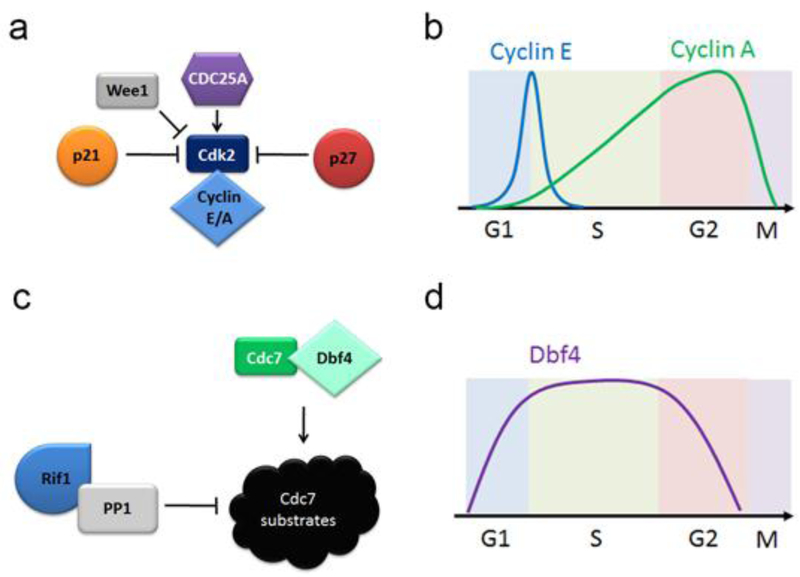
The kinases Cdc7 [85, 86] and Cdk2 [72] play critical roles in the assembly and activation of pre-replication complexes in human cells (figure 1c) and as such their regulation determines the initiation of DNA replication. CMG helicase activation in yeast and human cells requires MCM phosphorylation by Cdc7/DDK. Dbf4-dependent kinase (DDK) (Figure 2c, d) is a complex of Cdc7 and either Dbf4 or Dbf4b (Drf1) [87]. Dbf4 or Dbf4b is required for maximum Cdc7 kinase activity and substrate recognition. In Xenopus, Dbf4b is proposed to play a critical role in the initiation of DNA replication that is more important than that of the canonical Dbf4 [88]. Human Dbf4b, unlike its Xenopus homolog, doesn’t bind chromatin, but appears to play an important role in the S and G2/M phases of the cell cycle [89, 90].
Figure 2.
Replication initiation signaling. 2a. Cdk2 regulation; 2b. Cyclins’ regulation. 2c. Cdc7 regulation. 2d. Dbf4 regulation.
It has been demonstrated that Treslin stimulates MCM2 phosphorylation by Cdc7 and that this may contribute to MCM gate opening [91]. In addition to Treslin, at least two more proteins appear to be important for Cdc7 recruitment to CMG in human cells: both Claspin [26] and Cdt1 [92] have been shown to directly interact with Cdc7 and recruit it to CMG helicase. Recent data [33] indicate that efficient MCM4 phosphorylation by Cdc7 requires the presence of And-1, however there’s no evidence of a direct interaction between And-1 and either Cdc7 or Dbf4. And-1 has been demonstrated to interact with Claspin [93], so one possible explanation is that And-1 is required to recruit Claspin, which in turn brings Cdc7 to the replication origins. However, there’s a controversy in Cdc7 activity being crucial for the recruitment of And-1 [72]. One possible explanation would be that both Claspin and Cdc7 interact with RecQ4-MCM10-And-1-polymerase alpha complex and Cdc7 phosphorylates its components before the recruitment resulting in stronger affinity for the CMG, however more research is required to investigate this process as it is inconsistent with Cdc7-dependent phosphorylation of DUE-B at the CDC45 recruitment step.
Cdk2 (Figure 2a, b) is required for both the assembly and activation of CMG and RecQ4-MCM10-And-1 recruitment during the initiation of DNA replication (discussed above). Cdk2 is a cyclin dependent kinase that is associated with either Cyclin E (in G1/S transition) or Cyclin A (during S-phase) [94] and the active kinase is phosphorylated on Thr160 and dephosphorylated on Tyr15 [95]. Cdk2 Tyr15 is phosphorylated by Wee1 kinase and dephosphorylated by Cdc25A phosphatase. This dephosphorylation appears to be the critical step in Cdk2 activation [96]. Cdk2 is an abundant protein in human cell (approximately 8.8×105 Cdk2 copies per HeLa cell compared to 2.1×104 copies of Cdc7 [97]) and with increased levels of cyclins during G1/S phase Cdk2 activity is unlikely to be rate-limiting for the initiation of DNA replication. Surprisingly, Cdk2 knockout mice are viable [98]. Cdk1 appears to be able to substitute for the absent Cdk2, in most of its functions, however, a study with analog sensitive Cdk2 mutant [99] showed an essential and non-redundant role for Cdk2 in G1/S transition, that includes outcompeting Cdk1 for binding to cyclins.
Recent observations that localized PP1 phosphatase activity is required for the spatiotemporal sequence of origin firing [100] suggests that the assembly and activation of CMG and the replisome is controlled by the gradients of kinase and phosphatase activities across the nucleus. While Cdc7 is localized to origins in a Claspin- or Cdt1-dependent manner, the opposing activities of PP1 family phosphatases, which removes Cdc7- dependent phosphorylations, is localized to origins though an interaction with Rif1 [100, 101]. The stochastic model of origin firing discussed above requires a unidentified mechanism that inhibits origin firing at a specific distance (7 to 120 kbp) around replication fork, a distance that corresponds to the chromatin loop size [16]. Since Rif1 is the main structural determinant of chromatin loops, the following model seems feasible: Rif1 recruits PP1 family phosphatases to the actively replicating loops to repress the local origin firing. While many questions remain, including the mechanism of PP1 recruitment to replicating but not replication loops that have been replicated or are yet to replicate, the Rif1-PP1 interaction may be the unidentified mechanism that inhibits origin firing at a specific distance predicted by the functional modeling of replication.
Regulation of replication initiation after DNA damage
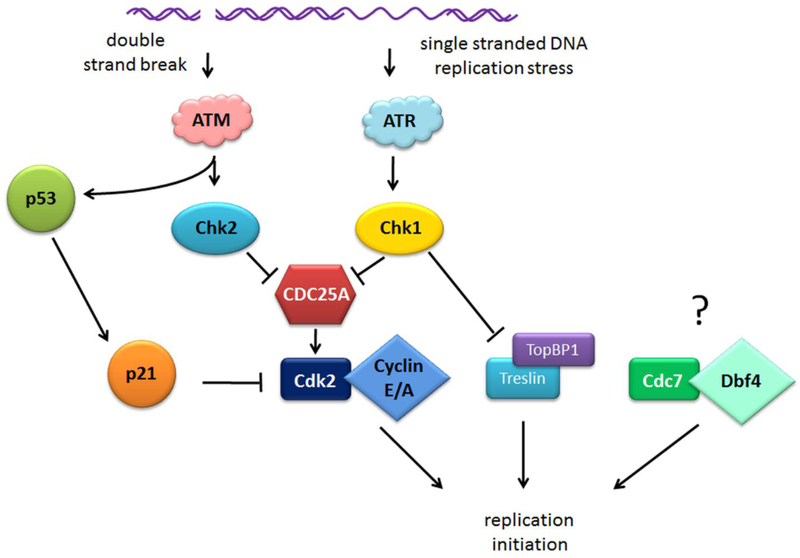
Key elements of the DNA damage response include cell cycle arrest and inhibition of the initiation of DNA replication (Figure 3). The regulation of DNA replication after DNA damage is a very complex process. While mild replication stress activates the initiation of replication from dormant origins, higher levels of damage completely block origin firing. Current models for this phenomenon suggests that low levels of ATR/Chk1 activity block the activation of replication in the new “replication factories” while allowing dormant origins to fire within active factories [102]. The main target of the replication checkpoint is Cdk2 kinase. Cdc25A phosphatase, that is critical for Cdk2 dephosphorylation and activation, is a known target of Chk1 and Chk2 kinases. Cdc25A is phosphorylated by Chk2 [103] and/or Chk1 [104] at serines 123, 178, 278, and 292 in response to DNA damage, and these phosphorylations lead to Cdc25A ubiquitylation and proteasomal degradation. In the absence of Cdc25A, Cdk2 remains in its Y15 phosphorylated inactive form and is unable to facilitate replication initiation.
Figure 3.
Regulation of DNA replication initiation after DNA damage
Another pathway that contributes to Cdk2 inactivation after DNA damage is p53-dependent. The tumor suppressor protein p53 is stabilized and activated in response to DNA damage in ATM/ATR and Chk1/Chk2 dependent manner. Active p53 acts as a transcription factor, inducing the expression of multiple DNA damage response genes which impact cell cycle arrest, senescence and apoptosis. One of these genes is the Cdk inhibitor p21 which directly interacts with and inhibits Cdk2 kinase activity [105] which blocks its ability to initiate DNA replication. Cdk2 can also be inhibited by p27 [106], but there is no evidence to suggest that p27 is regulated in response to replication stress.
While the Chk1/2-Cdc25A-Cdk2 axis signaling is considered the main mechanism that blocks the initiation of DNA replication after DNA damage, additional pathways have emerged. It has been demonstrated that in response to the replication stress induced by benzo[a]pyrene dihydrodiol epoxide (BPDE) the initiation of DNA replication is blocked in the absence of any noticeable Cdc25A degradation. In this instance, the block of replication initiation was found to be due to Chk1-dependent inhibition of CDC45 recruitment to licensed origins [107]. One possible explanation of this phenomenon is that Chk1-dependent phosphorylation of Treslin in response to replication stress disrupts the interaction of Treslin with TopBP1 that is known to be critical for CDC45 recruitment to CMG from yeast to Xenopus and human [61, 108].
Many studies have investigated the regulation of Cdc7-Dbf4 activity in response to DNA damage, but the results are largely inconclusive. While in yeast Rad53-dependent phosphorylation of Dbf4 [109] has been demonstrated to contribute to the replication checkpoint, ATR-dependent regulation of DDK activity in human cells and Xenopus cell free extract remains highly controversial [110]. Some studies claim that the DDK complex is dissociated, and Cdc7 activity is decreased in response to replication checkpoint signaling induced by etoposide in Xenopus cell-free extracts [60], and even in human cells [111], while others detect no effect of ATR/Chk1 activation on Cdc7-Dbf4 interaction in human cells after UVC irradiation [112], and no reduction in Cdc7 activity after treatments with hydroxyurea or etoposide [113]. However, Dbf4 overexpression appeared to abrogate the UVC-induced (but not IR-induced) replication checkpoint [112]. These overexpression experiments are hard to interpret as Dbf4 appears to be one of the limiting factors in the regulation of the initiation of DNA replication that is stabilized at the G1/S transition [114], so its overexpression may have affected the cell cycle and DNA synthesis rate in general. Dbf4 has been demonstrated to be a direct substrate of both ATM/ATR [110] and Chk1 [112] kinases in human cells, and mutant unphosphorylatable Dbf4 was able to partially suppress IR-induced replication arrest [110]. However the role of ATM/ATR-dependent phosphorylations on Dbf4 in inhibiting the initiation of DNA replication after DNA damage is not clear, as they do not affect DDK activity or stability, and appear to only prevent rereplication [110]. One study suggests that DDK activity during replication stress acts upstream of ATR/Chk1 signaling as it is shown that Dbf4 overexpression inhibits replication checkpoint in both human and Xenopus systems [115].
The variability in the type of damage as well as the DNA synthesis assays used to study the impact of DDK on the replication checkpoint and the frequent use of Dbf4 overexpression makes it very difficult to compare and interpret different studies. Further investigation is needed to establish the role of the ATR-DDK axis in the replication checkpoint. With recent observations that the PP1 family of phosphatases contribute to reversing Cdc7-dependent phosphorylations at dormant origins [100, 101], regulation of these phosphatases in response to DNA damage should also be investigated.
Conclusions and future directions
The initiation of DNA replication in humans is an extremely complex process, and caution is necessary while attempting to translate findings from relatively simple yeast and Xenopus extract systems into mammalian cells. Major gaps in knowledge about origin firing in human cells include the mechanism though which the GINS complex and DNA polymerase epsilon are recruited to the MCM helicase and mechanisms that regulate DDK after DNA damage. Also, while the requirement for Cdk2 and Cdc7 activities for certain steps in replication initiation is clear, the targets of these kinases and the functions of the phosphorylated targets are yet to be elucidated in detail. It is very likely that kinase-phosphatase gradients play critical roles in the regulation of the initiation of DNA replication, and that balancing Cdc7- Cdk2- Chk1 and ATR-dependent phosphorylation systems determines the spatiotemporal pattern of origin firing that is conserved from one cell division to the next. Identifying the mechanisms that underlie these gradients is of fundamental and translational interest.
Table 1:
Key players of replication initiation in various organisms:
| Step | S. cerevisiae | Xenopus laevis | Homo sapiens |
|---|---|---|---|
| MCM loading | MCM2-7; Cdt1; Cdc6; ORC1-6 |
MCM2-7; Cdt1; Cdc6; ORC1-6 |
MCM2-7; Cdt1; Cdc6; ORC1-6; HBO1; SNF2H; GRWD1; WDR18 |
| CMG assembly | Cdc45; GINS 1-4; Sld3; Sld7; polymerase epsilon; CDK; DDK, |
Cdc45; GINS 1-4; Treslin; DUE-B; MTBP; TopBP1; GEMC1; Cdk2; DDK |
CDC45; GINS 1-4; Treslin; DUE-B; MTBP; TopBP1; Cdk2; DDK |
| CMG activation | DDK | DDK | DDK |
| Replication initiation |
Ctf4, MCM10, polymerase alpha |
Ctf4, MCM10, polymerase alpha |
And-1, MCM10, RecQ4, polymerase alpha, DDK, Cdk2 |
Table 2.
Modifications, required for origin firing in various organisms (X – unidentified substrate)
| Step | S. cerevisiae | Xenopus laevis | Homo sapiens |
|---|---|---|---|
| MCM loading | Acetyl-H3K14 Acetyl-H4 |
||
| CMG assembly | Phospho-S1d3 (CDK) Phospho-MCM (DDK) |
Phospho-Treslin (CDK) | Phospho-Treslin (Cdk2) Phospho-Due-B (DDK) |
| CMG activation | Phospho-MCM (DDK) | Phospho-MCM (DDK) | Phospho-MCM (DDK) |
| Replication initiation |
Phospho-MCM10(CDK) | Phospho-GIN S4(DDK) Phospho-X (Cdk2) |
Acknowledgements:
This work was supported by the NIH Grant RO1 CA204173
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- 1.Bell SP and Labib K, Chromosome Duplication in Saccharomyces cerevisiae. Genetics, 2016. 203(3): p. 1027–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blow JJ and Laskey RA, Xenopus cell-free extracts and their contribution to the study of DNA replication and other complex biological processes. Int J Dev Biol, 2016. 60(7-8-9): p. 201–207. [DOI] [PubMed] [Google Scholar]
- 3.Jackson DA and Pombo A, Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol, 1998. 140(6): p. 1285–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chagin VO, et al. , 4D Visualization of replication foci in mammalian cells corresponding to individual replicons. Nat Commun, 2016. 7: p. 11231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O'Donnell M, Langston L, and Stillman B, Principles and concepts of DNA replication in bacteria, archaea, and eukarya. Cold Spring Harb Perspect Biol, 2013. 5(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira J, et al. , Spatial organization of large-scale chromatin domains in the nucleus: a magnified view of single chromosome territories. J Cell Biol, 1997. 139(7): p. 1597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerhardt J, et al. , Identification of new human origins of DNA replication by an origintrapping assay. Mol Cell Biol, 2006. 26(20): p. 7731–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petryk N, et al. , Replication landscape of the human genome. Nat Commun, 2016. 7: p. 10208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Picard F, et al. , The spatiotemporal program of DNA replication is associated with specific combinations of chromatin marks in human cells. PLoS Genet, 2014. 10(5): p. e1004282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mahbubani HM, et al. , Cell cycle regulation of the replication licensing system: involvement of a Cdk-dependent inhibitor. J Cell Biol, 1997. 136(1): p. 125–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donovan S, et al. , Cdc6p-dependent loading of Mcm proteins onto pre-replicative chromatin in budding yeast. Proc Natl Acad Sci U S A, 1997. 94(11): p. 5611–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Burkhart R, et al. , Interactions of human nuclear proteins P1Mcm3 and P1Cdc46. Eur J Biochem, 1995. 228(2): p. 431–8. [PubMed] [Google Scholar]
- 13.Ibarra A, Schwob E, and Mendez J, Excess MCM proteins protect human cells from replicative stress by licensing backup origins of replication. Proc Natl Acad Sci U S A, 2008. 105(26): p. 8956–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ge XQ, Jackson DA, and Blow JJ, Dormant origins licensed by excess Mcm2–7 are required for human cells to survive replicative stress. Genes Dev, 2007. 21(24): p. 3331–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Macheret M and Halazonetis TD, Intragenic origins due to short G1 phases underlie oncogene-induced DNA replication stress. Nature, 2018. 555(7694): p. 112–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lob D, et al. , 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression. Nat Commun, 2016. 7: p. 11207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gindin Y, et al. , A chromatin structure-based model accurately predicts DNA replication timing in human cells. Mol Syst Biol, 2014. 10: p. 722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Miotto B, Ji Z, and Struhl K, Selectivity of ORC binding sites and the relation to replication timing, fragile sites, and deletions in cancers. Proc Natl Acad Sci U S A, 2016. 113(33): p. E4810–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hyrien O, How MCM loading and spreading specify eukaryotic DNA replication initiation sites. F1000Res, 2016. 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang SC, Rhind N, and Bechhoefer J, Modeling genome-wide replication kinetics reveals a mechanism for regulation of replication timing. Mol Syst Biol, 2010. 6: p. 404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das SP, et al. , Replication timing is regulated by the number of MCMs loaded at origins. Genome Res, 2015. 25(12): p. 1886–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamazaki S, et al. , Rif1 regulates the replication timing domains on the human genome. EMBO J, 2012. 31(18): p. 3667–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dixon JR, et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature, 2012. 485(7398): p. 376–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pope BD, et al. , Topologically associating domains are stable units of replicationtiming regulation. Nature, 2014. 515(7527): p. 402–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eser U, et al. , Form and function of topologically associating genomic domains in budding yeast. Proc Natl Acad Sci U S A, 2017. 114(15): p. E3061–E3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng Y, et al. , BRPF3-HBO1 regulates replication origin activation and histone H3K14 acetylation. EMBO J, 2016. 35(2): p. 176–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brustel J, et al. , Histone H4K20 tri-methylation at late-firing origins ensures timely heterochromatin replication. EMBO J, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sansam CG, et al. , A mechanism for epigenetic control of DNA replication. Genes Dev, 2018. 32(3–4): p. 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, et al. , A replicator-specific binding protein essential for site-specific initiation of DNA replication in mammalian cells. Nat Commun, 2016. 7: p. 11748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Utani K, et al. , Phosphorylated SIRT1 associates with replication origins to prevent excess replication initiation and preserve genomic stability. Nucleic Acids Res, 2017. 45(13): p. 7807–7824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Couch FB, et al. , ATR phosphorylates SMARCAL1 to prevent replication fork collapse. Genes Dev, 2013. 27(14): p. 1610–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kwok M, et al. , Synthetic lethality in chronic lymphocytic leukaemia with DNA damage response defects by targeting the ATR pathway. Lancet, 2015. 385 Suppl 1: p. S58. [DOI] [PubMed] [Google Scholar]
- 33.Moiseeva T, et al. , ATR kinase inhibition induces unscheduled origin firing through a Cdc7-dependent association between GINS and And-1. Nat Commun, 2017. 8(1): p.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takeda DY, et al. , Recruitment of ORC or CDC6 to DNA is sufficient to create an artificial origin of replication in mammalian cells. Genes Dev, 2005. 19(23): p. 2827–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Giordano-Coltart J, et al. , Studies of the properties of human origin recognition complex and its Walker A motif mutants. Proc Natl Acad Sci U S A, 2005. 102(1): p. 69–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rialland M, Sola F, and Santocanale C, Essential role of human CDT1 in DNA replication and chromatin licensing. J Cell Sci, 2002. 115(Pt 7): p. 1435–40. [DOI] [PubMed] [Google Scholar]
- 37.Cook JG, et al. , Analysis of Cdc6 function in the assembly of mammalian prereplication complexes. Proc Natl Acad Sci U S A, 2002. 99(3): p. 1347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimoto N, et al. , Redundant and differential regulation of multiple licensing factors ensures prevention of re-replication in normal human cells. J Cell Sci, 2009. 122(Pt 8): p. 1184–91. [DOI] [PubMed] [Google Scholar]
- 39.Tatsumi Y, et al. , The ORC1 cycle in human cells: I. cell cycle-regulated oscillation of human ORC1. J Biol Chem, 2003. 278(42): p. 41528–34. [DOI] [PubMed] [Google Scholar]
- 40.Li CJ, Vassilev A, and DePamphilis ML, Role for Cdk1 (Cdc2)/cyclin A in preventing the mammalian origin recognition complex’s largest subunit (Orc1) from binding to chromatin during mitosis. Mol Cell Biol, 2004. 24(13): p. 5875–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saha P, et al. , Human CDC6/Cdc18 associates with Orc1 and cyclin-cdk and is selectively eliminated from the nucleus at the onset of S phase. Mol Cell Biol, 1998. 18(5): p. 2758–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jiang W, Wells NJ, and Hunter T, Multistep regulation of DNA replication by Cdk phosphorylation of HsCdc6. Proc Natl Acad Sci U S A, 1999. 96(11): p. 6193–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petersen BO, et al. , Phosphorylation of mammalian CDC6 by cyclin A/CDK2 regulates its subcellular localization. EMBO J, 1999. 18(2): p. 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nishitani H, Lygerou Z, and Nishimoto T, Proteolysis of DNA replication licensing factor Cdt1 in S-phase is performed independently of geminin through its N-terminal region. J Biol Chem, 2004. 279(29): p. 30807–16. [DOI] [PubMed] [Google Scholar]
- 45.Kim Y and Kipreos ET, Cdt1 degradation to prevent DNA re-replication: conserved and non-conserved pathways. Cell Div, 2007. 2: p. 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wohlschlegel JA, et al. , Inhibition of eukaryotic DNA replication by geminin binding to Cdt1. Science, 2000. 290(5500): p. 2309–12. [DOI] [PubMed] [Google Scholar]
- 47.Maiorano D, Rul W, and Mechali M, Cell cycle regulation of the licensing activity of Cdt1 in Xenopus laevis. Exp Cell Res, 2004. 295(1): p. 138–49. [DOI] [PubMed] [Google Scholar]
- 48.Miotto B and Struhl K, HBO1 histone acetylase activity is essential for DNA replication licensing and inhibited by Geminin. Mol Cell, 2010. 37(1): p. 57–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugimoto N, et al. , Chromatin remodeler sucrose nonfermenting 2 homolog (SNF2H) is recruited onto DNA replication origins through interaction with Cdc10 protein-dependent transcript 1 (Cdt1) and promotes pre-replication complex formation. J Biol Chem, 2011. 286(45): p. 39200–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sugimoto N, et al. , Cdt1-binding protein GRWD1 is a novel histone-binding protein that facilitates MCM loading through its influence on chromatin architecture. Nucleic Acids Res, 2015. 43(12): p. 5898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huang Y, et al. , A Role of hIPI3 in DNA Replication Licensing in Human Cells. PLoS One, 2016. 11(4): p. e0151803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ritzi M, et al. , Human minichromosome maintenance proteins and human origin recognition complex 2protein on chromatin. J Biol Chem, 1998. 273(38): p. 24543–9. [DOI] [PubMed] [Google Scholar]
- 53.Symeonidou IE, et al. , Multi-step loading of human minichromosome maintenance proteins in live human cells. J Biol Chem, 2013. 288(50): p. 35852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Das M, et al. , MCM Paradox: Abundance of Eukaryotic Replicative Helicases and Genomic Integrity. Mol Biol Int, 2014. 2014: p. 574850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szambowska A, et al. , Cdc45-induced loading of human RPA onto single-stranded DNA. Nucleic Acids Res, 2017. 45(6): p. 3217–3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kohler C, et al. , Cdc45 is limiting for replication initiation in humans. Cell Cycle, 2016. 15(7): p. 974–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yeeles JT, et al. , Regulated eukaryotic DNA replication origin firing with purified proteins. Nature, 2015. 519(7544): p. 431–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kumagai A, et al. , Direct regulation of Treslin by cyclin-dependent kinase is essential for the onset of DNA replication. J Cell Biol, 2011. 193(6): p. 995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schmidt U, et al. , Characterization of the interaction between the human DNA topoisomerase IIbeta-binding protein 1 (TopBP1) and the cell division cycle 45 (Cdc45) protein. Biochem J, 2008. 409(1): p. 169–77. [DOI] [PubMed] [Google Scholar]
- 60.Kumagai A, et al. , Treslin collaborates with TopBP1 in triggering the initiation of DNA replication. Cell, 2010. 140(3): p. 349–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Boos D, et al. , Regulation of DNA replication through Sld3-Dpb11 interaction is conserved from yeast to humans. Curr Biol, 2011. 21(13): p. 1152–7. [DOI] [PubMed] [Google Scholar]
- 62.Boos D, Yekezare M, and Diffley JF, Identification of a heteromeric complex that promotes DNA replication origin firing in human cells. Science, 2013. 340(6135): p. 981–4 [DOI] [PubMed] [Google Scholar]
- 63.Im JS, et al. , Assembly of the Cdc45-Mcm2-7-GINS complex in human cells requires the Ctf4/And-1, RecQL4, and Mcm10 proteins. Proc Natl Acad Sci U S A, 2009. 106(37): p. 15628–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jeon Y, et al. , Human TopBP1 participates in cyclin E/CDK2 activation and preinitiation complex assembly during G1/S transition. J Biol Chem, 2007. 282(20): p. 14882–90. [DOI] [PubMed] [Google Scholar]
- 65.Chowdhury A, et al. , The DNA unwinding element binding protein DUE-B interacts with Cdc45 in preinitiation complex formation. Mol Cell Biol, 2010. 30(6): p. 1495–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Balestrini A, et al. , GEMC1 is a TopBP1-interactingprotein required for chromosomal DNA replication. Nat Cell Biol, 2010. 12(5): p. 484–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gao Y, et al. , Protein phosphatase 2A and Cdc7 kinase regulate the DNA unwinding element-binding protein in replication initiation. J Biol Chem, 2014. 289(52): p. 35987–6000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Caillat C, et al. , The structure of the GemC1 coiled coil and its interaction with the Geminin family of coiled-coil proteins. Acta Crystallogr D Biol Crystallogr, 2015. 71(Pt 11): p. 2278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wu R, et al. , H3K9me3 demethylase Kdm4d facilitates the formation of pre-initiative complex and regulates DNA replication. Nucleic Acids Res, 2017. 45(1): p. 169–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Aparicio T, et al. , The human GINS complex associates with Cdc45 and MCM and is essential for DNA replication. Nucleic Acids Res, 2009. 37(7): p. 2087–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Muramatsu S, et al. , CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev, 2010. 24(6): p. 602–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Im JS, et al. , RecQL4 is required for the association of Mcm10 and Ctf4 with replication origins in human cells. Cell Cycle, 2015. 14(7): p. 1001–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Perez-Arnaiz P and Kaplan DL, An Mcm10 Mutant Defective in ssDNA Binding Shows Defects in DNA Replication Initiation. J Mol Biol, 2016. 428(23): p. 4608–4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Izumi M, et al. , The Mcm2-7-interacting domain of human mini-chromosome maintenance 10 (Mcm10) protein is important for stable chromatin association and origin firing. J Biol Chem, 2017. 292(31): p. 13008–13021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Di Perna R, et al. , The physical interaction of Mcm10 with Cdc45 modulates their DNA-binding properties. Biochem J, 2013. 454(2): p. 333–43. [DOI] [PubMed] [Google Scholar]
- 76.Zhu W, et al. , Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev, 2007. 21(18): p. 2288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bermudez VP, et al. , Studies on human DNA polymerase epsilon and GINS complex and their role in DNA replication. J Biol Chem, 2011. 286(33): p. 28963–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mimura S, et al. , Central role for cdc45 in establishing an initiation complex of DNA replication in Xenopus egg extracts. Genes Cells, 2000. 5(6): p. 439–52. [DOI] [PubMed] [Google Scholar]
- 79.Sangoram AM, et al. , Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron, 1998. 21(5): p. 1101–13. [DOI] [PubMed] [Google Scholar]
- 80.Yoshizawa-Sugata N and Masai H, Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem, 2007. 282(4): p. 2729–40. [DOI] [PubMed] [Google Scholar]
- 81.Lewis JS, et al. , Single-molecule visualization of Saccharomyces cerevisiae leading-strand synthesis reveals dynamic interaction between MTC and the replisome. Proc Natl Acad Sci U S A, 2017. 114(40): p. 10630–10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Katou Y, et al. , S-phase checkpoint proteins Tof1 and Mrcl form a stable replicationpausing complex. Nature, 2003. 424(6952): p. 1078–83. [DOI] [PubMed] [Google Scholar]
- 83.Leman AR, et al. , Human Timeless and Tipin stabilize replication forks and facilitate sister-chromatid cohesion. J Cell Sci, 2010. 123(Pt 5): p. 660–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu X, et al. , TIMELESS Suppresses the Accumulation of Aberrant CDC45.MCM2-7.GINS Replicative Helicase Complexes on Human Chromatin. J Biol Chem, 2016. 291(43): p. 22544–22558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Jiang W, et al. , Mammalian Cdc7-Dbf4 protein kinase complex is essential for initiation of DNA replication. EMBO J, 1999. 18(20): p. 5703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsuji T, Ficarro SB, and Jiang W, Essential role of phosphorylation of MCM2 by Cdc7/Dbf4 in the initiation of DNA replication in mammalian cells. Mol Biol Cell, 2006. 17(10): p. 4459–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kitamura R, et al. , Molecular mechanism of activation of human Cdc7 kinase: bipartite interaction with Dbf4/activator of S phase kinase (ASK) activation subunit stimulates ATP binding and substrate recognition. J Biol Chem, 2011. 286(26): p. 23031–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Silva T, et al. , Xenopus CDC7/DRF1 complex is required for the initiation of DNA replication. J Biol Chem, 2006. 281(17): p. 11569–76. [DOI] [PubMed] [Google Scholar]
- 89.Montagnoli A, et al. , Drf1, a novel regulatory subunit for human Cdc7 kinase. EMBO J, 2002. 21(12): p. 3171–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yoshizawa-Sugata N, et al. , A second human Dbf4/ASK-related protein, Drf1/ASKL1, is required for efficient progression of S and M phases. J Biol Chem, 2005. 280(13): p. 13062–70. [DOI] [PubMed] [Google Scholar]
- 91.Bruck I and Kaplan DL, Conserved mechanism for coordinating replication fork helicase assembly with phosphorylation of the helicase. Proc Natl Acad Sci U S A, 2015. 112(36): p. 11223–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ballabeni A, et al. , Human CDT1 associates with CDC7 and recruits CDC45 to chromatin during S phase. J Biol Chem, 2009. 284(5): p. 3028–36. [DOI] [PubMed] [Google Scholar]
- 93.Hao J, et al. , And-1 coordinates with Claspin for efficient Chk1 activation in response to replication stress. EMBO J, 2015. 34(15): p. 2096–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Merrick KA, et al. , Distinct activation pathways confer cyclin-binding specificity on Cdk1 and Cdk2 in human cells. Mol Cell, 2008. 32(5): p. 662–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Gu Y, Rosenblatt J, and Morgan DO, Cell cycle regulation of CDK2 activity by phosphorylation of Thr160 and Tyr15. EMBO J, 1992. 11(11): p. 3995–4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Welburn JP, et al. , How tyrosine 15 phosphorylation inhibits the activity of cyclin-dependent kinase 2-cyclin A. J Biol Chem, 2007. 282(5): p. 3173–81. [DOI] [PubMed] [Google Scholar]
- 97.Kulak NA, et al. , Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat Methods, 2014. 11(3): p. 319–24. [DOI] [PubMed] [Google Scholar]
- 98.Berthet C, et al. , Cdk2 knockout mice are viable. Curr Biol, 2003. 13(20): p. 1775–85. [DOI] [PubMed] [Google Scholar]
- 99.Merrick KA, et al. , Switching Cdk2 on or off with small molecules to reveal requirements in human cell proliferation. Mol Cell, 2011. 42(5): p. 624–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hiraga SI, et al. , Human RIF1 and protein phosphatase 1 stimulate DNA replication origin licensing but suppress origin activation. EMBO Rep, 2017. 18(3): p. 403–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Alver RC, et al. , Reversal of DDK-Mediated MCM Phosphorylation by Rif1-PP1 Regulates Replication Initiation and Replisome Stability Independently of ATR/Chk1. Cell Rep, 2017. 18(10): p. 2508–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ge XQ and Blow JJ, Chk1 inhibits replication factory activation but allows dormant origin firing in existing factories. J Cell Biol, 2010. 191(7): p. 1285–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Falck J, et al. , The ATM-Chk2-Cdc25A checkpoint pathway guards against radioresistant DNA synthesis. Nature, 2001. 410(6830): p. 842–7. [DOI] [PubMed] [Google Scholar]
- 104.Sorensen CS, et al. , Chk1 regulates the S phase checkpoint by coupling the physiological turnover and ionizing radiation-induced accelerated proteolysis of Cdc25A. Cancer Cell, 2003. 3(3): p. 247–58. [DOI] [PubMed] [Google Scholar]
- 105.Harper JW, et al. , Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell, 1995. 6(4): p. 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Soos TJ, et al. , Formation of p27-CDK complexes during the human mitotic cell cycle. Cell Growth Differ, 1996. 7(2): p. 135–46. [PubMed] [Google Scholar]
- 107.Liu P, et al. , The Chk1-mediated S-phase checkpoint targets initiation factor Cdc45 via a Cdc25A/Cdk2-independent mechanism. J Biol Chem, 2006. 281(41): p. 30631–44. [DOI] [PubMed] [Google Scholar]
- 108.Guo C, et al. , Interaction of Chk1 with Treslin negatively regulates the initiation of chromosomal DNA replication. Mol Cell, 2015. 57(3): p. 492–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zegerman P and Diffley JF, Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation. Nature, 2010. 467(7314): p. 474–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee AY, et al. , Dbf4 is direct downstream target of ataxia telangiectasia mutated (ATM) and ataxia telangiectasia and Rad3-related (ATR) protein to regulate intra-S-phase checkpoint. J Biol Chem, 2012. 287(4): p. 2531–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Dierov J, Dierova R, and Carroll M, BCR/ABL translocates to the nucleus and disrupts an ATR-dependent intra-S phase checkpoint. Cancer Cell, 2004. 5(3): p. 275–85. [DOI] [PubMed] [Google Scholar]
- 112.Heffernan TP, et al. , Cdc7-Dbf4 and the human S checkpoint response to UVC. J Biol Chem, 2007. 282(13): p. 9458–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Tenca P, et al. , Cdc7 is an active kinase in human cancer cells undergoing replication stress. J Biol Chem, 2007. 282(1): p. 208–15. [DOI] [PubMed] [Google Scholar]
- 114.Pasero P, et al. , A role for the Cdc7 kinase regulatory subunit Dbf4p in the formation of initiation-competent origins of replication. Genes Dev, 1999. 13(16): p. 2159–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Tsuji T, et al. , The role of Dbf4/Drf1-dependent kinase Cdc7 in DNA-damage checkpoint control. Mol Cell, 2008. 32(6): p. 862–9. [DOI] [PMC free article] [PubMed] [Google Scholar]