Abstract
Lutein is a dietary carotenoid of particular nutritional interest as it is preferentially taken up by neural tissues. Often linked with beneficial effects on vision, a broader role for lutein in neuronal differentiation has emerged recently, although the underlying mechanisms for these effects are not yet clear. The purpose of this study was to investigate the effect of lutein on neuronal differentiation and explore the associated underpinning mechanisms. We found that lutein treatment enhanced the differentiation of SH-SY5Y cells, specifically increasing neuronal arborization and expression of the neuronal process filament protein microtubule-associated protein 2. This effect was mediated by the intracellular phosphoinositide-3-kinase (PI3K) signaling pathway. While PI3K activity is a known trigger of neuronal differentiation, more recently it has also been shown to modulate the metabolic state of cells. Our analysis of bioenergetics found that lutein treatment increased glucose consumption, rates of glycolysis and enhanced respiratory activity of mitochondrial complexes. Concomitantly, the generation of reactive oxygen species was increased (consistent with previous reports that reactive oxygen species promote neuronal differentiation), as well as the production of the key metabolic intermediate acetyl-CoA, an essential determinant of epigenetic status in the cell. We suggest that lutein-stimulated neuronal differentiation is mediated by PI3K-dependent modulation of mitochondrial respiration and signaling, and that the consequential metabolic shifts initiate epigenetically dependent transcriptomic reprogramming in support of this morphogenesis. These observations support the potential importance of micronutrients supplementation to neurogenesis, both during normal development and in regenerative repair.
Keywords: lutein, micronutrient, neuronal differentiation, metabolism, PI3K-AKT pathway, glycolysis, mitochondria, gene expression
Introduction
Lutein is a carotenoid vitamin which has received particular interest because comparison across tissues indicates that it is preferentially accumulates in the human brain and readily crosses the blood-retina barrier to form macular pigments (Johnson, 2012). It is the most abundant carotenoid in the postnatal infant brain, accounting for more than half of total carotenoids. Moreover, lutein concentrations are significantly lower in the brain of pre-term infants, which suggests that the uptake of carotenoids from nutritional inputs is dramatic during the third trimester of pregnancy (Mostofsky et al., 2001; Vishwanathan et al., 2014). Supplementation of lutein may thus help to mitigate the recognized neurocognitive deficits measured in pre-term infants. Indeed, dietary supplementation of lutein to infant rhesus macaques results in higher lutein levels in serum, and brain tissues, the occipital cortex exhibiting the highest accumulation of lutein among several different brain areas including prefrontal and superior temporal cortex, striatum, occipital cortex, hippocampus and cerebellum (Jeon et al., 2016). Further, also in rhesus macaques it has been shown that lutein is the only carotenoid accumulating in frontal cortex tissue, and is incorporated into all neural membranes, but particularly in myelin, when comparing to the neuronal cellular, mitochondrial and nuclear membrane (Mohn et al., 2015). Significantly, the post-mortem metabolic analysis of human infant brain tissues demonstrates that lutein uptake is positively correlated with the metabolism of lipids, amino acid neurotransmitters and antioxidant homocarnosine (Lieblein-Boff et al., 2015a). These in vivo studies indicate that lutein may function to modulate metabolic pathways which are necessary for neuronal development. A high rate of metabolic flux arising from relatively inefficient glycolysis is a common feature of undifferentiated neurons, consistent with the relatively low levels of bioenergetics and biosynthesis required for protracted cycles of cellular proliferation (O’Brien et al., 2015). The glycolytic metabolism of stem cells favors the utilization of extracellular nutrients and glucose to produce ATP and the intermediates needed for biosynthetic pathways, including ribose sugars, glycerol and citrate (DeBerardinis et al., 2008). A further benefit of anaerobic glycolysis is the lower generation of reactive oxygen species in the hypoxic state, and thus greater protection of cellular DNA from mutation and damage (Kim et al., 2006). In contrast, mature neurons have a higher energy requirement, for the production of ATP, maintenance and restoration of the conserved energy along the ion gradient, and the generation of neurotransmitters (Mergenthaler et al., 2013) (Alle et al., 2009). Notably, levels of lutein are lower in elderly people than infants (Johnson et al., 2010), and in the brains of individuals who have mild cognitive impairment, supplementation with lutein can significantly improve older women’s verbal fluency, while simultaneously stimulating regenerative processes (Hammond et al., 2017). Apart from these, lutein also appears to have a direct differentiation-promoting effect on human stem cells (Kuchan et al., 2013). Although lutein is seemingly necessary in the optimal development of the infant brain, and the neuronal system specifically, the mechanism by which it achieves this benefit is still poorly defined. As lutein cannot be synthesized de novo, the link between its dietary supply and the mechanism by which it affects neuronal development needs to be more fully elucidated.
The fatty acid docosahexaenoic acid (DHA) has long been used as an additive in infant formula because of perceived benefit to neuronal development and plasticity (Rogers et al., 2013). Specifically, DHA is reported to promote neurogenesis, by stimulating neuronal differentiation in rat fetal NSCs (Kawakita et al., 2006). Moreover, DHA can stimulate damaged nerve regeneration after injury (He et al., 2017). DHA decreases Hes1 expression and increases p27kip1 expression (Katakura et al., 2013), activating G-protein coupled protein 40 (Ma et al., 2010) and increasing phosphatidylserine accumulation in cell membrane (Kim, 2007). Thus, DHA has been used as “gold standard” measure and comparison for assessing the potential benefits of lutein supplementation on neurogenesis. In this study, we firstly studied whether lutein affect the differentiation of neuronal cells by comparing neurite number, neurite length, expression of neuronal marker with DHA treated cells. Then we explored the involvement of AKT signaling pathway, metabolic reprogramming and the associated epigenetic changes during lutein-driven differentiation process.
Materials and Methods
Cell culture
SH-SY5Y cells were purchased from American Type Culture Collection (Manassas, VA, USA) and cultured in DMEM/F12 (Thermo Fisher Scientific, Waltham, MA, USA), with 10% fetal bovine serum (Thermo Fisher Scientific), penicillin (50 U/mL), streptomycin (50 μg/mL) in a humidified 5% CO2 incubator at 37°C. For micronutrient treatment, undifferentiated cells (less than 10 passages) were seeded at 3 × 105 cells/well in 6-well plates. Twenty-four hours after seeding, 5 μM lutein (Royal DSM, Heerlen, Netherlands) or 10 μM DHA (Cayman Chemical, Ann Arbor, MI, USA) was added (Shimazawa et al., 2009; Gao et al., 2011), and culture medium was renewed at 72 hours post-seeding. At 96 hours post-seeding, cells were harvested for analysis.
Morphological analysis
Cells were cultured at 1 × 105 cells/well in 8-well chamber slides (Thermo Fisher Scientific). After micronutrient treatment, cells were fixed with 4% formaldehyde (Sigma Aldrich, St. Louis, MO, USA) for 15 minutes, and then sequentially stained with 5 μg/mL WGA488 (Thermo Fisher Scientific) for 7 minutes and a further 3 minutes with 1 μg/mL DAPI (Thermo Fisher Scientific). Stained cells were rinsed with phosphate-buffered saline pH 7.4 (PBS) twice and then mounted onto glass slides with mounting medium. Cells were visualized, and photos were taken by Leica DMR fluorescence microscope (Leica, Wetzlar, Germany). Images were further analyzed by using NeuronJ and Sholl Analysis plugins in ImageJ version 1.5 (https://imagej.nih.gov). Forty randomly selected images (~150 cells in total for each group) were traced semi-manually using NeuronJ and the number of neurites developed per cell was manually counted (Higgins et al., 2013). Average of intersections over the whole area occupied by the arbor (Nav) from 40 cells was measured using the Sholl technique (Langhammer et al., 2010).
Measurement of cell density and viability
SH-SY5Y cells were seeded in 24-well plates at a density of 3 × 104 cells/well, After 72-hour treatment of micronutrients as described above, cell density and viability were then determined by Trypan blue (Thermo Fisher Scientific) exclusion: floating SH-SY5Y cells in the supernatant were collected to an Eppendorf tube, adherent cells in each well were digested by 50 μL 0.25% trypsin Ethylenediaminetetraacetic acid (EDTA) for 2 minutes, then floating cells with culture medium were added back and mixed with the digested cells. To prepare 2-fold diluted samples for cell counting, cell samples (20 μL) were mixed with 20 μL 0.4% trypan blue solution. Cell counting results were calculated as percentage of (untreated) control.
RNA isolation and quantitative polymerase chain reaction (qPCR)
Total RNA from cells was isolated using TRIzol® reagent and PureLink® RNA mini kit (Thermo Fisher Scientific) according to manufacturer’s protocol. Briefly, 1 μg of RNA was used for cDNA synthesis using High Capacity cDNA Reverse Transcription kit (Thermo Fisher Scientific). PCR was carried out with LightCycler® 480 SYBR Green I Master Mix and assayed on LightCycler® 480 (Roche Diagnostics, Bazel, Switzerland). Primer sequences used are: Peptidylprolyl isomerase A (PPIA; reference gene), forward: 5′-TCT TGA GGG AAG CAT ATT GG-3′; reverse: 5′-CAG GGA GAC TGA CTG TAG CAC-3′. Superoxide dismutase 2 (SOD2), forward: 5′-AGA AGT ACC AGG AGG CGT TG-3′; reverse: 5′-TTG ATA TGA CCA CCA CCA TTG-3′. Glutathione peroxidase 1 (GPX1), forward: 5′-GAA TGT GGC GTC CCT CTG AG-3′; reverse: 5′-TTC GTT CTT GGC GTT CTC CTG-3′. Catalase (CAT), forward: 5′-TTT GGC CTC ACA AGG ACT AC-3′; reverse: 5′-GGT CGA AGG CTA TCT GTT CAA-3′. Mitochondrial Encoded NADH Dehydrogenase (MT-ND2), forward: 5′-CTA CTC CAC CTC AAT CAC ACT AC-3′; reverse: 5′-AGG TAG GAG TAG CGT GGT AAG-3′. MT-ND5, forward: 5′-CTA CCT AAA ACT CAC AGC CCT C-3′; reverse: 5′-GGG TAG AAT CCG AGT ATG TTG G-3′. Mitochondrial encoded ATP synthase 6 (MT-ATP6), forward: 5′-ACA CCC CTT ATC CCC ATA CTA G-3′; reverse: 5′-ATG GTT GAT ATT GCT AGG GTG G-3′. Glucose transpoter 3 (Glut3), forward: 5′-CAGGTTTTGTGCCCATGTAC-3′; reverse: 5′-CCA TAG CTC TTC AGA CCC AAG-3′. Hexokinase 1 (HK1), forward: 5′-ACA TTG TCT CCT GCA TCT CTG-3′; reverse: 5′-GCC TTA AAA CCC TTT GTC CAC-3′. Hexokinase 2 (HK2), forward: 5′-AAG CCC TTT CTC CAT CTC CT-3′; reverse: 5′-CTT CTT CAC GGA GCT CAA CC-3′. Glucose-6-phosphate isomerase (GPI), forward: 5′-TAG ACG GCA AGG ATG TGA TG-3′; reverse: 5′-CGC CAA TGC CAA TGT TGA TG-3′. Phosphofructokinase, muscle (PFKM), forward: 5′-TGA CCA AAG ATG TGA CCA AGG-3′; reverse: 5′-GCG AAC CAC TCT TAG ATA CCG-3′.
Whole genome mRNA profiles of SY5Y cells treated by lutein or DHA were measured by Affemetrix HuGene 4.0 array (Affymetrix, Inc, Santa Clara, CA, USA) as described by the manufacturer. Hybridisation, washing, scanning and initial data analyzing of the microarrays were performed by the Adelaide microarray center (University of Adelaide, Australia).
Measurement of mitochondrial membrane potential
Mitochondrial membrane potential was monitored with the potentiometric dye JC-1 using the Mitoprobe assay kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. JC-1 dye exhibits potential-dependent accumulation in mitochondria, indicated by a fluorescence emission shift from green (~529 nm) to red (~590 nm). JC-1 accumulates in polarized mitochondria with a resting membrane potential and fluoresces red. However, on loss of ΔΨm, JC-1 aggregates are released from the mitochondria and fluoresce green. Thus, to assess mitochondrial potential after the treatment of micronutrients, cells were stained with JC-1 (2μM) for 20 minutes at 37°C 5% CO2, after washing and resuspension in PBS, fluorescence was monitored by flow cytometry (BD LSRII, San Jose, CA, USA).
Measurement of the generation of reactive oxygen species (ROS)
Intracellular ROS was measured as previously (Ding et al., 2008; Lee et al., 2011). Cells were cultured in 48-well plates. After micronutrients treatment, cells were incubated with 5 μM 2′,7′-dichlorodihydrofluorescein diacetate (H2DCFDA, Thermo Fisher Scientific) in DMEM/F12 (phenol red-free, Thermo Fisher Scientific) at 37°C for 30 minutes. Fluorescence was then read by micro-plate reader Synergy 2 (BioTek Instruments, Inc., Winooski, VT , USA) at excitation/emission 485/530 nm. After the measurement of ROS, the cells were washed with pre-warmed PBS twice and the cell density then measured in situ by Alamar Blue (Thermo Fisher Scientific) following manufacturer’s instructions, Briefly, cells were incubated in fresh medium (mixed with 10% alamar blue) for 2–3 hours in a humidified incubator at 37°C before measuring the fluorescence at Ex/Em 560/590 nm. Cellular ROS measurements were normalized to cell density.
Measurement of glucose consumption and lactate generation
Cells were cultured in 24-well plates with a working volume of 500 μL. After micronutrient treatment, 200 μL spent medium were collected. Cell density was measured in situ by Alamar Blue by following manufacturer’s instruction. Glucose and lactate were quantified using GLU and LACT2 kits (Roche Diagnostics, Basel, Switzerland) respectively, on Cobas c311 analyzer (Roche Diagnostics), and then normalized by cell density. The consumption of glucose was calculated by:
Consumption = (G0–Gt)/cell density
G0 represents glucose concentration in the fresh medium while Gt means glucose concentration remaining at the time of testing.
The generation of lactate was calculated by:
Generation = (L0–Lt)/cell density
L0 represents lactate concentration in the fresh medium while Lt means lactate concentration remaining at the time of testing.
Cellular bioenergetics
Cells were seeded in XF cell culture micro-plate (Seahorse Bioscience, Copenhagen, Denmark) in triplicate at 3 × 104 cells/well in 200 μL of growth medium and then cultured at 37°C in a 5% CO2 incubator. After micronutrient treatment, assays were initiated by removing the growth medium from each well and replacing with 175 μL of pre-warmed assay medium (pH 7.4, containing XF Base medium, 10 mM glucose and 1 mM sodium pyruvate). The cells were incubated at 37°C in a non-CO2 incubator for 1 hour to allow media temperature and pH to reach equilibrium before measurement. MitoStress Assay Kit (Seahorse Bioscience, Copenhagen, Denmark) was used to measure extracellular oxygen consumption rate (OCR) according to manufacturer’s protocol. After initial baseline measurements, the following compounds were used: ATP synthase inhibitor oligomycin (2 μM) for measuring ATP-linked respiration, electron transport chain accelerator p-trifluoromethoxy carbonyl cyanide phenyl hydrazone at 2 μM for measuring maximum respiration capacity, and lastly, mitochondrial complex I/III inhibitors rotenone/antimycin mix (1 μM each) for measuring non-mitochondrial respiration. Proton leak is calculated by subtracting OCR (after injecting rotenone/antimycin) from OCR (after injecting oligomycin). For extracellular acidification rate of glycolysis, oligomycin was added to generate the maximum glycolytic function while 50 mM 2-DG was added later to calculate the non-glycolytic acidification. Data was normalized to total cellular protein content, which was determined using Pierce™ BCA Protein Assay (Thermo Fisher Scientific).
Measurement of cellular enzymic activity and metabolic intermediates
Micronutrients treated cells were harvested and snap frozen in liquid nitrogen before store in –80°C. Upon analysis, cells were diluted in proper assay buffer according to manufacturer’s instruction and then activities were quantified by using microplate assay kits (Abcam, Cambridge, United Kingdom). For HK activity measurement, glucose was catalyzed to glucose-6-phosphate by HK, along with the reduction of probe and generating an intense fluorescence as Ex/Em: 535/587 nm; For the measurement of PDH, a 96 wells plate was pe-coated with anti-PDH antibody. PDH complex in cell samples can be captured by the antibody and then converting NAD+ to NADH, coupling to the reduction of a reporter dye to yield a color product at OD450 nm. For pyruvate analysis, pyruvate was oxidized by pyruvate oxidase and generating a color product at OD579 nm. For acetyl CoA analysis, acetyl CoA was converted to free CoA and the resulting NADH captured by probe as indicated by fluorescence at Ex/Em 535/587 nm.
DPPH free radical scavenging assay
The ability of tested micronutrients to scavenge 2,2-diphenyl-1-picrylhydrazyl (DPPH, Sigma Aldrich) radical were determined according to the method of reference (Bertalanic et al., 2012). DPPH• is one of the few stable and commercially available organic nitrogen radicals, this purple radical turns to yellow followed by the formation of DPPH upon absorption of hydrogen from an antioxidant. The antioxidant effect can be easily evaluated by following the decrease of UV absorption at 517 nm. The reaction mixture contains 0.1 mL of DPPH radical solution (0.1 mM) and 5 μM lutein or 10 μM DHA. Total volume of the reaction mixture was 0.3 mL. Absorption of DPPH radical at 517 nm was determined after 30 min against a blank solution that contained only methanol. 50 µM L-ascorbic acid was used as a positive control.
Scavenging rate = (1 – absorbance of sample)/absorbance of blank × 100%
Western blotting
Cells were treated with micronutrients with or without PI3K inhibitor LY294002 (Abcam), and harvested by RIPA Lysis and Extraction Buffer (Thermo Fisher Scientific). An aliquot of 20 μg protein was resolved on 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and blotted onto a piece of the polyvinylidene fluoride membrane. After blocking with 5% skim milk in tris-buffered saline and tween 20 buffer (50 mM Tris-Cl, 150 mM NaCl, 0.1% Tween 20, pH 7.4) for 1 hour, the membrane was incubated with relevant primary antibody at 4°C overnight. The blot was then incubated with horseradish peroxidase (HRP)-conjugated secondary antibody for 1 hour before washing three times with PBS at room temperature (25°C). Bands on the immunoblot were visualized using Pierce™ ECL enhanced chemiluminescence detection kit (Thermo Fisher Scientific) by following manufacturer’s instructions, and chemiluminescent bands were quantified by ImageJ version 1.5 and normalized by loading control of β-actin (relative expression = band density of target/ band density of loading control). The primary antibodies are AKT (catalog #9272, Cell Signaling Technology, rabbit-origin, 1:1000 dilution), pAKT (catalog #4058, Cell Signaling Technology, rabbit-origin, 1:1000 dilution), microtubule-associated protein 2 (MAP2) (catalog #4542, Cell Signaling Technology, rabbit-origin, 1:1000 dilution), β-actin (catalog #58673, Santa Cruz Biotechnology, mouse-origin, 1:500 dilution). HRP-conjugated goat anti-rabbit IgG (H + L) (catalog #31460, 1:10,000 dilution) was purchased from Thermo Fisher Scientific, and HRP-conjugated rabbit anti-mouse IgG (H + L) (catalog #AB97046, 1:10,000 dilution) was purchased from Abcam.
Statistical analysis
Data are presented as mean ± SEM from at least 3 independent experiments unless otherwise indicated in the figure legend. To assess statistical significance, one-way analysis of variance with a least significant difference (LSD) post hoc test was performed in Prism GraphPad version 6.0 (GraphPad Software, La Jolla, CA, USA). P < 0.05 was considered statistically significant.
Results
Lutein treatment promotes neuronal differentiation
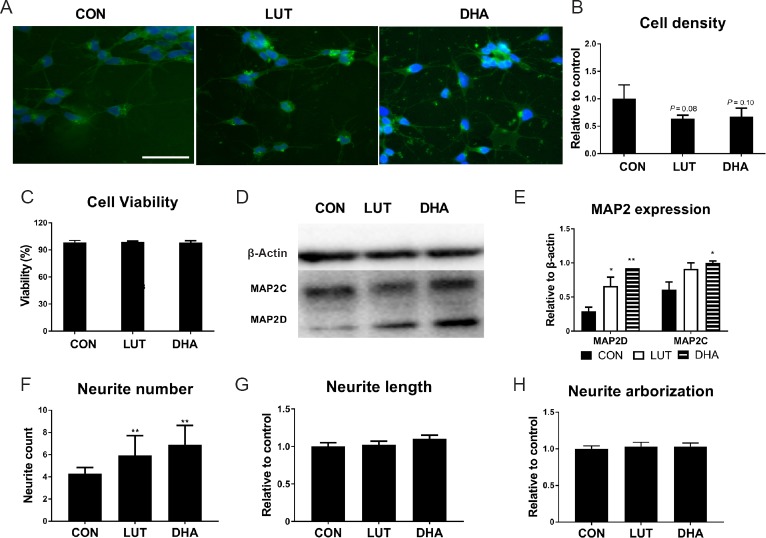
It is recognized that cells exit the cell cycle and decrease cellular proliferation during differentiation (Walsh and Perlman, 1997; Ruijtenberg and van den Heuvel, 2016), leading to reduced cell density. Our trypan blue exclusion results indicated that 72-hour lutein treatment results in a reduced cell density of 36%, similar to the 33% reduction seen with our ‘positive control’ DHA. Meanwhile, cell viability in both treatment groups was maintained at higher than 98% (Figure 1A–C).
Figure 1.
Lutein and DHA promote neuronal differentiation
(A) SH-SY5Y cells cultured with micronutrients for 3 days (40× magnification). The nuclei were counterstained with DAPI (blue); cell membrane was stained with Alexa Fluor® 488 (WGA488) (green). Scale bar: 50 µm. (B) Cell density measured by trypan blue exclusion assay (relative to control, F(2,6) = 3.9, P = 0.08). (C) Cell viability measured by trypan blue exclusion assay (F(2,6) = 0.25, P = 0.79). (D & E) The protein expression of neuron marker MAP-2 measured by Western blot (F(2,3) = 29.7, P < 0.05). (F) Quantification of neurite number using ImageJ (F(2,108) = 30.5, P < 0.01). (G) Average neurite length measured by NeuronJ (relative to control, F(2,437) = 166, P < 0.01). (H) Neuronal arborization measured by Sholl analysis of ImageJ (relative to control, F(2,101) = 4.5, P < 0.05). Statistical significance: *P < 0.05 and **P < 0.01, vs. control (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, n = 4 measurements per group. For image quantification, about 100 cells were counted for each group. CON: Control; LUT: lutein (5 μM); DHA: docosahexaenoic acid (10 μM). DAPI: 4′,6-diamidino-2-phenylindole.
MAP2 is a neuron-specific protein which regulates the structure and stability of microtubules, neuronal morphogenesis, cytoskeleton dynamics, and organelle trafficking in axons and dendrites (Chamak et al., 1987). It is widely used as an early-stage neuron differentiation marker (Maddodi et al., 2010). Lutein treatment increased expression of MAP2D isoform more than the MAP2C, 2-fold (P < 0.05) and 1.4-fold respectively. DHA treatment also increased MAP2D more than the MAP2C isoform, 3-fold (P < 0.01) and 1.6-fold respectively (P < 0.05) (Figure 1D & E).
Neurite outgrowth is the initiation of neuron development and proceeds by dynamic behavior of growth cones. It is the defining morphological and functional feature of neuronal cells. The importance of this process for optimal brain growth and neurocognitive ability has been well recognized (Riederer et al., 1997; Wu et al., 2009). In order to investigate whether lutein promotes neurite outgrowth, we used NeuronJ and Sholl analysis to analysis WGA488-stained cells with different treatments. According to the results, neuronal cells displayed more neurites following lutein treatment as well as DHA (Figure 1F), although both neurite length (Figure 1G) and neuronal arborization (data not shown) remained unchanged.
PI3K-AKT pathway and lutein-induced differentiation
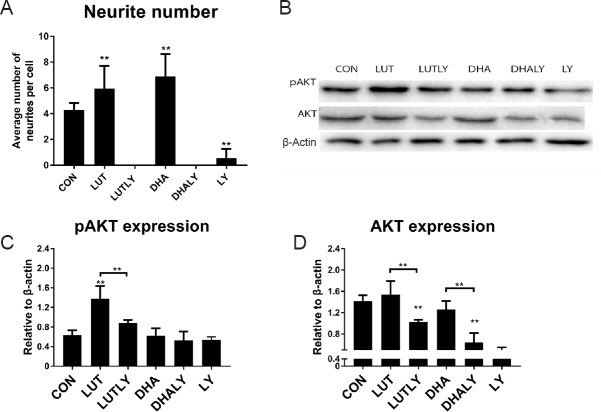
Activation of PI3K signaling is not only required for initiation of neurite elongation during SH-SY5Y differentiation, it is also essential for maintenance of the neuronal morphology (Sanchez et al., 2004). A significant increase in phosphorylated AKT has been documented with SH-N-SH (the original cell line of SH-SY5Y) cell differentiation (Qiao et al., 2012). To explore if PI3K/AKT pathway is involved in lutein-induced neuronal differentiation and metabolic reprogramming, we measured the expression of both phosphorylated and pan AKT (Lopez-Carballo et al., 2002; Amengual et al., 2011).
As shown in Figure 2, lutein significantly increased level of phosphorylated AKT (pAKT) by 2-fold (P < 0.01) compared to cells cultured in the absence of lutein. However, the level of pan AKT remained unchanged. Co-treatment with PI3K inhibitor LY294002 abolished the lutein-mediated increase in pAKT (Figure 2C) and significantly decreased the expression of pan AKT (Figure 2D). Co-treatment with LY294002 also completely blocked neurite outgrowth (P < 0.01, Figure 2A). However, as the positive control of neural differentiation inducer, distinct from lutein, DHA treatment did not increase the expression of either pAKT (Figure 2C), nor pan AKT (Figure 2D). This result indicates that DHA mediated neuronal differentiation is not via activating AKT kinase, which is consistent with previous researches (Kamata et al., 2007; Zamarbide et al., 2014). However, co-treatment with DHA and LY294002 did lead to reduced pan AKT (P < 0.01; Figure 2D), blocked neurite outgrowth (P < 0.01; Figure 2A). These results suggest that though PI3K/AKT is not the pathway which DHA regulate neuronal differentiation, a physiological activity of AKT is still needed to support the differentiation process (see discussion).
Figure 2.
Lutein treatment alters PI3K-AKT protein expression.
(A) Neurite outgrowth was abolished by PI3K inhibitor LY294002 (F(5,162) = 175.9, P < 0.01). (B) Western blot analysis of pan and phospho-AKT (S473) expression in the presence of lutein (5 μM) or DHA (10 μM) with (or without) LY294002 (10 μM) co-treatment (represented results of eight replications). (C) Quantification of pAKT expression. Lutein-mediated increase in pAKT expression was reversed by LY294002 co-treatment (F(5, 28) = 25, P < 0.01). (D) Quantification of pan AKT expression. Sole treatment with either micronutrient had no effect on AKT expression, while co-treatment with LY294002 significantly reduced AKT expression (F(5,29) = 23, P < 0.01); Statistical significance: *P < 0.05 and **P < 0.01, vs. control (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, n = 8 measurements per group). CON: Control; LUT: lutein (5 μM); LUTLY: lutein (5 μM) + LY294002 (10 μM); DHA: docosahexaenoic acid (10 μM); DHALY: DHA (10 μM) + LY294002 (10 μM); LY: LY294002 (10 μM).
Lutein and DHA influences glycolytic and mitochondrial metabolism
Glycolysis and mitochondrial respiration are two main cellular energy resources. In undifferentiated pluripotent cells, anaerobic glycolysis serves as their main energy resource, and an elevated oxidative phosphorylation is a necessary process for differentiation (O’Brien et al., 2015; Agostini et al., 2016; Zheng et al., 2016). Micronutrients such as DHA have long been recognized to increase mitochondrial membrane fluidity and function (Sherratt et al., 2016), which is critical for mitochondrial respiration. To study the effects of lutein treatment on glycolysis and mitochondrial respiration, we measured intracellular glycolytic flux and mitochondrial respiration in cells in the presence (or absence) of lutein and DHA treatment.
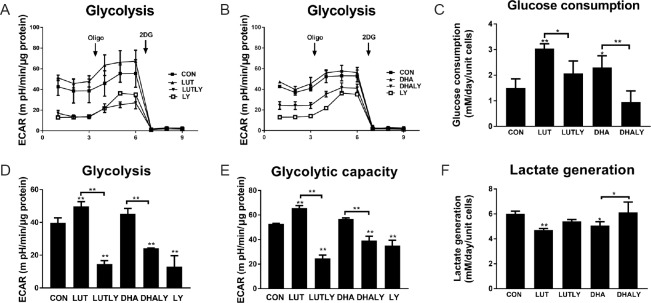
Our real time metabolic flux results show that lutein treatment increased cellular glycolytic flux, while DHA increased the rates of glycolysis (Figure 3A, B & D). Specifically, the basal glycolytic function was increased by 1.2-fold following lutein treatment (P < 0.05; Figure 3D). Moreover, lutein treatment resulted in a 1.25-fold increase in glycolytic maximal capacity (P < 0.01; Figure 3E), compared to non-significant increase of DHA treatment. LY294002 co-treatment reversed the beneficial effects of lutein on glycolysis, reducing both basal and maximal glycolysis to levels which are significantly lower than control (Figure 3D & E). As the primary resource for glycolysis, glucose consumption was also measured. As shown in Figure 3C, lutein treatment doubled the consumption of glucose to 3 mM/d (P < 0.01), but this elevating consumption was inhibited by LY294002 co-treatment. Similarly, DHA treatment increased glucose consumption by 1.5-fold (P < 0.05) and was reversed to baseline level by LY294002 co-treatment. In contrast, lactate production did not show a pattern of increase as that for glycolysis; it was significantly decreased by about 25% (P < 0.01; Figure 3F) in the presence of lutein or DHA, suggesting that increased glucose consumption is not necessarily catalyzed to lactate, and may enter other metabolic pathways. Under LY co-treatment, we measured a significant increase of about 1.2-fold in lactate production in lutein- or DHA-supplemented cells co-treated with LY294002 when compared to their no-inhibitor counterparts (Figure 3F).
Figure 3.
Lutein or DHA treatment affects cellular glycolysis.
(A) Lutein increased glycolytic function and it was reversed by PI3K inhibitor LY294002 co-treatment. (B) DHA increased glycolytic and was reversed by LY294002 co-treatment. (C) Glucose consumption increased by treatment of lutein or DHA. LY294002 co-treatment with lutein or DHA reversed this increasing (F(4,15) = 15.7, P < 0.01). (D) Quantification of basal glycolysis by subtracting non-glycolytic ECAR (after adding 2DG) from ECAR before adding oligomycin (F(5,11) = 49.5, P < 0.01). (E) Quantification of maximum glycolytic capacity by subtracting non-glycolytic ECAR (after adding 2DG) from ECAR after adding oligomycin (F(5,11) = 9.8, P < 0.01). (F) Generation of lactate reduced by treatment of DHA or lutein. LY294002 co-treatment with DHA or lutein reversed this reduction (F(4,15) = 8.4, P < 0.01). Statistical significance: *P < 0.05 and **P < 0.01, vs. CON (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, three replications in each group. ECAR: Extracellular acidification rate; Oligo: oligomycin (2 μM); 2DG: 2-Deoxy-D-glucose (50 mM); CON: control; LUT: lutein (5 μM); LUTLY: lutein (5 μM) + LY294002 (10 μM); DHA: docosahexaenoic acid (10 μM); DHALY: DHA (10 μM) + LY294002 (10 μM); LY: LY294002 (10 μM).
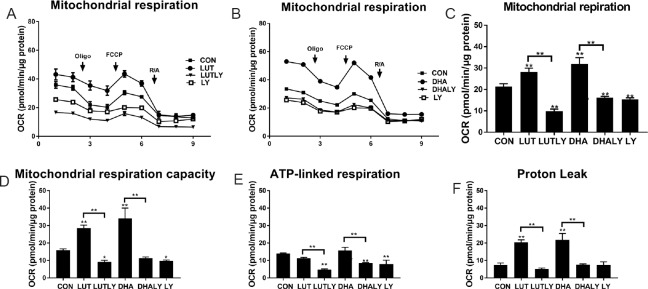
As shown in Figure 4, lutein, similar as DHA, increased mitochondrial respiration (respiration chain activity) when compared with control cells. In the presence of lutein, mitochondrial respiration showed a 36% increase (P < 0.01; Figure 4C), while DHA significantly increased mitochondrial respiration for 50% (P < 0.01; Figure 4C). Moreover, lutein increased mitochondrial respiration capacity by 1.8-fold when compared with control cells (P < 0.01; Figure 4D), but ATP-linked respiration was unchanged either by the treatment of lutein or DHA (Figure 4E). This lutein-mediated increase on mitochondrial function was reversed by co-treatment with LY294002, which made the mitochondrial respiration, ATP-linked respiration and maximum capacity to a minimum level when compared to control. Moreover, this reduction is not because of a LY294002 cytotoxic effect, given that trypan blue exclusion assays were not affected, and confirming that cell viability is maintained at 98% Figure 1C). Similarly, DHA increased mitochondrial function and maximal mitochondrial respiration capacity by 1.7-fold (P < 0.01) and 2.1-fold (P < 0.05) respectively, but had no significant effect on ATP production when compared to control (Figure 4C–E). LY294002 co-treatment abolished the DHA-mediated increase in basal and maximal mitochondrial respiration, resulting in baseline (control) levels. It is worthy to note out that proton leak, an odd coupler of cellular reactive oxidative stress species (ROS) (Brookes, 2005), was doubled by the treatment of lutein (Figure 4F). DHA differentiated cells also showed a similar level of increased proton leak. However, co-treatment with LY294002 completely blocked this increase in proton leak (Figure 4F).
Figure 4.
Lutein or DHA treatment increases mitochondrial function
(A) Lutein-mediated increased mitochondrial respiration and was reversed by PI3K inhibitor LY294002 co-treatment. (B) DHA-mediated increased mitochondrial respiration and was reversed by LY294002 co-treatment. (C) Quantification of mitochondrial respiration by subtracting non-mitochondrial OCR (after adding R/A) from OCR before adding oligomycin (F(5,11) = 88.9, P < 0.01). (D) Quantification of mitochondrial maximum capacity by subtracting non-mitochondrial OCR (after adding R/A) from OCR after adding FCCP (F(5,11) = 68, P < 0.01). (E) Quantification of ATP linked respiration by subtracting non-mitochondrial OCR (after adding R/A) from OCR after adding oligomycin (F(5,11) = 32.5, P < 0.01). (F) Quantification of proton leak by subtracting non-mitochondrial OCR (after adding R/A) from OCR before adding FCCP (F(5,11) = 55.5, P < 0.01). Statistical significance: *P < 0.05 and **P < 0.01, vs. CON (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, three replications in each group. OCR: Oxygen consumption rate; FCCP: carbonyl cyanide-4-(trifluoromethoxy)phenylhydrazone; Oligo: oligomycin (2 μM); R/A: rotenone and antiomycin (1 μM); CON: control; LUT: lutein (5 μM); LUTLY: lutein (5 μM) + LY294002 (10 μM); DHA: docosahexaenoic acid (10 μM); DHALY: DHA (10 μM) + LY294002 (10 μM); LY: LY294002 (10 μM).
Micronutrients increase ROS generation during neuronal differentiation
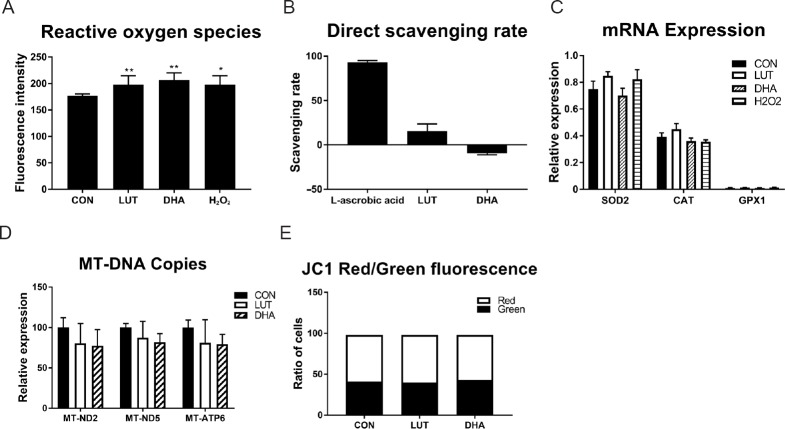
Elevated levels of ROS (mainly produced through mitochondrial I/III respiration process) was also reported during neuronal differentiation (Tsatmali et al., 2006; Murphy, 2009; Ji et al., 2010). To investigate the levels of ROS during lutein or DHA induced neuronal differentiation, cells were incubated with the ROS indicator H2DCFDA. We found a significant increase in ROS generation (P < 0.01; Figure 5A) in the presence of lutein or DHA, compared to untreated control. As lutein and DHA have been seen as antioxidant previously (Shimazawa et al., 2009; Erdman et al., 2015), we then measured the free radical scavenging ability of lutein and DHA, the results showed that radical scavenging rate of lutein was only 15%, while that of DHA was –9%, both of which were far less than 93% of L-ascrobic acid (positive control, Figure 5B), indicating that lutein and DHA have poor radical scavenging ability at the concentration we tested. Despite an increase in cellular ROS following lutein treatment, however, we found unchanged expression of the oxidant dismutase SOD2/GPX1/CAT after lutein treatment (Figure 4C). Moreover, we also found the expression of these genes were not changed by 100 μM H2O2 exposure, though ROS species were significantly increased (Figure 5A & C).
Figure 5.
Lutein or DHA treatment increases cellular ROS.
Lutein (5 μM) or DHA (10 μM) treatment: (A) increased the generation of ROS during neuronal differentiation (F(3,27) = 11.1, P < 0.01); (B) showed a low activity of DPPH free radical scavenging when compared with 50 μM L-ascrobic acid, indicating that 5 μM lutein or 10 μM DHA does not show any antioxidant effect (F(2,9) = 468, P < 0.01); (C) had no effect on the expression of antioxidant enzymes, indicating lutein or DHA treatment doesn’t trigger cellular antioxidant pathway; (D) had no effect on copy number of mt-ND2, mt-ND5 and mt-ATP6 of the mitochondrial genome, indicating lutein or DHA does not change mitochondrial mass; (E) had no effect on the ratio of cells with high mitochondrial membrane potential (MMP; red fluorescence) and low MMP (green fluorescence) measured by JC1 staining, indicating lutein or DHA does not change mitochondrial membrane potential. Statistical significance: *P < 0.05 and **P < 0.01, vs. control (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, six replications in each group. CON: Control; LUT: lutein (5 μM); DHA: docosahexaenoic acid (10 μM).
Mitochondrial biogenesis has been reported to occur during the process of neuronal differentiation and contributes to the increase in mitochondrial respiration and neuronal differentiation (Agostini et al., 2016). To assess whether an increase in mitochondrial number is involved in the micronutrient-mediated elevation of mitochondrial function, three mitochondrial DNA (mt-DNA) encoded genes (mt-ND2, mt-ND5 and mt-ATP6) were measured by qPCR as indications of mitochondrial copy number. We found no change in mitochondrial DNA copy number following lutein or DHA treatment (Figure 5D), which was consistent with previous report of unchanged mitochondrial copies during SY5Y differentiation (Schneider et al., 2011), suggesting that lutein- and DHA-mediated increases in mitochondrial respiration was a result of increased physiologic function of pre-existing mitochondria rather than increased mitochondrial mass.
Mitochondrial respiration pumps protons from matrix to mitochondrial inter-membrane space and therefore generating mitochondrial membrane potential (MMP), of which the central function is to drive ATP synthesis. As our results showed mitochondrial respiration was increased by lutein or DHA treatment, but without a concomitant increase in ATP production, we then investigated the level of MMP in the presence of micronutrients. As Figure 5E showed, the ratio of polarized cells (red) in the presence of micronutrients were similar as untreated control cells, indicating that MMP was not changed during micronutrients mediated increase of mitochondrial respiration.
Activity of glycolytic enzymes increased by lutein treatment
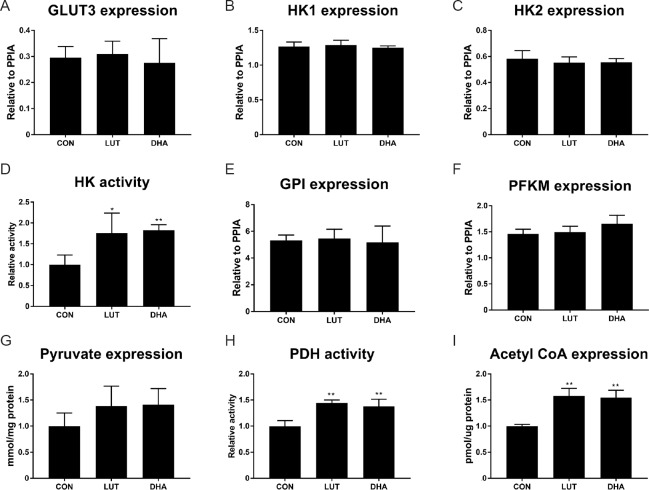
As our results showed that cellular glycolytic function was elevated by micronutrients, we next analysed whether these metabolic effects were caused by changed function of relevant enzymes. Interestingly, qPCR showed that the gene expression of several glycolytic enzymes, namely neuronal specific glucose transporter 3 (GLUT3), hexokinase 1/2 (HK1/2), glucose-6-phosphate isomerase (GPI), phosphofructokinase (PFKM) remained unchanged following micronutrient treatments (Figure 6A–C, E & F).
Figure 6.
Effect of lutein on glycolytic enzymes and intermediates.
After treatment of 5 μM lutein or 10 μM DHA for 3 days, total RNA was extracted from SH-SY5Y cells and mRNA levels of glucose transporter 3 (A, GLUT3), hexokinase 1 (B, HK1), hexokinase 2 (C, HK2), glucose-6-phosphate isomerase (E, GPI), phosphofructokinase, muscle (F, PFKM) were assessed by qPCR. For the measurement of enzymatic activity and metabolic intermediates, SH-SY5Y cells were harvested after treatment, and cell lysate were used to measure intracellular enzymatic activities (relative to control) as described in methods: (D) The activity of HK (F(2,9) = 8.2, P < 0.01); (H) the activity of pyruvate dehydrogenase (PDH) (F(2,9) = 20.9, P < 0.01); (G) concentration of pyruvate (F(2,9) = 2.1, P = 0.18); (I) concentration of acetyl-CoA (F(2,9)=29.1, P < 0.01). *P < 0.05, **P < 0.01, vs. untreated control cells (one-way analysis of variance followed by the least significant difference post hoc test), data are expressed as the mean ± SEM, four replications in each group. DHA: Docosahexaenoic acid; qPCR: quantitative polymerase chain reaction; CON: control; LUT: lutein.
We next measured the activities of two rate-limiting glycolytic enzymes, HK and PDH. The results showed that lutein or DHA significantly increased the activity of hexokinase by 1.7-fold compared to untreated control (Figure 6D), while activity of pyruvate dehydrogenase was also elevated by 1.4-fold (P < 0.01; Figure 6H) in the presence of lutein or DHA, compared to untreated control.
Activity of these enzymes are critical for both cellular glycolytic and mitochondrial metabolism, as they determine the availability of metabolic substrates for the TCA cycle and mitochondrial respiration. In light of this, we then measured the concentration of pyruvate and acetyl-CoA, which link glycolysis and mitochondrial metabolism. As the results showed, lutein or DHA only moderately increased the level of pyruvate by 1.3-fold without statistically significant, compared to untreated control cells (Figure 6G). However, the concentration of acetyl-CoA was significantly increased for 40% (P < 0.01; Figure 5I) following treatment with lutein or DHA, relative to untreated control.
Micronutrients affect cellular gene expression
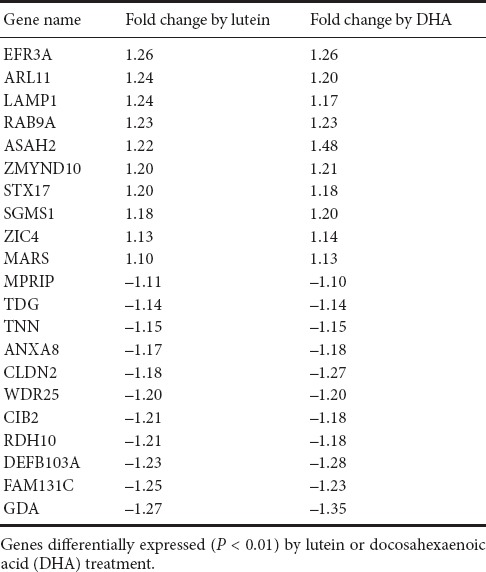
As our results showed a significant increase in the level of acetyl-CoA, a key epigenetic regulator, in the presence of lutein and DHA. We then investigate whether there are common genes regulated by treatment of lutein and DHA by Affymetrix mRNA array. As shown in Table 1, a total of 21 genes were significantly regulated by lutein and DHA. Among these 21 genes, LAMP1, TNN and RDH10 were proven neuronal-related, GDA, ASAH2, RDH10 were metabolism related and TNN was cell growth related.
Table 1.
Gene expression co-regulated by lutein and DHA
Discussion
The importance of nutrition for brain development during fetal early life and neural regeneration is now well established (Georgieff, 2007; Altun and Kurutas, 2016). In this context and given the structural and functional significance of fatty acids to neural tissue, much attention has focussed on the importance of dietary sourced micronutrients like PUFAs, tocopherols and carotenoids for optimal neuronal differentiation (Rubin et al., 2012; Bazinet and Laye, 2014; Vishwanathan et al., 2014; Deng et al., 2015). As might be expected then, epidemiological evidence links deficiencies in these micronutrients with sub-optimal neurodevelopment outcomes (Erdman et al., 2015). The supplementation of key nutrients such as DHA, Vitamin E and lutein, has been shown to have health benefits for preterm or nutrient-deficient infants (Rubin et al., 2012; Makrides, 2013). DHA has long been indicated as an essential additive for optimal neuronal function and regeneration. In addition to direct incorporation as a physical component of the neural membrane, DHA can also act as an endogenous ligand to activate the retinoid X receptor and thus potential promote neuronal differentiation (Kim, 2007; Mounier et al., 2015). Further, DHA enhanced neurite growth has been shown to occur by cellular internalization, allowing DHA to be converted to acetyl-CoA and metabolized within the TCA to yield NADH (Marszalek et al., 2004; Pietrocola et al., 2015). Meanwhile, lutein is preferentially accumulated in pediatric brain tissues and has recently been linked with beneficial effects on cognitive ability (Erdman et al., 2015; Panagos et al., 2016). Although often discussed as a potential antioxidant (by absorbing harmful blue light) in the context of ocular pathologies (Bian et al., 2012), the mechanistic basis of lutein effects in other regions of the nervous system is yet to be defined. It has been shown that lutein and its metabolites can function as exogenous ligands for retinoic acid receptor stimulating downstream signaling (Sayo et al., 2013), which is functionally similar to the tropic effect of DHA binding to retinoid X receptor (Kim, 2007). In primate, lutein is the only carotenoid to accumulate in all membranes and with a similar distribution to that of the DHA (Mohn et al., 2015). Further, post-mortem metabolic analysis of human infants indicates that the accumulation of lutein in the brain is strongly aligned to that of various lipid pathway metabolites, energy metabolites, brain osmolytes, amino acid neurotransmitters and the antioxidant homocarnosine (Lieblein-Boff et al., 2015b). In particular, lutein levels are positively correlated with levels of the neurotransmitter GABA, which is thought to regulate neuronal maturation and neurite outgrowth (Barbin et al., 1993; Represa and Ben-Ari, 2005; Ben-Ari et al., 2007).
The general finding of our current study is that lutein promotes neuronal differentiation as evidenced by enhanced neurite outgrowth and increased expression of the lineage and functionally specific marker MAP2. More significantly, we present evidence that this beneficial effect occurs through the modulation of metabolism and up-regulation of mitochondrial function, which have been well recognized in cell differentiation (Lee et al., 2011; Tormos et al., 2011; Pereira et al., 2013). The key regulatory role of metabolic reprogramming in cell commitment and differentiation is an emerging paradigm and has been linked with the increasing of aerobic respiration during neuronal differentiation (Agostini et al., 2016; Zheng et al., 2016; Xie and Sheppard, 2018). As the obligatory energy source of the brain, glucose consumption under normal conditions in an adult human accounts for approximately 25% of the systemic glucose available (Belanger et al., 2011). Not surprisingly, we found that lutein treatment led to a significant increase in glucose consumption and a reduction in lactate formation (presumably reflecting the need to generate pyruvate) alongside the increased metabolic activity associated with enhanced differentiation. The PI3K/Akt intracellular signaling pathway is key to glucose utilization in many cells, including differentiating neurons and neuronal regeneration (Zhao et al., 2012). Akt can directly regulate the differentiation of embryonal carcinoma cells, through phosphorylation of pluripotency/differentiation factors such as Stab1 (special AT-rich binding protein 1), nestin and other genes (Chen et al., 2013). Similarly, Akt signaling is essential for the differentiation of lineage precursors of bone, muscle and neuronal cells, following activation and/or translocation to the mitochondria (Sinor and Lillien, 2004; Mukherjee and Rotwein, 2009; Gardner et al., 2012; Qiao et al., 2012). By using small molecule inhibitor, we demonstrated that lutein dependent neuronal differentiation, the consequential increase in Akt phosphorylation, and the accompanying mitochondrial function are all reversed when PI3K signaling is inhibited. However, our results also showed that the treatment of DHA doesn’t change the activity of Akt, nor protect Akt activity against LY294002. This indicates DHA may drive neuronal differentiation by a distinct pathway, such as ERK/MAPK (Kamata et al., 2007; Zamarbide et al., 2014), though a physiological level of AKT is still needed for DHA mediated differentiation. As the central mediator of glucose, Akt promotes glycolysis, and also stimulates mitochondrial respiration by increasing substrate availability (Goo et al., 2012).
Recent studies suggest that ROS generation may rather be both necessary and an important regulator of cell fate commitment and cell differentiation (Sauer et al., 2000; Ji et al., 2010; Bigarella et al., 2014). These superoxides are mainly generated in complex I/III in mitochondria, and then dismutated into hydrogen peroxide by the SOD2 enzyme (Lee et al., 2011). Increased ROS levels (H2O2) not only induce G2/M cell cycle arrest (Guo et al., 2010), but also act as secondary messengers in growth factor signaling (Sundaresan et al., 1995), interacting directly with proteins, kinases, and transcription factors to regulate various aspects of cell cycle progression, apoptosis, quiescence and differentiation. ROS mediated signaling to amino acids can cause functional (activities) changes in a range of proteins, and therefore influences the target protein’s function, stability, subcellular localization, interactions with other proteins and other processes (Bigarella et al., 2014). During the differentiation of SH-SY5Y cells, ROS signaling is also associated to the increased phosphorylation of AKT and ERK1/2 (Kunzler et al., 2017). Importantly, ROS also influences nutrient-sensing pathways that direct metabolic flux (Bigarella et al., 2014). Our results showed that lutein elevated the levels of ROS following the increased activity of respiration chain complexes, and it is interesting that lutein induced ROS generation did not result in higher expression of the neuron-specific ROS dismutase-SOD2, GPX1 and CAT, suggesting that these effects do not trigger an obligatory free radical scavenger response and enabling the ROS to be directed toward promotion of cellular differentiation (Ji et al., 2010). Moreover, differentiation will be increased by increasing ROS through genetic deletion of SOD2, while the reduction of ROS by mitochondrial targeted antioxidants prohibited the differentiation of human stem cells (Owusu-Ansah and Banerjee, 2009; Tormos et al., 2011; Sena and Chandel, 2012).
An increase in ATP production during stimulated cell differentiation has been reported (Agostini et al., 2016). However, we do not observe such an increase in ATP following DHA or lutein supplementation, suggesting that such micronutrients enhance the earlier stages of the differentiation process (see below), specifically through the modulation of mitochondrial complex I/III, generating membrane potential and the accompanying production of ROS, rather than simply promoting energy production.
Derivative metabolites generated from the glycolytic intermediate, acetyl-CoA are essential for modulating the epigenetic status of cells, serving as substrates or co-factors for enzymes which orchestrate the chemical modification of histones and DNA (Janke et al., 2015) and thus alter gene expression patterns. More direct redox modification of transcription factor proteins, such as FOXOs and HIFs, would also impact on stem cell fate and commitment (Bigarella et al., 2014). The preliminary genome wide transcriptomic analysis we have carried out following micronutrient supplementation of SH-SY5Y cells identified some potentially key genes involved in metabolic regulation and/or early neuronal differentiation. Lysosome function (implicated by the LAMP1 gene) is involved in sensing of nutrient availability, and modulates energy metabolism by regulating the expression of fatty acid β-oxidation and neuronal differentiation through expression of PPARα (Settembre et al., 2013). Tenascin N (encoded by TNN gene) is the most highly expressed tenascin in neurons. It is known to mediate neurite dynamics in hippocampal cells, and a splice variant fragment of TNN influences neurite outgrowth (Neidhardt et al., 2003). Guanine deaminase (encoded by the GDA gene) removes amino acids from guanine and decreases global GTPase level in the cell, biochemical changes which are critical for all stages of axonogenesis and dendrite elaboration (Luo, 2000; Hall and Lalli, 2010). N-acylsphingosine amidohydrolase 2 (encoded by the ASAH2 gene) can hydrolyses the sphingolipid ceramide into free fatty acids and sphingosine and acting as a repressor of apoptosis. The increased expression of ASAH2 presumably protects against the possibility of apoptosis in committed cells. Collectively, these initial observations suggest that micronutrient supplementation supports the earliest stages of neuronal differentiation, encouraging the transition from proliferation to differentiation, and the initial morphological changes and cellular polarity which have been defined previously.
In summary, we have found that the lutein promotes neural differentiation potentially in a PI3K-AKT dependent manner and that this effect is accompanied by enhanced rates of glycolysis and mitochondrial function. Further, the increased metabolic activities drive production of the neuronal signaling molecule ROS, and of the TCA cycle intermediate Acetyl-CoA in particular, consistent with epigenetic-based changes in the transcriptome that would serve to encode and enhance neuronal differentiation. As many studies suggested that both mitochondrial function and ROS signaling are critical for neural regeneration to proceed (Han et al., 2016; Meda et al., 2017), the mechanistic findings presented here support a view that dietary intervention may have clinical potential for enhancing the processes of neural regeneration. Further studies to explore the molecular mechanisms of lutein function by using primary neural stem cells or clinical study will confirm this beneficial function to the process of neuronal differentiation.
Footnotes
Conflicts of interest: The authors declare that they have no competing interests.
Financial support: None.
Copyright license agreement: The Copyright License Agreement has been signed by all authors before publication.
Data sharing statement: Datasets analyzed during the current study are available from the corresponding author on reasonable request.
Plagiarism check: Checked twice by iThenticate.
Peer review: Externally peer reviewed.
C-Editor: Zhao M; S-Editor: Li CH; T-Editor: Liu XL
References
- 1.Agostini M, Romeo F, Inoue S, Niklison-Chirou MV, Elia AJ, Dinsdale D, Morone N, Knight RA, Mak TW, Melino G. Metabolic reprogramming during neuronal differentiation. Cell Death Differ. 2016;23:1502–1514. doi: 10.1038/cdd.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alle H, Roth A, Geiger JR. Energy-efficient action potentials in hippocampal mossy fibers. Science. 2009;325:1405–1408. doi: 10.1126/science.1174331. [DOI] [PubMed] [Google Scholar]
- 3.Altun I, Kurutas EB. Vitamin B complex and vitamin B12 levels after peripheral nerve injury. Neural Regen Res. 2016;11:842–845. doi: 10.4103/1673-5374.177150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amengual J, Lobo GP, Golczak M, Li HN, Klimova T, Hoppel CL, Wyss A, Palczewski K, von Lintig J. A mitochondrial enzyme degrades carotenoids and protects against oxidative stress. FASEB J. 2011;25:948–959. doi: 10.1096/fj.10-173906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbin G, Pollard H, Gaiarsa JL, Ben-Ari Y. Involvement of GABAA receptors in the outgrowth of cultured hippocampal neurons. Neurosci Lett. 1993;152:150–154. doi: 10.1016/0304-3940(93)90505-f. [DOI] [PubMed] [Google Scholar]
- 6.Bazinet RP, Laye S. Polyunsaturated fatty acids and their metabolites in brain function and disease. Nat Rev Neurosci. 2014;15:771–785. doi: 10.1038/nrn3820. [DOI] [PubMed] [Google Scholar]
- 7.Belanger M, Allaman I, Magistretti PJ. Brain energy metabolism: focus on astrocyte-neuron metabolic cooperation. Cell Metab. 2011;14:724–738. doi: 10.1016/j.cmet.2011.08.016. [DOI] [PubMed] [Google Scholar]
- 8.Ben-Ari Y, Gaiarsa JL, Tyzio R, Khazipov R. GABA: A pioneer transmitter that excites immature neurons and generates primitive oscillations. Physiol Rev. 2007;87:1215–1284. doi: 10.1152/physrev.00017.2006. [DOI] [PubMed] [Google Scholar]
- 9.Bertalanic L, Kosmerl T, Poklar Ulrih N, Cigic B. Influence of solvent composition on antioxidant potential of model polyphenols and red wines determined with 2, 2-diphenyl-1-picrylhydrazyl. J Agric Food Chem. 2012;60:12282–12288. doi: 10.1021/jf3041512. [DOI] [PubMed] [Google Scholar]
- 10.Bian Q, Gao S, Zhou J, Qin J, Taylor A, Johnson EJ, Tang G, Sparrow JR, Gierhart D, Shang F. Lutein and zeaxanthin supplementation reduces photooxidative damage and modulates the expression of inflammation-related genes in retinal pigment epithelial cells. Free Radic Biol Med. 2012;53:1298–1307. doi: 10.1016/j.freeradbiomed.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bigarella CL, Liang R, Ghaffari S. Stem cells and the impact of ROS signaling. Development. 2014;141:4206–4218. doi: 10.1242/dev.107086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brookes PS. Mitochondrial H(+) leak and ROS generation: an odd couple. Free Radic Biol Med. 2005;38:12–23. doi: 10.1016/j.freeradbiomed.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 13.Chamak B, Fellous A, Glowinski J, Prochiantz A. MAP2 expression and neuritic outgrowth and branching are coregulated through region-specific neuro-astroglial interactions. J Neurosci. 1987;7:3163–3170. doi: 10.1523/JNEUROSCI.07-10-03163.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen B, Xue Z, Yang GH, Shi BY, Yang B, Yan YM, Wang X, Han DS, Huang Y, Dong WJ. Akt-signal integration is involved in the differentiation of embryonal carcinoma cells. PLoS One. 2013;8:e64877. doi: 10.1371/journal.pone.0064877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBerardinis RJ, Lum JJ, Hatzivassiliou G, Thompson CB. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008;7:11–20. doi: 10.1016/j.cmet.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Deng S, Hou G, Xue Z, Zhang L, Zhou Y, Liu C, Liu Y, Li Z. Vitamin E isomer delta-tocopherol enhances the efficiency of neural stem cell differentiation via L-type calcium channel. Neurosci Lett. 2015;585:166–170. doi: 10.1016/j.neulet.2014.11.031. [DOI] [PubMed] [Google Scholar]
- 17.Ding Y, Choi KJ, Kim JH, Han X, Piao Y, Jeong JH, Choe W, Kang I, Ha J, Forman HJ, Lee J, Yoon KS, Kim SS. Endogenous hydrogen peroxide regulates glutathione redox via nuclear factor erythroid 2-related factor 2 downstream of phosphatidylinositol 3-kinase during muscle differentiation. Am J Pathol. 2008;172:1529–1541. doi: 10.2353/ajpath.2008.070429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erdman JW, Smith JW, Kuchan MJ, Mohn ES, Johnson EJ, Rubakhin SS, Wang L, Sweedler JV, Neuringer M. Lutein and Brain Function. Foods. 2015;4:547–564. doi: 10.3390/foods4040547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gao S, Qin T, Liu Z, Caceres MA, Ronchi CF, Chen CY, Yeum KJ, Taylor A, Blumberg JB, Liu Y, Shang F. Lutein and zeaxanthin supplementation reduces H2O2-induced oxidative damage in human lens epithelial cells. Mol Vis. 2011;17:3180–3190. [PMC free article] [PubMed] [Google Scholar]
- 20.Gardner S, Anguiano M, Rotwein P. Defining Akt actions in muscle differentiation. Am J Physiol Cell Physiol. 2012;303:C1292–1300. doi: 10.1152/ajpcell.00259.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgieff MK. Nutrition and the developing brain: nutrient priorities and measurement. Am J Clin Nutr. 2007;85:614S–620S. doi: 10.1093/ajcn/85.2.614S. [DOI] [PubMed] [Google Scholar]
- 22.Goo CK, Lim HY, Ho QS, Too HP, Clement MV, Wong KP. PTEN/Akt signaling controls mitochondrial respiratory capacity through 4E-BP1. PLoS One. 2012;7:e45806. doi: 10.1371/journal.pone.0045806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guo YL, Chakraborty S, Rajan SS, Wang R, Huang F. Effects of oxidative stress on mouse embryonic stem cell proliferation, apoptosis, senescence, and self-renewal. Stem Cells Dev. 2010;19:1321–1331. doi: 10.1089/scd.2009.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hall A, Lalli G. Rho and Ras GTPases in axon growth, guidance, and branching. Cold Spring Harb Perspect Biol. 2010;2:a001818. doi: 10.1101/cshperspect.a001818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hammond BR, Jr, Miller LS, Bello MO, Lindbergh CA, Mewborn C, Renzi-Hammond LM. Effects of lutein/zeaxanthin supplementation on the cognitive function of community dwelling older adults: a randomized, double-masked, placebo-controlled trial. Front Aging Neurosci. 2017;9:254. doi: 10.3389/fnagi.2017.00254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Han SM, Baig HS, Hammarlund M. Mitochondria localize to injured axons to support regeneration. Neuron. 2016;92:1308–1323. doi: 10.1016/j.neuron.2016.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, Neumann D, Kakazu A, Pham TL, Musarrat F, Cortina MS, Bazan HEP. PEDF plus DHA modulate inflammation and stimulate nerve regeneration after HSV-1 infection. Exp Eye Res. 2017;161:153–162. doi: 10.1016/j.exer.2017.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins S, Lee JS, Ha L, Lim JY. Inducing neurite outgrowth by mechanical cell stretch. Biores Open Access. 2013;2:212–216. doi: 10.1089/biores.2013.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Janke R, Dodson AE, Rine J. Metabolism and epigenetics. Annu Rev Cell Dev Biol. 2015;31:473–496. doi: 10.1146/annurev-cellbio-100814-125544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jeon S, Ranard KM, Neuringer M, Johnson EE, Kuchan M, Johnson EJ, Erdman JW. Lutein-supplemented formula enhances lutein accumulation in brain and other tissues in infant rhesus macaques. FASEB J. 2016;30(1_supplement):68911–68911. [Google Scholar]
- 31.Ji AR, Ku SY, Cho MS, Kim YY, Kim YJ, Oh SK, Kim SH, Moon SY, Choi YM. Reactive oxygen species enhance differentiation of human embryonic stem cells into mesendodermal lineage. Exp Mol Med. 2010;42:175–186. doi: 10.3858/emm.2010.42.3.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson EJ. A possible role for lutein and zeaxanthin in cognitive function in the elderly. Am J Clin Nutr. 2012;96:1161S–1165S. doi: 10.3945/ajcn.112.034611. [DOI] [PubMed] [Google Scholar]
- 33.Johnson EJ, Maras JE, Rasmussen HM, Tucker KL. Intake of lutein and zeaxanthin differ with age, sex, and ethnicity. J Am Diet Assoc. 2010;110:1357–1362. doi: 10.1016/j.jada.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 34.Kamata Y, Shiraga H, Tai A, Kawamoto Y, Gohda E. Induction of neurite outgrowth in PC12 cells by the medium-chain fatty acid octanoic acid. Neuroscience. 2007;146:1073–1081. doi: 10.1016/j.neuroscience.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 35.Katakura M, Hashimoto M, Okui T, Shahdat HM, Matsuzaki K, Shido O. Omega-3 polyunsaturated Fatty acids enhance neuronal differentiation in cultured rat neural stem cells. Stem Cells Int. 2013;2013:490476. doi: 10.1155/2013/490476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kawakita E, Hashimoto M, Shido O. Docosahexaenoic acid promotes neurogenesis in vitro and in vivo. Neuroscience. 2006;139:991–997. doi: 10.1016/j.neuroscience.2006.01.021. [DOI] [PubMed] [Google Scholar]
- 37.Kim HY. Novel metabolism of docosahexaenoic acid in neural cells. J Biol Chem. 2007;282:18661–18665. doi: 10.1074/jbc.R700015200. [DOI] [PubMed] [Google Scholar]
- 38.Kim JW, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 39.Kuchan MWF, Geng Y, Feng B. Lutein stimulates the differentiation of human stem cells to neural progenitor cells in vitro. Paper presented at: Advances and Controversies in Clinical Nutrition Abstract no.23. 2013 [Google Scholar]
- 40.Kunzler A, Zeidan-Chulia F, Gasparotto J, Girardi CS, Klafke K, Petiz LL, Bortolin RC, Rostirolla DC, Zanotto-Filho A, de Bittencourt Pasquali MA, Dickson P, Dunkley P, Moreira JCF, Gelain DP. Changes in cell cycle and up-regulation of neuronal markers during SH-SY5Y neurodifferentiation by retinoic acid are mediated by reactive species production and oxidative stress. Mol Neurobiol. 2017;54:6903–6916. doi: 10.1007/s12035-016-0189-4. [DOI] [PubMed] [Google Scholar]
- 41.Langhammer CG, Previtera ML, Sweet ES, Sran SS, Chen M, Firestein BL. Automated Sholl analysis of digitized neuronal morphology at multiple scales: whole cell Sholl analysis versus Sholl analysis of arbor subregions. Cytometry Part A. 2010;77A:1160–1168. doi: 10.1002/cyto.a.20954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lee S, Tak E, Lee J, Rashid MA, Murphy MP, Ha J, Kim SS. Mitochondrial H2O2 generated from electron transport chain complex I stimulates muscle differentiation. Cell Res. 2011;21:817–834. doi: 10.1038/cr.2011.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lieblein-Boff J, Johnson E, Kennedy A, Lai CS, Kuchan M. Lutein Accretion Corresponds to Activity of Metabolic Pathways during Brain Development. FASEB J. 2015a;29(1_supplement):6036. [Google Scholar]
- 44.Lieblein-Boff JC, Johnson EJ, Kennedy AD, Lai CS, Kuchan MJ. Exploratory metabolomic analyses reveal compounds correlated with lutein concentration in frontal cortex, hippocampus, and occipital cortex of human infant brain. PLoS One. 2015b;10:e0136904. doi: 10.1371/journal.pone.0136904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez-Carballo G, Moreno L, Masia S, Perez P, Barettino D. Activation of the phosphatidylinositol 3-kinase/Akt signaling pathway by retinoic acid is required for neural differentiation of SH-SY5Y human neuroblastoma cells. J Biol Chem. 2002;277:25297–25304. doi: 10.1074/jbc.M201869200. [DOI] [PubMed] [Google Scholar]
- 46.Luo L. Rho GTPases in neuronal morphogenesis. Nat Rev Neurosci. 2000;1:173–180. doi: 10.1038/35044547. [DOI] [PubMed] [Google Scholar]
- 47.Ma D, Zhang M, Larsen CP, Xu F, Hua W, Yamashima T, Mao Y, Zhou L. DHA promotes the neuronal differentiation of rat neural stem cells transfected with GPR40 gene. Brain Res. 2010;1330:1–8. doi: 10.1016/j.brainres.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 48.Maddodi N, Bhat KM, Devi S, Zhang SC, Setaluri V. Oncogenic BRAFV600E induces expression of neuronal differentiation marker MAP2 in melanoma cells by promoter demethylation and down-regulation of transcription repressor HES1. J Biol Chem. 2010;285:242–254. doi: 10.1074/jbc.M109.068668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Makrides M. DHA supplementation during the perinatal period and neurodevelopment: Do some babies benefit more than others. Prostag Leukotr Ess. 2013;88:87–90. doi: 10.1016/j.plefa.2012.05.004. [DOI] [PubMed] [Google Scholar]
- 50.Marszalek JR, Kitidis C, Dararutana A, Lodish HF. Acyl-CoA synthetase 2 overexpression enhances fatty acid internalization and neurite outgrowth. J Biol Chem. 2004;279:23882–23891. doi: 10.1074/jbc.M313460200. [DOI] [PubMed] [Google Scholar]
- 51.Meda F, Joliot A, Vriz S. Nerves and hydrogen peroxide: how old enemies become new friends. Neural Regen Res. 2017;12:568–569. doi: 10.4103/1673-5374.205088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mergenthaler P, Lindauer U, Dienel GA, Meisel A. Sugar for the brain: the role of glucose in physiological and pathological brain function. Trends Neurosci. 2013;36:587–597. doi: 10.1016/j.tins.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mohn E, Matthan N, Neuringer M, Crivello N, Erdman J, Kuchan M, Johnson E. Distribution of lutein in membranes of rhesus macaque brain. FASEB J. 2015;29(1_supplement):6036. [Google Scholar]
- 54.Mostofsky DI, Yehuda S, Salem N. Totowa, NJ, USA: Humana Press; 2001. Fatty acids : physiological and behavioral functions. [Google Scholar]
- 55.Mounier A, Georgiev D, Nam KN, Fitz NF, Castranio EL, Wolfe CM, Cronican AA, Schug J, Lefterov I, Koldamova R. Bexarotene-activated retinoid X receptors regulate neuronal differentiation and dendritic complexity. J Neurosci. 2015;35:11862–11876. doi: 10.1523/JNEUROSCI.1001-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mukherjee A, Rotwein P. Akt promotes BMP2-mediated osteoblast differentiation and bone development. J Cell Sci. 2009;122:716–726. doi: 10.1242/jcs.042770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009;417:1–13. doi: 10.1042/BJ20081386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Neidhardt J, Fehr S, Kutsche M, Lohler J, Schachner M. Tenascin-N: characterization of a novel member of the tenascin family that mediates neurite repulsion from hippocampal explants. Mol Cell Neurosci. 2003;23:193–209. doi: 10.1016/s1044-7431(03)00012-5. [DOI] [PubMed] [Google Scholar]
- 59.O’Brien LC, Keeney PM, Bennett JP., Jr Differentiation of human neural stem cells into motor neurons stimulates mitochondrial biogenesis and decreases glycolytic flux. Stem Cells Dev. 2015;24:1984–1994. doi: 10.1089/scd.2015.0076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Owusu-Ansah E, Banerjee U. Reactive oxygen species prime Drosophila haematopoietic progenitors for differentiation. Nature. 2009;461:537–541. doi: 10.1038/nature08313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Panagos PG, Vishwanathan R, Penfield-Cyr A, Matthan NR, Shivappa N, Wirth MD, Hebert JR, Sen S. Breastmilk from obese mothers has pro-inflammatory properties and decreased neuroprotective factors. J Perinatol. 2016;36:284–290. doi: 10.1038/jp.2015.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pereira SL, Graos M, Rodrigues AS, Anjo SI, Carvalho RA, Oliveira PJ, Arenas E, Ramalho-Santos J. Inhibition of mitochondrial complex III blocks neuronal differentiation and maintains embryonic stem cell pluripotency. PLoS One. 2013;8:e82095. doi: 10.1371/journal.pone.0082095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, Madeo F, Kroemer G. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014. [DOI] [PubMed] [Google Scholar]
- 64.Qiao J, Paul P, Lee S, Qiao L, Josifi E, Tiao JR, Chung DH. PI3K/AKT and ERK regulate retinoic acid-induced neuroblastoma cellular differentiation. Biochem Biophys Res Commun. 2012;424:421–426. doi: 10.1016/j.bbrc.2012.06.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Represa A, Ben-Ari Y. Trophic actions of GABA on neuronal development. Trends Neurosci. 2005;28:278–283. doi: 10.1016/j.tins.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 66.Riederer BM, Pellier V, Antonsson B, Di Paolo G, Stimpson SA, Lutjens R, Catsicas S, Grenningloh G. Regulation of microtubule dynamics by the neuronal growth-associated protein SCG10. Proc Natl Acad Sci U S A. 1997;94:741–745. doi: 10.1073/pnas.94.2.741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rogers LK, Valentine CJ, Keim SA. DHA supplementation: current implications in pregnancy and childhood. Pharmacol Res. 2013;70:13–19. doi: 10.1016/j.phrs.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rubin LP, Chan GM, Barrett-Reis BM, Fulton AB, Hansen RM, Ashmeade TL, Oliver JS, Mackey AD, Dimmit RA, Hartmann EE, Adamkin DH. Effect of carotenoid supplementation on plasma carotenoids, inflammation and visual development in preterm infants. J Perinatol. 2012;32:418–424. doi: 10.1038/jp.2011.87. [DOI] [PubMed] [Google Scholar]
- 69.Ruijtenberg S, van den Heuvel S. Coordinating cell proliferation and differentiation: Antagonism between cell cycle regulators and cell type-specific gene expression. Cell Cycle. 2016;15:196–212. doi: 10.1080/15384101.2015.1120925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sanchez S, Jimenez C, Carrera AC, Diaz-Nido J, Avila J, Wandosell F. A cAMP-activated pathway, including PKA and PI3K, regulates neuronal differentiation. Neurochem Int. 2004;44:231–242. doi: 10.1016/s0197-0186(03)00150-5. [DOI] [PubMed] [Google Scholar]
- 71.Sauer H, Rahimi G, Hescheler J, Wartenberg M. Role of reactive oxygen species and phosphatidylinositol 3-kinase in cardiomyocyte differentiation of embryonic stem cells. FEBS Lett. 2000;476:218–223. doi: 10.1016/s0014-5793(00)01747-6. [DOI] [PubMed] [Google Scholar]
- 72.Sayo T, Sugiyama Y, Inoue S. Lutein, a nonprovitamin A, activates the retinoic acid receptor to induce HAS3-dependent hyaluronan synthesis in keratinocytes. Biosci Biotechnol Biochem. 2013;77:1282–1286. doi: 10.1271/bbb.130124. [DOI] [PubMed] [Google Scholar]
- 73.Schneider L, Giordano S, Zelickson BR, M SJ, G AB, Ouyang X, Fineberg N, Darley-Usmar VM, Zhang J. Differentiation of SH-SY5Y cells to a neuronal phenotype changes cellular bioenergetics and the response to oxidative stress. Free Radic Biol Med. 2011;51:2007–2017. doi: 10.1016/j.freeradbiomed.2011.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sena LA, Chandel NS. Physiological roles of mitochondrial reactive oxygen species. Mol Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Settembre C, Fraldi A, Medina DL, Ballabio A. Signals from the lysosome: a control centre for cellular clearance and energy metabolism. Nat Rev Mol Cell Biol. 2013;14:283–296. doi: 10.1038/nrm3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sherratt SCR, Shrivastava S, Jacob RF, Chattopadhyay A, Mason RP. Docosahexaenoic acid (DHA), but not eicosapentaenoic acid (EPA), increases both membrane fluidity and cholesterol crystalline domain formation in lipid vesicles. Biophys J. 2016;110:583a–583a. [Google Scholar]
- 77.Shimazawa M, Nakajimaa Y, Mashima Y, Hara H. Docosahexaenoic acid (DHA) has neuroprotective effects against oxidative stress in retinal ganglion cells. Brain Res. 2009;1251:269–275. doi: 10.1016/j.brainres.2008.11.031. [DOI] [PubMed] [Google Scholar]
- 78.Sinor AD, Lillien L. Akt-1 expression level regulates CNS precursors. J Neurosci. 2004;24:8531–8541. doi: 10.1523/JNEUROSCI.1470-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–299. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 80.Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metab. 2011;14:537–544. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tsatmali M, Walcott EC, Makarenkova H, Crossin KL. Reactive oxygen species modulate the differentiation of neurons in clonal cortical cultures. Mol Cell Neurosci. 2006;33:345–357. doi: 10.1016/j.mcn.2006.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Vishwanathan R, Kuchan MJ, Sen S, Johnson EJ. Lutein and preterm infants with decreased concentrations of brain carotenoids. J Pediatr Gastroenterol Nutr. 2014;59:659–665. doi: 10.1097/MPG.0000000000000389. [DOI] [PubMed] [Google Scholar]
- 83.Walsh K, Perlman H. Cell cycle exit upon myogenic differentiation. Curr Opin Genet Dev. 1997;7:597–602. doi: 10.1016/s0959-437x(97)80005-6. [DOI] [PubMed] [Google Scholar]
- 84.Wu H, Ichikawa S, Tani C, Zhu B, Tada M, Shimoishi Y, Murata Y, Nakamura Y. Docosahexaenoic acid induces ERK1/2 activation and neuritogenesis via intracellular reactive oxygen species production in human neuroblastoma SH-SY5Y cells. Biochim Biophys Acta. 2009;1791:8–16. doi: 10.1016/j.bbalip.2008.10.004. [DOI] [PubMed] [Google Scholar]
- 85.Xie K, Sheppard A. Dietary micronutrients promote neuronal differentiation by modulating the mitochondrial-nuclear dialogue. Bioessays. 2018;40:e1800051. doi: 10.1002/bies.201800051. [DOI] [PubMed] [Google Scholar]
- 86.Zamarbide M, Etayo-Labiano I, Ricobaraza A, Martinez-Pinilla E, Aymerich MS, Luis Lanciego J, Perez-Mediavilla A, Franco R. GPR40 activation leads to CREB and ERK phosphorylation in primary cultures of neurons from the mouse CNS and in human neuroblastoma cells. Hippocampus. 2014;24:733–739. doi: 10.1002/hipo.22263. [DOI] [PubMed] [Google Scholar]
- 87.Zhao T, Qi Y, Li Y, Xu K. PI3 Kinase regulation of neural regeneration and muscle hypertrophy after spinal cord injury. Mol Biol Rep. 2012;39:3541–3547. doi: 10.1007/s11033-011-1127-1. [DOI] [PubMed] [Google Scholar]
- 88.Zheng X, Boyer L, Jin M, Mertens J, Kim Y, Ma L, Ma L, Hamm M, Gage FH, Hunter T. Metabolic reprogramming during neuronal differentiation from aerobic glycolysis to neuronal oxidative phosphorylation. Elife. 2016;5:e13374. doi: 10.7554/eLife.13374. [DOI] [PMC free article] [PubMed] [Google Scholar]