Abstract
Selenocysteine (Sec) lacks a cognate aminoacyl-tRNA synthetase. Instead, seryl-tRNA synthetase (SerRS) produces Ser-tRNASec, which is subsequently converted by selenocysteine synthase to Sec-tRNASec. Escherichia coli SerRS serylates tRNASec poorly; this may hinder efficient production of designer selenoproteins in vivo. Guided by structural modeling and selection for chloramphenicol acetyltransferase activity, we evolved three SerRS variants capable of improved Ser-tRNASec synthesis. They display 10-, 8-, and 4-fold increased kcat/KM values compared to wild-type SerRS using synthetic tRNASec species as substrates. The enzyme variants also facilitate in vivo read-through of a UAG codon in the position of the critical serine146 of chloramphenicol acetyltransferase. These results indicate that the naturally evolved SerRS is capable of further evolution for increased recognition of a specific tRNA isoacceptor.
Keywords: Selenoproteins, synthetic biology, seryl-tRNA synthetase, tRNA, protein engineering
Introduction
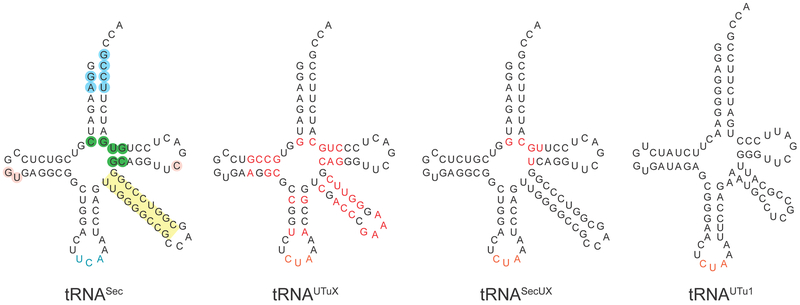
Selenocysteine (Sec), genetically encoded by UGA, is the only naturally occurring amino acid that lacks a cognate aminoacyl-tRNA synthetase. Instead, seryl-tRNA synthetase (SerRS) acylates tRNASec with serine (Ser); subsequently, the selenocysteine synthase SelA transforms the RNA-bound Ser to Sec. The resulting Sec-tRNASec is delivered to the ribosome by the dedicated elongation factor SelB, which in the presence of a specific mRNA structure aids accurate recoding of UGA for Sec insertion into proteins [1]. Thus, E. coli tRNASec evolved to optimally interact with SerRS, SelA, and SelB, and to reject the general elongation factor EF-Tu. Yet, strategies for in vivo synthesis of selenoproteins in E. coli rely on tRNASec variants that interact with EF-Tu and SelA. For this reason a number of tRNASec variants were designed [2–4]. Secondary structures and altered nucleotides (compared to tRNASec) of tRNAUTuX, tRNASecUX and tRNAUTu1 are presented in Fig. 1. Significantly, the three base pairs (green) in the center of tRNASec are identity elements for SelB recognition and reject EF-Tu. Replacing these base pairs with G7:C66, C/U49:G65 and A/C50:U64 allows these tRNAs to be recognized by EF-Tu. Sec insertion by these variant tRNAs may be feasible for virtually any codon [5].
Figure 1.
Wild-type and synthetic selenocysteine tRNAs. Identity nucleotide determinants of wild-type E. coli tRNASec are shaded blue, and the variable arm, whose length and orientation are specifically recognized by SerRS, is highlighted in yellow. Nucleotides that govern the tertiary fold of tRNASec are given in pink. Base pairs acting as EF-Tu antideterminants are shown in green. Synthetic selenocysteine tRNAs employed in this study are given in black, except the amber anticodon (in purple). Mutations introduced into tRNAUTuX and tRNASecUX are indicated in red. tRNAUTu1, derived from a metagenome sequence [10,15] is too dissimilar to E. coli tRNASec to highlight nucleotide differences.
E. coli SerRS charges five tRNASer isoacceptors with equal efficiency [6]. However, compared to the tRNASer species, tRNASec [7] is serylated 100 times more poorly (as judged by kcat/KM) [8]. This is plausible; given that there is only one selenoprotein in E. coli [9], efficient genome translation requires very much less Sec-tRNASec than Ser-tRNASer (for translation of >78,000 codons). In light of our synthetic biology efforts to develop an efficient EF-Tu-dependent translation system for UAG directed, site-specific Sec incorporation into proteins [10], we decided to design a variant SerRS enzyme endowed with improved serylation of the tRNASec variants tRNAUTuX [3], tRNAUTu1 [10]), and tRNASecUX [4].
We based our E. coli SerRS mutagenesis strategy on the examination of Thermus thermophilus SerRS•tRNASer [11] and Homo sapiens SerRS•tRNASec [12] co-crystal structures, corresponding library construction, and variant selection by an in vivo chloramphenicol acetyltransferase (CAT) assay [13]. This approach led to the discovery of three desired SerRS variants.
Materials and Methods
2.1. Reagents and strains
Pfu Ultra II DNA polymerase was obtained from Agilent Technologies; restriction enzymes, T4 DNA ligase, and the Gibson Assembly mix were from New England Biolabs. In-Fusion cloning kit was from Clontech. The thermosensitive E. coli strain KL235 [14] was obtained from the Coli Genetic Stock Center (Yale University).
2.2. Plasmid construction
For in vivo serylation assays plasmids pXF113, pXF114 and pXF114 were created by inserting a lpp-tRNA-rrnC cassette into a modified pACYC184 backbone containing a cat gene where an amber stop codon was introduced at position 146 [15]. The cassette and pACYC184 backbone were amplified using primers listed in Table S1 and ligated using In-Fusion cloning. pXF113, pXF114 and pXF115 contained genes from tRNAUTuX, tRNASecUX and tRNAPyl, respectively. Plasmids pXF113 containing coding sequences for tRNAUTu1, SupD, and SelC(amb) were created by Gibson Assembly, using sets of primer listed in Table S1. The gene for E. coli tRNASec with an amber anticodon (selC(amb)) was amplified using a previously published template [2]. To generate pBAD-SerRS plasmids oligonucleotides EcSerSf and EcSerSr (Table S1) were used to amplify the SerRS gene. The product was then digested with NcoI and KpnI and inserted into a modified pKTS vector containing araC gene and PBAD promoter. SerRS variants containing S55A mutation or a GGGGS linker were constructed by In-Fusion cloning, using primer sets listed in Table S1. Plasmids constitutively expressing tRNAs and wild-type or SerRS variants were created by inserting a lpp-tRNA-rrnC cassette into pBAD-SerRS plasmids by In-Fusion cloning (Table S1). pTrc99A-SerRS plasmids were generated by In-Fusion using the primer sets listed in Table S1. To generate pTrc99A-SerRS.V20-A55S the plasmid was mutagenized using primers A55S.F and A55S.R (Table S1). For protein isolation purposes, SerRS variants were cloned into pET15 vector (Novagen) using Gibson Assembly (NEB). The open reading frames of E. coli SerRS genes and the pET15 backbone were amplified using primer pairs listed in Table S1. Artificial tRNASec genes (tRNAUTuX, tRNASecUX, tRNAUTu1, [3,4,10]) and wild-type tRNASerCGA were amplified using primer pairs listed in Table S1 and cloned using Gibson Assembly (NEB) into a modified pUC18 vector (T7 promotor adjacent to the BamHI restriction site). Construction of pUC19 with wild-type E. coli tRNASec was described in [16].
2.3. Construction of E. coli knock-out strains
Strains DH10B ΔselAB::zeo strain and WL30153 were used to test serylation of different tRNAs in vivo and for selection purposes. DH10B ΔselAB::zeo was created by λ-Red recombination [17] using primers EM7-zeo FW and EM7-zeo RV (Table S1) to amplify the EM7 - zeocin cassette. E. coli strain WL30153 (MC4100 ΔselAB) was described [18].
2.4. Library creation and chloramphenicol-resistance selection
Four fragments of serS gene were amplified by primers listed in Table S1 to generate SerRS coding sequences that are fully randomized at given positions (residues K86, A87, D90, S160 and D228 and T229) by overlap PCR. The mixture of pBAD-SerRS plasmids expressing the wild-type and the S55A variant with 1 to 1 ratio was used as the template to amplify serRS gene fragments. Inserts were then digested with NcoI and NdeI and ligated into pRSF-based vector, under the control of a PBAD promoter. The SerRS library transformation resulted with 8.5 × 108 clones, oversampling the theoretical diversity of 3.5 × 108. For selection purposes, the library was co-transformed with pXF113 into DH10B ΔselAB::zeo, and plated onto LB agar supplemented with 55 mg/l Cm.
2.5. Protein purification
E. coli BL21(DE3) cells harboring pET15b-serS plasmids were grown to OD600 0.4 at 37 °C, after which 0.3 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) was added. Following induction, cells were grown for 3 hr, pelleted and resuspended in buffer A (0.1 M Tris-Cl pH 7.4, 0.1 M NaCl, 5 mM 2-mercaptoethanol, 0.3 mM PMSF). Cells were then lysed by ultrasonic treatment and soluble proteins collected by centrifugation at 12,000 × g. Wild-type SerRS and SerRS variants were then purified by two rounds of anionic exchange, first on HiTrap DEAE FF Sepharose (GE Healthcare) and then HiTrap Q (GE Healthcare). Purification was executed in a linear gradient of salt using buffers A and B (0.1 M Tris-Cl pH 7.4, 1 M NaCl, 5 mM 2-mercaptoethanol). Eluted proteins were concentrated and stored at −80°C in buffer S (0.1 M Tris-Cl pH 7.4, 0.15 M NaCl, 50% glycerol, 5 mM 2-mercaptoethanol). Prior to the aminoacylation assay, proteins were diluted in buffer D (50 mM Tris-Cl pH 8.0, 1 mM 1,4-dithiothreitol (DTT)) and their concentration was measured according to the theoretical extinction coefficients (68760 M−1 cm−1 for wild-type and SerRS v.20 and v.22, and 71740 M−1 cm−1 for SerRS v.8).
2.6. Transcript preparation and purification
pUC18 plasmids containing genes for tRNASer and tRNASec variants were transformed into E. coli DH5α cells, grown overnight and isolated using Plasmid Maxi kit (Qiagen). Following BstNI digestion to generate appropriate 3′-CCA ends, tRNAs were generated by in vitro transcription using recombinantly produced T7 RNA polymerase [19]. Transcripts were purified on a HiTrap DEAE FF column (GE Healthcare), as described [20]. Prior use tRNAs were heated to 92°C for 3 min and slowly cooled to room temperature in the presence of 5 mM MgCl2 to allow renaturation. tRNA concentration was estimated according to its absorbance at 260 nm and the percentage of active transcript was obtained from the plateau charging (see below).
2.7. In vitro aminoacylation assays
All aminoacylation reactions were performed at 37 °C, essentially as reported [21], and contained 100 mM Tris-Cl (pH 7.5), 10 mM KCl, 10 mM MgCl2, 1 mM DTT, 10 mM ATP and 0.4 mg/ml bovine serum albumin, 0.05 U thermostable inorganic pyrophosphatase (NEB) and 125 μM [14C]serine (Perkin Elmer). Aliquots of the reaction were removed at given intervals, spotted onto 3 MM filter papers and immersed in 10% trichloroacetic acid to precipitate aminoacyl-tRNA. For the KM and kcat determination in case of tRNAUTuX, tRNA concentration was varied over the range of ~0.1 – 5.6 μM (for SerRS V8 and V20) and ~0.1 – 8.6 μM (for wt and SerRS V22).
In case of tRNASecUX, tRNA concentration was varied over the range of ~0.6 – 11.1 μM and in case of tRNAUTu1 over the range of ~0.2 – 12 μM for all SerRS enzymes. To determine the plateau of any selenocysteine tRNA serylation, 2 μM wt SerRS was incubated with of 2 μM transcript for 30 min.
2.8. Complementation assay
A culture of E. coli strain KL235 (in which lack of growth at elevated temperatures is a result of a thermosensitive SerRS [14]) was grown to early exponential phase at 30°C. The cells were made electrocompetent and were transformed with plasmids pTrc99A, in which the genes for wild-type and SerRS variants were placed under the control of trc promoter. As a negative control, a batch of cells were transformed with pTrc99A lacking the insert. Individual clones of KL235 containing desired plasmids were grown overnight, diluted to OD600=1 and spotted in serial dilutions onto LB plates containing 0.05 and 0.1 mM IPTG. Colonies were then grown at 30, 37 and 42°C for 20 hr.
2.9. Model of the E. coli SerRS•tRNASec
E. coli SerRS was comparatively modeled by Phyre2 (Protein Homology/AnalogY Recognition Engine, [22]. Superposition of the model structure of E. coli SerRS onto SerRS•tRNASec complex was achieved by UCSF Chimera [23].
Results
Designing a SerRS variant library
SerRS recognizes tRNASec and tRNASer in a similar manner. The two tRNA species share a set of minor identity elements (discriminator base G73, C72, and acceptor-stem pairs G2:C71, A3:U70, Fig. 1) [24,25]. However, the most important element is the long variable arm, which is recognized by SerRS in a sequence-independent manner; the length and relative orientation have the greatest effect. The binding of the variable arm (of both tRNA species) is accompanied by the movement of the N-terminal tRNA binding domain (NTD) [11,12].
Another idiosyncratic feature of tRNASec is the G19:U20:C56 base triplet that defines its tertiary fold [12,26] (Fig. 1). Interestingly, U20 protrudes in the human SerRS•tRNASec structure and is solvent accessible [12,26], unlike the corresponding C20 in TttRNASer that is buried in the tRNA core [11]. The highly conserved S61 residue presumably allows SerRS to distinguish between tRNASec and tRNASer by interacting specifically with nucleotide determinant in position 20 [12]. Both S61 (S55 in E. coli SerRS) and U20 are present in EcSerRS and EctRNASec, respectively. Furthermore, all tRNASec derivatives (tRNASecUX, tRNAUTu1 and tRNAUTuX, Fig. 1) retain this uridine. However, our attempts to improve tRNASec serylation via S55 mutagenesis, or by increasing NTD flexibility in order to accommodate the longer acceptor-TψC arm of tRNASec were unsuccessful (see Supplementary Information for detailed explanations).
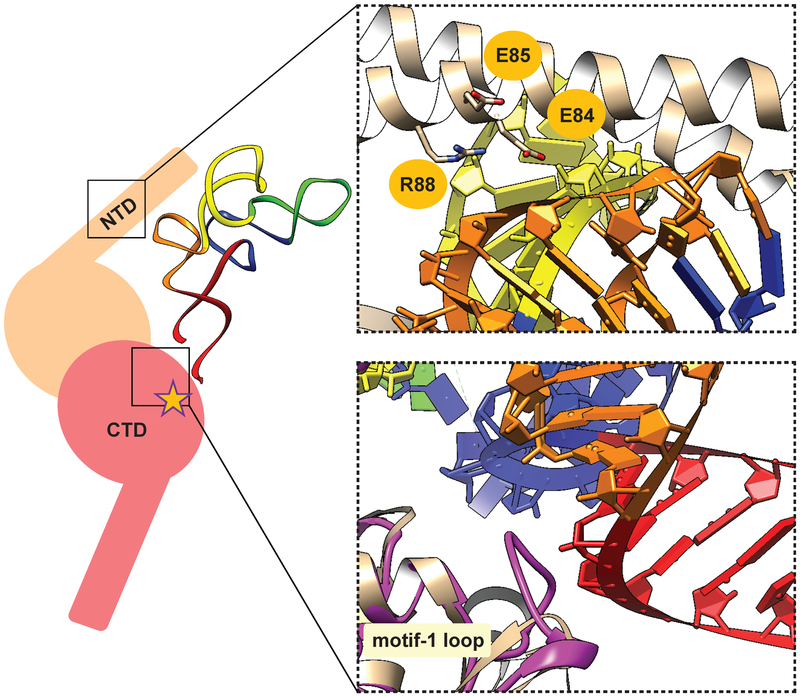
Next, we decided to simultaneously target multiple residues that, in the T. thermophilus SerRS•tRNASer complex, are in close contact (Fig. 2) [11]. We decided to use this structure because tRNASec and tRNASer share some common features responsible for their recognition by SerRS [25], and because TtSerRS and EcSerRS are more closely related than HsSerRS and EcSerRS [27]. While the discriminator base G73 and the orientation of the variable arm constitute the minimal set of recognition parameters in tRNASer and tRNASec [8,28,29], the NTD of TtSerRS also appears to interact with the T loop of tRNASer (Fig. 2, [11]). Similarly, T loop of tRNASec is also in proximity of NTD in the human SerRS•tRNASec complex [12]. Given the difference in nucleotide composition of TψC arms of tRNASer and tRNASec, we hypothesized that certain residues in NTD might account for different mode of recognition of these tRNAs. Most closely placed to tRNASer, charged residues E84, E85, and R88 of T. thermophilus SerRS are likely to form direct interactions with the tRNA (Fig. 2). Thus, we decided to randomize the corresponding residues of EcSerRS (K86, A87, and D90) to generate a SerRS variant capable of improved serylation of tRNASec.
Fig. 2.
Key interactions between SerRS and tRNASer as a guide for SerRS engineering. (left) Cartoon representation of SerRS•tRNASec productive complex. SerRS is given in two shades of pink, for each monomer of the homodimer. The N-terminal tRNA binding domain (NTD), C-terminal catalytic domain (CTD) and the active site (star) are indicated. Cognate tRNA binds across the dimer. Acceptor, TψC, variable, anticodon, and D arms of tRNASec are shown in red, orange, yellow, green and blue, respectively. (right) Magnified view of the two regions targeted for mutagenesis. (up) In the crystal structure of T. thermophilus SerRS•tRNASer complex (PDB ID: 1SER) NTD residues E84, E85, and R88 are in the vicinity and/or form contacts with the TψC arm. These residues correspond to K86, A87, and D90 of E. coli SerRS. (down) Compared to T. thermophilus SerRS (light pink), E. coli SerRS (purple) has a longer motif 1 loop, which may interact with the base of the acceptor arm.
In addition to NTD residues, we wanted to simultaneously target the amino acids located in the catalytic domain of the enzyme. Compared to T. thermophilus, E. coli SerRS has a much longer motif 1 loop (residues R221-N231) [30]. This exposed motif 1 loop and S160 of EcSerRS are in proximity to the base of the acceptor arm according to our structural model (Fig. 2). Nucleotides in the tRNA that are likely to form interactions with SerRS occupy positions 65 and 66; they differ in tRNASer and tRNASec (tRNASer has C65 and G66 while tRNASec contains G65 and A66). Thus, to examine the role of catalytic domain residues in differential recognition of these two tRNA species we targeted D228 and T229 in the motif 1 loop and S160.
Read-through of UAG codons in chloramphenicol acetyltransferase
The synthetic selenocysteine tRNAs tRNAUTuX, tRNASecUX and tRNAUTu1 (Fig. 1) charged with Ser are recognized by EF-Tu, as shown by minor mis-incorporation of Ser instead of Sec into reporter proteins [2,3,10]. Therefore, the capacity of SerRS to aminoacylate these tRNASec variants can be monitored in vivo via stop-codon read-through in a selA deficient strain.
Using the CAT with a TAG stop codon at position 112 as reporter, tRNAUTuX and tRNASecUX facilitated production of the full-length CAT protein at moderate Cm levels (50 mg/l and 25 mg/l for the respective tRNAs, Fig. S1). Since a number of different amino acids in position 112 lead to CAT activity, we switched to a cat gene TAG variant (position 146) that required Ser for activity [31]. Ser-tRNAUTuX facilitated UAG suppression that allowed growth at ≤ 20 mg/l, while no growth was observed with tRNASecUX (Fig. S2). Both types of assay were conducted in DH10B ΔselAB::zeo in which there was no observable near-cognate suppression (Fig. S1).
Selection of desirable variants (a) recognition of tRNAUTuX
A SerRS library containing fully randomized positions 84, 85, 88, 160, 228 and 229 was created using both wt and SerRS-S55A coding sequences as templates. SerRS coding sequence was placed under the control of the PBAD promoter to regulate its expression and reduce its toxicity on cell growth. After confirming the library quality, library plasmids were re-transformed into the DH10B ΔselAB::zeo strain harboring the selection plasmid (pXF113 that carries the cat-146TAG and tRNAUTuX genes). The number of apparent cat revertants was determined with pXF115 carrying tRNAPyl [15] (see above).
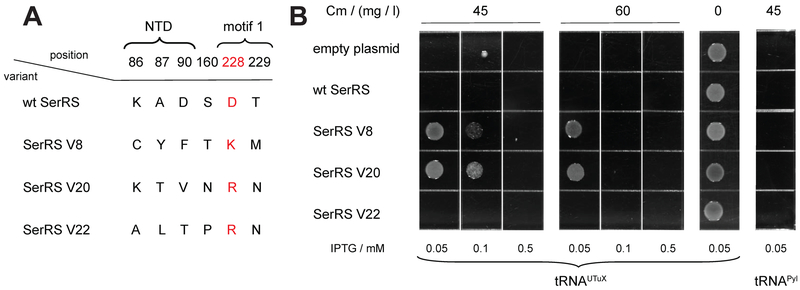
To identify SerRS variants with improved activity toward tRNAUTuX we selected those clones that facilitated cell growth at 55 mg/l Cm. About 100 colonies grew on an LB plate supplemented with 55 mg/l Cm when tRNAUTuX was expressed, while only a few colonies appeared when tRNAPyl was provided. Repeated testing of 22 positives showed that four SerRS variants confer Cm resistance up to 65 mg/l in a tRNAUTuX-dependent manner (Fig. S4). After recloning only variants V8, V20 and V22 showed reproducible improvement in CAT read-through assay. Relevant SerRS sequences of these three variants are shown in Fig. 3A. All three variants possess different mutations, except for D228. In all cases D228 is converted to a positively charged K or R, implicating the importance of this motif 1 residue in the interaction with tRNAUTuX. Furthermore, S55 is mutated to Ala in SerRS V20. However, this sequence heterogeneity is not accidental. After mutating Ala55 to wild-type serine, SerRS V20 loses its ability to efficiently charge tRNAUTuX and other selenocysteine tRNAs (Fig. S5). Thus, the S55A mutation is advantageous when accompanied by other specific mutations.
Fig. 3.
SerRS variants improve tRNAUTuX-mediated amber stop-codon suppression. (A) Three most proficient SerRS variants contain different mutations, except for D228; this negatively charged motif-1 residue is converted into a basic (K or R) amino acid in all variants (highlighted). Locations of the residues are indicated (NTD or motif 1). In addition to the mutations listed here, SerRS V20 also contains a S55A mutation. (B) Comparison of in vivo acylation activity between wt and mutant SerRS for tRNAUTuX. E. coli cells DH10B ΔselAB::zeo expressing tRNAUTuX and SerRS variants were grown on LB plates with different concentration of Cm (0, 45, 60 mg/l). tRNAUTuX was under the control of lpp promoter; a CAT gene with a TAG codon at position 146 was on the same plasmid. SerRS expression was controlled by the trc promoter on the second plasmid. The read-through facilitated by the variants is tRNAUTuX dependent, as no resistance is conferred in the presence of tRNAPyl.
Sequencing of the entire plasmids that encode the variants revealed a partial deletion of the araC gene. As mutations in the AraC protein may give rise to various induction/repression phenotypes [32] we did not want to take a guess on the SerRS variant expression levels (Fig. S4 and discussion therein). To unambiguously demonstrate that increased Cm resistance correlates with the activity of evolved SerRS variants, and not caused by unintended mutations in the vector, we recloned the SerRS coding sequences under the control of a trc promoter into another low-copy plasmid (Fig. 3B).
The CAT-146TAG read-through assay by the SerRS variants placed under the trc inducible promoter revealed that SerRS V8 and V20 outperform wt SerRS and allow growth at higher Cm levels (Fig. 3B). However, high Cm resistance (45 and 60 mg/l) could only be achieved if expression of the variants was induced with 0.05 and 0.1 mM IPTG, presumably allowing only modest level of expression (Fig. 3B). Higher IPTG concentrations led to decreased cell viability suggesting that SerRS overexpression is toxic to the cell [33]. This may explain why AraC deletion variants accumulated during selection of the SerRS library. Our results indicate that the efficiency of CAT stop-codon suppression is related to increased levels of Ser-tRNAUTuX.
To examine the biochemical properties of the evolved variants with tRNAUTuX as substrate, we tested their activity in vitro (Table 1). Using an established protocol [29] and similar active enzyme concentrations (determined by active site titration), SerRS V8 and V20 serylate tRNAUTuX 10- and 8-times better than wild-type SerRS (Table 1). SerRS V22 provides a more moderate improvement (about 4-times), yet this is not sufficient to allow higher suppression levels in vivo (Fig. 3B). Interestingly, the major contribution to improved activity is the consequence of increased turnover (8- to 19-increased kcat for V20 and V8, respectively), rather than increased affinity (Table 1). Taken together, SerRS variants V8 and V20 aminoacylate tRNAUTuX better than the wild-type enzyme, under in vitro and in vivo conditions.
Table 1.
Steady-state kinetic parameters for tRNAUTuX for wild-type SerRS and evolved variants. The tRNAUTuX concentrations were corrected for plateau charging (48 %). Enzyme concentrations were: 650 nM wild-type SerRS, 187 nM SerRS V8, 92 nM SerRS V20, 224 nM SerRS V22.
| enzyme | KM (μM) | kcat (min−1) | kcat / KM (min−1 μM−1) | relative improvement |
|---|---|---|---|---|
| wt | 1.3 | 0.075 | 0.057 | 1 |
| V8 | 2.4 | 1.4 | 0.58 | 10 |
| V20 | 1.2 | 0.56 | 0.48 | 8.4 |
| V22 | 7.2 | 1.5 | 0.21 | 3.6 |
Selection of desirable variants (b) recognition of tRNASecUX and tRNAUTu1
As the different synthetic tRNASec species displayed differences in selenoprotein formation we tested the substrate properties of tRNASecUX and tRNAUTu1 (Fig. 1) [4,10,29] with our SerRS variants in vitro and in vivo. In enzyme-kinetic assays (described above) we determined the steady-state parameters. The values for tRNASecUX reveal significant serylation improvement by SerRS V8, while the kinetic parameters of SerRS V20 were only modestly improved (Table 2). As was the case with tRNAUTuX, the improvement of SerRS V8 activity arises from a kcat increase (Table 1, 2). This is plausible, as both tRNAUTuX and tRNASecUX are highly similar in both sequence and structure. Interestingly, SerRS V8 and V20 aminoacylate the more dissimilar tRNAUTu1 [10] 11- and 6-times better than wild-type SerRS. Although tRNAUTu1, as allo-tRNA [15], differs significantly from the tRNAUTuX species used in SerRS V8 and V20 evolution, both of these enzyme variants have higher affinity toward tRNAUTu1 than the wild-type SerRS. As with tRNAUTuX and tRNASecUX, both variants have significantly higher turnover numbers (~3 – 6-times for SerRS V20 and V8, respectively, Table 2).
Table 2.
Steady-state kinetic parameters for tRNASecUX and tRNAUTu1 for wild-type SerRS and variants V8 and V20. The tRNASecUX and tRNAUTu1 concentrations were corrected for plateau charging (39 % and 55 %, respectively). For tRNASecUX serylation the enzyme concentrations were: 500 nM wild-type SerRS, 187 nM SerRS V8, 92 nM SerRS V20; for tRNAUTu1 the enzyme concentrations were: 150 nM wild-type SerRS, 34 nM SerRS V8, 34 nM SerRS V20, 224 nM SerRS V22.
| enzyme | KM (μM) | kcat (min−1) | kcat / KM (min−1 μM−1) | relative improvement | tRNA |
|---|---|---|---|---|---|
| wt | 3.7 | 0.14 | 0.038 | 1 | tRNASecUX |
| V8 | 4.9 | 1 | 0.21 | 5.5 | |
| V20 | 3.7 | 0.26 | 0.071 | 1.9 | |
| wt | 5.7 | 4.9 | 0.86 | 1 | tRNAUTu1 |
| V8 | 3.1 | 29 | 9.3 | 11 | |
| V20 | 2.7 | 13 | 4.9 | 5.7 |
The CAT-146TAG read-through assay with tRNASecUX and tRNAUTu1 corroborated the above in vitro data. Both SerRS V8 and V20 confer significantly higher Cm resistance compared to wt SerRS (Fig. 4). In case of tRNASecUX, both SerRS variants allow growth at 20 mg/l Cm, while the wt enzyme does not. Using the much stronger UAG suppressor tRNAUTu1 [10], SerRS V8 and V20 allow robust growth at Cm 400 mg/l. Wild-type SerRS can also facilitate growth at these Cm levels albeit modestly, and only if expressed at a very low level (0.05 mM IPTG, Fig. 4). Thus, in case of all three selenocysteine tRNAs, the evolved SerRS variants allow growth at higher Cm concentrations but also operate in a larger dynamic range of expression.
Fig. 4.
In addition to tRNAUTuX, SerRS V8 and V20 improve tRNASecUX and tRNAUTu1-mediated TAG codon suppression. WL30153 cells expressing tRNAUTu1 and SerRS variants were grown on LB plates with 400 mg/l Cm. The same strain expressing tRNASecUX and SerRS variants was grown on LB plates with 20 mg/l Cm; both tRNAs were under the control of lpp promoter, and the cat gene with TAG at codon position 146 was on the same plasmid. SerRS expression was controlled by the trc promoter on the second plasmid.
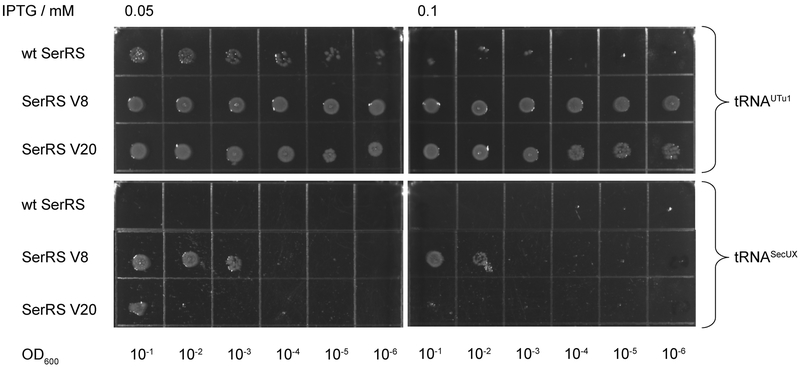
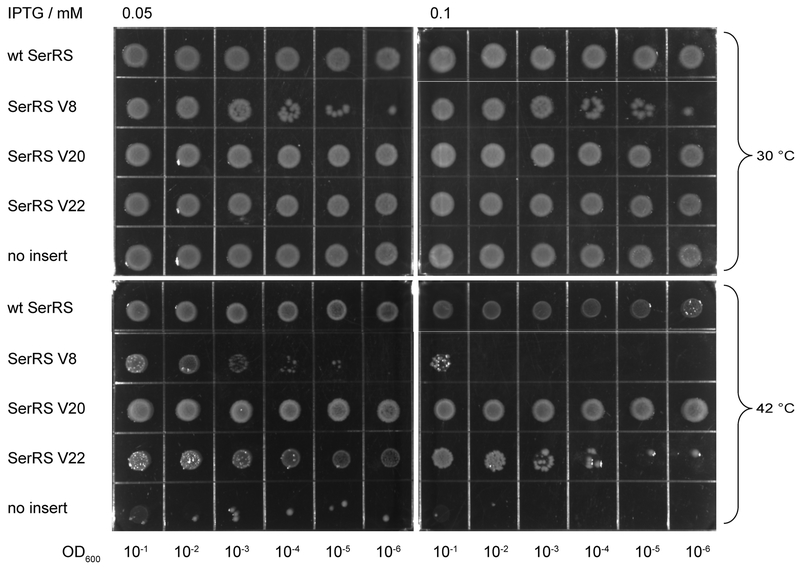
SerRS variant V20 supports E. coli growth in the absence of other active SerRS enzymes
To examine if the mutations, that gave rise to our SerRS variants, influenced their ability to provide Ser-tRNASer sufficient for active cell growth, we determined whether the variant enzymes could restore growth of E. coli strain KL235 that carries a temperature-sensitive SerRS [1]. This strain contains a genomic copy of serS15, encoding a thermolabile SerRS enzyme; as a result the host strain is unable to grow at 42°C. The SerRS variants were induced with low amounts of IPTG (0.05 and 0.1 mM) for 20 hr at 42°C. Robust growth was detected for KL235 cells expressing either wild-type SerRS or variant V20 (Figure 5). Toxicity of SerRS overexpression was more apparent in case of SerRS V22 that can replace SerS15 only when expressed at a very low level (0.05 mM IPTG). Similarly, SerRS V8, which can (albeit pretty inefficiently) rescue growth when induced with 0.05 mM IPTG, loses this capacity altogether when 0.1 mM IPTG is added. This indicates that SerRS V20 is able to supply sufficient serylated tRNA species for vigorous growth.
Fig. 5.
SerRS V20 rescues the thermolabile SerRS at nonpermissive temperature. Plasmids pTrc99A containing wt or SerRS variants were transformed into E. coli strain KL235 [1]; as a negative control, pTrc99A without an insert was also used. Plates were removed from the incubator after 20 hr at 30°C (upper panels) and 42°C (lower panels).
Additional information on the serylation properties of SerRS came from a serine-specific CAT-146TAG read-through assay. In order to decode the 146 UAG codon, we used the natural E. coli tRNASer suppressor SupD [34]. The suppression efficiencies mediated by the SerRS V8•SupD and V20•SupD pairs were comparable, and even higher than in case of wt SerRS. SupD allowed moderate to good growth at 300 mg/l Cm (Supplementary Fig. S6). However, in vitro aminoacylation reveals somewhat reduced affinity of SerRS V8 and V20 for SupD; both of these enzymes have about 2-times higher KM, while kcat is 2–3-times lower, compared to the wt SerRS (Table S2). This suggests that certain mutations improving selenocysteine tRNA recognition may have adverse effects on tRNASer aminoacylation by SerRS variants (Figure 5).
Discussion
E. coli SerRS catalyzes serylation of tRNASer and tRNASec with a 100-fold difference in kcat/KM [8]. One of the requirements for effective SelA recognition, the 8-bp long acceptor stem of tRNASec, negatively influences recognition by SerRS, as demonstrated by the improved activity of tRNASec variants lacking the 5A:67A bp [8]. These differences are also reflected by the extent of CAT synthesis in vivo, where the conferred resistance matches relative serylation levels of the SupD tRNA suppressor and the synthetic tRNASec species (see above). Enzyme-kinetic analyses of these tRNAs with wild-type SerRS reveal tRNAUTuX to be a marginally better substrate than tRNASecUX (kcat/KM is 1.5-time higher, Table 1, 2), and that tRNAUTu1 is 15-times better aminoacylated than tRNAUTuX (Table 1, 2). However, even tRNAUTu1 has about 9-times lower kcat/KM value than SupD (Table 2, S2). The improved serylation of the synthetic tRNASec species by the designed SerRS V8 and V20 variants (Table 1, 2) proves that SerRS can be adapted to better recognize atypical tRNASec structures. Compared to wt SerRS, these variants are inferior in the SupD tRNASer acylation (4–5 fold), but superior for synthetic tRNASec species (up to 11-fold).
Given the central role of aminoacyl-tRNA synthetases in protein synthesis, the last three decades saw much work directed toward ‘improvement’ of these enzymes. This effort led to tRNA synthetases endowed with an altered or expanded substrate spectrum for either tRNAs [35] or amino acids [36], or unnatural amino acid substrates for synthetic biology applications [37]. In this work we addressed a different challenge, to design variants of a normal cellular tRNA synthetase that aminoacylates multiple cognate isoacceptors. While wt E. coli SerRS serylates its cognate tRNASer species 100-fold better than its tRNASec isoacceptor [8], we created an enzyme variant with improved serylation properties only for tRNASec species; yet this enzyme fully maintains its essential role in providing Ser-tRNASer for cellular protein synthesis. This is another illustration of the great plasticity of the translation machinery.
Supplementary Material
Acknowledgments
The authors are grateful to Takahito Mukai, Li-Tao Guo, and Yuchen Liu for enlightened discussions and Anusha Manglik for help with experiments. This work was supported by the National Institute of General Medical Sciences (R35GM122560 to DS), and the Division of Chemical Sciences, Geosciences and Biosciences, Office of Basic Energy Sciences of the Department of Energy (DE-FG02–98ER20311 to DS) for the genetic experiments.
Abbreviations:
- aaRS
aminoacyl-tRNA synthetase
- SerRS
seryl-tRNA synthetase
- SelA
selenocysteine synthase
- CAT
chloramphenicol acetyltransferase
- NTD
N-terminal tRNA binding domain
References
- [1].Yoshizawa S and Böck A (2009) The many levels of control on bacterial selenoprotein synthesis. Biochim Biophys Acta 1790: 1404–1414 [DOI] [PubMed] [Google Scholar]
- [2].Aldag C, Bröcker MJ, Hohn MJ, Prat L, Hammond G, Plummer A and Söll D (2013) Rewiring translation for elongation factor Tu-dependent selenocysteine incorporation. Angew Chem Int Ed Engl 52: 1441–1445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Miller C, Bröcker MJ, Prat L, Ip K, Chirathivat N, Feiock A, Veszpremi M and Söll D (2015) A synthetic tRNA for EF-Tu mediated selenocysteine incorporation in vivo and in vitro. FEBS Lett 589: 2194–2199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Thyer R, Robotham SA, Brodbelt JS and Ellington AD (2015) Evolving tRNASec for efficient canonical incorporation of selenocysteine. J Am Chem Soc 137: 46–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Bröcker MJ, Ho JM, Church GM, Söll D and O’Donoghue P (2014) Recoding the genetic code with selenocysteine. Angew Chem Int Ed Engl 53: 319–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Roy KL and Söll D (1970) Purification of five serine transfer ribonucleic acid species from Escherichia coli and their acylation by homologous and heterologous seryl transfer ribonucleic acid synthetases. J Biol Chem 245: 1394–1400 [PubMed] [Google Scholar]
- [7].Schön A, Böck A, Ott G, Sprinzl M and Söll D (1989) The selenocysteine-inserting opal suppressor serine tRNA from E. coli is highly unusual in structure and modification. Nucleic Acids Res 17: 7159–7165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Baron C and Böck A (1991) The length of the aminoacyl-acceptor stem of the selenocysteine-specific tRNASec of Escherichia coli is the determinant for binding to elongation factors SELB or Tu. J Biol Chem 266: 20375–20379 [PubMed] [Google Scholar]
- [9].Böck A and Thanbichler M (2004) Selenocysteine. EcoSal Plus 1: doi: 10.1128/ecosalplus.1123.1126.1121.1121 [DOI] [PubMed] [Google Scholar]
- [10].Mukai T, Sevostyanova A, Suzuki T, Fu X and Söll D (2018) A facile method for producing selenocysteine-containing proteins. Angew Chem Int Ed Engl 57: 7215–7219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Biou V, Yaremchuk A, Tukalo M and Cusack S (1994) The 2.9 Å crystal structure of T. thermophilus seryl-tRNA synthetase complexed with tRNASer. Science 263: 1404–1410 [DOI] [PubMed] [Google Scholar]
- [12].Wang C, Guo Y, Tian Q, Jia Q, Gao Y, Zhang Q, Zhou C and Xie W (2015) SerRS-tRNASec complex structures reveal mechanism of the first step in selenocysteine biosynthesis. Nucleic Acids Res 43: 10534–10545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Liu DR and Schultz PG (1999) Progress toward the evolution of an organism with an expanded genetic code. Proc Natl Acad Sci U S A 96: 4780–4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Low B, Gates F, Goldstein T and Söll D (1971) Isolation and partial characterization of temperature-sensitive Escherichia coli mutants with altered leucyl- and seryl-transfer ribonucleic acid synthetases. J Bacteriol 108: 742–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mukai T, Vargas-Rodriguez O, Englert M, Tripp HJ, Ivanova NN, Rubin EM, Kyrpides NC and Söll D (2017) Transfer RNAs with novel cloverleaf structures. Nucleic Acids Res 45: 2776–2785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Sherrer RL, Ho JM and Söll D (2008) Divergence of selenocysteine tRNA recognition by archaeal and eukaryotic O-phosphoseryl-tRNASec kinase. Nucleic Acids Res 36: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Datsenko KA and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Leinfelder W, Forchhammer K, Zinoni F, Sawers G, Mandrand-Berthelot MA and Böck A (1988) Escherichia coli genes whose products are involved in selenium metabolism. J Bacteriol 170: 540–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Korencic D, Söll D and Ambrogelly A (2002) A one-step method for in vitro production of tRNA transcripts. Nucleic Acids Res 30: e105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lukavsky PJ and Puglisi JD (2004) Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA 10: 889–893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Borel F, Vincent C, Leberman R and Härtlein M (1994) Seryl-tRNA synthetase from Escherichia coli: implication of its N-terminal domain in aminoacylation activity and specificity. Nucleic Acids Res 22: 2963–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kelley LA, Mezulis S, Yates CM, Wass MN and Sternberg MJ (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10: 845–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC and Ferrin TE (2004) UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- [24].Giegé R, Sissler M and Florentz C (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Fu X, Söll D and Sevostyanova A (2018) Challenges of site-specific selenocysteine incorporation into proteins by Escherichia coli. RNA Biol 15: 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Holman KM, Puppala AK, Lee JW, Lee H and Simonović M (2017) Insights into substrate promiscuity of human seryl-tRNA synthetase. RNA 23: 1685–1699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Lenhard B, Orellana O, Ibba M and Weygand-Đurašević I (1999) tRNA recognition and evolution of determinants in seryl-tRNA synthesis. Nucleic Acids Res 27: 721–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Asahara H, Himeno H, Tamura K, Nameki N, Hasegawa T and Shimizu M (1994) Escherichia coli seryl-tRNA synthetase recognizes tRNASer by its characteristic tertiary structure. J Mol Biol 236: 738–748 [DOI] [PubMed] [Google Scholar]
- [29].Sampson JR and Saks ME (1993) Contributions of discrete tRNASer domains to aminoacylation by E. coli seryl-tRNA synthetase: a kinetic analysis using model RNA substrates. Nucleic Acids Res 21: 4467–4475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Fujinaga M, Berthet-Colominas C, Yaremchuk AD, Tukalo MA and Cusack S (1993) Refined crystal structure of the seryl-tRNA synthetase from Thermus thermophilus at 2.5 Å resolution. J Mol Biol 234: 222–233 [DOI] [PubMed] [Google Scholar]
- [31].Lewendon A, Murray IA, Shaw WV, Gibbs MR and Leslie AG (1990) Evidence for transition-state stabilization by serine-148 in the catalytic mechanism of chloramphenicol acetyltransferase. Biochemistry 29: 2075–2080 [DOI] [PubMed] [Google Scholar]
- [32].Ross JJ, Gryczynski U and Schleif R (2003) Mutational analysis of residue roles in AraC function. J Mol Biol 328: 85–93 [DOI] [PubMed] [Google Scholar]
- [33].Swanson R, Hoben P, Sumner-Smith M, Uemura H, Watson L and Söll D (1988) Accuracy of in vivo aminoacylation requires proper balance of tRNA and aminoacyl-tRNA synthetase. Science 242: 1548–1551 [DOI] [PubMed] [Google Scholar]
- [34].Eggertsson G and Söll D (1988) Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev 52: 354–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Inokuchi H, Hoben P, Yamao F, Ozeki H and Söll D (1984) Transfer RNA mischarging mediated by a mutant Escherichia coli glutaminyl-tRNA synthetase. Proc Natl Acad Sci U S A 81: 5076–5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Döring V, Mootz HD, Nangle LA, Hendrickson TL, de Crecy-Lagard V, Schimmel P and Marliere P (2001) Enlarging the amino acid set of Escherichia coli by infiltration of the valine coding pathway. Science 292: 501–504 [DOI] [PubMed] [Google Scholar]
- [37].Liu CC and Schultz PG (2010) Adding new chemistries to the genetic code. Annu Rev Biochem 79: 413–444 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.