Abstract
We are currently witnessing a decline in the development of efficient new anticancer drugs, despite the salient efforts made on all fronts of cancer drug discovery. This trend presumably relates to the substantial heterogeneity and the inherent biological complexity of cancer, which hinder drug development success. Protein-protein interactions (PPIs) are key players in numerous cellular processes and aberrant interruption of this complex network provides a basis for various disease states, including cancer. Thus, it is now believed that cancer drug discovery, in addition to the design of single-targeted bioactive compounds, should also incorporate diversity-oriented synthesis (DOS) and other combinatorial strategies in order to exploit the ability of multi-functional scaffolds to modulate multiple protein-protein interactions (biological hubs). Throughout the review, we highlight the chemistry driven approaches to access diversity space for the discovery of small molecules that disrupt oncogenic PPIs, namely the p53-Mdm2, Bcl-2/Bcl-xL-BH3, Myc-Max, and p53-Mdmx/Mdm2 interactions.
Keywords: diversity-oriented synthesis, combinatorial chemistry, protein-protein interactions, cancer
1. Introduction
Cancer develops through a multistep complex process that involves a series of genetic and epigenetic alterations which in turn, ultimately, lead to malignant phenotypes [1,2,3,4]. Deciphering of the molecular mechanisms of cancer stirred high expectations for the development of smart drugs that could efficiently inhibit aberrantly functioning cancer-driving oncoproteins [5]. Indeed, targeted drugs substantially improved the therapy of chronic myeloid leukemia, acute promyelocytic leukemia and gastrointestinal stromal tumors, in which cases tumor specific driver oncoproteins were identified and successfully targeted [6,7,8]. However, these tumors make up a minor fraction of worldwide cancer. In the settings of major killing cancers, such as lung, gastric, pancreatic, colon, breast and prostate cancer, the success has been disappointingly limited, not affecting survival rates [9]. Examples of effective targeted therapies that have entered clinical use are: proteasome inhibitor bortezomib which has improved the median survival of multiple myeloma patients [10,11], several tyrosine kinase inhibitors effective against renal and lung cancer [12], mTOR inhibitors [13], and lately, DNA methylation and chromatin modifying epigenetic drugs [14,15]. All this new therapeutic armamentarium has had, unfortunately, little effect on the ultimate clinical outcome of most cancers [16].
The limited efficacy of many first generation targeted drugs, designed to attack specific molecular alterations, is now understood to be a consequence of the complexity of aberrantly functioning of biomolecules in cancer [17,18]. It is therefore imperative to devise and develop novel drug discovery strategies in order to have chances to address the hard kernel of cancer complexity and improve cancer therapy [19,20,21]. Recently, advances towards the understanding of cancer system biology led to consider cancer-related protein-protein interaction networks as appropriate therapeutic targets, although the druggability of this approach is questioned [22,23,24]. Nevertheless, positive results of early studies provided encouraging evidence of selective and efficient interruption of aberrant protein-protein interactions in cancer, opening up a new avenue in cancer drug development [25,26,27].
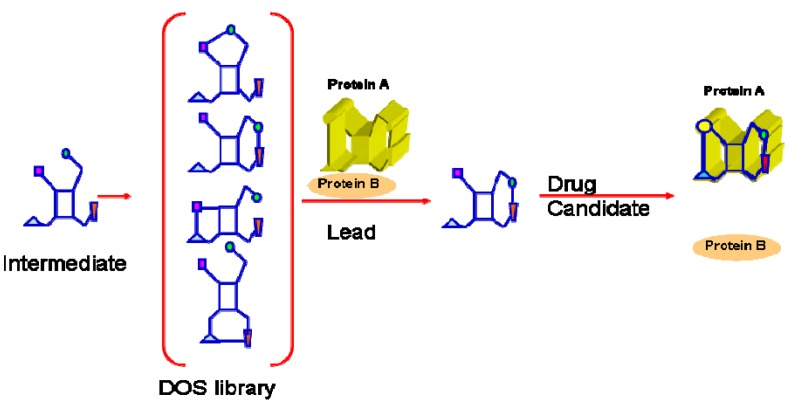
Combinatorial Chemistry and Diversity Oriented Synthesis (DOS) [28,29,30,31] are chemical technologies that have been used to generate screening collections which contain various aspects of structural diversity. The main intent of both approaches is to exploit state of the art synthesis and technological advancements. DOS is an evolution of combinatorial synthesis which leverages a forward synthetic planning strategy in order to obtain the most diverse set of molecules in an efficient manner (Figure 1). As an approach, DOS is uniquely different from traditional target oriented synthesis (TOS). TOS requires retrosynthetic planning strategies to manage the sometimes monumental synthetic challenges as well as prioritize the numerous synthetic options en route to a single and usually complex target molecule.
Figure 1.
In DOS, libraries of structurally diverse compounds are derived from common intermediates (reaction of the same starting material with different reagents or reaction of different starting materials with common reagents). Different chemical groups are presented with different colors. The binding cavity that is targeted by DOS compounds in a protein-protein interaction is colored in yellow. Optimized DOS compounds can dissociate such PPIs.
The hypothesis at the conception of DOS was that large collections of molecules derived from combinatorial chemistry were too similar to each other and not novel, diverse or complex enough to be probes for challenging difficult biological targets such as protein-protein interactions (PPIs). DOS evolved to encompass the rapid synthesis of complex small molecules as a compromise between total synthesis and combinatorial chemistry techniques. In the design of DOS libraries, relevant parameters that define compound properties are also taken into consideration, such as the Lipinski rules [32]. Certain deviations from these traditional parameters such as the incorporation of higher molecular weight (MW) compounds are included. The resulting compounds derived from DOS strategies have helped to launch an area of research known as chemical genomics [33]. Several other approaches similar to DOS such as Biology Oriented Synthesis (BiOS) [34,35], Functional Oriented Synthesis (FOS) [36], and Diverted Total Synthesis (DVT) [37] have also emerged with the intent to capture and leverage “natural product like (NPL) features”. There are no generally accepted parameters for what NPL is. However, notable attempts to compare NP collections with other synthetic collections have been described [38,39], providing empirical evidence of higher polarity, decreased hydrophobicity, higher molecular weights, increased stereochemical features and therefore higher sp3 content [40], unique molecular architectures and fewer aromatic rings [31]. These strategies are a clever way to address the shortcomings of using unguided combinatorial chemistry techniques [41,42] in the absence of other traditional structure-guided approaches.
There are a few salient structural characteristics regarding various diversity aspects of DOS that have been previously reviewed [43,44], such as appendage, substitutional, stereochemical, or scaffold diversity. For the most part, each descriptor contributes to the overall shape of the library. Recently, principle moments of inertia (PMI) plots [45] have been used to generally differentiate the shapes of molecules, and progress has been made in evaluating multiple conformations of the same molecule. Frustratingly, little is still known on how to predict or measure the diversity of stereoisomers despite the general acceptance that stereochemistry is a differentiating feature of many drugs [40]. Often, novelty or general characteristics such as NP likeness and/or complexity of the final compounds are considered. In order to steer the finite resources available to synthesize screening collections, there is usually a guiding principle or inspiration used as a nucleus for the ideas. Some more popular strategies are guided by the specific shape of a target enzyme or protein, a privileged structural motif [46], or a chemical methodology that enables access to novel chemical structures. In the following sections, molecules that target oncogenic protein-protein interactions and have been derived directly from DOS libraries or through the use of combinatorial techniques, in confluence with other technologies, are described (Figure 2).
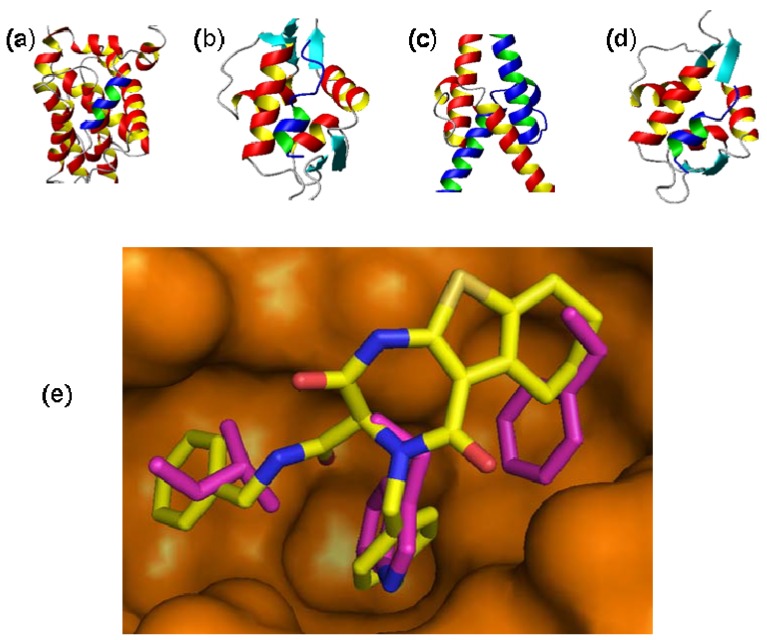
Figure 2.
Several three dimensional structures of protein-protein interactions that have been targeted by DOS have been solved by X-ray crystallography or Nuclear Magnetic Resonance spectroscopy (NMR): (a) Structure of a helical peptide from the death-promoting region of the Bcl-2-related protein Bak (colored in blue/green) bound to the survival protein Bcl-xL (colored in red/yellow) (pdbid:1BXL); (b) Structure of a p53 helical peptide (colored in blue/green) bound to Mdm2 (colored in red/yellow) (pdbid: 1YCR); (c) Structure of the basic/helix-loop-helix/leucine zipper (bHLHZ) domains of Myc (colored in red/yellow) and Max (colored in blue/green) heterodimers (pdbid: 1NKP); (d) Structure of a 15-residue transactivation domain peptide of human p53 (colored in blue/green) bound to the N-terminal domain of human Mdmx (colored in red/yellow) (pdbid: 3DAB). (e) Docking of an inhibitor of p53-Mdm2 interaction, developed from a small focused compound library of 1,4-thienodiazepine-2,5-diones, into the p53-binding site of Mdm2 (pdbid: 1YCR). The inhibitor (colored in yellow) is able to mimic hot spot residues (colored in purple) implicated in the p53-Mdm2 interface [47].
2. Targeting Oncogenic Protein-Protein Interactions with Small Molecules
Traditionally, drug development has focused on a small number of protein classes, (i.e., enzymes and receptors), not exceeding more than 1% of the roughly 30,000 unique protein sequences that comprise the human proteome [48]. Protein-protein interactions (PPIs) are key players in numerous cellular processes and it has been estimated that a very high number of them (40,000 to 200,000) exist in the human interactome. Since aberrant interruption of this complex network provides a basis for various disease states, a fine-tuning of these binding events via small molecule interactions has emerged as a rather important strategy for human therapeutics. Given the biological complexity of PPIs, the discovery and optimization of small molecules provides a significant challenge for drug development. A recent analysis of the network characteristics and interface properties of cancer-related proteins revealed that these are distinct from non-cancer proteins [49,50]. Specifically, it was shown that cancer-related proteins tend to interact with their partners through distinct interfaces, corresponding mostly to multi-interface hubs [49]. In addition, it was shown that they possess more planar, more hydrophilic, but smaller binding sites compared to non-cancer proteins, indicating low affinity and high specificity of the cancer-related interactions [49]. Such decoding is of importance only to reveal the details of specific binding regions for cancer-related protein interactions and may be utilized to formulate the drug development process accordingly. An in vivo proof of principle on the efficacy of protein-protein interaction inhibitors as anticancer drugs exists [26,51,52].
Although the importance of PPIs in drug development is well documented, PPIs have been extremely challenging targets. However, it should be noted that traditional approaches, such as high-throughput screening, have been successfully exploited in developing potent selective PPI antagonists. For instance, the discovery of Nutlins, the cis-imidazoline analogs that target the MDM2-p53 protein-protein interaction on the intent to reactivate p53, as well as the discovery of potent small molecules inhibitors that interfere with bcl-2 protein-protein antiapoptotic interactions, constitute such examples [51,53,54,55,56,57,58]. Navitoclax, a targeted high affinity inhibitor of Bcl-2 has already been evaluated in phase I and Nutlin-3 is currently about to enter early clinical evaluation [59,60].
Numerous factors have hindered a fruitful exploitation of PPIs as potential intervention points for the development of anticancer agents. For instance, PPI surfaces are large (750 Å–1,500 Å) [61] and devoid of deep interventions [22]. Affinity is achieved from the accumulation of numerous weak interactions. Therefore, it is inherently difficult for a small molecule to compete for binding on such an extensive interface composed of a large number of individual and complimentary interactions. To complicate the situation further, the inherent malleability of proteins to accommodate surface complementarity significantly handicaps structure-guided approaches. Furthermore, the small number of available assays to discriminate real from artifactual binding could hinder the development of small molecule antagonists for PPIs. However, despite the aforementioned difficulties, important progress has nonetheless been achieved towards the discovery of PPI antagonists. Indeed, this became evident upon analysis of protein-protein interfaces which showed that a centralized region of residues, the so called “hot-spots” [62], mediate all the key interactions that contribute to the binding affinity and presents comparable dimensions to the size of the small organic molecule. Such observations have recently challenged the traditional thought that PPIs are “undruggable” targets and numerous small-molecule inhibitors of PPIs are now in clinical trials [48,61]. Additionally, several strategies have surfaced regarding the discovery of small-molecule modulators of PPIs (for an insightful review see ref. [63]). Thus, disruption of oncogenic PPIs with small molecules might lead to a new class of anticancer therapeutics. Since α-helix-mediated PPIs are involved in a wide array of cellular signaling pathways, discovery of cell permeable and bioavailable small molecule inhibitors of these interactions could pave the way in the field. In many PPIs, short helical peptides play an important role as a recognition motif, where side chains at i, i+3 or i+4, and i+7 positions often become a critical determinant for PPIs [64,65]. Indeed, DOS has been successful in the discovery of lead compounds targeting α-helix-mediated PPIs in numerous cases such as: the complex between the Bcl-2-related proapoptotic protein Bak bound to the survival protein Bcl-xL (Figure 2a), the p53 derived helical peptide bound to murine double minute 2 (Mdm2) (Figure 2b), the complex of the basic/helix-loop-helix/leucine zipper (bHLHZ) domains of Myc and Max heterodimers (Figure 2c), and the complex of a 15-residue transactivation domain peptide of human p53 bound to the N-terminal domain of human Mdmx (Figure 2d). These applications will be analyzed in the following sections.
3. Antagonists of p53-Mdm2 Interactions
The tumor suppressor p53 is a well recognized target in cancer drug discovery which could offer new therapeutic opportunities [66]. The activation of wild-type p53 in human tumors with small molecules that antagonize Mdm2 appears to be a promising strategy in the treatment of cancer [67]. The disruption of the p53-Mdm2 interaction is usually accomplished by peptides, foldamers and peptoids (α-helical transactivation domain), chemical entities aiming mainly to mimic the p53 fragment and the Mdm2-binding site [68]. Also, the design of small molecules with appropriate physicochemical properties (i.e., bioavailability, aqueous solubility, stability) could enable the discovery of efficient Mdm2 antagonists [69].
3.1. Antagonists of p53-Mdm2 Interactions Based on 1,4-thienodiazepine-2,5-dione Based Core Structures
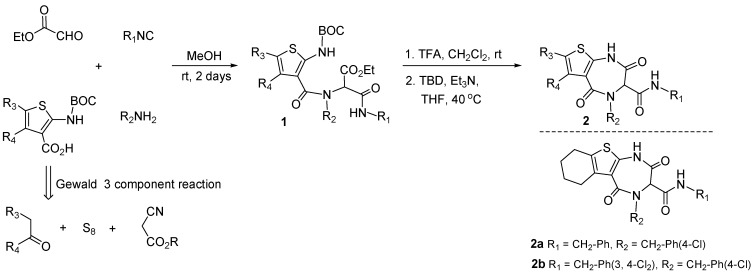
A peptidomimetic strategy was employed to synthesize small molecule p53-Mdm2 antagonists taking advantage of an Ugi-deprotection-cyclization sequence. The Ugi reaction has been exploited in combinatorial chemistry because it combines four separate components to make one scaffold. This provides easy access to appendage diversity around one single scaffold. In this case, the scaffold is a peptidomimetic 1,4-thienodiazepine-2,5-dione and was envisioned to act as an α-helix mimetic and to disturb the p53-Mdm2 interaction. A small library of 18 diverse thienodiazepine-2,5-diones with general structure 2, selected from a large virtual library, was prepared in one pot by solution phase synthesis via an Ugi-deprotection-cyclization strategy [47]. Condensation of 2-aminothiophene carboxylic acids, ethyl glyoxalate, amines and isonitriles, gave the Ugi-4CR product 1. Boc deprotection followed by TBD (1,5,7-triazabicyclo [4,4,0]dec-5-ene) mediated cyclization produced azepinediones 2. Access to a diverse set of 2-aminothiophene carboxylic acids can result from the Gewald three component reactions (Figure 3).
Figure 3.
Application of an Ugi-4CR in the discovery of p53-Mdm2 antagonists.
Library screening, following two complimentary techniques, found 1,4-thienodiazepine-2,5-dione 2a and 2b to antagonize the p53-Mdm2 protein interaction. Both compounds were found to antagonize the p53-Mdm2 interaction in a fluorescence polarization assay, exhibiting a dose dependent effect to compete with a p53-like peptide. Diazepinediones 2a and 2b inhibited Mdm2 with inhibition constant (Ki) values of 40 μM and 45 μM, respectively. Also, in a Nuclear Magnetic Resonance (NMR) competition assay performed on the Mdm2⁄p53 complex, compounds 2a and 2b were found to dissociate the Mdm2⁄p53 complex with Kd values of 30 ± 20 μM and 10 ± 6 μM, respectively.
This study demonstrated that the 1,4-thienodiazepine-2,5-dione scaffold is bioisosteric to the well known benzodiazepinediones and perhaps by correlation, may be a bioisosteric privileged structure. Enumeration of the scaffold followed by plotting MW vs. TPSA, compared to available benzodiazepine compounds through eMolecules [70], suggests that there is a large potential diversity which can be accessed based on the developed chemistry.
4. Targeting Anti-Apoptotic Members of the Bcl-2 Family Proteins
The Bcl-2 (B-cell lymphoma) family proteins regulate the equilibrium between cell proliferation and cell death (apoptosis) through complex protein-protein interactions. This family is composed of antiapoptotic and proapoptotic members. The antiapoptotic members contain four Bcl homology (BH) domains (BH1−BH4) and include Bcl-xL, Bcl-w, Bcl-2, Bcl2-A1 and Mcl-1, whereas the proapoptotic members contain either a single BH3 domain (BH3-only) (Puma, Bad, Bik, Bid, Bim) or three (BH) domains (BH1−BH3) (Bak, Bax). Apoptosis, or programmed cell death, is a highly controlled biological mechanism regulating the removal of aged, damaged, and unnecessary cells [71,72,73,74,75]. Aberrations in this equilibrium circuit can allow transformed cells to evade death and become resistant to cytotoxic therapies. Hence, the Bcl-2 pathway has been a compelling target for drug development for more than two decades. The critical event in Bcl-2 family signal propagation is the direct association of a protein containing a BH3 death domain with a multi-domain Bcl-2 family member. The antiapoptotic proteins bind their proapoptotic counterparts and sequester them from the cellular environment, thus inhibiting the apoptosis process. The up regulation of antiapoptotic members of this family (Bcl-2, Bcl-xL) is observed in many cancers. This overexpression prevents the activation of apoptosis and can protect cancer cells, favoring their proliferation and survival when exposed to anticancer compounds [76,77,78]. Therefore, the design of small molecules that bind the BH3 domain of antiapoptotic proteins and inhibit PPIs, can offer new strategies in cancer therapy [79]. Analysis of the three-dimensional structures of antiapoptotic Bcl-2 family proteins showed how these specific proteins interact with their proapoptotic counterparts [76,77,78]. It was revealed that the binding cavity for the proapoptotic molecules was an elongated hydrophobic crevice of approximately 20 Å, called BH3 binding groove. The understanding of these protein-protein interactions has opened new directions for rational design of novel inhibitors.
4.1. Discovery of Novel Bcl-2 Inhibitors Based on Rigid Pyridone Scaffolds
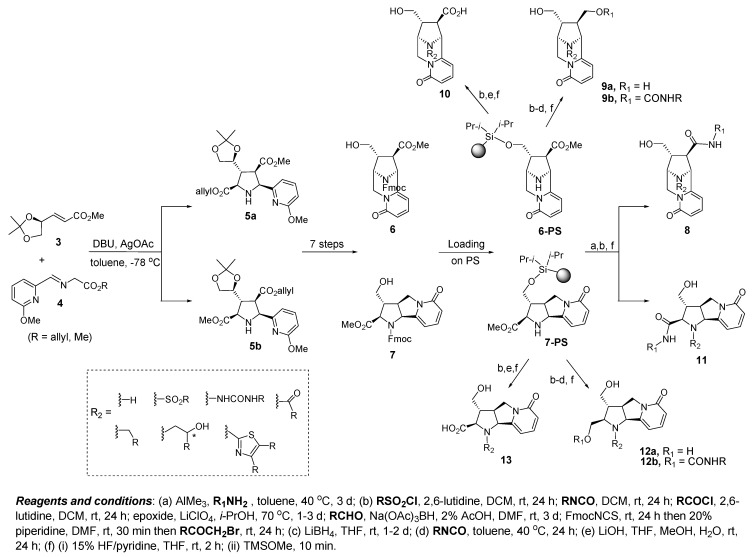
Screening of a DOS library, containing 15,000 compounds inspired by the tricyclic alkaloid natural product cytisine containing the privileged structural pyridone motif, led to the identification of novel inhibitors of Bcl-2 [80]. The stereochemical and skeletal diversity is accomplished by taking advantage of highly substituted pyrrolidines 5a and 5b, accessed from a stereoselective [3+2] dipolar cycloaddition that then diverges into two distinct and novel tricyclic scaffolds 6 and 7 (Figure 4).
Figure 4.
Discovery of Bcl-2 inhibitors based on DOS of pyridone core structures.
Appendage diversity was exploited by loading the scaffolds 6 and 7 onto a solid phase support to provide 6-PS and 7-PS and then employing a split and pool strategy. In this way, a sparse matrix could be utilized by exploring all combinations of capping strategies including changing the oxidation state of the handles. For example, amidation of the methyl ester of 6-PS under Weinreb conditions followed by capping of the secondary pyrrolidine nitrogen with a series of electrophiles, gave access to a diverse set of pyridones with general structure 8 after fluoride mediated resin cleavage. Alternatively, capping of the secondary pyrrolidine nitrogen first followed by ester reduction, resulted in alcohol 9a after resin cleavage. Also, treatment of the intermediate resin-bound primary hydroxyl with a series of isocyanates, produced carbamates 9b after release from the resin. Furthermore, a number of compounds with general structure 10 were prepared from 6-PS after derivatization of the pyrrolidine nitrogen and ester hydrolysis. Pyridone 7-PS gave access to chemotypes 11, 12a,b and 13 as well, by application of the same protocols.
The library compounds were screened for binding affinity against Bcl-2 and Bcl-xL in traditional solution based competition binding assays, formatted for HTS analysis, against a fluorescently tagged BH3 peptide. The hit rate from this library screen was 1.1% and 0.2% against Bcl-2 and Bcl-xL respectively, with the best inhibitors having single digit micromolar activities [80]. The most potent compounds identified in the bridged bicyclic pyridone chemotype 8 were those containing a diamine at R1 and a chloro-substituted diphenyl 2-aminothiazole at R2 (Figure 5). Both enantiomers in this series were equally active against Bcl-2, perhaps indicative of non-specific binding. They also displayed inhibitory activity against Bcl-xL except for compounds which lacked the diamine at R1. The most active compounds derived from the tricyclic pyridone chemotype 11 were the products of reductive alkylation and Weinreb amidation. For one enantiomeric series there was a distinct preference at R2 for cyclohexyl carboxaldehyde in combination with primary amines containing a hydrophobic aromatic ring at R1 (Figure 5). For the enantiomeric series (ent-11), 3,4-dichlorobenzaldehyde as well as 4-phenoxybenzaldehyde were preferred at R2 in combination with benzyl or phenyl substituted piperidines at R1 (Figure 5). This difference in activity for the two enantiomeric series suggests more specific binding as compared to the 2-aminothiazole compounds. In both cases the active compounds in this series were selective for Bcl-2 over Bcl-xL. Notably, this example highlights that a purely chemistry driven approach to novel alkaloid scaffolds can lead to the discovery of inhibitors of protein-protein interactions. Furthermore, this chemistry approach provides the flexibility to design specific appendage diversity enabling a quick understanding of the SAR.
Figure 5.
Discovery of selective inhibitors of Bcl-2 based on tricyclic pyridone scaffolds.
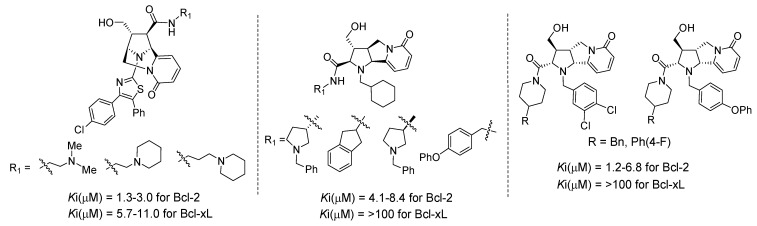
4.2. Discovery of Bcl-2 Inhibitors from an Isoxazolidine Library
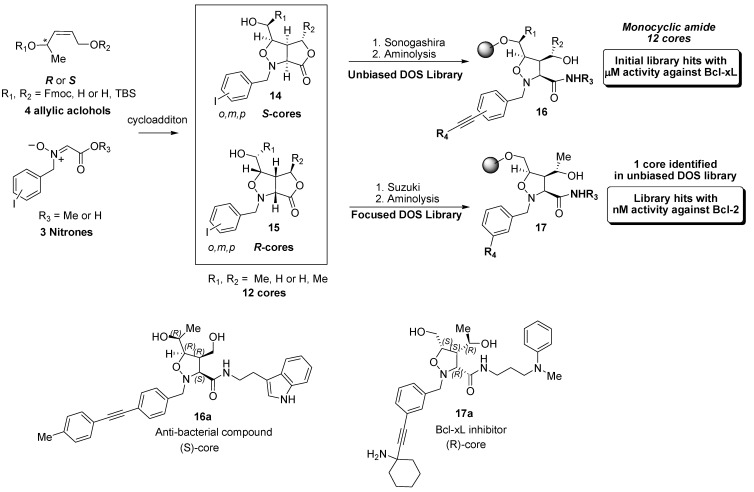
An unbiased DOS library which incorporated appendage, scaffold, and stereochemical diversity has previously been reviewed [81] (Figure 6). Notably, the library was screened in multiple assays and in different therapeutic areas which resulted in the discovery of low micromolar hits in both Bcl-xL 17a as well as an antibacterial hit 16a. The stereochemical importance is underscored by the fact that the respective hits were derived from the enantiomers of related cores. In order to access the structural and stereochemical diversity of this library, a diastereoselective 1,3-dipolar cycloaddition/acyclation reaction between each enantiomer of the allylic alcohols and various nitrone carboxylic acids was employed to generate 12 scaffolds represented by the general bicyclic structures 14 and 15. A small library of bicyclic compounds was synthesized and a larger monocyclic isoxazolidine library 16 was obtained from Sonogashira coupling of the aromatic iodides and substituted terminal alkynes, followed by aminolysis of the lactone. Notably, the lead compound was identified from only this subset of the entire library. Further structural diversity was obtained via cleavage of the N-O bond to provide a library of α-amino amide compounds (not shown). Despite the similarity of the appendage diversity, this subtle change resulted in no Bcl-2 activity.
Figure 6.
Isoxazolidine based Bcl-2 inhibitors.
Interestingly, a recent patent application has been published [82] which indicates that hit to lead activities have identified biaryl analogs 17 where potency has been pushed to nanomolar levels and optimized for Bcl-2 instead of Bcl-xL inhibition. These biaryl compounds were not part of the initial screening library and exemplify how the library design can be exploited quickly to optimize hit to lead efforts by simply replacing the sonogashira couling with the Suzuki reaction.
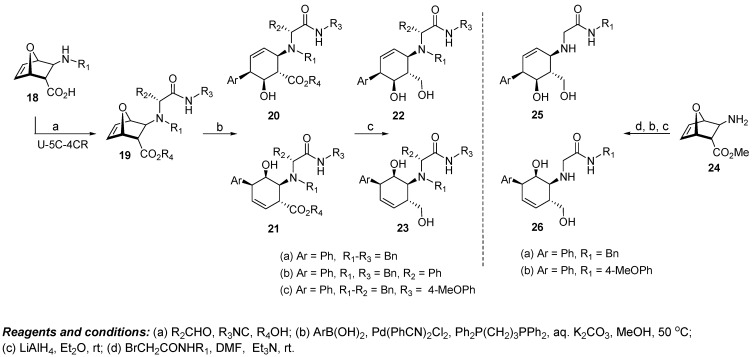
4.3. Discovery of Bcl-xL Antagonists Resulting from Oxabicyclic Scaffolds
A strategy employing iterative molecular docking of unique oxabicyclic scaffolds, combined with NMR studies, was employed in an attempt to identify antagonists of the antiapoptotic protein Bcl-xL, a member of the Bcl-2 family of survival proteins [83]. The pluripotent scaffold 19, derived from chiral β-amino acid 18 via an Ugi-five center-four component reaction (U-5C-4CR), was selected for this study because of its ability to provide easy access to new cyclohexene cores where the olefin and the substitution pattern of the ring can be varied. For this example, although the actual scaffold (cyclohexene ring) was not changed, the shape of the core is varied by the regio and appendage diversity to provide enough differentiation in structure. A set of virtual libraries were designed based on these scaffolds and then tested in silico for their capacity to bind to the BH3 binding groove of Bcl-xL. Analysis of docking calculations and comparison of all tested compounds revealed that scaffolds 20 and 21 can be used in the design of promising molecules, particularly the alcohol series 22 and 23. Therefore, in this particular case, the potential diversity of the oxabicyclic scaffold as it relates to the BH3 binding groove was narrowed on the basis of computational results, and compounds 22a,c and 23a-c as well as their enantiomers (ent-22a,c and ent-23a-c) were identified as potential antagonists (Figure 7). Cyclohexenols 20 and 21 were prepared as a regioisomeric mixture from the palladium catalyzed ring opening of the chiral oxabicyclic scaffold 19 in the presence of aryl/heteroaryl boronic acids. Ester reduction resulted in diols 22 and 23, respectively.
Figure 7.
Design of Bcl-xL antagonists through structural modification of oxabicyclic scaffolds.
When compounds 22a,c, 23a-c, ent-22a,c, and ent-23a-c were tested for their ability to bind Bcl-xL in NMR-based binding assays, the results were inconclusive due to the low solubility of the compounds in the solvent used for the NMR experiments. Therefore, the more hydrophilic analogs 25 and 26 were chosen for the NMR based screening. Computational screening identified compounds 25a,b, 26a,b as well as their enantiomers ent-25a,b and ent-26a,b to possess the structural elements required for binding. Access to chemotypes 25 and 26 was obtained through conversion of amino ester 24 via an alkylation reaction sequence with bromoacetamides. Experimental NMR data showed weak binding of this group of compounds with Bcl-xL. Unfortunately, the corresponding fluorescence polarization assay, using a fluoresceinated Bak peptide with the full length protein, did not show any appreciable displacement at 200 μM concentration. Therefore, despite the unique access to the oxabicyclic scaffold via the Ugi reaction, promising computational studies and NMR work, the oxabicyclic scaffold did not provide the intended results. An intriguing experiment would be to exploit the developed methodology to provide a more diverse set of compounds to see if a better hit would be derived. Moreover, the resulting compounds should be tested in other biochemical assays to see if the oxabicyclic scaffold is more suited for a different target.
5. Isoindoline Based Antagonists of the Myc-Max Protein-Protein Interaction
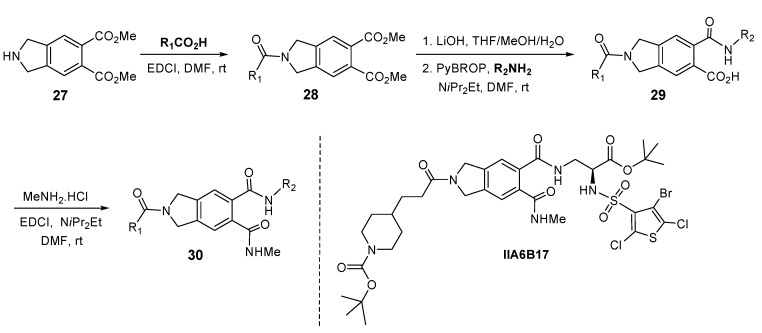
Disruption of the oncogenic PPIs between the Myc and Max transcription factors by small molecules, should enable the discovery of valuable probes for dissecting the roles of these transcription factors in cancer and for evaluating their potential as new therapeutic targets. Myc is aberrantly activated in a number of human cancers [84,85,86,87,88,89,90] and acts by heterodimerization with Max via their helix-loop-helix leucine zipper domains, a process that leads to the transcription of Myc target genes. In an effort to identify small molecules that antagonize the Myc-Max heterodimerization process, a screening collection of approximately 7,000 small organic molecules were used in a FRET assay. A subset of the 7,000 compounds contained a 240 membered privileged structure library based on an isoindoline scaffold: four isoindoline compounds were identified as hits. One hit originating from the diamide-acid chemotype 29 and three compounds from the triamide 30, were identified as PPI inhibitors between the Myc and Max transcription factors [91]. The library design is mainly focused on introduction of appendage diversity through sequential elaboration of the trifunctional isoindoline scaffold 27 by solution phase parallel synthesis. The objective was to identify nonpeptide RGD-based antagonists [92]. The appendage diversity was limited to amine and acid building blocks to make amides. Amidation of 27 with a set of carboxylic acids gave access to amide 28. Ester hydrolysis to diacid, followed by amidation with a set of amines, resulted in 120 diverse diamides 29. Sequential amidation with MeNH2.HCl produced 120 triamides of general structure 30 (Figure 8).
Figure 8.
Isoindoline-based antagonists of the Myc-Max protein-protein interaction.
The initial hits were found to be active in ELISA and electrophoretic mobility shift assays (EMSA) as well, where isoindoline IIA6B17 stood out as the most active compound (ELISA IC50 ≈ 125 μM; EMSA IC50 ≈ 50 μM). Also, two of these hits inhibited cell focus formation in Myc-transformed chicken embryo fibroblasts (IIA6B17 IC50 = 15–20 μM). Although low micromolar inhibitors were identified in this study, this work highlighted the feasibility of inhibiting oncogenic transcription factor PPIs with small molecules [93,94]. In addition, a few diamide and triamide compounds 29 and 30 have also exhibited cytotoxic activity in a leukemia mouse L-1210 assay [92]. Further elaboration of the appendage diversity could be achieved on this particular isoindoline scaffold by incorporating other functionality besides amides, thus leading to a wider selection of chemotypes for screening in other therapeutic areas.
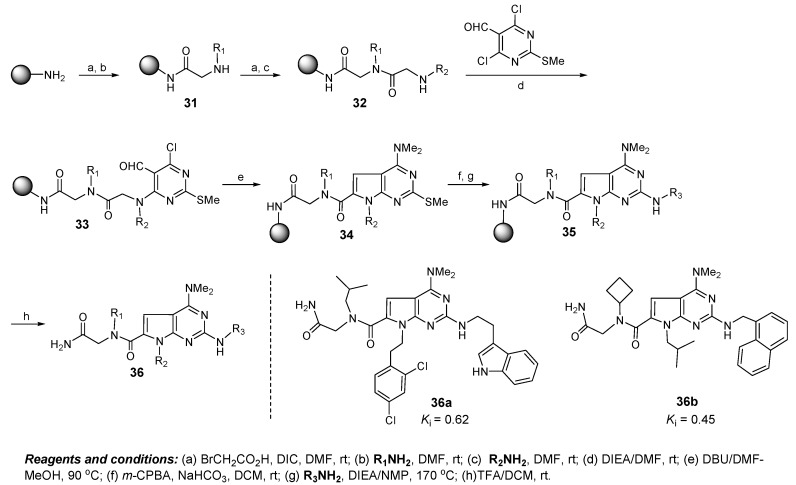
6. Discovery of Dual Mdmx/Mdm2 Inhibitors Based on Pyrrolopyrimidine Scaffolds as A-Helix Mimetics
A library of 900 compounds based on a pyrrolopyrimidine scaffold as an α-helix mimetic, was prepared by solid phase parallel synthesis in the hope to discover small molecules able to disrupt the interaction between p53 and Mdmx/Mdm2 [95]. Mdmx is overexpressed in many cancers and functions as a major regulator of p53 activity (both independently and synergistically with Mdm2). Thus, the development of Mdmx inhibitors that could act solely on Mdmx or on both Mdmx and Mdm2 is highly desirable but still remains challenging [96,97,98].
The pyrrolopyrimidine based diversity library was inspired by a natural α-helical motif and was designed to explore the appendage diversity of specific vectors around the scaffold. The library synthesis commenced with the efficient preparation on Rink resin of the dimeric peptoid 32 to incorporate diversity elements R1 and R2 through an iterative bromacetylation followed by displacement of the bromide with primary amines (Figure 9).
Figure 9.
Design of dual Mdmx/Mdm2 inhibitors based on pyrrolopyrimidine scaffolds.
Coupling of 32 with 4,6-dichloro-2-(methylthio)-pyrimidine-5-carbaldehyde gave pyrimidine 33 which was cyclized to pyrrolopyrimidine 34 with a concomitant dimethylamination. Oxidation of the thiomethyl ether group to sulfone, followed by substitution with a wide range of amines, gave a diverse set of pyrrolopyrimidines 36 after cleavage from the resin. The library molecules were screened at ~40 μM concentration for their ability to displace a Rhodamine-labeled 15-mer p53 peptide from Mdmx protein by a fluorescence polarization assay. Compounds 36a and 36b were the two most active agents identified in the screen and effectively inhibited the binding of p53-Mdmx with Ki = 0.62 and 0.45 μM respectively, comparable with that of a 15-mer p53 peptide (Ki = 0.8 μM). They were also found to inhibit the p53-Mdm2 interaction with Ki = 0.62 and 0.84 μM respectively, similar to the binding affinities for Mdmx, suggesting that pyrrolopyrimidines 36a and 36b act as dual inhibitors of Mdmx- and Mdm2-p53 interactions.
This library highlights the importance of a specific privileged scaffold or motif. Furthermore, stereochemistry or scaffold diversity was not a defining diversity aspect. Using a specifically designed and very rigid scaffold enabled the exploration of the appendage diversity, resulting in the successful discovery of an inhibitor of Mdmx/Mdm2. Potentially, this approach could be utilized to generate similar libraries and could serve as a useful tool in the discovery of inhibitors of other α-helix-mediated PPIs.
7. Future Directions
Today the combined efforts of the biotech and pharmaceutical industry as well as governments in drug discovery have yielded improved technologies in several domains: automation, stereoselective methodologies in organic synthesis, assays development, analysis of genetic targets, computational strategies, structural biology, etc. However, they have been unable to capitalize and integrate these technologies effectively enough to improve the success of drug discovery in targeting oncogenic protein-protein interactions. For instance, various problems can appear in biochemical screening assays that may hamper the correct validation of PPI inhibitors, including aggregator agents [99,100], reactive false positives [101], frequent hitters [102], and warhead-containing agents [103]. Thus, despite the fact that combinatorial chemistry and diversity oriented synthesis have provided a few hits and leads, there is still a need for novel advancements in order to diagnose artifact pitfalls early on [99,100,101].
DOS is training us to think about diversity and to think forward on how we can access unique chemical space. Combining this with combinatorial know how has provided an interesting approach in building screening decks for drug discovery. This is an important objective since traditional approaches alone are falling short on providing small molecules that modulate key protein-protein interactions important for the regulation of cancer. However, the development of such libraries is still costly and time consuming and it would be a notable improvement to direct our collective synthetic resources towards the exploration of chemical space around specific scaffolds which regulate cancer-related proteins. Recent work on network characteristics and interface properties of cancer-related proteins revealed a distinct trend in comparison to non-cancer proteins [49,50]. Novel cheminformatic tools have been put into place to facilitate the analysis of protein-protein interfaces with regard to their suitability for small molecule drug design [104,105]. Nonetheless, knowing specifically what chemical matter is most relevant is still a far reaching objective. Such discriminative capabilities could be used to rationally design focused libraries. Until then, exploring chemical space to find unique and novel starting points may be our best chance to find druggable chemical matter for these important and challenging protein-protein interactions.
Acknowledgements
We would like to thank Alexander Dömling (University of Pittsburgh, Pittsburgh, PA, USA) for providing the coordinates to construct Figure 2e.
References and Notes
- 1.Jones P.A., Baylin S.B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 2002;3:415–428. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Rosen J.M., Jordan C.T. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.The Cancer Genome Atlas (TCGA) Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson K., Lutz C., van Delft F.W., Bateman C.M., Guo Y., Colman S.M., Kempski H., Moorman A.V., Titley I., Swansbury J., Kearney L., Enver T., Greaves M. Genetic variegation of clonal architecture and propagating cells in leukaemia. Nature. 2011;469:356–361. doi: 10.1038/nature09650. [DOI] [PubMed] [Google Scholar]
- 5.Sawyers C. Targeted cancer therapy. Nature. 2004;432:294–297. doi: 10.1038/nature03095. [DOI] [PubMed] [Google Scholar]
- 6.Capdeville R., Buchdunger E., Zimmermann J., Matter A. Glivec (STI571, imatinib), a rationally developed, targeted anticancer drug. Nat. Rev. Drug Discov. 2002;1:493–502. doi: 10.1038/nrd839. [DOI] [PubMed] [Google Scholar]
- 7.de Bono J.S., Ashworth A. Translating cancer research into targeted therapeutics. Nature. 2010;467:543–549. doi: 10.1038/nature09339. [DOI] [PubMed] [Google Scholar]
- 8.Grignani F., Fagioli M., Alcalay M., Longo L., Pandolfi P.P., Donti E., Biondi A., Lo Coco F., Pelicci P.G. Acute promyelocytic leukemia: From genetics to treatment. Blood. 1994;83:10–25. [PubMed] [Google Scholar]
- 9.Burris H.A., III. Shortcomings of current therapies for non-small-cell lung cancer: Unmet medical needs. Oncogene. 2009;28 (Suppl. 1):S4–S13. doi: 10.1038/onc.2009.196. [DOI] [PubMed] [Google Scholar]
- 10.Harousseau J.L., Palumbo A., Richardson P.G., Schlag R., Dimopoulos M.A., Shpilberg O., Kropff M., Kentos A., Cavo M., Golenkov A., et al. Superior outcomes associated with complete response in newly diagnosed multiple myeloma patients treated with nonintensive therapy: analysis of the phase 3 VISTA study of bortezomib plus melphalan-prednisone versus melphalan-prednisone. Blood. 2010;116:3743–3750. doi: 10.1182/blood-2010-03-275800. [DOI] [PubMed] [Google Scholar]
- 11.McConkey D.J. Bortezomib paradigm shift in myeloma. Blood. 2009;114:931–932. doi: 10.1182/blood-2009-06-223230. [DOI] [PubMed] [Google Scholar]
- 12.Saijo N. Targeted therapies: Tyrosine-kinase inhibitors--new standard for NSCLC therapy. Nat. Rev. Clin. Oncol. 2010;7:618–619. doi: 10.1038/nrclinonc.2010.168. [DOI] [PubMed] [Google Scholar]
- 13.Meric-Bernstam F., Gonzalez-Angulo A.M. Targeting the mTOR signaling network for cancer therapy. J. Clin. Oncol. 2009;27:2278–2287. doi: 10.1200/JCO.2008.20.0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sausville E.A., Carducci M.A. Making bad cells go good: the promise of epigenetic therapy. J. Clin. Oncol. 2005;23:3875–3876. doi: 10.1200/JCO.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Karberg S. Switching on epigenetic therapy. Cell. 2009;139:1029–1031. doi: 10.1016/j.cell.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 16.Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 17.Check Hayden E. Cancer complexity slows quest for cure. Nature. 2008;455:148. doi: 10.1038/455148a. [DOI] [PubMed] [Google Scholar]
- 18.Stewart D.J., Kurzrock R. Cancer: the road to Amiens. J. Clin. Oncol. 2009;27:328–333. doi: 10.1200/JCO.2008.18.9621. [DOI] [PubMed] [Google Scholar]
- 19.Gupta P.B., Onder T.T., Jiang G., Tao K., Kuperwasser C., Weinberg R.A., Lander E.S. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatenby R.A. A change of strategy in the war on cancer. Nature. 2009;459:508–509. doi: 10.1038/459508a. [DOI] [PubMed] [Google Scholar]
- 21.Kamb A., Wee S., Lengauer C. Why is cancer drug discovery so difficult? Nat. Rev. Drug Discov. 2007;6:115–120. doi: 10.1038/nrd2155. [DOI] [PubMed] [Google Scholar]
- 22.Fry D.C., Vassilev L.T. Targeting protein-protein interactions for cancer therapy. J. Mol. Med. 2005;83:955–963. doi: 10.1007/s00109-005-0705-x. [DOI] [PubMed] [Google Scholar]
- 23.Sugaya N., Ikeda K. Assessing the druggability of protein-protein interactions by a supervised machine-learning method. BMC Bioinformatics. 2009;10:263. doi: 10.1186/1471-2105-10-263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yao C., Li H., Zhou C., Zhang L., Zou J., Guo Z. Multi-level reproducibility of signature hubs in human interactome for breast cancer metastasis. BMC Syst. Biol. 2010;4:151. doi: 10.1186/1752-0509-4-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fujii N., You L., Xu Z., Uematsu K., Shan J., He B., Mikami I., Edmondson L.R., Neale G., Zheng J., Guy R.K., Jablons D.M. An antagonist of dishevelled protein-protein interaction suppresses beta-catenin-dependent tumor cell growth. Cancer Res. 2007;67:573–579. doi: 10.1158/0008-5472.CAN-06-2726. [DOI] [PubMed] [Google Scholar]
- 26.Harris C.C. Protein-protein interactions for cancer therapy. Proc. Natl. Acad. Sci. USA. 2006;103:1659–1660. doi: 10.1073/pnas.0510948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Araujo R.P., Liotta L.A., Petricoin E.F. Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nat. Rev. Drug Discov. 2007;6:871–880. doi: 10.1038/nrd2381. [DOI] [PubMed] [Google Scholar]
- 28.Dandapani S., Marcaurelle L.A. Current strategies for diversity-oriented synthesis. Curr. Opin. Chem. Biol. 2010;14:362–370. doi: 10.1016/j.cbpa.2010.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Schreiber S.L. Target-oriented and diversity-oriented organic synthesis in drug discovery. Science. 2000;287:1964–1969. doi: 10.1126/science.287.5460.1964. [DOI] [PubMed] [Google Scholar]
- 30.Thomas G.L., Wyatt E.E., Spring D.R. Enriching chemical space with diversity-oriented synthesis. Curr. Opin. Drug Discov. Devel. 2006;9:700–712. [PubMed] [Google Scholar]
- 31.Bauer R.A., Wurst J.M., Tan D.S. Expanding the range of 'druggable' targets with natural product-based libraries: an academic perspective. Curr. Opin. Chem. Biol. 2010;14:308–314. doi: 10.1016/j.cbpa.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lipinski C.A., Lombardo F., Dominy B.W., Feeney P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 2001;46:3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 33.Stockwell B.R. Chemical genetics: ligand-based discovery of gene function. Nat. Rev. Genet. 2000;1:116–125. doi: 10.1038/35038557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilk W., Zimmermann T.J., Kaiser M., Waldmann H. Principles, implementation, and application of biology-oriented synthesis (BIOS) Biol. Chem. 2010;391:491–497. doi: 10.1515/BC.2010.013. [DOI] [PubMed] [Google Scholar]
- 35.Kombarov R., Altieri A., Genis D., Kirpichenok M., Kochubey V., Rakitina N., Titarenko Z. BioCores: identification of a drug/natural product-based privileged structural motif for small-molecule lead discovery. Mol. Divers. 2010;14:193–200. doi: 10.1007/s11030-009-9157-5. [DOI] [PubMed] [Google Scholar]
- 36.Wender P.A., Verma V.A., Paxton T.J., Pillow T.H. Function-oriented synthesis, step economy, and drug design. Acc. Chem. Res. 2008;41:40–49. doi: 10.1021/ar700155p. [DOI] [PubMed] [Google Scholar]
- 37.Wilson R.M., Danishefsky S.J. Small molecule natural products in the discovery of therapeutic agents: the synthesis connection. J. Org. Chem. 2006;71:8329–8351. doi: 10.1021/jo0610053. [DOI] [PubMed] [Google Scholar]
- 38.Newman D.J., Cragg G.M., Snader K.M. Natural products as sources of new drugs over the period 1981-2002. J. Nat. Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- 39.Harvey A.L. Natural products in drug discovery. Drug Discov. Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 40.Lovering F., Bikker J., Humblet C. Escape from flatland: increasing saturation as an approach to improving clinical success. J. Med. Chem. 2009;52:6752–6756. doi: 10.1021/jm901241e. [DOI] [PubMed] [Google Scholar]
- 41.Ortholand J.Y., Ganesan A. Natural products and combinatorial chemistry: back to the future. Curr. Opin. Chem. Biol. 2004;8:271–280. doi: 10.1016/j.cbpa.2004.04.011. [DOI] [PubMed] [Google Scholar]
- 42.Feher M., Schmidt J.M. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 43.Burke M.D., Schreiber S.L. A planning strategy for diversity-oriented synthesis. Angew. Chem. Int. Ed. Engl. 2004;43:46–58. doi: 10.1002/anie.200300626. [DOI] [PubMed] [Google Scholar]
- 44.Spandl R.J., Bender A., Spring D.R. Diversity-oriented synthesis; a spectrum of approaches and results. Org. Biomol. Chem. 2008;6:1149–1158. doi: 10.1039/b719372f. [DOI] [PubMed] [Google Scholar]
- 45.Sauer W.H., Schwarz M.K. Molecular shape diversity of combinatorial libraries: a prerequisite for broad bioactivity. J. Chem. Inf. Comput. Sci. 2003;43:987–1003. doi: 10.1021/ci025599w. [DOI] [PubMed] [Google Scholar]
- 46.Welsch M.E., Snyder S.A., Stockwell B.R. Privileged scaffolds for library design and drug discovery. Curr. Opin. Chem. Biol. 2010;14:347–361. doi: 10.1016/j.cbpa.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., Wolf S., Bista M., Meireles L., Camacho C., Holak T.A., Domling A. 1,4-Thienodiazepine-2,5-diones via MCR (I): synthesis, virtual space and p53-Mdm2 activity. Chem. Biol. Drug Des. 2010;76:116–129. doi: 10.1111/j.1747-0285.2010.00989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Whitty A., Kumaravel G. Between a rock and a hard place? Nat. Chem. Biol. 2006;2:112–118. doi: 10.1038/nchembio0306-112. [DOI] [PubMed] [Google Scholar]
- 49.Kar G., Gursoy A., Keskin O. Human cancer protein-protein interaction network: A structural perspective. PLoS Comput. Biol. 2009;5:e1000601. doi: 10.1371/journal.pcbi.1000601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sun J., Zhao Z. A comparative study of cancer proteins in the human protein-protein interaction network. BMC Genomics. 2010;11 (Suppl. 3):S5. doi: 10.1186/1471-2164-11-S3-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vassilev L.T., Vu B.T., Graves B., Carvajal D., Podlaski F., Filipovic Z., Kong N., Kammlott U., Lukacs C., Klein C., Fotouhi N., Liu E.A. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 52.Tovar C., Rosinski J., Filipovic Z., Higgins B., Kolinsky K., Hilton H., Zhao X., Vu B.T., Qing W., Packman K., Myklebost O., Heimbrook D.C., Vassilev L.T. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc. Natl. Acad. Sci. USA. 2006;103:1888–1893. doi: 10.1073/pnas.0507493103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhang W., Konopleva M., Burks J.K., Dywer K.C., Schober W.D., Yang J.Y., McQueen T.J., Hung M.C., Andreeff M. Blockade of mitogen-activated protein kinase/extracellular signal-regulated kinase kinase and murine double minute synergistically induces Apoptosis in acute myeloid leukemia via BH3-only proteins Puma and Bim. Cancer Res. 2010;70:2424–2434. doi: 10.1158/0008-5472.CAN-09-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonnemann J., Palani C.D., Wittig S., Becker S., Eichhorn F., Voigt A., Beck J.F. Anticancer effects of the p53 activator nutlin-3 in Ewing's sarcoma cells. Eur. J. Cancer. 2011;47:1432–1441. doi: 10.1016/j.ejca.2011.01.015. [DOI] [PubMed] [Google Scholar]
- 55.Petros A.M., Huth J.R., Oost T., Park C.M., Ding H., Wang X., Zhang H., Nimmer P., Mendoza R., Sun C., et al. Discovery of a potent and selective Bcl-2 inhibitor using SAR by NMR. Bioorg. Med. Chem. Lett. 2010;20:6587–6591. doi: 10.1016/j.bmcl.2010.09.033. [DOI] [PubMed] [Google Scholar]
- 56.Ackler S., Mitten M.J., Foster K., Oleksijew A., Refici M., Tahir S.K., Xiao Y., Tse C., Frost D.J., Fesik S.W., et al. The Bcl-2 inhibitor ABT-263 enhances the response of multiple chemotherapeutic regimens in hematologic tumors in vivo. Cancer Chemother. Pharmacol. 2010;66:869–880. doi: 10.1007/s00280-009-1232-1. [DOI] [PubMed] [Google Scholar]
- 57.Oltersdorf T., Elmore S.W., Shoemaker A.R., Armstrong R.C., Augeri D.J., Belli B.A., Bruncko M., Deckwerth T.L., Dinges J., Hajduk P.J., et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 58.Ezzoukhry Z., Louandre C., Francois C., Saidak Z., Godin C., Maziere J.C., Galmiche A. The BH3 mimetic ABT-737 Reveals the Dynamic Regulation of BAD, a Pro-apoptotic Protein of the BCL2 family, by BCL-XL. Mol. Pharmacol. 2011;79:997–1004. doi: 10.1124/mol.110.070565. [DOI] [PubMed] [Google Scholar]
- 59.Wilson W.H., O'Connor O.A., Czuczman M.S., LaCasce A.S., Gerecitano J.F., Leonard J.P., Tulpule A., Dunleavy K., Xiong H., Chiu Y.L., et al. Navitoclax, a targeted high-affinity inhibitor of BCL-2, in lymphoid malignancies: A phase 1 dose-escalation study of safety, pharmacokinetics, pharmacodynamics, and antitumour activity. Lancet Oncol. 2011;11:1149–1159. doi: 10.1016/S1470-2045(10)70261-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Secchiero P., Bosco R., Celeghini C., Zauli G. Recent advances in the therapeutic perspectives of nutlin-3. Curr. Pharm. Des. 2011;17:569–577. doi: 10.2174/138161211795222586. [DOI] [PubMed] [Google Scholar]
- 61.Arkin M.R., Wells J.A. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat. Rev. Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 62.Arkin M.R., Randal M., DeLano W.L., Hyde J., Luong T.N., Oslob J.D., Raphael D.R., Taylor L., Wang J., McDowell R.S., Wells J.A., Braisted A.C. Binding of small molecules to an adaptive protein-protein interface. Proc. Natl. Acad. Sci. USA. 2003;100:1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meireles L.M., Mustata G. Discovery of modulators of protein-protein interactions: current approaches and limitations. Curr. Top. Med. Chem. 2011;11:248–257. doi: 10.2174/156802611794072632. [DOI] [PubMed] [Google Scholar]
- 64.Davis J.M., Tsou L.K., Hamilton A.D. Synthetic non-peptide mimetics of alpha-helices. Chem. Soc. Rev. 2007;36:326–334. doi: 10.1039/b608043j. [DOI] [PubMed] [Google Scholar]
- 65.Cummings C.G., Hamilton A.D. Disrupting protein-protein interactions with non-peptidic, small molecule alpha-helix mimetics. Curr. Opin. Chem. Biol. 2010;14:341–346. doi: 10.1016/j.cbpa.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 66.Vazquez A., Bond E.E., Levine A.J., Bond G.L. The genetics of the p53 pathway, apoptosis and cancer therapy. Nat. Rev. Drug Discov. 2008;7:979–987. doi: 10.1038/nrd2656. [DOI] [PubMed] [Google Scholar]
- 67.Vassilev L.T. MDM2 inhibitors for cancer therapy. Trends Mol. Med. 2007;13:23–31. doi: 10.1016/j.molmed.2006.11.002. [DOI] [PubMed] [Google Scholar]
- 68.Robinson J.A. Design of protein-protein interaction inhibitors based on protein epitope mimetics. ChemBioChem. 2009;10:971–973. doi: 10.1002/cbic.200900055. [DOI] [PubMed] [Google Scholar]
- 69.Domling A. Small molecular weight protein-protein interaction antagonists: An insurmountable challenge? Curr. Opin. Chem. Biol. 2008;12:281–291. doi: 10.1016/j.cbpa.2008.04.603. [DOI] [PubMed] [Google Scholar]
- 70.eMolecules, Inc. [accessed on 8 January 2011]. Available online: http://www.emolecules.com/
- 71.Adams J.M., Cory S. The Bcl-2 protein family: Arbiters of cell survival. Science. 1998;281:1322–1326. doi: 10.1126/science.281.5381.1322. [DOI] [PubMed] [Google Scholar]
- 72.Reed J.C. Bcl-2 family proteins: Strategies for overcoming chemoresistance in cancer. Adv. Pharmacol. 1997;41:501–532. doi: 10.1016/S1054-3589(08)61070-4. [DOI] [PubMed] [Google Scholar]
- 73.Chao D.T., Korsmeyer S.J. BCL-2 family: Regulators of cell death. Annu. Rev. Immunol. 1998;16:395–419. doi: 10.1146/annurev.immunol.16.1.395. [DOI] [PubMed] [Google Scholar]
- 74.Minn A.J., Swain R.E., Ma A., Thompson C.B. Recent progress on the regulation of apoptosis by Bcl-2 family members. Adv. Immunol. 1998;70:245–279. doi: 10.1016/S0065-2776(08)60388-0. [DOI] [PubMed] [Google Scholar]
- 75.Storey S. Targeting apoptosis: Selected anticancer strategies. Nat. Rev. Drug Discov. 2008;7:971–972. doi: 10.1038/nrd2662. [DOI] [PubMed] [Google Scholar]
- 76.Sattler M., Liang H., Nettesheim D., Meadows R.P., Harlan J.E., Eberstadt M., Yoon H.S., Shuker S.B., Chang B.S., Minn A.J., et al. Structure of Bcl-xL-Bak peptide complex: Recognition between regulators of apoptosis. Science. 1997;275:983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 77.Petros A.M., Nettesheim D.G., Wang Y., Olejniczak E.T., Meadows R.P., Mack J., Swift K., Matayoshi E.D., Zhang H., Thompson C.B., Fesik S.W. Rationale for Bcl-xL/Bad peptide complex formation from structure, mutagenesis, and biophysical studies. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Day C.L., Chen L., Richardson S.J., Harrison P.J., Huang D.C., Hinds M.G. Solution structure of prosurvival Mcl-1 and characterization of its binding by proapoptotic BH3-only ligands. J. Biol. Chem. 2005;280:4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 79.Becattini B., Kitada S., Leone M., Monosov E., Chandler S., Zhai D., Kipps T.J., Reed J.C., Pellecchia M. Rational design and real time, in-cell detection of the proapoptotic activity of a novel compound targeting Bcl-X(L) Chem. Biol. 2004;11:389–395. doi: 10.1016/j.chembiol.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 80.Marcaurelle L.A., Johannes C., Yohannes D., Tillotson B.P., Mann D. Diversity-oriented synthesis of a cytisine-inspired pyridone library leading to the discovery of novel inhibitors of Bcl-2. Bioorg. Med. Chem. Lett. 2009;19:2500–2503. doi: 10.1016/j.bmcl.2009.03.037. [DOI] [PubMed] [Google Scholar]
- 81.Marcaurelle L.A., Johannes C.W. Application of natural product-inspired diversity-oriented synthesis to drug discovery. Prog. Drug Res. 2008;66:187–216. doi: 10.1007/978-3-7643-8595-8_3. [DOI] [PubMed] [Google Scholar]
- 82.Castro C. Compounds and methods for inhibiting the interaction of BCL proteins with binding partners. US Patent 7,851,637. 2010 Dec 14 ;
- 83.Di Micco S., Vitale R., Pellecchia M., Rega M.F., Riva R., Basso A., Bifulco G. Identification of lead compounds as antagonists of protein Bcl-xL with a diversity-oriented multidisciplinary approach. J. Med. Chem. 2009;52:7856–7867. doi: 10.1021/jm9010687. [DOI] [PubMed] [Google Scholar]
- 84.Bange J., Zwick E., Ullrich A. Molecular targets for breast cancer therapy and prevention. Nat. Med. 2001;7:548–552. doi: 10.1038/87872. [DOI] [PubMed] [Google Scholar]
- 85.Escot C., Theillet C., Lidereau R., Spyratos F., Champeme M.H., Gest J., Callahan R. Genetic alteration of the c-myc protooncogene (MYC) in human primary breast carcinomas. Proc. Natl. Acad. Sci. USA. 1986;83:4834–4838. doi: 10.1073/pnas.83.13.4834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Liao D.J., Dickson R.B. c-Myc in breast cancer. Endocr. Relat. Cancer. 2000;7:143–164. doi: 10.1677/erc.0.0070143. [DOI] [PubMed] [Google Scholar]
- 87.Little C.D., Nau M.M., Carney D.N., Gazdar A.F., Minna J.D. Amplification and expression of the c-myc oncogene in human lung cancer cell lines. Nature. 1983;306:194–196. doi: 10.1038/306194a0. [DOI] [PubMed] [Google Scholar]
- 88.Erisman M.D., Rothberg P.G., Diehl R.E., Morse C.C., Spandorfer J.M., Astrin S.M. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol. Cell. Biol. 1985;5:1969–1976. doi: 10.1128/mcb.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.He T.C., Sparks A.B., Rago C., Hermeking H., Zawel L., da Costa L.T., Morin P.J., Vogelstein B., Kinzler K.W. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 90.Pavlidis N., Briassoulis E., Bai M., Fountzilas G., Agnantis N. Overexpression of C-myc, Ras and C-erbB-2 oncoproteins in carcinoma of unknown primary origin. Anticancer Res. 1995;15:2563–2567. [PubMed] [Google Scholar]
- 91.Berg T., Cohen S.B., Desharnais J., Sonderegger C., Maslyar D.J., Goldberg J., Boger D.L., Vogt P.K. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc. Natl. Acad. Sci. USA. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Boger D.L., Lee J.K., Goldberg J., Jin Q. Two comparisons of the performance of positional scanning and deletion synthesis for the identification of active constituents in mixture combinatorial libraries. J. Org. Chem. 2000;65:1467–1474. doi: 10.1021/jo9916481. [DOI] [PubMed] [Google Scholar]
- 93.Boger D.L., Desharnais J., Capps K. Solution-phase combinatorial libraries: modulating cellular signaling by targeting protein-protein or protein-DNA interactions. Angew. Chem. Int. Ed. Engl. 2003;42:4138–4176. doi: 10.1002/anie.200300574. [DOI] [PubMed] [Google Scholar]
- 94.Yin X., Giap C., Lazo J.S., Prochownik E.V. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 95.Lee J.H., Zhang Q., Jo S., Chai S.C., Oh M., Im W., Lu H., Lim H.S. Novel Pyrrolopyrimidine-Based alpha-Helix Mimetics: Cell-Permeable Inhibitors of Protein-Protein Interactions. J. Am. Chem. Soc. 2011;133:676–679. doi: 10.1021/ja108230s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Laurie N.A., Donovan S.L., Shih C.S., Zhang J., Mills N., Fuller C., Teunisse A., Lam S., Ramos Y., Mohan A., et al. Inactivation of the p53 pathway in retinoblastoma. Nature. 2006;444:61–66. doi: 10.1038/nature05194. [DOI] [PubMed] [Google Scholar]
- 97.Toledo F., Wahl G.M. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat. Rev. Cancer. 2006;6:909–923. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 98.Marine J.C., Dyer M.A., Jochemsen A.G. MDMX: from bench to bedside. J. Cell. Sci. 2007;120:371–378. doi: 10.1242/jcs.03362. [DOI] [PubMed] [Google Scholar]
- 99.McGovern S.L., Helfand B.T., Feng B., Shoichet B.K. A specific mechanism of nonspecific inhibition. J. Med. Chem. 2003;46:4265–4272. doi: 10.1021/jm030266r. [DOI] [PubMed] [Google Scholar]
- 100.Seidler J., McGovern S.L., Doman T.N., Shoichet B.K. Identification and prediction of promiscuous aggregating inhibitors among known drugs. J. Med. Chem. 2003;46:4477–4486. doi: 10.1021/jm030191r. [DOI] [PubMed] [Google Scholar]
- 101.Huth J.R., Mendoza R., Olejniczak E.T., Johnson R.W., Cothron D.A., Liu Y., Lerner C.G., Chen J., Hajduk P.J. ALARM NMR: A rapid and robust experimental method to detect reactive false positives in biochemical screens. J. Am. Chem. Soc. 2005;127:217–224. doi: 10.1021/ja0455547. [DOI] [PubMed] [Google Scholar]
- 102.Roche O., Schneider P., Zuegge J., Guba W., Kansy M., Alanine A., Bleicher K., Danel F., Gutknecht E.M., Rogers-Evans M., et al. Development of a virtual screening method for identification of "frequent hitters" in compound libraries. J. Med. Chem. 2002;45:137–142. doi: 10.1021/jm010934d. [DOI] [PubMed] [Google Scholar]
- 103.Rishton G.M. Nonleadlikeness and leadlikeness in biochemical screening. Drug Discov. Today. 2003;8:86–96. doi: 10.1016/S1359644602025722. [DOI] [PubMed] [Google Scholar]
- 104.ANCHOR. [accessed on 8 January 2011]. Available online: http://structure.pitt.edu/anchor/
- 105.AnchorQuery. [accessed on 8 January 2011]. Available online: http://anchorquery.ccbb.pitt.edu/