Abstract
Mutations in CNGA3 and CNGB3, the genes encoding the subunits of the tetrameric cone photoreceptor cyclic nucleotide–gated ion channel, cause achromatopsia, a congenital retinal disorder characterized by loss of cone function. However, a small number of patients carrying the CNGB3/c.1208G>A;p.R403Q mutation present with a variable retinal phenotype ranging from complete and incomplete achromatopsia to moderate cone dysfunction or progressive cone dystrophy. By exploring a large patient cohort and published cases, we identified 16 unrelated individuals who were homozygous or (compound-)heterozygous for the CNGB3/c.1208G>A;p.R403Q mutation. In-depth genetic and clinical analysis revealed a co-occurrence of a mutant CNGA3 allele in a high proportion of these patients (10 of 16), likely contributing to the disease phenotype. To verify these findings, we generated a Cngb3R403Q/R403Q mouse model, which was crossbred with Cnga3-deficient (Cnga3–/–) mice to obtain triallelic Cnga3+/– Cngb3R403Q/R403Q mutants. As in human subjects, there was a striking genotype-phenotype correlation, since the presence of 1 Cnga3-null allele exacerbated the cone dystrophy phenotype in Cngb3R403Q/R403Q mice. These findings strongly suggest a digenic and triallelic inheritance pattern in a subset of patients with achromatopsia/severe cone dystrophy linked to the CNGB3/p.R403Q mutation, with important implications for diagnosis, prognosis, and genetic counseling.
Keywords: Genetics, Ophthalmology
Keywords: Molecular genetics, Retinopathy
Introduction
Mutations in 6 genes (CNGA3, CNGB3, GNAT2, PDE6C, PDE6H, and ATF6) are so far known to cause autosomal recessive achromatopsia (ACHM) (1–8), a rare congenital disorder characterized by poor visual acuity, reduced or complete lack of color discrimination, photophobia, and nystagmus. Patients have small central scotoma and eccentric fixation. Most present with the complete form of the disease, few with incomplete ACHM in which symptoms are milder. ACHM patients lack or have strongly diminished cone photoreceptor function from birth. In addition, variable progressive foveomacular degeneration is observed in most patients upon optical coherence tomography (OCT) (9–17). Biallelic mutations in either CNGA3 or CNGB3 encoding the ion-conducting and the modulatory subunits of the cone-specific cyclic nucleotide–gated (CNG) ion channel, respectively, are by far the most common cause of ACHM and account for the vast majority of cases in the patient population (18–20). CNG channels are key components of the phototransduction cascade and mediate membrane hyperpolarization upon a light-triggered decrease in the level of the channel’s ligand cGMP (21). Native cone CNG channels are composed of 3 CNGA3 subunits and 1 CNGB3 subunit (22). In heterologous systems, the sole expression of CNGA3 but not of CNGB3 yields functional channels, defining CNGB3 as an accessory subunit. However, heteromeric A3/B3 channels differ from homomeric A3 channels in some important aspects, such as ligand selectivity and gating properties (23). Moreover, formation of heteromeric channels seems to be important for maintaining protein stability and effectively targeting CNG channels to the outer segment of photoreceptors (24, 25).
The majority of reported mutations in CNGB3 are nonsense, splicing, or frameshift mutations that most likely represent null alleles (2, 3, 12, 19, 26) and are associated with typical clinical findings of ACHM or severe cone dystrophy, which differs from ACHM in its progressive course and some minute residual cone function, depending on the stage of the disease (12). A notable exception is the c.1208G>A missense mutation, which causes an arginine-to-glutamine substitution in the evolutionarily conserved pore helix of CNGB3 at amino acid position 403 (p.Arg403Gln). This substitution was reported in patients presenting with a rather variable retinal phenotype described as progressive macular dystrophy, macular degeneration, or cone dystrophy (12, 26, 27). The clinical expression in these patients is distinct from that in patients with ACHM, since visual acuity, photopic electroretinographic responses, and color vision are much less impaired. Bright and coworkers found that coexpression of WT CNGA3 and mutant CNGB3/p.R403Q in Xenopus oocytes resulted in formation of heterotetrameric CNG channels with normal surface expression, but increased apparent ligand sensitivity and increased outward rectification (23, 28).
Targeted knockout mice are available for Cnga3 and Cngb3 as models for human ACHM, and to investigate and dissect the function of both subunits (29, 30). Cnga3-KO mice show a visual phenotype that reproduces the complete loss of cone function in human ACHM. Moreover, as in ACHM patients, perturbed morphology of cone somata, cone opsin mislocalization, and cGMP accumulation in the cone outer segments were observed in these animals (31, 32). Interestingly, the disease phenotype in Cngb3-KO mice is attenuated compared with that in human patients bearing CNGB3 mutations. The latter present with a typical ACHM phenotype, while Cngb3-knockout mice show robust residual cone function with a photopic electroretinographic response amplitude of approximately 50% of that observed in WT littermates at postnatal day 15 that declines to 20% at the age of 12 months (33). The reduced function is accompanied by a slowly progressive loss of cones, with cone densities that drop to about 50% by 12 months of age (30, 33).
Biallelic mutations in one of the known ACHM genes explain the disease in the vast majority of patients. However, there is a small fraction of patients with unexplained variable clinical phenotypes, suggesting the existence of more complex genetic scenarios.
Here we report strong evidence for a digenic triallelic cause of ACHM involving biallelic CNGB3 mutations combined with an additional monoallelic CNGA3 mutation that explains the variable clinical phenotype in patients carrying the CNGB3/p.R403Q mutation. We corroborated these findings by modeling this digenic triallelic inheritance in mice. In particular, we generated a Cngb3R403Q/R403Q knockin mouse and crossbred it with Cnga3-KO mice (29). In line with the human data, the resulting triallelic Cnga3+/– Cngb3R403Q/R403Q mutant mice also showed a more severe retinal disease phenotype than their homozygous Cngb3R403Q/R403Q littermates.
Results
Accessory mutations in CNGA3 exacerbate disease phenotype in carriers of CNGB3/p.R403Q.
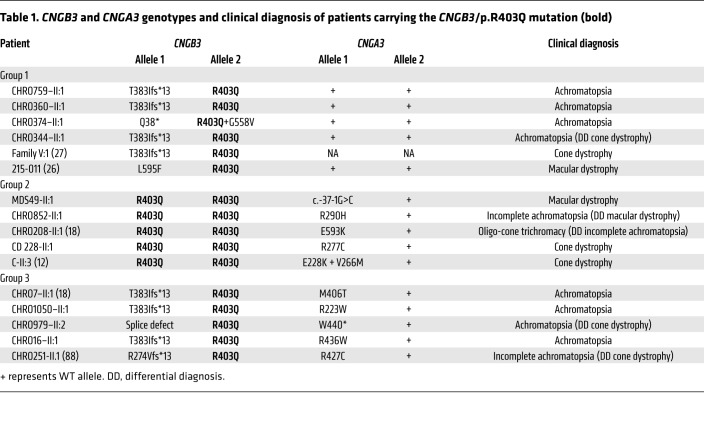
Given the unexplained clinical heterogeneity in patients with the CNGB3/p.R403Q mutation reported in the literature (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI96098DS1) (12, 26, 27), we retrospectively screened the Tübingen ACHM DNA sample registry for patients carrying the c.1208G>A;p.R403Q mutation in CNGB3. We furthermore selectively screened patients with a clinical diagnosis of cone (n = 316), cone-rod (n = 450), and macular (n = 408) dystrophy for this distinct CNGB3 mutation. All patients carrying the CNGB3/p.R403Q mutation were also screened for additional mutations in CNGA3. In total, we identified 13 patients who were either homozygous for CNGB3/p.R403Q (n = 4) or (compound-)heterozygous for CNGB3/p.R403Q and one of various, most likely pathogenic CNGB3 mutations (n = 9) (Supplemental Tables 1 and 2), as well as 4 single-heterozygous cases (data not shown).
Apart from screening our own cohorts, we performed an extensive literature review for cases reported with CNGB3/p.R403Q (Supplemental Table 1), and contacted the authors of these publications to learn to what extent CNGB3 and CNGA3 had been analyzed or to request DNA for such an analysis. We obtained DNA of the individuals (078-053 and 215-011) reported by Nishiguchi and coworkers (26), and resequencing of CNGA3 and CNGB3 resulted in the following observations: patient 078-053, who had been reported to be homozygous for p.R403Q, turned out to be heterozygous, and no additional variants in CNGA3 were identified. In patient 215-011, the CNGB3 genotype p.[R403Q](;)[L595F] was confirmed, while sequencing of CNGA3 did not reveal any additional variant in this gene.
Strikingly, we observed that the majority of patients (10 of 16; 62.5%) carried an additional heterozygous variant in CNGA3 (Table 1, Figure 1, and Supplemental Tables 1 and 2). Eight of these CNGA3 variants represent already-reported mutations, including 7 that have already been functionally validated in heterologous expression systems (p.R223W, p.E228K, p.R277C, p.M406T, p.R427C, p.R436W, and p.E593K) (18, 34, 35). The eighth known mutation is a nonsense mutation (p.W440*) that is predicted to result in a truncated CNGA3 protein lacking the entire cGMP-binding domain. In addition, 2 novel CNGA3 variants, the missense substitution p.R290H and a putative splicing mutation, c.-37-1G>C, were identified in families CHRO852 and MDS49, respectively (Figure 1). When expressed heterologously, the CNGA3/p.R290H mutant gave rise to a functional CNG channel interacting with the CNGB3 subunit. However, the apparent cGMP affinity of CNGA3/p.R290H was 3-fold lower compared with that of WT CNGA3 channels, suggesting that the pathology of CNGA3/p.R290H is related to a lower affinity to its ligand (Supplemental Figure 1). Finally, we tested the c.-37-1G>C variant using a minigene splicing assay in human embryonic kidney 293T (HEK293T) and murine cone photoreceptor–like 661W cells. We found that the c.-37-1G>C variant induces the use of an alternative splice acceptor, resulting in the loss of the first 75 bp of exon 1, including the start codon ATG (Supplemental Figure 2). Therefore, protein translation of the misspliced transcript is likely abolished, unless an alternative start codon in exon 2 is used (e.g., M52). However, we also observed small amounts of correctly spliced transcript in the minigene assay, suggesting that small amounts of WT CNGA3 protein can be translated from this mutant allele. The hypomorphic nature of this mutant is compatible with the mild phenotype in the patient (MDS49-II:1). According to their genotypes, patients carrying the CNGB3/p.R403Q mutation were divided into 3 groups, as follows (see Supplemental Table 1 for a compilation of patients’ clinical data and related genotypes; and Supplemental Figure 3 for retinal imaging data of selected patients).
Table 1. CNGB3 and CNGA3 genotypes and clinical diagnosis of patients carrying the CNGB3/p.R403Q mutation (bold).
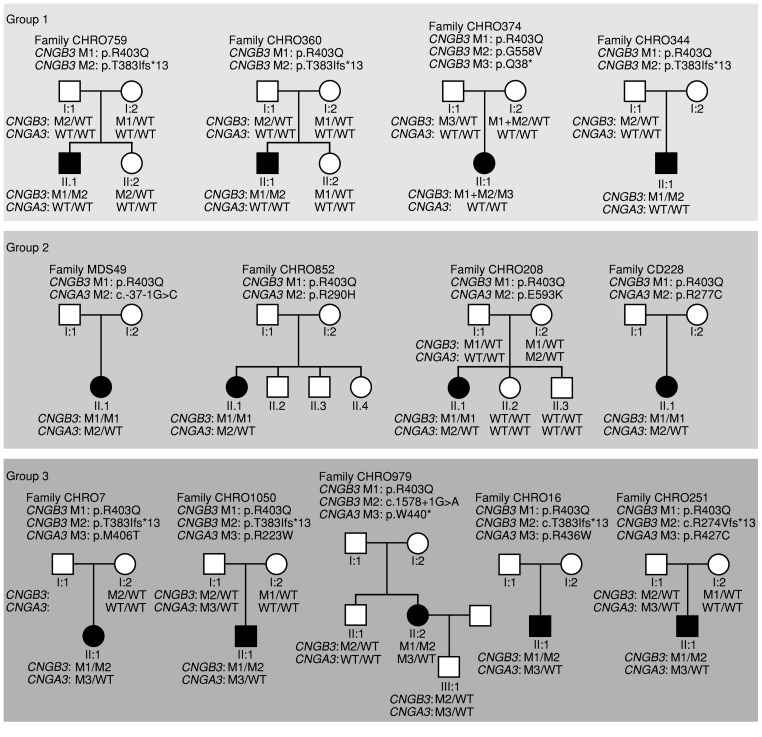
Figure 1. Pedigrees of patients and segregation of CNGB3 and CNGA3 disease alleles.
Pedigrees are subdivided into 3 groups according to genotypes and correlated disease severity. Group 1: Compound heterozygosity for CNGB3/p.R403Q and a protein truncation mutation in CNGB3 and no mutation in CNGA3. Group 2: Homozygosity for the CNGB3/c.1208G>A;p.R403Q mutation and an additional heterozygous CNGA3 mutation. Group 3: Compound heterozygosity for CNGB3/c.1208G>A;p.R403Q and a protein truncation or splicing mutation in CNGB3 and an additional heterozygous CNGA3 mutation. The genotypes for CNGB3 and CNGA3 of all tested individuals are provided. The presence of 2 mutant CNGB3 alleles with homozygosity for the CNGB3/c.1208G>A;p.R403Q mutation was confirmed by qRT-PCR. Notably, siblings and parents carrying single heterozygous mutations in both CNGB3 and CNGA3 (CHRO208-I:2; CHRO251-I:1; CHRO979-III:1, and CHRO1050-I:1) are unaffected. The different mutations (M1-M3) in each family are defined above the respective pedigree. M1 was always selected for the p.R403Q mutation.
Group 1: compound heterozygosity for CNGB3/p.R403Q and a protein truncation mutation in CNGB3 and no mutation in CNGA3.
Four of the 6 patients in this group (CHRO759-II:1, CHRO360-II:1, CHRO344-II:1, family V:1) actually had the same genotype, p.[R403Q];[T383Ifs*13], but were clinically examined at different ages (range, 5.5–56 years), providing quasi-longitudinal phenotypic data. Clinical diagnosis ranged from macular and cone dystrophy to ACHM, and in one case progression of symptoms was described (27). Patient CHRO374-II:1 in this group had a different genotype, p.[Q38*];[p.R403Q;p.G558V], including a complex allele with p.R403Q and p.G558V mutations in cis and the nonsense mutation in trans. Patient 215-011 carries 2 heterozygous mutations, c.[1208G>A(;)c.1783C>T] and p.[R403Q(;)L595F], yet segregation analysis was not done to assure compound heterozygosity. Nishiguchi and coworkers describe this case with a clinical diagnosis of macular dystrophy (26).
Group 2: homozygosity for the CNGB3/p.R403Q mutation and an additional heterozygous CNGA3 mutation.
Patient MDS49-II:1 had late-onset macular dystrophy, with slightly reduced visual acuity and reduced central responses in the multifocal electroretinogram (ERG) but normal full-field photopic ERG responses at 48 years of age. Compared with that of the other patients in this genotype group, this mild phenotype is probably due to the hypomorphic nature of the CNGA3 mutation (i.e., leaky splice defect). Two patients in this group (CHRO852-II:1, CHRO208-II:1) presented with typical features of ACHM (visual acuity ≤0.2, nonrecordable photopic ERG responses, photophobia, and nystagmus) but with some residual color vision, justifying a clinical diagnosis of oligocone trichromacy and incomplete ACHM, respectively. Patient CD228-II:1 was diagnosed with cone dystrophy, highlighting that the additional CNGA3 allele plays an important role for the resulting phenotype. Progression was reported in 2 cases.
Group 3: compound heterozygosity for CNGB3/p.R403Q and a protein truncation or splicing mutation in CNGB3, and an additional heterozygous CNGA3 mutation.
Four of the 5 patients in this group (CHRO7-II:1, CHRO1050-II:1, CHRO979-II:2, and CHRO16-II:1) presented with clinical findings typical for complete ACHM, and 2 of the oldest patients (CHRO251-II:1 and CHRO979-II:2, aged 53 and 57, respectively) showed pronounced foveal or macular lesions and retinal pigment irregularities. The fifth patient in this group (CHRO1050-II:1, 17 years old) had photophobia, nystagmus, and a nonrecordable photopic ERG response in common with the other patients in this group. Nevertheless, color vision and visual acuity (oculus dexter [OD], 0.4; oculus sinister [OS], 0.25) were impaired to a lesser extent in this patient.
Notably, none of the double-heterozygous family members (i.e., parents) of group 2 and group 3 patients who were identified during the segregation analysis reported visual impairment (Figure 1). However, detailed phenotyping was not performed in these double-heterozygous individuals.
Collectively, these findings suggest that CNGB3/p.R403Q is a hypomorphic mutation that possibly causes a relatively mild and late-onset or a subclinical retinal disease in the homozygous state. Furthermore, these data show a correlation of disease severity with the actual CNGB3 and CNGA3 genotype, and provide evidence for a novel digenic triallelic cause of ACHM involving biallelic CNGB3 mutations and single heterozygous CNGA3 mutations.
Enhanced photoreceptor function loss in Cnga3+/– Cngb3R403Q/R403Q compared with Cngb3R403Q/R403Q mutant mice.
To verify our hypothesis of a digenic triallelic cause of ACHM, we generated Cngb3R403Q/R403Q mice and consecutively crossed them with Cnga3–/– mice to produce triallelic Cnga3+/– Cngb3R403Q/R403Q animals to model the condition in triallelic mutant patients (Figure 1). Both mouse lines were established on 2 different genetic backgrounds (129/Sv × C57BL/6N and 129/Sv) to evaluate the effects of inbred and hybrid backgrounds on the expression of the mutants’ phenotype.
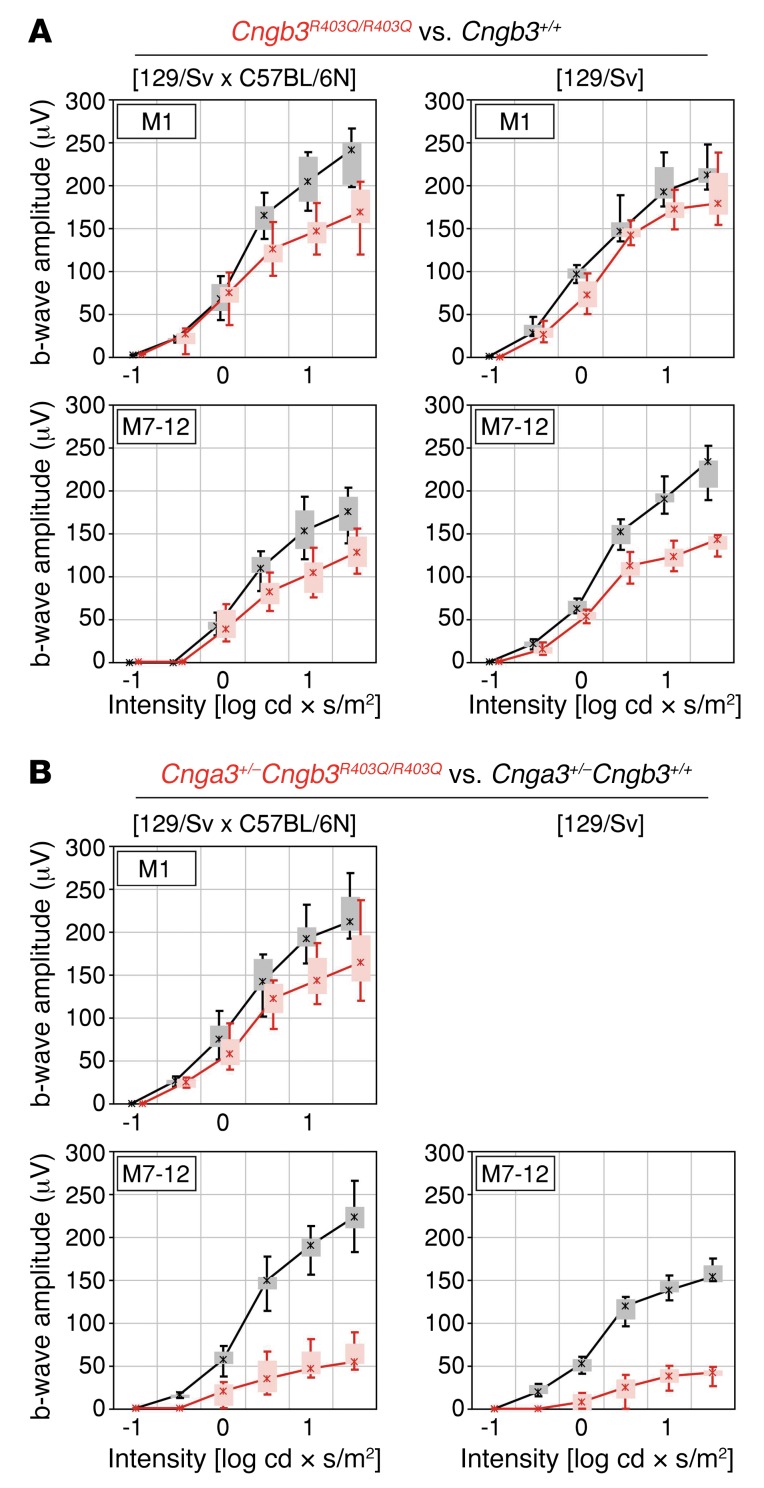
We first analyzed retinal photoreceptor function by means of ERG recordings in Cngb3R403Q/R403Q mice and litter-matched controls at 4–5 weeks and 7–12 months. At young ages, Cngb3R403Q/R403Q mice did not show substantially altered ERG responses, although a tendency toward reduced amplitudes under light-adapted conditions was observed in mixed-background animals (Figure 2A). However, at the age of 7–12 months, Cngb3R403Q/R403Q mutants on the 129/Sv and to a lesser extent on the 129/Sv × C57BL/6N background displayed reduced photopic single-flash ERG responses to 60% of the WT control b-wave amplitudes (Figure 2A). In addition, we also analyzed ERG responses in 4- to 5-week-old Cngb3–/– mice, which have been reported to retain residual cone function (33). Under the same experimental conditions, the Cngb3–/– mice showed clearly reduced photopic ERG amplitudes (Supplemental Figure 4) when compared with the amplitudes obtained from Cngb3R403Q/R403Q mutants. These data demonstrate the pathogenic impact of the p.R403Q mutation on cone photoreceptor function and confirm its hypomorphic effect compared with full ablation of Cngb3. We then analyzed ERG responses in Cnga3+/– Cngb3R403Q/R403Q mice and Cnga3+/– Cngb3+/+ controls at 1 month (for animals on the hybrid background) and at 7–12 months of age (for animals on hybrid and inbred backgrounds). While in young mice no substantial differences in ERG responses between genotypes were detected, we observed a distinct reduction of b-wave amplitudes in aged mice under mesopic and photopic light stimuli for the triallelic mutant irrespective of genetic background (Figure 2B). This effect was so strong that ERG b-wave amplitude of the triallelic mutant was only about 25% of that in Cnga3+/– Cngb3+/+ controls for the brightest light stimuli, while the differences in ERG b-wave amplitude between WT and Cngb3R403Q/R403Q mice under the same condictions were only minor (Figure 2). This finding confirms the additive negative effect of reduced Cnga3 dosage on cone function in this mutant. Of note, rod ERG responses remained unchanged among all mice tested (Supplemental Figures 5 and 6), which indicates pure cone disease with no effects on rod survival and activity. No gross alterations in retinal morphology were detected in the Cnga3+/– Cngb3R403Q/R403Q mutant mice and WT controls by in vivo scanning laser ophthalmoscopy (SLO) and ophthalmic coherence tomography (OCT) at the age of 7–12 months (Supplemental Figure 7).
Figure 2. Cone function loss in aged Cngb3R403Q/R403Q and Cnga3+/– Cngb3R403Q/R403Q mutants.
(A) Results of the ERG recordings of WT (black) and Cngb3R403Q/R403Q mice (red) at 1 month (M1) and 7–12 months (M7–12) of age (n = 3–5). Furthermore, mice were tested on 2 genetic backgrounds (129/Sv × C57BL/6N [n = 4] and 129/Sv [n = 3]). (B) Results of the ERG recordings of Cnga3+/– (black) and Cnga3+/– Cngb3R403Q/R403Q mice (red) (n = 3). ERG data are presented as box-and-whisker plots (boxes: 25%–75% quantile range, whiskers: 5% and 95% quantiles, asterisks: median).
Enhanced photoreceptor degeneration in Cnga3+/– Cngb3R403Q/R403Q mice.
Western blot analysis revealed that CNGB3 protein expression was strongly decreased in retinas of Cngb3R403Q/R403 mice compared with controls (Supplemental Figure 8A). A similar decrease in CNGB3 was observed in Cnga3–/– mice. In line with this, the CNGA3 protein level was also reduced in Cngb3R403Q/R403Q mutants. These results suggest that Cngb3R403Q/R403Q markedly reduces the number of functional CNG channels in the cone outer segment. Cngb3 transcripts, however, remained unchanged in Cngb3R403Q/R403Q as well as in Cnga3+/– Cngb3R403Q/R403Q mutants (Supplemental Figure 8, B and C).
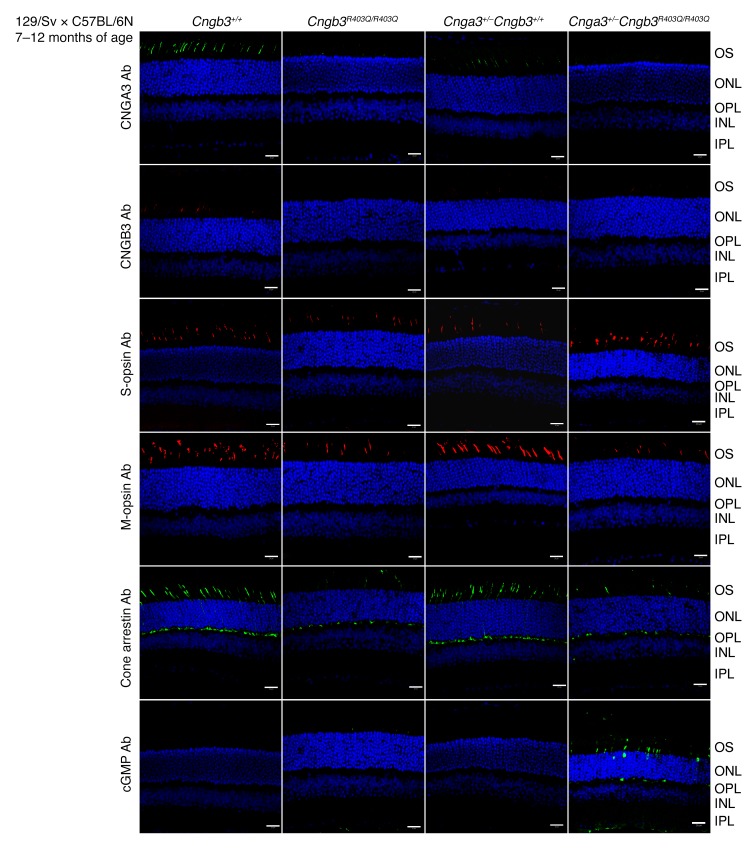
The abnormal ERG of the 7- to 12-month-old mutants prompted us to analyze CNG channel protein abundances and to seek evidence of emerging degenerative processes in photoreceptors. For this purpose, retinas of 7- to 12-month-old mice used in the ERG recordings were investigated by immunohistochemistry (Figure 3). Immunostaining with an anti-CNGA3 antibody and anti-CNGB3 antiserum revealed almost complete absence of both CNG channel subunits in Cngb3R403Q/R403Q and Cnga3+/+ Cngb3R403Q/R403Q mutants. Further immunostainings with cone markers showed apparently reduced levels of M-opsin in both Cngb3R403Q/R403Q and Cnga3+/+ Cngb3R403Q/R403Q mice, whereas S-opsin staining remained unchanged, suggesting a preferential loss of M-opsin–positive cones in homozygous Cngb3R403Q/R403Q mutants (Figure 3). Moreover, cone arrestin was also found to be reduced in both mutants. A decline in arrestin expression has been suggested as an early marker of cone degeneration (36). Most strikingly, immunolabeling with an anti-cGMP antibody revealed that intracellular cGMP levels were profoundly elevated and could be detected by immunolabeling with an anti-cGMP antibody (32) in the retina of Cnga3+/– Cngb3R403Q/R403Q and, to a lesser extent, Cngb3R403Q/R403Q animals. Accumulated cGMP was localized in photoreceptor outer segments and the outer nuclear layer as well as the outer plexiform layer, representing cone photoreceptor axon terminals and somata, respectively. In contrast, the anti-cGMP antibody failed to detect the low cGMP levels in the retina of Cngb3+/+ and Cnga3+/– Cngb3+/+ control animals. cGMP accumulation has been observed in homozygous Cnga3–/– and Cngb3–/– mice (37, 38), and is a common finding in many models of retinal degeneration (39), reflecting an advanced state of metabolic dysregulation in photoreceptors prior to cell death (37). Taken together, the results indicate that triallelic Cnga3+/– Cngb3R403Q/R403Q mice present with an exacerbated disease phenotype when compared with diallelic Cngb3R403Q/R403Q mutants, supporting the findings in patients carrying the CNGB3/p.R403Q mutation with or without an additional CNGA3-mutant allele.
Figure 3. Cone degeneration and reduced CNGA3 and CNGB3 immuno staining in aged Cngb3R403Q/R403Q and Cnga3+/– Cngb3R403Q/R403Q mutants.
Immunofluorescence of cryosections from murine retinas (n = 3 per genotype) at 7–12 months of age (WT, Cnga3+/– Cngb3+/+, Cngb3R403Q/R403Q, and Cnga3+/– Cngb3R403Q/R403Q). Scale bars: 20 μm. S-opsin and cGMP staining with the respective antibodies was performed simultaneously on the same slices and imaged by multicolor laser scanning confocal micrography. Antibodies were diluted as follows: cGMP: 1:3,000; cone arrestin: 1:500; S-opsin: 1:300; M-opsin: 1:300; and CNGB3-antiserum: 1:5,000. INL, inner nuclear layer; IPL, inner plexiform layer; ONL, outer nuclear layer; OPL, outer plexiform layer; OS, outer segment.
Discussion
Biallelic mutations in CNGB3 cause autosomal recessive ACHM or a clinically closely related form of early-onset severe cone dystrophy that differs from the former in having some residual minor cone function that is progressively lost (12, 19, 26). A notable exception are phenotypes associated with the CNGB3/c.1208G>A;p.R403Q mutation, which has been reported in patients who present with highly variable clinical findings ranging from mild macular dystrophy to severe cone dystrophy with near complete loss of cone function (12, 26, 27).
Our in-depth exploration of a large cohort of patients with ACHM, cone dystrophy, and macular dystrophy and an extensive literature search corroborated these individual reports (Supplemental Table 1). Moreover, our compiled data provide an explanatory model for this phenotypic variability based on the allelic composition of CNGB3 mutations and the presence or absence of additional heterozygous mutations in CNGA3 that constitute digenic triallelic inheritance of the observed phenotypes. In fact, we can differentiate 3 distinct groups of patients (groups 1–3; Table 1 and Supplemental Table 1) who show a tendency toward phenotypic outcome and severity.
Our study provides compelling evidence that the CNGB3/p.R403Q variant is in fact a hypomorphic mutation. First, all subjects with biallelic mutations in CNGB3 involving p.R403Q either in homozygous or compound heterozygous state in trans with a known pathogenic CNGB3 mutation exhibit a retinal disease phenotype. In none of these cases, we found biallelic CNGA3 mutations which could explain the disease irrespective of the CNGB3/p.R403Q variant. Second, the arginine residue at amino acid position 403 in CNGB3 is evolutionarily fully conserved in vertebrates, and common prediction algorithms classify the p.R403Q substitution as deleterious, damaging, or disease-causing (Supplemental Table 2). Third, heterologous expression of the human CNGB3/p.R403Q variant or mutants of the homologous residue in bovine CNGB1 — together with the respective CNGA subunit — showed significantly elevated cGMP affinity and strong outward rectification behavior (28, 40). Fourth, reduced cone function (Figure 2 and Supplemental Figure 6) and strongly decreased CNG channel expression in the Cngb3R403/R403 mouse mutant (Figure 3 and Supplemental Figure 8) demonstrate the mild but deleterious effect of the CNGB3/p.R403Q mutation in vivo that is further aggravated by a heterozygous mutation in CNGA3, modeled by the crossing of the Cngb3R403Q/R403Q and Cnga3-KO mice.
The macular dystrophy in patients MDS49-II:1 and 215-011 and variable other phenotypes in human patients, and the intermediate phenotype in the Cngb3R403Q/R403Q mouse mutant in terms of photopic ERG responses, strongly suggests that the CNGB3/p.R403Q mutation represents a hypomorphic CNGB3 allele with residual cone function and a slowly progressive deterioration of cone function and integrity. Consistent with these findings, we generally observed more severe clinical phenotypes but still some minor residual cone function in patients compound heterozygous for the CNGB3/p.R403Q mutation and with a truncating mutation in CNGB3 (group 1 patients; Table 1 and Supplemental Table 1).
Intriguingly, we documented a large proportion of CNGB3/p.R403Q-positive patients (10 of 16) carrying an additional heterozygous CNGA3 mutation (Supplemental Table 1). These CNGA3 mutations are truly pathogenic (Supplemental Table 2), since they (i) constitute a null allele (p.W440*), (ii) have been recurrently reported in ACHM patients (p.R223W, p.E228K, p.R277C, p.R427C, p.R436W) (18, 41–44), or (iii) have been functionally validated in heterologous expression systems in prior studies (p.R223W, p.E228K, p.R277C, p.M406T, p.R427C, p.R436W, p.E593K) (34, 35) or in this work (p.R290H; Supplemental Figure 1). The probability of the simultaneous presence of a heterozygous CNGA3 mutation just by chance in 10 of 16 CNGB3/p.R403Q-positive subjects is exceedingly low given the estimated carrier frequency of 0.0063 for CNGA3 mutations among populations of European descent. Moreover, there was a tendency toward a milder clinical phenotype of incomplete ACHM with minimal cone function observed in the group 2 patients (homozygous for CNGB3/p.R403Q and heterozygous for additional CNGA3 mutations) (Supplemental Table 1).
To further substantiate this digenic triallelic nature of the disease in group 2 and group 3 patients, we specifically generated a CNGB3/p.R403Q-knockin mouse mutant (Cnga3+/+ Cngb3R403Q/R403Q) and modeled the triallelic state through crossbreeding with the well-characterized Cnga3–/– mouse line (Cnga3+/– Cngb3R403Q/R403Q). Importantly, most mouse ACHM models closely recapitulate the principal defect in terms of cone function and cone photoreceptor survival — e.g., Cnga3–/– and cpfl5(Cnga3) (29, 45), Cngb3–/– (30), cpfl3(Gnat2) (46), and cpfl1(Pde6c) (5).
Cone function in 4-week-old Cnga3+/+ Cngb3R403Q/R403Q and Cnga3+/– Cngb3R403Q/R403Q mutants was only slightly reduced, as revealed by photopic flash and flicker ERG, whereas both CNG subunits were hardly detectable at the protein level (Figure 3 and Supplemental Figure 8A). Cngb3–/– mice display early cone function loss with slow progression (30, 33). Cnga3+/+ Cngb3R403Q/R403Q as well as Cnga3+/– Cngb3R403Q/R403Q mutants, however, displayed late-onset cone function loss, with milder cone degeneration. The moderate reduction in cone function in Cnga3+/+ Cngb3R403Q/R403Q mice is comparable to a mild to subclinical phenotype, while the pronounced functional decline at age 7–12 months of the triallelic Cnga3+/– Cngb3R403Q/R403Q mutants reflects the phenotype of patient groups 2 and 3. Obviously, an additional heterozygous CNGA3 mutation exacerbates the Cngb3R403Q/R403Q disease phenotype in both humans and mice. Of note, in vivo retinal imaging and retinal histopathology revealed no gross retinal abnormalities in either mouse mutant. The foveomacular lesions often observed in patients with CNGA3- and CNGB3-linked ACHM (9) are likely due to the loss of cone photoreceptors in the primate macula and fovea, the anatomical feature with the highest cone density, which is not present in the murine retina.
Interestingly, M-opsin–positive cell counts were found to be reduced in aged triallelic Cnga3+/– Cngb3R403Q/R403Q mutants. Differences in the expression, distribution, and/or susceptibility to degeneration of both M- and S-cones have been described in several studies (47–50). Also, arrestin levels in Cngb3R403Q/R403Q genotypes were diminished beginning at 4–5 weeks of age. Altered expression of cone arrestin was indeed observed in mice during cone degeneration (36, 51–53).
Intriguingly, Cnga3+/– Cngb3R403Q/R403Q mutants showed pronounced cGMP accumulation at 7–12 months of age. cGMP was described as a biomarker for photoreceptor dysfunction in a treatment study on the murine ACHM model,indicating an attempt of the cell to overcome the lack of channel activity by a reversible upregulation of the driving factor (31). Thus, the observed differences in cGMP accumulation between the different mutants very probably reflect the degree of overall CNG channel functionality in photoreceptors in the in vivo situation, incorporating factors like trafficking which are hard to model ex vivo. Also, in other, less specific retinal degeneration mouse mutants, affected photoreceptors accumulate vast amounts of cGMP (32). This accumulation was absent in respective controls and hardly detectable in Cnga3+/+ Cngb3R403Q/R403Q mice, underpinning the important aggravation of cone impairment due to the loss of 1 Cnga3 allele, which on its own (i.e., in Cnga3+/– mice) does not cause an impaired visual or retinal phenotype (29).
Taken together, the results show that Cnga3+/+ Cngb3R403Q/R403Q and Cnga3+/– Cngb3R403Q/R403Q mutants recapitulate the principal defect in cone function and morphology observed in patients with homologous genotypes, although some differences exist due to (i) lack of a cone-rich macula and therefore attenuation of the murine phenotype compared with human patients carrying the same mutation; and (ii) formation of some functional homomeric CNGA3 channels in Cngb3–/– mice, which properly traffic to the cone outer segment membrane and result in an attenuated retinal phenotype in Cngb3–/– mice compared with human patients with CNGB3-linked ACHM (30, 54–56). The indistinguishable clinical phenotype of CNGA3-linked ACHM and ACHM caused by loss-of-function mutations in CNGB3 suggests that homomeric cone CNG channels are not formed or do not traffic to the outer segment in humans.
With comprehensive genetic testing now being practiced in medical genetics, the number of reported instances of digenic inheritance of disease in human patients increases. However, only a fraction of those reports withstand thorough validation, such as (i) replication of the findings in multiple independent patients or families, as shown in Table 1 and Supplemental Table 1; (ii) validation of the pathogenicity of the variants involved by means of genetic evidence (e.g., recurrent mutation in affected patients or cosegregation of the variant with disease in families, as shown in Figure 1) or preferably by phenotypical analysis of corresponding knockin mouse models, as realized in our triallelic Cnga3+/– Cngb3R403Q/R403Q mutant; (iii) direct or indirect physical interaction of the 2 involved gene products as part of a multimeric protein complex or their participation in the same biochemical or signaling pathway that is suggested for CNGA3 and CNGB3; and (iv) modeling of the digenic interaction in an appropriate cellular or animal model with adequate readout (e.g., comparative analysis of Cnga3+/+ Cngb3R403Q/R403Q, Cnga3+/– Cngb3+/+, and Cnga3+/– Cngb3R403Q/R403Q mice). A classic and commonly cited example of digenic inheritance in inherited retinal dystrophies is retinitis pigmentosa caused by simultaneous mutations in the PRPH2 (RDS/peripherin) and the ROM1 gene (57, 58). Functional defects in the assembly of mutant RDS/ROM1 hetero-oligomers have been shown (59–61). However, no confirmation of the digenic interaction in an in vivo model has been reported so far. Digenic triallelic inheritance — such as the mode of inheritance for CNGB3/p.R403Q-associated ACHM substantiated in this study — has been proposed in patients with Bardet-Biedl syndrome (BBS). BBS is a rare multiorgan disorder belonging to the ciliopathies, featuring retinal dystrophy, polydactyly, obesity, renal malformations, intellectual disability, and hypogonadism (62), and is genetically highly heterogeneous, with almost 20 disease genes described so far (62, 63). The majority of those gene products are part of a multiprotein complex, the BBSome, and BBS proteins are involved in cargo shuttling in photoreceptors (64, 65). Protein defects result in ciliopathies associated with defective ciliary trafficking and photoreceptor degeneration. Digenic triallelic interactions that are either obligatory for disease expression or that worsen the disease phenotype have been reported between multiple pairs of BBS genes (i.e., refs. 66–70; for an overview, see the Digenic Disease Database (DIDA), http://dida.ibsquare.be/). However, it should be noted that triallelism may only characterize a minor fraction of BBS patients (71–73).
The evidence of a digenic triallelic interaction between the CNGB3/p.R403Q mutation and single-heterozygous CNGA3 mutations is compelling and fulfills all criteria listed above, supporting the following conclusions: (i) triallelism was present in 10 of 16 subjects carrying the CNGB3/p.R403Q allele in this study; (ii) all CNGB3 and CNGA3 variants observed in the triallelic genotypes were validated as pathogenic mutations by genetic criteria (Supplemental Table 2; i.e., recurrently observed in ACHM patients, concordant segregation in families), and their obvious deleterious consequences were shown at the transcript or protein level (c.1578+1G>A and p.Thr383Ifs*13 in CNGB3; p.W440* and c.-37-1G>C in CNGA3) and/or by functional analysis in heterologous expression systems or in an animal model (Cngb3R403Q/R403Q in this study); (iii) a direct physical interaction of CNGB3 and CNGA3 is required for the formation of the native heteromeric cone CNG channel, for mutual protein stability, and likely also for transport to the photoreceptor outer segment (29, 30, 74, 75); and (iv) we modeled the digenic interaction in the Cnga3+/– Cngb3R403/R403Q mouse mutant and demonstrated the additive effect on visual function and cone photoreceptor integrity by means of ERG recordings and retinal histopathology. In summary, we describe a digenic triallelic inheritance pattern for cone retinopathies supported by data from patients and targeted knockin mice, with strong clinical implications for prognosis.
Methods
Molecular genetic analysis.
DNA was isolated from peripheral blood according to standard procedures at the different centers, and banked at the Institute for Ophthalmic Research, Tübingen, as per the standard protocol. All coding exons and flanking intronic and UTR sequences of CNGB3 (RefSeq NM_019098) and CNGA3 (RefSeq NM_001298) were analyzed as reported previously (1, 2, 18, 19). For primer sequences, as well as PCR and sequencing conditions, see Supplemental Methods. All variants and genotypes were deposited to the ClinVar database (76), with accession codes for the single variants (SCV000700209–SCV000700226) listed in Supplemental Table 2.
Segregation analysis within the families was performed by PCR/RFLP or Sanger sequencing (Figure 1). For patients MDS49-II:1, CHRO852-II:1, CHRO16-II:1, and CD228-II:1, no family members were available. Therefore, homozygosity for the CNGB3/p.R403Q mutation was confirmed by quantitative real-time PCR (qRT-PCR), applying the QuantiTect SYBR GreenPCR Kit (QIAGEN) for MDS49-II:1 and CHRO852-II:1 on an Applied Biosystems 7500 Real-Time PCR system following the manufacturer’s instructions and primers for CNGB3 exons 10 (forward: 5′-GGCTGTATTTCAGAAACAACATGA-3′, reverse: 5′-CACAGGCATTAATGTGCAGAA-3′), 11 (forward: 5′-CCTTCCAGAACCACAAACTTTA-3′, reverse: 5′-CAAAATGACAGCACTGTGTATCC-3′), and 12 (forward: 5′-CATGTGGAAAACATGTAATTTGACT-3′, reverse: 5′-TTGGCTGTAGCTGCTCCAAT-3′) and the reference gene SDC4 (forward: 5′-CAGGGTCTGGGAGCCAAG-3′, reverse: 5′-GCACAGTGCTGGACATTGACA-3′). In addition, for the p.R403Q-homozygous patients MDS49-II:1, CHRO852-II:1, and CHRO16-II:1, exome sequencing was performed to exclude mutations in other genes known to be associated with inherited retinal dystrophies as listed in RetNet (https://sph.uth.edu/Retnet/), as previously described (77).
To test the effect of the variant c.-37-1G>C on splicing, a 714-bp fragment comprising the first coding CNGA3 exon with 295 bp of upstream and 419 bp of downstream sequence was amplified using a proofreading polymerase and genomic DNA from patient MDS49-II:1, thereby co-amplifying the normal and the mutant allele. Cloning into the exon-trapping vector pSPL3, transfection of HEK293T cells as well as murine 661W photoreceptor-derived cells, RNA isolation, and cDNA synthesis were performed as described previously (78, 79). The HEK293T cell line was purchased from ATCC. The murine cone photoreceptor–derived 661W cells were provided by Muayyad R. Al-Ubaidi (University of Oklahoma Health Sciences Center).
Cell culture and heterologous expression of CNGA3/p.R290H.
HEK293 cells (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DMSZ]) were maintained in DMEM medium (Thermo Fisher Scientific) supplemented with 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin and incubated at 37°C with 5% CO2. For transfection experiments HEK293 cells were seeded on 35-mm petri dishes at a density of 200,000 cells per dish. After 6 hours, cells were transfected with the expression plasmid DNA (1.5 μg of each plasmid per dish) using the TurboFect transfection reagent (Thermo Fisher Scientific). For electrophysiological measurements, transfected cells were detached using 0.05% trypsin/0.53 mM EDTA (Thermo Fisher Scientific) and replated onto 12-mm poly-l–lysine-coated coverslips in 24-well plates (23).
Electrophysiological measurement of channels containing CNGA3/p.R290H.
Currents of heterologously expressed CNG channels were measured at room temperature 2–3 days after transfection using excised inside-out voltage clamp configuration. Transfected cells were identified by green fluorescence due to EGFP expression encoded by an IRES2-EGFP sequence within the CNG channel expression cassette of the plasmid. The bath and pipette solutions were composed of (in mM): 140 NaCl, 5 KCl, 10 HEPES, 1 EGTA, pH 7.4 adjusted with NaOH. Pipettes were pulled from borosilicate glass capillaries (GC150TF, Harvard Apparatus) and had resistances of 2–3 Ωm when filled with the pipette solution. cGMP and cAMP were purchased from Sigma-Aldrich. L-(-)-cis-diltiazem hydrochloride was purchased from Enzo Life Sciences. cGMP and cAMP solutions were dissolved in bath solution directly. Stock solution of L-cis-diltiazem was prepared in water and was freshly diluted in bath solution before use in experiments. Data were acquired at 10 kHz using an Axopatch 200B amplifier and pClamp 10 (Axon Instruments). Voltage clamp data were stored on a computer hard disk and analyzed off-line by using Clampfit 10 (Axon Instruments). Leakage current was removed by subtraction of current without cGMP or cAMP. Dose-response curves were determined by fitting with the Hill function unless otherwise specified.
Generation of the CNGB3/p.R403Q-knockin mouse model.
The p.R403Q mutation results in an exchange of arginine into glutamine, which could be achieved with 2 different codons: CAG and CAA. Corresponding to the preferred codon usage in mice, we chose the CAG codon for construction of the CNGB3/p.R403Q targeting vector.
The primers Cngb3_FM: 5′-CTCTCACTGAGAGATGGAGCCT-3′, Cngb3_RM: 5′-AAAGCTAACCATAAGTTTGAGG-3′, Cngb3_CAG: 5′-TATTGGGCAGTTCAGACTTTAAT-3′, and Cngb3_R_CAG: 5′-ATTAAAGTCTGAACTGCCCAATA-3′ were used to introduce the p.R403Q mutation. Moreover, the use of the CAG codon enabled subsequent analysis of the p.R403Q mutation via PCR/RFLP due to a loss of a TaqI restriction site and generation of a novel Hpy188I restriction site compared with the WT sequence.
The Cngb3 homology arm fragments for the assembly of the targeting construct were subcloned from a BAC isolated from a 129/Sv mouse BAC library (Deutsches Ressourcenzentrum für Genomforschung [RZPD]). The targeting vector contained a Neo/Tk selection cassette flanked by 2 loci of loxP sites located upstream of exon 11 and a single loxP site together with a new SphI restriction site (for subsequent identification of the homologous recombination by Southern blot analysis) in intron 10 (Supplemental Figure 9). R1 embryonic stem (ES) cells were electroporated with the linearized targeting construct and screened for G418-resistant clones. Homologous recombination was confirmed by Southern blot analysis upon MscI-, SphI-, and BamHI-digested ES cell DNA. Two correctly targeted clones were injected into C57BL/6 blastocysts. The resulting chimeras were mated with C57BL/6N and 129/Sv mice to obtain germline transmission, resulting in heterozygous CNGB3/p.R403Q offspring. Heterozygous offspring (genotype: Cngb3R403Q/+) were intercrossed to establish homozygous Cngb3-mutant mice (Cngb3R403Q/R404Q) and littermate control animals for the experiments. Cngb3R403Q/+ mice were also crossed with Cnga3+/– mice to obtain Cnga3+/– Cngb3+/R403Q mice, which were intercrossed to obtain Cnga3+/– Cngb3R403Q/R403Q mutant mice and the corresponding Cnga3+/– and WT controls.
Genotyping of CNGB3/p.R403Q mutants.
Animals were genotyped by PCR using genomic DNA extracted from mouse tails with 2 primers for Cngb3 (F1: 5′-GTCGACTAGAGCTTGCGGAAC-3′; R: 5′-AATATTGTAGTCTCTTGCCTT-3′) that amplified either the WT (220 bp) or the Cngb3-knockin allele (366 bp) (Supplemental Figure 9F), and 3 primers for Cnga3 (F1: 5′-CTTAGGTTTCCTTGAGGCAAG-3′; F2: 5′-GCCTGCTCTTTACTGAAGGCT-3′; R: 5′-CAAGTTCCCTATCCTGAACAC-3′) that yielded either the WT (248 bp) or Cnga3-KO allele (340 bp). Furthermore, animals on a hybrid background (129/Sv × C57BL6/N) were tested for the presence of the rd8 allele to exclude tampering or aggravation of investigated phenotypes with primers (MM_RD8_F: 5′-GCCCCTGTTTGCATGGAGGAAACTTGGAAGACAGCTACAGTTCATAT-3′; MM_RD8_R: 5′-GCCCCATTTGCACACTGATGAC-3′), resulting in a 244-bp amplicon that can be cleaved by Ndel into a 45-bp and a 199-bp fragment if the rd8 mutation is present.
Electroretinography.
Full-field ERGs were recorded from Cngb3+/+, Cnga3+/–, Cngb3R403Q/R403Q, and Cnga3+/– Cngb3R403Q/R403Q littermate mice at the age of 4–5 weeks or 8–12 months according to procedures described previously (80). In brief, mice were dark-adapted overnight before the experiments and anesthetized with s.c. injection of a mixture of ketamine (66.7 mg/kg body weight) and xylazine (11.7 mg/kg body weight). The pupils were dilated, and single-flash ERGs were obtained under dark-adapted (no background illumination, 0 cd/m2) and light-adapted (background illumination of 30 cd/m2 starting 10 minutes before recording) conditions. Single white-flash stimuli ranged from –4.0 to 1.5 log cd×s/m2 under dark-adapted and from –2.0 to 1.5 log cd×s/m2 under light-adapted conditions. Ten responses were averaged with inter-stimulus intervals of 5 seconds (for –4 to –0.5 log cd×s/m2) or 17 seconds (for 0 to 1.5 log cd×s/m2). Responses to series of brief flashes (flicker) at a fixed intensity (0.5 log cd×s/m2) with 12 frequencies (0.5, 1, 2, 3, 5, 7, 10, 12, 15, 18, 20, and 30 Hz) were obtained without any background illumination (0 cd/m2) and were averaged either 20 times (for 0.5 to 3 Hz) or 30 times (for 5 Hz and above) (81). Band-pass filter cutoff frequencies were 0.3 and 300 Hz for all ERG recordings.
SLO and OCT.
SLO and OCT imaging was done in the same session as the ERGs in Cngb3+/+, Cnga3+/–, Cngb3R403Q/R403Q, and Cnga3+/– Cngb3R403Q/R403Q mice. SLOs and OCTs were obtained as reported previously (82). Analysis were performed using an s.c. injection of 75 mg/kg body weight fluorescein-Na (University Pharmacy, University of Tübingen) and ICGA following an s.c. injection of 50 mg/kg body weight ICG (ICG-Pulsion, Pulsion Medical Systems).
SD-OCT imaging was performed with a Spectralis HRA+OCT device from Heidelberg Engineering featuring a broadband superluminescent diode at I 1/4 870 nm as low coherent light source. Each 2-dimensional B-scan recorded with the equipment set to 308 field of view consisted of 1,536 A-scans acquired at a speed of 40,000 scans per second. Optical depth resolution was 7 μm, with digital resolution reaching 3.5 mm. Image data were analyzed using the proprietary software package Eye Explorer from Heidelberg Engineering.
Generation of anti-CNGB3 rabbit antiserum.
CNGB3 antigen was obtained from the N-terminal (208 amino acid residues) part of the murine CNGB3 WT protein. The cDNA was cloned into a pRSET expression vector (Invitrogen/Thermo Fisher Scientific) containing a 6xHis-tag. The expression vector was used to transform E. coli BL21-Gold(DE3)pLysS cells, enabling stable and reliable expression controlled by the IPTG-inducible lacUV5 promoter. Transformed BL21-Gold(DE3)pLysS cells were cultured in 200 ml super optimal broth media with additional 2 ml MgCl2 solution (1 mol/l), 400 μl ampicillin, and 300 μl chloramphenicol. After reaching an OD600 of 0.5, protein expression was induced by addition of IPTG (1 mM) for 3 hours. Bacteria were harvested with an OD600 of 1.0, and the CNGB3 protein fragment was purified with Ni2+-NTA columns (QIAGEN). Protein size was determined using denaturing discontinuous gel electrophoresis (83). For subsequent immunization of rabbits, a total of 2 mg purified Cngb3 fragment was used (~500 μg/rabbit). Immunization was performed by PINEDA antibody service, and serum was taken 1 day before immunization (preimmune serum) and on days 61, 90, and 120. Antiserum was tested with material from WT mice by means of Western blotting and immunohistochemical methods.
Immunoblotting of proteins from murine retina.
Mouse eyes were enucleated and homogenized (Ultra-Turrax) in homogenization buffer (20 mM Tris-HCl pH 8.3, 0.67% SDS, 238 mM β-mercaptoethanol, 0.2 mM PMSF). Protein extraction was performed with pooled mouse retinas (n = 10 per genotype). Western blot analysis was done according to a previously established protocol (74). WT retinas were used as positive controls, and retinas from Cnga3–/– mice were used as negative controls. Extracted proteins were separated by their molecular weight using denaturing 17.5% SDS-PAGE electrophoresis.
qRT-PCR.
Total RNA extraction was performed using the RNeasy-Mini Kit (QIAGEN) according to the manufacturer’s protocol. RNA quality was analyzed on an Agilent Bioanalyzer (Agilent Technologies). Reverse transcription–PCR (RT-PCR) was performed using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). qRT-PCR was performed on a LightCycler 480 System (Roche Applied Science) using GoTaq qPCR Master Mix (Promega). Six different biological samples were analyzed in duplicate and normalized to the expression of the housekeeping gene aminolevulinic acid synthase (Alas). Relative quantification was determined by the method described by Pfaffl (84). The following primers were used (5′→3′orientation): Cnga3 forward: CCACCCCCCGTGAAAGAGTA, Cnga3 reverse: GGAGTCGATCTTAGCCTGGA, Cngb3 forward: GTTGTGGCCCACGGATTTGC, Cngb3 reverse: GTTGTGGCCCACGGATTTGC, ALAS forward: TCGCCGATGCCCATTCTTATC, ALAS reverse: GGCCCCAACTTCCATCATCT.
Immunohistochemistry of the murine retina.
Immunohistochemical staining was performed on retinal cryosections according to procedures described previously (74). In brief, enucleated eyes were punctured with a needle (ora serrata) and fixed with 4% PFA in 0.1 M phosphate buffer (PB) for 5 minutes. Cornea and lens were removed, and the residual eye cup was fixed for 45 minutes in 4% PFA/PB and washed 3 times (0.1 M PB). Subsequently, the eye cup was incubated overnight in 30% sucrose/PB. After embedding in tissue freezing medium (Tissue-Tek O.C.T Compound, Sakura Finetech), vertical cryosections were cut at 10 μm and stored at –20°C until use. The retina slices were rehydrated with 0.1 M PB and then fixed for 10 minutes with 4% PFA. After 3 washing steps (0.1 M PB), slices were incubated with primary antibody overnight at 4°C in a solution of 0.1 M PB, 5% chemiBLOCKER (Merck Millipore), and 0.3% Triton X-100. Subsequently, the slices were washed 3 times in 0.1 M PB, before proceeding with secondary detection using Alexa Fluor 488 anti-mouse or rabbit IgG F(ab′)2 fragments (Cell Signaling Technology) or anti–guinea pig IgG (Mobitec) or Cy3 anti-mouse or anti-rabbit IgG (Jackson ImmunoResearch Laboratories Inc.). The cell nuclei were stained with Hoechst 33342. Finally, the sections were washed (0.1 M PB) and covered with coverslips. Multicolor laser scanning confocal micrographs were taken using an LSM 510 Meta microscope (Zeiss). The following primary antibodies were used: rabbit anti-CNGA3 (85) (1:3,000), rabbit anti–M-OPSIN (1:300; AB5405, Merck Millipore), rabbit anti–S-OPSIN (1:300; AB5407, Merck Millipore), rabbit anti–cone arrestin (86) (1:500), and sheep anti-cGMP (31, 87) (1:3,000), as well as rabbit antiserum against CNGB3 (see Generation of anti-CNGB3 rabbit antiserum) (1:5,000).
Study approval: human subjects and clinical examination.
Study participants were recruited over 20 years from international collaborating centers (Centre for Ophthalmology, Tübingen, Germany; Augenzentrum Siegburg, Germany; University Eye Hospital Bonn, Germany; McGill Ocular Genetics Centre, Montreal, Quebec, Canada; University Eye Hospital Munich, Germany; Kellogg Eye Center, Ann Arbor, Michigan, USA; The National Eye Institute, Bethesda, Maryland, USA; Casey Eye Institute, Portland, Oregon, USA; and Institut des Neurosciences, Montpellier, France) specializing in inherited retinal diseases. Blood or DNA samples were sent to the Centre for Ophthalmology, University of Tübingen, for genetic investigation. Clinical and genetic studies were performed according to the tenets of the Declaration of Helsinki and approved by the respective local research and ethical review boards (Ethics Committee, Medical Faculty, University Tübingen, Germany; Ethics Committee, Medical Faculty, University of Bonn, Germany; Ethics Committee, Medical Faculty, University of Munich, Germany; University of Michigan Medical School Institutional Review Board, Ann Arbor, MI, USA; Oregon Health & Science University Institutional Review Board, Portland, OR, USA; McGill University Institutional Review Board, Montreal, Canada; the Ministry of Public Health of France.), and all participants provided written informed consent.
Patients underwent comprehensive ophthalmological examination including — varying and depending on the different centers — psychophysical testing (best-corrected visual acuity, color vision), electrophysiological assessment (i.e., full-field and multifocal ERG), and retinal imaging (i.e., color fundus photography, fundus autofluorescence [FAF], and OCT). Color vision was assessed with various tests, including Ishihara plates, Lanthony Tritan album (LTA), American Optical Hardy-Rand-Rittler (AOHRR), saturated Roth 28-hue test, Farnsworth Munsell 100-hue tests, and the Nagel anomaloscope. Full-field ERG and multifocal ERG recordings were measured in accordance with International Society for Clinical Electrophysiology of Vision (ISCEV) recommendations.
Study approval: animal welfare and genetic background.
Experimental mice were bred and maintained at the animal facility of the Institute of Pharmacy, Department of Pharmacology, Toxicology, and Clinical Pharmacy, University of Tübingen. All procedures with respect to mice were performed with permission of local authorities (Regierungspräsidium Tübingen, Tübingen, Germany and Regierungspräsidium Karlsruhe, Karlsruhe, Germany) and conducted in accordance with German legislation on the protection of animals. The mice were housed in temperature- and humidity-controlled cages with unrestricted access to food and water in a standard 12-hour light/12-hour dark cycle. For experiments, Cngb3R403/R403Q and Cnga3+/– b3R403Q/R403Q mutant mice were compared with age- or age- and litter-matched WT mice or Cnga3+/– mice on a hybrid 129/Sv × C57BL/6 or pure 129/Sv genetic background. Mice were used irrespective of their sex at 4–52 weeks of age. Cnga3+/– mice (29) were on a 129/Sv genetic background. The Cngb3R403/R403Q mice that were generated for this study were either on a 129/Sv background or on a hybrid background (C57BL/6N × 129/Sv) as specified in the corresponding figure legends. Cngb3–/– mice (30) (Supplemental Figure 4) were on C57BL/6J background.
Statistics.
Electrophysiological data from HEK293 experiments are presented as mean ± SEM (n indicates number of recorded cells). Student’s 2-tailed t test or 1-way ANOVA was calculated with Origin6.1 (OriginLab). qRT-PCR data are presented as mean ± SEM (n indicates number of animals). One-way ANOVA was were performed with GraphPad Prism 5 (GraphPad Software). Box-and-whisker plots were used to present ERG data distribution of VlogI (amplitude versus log intensity) graph, where boxes indicate 25%–75% quantile range, whiskers 5% and 95% quantiles, and asterisks the median of data.
Author contributions
BW and PR originally developed the concept and designed the experiments together with M Burkard, SK, TK, CB, PR, and RL. SK, AKM, BB, and NW generated, analyzed, and interpreted the genetic data. SK, DZ, and GAH compiled the clinical data from patients. M Burkard and TK generated the Cngb3R403Q/R403Q knockin mouse. M Burkard characterized the mutant mouse line. M Burkard, SK, NT, XZ, CB, TK, AEB, PR, VS, SCB, GH, AKM, KJ, and BB performed the experiments. MS, NT, VS, SCB, and GH planned, established, and performed the ERG, SLO, and OCT animal experiments. FK, EB, and SM planned, established, and performed the immunofluorescence, qRT-PCR, and Western blot experiments. XQD and SV analyzed and interpreted experimental data. M Biel and XZ planned and performed the experiments on the CNGA3/p.R290H variant. DZ, GAH, UK, PCI, RKK, GR, JH, PS, RGW, and CH clinically examined the patients and provided clinical data. M Burkard, SK, SM, MWS, BW, and PR interpreted the data and wrote the manuscript. All authors read, revised and approved the final manuscript.
Supplementary Material
Acknowledgments
We thank Sui Mei Chiu for patient referral, Kinga Bujakowska and Eric Pierce for supplying DNA of the probands published in ref. 26, Michel Michaelides for his willingness to discuss the case presented in ref. 27, and Fred Koch for his help in phenotypical mouse analysis. The study was supported by grants from the Deutsche Forschungsgemeinschaft (KFO134) to SK, BW, and PR, and BMBF grant 01GM1108A from the German Federal Ministry of Research and Education to SK and BW. M Burkard and SV were further supported by grants from the Institutional Strategy of the Eberhard Karls University of Tuebingen (Deutsche Forschungsgemeinschaft [DFG], ZUK 63), from the Wissenschaftsfoerderung der Deutschen Brauwirtschaft e.V. project B103, and the European Foundation for Alcohol Research (ERAB; ref. EA 15 28), and grants from Wissenschaftsfoerderung der Deutschen Brauwirtschaft e.V. project B103 and the European Foundation for Alcohol Research (ERAB; ref. EA 15 28). PCI was supported by the National Institute for Health Research (NIHR) Oxford Biomedical Research Centre (BRC). Finally, we acknowledge support from Deutsche Forschungsgemeinschaft and the Open Access Publishing Fund of University of Tübingen. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR or the Department of Health.
Version 1. 11/12/2018
Electronic publication
Version 2. 12/03/2018
Print issue publication
Footnotes
Conflict of interest: RW serves on Scientific Advisory boards for the Foundation Fighting Blindness. This relationship has been reviewed and managed by OHSU.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5663–5675.https://doi.org/10.1172/JCI96098.
Contributor Information
Markus Burkard, Email: markus.burkard@uni-tuebingen.de.
Susanne Kohl, Email: Susanne.Kohl@med.uni-tuebingen.de.
Timm Krätzig, Email: timmkraetzig@gmx.de.
Naoyuki Tanimoto, Email: Naoyuki.Tanimoto@uksh.de.
Christina Brennenstuhl, Email: cbrennenstuhl@aol.com.
Katrin Junger, Email: katrinjunger@yahoo.de.
Peggy Reuter, Email: peggy.reuter@med.uni-tuebingen.de.
Vithiyanjali Sothilingam, Email: vithiyanjali.sothilingam@med.uni-tuebingen.de.
Susanne C. Beck, Email: su.beck@uni-tuebingen.de.
Gesine Huber, Email: gesine.huber@gmx.de.
Xi-Qin Ding, Email: xi-qin-ding@ouhsc.edu.
Britta Baumann, Email: Britta.Baumann@med.uni-tuebingen.de.
Nicole Weisschuh, Email: nicole.weisschuh@uni-tuebingen.de.
Ditta Zobor, Email: ditta.zobor@med.uni-tuebingen.de.
Gesa-Astrid Hahn, Email: gesa-astrid.hahn@med.uni-tuebingen.de.
Ulrich Kellner, Email: kellneru@mac.com.
Sascha Venturelli, Email: sascha.venturelli@gmx.de.
Elvir Becirovic, Email: elvir.becirovic@cup.uni-muenchen.de.
Peter Charbel Issa, Email: peter.issa@ouh.nhs.uk.
Robert K. Koenekoop, Email: robkoenekoop@hotmail.com.
Günther Rudolph, Email: guenther.rudolph@med.uni-muenchen.de.
Christian Hamel, Email: christian.hamel@inserm.fr.
Xiangang Zong, Email: xiangang.zong@cup.uni-muenchen.de.
Martin Biel, Email: mbiel@cup.uni-muenchen.de.
Robert Lukowski, Email: robert.lukowski@uni-tuebingen.de.
Stylianos Michalakis, Email: stylianos.michalakis@cup.uni-muenchen.de.
Bernd Wissinger, Email: wissinger@uni-tuebingen.de.
Peter Ruth, Email: peter.ruth@uni-tuebingen.de.
References
- 1.Kohl S, et al. Total colourblindness is caused by mutations in the gene encoding the alpha-subunit of the cone photoreceptor cGMP-gated cation channel. Nat Genet. 1998;19(3):257–259. doi: 10.1038/935. [DOI] [PubMed] [Google Scholar]
- 2.Kohl S, et al. Mutations in the CNGB3 gene encoding the beta-subunit of the cone photoreceptor cGMP-gated channel are responsible for achromatopsia (ACHM3) linked to chromosome 8q21. Hum Mol Genet. 2000;9(14):2107–2116. doi: 10.1093/hmg/9.14.2107. [DOI] [PubMed] [Google Scholar]
- 3.Sundin OH, et al. Genetic basis of total colourblindness among the Pingelapese islanders. Nat Genet. 2000;25(3):289–293. doi: 10.1038/77162. [DOI] [PubMed] [Google Scholar]
- 4.Kohl S, et al. Mutations in the cone photoreceptor G-protein alpha-subunit gene GNAT2 in patients with achromatopsia. Am J Hum Genet. 2002;71(2):422–425. doi: 10.1086/341835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang B, et al. A homologous genetic basis of the murine cpfl1 mutant and human achromatopsia linked to mutations in the PDE6C gene. Proc Natl Acad Sci U S A. 2009;106(46):19581–19586. doi: 10.1073/pnas.0907720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Thiadens AA, et al. Homozygosity mapping reveals PDE6C mutations in patients with early-onset cone photoreceptor disorders. Am J Hum Genet. 2009;85(2):240–247. doi: 10.1016/j.ajhg.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohl S, et al. A nonsense mutation in PDE6H causes autosomal-recessive incomplete achromatopsia. Am J Hum Genet. 2012;91(3):527–532. doi: 10.1016/j.ajhg.2012.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohl S, et al. Mutations in the unfolded protein response regulator ATF6 cause the cone dysfunction disorder achromatopsia. Nat Genet. 2015;47(7):757–765. doi: 10.1038/ng.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiadens AA, et al. Progressive loss of cones in achromatopsia: an imaging study using spectral-domain optical coherence tomography. Invest Ophthalmol Vis Sci. 2010;51(11):5952–5957. doi: 10.1167/iovs.10-5680. [DOI] [PubMed] [Google Scholar]
- 10.Thomas MG, McLean RJ, Kohl S, Sheth V, Gottlob I. Early signs of longitudinal progressive cone photoreceptor degeneration in achromatopsia. Br J Ophthalmol. 2012;96(9):1232–1236. doi: 10.1136/bjophthalmol-2012-301737. [DOI] [PubMed] [Google Scholar]
- 11.Thomas MG, Kumar A, Kohl S, Proudlock FA, Gottlob I. High-resolution in vivo imaging in achromatopsia. Ophthalmology. 2011;118(5):882–887. doi: 10.1016/j.ophtha.2010.08.053. [DOI] [PubMed] [Google Scholar]
- 12.Thiadens AA, et al. Comprehensive analysis of the achromatopsia genes CNGA3 and CNGB3 in progressive cone dystrophy. Ophthalmology. 2010;117(4):825–830.e1. doi: 10.1016/j.ophtha.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Sundaram V, et al. Retinal structure and function in achromatopsia: implications for gene therapy. Ophthalmology. 2014;121(1):234–245. doi: 10.1016/j.ophtha.2013.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aboshiha J, et al. A prospective longitudinal study of retinal structure and function in achromatopsia. Invest Ophthalmol Vis Sci. 2014;55(9):5733–5743. doi: 10.1167/iovs.14-14937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scoles D, et al. In vivo imaging of human cone photoreceptor inner segments. Invest Ophthalmol Vis Sci. 2014;55(7):4244–4251. doi: 10.1167/iovs.14-14542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carroll J, Choi SS, Williams DR. In vivo imaging of the photoreceptor mosaic of a rod monochromat. Vision Res. 2008;48(26):2564–2568. doi: 10.1016/j.visres.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubis AM, et al. Genotype-dependent variability in residual cone structure in achromatopsia: toward developing metrics for assessing cone health. Invest Ophthalmol Vis Sci. 2014;55(11):7303–7311. doi: 10.1167/iovs.14-14225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wissinger B, et al. CNGA3 mutations in hereditary cone photoreceptor disorders. Am J Hum Genet. 2001;69(4):722–737. doi: 10.1086/323613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kohl S, et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur J Hum Genet. 2005;13(3):302–308. doi: 10.1038/sj.ejhg.5201269. [DOI] [PubMed] [Google Scholar]
- 20.Mayer AK, et al. CNGB3 mutation spectrum including copy number variations in 552 achromatopsia patients. Hum Mutat. 2017;38(11):1579–1591. doi: 10.1002/humu.23311. [DOI] [PubMed] [Google Scholar]
- 21.Kaupp UB, Seifert R. Cyclic nucleotide-gated ion channels. Physiol Rev. 2002;82(3):769–824. doi: 10.1152/physrev.00008.2002. [DOI] [PubMed] [Google Scholar]
- 22.Shuart NG, Haitin Y, Camp SS, Black KD, Zagotta WN. Molecular mechanism for 3:1 subunit stoichiometry of rod cyclic nucleotide-gated ion channels. Nat Commun. 2011;2:457. doi: 10.1038/ncomms1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerstner A, Zong X, Hofmann F, Biel M. Molecular cloning and functional characterization of a new modulatory cyclic nucleotide-gated channel subunit from mouse retina. J Neurosci. 2000;20(4):1324–1332. doi: 10.1523/JNEUROSCI.20-04-01324.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Michalakis S, et al. Loss of CNGB1 protein leads to olfactory dysfunction and subciliary cyclic nucleotide-gated channel trapping. J Biol Chem. 2006;281(46):35156–35166. doi: 10.1074/jbc.M606409200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, et al. Knockout of GARPs and the β-subunit of the rod cGMP-gated channel disrupts disk morphogenesis and rod outer segment structural integrity. J Cell Sci. 2009;122(pt 8):1192–1200. doi: 10.1242/jcs.042531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishiguchi KM, Sandberg MA, Gorji N, Berson EL, Dryja TP. Cone cGMP-gated channel mutations and clinical findings in patients with achromatopsia, macular degeneration, and other hereditary cone diseases. Hum Mutat. 2005;25(3):248–258. doi: 10.1002/humu.20142. [DOI] [PubMed] [Google Scholar]
- 27.Michaelides M, et al. Progressive cone dystrophy associated with mutation in CNGB3. Invest Ophthalmol Vis Sci. 2004;45(6):1975–1982. doi: 10.1167/iovs.03-0898. [DOI] [PubMed] [Google Scholar]
- 28.Bright SR, Brown TE, Varnum MD. Disease-associated mutations in CNGB3 produce gain of function alterations in cone cyclic nucleotide-gated channels. Mol Vis. 2005;11:1141–1150. [PubMed] [Google Scholar]
- 29.Biel M, et al. Selective loss of cone function in mice lacking the cyclic nucleotide-gated channel CNG3. Proc Natl Acad Sci U S A. 1999;96(13):7553–7557. doi: 10.1073/pnas.96.13.7553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding XQ, Harry CS, Umino Y, Matveev AV, Fliesler SJ, Barlow RB. Impaired cone function and cone degeneration resulting from CNGB3 deficiency: down-regulation of CNGA3 biosynthesis as a potential mechanism. Hum Mol Genet. 2009;18(24):4770–4780. doi: 10.1093/hmg/ddp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Michalakis S, et al. Restoration of cone vision in the CNGA3-/- mouse model of congenital complete lack of cone photoreceptor function. Mol Ther. 2010;18(12):2057–2063. doi: 10.1038/mt.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Michalakis S, Xu J, Biel M, Ding XQ. Detection of cGMP in the degenerating retina. Methods Mol Biol. 2013;1020:235–245. doi: 10.1007/978-1-62703-459-3_16. [DOI] [PubMed] [Google Scholar]
- 33.Xu J, Morris L, Fliesler SJ, Sherry DM, Ding XQ. Early-onset, slow progression of cone photoreceptor dysfunction and degeneration in CNG channel subunit CNGB3 deficiency. Invest Ophthalmol Vis Sci. 2011;52(6):3557–3566. doi: 10.1167/iovs.10-6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muraki-Oda S, et al. Functional analysis of rod monochromacy-associated missense mutations in the CNGA3 subunit of the cone photoreceptor cGMP-gated channel. Biochem Biophys Res Commun. 2007;362(1):88–93. doi: 10.1016/j.bbrc.2007.07.152. [DOI] [PubMed] [Google Scholar]
- 35.Koeppen K, et al. Functional analysis of human CNGA3 mutations associated with colour blindness suggests impaired surface expression of channel mutants A3(R427C) and A3(R563C) Eur J Neurosci. 2008;27(9):2391–2401. doi: 10.1111/j.1460-9568.2008.06195.x. [DOI] [PubMed] [Google Scholar]
- 36.Venkatesh A, Ma S, Le YZ, Hall MN, Rüegg MA, Punzo C. Activated mTORC1 promotes long-term cone survival in retinitis pigmentosa mice. J Clin Invest. 2015;125(4):1446–1458. doi: 10.1172/JCI79766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu J, et al. cGMP accumulation causes photoreceptor degeneration in CNG channel deficiency: evidence of cGMP cytotoxicity independently of enhanced CNG channel function. J Neurosci. 2013;33(37):14939–14948. doi: 10.1523/JNEUROSCI.0909-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bissinger AL, Fehrle C, Werner CR, Lauer UM, Malek NP, Berg CP. Epidemiology and genotyping of patients with chronic hepatitis B: genotype shifting observed in patients from Central Europe. Pol J Microbiol. 2015;64(1):15–21. [PubMed] [Google Scholar]
- 39.Arango-Gonzalez B, et al. Identification of a common non-apoptotic cell death mechanism in hereditary retinal degeneration. PLoS One. 2014;9(11):e112142. doi: 10.1371/journal.pone.0112142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martínez-François JR, Xu Y, Lu Z. Mutations reveal voltage gating of CNGA1 channels in saturating cGMP. J Gen Physiol. 2009;134(2):151–164. doi: 10.1085/jgp.200910240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fahim AT, et al. Diagnostic fundus autofluorescence patterns in achromatopsia. Am J Ophthalmol. 2013;156(6):1211–1219.e2. doi: 10.1016/j.ajo.2013.06.033. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Huang L, Xiao X, Jia X, Guo X, Zhang Q. Identification of CNGA3 mutations in 46 families: common cause of achromatopsia and cone-rod dystrophies in Chinese patients. JAMA Ophthalmol. 2014;132(9):1076–1083. doi: 10.1001/jamaophthalmol.2014.1032. [DOI] [PubMed] [Google Scholar]
- 43.Saqib MA, et al. Homozygosity mapping reveals novel and known mutations in Pakistani families with inherited retinal dystrophies. Sci Rep. 2015;5:9965. doi: 10.1038/srep09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reuter P, et al. Mutations in CNGA3 impair trafficking or function of cone cyclic nucleotide-gated channels, resulting in achromatopsia. Hum Mutat. 2008;29(10):1228–1236. doi: 10.1002/humu.20790. [DOI] [PubMed] [Google Scholar]
- 45.Pang JJ, et al. AAV-mediated cone rescue in a naturally occurring mouse model of CNGA3-achromatopsia. PLoS ONE. 2012;7(4):e35250. doi: 10.1371/journal.pone.0035250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jobling AI, Vessey KA, Waugh M, Mills SA, Fletcher EL. A naturally occurring mouse model of achromatopsia: characterization of the mutation in cone transducin and subsequent retinal phenotype. Invest Ophthalmol Vis Sci. 2013;54(5):3350–3359. doi: 10.1167/iovs.13-11831. [DOI] [PubMed] [Google Scholar]
- 47.Veleri S, Lazar CH, Chang B, Sieving PA, Banin E, Swaroop A. Biology and therapy of inherited retinal degenerative disease: insights from mouse models. Dis Model Mech. 2015;8(2):109–129. doi: 10.1242/dmm.017913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swaroop A, Kim D, Forrest D. Transcriptional regulation of photoreceptor development and homeostasis in the mammalian retina. Nat Rev Neurosci. 2010;11(8):563–576. doi: 10.1038/nrn2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cunea A, Powner MB, Jeffery G. Death by color: differential cone loss in the aging mouse retina. Neurobiol Aging. 2014;35(11):2584–2591. doi: 10.1016/j.neurobiolaging.2014.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Ng L, et al. A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat Genet. 2001;27(1):94–98. doi: 10.1038/83829. [DOI] [PubMed] [Google Scholar]
- 51.Chan S, et al. Functional comparisons of visual arrestins in rod photoreceptors of transgenic mice. Invest Ophthalmol Vis Sci. 2007;48(5):1968–1975. doi: 10.1167/iovs.06-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nikonov SS, et al. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008;59(3):462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Weiss ER, Ducceschi MH, Horner TJ, Li A, Craft CM, Osawa S. Species-specific differences in expression of G-protein-coupled receptor kinase (GRK) 7 and GRK1 in mammalian cone photoreceptor cells: implications for cone cell phototransduction. J Neurosci. 2001;21(23):9175–9184. doi: 10.1523/JNEUROSCI.21-23-09175.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ding XQ, Fitzgerald JB, Matveev AV, McClellan ME, Elliott MH. Functional activity of photoreceptor cyclic nucleotide-gated channels is dependent on the integrity of cholesterol- and sphingolipid-enriched membrane domains. Biochemistry. 2008;47(12):3677–3687. doi: 10.1021/bi7019645. [DOI] [PubMed] [Google Scholar]
- 55.Yu WP, Grunwald ME, Yau KW. Molecular cloning, functional expression and chromosomal localization of a human homolog of the cyclic nucleotide-gated ion channel of retinal cone photoreceptors. FEBS Lett. 1996;393(2-3):211–215. doi: 10.1016/0014-5793(96)00889-7. [DOI] [PubMed] [Google Scholar]
- 56.Faillace MP, Bernabeu RO, Korenbrot JI. Cellular processing of cone photoreceptor cyclic GMP-gated ion channels: a role for the S4 structural motif. J Biol Chem. 2004;279(21):22643–22653. doi: 10.1074/jbc.M400035200. [DOI] [PubMed] [Google Scholar]
- 57.Kajiwara K, Berson EL, Dryja TP. Digenic retinitis pigmentosa due to mutations at the unlinked peripherin/RDS and ROM1 loci. Science. 1994;264(5165):1604–1608. doi: 10.1126/science.8202715. [DOI] [PubMed] [Google Scholar]
- 58.Dryja TP, Hahn LB, Kajiwara K, Berson EL. Dominant and digenic mutations in the peripherin/RDS and ROM1 genes in retinitis pigmentosa. Invest Ophthalmol Vis Sci. 1997;38(10):1972–1982. [PubMed] [Google Scholar]
- 59.Goldberg AF, Molday RS. Subunit composition of the peripherin/rds-rom-1 disk rim complex from rod photoreceptors: hydrodynamic evidence for a tetrameric quaternary structure. Biochemistry. 1996;35(19):6144–6149. doi: 10.1021/bi960259n. [DOI] [PubMed] [Google Scholar]
- 60.Goldberg AF, Molday RS. Defective subunit assembly underlies a digenic form of retinitis pigmentosa linked to mutations in peripherin/rds and rom-1. Proc Natl Acad Sci U S A. 1996;93(24):13726–13730. doi: 10.1073/pnas.93.24.13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Loewen CJ, Moritz OL, Molday RS. Molecular characterization of peripherin-2 and rom-1 mutants responsible for digenic retinitis pigmentosa. J Biol Chem. 2001;276(25):22388–22396. doi: 10.1074/jbc.M011710200. [DOI] [PubMed] [Google Scholar]
- 62. Forsythe E, Beales PL. Bardet-Biedl syndrome. In: Adam MP, et al, eds. GeneReviews. Seattle, Washington, USA: University of Washington; 1993–2018. [Google Scholar]
- 63.Khan SA, Muhammad N, Khan MA, Kamal A, Rehman ZU, Khan S. Genetics of human Bardet-Biedl syndrome, an updates. Clin Genet. 2016;90(1):3–15. doi: 10.1111/cge.12737. [DOI] [PubMed] [Google Scholar]
- 64.Nachury MV, et al. A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell. 2007;129(6):1201–1213. doi: 10.1016/j.cell.2007.03.053. [DOI] [PubMed] [Google Scholar]
- 65.Seo S, et al. BBS6, BBS10, and BBS12 form a complex with CCT/TRiC family chaperonins and mediate BBSome assembly. Proc Natl Acad Sci U S A. 2010;107(4):1488–1493. doi: 10.1073/pnas.0910268107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katsanis N, et al. Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science. 2001;293(5538):2256–2259. doi: 10.1126/science.1063525. [DOI] [PubMed] [Google Scholar]
- 67.Beales PL, et al. Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet. 2003;72(5):1187–1199. doi: 10.1086/375178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hjortshøj TD, et al. Bardet-Biedl syndrome in Denmark--report of 13 novel sequence variations in six genes. Hum Mutat. 2010;31(4):429–436. doi: 10.1002/humu.21204. [DOI] [PubMed] [Google Scholar]
- 69.Chen J, et al. Molecular analysis of Bardet-Biedl syndrome families: report of 21 novel mutations in 10 genes. Invest Ophthalmol Vis Sci. 2011;52(8):5317–5324. doi: 10.1167/iovs.11-7554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Billingsley G, et al. Mutations in chaperonin-like BBS genes are a major contributor to disease development in a multiethnic Bardet-Biedl syndrome patient population. J Med Genet. 2010;47(7):453–463. doi: 10.1136/jmg.2009.073205. [DOI] [PubMed] [Google Scholar]
- 71.Smaoui N, et al. Screening of the eight BBS genes in Tunisian families: no evidence of triallelism. Invest Ophthalmol Vis Sci. 2006;47(8):3487–3495. doi: 10.1167/iovs.05-1334. [DOI] [PubMed] [Google Scholar]
- 72.Abu-Safieh L, et al. In search of triallelism in Bardet-Biedl syndrome. Eur J Hum Genet. 2012;20(4):420–427. doi: 10.1038/ejhg.2011.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ece Solmaz A, et al. Targeted multi-gene panel testing for the diagnosis of Bardet Biedl syndrome: identification of nine novel mutations across BBS1, BBS2, BBS4, BBS7, BBS9, BBS10 genes. Eur J Med Genet. 2015;58(12):689–694. doi: 10.1016/j.ejmg.2015.10.011. [DOI] [PubMed] [Google Scholar]
- 74.Matveev AV, Quiambao AB, Browning Fitzgerald J, Ding XQ. Native cone photoreceptor cyclic nucleotide-gated channel is a heterotetrameric complex comprising both CNGA3 and CNGB3: a study using the cone-dominant retina of Nrl-/- mice. J Neurochem. 2008;106(5):2042–2055. doi: 10.1111/j.1471-4159.2008.05548.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hüttl S, et al. Impaired channel targeting and retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGB1. J Neurosci. 2005;25(1):130–138. doi: 10.1523/JNEUROSCI.3764-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Landrum MJ, et al. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014;42(Database issue):D980–D985. doi: 10.1093/nar/gkt1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Weisschuh N, et al. Mutation detection in patients with retinal dystrophies using targeted next generation sequencing. PLoS One. 2016;11(1):e0145951. doi: 10.1371/journal.pone.0145951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Weisschuh N, Wissinger B, Gramer E. A splice site mutation in the PAX6 gene which induces exon skipping causes autosomal dominant inherited aniridia. Mol Vis. 2012;18:751–757. [PMC free article] [PubMed] [Google Scholar]
- 79.Mayer AK, et al. Homozygosity mapping and whole-genome sequencing reveals a deep intronic PROM1 mutation causing cone-rod dystrophy by pseudoexon activation. Eur J Hum Genet. 2016;24(3):459–462. doi: 10.1038/ejhg.2015.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tanimoto N, et al. Vision tests in the mouse: Functional phenotyping with electroretinography. Front Biosci (Landmark Ed) 2009;14:2730–2737. doi: 10.2741/3409. [DOI] [PubMed] [Google Scholar]
- 81.Tanimoto N, Sothilingam V, Kondo M, Biel M, Humphries P, Seeliger MW. Electroretinographic assessment of rod- and cone-mediated bipolar cell pathways using flicker stimuli in mice. Sci Rep. 2015;5:10731. doi: 10.1038/srep10731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Seeliger MW, et al. In vivo confocal imaging of the retina in animal models using scanning laser ophthalmoscopy. Vision Res. 2005;45(28):3512–3519. doi: 10.1016/j.visres.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 83.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 84.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29(9):e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Michalakis S, Geiger H, Haverkamp S, Hofmann F, Gerstner A, Biel M. Impaired opsin targeting and cone photoreceptor migration in the retina of mice lacking the cyclic nucleotide-gated channel CNGA3. Invest Ophthalmol Vis Sci. 2005;46(4):1516–1524. doi: 10.1167/iovs.04-1503. [DOI] [PubMed] [Google Scholar]
- 86.Zhang T, Baehr W, Fu Y. Chemical chaperone TUDCA preserves cone photoreceptors in a mouse model of Leber congenital amaurosis. Invest Ophthalmol Vis Sci. 2012;53(7):3349–3356. doi: 10.1167/iovs.12-9851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Michalakis S, et al. Mosaic synaptopathy and functional defects in Cav1.4 heterozygous mice and human carriers of CSNB2. Hum Mol Genet. 2014;23(6):1538–1550. doi: 10.1093/hmg/ddt541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Khan NW, Wissinger B, Kohl S, Sieving PA. CNGB3 achromatopsia with progressive loss of residual cone function and impaired rod-mediated function. Invest Ophthalmol Vis Sci. 2007;48(8):3864–3871. doi: 10.1167/iovs.06-1521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.