Abstract
Chronic lymphocytic leukemia (CLL) is characterized by clonal proliferation and progressive accumulation of mature B lymphocytes in the peripheral blood, lymphoid tissues, and bone marrow. CLL is characterized by profound immune defects leading to severe infectious complications. T cells are numerically, phenotypically, and functionally highly abnormal in CLL, with only limited ability to exert antitumor immune responses. Exhaustion of T cells has also been suggested to play an important role in antitumor responses. CLL-mediated T cell exhaustion is achieved by the aberrant expression of several inhibitory molecules on CLL cells and their microenvironment, prominently the programmed cell death ligand 1/programmed cell death 1 (PD-L1/PD-1) receptors. Previously, we showed that CD84, a member of the SLAM family of receptors, bridges between CLL cells and their microenvironment. In the current study, we followed CD84 regulation of T cell function. We showed that cell-cell interaction mediated through human and mouse CD84 upregulates PD-L1 expression on CLL cells and in their microenvironment and PD-1 expression on T cells. This resulted in suppression of T cell responses and activity in vitro and in vivo. Thus, our results demonstrate a role for CD84 in the regulation of immune checkpoints by leukemia cells and identify CD84 blockade as a therapeutic strategy to reverse tumor-induced immune suppression.
Keywords: Immunology, Oncology
Keywords: Cellular immune response, Leukemias
Introduction
Chronic lymphocytic leukemia (CLL) is the most common leukemia in the Western world. The disease is characterized by clonal proliferation and continuous accumulation of mature CD5+ B lymphocytes in the peripheral blood (PB), lymphoid tissues, and bone marrow (BM) (1). This accumulation of cells results from decreased apoptosis rather than overproliferation.
Malignant cells can evade the host immune system and its tumor surveillance mechanisms by inhibiting T cell immunity. Immunosuppression is a prevailing clinical feature in patients with CLL, with many of these patients demonstrating increased susceptibility to infections, as well as increased failure to induce an antitumor immune response. T cell exhaustion represents a continuous spectrum of cellular dysfunction induced during chronic viral infection, autoimmunity, or cancer. Exhaustion has also been implicated in playing an important role in antitumor responses, whereby exhausted tumor-infiltrating lymphocytes fail to control tumor progression (2). Programmed cell death 1 (PD-1, also known as CD279) and its ligands programmed cell death ligand 1 (PD-L1, also known as B7-H1 and CD274) and PD-L2 (also known as B7-DC and CD273) have been suggested to be the most important axis in the maintenance of a malignant prosurvival microenvironment (3). Exhausted T cells also express cytotoxic T lymphocyte antigen-4 (CTLA-4), lymphocyte activation gene 3 (LAG-3), Tim-3, 2B4, CD160, and other markers (4).
TCL1-transgenic mice are a commonly used murine model for CLL studies. These transgenic mice express the human TCL1 gene under the control of a VH chain promoter-IgH-Eμ enhancer, thereby targeting its expression to B cells. Mice overexpressing TCL1 develop a CLL-like disease that resembles a more advanced-stage disease and occurs at a rather old age, much like the human pathology (5).
Dynamic interactions between cell-surface molecules orchestrate the immune response. The signaling lymphocyte activation molecule (SLAM) family includes 9 receptors that modulate immune responses through homophilic and heterophilic interactions (6). CD84 is a member of the SLAM family. It is a cell-surface protein that forms homophilic dimers by self-association (7–9). Our studies have previously characterized a survival pathway in CLL regulated by CD84 (10). In addition, we recently showed that CD84 serves as an important bridge mediating the interaction between CLL cells and the various cells in their microenvironment in vitro and in vivo (11).
In the current study, we examined downstream events following CD84 ligation on CLL cells and their stroma. Our results showed an elevation of PD-L1 expression in CD84-activated CLL and stromal cells. Downregulation of CD84 expression reduced PD-L1 expression levels on CLL cells and in the CLL microenvironment and also reduced the expression of PD-1 and additional exhaustion marker on T cells. This led to an increase in antitumor T cell activity. Thus, our results reveal a role for CD84 in the regulation of immune checkpoint expression by leukemia cells and provide a therapeutic strategy for blocking CD84 and thus restoring T cell function.
Results
CD84 activation upregulates PD-L1 expression on CLL cells and in their microenvironment.
To analyze the mechanism of action of CD84 in regulating crosstalk between CLL cells and their microenvironment, we used genome-wide gene expression profiling to search for target genes induced by CD84 engagement in primary CLL and M210B4 stromal cells, which are known to support CLL cell survival (11, 12). We identified a set of genes differentially expressed between the control and CD84-activated fractions (Gene Expression Omnibus [GEO] number GSE107140) (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI96610DS1). PD-L1 was one of the genes that was upregulated in both cell types.
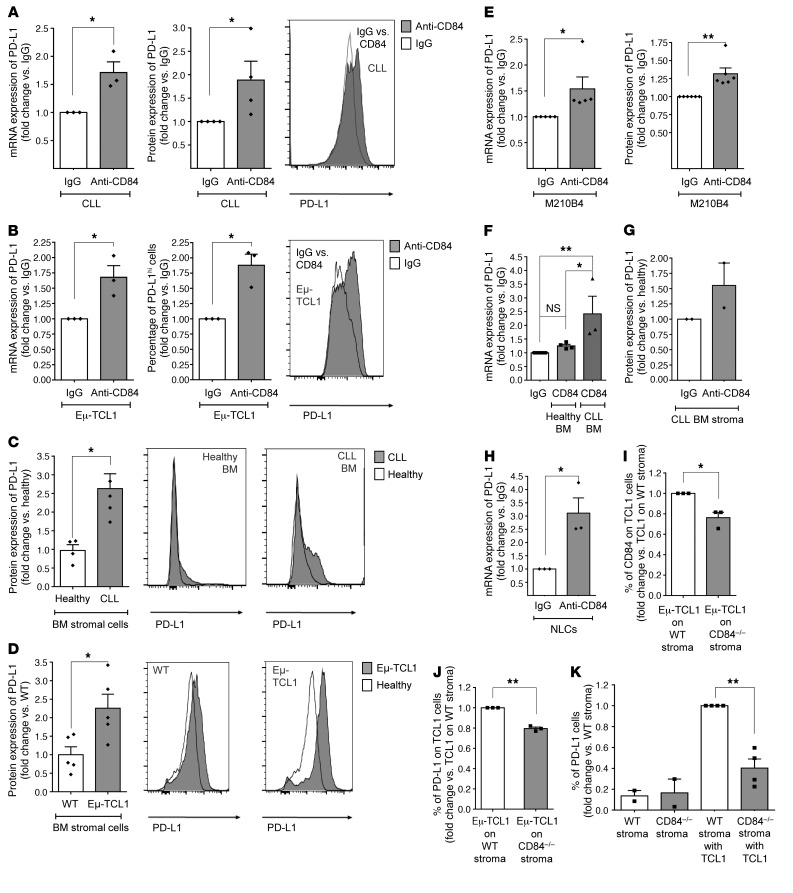
As previously described, PD-L1 cell-surface levels are significantly upregulated on CLL cells compared with expression on healthy B cells (ref. 13 and Supplemental Figure 2A). To directly show the regulation of PD-L1 expression by CD84, human (Figure 1A) and murine (Eμ-TCL1) (Figure 1B) CLL cells were stimulated with anti-CD84–activating antibody (10, 11). We observed that PD-L1 mRNA and protein levels were significantly elevated in both human and murine CLL cells following CD84 activation. We next examined the effect of CD84 on PD-L1 expression in stromal cells. First, we compared PD-L1 expression levels on healthy and CLL-derived BM stromal cells (CD34–CD45–) (Supplemental Figure 2, B and C). We detected elevated levels of PD-L1 on CLL-derived stromal cells (Figure 1C), which have previously been shown to express high levels of CD84 (11). We also detected an increase in PD-L1 levels on BM stromal cells derived from Eμ-TCL1–transgenic mice (a model of aggressive CLL that overexpresses the human TCL1 gene) (Figure 1D), which express higher levels of CD84 compared with stromal cells derived from healthy mice (11).
Figure 1. CD84 regulates PD-L1 expression on human and murine CLL cells and cells in their microenvironment.
(A and B) CLL cells derived from patients (at different stages of disease: n = 3 Binet A, n = 1 Binet B, n = 1 Rai II, n = 1 Binet C, and n = 1 Rai III) (A) or from Eμ-TCL1 CLL mice (B) were stimulated with anti-CD84 or control IgG (5 μg/ml) antibodies, and PD-L1 mRNA and protein levels were determined by qRT-PCR and flow cytometry, respectively. *P < 0.05, 1-tailed, paired t test (A, right), 2-tailed, paired t test (A, left, and B). n = 3 (A) and n = 4 (B). Representative histograms are shown. (C and D) BM stromal cells derived from human CLL samples (n = 4) and healthy patients (n = 4) (C), or Eμ-TCL1 (n = 5) and healthy (n = 5) mice (D) were analyzed by FACS for PD-L1 expression levels. Representative histograms are shown. IgG is shown in white and anti-CD84 in gray. *P < 0.05, 2-tailed, paired t test (C and D). (E) M210B4 stromal cells were stimulated with IgG or anti-CD84 (4 μg/ml) antibody for 24 or 48 hours. PD-L1 mRNA levels were analyzed by qRT-PCR (n = 5), and protein levels were analyzed by flow cytometry (n = 6). *P < 0.05, 2-tailed, paired t test and **P < 0.01, 1 tailed, paired t test. (F–H) BM stromal cells derived from healthy volunteers or CLL patients (F and G) or human NLCs (H) were stimulated with 4 μg/ml anti-CD84 or control antibodies for 24 hours for mRNA expression or 48 hours for protein expression. PD-L1 mRNA levels were analyzed by qRT-PCR using PSMB as a housekeeping control gene (n = 3), and protein levels were determined by flow cytometry (n = 2). *P < 0.05 and **P < 0.01, 1-way ANOVA with Holm-Sidak–corrected multiple comparisons (F) and 2-tailed, paired t test (H). (I–K) Eμ-TCL1 splenocytes were seeded on near-confluent WT or CD84–/– stroma. Forty-eight hours later, protein levels of CD84 and PD-L1 were measured on TCL1 (gated on CD5+CD19+) cells (I and J) and on stroma before and after addition of TCL1 cells (K). n = 3–4 TCL1 mice. *P < 0.05 and **P < 0.01, 2-tailed, paired t test.
To corroborate the regulation of PD-L1 expression by CD84 in the tumor microenvironment, we analyzed PD-L1 expression on the CD84-stimulated stromal cell line M210B4 (Figure 1E), on stromal cells derived from CLL patients and healthy volunteers (Figure 1, F and G), and on monocyte-derived nurse-like cells (NLCs) (Figure 1H). We detected a significant increase in the levels of PD-L1 mRNA (Figure 1, F and H) and protein (Figure 1G) in all 3 cell types following stimulation. Next, we examined the expression of CD84 and PD-L1 on additional cells derived from the tumor microenvironment. Macrophages (MQs) and DCs expressed basal levels of CD84 (Supplemental Figure 2, D and E, and ref. 11). Furthermore, healthy CD84–/– DCs and MQs expressed levels of PD-L1 similar to those in WT cells (Supplemental Figure 2, G and H). Activation of CD84 expressed on healthy cells induced only a modest elevation of PD-L1 expression (Supplemental Figure 2, I–K) compared with that seen on cells derived from the tumor microenvironment. These results suggest that the low expression levels of CD84 on healthy cells induced only a minor change in PD-L1 levels, indicating an effect of the microenvironment on expression of CD84 and regulation of the immune response.
Next, to determine whether the malignant cells regulate PD-L1 expression on the stroma in a CD84-dependent manner, we incubated TCL1 splenocytes alone or in the presence of WT or CD84–/– stromal cells. Forty-eight hours later, CD84 (Figure 1I) and PD-L1 (Figure 1J) protein levels were measured on TCL1 and stromal cells (Figure 1K). PD-L1 expression was significantly reduced on both cell types, whereas a more significant decrease (~60%) in PD-L1 expression was detected on the stroma. This decrease did not result from the lack of CD84 on these cells, but rather from the lack of CD84-dependent contact with the malignant cells, since the difference was not detected on stromal cells cultured alone (Figure 1K). Thus, CD84 controls PD-L1 expression on CLL cells and their stroma.
CD84 ligation upregulates PD-L1 expression through the AKT/mTOR pathway.
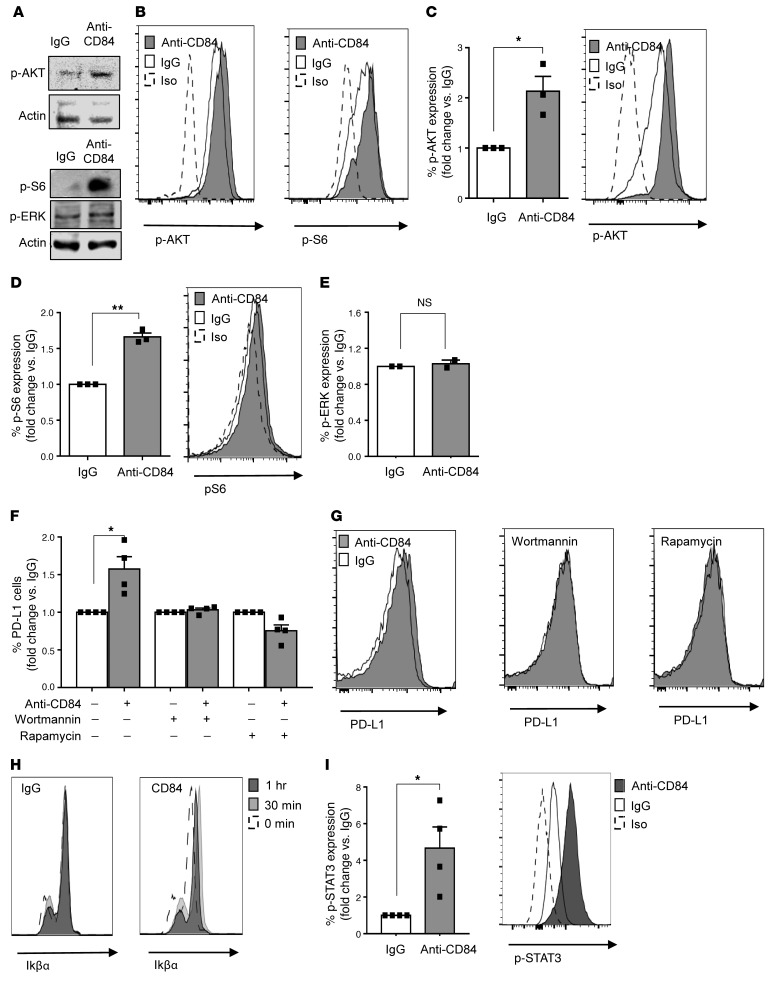
We next identified the CD84-induced cascades that elevate PD-L1 expression. AKT/mTOR and the MAPK pathways were previously shown to regulate PD-L1 expression (14). Phosphorylation of S6 is a downstream event following mTOR activation, a known regulator of PD-L1 expression in lung cancer (15). We detected an increase in the phosphorylation of AKT (Supplemental Figure 2I), ERK (Supplemental Figure 2J), and S6 (Supplemental Figure 2K) in the PD-L1hi–expressing Eμ-TCL1 cells. Therefore, we analyzed the activation of these pathways in CD84-stimulated M210B4 cells. While no change in the MAPK pathway activation was detected (Figure 2A), we found that AKT activation was markedly increased upon CD84 stimulation and that the levels of phosphorylated AKT (p-AKT) as well as p-S6 were elevated (Figure 2A). These results were confirmed by flow cytometry, which showed elevation of p-AKT and p-S6 levels in CD84-activated cells (Figure 2B). Next, we analyzed activation of the AKT/mTOR pathway in primary CLL cells. As with our results in stromal cells, CD84 activation induced the phosphorylation of AKT (Figure 2C) and S6 (Figure 2D), while no change was observed in ERK phosphorylation (Figure 2E). These results suggest that CD84 increases PD-L1 protein expression through activation of the AKT/mTOR pathway and p-S6.
Figure 2. Activation of cell-surface CD84 elevates PD-L1 levels through the p-AKT/mTOR pathway.
(A and B) M210B4 cells (1 × 105) were stimulated with anti-CD84 (4 μg/ml) for 15 minutes, followed by anti-FAB cross-linking for 5 minutes. (A) Cells were lysed, and lysates were separated on 12% (wt/vol) SDS-PAGE and blotted with anti–p-S6, anti–p-AKT, anti–p-ERK, or actin. Blots are representative of 3 independent experiments. (B) Cells were fixed, permeabilized, and subsequently stained with anti–p-AKT and anti–p-S6 antibodies followed by a secondary anti-rabbit allophycocyanin-conjugated (APC-conjugated) antibody. Histograms are representative of 2 independent experiments. (C–E) Primary CLL cells were stimulated with control (IgG) or anti-CD84–activating (5 μg/ml) antibodies for 5 minutes, followed by anti-FAB cross-linking for an additional 5 minutes. Then, the cells were fixed, and p-ERK, p-AKT, and p-S6 levels were analyzed by flow cytometry (n = 3; *P < 0.05 and **P < 0.01, 2-tailed, paired t test). (F and G) CLL cells were incubated with the inhibitors wortmannin and rapamycin (0.1 μM) for 20 minutes. The cells were then stimulated with anti-CD84–activating (5 μg/ml) antibodies, or IgG control. After 48 hours, the cells were analyzed by FACS for PD-L1 cell-surface expression (n = 4; *P < 0.05, 2-tailed, paired t test). (H and I) CLL cells were stimulated with IgG or anti-CD84–activating (5 μg/ml) antibodies for 0, 15, and 30 minutes, followed by anti-FAB cross-linking for 0, 15, and 30 minutes. Cells were then fixed, permeabilized, and stained with IkBα (n = 2) (H), or p-STAT3 (n = 4) (I) antibodies. *P < 0.05, 2-tailed, paired t test.
To directly prove that PD-L1 upregulation is dependent on the AKT/mTOR pathway, CD84 expression on CLL cells, which were incubated in the presence or absence of AKT (0.1 μM Wortmannin) or mTOR (0.1 μM rapamycin) inhibitors, was activated as previously described (11). As demonstrated in Figure 2, F and G, the 2 inhibitors completely abrogated the CD84-induced expression of PD-L1, showing that CD84 upregulates PD-L1 through the activation of AKT and mTOR.
To gain additional insight into the transcriptional consequences that might promote PD-L1 expression, we analyzed possible transcription factors previously shown to regulate PD-L1 expression (16) following CD84 activation. NF-κB has been shown to regulate PD-L1, and is a major mediator of IFN-γ–induced PD-L1 expression (17). To determine whether CD84 activation involves the NF-κB pathway, we examined the degradation of its inhibitor, IκB, at different time points following CD84 activation. As shown in Figure 2H, no degradation (and even a slight elevation) of IκB was detected following activation, suggesting that this pathway is not activated by CD84.
STAT3 binds the PD-L1 promoter and regulates its expression (16). We therefore next analyzed p-STAT3 levels following CD84 activation. As shown in Figure 2I, p-STAT3 levels significantly increased upon CD84 activation, suggesting the involvement of STAT3 in the CD84-induced cascade.
CD84 regulates PD-L1 expression in vivo.
To analyze CD84-mediated regulation of PD-L1 expression in vivo, we used a transfer model in which splenocytes derived from sick Eμ-TCL1 mice were injected into coisogenic WT or CD84–/– mice, as previously reported (11). As previously shown, lack of CD84 in the tumor environment resulted in a reduced number of malignant cells 2 weeks (11) and 5 weeks (Supplemental Figure 3, A and B) after cell transfer and prolonged survival of the tumor-bearing mice (Supplemental Figure 3C).
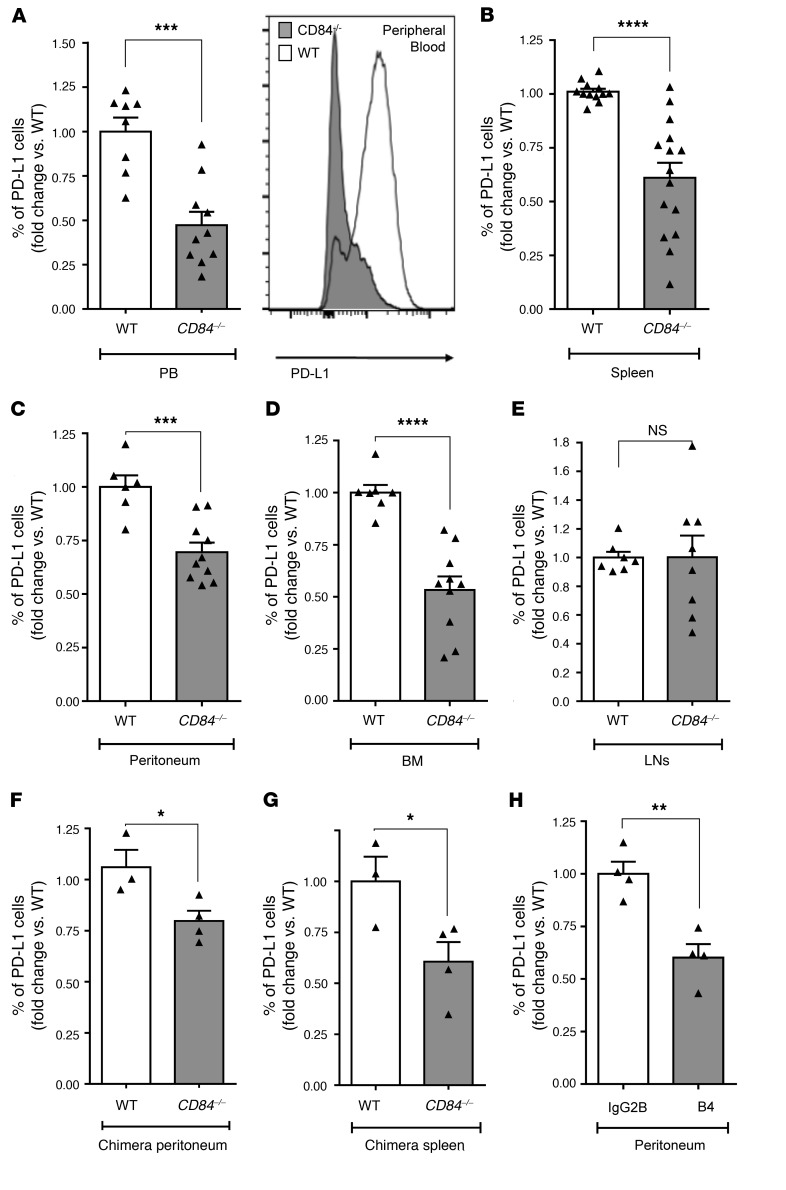
CD84 regulation of PD-L1 expression was specific to the tumor microenvironment, since we detected similar PD-L1 levels on the cell surface of B cells derived from WT and CD84-deficient mice (Supplemental Figure 3D). Next, we analyzed the expression of PD-L1 on the Eμ-TCL1 cells retrieved from PB (Figure 3A and Supplemental Figure 3E), spleen (Figure 3B and Supplemental Figure 3F), peritoneum (Figure 3C and Supplemental Figure 3G), and BM (Figure 3D and Supplemental Figure 3H) two to three weeks after cell transfer. In these compartments, we detected a significant reduction in PD-L1 expression on cells derived from the CD84-deficient, as compared with the CD84-positive, environments. However, no change in PD-L1 expression was detected on Eμ-TCL1 cells in lymph node (LN) cells (Figure 3E). The reduction of PD-L1 expression did not result from cell death, as a similar decrease in PD-L1 expression was detected on a gated population of annexin V– and 7AAD– live Eμ-TCL1 cells (Supplemental Figure 3I). In addition, the reduction in PD-L1 expression did not result from a reduced tumor load in the various compartments in mice lacking CD84, because although the tumor load in the LNs was higher compared with the load in the BM (Supplemental Figure 3J), no change in PD-L1 expression was detected in this organ. These results indicate a role for CD84 in the regulation of PD-L1 expression on tumor cells.
Figure 3. Decreased PD-L1 expression in vivo on Eμ-TCL1 cells derived from CD84–/– mice.
(A–E) Eμ-TCL1 splenocytes (4 × 107) were injected i.v. into the tail vein of C57BL/6 WT or CD84–/– mice. After 14 to 21 days, the mice were sacrificed, and expression of PD-L1 was determined by flow cytometry on CD5/CD19 Eμ–TCL1 cells in (A) PB (n = 8–10), (B) spleen (n = 12–14), (C) peritoneum (n = 6–10), (D) BM (n = 7–10), and (E) LNs (n = 7–8). A representative histogram of PD-L1 expression on PB cells is shown in A. ***P < 0.001, 2-tailed t test (A), ****P < 0.0001, 2-tailed t test (B–D), and P = 0.9893 (NS), 2-tailed t test (E). (F and G) BM cells (5 × 106) derived from 8-week-old Eμ-TCL1 mice or negative control littermates (WT) were injected into lethally irradiated C57BL/6 (WT) or CD84-deficient (CD84–/–) mice. After 6 months, the mice were euthanized, and the expression of PD-L1 was determined on Eμ-TCL1 cells in the (F) peritoneum (n = 3–4) and (G) spleen (n = 3–4). *P < 0.05, 2-tailed t test. (H) C57BL/6 mice were injected with 4 × 107 Eμ-TCL1 splenocytes and, starting from the second day, were treated i.v. with 1 mg/kg body weight B4 or isotype control antibodies. After 14 days, the mice were sacrificed, and PD-L1 expression was determined on CD5+CD19+ peritoneal cells (n = 4; **P < 0.01, 1-tailed t test).
Next, we examined the regulation of PD-L1 expression by CD84 in long-term experiments in BM chimeric mice. Lethally irradiated WT or CD84−/− mice were injected with BM cells derived from age-matched Eμ-TCL1 or WT mice. After 6 months, we analyzed the Eμ-TCL1 cell populations in each compartment. A significant decrease in PD-L1 expression was detected on Eμ-TCL1 cells derived from the CD84– microenvironment in the peritoneum and spleen (Figure 3, F and G). These results suggest that CD84 expressed within the microenvironment regulates PD-L1 expression on malignant cells and that, in the absence of CD84, PD-L1 levels are reduced.
Finally, to further determine the role of CD84 in regulating PD-L1 expression in vivo, CD84 was blocked on Eμ-TCL1 cells using the anti-CD84–antagonistic B4 antibody (10, 11). As shown in Figure 3H, blocking CD84 lowered PD-L1 expression levels on Eμ-TCL1 cells derived from the peritoneal cavity.
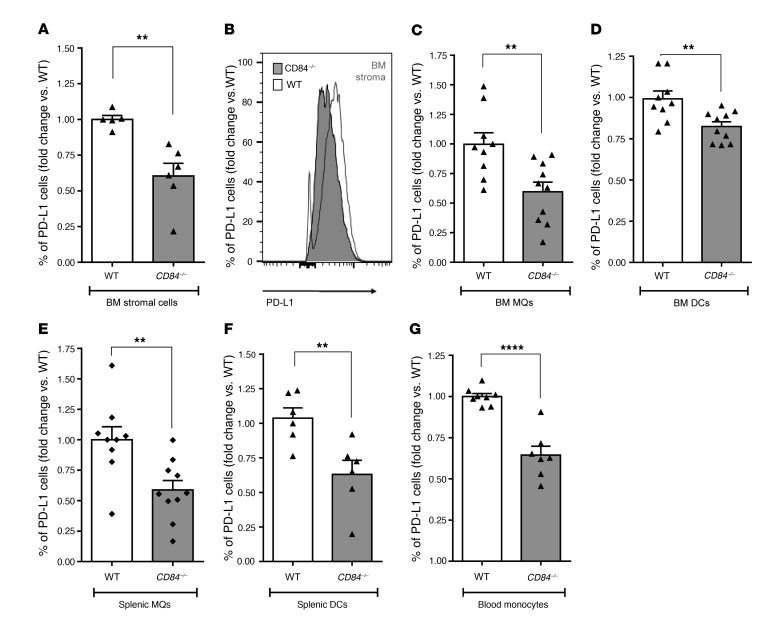
To investigate cells within the tumor microenvironment, we analyzed PD-L1 expression on cells retrieved following transfer of Eμ-TCL1 cells into CD84-proficient and -deficient animals. As shown in Figure 4, compared with controls, a significant decrease in PD-L1 expression was detected on BM stromal cells (Figure 4, A and B), BM MQs (Figure 4C), BM DCs (Figure 4D), splenic MQs (Figure 4E), splenic DCs (Figure 4F), and PB monocytes (Figure 4G) derived from the CD84-deficient environment, while no difference in PD-L1 expression was detected on these cells derived from the microenvironment of mice prior to TCL1 injection (Supplemental Figure 3, K and L). These results indicate a CD84-mediated global regulation of PD-L1 expression in the tumor microenvironment.
Figure 4. Decreased PD-L1 expression on cells in the microenvironment in the absence of CD84.
(A–G) Eμ-TCL1 splenocytes (4 × 107) were injected i.v. into the tail vein of C57BL/6 WT or CD84–/– mice. After 14 to 21 days, the mice were sacrificed, and expression of PD-L1 was determined on (A and B) BM stromal cells (a representative histogram is shown in B) (n = 9–10); (C) BM MQs (n = 9–10); (D) BM DCs (n = 9–10); (E) splenic MQs (n = 5–6); (F) splenic DCs (n = 6); and (G) PB inflammatory monocytes (n = 7–9). **P < 0.01 (A and C–F) and ****P < 0.0001 (G), 2-tailed t test.
T cells retrieved from the CD84-deficient CLL microenvironment show a decrease in PD-1 expression and induced T cell functionality.
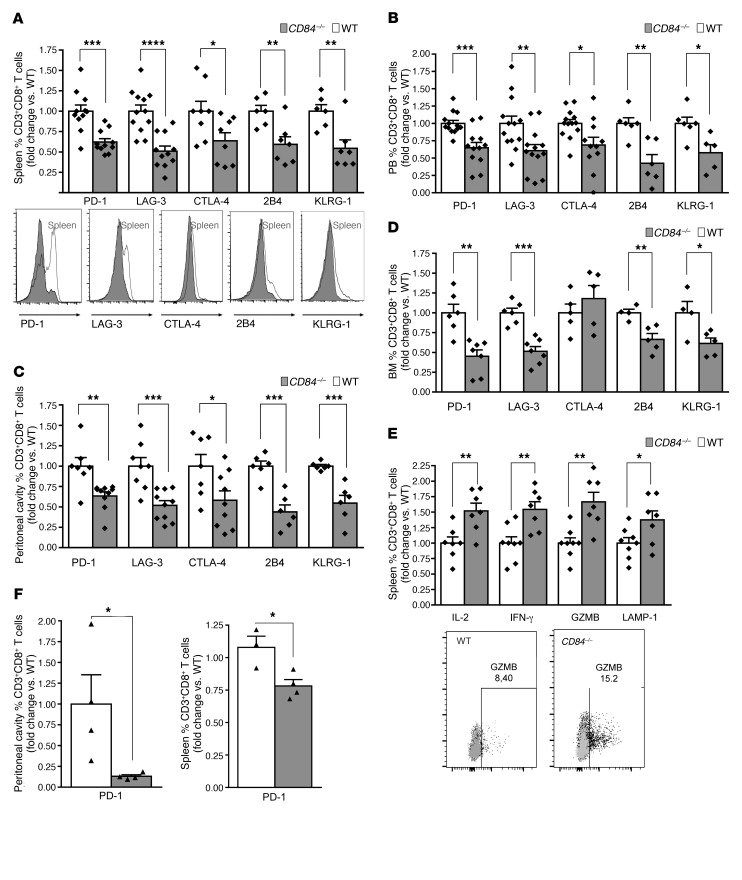
Given that PD-L1 is known as a negative regulator of T cell activation following its binding to PD-1 (18), we next analyzed PD-1 expression on T cells in the Eμ-TCL1 cells transferred into CD84–/– versus WT environments. We analyzed expression levels of PD-1 and additional T cell exhaustion markers on T cells retrieved from spleen (Figure 5A and Supplemental Figure 4A), PB (Figure 5B), peritoneum (Figure 5C), and BM (Figure 5D). A significant reduction in the expression of these exhaustion markers was detected on CD8+ T cells derived from these organs (Figure 5, A–D). Interestingly, we detected no change in the expression of these markers on cells derived from LN (Supplemental Figure 4B), further suggesting that CD84 does not play a role in this environment. We detected less dramatic changes on CD4+ T cells derived from Eμ-TCL1–injected mice (Supplemental Figure 4, C–I), suggesting a less significant effect of CD84 on this cell population.
Figure 5. Reduced abundance of exhausted CD8+ T cells in the CD84–/– environment.
Eμ-TCL1 splenocytes (4 × 107) were injected i.v. into the tail vein of C57BL/6 WT or CD84–/– mice. (A–D) After 14 to 21 days, the mice were sacrificed, and expression of the exhaustion markers PD-1, LAG-3, CTLA-4, 2B4, and KLRG1 were determined on CD8+ T cells in (A) spleen (n = 6–11) histograms show representative results; (B) PB (n = 5–11); (C) the peritoneal cavity (n = 6–10); and (D) BM (n = 4–7). (E) After 14 to 21 days, the mice were sacrificed, and splenic cells were cultured for 24 hours with anti-CD3 and then with brefeldin A for the last 2 hours of culture. CD8+ T cells were then analyzed for IFN-γ, IL-2, GZMB, and LAMP-1 expression (n = 7–8). Dot plot show representative results of granzyme b expression. (F and G) BM cells (5 × 106) derived from 8-week-old Eμ-TCL1 mice or negative control littermates (WT) were injected into lethally irradiated C57BL/6 (WT) or CD84-deficient (CD84−/−) mice. After 6 months, the mice were euthanized, and the expression of PD-1 was determined on CD8+ T cells in the peritoneum (F) and spleen (G) (n = 3–4). *P < 0.05 (A–G), **P < 0.01 (A–E), ***P < 0.001 (A–D), and ****P < 0.0001 (A), 2-tailed t test. P = 0.3172 (NS), 2-tailed t test (E).
Next, we compared the differentiation and functionality of the CD8+ T cells derived from the transferred models. We analyzed T central memory (TCM) (CD44+CD62L+) and T effector memory (TEM) (CD44+CD62L–) cell populations in the TCL1 transfer model. As shown in Supplemental Figure 5, A–D, we detected an elevation in these cell populations in CD84–/– mice transferred with Eμ-TCL1 cells, suggesting an increased functionality of cells in an environment deficient in CD84. In addition, we detected higher levels of IL-2, IFN-γ, granzyme B (GZMB), and LAMP-1 in CD8+ splenic T cells derived from the Eμ-TCL1 cells transferred into the CD84–/– environment, confirming that in the absence of CD84, CD8+ T cells display a more activated phenotype (Figure 5E).
To study CD84 regulation of PD-1 expression on T cells in a long-term model, we analyzed PD-1 expression levels on CD8+ T cells derived from chimeric mice, whereby BM derived from Eμ-TCL1 mice was injected into WT or CD84-deficient environments. As shown in Figure 5, F and G, we detected a significant decrease in PD-1 expression levels on CD8+ T cells derived from the CD84–/– chimeras.
To exclude the possibility that changes in T cell activity result from intrinsic differences caused by CD84, we activated WT and CD84–/– T cells in whole splenic cultures with anti-CD3 and analyzed their exhaustion markers. As shown in Supplemental Figure 6, we observed no significant differences in the expression of PD-1, LAG-3, CTLA-4, 2B4, or KLRG-1 (Supplemental Figure 6, A and B), or in IL-2, IFN-γ, GZMB, or LAMP-1 (Supplemental Figure 6C), findings that agree with previous studies showing similar levels of cytokine secretion in WT and CD84–/– T cells (19).
The regulation of PD-1 expression on T cells and their functionality may result from a direct effect of CD84 expression on these cells or, alternatively, from the regulation of PD-L1 expression and cytokine secretion on the malignant cells and in their microenvironment, affecting T cell function. To determine whether CD84 activation on T cells directly affects their functionality, we stimulated CD84 expression on T cells derived from Eμ-TCL1 mice and analyzed the expression of PD-1, LAG-3, CTLA-4, 2B4 and KLRG-1. As shown in Supplemental Figure 6D, a significant, but modest, elevation in the expression of these molecules was detected following CD84 activation, suggesting that the exhausted phenotype of TCL1-derived T cells results, at least in part, from a direct effect of T cell–expressed CD84 on T cell functionality.
Regulation of CLL-derived human T cell activity.
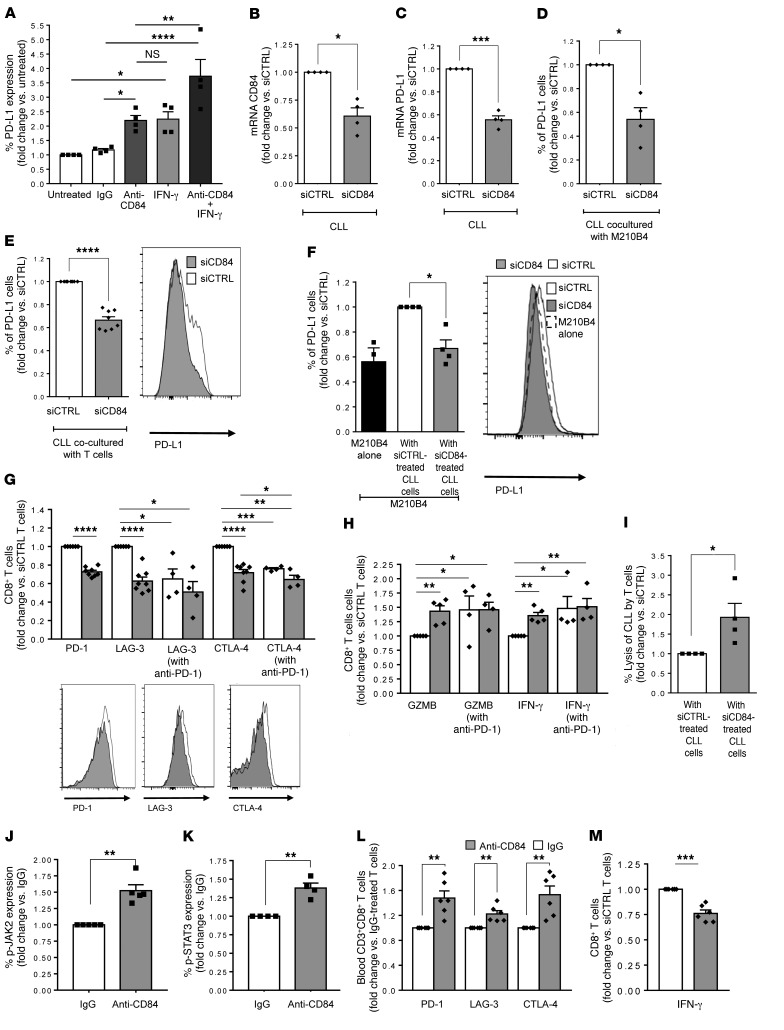
Our murine models demonstrated the regulation of PD-L1/PD-1 expression by CD84. We next sought to determine whether CD84 regulates PD-L1 expression and T cell function in cells derived from patients with CLL. To this end, purified PB CLL cells (Supplemental Figure 7A) were stimulated with anti-CD84–activating antibody (11), and PD-L1 cell-surface expression levels were analyzed. As shown in Figure 6A, cell-surface levels of PD-L1 were significantly elevated following CD84 activation. To compare the induction of PD-L1 expression following ligation of CD84 with IFN-γ–induced activation, we incubated purified CLL cells with either activating anti-CD84 antibody or IFN-γ (500 IU/ml) for 48 hours. We found that both inducers upregulated the expression of PD-L1 to similar levels. Furthermore, incubation of CLL cells with both inducers together resulted in an even greater elevation of PD-L1 protein expression, suggesting that these 2 inducers affect different pathways and can work additively.
Figure 6. Downregulation of CD84 expression in human CLL reduces PD-L1 on CLLs and stroma and induces T cell activity.
(A) Primary human CLL cells were incubated for 48 hours in the presence or absence of anti-CD84–activating or control (IgG) (5 μg/ml) antibodies, IFN-γ (500 IU/ml), or both treatments together. Cells were then analyzed by FACS for PD-L1 cell-surface expression (n = 4; *P < 0.05 , **P < 0.01, and ****P < 0.0001, 1-way ANOVA with Holm-Sidak–corrected multiple comparisons). (B and C) CLL cells were treated with siCTRL or siCD84 for 24 hours, and levels of (B) CD84 (n = 4; *P < 0.05) and (C) PD-L1 (n = 4–5; ***P < 0.001, 2-tailed, paired t test) were analyzed. (D–F) The siRNA-treated cells were then incubated for 48 hours with M210B4 (D and F), or T cells (E, G, and H) derived from the same patient. (D and E) PD-L1 cell-surface expression on CLL cells derived from the coculture was analyzed by FACS. E shows a representative histogram. n = 4, *P < 0.05 (D) and n = 11, ****P < 0.0001 (E), 2-tailed, paired t test. (F) M210B4 cells were stained for PD-L1 cell-surface expression. A representative histogram is shown (n = 4; *P < 0.05, 2-tailed, paired t test). (G and H) CLL cells were treated with siCTRL or siCD84. The siRNA-treated cells were then incubated in the presence or absence T cells treated with anti–PD-1–blocking antibody (5 μg/ml) for 24 hours prior to coculture. (G) CD8+ T cells were analyzed for expression of the exhaustion markers PD-1, LAG-3, and CTLA-4 (n = 4). Representative histograms are shown in G. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001, 2-tailed, paired t test for siCTRL and siCD84, and 1-way ANOVA with Fisher’s least significant difference (LSD) for the others. (H) Cultures were incubated with anti-CD3 (0.5 μg/ml) and brefeldin A for the last 2 hours. T cells were then analyzed for GZMB and IFN-γ expression. Graph show the percentage of CD8+ T cells that expressed GZMB and IFN-γ with and without PD-1 inhibition (5 μg/ml, clone EH12.2H7) (n = 4; *P < 0.05 and **P < 0.01, 2-tailed, paired t test for siCTRL and siCD84, and 1-way ANOVA with Fisher’s LSD for the others). (I) CLL cell lysis by the T cells in B was determined with the CytoTox-ONE Kit (Promega) (n = 4; *P < 0.05, 2-tailed, paired t test). (J–M) Purified human T cells were treated with anti-CD84 (5 μg/ml) and analyzed for p-JAK2 (I), p-STAT3 (K), PD-1, LAG-3, and CTLA-4 (L), and IFN-γ expression (M) (n = 4–6; **P < 0.01 and ***P < 0.001, 2-tailed, paired t test).
Next, CD84 expression levels were downregulated with siCD84 or siControl (siCTRL). Reduced expression of CD84 in CLL cells (Figure 6B) resulted in the downregulation of PD-L1 mRNA (Figure 6C) and protein expression on CLL cells cultured with M210B4 cells (Figure 6D) or non-CLL PB cells (which contained the T cells) (Figure 6E). Interestingly, downregulation of CD84 expression in CLL cells also reduced CD84 expression levels on stromal cells (Supplemental Figure 7B). The reduced levels of CD84 in CLL cells lowered the levels of PD-L1 expressed on M210B4 to levels detected on M210B4 cells cultured without CLL cells (Figure 6F). These results suggest that CD84 mediates the interaction between CLL cells and their stromal cells and that this interaction induces PD-L1 expression on these cells.
Interestingly, T cells incubated with the siCD84-treated CLL cells showed reduced expression levels of the exhaustion markers PD-1, LAG-3, and CTLA-4 (Figure 6G). Moreover, downregulation of CD84 expression in CLL cells resulted in elevated expression of IFN-γ and GZMB (Figure 6H), further indicating a role for CD84 in the regulation of T cell functionality. To compare the CD84-dependent regulation of exhaustion marker expression on T cells with anti–PD-1 treatment, we incubated T cells with siCD84-treated CLL cells in the presence or absence of anti–PD-1 antibody. As shown in Figure 6, G and H, reduced expression of CD84 or blockage of PD-1 activity lowered the expression of T cell exhaustion markers and elevated the expression of GZMB and IFN-γ to similar levels. Interestingly, blocking PD-1 did not have an additive effect on the expression of exhaustion markers and functionality of T cells. Thus, CD84 expressed on CLL cells regulates the functionality of T cells. As shown in the murine model, the effect on CD8+ T cells was more striking than that observed on the CD4+ T cell population (Figure 6G and Supplemental Figure 7C).
Next, to directly demonstrate that CD84 deficiency induces T cell activity in killing CLL cells, we performed a cytotoxicity assay comparing the activity of T cells cultured with siCTRL or siCD84-treated CLL cells. To exclude the direct effect of siCD84 treatment on CLL cell survival, the killing activity of T cells started 24 hours after the introduction of siCD84, since CD84 has a direct effect on CLL survival during the first 24 hours. We treated purified CLL cells from PB with siCTRL or siCD84. After 24 hours, we replaced the conditioned medium with fresh medium. We added the rest of the cells from the CLL blood sample to the culture for 48 hours and monitored lactate dehydrogenase (LDH) release. As shown in Supplemental Figure 7D, we observed that LDH release was above the background level only in the presence of cells from the CLL microenvironment. Interestingly, we detected a significant increase of approximately 2-fold in the lysis of siCD84-treated CLLs in the presence of T cells cocultured with CD84-deficient CLL cells compared with CLL cells expressing CD84 (Figure 6I).
We detected similar levels of CD84 on naive, activated (cultured with CD3 and CD28), and exhausted T cells (obtained from CLL patients) (Supplemental Figure 7E). To elucidate whether CD84 directly affects chronic TCR activation, which in turn drives T cell exhaustion, or whether the exhaustion is mediated through the PD-1/PD-L1 pathway, we analyzed the CD84-induced downstream cascade in human T cells. Ligation of CD84 on CLL-derived human T cells did not induce a change in IκB degradation, indicating that the NF-κB pathway was not activated (Supplemental Figure 7F). However, we observed that CD84 activation increased the levels of p–JAK-2, p-STAT3, and exhaustion markers and reduced the levels of IFN-γ (Figure 6, J–M).
To determine whether CD84 directly regulates T cell exhaustion by inhibition of the T cell receptor (TCR) downstream cascade, we examined the downstream TCR activation pathway by analyzing CD69 expression. CD69 receptor expression is upregulated in activated T cells (20) and serves as a costimulatory molecule that promotes cellular proliferation, cytokine production, and/or cytotoxicity (21). As shown in Supplemental Figure 7G, we observed no difference in CD69 expression in the CD84-activated CD8+ T cells, indicating that CD84 has no direct effect on the TCR-induced cascade. This suggests that the CD84-mediated pathway enhances PD-1 activity and T cell exhaustion and that reduced expression of PD-1 results in elevated T cell activity.
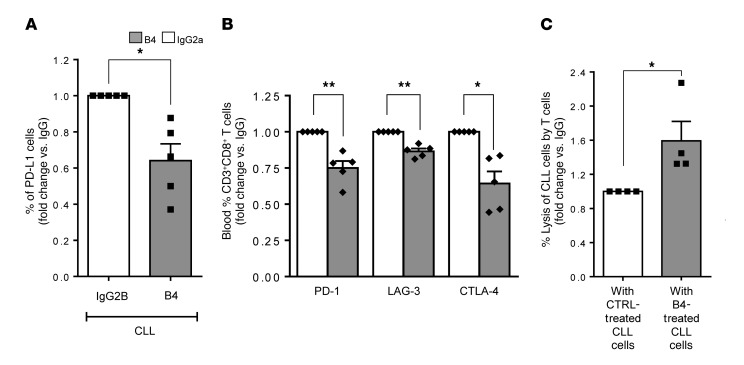
Finally, to further confirm the role of CD84 in regulating T cell activity, we blocked CD84 expression on CLL and T cells using the anti-CD84–blocking antibody B4 and assessed T cell activity.
We found that blocking CD84 significantly reduced the expression of PD-L1 on the surface of CLL cells (Figure 7A) and reduced expression of the exhaustion markers PD-1, LAG-3, and CTLA-4 on T cells (Figure 7B and Supplemental Figure 7H). The reduced expression of PD-L1 and T cell exhaustion markers resulted in a significant increase in CLL cell lysis (Figure 7C).
Figure 7. Reduction of PD-L1 on CLL cells and activation of T cells can be induced by treatment with the B4 anti-CD84 antibody.
Purified CLL cells from fresh PB samples obtained from patients were incubated with 20 μg anti-CD84 (B4) or anti-IgG2a (BioLegend) antibodies for 24 hours and were then incubated with T cells from the same sample for 48 hours. (A) CLLs cells were analyzed for PD-L1 expression (n = 5; P < 0.05, 2-tailed, paired t test). (B) T cells were examined for PD-1, LAG-3, and CTLA-4 expression (n = 5; *P < 0.05 and **P < 0.01, 2-tailed, paired t test). (C) CLL cell lysis by the T cells in B was determined with the CytoTox-ONE Kit (n = 4; *P < 0.05, 2-tailed, paired t test).
Together, these results show that CD84 regulates the activity of human T cells and suggest that downregulation of CD84 expression can induce T cell activation, possibly resulting in the killing of CLL cells.
Discussion
Cancers often circumvent immune surveillance by utilizing mechanisms of tolerance, such as the expression of PD-L1, a ligand whose inhibition results in potent antitumor immunity (22, 23). T cell dysfunction through the PD-L1 pathway has been extensively described in CLL (24–28). Prior studies have shown that PD-L1 expressed on nonhematopoietic cells in the tumor microenvironment regulates immune function in both homeostatic and malignant states (29, 30). Thus, given the important role of PD-L1 and PD-1 as immune checkpoints of T cell function and the fact that PD-1 inhibition is already used in cancer therapy, it is of great importance to understand the regulation of PD-L1 and PD-1 expression.
Several studies have identified the signaling pathways required for PD-L1 expression. Both type I and II IFNs upregulate PD-L1 expression (31, 32). PD-L1 expression in cell lines is decreased when MyD88, TRAF6, and MEK are inhibited (33). JAK2 has also been shown to induce PD-L1 expression (33, 34). Loss or inhibition of PTEN, a modifier of PI3K and AKT signaling, increases posttranscriptional PD-L1 expression in many cancers (35). Cyclin-dependent kinase 5 (Cdk5) allows medulloblastoma to evade immune elimination by IFN-γ–induced PD-L1, and disruption of Cdk5 expression in a mouse model of medulloblastoma resulted in potent CD4+ T cell–mediated tumor rejection (36).
The regulation of PD-1 expression is complex. Many factors have been reported to control PD-1 expression including cis-DNA elements, transcription factors, and epigenetic components such as DNA methylation and histone modifications. In addition, in T cells, PD-1 expression is elevated in response to the vast majority of immune challenges that fine-tune PD-1 expression in different inflammatory environments and across numerous cell types to modulate the immune response. Expression of this inhibitory pathway is rapidly downregulated in acute settings, allowing for a normal immune response. However, in the case of chronic stimulation, antigen-specific T cells retain high PD-1 expression, resulting in an impaired response to stimuli (37).
In the current study, we show that a direct interaction between CLL cells and their microenvironment, mediated through CD84, regulates T cell function in CLL. CD84 was expressed at low levels on healthy B and T cells and their microenvironment and only minimally affected PD-1 and PD-L1 expression. Tumor cells and their microenvironment significantly elevated CD84 expression. Activation of CD84 on tumor cells significantly upregulated the expression of PD-L1 on the tumor cells themselves and in their microenvironment, as well as expression of PD-1 on T cells. T cells lacking CD84 were functionally similar to WT T cells, suggesting that only in the malignant environment does activation of CD84 induce PD-1 expression and T cell exhaustion. This elevation in T cell exhaustion was not dependent on TCR activation, suggesting that the CD84-induced cascade in T cells directly upregulates T cell exhaustion.
Our results show that CD84 plays an important role in T cell–mediated antitumor immunity in CLL. Activation of CD84 induced a cascade involving S6 and AKT and p-STAT3 levels, which together elevated PD-L1 expression on murine and human CLL cells and in their microenvironment. Downregulation of CD84 expression or its inhibition reduced PD-L1 expression, resulting in decreased expression of T cell exhaustion markers and activated T cell cytotoxicity. Thus, we propose that CD84 has an essential role in inhibiting the T cell antitumor response in patients with CLL. It may allow tumor progression by upregulating the expression of PD-1 and PD-L1, which suppress the T cell–mediated antitumor response.
We further demonstrate that disruption of CD84-mediated cell-cell contact decreased the levels of PD-L1 expression on CLL cells and their microenvironment, resulting in reduced PD-1 expression on T cells and allowing elevated activity of these cells. Thus, the exhaustive phenotype of T cells is reversible, and these cells can gain back their activity. Currently, there are many cytostatic agents and kinase inhibitors in clinical trials for the treatment of CLL. However, the development of drug resistance and the persistence of minimal residual disease in the BM make this cancer difficult to cure. Therefore, a combination of treatments targeting both the malignant cell population and its interaction with the microenvironment by inducing the immune system to eradicate malignant cells might provide a better approach for treating CLL.
Methods
Mice.
C57BL/6 WT were purchased from Harlan Biotech Israel. Eμ–TCL1 (5) mice were provided by C.M. Croce (The Ohio State University, Columbus, Ohio, USA). TCL1 mice were backcrossed with C57BL/6 mice. CD84-KO (CD84–/–) mice were previously described (19). All mice were used at 6 to 8 weeks of age.
Primary human CLL samples.
Primary human CLL cells and patients’ own T cells were provided in compliance with the hospitals’ IRBs, as previously described (38). The diagnosis of CLL was based on standard criteria (39). CLL was staged according to the Rai and Binet staging system (ref. 40 and Supplemental Table 1).
Primary human BM cells.
Primary human BM stem cells (BMSCs) were provided in compliance with the IRBs of the hospitals, as previously described (38). The diagnosis of CLL was based on standard criteria (39). Patients were staged according to the Rai and Binet (40) staging system (Supplemental Table 1).
Murine Eμ-TCL1 cells.
B cells were isolated from Eμ-TCL1 mice by harvesting spleens and sorting for CD5+ and CD19+ TCL1 cells.
Stromal cell line.
The murine BM cell line M210B4 (CRL-1972) was purchased from the American Type Culture Collection (ATCC) and maintained and passaged every third day in RPMI 1640 medium containing 10% (v/v) FCS.
Primary mouse BM cells.
Tibia and femur bones were flushed out, and cells were seeded on 24-well plates. Full medium was added, and the cells were maintained by washing away nonadherent cells and changing the medium (10% RPMI) weekly.
NLCs.
Peripheral blood mononuclear cells (PBMCs) from patients with CLL were suspended in RPMI 1640 medium containing 10% FCS in culture for 2 weeks until outgrowth of NLCs was observed (41).
RNA isolation and quantitative real-time PCR.
Total RNA from CLL or M210B4 cells was isolated as previously described (38, 42). mRNA from primary human BM and T cell samples was isolated using the PerfectPure RNA Cultured Cell Kit (5 Prime) according to the manufacturer’s instructions. mRNA levels were analyzed by quantitative real-time PCR (qRT-PCR) as previously described (43). Primers were purchased from MilliporeSigma, and the primer sequences for PCR are listed in Supplemental Table 2.
Microarray analysis.
Affymetrix GeneChip 2.0 ST mouse gene analysis was performed as previously described (44). Microarray data were deposited in the NCBI’s GEO database (GEO GSE107140). The resulting genes were compared using Python, version 3.6, and only genes found in both data sets were selected. Thereafter, the change in gene expression was compared. Heatmaps displaying these results were made using the Seaborn library in Python, version 3.6, and were based on either the highest differentiated genes upon treatment or on proteins known to interact with PD-L1 (CD274) (STRING protein-protein interaction database; https://string-db.org/).
Staining for flow cytometry.
Cells were stained using specific antibodies (see Supplemental Table 3), as previously described (11). For intracellular cytokine staining, cells were treated for 2 hours with brefeldin A (BioLegend) and then harvested, fixed, and permeabilized using a commercial kit from BD Biosciences, followed by staining. For transcription factor staining, the cells were treated for 2 hours with brefeldin A (BioLegend) and then harvested, fixed, and permeabilized using an eBioscience commercial kit, followed by staining.
Activation of T cells.
Whole spleens were incubated on CD3 plates for 24 hours. The cells were harvested and stained for flow cytometry after 24 hours, as this was the closest to the time point at which cytokine expression is shown in vivo.
CD84 activation.
Stimulation of CD84 on CLL cells, T cells, and stroma with an agonistic antibody was performed as previously described (10, 11). mRNA was analyzed 24 hours after activation and protein after 48 hours.
Stimulation with IFN-γ.
Stimulation of IFN-γ on CLL cells was performed by incubating cells with 500 IU/ml IFN-γ (Peprotech) in 10% RPMI medium.
MTOR and AKT inhibitors.
In order to inhibit the MTOR/AKT pathway, CLL cells were incubated for 20 minutes with 0.1 μM rapamycin and wortmannin (MilliporeSigma), respectively, in 10% RPMI medium prior to any treatment.
PD-1 blocking.
In order to block PD-1, T cells were incubated with anti–PD-1 antibody (clone EH12.2H7, BioLegend, 5 μg/ml) for 24 hours prior to culturing with CLL cells.
CD84 downregulation by siRNA on human samples.
CLL cells were purified with anti CD19 beads (Miltenyi Biotec). Thereafter, downregulation of CD84 was performed using siCD84 or siCTRL (Dharmacon) introduced by electroporation as previously described (11).
CD84 blocking with the B4 antibody in human samples.
Purified CLL cells (anti-CD19 beads, Miltenyi Biotec) were incubated with 20 μg/ml B4 antibody (11) or isotype control IgG2a (BioLegend) for 24 hours in 10% RPMI medium; thereafter, fresh 10% RPMI medium was added (for the cytotoxic assay) prior to coculture with T cells.
Cocultures with M210B4 and mouse stromal cells.
CLL/Eμ-TCL1 and M210B4/mouse stromal cells were cocultured at a ratio of 1:16 (M210B4/CLL), as previously described (12).
Cocultures with human T cells.
CLL cells were cocultured for 48 hours with the non-CLL fraction from the purification (after pure CLL cells were manipulated) at the same ratio as the original blood sample from the patient, and T cells were then stained with antibodies and analyzed by flow cytometry.
Cytotoxicity assays.
siCTRL- or siCD84-treated CLL cells were divided into 2 fractions, one cultured alone and the other cultured with the non-CLL fraction. A cytotoxic assay was conducted using the CytoTox-ONE detection system, according to the manufacturer’s instructions (Promega). Twenty-four hours after treatment with siRNA or B4, the conditioned medium was replaced with fresh medium. Next, the remaining cells in the CLL samples were added to the CLL culture for a 48-hour coculture. The amount LDH in the medium was compared with the medium without any cells (background), cells without CLL cells (negative control), lysed CLL cells (positive control), and CLL cells alone (negative control).
Western blot analysis.
RIPA lysis buffer was added with a protease inhibitor cocktail (MilliporeSigma), and a phosphatase inhibitor was added to the CD84-stimulated cells. The cells were then loaded onto 12% SDS-polyacrylamide gels, subjected to electrophoresis, and transferred onto nitrocellulose membranes. The blots were probed with antibodies against actin (MilliporeSigma) as a housekeeping gene, anti–p-ERK, anti p-AKT, and anti–p-S6 (Cell Signaling Technology), and the bands were detected by Odyssey system (Li-Cor).
Eμ-TCL1 adoptive transfer.
Eμ-TCL1 adoptive transfer was performed as previously described (11).
In vivo CD84 inhibition.
Blocking of CD84 in vivo was performed using the B4 antibody or IgG2a isotype control (eBioscience). In adoptive transfer experiments, treatment with the B4 or IgG2a isotype control antibody started the second day after Eμ-TCL1 splenocyte injection and continued every second day with 1 mg antibody/kg body weight injected into the tail vein.
Chimeric mice.
Six- to eight-week-old C57BL/6 WT or CD84–/– mice were lethally irradiated (10.5 Gy). The following day, donor animals (WT and Eμ-TCL1 mice) were sacrificed and their BM was isolated. From these isolated cells, 5 × 106 cells were injected into the tail vein of recipient mice.
Statistics.
Data analysis was performed with GraphPad Prism, version 6.0 (GraphPad Software), Python, version 3.6, or MATLAB R2017b. For most experiments, the mean with the SEM is provided. To test for significance, a 2-tailed Student’s t test was used and a paired t test for same patient or mouse cells in different treatments unless stated otherwise in the figure legends. For normalized data, a ratio t test was performed to correct for non-normally distributed data points. Results were deemed significant with a P value of 0.05 or less.
Study approval.
All animal procedures were reviewed and approved by the Animal Research Committee of the Weizmann Institute of Science. Samples from CLL patients and healthy controls were provided by the Hematology Institute at the Kaplan Medical Center, the Laniadio Medical Center, and the Tel Aviv Sourasky Medical Center, in accordance with the IRBs of these hospitals. CLL staging was done using the Rai and Binet systems.
Author contributions
HL designed research, performed research, analyzed data, and wrote the manuscript. AFB, VH, MPK, LR, LS, and SBH designed research, performed research, and analyzed data. IO performed research and analyzed data. VM, ND, MS, and LS contributed vital reagents. YC performed research and contributed vital reagents. MS designed research and analyzed data. YH contributed vital reagents and designed research. IS designed research, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors wish to thank the members of the Shachar laboratory for fruitful discussions and support. IS is the incumbent of the Dr. Morton and Ann Kleiman Professorial Chair. This research was supported by DKFZ-MOST (German Cancer Research Center–Israeli Ministry of Science and Technology) Cooperation in Cancer Research; the European Research Area Network (ERA-NET) TRANSCAN-2 program (JTC 2014, Fighting Resistance in CLL [FIRE-CLL] project); the Quinquin Foundation; and the Binational Science Foundation (BSF) (grant 711979).
Version 1. 10/02/2018
In-Press Preview
Version 2. 11/05/2018
Electronic publication
Version 3. 12/03/2018
Print issue publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2018, American Society for Clinical Investigation.
Reference information: J Clin Invest. 2018;128(12):5465–5478.https://doi.org/10.1172/JCI96610.
Contributor Information
Hadas Lewinsky, Email: hadas.lewinsky@weizmann.ac.il.
Avital F. Barak, Email: avital.barak@weizmann.ac.il.
Victoria Huber, Email: victoria.huber@weizmann.ac.il.
Matthias P. Kramer, Email: matthias.kramer@weizmann.ac.il.
Lihi Radomir, Email: lihi.radomir@weizmann.ac.il.
Lital Sever, Email: lital.sever@weizmann.ac.il.
Irit Orr, Email: irit.orr@weizmann.ac.il.
Vita Mirkin, Email: VitaMi@clalit.org.il.
Nili Dezorella, Email: nili.dezo@gmail.com.
Mika Shapiro, Email: mikash@tlvmc.gov.il.
Yosef Cohen, Email: yocohen@laniado.org.il.
Lev Shvidel, Email: Levsh@clalit.org.il.
Martina Seiffert, Email: m.seiffert@dkfz.de.
Yair Herishanu, Email: herishanu@gmail.com.
Shirly Becker-Herman, Email: shirly.herman@weizmann.ac.il.
Idit Shachar, Email: idit.shachar@weizmann.ac.il.
References
- 1.Rozman C, Montserrat E. Chronic lymphocytic leukemia. N Engl J Med. 1995;333(16):1052–1057. doi: 10.1056/NEJM199510193331606. [DOI] [PubMed] [Google Scholar]
- 2.McKinney EF, Smith KG. T cell exhaustion and immune-mediated disease-the potential for therapeutic exhaustion. Curr Opin Immunol. 2016;43:74–80. doi: 10.1016/j.coi.2016.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Greaves P, Gribben JG. The role of B7 family molecules in hematologic malignancy. Blood. 2013;121(5):734–744. doi: 10.1182/blood-2012-10-385591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pauken KE, et al. Cutting edge: identification of autoreactive CD4+ and CD8+ T cell subsets resistant to PD-1 pathway blockade. J Immunol. 2015;194(8):3551–3555. doi: 10.4049/jimmunol.1402262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bichi R, et al. Human chronic lymphocytic leukemia modeled in mouse by targeted TCL1 expression. Proc Natl Acad Sci U S A. 2002;99(10):6955–6960. doi: 10.1073/pnas.102181599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calpe S, et al. The SLAM and SAP gene families control innate and adaptive immune responses. Adv Immunol. 2008;97:177–250. doi: 10.1016/S0065-2776(08)00004-7. [DOI] [PubMed] [Google Scholar]
- 7.Martin M, et al. CD84 functions as a homophilic adhesion molecule and enhances IFN-γ secretion: adhesion is mediated by Ig-like domain 1. J Immunol. 2001;167(7):3668–3676. doi: 10.4049/jimmunol.167.7.3668. [DOI] [PubMed] [Google Scholar]
- 8.Romero X, et al. CD229 (Ly9) lymphocyte cell surface receptor interacts homophilically through its N-terminal domain and relocalizes to the immunological synapse. J Immunol. 2005;174(11):7033–7042. doi: 10.4049/jimmunol.174.11.7033. [DOI] [PubMed] [Google Scholar]
- 9.Yan Q, et al. Structure of CD84 provides insight into SLAM family function. Proc Natl Acad Sci U S A. 2007;104(25):10583–10588. doi: 10.1073/pnas.0703893104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Binsky-Ehrenreich I, et al. CD84 is a survival receptor for CLL cells. Oncogene. 2014;33(8):1006–1016. doi: 10.1038/onc.2013.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marom A, et al. CD84 mediates CLL-microenvironment interactions. Oncogene. 2017;36(5):628–638. doi: 10.1038/onc.2016.238. [DOI] [PubMed] [Google Scholar]
- 12.Kurtova AV, et al. Diverse marrow stromal cells protect CLL cells from spontaneous and drug-induced apoptosis: development of a reliable and reproducible system to assess stromal cell adhesion-mediated drug resistance. Blood. 2009;114(20):4441–4450. doi: 10.1182/blood-2009-07-233718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grzywnowicz M, et al. Programmed death-1 and its ligand are novel immunotolerant molecules expressed on leukemic B cells in chronic lymphocytic leukemia. PLoS One. 2012;7(4):e35178. doi: 10.1371/journal.pone.0035178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ritprajak P, Azuma M. Intrinsic and extrinsic control of expression of the immunoregulatory molecule PD-L1 in epithelial cells and squamous cell carcinoma. Oral Oncol. 2015;51(3):221–228. doi: 10.1016/j.oraloncology.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 15.Lastwika KJ, et al. Control of PD-L1 expression by oncogenic activation of the AKT-mTOR pathway in non-small cell lung cancer. Cancer Res. 2016;76(2):227–238. doi: 10.1158/0008-5472.CAN-14-3362. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Jiang CC, Jin L, Zhang XD. Regulation of PD-L1: a novel role of pro-survival signalling in cancer. Ann Oncol. 2016;27(3):409–416. doi: 10.1093/annonc/mdv615. [DOI] [PubMed] [Google Scholar]
- 17.Gowrishankar K, Gunatilake D, Gallagher SJ, Tiffen J, Rizos H, Hersey P. Inducible but not constitutive expression of PD-L1 in human melanoma cells is dependent on activation of NF-κB. PLoS One. 2015;10(4):e0123410. doi: 10.1371/journal.pone.0123410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26:677–704. doi: 10.1146/annurev.immunol.26.021607.090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cannons JL, et al. Optimal germinal center responses require a multistage T cell:B cell adhesion process involving integrins, SLAM-associated protein, and CD84. Immunity. 2010;32(2):253–265. doi: 10.1016/j.immuni.2010.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Testi R, D’Ambrosio D, De Maria R, Santoni A. The CD69 receptor: a multipurpose cell-surface trigger for hematopoietic cells. Immunol Today. 1994;15(10):479–483. doi: 10.1016/0167-5699(94)90193-7. [DOI] [PubMed] [Google Scholar]
- 21.Moretta A, et al. CD69-mediated pathway of lymphocyte activation: anti-CD69 monoclonal antibodies trigger the cytolytic activity of different lymphoid effector cells with the exception of cytolytic T lymphocytes expressing T cell receptor α/β. J Exp Med. 1991;174(6):1393–1398. doi: 10.1084/jem.174.6.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou W, Chen L. Inhibitory B7-family molecules in the tumour microenvironment. Nat Rev Immunol. 2008;8(6):467–477. doi: 10.1038/nri2326. [DOI] [PubMed] [Google Scholar]
- 23.Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer. 2012;12(4):252–264. doi: 10.1038/nrc3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanna BS, et al. Depletion of CLL-associated patrolling monocytes and macrophages controls disease development and repairs immune dysfunction in vivo. Leukemia. 2016;30(3):570–579. doi: 10.1038/leu.2015.305. [DOI] [PubMed] [Google Scholar]
- 25.McClanahan F, et al. Mechanisms of PD-L1/PD-1-mediated CD8 T-cell dysfunction in the context of aging-related immune defects in the Eμ-TCL1 CLL mouse model. Blood. 2015;126(2):212–221. doi: 10.1182/blood-2015-02-626754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riches JC, et al. T cells from CLL patients exhibit features of T-cell exhaustion but retain capacity for cytokine production. Blood. 2013;121(9):1612–1621. doi: 10.1182/blood-2012-09-457531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brusa D, et al. The PD-1/PD-L1 axis contributes to T-cell dysfunction in chronic lymphocytic leukemia. Haematologica. 2013;98(6):953–963. doi: 10.3324/haematol.2012.077537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grzywnowicz M, et al. Expression of Programmed Death 1 Ligand in different compartments of chronic lymphocytic leukemia. Acta Haematol. 2015;134(4):255–262. doi: 10.1159/000430980. [DOI] [PubMed] [Google Scholar]
- 29.Augello A, et al. Bone marrow mesenchymal progenitor cells inhibit lymphocyte proliferation by activation of the programmed death 1 pathway. Eur J Immunol. 2005;35(5):1482–1490. doi: 10.1002/eji.200425405. [DOI] [PubMed] [Google Scholar]
- 30.Nazareth MR, Broderick L, Simpson-Abelson MR, Kelleher RJ, Yokota SJ, Bankert RB. Characterization of human lung tumor-associated fibroblasts and their ability to modulate the activation of tumor-associated T cells. J Immunol. 2007;178(9):5552–5562. doi: 10.4049/jimmunol.178.9.5552. [DOI] [PubMed] [Google Scholar]
- 31.Eppihimer MJ, et al. Expression and regulation of the PD-L1 immunoinhibitory molecule on microvascular endothelial cells. Microcirculation. 2002;9(2):133–145. doi: 10.1080/713774061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schreiner B, et al. Interferon-β enhances monocyte and dendritic cell expression of B7-H1 (PD-L1), a strong inhibitor of autologous T-cell activation: relevance for the immune modulatory effect in multiple sclerosis. J Neuroimmunol. 2004;155(1–2):172–182. doi: 10.1016/j.jneuroim.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Liu J, et al. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1) and increase expression after stimulation with IFN-{gamma} and TLR ligands via a MyD88-, TRAF6-, and MEK-dependent pathway. Blood. 2007;110(1):296–304. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 34.Lee SJ, et al. Interferon regulatory factor-1 is prerequisite to the constitutive expression and IFN-gamma-induced upregulation of B7-H1 (CD274) FEBS Lett. 2006;580(3):755–762. doi: 10.1016/j.febslet.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 35.Parsa AT, et al. Loss of tumor suppressor PTEN function increases B7-H1 expression and immunoresistance in glioma. Nat Med. 2007;13(1):84–88. doi: 10.1038/nm1517. [DOI] [PubMed] [Google Scholar]
- 36.Dorand RD, et al. Cdk5 disruption attenuates tumor PD-L1 expression and promotes antitumor immunity. Science. 2016;353(6297):399–403. doi: 10.1126/science.aae0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bally AP, Austin JW, Boss JM. Genetic and epigenetic regulation of PD-1 expression. J Immunol. 2016;196(6):2431–2437. doi: 10.4049/jimmunol.1502643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Binsky I, et al. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc Natl Acad Sci U S A. 2007;104(33):13408–13413. doi: 10.1073/pnas.0701553104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hallek M, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Binet JL, et al. A clinical staging system for chronic lymphocytic leukemia: prognostic significance. Cancer. 1977;40(2):855–864. doi: 10.1002/1097-0142(197708)40:2<855::AID-CNCR2820400239>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 41.Burger JA, Tsukada N, Burger M, Zvaifler NJ, Dell’Aquila M, Kipps TJ. Blood-derived nurse-like cells protect chronic lymphocytic leukemia B cells from spontaneous apoptosis through stromal cell-derived factor-1. Blood. 2000;96(8):2655–2663. [PubMed] [Google Scholar]
- 42.Binsky I, et al. TAp63 regulates VLA-4 expression and chronic lymphocytic leukemia cell migration to the bone marrow in a CD74-dependent manner. J Immunol. 2010;184(9):4761–4769. doi: 10.4049/jimmunol.0904149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordin M, et al. c-Met and its ligand hepatocyte growth factor/scatter factor regulate mature B cell survival in a pathway induced by CD74. J Immunol. 2010;185(4):2020–2031. doi: 10.4049/jimmunol.0902566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lantner F, et al. CD74 induces TAp63 expression leading to B-cell survival. Blood. 2007;110(13):4303–4311. doi: 10.1182/blood-2007-04-087486. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.