Abstract
Natural killer (NK) cells are innate lymphocytes that have features of adaptive immunity such as clonal expansion and generation of long-lived memory. Interleukin-12 (IL-12) signaling through its downstream transcription factor signal transducer and activator of transcription 4 (STAT4) is required for the generation of memory NK cells after expansion. We identify gene loci that are highly enriched for STAT4 binding using chromatin immunoprecipitation sequencing for STAT4 and the permissive histone mark H3K4me3 in activated NK cells. We found that promoter regions of Runx1 and Runx3 are targets of STAT4 and that STAT4 binding during NK cell activation induces epigenetic modifications of Runx gene loci resulting in increased expression. Furthermore, specific ablation of Runx1, Runx3, or their binding partner Cbfb in NK cells resulted in defective clonal expansion and memory formation during viral infection, with evidence for Runx1-mediated control of a cell cycle program. Thus, our study reveals a mechanism whereby STAT4-mediated epigenetic control of individual Runx transcription factors promotes the adaptive behavior of antiviral NK cells.
INTRODUCTION
Although natural killer (NK) cells are generally thought to represent the cytolytic arm of the innate immune system, recent findings in mice and humans have demonstrated that these innate lymphocytes can have features of adaptive immunity, including clonal expansion and generation of memory (1–4). In certain strains of mice, NK cells bearing the Ly49H receptor recognize the viral glycoprotein m157 expressed by mouse cytomegalovirus (MCMV)–infected cells and undergo prolific expansion (100- to 1000-fold), resulting in a long-lived pool of self-renewing memory NK cells able to be recalled (5). Proinflammatory cytokines (6–9) and downstream transcription factors (7, 9, 10) can promote these adaptive NK cell responses via distinct mechanisms (2); however, how transcriptional and epigenetic regulation of NK cell expansion and memory are initiated and maintained are not fully understood.
Interleukin-12 (IL-12) binding to its heterodimeric receptor on NK cells results in a signaling cascade leading to Janus kinase–mediated phosphorylation and homodimerization of signal transducer and activator of transcription 4 (STAT4) (11), which translocates into the nucleus, where it binds to target sequences in IL-12-responsive loci and activates transcription of effector cytokine genes such as Ifng (12). In addition, IL-12 and STAT4 induction of the transcription factor Zbtb32 was found to promote the expansion of Ly49H+ NK cells after MCMV infection, involving a mechanism where the antiproliferative factor BLIMP-1 is repressed (10). Additional genes targeted by STAT4 in activated NK cells during virus infection remain unknown.
Here, we used STAT4 and H3K4me3 chromatin immunoprecipitation sequencing (ChIP-seq) to analyze the transcriptional and global epigenetic mechanisms that regulate IL-12–mediated pathways during NK cell activation. Using this approach, we found that Runx family transcription factors were among the genes highly associated with STAT4 binding in activated NK cells. Runx transcription factors are a family of evolutionarily conserved proteins that are crucial for hematopoiesis, neurogenesis, and osteogenesis (13). The Runt domain possessed by all three Runx transcription factors (Runx1, Runx2, and Runx3) mediates heterodimerization with the non–DNA binding core-binding factor β subunit (CBF-β) to regulate gene transcription. Dimerization with CBF-β enhances the DNA binding affinity of Runx proteins and results in activation and repression of a wide variety of target genes by interacting with other transcription factors, histone deacetylases, or histone acetyltransferases (14–16). Runx1 and Runx3 play an important role in T cell development, lineage specification, differentiation, and function (14, 17–22). During MCMV infection, Runx1 and Runx3 were both up-regulated in NK cells as a consequence of epigenetic modifications. Thus, we engineered mice containing specific deletions of Runx1, Runx3, or Cbfb in NK cells to investigate the influence of this family of transcription factors on NK cell activation, expansion, and response against MCMV infection.
RESULTS
STAT4 targets promoter and intronic regions of Runx1 and Runx3 in activated NK cells
STAT4, a signal transducer and activator of transcription downstream of the IL-12 receptor, has previously been demonstrated to be critical in the generation of memory NK cells during MCMV infection (9). To investigate the global occupancy of STAT4 across the genome, we stimulated primary mouse NK cells with proinflammatory cytokines (IL-12 plus IL-18) and performed STAT4 ChIP-seq. A total of 1196 reproducible peaks were identified within promoter, intronic, exonic, and intergenic regions (using cytokine-stimulated STAT4-deficient NK cells as a negative control for nonspecific antibody binding). This analysis revealed a majority of STAT4 occupancy within introns (35%) and intergenic regions (40%; Fig. 1A), the latter of which suggests that STAT4 binding can potentially function as a distal enhancer (23). STAT4 also localized to promoters (20%), defined as 2000 base pairs (bp) upstream to 500 bp downstream from transcriptional start sites (TSS), and a minority of binding occurred within exons (5%; Fig. 1A).
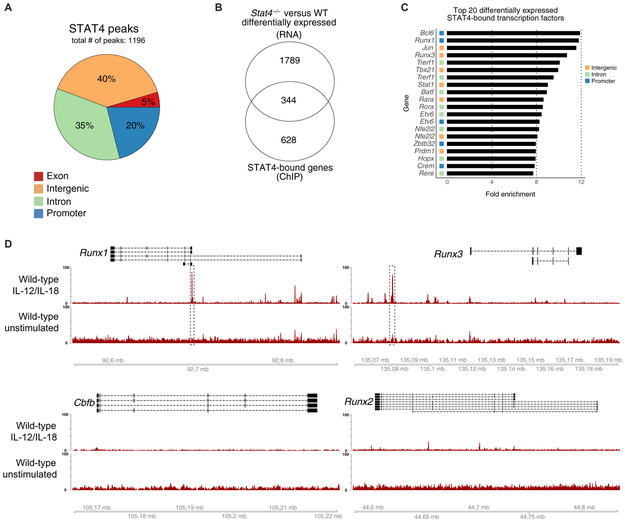
Fig. 1. STAT4 directly targets Runx transcription factors in activated NK cells.
Splenic NK cells (TCRβ−CD19−CD3ε−Ly6G−TER119−TCRγδ−NK1.1+) were sorted from WT mice and stimulated with IL-12 and IL-18 or media alone as a control (unstimulated). STAT4 ChIP was performed, followed by high-throughput DNA sequencing. (A) Proportions of STAT4 genome-wide occupancy at promoter (2 kb upstream and 0.5 kb downstream from TSS), intronic, exonic, or distal intergenic regions in cytokine-stimulated NK cells are shown. (B) RNA-seq was performed on splenic Ly49H+ WT NK cells and Stat4−/− NK cells sorted from mixed chimeras 2 days after MCMV infection. Venn diagram of overlap between differentially expressed genes (top; Padj < 0.05) identified through RNA-seq and reproducible STAT4-bound regions identified through ChIP-seq (bottom; IDR < 0.05). RNA-seq data were performed on n = 3 per condition. (C) Bar graphs depict the top 20 genes with greatest fold enrichment of STAT4 binding over input calculated by MACS2 in transcription factors that show differential expression in RNA-seq data. (D) Representative gene tracks for indicated core-binding factors from STAT4 ChIP-seq. ChIP-seq data are representative of three independent experiments with n = 15 to 20 pooled mice per group per experiment.
To determine which STAT4-bound genes were dependent on STAT4 for their expression, we performed RNA sequencing (RNA-seq) of Ly49H+ NK cells from mixed WT:Stat4−/− bone marrow chimeric (BMC) mice 2 days after MCMV infection. Among all differentially expressed genes, 344 harbored STAT4-bound regions either within or near the gene locus (Fig. 1B). We then ranked these genes on the basis of the strength of STAT4 binding, as measured by the fold enrichment over input. As a strong validation of our STAT4 ChIP-seq approach, Ifng was among the top hits of all STAT4-bound genes (fig. S1A), and a previously identified STAT4 target locus, the promoter of Zbtb32 (10), was a top hit among transcription factors (Fig. 1C). We found that the promoter and intronic regions of Runx transcription factors were also highly enriched for STAT4 binding (Fig. 1C). Individual analyses of the genomic region of all Runx family members and CBF-β in stimulated NK cells indicated substantial STAT4 binding of Runx1 and Runx3 gene loci at promoter and intergenic regions, respectively (Fig. 1D). In contrast, STAT4 binding at the Runx2 and Cbfb loci was minimal or not readily detected in activated NK cells (Fig. 1D). Together, these results suggest that cytokine stimulation of NK cells induces rapid and distinct STAT4 binding at gene loci encoding Runx1 and Runx3.
STAT4 binding increases trimethylation of histone H3 lysine 4 (H3K4me3) at Runx gene promoters during NK cell activation
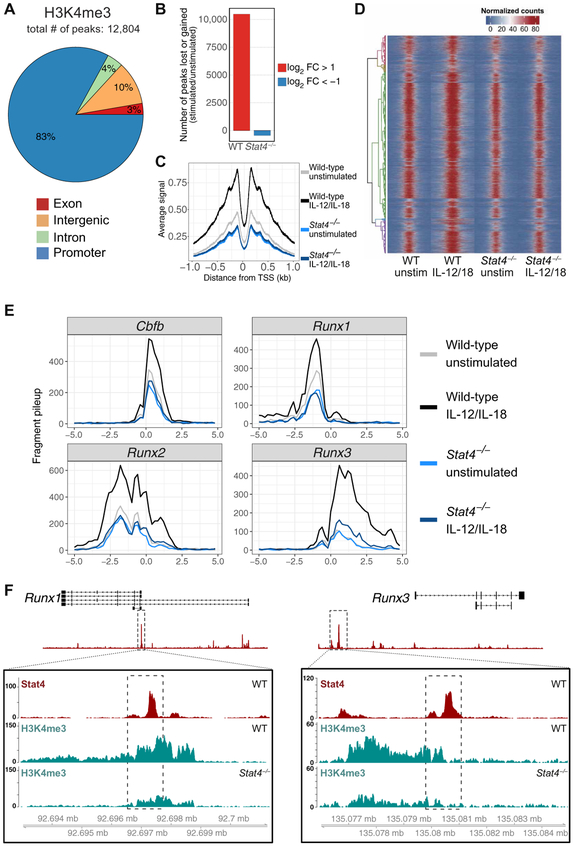
To investigate the epigenetic regulation of Runx family members and CBF-β during NK cell activation, we performed ChIP-seq on stimulated NK cells using an antibody against H3K4me3, a histone modification widely used to identify active gene promoters that, in certain settings, can predict the transcription of more than 80% of the genes carrying this modification (24). In cytokine-stimulated NK cells, the majority of the H3K4me3 modifications were detected at sites localized to promoters (83%), whereas 7 and 10% of the permissive marks were found within intragenic (4% introns and 3% exons) and distal intergenic regions, respectively (Fig. 2A). We observed global increases in promoter regions (Fig. 2C) and nonpromoter regions (fig. S1B), marked by H3K4me3 in stimulated wild-type (WT) NK cells compared with resting NK cells but not in stimulated STAT4-deficient NK cells, suggesting that the transcription factor STAT4 actively shapes the epigenetic landscape across the genome of NK cells during their activation (Fig. 2, B and D). Consistent with this, we detected an increase of H3K4me3 peaks at the promoters of all three Runx factors and CBF-β in stimulated NK cells (Fig. 2E), indicating a cytokine-induced permissiveness of transcription specifically at these genes but not all transcription factor genes (fig. S1C). In STAT4-deficient NK cells stimulated with proinflammatory cytokines, we found that the increase observed in H3K4me3 at Runx loci was largely ablated (Fig. 2E). STAT4 binding at the Runx1 promoter overlapped well with STAT4-dependent H3K4me3 activity (Fig. 2F). We observed that, whereas STAT4 deficiency leads to decreased activity at the Runx1 promoter, STAT4 binds to Runx3 at a distal region 40.8 kb upstream from the promoter site (Fig. 2F), suggesting that STAT4 may bind to an active enhancer region for Runx3 but not for Runx1.
Fig. 2. STAT4 facilitates a permissive epigenetic landscape (H3K4me3) in activated NK cells.
Splenic NK cells (TCRβ−CD19−CD3ε−Ly6G−TER119−TCRγδ−NK1.1+) were isolated from WT and Stat4−/− mice and stimulated with IL-12 and IL-18 or media alone as a control (unstimulated; unstim). H3K4me3 ChIP was performed, followed by high-throughput DNA sequencing. (A) Global proportions of H3K4me3 permissive marks at promoter, intronic, exonic, or distal inter-genic regions in cytokine-stimulated NK cells are shown. (B) Bar plots depict number of peaks that change on the basis of fold change (FC) of stimulated versus unstimulated NK cells. FC was calculated by taking the difference between log2-transformed normalized counts for each condition. Only peaks that showed a log2 FC greater than a magnitude of 1 were counted. (C) Meta-peak of all H3K4me3 promoter regions. Overlap of midpoints of ChIP fragments (defined as regions between properly paired sequence reads) for each TSS region was counted for each base pair ± 1 kb from the transcriptional start site. Line plot depicts average signal for all regions for each base pair. (D) Heat map of all H3K4me3 binding regions, with each row representing a single-peak region, row-clustered by normalized peak counts. Signal is displayed as normalized read counts over 5 kb centered at the peak region and is binned at 100-bp windows. (E) H3K4me3 signals from Cbfb, Runx1, Runx2, and Runx3 loci plotted as normalized fragment counts binned at 200 bp across a 10-kb window centered on the transcriptional start site. (F) Zoomed-in histograms of STAT4 ChIP and H3K4me3 ChIP reads mapped to Runx1 and Runx3 loci. Dashed box within boxed tracks indicate STAT4 ChIP called peak region. Data are representative of two independent experiments with n = 15 to 20 pooled mice per group per experiment.
MCMV infection up-regulates Runx1 and Runx3 expression in virus-specific NK cells
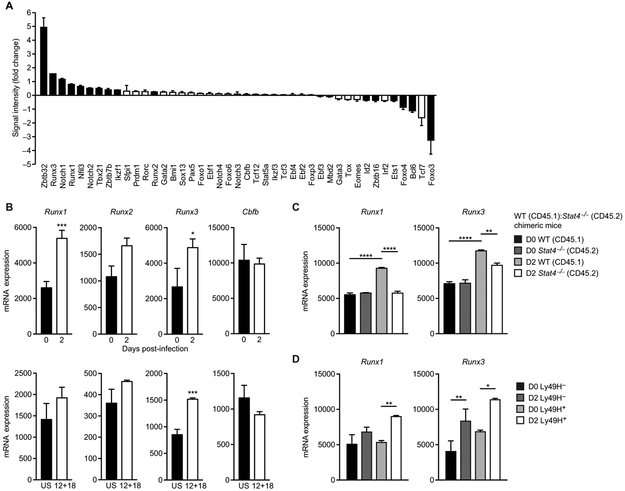
Next, we investigated whether STAT4 binding and increased H3K4me3 at Runx loci coincided with greater transcription of these genes in activated NK cells after MCMV infection. We compared mRNA expression of known T, B, NK, and NK T cell-regulating transcription factors in sorted Ly49H+ NK cells from infected and uninfected mice by microarray (Fig. 3A). The most highly up-regulated transcription factor in NK cells on day 2 after MCMV infection was Zbtb32 [as reported previously (10)], followed by Runx3, Notch1, and Runx1 (Fig. 3A). Consistent with the microarray data, Runx1 and Runx3 mRNAs were increased by RNA-seq of NK cells at day 2 post-infection (PI) compared with uninfected controls, whereas no significant change in Cbfb and Runx2 transcript levels was observed (Fig. 3B). Furthermore, ex vivo stimulation with proinflammatory cytokines IL-12 plus IL-18 up-regulated Runx1 and Runx3 in NK cells (Fig. 3B), albeit not to the extent of virus infection, suggesting that additional signals are required for optimal expression of Runx transcription factors during MCMV exposure. MCMV infection in mixed WT:Stat4−/− chimeric mice provided evidence that up-regulation of Runx1 and Runx3 in NK cells is mostly dependent on STAT4 (Fig. 3C) and less on antigen receptor Ly49H signaling (Fig. 3D). Bc16 was among the genes that showed down-regulation (Fig. 3A) despite being the top transcription factor locus bound by STAT4 (Fig. 1C), suggesting that factors other than STAT4 may play the dominant role in the regulation of this locus in activated NK cells. This finding also demonstrates that STAT4 binding to a given gene locus does not necessarily always equate to enhanced transcription. Given the enrichment of STAT4 and H3K4me3 at the TSS of Runx1 and Runx3 in cytokine-stimulated NK cells and that Runx1 and Runx3 mRNA were up-regulated after viral infection, we aimed to further investigate the role of this transcription factor family for NK cell function.
Fig. 3. Runx1 and Runx3 are up-regulated during MCMV infection.
(A) Graph shows mRNA expression of highest induced lymphocyte-specific transcription factors in splenic Ly49H+ NK cells sorted from uninfected and MCMV-infected animals on day 2 PI, as assessed by microarray [data provided by the Immunological Genome Consortium (41)]. Data are shown as fold change in microarray signal intensity for the infected versus uninfected samples (n = 3 biological replicates per group and representative of three separate experiments). Solid black bars denote significant up-regulation or down-regulation as compared with uninfected controls (P < 0.05, two-tailed unpaired Student’s t test). (B) Normalized counts of Runx1, Runx2, Runx3, and Cbfb in splenic Ly49H+ NK cells sorted from MCMV-infected mice on day 2 PI and uninfected mice (top) or in unstimulated (US) or IL-12 plus IL-18-treated (12 + 18; 16-hour stimulation) splenic NK cells (bottom), as assessed by RNA-seq (n = 2 to 3 biological replicates per group). (C) RNA-seq was performed on purified Ly49H+ WT NK cells and Stat4−/− NK cells from uninfected and MCMV-infected mixed BMC mice (day 2 PI). Normalized counts of Runx family members are shown (n = 2 to 3 biological replicates per group). (D) RNA-seq was performed on purified Ly49H+ and Ly49H− WT NK cells from uninfected and MCMV-infected mice (day 2 PI). Normalized counts of Runx1 and Runx3 are shown (n = 2 to 3 biological replicates per group). Data are presented as means ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001).
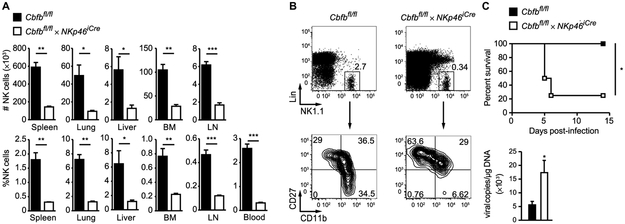
We first generated mice with NK cell–specific deletion of the gene encoding CBF-β (Cbfbfl/fl × NKp46iCre) because the function of all three Runx transcription factors depends on heterodimerization with CBF-β. Thus, Cbfbfl/fl × NKp46iCre mice represent a mouse model in which all Runx activity is ablated at the immature NK cell stage, when NKp46 is first expressed. Consistent with a previous report (25), we observed a profound reduction in the absolute numbers and percentages of NK cells in the bone marrow and peripheral organs of Cbfbfl/fl × NKp46Cre mice compared with littermate control mice (Fig. 4A), and the remaining NK cells in Cbfbfl/fl × NKp46iCre mice showed an immature phenotype (Fig. 4B). Moreover, lethally irradiated mice co-reconstituted with bone marrow from congenically distinct WT and Cbfbfl/fl × NKp46iCre mice revealed a cell-intrinsic requirement of Cbfb in NK cell development (fig. S2). To test whether the few remaining NK cells can provide a protective antiviral immune response, we infected Cbfbfl/fl × NKp46iCre and littermate control mice with MCMV. Cbfbfl/fl × NKp46icre mice were found to be highly susceptible to MCMV infection, with significantly increased viral titers and reduced overall survival compared with littermate controls (Fig. 4C). These data demonstrate that CBF-β and likely individual Runx transcription factors are required for NK cell development and protection against viral infection.
Fig. 4. CBF-β is required for NK cell development and host protection against MCMV infection.
(A) Percentage and absolute numbers of NK cells (Lin−NK1.1+) in the bone marrow (BM) and in peripheral organs of Cbfbfl/fl × NKp46iCre mice and Cbfbfl/fl littermate controls. (B) Representative flow cytometric plots show CD27 and CD11b expression of Lin− NK1.1+ NK cells in the blood of Cbfbfl/fl × NKp46iCre mice and Cbfbfl/fl littermate controls. Lin− refers to TCRβ−CD19−CD3ε−Ly6G−TER119−TCRγδ− cells. Data are representative of four independent experiments, with n = 3 to 5 mice. Samples were compared using two-tailed unpaired Student’s t test, and data are presented as means ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001). (C) Top graph shows survival of Cbfbfl/fl × NKp46iCre and Cbfbfl/fl littermate control mice infected with a lethal dose of MCMV. Bottom graph shows viral titers in the blood at 4 days PI. Data are representative of two to four independent experiments, with n = 3 to 5 mice. Samples were compared using two-tailed unpaired Student’s t test, and data are presented as means ± SEM (*P < 0.05).
Cbfb is essential for antiviral NK cell expansion and survival
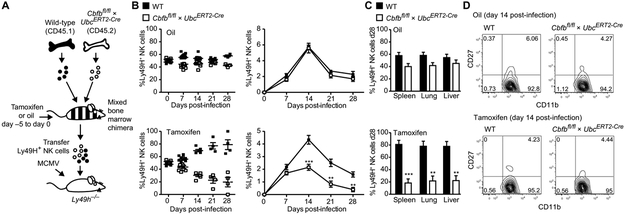
Because of the developmental defect, to investigate whether CBF-β is required specifically in mature NK cells during the antiviral response, we engineered mice in which the floxed Cbfb allele (Cbfbfl/fl) is crossed to a tamoxifen-inducible Cre cassette driven by the Ubiquitin promoter (UbcERT2-Cre). In mixed WT (CD45.1):Cbfbfl/fl × UbcERT2-Cre (CD45.2) BMC mice, tamoxifen treatment results in the Cre-mediated excision of the floxed Cbfb gene in all CD45.2+ cells, permitting us to investigate the function of CBF-β in peripheral NK cells while excluding the developmental defects observed in the Cbfbfl/fl × NKp46iCre mice. We adoptively transferred Ly49H+ NK cells from mixed WT (CD45.1):Cbfbfl/fl × UbcERT2-Cre (CD45.2) chimeras treated with either oil (as a control) or tamoxifen for 5 days into Ly49H-deficient mice (Fig. 5A). One day later, the recipients were infected with MCMV, and the ability of transferred Ly49H+ NK cells to undergo expansion and to generate long-lived memory cells was evaluated (Fig. 5A).
Fig. 5. Inducible deletion of CBF-β in NK cells results in defective expansion and generation of memory cells after viral infection.
(A) Schematic of experiment: WT:Cbfbfl/fl × UbcERT2-Cre–mixed bone marrow chimeras were treated with 1 mg of tamoxifen or oil daily for 5 days before adoptive transfer of Ly49H+ NK cells into Ly49H-deficient hosts. Recipient mice were infected with MCMV 1 day later. (B) Graphs depict the relative and absolute percentage of transferred WT and Cbfbfl/fl × UbcERT2-Cre NK cells in the blood at various time points after MCMV infection. (C) Percentage of transferred WT and Cbfbfl/fl × UbcERT2-Cre NK cells in indicated organs at day 28 PI. (D) CD27 versus CD11b expression of WT and Cbfbfl/fl × UbCERT2-Cre Ly49H+ NK cells at day 14 PI. Error bars show SEM, and graphs are representative of three independent experiments, with n = 4 to 6 mice per group (**P < 0.01, ***P < 0.001, two-tailed unpaired Student’s t test).
In this experiment, WT Ly49H+ NK cells rapidly expanded and reached peak percentages in tamoxifen-treated mice by day 14 PI, whereas expansion of Cbfb-deficient Ly49H+ NK cells was significantly reduced (Fig. 5B). This difference between the groups was not observed in control mice with adoptively transferred NK cell from WT:Cbfbfl/fl × UbcERT2-Cre chimeras treated with oil alone (Fig. 5B). Furthermore, a significant reduction in the percentage of Cbfb-deleted NK cells was detected in the spleen, lung, and liver 28 days after MCMV infection (Fig. 5C), revealing a crucial requirement for CBF-β in the generation of NK cell memory. Despite the defect in cell numbers, the maturation and activation of Cbfb-deleted NK cells were similar to WT controls during MCMV infection, as evidenced by the down-regulation of CD27 and up-regulation of CD11b (Fig. 5D). These findings suggest that activated NK cells lacking CBF-β may have a proliferation or survival defect. Transfer of NK cells from WT:Cbfbfl/fl × UbcERT2-Cre chimeras into Rag2−/− × Il2rg−/− mice followed by tamoxifen treatment resulted in reduced homeostatic expansion of Cbfb-deficient NK cells compared with WT NK cells (fig. S3, A and B), as IL-15 (required for NK cell homeostasis) was also shown to induce up-regulation of Runx1 and Runx3 expression in vitro (fig. S3C). Furthermore, Cbfb-deficient NK cells showed increased fluorochrome inhibitor of caspases (FLICA) incorporation when transferred into Rag2−/− × Il2rg−/− mice (fig. S3D), indicating that CBF-β and Runx family members may play an essential role in shielding peripheral NK cells from apoptosis during both virus- and lymphopenia-driven proliferation.
Nonredundant role for Runx1 and Runx3 in virus-specific NK cell expansion and generation of memory
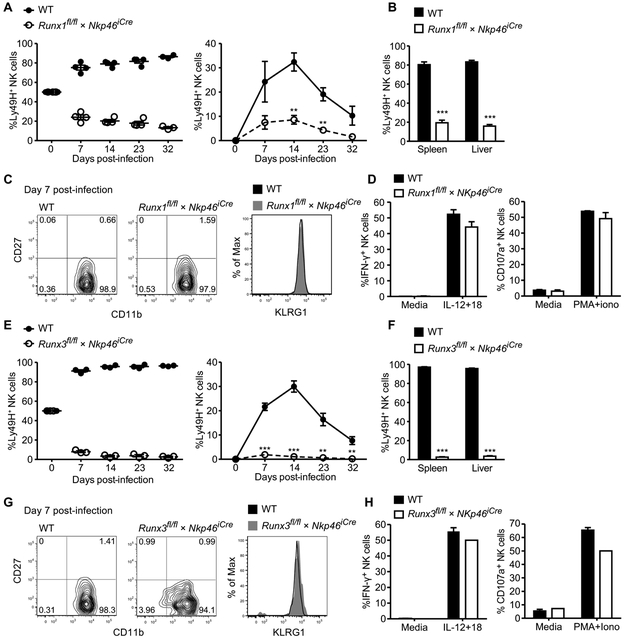
Given the observed expansion defect of Cbfb-deleted NK cells during viral infection, we sought to determine whether specific CBF-β-binding partners Runx1 or Runx3 were responsible for promoting NK cell expansion in response to MCMV. We generated mice with NK cell–specific deletion of Runx1 (Runx1fl/fl × NKp46iCre) or Runx3 (Runx3fl/fl × NKp46iCre). In contrast to Cbfbfl/fl × NKp46iCre mice, no significant changes were observed in NK cell percentages in mice with specific deletion of Runx1 or Runx3 compared with littermate controls (fig. S4) (25), suggesting that one Runx factor may be able to compensate for the loss of another during NK cell development or homeostasis. To test their responsiveness to MCMV infection, we cotransferred an equal number of WT (CD45.1) and Runx1fl/fl × NKp46iCre (CD45.2) NK cells into Ly49H-deficient mice followed by virus injection. WT NK cells robustly expanded in peripheral blood in response to MCMV infection, whereas Runx1fl/fl × NKp46iCre NK cells failed to expand (Fig. 6A). To determine whether a defect in trafficking of Runx1-deficient NK cells was responsible for the decreased numbers in peripheral blood, we harvested the spleen and liver of infected mice. Substantially higher numbers of WT NK cells were observed in the spleen and liver at 32 days PI (Fig. 6B), indicating a crucial requirement of Runx1 for NK cell expansion and memory formation in both lymphoid and nonlymphoid tissues. Runx1-deficient NK cells were also able to undergo maturation and activation similarly to WT NK cells during MCMV infection, as evidenced by the down-regulation of CD27 and up-regulation of CD11b and killer cell lectin-like receptor subfamily G member 1 (KLRG1; Fig. 6C). Furthermore, the few remaining memory NK cells in the Runx1-deficient population retained the ability to produce interferon-γ (IFN-γ) and degranulate after proinflammatory cytokine stimulation ex vivo (Fig. 6D), suggesting that, although Runx1 may mediate expansion of activated NK cells, it does not govern all NK cell effector functions.
Fig. 6. Runx1 and Runx3 are critical and nonredundant in promoting NK cell expansion and survival during MCMV infection.
WT and Runx1fl/fl × Nkp46iCre, or WT and Runx3fl/fl × Nkp46iCre Ly49H+ NK cells were cotransferred into Ly49H-deficient hosts and infected with MCMV. (A) Relative and absolute percentage of WT and Runx1fl/fl × Nkp46iCre Ly49H+ NK cells. (B) Percentage of transferred WT and Runx1fl/fl × Nkp46iCre NK cells in the spleen and liver at day 32 PI. (C) CD27 versus CDIIb and KLRG1 expression of WT and Runx1fl/fl × Nkp46iCre transferred NK cells in the blood at day 7 PI. (D) Memory NK cells isolated at day 28 PI were stimulated with IL-12 plus IL-18 (IL-12+18) or PMA plus ionomycin (PMA+iono) for 4 hours or were untreated. Percentages of memory WT and Runx1fl/fl × Nkp46iCre NK cells producing IFN-γ (left) or degranulating (right) are shown. (E) Graphs show percentage of WT and Runx3fl/fl × Nkp46iCre transferred NK cells in the blood at depicted time points as well as (F) in the spleen and liver at day 32 PI. (G) CD27 versus CD11b and KLRG1 expression of transferred NK cells in the blood for each group at day 7 PI. (H) Memory NK cells isolated at day 28 PI were stimulated as described in (D) and percentages of IFN-γ (left) or degranulating (right) memory NK cells are shown. Data are representative of two independent experiments, with n = 3 to 4 mice. Samples were compared using two-tailed unpaired Student’s t test, and data are presented as means ± SEM (**P < 0.01, ***P < 0.001).
Next, we tested the role of Runx3 in antiviral NK cells by cotransferring WT and Runx3fl/fl × NKp46iCre NK cells into Ly49H-deficient mice followed by MCMV infection. The results observed for Runx3-deficient NK cells were similar to the data obtained for Runx1-deficient NK cells in response to viral infection because Runx3fl/fl × NKp46iCre NK cells failed to expand (Fig. 6E) and were almost completely absent in the spleen and liver 32 days PI (Fig. 6F). As with the Runx1 deficiency, Runx3-deficient NK cells showed no defects in maturation and activation (Fig. 6G) and retained functional competence in memory populations (Fig. 6H). Together, these findings demonstrate a crucial and nonredundant requirement of Runx1 and Runx3 for the robust clonal expansion of NK cells during MCMV infection.
Runx1 regulates cell cycle during MCMV-driven NK cell proliferation
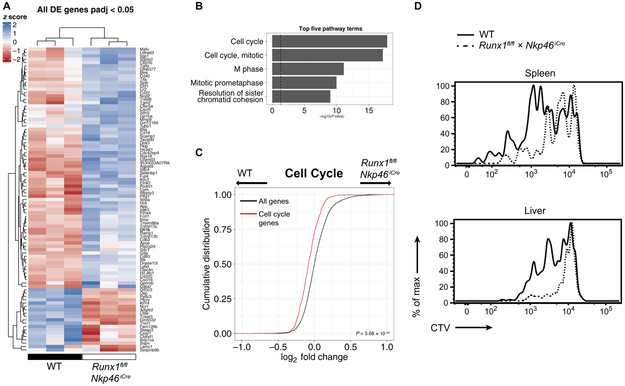
Last, we sought to determine the genes and cellular programs that Runx family transcription factors control in NK cells responding to MCMV infection. We performed RNA-seq on Ly49H+ WT and Runx1fl/fl × NKp46iCre NK cells isolated on day 3 PI, and our analysis revealed a subset of genes that were differentially up- or down-regulated (Fig. 7A). Although only a handful of genes were differentially expressed (Padj < 0.05; Fig. 7A), gene set analysis on moderated fold changes of all genes revealed a significant down-regulation of gene programs associated with cell cycle processes and cellular division in the Runx1-deficient NK cells compared with WT NK cells (Fig. 7, B and C). Consistent with this transcriptome analysis, CellTrace Violet (CTV) labeling of NK cells before adoptive cotransfer revealed that Runx1-deficient NK cells demonstrated decreased rates of cell division compared with their WT counterparts during MCMV infection (Fig. 7D). Together, these findings uncover a critical role for Runx1 in regulating a transcriptional program that controls cell cycle and proliferation during the adaptive NK cell response against virus infection.
Fig. 7. Runx1 promotes a cell cycle program during MCMV infection.
Splenic NK cells harvested from WT or Runx1fl/fl × Nkp46iCre mice were adoptively transferred into Ly49H-deficient recipients and infected with MCMV. RNA-seq was performed on WT and Runx1-deficient Ly49H+ NK cells harvested from the spleen 3 days PI. (A) Heat map depicting row-normalized blind rlog counts calculated by DESeq2. Genes that show differential expression at a cutoff of Padj < 0.05 are shown. (B) Top five enriched pathways as calculated by GAGE. Bar plots depict −log10 P values calculated by GAGE. (C) Cumulative distribution function plot of moderated log2 fold changes calculated by DESeq2 comparing cell cycle genes (red) and all genes (black). P value was calculated by one-sided Kolmogorov-Smirnov test. (D) WT and Runx1-deficient NK cells were labeled with CTV before adoptive transfer and MCMV infection. Representative histograms of CTV expression in Ly49H+ NK cells of indicated genotypes at 3.5 days PI are shown, and data are representative of three independent experiments, with n = 3 to 4 mice.
DISCUSSION
Here, we have found a role for the transcription factors Runx1 and Runx3 in driving the adaptive NK cell response during MCMV infection. Through STAT4 ChIP-seq analysis, we identified these two members of the Runx family among the top 20 transcription factor genes that show enrichment in STAT4 occupancy after stimulation of NK cells. Consistent with the known mechanism of STATs (26), our data demonstrate that proinflammatory cytokines (such as IL-12) that induce STAT4 phosphorylation and nuclear translocation in NK cells will initiate STAT4 binding to regulatory gene regions and subsequently induce gene expression, perhaps by modifying chromatin accessibility at specific gene loci.
Before the current study, relatively little was known about STAT4-dependent gene regulation within NK cells. In T cells, STAT4 ChIP has revealed many genes bound and regulated by STAT4, including Ifng, Il12rb2, and Il2ra (27, 28). Consistent with a broad role for STAT4 in the initiation of type 1 immune responses, IL-12, together with IFN-α and IFN-β, induces IFN-γ production in antiviral T cells via STAT4 (29). We previously demonstrated a similar mechanism of STAT4 regulation in NK cells, where Il12rb2−/− NK cells do not efficiently phosphorylate STAT4 and produce less IFN-γ than WT NK cells during viral infection (9). Furthermore, cotransfer of WT and Stat4−/− Ly49H+ NK cells into Ly49H-deficient hosts followed by MCMV infection revealed a crucial role of STAT4 for NK cell expansion and generation of long-lived memory NK cells (9). The expansion defect of IL-12 receptor– and STAT4-deficient NK cells closely resembles the phenotype observed in Runx1- and Runx3-deficient NK cells. In addition to the Runx transcription factors, our analysis revealed a wide variety of transcription factor genes, whose expression appears to be dependent on the presence of STAT4. Future studies will endeavor to determine whether additional factors synergize with Runx1 and Runx3 in driving NK cell expansion.
STAT transcription factor–induced expression of a particular gene is dependent on the status of chromatin structure, which is altered by adenosine triphosphate–induced nucleosome remodeling, histone incorporation, posttranslational histone modification, and DNA methylation (30). Little is known about the interaction of STATs and histone epigenetic marks in orchestrating gene expression in NK cells. During T cell differentiation, STAT transcription factors can induce epigenetic modifications of Ifng, Il4, Il3, and Il5 genes (30–33). More recently, it was reported that, in helper T cell differentiation, STATs can play a dominant role in promoting epigenetic changes associated with transcriptional activation (34). Our data support such an epigenetic mechanism and provide further evidence that STAT4 binding at gene loci induces a permissive epigenetic landscape (i.e., increase in H3K4me3) essential for increased gene expression, observed in activated NK cells after proinflammatory cytokine exposure or MCMV infection. Our study does not preclude the possibility that the H3K4me3 can reciprocally mediate recruitment of STAT4, and further studies using complementary techniques such as assay for transposase-accessible chromatin sequencing will lead to a more intricate view of the hierarchy and kinetics of chromatin accessibility events. However, on the basis of our current findings, we propose a model whereby STAT4 is a major regulator of H3K4me3 at Runx family gene loci and consequently controls expression of individual Runx genes during NK cell activation.
The role of CBF-β and Runx transcription factors has largely been studied in the context of T cell development and lineage specification (14). Previous reports have described how Runx1 is predominantly expressed in NK T and CD4+ T cells, whereas Runx3 is expressed in NK cells and CD8+ T cells. Runx3 knockout mice are viable but show severe defects in neurogenesis and thymopoiesis (35). CD4-targeted depletion of Runx3 results in derepression of Cd4 and decreased single-positive thymic CD8+ T cell numbers (17). Although previous groups reported that defects in Runx3 resulted in impaired NK cell development (36, 37), we only observed a minor decrease in NK cell numbers using Runx3fl/fl × NKp46iCre mice. In addition, Runx3 has been reported to regulate transcriptional programs in NK cells by initiating the expression of the human NK cell–specific killer cell immunoglobulin-like receptors and NKp46 (38, 39). Our findings in the Runx3fl/fl × NKp46iCre mice are also not consistent with this study because we find no defects in Ly49 repertoire or NKp46 expression. The discrepancies between our observations and existing data may be due to the nature of the models (genetic ablation versus overexpression of mutants) or the timing of Runx3 ablation in lymphocytes (at the immature NK cell stage versus germline mutations affecting T and NK cells). More recently, Runx3 has been shown to promote IL-15 signaling in NK cells by cooperating with E26 transformation-specific (ETS) and T-box transcription factors (36), and ChIP-seq analysis demonstrated specific gene regulation by Runx3 in IL-2-activated cytotoxic cells (40), together suggesting a general role for Runx3 in the response of lymphocytes to common γ-chain family cytokines. Because IL-15 is required for the homeostasis and survival of NK cells, these data are consistent with our observations that NK cell–specific ablation of Runx3 resulted in moderately decreased peripheral NK cell numbers. Last, Runx3 was recently found to affect the development and function of group 3 innate lymphoid cells (ILC3), where it specifically promoted retinoic acid receptor-related orphan receptor γ t (RORγt) expression in a lineage-specific manner to drive ILC3 development (25).
Beyond NK cell development, little was previously known about the requirement of Runx family members and CBF-β during activation and antiviral effector responses of mature NK cells. The expression of Runx and CBF-β transcription factors at all maturation stages during the life span of NK cells suggested a crucial and continuous role of these proteins for NK cell activation and function (41). Consistent with our observation of a loss in NK cell numbers during NK cell–specific or tamoxifen-inducible deletion of Cbfb, germline CBF-β deficiency (which abrogates the function of all Runx proteins) results in embryonic lethality (42). Similarly, Runx1-deficient mice showed a loss in hematopoietic stem cells resulting in embryonic lethality between days 12.5 and 13.5 (42). A conditional knockout of Cbfb in CD4-expressing cells induces derepression of Zbtb7b in CD4+ CD8low T cells (43), whereas a CD4-specific knockout of Runx1 disabled positive selection and maturation of single-positive thymic CD4+ T cells and resulted in a loss of NK T cells (17). Our current findings demonstrate that Cbfb, Runx1, and Runx3 are all essential factors throughout NK cell development and response against pathogens, in contrast to Nfil3, a transcription factor that is critical for NK cell and ILC development but dispensable for the homeostasis of mature NK cells and ILCs, and during pathogen infection (44–48).
Although our work would have been technically challenging without the use of specific Cre and loxP systems, we acknowledge the experimental limitations associated with these genetic tools. Despite the inherent caveats found in our mouse models, it will still be interesting to address whether specific and tamoxifen-inducible deletion of Runx1 or Runx3 at later time points will affect generation or maintenance of long-lived memory NK cells in infection or homeostasis settings; such inducible ablation of individual Runx family members will allow us to investigate whether these transcription factors have an enduring or sustained impact beyond when they are first up-regulated. A second caveat of the study can be found in the in vivo versus in vitro comparisons, where we have drawn some conclusions based on NK cells from viral infection (RNA-seq) and NK cells stimulated with proinflammatory cytokines (ChIP-seq). Because existing ChIP-seq techniques require large numbers of cells (in the millions), it is not feasible to assess STAT4 binding or H3K4me3 levels in NK cells from infected mice (numbering in the thousands) at the current time. As improved protocols are developed where specific transcription factor binding or chromatin modifications can be assessed using low numbers of cells, we will be able to determine the precise nature of the signals and transcriptional requirements for mediating epigenetic control of gene transcription and cellular processes such as clonal proliferation and immune memory.
Here, we have demonstrated the mechanistic induction and epigenetic regulation of CBF-β and Runx family members in NK cells exposed to the proinflammatory cytokine milieu generated during virus challenge. In mice infected with MCMV, we observed a pronounced expansion and memory defect in NK cells deficient in either CBF-β or one of its binding partners Runx1 or Runx3 and ascribe an indispensible role for these transcription factors in promoting the adaptive responses of virus-specific NK cells. Furthermore, we find that Runx1 controls a broad transcriptional program associated with cell cycle and cellular proliferation. Future transcriptome and functional studies will address whether Runx3 contributes in a similar fashion or plays a different and unique role (e.g., preventing apoptosis) from Runx1 in driving the clonal expansion of antiviral NK cells. Nonetheless, our current work highlights the importance of core-binding transcription factors for NK cell proliferation and survival during their adaptive response against viral infection and may inform current therapeutic strategies being developed to harness these potent cytotoxic innate lymphocytes toward the treatment of human diseases.
MATERIALS AND METHODS
Mice and viral infection
All mice used in this study were bred at the Memorial Sloan Kettering Cancer Center in accordance with the guidelines of the Institutional Animal Care and Use Committee. The following strains were used, all on the C57BL/6 genetic background: C57BL/6 (CD45.2; the Jackson Laboratory), B6.SJL (CD45.1; Taconic), NKp46iCre (49), Runx1fl/fl (50), Runx3fl/fl and Cbfbfl/fl (51), Stat4−/− (52), Klra8−/− [Ly49h−/− (53)], UbcERT2-Cre, and Rag2−/− × Il2rg−/− (the Jackson Laboratory). Generation of mixed BMC mice and adoptive transfer studies were performed as previously described (54).
In some experiments, Rag2−/− × Il2rg−/− recipient mice or BMC mice were injected intraperitoneally with 4-hydroxytamoxifen (1 mg per day) dissolved in corn oil (Sigma) or corn oil alone as a control for five consecutive days. Experimental mice were infected by intraperitoneal injection of 7.5 × 102 plaque-forming units (PFU) of in vivo passaged, salivary gland–derived MCMV (Smith Strain). Mice infected with a lethal dose of MCMV were intraperitoneally injected with 4 × 104 PFU of MCMV. Experiments were conducted using age- and gender-matched mice in accordance with approved institutional protocols.
Determining viral titers
Viral titers were determined as previously described (55). Briefly, DNA was isolated from indicated organs using a genomic purification kit (Qiagen). After isolation, DNA concentration was measured using NanoDrop for each sample, and 3 μl was added into Master Mix containing iQ SYBR Green (Bio-Rad) and primers specific to MCMV DNA (forward: 5′-TCGCCCATCGTTTCGAGA-3′ and reverse: 5′-TCTCGTAGGTCCACTGACCGA-3′). Copy number was determined by comparing Cq values with a standard curve of known dilutions of a MCMV plasmid.
Flow cytometry and cell sorting
Cell surface staining of single-cell suspensions from the indicated organs was performed using fluorophore-conjugated antibodies (BD Biosciences, eBioscience, BioLegend, Tonbo Biosciences, and R&D Systems). For intracellular cytokine staining, cells were fixed and permeabilized with the transcription factor staining kit (eBioscience). Apoptosis was evaluated by caspase activity staining using the carboxy-fluorescein FLICA Poly Caspase Assay kit (ImmunoChemistry Technologies). NK cell proliferation was analyzed by labeling cells with 5 μM CTV (Invitrogen) before transfer into recipient mice, and CTV labeling was performed according to the manufacturer’s protocol. Flow cytometry and cell sorting were performed on the LSR II and Aria II cytometers (BD Biosciences), respectively. The data were analyzed using FlowJo software (Tree Star).
Chromatin immunoprecipitation and DNA sequencing or RNA-seq
NK cells (5 × 106 to 10 × 106; TCRβ−CD19−CD3ε−Ly6G−TER119−T-CRγδ−NK1.1+) were sorted from spleens of pooled C57BL/6 mice and incubated with or without IL-12 (20 ng/ml) and IL-18 (10 ng/ml). NK cells were stimulated for 30 min or 18 hours, followed by H3K4me3 or STAT4 ChIP, respectively. DNA and proteins were cross-linked for 7.5 min using 0.75% formaldehyde. ChIP was performed as previously described (10, 16), using 10 μg of rabbit polyclonal anti-STAT4 antibody (Santa Cruz Biotechnology, sc-468, clone C-20) or 1 μg of rabbit polyclonal anti-trimethyl-histone H3 (Lys4) antibody (H3K4me3; Millipore, 07473) followed by Illumina next-generation sequencing. For RNA-seq, RNA was isolated from sorted cell populations using TRIzol (Invitrogen), followed by SMARTer amplification and paired-end Illumina next-generation sequencing.
ChIP-seq and RNA-seq analyses
ChIP-seq reads from all samples were trimmed to remove low-quality reads using Trimmomatic (version 0.36) (56). Trimmed reads were mapped to the Mus musculus genome (mm10 assembly) using Bowtie2 (version 2.2.9) (57). Concordantly aligned paired mates were used for peak calling by MACS2 (version 2.1.1.20160309) (58) at P = 0.01. For STAT4 ChIP samples, irreproducible discovery rate (IDR) calculations using Python3 scripts (version 2.0.2) (59) were performed on three independent replicates, and a union of peaks passing a threshold of 0.05 was used as a reference peak list for read counts. This list was further filtered by selecting only peaks that showed more than a 1.5-fold increase in counts over a stimulated Stat4−/− ChIP sample. Peaks were annotated using the Bioconductor ChIPpeakAnno package (version 3.6.5) (60, 61) with the University of California Santa Cruz mm10 knownGene gene model. List of transcription factors was obtained from the online Animal Transcription Factor Database (62). For H3K4me3 ChIP samples, peaks were called using MACS2 arguments --broad --broad-cutoff 0.1 --nomodel --extsize 146 and further filtered using a more stringent cutoff of q Value > 10 (or q < 1 × 10−10) before creating a union of peaks. To normalize for the sequencing depth of the H3K4me3 samples, reads were counted within regions equal in width to called peaks located at the midpoint between the called peak and its closest neighbor. These numbers were used to calculate size factors using the DESeq2 (version 1.14.1) method (60, 61) and subsequently used to scale raw values. All genomic tracks were generated using the Gvis R package (version 1.18.2) (63).
RNA-seq samples were trimmed and mapped as above. Counts were normalized for sequencing depth using size factors calculated by DESeq2 in R (versions 3.3.2 and 3.3.3). For gene set analysis, log2 fold changes estimated by the DESeq2 pipeline were used as input for gene set enrichment analysis by Generally Applicable Gene Set Enrichment (GAGE; version 2.24.0) (64). Pathways were retrieved from the Reactome database available on Bioconductor (65). The M. musculus cell cycle gene set plotted in Fig. 7 was Reactome ID 5990980.
Statistical analyses
Data are shown as means ± SEM in all graphs, and statistical differences were calculated using two-tailed unpaired Student’s t test or as indicated. P < 0.05 was considered significant. All statistical analyses and plots were produced in GraphPad Prism or R.
Supplementary Material
Fig. S1. Epigenetic regulation by STAT4 in cytokine-stimulated NK cells.
Fig. S2. Cell-intrinsic requirement of CBF-β for the development of mature NK cells.
Fig. S3. CBF-β is critical for NK cell survival after homeostatic proliferation.
Fig. S4. Runx1- or Runx3-deficient mice have normal NK cell numbers.
Acknowledgments:
We thank members of the Sun laboratory for comments, discussions, technical support, and experimental assistance. We thank the Immunological Genome Consortium for providing the microarray data used in this study. We thank D. Littman, S. Vidal, E. Vivier, S. Rudensky, and T. Egawa for providing mice used in this study.
Funding: M.R. was supported by a fellowship from the German Academic Exchange Service (DAAD; Germany). C.M.L. was supported by a T32 award from the NIH (CA009149). N.M.A. was supported by a Medical Scientist Training Program grant from the National Institute of General Medical Sciences of the NIH under award number T32GM007739 to the Weill Cornell/Rockefeller/Sloan-Kettering Tri-Institutional MD-PhD Program. T.E.O. was supported by a fellowship from the American Cancer Society. J.C.S. was supported by the Ludwig Center for Cancer Immunotherapy, the Burroughs Wellcome Fund, the American Cancer Society, and grants from the NIH (AI100874, AI130043, and P30CA008748).
Footnotes
SUPPLEMENTARY MATERIALS
immunology.sciencemag.org/cgi/content/full/2/18/eaan3796/DC1
Competing interests: The authors declare that they have no competing interests.
REFERENCES AND NOTES
- 1.Cerwenka A, Lanier LL, Natural killer cell memory in infection, inflammation and cancer. Nat. Rev. Immunol 16, 112–123 (2016). [DOI] [PubMed] [Google Scholar]
- 2.O’Sullivan TE, Sun JC, Lanier LL, Natural killer cell memory. Immunity 43, 634–645 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun JC, Lanier LL, NK cell development, homeostasis and function: Parallels with CD8+ T cells. Nat. Rev. Immunol 11, 645–657 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vivier E, Raulet DH, Moretta A, Caligiuri MA, Zitvogel L, Lanier LL, Yokoyama WM, Ugolini S, Innate or adaptive immunity? The example of natural killer cells. Science 331, 44–49 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun JC, Lopez-Verges S, Kim CC, DeRisi JL, Lanier LL, NK cells and immune “memory”. J. Immunol 186, 1891–1897 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper MA, Elliott JM, Keyel PA, Yang L, Carrero JA, Yokoyama WM, Cytokine-induced memory-like natural killer cells. Proc. Natl. Acad. Sci. U.S.A 106, 1915–1919 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Madera S, Rapp M, Firth MA, Beilke JN, Lanier LL, Sun JC, Type I IFN promotes NK cell expansion during viral infection by protecting NK cells against fratricide. J. Exp. Med 213, 225–233 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Madera S, Sun JC, Cutting edge: Stage-specific requirement of IL-18 for antiviral NK cell expansion. J. Immunol 194, 1408–1412 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun JC, Madera S, Bezman NA, Beilke JN, Kaplan MH, Lanier LL, Proinflammatory cytokine signaling required for the generation of natural killer cell memory. J. Exp. Med 209, 947–954 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beaulieu AM, Zawislak CL, Nakayama T, Sun JC, The transcription factor Zbtb32 controls the proliferative burst of virus-specific natural killer cells responding to infection. Nat. Immunol 15, 546–553 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morinobu A, Gadina M, Strober W, Visconti R, Fornace A, Montagna C, Feldman GM, Nishikomori R, O’Shea JJ, STAT4 serine phosphorylation is critical for IL-12-induced IFN-γ production but not for cell proliferation. Proc. Natl. Acad. Sci. U.S.A 99, 12281–12286 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchieri G, Interleukin-12 and the regulation of innate resistance and adaptive immunity. Nat. Rev. Immunol 3, 133–146 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Ito Y, Oncogenic potential of the RUNX gene family: ‘Overview’. Oncogene 23, 4198–4208 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Collins A, Littman DR, Taniuchi I, RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol 9, 106–115 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taniuchi I, Littman DR, Epigenetic gene silencing by Runx proteins. Oncogene 23, 4341–4345 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Zheng Y, Josefowicz SZ, Kas A, Chu T-T, Gavin MA, Rudensky AY, Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR, The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J. Exp. Med 204, 1945–1957 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grueter B, Petter M, Egawa T, Laule-Kilian K, Aldrian CJ, Wuerch A, Ludwig Y, Fukuyama H, Wardemann H, Waldschuetz R, Möröy T, Taniuchi I, Steimle V, Littman DR, Ehlers M, Runx3 regulates integrin αE/CD103 and CD4 expression during development of CD4−/CD8+ T cells. J. Immunol 175, 1694–1705 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Kitoh A, Ono M, Naoe Y, Ohkura N, Yamaguchi T, Yaguchi H, Kitabayashi I, Tsukada T, Nomura T, Miyachi Y, Taniuchi I, Sakaguchi S, Indispensable role of the Runx1-Cbfβ transcription complex for in vivo-suppressive function of FoxP3+ regulatory T cells. Immunity 31, 609–620 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Komine O, Hayashi K, Natsume W, Watanabe T, Seki Y, Seki N, Yagi R, Sukzuki W, Tamauchi H, Hozumi K, Habu S, Kubo M, Satake M, The Runx1 transcription factor inhibits the differentiation of naive CD4+ T cells into the TH2 lineage by repressing GATA3 expression. J. Exp. Med 198, 51–61 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rudra D, Egawa T, Chong MMW, Treuting P, Littman DR, Rudensky AY, Runx-CBFβ complexes control expression of the transcription factor Foxp3 in regulatory T cells. Nat. Immunol 10, 1170–1177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taniuchi I, Osato M, Egawa T, Sunshine MJ, Bae S-C, Komori T, Ito Y, Littman DR, Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell 111, 621–633 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Vahedi G, Takahashi H, Nakayamada S, Sun H-W, Sartorelli V, Kanno Y, O’Shea JJ, STATs shape the active enhancer landscape of T cell populations. Cell 151, 981–993 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung W-K, Shahab A, Kuznetsov VA, Bourque G, Oh S, Ruan Y, Ng H-H, Wei C-L, Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell 1, 286–298 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, Groner Y, Bern MD, Stappenbeck TS, Colonna M, Egawa T, Yokoyama WM, Runx3 specifies lineage commitment of innate lymphoid cells. Nat. Immunol 16, 1124–1133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy DE, Darnell JE Jr., Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol 3, 651–662 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Letimier FA, Passini N, Gasparian S, Bianchi E, Rogge L, Chromatin remodeling by the SWI/SNF-like BAF complex and STAT4 activation synergistically induce IL-12Rβ2 expression during human Th1 cell differentiation. EMBO J 26, 1292–1302 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O’Sullivan A, Chang H-C, Yu Q, Kaplan MH, STAT4 is required for interleukin-12-induced chromatin remodeling of the CD25 locus. J. Biol. Chem 279, 7339–7345 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA, Critical role for STAT4 activation by type 1 interferons in the interferon-γ response to viral infection. Science 297, 2063–2066 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Bernstein BE, Meissner A, Lander ES, The mammalian epigenome. Cell 128, 669–681 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA, T helper cell differentiation: Regulation by cis elements and epigenetics. Immunity 24, 369–379 (2006). [DOI] [PubMed] [Google Scholar]
- 32.Wilson CB, Rowell E, Sekimata M, Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol 9, 91–105 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Zhang F, Boothby M, T helper type 1–specific Brg1 recruitment and remodeling of nucleosomes positioned at the IFN-γ promoter are Stat4 dependent. J. Exp. Med 203, 1493–1505 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wei L, Vahedi G, Sun H-W, Watford WT, Takatori H, Ramos HL, Takahashi H, Liang J, Gutierrez-Cruz G, Zang C, Peng W, O’Shea JJ, Kanno Y, Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity 32, 840–851 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levanon D, Bettoun D, Harris-Cerruti C, Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C, Fliegauf M, Kremer E, Otto F, Brenner O, Lev-Tov A, Groner Y, The Runx3 transcription factor regulates development and survival of TrkC dorsal root ganglia neurons. EMBO J 21, 3454–3463 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levanon D, Negreanu V, Lotem J, Bone KR, Brenner O, Leshkowitz D, Groner Y, Transcription factor Runx3 regulates interleukin-15-dependent natural killer cell activation. Mol. Cell. Biol 34, 1158–1169 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ohno S-I, Sato T, Kohu K, Takeda K, Okumura K, Satake M, Habu S, Runx proteins are involved in regulation of CD122, Ly49 family and IFN-γ expression during NK cell differentiation. Int. Immunol 20, 71–79 (2008). [DOI] [PubMed] [Google Scholar]
- 38.Anderson SK, Transcriptional regulation of NK cell receptors. Curr. Top. Microbiol. Immunol 298, 59–75 (2006). [DOI] [PubMed] [Google Scholar]
- 39.Lai CB, Mager DL, Role of runt-related transcription factor 3 (RUNX3) in transcription regulation of natural cytotoxicity receptor 1 (NCR1/NKp46), an activating natural killer (NK) cell receptor. J. Biol. Chem 287, 7324–7334 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lotem J, Levanon D, Negreanu V, Leshkowitz D, Friedlander G, Groner Y, Runx3-mediated transcriptional program in cytotoxic lymphocytes. PLOS ONE 8, e80467 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bezman NA, Kim CC, Sun JC, Min-Oo G, Hendricks DW, Kamimura Y, Best JA, Goldrath AW, Lanier LL, The Immunological Genome Project Consortium, Molecular definition of the identity and activation of natural killer cells. Nat. Immunol 13, 1000–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang Q, Stacy T, Miller JD, Lewis AF, Gu T-L, Huang X, Bushweller JH, Bories J-C, Alt W, Ryan G, Liu PP, Wynshaw-Boris A, Binder M, Marín-Padilla M, Sharpe AH, Speck NA, The CBFβ subunit is essential for CBFα2 (AML1) function in vivo. Cell 87, 697–708 (1996). [DOI] [PubMed] [Google Scholar]
- 43.Setoguchi R, Tachibana M, Naoe Y, Muroi S, Akiyama K, Tezuka C, Okuda T, Taniuchi I, Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science 319, 822–825 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Firth MA, Madera S, Beaulieu AM, Gasteiger G, Castillo EF, Schluns KS, Kubo M, Rothman PB, Vivier E, Sun JC, Nfil3-independent lineage maintenance and antiviral response of natural killer cells. J. Exp. Med 210, 2981–2990 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, O’Sullivan TE, van den Brink MR, Pamer EG, Hanash AM, Sun JC, Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J. Exp. Med 211, 1723–1731 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, Huntington ND, Belz T, Carotta S, Nfil3 is required for the development of all innate lymphoid cell subsets. J. Exp. Med 211, 1733–1740 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xu W, Domingues RG, Fonseca-Pereira D, Ferreira M, Ribeiro H, Lopez-Lastra S, Motomura Y, Moreira-Santos L, Bihl F, Braud V, Kee B, Brady H, Coles MC, Vosshenrich C, Kubo M, Di Santo JP, Veiga-Fernandes H, NFIL3 orchestrates the emergence of common helper innate lymphoid cell precursors. Cell Rep 10, 2043–2054 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Yu X, Wang Y, Deng M, Li Y, Ruhn KA, Zhang CC, Hooper LV, The basic leucine zipper transcription factor NFIL3 directs the development of a common innate lymphoid cell precursor. Elife 3, e04406 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Narni-Mancinelli E, Chaix J, Fenis A, Kerdiles YM, Yessaad N, Reynders A, Gregoire C, Luche S Ugolini, E. Tomasello, T. Walzer, E. Vivier, Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proc. Natl. Acad. Sci. U.S.A 108, 18324–18329 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Growney JD, Shigematsu H, Li Z, Lee BH, Adelsperger J, Rowan R, Curley DP, Kutok JL, Akashi K, Williams IR, Speck NA, Gilliland DG, Loss of Runx1 perturbs adult hematopoiesis and is associated with a myeloproliferative phenotype. Blood 106, 494–504 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Naoe Y, Setoguchi R, Akiyama K, Muroi S, Kuroda M, Hatam F, Littman DR, Taniuchi I, Repression of interleukin-4 in T helper type 1 cells by Runx/Cbfβ binding to the Il4 silencer. J. Exp. Med 204, 1749–1755 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kaplan MH, Sun Y-L, Hoey T, Grusby MJ, Impaired IL-12 responses and enhanced development of TH2 cells in Stat4-deficient mice. Nature 382, 174–177 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Fodil-Cornu N, Lee S-H, Belanger S, Makrigiannis AP, Biron CA, Buller RM, Vidal SM, Ly49h-deficient C57BL/6 mice: A new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J. Immunol 181, 6394–6405 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sun JC, Beilke JN, Lanier LL, Adaptive immune features of natural killer cells. Nature 457, 557–561 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Johnson LR, Weizman O-E, Rapp M, Way SS, Sun JC, Epitope-specific vaccination limits clonal expansion of heterologous naive T cells during viral challenge. Cell Rep 17, 636–644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bolger AM, Lohse M, Usadel B, Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Langmead B, Salzberg SL, Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, Liu XS, Model-based analysis of ChIP-Seq (MACS). Genome Biol 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Li Q, Brown JB, Huang H, Bickel PJ, Measuring reproducibility of high-throughput experiments. Ann. Appl. Stat 5, 1752–1779 (2011). [Google Scholar]
- 60.Zhu LJ, Gazin C, Lawson ND, Pagès H, Lin SM, Lapointe DS, Green MR, ChIPpeakAnno: A bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang H-M, Chen H, Liu W, Liu H, Gong J, Wang H, Guo A-Y, AnimalTFDB: A comprehensive animal transcription factor database. Nucleic Acids Res 40, D144–D149 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hahne F, Ivanek R, Visualizing genomic data using Gviz and bioconductor. Methods Mol. Biol 1418, 335–351 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ, GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinformatics 10, 161 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yu G, He Q-Y, ReactomePA: An R/Bioconductor package for reactome pathway analysis and visualization. Mol. Biosyst 12, 477–479 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Epigenetic regulation by STAT4 in cytokine-stimulated NK cells.
Fig. S2. Cell-intrinsic requirement of CBF-β for the development of mature NK cells.
Fig. S3. CBF-β is critical for NK cell survival after homeostatic proliferation.
Fig. S4. Runx1- or Runx3-deficient mice have normal NK cell numbers.