Abstract
Rapid amplification of cDNA ends (RACE) is a prevalent technique used to obtain the 5′ ends of transcripts. Several different 5′ RACE methods have been developed, and one particularly simple and efficient approach called CapFinder relies on the 5′ cap-dependent template-switching that occurs in eukaryotes. However, most prokaryotic transcripts lack a 5′ cap structure. Here, we report a procedure to capture primary transcripts based on capping the 5′ triphosphorylated RNA in prokaryotes. Primary transcripts were first treated with vaccinia capping enzyme to add a 5′ cap structure. First-strand cDNA was then synthesized using Moloney murine leukaemia virus reverse transcriptase. Finally, a template-switching oligonucleotide with a tail containing three ribonucleic acid guanines was hybridized to the cDNA 3′ poly(C) and further used as template for reverse transcriptase. It is oligonucleotide sequence independent and is more sensitive compared to RLM-RACE. This approach specifically identified the transcription start sites of ompA, sodB and shiA in Escherichia coli and of ompA, rne and rppH in Brucella melitensis. Furthermore, we also successfully identified the transcription start sites of small noncoding genes ryhB and micC in E. coli and bsnc135 and bsnc149 in B. melitensis. Our findings suggest that Capping-RACE is a simple, accurate, and sensitive 5′ RACE method for use in prokaryotes.
INTRODUCTION
Bacteria respond to a variety of environmental conditions by manipulating the expression of related genes, and the regulation of transcripts plays an important role in this process. The 5′ untranslated region (5′ UTR) extending from the transcription start site (TSS) to the start codon is crucial for mRNA stability and translational efficiency (1). Therefore, the accurate identification of TSSs is indispensable for characterizing the structure and function of genes.
Rapid amplification of cDNA ends (RACE) is a novel technique used to determine the 5′ end sequences of transcripts (2). In classic RACE, a homopolymer tail that acts as an anchor sequence can be added to the 3′ end of cDNA by terminal deoxynucleotidyl transferase (TdT). The 5′ terminal information is then obtained by PCR using a gene-specific primer (GSP) and an anchored primer complementary to the anchored sequence (2–4). Sequence- and ligation-independent cloning PCR (SLIC-PCR) and ligation-anchored PCR (LA-PCR) were subsequently developed to circumvent the nonspecific amplifications caused by using primers containing the anticomplementary sequence of the homopolymeric tail (5,6). These two methods are based on the ligation of a single-stranded modified oligonucleotide to the 3′ end of first-strand cDNA, catalysed by T4 RNA ligase. However, the 5′ ends determined with these methods might not be the actual transcription start sites because reverse transcriptases are subject to premature termination (7).
Another strategy for reducing nonspecific amplications, known as RNA ligase-mediated amplification of cDNA ends (RLM-RACE), was also developed (7,8). In RLM-RACE, the full-length RNA strands are treated by tobacco acid pyrophosphatase (TAP) to remove the 5′ cap structure and pyrophosphate, and then a RNA adaptor is directly ligated to the RNA 5′ ends using T4 RNA ligase prior to reverse transcription. However, the RNA truncated at the 5′ end can also be ligated to the RNA adaptor in this method. To address this limitation, an alternate method, oligo-capping, was developed. This approach works by removing the 5′ pyrophosphate of non-capped RNAs to leave a hydroxyl before TAP treatment; thus, only the 5′ ends of capped RNAs can be obtained using this method (9).
The methods described above all have multiple enzymatic steps, making them very time-consuming to perform, and also require quite large amounts of starting RNA. The CapFinder RACE method was designed to address these limitations (10,11). This procedure relies on the ability of the Moloney murine leukaemia virus (M-MLV) reverse transcriptase to add two to four cytosine residues to the 3′ end of newly synthesized cDNA upon reaching the 5′ cap structure of mRNAs. A template-switching oligonucleotide (TSO) containing a 3′ poly(rG) tail is added to base pair with the extra cytosine residues, and then the TSO serves as a template for reverse transcription. As a result, a known adaptor is attached to the 3′ terminus of the cDNA. To enhance the specificity and efficiency of this approach, various additional techniques, such as step-out PCR, inverse PCR and T-RACE, have been devised (12–14).
Notably, most of the methods mentioned above were developed for the mapping of eukaryotic transcripts. Prokaryotic RNA generally lack the 5′ cap structure, which is essential for the application of CapFinder RACE (10,11). Furthermore, prokaryotic transcripts also lack a 3′ poly(A) tail, which is used as a reverse transcription primer in eukaryotic RACE (15). Thus, the enrichment of primary transcripts in prokaryotes has been a great challenge. Recently, a novel approach called dRNA-seq was established (16,17). This protocol can enrich primary transcripts containing 5′ triphosphates by utilizing terminator exonuclease (TEX), which specifically eliminates 5′ monophosphorylated RNA. dRNA-seq has been widely applied to the genome-wide mapping of TSSs in different organisms and has even been applied to TSS mapping of the barley chloroplast (18–25). In addition, the Cappable-seq method has been reported as an alternative strategy to isolate primary transcripts in high-throughput sequencing (26,27). Unlike dRNA-seq, which removes unnecessary RNAs, in Cappable-seq, the 5′ triphosphorylated RNAs are capped with biotinylated GTP using the vaccinia capping enzyme (VCE); the capped RNAs are then captured on streptavidin-coated magnetic beads and separated from other RNA.
Here, we describe a simplified 5′ RACE method to precisely determine the TSSs of specific genes in prokaryotes that is based on the addition of a 7-methylguanylate cap structure to the 5′ end of triphosphorylated RNA using VCE. In this approach, the 3′ ends of cDNAs are obtained by template-switching as in CapFinder (10,11) and are then amplified by nest-PCR. Using this new method, we successfully identified the TSSs of protein-coding genes and non-coding genes in Escherichia coli and Brucella melitensis, suggesting that the 5′ end sequences of other prokaryotic transcripts could be readily mapped by this Capping-RACE procedure.
MATERIALS AND METHODS
Primers
The complete list of primers used in this study is provided as Supplementary Material (Supplementary Table S1).
Bacterial strains and growth conditions
Escherichia coli strain MG1655 was cultured in Luria–Bertani (LB) medium. Brucella melitensis strain 16M was grown in tryptic soy broth (TSB) in a Biosafety Level 3 laboratory as previously described (28). Bacteria were grown at 37°C, 180 rpm to an OD600 of 0.5–0.6. Growth of bacterial culture (5 ml per sample) was stopped by adding 0.625 ml of pre-chilled stop solution (5% phenol/95% ethanol). Cells were harvested by centrifugation for 10 min at 6000 × g, and the resulting pellet was stored at −80°C.
RNA extraction and DNase I treatment
Frozen cell pellets were thawed on ice and resuspended in 100 μl per pellet of TE buffer (10 mM Tris–HCl, 1 mM EDTA, pH8.0). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). The RNA samples were then subjected to treatment with DNase I (TaKaRa, Dalian, China) to remove genomic DNA. The concentration and quality of each sample was evaluated using a Nano Drop (Thermo Fisher Scientific, Waltham, MA, USA) and agarose gel electrophoresis.
In vitro RNA transcription and purification
The template for the 111 nt in vitro transcription RNA was amplified from pwtCas9-bacteria obtained from Addgene (#44250) by forward primer 5′-CAGTAATACGACTCACTATTAGAAGCGACTCGTCTCAAACGG-3′, which contains a T7 φ2.5 promoter, and reverse primer 5′-AGGATGACGTTCATGCTTCTTGT-3′. The templates for A8AGAA, G8GGAA, G8GAAA, G8GUAA, G8GCAA, G0, G1, G2, G3 and G4 were amplified by pairs of partially complementary oligodeoxynucleotides (Supplementary Table S1). The in vitro transcribed RNAs were generated with a T7 High Efficiency Transcription Kit (TransGen Biotech, Beijing, China). The 111 nt in vitro transcription RNA was purified using a Zymo Concentrator-5 kit (Zymo Research, Irvine, CA, USA), all other RNAs were purified by ethanol precipitation.
Different treatments of the 111 nt in vitro transcribed RNA
The 111 nt in vitro transcribed RNA was pre-treated to modify the 5′ ends as follows: (i) 1 μg of RNA was incubated with 1 μl of RNA 5′ pyrophosphohydrolase (RppH) (NEB, Ipswich, MA, USA) for 1 h at 37°C; (ii) 1 μg of RNA was incubated with 1 μl of VCE (NEB) for 30 min at 37°C; or (iii) 1 μg of RNA was treated with RppH for 1 h at 37°C and purified by ethanol precipitation, followed by incubation with VCE for 30 min at 37°C. All samples were purified by ethanol precipitation and evaluated using a Nano Drop (Thermo Fisher Scientific).
Capping of total bacterial RNA
A total of 1–10 μg of bacterial RNA was treated with 1 μl of VCE (NEB) as described above for 5′ RACE, and the treated RNA was then purified by ethanol precipitation.
Capping-RACE cDNA synthesis
The cDNA synthesis was performed in two steps as previously described (29). A total of 0.5 μg of RNA and 2 pmol of gene-specific primer 1 (GSP1) were incubated at 65°C for 5 min and then quickly chilled on ice for 2 min before the addition of the following remaining components in a total volume of 10 μl: 1 μl of 10 mM dNTPs, 2 μl of 5 × buffer, 0.5 μl of DTT and 0.5 μl of 200 U SuperScript III Reverse Transcriptase (Invitrogen). After incubation at 50°C for 60 min, 10 μl of a mixture of 2 μl of 0.1% bovine serum albumin (BSA), 0.8 μl of 50 mM MnCl2, 1 μl of TSO, 2 μl of 5× buffer, 0.5 μl of DTT, 0.5 μl of 200 U SuperScript III Reverse Transcriptase, and 3.2 μl DEPC-treated water was added to each sample. The combined reaction was then incubated at 42°C for an additional 90 min. Finally, the reverse transcriptase was inactivated by heating the samples at 70°C for 10 min.
RLM-RACE
RLM-RACE was performed as described previously with modification (7–9). Briefly, bacterial RNA was treated with 1 μl of RNA 5′ pyrophosphohydrolase (RppH) (NEB, Ipswich, MA, USA) for 1 h at 37°C, and purified by ethanol precipitation. The treated RNA was ligated to a synthetic RNA adaptor with T4 RNA ligase 1 (Thermo Fisher Scientific) for 30 min at 37°C. The ligation product was reverse transcribed with 2 pmol of gene-specific primer 1 (GSP1) by SuperScript III Reverse Transcriptase (Invitrogen) for 1 h at 55°C and 10 min at 70°C. The cDNA was amplified by nest-PCR. Finally, the PCR products were detected by agarose gel electrophoresis.
Nest-PCR and sequencing
For amplification of the 5′ end sequences, nest-PCR was performed using multiple sets of primers. Briefly, the first-round PCR was carried out using the outer primer and GSP1. The outer primer contains a part of the TSO. The resulting product was then diluted 100-fold before being used as the template for the second-round PCR, which was performed using the inner primer and GSP2. The inner primer is identical to the 3′ end of the outer primer. The final PCR products were separated by agarose gel electrophoresis and purified from the gel using a Universal DNA Purification Kit (Tiangen, Beijing, China). Finally, the purified product was cloned into pMD18-T vector (Takara), and the resulting clones were sequenced.
RESULTS
General strategy for Capping-RACE
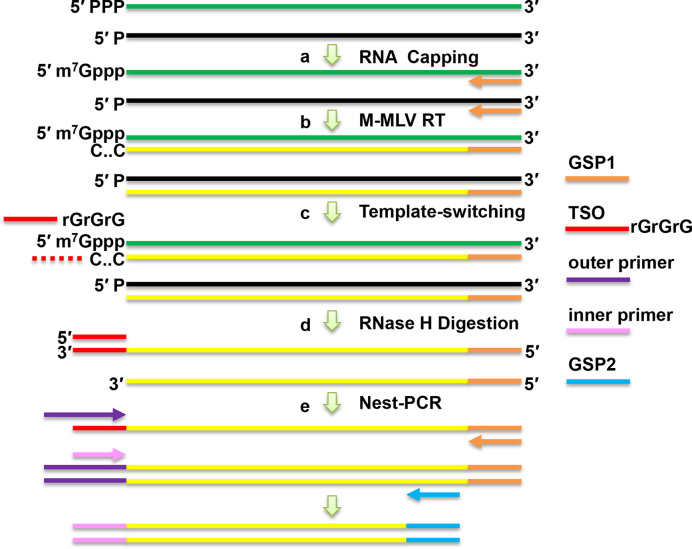
The overall strategy for the Capping-RACE approach is illustrated in Figure 1. The basic principle of this method is the treatment of total RNA with VCE to add a cap structure to the end of 5′ triphosphorylated RNA (Figure 1A). The cDNA synthesis by M-MLV reverse transcriptase begins with a gene-specific primer (GSP1) (Figure 1B). In the presence of MnCl2 and BSA, the reverse transcriptase adds a few cytosines to the 3′ end of the cDNA when reaching the cap structure; this allows cDNA generated by the capped RNAs to be hybridized to a TSO adaptor containing a 3′ poly(rG) tail (Figure 1C). The cDNA polycytosine end synthesized by RT continues to extend in response to the TSO. RNase H is then used for the removal of complementary RNA to enhance the PCR efficiency (Figure 1D). Next, nest-PCR is performed using two sets of primers (Figure 1E). Finally, the PCR products are cloned into a pMD18-T vector, and the products are sent away for sequencing.
Figure 1.
Schematic representation of the Capping-RACE procedure. (A) Total RNA is treated with vaccinia capping enzyme (VCE), which caps only the 5′ triphosphorylated RNA. (B) First-strand cDNA synthesis is performed using M-MLV reverse transcriptase with a gene-specific primer (GSP1). (C) A template switching oligonucleotide (TSO) with a 3′ end composed of three ribonucleic acid guanines hybridizes to the cDNA 3′ poly(C) and is used as a template for reverse transcriptase. (D) Hybrid RNA is removed by RNase H. (E) Nest-PCR is performed using two sets of primers. The PCR products are cloned into pMD18-T vectors, which are sent out for commercial sequencing.
Verification of the feasibility of Capping-RACE
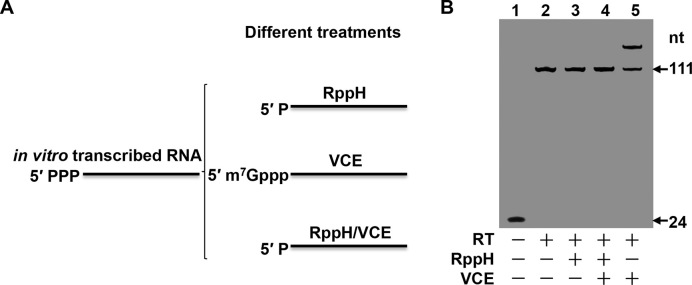
To determine whether the Capping-RACE method can be used to map primary transcripts, we conducted experiments on a 111 nt in vitro transcribed RNA that has a 5′ triphosphorylated end. The in vitro transcribed RNA was pre-treated to modify the 5′ end in one of three ways (Figure 2A). In pre-treatment 1, the RppH treatment removes pyrophosphate from the RNA to leave a 5′ monophosphate RNA. In pre-treatment 2, the VCE treatment adds a cap structure to the 5′ end of the RNA. Pre-treatment 3 was a dual-treatment in which RppH treatment was performed prior to VCE treatment; this served as a control. The first-strand cDNA synthesis was then initiated with the FAM-RT primer 1 modified on the 5′ end using 6-carboxy fluorescein phosphoramidite. As expected, template-switching did not occur for the 5′ triphosphorylated and 5′ monophosphorylated RNAs that lacked cap structures (Figure 2B, lanes 2 and 3). Since RppH-treated RNA cannot be capped, the dual-treated RNA was also not attached to a TSO adaptor (Figure 2B, lane 4). About 60% of the capped RNA was attached by the TSO adaptor (Figure 2B, lane 5). These results suggest that Capping-RACE is feasible for mapping the 5′ end of transcribed RNAs.
Figure 2.
Experimental verification of Capping-RACE with in vitro transcribed RNA. (A) Different treatments of in vitro transcribed RNA. (B) The cDNA products from the differently treated RNA described in (A). Lane 1, the negative control, performed without adding reverse transcriptase (RT). Lane 2, in vitro transcribed RNA that was not subjected to any treatment. Lane 3, in vitro transcribed RNA subjected to RppH treatment. Lane 4, in vitro transcribed RNA subjected to dual treatment, i.e. RppH treatment prior to VCE treatment. Lane 5, in vitro transcribed RNA subjected to vaccinia capping enzyme (VCE) treatment. The reaction products were analysed on a 12% non-denaturing polyacrylamide gel and detected by a fluorescence image analyser (FUJIFILM, FLA-5100).
Capping-RACE is sequence independent
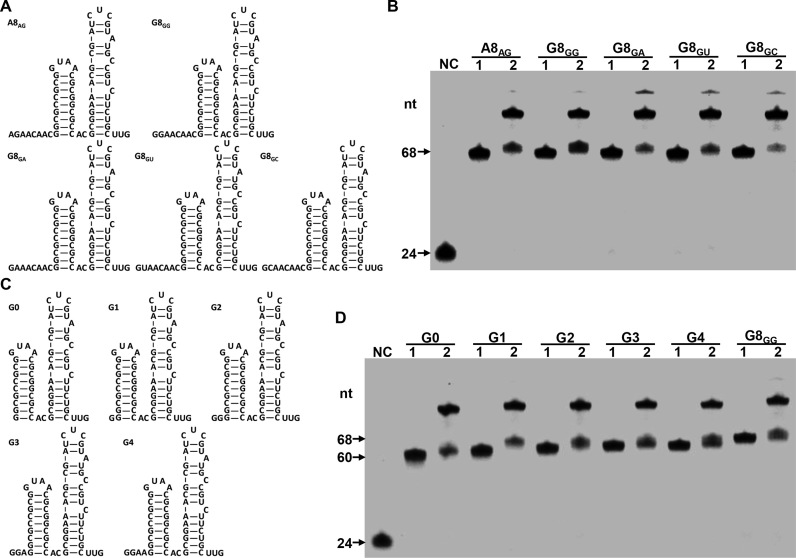
The data described above (Figure 2) indicated that Capping-RACE could be a simplified 5′ RACE method. To investigate whether Capping-RACE has a bias for 5′ terminal sequence of transcripts, we designed several in vitro transcribed RNAs which had known nucleic acid sequence and secondary structure, referring to reported oligonucleotides with modifications (30). All the RNAs comprised an 8-nucleotide single-stranded segment and two stem-loop structures (Figure 3A). We tested the efficiency of Capping-RACE for these RNAs. The first-strand cDNA synthesis was initiated with the FAM-RT primer 2 modified on the 5′ end using 6-carboxy fluorescein phosphoramidite. As shown in Figure 3B, all the RNAs were capped and subsequently ligated to the TSO with a high yield (>50%).
Figure 3.
Determination of the Capping-RACE efficiency for transcripts with different 5′ terminal sequence. (A and C) The sequence and secondary structure of in vitro transcribed RNAs are shown. The letter indicates the 5′ terminal nucleotide, and the numeral indicates the number of unpaired nucleotides at the 5′ end. (B and D) The cDNA products from in vitro transcribed RNAs. Lane NC, the negative control, FAM-RT primer 2 on the 5′ end using 6-carboxy fluorescein phosphoramidite. Lane 1 and lane 2, Capping-RACE for the in vitro transcribed RNAs without or with TSO.
Next, to determine the effect of complex structures on Capping-RACE, we synthesized a set of G8GG variants by truncating the length of single-stranded segment (G0, G1, G2, G3 and G4; Figure 3C). These variants have only 0 to 4 unpaired nucleotides at the 5′ end. Then we applied Capping-RACE to these variants. As anticipated, the effectiveness of Capping-RACE still exceeded 50% for each RNA (Figure 3D). These results gave further support to the utility of Capping-RACE for prokaryotic transcripts.
The application and reproducibility of Capping-RACE for selected examples
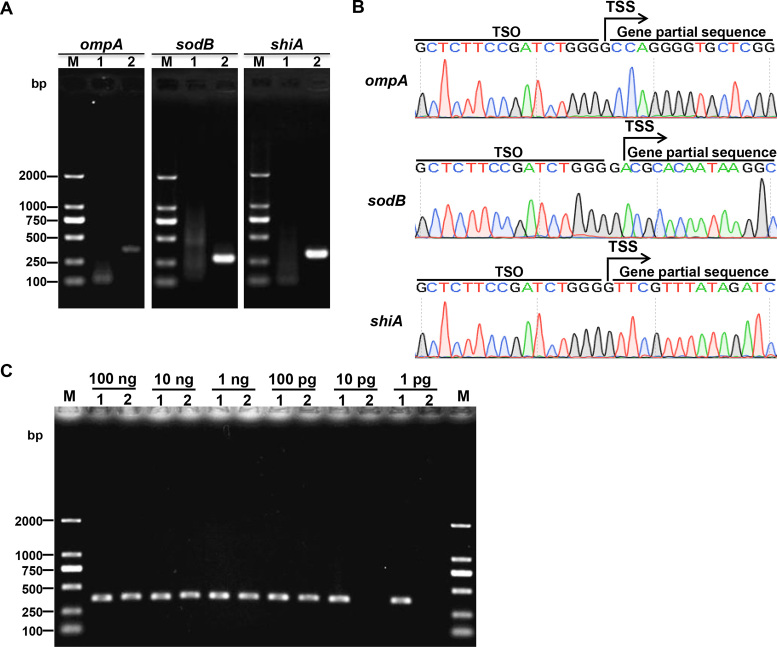
To test Capping-RACE in prokaryotes, we selected three protein-coding genes, ompA, sodB and shiA, of the model organism E. coli MG1655 as targets. The resulting PCR products are shown in Figure 4A. The specific second-round PCR products were purified from the gel and sequenced (Figure 4B). The TSS information acquired by Capping-RACE is in accordance with the experimental data published previously (31). To ascertain the sensitivity of Capping-RACE, we used a variety of starting input of total RNA to identify the TSS of ompA. Simultaneously, RLM-RACE was carried out for comparison. The result showed that only 1 pg of total RNA was required for Capping-RACE, however, RLM-RACE need at least 100 pg of total RNA (Figure 4C). The data suggest that Capping-RACE is more sensitive than RLM-RACE.
Figure 4.
5′ RACE mapping of E. coli MG1655 protein-coding genes. (A) Nest-PCR products of 5′ RACE starting from E. coli MG1655 total RNA. ompA, outer membrane protein A (371 bp); sodB, superoxide dismutase (294 bp); shiA, shikimate transporter (320 bp). Lane M, DL2000 DNA Marker (Takara); lane 1, first-round PCR products; lane 2, second-round PCR products. (B) The sequencing result of the second-round PCR products. (C) The second-round PCR products of Capping-RACE and RLM-RACE for ompA, starting from different input of total RNA. Lane M, DL2000 DNA Marker (Takara); lane1, Capping-RACE; lane2, RLM-RACE.
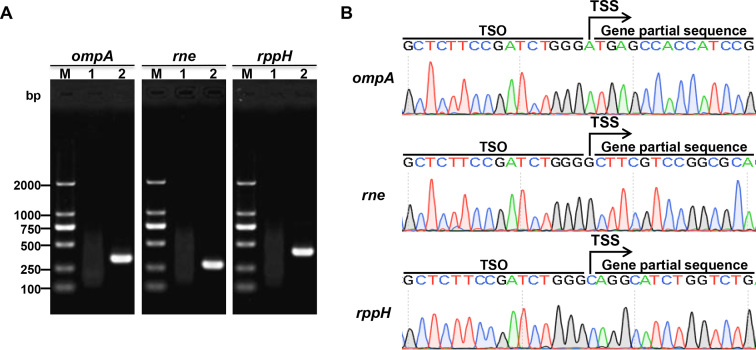
For further validation of this method in another prokaryotic species, we applied Capping-RACE to identifying the TSSs of protein-coding genes of B. melitensis 16M, which is an important facultative intracellular pathogen. As shown in Figure 5, accurate TSSs were identified by the Capping-RACE data. These results confirm that Capping-RACE could be used for TSS identification of prokaryotic protein-coding genes.
Figure 5.
5′ RACE mapping of B. melitensis 16M protein-coding genes. (A) Nest-PCR products of 5′ RACE starting from B. melitensis 16M total RNA. ompA, outer membrane protein A (337 bp); rne, an endoribonuclease (286 bp); rppH, RNA pyrophosphohydrolase (400 bp). Lane M, DL2000 DNA Marker (Takara); lane 1, first-round PCR products; lane 2, second-round PCR products. (B) The sequencing result of the second-round PCR products.
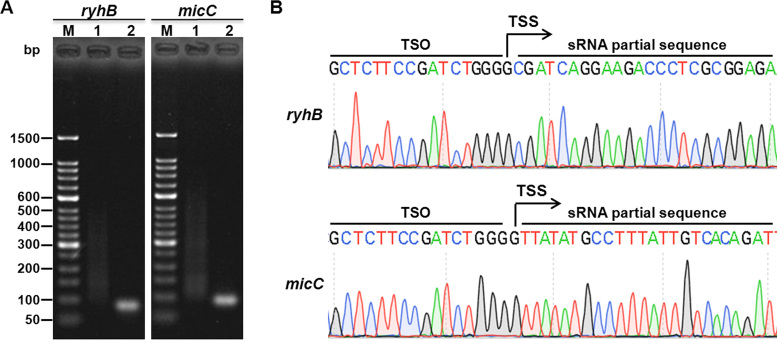
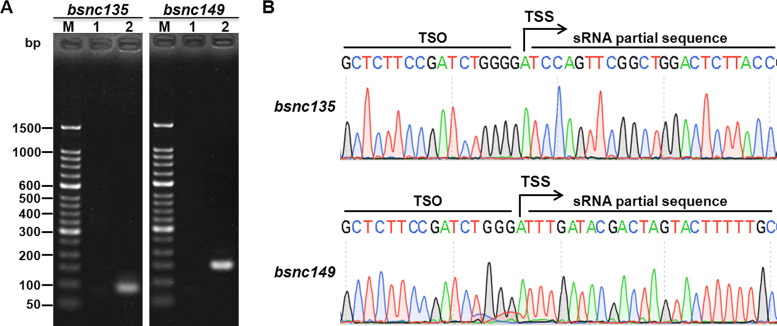
Capping-RACE of prokaryotic non-coding genes
We also selected two sRNAs from each of the above two bacterial species. RyhB and micC are two of the most important sRNAs of E. coli, and bsnc135 and bsnc149 are two sRNAs of B. melitensis 16M that were identified previously (32). As expected, the 5′ ends were detected successfully (Figures 6 and 7), and all the TSS information obtained was in accordance with published data (30,32). The results reveal that Capping-RACE can be applied not only to protein-coding genes but also to non-coding genes.
Figure 6.
5′ RACE mapping of E. coli MG1655 non-coding genes. (A) Nest-PCR products of 5′ RACE of two E. coli MG1655 sRNAs, ryhB (85 bp) and micC (93 bp). Lane M, 50 bp DNA Ladder (Takara); lane 1, first-round PCR products; lane 2, second-round PCR products. (B) The sequencing result of the second-round PCR products. The positions of the TSSs are indicated by black arrows.
Figure 7.
5′ RACE mapping of B. melitensis 16M non-coding genes. (A) Nest-PCR products of 5′ RACE of two B. melitensis 16M sRNAs, bsnc135 (92 bp) and bsnc149 (155 bp). Lane M, 50 bp DNA Ladder (Takara); lane 1, first-round PCR products; lane 2, second-round PCR products. (B) The sequencing result of the second-round PCR products. The positions of the TSSs are indicated by black arrows.
DISCUSSION
RNA-seq has rapidly become a powerful tool for understanding gene sequence and structure as well as RNA-based regulation in diverse organisms. However, RNA-seq is an extremely complex and time-consuming method because it requires the construction of a cDNA library followed by sequencing and data analysis. This technique is mainly suitable for comprehensive transcriptome analyses. Currently, dRNA-seq and Cappable-seq provide approaches for genome-wide mapping of TSSs (16,17,26,27). However, RACE is still an efficient technique for precisely obtaining the 5′ end sequence of specific transcripts. Since most prokaryotic transcripts lack a 5′ cap structure and a 3′ poly(A) tail, the previous RACE methods that were developed for use with eukaryotic transcripts cannot be used effectively for prokaryotes.
In this study, we present a simplified 5′ RACE strategy, i.e. Capping-RACE, for targeting genes in prokaryotes. This approach adds a cap structure to any transcripts containing a 5′ terminal triphosphate. The 5′ ends of cDNAs are then obtained by template-switching in the same manner as in CapFinder (10,11). Our data show that this new method can be successfully applied to determining the 5′ ends of both protein-coding and non-coding genes (Figures 4–7). Since this approach cannot add cap structure to transcripts terminated with 5′ monophosphate, 5′ hydroxyl or 5′ NAD cap, the method is not suitable for these transcrips. Compared with previous procedures, our method has several advantages. It is more accurate and sensitive because it avoids many non-specific effects by capping the 5′ triphosphorylated RNA. This method has been confirmed to be sequence independent (Figure 3). Furthermore, only a minimal amount of total RNA is required for template-switching, so this method requires less starting material than alternative techniques (Figure 4C). Our results suggest that Capping-RACE has significant application potential for the identification of TSSs of prokaryotic transcripts.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Katie Oakley, PhD, from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Technique Innovation Program of Hubei Province [2016ABA124]; Fundamental Research Funds for the Central Universities [2016PY052 to Z.-F.L.]. Funding for open access charge: Technique Innovation Program of Hubei Province.
Conflict of interest statement. None declared.
REFERENCES
- 1. Waters L.S., Storz G.. Regulatory RNAs in bacteria. Cell. 2009; 136:615–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Frohman M.A., Dush M.K., Martin G.R.. Rapid production of full-length cdnas from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc. Natl. Acad. Sci. U.S.A. 1988; 85:8998–9002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ohara O., Dorit R.L., Gilbert W.. One-sided polymerase chain reaction: the amplification of cdna. Proc. Natl. Acad. Sci. U.S.A. 1989; 86:5673–5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Loh E.Y., Elliott J.F., Cwirla S., Lanier L.L., Davis M.M.. Polymerase chain reaction with single-sided specificity: analysis of t cell receptor delta chain. Science. 1989; 243:217–220. [DOI] [PubMed] [Google Scholar]
- 5. Edwards J.B., Delort J., Mallet J.. Oligodeoxyribonucleotide ligation to single-stranded cdnas: a new tool for cloning 5′ ends of mrnas and for constructing cdna libraries by in vitro amplification. Nucleic Acids Res. 1991; 19:5227–5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Troutt A.B., Mcheyzer-Williams M.G., Pulendran B., Nossal G.J.. Ligation-anchored pcr: a simple amplification technique with single-sided specificity. Proc. Natl. Acad. Sci. U.S.A. 1992; 89:9823–9825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Liu X., Gorovsky M.A.. Mapping the 5′ and 3′ ends of tetrahymena thermophila mrnas using rna ligase mediated amplification of cdna ends (rlm-race). Nucleic Acids Res. 1993; 21:4954–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fromont-Racine M., Bertrand E., Pictet R., Grange T.. A highly sensitive method for mapping the 5′ termini of mrnas. Nucleic Acids Res. 1993; 21:1683–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Maruyama K., Sugano S.. Oligo-capping: a simple method to replace the cap structure of eukaryotic mrnas with oligoribonucleotides. Gene. 1994; 138:171–174. [DOI] [PubMed] [Google Scholar]
- 10. Schmidt W.M., Mueller M.W.. Capselect: a highly sensitive method for 5′cap-dependent enrichment of full-length cdna in pcr-mediated analysis of mrnas. Nucleic Acids Res. 1999; 27:e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schramm G., Bruchhaus I., Roeder T.. A simple and reliable 5′-race approach. Nucleic Acids Res. 2000; 28:E96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matz M., Shagin D., Bogdanova E., Britanova O., Lukyanov S., Diatchenko L., Chenchik A.. Amplification of cdna ends based on template-switching effect and step-out pcr. Nucleic Acids Res. 1999; 27:1558–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huang J.-C., Chen F.. Simultaneous amplification of 5′ and 3′ cDNA ends based on template-switching effect and inverse PCR. BioTechniques. 2006; 40:187–189. [DOI] [PubMed] [Google Scholar]
- 14. Bower N.I., Johnston I.A.. Targeted rapid amplification of cDNA ends (T-RACE)–an improved RACE reaction through degradation of non-target sequences. Nucleic Acids Res. 2010; 38:e194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sorek R., Cossart P.. Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat. Rev. Genet. 2009; 11:9–16. [DOI] [PubMed] [Google Scholar]
- 16. Sharma C.M., Hoffmann S., Darfeuille F., Reignier J., Findeiß S., Sittka A., Chabas S., Reiche K., Hackermüller J., Reinhardt R. et al. . The primary transcriptome of the major human pathogen Helicobacter pylori. Nature. 2010; 464:250–255. [DOI] [PubMed] [Google Scholar]
- 17. Sharma C.M., Vogel J.. Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014; 19:97–105. [DOI] [PubMed] [Google Scholar]
- 18. Bohn C., Rigoulay C., Chabelskaya S., Sharma C.M., Marchais A., Skorski P., Borezée-Durant E., Barbet R., Jacquet E., Jacq A. et al. . Experimental discovery of small RNAs in Staphylococcus aureus reveals a riboregulator of central metabolism. Nucleic Acids Res. 2010; 38:6620–6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhelyazkova P., Sharma C.M., Forstner K.U., Liere K., Vogel J., Borner T.. The primary transcriptome of barley chloroplasts: numerous noncoding RNAs and the dominating role of the Plastid-Encoded RNA polymerase. Plant Cell. 2012; 24:123–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cortes T., Schubert Olga T., Rose G., Arnvig Kristine B., Comas I., Aebersold R., Young Douglas B.. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in mycobacterium tuberculosis. Cell Rep. 2013; 5:1121–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Voigt K., Sharma C.M., Mitschke J., Joke Lambrecht S., Voß B., Hess W.R., Steglich C.. Comparative transcriptomics of two environmentally relevant cyanobacteria reveals unexpected transcriptome diversity. ISME J. 2014; 8:2056–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Babski J., Haas K.A., Näther-Schindler D., Pfeiffer F., Förstner K.U., Hammelmann M., Hilker R., Becker A., Sharma C.M., Marchfelder A. et al. . Genome-wide identification of transcriptional start sites in the haloarchaeon Haloferax volcanii based on differential RNA-Seq (dRNA-Seq). BMC Genomics. 2016; 17:629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhukova A., Fernandes L.G., Hugon P., Pappas C.J., Sismeiro O., Coppée J.-Y., Becavin C., Malabat C., Eshghi A., Zhang J.-J. et al. . Genome-Wide transcriptional start site mapping and sRNA identification in the pathogen leptospira interrogans. Front. Cell. Infect. Microbiol. 2017; 7:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heidrich N., Bauriedl S., Barquist L., Li L., Schoen C., Vogel J.. The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res. 2017; 45:6147–6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hilzinger J.M., Raman V., Shuman K.E., Eddie B.J., Hanson T.E., Drake H.L.. Differential RNA sequencing implicates sulfide as the master regulator of S0 metabolism in chlorobaculum tepidum and other green sulfur bacteria. Appl.Environ.Microbiol. 2018; 84:e01966-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ettwiller L., Buswell J., Yigit E., Schildkraut I.. A novel enrichment strategy reveals unprecedented number of novel transcription start sites at single base resolution in a model prokaryote and the gut microbiome. BMC Genomics. 2016; 17:199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boutard M., Ettwiller L., Cerisy T., Alberti A., Labadie K., Salanoubat M., Schildkraut I., Tolonen A.C.. Global repositioning of transcription start sites in a plant-fermenting bacterium. Nat. Commun. 2016; 7:13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Zheng K., Chen D.-S., Wu Y.-Q., Xu X.-J., Zhang H., Chen C.-F., Chen H.-C., Liu Z.-F.. MicroRNA expression profile in RAW264.7 cells in response to brucella melitensis infection. Int. J. Biol. Sci. 2012; 8:1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pinto F.L., Lindblad P.. A guide for in-house design of template-switch-based 5′ rapid amplification of cDNA ends systems. Anal. Biochem. 2010; 397:227–232. [DOI] [PubMed] [Google Scholar]
- 30. Hsieh P.K., Richards J., Liu Q., Belasco J.G.. Specificity of RppH-dependent RNA degradation in Bacillus subtilis. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:8864–8869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gama-Castro S., Salgado H., Santos-Zavaleta A., Ledezma-Tejeida D., Muñiz-Rascado L., García-Sotelo J.S., Alquicira-Hernández K., Martínez-Flores I., Pannier L., Castro-Mondragón J.A. et al. . RegulonDB version 9.0: high-level integration of gene regulation, coexpression, motif clustering and beyond. Nucleic Acids Res. 2015; 44:D133–D143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saadeh B., Caswell C.C., Chao Y., Berta P., Wattam A.R., Roop R.M., O′Callaghan D., Zhulin I.B.. Transcriptome-wide identification of Hfq-associated RNAs in Brucella suis by deep sequencing. J. Bacteriol. 2016; 198:427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.