Abstract
Living organisms possess two types of tRNAs for methionine. Initiator tRNAs bind directly into the ribosomal P-site to initiate protein synthesis, and the elongators bind to the A-site during the elongation step. Eubacterial initiators (tRNAfMet) are unique in that the methionine attached to them is formylated to facilitate their binding to initiation factor 2 (IF2), and to preclude them from binding to elongation factor Tu (EFTu). However, in mammalian mitochondria, protein synthesis proceeds with a single dual function tRNAMet. Escherichia coli possesses four tRNAfMet (initiator) and two tRNAMet (elongator) genes. Free-living organisms possessing the mitochondrion like system of single tRNAMet are unknown. We characterized mutants of E. coli tRNAfMet that function both as initiators and elongators. We show that some of the tRNAfMet mutants sustain E. coli lacking all four tRNAfMet and both tRNAMet genes, providing a basis for natural occurrence of mitochondria like situation in free living organisms. The tRNA mutants show in vivo binding to both IF2 and EFTu, indicating how they carry out these otherwise mutually exclusive functions by precise regulation of their in vivo formylation. Our results provide insights into how distinct initiator and elongator methionine tRNAs might have evolved from a single ‘dual function’ tRNA.
INTRODUCTION
Living organisms have evolved with two essential and distinct forms of methionine decoding tRNAs. The initiator tRNA (i-tRNA or tRNAfMet) inserts methionine at the first position, and the elongator tRNAs (tRNAMet) at the downstream positions in proteins (1). In bacteria, while tRNAfMet is recognized by initiation factor 2 (IF2) for its binding to the ribosomal P-site, tRNAMet binds elongation factor Tu (EFTu) to enter the A-site (2,3). The exclusive role of tRNAfMet in initiation is facilitated by its unique structural and sequence characteristics (4). The three consecutive G:C base pairs found in the anticodon stems of i-tRNAs are evolutionarily conserved in all domains of life, and facilitate i-tRNA binding to the ribosomal P-site. In addition, tRNAfMet possesses a Watson Crick mismatch at position 1 × 72, which together with the 2:71 and 3:70 base pairs (Supplementary Figure S1), constitutes a major element for its recognition by methionine-tRNA formyltransferase (Fmt) (5,6). Formylation of the amino acid attached to tRNAfMet plays a critical role in its binding to IF2 (3,7,8).
In contrast, mammalian mitochondria possess a single tRNAMet (9) which binds to the mitochondrial IF2 (IF2mt) to take part in initiation (when formylated), and to EFTumt to take part in elongation (when not formylated). Competition between Fmtmt and EFTumt for binding to Met-tRNAMet is assumed to ensure the dual function of the tRNAMet in mammalian mitochondria (10). The dual-nature of this tRNAMet is aided, at least in part, by the fact that Fmtmt primarily recognizes the amino acid attached to the tRNA as opposed to eubacterial Fmt, whose primary determinants are situated in the acceptor stem of the tRNA itself, necessitating two distinct tRNAs for initiation and elongation functions (10). In yet another example of dual function tRNAs, Trypanosoma brucei, a protist that lacks mitochondrially encoded tRNAs and relies on tRNAs imported from the cytosol to sustain translation in the organelle, utilizes the eukaryotic-type elongator tRNAMet imported from cytosol for initiation after its formylation in mitochondria (11). Understanding the origin of dedicated tRNAs for initiation and elongation from such dual function tRNAs may provide evolutionary insights. In addition, it is of interest to understand the mechanism by which the single mitochondrial tRNAMet is distributed into the initiation and elongation steps of protein synthesis.
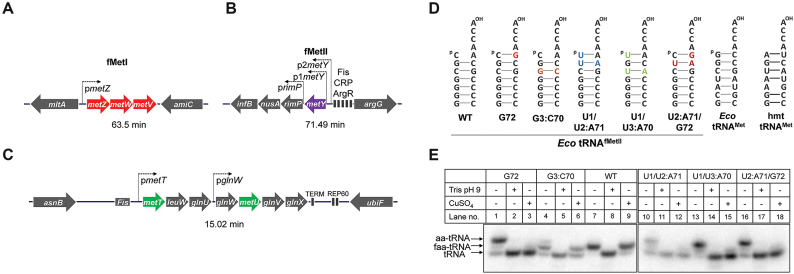
In E. coli, of the four tRNAfMet genes, three (metZ, metW and metV) encoding tRNAfMetI are in an operon at 63.5 min (metZWV), and the fourth (metY) encoding tRNAfMetII is present at 71.49 min (Figure 1A and B). The tRNAfMetI and tRNAfMetII differ by a single nucleotide at position 46 (12). Interestingly, this change allows separation of the two tRNAs on the native gels (13). As for the elongators, two genes (metT and metU) encoding identical tRNAMet are present in the metT-leuW-glnU-glnW-metU-glnV-glnX operon at 15.02 min along with other essential elongator tRNA genes (Figure 1C). The metZWV and metY genes can be independently deleted without significantly affecting growth at 37°C (14–16) (Supplementary Figure S2). Because the function of tRNAfMet is essential in E. coli, simultaneous deletion of both loci can only be achieved in the presence of supporting tRNAfMet gene(s) elsewhere in the chromosome or on a plasmid (16). Likewise, it should be possible to delete both the chromosomal copies of the tRNAMet genes in the presence of tRNAMet gene support. However, generating a mitochondrion like system for tRNAMet in E. coli would demand deletion of all four of the tRNAfMet (metZWV and metY) and both the tRNAMet (metT and metU) genes from the chromosome and sustaining the strain on a single gene encoding methionine tRNA with dual functions as initiator and elongator.
Figure 1.
Mutants of the E. coli initiator tRNA and their in vivo status. Schematic of organization of the initiator (A and B) and elongator (C) methionine tRNA gene loci in E. coli. fMetI encoding tRNA genes (metZWV) are shown in red, the fMetII encoding gene (metY) is shown in purple and the methionine elongator tRNA genes (metT and metU) are shown in green. Only the promoters relevant to tRNA gene transcription have been depicted. (D) Structures of the acceptor stems of the E. coli tRNAfMet, tRNAfMet mutants used, and tRNAMet. Mutations are shown in colour. (E) In vivo status of the tRNAfMet mutants as analyzed by acid urea PAGE followed by northern blot analysis using 32P-5′-end labeled oligomers complementary to nucleotides 39–55 of metY. Total tRNA was isolated from ΔmetY strains overexpressing the mutant tRNAs from plasmid. Aminoacylated tRNA population is depicted as aa-tRNA, formylated tRNA population as faa-tRNA and decylated tRNA population as tRNA.
MATERIALS AND METHODS
Strains, plasmids and DNA oligomers
Lists of strains/plasmids and DNA oligomers used are provided in Supplementary Tables S1 and S2.
Generation of plasmid constructs
pT6
The 881 bp genomic region containing the leuW-glnU-glnW-metU-glnV-glnX operon was PCR amplified from E. coli KL16 genomic DNA using the primers leuW_NcoIfp and glnX_HindIIIrp. The amplicon was digested with NcoI and HindIII and ligated into similarly digested pACDH (TetR) to obtain pT6. Expression of the tRNAs from the construct was verified by northern blot analysis (Supplementary Figure S3).
pA5
The regions containing the genes leuW-glnU-glnW and glnV-glnX were PCR amplified from E. coli KL16 genomic DNA using the primers leuW_NcoIfp, glnW_EcoRIrp, glnV_EcoRIfp and glnX_HindIIIrp respectively. The PCR amplicons were sequentially ligated into the NcoI EcoRI and EcoRI HindIII sites in pACDH (TetR) to obtain pT5. The Tet marker in pT5 was then disrupted by NruI EcoRV digestion and a blunt ended 1369 bp ampicillin resistance (AmpR) cassette obtained by SnaBI/MscI digestion of pKD4 was ligated at this site to obtain pA5. The resulting plasmid contains 5 genes of the 7 gene operon excluding metT and metU. The constructs were verified by restriction digestion followed by DNA sequencing.
pIF2 and pEFTu
The genes encoding IF2 and EFTu (infB and tufA) were PCR amplified from E. coli KL16 genomic DNA with the primers IF2_Hisfp, IF2_rp, EFTu_fp and EFTu_Hisrp designed to incorporate N-terminal and C-terminal His tags, respectively. The amplicons of 2.7 and 1.2 kb, respectively were cloned into NcoI/HindIII sites of pA5, replacing the tRNA genes in the process. The clones were screened by restriction digestion and confirmed by DNA sequencing.
Generation of strains
ΔmetZWV::cm
E. coli KL16/pKD46 was grown at 30°C to 0.2 OD600, induced with 1 mM arabinose and made electrocompetent at 0.7 OD600 by three successive washes in sterile 10% glycerol, obtaining a 100-fold concentration of cells in the process. A 50 μl aliquot of the cells was used for electroporation. The linear DNA substrate for recombination was made by PCR amplification of the chloramphenicol (cm)R cassette from pKD3 with 36 bp flank sequences corresponding to the metZWV locus using the primers metZWV_KOfp and metZWV_KOrp. About 300 ng of the 1 kb amplicon was electroporated into the cells at 1.8 kV, 200 Ω and 25 μF in a 0.1 cm cuvette. The cells were allowed to recover in LB for 4 h at 37°C and selected on LB agar containing 30 μg/ml chloramphenicol. Knockouts were confirmed by colony PCR with metZWV_fp and metZWV_rp where knockout strains give an amplicon of 1087 bp, while the parental strain gives an amplicon of 331 bp (Supplementary Figure S4). The knockout strains were further confirmed by native PAGE of total tRNA followed by northern blot analysis to verify the absence of tRNAs derived from the metZWV locus (Supplementary Figure S2).
ΔmetY
An E. coli KL16 strain with the metY gene replaced by a chloramphenicol cassette, ΔmetY::cm, has been reported previously (16). The chloramphenicol cassette used to replace metY was removed by pCP20 mediated expression of Flp recombinase to make an unmarked deletion of metY.
Δ7::kan/pT6
The strain Δ7::kan/pT6 where the entire operon of the 7 tRNA genes (metT-leuW-glnU-glnW-metU-glnV-glnX) is deleted and supported by six of the genes (excluding metT) on a plasmid copy was generated by lambda Red mediated recombination. The knockout cassette was PCR amplified using pKD4 as template with the primers metT_KOfp2 and metT_KOrp2. The 1571 bp PCR product was electroporated into E. coli KL16/pT6/pKD46 as above. Selection was carried out on LB agar containing 25 μg/ ml kanamycin and the knockout strains were confirmed by colony PCR using metT_KO_conf_fp and metT_KO_conf_rp where knockout strains give a 2 kb amplicon while the parental strain gives a 1 kb amplicon (Supplementary Figure S5).
Strains sustained on mutant tRNAs for initiator function
Strains deleted for all four genomic copies of the initiator tRNA and supported by mutant tRNAs on plasmid (ΔmetZWV::cm ΔmetY/pmetY*; also referred to as DK/pmetY*) were generated by P1 phage transduction mediated transfer of the ΔmetZWV::cm locus from E. coli KL16 ΔmetZWV::cm to ΔmetY/pmetY*. Strains were selected on LB agar containing 30 μg/ml chloramphenicol and 7.5 μg/ml tetracycline. Transductants were verified by colony PCR using metZWV_fp and metZWV_rp as above. Deletion of all wild type initiator tRNA gene copies was further verified by native PAGE of total tRNA followed by northern blot analysis using the met33 probe, which binds to initiator tRNA molecules derived from both metZWV and metY locus. Reversion of the mutant tRNA genes supplied on plasmid to the wild type sequence was ruled out by DNA sequencing of the plasmid isolated from the knockout strains.
Strains sustained on mutant tRNAs for elongator tRNAMet function
Strains deleted for both copies of elongator tRNAMet (Δ7::kan ΔmetY::cm/pmetY*/pA5; renamed as EK/pmetY*) were generated by P1 transduction mediated transfer of the Δ7::kan locus from Δ7::kan/pT6 strain to ΔmetY::cm/pA5/pmetY* strain. Strains were selected on LB agar containing 25 μg/ml kanamycin, 100 μg/ml ampicillin and 7.5 μg/ml tetracycline. Transductants were verified by colony PCR using metT_KO_conf_fp and metT_KO_conf_rp as above. Deletion of all elongator tRNAMet gene copies was further verified by native PAGE of total tRNA followed by northern blot analysis using met-elongator probe. Reversion of the mutant tRNAs to the wild type sequence was ruled out by DNA sequencing of the plasmid isolated from the knockout strains.
Strains sustained on mutant tRNAfMet for initiator and elongator functions
Strains deleted for all copies of the native initiator and elongator methionine tRNA genes (ΔmetZWV::cm ΔmetY Δ7::kan/pmetY*/pA5; referred to as TK/pmetY*) were generated using the ΔmetZWV::cm ΔmetY strain constructed as above. The double knockout strains harboring pA5 were transduced with P1 phage raised on Δ7::kan/pT6. The resulting transductants are deleted for all genomic copies of methionine initiator and elongator tRNAs and are sustained exclusively on mutant tRNAfMet supplied on plasmid. Transductants were selected on LB agar containing 100 μg/ml ampicillin, 25 μg/ml kanamycin and 7.5 μg/ml tetracycline and were verified by colony PCR using metT_KO_conf_fp and metT_KO_conf_rp as above. Deletion of all elongator and initiator methionine tRNA gene copies was further verified by native PAGE of total tRNA followed by northern blot analysis using both met33 and met-elongator probes. Reversion of the mutant tRNAs to the wild type sequence was ruled out by DNA sequencing of the plasmid isolated from the knockout strains.
ΔmetY tufB-His:kan
EFTu is encoded by two near-identical genes, tufA and tufB in E. coli. A C-terminal His-tag was added to the genomic copy of tufB by lambda Red mediated recombination. A 1.25 kb kanamycin cassette was PCR amplified from pST-K (17) using the primers Kan_HindIIIfp and Kan_HindIIIrp and cloned downstream of the EFTu gene at the HindIII site in pEFTu to obtain pEFTu-K. The C-terminal region of EFTu including the His-tag and its 100 bp upstream region were PCR amplified along with the kanamycin cassette from pEFTu-K incorporating a 40 bp flank sequence (corresponding to the genomic sequence downstream of tufB) at the 3′ end using the primers tufB_KI_fp and tufB_KI_rp. The 1454 bp PCR product was electroporated into KL16/ pKD46 cells as above and recombinants were selected on LB agar containing 25 μg/ ml kanamycin. The strains were verified by colony PCR using tufB_flankfp and tufB_flankrp. The tufB-His:kan locus was moved into KL16 ΔmetY by P1 mediated transduction to obtain KL16 ΔmetY tufB-His:kan.
Growth analysis and doubling times
Growth analysis was performed in a BioScreen C growth analyzer using 100-well honeycomb plates. Thousand-fold dilutions of the strains in 5 biological replicates were grown in LB at either 37°C or 22°C to saturation and OD600 was measured at 1 h intervals. Growth curves were plotted using GraphPad Prism v6 with SEM. Doubling times were calculated from manual growth curves performed in duplicates. Cells were grown in 100 ml conical flasks containing 20 ml of LB at 37°C and 180 rpm. Aliquots (100 μl) of cells were taken at hourly intervals and OD600 was measured. The linear part of the curve was identified by plotting the natural log of OD600 against time. Doubling times were calculated from the linear part of the curve using the formula: d = ln 2 * (t2 – t1)/ln (OD2 – OD1), where OD2 and OD1 are the OD600 values at times t2 and t1.
IF2 and EFTu pulldowns
IF2 and EFTu pulldowns were performed in DK/pmetY*/pIF2 and either E. coli KL16/pmetY*/pEFTu or KL16 ΔmetY tufB-His:kan, respectively. About 300 ml of each culture was grown to about 1.0 OD600 at 37°C without induction. All further steps were performed at 4°C. The cultures were chilled on ice, harvested, washed and resuspended in 4 ml of lysis buffer (20 mM Tris–Cl pH 7.8, 500 mM NaCl, 2 mM β-mercaptoethanol, 1 mM GTP). Cells were incubated for 1 h on ice after addition of 100 μl of 40 mg/ml lysozyme and subsequently lysed by freeze-thaw in liquid nitrogen. The lysate was supplemented with 20 U of RNase-free DNase I and 0.2 mM CaCl2 during the last 15 min of incubation. The clear supernatant obtained after centrifugation at 20 000g for 30 min at 4°C was mixed with 100 μl Ni-NTA agarose beads (Qiagen) pre-equilibriated with lysis buffer containing 10 mM imidazole. Binding was allowed to occur for 1 h at 4°C with shaking. The beads were harvested by centrifugation at 3000g for 1 min and washed thrice with 1 ml lysis buffer containing 10 mM imidazole. Proteins were eluted from the beads using 300 μl lysis buffer containing 1 M imidazole. About 20 μl of this eluted fraction was loaded on a 12% SDS-PAGE to visualize the pulled down proteins. About 50 μl of the fraction was resolved on a 15% native PAGE to visualize the tRNA bound to IF2 or EFTu. Northern blot analysis was performed using a mixture of 32P end-labelled met33 and met-elongator probes.
Phenotypic microarray profiling
Phenotypic microarrays were performed using strains freshly streaked out on Biolog Universal Growth (BUG) agar. Briefly, the colonies were resuspended in the manufacturer supplied ‘IF0 inoculation fluid’ (Biolog Inc., Hayward CA, USA) containing ‘Redox dye mix A’ to obtain a turbidity of 85% T. Aliquots (100 μl) of this suspension were added into each well of the phenotypic microarray plates PM1 (carbon sources), PM3 (nitrogen sources), or PM5 (nutrient supplements). For PM3 and PM5, 20 mM sodium pyruvate was added as the carbon source. The plates were incubated at 37°C for 48 h and reduction of the dye was monitored. Pairwise comparisons of the growth profiles of strains were performed using the manufacturer supplied software.
Identification of natural tRNAs similar to the dual-function mutants
To identify tRNA genes with characteristics similar to the dual-function mutant tRNAs generated, the list of predicted tRNAs from all sequenced bacterial genomes was downloaded from tRNADB-CE (http://trna.ie.niigata-u.ac.jp) (18), a manually curated database of tRNAs predicted by multiple analyses. The sequences corresponding to initiator tRNAs were filtered out and tRNAs containing a 1:72 pair were identified using a regular expression search. The identified sequences were manually verified by determining the most stable secondary structure using RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi) (19).
RESULTS
Formylation deficient mutants of the E. coli tRNAfMet function as initiators and elongators
Mutants of tRNAfMet harboring substitutions in its acceptor stem (G72, G72G73, G3:C70, U1, U1/U2:A71, U1/U3:A70 and U2:A71/G72) are known, which are capable of functioning as elongators or both as initiators and elongators at least in an in vitro translation system or in a reporter based system in E. coli (20). These tRNAfMet mutants are defective in formylation (by Fmt) to varying degrees, and have become substrates for peptidyl-tRNA hydrolase (Pth) which hydrolyses formyl-aminoacyl-tRNAfMet (with a Watson:Crick base pair at 1:72) to free tRNA (20–22). However, it is unknown whether these tRNAs are capable of sustaining the entire initiation or elongation load of the cell. Could these tRNAs sustain the cellular demand for all the six genes that encode the initiators (tRNAfMet) and elongators (tRNAMet) and allow E. coli to survive?
Of the above tRNAfMet acceptor stem mutants, the U1 mutant is not significantly compromised for its formylation by Fmt and serves as a poor elongator even in a reporter based assay (20). And, the G72G73 mutant is an extremely poor substrate for Fmt (20). Thus, we excluded both these mutants from our analyses. Analyses of the remaining mutants (G72, G3:C70, U1/U2:A71, U1/U3:A70 and U2:A71/G72, Figure 1D) on acid urea gels for their in vivo aminoacylation/formylation revealed their cellular accumulation both in the aminoacylated and formyl-aminoacylated forms (Figure 1E). As a control, the wild type tRNAfMet accumulated in completely formylated form (Figure 1E, lane 7). The G72, U1:U3/A70 and U2:A71/G72 mutants, existed predominantly in aminoacylated form with ∼20–30% of the tRNAs present in formylated form (Figure 1E and Supplementary Figure S6). The G3:C70 mutant existed predominantly in the formylated form (Figure 1E, lane 4), and the U1/U2:A71 mutant had higher levels of the aminoacylated form (Figure 1E, lane 10). A small fraction of most tRNAfMet mutants was also present in the deacylated form as, owing to the Watson:Crick base pair at the 1:72 position, they are substrates for Pth (20). The U1/U2:A71 mutant, most likely due to its structural instability (22), accumulates to lower cellular levels than the other mutants. Nonetheless, the analysis endorsed the suitability of the tRNAfMet mutants for further investigation.
Mutants of tRNAfMet that sustain E. coli for initiation
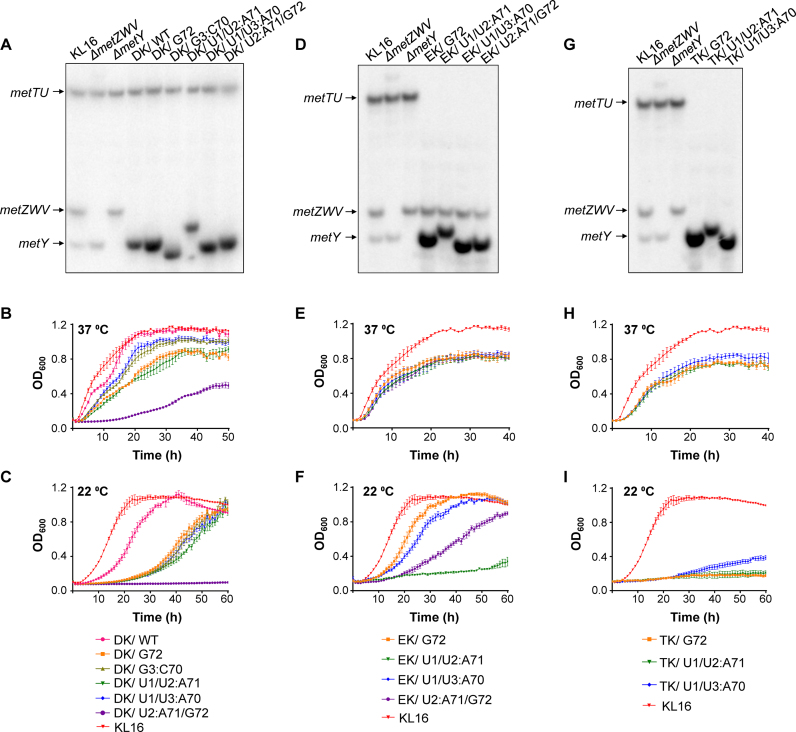
To test if the mutant tRNAfMet sustained the entire load of initiation in E. coli to allow deletion of all four tRNAfMet genes from the chromosome, we used a transduction based genetic strategy (16) (Supplementary Figure S7a) wherein P1 phage lysate raised on ΔmetZWV::cm donor strain (deleted for the metZWV locus) was used to transduce the ΔmetY recipient strains harboring plasmid borne genes for the wild type or mutant tRNAfMet (indicated as pmetY*). The resulting transductants (ΔmetZWV::cm ΔmetY/pmetY*) referred to as double locus knockout strains (DK/pmetY*) were deleted for all four tRNAfMet genes (metZWV and metY) and supported entirely by a plasmid borne tRNAfMet (wild type or mutant) gene (Figure 2A). Growth curve analysis at 37°C (Figure 2B) revealed that the deletion strain supported by wild type tRNAfMet (DK/pmetY) grew the best (pink curve). Consistent with the significant fraction of their accumulation in formylated form, the G3:C70 and U1/U3:A70 were most capable of initiation (olive and blue curves). Growth of the strain supported by the U1/U2:A71 mutant (dark green) was comparable to that of the G72 mutant supported strain (orange). The strain supported by the U2:A71/G72 mutant (purple) and which accumulated very little in the formylated form, grew the poorest. All the strains (supported by the mutant tRNAfMet) were cold sensitive at 22°C (Figure 2c) like the ΔmetZWV strain (15,23). Importantly, all the mutants (G72, G3:C70, U1/U2:A71, U1/U3:A70 or U2:A71/G72) or the wild type tRNAfMet are capable of sustaining E. coli for the cellular load of initiation. However, we do not discount that in the cases of the mutant tRNAs which are suboptimal in their function, their increased expression (from the plasmid borne genes) might have also contributed to the sustenance of the strain.
Figure 2.
Generation and growth analysis of the knockout strains. (A) Confirmation of the double locus knockout (DK) strains (ΔmetZWV and ΔmetY) by northern blot analysis of tRNAs prepared from flask cultures and separated on native PAGE. The tRNAMet (metT/metU), tRNAfMetI (metZWV) and tRNAfMetII (metY) are indicated by arrows. Lanes 1, 2 and 3 represent RNA from KL16 (wild type for both the metZWV and metY) its ΔmetZWV::cm and ΔmetY derivatives, respectively. The plasmid borne tRNAfMet, pmetY* (harboring WT or the mutant tRNAfMet, as indicated) were introduced into ΔmetY background and subjected to deletion of the metZWV locus (lanes 4–9). (B and C) Growth (in microtiter plates) of the DK strains supported by tRNAfMet or its mutants (as indicated) at 37°C and 22°C, respectively. (D) Confirmation of the ΔmetT and ΔmetU strains (EK) by northern blot analysis of tRNAs prepared from flask cultures and separated on native PAGE. Lanes 1, 2, and 3 are as in (A). The plasmids pmetY* (harboring tRNAfMet mutants as indicated) and pA5 (harboring leuW-glnU-glnW-glnV-glnX) were introduced into ΔmetY background and subjected to deletion of the metT/metU locus (Δ7::kan) (lanes 4–7). (E and F) Growth (in microtiter plates) of the tRNAMet gene knockout (EK) strains (as indicated) supported by the mutant tRNAs (EK/pmetY*) at 37°C and 22°C, respectively. (G) Confirmation of the triple locus knockout (ΔmetZWV, ΔmetY, ΔmetT and ΔmetU shown as TK) strains by northern blot analysis of tRNAs prepared from flask cultures and separated on native PAGE. Lanes 1, 2, and 3 are as in (A). (H and I) Growth (in microtiter plates) of the TK strains supported by tRNAfMet mutants (as indicated) at 37°C and 22°C, respectively. For (B)–(I), error bars indicate standard error of mean (SEM). All growth curve analyses were done at the same time. Data for the parent strain (KL16) have been shown in all panels (B, E, and H at 37°C; and C, F and I at 22°C) for comparison.
Mutants of tRNAfMet that sustain E. coli for elongation
Next, we designed a strategy to test if the tRNAfMet mutants support E. coli for its elongator tRNAMet function. Both metT and metU genes encode identical tRNAMet, and are in the metT-leuW-glnU-glnW-metU-glnV-glnX operon with five other essential tRNA genes. Using standard genetic engineering techniques, we first cloned two versions of the operon segments into plasmids, pT6 (leuW-glnU-glnW-metU-glnV-glnX) and pA5 (leuW-glnU-glnW-glnV-glnX), such as to lack either one (metT) or both (metT and metU) tRNAMet genes, respectively (Supplementary Figure S8). The pT6 (TetR) and pA5 (AmpR) are pACYC based plasmids compatible with pmetY* (tRNAfMet harboring plasmids). We then followed a two-step protocol. In the first step, we generated Δ(metT-leuW-glnU-glnW-metU-glnV-glnX)::kan/pT6 strain (referred to as Δ7::kan/pT6), deleted for all seven tRNA genes of the operon from the chromosome with pT6 support (Supplementary Figure S8). The resulting strain has no tRNAMet genes on the chromosome and is sustained by a single tRNAMet gene, metU, on pT6. In the second step, we transduced the ΔmetY::cm/pA5/pmetY* strain (recipient) with the P1 raised on Δ7::kan/pT6 (donor) strain (Supplementary Figure S7b). The resulting transductants (ΔmetY::cm Δ7::kan/pA5/pmetY*; referred to as elongator knockout strains, EK/pmetY*) are devoid of all copies of the elongator tRNAMet genes and are sustained for the elongation function entirely by the plasmid borne mutant tRNAfMet. In this experiment, we obtained transductants for the strains harboring G72, U1/U2:A71, U1/U3:A70 and U2/A71:G72 metY genes (Figure 2D). However, the strain EK/pmetY* harboring wild type tRNAfMet gene did not yield transductants, suggesting, not unexpectedly, that it is unable to sustain elongation function. Also, the strain EK/pmetY* harboring the G3:C70 mutant tRNAfMet gene failed to produce any transductants. In fact, only a minor fraction of the G3:C70 tRNAfMet accumulated in aminoacylated form explaining why this mutant might have failed to substitute for the elongator tRNAMet function. In the growth curve experiment, the strains supported by either of the G72, U1/U2:A71, U1/U3:A70 and U2/A71:G72 mutants grew comparably at 37°C (Figure 2E). However, at 22°C (Figure 2F) the strains supported by G72 (Figure 2f, orange) and U1/U3:A70 (Figure 2F, blue) were the healthiest, followed by U2:A71/G72 (Figure 2F, purple). The strain supported by U1/U2:A71 (Figure 2F, dark green) was highly compromised for growth at 22°C, although it grew at 37°C. These data showed that at least some of the tRNAfMet mutants could sustain E. coli for the elongator tRNAMet function. It may be noted that while the EK/pmetY* strains are deleted for metY, they possess the metZWV locus encoding three of the four tRNAfMet genes.
Mutants of tRNAfMet that sustain E. coli for both initiation and elongation
From the above experiments, we obtained four mutants of tRNAfMet (G72, U1/U2:A71, U1/U3:A70 and U2:A71/G72), which sustained E. coli for the independent functions of initiation and elongation. As the next step, we wished to check (by deletion of the metZWV locus from the EK/pmetY* strains) if any of the four tRNAfMet mutants, supported E. coli for the tRNAfMet and tRNAMet functions, simultaneously. To do this, we transduced the double knockout strains (DK/pA5/pmetY*) supported by any of the four tRNAfMet mutants with the P1 raised on the Δ7::kan strain to obtain triple locus knockout (TK) strains deleted for all copies of the chromosomal tRNAfMet and tRNAMet genes (ΔmetY ΔmetZWV::cm Δ7::kan/pA5/pmetY* (or TK/pmetY*) (Supplementary Figure S7c). Interestingly, we were able to obtain triple knockout strains with three of the tRNAfMet mutants G72, U1/U2:A71 and U1/U3:A70 (Figure 2G). Under the conditions used, the U2:A71/G72 mutant did not support a triple knockout even though it supports independent functions of either initiation or elongation. The strains grew comparably at 37°C (Figure 2H) or at 22°C (Figure 2I), with the strain supported by U1/U3:A70 tRNAfMet growing slightly better than the other two mutants.
To characterize the knockout strains further, we assessed the growth of KL16 (parent strain), ΔmetY, ΔmetZWV::cm, DK/U1/U3:A70, TK/G72 and TK/U1/U3:A70 on various carbon sources, nitrogen sources and nutrient supplements using the Biolog phenotypic microarray platform. The single knockout strains did not show any appreciable change in growth profile compared to the parent strain (Supplementary Figure S9a and b), but the DK/U1/U3:A70 strain showed mild growth defects on some carbon and nitrogen sources (Supplementary Figure S9c). The triple knockout strains, TK/G72 and TK/U1/U3:A70, showed stronger growth defects on many carbon and nitrogen sources (Supplementary Figure S9d and e), primarily with respect to growth on metabolites which are direct intermediates of the Kreb's cycle, with the defects seen for TK/G72 being larger compared to TK/U1/U3:A70, indicating that the differences observed may be directly related to the growth rate of the strains. Doubling times of the triple knockout strains calculated from growth in LB in flask cultures show that they grow about 1.5–2-fold slower than the vector control (Supplementary Figure S10) with the strains supported by G72, U1/U2:A71, and U1/U3:A70 tRNAs having a doubling time of 61.7, 67.8 and 55 min compared to 35.2 min for the vector control.
Dual-function mutants bind to both IF2 and EFTu in vivo
In mitochondrial protein synthesis where the single tRNAMet is partitioned into initiation and elongation steps, the tRNAMet either binds IF2mt or EFTumt depending on its formylation status. We wondered whether a similar phenomenon operated in the TK strains. To analyze this, we checked for binding of tRNAfMet mutants to N-terminally or C-terminally His6 appended IF2 or EFTu, respectively following a pulldown with Ni-NTA beads. Expectedly, we detected binding of wild type tRNAfMet (both the fMetI and fMetII) to IF2 in wild type cells (Figure 3B, lane 1). There was negligible binding of tRNAMet to IF2 (Figure 3B, top panel). The amounts of mutant tRNAfMet bound to IF2 were roughly in the order of their efficiency of initiation as analyzed by the growth of the double knockout (DK) strains supported by the mutants. The binding of these tRNAs to EFTu was also checked. Overexpression of a plasmid encoded EFTu-His gene led to significant binding to even wild type initiator tRNA (Supplementary Figure S11). Thus, a His-tagged copy of EFTu was knockedin into the tufB locus using lambda Red mediated recombination (Supplementary Figure S12). Pulldowns performed in this strain background were significantly more specific and indicative of in vivo binding. As opposed to wild type tRNAfMet which bound poorly to EFTu, the tRNAfMet mutants capable of supporting elongation function, bound strongly to EFTu (Figure 3E, lane 3, 6 and 7). The only exception was U1/U2:A71, which bound poorly to EFTu (Figure 3E, lane 5). However, it may be noted that this mutant accumulated to low levels in the cell.
Figure 3.
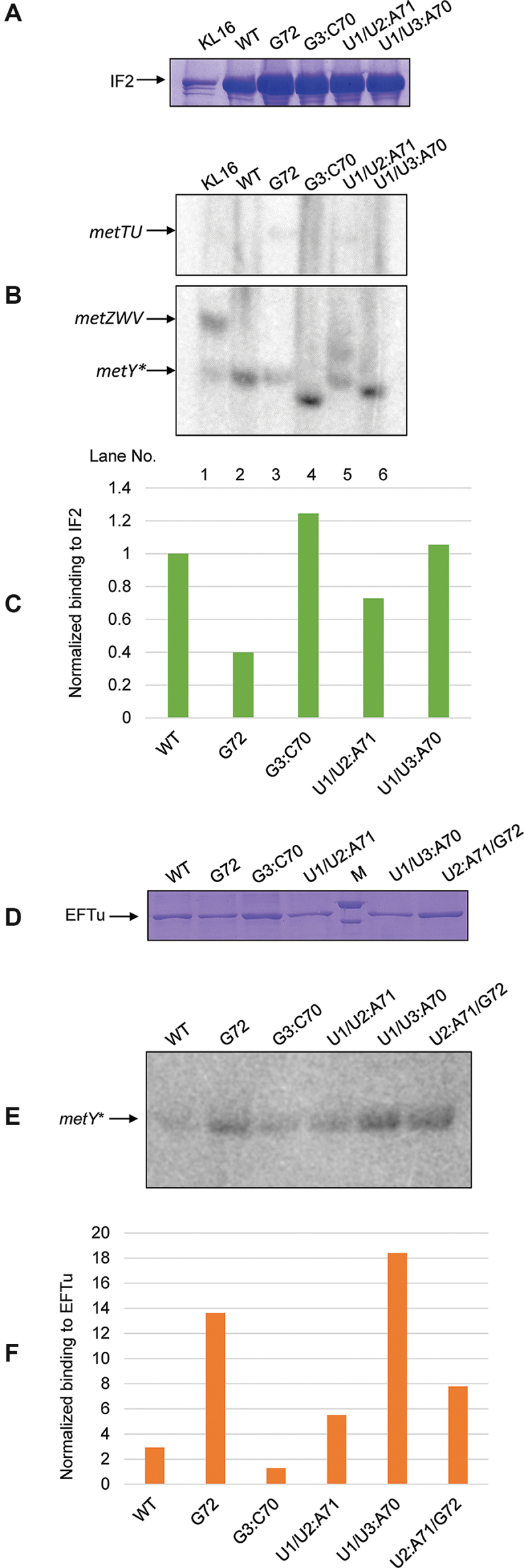
Binding of tRNAfMet or its mutants to IF2 and EFTu. (A) Analysis of IF2 pulldown fractions by 12% SDS-PAGE. Pulldowns were performed in ΔmetY ΔmetZWV (DK) strains supported by the mutant indicated, and harboring a His-tagged plasmid encoded copy of IF2. The band corresponding to full length His-IF2 (∼98.5 kDa) is indicated. (B) Northern blot analysis of the pulldown samples separated on 15% native PAGE. IF2 bound RNA fraction was probed with 32P-5′-end labeled oligomers capable of binding to both fMetI and fMetII tRNAs. Quantification of binding from the northern blot is shown in (C). (D) Analysis of EFTu pulldown fractions by 12% SDS-PAGE. Pulldowns were performed in ΔmetY tufB-His:kan strains harboring the mutant tRNAs indicated. The band corresponding to full length EFTu-His (∼44 kDa) is indicated. (E) Northern blot analysis of the EFTu pulldown fractions separated on 15% native PAGE. Northern blot analysis was performed as in (b). Quantification of binding from the northern blot is shown in (F).
Naturally occurring variants of i-tRNAs with a base pair at the 1:72 position
The results shown above provide a basis for the possibility of occurrence of a phenomenon observed in mitochondria, i.e. for the natural occurrence of a single tRNAMet in bacterial systems. To test whether variants of i-tRNA which could function in elongation exist naturally, we analyzed the structures of i-tRNAs from all sequenced bacterial genomes to date. We find that there are naturally occurring i-tRNAs which possess the characteristic three consecutive G:C base pairs (or their functional variants) (24) along with a U1:A72 pair (Supplementary Figure S13a–e), U1:G72 wobble pair (Supplementary Figure S13f–k), and even a G1:C72 pair (Supplementary Figure S13l–m) in the acceptor stem. Although many of these organisms do possess separate methionine elongator tRNAs, it may well be that the i-tRNAs with the U1:G72 and G1:C72 base pairs could also carry out elongator tRNA functions, at least under some conditions.
DISCUSSION
Overall, our studies show that by fine tuning formylation efficiencies (by introducing mutations in the acceptor stem of tRNAfMet), it is possible to direct a tRNA into either the initiation or the elongation pathways, providing an insight into the evolution of distinct methionine tRNA species. As Fmtmt primarily recognizes the amino acid attached to the tRNA, mitochondrial tRNAMet could be driven either into the initiation or elongation by regulating the levels of formylase enzyme. As opposed to this, eubacterial Fmt recognizes sequence elements on the tRNAfMet body itself. Although this necessitates the presence of two distinct tRNAs for initiation and elongation functions, we have shown that it is possible to change the fate of the tRNA by altering the formylatability of the tRNAfMet. However, instead of regulation at the level of Fmt expression, shifting the balance of fate in bacterial systems require sequence changes in the determinants of the Fmt enzyme, a mechanism that could have been adopted during the evolution of distinct tRNAs for initiation and elongation. Interestingly, the mutant that is most efficient at sustaining initiation and elongation together, U1/U3:A70, is also the one that is most similar to the human mitochondrial methionine tRNA (Supplementary Figure S1), supporting such an idea.
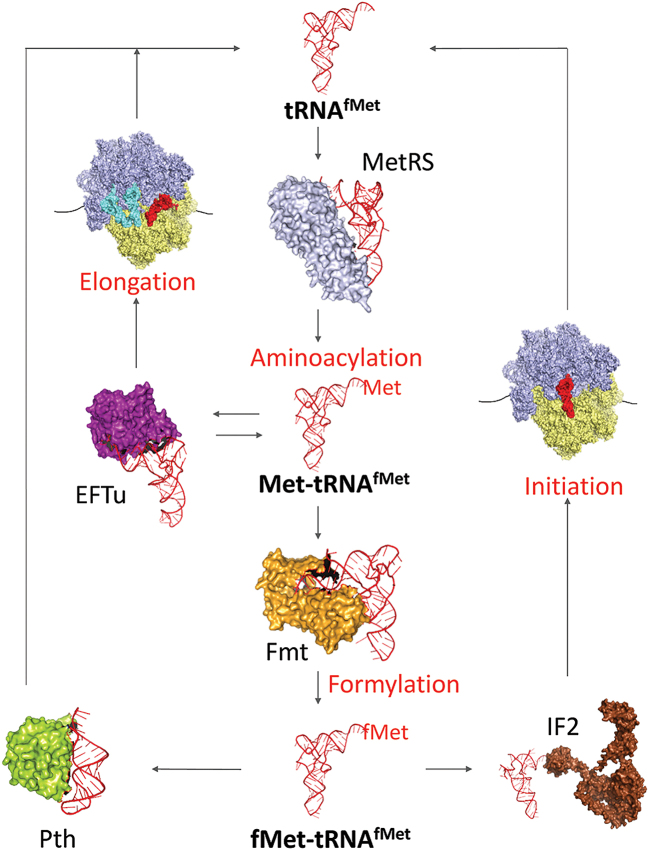
Mutations in the MTFMT gene encoding human mitochondrial formylase cause Leigh syndrome and combined OXPHOS deficiency (25). Such patients show a significant reduction in the levels of fMet-tRNAMet, with a severe defect in mitochondrial translation. Interestingly, the triple knockout strains supported by the U1/U2:A71 mutant (which has the least amount of formylated species at steady state, Figure 1E) shows slowest growth among the three triple knockout strains in our E. coli knockout system. In addition, our study provides insights into bacterial protein synthesis and the delicate balance of protein factors and sequence elements in the acceptor stem of tRNAfMet that decide the fate of the bacterial i-tRNA (Figure 4).
Figure 4.
Model for alternate fates of the bacterial initiator tRNA. Following aminoacylation by MetRS, the formylability of the initiator tRNA by Fmt determines its entry into either initiation (by binding of the formylated species to IF2) or elongation (by binding of the unformylated species to EFTu). Partitioning of tRNAfMet between various factors (Fmt/IF2 or EFTu) determines its fate in protein synthesis.
We noted the existence of i-tRNA variants that harbor U1:A72 pair, U1:G72 wobble pair, or a G1:C72 pair in the acceptor stem in many of the sequenced bacterial genomes. Although not known so far, our observation provides a basis for the possibility of natural occurrence of a single tRNA system for initiation and elongation in at least some bacteria. Importantly, together with our earlier studies (26,27), our current finding reinforces the use of E. coli as a model to investigate the evolutionary and mechanistic aspects of the mitochondrial protein synthesis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank their laboratory colleagues for critical review of the work and manuscript. We thank Prof. Dipankar Chatterji and Dr. Neerupma Bhardwaj of the Molecular Biophysics Unit, IISc for the use of the Biolog machine and their help in carrying out the phenotypic microarray analyses. The authors acknowledge the DBT-IISc partnership programme, University Grants Commission, New Delhi for the Centre of Advanced Studies, and the DST-FIST level II infrastructure supports to carry out this work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Biotechnology (DBT), Ministry of Science and Technology; Department of Science and Technology (DST), Ministry of Science and Technology; J.C. Bose Fellowship (to U.V.). Funding for open access charge: Government of India, through DBT and DST.
Conflict of interest statement. None declared.
REFERENCES
- 1. Kozak M. Comparison of initiation of protein synthesis in procaryotes, eucaryotes, and organelles. Microbiol. Rev. 1983; 47:1–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu. Rev. Biochem. 1988; 57:199–233. [DOI] [PubMed] [Google Scholar]
- 3. Gualerzi C.O., Pon C.L.. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990; 29:5881–5889. [DOI] [PubMed] [Google Scholar]
- 4. Sprinzl M., Hartmann T., Weber J., Blank J., Zeidler R.. Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 1989; 17:r1–r172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lee C.P., Seong B.L., RajBhandary U.L.. Structural and sequence elements important for recognition of Escherichia coli formylmethionine tRNA by methionyl-tRNA transformylase are clustered in the acceptor stem. J. Biol. Chem. 1991; 266:18012–18017. [PubMed] [Google Scholar]
- 6. Guillon J.M., Mechulam Y., Schmitter J.M., Blanquet S., Fayat G.. Disruption of the gene for Met-tRNA(fMet) formyltransferase severely impairs growth of Escherichia coli. J. Bacteriol. 1992; 174:4294–4301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guillon J.M., Heiss S., Soutourina J., Mechulam Y., Laalami S., Grunberg-Manago M., Blanquet S.. Interplay of methionine tRNAs with translation elongation factor Tu and translation initiation factor 2 in Escherichia coli. J. Biol. Chem. 1996; 271:22321–22325. [DOI] [PubMed] [Google Scholar]
- 8. Wu X.Q., RajBhandary U.L.. Effect of the amino acid attached to Escherichia coli initiator tRNA on its affinity for the initiation factor IF2 and on the IF2 dependence of its binding to the ribosome. J. Biol. Chem. 1997; 272:1891–1895. [DOI] [PubMed] [Google Scholar]
- 9. Anderson S., de Bruijn M.H., Coulson A.R., Eperon I.C., Sanger F., Young I.G.. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J. Mol. Biol. 1982; 156:683–717. [DOI] [PubMed] [Google Scholar]
- 10. Takeuchi N., Vial L., Panvert M., Schmitt E., Watanabe K., Mechulam Y., Blanquet S.. Recognition of tRNAs by Methionyl-tRNA transformylase from mammalian mitochondria. J. Biol. Chem. 2001; 276:20064–20068. [DOI] [PubMed] [Google Scholar]
- 11. Tan T.H., Bochud-Allemann N., Horn E.K., Schneider A.. Eukaryotic-type elongator tRNAMet of Trypanosoma brucei becomes formylated after import into mitochondria. PNAS. 2002; 99:1152–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kenri T., Imamoto F., Kano Y.. Three tandemly repeated structural genes encoding tRNA(f1Met) in the metZ operon of Escherichia coli K-12. Gene. 1994; 138:261–262. [DOI] [PubMed] [Google Scholar]
- 13. Seong B.L., RajBhandary U.L.. Escherichia coli formylmethionine tRNA: mutations in GGGCCC sequence conserved in anticodon stem of initiator tRNAs affect initiation of protein synthesis and conformation of anticodon loop. PNAS. 1987; 84:334–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kenri T., Imamoto F., Kano Y.. Construction and characterization of an Escherichia coli mutant deficient in the metY gene encoding tRNA(f2Met): either tRNA(f1Met) or tRNA(f2Met) is required for cell growth. Gene. 1992; 114:109–114. [DOI] [PubMed] [Google Scholar]
- 15. Kenri T., Kohno K., Goshima N., Imamoto F., Kano Y.. Construction and characterization of an Escherichia coli mutant with a deletion of the metZ gene encoding tRNA (f1Met). Gene. 1991; 103:31–36. [DOI] [PubMed] [Google Scholar]
- 16. Samhita L., Shetty S., Varshney U.. Unconventional initiator tRNAs sustain Escherichia coli. PNAS. 2012; 109:13058–13063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Parikh A., Kumar D., Chawla Y., Kurthkoti K., Khan S., Varshney U., Nandicoori V.K.. Development of a new generation of vectors for gene expression, gene replacement, and protein-protein interaction studies in mycobacteria. Appl. Environ. Microbiol. 2013; 79:1718–1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Abe T., Ikemura T., Sugahara J., Kanai A., Ohara Y., Uehara H., Kinouchi M., Kanaya S., Yamada Y., Muto A. et al. . tRNADB-CE 2011: tRNA gene database curated manually by experts. Nucleic Acids Res. 2011; 39:D210–D213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lorenz R., Bernhart S.H., Honer Zu Siederdissen C., Tafer H., Flamm C., Stadler P.F., Hofacker I.L.. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011; 6:26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Varshney U., Lee C.P., Seong B.L., RajBhandary U.L.. Mutants of initiator tRNA that function both as initiators and elongators. J. Biol. Chem. 1991; 266:18018–18024. [PubMed] [Google Scholar]
- 21. Thanedar S., Kumar N.V., Varshney U.. The fate of the initiator tRNAs is sensitive to the critical balance between interacting proteins. J. Biol. Chem. 2000; 275:20361–20367. [DOI] [PubMed] [Google Scholar]
- 22. Varshney U., Lee C.P., RajBhandary U.L.. Direct analysis of aminoacylation levels of tRNAs in vivo. Application to studying recognition of Escherichia coli initiator tRNA mutants by glutaminyl-tRNA synthetase. J. Biol. Chem. 1991; 266:24712–24718. [PubMed] [Google Scholar]
- 23. Shetty S., Varshney U.. An evolutionarily conserved element in initiator tRNAs prompts ultimate steps in ribosome maturation. PNAS. 2016; 113:E6126–E6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Shetty S., Shah R.A., Chembazhi U.V., Sah S., Varshney U.. Two highly conserved features of bacterial initiator tRNAs license them to pass through distinct checkpoints in translation initiation. Nucleic Acids Res. 2017; 45:2040–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tucker E.J., Hershman S.G., Kohrer C., Belcher-Timme C.A., Patel J., Goldberger O.A., Christodoulou J., Silberstein J.M., McKenzie M., Ryan M.T. et al. . Mutations in MTFMT underlie a human disorder of formylation causing impaired mitochondrial translation. Cell Metab. 2011; 14:428–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gaur R., Grasso D., Datta P.P., Krishna P.D., Das G., Spencer A., Agrawal R.K., Spremulli L., Varshney U.. A single mammalian mitochondrial translation initiation factor functionally replaces two bacterial factors. Mol. Cell. 2008; 29:180–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ayyub S.A., Aswathy S.L., Dobriyal D., Aluri S., Spremulli L.L., Varshney U.. Fidelity of translation in the presence of mammalian mitochondrial initiation factor 3. Mitochondrion. 2018; 39:1–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.