Abstract
Skeletal muscle atrophy due to excessive protein degradation is the main cause for muscle dysfunction, fatigue, and weakening of athletic ability. Endurance exercise is effective to attenuate muscle atrophy, but the underlying mechanism has not been fully investigated. α-Ketoglutarate (AKG) is a key intermediate of tricarboxylic acid cycle, which is generated during endurance exercise. Here, we demonstrated that AKG effectively attenuated corticosterone-induced protein degradation and rescued the muscle atrophy and dysfunction in a Duchenne muscular dystrophy mouse model. Interestingly, AKG also inhibited the expression of proline hydroxylase 3 (PHD3), one of the important oxidoreductases expressed under hypoxic conditions. Subsequently, we identified the β2 adrenergic receptor (ADRB2) as a downstream target for PHD3. We found AKG inhibited PHD3/ADRB2 interaction and therefore increased the stability of ADRB2. In addition, combining pharmacologic and genetic approaches, we showed that AKG rescues skeletal muscle atrophy and protein degradation through a PHD3/ADRB2 mediated mechanism. Taken together, these data reveal a mechanism for inhibitory effects of AKG on muscle atrophy and protein degradation. These findings not only provide a molecular basis for the potential use of exercise-generated metabolite AKG in muscle atrophy treatment, but also identify PHD3 as a potential target for the development of therapies for muscle wasting.—Cai, X., Yuan, Y., Liao, Z., Xing, K., Zhu, C., Xu, Y., Yu, L., Wang, L., Wang, S., Zhu, X., Gao, P., Zhang, Y., Jiang, Q., Xu, P., Shu, G. α-Ketoglutarate prevents skeletal muscle protein degradation and muscle atrophy through PHD3/ADRB2 pathway.
Keywords: tricarboxylic acid cycle, metabolite, Duchenne muscular dystrophy, metabolism
Muscle atrophy is characterized as the accumulation of damaged or dysfunctional fiber components and muscle weakness caused by structural or functional impairment (1, 2). Human muscle atrophy generally occurs with many diseases including cancers (3), sepsis (4), renal or heart failure (4), muscle genetic diseases, and neurodegenerative disorders (5). Excessive muscle atrophy aggravates illness and leads to death. These highlight the urgent need to better understand the etiology of muscle atrophy and to develop more effective therapies to combat muscle atrophy. Although muscle atrophy has received increasing attention in the literature, little is known about the triggers or the molecular mechanism underlying this process. Muscle atrophy is attributed to the imbalance of protein degradation and synthesis. There are two major signaling pathways controlling protein degradation, the ubiquitin–proteasome (6–9) and the autophagic lysosomal pathways (1). Both pathways are activated during muscle atrophy and contribute to the loss of muscle mass. These pathways involve a variety of atrophy-related genes or atrogenes, which are controlled by specific transcription factors, such as FoxOs and their downstream target genes, muscle atrophy F-box (MAFbx) and muscle RING-finger protein 1 (MuRF1) (10–12). Our goal was to find potential therapeutic compounds that specifically target these atrophy-related genes or transcription factors to ameliorate muscle atrophy caused by protein degradation.
α-Ketoglutarate (AKG), a product of glutamine deamination (glutaminolysis), is a crucial intermediate in the tricarboxylic acid cycle (13, 14). It has been reported that AKG plays a key role in protein turnover. For example, intracellular AKG levels are strictly dependent on amino acid availability (15). AKG treatment in vitro inhibits glutamine degradation and enhances protein synthesis in intestinal porcine epithelial cells (16). Our previous study consistently found that AKG promotes skeletal muscle protein synthesis and muscle hypertrophy via PI3K/Akt signaling pathway (17). Importantly, the key protein degradation transcript, forkhead box O1 (FoxO1), is suppressed by AKG in a G-protein–coupled receptor 91 (GPR91)-independent manner (17). Thus, we speculate that besides the stimulatory effects on protein synthesis, AKG also inhibits protein degradation and may have potential to be used in muscle atrophy therapies.
In this study, by using a corticosterone-induced protein degradation model, we found that AKG prevents protein degradation and skeletal muscle atrophy both in vitro and in vivo. Combining pharmacologic, lentivirus-mediated knockdown and vector-mediated overexpression approaches, we further demonstrated that the inhibitory effects of AKG on protein degradation and skeletal muscle atrophy are mediated by suppressing prolyl hydroxylase 3 (PHD3) and subsequently stimulating β-2 adrenergic receptor (ADRB2). Importantly, the same inhibitory effects of AKG on muscle atrophy were consistently observed in a Duchenne muscular dystrophy (DMD) mouse model. Thus, our data support a model that AKG inhibits protein degradation through a PHD3/ADRB2-dependent mechanism and provide preclinical evidence that AKG could to be used to treat inactivity-induced muscle fiber atrophy.
MATERIALS AND METHODS
Cell culture
Mouse myoblast C2C12 cells were purchased from China Infrastructure of Cell Line Researcher (Beijing, China). The cells were cultured in high-glucose DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific) at 37°C in a humidified atmosphere that contained 5% CO2. The C2C12 myoblasts were induced differently than myotubes by a differentiation medium that contained 2% horse serum (Thermo Fisher Scientific) for 4 d.
Animals
C57BL/10ScSn-Dmdmdx/Nju (mdx) mice in a C57BL/10ScSnNju background were purchased from the Nanjing Biomedical Research Institute of Nanjing University [permission SCXK (Su) 2015-0001]. Wild-type mice (C57BL/6J mice) were purchased from the Animal Experiment Center of Guangdong province [permission SYXK (Yue) 2014-0136]. The mice were divided into 3 groups of 10 animals each: control, mdx, and mdx supplemented with AKG (α-ketoglutarate sodium). AKG (2%) was provided in drinking water for 9 wk in the mdx supplemented with AKG group, and the other 2 groups were provided sterile water. The mice were housed under a 12-light–dark cycle (6 am and 6 pm, 23°C and 70 ± 80% humidity). Body composition was analyzed by NMR (MesoQMR23-060H; Niumag, Shanghai, China). At the end of the experiment, the mice were humanely killed to collect blood and gastrocnemius tissue samples for further testing. The tissues samples were stored at −80°C for real-time quantitative PCR (qPCR) and Western blot analysis. All experimental protocols and methods were approved by the College of Animal Science, South China Agricultural University. All experiments were conducted in accordance with The Instructive Notions with Respect to Caring for Laboratory Animals (Ministry of Science and Technology, Beijing, China).
Strength and exercise endurance
Mouse maximum limb muscle force was measured by a grip strength meter (BIO-GS3; Bioseb, Vitrolles, France). For assessing limb muscle force, we conducted 3 sets of 10 successive experiments. The mean maximum limb muscle strength in each set of measurements was used for data analysis. Mouse exercise endurance was evaluated by a treadmill running test. The mice were placed on an FT200 treadmill (Chengdu Techman Software, Chengdu, China) with a 10-degree incline at an initial velocity of 10 m/min for 10 min. After a rest of 10 min, the velocity was increased by 5 m/min per 2 min up to 40 m/min as a high-speed running test. In the slow-speed running test, the velocity was increased by 1 m/min per 3 min up to 20 m/min. The end time was used for data analysis.
ADRB2 knockdown and inhibition
The shADRB2 lentivirus was generated from Hanbio Biotechnology (Shanghai, China). Forty 6-wk-old C57BL/6J mice were randomly divided into 4 groups of 10 animals each: LV-shScrambled group, LV-shScrambled + AKG group, LV-shADRB2 group, and LV-shADRB2 + AKG group. After the interference efficiency was verified, the lentivirus was injected intramuscularly at 60 μl (10 × 7 titers) in 3 different sites of the gastrocnemius. The groups containing AKG received 2% AKG supplementation in drinking water, and the other groups were provided sterile water for 8 wk. At the end of the experiment, we measured the animals’ body composition, muscle strength, and exercise endurance. Blood and muscle samples were collected for further testing.
We also used ICI-118,551 hydrochloride [ICI; a highly selective β2 adrenergic receptor (ADRB2) antagonist, I127; Sigma-Aldrich, St. Louis, MO, USA] to block the ADRB2 pathway in C2C12 myotubes and mouse gastrocnemius. C2C12 myotubes were cotreated with 10 μM inhibitor and 2 mM AKG for 48 h to analyze the roles of ADRB2 pathway on protein degradation and fiber size. To examine the effect of ADRB2, male C57BL/6J mice were subjected to 5 mg/kg inhibitor or 1 g/kg AKG by intraperitoneal injection for 3 h. Mouse gastrocnemius samples were collected for Western blot and immunohistochemical analyses.
PHD3 overexpression
To investigate the underlying mechanisms of PHD3-mediated AKG on protein degradation and skeletal muscle atrophy, we overexpressed PHD3 by using the FLAG-EglN3-pLenti6 plasmid, which was a gift from W. Kaelin (36951; Addgene, Cambridge, MA, USA). The EglN3-pLenti6 plasmid was transfected into C2C12 myoblasts along with control vector. After 24 h, the myoblasts were introduced into myotubes with differentiation medium. Then the myotubes were treated with 2 mM AKG for 48 h. The samples were collected for further analysis.
Flow cytometric analysis
We cultured C2C12 cells in 24-well plates with differentiation medium. In 24-well plates, 50 μl of 10 μg/ml puromycin solution was added, and the myotubes were incubated for 10 min in a CO2 incubator. The cells were washed 2 times with 200 μl prewarmed differentiation medium, incubated for 50 min at 37°C and 5% CO2, and washed 2 times. Cells were collected and washed with cold PBS containing 0.1% bovine serum albumin (BSA). Then the cells were resuspended with cold PBS including BSA and 1 μg 12D10 (puromycin antibody) and incubated for 30 min. The cells were then washed 2 times, fixed with 50 μl 2% paraformaldehyde solution (PBS/BSA), and stored at 4°C until fluorescence-activated cell sorting analysis. We used a similar procedure for the ADRB2 analysis.
Coomassie Brilliant Blue staining
C2C12 cells underwent induced differentiation for 4 d, and the anti-PHD3 was used to pull down PHD3 binding protein. The precipitated samples were used for SDS-PAGE; then the strips were stained with Coomassie Brilliant Blue. The samples were also measured by mass spectrometry at the Beijing Genomics Institute (Shenzhen, China).
Protein degradation assessment
Protein degradation of long-lived proteins was determined by using Click-It metabolic labeling for proteins (C10102; Thermo Fisher Scientific) as previously described (18). C2C12 myotubes were washed with PBS, and the cells were incubated at 37°C for 1 h to deplete methionine reserves. Then the cells were incubated for 12 h with 50 μM Click-It AHA. Cells were washed with PBS, fixed with 4% paraformaldehyde for 15 min, and permeabilized with 0.25% Triton X-100 in PBS. Finally, the cells were washed in 3% BSA in PBS. The detection click reaction was obtained by incubating fixed cells for 2 h with Click-It Tamra Dibenzocyclooctyne (DIBO) Alkyne (C10410; Thermo Fisher Scientific). The myotubes were washed with 3% BSA and used for imaging.
3-Methylhistidine level assay
3-Methylhistidine (3-MeHis) is mainly located in myosin and actin in myofibrillar protein and is not reused for protein synthesis (19). Therefore, 3-MeHis release from skeletal muscle directly shows skeletal muscle and myofibrillar protein degradation. To measure skeletal muscle and C2C12 myotube protein degradation, the amount of 3-MeHis was measured by HPLC as previously described (20).
Western blot assay
Protein expression was tested by Western blot analysis. Cells and gastrocnemius samples were lysed in RIPA lysis buffer that contained 1 mM PMSF. Total protein concentration was determined using bicinchoninic acid protein assays (Thermo Fisher Scientific). After SDS-PAGE gels, the proteins were reacted with the following primary antibodies: rabbit anti–β-actin (bs-0061R; Bioss Antibodies, Woburn, MA, USA) and mouse puromycin antibody 12D10 (MABE343; EMD Millipore, Billerica, MA, USA); rabbit anti–cAMP-response element binding protein (CREB; 9197; Cell Signaling Technology, Danvers, MA, USA), rabbit anti–phospho-CREB (Ser133, 9198; Cell Signaling Technology), mouse anti-MuRF1 (sc-32920; Santa Cruz Biotechnology, Santa Cruz, CA, USA) and anti-MAFbx (sc-33782; Santa Cruz Biotechnology), rabbit anti-FoxO1 (2880; Cell Signaling Technology), and rabbit anti–phospho-FoxO1 (Ser256; 2880, Cell Signaling Technology); or ADRB2 (bs-0947R; Bioss Antibodies), PHD3 (bs-0532R; Bioss Antibodies), p-PKA (Ser339, bs-4024R; Bioss Antibodies), and PKA (bs-0520R; Bioss Antibodies). The primary antibodies were incubated at 4°C overnight, followed by incubation with the appropriate secondary antibody (Bioss Antibodies) for 1 h at room temperature. Protein expression was measured and analyzed using a FluorChem M fluorescent imaging system (ProteinSimple, Santa Clara, CA, USA) and ImageJ software (Image Processing and Analysis in Java; National Institutes of Health, Bethesda, MD, USA; http://imagej.nih.gov/).
RNA extraction, reverse transcription, and quantitative PCR
Total RNAs were extracted from C2C12 myotubes and mouse gastrocnemius using Trizol reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. After treatment with DNase I (Takara, Kyoto, Japan), total RNA (2 μg) was reverse transcribed to cDNA in a final 20 μl using M-MLV Reverse Transcriptase (Promega, Madison, WI, USA) and Random Primer 9 (Takara) according to the manufacturer’s instructions. β-Actin was used as a candidate housekeeping gene. SYBR Green Real-Time PCR Master Mix reagents (Toyobo, Osaka, Japan) and sense and antisense primers (200 nM for each gene) were used for quantitative PCR. PCR reactions were performed with an Mx3005p instrument (Stratagene, San Diego, CA, USA). The primer sequences are presented in Table 1.
TABLE 1.
PCR primer sequences and amplification parameters
| Gene | Primer sequence, 5′–3′ |
Product size (bp) | Temperature (°C) | |
|---|---|---|---|---|
| Sense | Antisense | |||
| β-Actin | GGTCATCACTATTGGCAACGAG | GAGGTCTTTACGGATGTCAACG | 142 | 57 |
| MAFbx | TCAGAGAGGCAGATTCGCAA | TCCAGGAGAGAATGTGGCAG | 154 | 59 |
| MuRF1 | TTTGACACCCTCTACGCCAT | TTGGCACTTGAGAGGAAGGT | 203 | 59 |
| PHD1 | TGTCACTGTGGTGTTTGGCTAC | GCATTTATCAGGATGGGAAGG | 123 | 61 |
| PHD2 | CCTGCCATTGGTGATAGTGAC | GGGTGGAAGGGTAAGAAACAT | 153 | 63 |
| PHD3 | TCAAGGCTGTGAGGTAGTCT | CTTGCATGGGAGGCTCATC | 186 | 61 |
| ADRB2 | TCAAGGCTGTGAGGTAGTCT | CTTGCATGGGAGGCTCATC | 192 | 59 |
Immunofluorescence and immunocytochemistry
C57BL/6J mouse gastrocnemius was sliced to 10 µm with a cryostat (CM 1850; Leica Microsystems, Buffalo Grove, IL, USA). The sections and C2C12 cells were rinsed 3 times in PBS and washed in 0.3% H2O2 for 30 min, then blocked for 1 h at room temperature. The sections were incubated in anti-MuRF1 (sc-32920; Santa Cruz Biotechnology) overnight at room temperature. Upright microscopes were used to take photographs. Image-Pro Plus software (Media Cybernetics, Rockville, MD, USA) was used for quantifying grayscale. Up to 6 fields of view were captured from the same location within each gastrocnemius muscle. Six hundred myofibers were measured per muscle. C2C12 cells were ncubated overnight in rabbit anti-PHD3 (bs-0532R; Bioss Antibodies) and rabbit anti-ADRB2 (bs-0947R; Bioss Antibodies) at 4°C. The next day, the sections were transferred to biotin secondary antibody (Bioworld Technology, St. Louis Park, MN, USA) for 1 h, and the C2C12 cells were incubated in FITC secondary antibody (Bioss Antibodies). C2C12 cells were then observed, and the fluorescence was quantified via Nikon Eclipse Ti-s microscopy with Nis-Elements BR software (Nikon, Tokyo, Japan). Up to 6 fields of view were captured from every group.
Hematoxylin and eosin staining
The mouse gastrocnemius samples were sliced to 10 µm with a cryostat. Cross sections were fixed in 4% formaldehyde at room temperature for 20 min and stained with hematoxylin and eosin (21). The myofiber area was quantified by Image-Pro Plus. Up to 6 fields of view were captured from the same location within each gastrocnemius muscle. Six hundred myofibers were measured per muscle.
Statistical analysis
Statistical analyses were performed by SPSS 18.0 (IBM SPSS, Chicago, IL, USA). Methods of statistical analyses were chosen on the basis of the design of each experiment and are indicated in the figure captions. The data were presented as means ± sem. A value of P ≤ 0.05 was considered statistically significant.
RESULTS
AKG reduces corticosterone-induced skeletal muscle protein degradation
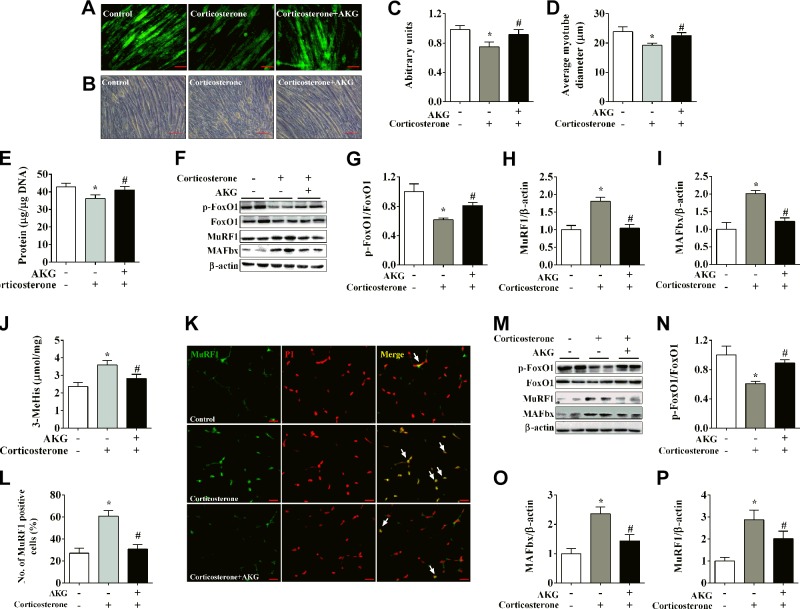
Corticosterone was used to induce skeletal muscle protein degradation both in vitro and in vivo. As an in vitro model of skeletal muscle development, C2C12 myotube was incubated with 10 μM corticosterone for 48 h to induce protein degradation. We found significant decreases of Click-It AHA (a muscle long-lived protein), myotube size, and total protein levels (Fig. 1A–E), all of which are consistent with previous observations (22) and validated our protein degradation model. Importantly, these inhibitory effects of corticosterone were abolished by AKG treatment (Fig. 1A–E), suggesting an antiprotein degradation role of AKG. Consistently, corticosterone-induced protein degradation was associated with decreased phosphorylation of the forkhead transcription factor FoxO1 and increased the E3 ubiquitin ligases MuRF1 and MAFbx. These modulations are common mechanisms for muscle atrophy and were diminished by AKG treatment (Fig. 1F–I). This protective role of AKG is further supported by the evidence from in vivo mouse model. Protein degradation was similarly induced by intraperitoneal injection of corticosterone, as demonstrated by the increases of 3-MeHis, MuRF1, MAFbx, and the nucleus shuttles of MuRF1 (Fig. 1J–M, O, P), and by the decreases of FoxO1 phosphorylation (Fig. 1M, N) in gastrocnemius of C57BL/6J mice. All of these corticosterone-induced effects were abolished by intraperitoneal coinjection of AKG (Fig. 1J–P). Both in vitro and in vivo data suggested that AKG inhibits corticosterone-induced skeletal muscle protein degradation.
Figure 1.
AKG inhibits corticosterone-induced protein degradation in skeletal muscle. A, C) Representative images (A) and fluorescence intensity quantification (C) of long-lived protein Click-It AHA in C2C12 myotubes treated with vehicle, 10 μM corticosterone, or 10 μM corticosterone + 2 mM AKG for 48 h. B, D) Representative images (B) and quantification (D) of C2C12 myotubes size treated for 48 h. E) Total protein levels of C2C12 myotubes treated for 48 h. F–I) Immunoblots (F) and quantification (G–I) of pFoxO1 and FoxO1 (G), MuRF1 (H), and MAFbx (I) in C2C12 myotubes treated for 48 h. J) 3-MeHis levels in gastrocnemius muscle from male C57BL/6J mice injected intraperitoneally with vehicle, 50 μg/kg corticosterone, or 50 μg/kg corticosterone + 1.0 g/kg AKG. 3-MeHis levels were detected by HPLC to measure protein degradation. K, L) Representative images (K) and quantification (L) of propidium iodide (PI, red)–positive MuRF1 (green) cells (yellow nucleus indicated by white arrows) in gastrocnemius muscle from male C57BL/6J mice injected intraperitoneally with vehicle, corticosterone, or corticosterone + AKG. M–P) Immunoblots (M) and quantification (N–P) of pFoxO1 and FoxO1 (N), MuRF1 (O), and MAFbx (P) in gastrocnemius muscle from male C57BL/6J mice injected intraperitoneally with vehicle, corticosterone, or corticosterone + AKG. All results contain 6 replicates per group (n = 6/group). Data are presented as means ± sem and were analyzed by 1-way ANOVA, followed by post hoc Bonferroni tests. β-Actin served as housekeeping gene. *P < 0.05 compared to control, #P < 0.05 compared to corticosterone group.
AKG rescues skeletal muscle atrophy in DMD mice
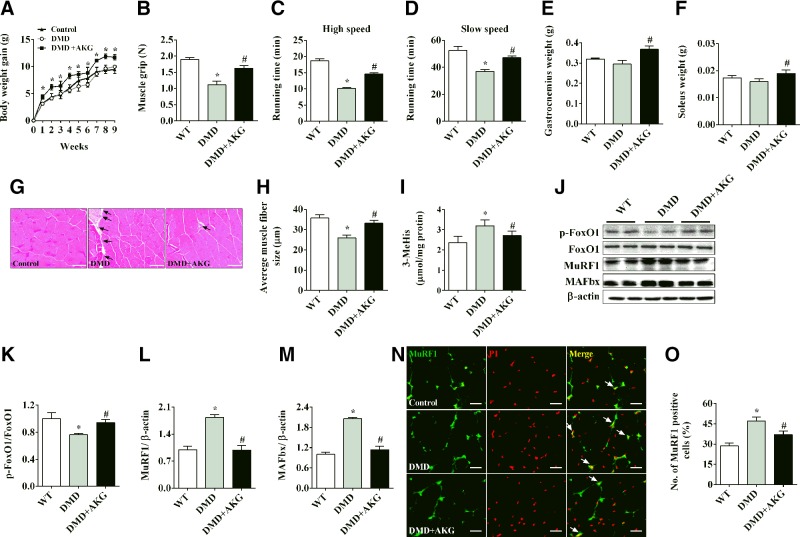
DMD is an X-linked disease caused by mutations in the DMD gene and loss of the protein dystrophin (23). Here we used the most commonly used mdx mouse, which has a point mutation in the dystrophin gene, as a model to study the effects of AKG on DMD muscle-wasting disorder. Consistent with previous descriptions of the mdx mouse model (24), we found that mdx mice showed normal body weight gain (Fig. 2A) and the same gastrocnemius and soleus muscle weight (Fig. 2E, F), but decreased limb strength (Fig. 2B) and exercise endurance (Fig. 2C, D). Interestingly, mdx mice fed with AKG supplements had significantly increased body weight gain, as well as weight of gastrocnemius and soleus muscle, compared to control mdx mice (Fig. 2A, E, F), suggesting a role of AKG in growth and muscle mass increase. More importantly, AKG also improved the limb strength and exercise endurance in mdx mice (Fig. 2B–D), indicating a protective function of AKG on muscle weakness induced by the dystrophin mutation in mdx mice. This point is further supported by the evidence that AKG attenuated the inhibition on muscle fiber size (Fig. 2G, H) and the stimulation on protein degradation (Fig. 2I) in the gastrocnemius muscle of the mdx mice. Consistently, the mdx mice showed decreased FoxO1 phosphorylation (Fig. 2J, K) and increased MuRF1, MAFbx, and nucleus shuttles of MuRF1 (Fig. 2J, L–O), while AKG diminished all these effects (Fig. 2J–O). Thus, similar to the observations in our corticosterone-induced muscle atrophy model, our data support a protective role of AKG in DMD muscle atrophy.
Figure 2.
AKG rescues skeletal muscle atrophy in mdx mice. A) Body weight of male C57BL/6J mice receiving tap water, mdx mice receiving water, or mdx mice receiving water supplemented with 2% AKG for 8 wk. B) Muscle grip of male mice provided water or 2% AKG for 8 wk. C, D) Treadmill running time of male mice provided water or 2% AKG for 8 wk at high-speed (C) and slow-speed (D) settings. E, F) Gastrocnemius (E) and soleus (F) muscle weight of male mice provided water or 2% AKG for 8 wk. G, H) Representative images (G) and quantification (H) of gastrocnemius muscle hematoxylin and eosin staining from male mice provided water or 2% AKG for 8 wk. I) Levels of 3-MeHis in gastrocnemius muscle from male mice provided water or 2% AKG for 8 wk. J–M) Immunoblots (J) and quantification (K–M) of pFoxO1 and FoxO1 (K), MuRF1 (L), and MAFbx (M) in gastrocnemius muscle from male mice provided water or 2% AKG for 8 wk. N, O) Representative images (N) and quantification (O) of propidium iodide (PI, red)-positive MuRF1 (green) cells (yellow nucleus indicated by white arrows) in gastrocnemius muscle from male mice provided water or 2% AKG for 8 wk. All results contain 10 replicates per group (n = 10/group). Data are presented as means ± sem and were analyzed by 1-way ANOVA, followed by post hoc Bonferroni tests. β-Actin served as housekeeping gene. *P < 0.05 compared to control; #P < 0.05 compared to MDX group.
PHD3 mediates inhibitory effects of AKG on protein degradation and myotube atrophy
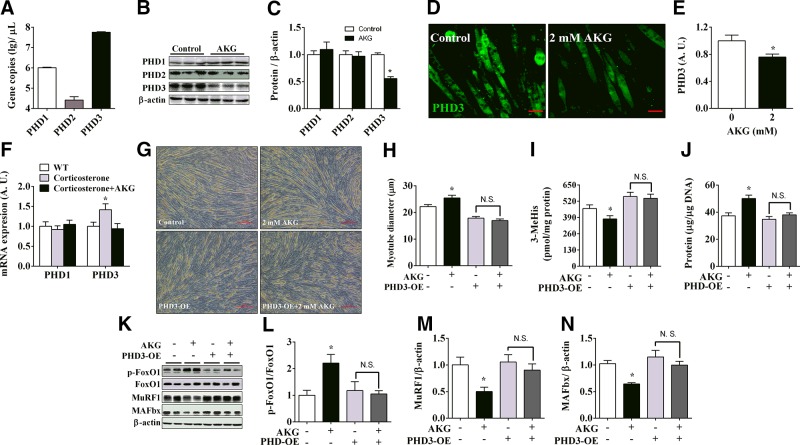
PHDs, also referred to as the hypoxia-inducible factor prolyl-hydroxylase, previously have been shown to mediate the stimulatory effects of AKG on rapamycin (mammalian target of rapamycin) signaling pathway (15). Given the fact that mammalian target of rapamycin enhances protein synthesis and inhibits protein degradation (25), we speculated that AKG inhibits protein degradation through PHD-mediated pathways. Because PHDs are a heterogeneous enzyme family composed of 3 different proteins including PHD1, PHD2, and PHD3, we first compared the expression levels of different PHDs in C2C12 myotubes. Although 3 forms of PHDs were all expressed, PHD3 had much higher expression levels than the other 2 forms in C2C12 myotubes (Fig. 3A). In addition, as demonstrated by both Western blot analysis (Fig. 3B, C) and immunofluorescence staining (Fig. 3D, E), PHD3 protein expression was significantly down-regulated by in vitro AKG treatment, while PHD1 and -2 did not respond to AKG. Importantly, in vivo corticosterone treatment significantly increased mRNA expression of PHD3, while AKG blocked this corticosterone-induced PHD3 increase. Notably, unlike PHD3, the mRNA expression of PHD1 was not changed by either corticosterone or AKG (Fig. 3F). These results suggest that PHD3 may play an important role in AKG’s antiprotein degradation effects. Consistent with this view, we found that overexpression of PHD3 blocked AKG’s antiprotein degradation effects in the C2C12 myotubes, including stimulation on myotube diameter (Fig. 3G, H), total protein content (Fig. 3J), and phosphorylation of FoxO1 (Fig. 3K, L), as well as inhibition on medium 3-MeHis concentration (Fig. 3I) and protein expression of MuRF1 and MAFbx (Fig. 3K, M, N). These observations support a model that AKG reduces protein degradation by inhibiting PHD3.
Figure 3.
PHD3 mediates inhibitory effects of AKG on protein degradation and myotube atrophy. A) C2C12 cells were induced for 4 d to differentiate into myotubes. Then C2C12 myotubes were collected for examining expression of 3 PHDs subtypes by absolute quantification PCR (n = 6/group). B, C) Immunoblots (B) and quantification (C) of PHDs in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). D, E) Representative images (D) and quantification (E) of PHD3 in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). F) mRNA expression of PHD1 and 3 in gastrocnemius muscle from 8 wk old male C57BL/6J mice injected intraperitoneally with vehicle, 50 μg/kg corticosterone, or 50 μg/kg corticosterone + 1.0 g/kg AKG (n = 6/group). G, H) Representative images (G) and quantification (H) of diameters of normal or PHD3-overexpressing C2C12 myotubes treated with 2 mM AKG for 48 h (n = 6/group). I) 3-MeHis levels in culture medium from normal or PHD3-overexpressing C2C12 myotubes treated with 2 mM AKG for 48 h (n = 6/group). J) Total protein levels in normal or PHD3-overexpressing C2C12 myotubes treated with 2 mM AKG for 48 h (n = 6/group). K–N) Immunoblots (K) and quantification (L–N) of pFoxO1 and FoxO1 (L), MuRF1 (M), and MAFbx (N) in normal or PHD3-overexpressing C2C12 myotubes treated with 2 mM AKG for 48 h (n = 6/group). Data are presented as means ± sem and were analyzed by 1-way ANOVA, followed by post hoc Bonferroni tests. β-Actin served as housekeeping gene. *P < 0.05 compared to control.
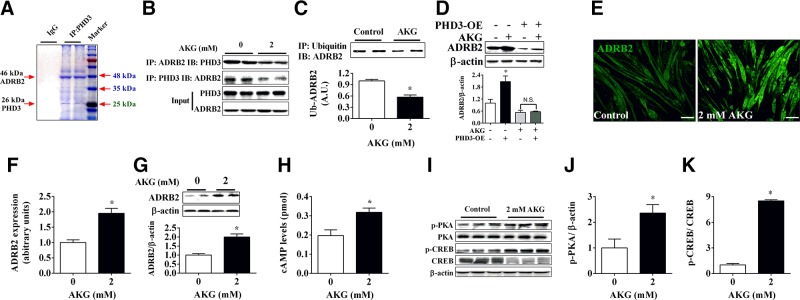
AKG inhibits interaction between PHD3 and its target ADRB2
As a member of the proline hydroxylase family, PHD3 has been reported to have many important downstream targets, but it is still not clear which target plays a crucial role in the inhibitory effects of PHD3 on protein degradation. To identify this essential target, we performed immunoprecipitation to screen for proteins that bind with PHD3. We found that in C2C12 myotubes, PHD3 interacted with many different proteins, and the primary binding protein had a molecular mass of approximately 48 kDa (Fig. 4A). Accordingly, the most likely protein is ADRB2, with a molecular mass of 46 kDa, which is hydroxylated and degraded by PHD3 (26). Subsequent coimmunoprecipitation results further support this point. We found that PHD3 bound with ADRB2 and that AKG significantly inhibited the interaction between PHD3 and ADRB2 (Fig. 4B). Because PHD3 also promotes the degradation of ADRB2 through ubiquitination (26), we tested the ubiquitination of ADRB2 and found that it was significantly reduced by AKG (Fig. 4C), which may result in up-regulation of ADRB2. Consistent with this, both Western blot and immunofluorescence results demonstrated that AKG significantly promoted the expression of ADRB2 in C2C12 myotubes (Fig. 4E–G). More importantly, this stimulatory effect of AKG on ADRB2 expression was totally blocked by PHD3 overexpression (Fig. 4D), suggesting a mediating role for PHD3. We next investigated the effects of AKG on ADRB2’s downstream cAMP/PKA pathway. Consistent with the stimulatory effects on ADRB2, AKG significantly increased cAMP levels in C2C12 myotubes (Fig. 4H) and promoted the phosphorylation of PKA and CREB (Fig. 4I–K). These results indicated that AKG inhibits PHD3/ADRB2 interaction to stimulate ADRB2 signaling pathway in skeletal muscle.
Figure 4.
ADRB2 is target of PHD3 in response to AKG. A) C2C12 cells were induced differentiation for 4 d, and anti-PHD3 was used to pull down PHD3 binding protein. Precipitated samples were used for SDS-PAGE, and then strips were stained by Coomassie Brilliant Blue. B) C2C12 myotubes were treated by 2 mM AKG for 48 h, and anti-PHD3 and anti-ADRB2 were used to precipitate PHD3 and ADRB2, respectively. Precipitated samples was subjected to immunoblotting of ADRB2 or PHD3 (n = 3 group). C) C2C12 myotube samples from control or 2 mM AKG–treated group were pulled down by anti-ubiquitin antibody and subjected to immunoblotting of ADRB2 (n = 3/group). D) Immunoblots and quantification of ADRB2 in normal or PHD3-overexpressing C2C12 myotubes treated with 2 mM AKG for 48 h (n = 6/group). E, F) Representative images (E) and quantification (F) of ADRB2 in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). G) Immunoblots and quantification of ADRB2 in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). H) cAMP levels in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). I–K) Immunoblots (I) and quantification (J, K) of p-PKA (J), PKA, p-CREB (K), and CREB (K) in C2C12 myotubes treated with 0 or 2 mM AKG for 48 h (n = 6/group). Data are presented as means ± sem and were analyzed by 1-way ANOVA followed by post hoc Bonferroni tests for panel D, and nonpaired Student’s t test for all others. β-Actin served as housekeeping gene. *P < 0.05 compared to control.
ADRB2 mediates the inhibitory effects of AKG on skeletal muscle atrophy and protein degradation
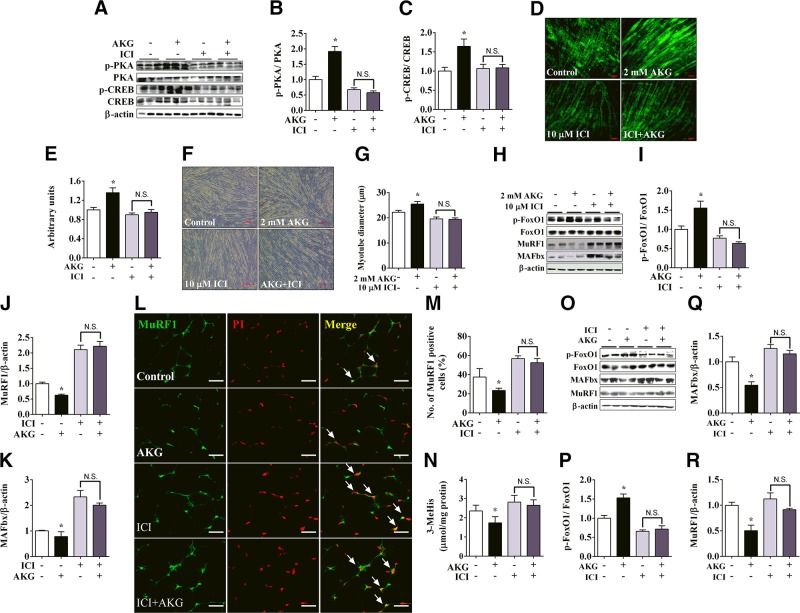
Given the facts that AKG increases ADRB2 expression and activates its downstream signaling pathway, and that ADRB2 plays an important role in skeletal muscle hypertrophy and protein synthesis (27), we speculate that AKG inhibits skeletal muscle atrophy and protein degradation by activating ADRB2. To test this hypothesis, we generated models with ADRB2 suppressed by the antagonist or knocked down by the specific lentivirus both in vitro and in vivo. We found that in C2C12 myotubes, the antiprotein degradation effects of AKG were abolished by cotreatment with ICI, an inhibitor of ADRB2 (Fig. 5A–K). Specifically, not only the stimulation of phosphorylation of PKA, CREB, and FoxO1 (Fig. 5A–C, H, I), expression of Click-It AHA (Fig. 5D, E), and diameter of myotubes (Fig. 5F, G) but also the inhibition of expression of MuRF1 and MAFbx (Fig. 5H, J, K) were diminished by ICI. These results clearly suggest a mediating role of ADRB2 for AKG’s antiprotein degradation effects. Consistently, a similar in vivo response was observed in the gastrocnemius muscle from male C57BL/6J mice. We found AKG intraperitoneal administration inhibited nucleus shuttles of MuRF1 (Fig. 5L, M), 3-MeHis contents (Fig. 5N), and protein expression of MuRF1 and MAFbx (Fig. 5O, Q, R), while ICI coinjection blocked these effects. Additionally, the stimulatory effect of AKG on phosphorylation of FoxO1 was also abolished by ICI (Fig. 5O, P). These pharmacologic results indicated that ADRB2 mediates the inhibitory effects of AKG on skeletal muscle atrophy and protein degradation.
Figure 5.
Pharmacologic inhibition of ADRB2 blocked inhibitory effects of AKG on skeletal muscle atrophy and protein degradation. A–C) Immunoblots (A) and quantification (B, C) of p-PKA and PKA (B) and p-CREB and CREB (C) in C2C12 myotubes treated with vehicle, 2 mM AKG, 10 μM ICI, or 2 mM AKG + 10 μM ICI for 48 h (n = 6/group). D, E) Representative images (D) and quantification (E) of long-life protein Click-It AHA in C2C12 myotubes treated with vehicle, 2 mM AKG, 10 μM ICI, or 2 mM AKG + 10 μM ICI for 48 h (n = 6/group). F, G) Representative images (F) and quantification (G) of fiber diameter of C2C12 myotubes treated with vehicle, 2 mM AKG, 10 μM ICI, or 2 mM AKG + 10 μM ICI for 48 h (n = 6/group). H–K) Immunoblots (H) and quantification (I–K) of pFoxO1 and FoxO1 (I), MuRF1 (J), and MAFbx (K) in C2C12 myotubes treated with vehicle, 2 mM AKG, 10 μM ICI, or 2 mM AKG + 10 μM ICI for 48 h (n = 6/group). L, M) Representative images (L) and quantification (M) of propidium iodide (PI, red)-positive MuRF1 (green) cells (yellow nucleus indicated by white arrows) in gastrocnemius muscle from male C57BL/6J mice 3 h after intraperitoneal injection with vehicle, 1 g/kg AKG, 5 μg/kg ICI, or 1 g/kg AKG + 5 μg/kg ICI (n = 6). N) 3-MeHis in gastrocnemius muscle from male C57BL/6J mice 3 h after intraperitoneal injection with vehicle, 1 g/kg AKG, 5 μg/kg ICI, or 1 g/kg AKG + 5 μg/kg ICI (n = 6). O–R) Immunoblots (O) and quantification (P–R) of pFoxO1 and FoxO1 (P), MuRF1 (Q), and MAFbx (R) in gastrocnemius muscle from male C57BL/6J mice 3 h after injected intraperitoneally with vehicle, 1 g/kg AKG, 5 μg/kg ICI, or 1 g/kg AKG + 5 μg/kg ICI (n = 6). Data are presented as means ± sem and were analyzed by 1-way ANOVA, followed by post hoc Bonferroni tests. β-Actin served as housekeeping gene. *P < 0.05 compared to control.
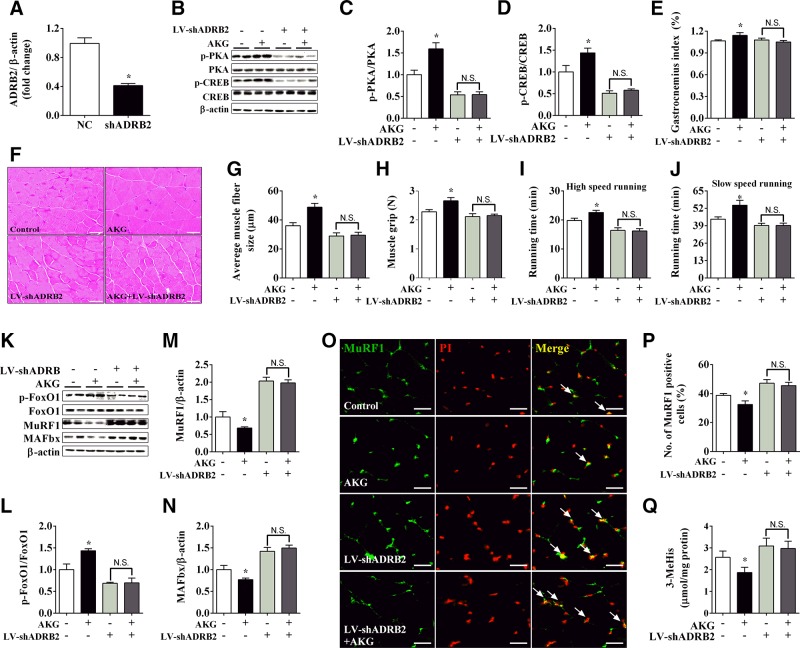
Besides pharmacologic methods, we also generated a mouse model with ADRB2 specifically knocked down in the gastrocnemius muscle by lentivirus to test our hypothesis. The interference efficiency of the lentivirus was first tested in the gastrocnemius muscle. We found that the mRNA expression of ADRB2 dropped by 70% in the shADRB2-treated group (Fig. 6A), which validated our knockdown model. Consistent with the pharmacologic data, ADRB2 knockdown blocked the increases of PKA, CREB, and FoxO1 phosphorylation (Fig. 6B–D, K, L) as well as the up-regulation of gastrocnemius weight and fiber diameter (Fig. 6E–G) induced by 8 wk of AKG supplementation. Additionally, muscle strength and exercise endurance, which are important indicators of skeletal muscle atrophy and protein degradation, were significantly improved by AKG treatment, while ADRB2 knockdown prevented these improvements (Fig. 6H–J). Similarly, ADRB2 knockdown also diminished AKG-induced inhibition on protein expression of MuRF1 and MAFbx (Fig. 6K, M, N), nucleus shuttles of MuRF1 (Fig. 6O, P), and 3-MeHis contents (Fig. 6Q) in the gastrocnemius muscle. Taken together, our pharmacologic and knockdown results support the notion that AKG stimulates ADRB2 to inhibit skeletal muscle atrophy and protein degradation.
Figure 6.
ADRB2 knockdown abolished inhibitory effects of AKG on muscle atrophy and protein degradation. A) mRNA expression of ADRB2 in gastrocnemius muscle from male C57BL/6J mice injected intramuscularly with LV-shScrambled or LV-shADRB2. B–D) Immunoblots (B) and quantification (C, D) of p-PKA and PKA (C) and p-CREB and CREB (D) in gastrocnemius muscle from male C57BL/6J mice receiving LV-shScrambled, LV-shScrambled + AKG, LV-shADRB2, or LV-shADRB2 +AKG. E–J) Gastrocnemius weight (E), gastrocnemius muscle fiber size (F, G), muscle grip (H), high-speed running time (I), and slow-speed running time (J) of male C57BL/6J mice receiving LV-shScrambled, LV-shScrambled + AKG, LV-shADRB2, or LV-shADRB2 + AKG. K–N) Immunoblots (K) and quantification (L–N) of pFoxO1 and FoxO1 (L), MuRF1 (M), and MAFbx (N) in gastrocnemius muscle from male C57BL/6J mice receiving LV-shScrambled, LV-shScrambled + AKG, LV-shADRB2, or LV-shADRB2 + AKG. O, P) Representative images (O) and quantification (P) of propidium iodide (PI, red) positive MuRF1 (green) cells (yellow nucleus indicated by white arrows) in gastrocnemius muscle from male C57BL/6J mice receiving LV-shScrambled, LV-shScrambled + AKG, LV-shADRB2, or LV-shADRB2 + AKG. Q) Levels of 3-MeHis in gastrocnemius muscle from male C57BL/6J mice receiving LV-shScrambled, LV-shScrambled + AKG, LV-shADRB2, or LV-shADRB2 + AKG. Data are presented as means ± sem and were analyzed by nonpaired Student’s t test for panel A, and 1-way ANOVA, followed by post hoc Bonferroni tests for all others. β-Actin served as housekeeping gene. *P < 0.05 compared to control.
DISCUSSION
Excessive protein degradation is the main cause for skeletal muscle atrophy, which is harmful to human health and leads to muscle fatigue and reduced life span (28–31). Abnormal protein degradation and muscle atrophy generally occur with genetic mutation (DMD) (32), hormone exposure (glucocorticoid) (33), or disease (cachexia) (34). Many studies have shown that appropriate exercise can induce muscle hypertrophy and is also an effective way to rescue muscle atrophy (35–37). Interestingly, clinical studies demonstrated that exercise is associated with increased serum AKG (38), which previously has been shown to down-regulate protein degradation–related genes in the skeletal muscle of mice (17). These studies indicate a potential mediating role of AKG in anti–muscle atrophy function of exercise. Here, we found that AKG improved corticosterone-induced skeletal muscle protein degradation both in vitro and in vivo. Moreover, AKG not only enhanced muscle strength and exercise endurance but also ameliorated skeletal muscle atrophy and fatigue in DMD mice. These data suggest that AKG plays a key role in skeletal muscle atrophy and protein degradation.
PHDs, a family of hypoxia-induced hydroxylases (39), activate the cell proliferative signaling (40), participate in cancer metabolism (41), and regulate the development of regulatory T cells (42) through downstream target proteins, including hypoxia-inducible factor 1α (43), epidermal growth factor receptor (40), TNF-α (44), and IKK (45). As an important intermediate product in tricarboxylic acid cycles, AKG acts not only as the ligand for GPR99 receptor but also as a sensor of PHDs. Because only PHDs but not GPR99 express in the skeletal muscle (46), we wondered if PHDs act as downstream targets for AKG to mediate its antiprotein degradation effects. PHDs are a family of AKG-dependent dioxygenases, including PHD1–3 (47–49). We found that PHD3 has the highest expression in the skeletal muscle tissues, which is consistent with a previous study (50). More interestingly, we found that AKG down-regulated the expression of PHD3, but not PHD1 and -2, and that PHD3 overexpression blocked the stimulatory effects of AKG on skeletal muscle atrophy and protein degradation. These results suggest that PHD3 is a primary mediator for AKG’s antiprotein degradation effects.
To identify the essential PHD3-targeting proteins that mediate AKG’s antiprotein degradation effects, we screened the proteins that interacted with PHD3 in the skeletal muscle. We found that PHD3 has the most abundant binding with ADRB2, an anti–muscle atrophy GPCR (51), which is consistent with previous reports in cardiac and HEK293 cells (26, 52). PHD3 inhibits downstream target proteins by hydroxylation and ubiquitylation (26, 53). Here we found that AKG significantly inhibited the interaction between PHD3 and ADRB2, and reduced the ubiquitination of ADRB2, which in return increased the expression of ADRB2. More importantly, AKG-induced up-regulation of ADRB2 was blocked by PHD3 overexpression, suggesting a mediating role of PHD3 in regulatory effects of AKG on ADRB2. ADRB2 has been shown to be activated by two different mechanisms, binding its ligands or accumulating in cell membranes, both of which activate the downstream signaling pathway (54). Consistently, we found that AKG not only increased the stability and up-regulated the expression of ADRB2 but also activated downstream cAMP/PKA signaling pathway. These results clearly demonstrate an AKG-regulated PHD3/ADRB2 pathway.
ADRB2, mainly activating Gαs stimulatory protein (55), previously has been shown to inhibit muscle atrophy. For example, ADRB2 agonists reduce muscle atrophy induced by cancer cachexia (56, 57), and efficacy depends on the expression levels of ADRB2 in skeletal muscle. Additionally, ADRB2 global knockout increases the expression levels of proteins related to the ubiquitin–proteasome system and skeletal muscle atrophy (58). These raise the possibility that ADRB2 mediates the inhibitory effects of AKG on muscle atrophy. This view is further supported by the pharmacologic evidence that the ADRB2 inhibitor blocked the inhibitory effects of AKG on protein degradation. In addition, genetic knockdown of ADRB2 in vivo diminished AKG-induced anti–muscle atrophy effects, including inhibited protein degradation, improved muscle strength and exercise endurance, and increased muscle fiber and mass. Both pharmacologic and genetic evidence argue for a mediating role of ADRB2 in AKG’s anti–muscle atrophy effects.
Consistent with this view is our finding that AKG activates ADRB2’s downstream cAMP/PKA pathway, which previously has been shown to be involved in skeletal muscle hypertrophy, metabolism, and regeneration (55, 59, 60). Early studies demonstrated that cAMP and PKA activity increase during embryonic muscle development and myoblast differentiation (61–63), suggesting a myogenic role of cAMP/PKA pathway. Consistently, sustained activation of cAMP signaling promotes skeletal muscle hypertrophy and prevents muscle atrophy in rodents (64, 65). Notably, we also observed AKG induced an ADRB2-mediated stimulation on phosphorylation of CREB, the direct downstream targeting transcription factor of the cAMP/PKA pathway (66). CREB has been shown to suppress the transcription of FoxO1/3a and to decrease the expression of the atrophy-related genes MuRF1 and MAFbx (67–70). Collectively, these observations support a model that AKG up-regulates ADRB2 and subsequently activates cAMP/PKA to inhibit skeletal muscle atrophy.
In summary, we demonstrated that AKG ameliorates skeletal muscle atrophy and protein degradation by inhibiting PHD3 to increase the stability of ADRB2. Our findings identified AKG as a potential treatment for muscle atrophy and AKG may be developed as a muscle-targeted “exercise mimetics” to combat the increase in a sedentary lifestyle.
ACKNOWLEDGMENTS
This work was supported by the National Basic Research Program of China (Grant 2013CB127306 to G.S.), National Key Point Research and Invention Program (Grant 2016YFD0501205 to G.S.), Training Program for Outstanding Young Teachers in the Universities of Guangdong Province (G.S.), National Natural Science Foundation of China (Grants 31572480 to G.S. and 31472105 to Q.J.), and the U.S. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases (Grant K99DK107008 to P.X.). The authors declare no conflicts of interest.
Glossary
- ADRB2
β2 adrenergic receptor
- AKG
α-ketoglutarate
- BSA
bovine serum albumin
- CREB
cAMP-response element binding protein
- DMD
Duchenne muscular dystrophy
- FoxO1
forkhead box O1
- GPR91/99
G protein–coupled receptor 91/99
- ICI
ICI-118,551 hydrochloride
- MAFbx
muscle atrophy F-box
- MuRF1
muscle RING-finger protein 1
- PHD1/2/3
proline hydroxylase 1/2/3
- qPCR
quantitative PCR
- 3-MeHis
3-methylhistidine
AUTHOR CONTRIBUTIONS
X. Cai and Y. Yuan were the main contributors to the conduct of the study, data collection and analysis, data interpretation, and article writing; Z. Liao, K. Xing, C. Zhu, Y. Xu, and L. Yu contributed to the conduct of the study; L. Wang, S. Wang, X. Zhu, P. Gao, and Y. Zhang contributed to the study design and data interpretation; and Q. Jiang, P. Xu, and G. Shu contributed to the study design, data interpretation, and article writing.
REFERENCES
- 1.Bonaldo P., Sandri M. (2013) Cellular and molecular mechanisms of muscle atrophy. Dis. Model. Mech. 6, 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malicdan M. C., Noguchi S., Nonaka I., Saftig P., Nishino I. (2008) Lysosomal myopathies: an excessive build-up in autophagosomes is too much to handle. Neuromuscul. Disord. 18, 521–529 [DOI] [PubMed] [Google Scholar]
- 3.Stephens N. A., Gallagher I. J., Rooyackers O., Skipworth R. J., Tan B. H., Marstrand T., Ross J. A., Guttridge D. C., Lundell L., Fearon K. C., Timmons J. A. (2010) Using transcriptomics to identify and validate novel biomarkers of human skeletal muscle cancer cachexia. Genome Med. 2, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gordon B. S., Kelleher A. R., Kimball S. R. (2013) Regulation of muscle protein synthesis and the effects of catabolic states. Int. J. Biochem. Cell Biol. 45, 2147–2157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Verdijk L. B., Dirks M. L., Snijders T., Prompers J. J., Beelen M., Jonkers R. A., Thijssen D. H., Hopman M. T., Van Loon L. J. (2012) Reduced satellite cell numbers with spinal cord injury and aging in humans. Med. Sci. Sports Exerc. 44, 2322–2330 [DOI] [PubMed] [Google Scholar]
- 6.Bonifacio A., Sanvee G. M., Bouitbir J., Krähenbühl S. (2015) The AKT/mTOR signaling pathway plays a key role in statin-induced myotoxicity. Biochim. Biophys. Acta 1853, 1841–1849 [DOI] [PubMed] [Google Scholar]
- 7.White J. P., Gao S., Puppa M. J., Sato S., Welle S. L., Carson J. A. (2013) Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol. Cell. Endocrinol. 365, 174–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbé C., Kalista S., Loumaye A., Ritvos O., Lause P., Ferracin B., Thissen J. P. (2015) Role of IGF-I in follistatin-induced skeletal muscle hypertrophy. Am. J. Physiol. Endocrinol. Metab. 309, E557–E567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dibble C. C., Cantley L. C. (2015) Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25, 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A., Walsh K., Schiaffino S., Lecker S. H., Goldberg A. L. (2004) Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell 117, 399–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stitt T. N., Drujan D., Clarke B. A., Panaro F., Timofeyva Y., Kline W. O., Gonzalez M., Yancopoulos G. D., Glass D. J. (2004) The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophy–induced ubiquitin ligases by inhibiting FOXO transcription factors. Mol. Cell 14, 395–403 [DOI] [PubMed] [Google Scholar]
- 12.Lee S. W., Dai G., Hu Z., Wang X., Du J., Mitch W. E. (2004) Regulation of muscle protein degradation: coordinated control of apoptotic and ubiquitin–proteasome systems by phosphatidylinositol 3 kinase. J. Am. Soc. Nephrol. 15, 1537–1545 [DOI] [PubMed] [Google Scholar]
- 13.Hou Y., Wang L., Ding B., Liu Y., Zhu H., Liu J., Li Y., Kang P., Yin Y., Wu G. (2011) Alpha-ketoglutarate and intestinal function. Front. Biosci. (Landmark Ed.) 16, 1186–1196 [DOI] [PubMed] [Google Scholar]
- 14.Kristensen N. B., Jungvid H., Fernández J. A., Pierzynowski S. G. (2002) Absorption and metabolism of alpha-ketoglutarate in growing pigs. J. Anim. Physiol. Anim. Nutr. (Berl.) 86, 239–245 [DOI] [PubMed] [Google Scholar]
- 15.Durán R. V., MacKenzie E. D., Boulahbel H., Frezza C., Heiserich L., Tardito S., Bussolati O., Rocha S., Hall M. N., Gottlieb E. (2013) HIF-independent role of prolyl hydroxylases in the cellular response to amino acids. Oncogene 32, 4549–4556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yao K., Yin Y., Li X., Xi P., Wang J., Lei J., Hou Y., Wu G. (2012) Alpha-ketoglutarate inhibits glutamine degradation and enhances protein synthesis in intestinal porcine epithelial cells. Amino Acids 42, 2491–2500 [DOI] [PubMed] [Google Scholar]
- 17.Cai X., Zhu C., Xu Y., Jing Y., Yuan Y., Wang L., Wang S., Zhu X., Gao P., Zhang Y., Jiang Q., Shu G. (2016) Alpha-ketoglutarate promotes skeletal muscle hypertrophy and protein synthesis through Akt/mTOR signaling pathways. Sci. Rep. 6, 26802. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Polletta L., Vernucci E., Carnevale I., Arcangeli T., Rotili D., Palmerio S., Steegborn C., Nowak T., Schutkowski M., Pellegrini L., Sansone L., Villanova L., Runci A., Pucci B., Morgante E., Fini M., Mai A., Russo M. A., Tafani M. (2015) SIRT5 regulation of ammonia-induced autophagy and mitophagy. Autophagy 11, 253–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Young V. R., Munro H. N. (1978) Ntau-methylhistidine (3-methylhistidine) and muscle protein turnover: an overview. Fed. Proc. 37, 2291–2300 [PubMed] [Google Scholar]
- 20.Sato T., Ito Y., Nedachi T., Nagasawa T. (2014) Lysine suppresses protein degradation through autophagic-lysosomal system in C2C12 myotubes. Mol. Cell. Biochem. 391, 37–46 [DOI] [PubMed] [Google Scholar]
- 21.Gianelo M. C. S., Polizzelo J. C., Chesca D., Mattiello-Sverzut A. C. (2016) Three days of intermittent stretching after muscle disuse alters the proteins involved in force transmission in muscle fibers in weanling rats. Braz. J. Med. Biol. Res. 49, e4118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohtsuka A., Hayashi K., Noda T., Tomita Y. (1992) Reduction of corticosterone-induced muscle proteolysis and growth retardation by a combined treatment with insulin, testosterone and high-protein–high-fat diet in rats. J. Nutr. Sci. Vitaminol. (Tokyo) 38, 83–92 [DOI] [PubMed] [Google Scholar]
- 23.Kornegay J. N. (2017) The golden retriever model of Duchenne muscular dystrophy. Skelet. Muscle 7, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ng R., Banks G. B., Hall J. K., Muir L. A., Ramos J. N., Wicki J., Odom G. L., Konieczny P., Seto J., Chamberlain J. R., Chamberlain J. S. (2012) Animal models of muscular dystrophy. Prog. Mol. Biol. Transl. Sci. 105, 83–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao J., Zhai B., Gygi S. P., Goldberg A. L. (2015) mTOR inhibition activates overall protein degradation by the ubiquitin proteasome system as well as by autophagy. Proc. Natl. Acad. Sci. USA 112, 15790–15797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xie L., Xiao K., Whalen E. J., Forrester M. T., Freeman R. S., Fong G., Gygi S. P., Lefkowitz R. J., Stamler J. S. (2009) Oxygen-regulated beta(2)-adrenergic receptor hydroxylation by EGLN3 and ubiquitylation by pVHL. Sci. Signal. 2, ra33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Glass D. J. (2003) Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 5, 87–90 [DOI] [PubMed] [Google Scholar]
- 28.Atherton P. J., Greenhaff P. L., Phillips S. M., Bodine S. C., Adams C. M., Lang C. H. (2016) Control of skeletal muscle atrophy in response to disuse: clinical/preclinical contentions and fallacies of evidence. Am. J. Physiol. Endocrinol. Metab. 311, E594–E604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Percival J. M. (2011) nNOS regulation of skeletal muscle fatigue and exercise performance. Biophys. Rev. 3, 209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anthony T. G. (2016) Mechanisms of protein balance in skeletal muscle. Domest. Anim. Endocrinol. 56(Suppl), S23–S32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossetti M. L., Steiner J. L., Gordon B. S. (2017) Androgen-mediated regulation of skeletal muscle protein balance. Mol. Cell. Endocrinol. 447, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Escobar R. G., Lucero N., Solares C., Espinoza V., Moscoso O., Olguín P., Muñoz K. T., Rosas R. (2017) Upper limb functional assessment scale for children with Duchenne muscular dystrophy and spinal muscular atrophy. Rev. Chil. Pediatr. 88, 92–99 [DOI] [PubMed] [Google Scholar]
- 33.Kang S. H., Lee H. A., Kim M., Lee E., Sohn U. D., Kim I. (2017) Forkhead box O3 plays a role in skeletal muscle atrophy through expression of E3 ubiquitin ligases Murf-1 and atrogin-1 in Cushing’s syndrome. Am. J. Physiol. Endocrinol. Metab. 312, E495–E507 [DOI] [PubMed] [Google Scholar]
- 34.Lee D. E., Brown J. L., Rosa-Caldwell M. E., Blackwell T. A., Perry R. A. Jr., Brown L. A., Khatri B., Seo D., Bottje W. G., Washington T. A., Wiggs M. P., Kong B. W., Greene N. P. (2017) Cancer cachexia-induced muscle atrophy: evidence for alterations in microRNAs important for muscle size. Physiol. Genomics 49, 253–260 [DOI] [PubMed] [Google Scholar]
- 35.Bamman M. M., Roberts B. M., Adams G. R. (2017) Molecular regulation of exercise-induced muscle fiber hypertrophy. [E-pub ahead of print] Cold Spring Harb. Perspect. Med. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim J. Y. (2016) Therapeutic potential of eccentric exercises for age-related muscle atrophy. Integr. Med. Res. 5, 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fan J., Yang X., Li J., Shu Z., Dai J., Liu X., Li B., Jia S., Kou X., Yang Y., Chen N. (2017) Spermidine coupled with exercise rescues skeletal muscle atrophy from d-gal-induced aging rats through enhanced autophagy and reduced apoptosis via AMPK-FOXO3a signal pathway. Oncotarget 8, 17475–17490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leibowitz A., Klin Y., Gruenbaum B. F., Gruenbaum S. E., Kuts R., Dubilet M., Ohayon S., Boyko M., Sheiner E., Shapira Y., Zlotnik A. (2012) Effects of strong physical exercise on blood glutamate and its metabolite 2-ketoglutarate levels in healthy volunteers. Acta Neurobiol. Exp. (Warsz.) 72, 385–396 [DOI] [PubMed] [Google Scholar]
- 39.Schofield C. J., Ratcliffe P. J. (2004) Oxygen sensing by HIF hydroxylases. Nat. Rev. Mol. Cell Biol. 5, 343–354 [DOI] [PubMed] [Google Scholar]
- 40.Garvalov B. K., Foss F., Henze A. T., Bethani I., Gräf-Höchst S., Singh D., Filatova A., Dopeso H., Seidel S., Damm M., Acker-Palmer A., Acker T. (2014) PHD3 regulates EGFR internalization and signalling in tumours. Nat. Commun. 5, 5577. [DOI] [PubMed] [Google Scholar]
- 41.German N. J., Yoon H., Yusuf R. Z., Murphy J. P., Finley L. W., Laurent G., Haas W., Satterstrom F. K., Guarnerio J., Zaganjor E., Santos D., Pandolfi P. P., Beck A. H., Gygi S. P., Scadden D. T., Kaelin W. G. Jr., Haigis M. C. (2016) PHD3 loss in cancer enables metabolic reliance on fatty acid oxidation via deactivation of ACC2. Mol. Cell 63, 1006–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Singh Y., Shi X., Zhang S., Umbach A. T., Chen H., Salker M. S., Lang F. (2016) Prolyl hydroxylase 3 (PHD3) expression augments the development of regulatory T cells. Mol. Immunol. 76, 7–12 [DOI] [PubMed] [Google Scholar]
- 43.Schoepflin Z. R., Silagi E. S., Shapiro I. M., Risbud M. V. (2017) PHD3 is a transcriptional coactivator of HIF-1α in nucleus pulposus cells independent of the PKM2-JMJD5 axis. FASEB J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujita N., Gogate S. S., Chiba K., Toyama Y., Shapiro I. M., Risbud M. V. (2012) Prolyl hydroxylase 3 (PHD3) modulates catabolic effects of tumor necrosis factor-α (TNF-α) on cells of the nucleus pulposus through co-activation of nuclear factor κB (NF-κB)/p65 signaling. J. Biol. Chem. 287, 39942–39953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xue J., Li X., Jiao S., Wei Y., Wu G., Fang J. (2010) Prolyl hydroxylase-3 is down-regulated in colorectal cancer cells and inhibits IKKbeta independent of hydroxylase activity. Gastroenterology 138, 606–615 [DOI] [PubMed] [Google Scholar]
- 46.Diehl J., Gries B., Pfeil U., Goldenberg A., Mermer P., Kummer W., Paddenberg R. (2016) Expression and localization of GPR91 and GPR99 in murine organs. Cell Tissue Res. 364, 245–262 [DOI] [PubMed] [Google Scholar]
- 47.Taylor M. S. (2001) Characterization and comparative analysis of the EGLN gene family. Gene 275, 125–132 [DOI] [PubMed] [Google Scholar]
- 48.West C. M., van der Wel H., Wang Z. A. (2007) Prolyl 4-hydroxylase-1 mediates O2 signaling during development of Dictyostelium. Development 134, 3349–3358 [DOI] [PubMed] [Google Scholar]
- 49.Epstein A. C., Gleadle J. M., McNeill L. A., Hewitson K. S., O’Rourke J., Mole D. R., Mukherji M., Metzen E., Wilson M. I., Dhanda A., Tian Y. M., Masson N., Hamilton D. L., Jaakkola P., Barstead R., Hodgkin J., Maxwell P. H., Pugh C. W., Schofield C. J., Ratcliffe P. J. (2001) C. elegans EGL-9 and mammalian homologs define a family of dioxygenases that regulate HIF by prolyl hydroxylation. Cell 107, 43–54 [DOI] [PubMed] [Google Scholar]
- 50.Lieb M. E., Menzies K., Moschella M. C., Ni R., Taubman M. B. (2002) Mammalian EGLN genes have distinct patterns of mRNA expression and regulation. Biochem. Cell Biol. 80, 421–426 [DOI] [PubMed] [Google Scholar]
- 51.Wannenes F., Magni L., Bonini M., Dimauro I., Caporossi D., Moretti C., Bonini S. (2012) In vitro effects of beta-2 agonists on skeletal muscle differentiation, hypertrophy, and atrophy. World Allergy Organ. J. 5, 66–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Xie L., Pi X., Townley-Tilson W. H., Li N., Wehrens X. H., Entman M. L., Taffet G. E., Mishra A., Peng J., Schisler J. C., Meissner G., Patterson C. (2015) PHD2/3-dependent hydroxylation tunes cardiac response to β-adrenergic stress via phospholamban. J. Clin. Invest. 125, 2759–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grosfeld A., Stolze I. P., Cockman M. E., Pugh C. W., Edelmann M., Kessler B., Bullock A. N., Ratcliffe P. J., Masson N. (2007) Interaction of hydroxylated collagen IV with the von Hippel–Lindau tumor suppressor. J. Biol. Chem. 282, 13264–13269 [DOI] [PubMed] [Google Scholar]
- 54.Bowman S. L., Shiwarski D. J., Puthenveedu M. A. (2016) Distinct G protein–coupled receptor recycling pathways allow spatial control of downstream G protein signaling. J. Cell Biol. 214, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Berdeaux R., Stewart R. (2012) cAMP signaling in skeletal muscle adaptation: hypertrophy, metabolism, and regeneration. Am. J. Physiol. Endocrinol. Metab. 303, E1–E17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carbó N., López-Soriano J., Tarragó T., González O., Llovera M., López-Soriano F. J., Argilés J. M. (1997) Comparative effects of beta2-adrenergic agonists on muscle waste associated with tumour growth. Cancer Lett. 115, 113–118 [DOI] [PubMed] [Google Scholar]
- 57.Fuster G., Busquets S., Ametller E., Olivan M., Almendro V., de Oliveira C. C., Figueras M., López-Soriano F. J., Argilés J. M. (2007) Are peroxisome proliferator-activated receptors involved in skeletal muscle wasting during experimental cancer cachexia? Role of beta2-adrenergic agonists. Cancer Res. 67, 6512–6519 [DOI] [PubMed] [Google Scholar]
- 58.Voltarelli V. A., Bechara L. R., Bacurau A. V., Mattos K. C., Dourado P. M., Bueno C. R. Jr., Casarini D. E., Negrao C. E., Brum P. C. (2014) Lack of β2-adrenoceptors aggravates heart failure–induced skeletal muscle myopathy in mice. J. Cell. Mol. Med. 18, 1087–1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Minetti G. C., Feige J. N., Rosenstiel A., Bombard F., Meier V., Werner A., Bassilana F., Sailer A. W., Kahle P., Lambert C., Glass D. J., Fornaro M. (2011) Gαi2 signaling promotes skeletal muscle hypertrophy, myoblast differentiation, and muscle regeneration. Sci. Signal. 4, ra80. [DOI] [PubMed] [Google Scholar]
- 60.Gonçalves D. A., Silveira W. A., Lira E. C., Graça F. A., Paula-Gomes S., Zanon N. M., Kettelhut I. C., Navegantes L. C. (2012) Clenbuterol suppresses proteasomal and lysosomal proteolysis and atrophy-related genes in denervated rat soleus muscles independently of Akt. Am. J. Physiol. Endocrinol. Metab. 302, E123–E133 [DOI] [PubMed] [Google Scholar]
- 61.Le Peuch C. J., Ferraz C., Walsh M. P., Demaille J. G., Fischer E. H. (1979) Calcium and cyclic nucleotide dependent regulatory mechanisms during development of chick embryo skeletal muscle. Biochemistry 18, 5267–5273 [DOI] [PubMed] [Google Scholar]
- 62.Rogers J. E., Narindrasorasak S., Cates G. A., Sanwal B. D. (1985) Regulation of protein kinase and its regulatory subunits during skeletal myogenesis. J. Biol. Chem. 260, 8002–8007 [PubMed] [Google Scholar]
- 63.Zalin R. J., Montague W. (1974) Changes in adenylate cyclase, cyclic AMP, and protein kinase levels in chick myoblasts, and their relationship to differentiation. Cell 2, 103–108 [DOI] [PubMed] [Google Scholar]
- 64.Hinkle R. T., Dolan E., Cody D. B., Bauer M. B., Isfort R. J. (2005) Phosphodiesterase 4 inhibition reduces skeletal muscle atrophy. Muscle Nerve 32, 775–781 [DOI] [PubMed] [Google Scholar]
- 65.Navegantes L. C., Resano N. M., Migliorini R. H., Kettelhut I. C. (2000) Role of adrenoceptors and cAMP on the catecholamine-induced inhibition of proteolysis in rat skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 279, E663–E668 [DOI] [PubMed] [Google Scholar]
- 66.Delghandi M. P., Johannessen M., Moens U. (2005) The cAMP signalling pathway activates CREB through PKA, p38 and MSK1 in NIH 3T3 cells. Cell. Signal. 17, 1343–1351 [DOI] [PubMed] [Google Scholar]
- 67.Chung Y. W., Kim H. K., Kim I. Y., Yim M. B., Chock P. B. (2011) Dual function of protein kinase C (PKC) in 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced manganese superoxide dismutase (MnSOD) expression: activation of CREB and FOXO3a by PKC-alpha phosphorylation and by PKC-mediated inactivation of Akt, respectively. J. Biol. Chem. 286, 29681–29690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Van der Heide L. P., Smidt M. P. (2005) Regulation of FoxO activity by CBP/p300-mediated acetylation. Trends Biochem. Sci. 30, 81–86 [DOI] [PubMed] [Google Scholar]
- 69.Zhang X., Odom D. T., Koo S. H., Conkright M. D., Canettieri G., Best J., Chen H., Jenner R., Herbolsheimer E., Jacobsen E., Kadam S., Ecker J. R., Emerson B., Hogenesch J. B., Unterman T., Young R. A., Montminy M. (2005) Genome-wide analysis of cAMP-response element binding protein occupancy, phosphorylation, and target gene activation in human tissues. Proc. Natl. Acad. Sci. USA 102, 4459–4464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Silveira W. A., Goncalves D. A., Graca F. A., Andrade-Lopes A. L., Bergantin L. B., Zanon N. M., Godinho R. O., Kettelhut I. C., Navegantes L. C. (2014) Activating cAMP/PKA signaling in skeletal muscle suppresses the ubiquitin–proteasome–dependent proteolysis: implications for sympathetic regulation. J. Appl. Physiol. (1985) 117, 11–19 [DOI] [PubMed] [Google Scholar]