Abstract
CatSper is a voltage-dependent calcium channel located in the plasma membrane of the sperm flagellum and is responsible for triggering hyperactive motility. A homology model for the transmembrane region was built in which the arrangement of the subunits around the pseudo-four-fold symmetry axis was deduced by the pairing of conserved transmembranal cysteines across mammals. Directly emergent of the predicted quaternary structure is an architecture in which tetramers polymerize through additional, highly conserved cysteines, creating one or more double-rows channels extending the length of the principal piece of the mammalian sperm tail. The few species that are missing these cysteines are eusocial or otherwise monogamous, suggesting that sperm competition is selective for a disulfide-crosslinked macromolecular architecture. The model suggests testable hypotheses for how CatSper channel opening might behave in response to pH, 2-arachidonoylglycerol, and mechanical force. A flippase function is hypothesized, and a source of the concomitant disulfide isomerase activity is found in CatSper-associated proteins β, δ and ε.
Keywords: Electrophysiology, Integral membrane protein, Sperm, Contraception, Hyperactive motility, sperm competition, Disulfide linkage, Bioinformatics, Mechanotransduction, Disulfide isomerase
1. Introduction
Successful transmission of genetic material during sexual reproduction in mammals provides strong selective pressure for sperm motility, especially in species where sperm competition takes place. CatSper is a sperm-associated voltage-dependent calcium channel that is required for the hyperactive motility that is required to drive the sperm through the outer layers of the oocyte [1]. Knock-out of any of the four subunits of CatSper (CatSper1–4) in mouse produces infertile males but has no phenotypic effect on the female and no additional effect on the male [2]. CatSper is a member of the ion transport protein family (pfam00520), having six transmembrane helices and an ion selective pore. Several atomic-resolution structures exist for the homo-logous voltage-sensitive cation channel sub-families NaV, KV and CaV [3]. Each consists of four transmembrane helical segments of the voltage-sensing (VS) domain orbiting like satellites around the central pore domain (P). The P-domain is made up of four copies of the two C-terminal transmembrane segments with an intervening, pore-defining “P-loop”. The P-loop of CatSper contains a conserved sequence motif, TxDxW, that is associated with a calcium ion selective channel [4].
CatSper is the only known natively heterotetrameric CaV. All others are homotetramers or a single chain fusion of four transmembrane domains [5], with the special exception of Xenopus epithelial Ca2+ channels TRPV5 and TRPV6 which, when co-expressed, assemble into functional but non-native heteromeric calcium channels with different ion selectivities [6]. The requirement for four CatSper genes in mammals suggests that there is a structural selective pressure that goes beyond the form and function of the individual channel, since one gene would do if sperm required only a voltage-dependent and ion selective channel. The arrangement of CatSper into four lines extending the length of the principal piece of the sperm flagellum [60], and the observed association of defects in this architecture with reduced motility [7], suggests a connection between the CatSper macromolecular architecture and hyperactive motility.
CatSper is a target for immune-based contraception [65,66]. A reliable model will help to identify antigenic sites. Also, a model for the architecture of CatSper may help us to understand the relationship between its macromolecular form and its function in capacitation-induced hyperactive motility. Subunit-specific sequence conservation in the context of structure provides hints as to the basis of the selective pressure. Here we discuss conserved cysteine residues in mammalian CatSpers. Their cross-linking ability may serve to dictate the relative positioning of the four subunits. Here we describe a quaternary structure and an emergent higher-order (super-quaternary) structure that carries implications with regard to the mechanisms of voltage-dependent, pH-induced calcium channel opening in capacitated mammalian sperm.
2. Materials and methods
2.1. Bioinformatics
Homolog sequences for each CatSper subunit were identified by searching the RefSeq [11] database using BLASTp [43], restricting the search to mammals, and requiring the Entrez [44] search string “cation channel sperm-associated protein [n]” where n was 1,2,3,4 respectively in four BLASTp searches. Sequences with the annotation ‘LOW QUALITY SEQUENCE’ or which lacked any of the six transmembrane regions, or of species that were not represented by all four subunits, were removed. All four subunits of each of the remaining 41 species were concatenated in numerical order and the N- and C-terminal sequences were removed for further analysis. Sequences were aligned in UGENE [45] using MUSCLE [46]. Sequences were removed if they exceeded 92% sequence identity to any other sequence, leaving 24 species. Poorly aligned regions were omitted before calculating phylogenetic trees. Phylogenetic trees were computed in UGENE using the PhyML Maximum Likelihood [47] method, using the Blosum62 substitution matrix. Regions around conserved cysteines were dissected out using in-house fortran programs and presented along with the phylo-gram using the ETE python objects [48]. Modified “Zappo” coloring is used throughout [49].
For multiple sequence alignments of individual CatSper subunits, all species passing the above filters were included after removing those with 90% identity or greater, where the cutoff was based on all aligned positions. Any N- or C-terminal sequences that aligned poorly were left unaligned in Fig. 1.
Fig. 1.

Multiple sequence alignment for mammalian CatSper1–4. Ten representative mammal sequences were used for each of subunit in the TM region, comprising the VS and P-domains. N and C-terminal regions were aligned separately for each subunit using 55, 23, 39, and 36 mammalian sequences for CatSpers 1–4 respectively, based on a cutoff of 92% maximum sequence identity. Pie charts show amino acid composition of terminal domains color-coded as shown along the bottom. Names for transmembrane segments are indicated along the bottom.
The significance of finding four disulfide isomerase motif sequences CXC or CXXC within three out of five CatSper-associated sequences was calculated as the empirical probability of being conserved across mammals in CatSper-associated proteins (β:0.59,γ:0.51,δ:0.59,ε:0.46,ζ:0.45) times the frequency of cysteine respectively in those sequences (β:0.019,γ:0.019,δ:0.024,ε:0.022,ζ:0.0089) squared because there are two cysteines in each motif, times the respective length of the protein (β:994,γ:997,δ:781,ε:842,ζ:197). The expected number of CxC or CxxC motifs by chance in the five proteins is 0.12. The chance likelihood of four motifs was found by randomly sampling this distribution 10000 times and summing the number of times that four motifs or more was found across three or more proteins, p-value = 0.0009. Therefore, chance occurrence is rejected with 99.9% confidence.
Transmembrane helix prediction was done using TMHMM [50]. Secondary structure prediction for globular proteins was done using PsiPred [51]. Disordered regions were predicted using DISOPRED. Glycosylation sites were predicted using NetNGlyc [52]. Compositional analysis, tree pruning, pH and charge calculations were carried out using Microsoft Excel 2016 and in-house Fortran programs. Sequence motifs were identified using PrositeScan [53]. Charge calculations were done by summing the formal changes of the sidechains times the pH-dependent fraction charged using the Henderson-Hasselbach equation.
2.2. Molecular modeling
All molecular modeling was carried out using MOE (Chemical Computing Group, Montreal). All six possible subunit arrangements for the P-loop segment were built using MOE’s homology modeling tools. Side chains were repacked using MOE’s Gizmin function while holding backbone atoms fixed. Multiple tetramer models were docked manually to form dimers and higher-order oligomers by searching in the space of two translational and one rotational degree of freedom. Rotations and translations were constrained to the plane of the membrane. Conserved intramembranal cysteines that could come into contact were assumed to form disulfide bonds [54]. 2-Arachidonoylglycerol (2AG) was docked to CatSper interactively while energy minimizing using MOE’s Gizmin function. Backbone atoms were fixed.
Template-based remote homolog three-dimensional structure predictions for cytoplasmic CatSper domains and CatSper associated proteins were carried out using RaptorX [55]. In these cases, several mammalian homolog sequences were predicted independently and the predictions were compared to extract consensus features. No consensus structures were found.
2.3. Disulfide prediction
Evolutionary conservation of cysteines in structural context predicts disulfide formation [56,57]. Specifically, proteins that contain cysteines which find themselves within 6 Å of each other in the folded state readily crosslink when expressed in the lumen of the ER, depending on the presence of protein disulfide isomerases [58]. Disulfides do not dictate the protein topology, but they serve to lock the structure into place once formed. Moreover, insertion of cysteines into a sequence is a technique for the determination the transmembrane structure by identifying positions that support spontaneous crosslinking [59,54].
3. Results
3.1. Terminology and notation
Sequence positions denoted (Mm: < residue > < number >) refer to the Mus musculus sequences for Catsper n (n = 1–4,β,γ,δ,ε,ζ). Positions in sequences from other species are referenced using the aligned mouse sequence numbering. Transmembrane (TM) helical segments are denoted s1 through s6, where s1-s4 constitute the VS-domain and s5–s6 constitute the P-domain. Subunit arrangements for the CatSper tetramer around the pseudo-four-fold axis are denoted in clockwise order as viewed from the extracellular side. Thus, arrangements 1234, 2341, 3412 and 4123 all reference the same tetramer. We introduce the term “super-quaternary” to mean the arrangement of tetramers into higher-order macromolecular structures.
3.2. Homology modeling of CatSper monomers
The closest homolog of known structure to CatSper is an archeal voltage-gated calcium channel, (CaV) from Arcobacter butzleri [8]. The archeal and mammalian sequences share only “twilight zone” [9] similarity and then only in the transmembrane regions. But ancillary data makes the assignment of homology much more solid. Specifically, all mammalian CatSpers have six TM segments, a pore-forming motif between s5 and s6, and the presence of periodic arginines in s4 that are characteristic of a VS-domain [10].
The RefSeq database [11] contains between 100 and 130 mammalian CatSper sequences for each subunit. As a group, the four paralogs are characterized by conservation only in the TM region (Fig. 1). The N-and C-terminal domains of the individual subunits are conserved across mammals but no detectable homology was found between any of the terminal domains and any protein of known structure, therefore the homology models discussed here cover only the TM region. Interesting features of the terminal domains will be discussed later. The TM regions show intersubunit conservation in the TM helical segments, in the internal loops and in the P-loop. CatSpers also align confidently to the archeal CaV template within these segments (Supplemental Fig. S1). The spatial positioning of amino acids in sequences that are confidently predicted to be homologs is generally regarded as reliable for the aligned regions, sufficiently so that homology models are routinely used to solve crystal structures by molecular replacement. Expectation values for alignments of mouse CatSpers1–3 to arcobacter butzleri CaV (PDBID:4MS2) were highly significant (2e-18, 6e-19, 1e-7). Although CatSper 4 falls below BLAST-detectable similarity of 4MS2, it does align significantly (3e-9) to the rabbit CaV1.1 structure (PDBID:5GJW) [12] which superposes with 4MS2 with RMSD 3.44 Å. The two potential templates are remarkably similar in their TM structure despite having only 27.3% sequence identity (Supplemental Fig. S5). As expected, most of the external loops are highly variable and cannot be confidently modeled. In the discussions below, important inferences are confined to the TM helices and one of the internal loops, which are confidently aligned.
3.3. Tetrameric subunit arrangement
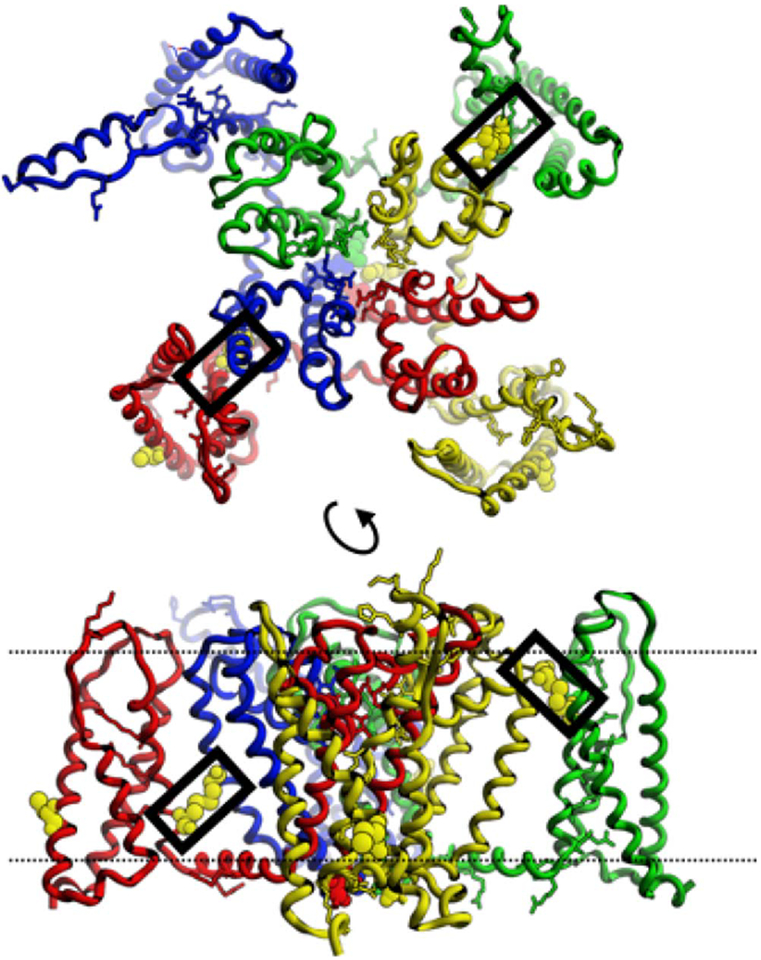
3.3.1. Disulfide 1 – Mm4:C170 to Mm1:C499
The template archeal CaV is homomeric but CatSper is heteromeric. There are six ways to organize four subunits clockwise around a pseudo-four-fold axis, abbreviated 1234, 1324, 1243, 1342, 1423, and 1432. We may assume that the energetic interactions between monomers dictate the selection of binding partners, but the prediction of the subunit arrangement is a prohibitively difficult computational docking problem given the uncertainties in energy calculations and the expected errors in the homology-based models. Conserved cysteines within the transmembrane segments provide the most compelling evidence for the correct arrangement of the subunits (Fig. 2). Mm1:C499 sits on s5 near the internal face, while Mm4:C170 sits on s4, also near the internal face. These two would form a disulfide if subunit 4 packed clockwise against subunit 1. The nature of the interaction is best described as the wrapping of subunit 4 P-domain and VS-domain around the P-domain of subunit 1 (Fig. 3). Both Mm4:C170 and Mm1:C499 are strictly conserved in mammals. Moreover, no other cysteine partners are available for Mm4:C170 on s5 of any of the other three CatSper subunits in any mammalian species, and no other cysteine partners are available for Mm1:C499 on s4 of any of the other three subunits in any mammalian species. Their strict conservation and the conspicuous absence of any alternative pairings for either subunit 1 or 4 in any species leads us to conclude that CatSper 4 (Mm4:C170) crosslinks with CatSper 1 (Mm1:C499) in a clockwise 41 arrangement. Crosslinking CatSper 4 and 1 eliminates four alternative arrangements, leaving just 1234 and 1324.
Fig. 2.
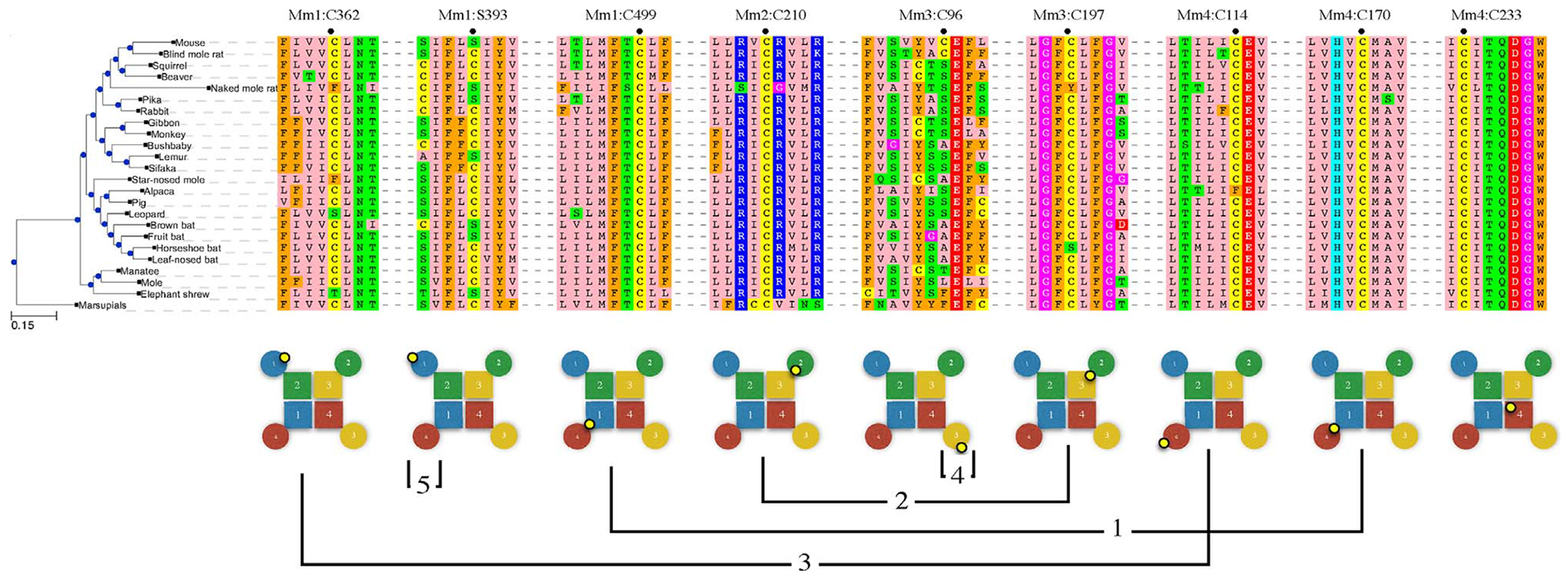
Blocks of multiple sequence alignment for 24 representative mammals were extracted from around the conserved cysteine positions that are discussed in the text. Mus musculus position labeled with a dot. Lower panel shows the location of the cysteine (small yellow circle) in the cartoon tetramers (circles: VS domain, squares: P domain, 1:blue, 2:green, 3:yellow, 4:red). Connecting lines indicate predicted disulfide crosslinks 1 through 4 as numbered in the text. “5” marks an alternative to Disulfide 4, a self-reacting crosslink. Mm4:C233 is strictly conserved but is not outward facing and cannot form a crosslink.
Fig. 3.
CatSper tetramer model (1: blue, 2: green, 3: yellow, 4: red) showing Disulfides 1 (left) and 2 (right) in black boxes. Lower panel shows the difference in depth of the two disulfides in the membrane. Dotted lines show approximate location of membranes surfaces (top: external, bottom: internal).
3.3.2. Disulfide 2 – Mm3:C197 to Mm2:C210
The only other conserved, outward-facing cysteine in the P-domain of any CatSper subunit is Mm3:C197, which sits on s5 near the extra-cellular surface. An approximate 10 Å difference in depth within the membrane makes Mm3:C197 unreachable by either of the Disulfide 1 cysteines Mm4:C170 or Mm1:C499. The only cysteine on the P-domain-facing surface of the VS domain on any subunit is Mm2:C210, which sits on s4 near the extracellular surface. Mm3:C197 and Mm2:C210 would come within 6 Å and crosslink if CatSper2 binds CatSper3 in a clockwise fashion. With 6 exceptions, no other cysteines are present at a position to pair with Mm3:C197 in any mammalian species. Those six exceptions (2 mol rats, guinea pig, common shrew, chinchilla, and degu) have a pairable cysteine at or near position Mm4:P160. However, if CatSper4 is already bound to CatSper1, then position Mm4:P160 is buried by the 41 dimerization interaction and unavailable for pairing with CatSper3. If we assume that the prior crosslinking of CatSpers 4 and 1 blocks access to Mm4:P160, then there are no alternative pairings for either Mm3:C197 or Mm2:C210 in any mammalian species. We conclude that Mm2:C210 crosslinks with Mm3:C197 in all mammals, and that therefore the subunit arrangement is 1234, not 1324. CatSpers 1 and 4 pair to form a 41 heterodimer, and CatSpers 2 and 3 simultaneously form a 23 dimer.
The final assembly of the channel consists of the specific association of the CatSper dimer 41 to the dimer 23. Since there are no additional conserved cysteines in positions to define the specificity of this tetramerization event, other reasons must be cited to account for selectivity of the 41 + 23 pairing over the 41 + 41 or 23 + 23 mispairings, which are also possible. We can safely say that both mispaired tetramers, 1414 and 2323 either do not form or are sub-functional, based on the CatSper-null mouse studies in which the knock-out of any subunit leads to hyperactive motility-impaired sperm [13]. Specific shape complimentarity interactions must account for the selective tetramerization of the correct and functional form. However, it cannot be ruled out that one of the CatSper associated proteins acts as a chaperone to block the formation of the mispaired tetramers.
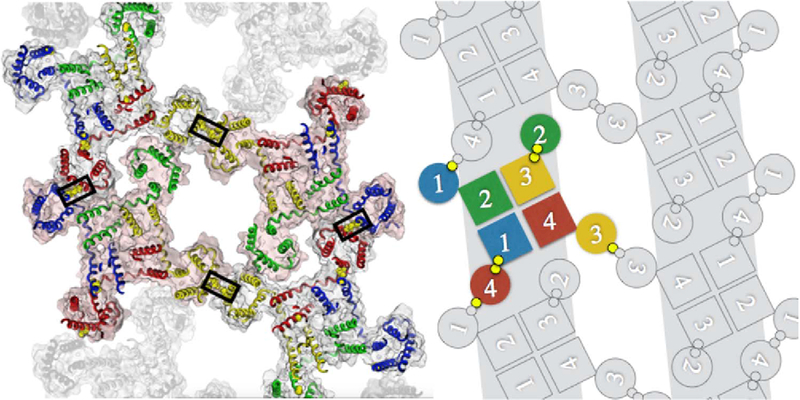
3.4. Superquaternary structure
The strong selective pressure for a heterotetrameric ion channel may be explained by considering the known organization of CatSper into four quadrilaterally-arranged rows along the principal piece of the flagellum [14]. If dictated by the CatSper structure itself, this organization would require asymmetry in the tetramer since specific interactions of a homomeric complex would be equally distributed in four directions, not just in two. Specific interactions between subunits of a heterotetrameric arrangement can form straight line polymer.
3.4.1. Disulfide 3 – Mm1:C362 to Mm4:C114
Upon inspection of the multiple sequence alignments in the context of the 1234 tetramer model, we find that cysteines Mm1:S393 and Mm3:C96 sit on the external, intramembranal surfaces of the VS-domains. Interestingly, these conserved cysteines vary across mammals in their sequence positions within the VS-domain. Another pair of cysteines, Mm1:C362 and Mm4:C114, are also externally-facing, intramembranal, and conserved across mammals, but these two occupy a fixed position throughout their evolutionary history (Fig. 2). A cysteine that pairs with itself on another tetramer (i.e. self-reacting) always sits at the same depth in the membrane as its crosslinking partner, which is itself, regardless of whether it has mutated to a new depth. However, if two different cysteines pair (i.e. cross-reacting) then there is no simple mutational pathway to a shifted depth within the membrane. Two cysteines would have to simultaneously appear in crosslinkable orientations, which is highly unlikely. Therefore, we expect that cysteines that vary in position over evolutionary time are probably self-reacting, while cysteines that remain fixed in their position are probably cross-reacting.
By assuming that the two fixed-position cysteines, Mm1:C362 and Mm4:C114, crosslink we create a linear polymer (Fig. 4). Although it is sterically possible that one of the variable-position cysteines, either Mm1:S393 or Mm3:C96 could pair with one of the fixed-position cysteines, such pairing would create a 90° rotation and further cross-linking would generate a tetramer of tetramers, not a linear polymer. Both the linear polymer and the tetramer of tetramers are possible, but only the linear polymer agrees with what we know about the macro-molecular architecture of CatSper. We assume once again that energetic packing interactions direct and determine the true super-quaternary structure, which is then locked into place by the actions of disulfide isomerases.
Fig. 4.
Zipper model in ribbon and cartoon representations. Two linear rows of CatSper tetramers are polymerized via cross-reacting Disulfide 3 linking CatSpers 1 (blue) and 4 (red). Antiparallel rows are linked at CatSper 3 (yellow) by self-reacting Disulfide 4. In cartoon version disulfide linkages are shown as small yellow circles.
3.4.2. Disulfide 4 – Mm3:C96 to Mm3:C96
If we accept the prediction of a linear polymer created by Disulfide 3, then the CatSper 4 VS-domain is docked to a notch between the VS-domains of CatSpers 1 and 2, and the VS-domains of CatSpers 1 and 3 remain exposed. The two variable-position cysteines Mm1:S393 and Mm3:C96 are positioned to self-react if two linear polymers are arranged antiparallel. Either the CatSper 1 side or the CatSper 3 side of the polymer may self-react. There is a no clear energetic difference between these two possibilities except the energetic ramifications of the positioning of the highly charged N-terminal domain of CatSper 1. Electrostatic repulsion would disfavor the self-association of CatSper 1’s. We tentatively propose a zipper structure in which CatSper polymers are cross-linked at Mm3:C96 (Fig. 4). It is not likely that the CatSper polymer rows associate in a parallel fashion by cross-reacting disulfides between Mm1:S393 and Mm3:C96 for the reasons described above regarding the evolutionary variability of the depth of these cysteines in the membrane, and because we do not see a correspondence across species between the depths of these cysteines in the membrane. In some mammals, this cross-link is possible but not in all.
4. Discussion
4.1. CatSper role in sperm competition
Swim speed is one of many phenotypic effector mechanisms selected by sperm competition [15], which also include sperm size, testes size, ejaculate volume, and seminal fluid proteins [16]. Additionally, many factors, not just CatSper function, influence swim speed in vertebrate sperm, including cell length and morphology [17] and the size of the mid-piece [18]. Thus no one factor explains success in sperm competition. But genetic adaptations in CatSper offer a uniquely adaptive advantage that they are expressed exclusively in the male and therefore suffer no conflicting manifestations in the female [19]. Combining strong selective pressure for motility with the absence of genetic interference may have focused evolution on the CatSper genes.
Three of the four cysteines proposed to form Disulfide 1 (Mm1:C499, Mm4:C170) and Disulfide 2 (Mm2:C210, Mm3:C197) are 100% conserved in mammals. Mm3:C197 is missing only in naked mole rat (Heterocephalus glaber) and chinese horseshoe bat (Rhinolophus sinicus). Naked mole rat is also missing Disulfide 3 cysteine Mm1:C362, and Disulfide 4 cysteine Mm3:C96, implying defects in subunit arrangement, polymerization, and cross-linking of rows in that species. Indeed, the swim speed for naked mole rat sperm has been quantified and characterized as “very slow”, and the slowness was attributed to the absence of sperm competition in that eusocial species [20]. Horseshoe bat has lost the proposed tetramer determinant Mm3:C197 from Disulfide 2, suggesting an impact on sperm motility. Unfortunately, there is no quantitative data on sperm motility in this or any bat. We can assert, however, a well-supported presence of sperm competition in bats, shown using comparative anatomy [21], and if taken as proof of sperm motility this suggests that loss of just one of the conserved disulfides is not enough to adversely affect hyperactive motility.
Disulfide 3 (Mm1:C362, Mm4:C114) is proposed to polymerize the CatSper tetramers into a row, but one or the other cysteine is absent in naked mole rat (Heterocephalus glaber), star-nosed mole (Condylura cristata), leopard (Panthera pandus), alpaca (Vicugna pacos) and elephant shrew (Elephantulus edwardii). Elephant shrews lack Mm1:C362, and also lack Disulfide 4 (Mm1:C393) which is hypothesized crosslink CatSper rows into a zipper. No data is available on the motility of elephant shrew sperm; however, it is known that elephant shrews are socially monogamous [22,23], which would appear to reduce the likelihood of sperm competition as a selective pressure. Some elephant shrews have sperm that are much shorter than those of such polyandrous species as rodents and primates [24]. Our model predicts that elephant shrew sperm motility, once it is measured, will be found to be slow.
The leopard (Panthera pardus) and other cats lack one cysteine of Disulfide 3 (Mm1:C362). But these animals are both promiscuous in their sexual behavior and show clear anatomical signs of sperm competition [25]. A closer look revealed that two partially-conserved cysteines (Mm2:C71, Mm3:C39) exist at positions just N-terminal to the start of the first TM helix of CatSpers 2 and 3 in cats and many other genera. This pair can form a disulfide in our model, which may serve as a “backup” in case Disulfide 3 is lost by mutation. Most mammals have both Disulfide 3 (Mm1:C362, Mm4:C114) and the “backup” (Mm2:C71, Mm3:C39) and both are compatible with the same polymer model. The “backup” crosslink is absent in star-nosed mole, alpaca, and elephant shrew. Alpaca and all other camelids also have the mutation Mm4:C114F, which would imply the absence of both Disulfide 3 and its “backup”. Although new world camelids (alpaca) have a cysteine at Mm4:Q48 which is spatially positionable to link to Mm1:C362 and rescue the Mm4:C114F mutation, old world camelids do not have this mutation. Thus, our model predicts that old world camelids may have slow swimming sperm. There are no studies to date on sperm motility in these species. Camelid’s ovulation is induced by seminal plasma [26,27] but induced ovulation has never been tied to the absence of sperm competition.
Sites on the outward facing edge of either CatSper 1 or CatSper 3 VS-domains are proposed to cross-link antiparallel CatSper polymers, but both Disulfide 4 (Mm3:C96) and its alternative self-reacting cysteine (Mm1:C393) are absent in two species: elephant shrew and lemur. Elephant shrew has been discussed above as probably lacking sperm competition. However, since lemur species probably do exhibit sperm competition [28,29], it would be premature to conclude that these zipper cross-bridges are required for sperm hyperactive motility.
4.2. The case for disulfide isomerase activity in CatSper associated proteins β, δ and ε
Five CatSper-associated proteins have been identified, CatSpers β [30], γ [31], δ [32], ε [7] and ζ [7]. CatSpers β, δ and ε each contain at least one strongly conserved instance of the CXC or CXXC motif, the presence of which is associated with disulfide isomerase activity [33]. The two cysteines in Mmβ:920-CNCT are 100% conserved and those in a second instance at Mmβ:187-CPC-189 are 80% conserved in a representative set of 27 mammalian sequences (Fig. S2). CatSperδ has a C-terminal region that is rich in cysteine. In this region, a CxC or CxxC motif occurs in 74% of the representative set of 38 mammalian sequences. Interestingly, African elephant is missing the cys-rich region but has a CXXC motif at another location, Mmδ305-CLAC, possibly suggestive of a rescue-of-function mutation. CatSper δ also contains a 100% conserved CCxxxxxxC motif of unknown function, but which is reminiscent of the SCCS motif that has been associated with mammalian selenoprotein thioredoxin reductase [34]. Selenium is known to up-regulate CatSper expression in mice [61]. CatSperε has a CXXC motif at Mmε:224 which is present in 27 out of 30 mammalian species. Two of the three that do not contain CXXC (ground squirrel and star-nosed mole) do contain CXXXC. Only mongolian gerbil is missing any disulfide isomerase motifs in CatSperε. Chance occurrence of four CxC or CxxC motifs occurring in three out of five CatSper associated proteins was rejected with 99.9% confidence using the resampling method.
No disulfide isomerase or any other enzyme activity has yet been experimentally determined for CatSper associated proteins. Moreover, the fragmentation of the CatSper stripes that was cited here as evidence for a defect in the disulfide crosslinking was seen in CatSperζ-null mutant sperm [7] even though CatSperζ has no disulfide isomerase motifs.
4.3. The case for a mechanotransduction function of CatSper zipper architecture
The presence of rheotaxis in sperm requires an explanation for how the forces generated by fluid flow manifest in a rotation of the tail [35]. Mechanotransdunction of calcium channel opening provided by the proposed zipper architecture provides such an explanation. Specifically, forces exerted on the membrane may stretch the zipper, pulling on the VS-domains where they are crosslinked. The VS-domains pull on the s4s5 linker helix, whose positioning is known to change upon opening of the cytoplasmic gate [36]. In superresolution images, CatSperζ-null mouse sperm labeled on CatSper1 show fragmentation [7] and are impaired in rheotaxis. Channel opening by mechanotransduction through disulfide cross-links would explain the coincidence a fragmented CatSper structure and the loss of efficient rheotaxis.
A common origin for mechano- and voltage-sensing has been previously noted, in which homology is found between voltage-gated cation channels and the bacterial large-conductance mechanosensitive channel [62]. A sequence database search using CatSper1 for sequences that contain the word “mechanosensitive” in the annotation string yielded several significant hits in bacteria and mammals. CatSper1 transmembrane domain has distant sequence homology with mouse CAV3.2 (NP_001157163) which has been shown to be associated with mechanotransduction in the dorsal root ganglia (DRG) cells of mouse hair follicles [63]. Specifically, the application of a high concentration of mibefradil, a selective T-type calcium channel antagonist, completely abolished mechanosensitivity within minutes of application, suggesting that the CAV3.2 calcium channel is responsible for mechanotransduction in DRG cells. Although the sequence identity between CAV3.2 and CatSpers 1–4 transmembrane domains is weak (27%), their structural homology is unambiguous, albeit indirectly confirmed. Mouse CAV3.2 is a close homolog of Nav1.4-beta1 From Electric Eel, which has a structure (PDBID: 5XSY) that has obvious homology to the CatSper model. Thus, we conclude that CatSper1–4 is a structural homolog of CAV3.2 and probably shares the mechanotransduction function.
4.4. The case for pH sensing in the N terminal domain of CatSper 1
While all of the N-terminal domains of CatSpers1–4 have a positive charge, the size and charge of CatSper1 N-terminal domain goes well beyond that required by the “positive-inside rule” of integral membrane proteins [37] and suggests that the domain has a role beyond simply dictating the topology of the TM helices. An abundance of histidines (Fig. 1) suggests a pH sensing function. In the epididymis, at pH 6.5, 24% of these histidines would be charged whereas upon ejaculation and mixing with the prostatic fluids to pH 8.0 [38] only 1% of the histidine sidechains would remain protonated. This pH shift represents an average shift of −12 in the ionic state of the domain. This domain is highly variable across mammals and the pH-induced shift varies from −38 charge units for Pacific Walrus to as little at −1.4 for Little Brown Bat and −1.2 for Alpaca. The charge shift suggests a possible “priming” mechanism for gate opening. A loss of positive charge, and in some species the acquisition of a negative charge, decreases or reverses the attractive force between the CatSper1 N-terminal domain and the negatively charged C-terminal domains, leading to a net repulsion of the C-terminal domains. The C-terminal domains are attached to the hydrophobic gate residues of s6. This hypothesis reiterates previously published suspicions about the role of the histidine-rich tail of CatSper1 [64].
4.5. The structural basis for a 2-AG inhibition
The mechanism for progesterone activation of CatSper during capacitation post coitus is believed to be the progesterone-mediated activation of α/β hydrolase domain–containing protein 2 (ABHD2), which hydrolyzes 2-arachidonoylglycerol (2AG) [67], which in turn inhibits CatSper. The model presented here provides insight as to how 2AG inhibits CatSper. The central channel has a wide central chamber that can fit one or two copies of 2AG if folded into a compact conformation. Large gaps (“lipid portals”) exist between the P-domains of each pair of CatSper subunits, wide enough to allow passage of 2AG. 2AG is an uncharged lipid ester, which is free to diffuse through the hydrophobic interior of the lipid bilayer, unlike charged lipids that are anchored to the bilayer surface. Also of critical import, 2AG is an all-cis leukotriene that can adopt a highly curved and compact conformation. 2AG in a compact conformation was able to pass through the lipid portals without disturbing protein backbone atoms and without unfolding. Its presence in the calcium channel would clearly block passage of ions. Hydrolysis of 2AG by ABHD2 would result in a negatively charged species which would not be able to diffuse freely in the lipid bilayer and thus would not be able to enter the CatSper channel. Supplementary Fig. S7 shows the lipid portal and the results of interactive docking of 2AG to CatSper.
4.6. The case for a flippase function in CatSper
Flippases, floppases and scramblases are transmembrane proteins that catalyze the translocation of membrane lipids from the inner to the outer leaf, from outer to inner, or both, respectively. Although the mechanism of translocation is still unknown, the crystal structure of PglK flippase contains a salt bridge cluster that suggests a hypothetical translocation intermediate containing a charge-stabilized buried lipid head group. A large conformational change is required for flippase activity, as demonstrated by the inhibition of its function by a cross-binding nanobody [39] In other studies, it was experimentally demonstrated that same side positioning of multiple Arg or His on a transmembrane α-helix was sufficient to generate flippase activity, using only short peptides [40]. The crystal structure of MurJ flippase (Supplementary Fig. S7) shows a mid-membrane cluster of changed sidechains with a net positive charge at neutral pH [68]. A similar cluster of structurally-aligned arginine and glutamate sidechains is found in the interface between CatSper VS- and P-domains where it may be subject to voltage-induced conformational changes. Mammalian sperm undergo capacitation-induced membrane reorganizations by hitherto unknown flippases (or floppases or scramblases) [41], and Cat-Sper’s voltage-dependent channel opening is capacitation-induced. This circumstantial evidence of a flippase-like structure, plus the experimentally observed membrane reorganization, does not together constitute proof that CatSper is a flippase, but it provides a plausible hypothesis. If so this may be an instance of “accidental” adaptation of a new function from an existing one (voltage dependence), absent any obvious selective advantage. It is notable that the coincidence of ion channel and flipping activities has been previously documented in the case of the Ca2+-activated chloride channel TMEM16F in mouse, a voltage-sensitive channel having scramblase activity; specifically, the knock-out of TMEM16F produces a defect in phospholipid scrambling in platelets [42].
Supplementary Material
Acknowledgements
This work was supported by the NIH grant R01 GM099827. Special thanks to Jean-Ju Chung (Cellular & Molecular Physiology, Yale University) for guidance and for critical reading of this manuscript.
Footnotes
Conflict of interest
I certify that I am the sole contributor to the work described in this paper and have no conflicts of interest.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.repbio.2018.01.005.
References
- [1].Ren D, Navarro B, Perez G. A sperm ion channel required for sperm motility and male fertility. Nature 2001;413(Oxctober (6856)):603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A 2007;104:1219–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Catterall WA. Structure and regulation of voltage-gated Ca2+ channels. Annu Rev Cell Dev Biol 2000;16(1):521–55. [DOI] [PubMed] [Google Scholar]
- [4].Tang SH, Mikala G, Bahinski A, Yatani A, Varadi G, Schwartz A. Molecular localization of ion selectivity sites within the pore of a human L-type cardiac calcium channel. J Biol Chem 1993;268(June (18)):13026–9. [PubMed] [Google Scholar]
- [5].Catterall WA. Voltage-gated calcium channels. Cold Spring Harb Perspect Biol 2011;3(8):a003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hoenderop JGJ, Voets T, Hoefs S, Weidema F, Prenen J, Nilius B, et al. Homo-and heterotetrameric architecture of the epithelial Ca 2+ channels TRPV5 and TRPV6. EMBO J 2003;224:776–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Chung JJ, Miki K, Kim D, Shim SH, Shi HF, Hwang JY, et al. CatSperζ regulates the structural continuity of sperm Ca2+ signaling domains and is required for normal fertility. eLife 2017;22(February (6)):e23082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Tang L, El-Din TMG, Payandeh J, Martinez GQ, Heard TM, Scheuer T, et al. Structural basis for Ca2+ selectivity of a voltage-gated calcium channel. Nature 2014;505(7481):56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Doolittle Russell F Proteins. Sci Am 1985;253(4):88–99. [DOI] [PubMed] [Google Scholar]
- [10].Cai X, Clapham DE. Evolutionary genomics reveals lineage-specific gene loss and rapid evolution of a sperm-specific ion channel complex: CatSpers and CatSperβ. PLoS One 2008;3(10):e3569 http://dx.doi.org/10.1371/journal.pone.0003569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Pruitt KD, Brown GR, Hiatt SM, Thibaud-Nissen F, Astashyn A, Ermolaeva O, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res 2013;42(November (D1)):D756–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu J, Yan Z, Li Z, Qian X, Lu S, Dong M, et al. Structure of the voltage-gated calcium channel Cav1. 1 at 3.6 Å resolution. Nature 2016;537(September (7619)):191–6. [DOI] [PubMed] [Google Scholar]
- [13].Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyper-activated motility. Proc Natl Acad Sci 2007;104(January (4)):1219–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chung JJ, Shim SH, Zhuang X, Clapham DE. Super-resolution imaging reveals a multi-array arrangement of catsper channel on the sperm tail. Biophys J 2012;102(January (3)):681a. [Google Scholar]
- [15].Montoto LG, Sánchez MV, Tourmente M, Martín-Coello J, Luque-Larena JJ, Gomendio M, et al. Sperm competition differentially affects swimming velocity and size of spermatozoa from closely related muroid rodents: head first. Reproduction 2011;142(6):819–30. [DOI] [PubMed] [Google Scholar]
- [16].Simmons Leigh W, Fitzpatrick John L. Sperm wars and the evolution of male fertility. Reproduction 2012;144(5):519–34. [DOI] [PubMed] [Google Scholar]
- [17].Gomendio M, Malo AF, Garde J, Roldan ER. Sperm traits and male fertility in natural populations. Reproduction 2007;134(July (1)):19–29. [DOI] [PubMed] [Google Scholar]
- [18].Firman RC, Simmons LW. Sperm midpiece length predicts sperm swimming velocity in house mice. Biol Lett 2010;6(August (4)):513–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Stockley P Sexual conflict resulting from adaptations to sperm competition. Trends Ecol Evol 1997;12(4):154–9. [DOI] [PubMed] [Google Scholar]
- [20].Van Der Horst G, Maree L, Kotzé SH, O’Riain MJ. Sperm structure and motility in the eusocial naked mole-rat, Heterocephalus glaber: a case of degenerative ortho-genesis in the absence of sperm competition? BMC Evol Biol 2011;11(1):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hosken DJ. Testes mass in megachiropteran bats varies in accordance with sperm competition theory. Behav Ecol Sociobiol 1998;44(3):169–77. [Google Scholar]
- [22].Rathbun GB, Rathbun CD. Social structure of the bushveld sengi (Elephantulus intufi) in Namibia and the evolution of monogamy in the Macroscelidea. J Zool 2006;269(3):391–9. [Google Scholar]
- [23].Gibbon CDF. The adaptive significance of monogamy in the golden-reumped elephantt-shrew. J Zool 1997;242(1):167–77. [Google Scholar]
- [24].Woodall PF. The male reproductive system and the phylogeny of elephant-shrews (Macroscelidea). Mammal Rev 1995;25(1–2):87–93. [Google Scholar]
- [25].Iossa G, Soulsbury CD, Baker PJ, Harris S. Sperm competition and the evolution of testes size in terrestrial mammalian carnivores. Funct Ecol 2008;22(4):655–62. [Google Scholar]
- [26].Ratto MH, Huanca W, Singh J, Adams GP. Local versus systemic effect of ovulation-inducing factor in the seminal plasma of alpacas. Reprod Biol Endocrinol 2005;3(1):29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Chen BX, Yuen ZX, Pan GW. Semen-induced ovulation in the Bactrian camel (Camelus bactrianus). J Reprod Fertil 1985;74(2):335–9. [DOI] [PubMed] [Google Scholar]
- [28].Parga JA. Copulatory plug displacement evidences sperm competition in Lemur catta. Int J Primatol 2003;24(4):889–99. [Google Scholar]
- [29].Hosken DJ. Sperm competition in bats. Proc R Soc Lond B: Biol Sci 1997;264(1380):385–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Liu J, Xia J, Cho KH, Clapham DE, Ren D. CatSperbeta, a novel transmembrane protein in the CatSper channel complex. J Biol Chem 2007;282:18945–52. [DOI] [PubMed] [Google Scholar]
- [31].Wang H, Liu J, Cho KH, Ren D. A novel, single, transmembrane protein CATSPERG is associated with CATSPER1 channel protein. Biol Reprod 2009;81(September (3)):539–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Chung JJ, Navarro B, Krapivinsky G, Krapivinsky L, Clapham DE. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat Commun 2011;2:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Woycechowsky KJ, Raines RT. The CXC motif: a functional mimic of protein disulfide isomerase. Biochemistry 2003;42(18):5387–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Johansson L, Arscott LD, Ballou DP, Williams CH, Arnér ES. Studies of an active site mutant of the selenoprotein thioredoxin reductase: the Ser-Cys-Cys-Ser motif of the insect orthologue is not sufficient to replace the Cys-Sec dyad in the mammalian enzyme. Free Radic Biol Med 2006;41(August (4)):649–56. [DOI] [PubMed] [Google Scholar]
- [35].Miki K, Clapham DE. Rheotaxis guides mammalian sperm. Curr Biol 2013;23(March (6)):443–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].McCusker EC, Bagnéris C, Naylor CE, Cole AR, D’avanzo N, Nichols CG, et al. Structure of a bacterial voltage-gated sodium channel pore reveals mechanisms of opening and closing. Nat Commun 2012;3:1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].von Heijne G, Gavel Y. Topogenic signals in integral membrane proteins. Eur J Biochem 1988;174(4):671–8. [DOI] [PubMed] [Google Scholar]
- [38].Harraway C, Berger NG, Dubin NH. Semen pH in patients with normal versus abnormal sperm characteristics. Am J Obstet Gynecol 2000;182(5):1045–7. [DOI] [PubMed] [Google Scholar]
- [39].Perez C, Köhler M, Janser D, Pardon E, Steyaert J, Zenobi R, et al. Structural basis of inhibition of lipid-linked oligosaccharide flippase PglK by a conformational nano-body. Sci Rep 2017;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nakao H, Ikeda K, Ishihama Y, Nakano M. Membrane-spanning sequences in endoplasmic reticulum proteins promote phospholipid flip-flop. Biophys J 2016;110(12):2689–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Visconti PE, Krapf D, De La Vega-beltrán JL, Acevedo JJ, Darszon A. Ion channels, phosphorylation and mammalian sperm capacitation. Asian J Androl 2011;13(3):395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yang H, Kim A, David T, Palmer D, Jin T, Tien J, et al. TMEM16F forms a Ca 2+-activated cation channel required for lipid scrambling in platelets during blood coagulation. Cell 2012;151(1):111–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 1997;25(September (17)):3389–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, et al. The NCBI biosystems database. Nucleic Acids Res 2009;38(suppl_1 (October)):D492–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Okonechnikov K, Golosova O, Fursov M. UGENE team. Unipro UGENE: a unified bioinformatics toolkit. Bioinformatics 2012;28(February (8)):1166–7. [DOI] [PubMed] [Google Scholar]
- [46].Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32(March (5)):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Guindon S, Lethiec F, Duroux P, Gascuel O. PHYML online—a web server for fast maximum likelihood-based phylogenetic inference. Nucleic Acids Res 2005;33(suppl_2 (July)):W557–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Huerta-Cepas J, Dopazo J, Gabaldón T. ETE: a python environment for tree exploration. BMC Bioinf 2010;11(1):24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49]. http://www.jalview.org/help/html/colourSchemes/zappo.html.
- [50].Krogh A, Larsson B, Von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 2001;305(January (3)):567–80. [DOI] [PubMed] [Google Scholar]
- [51].McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics 2000;16(April (4)):404–5. [DOI] [PubMed] [Google Scholar]
- [52].Blom N, Sicheritz-Pontén T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004;4(June (6)):1633–49. [DOI] [PubMed] [Google Scholar]
- [53].De Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, et al. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 2006;34(suppl_2):W362–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Lynch BA, Koshland DE. Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc Natl Acad Sci 1991;88(23):10402–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Morten Kallberg, Haipeng Wang, Sheng Wang, Jian Peng, Zhiyong Wang, Hui Lu, Jinbo Xu. Template-based protein structure modeling using the RaptorX web server. Nat Protoc 2012;7(8):1511–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Fariselli P, Riccobelli P, Casadio R. Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins: Struct Funct Bioinf 1999;36(3):340–6. [PubMed] [Google Scholar]
- [57].Fiser A, Simon I. Predicting the oxidation state of cysteines by multiple sequence alignment. Bioinformatics 2000;16(3):251–6. [DOI] [PubMed] [Google Scholar]
- [58].Frand AR, Kaiser CA. The ERO1gene of yeast is required for oxidation of protein dithiols in the endoplasmic reticulum. Mol Cell 1998;1(2):161–70. [DOI] [PubMed] [Google Scholar]
- [59].Pakula AA, Simon MI. Determination of transmembrane protein structure by disulfide cross-linking: the Escherichia coli Tar receptor. Proc Natl Acad Sci 1992;89(9):4144–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Chung JJ, Shim SH, Everley RA, Gygi SP, Zhuang X, Clapham DE. Structurally distinct Ca 2+ signaling domains of sperm flagella orchestrate tyrosine phosphorylation and motility. Cell 2014;157(4):808–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Mohammadi S, Movahedin M, Mowla SJ. Up-regulation of CatSper genes family by selenium. Reprod Biol Endocrinol 2009;7(1):126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kumánovics A, Levin G, Blount P. Family ties of gated pores: evolution of the sensor module. FASEB J 2002;16(12):1623–9. [DOI] [PubMed] [Google Scholar]
- [63].Shin JB, Martinez-Salgado C, Heppenstall PA, Lewin GR. A T-type calcium channel required for normal function of a mammalian mechanoreceptor. Nat Neurosci 2003;6(7). [DOI] [PubMed] [Google Scholar]
- [64].Navarro B, Kirichok Y, Chung JJ, Clapham DE. Ion channels that control fertility in mammalian spermatozoa. Int J Dev Biol 2008;52:607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Li H, Ding X, Guan H, Xiong C. Inhibition of human sperm function and mouse fertilization in vitro by an antibody against cation channel of sperm 1: the contraceptive potential of its transmembrane domains and pore region. Fertil Steril 2009;92(3):1141–6. [DOI] [PubMed] [Google Scholar]
- [66].Li H, Ding X, Guo C, Guan H, Xiong C. Immunization of male mice with B-cell epitopes in transmembrane domains of CatSper1 inhibits fertility. Fertil Steril 2012;97(2):445–52. [DOI] [PubMed] [Google Scholar]
- [67].Miller MR, Mannowetz N, Iavarone AT, Safavi R, Gracheva EO, Smith JF, et al. Unconventional endocannabinoid signaling governs sperm activation via the sex hormone progesterone. Science 2016;352(6285):555–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Kuk AC, Mashalidis EH, Lee SY. Crystal structure of the MOP flippase MurJ in an inward-facing conformation. Nat Struct Mol Biol 2017;24(2):171–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.