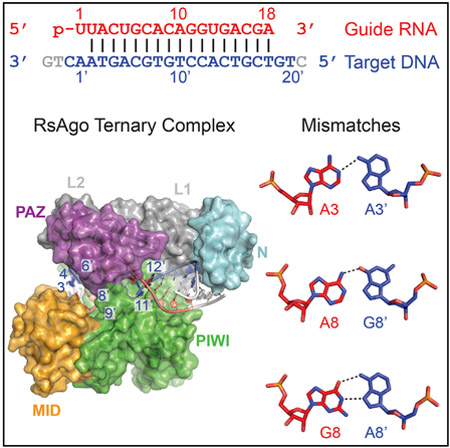
SUMMARY
Prokaryotic Argonaute (Ago) proteins were recently shown to target foreign genetic elements, thus making them a perfect model for studies of interference mechanisms. Here, we study interactions of Rhodobacter sphaeroides Ago (RsAgo) with guide RNA (gRNA) and fully complementary or imperfect target DNA (tDNA) using biochemical and structural approaches. We show that RsAgo can specifically recognize both the first nucleotide in gRNA and complementary nucleotide in tDNA, and both interactions contribute to nucleic acid binding. Non-canonical pairs and bulges on the target strand can be accommodated by RsAgo with minimal perturbation of the duplex but significantly reduce RsAgo affinity to tDNA. Surprisingly, mismatches between gRNA and tDNA induce dissociation of the guide-target duplex from RsAgo. Our results reveal plasticity in the ability of Ago proteins to accommodate helical imperfections, show how this might affect the efficiency of RNA silencing, and suggest a potential mechanism for guide release and Ago recycling.
In Brief
Prokaryotic Argonaute proteins (pAgos) target foreign genetic elements. Liu et al. structurally and functionally characterize interactions of pAgo with guide RNA and fully complementary or imperfectly targeted DNA, reveal plasticity in the ability of Ago proteins to accommodate helical imperfections, and suggest a potential mechanism for guide release.
Graphical Abstract
INTRODUCTION
The members of the Argonaute (Ago) protein family are key players in RNA-mediated gene silencing and are widely distributed in archaea, eubacteria, and eukaryotes (Hutvagner and Simard, 2008; Ipsaro and Joshua-Tor, 2015; Meister, 2013; Swarts et al., 2014b). In eukaryotes, Ago proteins form complexes with short interfering RNAs (siRNAs), microRNAs (miRNAs), and PIWI-interacting RNAs (piRNAs) that recognize complementary RNA targets resulting in mRNA degradation or translational and transcriptional inhibition (Bartel, 2004; Kuhn and Joshua-Tor, 2013; Parker, 2010). Structural biology studies on Ago proteins and their complexes with guide and target nucleic acids have greatly enhanced our understanding of gene silencing phenomena starting with studies of prokaryotic Agos (pAgos) and their complexes (Sheng et al., 2014; Song et al., 2004; Wang et al., 2009; Wang et al., 2008; Yuan et al., 2005) and subsequently extended to eukaryotic Agos and their complexes (Elkayam et al., 2012; Faehnle et al., 2013; Nakanishi et al., 2012, 2013; Schirle and MacRae, 2012; Schirle et al., 2014) and to PIWI proteins (Matsumoto et al., 2016).
Previous studies demonstrated that eukaryotic Ago proteins can recognize RNA targets containing multiple mismatches with Ago-associated RNA guides. This property plays a crucial role in the regulation of gene expression by RNA-guided complexes, which can thus recognize a wide range of RNA targets with various degrees of complementarity, and with various outcomes for the transcript fate. Analysis of target repression by miRNAs and siRNAs revealed that base pairing between the guide and the target is most important in the so-called seed region (positions 2–8 of guide RNA [gRNA]) and in the central guide segment (around positions 10–11 of gRNA, corresponding to the cleavage site in the RNA target), respectively (Bartel, 2004; Dahlgren et al., 2008; Du et al., 2005; Kloosterman et al., 2004). The ability to recognize mismatched RNA targets may be particularly important for piRNA-binding PIWI-clade Agos, which play key roles in silencing of multiple and diverse transposon copies in the germline of most metazoan species.
In vitro studies of eukaryotic Agos demonstrated that mismatches between guide and target RNAs in the seed region mostly interfere with target binding, while mismatches in the central part of the duplex decrease the catalytic rate of the complex (Salomon et al., 2015; Wee et al., 2012). Similarly, mismatches around the active center had the strongest effect on target cleavage by pAgos from Thermus thermophilus (TtAgo) and Marinitoga piezophila (MpAgo) (Doxzen and Doudna, 2017; Wang et al., 2008). However, the effects of mismatches on the architecture of ternary Ago complexes are poorly understood, and the molecular structures of mismatched complexes remain unknown.
While eukaryotic Agos use gRNAs to recognize and cleave target RNAs (Liu et al., 2004), pAgos were shown to use gRNAs (e.g., Rhodobacter sphaeroides RsAgo and MpAgo) or DNAs (e.g., TtAgo) to recognize target DNAs (tDNAs) (Kaya et al., 2016; Makarova et al., 2009; Olovnikov et al., 2013; Swarts et al., 2014a, 2014b; Yuan et al., 2005). Functional studies suggested that pAgos defend genomes against invasive DNA (Hur et al., 2014; Olovnikov et al., 2013; Schirle et al., 2014; Swarts et al., 2014a, 2015), a function similar to their eukaryotic counterparts. While some pAgos including TtAgo are active nucleases, amino acid residues in the PIWI domain that are required for catalytic endonuclease activity are not conserved in RsAgo. Indeed, RsAgo does not cleave the DNA target and cleavage seems to be mediated by another yet-to-be-identified nuclease that cuts it on either side of the region paired with the guide sequence (Olovnikov et al., 2013).
Structural studies of RsAgo in complex with gRNA and tDNA (Miyoshi et al., 2016) and of TtAgo in complex with gDNA and tDNA (Sheng et al., 2014) showed that both proteins accommodate nucleic acid duplexes between their N-PAZ and MID-PIWI lobes. Recent analysis of a PIWI-clade SIWI protein from silkworm revealed unexpected similarities with pAgos, including metal-dependent contacts with the RNA guide 5′ end in the MID pocket and similar organization of the active sites (Matsumoto et al., 2016). Further structural and functional studies of pAgos may shed light on the interference pathways mediated by these proteins and their eukaryotic counterparts and make a path to adopt pAgos as a tool for genome and epigenome engineering. This prompted us to undertake a structural study of RsAgo at the ternary complex level and extend these efforts to address how mismatches and bulges in the guide-target duplex are accommodated by the RsAgo scaffold.
RESULTS
Structure of the Ternary RsAgo Complex
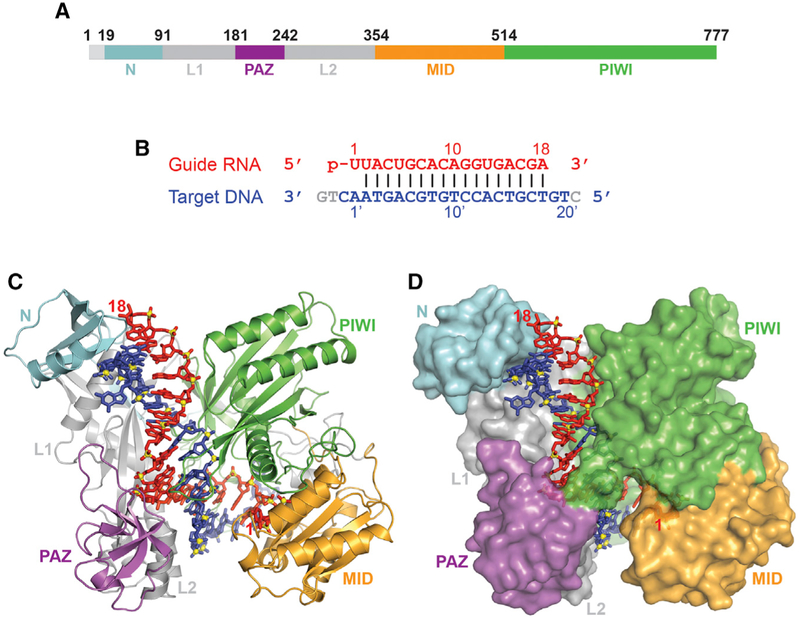
We successfully generated, purified, and crystallized a ternary complex of full-length RsAgo (Figure 1A) bound to 5′-phosphorylated 18-mer gRNA and complementary 24-mer tDNA (Figure 1B) and collected X-ray data at 2.1-Å resolution (X-ray statistics in Table S1). The structure was solved following Se-methionine labeling of RsAgo to solve the phase problem (Table S1). The structure of the ternary complex with bound nucleic acid (gRNA in red and tDNA in blue) in a stick representation and the color-coded protein either in a ribbon or surface representation are shown in Figures 1C and 1D, respectively. We can trace all domains and linkers of the RsAgo protein, the entire length of the 18-mer gRNA, and 21 out of 24 nt of the tDNA. In comparison with the previously published structure of the ternary RsAgo complex (Miyoshi et al., 2016), our structure contains a longer segment of the tDNA, which exactly corresponds to the length of short DNAs associated with RsAgo in vivo (Olovnikov et al., 2013).
Figure 1. Structure of RsAgo Bound to gRNA and tDNA.
(A) The domain architecture of RsAgo color-coded by domains.
(B) The sequence and pairing alignments of 5′-phosphorylated 18-mer gRNA (in red) and 24-mer tDNA (in blue); the nucleotides not observed in the structure are shown in gray.
(C and D) 2.1 Å structure of the ternary complex of RsAgo with gRNA and tDNA. The nucleic acid is in a stick representation, while the protein is in a ribbon (C) and a surface (D) representation.
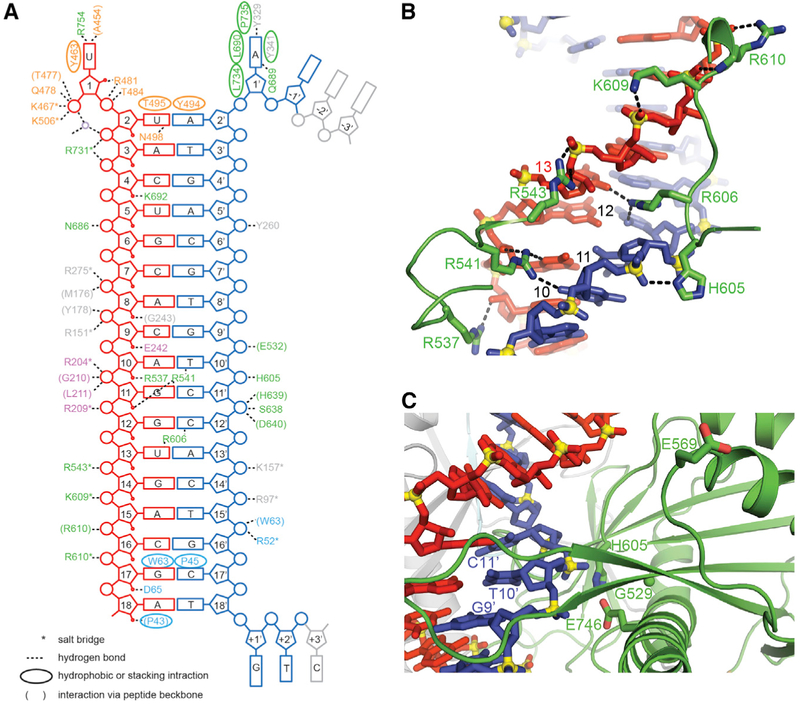
Similar to previous ternary Ago complex structures, the gRNA-tDNA duplex is positioned within a channel between the N-PAZ and MID-PIWI lobes of the RsAgo protein (Figures 1C and 1D). The bound duplex is fully Watson-Crick paired from position 2–2′ to 18–18′, with the binding channel traversing all the domains of the Ago scaffold. The ternary RsAgo complex is stabilized by a network of intermolecular hydrogen-bonding interactions, with the entire set shown schematically in Figure 2A, and a subset of which are shown in Figure 2B. These interactions are most prominent for the 5′ end of the phosphorylated gRNA and opposite it on the tDNA, but are also dispersed over the rest of the duplex. Except for intermolecular contacts to the base edges of U1 and A1′ (see below), the remainder of the intermolecular contacts are non-specific and involve the nucleic acid sugar-phosphate backbone primarily spanning the 5–17 positions on the guide strand and positions 9′–16′ on the target strand (Figure 2A). There are no intermolecular hydrogen bonding contacts for positions 2′–8′ of the seed segment of the tDNA strand (Figure 2A).
Figure 2. Intermolecular Interactions in the Structure of RsAgo Bound to gRNA and tDNA.
(A) Schematic listing of the intermolecular contacts in the complex. Hydrogen-bonding, electrostatic and hydrophobic or stacking interactions of protein residues with the bases (rectangles), backbone sugar (pentagons), and phosphate (circles) groups of the gRNA (in red) and tDNA (in blue) are indicated. Protein residues are color-coded by domains.
(B) Intermolecular contacts between the gRNA (in red) and tDNA (in blue) strands with residues within the Ago protein in the complex.
(C) Positioning of the target strand in the putative catalytic pocket of the PIWI domain in the complex. The putative catalytic residues Gly529, Glu569, His605, and Glu746 are labeled and shown in stick representation. The G9′-T10′-C11′ segment of the target strand is also labeled. Note the positioning of Glu569 in the “unplugged” conformation.
We have compared the structure of the bound gRNA-tDNA duplex in the ternary RsAgo complex with canonical A-form (Figures S1A–S1C) and B-form (Figures S1D–S1F) duplexes. Notably, the gRNA-tDNA duplex overlays poorly with canonical A-form and even more poorly with canonical B-form duplexes along both its entire 2–18 length (Figures S1A and S1D), as well over the 2–11 segment (Figures S1B and S1E) and the 12–18 (Figures S1C and S1F) segment. Thus, it appears that the RsAgo scaffold distorts the bound gRNA-tDNA away from canonical A- and B-helical duplex conformations. We have also compared the structures of individual strands to either A- or B-form (Figures S1G–S1L). Both the gRNA and the tDNA strands of the gRNA-tDNA duplex superimpose poorly with either canonical A-form or B-form (Figures S1G–S1L) segments. We conclude that the gRNA-tDNA duplex is not an A- and B-hybrid form.
Positioning of the 3′ End of the Guide Strand in the Ternary RsAgo Complex
Earlier studies had demonstrated that the 3′ end of RNA guide in binary Ago complexes containing bound guide strand (Elkayam et al., 2012; Schirle and MacRae, 2012; Wang et al., 2008) or ternary Ago complexes containing short target strands (Schirle et al., 2014; Sheng et al., 2014; Wang et al., 2009) are positioned in a binding pocket formed by the PAZ domain. The isolated PAZ domain was also shown to interact with RNA 3′ ends (Lingel et al., 2004; Ma et al., 2004). By contrast, the 3′ end of the guide is not positioned in the PAZ pocket in the RsAgo ternary complex and is indeed far from the PAZ pocket (Figures 1C and 1D), similar to the structure of ternary complexes of TtAgo containing > 15-bp guide-target duplex (Sheng et al., 2014; Wang et al., 2009). This conformational transition–release of gRNA 3′ end from its binding pocket in the PAZ domain–likely accompanies formation of the extended guide-target base pairing during target recognition. In addition, alignment of the PAZ domains of RsAgo, TtAgo, human Ago1, and human Dicer demonstrates that RsAgo lacks two (labeled 2 and 3) out of four segments that contribute to the PAZ pocket (Figure S2A). Thus, the PAZ domain of RsAgo is smaller in size than the RNA-bound domains of TtAgo, human Ago1, and human Dicer (Figure S2B), and may not form a 3′ end RNA-binding pocket even in binary complexes with gRNAs.
Positioning of Putative Catalytic Pocket Residues in the Ternary RsAgo Complex
The catalytic pockets of cleavage competent Agos contain a catalytic tetrad within the PIWI domain that is primarily composed of acidic residues (Nakanishi et al., 2012). RsAgo contains substitutions of two out of four acidic catalytic residues of the DEDX tetrad (Gly529, Glu569, His605, and Glu746 as shown in Figures2C and S3A). Previously, catalytically inactive human Ago1 protein was shown to be activated by restoration of a mutated tetrad residue (Faehnle et al., 2013; Nakanishi et al., 2013). To reveal whether the catalytically competent state may be also reconstituted in the RsAgo complex, we obtained a variant of RsAgo with the complete catalytic tetrad containing corresponding residues from TtAgo (substitutions G529D, A604R, H605D, and E746D; Figure S3A) and analyzed its cleavage activity. We detected no cleavage of different DNA targets (shown in Figure S4), even after prolonged incubations in different buffer solutions (Figure S3B), suggesting that additional changes might be necessary for correct positioning of amino acid residues involved in catalysis.
In TtAgo, a key Glu residue swings into the catalytic pocket during transition from a cleavage-incompetent (“unplugged”) state in complex with guide DNA to a cleavage-competent (“plugged-in”) state in the ternary complex with guide and tDNAs (Sheng et al., 2014). A similar conformational change is likely required for activation of catalysis by the eukaryotic PIWI-clade SIWI protein (Matsumoto et al., 2016). Plugged-in states are also observed in all available eukaryotic Ago2 complexes (Schirle et al., 2014). In contrast, RsAgo adopts an unplugged conformation even in the ternary complex with tDNA, with Glu569 positioned outside the catalytic pocket (Figure 2C), thus probably explaining the lack of its catalytic activity (see also Miyoshi et al., 2016).
Positioning of the Disrupted First RNA-DNA Base Pair in the Ternary RsAgo Complex
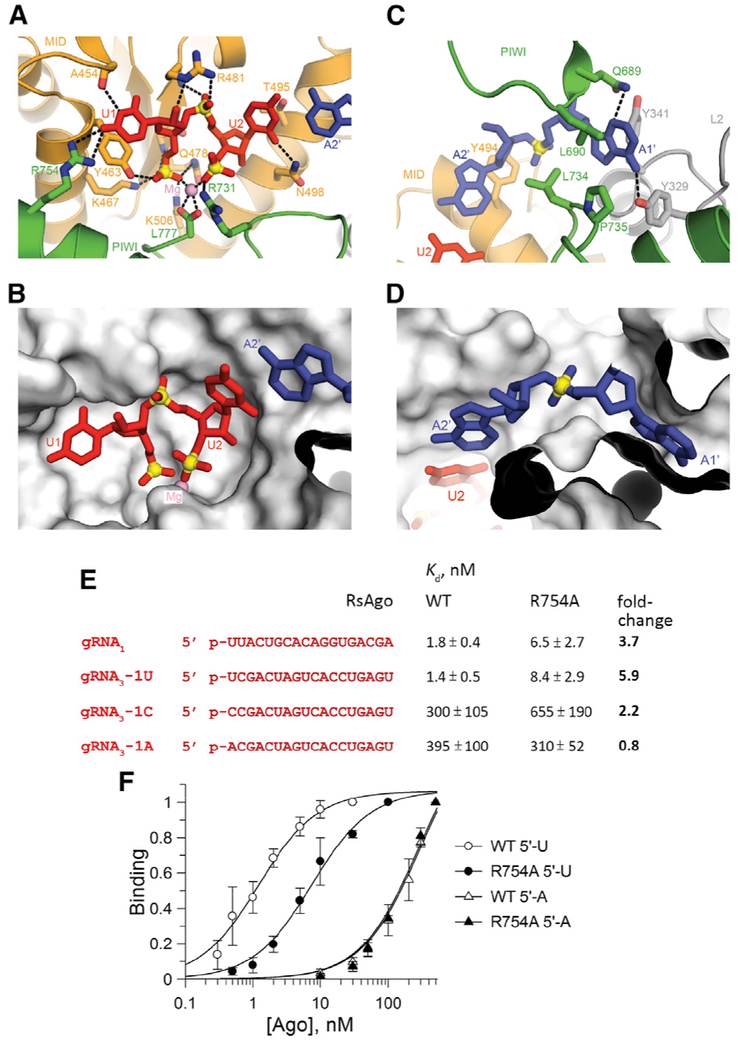
Similar to previously published structures of RsAgo (Miyoshi et al., 2016) and TtAgo (Sheng et al., 2014; Swarts et al., 2014a, 2017; Wang et al., 2009), the 5′-phosphate of the guide strand is positioned within the MID pocket and anchored in place through a network of direct hydrogen bonds from amino acid side chains originating from the MID (Tyr463, Lys467, Gln478, and Lys506) domain (Figures 3A and 3B). Indeed, mutation of Tyr463 and Lys467 residues make RsAgo unable to bind RNA guides in vivo (Olovnikov et al., 2013) and in vitro (Miyoshi et al., 2016). Furthermore, phosphates 1 and 3 of the U1-U2 step of the bound guide are coordinated to a bound Mg2+, which in turn is bonded to the carboxylate of C-terminal Ago residue Leu777 (Figures 3A and 3B). This makes RsAgo similar to the PIWI-clade SIWI protein (Matsumoto et al., 2016) but distinguishes it from Ago-clade Agos (exemplified by hAgo2), in which corresponding contacts are formed by a conserved lysine residue (Elkayam et al., 2012; Schirle and MacRae, 2012).
Figure 3. Recognition Features of MID-PIWI Pockets in the Structure of RsAgo Bound to gRNA and tDNA and Analysis of RsAgo Interactions with gRNA.
(A and B) Positioning of the 5′-pU1pU2 segment of the guide strand in the MID-PIWI domain in the ternary complex. The guide RNA segment is shown in a stick representation, while the protein is shown in a ribbon representation in (A) and in a surface representation in (B). A bound Mg2+ is shown by a purple ball. Hydrogen bonds are shown by dashed lines.
(C and D) Positioning of the A1′-A2′ segment of the target strand in the MID-PIWI domain of the complex. The target DNA segment is shown in a stick representation, while the protein is shown in a ribbon representation in (C) and in a surface representation in (D).
(E) Sequences of gRNAs used in the binding experiments with wild-type (WT) and R754A RsAgo variants. The apparent Kd values for each gRNA are shown on the right; fold-changes relative to the WT RsAgo are indicated in bold. Means and SDs from three to five independent experiments are shown.
(F) Binding of the 5′-U and 5′-A gRNA3 variants by WT and R754A RsAgo. In each experiment, the binding is shown relative to the binding observed at the maximal RsAgo concentration.
In the ternary RsAgo complex, residues U1 on the gRNA strand and A1′ on the tDNA strand are both aligned and anchored within their respective pockets. While residues U2-A2′ form a Watson-Crick pair, the U1-A1′ pair is disrupted by the splaying apart of the 1–2 step on the guide strand and 1′–2′ step on the target strand. Residue U1 in gRNA is specifically recognized through hydrogen bonding of its N3 to the backbone carbonyl of Ala454 and its C4 carbonyl to the side chain of Arg754 (Figure 3A). The 5′-pU1pU2 step is positioned within a surface-accessible pocket (Figure 3B). On the partner tDNA strand, the A1′–A2′ step is also splayed apart (Figures 3C and 3D), with the A1′ base anchored within its own pocket (Figures 2A and 3D) through hydrogen bonding of its 6-amino group with the side chain of Tyr329 and its N3 with the side chain of Gln689 (Figure 3C).
To explore the specificity of the first RNA-DNA base pair recognition by RsAgo, we measured apparent dissociation constants for the gRNA and tDNA binding using a filter binding assay (see Experimental Procedures for details). In agreement with published data (Miyoshi et al., 2016), 5′-U-containing gRNAs were bound by RsAgo with high affinity (Kd,app = 1.5–2 nM) (Figures 3E and 3F). Replacement of 5′-U with C or A dramatically decreased gRNA affinities (Kd,app = 300–400 nM). Alanine substitution of residue Arg754 that forms a contact with the base of 5′-U increased Kd,app for the binding of 5′-U-gRNA molecules ~4- to 6-fold, but had much lower or no effect on the binding of 5′-C and 5′-A gRNAs (Figures 3E and 3F). Thus, this residue contributes to specific recognition of the first gRNA position by RsAgo.
To reveal whether RsAgo has a preference for the adenine at the 1′ position of tDNA, we measured the interactions of gRNA-loaded RsAgo with either fully complementary tDNA, or tDNA variants containing substitutions of this residue (shown in Figure S4). In these experiments, gRNA was first incubated with a 2-fold molar excess of RsAgo, to prevent annealing of free gRNA to tDNA, followed by titration of labeled tDNA with increasing amounts of the gRNA-RsAgo complex. The apparent Kd for binding of fully complementary tDNA determined in this assay was an order of magnitude lower than the Kd for gRNA (0.13 versus 1.8 nM), demonstrating that the binary complex of gRNA with RsAgo has a very high affinity for specific DNA targets. In agreement with previous reports (Miyoshi et al., 2016), no significant DNA binding was observed in the absence of gRNA (Kd > 1,000 nM).
Replacement of A1′ in tDNA with either G, or C, or T measurably decreased apparent tDNA affinity (2- to 2.5-fold increase in the Kd,app values) (Figures 4 and S4). Furthermore, these substitutions significantly decreased the maximal tDNA binding observed at saturating gRNA-RsAgo concentrations (from ~70% for A1′ tDNA to 15%–30% for the other three variants; Figure S5A), an observation further discussed in the next section. Alanine substitution of residue Gln689, which forms a direct contact with the base of A1′, had only minor effects on tDNA binding irrespective of the 1′ nucleotide identity (Figure S5B), suggesting that other residues contribute to the specific tDNA recognition.
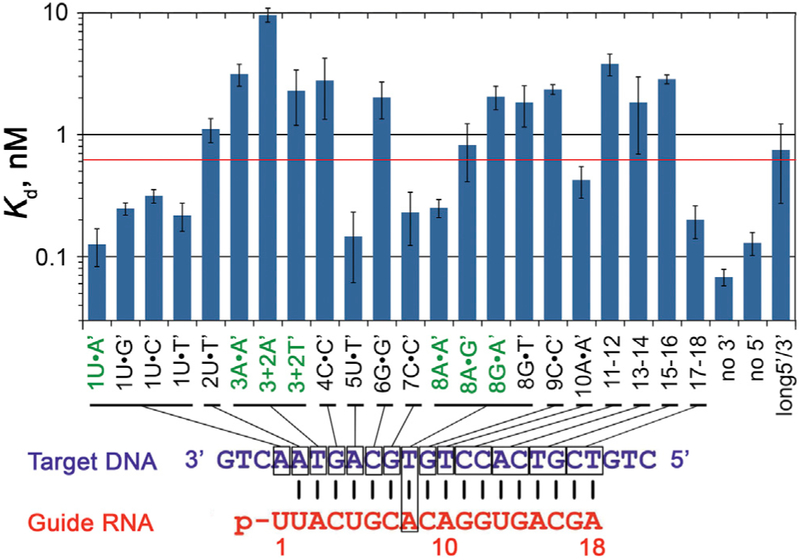
Figure 4. Apparent Affinities of gRNA-Loaded RsAgo to tDNAs Containing Mismatches at Various Positions.
The plot shows apparent Kd values for the binding of complementary and mismatched DNA targets, schematically shown below. The y axis is in the logarithmic scale, and the red line indicates the 5-fold level of the Kd,app value for fully complementary tDNA (1U-A′). The sequences of all oligonucleotides with corresponding Kd values are shown in Figure S4. The data are means and SDs from three to five independent measurements.
Effects of Mispairing between Guide and Target Strands on Target Recognition by RsAgo
To determine the effects of mismatches on target recognition by RsAgo, we used a series of tDNA molecules with substitutions at different positions, starting from the first (1′) to the last (18′) nucleotides of the corresponding gRNA (Figure S4). For positions 3′ and 8′, located in the proximal and distal parts of the seed region, we analyzed several possible substitutions: A3•A3′, A8•A8′, A8•G8′, G8•A8′, G8•T8′, and also 2-nt bulges (A′-A′ or T′-T′) between positions 3′ and 4′ in the tDNA strand. For positions from 11′ to 18′, outside of the seed region, we substituted two nucleotides at once (e.g., 11′–12′, 13′–14′) (Figure S4). For each tDNA, we measured its binding to RsAgo pre-loaded with the same gRNA that was used for structural analysis, except for the G8•A8′ and G8•T8′ pairs in which gRNA contained a substitution of G8 for A8.
Substitutions at most positions of the seed region significantly decreased apparent target affinity (Figures 4 and S4). The strongest effects were observed for positions 2′, 3′, 4′, 6′, and 8′; mismatches at these positions (e.g., A3•A3′, G8•A8′) and 2-nt bulges (3′+2A, 3′+2T) increased the apparent Kd values for tDNA binding 10-fold or more. The effects of mismatches at position 8′ depended on the nucleotide substitution; the A8•G8′, G8•A8′, and G8•T8′ pairs were bound with greatly decreased affinity while the A8•A8′ mismatch only moderately affected tDNA binding (Figure 4). Substitutions in tDNA outside of the seed region, including C9•C9′ and double mismatches at positions 11′–12′, 13′–14′, and 15′–16′, also strongly increased Kd,app values, with only the last two nucleotides (17′–18′) being unimportant for binding. Thus, pairing both within and downstream of the seed region in gRNA contributes to specific target recognition by RsAgo.
Removal of extra nucleotides from either 5′ or 3′ DNA ends (“no 3′” and “no 5′” targets) did not decrease apparent tDNA affinity to gRNA-loaded RsAgo. In contrast, increasing the length of the 5′ and 3′ ends by 5-nt (“long 5′/3′”) decreased tDNA affinity (Figures 4 and S4). Thus, additional DNA segments are unlikely to contribute to target binding by RsAgo.
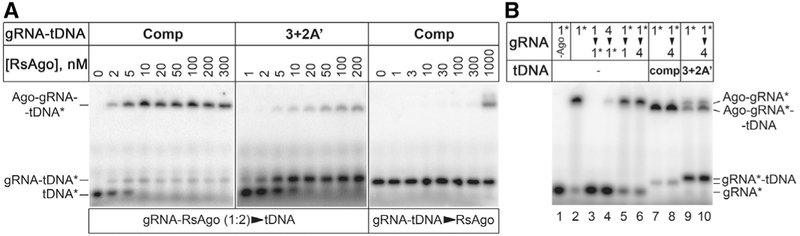
Certain changes in tDNA had a pronounced effect not only on the apparent Kd values but also on the maximal fraction of bound tDNA at saturating gRNA-RsAgo concentrations. While 70% of the fully complementary tDNA was bound in these conditions, the binding was decreased more than 2-fold for several tDNAs, including substitutions of residues 1′, 3′, 4′, and 9′, and 2-nt bulges between positions 3′ and 4′ (Figure S5A). This prompted us to analyze formation of the ternary complexes by native gel electrophoresis. Titration of fully complementary P32-labeled tDNA strand (tDNA*) with preformed gRNA-RsAgo complex led to the appearance of the low mobility ternary complex, with small amounts of free gRNA-tDNA* duplex (Figure 5A, left panel). In contrast, titration of preformed gRNA-tDNA* duplex with free RsAgo revealed essentially no ternary complex formation up to 300–1,000 nM RsAgo concentrations (Figure 5A, right panel). Similarly, apparent Kd value for duplex binding measured by the filter binding assay was >1,000 nM. Thus, RsAgo cannot interact with preformed RNA-DNA hybrids. Interestingly, previously published gel shift experiments revealed ternary complex formation only at high RsAgo concentrations (100 nM) (Miyoshi et al., 2016), suggesting that they were performed under sub-optimal conditions.
Figure 5. Analysis of Ternary Complex Formation by Native Gel Electrophoresis.
(A) Titration of labeled tDNA with increasing amounts of the preformed gRNA-RsAgo complex (1:2 ratio, RsAgo concentrations are shown at the top) for either fully complementary (“Comp,” left) or 3+2A′ (middle) targets. Titration of preformed gRNA-tDNA duplex with RsAgo (right).
(B) Formation of binary and ternary complexes containing labeled gRNA1*. The components were mixed as described in the text; gRNA1 and gRNA4 are indicated as “1” and “4.” The arrowheads indicate the order of addition of the labeled and unlabeled competitor gRNAs; the competitor gRNAs were added either before or 5 min after mixing of RsAgo and gRNA1*, followed by the addition of tDNA (when indicated) and incubation for 20 min at 30°C. The concentrations of gRNA1*, competitor gRNAs, and RsAgo were 20 nM, 1 μM, and 40 nM; tDNA was added to 40 nM. Positions of free nucleic acids and binary and ternary complexes are shown on the sides of the gels. The labeled components in each experiment are indicated with asterisks.
In the case of tDNA* containing a 2-nt bulge (3+2A′), only a minor fraction of it was observed in the ternary complex, with a majority found in the free gRNA-tDNA* duplex (Figure 5A, middle panel), in agreement with the filter binding assay (Figures 4 and S4). The free gRNA-tDNA duplex could be formed either in solution, as a result of incomplete gRNA binding to RsAgo, or as a result of duplex dissociation from the ternary complex after tDNA binding. To test which of these possibilities was more likely, we performed gel shift experiments with labeled gRNA strand (gRNA1*). Two competitor unlabeled gRNAs were used to prevent re-binding of dissociated nucleic acids to RsAgo: gRNA1 or unrelated gRNA4 that could not anneal to the tDNA strand and would not interfere with the ternary complex formation (Figure S4).
Free gRNA1* was efficiently bound by RsAgo, when mixed at the 1:2 ratio (Figure 5B, lane 2). Addition of excess phosphorylated unlabeled gRNA1 or gRNA4 prevented binary complex formation when the competitors were added before gRNA1* Figure 5B, lanes 3 and 4). Competitor gRNAs had only minor effect on the preformed binary complex (lanes 5 and 6), suggesting that gRNA1* does not dissociate during the course of experiment (t1/2 > 20 min). When fully complementary tDNA was added to the binary complex, the ternary complex was formed with high efficiency, and no release of gRNA1* was observed (lane 7). The presence of excess competitor gRNA4 (added after binary complex formation) did not affect ternary complex formation, suggesting that the complex is stable during measurements (lane 8). In contrast, when bulged tDNA (3+2A′) was added to the binary complex, a majority of gRNA1* was observed in free duplex with tDNA, while only a minor part of it remained in the binary and ternary complexes (lane 9). The same pattern was observed in the presence of competitor gRNA4, suggesting that once released, the duplex cannot rebind RsAgo even in the absence of competitor (compare lanes 9 and 10).
These observations favor the scenario in which mismatched tDNA is first bound by the binary gRNA-RsAgo complex to form the ternary complex, followed by dissociation of the gRNA-tDNA duplex. To further explore this mechanism, we analyzed the kinetics of dissociation of ternary complexes after the addition of tDNA (Figure S6). While fully complementary tDNA formed stable ternary complex, gRNA1*-tDNA duplexes were gradually released from the complexes with mismatches or bulges in the seed region (1U•T′, 3A•A′, 3+2A′). At the same time, tDNAs with mismatches at positions 8′ and 15′–16′ formed almost as stable complexes as the fully complementary target (Figure S6).
Alignment of Non-canonical Pairs and Bulges within the Seed Segment of Ternary RsAgo Complexes
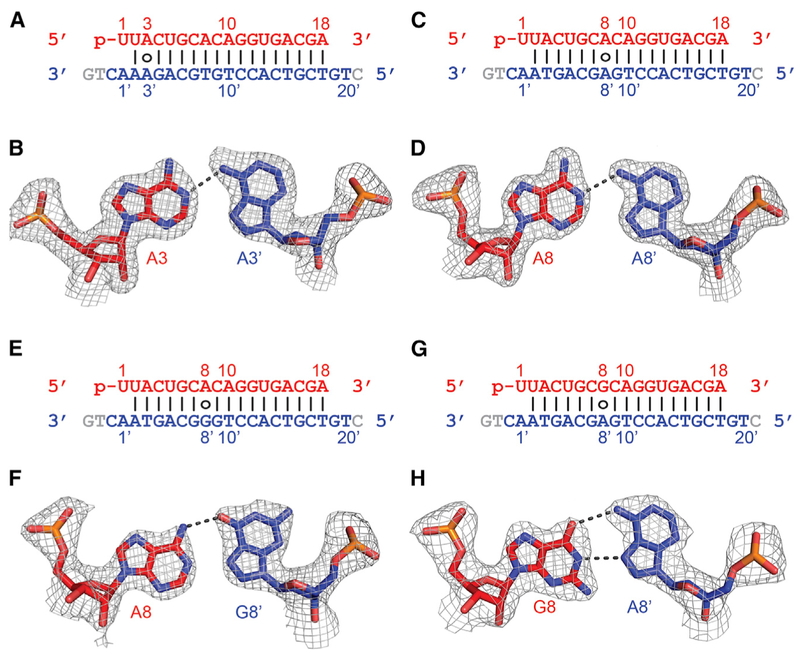
To get structural insight into the effects of mismatches between gRNA and tDNA on their interactions with RsAgo, we solved the structures of its ternary complexes with non-canonical pairs at positions 3′ (A3•A3′; Figures 6A and 6B) and 8′ (A8•A8′, A8•G8′, and G8•A8′; Figures 6C and 6D, 6E and 6F, and 6G and 6H, respectively) (X-ray statistics in Tables S2 and S3). Notably, the maximal resolution of some of these structures (1.81–1.85 Å for A•A non-canonical pairs) even exceeded the resolution of the complex containing fully complementary gRNA-tDNA.
Figure 6. Pairing Alignments of A•A′, A•G′, and G•A′ Non-canonical Pairs within the Seed Segment in the Complex of RsAgo with gRNA and tDNA.
(A) Sequence alignment of the gRNA-tDNA showing the position of the A3•A3′ non-canonical pair.
(B) Pairing alignment of the cis Watson-Crick A3(anti)•Hoogsteen A3′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(C) Sequence alignment of the gRNA-tDNA showing the position of the A8•A8′ non-canonical pair.
(D) Pairing alignment of the cis Watson-Crick A8(anti)•Hoogsteen A8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(E) Sequence alignment of the gRNA-tDNA showing the position of the A8•G8′ non-canonical pair.
(F) Pairing alignment of the cis Watson-Crick A8(anti)•Hoogsteen G8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
(G) Sequence alignment of the gRNA-tDNA showing the position of the G8•A8′ non-canonical pair.
(H) Pairing alignment of the cis Watson-Crick G8(anti)•Hoogsteen A8′(syn) pair in the ternary complex. The 2Fo-Fc omit electron density map is contoured at 1.0σ.
In the case of both A•A mismatches at positions 3′ and 8′, we observe formation of a cis Watson-Crick A3/A8-Hoogsteen A3′/A8′ non-canonical A•A′ pairs stabilized by a single hydrogen bond (Figures 6B and 6D). The adenine on the gRNA strand (A3 and A8) adopts an anti conformation, while adenine on the target strand (A3′ and A8′) adopts a syn conformation (Figures 6B and 6D). A comparison between the A•A′-containing (in color) and A•T′-containing (in silver) ternary RsAgo complexes shows minimal localized distortions for both the A3•A3′ (Figures S7A and S7B) and A8•A8′ (Figures S7C and S7D) non-canonical pairs, without disruption of the flanking Watson-Crick pairs.
In the case of A•G′ (Figure 6F) and G•A′ (Figure 6H) non-canonical pairs at position 8•8′, the purine on the gRNA strand adopts an anti conformation, while the purine on the tDNA strand adopts a syn conformation. Thus, for the A8•G8′ non-canonical pair, the alignment is cis Watson-Crick A8-Hoogsteen G8′ stabilized by one hydrogen bond (Figure 6F), while for the G8•A8′ non-canonical pair the alignment is cis Watson-Crick G8–Hoogsteen A8′ stabilized by two hydrogen bonds (Figure 6H). Once again the distortions are localized to the mismatch site for both the A8•G8′ (Figures S7E and S7F) and G8•A8′ (Figures S7G and S7H) non-canonical pairs, with minimal disruption of the flanking Watson-Crick pairs.
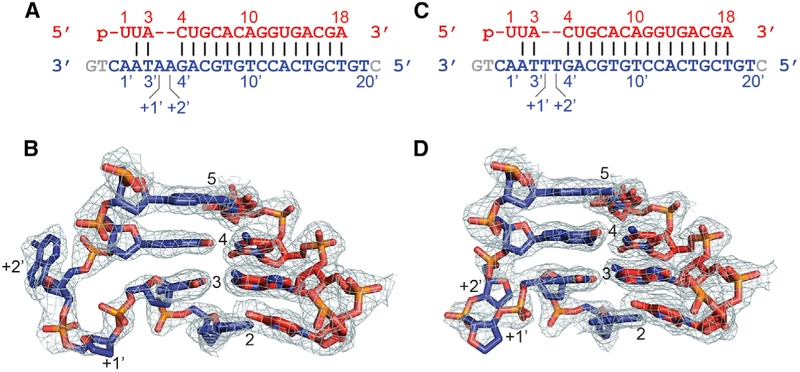
Finally, we also solved the structures of ternary RsAgo complexes with dual bulges, either A′-A′ (Figure 7A) or T′-T′ (Figure 7C) between positions 3′ and 4′ within the seed segment of the tDNA strand (X-ray statistics in Table S4). In each case, either the A′-A′ bulge (Figure 7B) or the T′-T′ bulge (Figure 7D) loops out of the duplex, while retaining flanking Watson-Crick pairs. The backbone of the looped-out segment can be traced in both bulges (Figures 7B and 7D). By contrast, for the A′-A′ bulge, the first looped-out A is disordered (Figure 7B), while for the T′-T′ bulge, both T bases are disordered (Figure 7D).
Figure 7. Looping Out of A-A and T-T Bulges on the DNA Target Strand within the Seed Segment in the Complex of RsAgo with gRNA and tDNA.
(A) Sequence of the gRNA-tDNA showing the position of the A′(+1)-A′(+2) bulge between residues 3′ and 4′ on the tDNA strand.
(B) Structure of the gRNA-tDNA duplex in the vicinity of the looped out A′(+1)-A′(+2) bulge in the ternary complex. The 2Fo-Fc electron density is contoured at 1.0σ.
(C) Sequence of the gRNA-tDNA showing the position of the T′(+1)-T′(+2) bulge between residues 3′ and 4′ on the tDNA strand.
(D) Structure of the gRNA-tDNA duplex in the vicinity of the looped out T′(+1)-T′(+2) bulge in the ternary complex. The 2Fo-Fc electron density is contoured at 1.0σ.
DISCUSSION
Our structural and biochemical studies report on formation of ternary RsAgo complex with the gRNA and tDNA strands. We find that helical imperfections between gRNA and tDNA can be tolerated in the ternary complex. However, they significantly reduce the apparent affinity of the RsAgo-gRNA complex to the tDNA, thus providing a molecular mechanism for highly specific target recognition. Furthermore, we found that the interaction with an imperfect target causes dissociation of gRNA from the RsAgo protein. These features make RsAgo and other pAgos attractive as a potential tool for highly specific genome manipulation. Below, we briefly discuss the interactions that contribute to specific recognition of the gRNA and tDNA by RsAgo and implications of these features for functional Ago activities.
Anchoring and Recognition of the Disrupted First Guide-Target Base Pair in the Ternary RsAgo Complex
The U1 base of gRNA in the ternary complex is stacked over a Tyr ring, with the 5′-phosphate positioned in the MID-PIWI pocket and coordinated to a bound Mg2+ cation, and the C-terminal carboxylate of RsAgo inserted into this pocket (Figure 3A). Similar features were observed previously in another RsAgo structure (Miyoshi et al., 2016), as well as in TtAgo (Sheng et al., 2014; Wang et al., 2008), archaeal Ago (Ma et al., 2005; Parker et al., 2005), and eukaryotic PIWI-clade Ago (Matsumoto et al., 2016) complexes.
Previous studies established that gRNA bound to RsAgo exhibited a strong bias for uracil at position 1 (Miyoshi et al., 2016; Olovnikov et al., 2013). This specificity is partially explained by formation of a specific intermolecular hydrogen bonding between the carbonyl group at position 4 of U1 and the Arg754 side chain in the RsAgo ternary complex (Figures 2A, 3A, 3E, and 3F). Similarly, sequence-specific recognition of the 5′-base in the guide strand were shown for several eukaryotic Agos (Boland et al., 2010; Frank et al., 2010; Matsumoto et al., 2016; Schirle and MacRae, 2012; Wang et al., 2009; Mi et al., 2008). On the contrary, TtAgo was proposed to have little specificity for the 5′-base binding (Swarts et al., 2014a, 2017).
tDNAs bound to RsAgo in vivo exhibited a strong bias for adenine at the position opposite 5′-U on the gRNA strand (Olovnikov et al., 2013). While this could be a mere consequence of their biogenesis from complementary genomic targets, replacement of A1′ with other nucleotides decreased tDNA binding, even though gRNA and tDNA are not paired at this position (Figures 4 and S5A). The interactions of RsAgo with A1′ on tDNA seem to be important for stabilization of the ternary complex, because replacements of this residue resulted in dissociation of the gRNA-tDNA duplex (Figures S5A and S6). We observe that A1′ in the RsAgo ternary complex is anchored within its own pocket (Figure 3D), with its 6-amino group forming specific intermolecular hydrogen bonds with the hydroxyl group of a Tyr residue (Figure 3C), possibly explaining the observed specificity.
Specific recognition of the A1′ residue in the target RNA strand was previously demonstrated for human Ago2 (Schirle et al., 2014), and a similar pocket was potentially identified in the SIWI protein (Matsumoto et al., 2016). By contrast, TtAgo preferentially binds G1′ in the target strand, which is also stabilized by specific hydrogen bonding (Sheng et al., 2014; Swarts et al., 2017). Thus, the ability to recognize the target nucleotide at position 1′ may be an ancient feature of Ago proteins present in the common ancestor of prokaryotic and eukaryotic lineages.
Accommodation of Non-canonical Pairs and Bulges within Seed Segment of Ternary RsAgo Complexes
In this work, we reveal how a subset of non-canonical pairs can be accommodated within the RsAgo-bound RNA-DNA heteroduplex. We observe the same cis Watson-Crick–Hoogsteen A•A′ non-canonical pairs at positions 3•3′ (Figure 6B) and 8•8′ (Figure 6D) in the RsAgo ternary complex. In each case, the A on the gRNA strand adopts an anti alignment, while the A′ on the tDNA strand adopts a syn alignment. The observed alignment is stabilized by only one N-H••N hydrogen bond and a potential non-linear C-H••N hydrogen bond. To our knowledge this type of cis Watson-Crick-Hoogsteen A•A non-canonical pair has not been reported previously, with most examples of A•A non-canonical pairs stabilized by two N-H••N hydrogen bonds (Leontis et al., 2002). It is conceivable that the Ago scaffold restricts alternate A•A pairing alignments in the RsAgo ternary complex.
We observe cis Watson-Crick–Hoogsteen A8(anti)•G8′(syn) (Figure 6F) and cis Watson-Crick–Hoogsteen G8(anti)•A8′(syn) (Figure 6H) pairing for A•G′ and G•A′ non-canonical pairs at position 8•8′ in the ternary RsAgo complex. The first pair is stabilized by one N-H••N hydrogen bond and would require protonation of A8 to form a second N-H••N hydrogen bond (Gao and Patel, 1988; Leontis et al., 2002). The second pair is stabilized by two N-H••N hydrogen bonds, a feature observed for G•A non-canonical pairs reported previously (Hunter et al., 1986; Leontis et al., 2002). Similarly to the A•A non-canonical pairs, the base on the gRNA strand adopts an anti alignment, while the base on the tDNA strand adopts a syn alignment (Figures 6F and 6H) for both A8•G8′ and G8•A8′ pairs. This may reflect the greater flexibility to adopt a syn alignment within the target strand of the seed segment (Figures 6B, 6D, 6F, and 6H), given that the sugar-phosphate backbone of this segment is not involved in intermolecular recognition (Figure 2A).
Systematic studies of nucleic acid bulges have shown that they can either stack into or alternately loop out of the duplex dependent on flanking sequence, the nature of tertiary interactions, and the role of cations (Hermann and Patel, 2000). In the present study, both bases of A′-A′ and T′-T′ bulges positioned between 3′ and 4′ nucleotides on the tDNA strand were found to loop out of the duplex in the ternary RsAgo complexes (Figures 7B and 7D). Although we can monitor the sugar-phosphate backbone at the bulge site, electron density for the looped-out bases was not observed except for one adenine residue (Figures 7B and 7D). There is minimal distortion in the duplex segment flanking the dual looped-out bases at the bulge site in the ternary RsAgo complexes. This contrasts recently reported effects of single-nucleotide bulges at positions 6′ and 7′ of the target strand in ternary complexes of TtAgo, which were shown to significantly affect the duplex conformation and its interactions with the protein (Sheng et al., 2017). This result highlights the ability of nucleic acid duplexes to accommodate helical imperfections without significant distortion of flanking base pair segments, depending on the position of the bulge relative to the nucleic acid/protein interface.
Implications for Functional Activities of Ago Proteins
Despite minimal perturbations of the overall ternary complex structure, distortions of the gRNA-tDNA interactions at different heteroduplex positions have significant effects on tDNA binding. In particular, mismatches or bulges at positions 2′, 3′, 4′, 6′, or 8′ strongly impair target binding (Figure 4), suggesting that correct guide-target alignment within the seed region play a critical role in the ternary complex formation. This is likely explained by the absence of protein-mediated interactions with the tDNA in the seed region (Figure 2A).
Previous studies of eukaryotic Agos demonstrated that mismatches in the seed region between siRNAs and miRNAs and their target mRNAs also have deleterious effects on the target repression in vivo and its cleavage in vitro (Bartel, 2009; Dahlgren et al., 2008; Du et al., 2005; Kloosterman et al., 2004; Salomon et al., 2015; Wee et al., 2012). In addition, a 3′-supplementary site in miRNAs was shown to contribute to the target mRNA recognition by eukaryotic Ago complexes (Bartel, 2009; Wee et al., 2012). Similarly, mismatches at several positions downstream of the seed region significantly reduced tDNA binding by the gRNA-RsAgo complex, suggesting that the 3′-site contributes to the tDNA recognition by RsAgo.
We found that mismatches between the gRNA and the tDNA can significantly destabilize the ternary complex, thus leading to its disassembly. In particular, mismatches and bulges at positions 3′ and 4′ in the seed region severely decreased the efficiency of ternary complex formation even at high RsAgo concentrations (Figures 5A and S5A). We demonstrated that this is likely explained by dissociation of the mismatched gRNA-tDNA heteroduplex from RsAgo (Figures 5B and S6). These experiments suggested an unexpected mechanism of gRNA release from RsAgo, triggered by binding of imperfect DNA targets. Intriguingly, recent analysis of human Ago2 interactions with non-canonical mRNA targets demonstrated that mismatches in the seed region of miRNAs resulted in their trimming and unloading from Ago2 in cell lysates (Park et al., 2017). Therefore, the mechanism for unloading of RNA guides from Ago proteins may be conserved in evolution and may facilitate guide exchange and Ago recycling, thus allowing fast re-programming of the interference systems toward various genetic targets.
Highly specific recognition of tDNA by RsAgo makes it and other pAgos attractive candidates to use as a potential tool for genome manipulation that will be orthogonal to widely used CRISPR/Cas9-genome editing systems. While RsAgo is inactive as an endonuclease and our attempts to restore its catalytic activity by point mutagenesis were unsuccessful (Figure S3), pAgos from other species have guide-dependent endonuclease activity both in vitro and in vivo (Kaya et al., 2016; Swarts et al., 2014a, 2015; Wang et al., 2008). In contrast to CRISPR/Cas9, pAgo-gRNA complex can potentially target virtually any sequence of interest as efficient recognition of the target does not require the presence of Protospacer adjacent motif (PAM), a short sequence following the DNA sequence targeted by Cas9. However, the use of pAgos as a genetic tool will depend on the development of approaches that would allow efficient formation of their binary and ternary complexes with guide nucleic acids inside eukaryotic cells, which will be an important goal for future studies.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
6×His-tagged full-length R. sphaeroides Ago (RsAgo) gene was inserted into pET-30a expression vector and transformed into E. coli strain BL21 (DE3). For detailed purification protocol, see Supplemental Experimental Procedures.
Crystallization, Data Collection, and Structure Determination
Diffraction data were collected at 100 K at beamline NE-CAT 24-IDC at the Advanced Photon Source (APS), Argonne National Laboratory. All data were processed with the HKL2000 suite (Otwinowski and Minor, 1997). Data-processing statistics are summarized in Tables S1-S4. See Supplemental Experimental Procedures for details.
Analysis of RsAgo Interactions with gRNA and tDNA
All RNA and DNA oligonucleotides used for analysis of RsAgo-nucleic acid interactions are shown in Figure S4. Determination of apparent Kd values was performed using nitrocellulose filtration method; binary and ternary complexes were analyzed by native gel electrophoresis; for detailed protocols, see Supplemental Experimental Procedures.
DATA AND SOFTWARE AVAILABILITY
The accession numbers for the ternary RsAgo complexes reported in this study are PDB: 6D8P (ternary RsAgo complex with fully paired gRNA-target DNA duplex), 6D92 (ternary RsAgo complex containing A3•A3′ pair), 6D95 (ternary RsAgo complex containing A8•A8′ pair), 6D9K (ternary RsAgo complex containing A8•G8′ pair), 6D9L (ternary RsAgo complex containing G8•A8′ pair), 6D8A (ternary RsAgo complex containing A-A bulge), and 6D8F (ternary RsAgo complex containing T-T bulge).
Supplementary Material
Highlights.
R. sphaeroides Ago ternary complex structures with mismatches/bulges
Dependence of target recognition by RsAgo on guide-target complementarity
Specific recognition of the first guide and target bases by prokaryotic Ago
Mismatched targets cause dissociation of guide RNA from ternary Ago complexes
ACKNOWLEDGMENTS
We thank the synchrotron beamline staff at the Argonne National Laboratory for their assistance. D.J.P. was supported by NIH Grant TR01 GM104962; A.A.A. was supported by NIH Grants R01 GM097369, R00 HD057233, and DP2 OD007371A and a Searle Scholar Award; and D.E. and A.K. were supported by Russian Science Foundation Grant 16-14-10377. D.J.P. was supported by NIH Grant GM104962 and the Memorial Sloan Kettering Cancer Center Core Grant (P30 CA008748).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at https://doi.org/10.10167j.celrep.2018.06.021.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Bartel DP (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281–297. [DOI] [PubMed] [Google Scholar]
- Bartel DP (2009). MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland A, Tritschler F, Heimstädt S, Izaurralde E, and Weichenrieder O (2010). Crystal structure and ligand binding of the MID domain of a eukaryotic Argonaute protein. EMBO Rep. 11, 522–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren C, Zhang HY, Du Q, Grahn M, Norstedt G, Wahlestedt C, and Liang Z (2008). Analysis of siRNA specificity on targets with double-nucleotide mismatches. Nucleic Acids Res. 36, e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxzen KW, and Doudna JA (2017). DNA recognition by an RNA-guided bacterial Argonaute. PLoS One 12, e0177097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Q, Thonberg H, Wang J, Wahlestedt C, and Liang Z (2005). A systematic analysis of the silencing effects of an active siRNA at all single-nucleotide mismatched target sites. Nucleic Acids Res. 33, 1671–1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkayam E, Kuhn CD, Tocilj A, Haase AD, Greene EM, Hannon GJ, and Joshua-Tor L (2012). The structure of human argonaute-2 in complex with miR-20a. Cell 150, 100–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle CR, Elkayam E, Haase AD, Hannon GJ, and Joshua-Tor L (2013). The making of a slicer: activation of human Argonaute-1. Cell Rep. 3, 1901–1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank F, Sonenberg N, and Nagar B (2010). Structural basis for 5′-nucleotide base-specific recognition of guide RNA by human AGO2. Nature 465, 818–822. [DOI] [PubMed] [Google Scholar]
- Gao X, and Patel DJ (1988). G(syn).cntdot.A(anti) mismatch formation in DNA dodecamers at acidic pH: pH-dependent conformational transition of G.cntdot.A mispairs detected by proton NMR. J. Am. Chem. Soc. 110, 5178–5182. [Google Scholar]
- Hermann T, and Patel DJ (2000). RNA bulges as architectural and recognition motifs. Structure 8, R47–R54. [DOI] [PubMed] [Google Scholar]
- Hunter WN, Brown T, and Kennard O (1986). Structural features and hydration of d(C-G-C-G-A-A-T-T-A-G-C-G); a double helix containing two G.A mispairs. J. Biomol. Struct. Dyn. 4, 173–191. [DOI] [PubMed] [Google Scholar]
- Hur JK, Olovnikov I, and Aravin AA (2014). Prokaryotic Argonautes defend genomes against invasive DNA. Trends Biochem. Sci 39, 257–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutvagner G, and Simard MJ (2008). Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 9, 22–32. [DOI] [PubMed] [Google Scholar]
- Ipsaro JJ, and Joshua-Tor L (2015). From guide to target: molecular insights into eukaryotic RNA-interference machinery. Nat. Struct. Mol. Biol 22, 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaya E, Doxzen KW, Knoll KR, Wilson RC, Strutt SC, Kranzusch PJ, and Doudna JA (2016). A bacterial Argonaute with noncanonical guide RNA specificity. Proc. Natl. Acad. Sci. USA 113, 4057–4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Wienholds E, Ketting RF, and Plasterk RH (2004). Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 32, 6284–6291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn CD, and Joshua-Tor L (2013). Eukaryotic Argonautes come into focus. Trends Biochem. Sci 38, 263–271. [DOI] [PubMed] [Google Scholar]
- Leontis NB, Stombaugh J, and Westhof E (2002). The non-Watson-Crick base pairs and their associated isostericity matrices. Nucleic Acids Res. 30, 3497–3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingel A, Simon B, Izaurralde E, and Sattler M (2004). Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nat. Struct. Mol. Biol 11, 576–577. [DOI] [PubMed] [Google Scholar]
- Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, and Hannon GJ (2004). Argonaute2 is the catalytic engine of mammalian RNAi. Science 305, 1437–1441. [DOI] [PubMed] [Google Scholar]
- Ma JB, Ye K, and Patel DJ (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature 429, 318–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, and Patel DJ (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434, 666–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS, Wolf YI, van der Oost J, and Koonin EV (2009). Prokaryotic homologs of Argonaute proteins are predicted to function as key components of a novel system of defense against mobile genetic elements. Biol. Direct 4, 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto N, Nishimasu H, Sakakibara K, Nishida KM, Hirano T, Ishitani R, Siomi H, Siomi MC, and Nureki O (2016). Crystal structure of silkworm PIWI-clade Argonaute Siwi bound to piRNA. Cell 167, 484–497.e9. [DOI] [PubMed] [Google Scholar]
- Meister G (2013). Argonaute proteins: functional insights and emerging roles. Nat. Rev. Genet 14, 447–459. [DOI] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell 133, 116–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi T, Ito K, Murakami R, and Uchiumi T (2016). Structural basis for the recognition of guide RNA and target DNA heteroduplex by Argonaute. Nat. Commun. 7, 11846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Weinberg DE, Bartel DP, and Patel DJ (2012). Structure of yeast Argonaute with guide RNA. Nature 486, 368–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, and Patel DJ (2013). Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 3, 1893–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olovnikov I, Chan K, Sachidanandam R, Newman DK, and Aravin AA (2013). Bacterial Argonaute samples the transcriptome to identify foreign DNA. Mol. Cell 51, 594–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Park JH, Shin SY, and Shin C (2017). Non-canonical targets destabilize microRNAs in human Argonautes. Nucleic Acids Res. 45, 1569–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS (2010). How to slice: snapshots of Argonaute in action. Silence 1, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Roe SM, and Barford D (2005). Structural insights into mRNA recognition from a PIWI domain-siRNA guide complex. Nature 434, 663–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomon WE, Jolly SM, Moore MJ, Zamore PD, and Serebrov V (2015). Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell 162, 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, and MacRae IJ (2012). The crystal structure of human Argonaute2. Science 336, 1037–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirle NT, Sheu-Gruttadauria J, and MacRae IJ (2014). Structural basis for microRNA targeting. Science 346, 608–613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Zhao H, Wang J, Rao Y, Tian W, Swarts DC, van der Oost J, Patel DJ, and Wang Y (2014). Structure-based cleavage mechanism of Thermus thermophilus Argonaute DNA guide strand-mediated DNA target cleavage. Proc. Natl. Acad. Sci. USA 111, 652–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng G, Gogakos T, Wang J, Zhao H, Serganov A, Juranek S, Tuschl T, Patel DJ, and Wang Y (2017). Structure/cleavage-based insights into helical perturbations at bulge sites within T. thermophilus Argonaute silencing complexes. Nucleic Acids Res. 45, 9149–9163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JJ, Smith SK, Hannon GJ, and Joshua-Tor L (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science 305, 1434–1437. [DOI] [PubMed] [Google Scholar]
- Swarts DC, Jore MM, Westra ER, Zhu Y, Janssen JH, Snijders AP, Wang Y, Patel DJ, Berenguer J, Brouns SJJ, and van der Oost J (2014a). DNA-guided DNA interference by a prokaryotic Argonaute. Nature 507, 258–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, and van der Oost J (2014b). The evolutionary journey of Argonaute proteins. Nat. Struct. Mol. Biol 21, 743–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Hegge JW, Hinojo I, Shiimori M, Ellis MA, Dumrongkulraksa J, Terns RM, Terns MP, and van der Oost J (2015). Argonaute of the archaeon Pyrococcus furiosus is a DNA-guided nuclease that targets cognate DNA. Nucleic Acids Res. 43, 5120–5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Szczepaniak M, Sheng G, Chandradoss SD, Zhu Y, Timmers EM, Zhang Y, Zhao H, Lou J, Wang Y, et al. (2017). Autonomous generation and loading of DNA guides by bacterial Argonaute. Mol. Cell 65, 985–998.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sheng G, Juranek S, Tuschl T, and Patel DJ (2008). Structure of the guide-strand-containing argonaute silencing complex. Nature 456, 209–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Juranek S, Li H, Sheng G, Wardle GS, Tuschl T, and Patel DJ (2009). Nucleation, propagation and cleavage of target RNAs in Ago silencing complexes. Nature 461, 754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee LM, Flores-Jasso CF, Salomon WE, and Zamore PD (2012). Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell 151, 1055–1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan YR, Pei Y, Ma JB, Kuryavyi V, Zhadina M, Meister G, Chen HY, Dauter Z, Tuschl T, and Patel DJ (2005). Crystal structure of A. aeolicus Argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell 19, 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.