Abstract
Four new citrinin derivatives, including two citrinin dimers and two citrinin monomer derivatives, were isolated and identified from a marine-derived fungal strain Penicillium sp. ML226 along with six known related compounds. Their structures were elucidated by spectroscopic and chemical methods. The new compounds showed modest cytotoxic activity against HepG-2 cell line and weak antimicrobial activity against Staphylococcus aureus.
Keywords: citrinin, Penicillium sp. ML226, new metabolites, marine fungus
1. Introduction
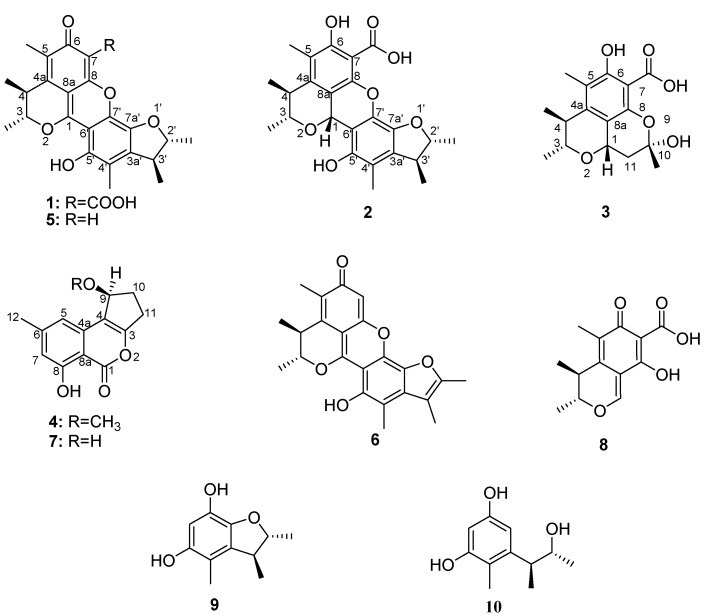
The search for new bioactive natural products is still the main way of discovering new drugs. Investigating the secondary metabolites of microorganisms isolated from specific ecological environments may increase the chance of finding compounds with novel skeletons and varied and unique bioactivities. It was reported that the specific situations that microorganisms live in might activate some silent genes and induce some unique biosynthetic pathways [1]. Marine microorganisms have attracted extensive attention in this context. Marine fungi are an important resource to find chemically and biologically diverse compounds due to their special living environment [2,3]. In order to search for new bioactive natural products, a marine-derived fungal strain, ML226, authenticated as Penicillium sp., was isolated from the Taiwan Strait, China. The EtOAc extract of Penicillium sp. ML226 exhibited cytotoxic and antimicrobial activity. Chemical investigation of the EtOAc extract of Penicillium sp. ML226 led to the isolation of two new citrinin dimers—penicitrinone E (1) and penicitrinol J (2)—two new citrinin monomer derivatives—penicitrinol K (3) and citrinolactone D (4)—together with six known compounds-penicitrinone A [4] (5), penicitrinone B [4] (6), citrinolactone B [5] (7), citrinin [6] (8), 2,3,4-trimethyl-5,7-dihydroxy-2,3-dihydrobenzofuran [7] (9) and phenol A [8] (10) (Figure 1). In this paper, we report the isolation and structural elucidation of compounds 1–10 and the cytotoxic and antimicrobial activity of 1–4. They all showed weak cytotoxicity against HepG-2 cell line in the concentration of 10 μg/mL with inhibition rate from 6% to 30%. Compounds 2 and 3 showed weak antimicrobial activity against Staphylococcus aureus.
Figure 1.
Structures of the isolated compounds 1–10.
2. Results and Discussion
2.1. Structural Elucidation of Compounds
Penicitrinone E (1) was obtained as a red, amorphous powder. 0 (0.002, MeOH). A molecular formula of C24H24O7 was assigned based on the interpretation of HRESIMS data at m/z 425.15965 [M + H]+ (calcd. 425.1600). The 1H-NMR data of 1 showed four tertiary methyl signals, two aromatic methyl signals, four sp3 methine protons (two oxygenated), and one hydroxyl proton (Table 1). The 13C-NMR and DEPT spectra for 1 displayed 24 carbon signals comprising four tertiary methyls, two aromatic methyls, four sp3 methines (two oxygenated), two carbonyl carbons, and 12 sp2 quaternary carbons (Table 1). Except for those of the benzopyran moiety, the NMR data were quite similar to those of 5 [4], indicating that they shared the same molecular skeleton. Compared with those of 5, the NMR spectra of 1 exhibited an additional carboxyl group (δc 165.4), two downfield shifts effect of C-1 (+4.6 ppm) and C-8 (+2.7 ppm) because of the inductive effect of the additional carboxyl group. The C-7 of 5 is a sp2 methine carbon but the C-7 of 1 is a sp2 quaternary carbon, indicating that the carboxyl group was linked to C-7.
Table 1.
1H- and 13C-NMR (600 and 150 MHz) data for compounds 1 and 2 (CDCl3, δ in ppm).
| No. | 1 | 2 | ||
|---|---|---|---|---|
| δH (mult., J [Hz], int.) | δc | δH (mult., J [Hz], int.) | δc | |
| 1 | - | 161.1s | 5.71 (s) | 66.3d |
| 3 | 5.17 (q, 6.7) | 83.2d | 4.10 (dq, 6.1, 6.8) | 79.0d |
| 3-CH3 | 1.49 (d, 6.7, 3H) | 19.04q | 1.47 (d, 6.8, 3H) | 21.9q |
| 4 | 3.29 (q, 7.2) | 34.8d | 3.03 (dq, 6.1, 7.0) | 37.7d |
| 4-CH3 | 1.38 (d, 7.2, 3H) | 18.97q | 1.34 (d, 7.0, 3H) | 19.6q |
| 4a | - | 132.7s | - | 144.7s |
| 5 | - | 130.8s | - | 120.6s |
| 5-CH3 | 2.22 (s, 3H) | 10.9q | 2.21 (s, 3H) | 11.2q |
| 6 | - | 183.8s | - | 161.9s |
| 6-OH | - | - | 12.52 (s) | - |
| 7 | - | 103.1s | - | 98.0s |
| 7-COOH | - | 165.4s | - | 170.6s |
| 8 | - | 160.9s | - | 145.3s |
| 8a | - | 99.5s | - | 108.9s |
| 2′ | 4.77 (dq, 4.3, 6.4) | 88.4d | 4.56 (m) | 88.3d |
| 2′-CH3 | 1.47 (d, 6.4, 3H) | 21.0q | 1.36 (d, 6.5, 3H) | 20.9q |
| 3′ | 3.25 (dq, 4.3, 7.1) | 45.0d | 3.09 (m) | 44.3d |
| 3′-CH3 | 1.36 (d, 7.1, 3H) | 18.82q | 1.35 (d, 7.2, 3H) | 19.3q |
| 3a′ | - | 142.4s | - | 133.0s |
| 4′ | - | 118.2s | - | 117.8s |
| 4′-CH3 | 2.28 (s, 3H) | 11.6q | 2.18 (s, 3H) | 11.6q |
| 5′ | - | 147.2s | - | 147.5s |
| 5′-OH | 8.25 (s) | - | 7.92 (s) | - |
| 6′ | - | 102.2s | - | 105.3s |
| 7′ | - | 136.2s | - | 130.7s |
| 7a′ | - | 139.2s | - | 138.1s |
a The assignments were based on DEPT, 1H-1H COSY, HMQC, and HMBC experiments, and chemical shift values are in ppm relative to TMS. “-”: no signal.
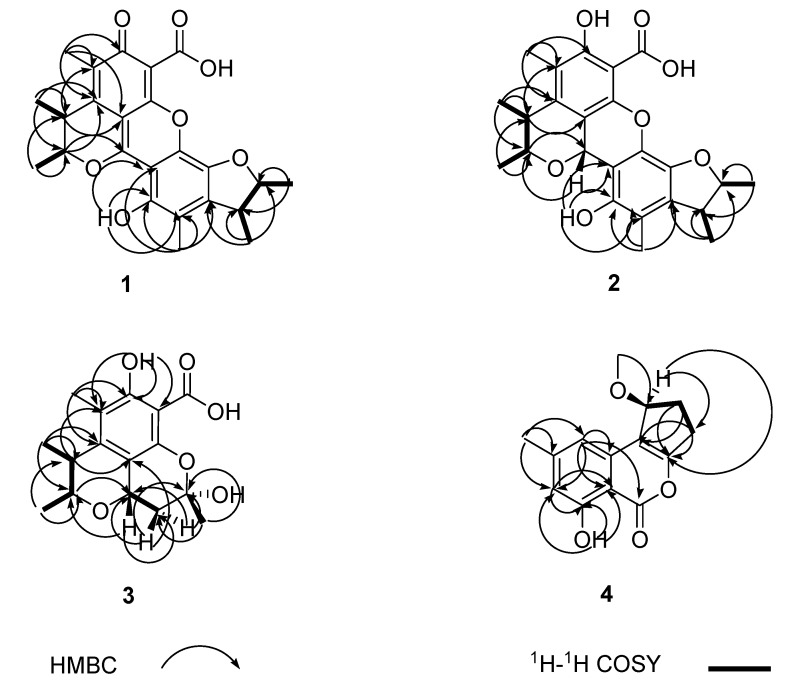
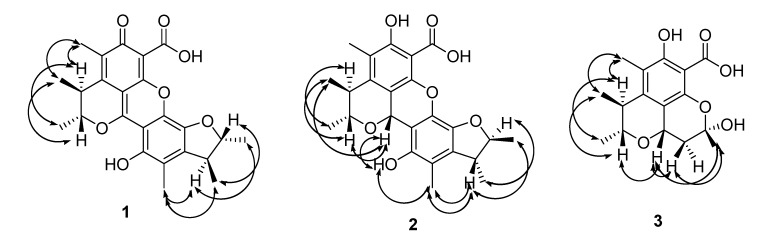
This deduction was further supported by analyses of the 2D (HMQC, 1H-1H COSY and HMBC) NMR spectra (Figure 2). The relative configuration of the two methyl residues in the benzopyran moiety was determined as trans based on the NOESY correlation of 4-CH3 with 3-H and J3,4 (<0.5 Hz); and the relative configuration of the two methyl residues in the benzofuran moiety was determined as trans based on the NOESY correlation of 3′-CH3 with 2′-H and J2′,3′ (=4.3 Hz) (Figure 3).
Figure 2.
1H-1H COSY and key HMBC correlations of compounds 1–4.
Figure 3.
Key NOESY effects of compounds 1–3.
Penicitrinol J (2) was isolated as a pale yellow, amorphous powder. −30.0 (0.001, MeOH). The molecular formula of 2 was determined as C24H26O7 by HRESIMS at m/z 427.17601 [M + H]+ (calcd. 427.1757). The 1H-NMR data of 2 showed four tertiary methyl signals, two aromatic methyl signals, five sp3 methine protons (three oxygenated), and two hydroxyl protons (Table 1). The 13C-NMR and DEPT spectra for 2 displayed 24 carbon signals including four tertiary methyls, two aromatic methyls, five sp3 methines (three oxygenated), one carbonyl carbon, and 12 sp2 quaternary carbons (Table 1). The NMR data were quite similar to those of 1 except for those of the benzopyran moiety. Compared with those of 1, the NMR spectra of 2 exhibited an additional oxygenated sp3 methine proton (δH 5.71) and an additional oxygenated sp3 methine carbon (δc 66.3), but missed one carbonyl carbon (δc 183.8 in 1). These indicated one of the two additional protons was linked to C-1, the other was the hydroxyl proton of 6-OH, which was further supported by the downfield shift effect of C-4a (+12 ppm) and the high-field shifts effect of H-3 (−1.07 ppm) and H-4 (−0.26 ppm) as a result of the missing of the double bond between C-1 and C-8a, and the 2D (HMQC, 1H-1H COSY and HMBC) NMR spectra (Figure 2). The NOESY correlation of 3-H with 4-CH3 and J3,4 (=6.1 Hz) established the trans of the two methyl residues in the benzopyran moiety; The NOESY correlation of 2′-H with 3′-CH3 and J2,3 (<0.5 Hz) demonstrated the trans of the two methyl residues in the benzofuran moiety; and the relative configuration of the 1-H and 3-H was determined as cis based on the NOESY correlations of 1-H with 3-H, 5′-OH and 4 -CH3 (Figure 3).
Penicitrinol K (3) was isolated as a white, amorphous powder. −125.2 (0.002, MeOH). The benzopyran moiety’s molecular formula of 3 was determined as C13H14O5 by HRESIMS at m/z 273.07299 [M + Na]+ (calcd. 273.0739) (Figure 4).
Figure 4.
The benzopyran moiety of compound 3.
The molecular formula of 3 was determined to be C16H20O6 by ESI-MS at m/z 331.2 [M + Na]+ and 291.4 [M − H2O + H]+. The 1H-NMR data of 3 showed three tertiary methyl signals, one aromatic methyl signal, one sp3 methylene signal, three sp3 methine protons (two being oxygenated), and one hydroxyl proton (Table 2). The 13C-NMR and DEPT spectra of 3 displayed signals for three tertiary methyls, one aromatic methyl, one sp3 methylene, three sp3 methines (two being oxygenated) and eight quaternary carbons (Table 2). Compared to those of 2, compound 3 shared the same benzopyran moiety with 2 which was further supported by the 2D (HMQC, 1H-1H COSY and HMBC) spectra (Figure 2), and the NMR spectra of 3 exhibited a high-field shift effect of C-1 (−1.01 ppm). The 1H-1H COSY correlations between 1-H and 11-Ha, 1-H and 11-Hb, and key HMBC correlations from 1-H, 11-H and 10-CH3 to corresponding carbons indicated that C-11 was linked to C-1, C-10 was linked to C-11, and 10-CH3 was linked to C-10 (Figure 2). Finally C-10 was linked to C-8 via O and 10-OH was linked to C-10, which established by the molecular formula of 3. The NOESY correlation of 3-H with 4-CH3 and J3,4 (=6.3 Hz) determined the trans of 3-CH3 and 4-CH3. The NOESY correlations between 1-H and 3-H and 1-H and 10-CH3 established the cis configurations of 1-H and 3-H and 1-H and 10-CH3 (Figure 3).
Table 2.
The NMR data for compound 3 (CDCl3, δ in ppm).
| No. | δH (mult., J [Hz], int.) | δc |
|---|---|---|
| 1 | 4.70 (dd, 6.1, 11.5) | 66.2d |
| 3 | 3.74 (dq, 6.3, 6.2) | 78.8d |
| 3-CH3 | 1.39 (d, 6.2, 3H) | 21.6q |
| 4 | 2.88 (dq, 6.3, 6.9) | 38.1d |
| 4-CH3 | 1.23 (d, 6.9, 3H) | 19.3q |
| 4a | - | 145.8s |
| 5 | - | 118.2s |
| 5-CH3 | 2.14 (s, 3H) | 11.1q |
| 6 | - | 161.3s |
| 6OH | 12.16 (s) | - |
| 7 | - | 97.6s |
| 7-COOH | - | 171.6s |
| 8 | - | 146.9s |
| 8a | - | 111.7s |
| 10 | - | 101.4s |
| 10-CH3 | 1.87 (s, 3H) | 29.3q |
| 11a | 2.53 (dd, 6.1, 12.8) | 37.3t |
| 11b | 1.84 (dd, 11.5, 12.8) |
Citrinolactone D (4) was obtained as a white, amorphous powder. +6.7 (0.004, MeOH). The molecular formula of 4 was determined as C14H14O4 by HRESIMS at m/z 269.07804 [M + Na]+ (calcd. 269.0790). The 1H-NMR data of 4 showed one aromatic methyl signal, one methoxyl group, two sp3 methylene signals, two aromatic protons, one oxygenated sp3 methine proton, and one hydroxyl proton (Table 3). The 13C-NMR spectrum of 4 displayed signals for two methyls (one of them oxygenated), two sp3 methylenes, one oxygenated sp3 methine, two sp2 methines, one carbonyl carbon, and 6 sp2 quaternary carbons (Table 3). The NMR data were quite similar to those of 7 [5]. By comparison with those of 7, the NMR spectra of 4 exhibited an additional methoxyl group (δH 3.50, δc 57.3) attached C-9, and which was further supported by HMBC correlations from 9-OCH3 to C-9 (Figure 2).
Table 3.
The NMR data of compound 4 (CDCl3, δ in ppm).
| No. | δH (mult., J [Hz], int.) | δc |
|---|---|---|
| 1 | - | 181.1s |
| 3 | - | 173.9s |
| 4 | - | 119.9s |
| 4a | - | 157.3s |
| 5 | 6.71 (s) | 107.6d |
| 6 | - | 146.7s |
| 7 | 6.63 (s) | 112.6d |
| 8 | - | 161.0s |
| 8-OH | 12.56 (s) | - |
| 8a | - | 109.0s |
| 9 | 4.95 (d, 6.8) | 79.4d |
| 10a | 2.35 (m) | 27.6t |
| 10b | 2.17 (m) | |
| 11a | 3.22 (m) | 30.2t |
| 11b | 2.81 (m) | |
| 12 | 2.40 (s, 3H) | 22.3q |
| 9-OCH3 | 3.50 (s, 3H) | 57.3q |
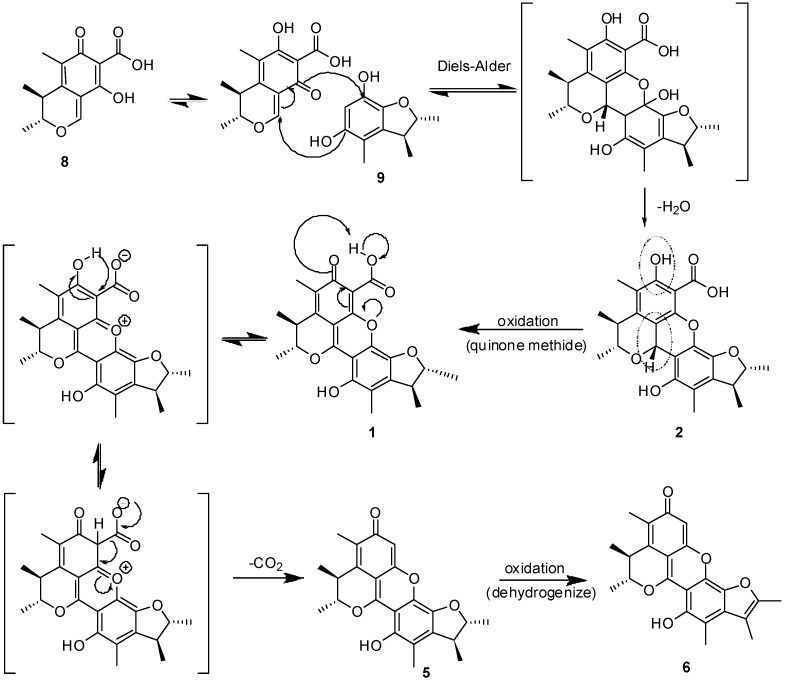
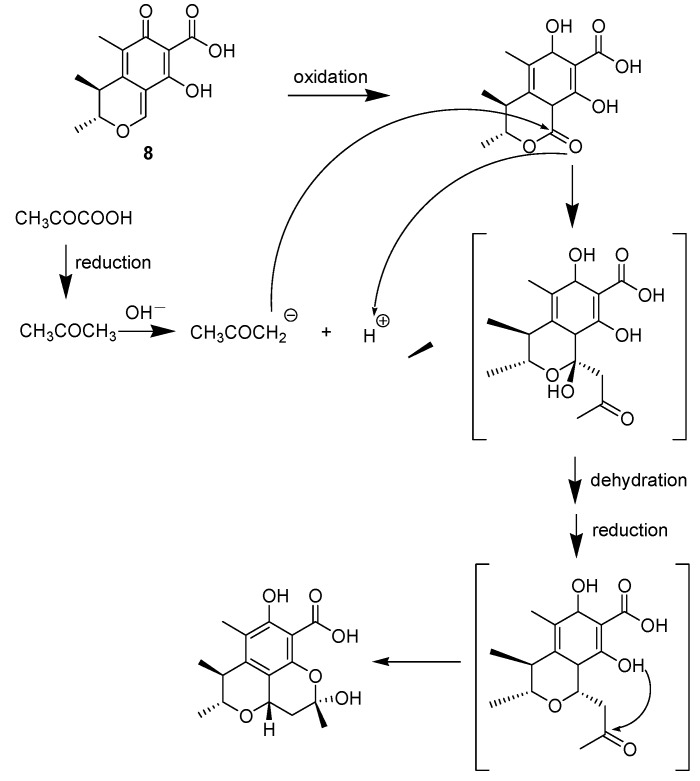
2.2. Biosynthesis
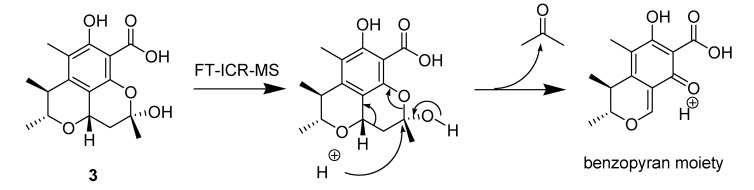
These compounds likely have the same biogenetic origin via the polyketide pathway [9]. citrinolactone B (7), citrinin (8), 2,3,4-trimethyl-5,7-dihydroxy-2,3-dihydrobenzofuran (9), phenol A (10) are biosynthesized from acetyl coenzyme A. Citrinolactone D (4) is the result of the methylation of citrinolactone B at 9-OH (7). To explain the biogenetic origin of penicitrinone E (1), penicitrinol J (2) and penicitrinol K (3), a postulated biosynthetic pathway is proposed in Scheme 1 and Scheme 2. Penicitrinone E (1) is postulated to derive from the oxidation of penicitrinol J (2), which results from the Diels-Alder reaction of 8 with 9 and then undergoes dehydration. Subsequent decarboxylation and the following dehydrogenation of penicitrinol J (2), leads successively to penicitrinone A (5) and penicitrinone B (6) [10] (Scheme 1). Oxidation of citrinin (8) forms dihydrocitrinone [11]. After undergoing aldol condensation of the carbonyl group at C-1 of dihydrocitrinone with an acetone anion, which is biosynthesized from pyruvic acid via enzymatic reduction [12], dihydrocitrinone changes to an intermediate structure. Further dehydration, reduction and the following aldol condensation of the intermediate structure yields penicitrinol K (3) (Scheme 2). Based on this biosynthetic scheme, the absolute configurations were deduced as 3R, 4S, 2′R, 3′S - for 1, 1R, 3R, 4S, 2′R, 3′S - for 2, 1S, 3R, 4S, 10S - for 3, 9S - for 4.
Scheme 1.
Postulated Biosynthesis of 1, 2, 5 and 6 resulting from 8 and 9.
Scheme 2.
Postulated Biosynthetic Pathway of 3.
To best of our knowledge, four penicitrinones have been reported: penicitrinones A–B [4], pennicitrinone C [9] and pennicitrinone D [13] and nine penicitrinols have been reported: Penicitrinol A [4], pennicitrinol B [9], penicitrinols C–E [12] and penicitrinols F–I [14]; Three citrinolactones have been reported: citrinolactones A–C [5]. Thus, the four new citrinin derivatives-penicitrinone E (1), penicitrinol J (2), penicitrinol K (3) and citrinolactone D (4) enrich the number of penicitrinones, penicitrinols and citrinolactones respectively. Markedly, the 7-COOH of penicitrinone E (1) and penicitrinol J-K (2–3) were reported in both penicitrinones and penicitrinols for the first time, which complemented the Diels-Alder reaction of citrinin [10,12].
2.3. Cytotoxic and Antimicrobial Activity
The new compounds 1–4 were tested for cytotoxic effects against the HeLa and HepG-2 cell lines using the MTT method [15]. However, they exhibited no remarkable cytotoxic activity against any of the cell lines in the concentration of 10 μg/mL (100% of cis-platinum as positive control). The results of cytotoxic tests of compounds 1–4 are shown in Table 4.
Table 4.
Biological Activities of Compounds 1–4.
| Compound | Inhibitory ratio (%) | |
| HeLa | HepG-2 | |
| 1 | - | 6.3 |
| 2 | - | 25.1 |
| 3 | - | 9.2 |
| 4 | 4.0 | 16.1 |
The antimicrobial activity of compounds 1–4 against Staphylococcus aureus (CMCC26003), Escherichia coli (CMCC44103), Candida albicans (AS2.538), and Aspergillus niger (ACCC30005) were also evaluated by paper diffusion method with concentration of 20 μg/6 mm paper disk. Only compounds 2 and 3 showed weak antimicrobial activity against Staphylococcus aureus CMCC26003 with inhibition zones of 10 and 9 mm diameter, respectively (18 mm of gentamicin as positive control).
3. Experimental
3.1. General Procedures
Optical rotations were obtained on a PerkinElmer 341 automatic polarimeter. UV spectra were recorded on a Persee TU-1901 spectrophotometer. IR spectra were recorded on a Nicolet Avatar 330FT spectrometer. 1H-NMR, 13C-NMR, and DEPT spectra and 2D NMR spectra were recorded on a Bruker Avance Ⅲ-600 NMR spectrometer using TMS as internal standard, and chemical shifts were recorded as values. HRESIMS data were measured on a Bruker FT-ICR-MS mass spectrometer. ESIMS was measured on a Finnigan mass spectrometer. TLC was carried out using glass-precoated silica gel GF254 (Qingdao Marine Chemical, Inc., Qingdao, China) and visualized under UV light or by spraying with vanillin (contains H2SO4) ethanol reagent. Sephadex LH-20 (40–70 μm, Amersham Pharmacia Biotech AB, Uppsala, Sweden), silica gel (200–300mesh, Qingdao Marine Chemical, Inc., Qingdao, China), and Lichroprep reversed-phase RP-18 silica gel (40–63 μm, Merck, Darmstadt, Germany) were used for column chromatography (CC).
3.2. Fungal Material
The fungal strain Penicillium sp. ML226 was isolated from the sediment of Fu Gong mangrove region, Long Hai, Taiwan Strait, China. It was identified according to its morphological characteristics and ITS sequence. It was identified as a sporulating fungus by traditional morphology. A BLAST search result showed that the internal transcribed spaces (ITS) sequence of ML226 was highly homologous (96% percent similarity) to that of a Penicillium species (JX192960), indicating that ML226 belongs to this genus. The voucher specimen is deposited in our laboratory at−80 °C. The producing strain was prepared on potato dextrose agar slants and stored at 4 °C.
3.3. Fermentation and Extraction
The fungus Penicillium sp. ML226 was inoculated on slope of YMG media (glucose 4.0, malt extract 10.0, yeast extract 4.0, pH 7.2) in a 250 mL solanad type flask containing solid media (25 mL/flask) at 28 °C for 4 days to afford spores. Then the spores were obtained by scraping and agitating from the slope of YMG media using 120 mL ddH2O. Solid media fermentation was performed with YMG media (12 L) at 28 °C for 7 days, and the spores was inoculated with inoculating loop. The cultured agar was chopped, diced and extracted with EtOAc-MeOH-AcOH (80:15:5, 3.5 liters) at room temperature overnight. The organic solution was collected through filtration, and the remaining agar residue was extracted several times more as described above until the filtrate was colourless. The combined filtrates were concentrated under vacuum to remove organic solvents. The aqueous solution was extracted five times with EtOAc to give an EtOAc solution, which was concentrated under vacuum to give a crude EtOAc extract. Then the EtOAc extract was dissolved with MeOH to give a MeOH solution. The MeOH solution was concentrated under vacuum to give a crude extract (5.00 g).
3.4. Purification
The crude MeOH extract (5.00 g) was subjected to MPLC over RP-18 silica gel (170 g) using a stepwise gradient of 30, 50, 70 and 100% (v/v) MeOH in H2O. Then we achieved Fr.B (244 mg) and Fr.C (251 mg) obtained from 30% MeOH and Fr.D (560 mg) and Fr.E (362 mg) obtained from 50% MeOH. These fractions were further purified by repeated column chromatography (CC) on Sephadex LH-20 and silica gel.
Fr.B (244 mg) was fractionated by Sephadex LH-20 CC (140 g, eluted with MeOH) to provide five fractions (Ba–Be). Fr.Bb (50.9 mg) was fractionated by Sephadex LH-20 CC (80 g, eluted with acetone) to provide four fractions (Bb1–Bb4). Fr.Bb2 (8.0 mg) was further purified by silica gel CC [step gradient, 0–3% EtOAc in petroleum ether (PE)] to yield 9 (3.4 mg). Fr.Bb4 (3.5 mg) was further purified by silica gel CC (step gradient, 0-10% EtOAc in PE) to afford 10 (1.8 mg).
Fr.C (251 mg) was fractionated by Sephadex LH-20 CC (140 g, eluted with MeOH) to provide seven fractions (Ca–Cg). Fr.Cb (36.6 mg) was further purified by silica gel CC (step gradient, 0–12.5% acetone in PE) to yield 1 (8.5 mg). Fr.Cd (60.0 mg) was fractionated by Sephadex LH-20 CC (80 g, eluted with acetone) to provide four fractions (Cd1–Cd4). Fr.Cd2 (10.0 mg) was further purified by silica gel CC (step gradient, 0–2.5% acetone in PE) to obtain 9 (3.2 mg). Fr.Cg (34, 2 mg) was further purified by silica gel CC (step gradient, 0-1.5% EtOAc in PE with 0.5% HCOOH) to produce 2 (4.0 mg) and 1 (2.2 mg).
After settling, some crystals of 8 (80.7 mg) appeared in Fr.D (560 mg) and the remaining mother liquor solution (463 mg) was fractionated by Sephadex LH-20 CC (140 g, eluted with MeOH) to provide seven fractions (Da–Dg). Fr.Dc (73.6 mg) was fractionated by Sephadex LH-20 CC [80 g, eluated with MeOH/acetone (1:4)] to provide four fractions (Dc1–Dc4). Fr.Dc3 (26.0 mg) was further purified by silica gel CC (step gradient, 0–50% EtOAc in PE) to yield 6 (3.7 mg).
Fr.E (362 mg) was fractionated by Sephadex LH-20 CC (140g, eluted with MeOH) to provide five fractions (Ec–Ee). Fr.Ec (55.1 mg) was fractionated by Sephadex LH-20 CC [80 g, eluted with MeOH/ acetone (1:4)] to provide three fractions (Ec1–Ec3). Fr.Ec2 (24.0 mg) was further purified by silica gel CC (step gradient, 0–25% acetone in PE) to yield 5 (3.0 mg). Fr.Ee (121.9 mg) was fractionated by Sephadex LH-20 CC (140g, eluted with acetone) to provide four fractions (Ee1–Ee4). Fr.Ee2 (12.9 mg) was further purified by silica gel CC (step gradient, 0–2.5% acetone in PE) to yield 3 (3.2 mg). Fr.Ee3 (43.0 mg) was further purified by silica gel CC (step gradient, 0-3.3% acetone in PE) to yield 4 (7.1 mg). Fr.Ee4 (73.0 mg) was fractionated by Sephadex LH-20 CC [80 g, eluted with MeOH/ acetone (1:4)] to provide three fractions ((Ee41–Ee43). Fr.Ee43 (36.9 mg) was further purified by silica gel CC (step gradient, 0-3.3% acetone in PE) to obtain 7 (2.6 mg).
3.5. Biological Assays
Cancer cell lines were derived from the cell bank of the Chinese Academy of Sciences. The cytotoxicities of the compounds 1–5 were measured by the MTT (Sigma) assay [15]. The cells in 100 μL of culture medium were plated in each well of 96-well plates (Falcon, CA). After 24 h of incubation for a density of 5 × 103/100 μL medium, the cells were treated in triplicate with the concentration of 10 μg/mL of every compound for 72 h at 37 °C. A 20 μL aliquot of MTT solution (5 mg/mL) was added directly to all wells and incubated for 4 h at 37 °C. To quench the reaction, 100 μL of triplex solution (10% SDS, 5% isobutanol, 12 mM HCl) was added to each well and incubated overnight at 37 °C. The optical density of each well was measured with a microplate reader (M-3350, Bio-Rad) at 595nm (excitation). Growth inhibition rates were calculated with the following equation:
3.6. Spectral Data
Penicitrinone E(1): red, amorphous powder; 0 (0.002, MeOH); UV (MeOH) λmax (log ε) 204 (2.74), 214 (2.76), 232 (2.58), 258 (2.37), 278 (2.49), 315 (2.34), 400 (2.49) nm; IR (KBr) νmax 3453, 2967, 2928, 2873, 2358, 2332, 1687, 1641, 1611, 1527, 1506, 1451, 1408, 1380, 1325, 1269, 1154, 1101, 1060, 1026, 993, 933, 899, 858, 822, 748, 700, 675, 652, 552, 505 cm−1; Rf = 0.318 (PE : acetone = 1:1), Rf = 0.611 (PE : acetone = 1:1, with formic acid); 1H- and 13C-NMR (see Table 1); HRESIMS m/z 425.15965 [M + H]+ (calcd. for C24H25O7, 425.1600).
Penicitrinol J(2): pale yellow, amorphous powder; −30.0 (0.001, MeOH); UV (MeOH) λmax (log ε) 203 (3.05), 215 (3.12), 257 (2.61), 277 (2.56), 315 (2.55), 400 (2.51) nm; IR (KBr) νmax 3343, 2967, 2925, 2869, 2364, 2338, 1684, 1636, 1586, 1527, 1504, 1451, 1421, 1385, 1328, 1274, 1239, 1165, 1136, 1074, 1022, 996, 974, 929, 907, 858, 815, 778, 754, 729, 704, 636, 550 cm−1; Rf = 0.591 (PE : acetone = 1:1), Rf = 1 (PE:acetone = 1:1, with formic acid); 1H- and 13C-NMR (see Table 1); HRESIMS m/z 427.17601 [M + H]+ (calcd. for C24H27O7, 427.1757).
Penicitrinol K (3): white, amorphous powder; −125.2 (0.002, MeOH); UV (MeOH) λmax (log ε) 213(2.68), 257 (2.34), 325 (1.90) nm; IR (KBr) νmax 3433, 3230, 2970, 2930, 2872, 2360, 2334, 1687, 1623, 1589, 1445, 1419, 1386, 1353, 1298, 1270, 1232, 1200, 1169, 1105, 1079, 1025, 905, 876, 809, 753, 729, 683, 582, 549 cm−1; Rf = 0.524 (CHCl3 : MeOH = 10:1); 1H- and 13C-NMR (see Table 2); The citrinin moiety’s HRESIMS m/z 273.07299 [M + Na]+ (calcd. for C13H14O5 Na, 273.0739); ESIMS m/z 331.2 [M + Na]+ and 291.4 [M − H2O + H]+.
Citrinolactone D(4): white, amorphous powder; +6.7 (0.004, MeOH); UV (MeOH) λmax (log ε) 204 (2.35), 229 (2.45), 237 (2.49), 257 (2.31), 325 (1.80) nm; IR (KBr) νmax 3437, 2965, 2924, 2824, 2365, 2337, 1653, 1622, 1599, 1490, 1451, 1364, 1296, 1253, 1206, 1141, 1083, 1031, 1011, 941, 922, 869, 822, 784, 757, 698, 656, 619, 570, 513, 471 cm−1; Rf = 0.675 (PE : acetone = 2:1); 1H- and 13C-NMR (see Table 3); HRESIMS m/z 269.07804 [M + Na]+ (calcd. for C14H14O4 Na, 269.0790).
4. Conclusions
Four new compounds, penicitrinone E (1), penicitrinol J (2), penicitrinol K (3) and citrinolactone D (4), were isolated together with six known compounds from the marine-derived Penicillium sp. ML226. Penicitrinone E (1), penicitrinol J (2) and penicitrinol K (3) showed modest selective cytotoxixity against HepG-2 cell line. Citrinolactone D (4) showed weak cytotoxixity against HepG-2 and HeLa cell lines. penicitrinol J (2) and penicitrinol K (3) showed mild antimicrobial activity against Staphylococcus aureus.
Acknowledgments
This work was financially supported by the “863” project (2006AA09Z410, 2007AA091503) of the Ministry of Science and Technology and the Fundamental Research Funds for the Central Universities (No.2011121037).
Supplementary Materials
The NMR spectra and HRESIMS spectra of compounds 1–4 can be accessed at: http://www.mdpi.com/1420-3049/18/5/5723/s1.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds 1–10 are available from the authors.
References
- 1.Tamburini E., Mastromei G. Do bacterial cryptic genes really exist? Res. Microbiol. 2000;151:179–182. doi: 10.1016/s0923-2508(00)00137-6. [DOI] [PubMed] [Google Scholar]
- 2.Liberra K., Lindequist U. Marine fungi—A prolific resource of biologically active natural products? Pharmazie. 1995;50:583–588. [PubMed] [Google Scholar]
- 3.Bugni T.S., Ireland C.M. Marine-derived fungi: a chemically and biologically diverse group of microorganisms. Nat. Prod. Rep. 2004;21:143–163. doi: 10.1039/b301926h. [DOI] [PubMed] [Google Scholar]
- 4.Wakana D., Hosoe T., Itabashi T., Okada K., Takaki G.M.D., Yaguchi T., Fukushima K., Kawai K. New citrinin derivatives isolated from Penicillium citrinum. J. Nat. Med. 2006;60:279–284. doi: 10.1007/s11418-006-0001-2. [DOI] [Google Scholar]
- 5.Kuramata M., Fujioka S., Shimada A., Kawano T., Kimura Y. Citrinolactones A, B and C, and Sclerotinin C, plant growth regulators from Penicillium citrinum. Biosci. Biotechnol. Biochem. 2007;71:499–503. doi: 10.1271/bbb.60538. [DOI] [PubMed] [Google Scholar]
- 6.Hetherington A.C., Raistrick H. Studies in the biochemistry of microorganisms. XI. On the production and chemical constitution of a new yellow coloring matter, citrinin, produced from glucose by Penicillium citrinum THOM. Philos. Trans. R. Soc. London Ser. B. 1931;220:226–297. [Google Scholar]
- 7.Chen C.H., Shaw C.Y., Chen C.C., Tsai Y.C. 2,3,4-Trimethyl-5,7-dihydroxy-2,3-dihydrobenzofuran, a novel antioxidant, from Penicillium citrinum F5. J. Nat. Prod. 2002;65:740–741. doi: 10.1021/np010605o. [DOI] [PubMed] [Google Scholar]
- 8.Rodel T., Gerlach H. Enantioselective Synthesis of the Polyketide Antibiotic (3R,4S)-(-)-Citrinin. Liebigs Ann. 1995;1995:885–888. doi: 10.1002/jlac.1995199505129. [DOI] [Google Scholar]
- 9.Lu Z.Y., Lin Z.J., Wang W.L., Du L., Zhu T.J., Fang Y.C., Gu Q.Q., Zhu W.M. Citrinin dimers from the halotolerant fungus Penicillium citrinum B-57. J. Nat. Prod. 2008;71:543–546. doi: 10.1021/np0704708. [DOI] [PubMed] [Google Scholar]
- 10.Clark B.R., Capon R.J., Lacey E., Tennant S., Gill J.H. Citrinin revisited: from monomers to dimers and beyond. Org. Biomol. Chem. 2006;4:1520–1528. doi: 10.1039/b600960c. [DOI] [PubMed] [Google Scholar]
- 11.Chien M.M., Schiff P.L., Jr., Slatkin D.J., Knapp J.E. Metabolites of aspergilli. III. The isolation of citrinin, dihydrocitrinone and sclerin from aspergilus carneus. Lloydia. 1977;40:301–302. [PubMed] [Google Scholar]
- 12.Chen L., Liu W., Hu X., Huang K., Wu J.L., Zhang Q.Q. Citrinin derivatives from the marine-derived fungus Penicillium citrinum. Chem. Pharm. Bull. (Tokyo) 2011;59:515–517. doi: 10.1248/cpb.59.515. [DOI] [PubMed] [Google Scholar]
- 13.Xin Z.H., Wang W.L., Zhang Y.P., Xie H., Gu Q.Q., Zhu W.M. Pennicitrinone D, a new citrinin dimer from the halotolerant fungus Penicillium notatum B-52. J. Antibiot. (Tokyo) 2009;62:225–227. doi: 10.1038/ja.2009.12. [DOI] [PubMed] [Google Scholar]
- 14.Chen L., Liu W., Huang K., Hu X., Fang Z.X., Wu J.L., Zhang Q.Q. Penicitrinols F-I, New Citrinin Derivatives from the Marine-Derived Fungus Penicillium citrinum. Heterocycles. 2011;83:1853–1858. doi: 10.3987/COM-11-12234. [DOI] [PubMed] [Google Scholar]
- 15.E Z. Tissue Culture And Molecular Biology Technology. Beijing Publishing House; Beijing, China: 1995. pp. 1–141. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.