Abstract
The human cerebellum plays an essential role in motor control, is involved in cognitive function (i.e., attention, working memory, and language), and helps to regulate emotional responses. Quantitative in-vivo assessment of the cerebellum is important in the study of several neurological diseases including cerebellar ataxia, autism, and schizophrenia. Different structural subdivisions of the cerebellum have been shown to correlate with differing pathologies. To further understand these pathologies, it is helpful to automatically parcellate the cerebellum at the highest fidelity possible. In this paper, we coordinated with colleagues around the world to evaluate automated cerebellum parcellation algorithms on two clinical cohorts showing that the cerebellum can be parcellated to a high accuracy by newer methods. We characterize these various methods at four hierarchical levels: coarse (i.e., whole cerebellum and gross structures), lobe, subdivisions of the vermis, and the lobules. Due to the number of labels, the hierarchy of labels, the number of algorithms, and the two cohorts, we have restricted our analyses to the Dice measure of overlap. Under these conditions, machine learning based methods provide a collection of strategies that are efficient and deliver parcellations of a high standard across both cohorts, surpassing previous work in the area. In conjunction with the rank-sum computation, we identified an overall winning method.
Keywords: Magnetic resonance imaging, cerebellar ataxia, attention deficit hyperactivity disorder, autism
1. Introduction
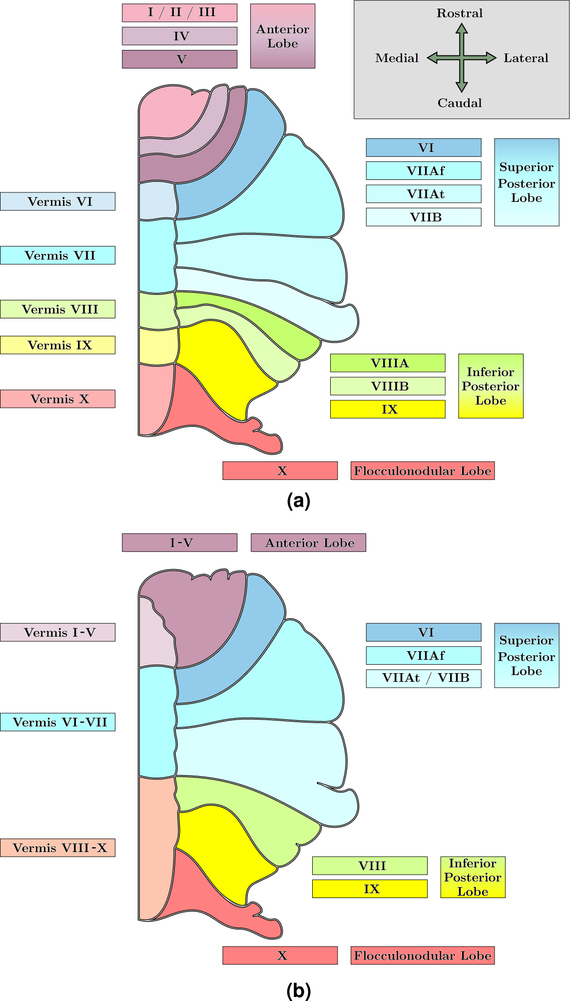
The cerebellum is a structure of great importance in the neuroanatomy of humans. It plays an essential role in motor coordination (Ito, 1984; Manto et al., 2013), as well as cognitive function such as attention (Schmahmann, 1991, 2004), working memory (Desmond and Fiez, 1998), and language (Silveri et al., 1994; Desmond and Fiez, 1998), regulates emotional responses (Schutter and Van Honk, 2005) including fear (Schmahmann and Caplan, 2006), and there is increasing understanding of perceptual processes in the cerebellum (Baumann et al., 2015). Anatomically, the cerebellum is nestled underneath the cerebral hemispheres behind the brainstem in the posterior cranial fossa. It is separated from the cerebrum by the tentorium cerebelli, a dura structure, and is connected to the brainstem at the pons. The cerebellum is divided into two hemispheres, like the cerebrum, and also has a midline zone which is known as the vermis. The cortical surface of the cerebellum is made up of finely spaced branches that radiate outwards from the cerebellar white matter (WM), which is known as the corpus medullare (CM). These WM branches conceal that the volume of the cerebellum is a tightly folded layer of gray matter (GM). Anatomists differentiate regions of the cerebellum hierarchically into groups of folds, known as lobes, and then into individual folds, referred to as lobules. The lobes are the anterior, superior posterior, inferior posterior, and the flocculonodular. The lobules are identified by Roman Numerals I through X (Schmahmann et al., 2000), however Lobules VII and VIII are further differentiated. This nomenclature comes from Schmahmann et al. (2000), derived in part from Larsell (1952); we refer to it as the Schmahmann nomenclature and note the differences between it and the classical nomenclature (Malacarne, 1776; Henle, 1879) in Table 1. Figure 1 shows the anatomical structure of the cerebellum, including the hierarchical breakdown of the lobes and lobules. Due to the importance of the cerebellum, any pathology can have serious consequences; however, the tightly folded structure of the cerebellum makes identifying specific structures challenging. Below we outline the clinical relevance of understanding the structure of the cerebellum and the various effects of cerebellar pathologies; we then provide an overview of the fully automated parcellation tools that exist in the literature.
Table 1:
A key to convert between the nomenclature of Schmahmann (Schmahmann et al., 2000), derived from Larsell (1952), and the classical nomenclature (Malacarne, 1776; Henle, 1879) of common cerebellar structures.
| Vermal Nomenclature | Hemisphere Nomenclature | ||
|---|---|---|---|
| Schmahmann | Classical | Schmahmann | Classical |
| Vermis I / II† | Lingula | L/R Lobule I/II | L/R Lingula (or Lingulae) |
| Vermis III† | Centralis | L/R Lobule III | L/R Centralis |
| Vermis IV† | Culmen I | L/R Lobule IV | L/R Quadrangularis |
| Vermis V† | Culmen II | L/R Lobule V | L/R Quadrangularis |
| Vermis VI | Declive | L/R Lobule VI | L/R Quadrangularis |
| Vermis VIIAf | Folium | L/R Lobule VIIAf (Crus I) | L/R Semi-Lunaris Superior |
| Vermis VIIAt | Tuber I | L/R Lobule VIIAt (Crus II) | L/R Semi-Lunaris Inferior |
| Vermis VIIB | Tuber II | L/R Lobule VIIB | L/R Semi-Lunaris Inferior |
| Vermis VIIIA | Pyramis I | L/R Lobule VIIIA | L/R Biventer I |
| Vermis VIIIB | Pyramis II | L/R Lobule VIIIB | L/R Biventer II |
| Vermis IX | Uvula | L/R Lobule IX | L/R Tonsilla (or Tonsil) |
| Vermis X | Nodulus | L/R Lobule X | L/R Flocculus |
It is widely acknowledged that there is no true vermis for the Anterior Lobe (Lobules I–V). The division in our Pediatric Cohort differentiates the midline portion of the Anterior Lobe from the body of the lobe.
Figure 1:
An illustration of a coronal view of one hemisphere of the human cerebellum (with orientation axes inset in (a)). Shown are the lobule labels for our (a) Adult and (b) Pediatric Cohort with their corresponding lobe groupings, based on the Schmahmann nomenclature (Schmahmann et al., 2000). Table 4 has a complete list of the provided labels for both cohorts. It is widely acknowledged that there is no true vermis for the Anterior Lobe (Lobules I–V). Thus the distinction between vermis and body in the Anterior Lobe differentiates the midline portion from the body of the lobe. Our Adult Cohort does not use this differentiation, whereas our Pediatric Cohort does.
Several anatomical studies have elucidated the view that the cerebellum is involved in more than just motor control (Baumann et al., 2015; Schmahmann, 2004; Schmahmann and Caplan, 2006; Strick et al., 2009). As such, the cerebellum projects to a diverse set of cortical areas via the thalamus, which then close the circuitry loop by reciprocating back to the cerebellum. It is now apparent that multiple cortical areas are the target of cerebellar output, including not only the primary motor cortex, but also subdivisions of premotor, oculomotor, prefrontal, and inferotemporal areas of cortex (Middleton and Strick, 2000); each of which form cerebello-thalamo-cortical circuits. Moreover, several clinical studies have shown that dysfunction in individual cerebellar loops with the cerebral cortex may underlie the development of specific neurological and psychiatric symptoms (Allen and Courchesne, 2003; Amaral et al., 2008; Andreasen and Pierson, 2008; Gottwald et al., 2004). Furthermore, clinical studies of cerebellum centric disorders, such as spinocerebellar ataxia (SCA), have been previously shown to have cerebellar shape (Yang et al., 2016a), clinical disability scores (Ying et al., 2006), and functional scores (Yang et al., 2014; Kansal et al., 2016) that correlate with SCA subtype in a region specific manner. More importantly, the cerebellum has been shown to be affected in diseases ranging from attention-deficit and hyperactivity disorder (ADHD) (Mostofsky et al., 1998b), schizophrenia (Nopoulos et al., 1999; Parker et al., 2014), Alzheimer’s disease (Thomann et al., 2008; Colloby et al., 2014), Parkinson’s disease (Lewis et al., 2013), to chronic alcoholism (Victor et al., 1959; Torvik and Torp, 1986; Cavanagh et al., 1997; Baker et al., 1999; Fitzpatrick et al., 2008). In patients with schizophrenia, a reduction in the volume of the vermis has been observed in multiple studies (Nopoulos et al., 1999; Okugawa et al., 2002, 2003) based on the manual parcellation of the cerebellum. Moreover, when the vermis has been further subdivided into the anterior and posterior portions, the volume differences are driven by changes in the posterior vermis (Womer et al., 2016) with a significant diagnosis-by-sex interaction. Several types of dementia exhibit correlations with the cerebellum; Alzheimer’s disease (AD) has shown a reduction in the volume of the posterior lobes (Thomann et al., 2008), whereas dementia with Lewy bodies has shown greater GM loss in Lobule VII than AD (Colloby et al., 2014). Several recent voxel based morphometry (VBM) studies have shown regional patterns of atrophy between AD and cerebellar GM and WM (Möller et al., 2013) and correlations between GM loss and the constructional praxis and constructional praxis recall test in the CERAD test battery (Dos Santos et al., 2011). However, older studies (Karas et al., 2003) that relied upon studying large regions—due to the FWHM size used in the VBM—showed no significant GM loss in the cerebellum suggesting that the effects of cerebellum/AD interaction can only be identified when smaller regions of interest are used. We highlight these studies and some of the other work done in improving our understanding, through neuroimaging, of the cerebellum and its role in health and disease in Table 2. However, Table 2 is far from a complete list of such studies, see Stoodley (2014) and Traut et al. (2018) for more comprehensive lists. There are two key points to take from this past work: 1) in-vivo assessment of the cerebellum through magnetic resonance (MR) imaging (MRI) is imperative to further our understanding and 2) manual parcellation or delineation remains a widely used approach for studying the cerebellum.
Table 2:
A summary of some cerebellar focused imaging studies exploring various pathologies. We include whether the study used manual delineations (MD) and the key cerebellar related findings. N (M/F) denotes the number of patients and the male/female ratio. Abbreviations: ADHD - Attention-deficit and hyperactivity disorder; AD - Alzheimer’s disease; AS - Asperger syndrone.
| Disease | Citation | N (M/F) | MD | Observations |
|---|---|---|---|---|
| ADHD | Mostofsky et al. (1998b) | 35 (35/0) | Y | Decreased inferior posterior vermis |
| Durston et al. (2004) | 90 (90/0) | N | Reduced right cerebellar volume | |
| Alcoholism | Torvik and Torp (1986) | 65 (65/0) | Y | Decreased vermis segments |
| Baker et al. (1999) | 19 (14/5) | Y | Non-significant loss in vermis and flocculus | |
| AD | Thomann et al. (2008) | 60 (29/31) | Y | Decreased superior and inferior posterior lobes |
| Möler et al. (2013) | 344 (175/169) | –† | Reduced GM throughout the cerebellum | |
| Colloby et al. (2014) | 127 (84/43) | –† | Bilateral reduction of Lobule VI | |
| AS | Catani et al. (2008) | 31 (31/0) | Y | Reduced cerebellar fractional anisotropy |
| Autism | Courchesne et al. (1994) | 103 (84/19) | Y | Reduced area in the vermis of Lobule VI and VII |
| Cleavinger et al. (2008) | 65 (65/0) | Y | No significant differences | |
| Webb et al. (2009) | 71 (56/15) | Y | Reduced area in various vermal labels | |
| D’Mello et al. (2015) | 70 (51/19) | N | Reduced GM in Lobule VII | |
| Dyslexia | Brambati et al. (2004) | 21 (10/11) | –† | Reduced cerebellar GM volume |
| Jednoróg et al. (2013) | 81 (39/42) | –† | Reduced GM volume in left Lobule I/II | |
| Fragile X Syndrome | Mostofsky et al. (1998a) | 188 (98/90) | Y | Decreased posterior vermis in males and females, though less significant in females. |
| Schizophrenia | Nopoulos et al. (1999) | 130 (130/0) | N‡ | Smaller vermis area and smaller anterior lobe |
| Okugawa et al. (2002) | 30 (30/0) | N‡ | Reduced posterior superior vermis | |
| Okugawa et al. (2003) | 116 (73/43) | N‡ | Reduced anterior vermis, posterior superior vermis, and posterior inferior vermis volumes | |
| Womer et al. (2016) | 104 (48/56) | Y | Decreased posterior vermis volumes in males |
The studies did not differentiate regions of the cerebellum and based assessment on an anatomists interpretation of the areas of change.
Automated processing for cerebellar volumes based on registration to a Talairach Atlas, augmented by manual tracings of the vermis.
Despite the continued use of manual delineation of the cerebellum in various studies (Womer et al., 2016) there has been work on both semi-automated (Pierson et al., 2002) and fully automated segmentation and parcellation of the cerebellum. Automating the parcellation, and consequently the volumetric analyses, of the cerebellum has benefits outside of the obvious efficiencies of speed and cost. Automated parcellation has been shown to remove manual bias, thus increasing consistency and comparability across studies and sites (Buckner et al., 2004; Hsu et al., 2002). We are only concerned with those methods that provide at a minimum the lobes of the cerebellum; hence methods like FreeSurfer (Dale et al., 1999; Fischl et al., 2004; Reuter and Fischl, 2011; Fischl, 2012), BrainSuite (Shattuck et al., 2001; Shattuck and Leahy, 2002; Shattuck et al., 2008), TOADS (Bazin and Pham, 2008; Shiee et al., 2010), CRUISE (Tosun et al., 2006; Landman et al., 2013), CRUISE+ (Shiee et al., 2014), MA-CRUISE (Huo et al., 2016a,b), and others (Carass et al., 2017c; Desikan et al., 2006; Guo et al., 2017; Ledig et al., 2015; Liu et al., 2012; Roy et al., 2015; Shao et al., 2018; Shiee et al., 2011; Tomas-Fernandez and Warfield, 2015; Van Leemput et al., 1999; Zhang et al., 2001; Zhao et al., 2017) that only provide tissue classes or coarse parcellations of the cerebellum are not directly relevant unless used in combination with other tools. The first published method that provided a fully automated parcellation of the cerebellar lobules was SUIT (Diedrichsen, 2006); the method used a spatially unbiased template of the human cerebellum that when registered with a subject image provided the parcellation. The method was later updated (Diedrichsen et al., 2009) to include a probabilistic atlas. As powerful as SUIT is in identifying the subdivisions of the cerebellum, it has primarily been used only for identifying cerebellar GM as a normalizing factor in functional MRI analysis. Prior to the introduction of the probabilistic version of SUIT, Powell et al. (2008) presented machine learning approaches for cerebellar parcellation that identified the lobes and vermis of the cerebellum. Bogovic et al. (2013a) presented ACCLAIM, a multi-object geometric deformable model (Bogovic et al., 2013c; Carass and Prince, 2016) approach that provides a parcellation of 28 labels of the cerebellum and included a comparison to SUIT. Price et al. (2014) presented the Cerebellar Analysis Toolkit (CATK) which used a Bayesian Appearance Modeling (Patenaude et al., 2011) with prior knowledge of shape, image intensity, and inter-shape relationships to provide five cerebellar labels. Weier et al. (2014) described the Rapid Automatic Segmentation of the human Cerebellum And its Lobules (RASCAL) which is a patch matching based approach that improved on the multi-atlas segmentation fusion technique presented in Coupe et al. (2011). Romero et al. (2017) presented CERES another patch-matching technique, that uses OPAL (Giraud et al., 2016; Ta et al., 2014) for its label fusion. Yang et al. (2016b) presented a multi-atlas labeling approach that used a graph-cut to help regularize the final segmentation. Several other methods have been reported in the literature (van der Lijn et al., 2009; Park et al., 2014; Plassard et al., 2016). A more detailed description of several of these methods is provided in Appendix A to help describe the approaches presented in this paper. To summarize, the previous work in this area includes: single and multi atlas registration, level sets, graph methods, a Bayesian framework, neural networks, support vector machines, and patch matching. Table 5 presents an overview of the methods presented and evaluated in this paper. It can be seen that deep learning, an important new class of algorithms in medical imaging, are represented among the methods tested in this paper.
Table 5:
An overview of the methods used in our comparison, with details of each method listed in the remainder of this Section.
| Name | Approach |
|---|---|
 SUIT*
SUIT*
|
Default SUIT v3.2 |
 C-SUIT*
C-SUIT*
|
C-SUIT is a customized SUIT, with Correction and Customized Atlas based on the Pediatric Cohort |
 FS-SUIT
FS-SUIT
|
FreeSurfer and SUIT in collaboration |
 LiviaNET
LiviaNET
|
A thirteen layer fully convolution network (FCN) |
 ConvNet
ConvNet
|
Convolution neural network |
 CERES2
CERES2
|
Updated version of CERES with improved intensity normalization and a new error correction method based on an ensemble of boosted patch-based neural networks |
 RASCAL
RASCAL
|
Updated patch-matching technique with cohort specific templates, improved intensity normalization, and non-linear registration |
 DeepNet
DeepNet
|
A U-net based FCN with ten layers |
– Denotes methods that only contributed results for the Pediatric Cohort.
There has been an increasing movement towards Grand Challenges (Styner et al., 2008; Schaap et al., 2009; Heimann et al., 2009; Menze et al., 2015; Mendrik et al., 2015; Maier et al., 2017; Carass et al., 2017a) in the medical imaging community in recent years. These challenges have helped to develop standards for evaluating the performance of different categories of medical imaging problems and for helping those on the peripheral of the community to understand the state-of-the-art and the general direction in which the technology is moving. In particular, the 2008 MICCAI MS Lesion challenge (Styner et al., 2008) was a significant step forward in the sharing of clinically relevant data. More recently, the 2015 Multimodal Brain Tumor Image Segmentation Benchmark (BRATS) (Menze et al., 2015) has been a disruptive step forward, having allowed groups without access to high-quality data with delineations to contribute innovative new solutions for segmenting brain tumors (Sauwen et al., 2015; Banerjee et al., 2016; Kamnitsas et al., 2017).
Thus in the spring of 2017, we invited colleagues from around the world to participate in a Cerebellum Parcellation Challenge as part of MICCAI 2017. As only eight groups responded to this call, it was decided that the workshop itself would not go forward due to lack of broad interest. Coming out of the discussions for this Cerebellum Parcellation Challenge (hereafter the Comparison), it was agreed that we would present the performance findings from seven of the research teams who participated in the Comparison (hereafter the Participants). In Section 2, we outline the two cohorts of data that were provided to the Participants and the evaluation used in comparing the submitted results from each of the Participants. One of the Participants submitted two methods, however two of the Participants contributed no results for our first cohort. Thus, that cohort has results from six algorithms, while our other cohort was processed by eight algorithms. Both cohorts are imaged using standard clinical protocols with an approximately 1 mm isotropic resolution, with complete details of the acquisition in Section 2. In our examination of these data and methods, we restrict our analyses to the Dice overlap; we outline our rationale behind this decision in Section 2.2. Section 3 provides a brief description of the methods contributed by the Participants for the Comparison, with a complete description of the presented methods in Appendix A. Section 4 includes the Comparison between the manual delineations for our two cohorts and the algorithms; it is broken down into hierarchical levels: 1) Coarse level including the whole cerebellum, whole vermis, and CM (3 labels); 2) Lobe level including the left and right of the four lobes (8 labels); 3) Vermis level which included the vermal subdivisions of the vermis (5 labels for our Adult Cohort, 3 labels for our Pediatric Cohort); 4) Lobule level (22 labels for our Adult Cohort, 14 labels for our Pediatric Cohort); and a5) Consolidated level, with further details in Sec. 4. In general the methods show agreement with the manual delineations of the cerebellar structures. However, the size of our cohorts restricted our statistical analyses, with rank-sum computations being used to determine an overall highest ranked method.
2. Materials and Metrics
2.1. Data
The Participants were given data from our Adult and Pediatric Cohorts. Our Adult Cohort is an expertly labeled data set collected by the Image Analysis and Communications Laboratory (IACL) at Johns Hopkins University (PI: J.L. Prince), complete details of the delineation protocol and the inter-rater variability are in Bogovic et al. (2013b). Our Pediatric Cohort comprises data collected at the Center for Neurodevelopmental Medicine and Imaging Research at the Kennedy Krieger Institute (PI: S.H. Mostofsky). The Pediatric Cohort was labeled using a cerebellar atlas developed at the Center for Neurodevelopmental Medicine and Imaging Research. The cerebellar atlas is based on a highly reliable manual parcellation protocol with interclass correlation coefficients ranging from .86–.99 across anatomically defined subdivisions as outlined in Table 4. The Participants were also encouraged to take advantage of other available data sets; in particular, they were made aware of data provided by Jörn Diedrichsen of the University of Western Ontario1. The Diedrichsen data comprises 20 normal adult subjects, each of which have 30 labeled cerebellar components.
Table 4:
The labeled cerebellar structures of both cohorts. For reference, we include a key to convert between the Schmahmann and classical nomenclature in Table 1.
| Adult Cohort (Healthy Controls and Ataxia Patients) | Pediatric Cohort (Healthy Controls, ADHD & HFA Patients) | ||
|---|---|---|---|
| Major Structure | Cerebellar Sub-components | Major Structure | Cerebellar Sub-components |
| Corpus Medullare | Corpus Medullare | ||
| Vermis | Vermis of Lobule VI | Vermis | Vermis of Lobule I–V |
| Vermis of Lobule VII | Vermis of Lobule VI–VII | ||
| Vermis of Lobule VIII | Vermis of Lobule VIII–X | ||
| Vermis of Lobule IX | L/R Anterior | L/R Lobule I–V | |
| Vermis of Lobule X | |||
| L/R Anterior | L/R Lobule I / II / III | L/R Superior Posterior | L/R Lobule VI |
| L/R Lobule IV | L/R Lobule VIIAf (Crus I) | ||
| L/R Lobule V | L/R Lobule VIIAt (Crus II) & VIIB | ||
| L/R Superior Posterior | L/R Lobule VI | L/R Inferior Posterior | L/R Lobule VIII |
| L/R Lobule VIIAf (Crus I) | L/R Lobule IX | ||
| L/R Lobule VIIAt(Crus II) | L/R Flocculonodular | L/R Lobule X | |
| L/R Lobule VIIB | |||
| L/R Inferior Posterior | L/R Lobule VIIIA | ||
| L/R Lobule VIIIB | |||
| L/R Lobule IX | |||
| L/R Flocculonodular | L/R Lobule X | ||
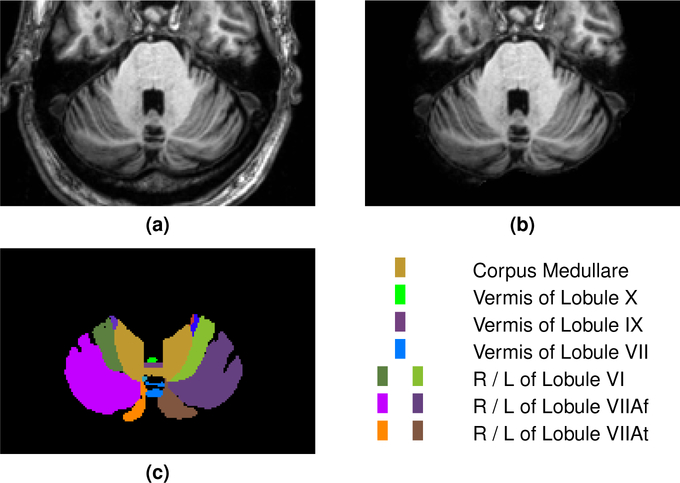
Our Adult Cohort contains 20 subjects, a mix of healthy controls and ataxia patients, each with 28 labeled cerebellar components (complete demographic information is provided in Table 3; see Fig. 2 for an example image and corresponding manual labels). Fifteen training examples were provided to the Participants, and the remaining five data sets were used for testing, with the goal being to label the cerebella of the test subjects to best agree with the expert labels. Magnetization prepared rapid gradient echo (MP-RAGE) images using a 3.0 T MR scanner (Intera, Phillips Medical Systems, Netherlands) were acquired with the following parameters: 1.1 mm slice thickness, 8° flip angle, TE = 3.9 ms, TR = 8.43 ms, FOV 21.2 × 21.2 cm, image matrix of 256 × 256. The images were resampled to have a 1.0 mm isotropic voxel; subsequently they were defaced using mri_deface from FreeSurfer (v5.3) (Fischl, 2012), a skull stripping mask was generated using SPECTRE (Carass et al., 2007, 2010), and the skull-stripped image was white matter (WM) peak normalized so that all images have a consistent WM peak intensity (Nyúl and Udupa, 1999a). For the training data, the defaced MR image, the WM peak skull stripped image, and the expert manual cerebellar parcellations were provided to the Participants. For the test subjects only the defaced MR image and the WM peak skull-stripped image were provided. All images in this cohort were acquired in an axial orientation. An example of both the defaced and WM peak skull stripped image for a data set are shown in Fig. 2 with the corresponding manual delineation.
Table 3:
Demographic details for the training and test data for both cohorts. The top line is the information of the entire data set, while subsequent lines within a section are specific to the patient diagnoses. N (M/F) denotes the number of patients and the male/female ratio, respectively. The Age column lists the mean, standard deviation, min, and max, in years, at scan time. The codes for the patient groups are: HC – Healthy controls; CB – Symptoms of cerebellar dysfunction without genetic diagnosis; SCA6 – Spinocerebellar ataxia type 6; ADHD – Attention-deficit and hyperactivity disorder; HFA – High-functioning Autism.
| Data Set | N (M/F) | Age | |
|---|---|---|---|
| Mean (SD) | [Min, Max] | ||
| Adult Cohort | |||
| Training | 15 (5/10) | 54.7(±11.97) | [30.0, 71.0] |
| HC | 6 (2/4) | 54.3(±14.69) | [30.0, 71.0] |
| CB | 3 (1/2) | 54.3(±8.02) | [46.0, 62.0] |
| SCA6 | 6 (2/4) | 55.3(±12.60) | [35.0, 70.0] |
| Testing | 5 (5/0) | 69.2(±5.81) | [62.0, 78.0] |
| CB | 5 (5/0) | 69.2(±5.81) | [62.0, 78.0] |
| Pediatric Cohort | |||
| Training | 20 (7/13) | 10.1(±1.36) | [8.3, 13.2] |
| HC | 10 (4/6) | 10.2(±1.33) | [8.4, 13.2] |
| ADHD | 7 (0/7) | 10.4(±1.61) | [8.3, 12.2] |
| HFA | 3 (3/0) | 9.2(±0.65) | [8.5, 9.7] |
| Testing | 10 (3/7) | 10.1(±1.29) | [8.4, 12.6] |
| HC | 5 (1/4) | 9.9(±1.04) | [8.4, 11.2] |
| ADHD | 3 (0/3) | 10.2(±1.06) | [9.2, 11.3] |
| HFA | 2 (2/0) | 10.6(±2.76) | [8.7, 12.6] |
Figure 2:
For our Adult Cohort, we show a cropped portion of a typical axial slice of (a) the defaced MP-RAGE, (b) the skull-stripped MP-RAGE, and (c) the manual labels with a corresponding color key for the prominent labels. The images are shown in radiological convention. A complete list of all the labels for the Adult Cohort is provided in Table 4. Results of the methods on the same data are shown in Fig. 4.
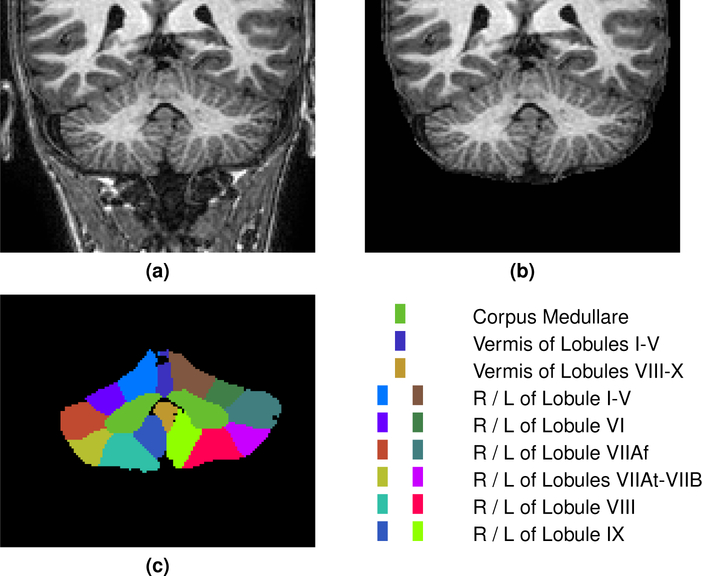
Our Pediatric Cohort comprises data collected at the Center for Neurodevelopmental and Imaging Research at the Kennedy Krieger Institute (PI: S.H. Mostofsky). These 30 expertly labeled data sets, with 18 labeled cerebellar components, are from 8–12 year old boys and girls with a mix of healthy controls, ADHD and high-functioning Autism (HFA) patients (complete demographic information is provided in Table 3). 20 of these were provided as training and 10 were reserved for testing. The objective was to label these cerebella to best agree with the expert labels. The provided MR images were MP-RAGE, acquired on a 3T Philips Gyroscan NT (Royal Philips Electronics) system with the following parameters: 1 mm slice thickness, 8° flip angle, TE = 3.0 ms TR = 7.0 ms, image matrix of 256 × 256. The Pediatric Cohort was preprocessed in an identical manner to our Adult Cohort; specifically, the images were defaced using mri_deface, skull-stripped using SPECTRE, and the skull-stripped image was WM peak normalized. For each of the 20 training images, the defaced MR image, the WM peak skull stripped image, and the expert manual cerebellar parcellation were provided to the Participants. For the test subjects only the defaced MR image and the WM peak skull stripped image were provided. All images in this cohort were acquired in a coronal orientation. An example of both the defaced and WM peak skull stripped image for a training data set are shown in Fig. 3 with the corresponding manual delineation. A complete list of the labels provided for the two cohorts is available in Table 4 and a key is provided in Table 1 to convert between the Schmahmann (Schmahmann et al., 2000) and classical (Malacarne, 1776; Henle, 1879) nomenclature.
Figure 3:
For our Pediatric Cohort, we show a cropped portion of a typical coronal slice of (a) the defaced MP-RAGE, (b) the skull-stripped MP-RAGE, and (c) the manual labels with a corresponding color key for the prominent labels. The images are shown in radiological convention. A complete list of all the labels for the Pediatric Cohort is provided in Table 4. Results of the methods on the same data are shown in Fig. 9.
2.2. Comparison Metric
To compare the results from the available methods with our expert delineations, we used the Dice overlap (Dice, 1945). The Dice overlap is a commonly used volume metric for comparing labels masks. If is the gold standard mask of a human rater and is the mask generated by a particular algorithm, then the Dice overlap for binary objects is computed as
where | · | is the cardinality (number of voxels). This overlap measure has values in the range [0,1], with 0 indicating no agreement between the two masks, and 1 meaning the two masks are identical. We have chosen to explicitly restrict our analysis to the Dice overlap for two reasons: 1) it is a widely reported and understood measure; 2) due to the large number of labels, the hierarchy of labels (from coarse to fine), the number of algorithms, and the two cohorts that we report on would make reporting multiple measures very lengthy. We note that in two recent challenge papers (Carass et al., 2017b; Maier et al., 2017) the final rankings of the methods—which used multiple metrics—were well correlated with the Dice overlap; see Table 7 in Maier et al. (2017) for example. A benefit of using a single measure in this manner is the clarity that is afforded in declaring a best method. We comment more on the pros and cons of this evaluation in Section 5.
Table 7:
For the Adult Cohort, we show the p-value for the two-sided Wilcoxon paired signed-rank test comparing the second ( LiviaNET) and third (
LiviaNET) and third ( DeepNet) placed teams to the top (
DeepNet) placed teams to the top ( CERES2) ranked team across the four hierarchies (Coarse, Lobe, Vermis, Lobule) of labeling and also the combination of all 38 labels (Consolidated). The mean Dice overlap for each method, at the respective hierarchy, is shown underneath the method’s name.
CERES2) ranked team across the four hierarchies (Coarse, Lobe, Vermis, Lobule) of labeling and also the combination of all 38 labels (Consolidated). The mean Dice overlap for each method, at the respective hierarchy, is shown underneath the method’s name.
| Hierarchy | p-value | ||
|---|---|---|---|
| Mean Dice Overlap | |||
| Coarse |
 CERES2
CERES20.9118 |
vs.  LiviaNET
LiviaNET0.8967 |
6.9 × 10−3 † |
vs.  DeepNet
DeepNet0.8908 |
6.1 × 10−5 ‡ | ||
| Lobe |
 CERES2
CERES20.8395 |
vs.  LiviaNET
LiviaNET0.8289 |
2.2 × 10−1 |
vs.  DeepNet
DeepNet0.8021 |
1.9 × 10−4 † | ||
| Vermis |
 CERES2
CERES20.8302 |
vs.  LiviaNET
LiviaNET0.8012 |
1.2 × 10−2 |
vs.  DeepNet
DeepNet0.8003 |
5.6 × 10−4 † | ||
| Lobule |
 CERES2
CERES20.7657 |
vs.  LiviaNET
LiviaNET0.7168 |
5.5 × 10−5 ‡ |
vs.  DeepNet
DeepNet0.7382 |
1.2 × 10−5 ‡ | ||
| Consolidated |
 CERES2
CERES20.8013 |
vs.  LiviaNET
LiviaNET0.7657 |
3.0 × 10−7 ‡ |
vs.  DeepNet
DeepNet0.7719 |
3.1 × 10−12 ‡ | ||
Denotes weak statistical significance (p-value < 0.001).
Denotes strong statistical significance (p-value < 0.0001).
3. Methods Overview
We introduce each method with a three line summary: the first line includes a colored square (that is used in subsequent plots and figures for quick reference) and the name of the method; second is a one line summary of the method; and finally in parentheses is the Participant(s) that contributed the method. Following each of the summaries is a brief overview of the respective method, a complete description of each method is available in Appendix A. A brief summary of each of the methods is provided in Table 5.
 SUIT
SUIT
Default SUIT v3.2
(Carlos H. Castillo)
Cerebellar isolation is performed using the unified segmentation (Ashburner and Friston, 2005) of SPM12. Then the cerebellar cortex is normalized into the spatially unbiased atlas template of the human cerebellum (SUIT) toolbox v3.2 (Diedrichsen et al., 2009), using a a fast-diffeomorphic normalization algorithm (DARTEL) (Ashburner, 2007). The probabilistic atlas included in the SUIT toolbox identify the cerebellar lobular boundaries. Then the inverse warp deformation field was calculated and then applied to map the SUIT atlas into a subject’s native space.
 C-SUIT
C-SUIT
Customized-SUIT (C-SUIT) with Corrections and Customized Atlas based on the Pediatric Cohort
(Paul Rasser)
The Pediatric data is preprocessed using FreeSurfer (v5.3) (FS) (Fischl, 2012) and then mapped into the SUIT space. In this space a customized pediatric atlas is constructed for application to the Pediatric Cohort. The ten test subjects from the Pediatric Cohort are also preprocessed with FreeSurfer and then diffeomorphically transformed into the customized atlas.
 FS-SUIT
FS-SUIT
FreeSurfer and SUIT in collaboration for Cerebellar Segmentation
(Melanie Ganz & Vincent Beliveau)
Whole brain and cerebellar GM and WM segmentation of structural MRI data was performed with FreeSurfer. FS-SUIT augments FreeSurfer with SUIT (v2.7) (Diedrichsen et al., 2009) to identify cerebellar lobules. To unify the results from FreeSurfer and SUIT into a coherent cerebellum lobule parcellation, a final segmentation is created by limiting the SUIT lobule parcellation to their intersection with the FreeSurfer cerebellar GM.
 LiviaNET
LiviaNET
Cerebellum parcellation from a deep learning perspective
(Jose Dolz, Ismail Ben Ayed, & Christian Desrosiers)
LiviaNET is built on the FCN described in Dolz et al. (2018) which had state-of-the-art performance for subcortical brain segmentation. To ensure that the network contains only convolutional layers, fully-connected layers are converted to a collection of 1 × 1 × 1 convolutions (Kamnitsas et al., 2017). As the structures in the cerebellum are often thinner than subcortical structures, to avoid losing small details when passing the target structures through several convolutional blocks, LiviaNET embeds the feature maps from all layers into the fully-connected layers.
 ConvNet
ConvNet
Cerebellum segmentation using convolutional neural networks
(Benjamin Thyreau)
ConvNet learns to segment the MRI using the expert labels as training data. ConvNet was intended to investigate whether whole-image input, as opposed to patch-based, could better capture high-level structure and human-expert variation. ConvNet took advantage of the overlap between the labeling schemes in both cohorts, which provided for data augmentation to train a base network that is then refined for the two cohorts separately.
 CERES2
CERES2
Cerebellum multi-atlas patch-based segmentation with a patch-based boosted neural network error corrector
(José E. Romero, Pierrick Coupé, & José V. Manjón)
A new version of CERES (Romero et al., 2017), which is a cerebellum lobule segmentation algorithm that is based on a recent method called Optimized Patch-Match Label fusion (OPAL) (Giraud et al., 2016; Ta et al., 2014) is presented. The method consists of a multi-atlas patch-based (Rousseau et al., 2011; Coupe et al., 2011) non-local label fusion technique that produces segmentations using a library of manually annotated cases. CERES2 improves on CERES by using a different intensity normalization method and by adding a systematic error correction step based on an ensemble of patch-based boosted neural networks.
 RASCAL
RASCAL
Patch-based label fusion
(Vladimir S. Fonov and D. Louis Collins)
The previously published RASCAL (Rapid Automatic Segmentation of the Human Cerebellum and its Lobules) (Weier et al., 2014) was adapted for use with the two cohorts. The data was preprocessed as follows: 1) linear registration to MNI-ICBM152 2009c stereotaxic space (Fonov et al., 2010); 2) linear intensity normalization based on quantile matching to normalize the intensity range to the MNI-ICBM152 2009c template; 3) extracted brain mask using thresholding of the provided SPECTRE brainmask; 4) created an unbiased population specific template (Fonov et al., 2010), the resultant template was used as a reference template for RASCAL.
 DeepNet
DeepNet
U-Net Parcellation of the Cerebellum
(Vladimir S. Fonov and D. Louis Collins)
DeepNet is an exploration of the potential of using an FCN based on U-net (Ronneberger et al., 2015; Çiçek et al., 2016) to parcellate the cerebellum.
4. Results
We present results using the Dice overlap measure to characterize the performance of the methods applied to both cohorts in our Comparison. Each Participating group provided a parcellation of the test data sets into lobules respecting the labeling scheme used in the respective cohort. To better characterize performance, we broke down the analysis using a hierarchical scheme. At the coarsest level we have the gross structures of the whole cerebellum, the whole vermis, and the corpus medullare (CM). We then have the subdivisions of the cerebellum into its left and right lobes; see Table 4 for the definitions of these structures for each cohort. The final two levels are the subdivisions of the vermis and the individual lobules, these are different for both cohorts—as the delineations draw distinctions between the vermis and the granularity with which the cerebellum compartments are identified. Specifically, for the Adult Cohort there are five subdivisions of the vermis and 22 lobule labels (11 per hemisphere), whereas for the Pediatric Cohort there are three vermal subdivisions and 14 lobule labels (seven per hemisphere). These levels are identified and defined as: 1) Coarse level which includes the whole cerebellum, whole vermis, and CM (3 labels); 2) Lobe level including the left and right of the four lobes (8 labels); 3) Vermis level which includes the vermal subdivisions of the vermis (5 labels for our Adult Cohort, 3 labels for our Pediatric Cohort); 4) Lobule level (22 labels for our Adult Cohort, 14 labels for our Pediatric Cohort); and a grouping listed as 5) Consolidated, which is a union of all the available labels (38 labels for the Adult Cohort, 28 labels for the Pediatric Cohort). These hierarchies have been generated (where necessary) based on the supplied parcellation of each algorithm by merging the appropriate labels; for example, the whole cerebellum label is given by merging all the labels. In Subsection 4.3, we summarize the Dice overlap results using the rank-sum to compare the performance of the various methods in a succinct manner. The rank-sum scoring assigns a score of 1 to the method with the highest mean Dice overlap measure, 2 to the second highest mean Dice overlap measure, et cetera, for each label. Table 6 provides a summary of the rank-sums for each of the hierarchies. The supplemental material includes details of the rank-sum calculation.
Table 6:
A summary of the rank-sum calculation for each of the hierarchies. The Coarse hierarchy includes three labels: whole cerebellum, whole vermis, and CM; the Lobe hierarchy includes eight labels: Left/Right Anterior Lobe, Left/Right Superior Posterior, Left/Right Inferior Posterior, and Left/Right Flocculonodular; the Vermis hierarchy is five labels for the Adult Cohort and three labels for the Pediatric Cohort (see Table 4 for details); the Lobule hierarchy contains 22 labels for the Adult Cohort and 14 labels for the Pediatric Cohort (see Table 4 for details). Complete rank-sum calculation is included in the supplemental material.
| 1st | 2nd | 3rd | 4th | 5th | 6th | 7th | 8th | ||
|---|---|---|---|---|---|---|---|---|---|
| Adult Cohort | Coarse |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 FS-SUIT
FS-SUIT
|
||
| Lobe |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 ConvNet
ConvNet
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 FS-SUIT
FS-SUIT
|
|||
| Vermis |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 FS-SUIT
FS-SUIT
|
|||
| Lobule |
 CERES2
CERES2
|
 Deep Net
Deep Net
|
 LiviaNET
LiviaNET
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 FS-SUIT
FS-SUIT
|
|||
| Consolidated |
 CERES2
CERES2
|
 Deep Net
Deep Net
|
 LiviaNET
LiviaNET
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 FS-SUIT
FS-SUIT
|
|||
| Pediatric Cohort | Coarse |
 LiviaNET
LiviaNET
|
 CERES2
CERES2
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 C-SUIT
C-SUIT
|
 FS-SUIT
FS-SUIT
|
 SUIT
SUIT
|
| Lobe |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 C-SUIT
C-SUIT
|
 ConvNet
ConvNet
|
 SUIT
SUIT
|
 FS-SUIT
FS-SUIT
|
|
| Vermis |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 ConvNet
ConvNet
|
 C-SUIT
C-SUIT
|
 SUIT
SUIT
|
 FS-SUIT
FS-SUIT
|
|
| Lobule |
 CERES2
CERES2
|
 Deep Net
Deep Net
|
 LiviaNET
LiviaNET
|
 RASCAL
RASCAL
|
 C-SUIT
C-SUIT
|
 ConvNet
ConvNet
|
 SUIT
SUIT
|
 FS-SUIT
FS-SUIT
|
|
| Consolidated |
 CERES2
CERES2
|
 LiviaNET
LiviaNET
|
 DeepNet
DeepNet
|
 RASCAL
RASCAL
|
 C-SUIT
C-SUIT
|
 ConvNet
ConvNet
|
 SUIT
SUIT
|
 FS-SUIT
FS-SUIT
|
|
4.1. Adult Cohort
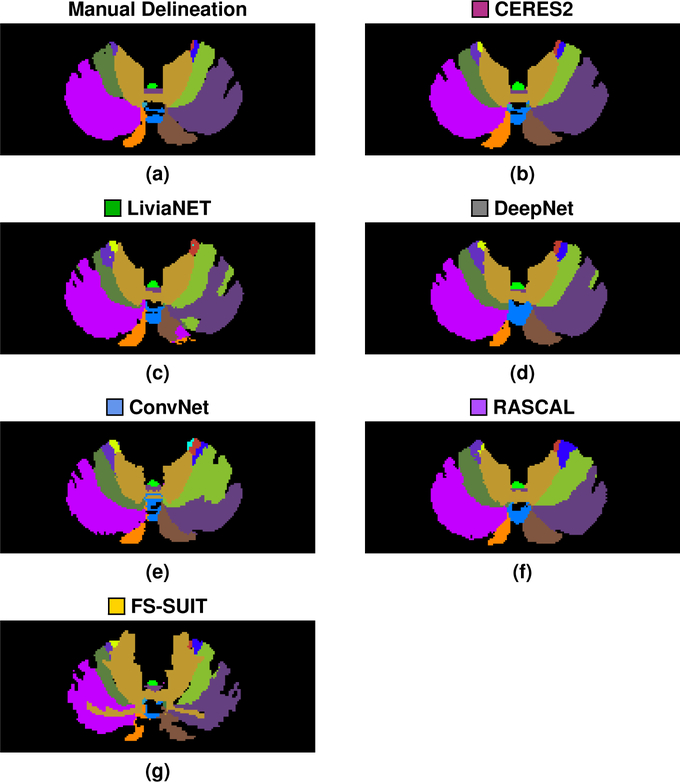
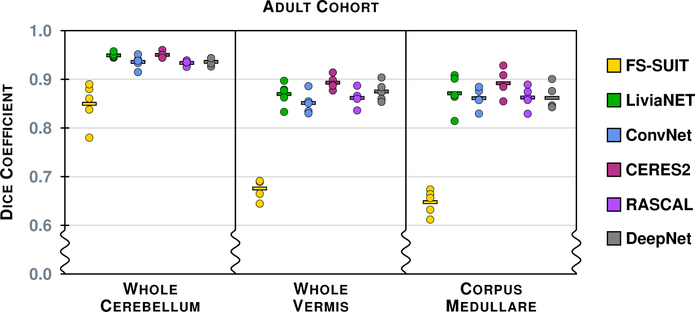
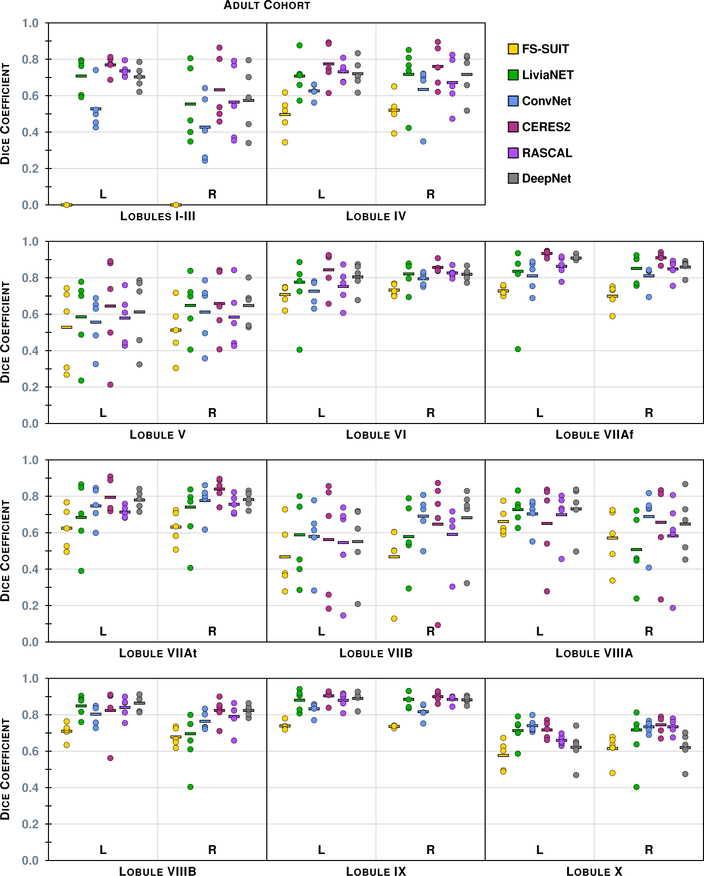
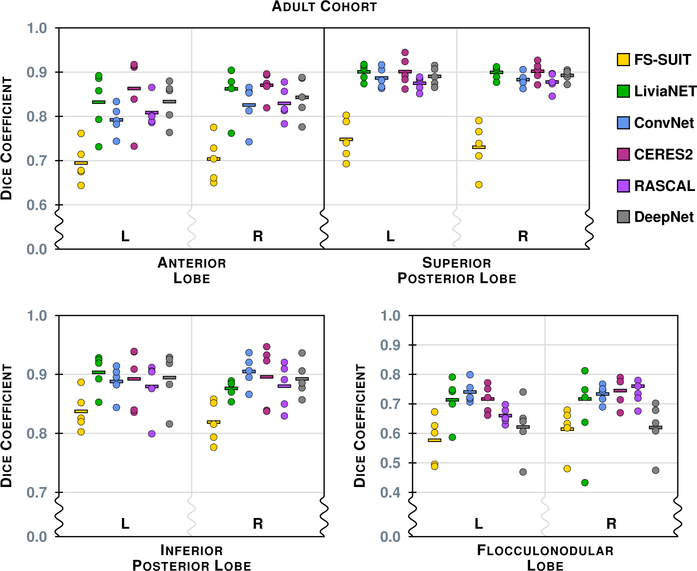
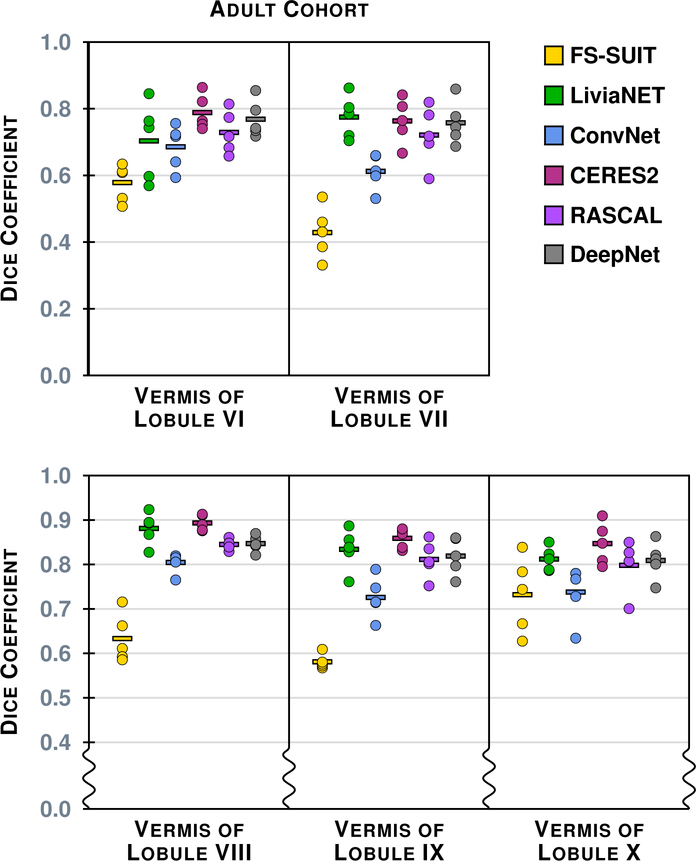
Figure 4 shows the results of the six methods on a typical axial slice from a test data set in the Adult Cohort: Fig. 2 shows the underlying MR data. Figures 5–8 show the Dice overlap for each of the methods across the various hierarchies; these plots show the individual data point for each of the five test data sets as well as showing the mean Dice overlap as a horizontal bar. Specifically, Fig. 5 shows the Dice overlap for the whole cerebellum, the whole vermis, and the CM. The mean Dice overlap of the methods on whole cerebellum was used to order the methods in Fig. 4. We can see that CERES2 has the highest mean Dice overlap for each of the Coarse labels; however, for the whole cerebellum label the difference between CERES2 and LiviaNET is quite small (0.950 vs. 0.949), though this is not the case for the other two Coarse Labels. This result sets the tone for many of the other labels in the Adult Cohort; in general for a given label the mean Dice overlap of CERES2 is the highest of the methods, with LiviaNET typically coming in second and on occasion the difference is negligible. Typical examples of this behavior are the Left and Right Anterior Lobe (Fig. 6), the Left and Right Superior Posterior Lobe (Fig. 6), Vermis of Lobules VIII through X (Fig. 7), and several cases in the Lobule hierarchy shown in Fig. 8. There are of course example of labels on which CERES2 does not achieve the maximum mean Dice overlap. See the Left and Right Inferior Posterior Lobe in Fig. 6, and the Vermis of Lobule VI in Fig. 7 for examples. In all 38 labels under consideration, there are 11 labels on which CERES2 is not ranked first; these 11 cases are split between LiviaNET (3 times), ConvNet (5 times), and DeepNet (3 times); see the supplemental material for complete details. We also observe in Figs. 6 and 8 that each algorithm has similar performance on both the left and right for each label. We make the observation that most of the methods have a mean Dice overlap above 0.8 for all the lobes except the Flocculonodular Lobe. For the vermal subdivisions, we see a slight degradation in results (mean Dice overlap in the range 0.7 to 0.9). Of course we see a further drop in performance when considering the lobe subdivisons, particularly for Lobules V, VIIB, and VIIIA. In fact, these lobules appear to be the most difficult to parcellate for all the methods; as each method has a large range of Dice overlap values for these regions.
Figure 4:
Shown for a test data set in the Adult Cohort are the (a) manual delineation, and the results for each of the methods: (b)
 CERES2; (c)
CERES2; (c)
 LiviaNET; (d)
LiviaNET; (d)
 DeepNet; (e)
DeepNet; (e)
 ConvNet; (f)
ConvNet; (f)
 RASCAL; and (g)
RASCAL; and (g)
 FS-SUIT, for the same axial slice shown in Fig. 2. The methods are ranked based on their mean whole cerebellum parcellation, see Fig. 5 for details.
FS-SUIT, for the same axial slice shown in Fig. 2. The methods are ranked based on their mean whole cerebellum parcellation, see Fig. 5 for details.
Figure 5:
The Dice overlap for the three labels associated with the Coarse hierarchy is shown for the Adult Cohort. Each column includes five data points, for the five test data sets in the Adult Cohort, showing the Dice overlap for a method-label pair (some of the data points are on top of one another and are thus occluded from view). The horizontal line in each column shows the mean Dice overlap for that particular method and label. We note that the scale has been zoomed to help appreciate the differences between the algorithms.
Figure 8:
The Dice overlap for the 22 labels (11 per hemisphere) associated with the Lobule hierarchy is shown for the Adult Cohort, see Table 4 for the list of lobule labels. See Fig. 5 for instructions on interpreting the plots.
Figure 6:
The Dice overlap for the eight labels associated with the Lobe hierarchy is shown for the Adult Cohort, see Table 4 for the list of lobe labels. We note that the scale has been zoomed to help appreciate the differences between the algorithms.
Figure 7:
The Dice overlap for the five labels associated with the Vermis hierarchy is shown for the Adult Cohort, see Table 4 for the list of vermis labels. See Fig. 5 for instructions on interpreting the plots. We note that some of the scale has been zoomed to help appreciate the differences between the algorithms.
4.2. Pediatric Cohort
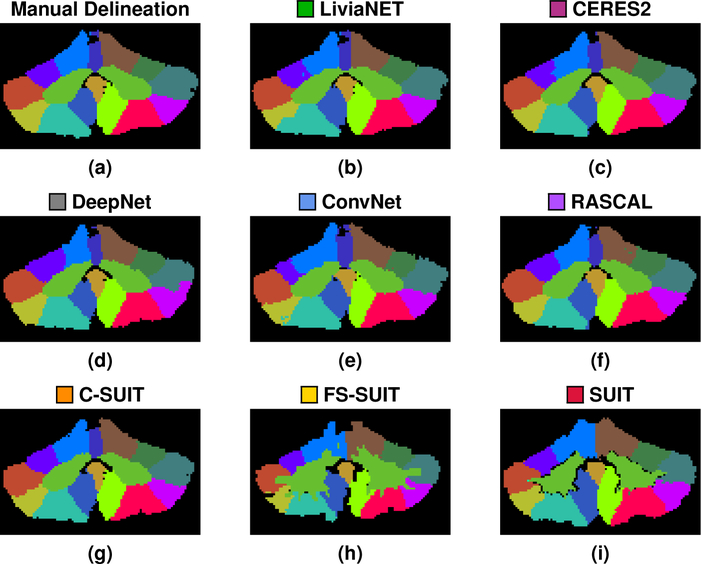
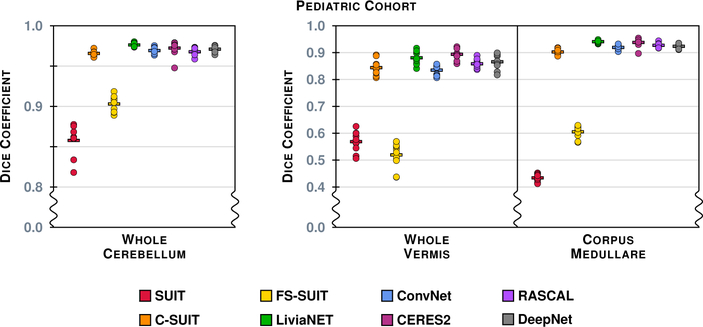
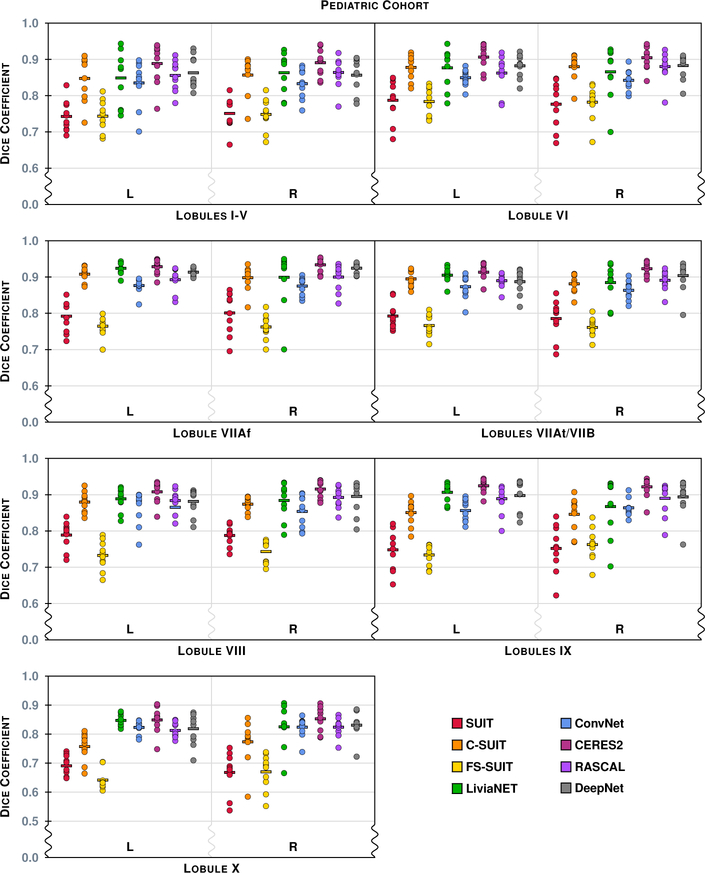
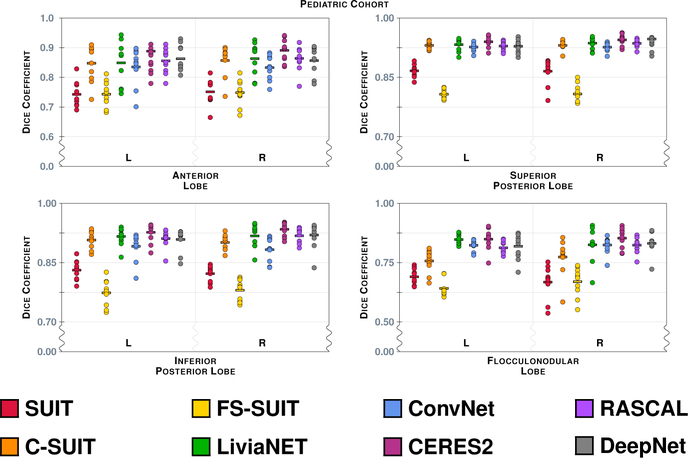
Figure 9 shows the results of the eight methods on a typical coronal slice from a test data set in the Pediatric Cohort, Fig. 3 shows the underlying MR data. Figures 10–13 show the Dice overlap for each of the methods across the various hierarchies; these plots show the individual data point for each of the ten test data sets as well as showing the mean Dice overlap as a horizontal bar. Specifically, Fig. 10 shows the Dice overlap for the whole cerebellum, the whole vermis, and the CM. The mean Dice overlap of the methods on the whole cerebellum was used to order the methods in Fig. 9. We can see that LiviaNET has the highest mean Dice overlap for the whole cerebellum and CM labels with CERES2 in second place; however, for the other coarse label the order of these two methods is reversed. In fact, unlike the Adult Cohort, where CERES2 was on top but definitely not unopposed, in the Pediatric Cohort CERES2 is quite dominant. The only labels for which it is not ranked first are the whole cerebellum and the CM. Similar to the Adult Cohort, we observe in Figs. 11 and 13 for the Pediatric Cohort that each algorithm performs consistently on both the left and right for each label.
Figure 9:
Shown for a test data set in the Pediatric Cohort are the (a) manual delineation, and the results for each of the methods: (b)
 LiviaNET; (c)
LiviaNET; (c)
 CERES2; (d)
CERES2; (d)
 DeepNet; (e)
DeepNet; (e)
 ConvNet; (f)
ConvNet; (f)
 RASCAL; (g)
RASCAL; (g)
 C-SUIT; (h)
C-SUIT; (h)
 FS-SUIT; and (i)
FS-SUIT; and (i)
 SUIT, for the same coronal slice shown in Fig. 3. The methods are ranked based on their mean whole cerebellum parcellation (see Fig. 10 for details).
SUIT, for the same coronal slice shown in Fig. 3. The methods are ranked based on their mean whole cerebellum parcellation (see Fig. 10 for details).
Figure 10:
The Dice overlap for the three labels associated with the Coarse hierarchy are shown for the Pediatric Cohort. Each column includes ten data points, for the ten test data sets in the Pediatric Cohort, showing the Dice overlap for a method-label pair (some of the data points are on top of one another and are thus occluded from view). The horizontal line in each column shows the mean Dice overlap for that particular method and label. We note that the scale has been zoomed to help appreciate the differences between the algorithms.
Figure 13:
The Dice overlap for the 14 labels (7 per hemisphere) associated with the Lobule hierarchy are shown for the Pediatric Cohort, see Table 4 for the list of lobule labels. See Fig. 10 for instructions on interpreting the plots. We note that the scale has been zoomed to help appreciate the differences between the algorithms.
Figure 11:
The Dice overlap for the eight labels associated with the Lobe hierarchy are shown for the Pediatric Cohort, see Table 4 for the list of lobe labels. See Fig. 10 for instructions on interpreting the plots. We note that the scale has been zoomed to help appreciate the differences between the algorithms.
4.3. Summary and Further Analysis
To create a readily interpretable representation of these results we computed the rank-sum for each method over the various hierarchies and both cohorts. These rank-sum results are presented in Table 6, with the details of the computation included in the supplemental material. Over both cohorts, we can easily discern some patterns in Table 6: clearly CERES2 is the overall winner, with LiviaNET and DeepNet trading back and forth between second and third place. We also see RASCAL is quite consistently fourth in both cohorts. Given the outcome of our rank-sum analysis, we identify the top three methods as CERES2, LiviaNET, and DeepNet. We next want to determine if there is a statistically significant difference between these top three methods. To this end, we use a two-sided Wilcoxon paired signed-rank test (Wilcoxon, 1945) between CERES2 & LiviaNET, and between CERES2 & DeepNet, to establish statistical significance. The Wilcoxon test is a nonparametric test of the null hypothesis that the two samples come from the same population against an alternative hypothesis. We tested using all the available Dice overlap values for a particular hierarchy; thus for the Coarse level on the Adult Cohort there are 15 values for each method (3 labels × 5 data sets). For the statistical comparisons we use an α level of 0.001 to note weak statistical significance and an α level of 0.0001 to denote strong statistical significance; we use these α values as we do not employ any multiple comparison correction techniques. The p-values for the Wilcoxon test and the mean values for the Dice overlap (for our top three methods) are shown in Table 7 for the Adult Cohort and Table 8 for the Pediatric Cohort. For the five hierarchies (Coarse, Lobe, Vermis, Lobule, and Consolidated) on the Adult Cohort CERES2 has the highest mean Dice overlap on all five hierarchies and is statistically significantly different on eight of the ten comparisons (with strong significance in five instances). The two cases where there is no statistically significant difference are between CERES2 and LiviaNET for the Lobe and Vermis hierarchies. For the Pediatric Cohort CERES2 has the highest mean Dice overlap on all five hierarchies and is statistically significantly different on nine of the ten comparisons (with strong significance in all nine cases). The single comparison for which there is not significance is between CERES2 and LiviaNET on the Coarse hierarchy.
Table 8:
For the Pediatric Cohort, we show the p-value for the two-sided Wilcoxon paired signed-rank test comparing the second ( LiviaNET) and third (
LiviaNET) and third ( DeepNet) placed teams to the top (
DeepNet) placed teams to the top ( CERES2) ranked team across the four hierarchies (Coarse, Lobe, Vermis, Lobule) of labeling and also the combination of all 28 labels (Consolidated). The mean Dice overlap for each method, at the respective hierarchy, is shown underneath the methods name.
CERES2) ranked team across the four hierarchies (Coarse, Lobe, Vermis, Lobule) of labeling and also the combination of all 28 labels (Consolidated). The mean Dice overlap for each method, at the respective hierarchy, is shown underneath the methods name.
| Hierarchy | p-value | ||
|---|---|---|---|
| Mean Dice Overlap | |||
| Coarse |
 CERES2
CERES20.9348 |
vs.  LiviaNET
LiviaNET0.9326 |
2.1 × 10−1 |
vs.  DeepNet
DeepNet0.9201 |
6.0 × 10−6 ‡ | ||
| Lobe |
 CERES2
CERES20.9033 |
vs.  LiviaNET
LiviaNET
0.8859 |
7.4 × 10−6 ‡ |
vs.  DeepNet
DeepNet0.8827 |
4.9 × 10−7 ‡ | ||
| Vermis |
 CERES2
CERES20.8763 |
vs.  LiviaNET
LiviaNET
0.8491 |
2.7 × 10−5 ‡ |
vs.  DeepNet
DeepNet0.8427 |
7.5 × 10−5 ‡ | ||
| Lobule |
 CERES2
CERES20.9043 |
vs.  LiviaNET
LiviaNET0.8776 |
1.6 × 10−11 ‡ |
vs.  DeepNet
DeepNet0.8808 |
1.4 × 10−12 ‡ | ||
| Consolidated |
 CERES2
CERES20.9043 |
vs.  LiviaNET
LiviaNET
0.8828 |
2.2 × 10−16 ‡ |
vs.  DeepNet
DeepNet0.8815 |
2.2 × 10−16 ‡ | ||
Denotes weak statistical significance (p-value < 0.001).
Denotes strong statistical significance (p-value < 0.0001).
To understand the inherent bias of any of the presented methods we have generated two bias plots (similar to Bland-Altman plots (Bland and Altman, 1986)), which are included in the supplemental material. Traditional Bland-Altman plots show the difference of two measurements vs. the mean for the same two measurements. To allow us to present the various methods on a single plot and to reflect our higher confidence in the manual delineation, we plot the volumetric difference between each method and the manual delineation vs. the volume identified by the manual delineation. In this way, all of the methods can be shown on a single plot and the differences of each method on a particular subject are directly comparable. The presented bias plots, included in supplemental material, are for the whole cerebellum label on the Adult and Pediatric Cohorts. These plots, included as Figs. 1 and 2 in the supplemental material, are indicative of the behavior of the methods across all the labels; in that FS-SUIT and C-SUIT have positive biases, while SUIT has a negative bias, and the other methods do not exhibit a consistent bias across labels.
5. Discussion and Conclusions
5.1. Ranking the Methods
The primary result of this Comparison is a ranking of the state-of-the-art methods for parcellating the cerebellum, which is summarized in Table 6 for both the Adult and Pediatric Cohorts. The different levels of labeling, which we have referred to as hierarchies, allows for some granularity in understanding the ranking of the various methods on our cohorts. Had all the Participants contributed results for the two cohorts it would have been feasible to merge the rankings; regardless of this, there is an obvious stratification that occurs across both cohorts that is almost independent of the hierarchy. We observe that the order of CERES2, LiviaNET, DeepNet, and RASCAL (as first through fourth) is very stable across both cohorts and the hierarchies. This is quite pleasing, as it points to a stability of both the algorithms and the labeling schemes used on both cohorts—even though the cohorts were labeled independently. We observe that these top methods all used spatial and intensity normalization to the MNI space.
Clearly improvements in the mean Dice overlap of 0.01 could be considered marginal, possibly even negligible, however the two-sided Wilcoxon paired signed-rank test establishes the results of CERES2 as being a statistical improvement over the second and third place methods of LiviaNET and DeepNet (see Tables 7 and 8). Other metrics may provide some subtle insight into the differences of these approaches that the Dice overlap cannot distinguish, however we note that recent work (Maier-Hein et al., 2018) has shown that the median Dice overlap is the most stable manner in which to evaluate challenge winners. The important point of this work is that all three of these methods provide a high level of accuracy in parcellating both the adult and pediatric cerebellum. This provides an opportunity for detailed analyses of the cerebellum on an unprecedented scale.
5.2. Criticisms
The current work has two major shortcomings: 1) flawed cohorts and 2) exclusive use of Dice overlap. The two cohorts are flawed in different ways. Firstly, the Adult Cohort while having a rich label set (CM label, five vermal labels, and 22 lobule labels) provided only five test data sets each of which showed signs of cerebellar dysfunction without a genetic diagnosis. In particular, the test data for the Adult Cohort had a mean age of 69.2 years of age, whereas the training data had a mean age of 54.7 years of age (see Table 3). A two-sided Wilcoxon signed-rank test (Wilcoxon, 1945) between the ages of the training and testing portions of the Adult Cohort has a p-value of 0.02, not significant but not a satisfactory situation either. The other issues with the Adult Cohort are its gender bias (all male test data versus training data that is only one third male) and the small size of the test data (N = 5). The effects of the gender bias are an unknown and the cohort size limits the statistical power of any tests. The cohort size also reduced the organizers’ willingness to report standard deviations for the Dice overlap, with such a small sample any reported standard deviations would be erroneous. In contrast, to the Adult Cohort, the Pediatric Cohort has a slightly smaller label set (CM label, three vermal labels, and 18 lobule labels), a larger training pool of 20 data sets and a larger testing pool with 10 data sets. The gender proportions are consistent throughout the training and testing data sets as well as throughout the disease classifications in both the training and testing data. When using a two-sided Wilcoxon signed-rank test to perform a comparison between the ages of the training and test data, we get a more pleasing p-value of 0.95. The unfortunate drawback of the Pediatric Cohort is that it is pediatric data. The pediatric cerebellum is an area of great potential research and the availability of these automated methods for future work is very promising. However, the pediatric cerebellum remains an understudied portion of the central nervous system. The organizers believe that the pooling of these two cohorts to validate these methods is still a comprehensive test for any cerebellum parcellation method.
Since both our data sets were acquired using Philips scanners, our study cannot be used to assess the robustness and stability across scanners. Additionally, our study did not explore the ability of any of these methods to identify group differences between populations; our cohorts’ size and composition did not permit this. We note that the top ranked method, LiviaNET, was validated on the Autism Brain Imaging Data Exchange (ABIDE) (Martino et al., 2014). LiviaNET used ABIDE I, which included 17 international sites consisting of 1112 individuals (539 with autism spectrum disorder and 573 healthy controls, individuals were between 6 and 64 years of age at scan time) and the authors claim that this demonstrated a robustness “to various acquisition protocols, demographics, and clinical factors” (Dolz et al., 2018). We further note that several of the included methods have been used on other data sources with high quality results (Romero et al., 2017; Weier et al., 2014), and studies based on the submitted methods have previously explored group differences (Bernard et al., 2015; Weier et al., 2016). However, we do not make any claims of robustness to data or efficacy for group comparisons for any of the reported methods.
The remaining concern is the exclusive use of the Dice overlap measure throughout the paper. If we ignore the hierarchical label evaluation we employed, there were 28 labels in the Adult Cohort and 22 labels in the Pediatric Cohort. Given this many labels it seemed impractical to the organizers to report multiple metrics. Moreover, it would have been quite difficult to develop a consensus as to how to combine such metrics in a meaningful and unbiased manner. We also note that the majority of papers comparing multiple algorithms, as this paper does, are focused on a small number of labels. In fact it is typical for there to be only one label under consideration: white matter lesions, for example (Styner et al., 2008). As organizers, we observed in Maier et al. (2017) (from Table 7) that the final ranking correlated with the mean Dice overlap; in fact, the mean Dice overlap correctly predicts the top three methods and only incorrectly ranks three of the fourteen methods under consideration. This occurs despite the fact that the Dice overlap is only one component of a multi-measure evaluation (Maier et al., 2017). Thus, we believe exclusive use of the Dice overlap is acceptable and that our analysis of this Comparison correctly represents the state-of-the-art in fully automated cerebellum parcellation.
There are three more (minor) concerns—image quality, subsequent analyses, and the use of FreeSurfer v5.3—each of which we comment on below. A limitation in both of our cohorts was quality assessment (QA) of the images, which was limited to ensuring images were free from artifacts. It is possible that other more subtle quality issues may have contributed to errors in generating the manual delineations. Our basic review was not a comprehensive QA, like MRI-QC (Esteban et al., 2017), which might identify an image that was not acquired in a manner consistent with the other data in the cohort. A further consideration, that is not covered by MRI-QC, would be a cerebellum specific processing assessment (Li et al., 2016; Zuo et al., 2018) that would highlight other issues due to preprocessing.
This paper has focused on the automated parcellation of the cerebellum to facilitate streamlined regional analyses. Such regional analyses can be used as a normalizing factor in functional MRI (Barrett et al., 2017) and positron emission tomography data (Murphy et al., 2013), and for studying the changing shape of the cerebellum in disease (Abulnaga et al., 2016; Kansal et al., 2016). However, there is continued interest in voxel based morphometry (VBM) analyses of the cerebellum (Colloby et al., 2014). In this regard several of the presented methods are deficient, as there is no convenient interpretation between the provided parcellation and a common atlas space that would lend itself to a VBM style study. This is an area for potential future research. Finally, two of the included methods used FreeSurfer v5.3 as part of their processing pipeline, however a newer version of FreeSurfer v6.0 is available. Based on the ChangeLog available for FreeSurfer, it was not immediately obvious of any improvement that would benefit either of these methods.
5.3. Comment on Inter-rater Performance
A portion of our Adult Cohort, along with other similarly acquired data, was used as part of an inter-rater comparison (Bogovic et al., 2013b). It is reassuring to see that the performance of the top methods in this Comparison have similar Dice overlap to that reported for the inter-rater analysis. In particular, the mean Dice overlap for CERES2, LiviaNET, and DeepNet, for the whole vermis are larger than those reported for the inter-rater values (Fig. 5 in Bogovic et al. (2013b)).
5.4. Impact of this Work
Several of the methods in this Comparison are either readily available for use through download or web interface. In particular, the top two methods are accessible to the community, CERES2 can be used through a web portal2 and LiviaNET is available for download3. Identifying the state-of-the-art in cerebellum parcellation is important for improving the robustness and speed with which cerebellum imaging studies can be completed. Although SUIT (Diedrichsen, 2006; Diedrichsen et al., 2009) has been available and widely used for over 10 years, our study clearly reveals that there are emerging methods with significantly better performance (given our performance criteria); we note that the probabilistic lobular segmentation generated by SUIT was meant to be informative and not definitive. As studies begin to emerge relating the volumes of cerebellar lobules to functional brain performance (cf. Kansal et al. (2016)), methods such as CERES2, LiviaNET, and DeepNet may offer a better alternative for identifying these volumes. As well, this study provides a baseline for future work on cerebellar parcellation, both in providing information on the best strategies to date and in providing Dice coefficients for comparison.
Supplementary Material
Figure 12:
The Dice overlap for the three labels associated with the Vermis hierarchy are shown for the Pediatric Cohort, see Table 4 for the list of vermis labels. See Fig. 10 for instructions on interpreting the plots.
Acknowledgments
The data collection and labeling of the cerebellum was supported in part by the NIH/NINDS grant R01 NS056307 (PI: J.L. Prince) and NIH/NIMH grants R01 MH078160 & R01 MH085328 (PI: S.H. Mostofsky). PMT is supported in part by the NIH/NIBIB grant U54 EB020403. CERES2 development was supported by grant UPV2016-0099 from the Universitat Politécnica de Valencia (PI: J.V. Manjón); the French National Research Agency through the Investments for the future Program IdEx Bordeaux (ANR-10-IDEX-03-02, HL-MRI Project; PI: P. Coupé) and Cluster of excellence CPU and TRAIL (HR-DTI ANR-10-LABX-57; PI: P. Coupé). Support for the development of LiviaNET was provided by the National Science and Engineering Research Council of Canada (NSERC), discovery grant program, and by the ETS Research Chair on Artificial Intelligence in Medical Imaging. The authors wish to acknowledge the invaluable contributions offered by Dr. George Fein (Dept. of Medicine and Psychology, University of Hawaii) in preparing this manuscript.
Appendix A. Methods
Here we provide detailed descriptions of all the methods used in the Comparison. We introduce each method with a three line summary: the first line includes a colored square (that is used in subsequent plots and figures for quick reference) and the name of the method; second is a one line summary of the method; and finally in parentheses is the Participant(s) that contributed the method.
 SUIT
SUIT
Default SUIT v3.2
(Carlos H. Castillo)
Data analysis were performed using MATLAB R2015b (The Mathworks Inc. Natick, MA), SPM12 (Ashburner et al., 2000), and the spatially unbiased atlas template of the human cerebellum (SUIT) toolbox v3.2 (Diedrichsen et al., 2009). To achieve the best performance from SUIT, all anatomical images were first reoriented into LPI (Neurological) orientation and then the origin of each T1-w image was assigned to the manually selected anterior commissure.
To ensure the correct normalization of the cerebellar cortex into the atlas template, SUIT first isolates the infra-tentorial structures from the rest of the brain. This is important because the occipital cortex has a similar intensity as the cerebellum and in most cases there is not a clearly visible separation between these two structures. SUIT v3.2 achieves this separation by using the unified segmentation (Ashburner and Friston, 2005) of SPM12; this segmentation procedure combines tissue classification and registration by means of both a mixture of Gaussians and tissue probability maps. Using this technique, the brain is segmented into eight tissue types: cerebral GM, cerebral WM, cerebellar GM, cerebellar WM, cerebrospinal fluid (CSF), bone, fat/skin, and air. Finally, a binary cerebellar mask is created by combining the cerebellar GM and WM segmentation maps including voxels with a tissue probability of greater than or equal to 90% of coming from either of those classes.
After the cerebellar isolation, SUIT uses a fast-diffeomorphic normalization algorithm (DARTEL) (Ashburner, 2007). DARTEL uses the probabilistic GM and WM segmentation maps to align the anatomy of the cerebellum of each participant to the SUIT atlas template. To increase the speed of the process, the non-linear registration is solved using a Levenberg-Marquardt strategy and a multigrid method; see Ashburner (2007) for complete details. The result is a non-linear deformed image coregistered to the SUIT atlas template and its respective deformation field.
To identify the cerebellar lobular boundaries, the probabilistic atlas of the cerebellum included in the SUIT toolbox was used. The SUIT atlas consists of a set of 34 probabilistic maps that indicates the likelihood that a certain voxel in the reference space belongs to each lobule. The SUIT atlas includes the cerebellar left and right lobules (I–IV, V, VI, Crus I, Crus II, VIIb, VIIIa, VIIIb, IX, X), vermis (VI, Crus I, Crus II, VIIb, VIIIa, VIIIb, IX, and X), and deep cerebellar nuclei. For this work, these compartments were combined to have only 18 labels (I–V, VI, Crus I, Crus II–VIIb, VIII, IX, X, Vermis I–V, Vermis VI–VII, Vermis VIII–X, and corpus medullare). For each subject, the inverse warp deformation field was calculated and then applied to the SUIT atlas using a nearest neighbor approach, so the values of each label were preserved. For each voxel one label was assigned depending on the maximum probability of the SUIT atlas, resulting in a lobular segmentation of the subject’s native space.
 C-SUIT
C-SUIT
Customized-SUIT (C-SUIT) with Corrections and Customized Atlas based on the Pediatric Cohort
(Paul Rasser)
FreeSurfer (v5.3) (FS) (Fischl, 2012) was used to preprocess the images from the Pediatric Cohort training set by performing bias correction, intensity normalization, and skull stripping on the provided defaced MP-RAGE images. A 6-paramerter transformation from their original defaced MP-RAGE to the conformed FreeSurfer space was found using minctracc (Collins et al., 1994) and was applied to both the defaced and the manually parcellated volumes. The parcellated volumes were used to correct differences in cerebellum GM as defined by the FreeSurfer subcortical segmentation output. SPM (Ashburner et al., 2000) was used to correct the coordinate system of the data sets to match the requirements of SUIT (Diedrichsen et al., 2009). The correction was followed by application of SUIT to provide an initial SUIT cerebellum mask that was subsequently corrected to conform to the cerebellar volume as defined by the corrected subcortical segmentation of FreeSurfer.
ANTs (Avants et al., 2008)—using the fast cross correlation metric—was used to find a symmetric diffeomorphic transformation between the normalized and bounded MP-RAGE to the SUIT space. All subject images (MP-RAGE, parcellated cerebellum, cerebellum binary mask) were transformed to the SUIT space. FreeSurfer was then used to create a bounded normalized atlas in SUIT space as well as a cerebellum mask in SUIT space by combining the output from 19 of the 20 training subjects from the Pediatric Cohort. The 20th subject was excluded from this atlas construction step due to its poor registration with the SUIT space. In the SUIT space, the 18 labels from the 19 subjects in the Pediatric Cohort were combined to create a customized parcellation using mri_concat from FreeSurfer.
To apply the customized atlas to the remaining ten subjects in the Pediatric Cohort, C-SUIT first preprocesses the ten subjects using FreeSurfer to perform bias correction, intensity normalization, and skull stripping. Preprocessing the data in this manner provided a consistency between the volumetric measures based on the cerebellar parcellation and the existing cerebral measures derived from FreeSurfer. This feature was included to provide a supplementary cerebellar parcellation for projects with existing measures of the cerebrum derived from FreeSurfer, such as the ENIGMA Project4.
Using SUIT the preprocessed MP-RAGED is bounded, then ANTs estimates a symmetric diffeomorphic transformation into the customized atlas. This is then applied to the normalized and bounded MP-RAGE. The inverse of this symmetric diffeomorphic transformation was applied to the average cerebellum hemisphere mask in SUIT space and used to create a binary SUIT style cerebellum mask. The SUIT command suit_normalize was applied to the bounded and normalized volume and its mask, with the inverse of this transformation applied to the customized cerebellum parcellation. As the cerebellum parcellation was required in the native space, the test subject’s cerebellum parcellation was transformed from the bounded space to their native space using fsl_rigid_register supplied with the FreeSurfer package. Finally, to remove any potential outliers in the cerebellum parcellation, FSL’s FAST tissue segmentation algorithm (Zhang et al., 2001) was used to create GM and WM masks that were applied to the final cerebellum parcellation.
 FS-SUIT
FS-SUIT
FreeSurfer and SUIT in collaboration for Cerebellar Segmentation
(Melanie Ganz & Vincent Beliveau)
The approach considered in this work combines FreeSurfer (v5.3) (Fischl, 2012) and SUIT (Diedrichsen et al., 2009), to overcome some of the limitations of these algorithms when used independently. Whole brain and cerebellar GM and WM segmentation of structural MRI data was performed with FreeSurfer. FreeSurfer processing included motion correction, removal of non-brain tissue, automated Talairach transformation, segmentation of subcortical WM and deep GM volumetric structures (including hippocampus, amygdala, caudate, putamen, ventricles) (Fischl et al., 2002, 2004), intensity normalization, and further cortical surface processing. FreeSurfer’s cerebellum segmentation is driven by a probabilistic atlas segmentation (Fischl et al., 2002). The procedure maintains tissue class statistics (e.g., means and variances of the MRI intensities of a given neuroanatomical structure) on a per-location per-class basis throughout an atlas space. In addition, local spatial relationships between structures are encoded in a Markov random field (MRF). FreeSurfer has been demonstrated to have good test-retest reliability across scanner manufacturers and field strengths (Han et al., 2006).
As FreeSurfer only provides a segmentation of the cerebellar GM and WM, FS-SUIT augments it with SUIT (v2.7) (Diedrichsen et al., 2009) to identify cerebellar lobules. SUIT is available as a compatible toolbox for SPM12 (Ashburner et al., 2000) which allows for the creation of a segmentation of the cerebellum into different lobules. SUIT consists of three steps: 1) cerebellum isolation; 2) normalization to SUIT atlas space; 3) reslicing into SUIT atlas space/individual subject space. SUIT uses SPM to perform the cerebellum isolation by segmenting the brain into tissue-types. The volume is cropped to include anything inferior to the tentorium cerebelli. The tissue-types are used to compute posterior probability for each voxel. The normalization to SUIT atlas space is performed by a nonlinear deformation map to the SUIT template using the cosine-basis function approach (Ashburner and Friston, 1999). Finally, SUIT applies the estimated deformation to map the subject into the SUIT atlas space. In SUIT the atlas is spatially unbiased.
To unify the results from FreeSurfer and SUIT into a coherent cerebellum lobule parcellation, a final segmentation is created by limiting the SUIT lobule parcellation to their intersection with the FreeSurfer cerebellar GM. While SUIT yields a segmentation of the cerebellar lobules, it largely ignores the individual WM and GM intensities. Whereas FreeSurfer provides a more accurate representation of the cerebellar WM and GM tissue classes, but it is also more sensitive in regions of low contrast between tissue types and tends to over-label peripheral tissue. Thus, the intersection provides a refinement of the GM and WM segmentation while reducing the over-labeling of peripheral tissue. Correspondingly, due to the restriction of the SUIT labels to FreeSurfer cerebellar GM, FS-SUIT uses the FreeSurfer WM to label the CM; otherwise there would be gaps between the lobule labels and the CM.
 LiviaNET
LiviaNET
Cerebellum parcellation from a deep learning perspective
(Jose Dolz, Ismail Ben Ayed, & Christian Desrosiers)
Originally designed for image recognition and classification tasks, convolutional neural networks (CNNs) are now commonly employed for semantic segmentation. The most naive approach follows a sliding-window strategy where regions defined by the window are processed one-by-one. This technique presents two main drawbacks: 1) processing image regions independently provides non-structured output, which reduces segmentation accuracy; and 2) due to many redundant convolution and pooling operations, the process is inefficient. To mitigate these limitations, the spatial map of class probabilities can be obtained in a single, dense inference step. This approach, known as a fully convolutional neural network (FCN) (Long et al., 2015), represents the network as a single non-linear convolution, which is trained end-to-end. Unlike the sliding-window approach, FCNs can avoid redundant convolution and pooling operations, making them computationally more efficient. Additionally, fully convolutional networks have an order of magnitude fewer coefficients, which makes them easier to train with fewer training samples.
The proposed method, which is built on top of DeepMedic (Kamnitsas et al., 2017), is based on the FCN described in Dolz et al. (2018) which had state-of-the-art performance for subcortical brain segmentation. This network is composed of 13 layers in total: 9 convolutional layers, followed by 3 fully-connected layers, and the classification layer. The number of kernels in each convolutional layer—from shallow to deep—is 25, 25, 25, 50, 50, 50, 75, 75, and 75, respectively. The kernel sizes are equal to 3 × 3 × 3 in all the convolutional layers. Three fully-connected layers, composed of 400, 200, and 150 hidden units each, are added after these convolutional layers for encoding semantic information. To ensure that the network contains only convolutional layers, fully-connected layers are converted to a collection of 1 × 1 × 1 convolutions (Kamnitsas et al., 2017). Dolz et al. (2018) described two intermediate-layer outputs (i.e., feature maps) that were embedded in the final predictions, encouraging consistency between features extracted at different scales, while injecting fine-grained information directly in the segmentation process. As the structures in the cerebellum are often thinner than subcortical structures, to avoid losing small details when passing the target structures through several convolutional blocks, LiviaNET embeds the feature maps from all layers into the fully-connected layers.
Due to computation and memory limitations, the LiviaNET network cannot apply dense training over the whole 3D input volume. Instead, LiviaNET sub-samples this volume into S smaller sub-volumes, which are then fed into the network. In this way: 1) LiviaNET avoids memory issues when the input is not down-sampled (as in this work), and 2) LiviaNET has a high number of samples from each image, removing the need for data augmentation. A Parametric Rectified Linear Unit (PReLU) (He et al., 2015), which applies an element-wise activation function, follows each convolutional layer. Let θ be the network trainable parameters, and as the set of ground-truth labels such that represents the label of voxel v in the sth sampled sub-volume for all the predicted voxels V. The cost function is
where is the output of the classification layer for voxel v in the segment s (i.e. softmax output) given their input feature maps, Xv. This cost function corresponds to the mean cross-entropy computed over all voxels and sub-volumes. Sample sizes were set to be larger for inference than in training (Dolz et al., 2018). In this particular application, the following combination was found to give satisfactory results: 27 × 27 × 27 for training and 35 × 35 × 35 for testing.
Optimization of network parameters was performed via the RMSprop optimizer (Tieleman and Hinton, 2015). Momentum was set to 0.6 and the initial learning rate was set to 0.001; the latter was reduced by a factor of 2 after every 5 epochs (starting from epoch 10). Weights in layer l were initialized based on a zero-mean Gaussian distribution with standard deviation , where nl denotes the number of connections to units in that layer. LiviaNET was trained for 35 epochs, with each epoch composed of 20 sub-epochs. At each sub-epoch, a total of 1000 samples were randomly selected from the training images, and processed in batches of size 10. The network architecture was developed using Theano (Bergstra et al., 2010), with modifications being made through LiviaNET 5. LiviaNET was used on a server with a NVIDIA Tesla P100 GPU and 16 GB of RAM; training took approximately 15 hours for LiviaNET (taking 25 minutes per epoch).
 ConvNet
ConvNet
Cerebellum segmentation using convolutional neural networks
(Benjamin Thyreau)
The basis of this work is a convolutional neural network (ConvNet) that learns to segment the MRI using the expert labels as training data. ConvNet was intended to investigate whether whole-image input, as opposed to patch-based, could better capture high-level structure and human-expert variation. ConvNet usually requires a large data set, so increasing the number of training examples available was important. As there is overlap between the labeling schemes in both cohorts, ConvNet uses a merged set of each provided expert delineation to have a common labeling scheme across the two cohorts. ConvNet is trained on this common set, followed by duplication and refinement of the ConvNets for each of the two cohorts.
A template image was created from all provided delineations using ANTs (Avants et al., 2008). A cerebellum mask is defined in this template space as the bounding box of the union of all labels in this template space. This cerebellum mask reduced the size of the computational domain for the processing. The general left/right symmetry was used to double the training-set size and further reduce the input FOV. Then each image was augmented 200× using random elastic distortions focused especially at label borders. The images were also intensity-normalized. Due to the memory constraints of the GPU platform (NVidia GTX 1080, 8Gb), the left/right resolution had to be halved which diminished some image details.
ConvNet was created with computational and time constraints in mind. Thus ConvNet structure has an alternating stack of 40- and 24-wide convolutional layers, with batch-normalization layers. No max-pooling nor other resolution changes were incorporated to reduce the model complexity. ConvNet also refrained from recalling low-level features within the final layers—usually a good practice in deep learning segmentation systems—as the goal is not pixel-perfect accuracy but rather expert imitation.
The loss function was a standard cross-entropy metric, weighted by each label size. When a label did not exist, which is a possibility since different label sets were merged, its weight was not accounted for in the loss function. Stochastic gradient decent (SGD) with momentum was used for the optimization of the initial ConvNet—as it tends to find better-quality minima with less over-fitting. The initial ConvNet model fitting took two days to train. The fine tuning of the separate ConvNets for each cohort used ADAM (Kingma and Ba, 2014) instead of SGD. This allowed for faster fine-tuning of the two ConvNets, taking approximately four hours per model. Mini-batches of size 6 were used. During development, one subject was left out of every data set for internal evaluation purpose, and later, as a final step the models were refitted on all available training data.
This ConvNet has limitations and room for improvement. For instance incorporation of multi-resolution features, tuning the number of parameters, and a different dropout scheme to ease convergence should all help improve the results. Additionally, as each of the ConvNets were trained on half-cerebella, the central voxels are arbitrary labeled according to their bounding-box side, which cause inaccuracies when the vermis is absent. Some post-processing, such as the use of an MRF, could be employed to enhance the results.
 CERES2
CERES2
Cerebellum multi-atlas patch-based segmentation with a patch-based boosted neural network error corrector
(José E. Romero, Pierrick Coupé, & José V. Manjón)
A new version of CERES (Romero et al., 2017), which is a cerebellum lobule segmentation algorithm that is based on a recent method called Optimized Patch-Match Label fusion (OPAL) (Giraud et al., 2016; Ta et al., 2014) is presented. The method consists of a multi-atlas patch-based (Rousseau et al., 2011; Coupe et al., 2011) non-local label fusion technique that produces segmentations using a library of manually annotated cases. CERES2 improves on CERES by using a different intensity normalization method and by adding a systematic error correction step based on an ensemble of patch-based boosted neural networks.
CERES2 preprocesses all the imaging data by denoising it using the spatially adaptive non-local means filter (Manjón et al., 2010) and corrects for intensity inhomogeneity using the N4 bias correction method (Tustison et al., 2010). The images were registered to the Montreal Neurological Institute (MNI) space using an affine transformation estimated using ANTs (Avants et al., 2008) and the MNI152 T1-w template; this is followed by intensity normalization (Nyúl and Udupa, 1999b) with the MNI152 images as the reference template. The images are then cropped to the cerebellum based on the manual segmentations of the subjects in the MNI space. Non-linear deformation was estimated using ANTs (Avants et al., 2008) using the cropped MNI152 template as reference. CERES2 completes the preprocessing by again applying intensity normalization (Nyúl and Udupa, 1999b) to the cropped images, further improving the intensity matching. After preprocessing, CERES2 has a library consisting of a set of cropped images (and segmentations) and their nonlinear transformations to the cropped MNI space. Similar to CERES, when given a new subject to segment a custom library is created online in the subject’s linear MNI space to avoid unwanted interpolation artifacts. This is done by concatenating the direct non-linear transformations of the library templates with the inverse non-linear transformation of the target case.
CERES2 employs a non-local patch-based label fusion, which is a multi-atlas segmentation fusion technique (Coupe et al., 2011). The resultant label for each image voxel is calculated by a weighted label fusion of multiple sample patches from a specific search area surrounding the target voxel for all the cases in the library, computed as
Here Vi is the search area around the ith voxel of the subject image, s iterates over the number of subjects in the library, and ys,j is the candidate label from voxel xs,j, the jth voxel in the sth subject. The function w(xi, xs,j) is the similarity between patches, which is defined as,
where P(·) is the patch around the respective voxel, ‖ · ‖2 is the normalized L2 norm (normalized by the number of elements), and h is a normalization factor calculated as the minimum of all patch distances from the search area.
The PatchMatch algorithm (Barnes et al., 2010) is an efficient way to find patch-wise correspondences between 2D images based on the approximated nearest neighbor field (ANNF) method. The core idea is that if two patches are a good match, then adjacent patches are likely to be a good match as well. Ta et al. (2014) presented the Optimized PatchMatch Label fusion (OPAL), an adaptation to 3D MR images, establishing correspondences between the input subject image and the library of L templates; the major benefit of OPAL is its run time is independent of the size of the library. A key difference between Barnes et al. (2010) and OPAL is the assumption, within OPAL, that both subject and library templates are located in the same coordinate frame (MNI space). A more complete description is available in Ta et al. (2014). Giraud et al. (2016) presented a multi-scale extension to OPAL, thus avoiding issues relating to fixed size patches. In this extension multiple label probability maps are computed and combined equally for each label before the label fusion step. Like CERES, CERES2 uses two scales and a label dependent weight as follows,
where p.(l) is the corresponding probability map for label l and α(l) ∈ [0, 1] is the mixing coefficient for label l. Details about the coefficient optimization are available in Romero et al. (2017). The final label for a voxel is simply the label with the highest probability.
Automatic segmentation methods suffer from random and systematic errors. Despite the fact that random error can be mitigated with aggregation techniques (such as multi-atlas techniques), systematic errors cannot be reduced using this strategy as they are not random. Nevertheless, their bias can be learned and used to correct the segmentations. Inspired by the work of Wang et al. (2013), in CERES2 a systematic error corrector using a patch-based ensemble of boosted neural networks to improve the segmentation accuracy is incorporated. The neural network ensemble is trained using samples from a region of interest of the label to correct as done in Wang et al. (2013). The feature vector was created by concatenating the following data: image patches of sizes 3 × 3 × 3 voxels (fully sampled); 7 × 7 × 7 and 11 × 11 × 11 voxels (subsampled by skipping two and four voxels at each dimension, respectively) from the T1-w image; the corresponding patches from the automatic segmentations; the Euclidean distance value of the voxel to the edge of the structure, and the coordinates in MNI space. Thus CERES2 has a feature vector of length 166 that is mapped to the corresponding manual segmentation patch of size 3 × 3 × 3 voxels. Features were extracted using an overcomplete scheme as done in Manjón et al. (2016). Such a structured prediction not only provides more accurate results than the voxel-wise version but also produces a more regular correction.
The neural network model used in CERES2 consists of a multilayer perceptron with two hidden layers of size 83 and 55 neurons resulting in a topology of 166 × 83 × 55 × 27 weights. CERES2 uses an ensemble comprising of 10 neural networks trained using a boosting strategy were wrongly classified training data samples were selected with higher probability than correctly classified ones. One ensemble per label is trained. At test time, the trained ensembles are used to correct the automatic segmentations produced by OPAL. The correction process takes a few seconds.
 RASCAL
RASCAL
Patch-based label fusion
(Vladimir S. Fonov and D. Louis Collins)
The previously published RASCAL (Rapid Automatic Segmentation of the Human Cerebellum and its Lobules) (Weier et al., 2014) was adapted for use with the two cohorts. The data was preprocessed as follows: 1) linear registration to MNI-ICBM152 2009c stereotaxic space (Fonov et al., 2010); 2) linear intensity normalization based on quantile matching to normalize the intensity range to the MNI-ICBM152 2009c template; 3) extracted brain mask using thresholding of the provided SPECTRE brainmask; 4) created an unbiased population specific template (Fonov et al., 2010), the resultant template was used as a reference template for RASCAL.
The RASCAL segmentation algorithm is an improved version of Coupe et al. (2011), which is also used by CERES2 and described above. The key differences are a change in how intensity normalization is done, majority voting to account for multiple labels, and the use of nonlinear registration. RASCAL (Weier et al., 2014) was then fine-tuned for the proposed data sets by employing a leave-one-out cross-validation. Linear registration, localized to the cerebellum, is performed using the affine registration mode of ANTs (Avants et al., 2008) with the Mattes cost function; which is followed by non-linear registration to the reference template using ANTs with the cross-correlation cost-function. After the registration steps are complete, all images are resampled to have the same resolution as the subject. Segmentations are then fused using the non-local patch-based algorithm.
 DeepNet
DeepNet
U-Net Parcellation of the Cerebellum
(Vladimir S. Fonov and D. Louis Collins)
DeepNet is an exploration of the potential of using an FCN based on U-net (Ronneberger et al., 2015; Çiçek et al., 2016) to parcellate the cerebellum. Recall that FCNs are CNNs designed for semantic segmentation. U-net has demonstrated state-of-the-art performance in several tasks (Chen et al., 2016; He et al., 2017; Zhao et al., 2017) while preserving the high resolution information throughout the contraction-expansion layers of the network. In brief, 3D U-net consists of a contracting (analysis) path and an expanding (synthesis) path. Each layer of the contracting portion consists of two 3×3×3 unpadded convolutions, followed by a ReLU and a 2×2×2 max pooling operation with a stride of 2 for downsampling in each dimension. Every step of the expansion consists of an upsampling convolution of 2×2×2 with a stride of 2 in each dimension followed by two 3×3×3 convolutions each followed by a ReLU. Shortcut connections from layers of equal resolution in the contracting path provide the high-resolution features to the expansion portion of the network. The last layer consists of a 1×1×1 convolution to reduce the number of output channels to the number of labels. Before each ReLU layer, batch normalization is performed during training with the mean, standard deviation, and global statistics updated using these values. This is followed by a layer to learn the scale and bias explicitly. At test time, normalization is done via the computed global statistics and the learned scale and bias.
As originally presented, 3D U-net used four analysis/synthesis steps; for the cerebellum parcellation task DeepNet uses five analysis/synthesis steps. DeepNet also modifies the default convolution kernels on a per-layer basis, with the details listed in Table A.1. The final layer of DeepNet contains two fully connected convolutional layers with 256 and 128 channels and a Dropout Layer. Thus the final layer creates a mapping from the 128 features into the segmentation labels—28 for the Adult Cohort and 18 for the Pediatric Cohort. Log Soft Max was used to calculate the negative log-likelihood error function, generalized kappa overlap metric was used to track performance on out-of-sample validation data. The total number of trainable parameters input into the model is 9,479,573.
Table A.1:
Layer parameters for  DeepNet.
DeepNet.
| U-net Layer | Input Channels | Output Channels | Convolution Kernels | Upsampling Kernel | |
|---|---|---|---|---|---|
| #1 | #2 | ||||
| 1 | 4 | 256 | 5×5×5 | 5×5×5 | 5×5×5 |
| 2 | 16 | 128 | 5×5×5 | 5×5×5 | 3×3×3 |
| 3 | 16 | 64 | 3×3×3 | 3×3×3 | 3×3×3 |
| 4 | 16 | 64 | 3×3×3 | 3×3×3 | 3×3×3 |
| 5 | 32 | 64 | 1×1×1 | 3×3×3 | 3×3×3 |
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Available from: http://www.diedrichsenlab.org/imaging/propatlas.htm
CERES2 can be used on http://www.volbrain.upv.es/
LiviaNET can be downloaded from https://github.com/josedolz/LiviaNET
References
- Abulnaga SM, Yang Z, Carass A, Kansal K, Jedynak BM, Onyike CU, Ying SH, Prince JL, 2016. A toolbox to visually explore cerebellar sub-structure shape changes in cerebellar ataxia, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27-March 3, 2016, pp. 9785–9785–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen G, Courchesne E, 2003. Differential effects of developmental cerebellar abnormality on cognitive and motor functions in the cerebellum: An fMRI study of Autism. Am. J. Psychiatry 160, 262–273. [DOI] [PubMed] [Google Scholar]
- Amaral DG, Schumann CM, Nordahl CW, 2008. Neuroanatomy of autism. Trends Neurosci. 31, 137–145. [DOI] [PubMed] [Google Scholar]
- Andreasen NC, Pierson R, 2008. The role of the cerebellum in schizophrenia. Biol. Psychiatry 64, 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, 2007. A fast diffeomorphic image registration algorithm. Neuroimage 38, 95–113. [DOI] [PubMed] [Google Scholar]
- Ashburner J, Andersson J, Friston KJ, 2000. Image registration using a symmetric prior - in three-dimensions. Human Brain Mapping 9, 212–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ, 1999. Nonlinear spatial normalization using basis functions. Human Brain Mapping 7, 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston KJ, 2005. Unified Segmentation. Neuroimage 26, 839–851. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Medical Image Analysis 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker KG, Harding AJ, Halliday GM, Kril JJ, Harper CG, 1999. Neuronal loss in functional zones of the cerebellum of chronic alcoholics with and without Wernicke’s encephalopathy. Neuroscience 91, 429–438. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Mitra S, Shankar BU, Hayashi Y, 2016. A Novel GBM Saliency Detection Model Using Multi-Channel MRI. PLoS ONE 11, e0146388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes C, Shechtman E, Golman DB, Finkelstein A, 2010. The generalized patchmatch correspondence algorithm, in: 2010 European Conference on Computer Vision (ECCV 2010), Springer Berlin Heidelberg; pp. 29–43. [Google Scholar]
- Barrett FS, Workman CI, Sair HI, Savonenko AV, Kraut MA, Sodums DJ, Joo JJ, Nassery N, Marano CM, Munro CA, Brandt J, Zhou Y, Wong DF, Smith GS, 2017. Association between serotonin denervation and resting-state functional connectivity in mild cognitive impairment. Human Brain Mapping 38, 3391–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann O, Borra RJ, Bower JM, Cullen KE, Habas C, Ivry RB, Leggio M, Mattingley JB, Molinari M, Moulton EA, Paulin MG, Pavlova MA, Schmahmann JD, Sokolov AA, 2015. Consensus Paper: The Role of the Cerebellum in Perceptual Processes. The Cerebellum 14, 197–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin PL, Pham DL, 2008. Homeomorphic brain image segmentation with topological and statistical atlases. Medical Image Analysis 12, 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergstra J, Breulex O, Bastien F, Lamblin P, Pascanu R, Desjardins G, Turian J, Warde-Farley B, Bengio Y, 2010. Theano: A CPU and GPU math compiler in Python, in: Proc. 9th Python in Science Conf., pp. 1–7. [Google Scholar]
- Bernard JA, Millman ZB, Mittal VA, 2015. Beat and metaphoric gestures are differentially associated with regional cerebellar and cortical volumes. Human Brain Mapping 36, 4016–4030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG, 1986. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 327, 307–310. [PubMed] [Google Scholar]
- Bogovic JA, Bazin PL, Ying SH, Prince JL, 2013a. Automated Segmentation of the Cerebellar Lobules Using Boundary Specific Classification and Evolution, in: 23rd Inf. Proc. in Med. Imaging (IPMI 2013), Springer Berlin Heidelberg; pp. 62–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic JA, Jedynak BM, Rigg R, Du A, Landman BA, Prince JL, Ying SH, 2013b. Approaching expert results using a hierarchical cerebellum parcellation protocol for multiple inexpert human raters. NeuroImage 64, 616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogovic JA, Prince JL, Bazin PL, 2013c. A multiple object geometric deformable model for image segmentation. Comput. Vis. Image Und 117, 145–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brambati SM, Termine C, Ruffino M, Stella G, Fazio F, Cappa SF, Perani D, 2004. Regional reductions of gray matter volume in familial dyslexia. Neurology 63,742–745. [DOI] [PubMed] [Google Scholar]
- Buckner RL, Head D, Parker J, Fotenos AF, Marcus D, Morris JC, Snyder Z, 2004. A unified approach for morphometric and functional data analysis in young, old, and demented adults using automated atlas-based head size normalization: reliability and validation against manual measurement of total intracranial volume. NeuroImage 23, 724–738. [DOI] [PubMed] [Google Scholar]
- Carass A, Cuzzocreo J, Wheeler MB, Bazin PL, Resnick SM, Prince JL, 2010. Simple paradigm for extra-cerebral tissue removal: Algorithm and analysis. NeuroImage 56, 1982–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carass A, Prince JL, 2016. An Overview of the Multi-Object Geometric Deformable Model Approach in Biomedical Imaging, in: Zhou SK (Ed.), Medical Image Recognition, Segmentation and Parsing. Academic Press, pp. 259–279. [Google Scholar]
- Carass A, Roy S, Jog A, Cuzzocreo JL, Magrath E, Gherman A, Button J, Nguyen J, Bazin PL, Calabresi PA, Crainiceanu C, Ellingsen LM, Reich DS, Prince JL, Pham DL, 2017a. Longitudinal multiple sclerosis lesion segmentation data resource. Data in Brief 12, 346–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carass A, Roy S, Jog A, Cuzzocreo JL, Magrath E, Gherman A, Button J, Nguyen J, Prados F, Sudre CH, Cardoso MJ, Cawley N, Ciccarelli O, Wheeler-Kingshott CAM, Ourselin S, Catanese L, Deshpande H, Maurel P, Commowick O, Barillot C, Tomas-Fernandez X, Warfileld SK, Vaidya S, Chunduru A, Muthuganapathy R, Krishnamurthi G, Jesson A, Arbel T, Maier O, Handels H, Iheme LO, Unay D, Jain S, Sima DM, Smeets D, Ghafoorian M, Platel B, Birenbaum A, Greenspan H, Bazin PL, Calabresi PA, Crainiceanu C, Ellingsen LM, Reich DS, Prince JL, Pham DL, 2017b. Longitudinal multiple sclerosis lesion segmentation: Resource & challenge. Neurolmage 148, 77–I02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carass A, Shao M, Li X, Dewey BE, Blitz AM, Roy S, Pham DL, Prince JL, Ellingsen LM, 2017c. Whole Brain Parcellation with Pathology: Validation on Ventriculomegaly Patients, in: Patch-MI 2017: Patch-Based Techniques in Medical Imaging, Springer Berlin Heidelberg; pp. 20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carass A, Wheeler MB, Cuzzocreo J, Bazin PL, Bassett SS, Prince JL, 2007. A joint registration and segmentation approach to skull stripping, in: 4th International Symposium on Biomedical Imaging (ISBI 2007), IEEE; pp. 656–659. [Google Scholar]
- Catani M, Jones DK, Daly E, Embiricos N, Deeley Q, Pugliese L Curran S, Robertson D, Murphy DGM, 2008. Altered cerebellar feedback projections in Asperger syndrome. NeuroImage 41,1184–1191. [DOI] [PubMed] [Google Scholar]
- Cavanagh JB, Holton JL, Nolan CC, 1997. Selective damage to the cerebellar vermis in chronic alcoholism: A contribution from neurotoxicology to an old problem of selective vulnerability. Neuropathology and applied neurobiology 23, 355–363. [PubMed] [Google Scholar]
- Çiçek O, Abdulkadir A, Lienkamp SS, Brox T, Ronneberger O, 2016. 3D U-Net: Learning Dense Volumetric Segmentation from Sparse Annotation, in: 19th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2016), Springer Berlin Heidelberg; pp. 424–432. [Google Scholar]
- Chen H, Qi X, Yu L, Heng PA, 2016. DCAN: Deep Contour-Aware Networks for Accurate Gland Segmentation, in: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 2487–2496. [Google Scholar]
- Cleavinger HB, Bigler ED, Johnson JL, Lu J, McMahon W, Lainhart JE, 2008. Quantitative magnetic resonance image analysis of the cerebellum in macrocephalic and normocephalic children and adults with autism. Journal of the International Neuropsychological Society 14, 401–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins DL, Neelin P, Peters TM, Evans AC, 1994. Automatic 3-D intersubject registration of MR volumetric data in standardized Talairach space. j. Comput. Assis. Tomogr 18, 192–205. [PubMed] [Google Scholar]
- Colloby SJ, O’Brien JT, Taylor JP, 2014. Patterns of cerebellar volume loss in dementia with Lewy bodies and Alzheimer’s disease: A VBM-DARTEL study. Psychiatry Research: Neuroimaging 223, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coupe P, Manjon JV, Fonov V, Pruessner j., Robles M, Collins DL, 2011. Patch-based segmentation using expert priors: application to hippocampus and ventricle segmentation. Neuroimage 54, 940–954. [DOI] [PubMed] [Google Scholar]
- Courchesne E, Saitoh O, Yeung-Courchesne R, Press GA, Lincoln AJ, Haas RH, Schreibman L, 1994. Abnormality of Cerebellar Vermian Lobules VI and VII in Patients with Infantile Autism: Identification of Hypoplastic and Hyperplastic with MR Imaging. Am. J. of Roentgenology 162, 123–130. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI, 1999. Cortical Surface-Based Analysis I: Segmentation and Surface Reconstruction. NeuroImage 9, 179–194. [DOI] [PubMed] [Google Scholar]
- Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, Hyman BT, Albert MS, Killiany RJ, 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. NeuroImage 31, 968–980. [DOI] [PubMed] [Google Scholar]
- Desmond JE, Fiez JA, 1998. Neuroimaging studies of the cerebellum: language, learning and memory. Trends Cogn. Sci 2, 355–362. [DOI] [PubMed] [Google Scholar]
- Dice LR, 1945. Measures of the Amount of Ecologic Association Between Species. Ecology 26, 297–302. [Google Scholar]
- Diedrichsen J, 2006. A spatially unbiased atlas template of the human cerebellum. NeuroImage 33, 127–18. [DOI] [PubMed] [Google Scholar]
- Diedrichsen J, Balsters JH, Flavell J, Cussans E, Ramnani N, 2009. A probabilistic MR atlas of the human cerebellum. NeuroImage 46, 39–46. [DOI] [PubMed] [Google Scholar]
- D’Mello AM, Crocetti D, Mostofsky SH Stoodley CJ, 2015. Cerebellar gray matter and lobular volumes correlate with core autism symptoms. NeuroImage: Clinical 7, 631–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolz J, Desrosiers C, Ayed IB, 2018. 3D fully convolutional networks for subcortical segmentation in MRI: A large-scale study. NeuroImage 170, 456–470. [DOI] [PubMed] [Google Scholar]
- Dos Santos V, Thomann PA, Wüstenberg T, Seidl U, Essig M, Schröder J, 2011. Morphological cerebral correlates of CERAD test performance in mild cognitive impairment and Alzheimer’s disease. Jrnl. Alzheimer’s Disease 23, 411–420 [DOI] [PubMed] [Google Scholar]
- Durston S, Hulshoff Pol HE, Schnack HG, Buitelaar JK, Steenhuis MP, Minderaa RB, Kahn RS, van engeland H, 2004. Magnetic Resonance Imaging of Boys With Attention-Deficit/Hyperactivity Disorder and Their Unaffected Siblings. Journal of the American Academy of Child & Adolescent Psychiatry 43, 332–340. [DOI] [PubMed] [Google Scholar]
- Esteban O, Birman D, Schaer M, Koyejo OO, Poldrack RA, Gorgolewski KJ, 2017. MRIQC: Advancing the automatic prediction of image quality in MRI from unseen sites. PLoS ONE 12, e0184661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B,2012. FreeSurfer. Neuroimage 62, 774–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, van der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A, Makris N, Rosen B, Dale AM, 2002. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33, 341–355. [DOI] [PubMed] [Google Scholar]
- Fischl B, Salat DH, van der Kouwe AJW, Makris N, Ségonne F, Quinn BT, Dale AM, 2004. Sequence-independent segmentation of magnetic resonance images. Neuroimage 23, S69–s84. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick LE, Jackson M, Crowe SF, 2008. The relationship between alcoholic cerebellar degeneration and cognitive and emotional functioning. Neuroscience & Biobehavioral Reviews 32, 466–485. [DOI] [PubMed] [Google Scholar]
- Fonov VS, Evans AC, Botteron K, McKinstry RC, Collins DL, the Brain Development Cooperative Group, 2010. Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54, 313–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraud R, Ta VT, Papadakis N, Manjón JV, Collins DL, Coupé P, Alzheimer’s Disease Neuroimaging Initiative, 2016. An optimized patchmatch for multi-scale and multi-feature label fusion. NeuroImage 124, 770–782. [DOI] [PubMed] [Google Scholar]
- Gottwald B, Wilde B, Mihajlovic Z, Mehdorn HM, 2004. Evidence for distinct cognitive deficits after focal cerebellar lesions. J. Neurol. Neurosurg. Psychiatry 75, 1524–1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Kashyap S, Sonka M, Oguz I, 2017. Machine learning in a graph framework for subcortical segmentation, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2017), Orlando, FL, February 11−16, 2017, pp. 101330H-101330H–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Jovicich J, Salat D, van der Kouwe A, Quinn B, Czanner S, Busa E, Pacheco J, Albert M, Killiany R, Maguire P, Rosas D, Makris N, Dale A, Dickerson B, Fischl B, 2006. Reliability of MRI-derived measurements of human cerebral cortical thickness: The effects of field strength, scanner upgrade and manufacturer. NeuroImage 32, 180–194. [DOI] [PubMed] [Google Scholar]
- He K, Zhang X, Ren S, Sun J, 2015. Delving deep into rectifiers: Surpassing humanlevel performance on imagenet classification, in: 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1026–1034. [Google Scholar]
- He Y, Carass A, Yun Y, Zhao C, Jedynak BM, Solomon SD, Saidha S, Calabresi PA, Prince JL, 2017. Towards Topological Correct Segmentation of Macular OCT from Cascaded FCNs, in: Fetal, Infant and Ophthalmic Medical Image Analysis: International Workshop, FIFI 2017, and 4th International Workshop, OMIA 2017, Held in Conjunction with MICCAI 2017, Québec City, QC, Canada, September 14, Proceedings, Springer Berlin Heidelberg; pp. 202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heimann T, van Ginneken B, Styner MA, Arzhaeva Y, Aurich V, Bauer C, Beck A, Becker C, Beichel R, Bekes G, Bello F, Binnig G, Bischof H, Bornik A, Cashman P, Ying C, Cordova A, Dawant BM, Fidrich M, Furst JD, Furukawa D, Grenacher L, Hornegger J, Kainmuller D, Kitney RI, Kobatake H, Lamecker H, Lange T, Lee J, Lennon B, Li R, Li S, Meinzer HP, Nemeth G, Raicu DS, Rau AM, van Rikxoort EM, Rousson M, Rusko L, Saddi KA, Schmidt G, Seghers D, Shimizu A, Slagmolen P, Sorantin E, Soza G, Susomboon R, Waite JM, Wimmer A, Wolf I, 2009. Comparison and evaluation of methods for liver segmentation from CT datasets. IEEE Trans. Med. Imag 28, 1251–1265. [DOI] [PubMed] [Google Scholar]
- Henle J, 1879. Handbuch der Nervenlehre des Menschen. Friedrich Vieweg und Sohn, Braunschweig. [Google Scholar]
- Hsu YY, Schuff N, Du AT, Mark K, Zhu X, Hardin D, Weiner MW, 2002. Comparison of automated and manual MRI volumetry of hippocampus in normal aging and dementia. Jrnl. of Magnetic Resonance Imaging 16, 305–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Carass A, Resnick SM, Pham DL, Prince JL, Landmann BA, 2016a. Combining multi-atlas segmentation with brain surface estimation, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27-March 3, 2016, pp. 9784–9784–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo Y, Plassard AJ, Carass A, Resnick SM, Pham DL, Prince JL, Landmann BA, 2016b. Consistent Cortical Reconstruction and Multi-atlas Brain Segmentation. NeuroImage 138, 197–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito M, 1984. The cerebellum and neural control. Raven, New York. [Google Scholar]
- Jednoróg K, Gawron N, Marchewka A, Heim S, Grabowska A, 2013. Cognitive subtypes of dyslexia are characterized by distinct patterns of grey matter volume. Brain Structure and Function 219, 1697–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menton DK, Rueckert D, Glocker B, 2017. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Medical Image Analysis 36, 61–78. [DOI] [PubMed] [Google Scholar]
- Kansal K, Yang Z, Fishman AM, Sair HI, Ying SH, Jedynak BM, Prince JL, Onyike CU, 2016. Structural cerebellar correlates of cognitive and motor dysfunctions in cerebellar degeneration. Brain 140, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karas GB, Burton EJ, Rombouts SARB, van Schijndel RA, O’Brien JT, Scheltens P, McKeith IG, Williams D, Ballard C, Barkhof F, 2003. A comprehensive study of gray matter loss in patients with Alzheimer’s disease using optimized voxel-based morphometry Neuroimage 18, 895–907. [DOI] [PubMed] [Google Scholar]
- Kingma DP, Ba J, 2014. Adam: A Method for Stochastic Optimization. CoRR abs/1412.6980. arXiv:1412.6980.
- Landman BA, Bogovic JA, Carass A, Chen M, Roy S, Shiee N, Yang Z, Kishore B, Pham D, Bazin PL, Resnick SM, Prince JL, 2013. System for Integrated Neuroimaging Analysis and Processing of Structure. Neuroinformatics 11, 91–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsell O, 1952. The morphogenesis and adult pattern of the lobules and tissues of the cerebellum of the white rat. J. Comp. Neuol 97, 281–356. [DOI] [PubMed] [Google Scholar]
- Ledig C, Heckemann RA, Hammers A, Lopez JC, Newcombe VF, Makropoulos A, Lotjonen J, Menon DK, Rueckert D, 2015. Robust whole-brain segmentation: application to traumatic brain injury. Medical Image Analysis 21, 40–58. [DOI] [PubMed] [Google Scholar]
- Lewis MM, Galley S, Johnson S, Stevenson J, Huang X, McKeown MJ, 2013. The Role of the Cerebellum in the Pathophysiology of Parkinson’s Disease. Canadian Journal of Neurological Sciences / Journal Canadien des Sciences Neurologiques 40, 299–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Ye C, Yang Z, Carass A, Ying SH, Prince JL, 2016. Quality Assurance using Outlier Detection on an Automatic Segmentation Method for the Cerebellar Peduncles, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27-March 3, 2016, pp. 9784–9784–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bazin PL, Carass A, Prince J, 2012. Topology Preserving Brain Tissue Segmentation Using Graph Cuts, in: IEEE Workshop on Mathematical Methods in Biomedical Image Analysis, pp. 185–190. [Google Scholar]
- Long J, Shelhamer E, Darrell T, 2015. Fully Convolutional Networks for Semantic Segmentation, in: 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- Maier O, Menze BH, von der Gablentz J, Häni L, Heinrich MP, Liebrand M, Winzeck S, Basit A, Bentley P, Chen L, Christiaens D, Dutil F, Egger K, Feng C, Glocker B, Götz M, Haeck T, Halme HL, Havaei M, Iftekharuddin KM, Jodoin PM, Kamnitsas K, Kellner E, Korvenoja A, Larochelle H, Ledig C, Lee JH, Maes F, Mahmood Q, Maier-Hein KH, McKinley R, Muschelli J, Pal C, Pei L, Rangarajan JR, Reza SMS, Robben D, Rueckert D, Salli E, Suetens P, Wang CW, Wilms M, Kirschke JS, Krämer UM, Münte TF, Schramm P, Wiest R, Handels H, Reyes M, 2017. ISLES 2015 - A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. Medical Image Analysis 35, 250–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier-Hein L, Eisenmann M, Reinke A, Onogur S, Stankovic M, Scholz P, Arbel T, Bogunovic H, Bradley AP, Carass A, Feldmann C, Frangi AF, Full PM, van Ginneken B, Hanbury A, Honauer K, Kozubek M, Landman BA, März K, Maier O, Maier-Hein K, Menze BH, Müller H, Neher PF, Niessen W, Rajpoot N, Sharp GC, Sirinukunwattana K, Speidel S, Stock C, Stoyanov D, Aziz Taha A, van der Sommen F, Wang CW, Weber MA, Zheng G, Jannin P, Kopp-Schneider A, 2018. Is the winner really the best? A critical analysis of common research practice in biomedical image analysis competitions. Nature Communications (In Review), arXiv:1806.02051<https://arxiv.org/abs/1806.02051>. [Google Scholar]
- Malacarne V, 1776. Nuova esposizione della vera struttura del cervelletto umano. Briolo, Torino. [Google Scholar]
- Manjón JV, Coupé P, Marti-Bonmati L, Collins DL, Robles M, 2010. Adaptive non-local means denoising of MR images with spatially varying noise levels. Mag. Reson. Im 31, 192–203. [DOI] [PubMed] [Google Scholar]
- Manjón JV, Coupé P, Raniga P, Xia Y, Fripp J, Salvado O, 2016. HIST: Hyperlntensity Segmentation Tool, in: Patch-MI 2016: Patch-Based Techniques in Medical Imaging, Springer Berlin Heidelberg. pp. 92–99. [Google Scholar]
- Manto M, Schmahmann JD, Koibuchi N, Rossi F, 2013. Handbook of the Cerebellum and Cerebellar Disorders. Springer, New York. [Google Scholar]
- Martino AD, Yan CG, Li Q, Denio E, Castellanos FX, Alaerts K, Anderson JS, Assaf M, Bookheimer SY, Dapretto M, Deen B, Delmonte S, Dinstein I, Ertl-Wagner B, Fair DA, Gallagher L, Kennedy DP, Keown CL, Keysers C, Lainhart JE, Lord C, Luna B, Menon V, Minshew NJ, Monk CS, Mueller S, Müller RA, Nebel MB, Nigg JT, O’Hearn K, Pelphrey KA, Peltier SJ, Rudie JD, Sunaert S, Thioux M, Tyszka JM, Uddin LQ, Verhoeven JS, Wenderoth N, Wiggins JL, Mostofsky SH, Milham MP, 2014. The autism brain imaging data exchange: towards a large-scale evaluation of the intrinsic brain architecture in autism. Molecular Psychiatry 19, 659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendrik AM, Vincken KL, Kuijf HJ, Breeuwer M, Bouvy W, de Bresser J, Alansary A, de Bruijn M, Carass A, El-Baz A, Jog A, Katyali R, Khan AR, van der Lijn F, Mahmood Q, Mukherjee R, van Opbroek A, Paneri S, Pereira S, Persson M, Rajchl M, Sarikayan D, Smedby O, Silva CA, Vrooman HA, Vyas S, Wang C, Zhaon L, Biessels GJ, Viergever MA, 2015. MRBrainS Challenge: Online Evaluation Framework for Brain Image Segmentation in 3T MRI Scans. Computational Intelligence and Neuroscience. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menze BH, Jakab A, Bauer S, Kalpathy-Cramer J, Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R, Lanczi L, Gerstner E, Weber MA, Arbel T, Avants BB, Ayache N, Buendia P, Collins DL, Cordier N, Corso JJ, Criminisi A, Das T, Delingette H, Demiralp Q, Durst CR, Dojat M, Doyle S, Festa J, Forbes F, Geremia E, Glocker B, Golland P, Guo X, Hamamci A, Iftekharuddin KM, Jena R, John NM, Konukoglu E, Lashkari D, Mariz JA, Meier R, Pereira S, Precup D, Price SJ, Riklin-Raviv T, Reza SMS, Ryan M, Sarikaya D, Schwartz L, Shin HC, Shotton J, Silva CA, Sousa N, Subbanna NK, Székely G, Taylor TJ, Thomas OM, Tustison NJ, Unal G, Vasseur F, Wintermark M, Ye DH, Zhao L, Zhao B, Zikic D, Prastawa M, Reyes M, Van Leemput K, 2015. The Multimodal Brain Tumor Image Segmentation Benchmark (bRaTS). IEEE Trans. Med. Imag 34, 1993–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middleton FA, Strick PL, 2000. Basal ganglia and cerebellar loops: motor and cognitive circuits. Brain Research Reviews 31, 236–250. [DOI] [PubMed] [Google Scholar]
- Möller C, Vrenken H, Jiskoot L, Versteeg A, Barkhof F, Scheltens P, van der Flier WM, 2013. Different patterns of gray matter atrophy in early- and late-onset Alzheimer’s disease. Neurobiology of Aging 34, 2014–2022. [DOI] [PubMed] [Google Scholar]
- Mostofsky SH, Mazzocco MMM, Aakalu G, Warsofsky IS, Denckla MB, Reiss AL, 1998a. Decreased cerebellar posterior vermis size in fragile X syndrome: Correlation with neurocognitive performance. Neurology 50, 121–130. [DOI] [PubMed] [Google Scholar]
- Mostofsky SH, Reiss AL, Lockhart P, Denckla MB, 1998b. Evaluation of cerebellarsize in attention-deficit hyperactivity disorder. j. Child Neurol. 13, 434–439. [DOI] [PubMed] [Google Scholar]
- Murphy KR, Landau SM, Choudhury KR, Hostage CA, Shpanskaya KS, Sair HI, Petrella JR, Wong TZ, Doraiswamy PM, 2013. Mapping the effects of ApoE4, age and cognitive status on 18F-florbetapir PET measured regional cortical patterns of beta-amyloid density and growth. NeuroImage 78, 474–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nopoulos PC, Ceilley JW, Gailis EA, Andreasen NC, 1999. An MRI study of cerebellar vermis morphology in patients with schizophrenia: Evidence in support of the cognitive dysmetria concept. Biological psychiatry 46, 703–711. [DOI] [PubMed] [Google Scholar]
- Nyúl LG, Udupa JK, 1999a. New Variants of a Method of MRI Scale Normalization, in: 16th Inf. Proc. in Med. Imaging (IPMI 1999), Springer Berlin Heidelberg; pp. 490–495. [Google Scholar]
- Nyúl LG, Udupa JK, 1999b. On Standardizing the MR Image Intensity Scale. Mag. Reson. Med 42, 1072–1081. [DOI] [PubMed] [Google Scholar]
- Okugawa G, Sedvall G, Nordström M, Andreasen N, Pierson R, Magnotta V, Agartz I, 2002. Selective reduction of the posterior superior vermis in men with chronic schizophrenia. Schizophrenia Research 55, 61–67. [DOI] [PubMed] [Google Scholar]
- Okugawa G, Sedvall GC, Agartz I, 2003. Smaller cerebellar vermis but not hemisphere volumes in patients with chronic schizophrenia. Am. J. of Psychiatry 160,1614–1617. [DOI] [PubMed] [Google Scholar]
- Park MTM, Pipitone J, Baer LH, Winterburn JL, Shah Y, Chavez S, Schira MM, Lobaugh NJ, Lerch JP, Voineskos AN, Chakravarty MM, 2014. Derivation of high-resolution MRI atlases of the human cerebellum at 3T and segmentation using multiple automatically generated templates. NeuroImage 95, 217–231 [DOI] [PubMed] [Google Scholar]
- Parker KL, Narayanan NS, Andreasen NC, 2014. The therapeutic potential of the cerebellum in schizophrenia. Front Syst Neurosci. 8, 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patenaude B, Smith SM, Kennedy DN, Jenkinson M, 2011. Bayesian model of shape and appearance for subcortical brain segmentation. NeuroImage 56, 907–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson R, Corson PW, Sears LL, Alicata D, Magnotta V, O’Leary D, Andreasen NC, 2002. Manual and semiautomated measurement of cerebellar subregions on MR images. NeuroImage 17, 61–76. [DOI] [PubMed] [Google Scholar]
- Plassard AJ, Yang Z, Rane S, Prince JL, Claassen DO, Landman BA, 2016. Improving cerebellar segmentation with statistical fusion, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27-March 3, 2016, pp. 9784–9784–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell S, Magnotta VA, Johnson H, Jammalamadaka VK, Pierson R, Andreasen NC, 2008. Registration and machine learning-based automated segmentation of subcortical and cerebellar brain structures. NeuroImage 39, 238–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price M, Cardenas V, Fein G, 2014. Automated MRI cerebellar size measurements using active appearance modeling. NeuroImage 103, 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Fischl B, 2011. Avoiding Asymmetry-Induced Bias in Longitudinal Image Processing. NeuroImage 57, 19–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero JE, Coupé P, Giraud R, Ta VT, Fonov V, Park MTM, Chakravarty MM, Voineskos AN, Manjón JV, 2017. CERES: A new cerebellum lobule segmentation method. NeuroImage 147, 916–924. [DOI] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P, Brox T, 2015. U-Net: Convolutional Networks for Biomedical Image Segmentation, in: 18th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2015), Springer Berlin Heidelberg; pp. 234–241. [Google Scholar]
- Rousseau F, Habas PA, Studholme C, 2011. A supervised patch-based approach for human brain labeling. IEEE Trans. Med. Imag 30, 1852–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, He Q, Sweeney E, Carass A, Reich DS, Prince JL, Pham DL, 2015. Subject-Specific Sparse Dictionary Learning for Atlas-Based Brain MRI Segmentation. IEEE Journal of Biomedical and Health Informatics 19, 1598–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauwen N, Sima DM, Van Cauter S, Veraart J, Leemans A, Maes F, Himmelreich U, Van Huffel S, 2015. Hierarchical non-negative matrix factorization to characterize brain tumor heterogeneity using multi-parametric MRI. NMR in Biomedicine 28, 1599–1624. [DOI] [PubMed] [Google Scholar]
- Schaap M, Metz CT, van Walsum T, van der Giessen AG, Weustink AC, Mollet NR, Bauer C, Bogunović H, Castro C, Deng X, Dikici E, O’Donnell T, Frenay M, Friman O, Hoyos MH, Kitslaar PH, Krissian K, Kuhnel C, Luengo-Oroz MA, Orkisz M, Smedby Ö, Styner M, Szymczak A, Tek H, Wang C, Warfield SK, Zambal S, Zhang Y, Krestin GP, Niessen WJ, 2009. Standardized evaluation methodology and reference database for evaluating coronary artery centerline extraction algorithms. Medical Image Analysis 13, 701–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmahmann JD, 1991. An emerging concept. The cerebellar contribution to higher function. Arch. Neurol 48, 1178–1187. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, 2004. Disorders of the cerebellum: ataxia, dysmetria of thought, and the cerebellar cognitive affective syndrome. The Journal of neuropsychiatry and clinical neurosciences 16, 367–378. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Caplan D, 2006. Cognition, emotion and the cerebellum. Brain 129, 290–292. [DOI] [PubMed] [Google Scholar]
- Schmahmann JD, Doyon J, Toga AW, Petrides M, Evans AC, 2000. MRI atlas of the human cerebellum. Academic Press, San Diego, CA. [DOI] [PubMed] [Google Scholar]
- Schutter DJLG, Van Honk J, 2005. The cerebellum on the rise in human emotion. The Cerebellum 4, 290–294. [DOI] [PubMed] [Google Scholar]
- Shao M, Carass A, Li X, Dewey BE, Blitz AM, Prince JL, Ellingsen LM, 2018. Multi-atlas segmentation of the hydrocephalus brain using an adaptive ventricle atlas, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2018), Houstan, TX, February 10 – 15, 2018, pp. 105780F-105780F–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Leahy RM, 2002. BrainSuite: An automated cortical surface identification tool. Medical Image Analysis 6, 129–142. [DOI] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW, 2008. Construction of a 3D probabilistic atlas of human cortical structures. NeuroImage 39, 1064–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Sandor-Leahy SR, Schaper KA, Rottenberg DA, Leahy RM, 2001. Magnetic resonance image tissue classification using a partial volume model. NeuroImage 13, 856–876. [DOI] [PubMed] [Google Scholar]
- Shiee N, Bazin PL, Cuzzocreo JL, Blitz A, Pham DL, 2011. Segmentation of brain images using adaptive atlases with application to ventriculomegaly, in: 22nd Inf. Proc. in Med. Imaging (IPMI 2011), Springer Berlin Heidelberg; pp. 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiee N, Bazin PL, Cuzzocreo JL, Ye C, Kishore B, Carass A, Calabresi PA, Reich DS, Prince JL, Pham DL, 2014. Reconstruction of the human cerebral cortex robust to white matter lesions: Method and validation. Human Brain Mapping 35, 3385–3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiee N, Bazin PL, Ozturk A, Reich DS, Calabresi PA, Pham DL, 2010. A topology-preserving approach to the segmentation of brain images with multiple sclerosis lesions. NeuroImage 49, 1524–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silveri MC, Leggeio MG, Molinari M, 1994. The cerebellum contributes to linguistic production. Neurology 44, 2047. [DOI] [PubMed] [Google Scholar]
- Stoodley CJ, 2014. Distinct regions of the cerebellum show gray matter decreases in autism, ADHD, and developmental dyslexia. Frontiers in Systems Neuroscience 8, 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strick PL, Dum RP, Fiez JA, 2009. Cerebellum and Nonmotor Function. Annual Review of Neuroscience 32, 413–434. [DOI] [PubMed] [Google Scholar]
- Styner M, Lee J, Chin B, Chin MS, Commowick O, Tran HH, Markovic-Plese S, Jewells V, Warfield S, 2008. 3D Segmentation in the Clinic: A Grand Challenge II: MS lesion segmentation, in: 11th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2008) 3D Segmentation in the Clinic: A Grand Challenge II, pp. 1–6. [Google Scholar]
- Ta VT, Giraud R, Collins DL, Coupé P, 2014. Optimized patch Match for near real time and accurate label fusion, in: 17th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2014), Springer Berlin Heidelberg; pp. 105–112. [DOI] [PubMed] [Google Scholar]
- Thomann PA, Schläfer C, Seidl U, Dos Santos V, Essig M, Schröder J, 2008. The cerebellum in mild cognitive impairment and Alzheimer’s disease - A structural MRI study. J. Psychiatric Res 42, 1198–1202. [DOI] [PubMed] [Google Scholar]
- Tieleman T, Hinton G, 2015. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude, in: COURSERA: Neural networks for machine learning. volume 4, pp. 26–31. [Google Scholar]
- Tomas-Fernandez X, Warfield SK, 2015. A Model of Population and Subject (MOPS) Intensities with Application to Multiple Sclerosis Lesion Segmentation. IEEE Trans. Med. Imag 34, 1349–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torvik A, Torp S, 1986. The prevalence of alcoholic cerebellar atrophy: A morphometric and histological study of an autopsy material. Journal of the Neurological Sciences 75, 43–51. [DOI] [PubMed] [Google Scholar]
- Tosun D, Rettmann ME, Naiman DQ, Resnick SM, Kraut MA, Prince JL, 2006. Cortical reconstruction using implicit surface evolution: Accuracy and precision analysis. NeuroImage 29, 838–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traut N, Beggiato A, Bourgeron T, Delorme R, Rondi-Reig L, P AL, Toro R, 2018. Cerebellar Volume in Autism: Literature Meta-analysis and Analysis ofthe Autism Brain Imaging Data Exchange Cohort. Biological Psychiatry 83, 579–588. [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA, Gee JC, 2010. N4ITK: Improved N3 Bias Correction. IEEE Trans. Med. Imag 29, 1310–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Lijn F, de Bruijne M, Hoogendam YY, Klein S, Hameeteman R, Breteler MMB, Niessen WJ, 2009. Cerebellum segmentation in MRI using atlas registration and local multi-scale image descriptors, in: 6th International Symposium on Biomedical Imaging (ISBI 2009), pp. 221–224. [Google Scholar]
- Van Leemput K, Maes F, Vandermeulen D, Suetens P, 1999. Automated modelbased tissue classification of MR images of the brain. IEEE Trans. Med. Imag 18, 897–908. [DOI] [PubMed] [Google Scholar]
- Victor M, Adams RD, Mancall EL, 1959. A Restricted Form of Cerebellar Cortical Degeneration Occurring in Alcoholic Patients. AMA Arch. Neurol 1, 579–688. [Google Scholar]
- Wang H, Suh JW, Das SR, Pluta JB, Craige C, Yushkevich PA, 2013. Multi-Atlas Segmentation with Joint Label Fusion. IEEE Trans. Patt. Anal. Mach. Intell 35, 611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SJ, Sparks BF, Friedman SD, Shaw DWW, Giedd J, Dawson G, Dager SR, 2009. Cerebellar vermal volumes and behavioral correlates in children with autism spectrum disorder. Psychiatry Research: Neuroimaging 172, 61–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier K, Fonov V, Lavoie K, Doyon J, Collins DL, 2014. Rapid automatic segmentation of the human cerebellum and its lobules (RASCAL)–Implementation and application of the patch-based label-fusion technique with a template library to segment the human cerebellum. Human Brain Mapping 35, 5026–5039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weier K, Till C, Fonov V, Yeh EA, Arnold DL, Banwell B, Collins D, 2016. Contribution of the cerebellum to cognitive performance in children and adolescents with multiple sclerosis. Multiple Sclerosis Journal 22, 599–607. [DOI] [PubMed] [Google Scholar]
- Wilcoxon F, 1945. Individual comparisons by ranking methods. Biometrics Bulletin 1, 80–83. [Google Scholar]
- Womer FY, Tang Y, Harms MP, Bai C, Chang M, Jiang X, Wei S, Wang F, Barch DM, 2016. Sexual dimorphism of the cerebellar vermis in schizophrenia. Schizophrenia Research 176, 164–170. [DOI] [PubMed] [Google Scholar]
- Yang Z, Abulnaga SM, Carass A, Kansal K, Jedynak BM, Onyike CU, Ying SH, Prince JL, 2016a. Landmark Based Shape Analysis for Cerebellar Ataxia Classification and Structural Change Pattern Visualization, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2016), San Diego, CA, February 27–March 3, 2016, pp. 9784 – 9784 – 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Ye C, Bogovic JA, Carass A, Jedynak BM, Ying SH, Prince JL, 2016b. Automated Cerebellar Lobule Segmentation with Application to Cerebellar Structural Analysis in Cerebellar Disease. NeuroImage 127, 435–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Zhong S, Carass A, Ying SH, Prince JL, 2014. Deep Learning for Cerebellar Ataxia Classification and Functional Score Regression, in: Machine Learning in Medical Imaging (MLMI 2014), pp. 68–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying SH, Choi SI, Perlman SL, Baloh RW, Zee DS, Toga AW, 2006. Pontine and cerebellar atrophy correlate with clinical disability in SCA2. Neurology 66, 424–426. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Brady M, Smith S, 2001. Segmentation of brain MR images through a hidden Markov random field model and the Expectation-Maximization algorithm. IEEE Trans. Med. Imag 20, 45–57. [DOI] [PubMed] [Google Scholar]
- Zhao C, Carass A, Lee J, He Y, Prince JL, 2017. Whole Brain Segmentation and Labeling from CT Using Synthetic MR Images, in: Machine Learning in Medical Imaging (MLMI 2017), Springer Berlin Heidelberg; pp. 291–298. [Google Scholar]
- Zuo L, Carass A, Han S, Prince JL, 2018. Automatic outlier detection using hidden Markov model for cerebellar lobule segmentation, in: Proceedings of SPIE Medical Imaging (SPIE-MI 2018), Houstan, TX, February 10 – 15, 2018, pp. 10578 – 10578 – 7. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.