Abstract
The primary deficiency in the membrane cytoskeletal protein dystrophin results in complex changes in dystrophic muscles. In order to compare the degree of secondary alterations in differently affected subtypes of skeletal muscles, we have conducted a global analysis of proteome-wide changes in various dystrophin-deficient muscles. In contrast to the highly degenerative mdx diaphragm muscle, which showed considerable alterations in 35 distinct proteins, the spectrum of mildly to moderately dystrophic skeletal muscles, including interosseus, flexor digitorum brevis, soleus, and extensor digitorum longus muscle, exhibited a smaller number of changed proteins. Compensatory mechanisms and/or cellular variances may be responsible for differing secondary changes in individual mdx muscles. Label-free mass spectrometry established altered expression levels for diaphragm proteins associated with contraction, energy metabolism, the cytoskeleton, the extracellular matrix and the cellular stress response. Comparative immunoblotting verified the differences in the degree of secondary changes in dystrophin-deficient muscles and showed that the up-regulation of molecular chaperones, the compensatory increase in proteins of the intermediate filaments, the fibrosis-related increase in collagen levels and the pathophysiological decrease in calcium binding proteins is more pronounced in mdx diaphragm as compared to the less severely affected mdx leg muscles. Annexin, lamin, and vimentin were identified as universal dystrophic markers.
Keywords: diaphragm, dystrophin, dystrophinopathy, Duchenne muscular dystrophy, extensor digitorum longus, flexor digitorum brevis, interosseus, muscle pathology, soleus, skeletal muscle proteome
1. Introduction
The full-length isoform of the membrane cytoskeletal actin-binding protein dystrophin, termed Dp427, represents the product of the largest gene in the human genome [1,2,3]. Although the Dp427 isoform is not an integral protein, it is tightly associated with the muscle sarcolemma via linkage to a large membrane-embedded assembly of glycoproteins [4,5,6]. The key trans-plasmalemma spanning protein within this dystrophin-associated complex was shown to be β-dystroglycan, a glycosylated protein of 43 kDa, which in turn binds to the extracellular laminin-receptor named α-dystroglycan of 156 kDa [7]. Since β-dystroglycan anchors dystrophin of 427 kDa to the surface membrane, the dystrophin-glycoprotein complex provides a connection between the actin membrane cytoskeleton of the fibre interior and the laminin network of the basal lamina and extended extracellular matrix that surrounds contractile cells [8]. The dystrophin lattice of the sarcolemma, in combination with the dystrophin-associated protein complex, is believed to stabilize muscle fibres during the repetitive physical strains of continuous excitation-contraction-relaxation cycles, and also functions as a binding partner for signalling molecules and ion channels [9,10]. It is therefore not surprising that the loss of function of such a crucial stabilizing component of the muscle fibre periphery causes severe secondary alterations in affected skeletal muscle fibres [6].
The disease grouping of dystrophinopathies is based on mutations or genetic rearrangements in the 79-exon spanning dystrophin gene [3]. Disorders of the skeletal musculature and the heart include highly progressive Duchenne muscular dystrophy, which is an early-onset and debilitating dystrophinopathy, Becker muscular dystrophy, which is a delayed-onset and milder dystrophinopathy, and X-linked dilated cardiomyopathy, which afflicts teenage men [11,12,13]. In muscular dystrophy, the absence of dystrophin isoform Dp427 was shown to cause a variety of physiological and biochemical changes, including a higher susceptibility to stretch-induced injury, impaired excitation-contraction coupling, lowered luminal calcium buffering, an elevated sarcolemmal influx of calcium ions and a concomitant increase in the proteolytic degradation of muscle proteins [14,15,16].
Model organisms have been instrumental in muscular dystrophy research, such as zebrafish, the mdx mouse and the grmd dog, including biomarker discovery studies [17,18,19,20]. Although genocopies of an inherited disorder often do not show the same clinical phenotype in animal models due to differences in down-stream pathophysiological changes, the mdx mouse model of Duchenne muscular dystrophy is highly suitable for biomedical studies and the initial evaluation of novel treatment strategies. Since the same primary abnormality, i.e., a single base substitution in exon 23 of the dystrophin gene that causes the premature termination of the Dp427 polypeptide chain, results in a great variety of changes in different subtypes of muscle tissue, the mdx mouse is an ideal system to test novel drugs or experimental treatments, such as myoblast transfer approaches or exon-skipping therapy [19]. While extraocular, laryngeal, and interosseus muscle only exhibit marginal changes, the limb musculature undergoes segmental necrosis and the diaphragm muscle shows severe fibre wasting and fibrosis in the mdx mouse. These differing degrees of histopathological changes are clearly reflected by the extent of proteome-wide alterations in dystrophic muscles [21,22,23,24,25].
Since these striking variations in the degree of muscle degeneration in the differing dystrophic phenotypes within the same organism must be due to dissimilar secondary effects and/or compensatory mechanisms, proteomics suggests itself as an ideal analytical tool to determine global differences between the various muscle subtypes in dystrophic animal models. Building on the findings from previous proteomic surveys of muscular dystrophy, as recently outlined in comprehensive reviews [26,27,28], we have determined here differential protein expression patterns in mildly vs. severely affected dystrophic mdx mouse skeletal muscles. The comparative label-free mass spectrometric analysis of the severely dystrophic and fibrotic diaphragm, the relatively mildly affected interosseus and flexor digitorum brevis muscles, and the moderately dystrophic soleus and extensor digitorum longus muscles identified annexin, lamin, and vimentin as universal dystrophic markers.
2. Results and Discussion
2.1. Comparative Label-Free Mass Spectrometric Analysis of mdx Skeletal Muscles
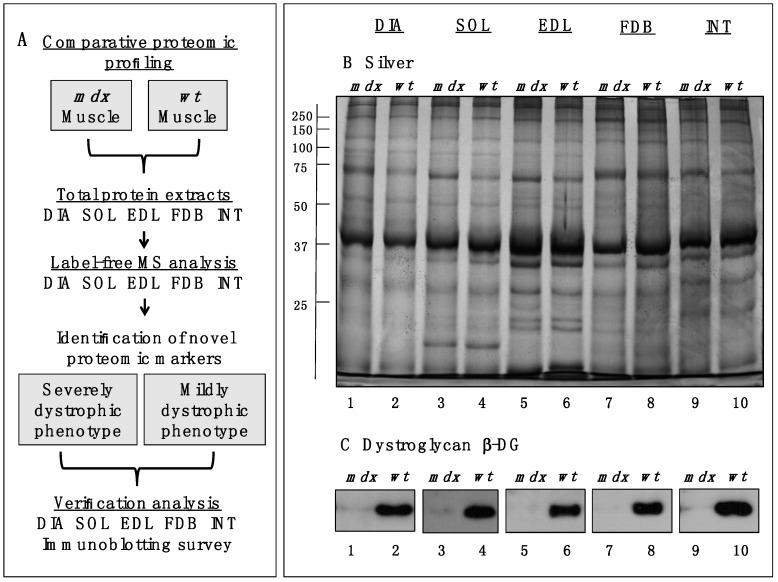
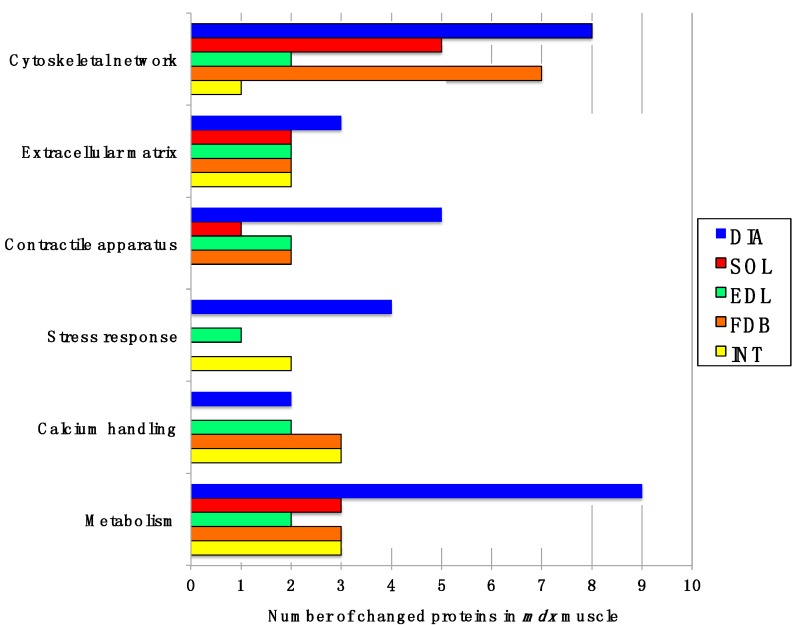
In order to identify common proteomic markers that exhibit a changed concentration in differently affected mdx skeletal muscles, total extracts from diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles were analysed by label-free mass spectrometry. Figure 1A,B outlines the workflow of this analytical strategy and shows a representative silver-stained gel of the various preparations used in this study. The immunoblot analysis of β-dystroglycan demonstrated a drastic reduction of this dystrophin-associated glycoprotein of 43 kDa in all mdx muscles investigated (Figure 1C) [29,30]. The below displayed Table 1 to Table 5 list the results for each analysed skeletal muscle detailing the accession numbers of identified proteins, the number of peptides used in the analysis, the MS scores, ANOVA values and the fold change. The comparative study revealed an altered abundance in 35, 16, 18, 23 and 14 proteins in DIA, SOL, EDL, FDB and INT preparations, respectively. The total number of positively identified proteins in wild type DIA, SOL, EDL, FDB, and INT was 296 ± 17, 270 ± 13, 202 ± 11, 207 ± 7 and 215 ± 4, respectively, and in mdx DIA, SOL, EDL, FDB and INT was 348 ± 20, 289 ± 4, 218 ± 6, 222 ± 11 and 235 ± 16, respectively. The comparison of the differences in the number of changes between individual subtypes of muscles and total numbers of identified proteins suggests, therefore, that the observed effects are mostly based on pathobiochemical variations and not technical issues associated with sample preparation. The direct comparison of proteomic findings related to the most severely affected DIA muscle vs. mildly dystrophic INT muscle illustrates this conclusion. Although 1.4-fold more protein species were overall identified in DIA preparations, the rate of changes in DIA muscle is 2.5 fold higher as compared to INT preparations (Table 1 and Table 5). This indicates that the variation in the number of changes between individual muscle subtypes is due to biological effects rather than technical matters. Figure 2 gives an overview of the number of changed proteins in individual mdx muscles and their association with distinct functional families, such as the cytoskeleton, the extracellular matrix, the contractile apparatus, the cellular stress response, Ca2+-homeostasis and metabolism. As can be deduced from Figure 2 and Table 1, the mdx DIA muscle showed both quantitatively and qualitatively the largest secondary changes due to deficiency in dystrophin. Changed expression levels were shown for proteins mostly involved in fibre contraction, energy metabolism, metabolite transportation, the cytoskeleton, the extracellular matrix and the cellular stress response.
Figure 1.
Comparative profiling of wild type (wt) muscles vs. dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx animal model of Duchenne muscular dystrophy. (A) Workflow of the comparative proteomic analysis of normal vs. dystrophic skeletal muscles; (B) Silver-stained gel of the mdx and wt preparations from DIA (lanes 1 and 2), SOL (lanes 3 and 4), EDL (lanes 5 and 6), FDB (lanes 7 and 8), and INT (lanes 9 and 10) muscles; (C) Comparative immunoblot analysis of β-dystroglycan showing antibody labelling of this dystrophin-associated glycoprotein of 43 kDa in normal vs. dystrophic muscles.
Table 1.
List of changed proteins in 100-day old mdx diaphragm muscle vs. age-matched wild type muscle as determined by label-free mass spectrometric analysis.
| Accession Number | Protein Name | Tissue Localization | Peptides | Score | ANOVA (p) | Fold Change |
|---|---|---|---|---|---|---|
| P13542 | Myosin-8 (perinatal MHC) | Myofibrils (myosin complex) | 3 | 518.76 | 1.88 × 10−5 | 798.68 |
| P13541 | Myosin-3 (embryonic MHC-3) | Myofibrils (myosin complex) | 2 | 345.71 | 1.13 × 10−5 | 84.52 |
| P09103 | Protein disulfide-isomerase | Endoplasmic reticulum | 2 | 154.39 | 1.39 × 10−5 | 3.35 |
| P48678 | Prelamin-A/C | Nuclear lamina | 8 | 511.85 | 2.87 × 10−7 | 3.31 |
| Q8CI43 | Myosin light chain 6B | Myofibrils (myosin complex) | 3 | 142.05 | 1.16 × 10−5 | 3.31 |
| P20152 | Vimentin | Intermediate filaments | 5 | 434.45 | 1.90 × 10−5 | 3.10 |
| A2AAJ9 | Obscurin | Contractile apparatus | 2 | 145.90 | 5.47 × 10−5 | 2.97 |
| P48036 | Annexin A5 | Sarcolemma region | 2 | 137.12 | 4.69 × 10−4 | 2.67 |
| Q91X72 | Hemopexin | Extracellular space | 2 | 94.70 | 1.03 × 10−4 | 2.62 |
| Q02788 | Collagen alpha-2(VI) chain | Extracellular matrix | 5 | 266.91 | 5.40 × 10−4 | 2.31 |
| P14733 | Lamin-B1 | Nuclear envelope | 2 | 127.83 | 5.32 × 10−5 | 2.23 |
| Q6LBE8 | Histone H3.2 | Nucleus | 2 | 149.87 | 1.23 × 10−3 | 2.20 |
| P26041 | Moesin | Cytoskeleton/sarcolemma | 2 | 113.60 | 4.09 × 10−4 | 2.18 |
| P07724 | Serum albumin | Extracellular space | 3 | 176.70 | 3.07 × 10−3 | 2.15 |
| Q04857 | Collagen alpha-1(VI) chain | Extracellular matrix | 5 | 305.12 | 4.35 × 10−4 | 2.12 |
| Q7TMM9 | Tubulin beta-2A chain | Microtubules | 3 | 188.58 | 2.56 × 10−4 | 2.05 |
| P20029 | 78 kDa glucose-regulated protein | Endoplasmic reticulum | 3 | 145.99 | 4.79 × 10−4 | 2.00 |
| Q921I1 | Serotransferrin | Extracellular space | 5 | 257.43 | 6.48 × 10−5 | 1.99 |
| Q8VHX6 | Filamin-C | Actin cytoskeleton | 16 | 1111.06 | 5.07 × 10−7 | 1.93 |
| P15864 | Histone H1.2 | Nucleus | 2 | 128.46 | 8.47 × 10−3 | 1.88 |
| P58252 | Elongation factor 2 | Cytoplasm/ribosome | 5 | 286.21 | 2.42 × 10−5 | 1.87 |
| P11499 | Heat shock protein Hsp90-beta (HSPAB1) | Cytoplasm | 5 | 285.81 | 6.42 × 10−5 | 1.86 |
| P63017 | Heat shock cognate 71 kDa protein (HSPA8, Hsc70) | Cytoplasm | 4 | 294.22 | 5.69 × 10−4 | 1.77 |
| P14602 | Heat shock protein beta-1 (HSPB1, Hsp27) | Cytoplasm | 2 | 114.96 | 2.04 × 10−4 | 1.77 |
| O55143 | SERCA2 Ca2+-ATPase | Sarcoplasmic reticulum | 2 | 106.96 | 3.43 × 10−2 | 1.72 |
| P31001 | Desmin | Intermediate filaments | 6 | 478.06 | 1.41 × 10−4 | 1.71 |
| Q64727 | Vinculin | Cytoskeleton | 2 | 145.57 | 7.51 × 10−4 | 1.68 |
| Q00897 | Alpha-1-antitrypsin 1-4 | Extracellular region | 3 | 148.95 | 1.47 × 10−6 | 1.63 |
| Q9WUB3 | Glycogen phosphorylase, muscle | Cytoplasm | 7 | 418.32 | 1.10 × 10−3 | −1.51 |
| Q8BWT1 | 3-ketoacyl-CoA thiolase, mitochondrial | Mitochondrion | 2 | 107.43 | 6.21 × 10−3 | −1.55 |
| P45952 | Medium-chain specific acyl-CoA dehydrogenase | Mitochondrial matrix | 2 | 112.72 | 4.60 × 10−4 | −1.56 |
| Q9JK37 | Myozenin-1 | Actin cytoskeleton | 2 | 96.43 | 1.24 × 10−3 | −1.97 |
| P04247 | Myoglobin | Cytoplasm | 3 | 144.52 | 3.35 × 10−4 | −2.73 |
| P32848 | Parvalbumin alpha | Cytoplasm | 4 | 255.00 | 6.93 × 10−4 | −2.83 |
| P16015 | Carbonic anhydrase CA3 | Cytoplasm | 3 | 192.85 | 3.38 × 10−4 | −3.76 |
Table 5.
List of changed proteins in 100-day old mdx interosseus muscle vs. age-matched wild type muscle as determined by label-free mass spectrometric analysis.
| Accession Number | Protein Name | Tissue Localization | Peptides | Score | ANOVA (p) | Fold Change |
|---|---|---|---|---|---|---|
| P13541 | Myosin-3 (perinatal MHC) | Myofibrils (myosin complex) | 2 | 299.48 | 6.78 × 10−4 | 15.70 |
| P13542 | Myosin-4 (MHC-IIB) | Myofibrils (myosin complex) | 3 | 502.66 | 1.99 × 10−4 | 8.88 |
| P07356 | Annexin A2 | Sarcolemma/basal lamina | 2 | 120.25 | 6.70 × 10−5 | 2.88 |
| P20152 | Vimentin | Intermediate filaments | 4 | 293.05 | 1.26 × 10−3 | 2.82 |
| P48678 | Prelamin-A/C | Nuclear lamina | 11 | 736.17 | 1.43 × 10−3 | 2.56 |
| Q99MQ4 | Asporin | Extracellular matrix | 2 | 93.48 | 3.76 × 10−3 | 2.53 |
| P10126 | Elongation factor 1-alpha 1 | Cytoplasm/ribosome | 2 | 87.24 | 3.21 × 10−3 | 2.27 |
| Q921I1 | Serotransferrin | Extracellular space | 3 | 167.69 | 7.26 × 10−3 | 2.05 |
| Q04857 | Collagen alpha-1(VI) chain | Extracellular matrix | 2 | 99.46 | 2.25 × 10−2 | 1.86 |
| P28654 | Decorin | Extracellular matrix | 2 | 162.96 | 1.49 × 10−2 | 1.79 |
| Q60854 | Serpin B6 | Cytoplasm | 2 | 113.51 | 3.31 × 10−3 | 1.66 |
| Q6PIE5 | Na+/K+-ATPase, alpha-2 subunit | Sarcolemma | 3 | 235.21 | 5.85 × 10−3 | −1.54 |
| Q8R429 | SERCA1 Ca2+-ATPase | Sarcoplasmic reticulum | 3 | 163.52 | 1.49 × 10−3 | −2.02 |
| Q91Z83 | Myosin-7 (cardiac MHC-β) | Myofibrils (myosin complex) | 6 | 528.44 | 1.84 × 10−2 | −2.88 |
Figure 2.
Diagrammatic presentation of the number of changed proteins in dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx animal model of Duchenne muscular dystrophy. The graph outlines affected functional protein families associated with the cytoskeleton, the extracellular matrix, the contractile apparatus, the cellular stress response, Ca2+-handling and metabolism.
2.2. Proteomic Analysis of mdx Diaphragm Muscle
As listed in Table 1, 28 proteins exhibited an increased concentration and 7 proteins a decreased abundance in dystrophic DIA muscle preparations. Overall, our analysis identified 103 changed DIA proteins including those that were recognized only by one peptide (not shown). The most drastically elevated expression levels with values of 2-fold or higher were established for myosin-8, myosin-3, protein disulphide isomerase, lamin-A/C, myosin light chain 6B, vimentin, obscurin, annexin A5, hemopexin, the collagen alpha-1(VI) and alpha-2(VI) chains, lamin-B1, histone H3.2, moesin, albumin, tubulin, and the 78 kDa glucose-regulated protein. Myosin heavy chain isoforms myosin-3 and myosin-8 are embryonic isoforms and important markers of skeletal muscle regeneration. Their substantially increased concentration suggests both fibre regeneration and the compensatory re-organization of myofibrils within dystrophic fibres. The perinatal myosin-8 isoform has been identified in mature skeletal muscles by shotgun proteomics [31] and its elevated levels in the mdx diaphragm suggests the potential recruitment of new myofibre populations containing embryonic isoforms of myosin heavy chains. Importantly, high levels of the intermediate filament protein vimentin in mdx diaphragm muscle were also previously described to occur in EDL, FDB, GAS (gastrocnemius), INT, SOL, and VL (vastus lateralis) muscles from various dystrophin-deficient mouse and dog models of X-linked muscular dystrophy [21,23,24,25,32,33,34,35,36]. This establishes this protein as an interesting biomarker candidate that might be extremely helpful to evaluate animal models of dystrophinopathy. Changes in annexins, such as isoforms A1, A2, A5, or A6, are also an established alteration in dystrophic muscle tissues [25,33,37,38]. The specific increase in annexin A5 in the 100-day old mdx diaphragm agrees with the proteomic profiling of the senescent mdx diaphragm muscle by two-dimensional fluorescence difference in-gel electrophoresis [37] and the six-month old mdx hind limb muscles using standard two-dimensional gel electrophoresis combined with silver staining [38].
Moderately increased levels, ranging from approximately 1.5- to 2-fold change, were shown for transferrin, filamin-C, histone H1.2, elongation factor EEF2, the heat shock proteins Hsp90-beta, Hsp71 cognate and Hsp beta-1, the SERCA2 isoform of the sarcoplasmic reticulum Ca2+-ATPase, desmin, vinculin, and anti-trypsin. Decreased proteins were identified as the muscle-specific isoform of glycogen phosphorylase, mitochondrial 3-ketoacyl-CoA thiolase, medium-chain specific acyl-CoA dehydrogenase, myozenin, myoglobin, parvalbumin, and carbonic anhydrase isoform CA3. In analogy to this study, a variety of molecular chaperones belonging to the Hsp70 and Hsp90 families of heat shock proteins [27] were also identified in gel-based studies of dystrophic muscles [23,37,39].
2.3. Proteomic Analysis of mdx Soleus Muscle
In the dystrophic SOL muscle, label-free mass spectrometric analysis revealed an increase of 2-fold or higher for vimentin, serpin B6, hemopexin, actin, annexin A2, and albumin (Table 2). A moderate elevation in expression levels was shown for lamin-A/C, elongation factors EEF1A1 and EEF2, antitrypsin, heat shock protein Hsp90-beta and the transitional endoplasmic reticulum ATPase. In contrast, dystrophin deficiency in SOL muscles was associated with decreases in lactate dehydrogenase, fatty acid-binding protein FABP3, myoglobin, and parvalbumin.
Table 2.
List of changed proteins in 100-day old mdx soleus muscle vs. age-matched wild type muscle as determined by label-free mass spectrometric analysis.
| Accession Number | Protein Name | Tissue Localization | Peptides | Score | ANOVA (p) | Fold Change |
|---|---|---|---|---|---|---|
| P20152 | Vimentin | Intermediate filaments | 5 | 462.30 | 3.75 × 10−5 | 5.44 |
| Q60854 | Serpin B6 | Cytoplasm | 2 | 136.46 | 8.01 × 10−5 | 3.34 |
| Q91X72 | Hemopexin | Extracellular space | 2 | 98.63 | 3.10 × 10−3 | 2.63 |
| P60710 | Actin, cytoplasmic 1 | Cytoplasm | 2 | 86.84 | 6.69 × 10−6 | 2.47 |
| P07356 | Annexin A2 | Sarcolemma/basal lamina | 3 | 184.47 | 8.10 × 10−5 | 2.41 |
| P07724 | Serum albumin | Extracellular space | 4 | 193.77 | 9.49 × 10−3 | 2.38 |
| P48678 | Prelamin-A/C | Nuclear lamina | 5 | 313.84 | 4.15 × 10−4 | 1.84 |
| P10126 | Elongation factor 1-alpha 1 | Cytoplasm/ribosome | 4 | 207.63 | 1.05 × 10−3 | 1.82 |
| Q00896 | Alpha-1-antitrypsin 1-3 | Extracellular region | 2 | 109.93 | 1.33 × 10−3 | 1.71 |
| P58252 | Elongation factor 2 | Cytoplasm/ribosome | 4 | 242.28 | 1.50 × 10−4 | 1.63 |
| P11499 | Heat shock protein HSP 90-beta (HSPAB1) | Cytoplasm | 6 | 373.77 | 1.98 × 10−3 | 1.61 |
| Q01853 | Transitional endoplasmic reticulum ATPase | Endoplasmic reticulum | 2 | 104.50 | 5.72 × 10−3 | 1.59 |
| P16125 | l-lactate dehydrogenase B chain | Cytoplasm | 2 | 129.92 | 8.59 × 10−4 | −1.71 |
| P11404 | Fatty acid-binding protein FABP3 | Cytoplasm | 2 | 83.44 | 8.28 × 10−4 | −1.84 |
| P04247 | Myoglobin | Cytoplasm | 2 | 117.20 | 7.47 × 10−4 | −2.10 |
| P32848 | Parvalbumin, alpha | Cytoplasm | 3 | 186.18 | 1.02 × 10−2 | −2.18 |
2.4. Proteomic Analysis of mdx Extensor Digitorum Longus Muscle
In the dystrophin-deficient EDL muscle, label-free mass spectrometric analysis showed a drastic increase of two-fold or higher for vimentin, albumin, annexin A2, lamin-A/C, serotransferrin, apolipoprotein A-I, histone H4, antitrypsin, heat shock proteins Hsp71 cognate, and Hsp90-beta. Moderate elevations were shown for the elongation factors EEF1A1 and EEF2, and phosphofructokinase (Table 3). The mass spectrometric analysis revealed decreases in myozenin-1, cytoplasmic aspartate aminotransferase, the alpha-1 and alpha-2 chains of collagen I, and myoglobin.
Table 3.
List of changed proteins in 100-day old mdx extensor digitorum longus muscle vs. age-matched wild type muscle as determined by label-free mass spectrometric analysis.
| Accession Number | Protein Name | Tissue Localization | Peptides | Score | ANOVA (p) | Fold Change |
|---|---|---|---|---|---|---|
| P20152 | Vimentin | Intermediate filaments | 8 | 561.29 | 4.53 × 10−4 | 7.32 |
| P07724 | Serum albumin | Extracellular space | 5 | 250.52 | 1.80 × 10−3 | 4.02 |
| P07356 | Annexin A2 | Sarcolemma/basal lamina | 2 | 124.10 | 1.77 × 10−4 | 3.93 |
| P48678 | Prelamin-A/C | Nuclear lamina | 4 | 267.71 | 7.35 × 10−6 | 3.50 |
| Q921I1 | Serotransferrin | Extracellular space | 2 | 89.28 | 2.42 × 10−2 | 2.81 |
| Q00623 | Apolipoprotein A-I | Cytoplasm/extracellular space | 3 | 234.45 | 1.82 × 10−2 | 2.55 |
| P62806 | Histone H4 | Nucleus | 2 | 123.52 | 2.26 × 10−3 | 2.41 |
| Q00896 | Alpha-1-antitrypsin 1-3 | Extracellular region | 2 | 93.70 | 3.09 × 10−2 | 2.35 |
| P63017 | Heat shock cognate 71 kDa protein (HSPA8, Hsc70) | Cytoplasm | 5 | 289.78 | 3.56 × 10−3 | 2.04 |
| P11499 | Heat shock protein Hsp90-beta (HSPAB1) | Cytoplasm | 2 | 105.43 | 5.75 × 10−5 | 2.02 |
| P58252 | Elongation factor 2 | Cytoplasm/ribosome | 3 | 185.26 | 1.63 × 10−2 | 1.85 |
| P10126 | Elongation factor 1-alpha 1 | Cytoplasm/ribosome | 2 | 91.27 | 4.50 × 10−3 | 1.53 |
| P47857 | 6-phosphofructokinase, muscle | Cytoplasm/glycolytic particle | 3 | 232.53 | 3.77 × 10−3 | 1.53 |
| Q9JK37 | Myozenin-1 | Actin cytoskeleton | 2 | 96.90 | 3.41 × 10−2 | −1.75 |
| P05201 | Aspartate aminotransferase | Cytoplasm | 2 | 123.54 | 2.64 × 10−2 | −1.92 |
| Q01149 | Collagen alpha-2(I) chain | Extracellular matrix | 3 | 142.26 | 1.99 × 10−2 | −2.62 |
| P11087 | Collagen alpha-1(I) chain | Extracellular matrix | 2 | 108.38 | 4.00 × 10−2 | −2.91 |
| P04247 | Myoglobin | Cytoplasm | 2 | 146.49 | 1.45 × 10−3 | −3.43 |
2.5. Proteomic Analysis of mdx Flexor Digitorum Brevis Muscle
In the FDB muscle from the mdx mouse, label-free mass spectrometric analysis revealed increases in vimentin, annexin A2 and A5, cytoplasmic actin, apolipoprotein A-I, alpha-1-antitrypsin 1-2, albumin, serotransferrin, heat shock protein Hsp90-beta, lamin-A/C, lumican, protein disulfide-isomerase, and the alpha-1 and alpha-2 chains of collagen VI (Table 4). A decreased abundance was observed for the glycolytic enzyme triosephosphate isomerase, protein NDRG2, glycogen phosphorylase, myozenin-1, the alpha-2 subunit of the Na+/K+-ATPase, myoglobin, perilipin-4, adenylate kinase isoform AK1, and parvalbumin. The reduction in the adenylate kinase isoform AK1 is an interesting finding and agrees with the first gel-based proteomic profiling of mdx hind limb muscles, published in 2003 by Ge et al. [38].
Table 4.
List of changed proteins in 100-day old mdx flexor digitorum brevis muscle vs. age-matched wild type muscle as determined by label-free mass spectrometric analysis.
| Accession Number | Protein Name | Tissue Localization | Peptides | Score | ANOVA (p) | Fold Change |
|---|---|---|---|---|---|---|
| P20152 | Vimentin | Intermediate filaments | 5 | 256.96 | 1.85 × 10−3 | 2.39 |
| P07356 | Annexin A2 | Sarcolemma/basal lamina | 3 | 194.99 | 1.79 × 10−3 | 2.29 |
| P60710 | Actin, cytoplasmic 1 | Cytoplasm | 2 | 191.53 | 7.90 × 10−3 | 2.27 |
| Q00623 | Apolipoprotein A-I | Cytoplasm/extracellular space | 2 | 140.56 | 2.04 × 10−3 | 2.17 |
| P22599 | Alpha-1-antitrypsin 1-2 | Extracellular region | 4 | 188.28 | 4.08 × 10−3 | 2.17 |
| P07724 | Serum albumin | Extracellular space | 2 | 98.97 | 2.57 × 10−3 | 2.16 |
| Q921I1 | Serotransferrin | Extracellular space | 4 | 216.10 | 3.21 × 10−3 | 2.01 |
| P11499 | Heat shock protein Hsp90-beta (HSPAB1) | Cytoplasm | 2 | 114.79 | 8.47 × 10−4 | 2.00 |
| P48678 | Prelamin-A/C | Nuclear lamina | 13 | 807.87 | 1.40 × 10−3 | 1.92 |
| P51885 | Lumican | Extracellular matrix | 3 | 175.81 | 6.93 × 10−3 | 1.91 |
| P48036 | Annexin A5 | Sarcolemma region | 2 | 116.73 | 1.34 × 10−2 | 1.80 |
| P09103 | Protein disulfide-isomerase | Endoplasmic reticulum | 2 | 142.68 | 5.43 × 10−4 | 1.67 |
| Q02788 | Collagen alpha-2(VI) chain | Extracellular matrix | 4 | 246.86 | 6.62 × 10−3 | 1.62 |
| Q04857 | Collagen alpha-1(VI) chain | Extracellular matrix | 3 | 192.60 | 3.43 × 10−3 | 1.61 |
| P17751 | Triosephosphate isomerase | Cytoplasm/glycolytic particle | 2 | 140.70 | 9.58 × 10−3 | −1.53 |
| Q9QYG0 | Protein NDRG2 | Cytoplasm/nucleoplasm | 2 | 168.53 | 5.98 × 10−3 | −1.61 |
| Q9WUB3 | Glycogen phosphorylase, muscle | Cytoplasm | 2 | 99.23 | 1.70 × 10−2 | −1.61 |
| Q9JK37 | Myozenin-1 | Actin cytoskeleton | 2 | 149.89 | 8.00 × 10−3 | −1.68 |
| Q6PIE5 | Na+/K+-ATPase, alpha-2 subunit | Sarcolemma | 4 | 216.92 | 8.65 × 10−3 | −1.71 |
| P04247 | Myoglobin | Cytoplasm | 3 | 193.88 | 4.83 × 10−3 | −1.81 |
| O88492 | Perilipin-4 | Cytoplasm | 2 | 138.76 | 5.52 × 10−4 | −2.04 |
| Q9R0Y5 | Adenylate kinase AK1 | Cytoplasm | 2 | 137.26 | 3.07 × 10−3 | −2.07 |
| P32848 | Parvalbumin, alpha | Cytoplasm | 2 | 113.41 | 8.62 × 10−3 | −2.69 |
2.6. Proteomic Analysis of mdx Interosseus Muscle
In the INT muscle, label-free mass spectrometric analysis identified a drastic increase in myosin-3 and myosin-4, as well as elevated levels of annexin A2, vimentin, lamin-A/C, aspirin, elongation factor EEF1A1, serotransferrin, collagen alpha-1(VI) chain, decorin, and serpin B6 (Table 5). In contrast, reductions were observed for the alpha-2 subunit of the Na+/K+-ATPase, the fast SERCA1 isoform of the sarcoplasmic reticulum Ca2+-ATPase and myosin-7.
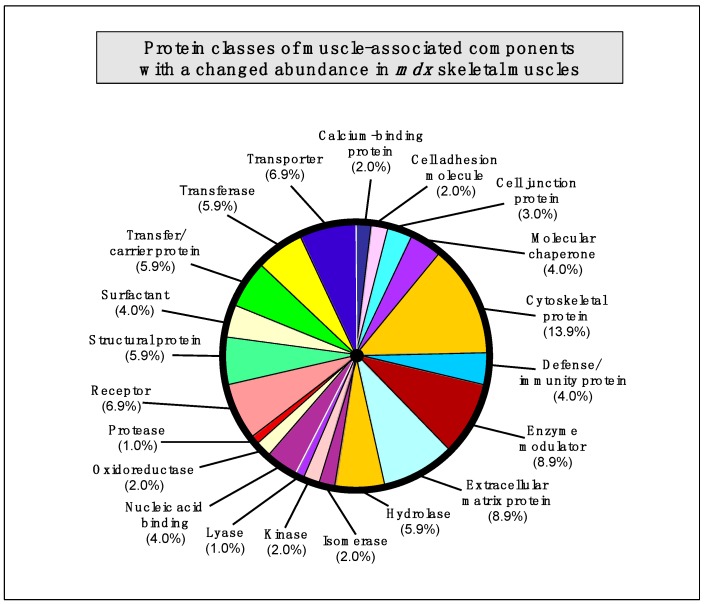
2.7. Summary of Protein Classes with a Changed Abundance in mdx Muscles
The bioinformatics analysis of altered proteins using the PANTHER database of protein families [40] resulted in the cataloguing of distinct muscle protein categories that had been identified by label-free mass spectrometric analysis in the various mdx tissues [41] (Figure 3).
Figure 3.
Bioinformatic summary of changed protein classes in mdx skeletal muscles, as determined by the software programme PANTHER [40,41]. The graph outlines the clustering of protein classes based on the label-free mass spectrometric analysis of normal vs. dystrophic muscles (Table 1, Table 2, Table 3, Table 4, Table 5).
The following protein classes exhibited a changed concentration: cytoskeletal protein (13.9%), extracellular matrix protein (8.9%), enzyme modulator (8.9%), enzyme modulator (8.9%), transporter (6.9%), receptor (6.9%), transferase (5.9%), structural protein (5.9%), hydrolase (5.9%), transfer/carrier protein (5.9%), nucleic acid binding protein (4.0%), molecular chaperone (4%), surfactant (4.0%), defense/immunity protein (4.0%), cell junction protein (3.0%), cell adhesion molecule (2.0%), calcium-binding protein (2.0%), cell adhesion molecule (2.0%), oxidoreductase (2.0%), kinase (2%), isomerase (2%), protease (1.0%), and lyase (1.0%).
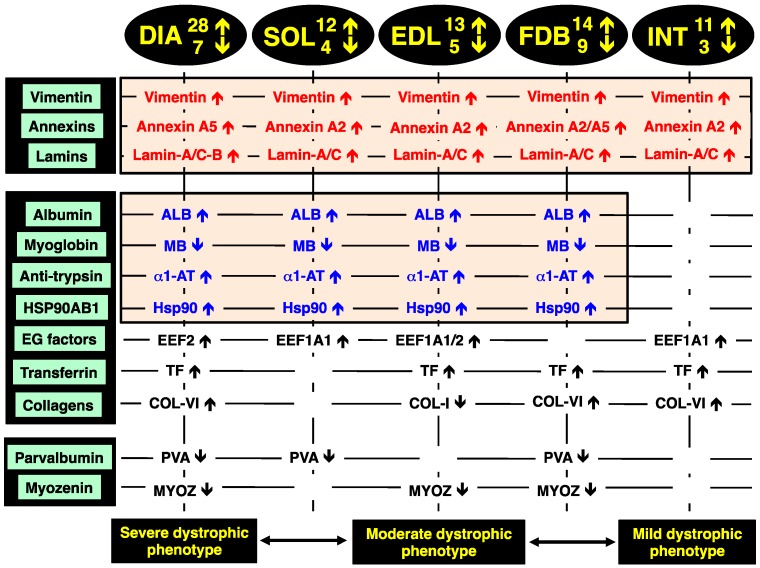
2.8. Identification of Universal Markers of Dystrophinopathy
The comparison of the proteomic data sets revealed that 3 protein species show similar changes in all dystrophic tissue specimens investigated in this study, i.e., a significant increase in the intermediate filament protein vimentin, the Ca2+-dependent membrane binding protein annexin (isoforms A2, A5), and the nuclear envelope protein lamin (isoforms A/C, B). These new universal biomarker candidates are summarised in Figure 4.
Figure 4.
Overview of the universal biomarker signature of dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx animal model of Duchenne muscular dystrophy, as revealed by label-free mass spectrometry.
In addition, all investigated mdx muscles, with the exception of INT preparations, showed analogous changes in four other muscle-associated proteins. Increases were demonstrated for albumin, anti-trypsin protein and the molecular chaperone Hsp90-beta (HSPAB1) and a decrease established for the cytoplasmic oxygen-carrier myoglobin. Additional proteins with a differential expression pattern in some of the analysed dystrophin-deficient muscles were identified as elongation factors EEF1A1 and EEF2, the iron-binding protein transferrin, various isoforms of collagen, the cytosolic Ca2+-binding protein parvalbumin, and the Z-line α-actinin binding protein myozenin (Figure 4).
The increase in the intermediate filament protein vimentin is probably a compensatory mechanism to stabilize the weakened cytoskeletal network and to rescue the load-bearing function of the fibre interior that lacks the dystrophin lattice in muscular dystrophy. Although a previous report has shown that vimentin is only transiently expressed during myotube maturation [42], vimentin can act synergistically to desmin and support the structural backbone of intermediate filaments [43]. The finding agrees with previous gel-based proteomic analyses of dystrophic mouse and dog skeletal muscles [23,24,25,32,34,36], as well as a comprehensive in vivo SILAC proteomic study of the mdx mouse [33] and a label-free mass spectrometric survey of the highly fibrotic mdx-4cv diaphragm [35]. This large number of corresponding proteomic results establishes this intermediate filament protein as a reliable and versatile muscle-associated biomarker candidate for evaluating animal models of dystrophinopathy [28].
The drastic increase in annexin isoforms A2 and A5 suggests impaired Ca2+-handling and an altered membrane organization in dystrophin-deficient fibres [44]. Since annexins are linked to the maintenance of the extracellular matrix and the actin-associated cytoskeletal network in muscle [45], their apparent up-regulation could also be an adaptive response and attempt to partially substitute for the loss of the dystrophin-actin axis [26]. The dystrophic grmd dog model of Duchenne muscular dystrophy also exhibited an increased level of annexins in the vastus lateralis muscle [36]. Since lamins are nuclear intermediate filament proteins that provide nuclear stability and support the structural linkage between muscle nuclei and the cytoskeleton [46], increased levels of lamin isoforms A/C and B probably enhance the assembly of lamin-based fibrous structures. This would maintain the inner nuclear membrane structure during degeneration-regeneration cycles and stabilize muscle fibres affected by inflammatory processes.
Altered albumin levels in dystrophic muscles may reflect disturbed oxidative metabolism and/or is connected to an increased permeability of the Dp427-deficient sarcolemma, which represents a major pathological feature in muscular dystrophies [47]. The up-regulation of alpha-1-antitrypin [48], which functions as a protective anti-protease and anti-inflammatory factor [49], might be an adaptation of dystrophic muscle tissues in response to fibre degeneration and inflammation [50]. Anti-trypsin, also known as serpina 1d protein, was also shown to be greatly increased in FDB muscles by a gel-based study [24]. The change in the Serpin B6 serine protease inhibitor was also reported by Ge et al. [38]. The loss of the cytoplasmic oxygen-carrier myoglobin, usually present at high levels in oxidative skeletal muscle fibres [51], could be due to leakage from dystrophic muscles and therefore be indicative of progressive disintegration of the dystrophic sarcolemma. Since elongation factors control muscle protein synthesis by delivering the aminoacyl-tRNA to the ribosome and thereby ensuring the proper elongation of the nascent polypeptide chain [52], increased levels of EEF1A1 and EEF2 may be involved in regenerative processes. Disturbed iron metabolism is indicated by the increased concentration of the iron-binding protein transferrin [53,54] in muscular dystrophy.
The drastic increase of collagen in the mdx diaphragm agrees with fibrosis-associated changes in dystrophinopathy [55,56,57] and confirms the results from previous analyses of dystrophic muscles [34,35]. The drastic decrease of the cytosolic Ca2+-binding protein parvalbumin [58,59] in dystrophic muscle indicates abnormal Ca2+-buffering and supports the calcium hypothesis of dystrophinopathy [60,61,62,63,64]. A preferential susceptibility of differing fibre populations may be linked to lowered levels of parvalbumin [24,35,37]. An interesting finding is the increase in the large heat shock protein Hsp90 that represents a major ATP-dependent molecular chaperone [27]. Hsp90 is involved in the activation and stabilization of many signalling proteins involved in cellular pathways. Changes in this molecular chaperone and its cyto-protective action indicate increased levels of cellular stress in muscular dystrophy [65,66] and might be a suitable marker of progressive dystrophic alterations [28]. Changes in the α-actinin binding protein myozenin of the Z-disc region [67] are also a potential new indicator of dystrophinopathy-related abnormalities within the complex arrangement of the contractile apparatus [68].
In order to relate the mass spectrometric analysis presented here with previous studies, significant proteomic hits were compared to major findings from already published reports on proteome-wide alterations in established model systems of dystrophinopathy. Table 6 and Table 7 summarize the list of major biomarker candidates identified in this report by label-free mass spectrometry and correlates the changes in 28 proteins with the proteomic results from 13 previous studies that have focused on the mdx mouse, the mdx-4cv mouse and the grmd dog models of Duchenne muscular dystrophy [21,22,23,24,25,32,33,34,35,36,37,38,39]. The tables outline the identified proteins, whereby certain proteomic hits exhibit dystrophy-related changes in more than one isoform [26,27,28], their subcellular localization and a list of analysed skeletal muscle types and animal models. Importantly, Table 6 illustrates that the observed increase in the intermediate filament component vimentin has also been shown in a considerable number of other studies [21,22,23,24,25,32,33,34,35,36] and Table 7 confirms that the significant decrease in the cytosolic Ca2+-binding protein parvalbumin was also identified in previous proteomic investigations [24,33,35,37] of dystrophin-deficient skeletal muscles. This establishes these two muscle-associated proteins with opposite changes in their concentration in dystrophic fibres as excellent analytical tools for the future assessment of animal models of X-linked muscular dystrophy.
Table 6.
Overview of increased biomarker candidates and correlation to previously published proteomic studies of secondary changes in animal models of dystrophinopathy.
| Protein Name | Tissue Localization | Animal Models | Skeletal Muscles | References |
|---|---|---|---|---|
| Vimentin | Intermediate filaments | mdx, mdx-4cv, grmd | DIA, EDL, FDB, GAS, INT, SOL, VL | [21,23,24,25,32,33,34,35,36] |
| Annexin A1, A2, A5, A6 | Sarcolemma region | mdx | DIA, EDL, FDB, INT, SOL | [25,33,37,38] |
| Lamin A/C, B | Nuclear lamina | mdx, mdx-4cv | DIA, EDL, FDB, INT, SOL | [35] |
| Myosin, embryonic MHC-3, MHC-8 | Myofibrils | mdx, mdx-4cv | DIA, GAS, INT | [33,35] |
| Obscurin | Contractile apparatus | mdx, mdx-4cv | DIA | [35] |
| Hemopexin | Extracellular space | mdx, grmd | DIA, SOL | [36] |
| Collagens, especially COL-VI | Extracellular matrix | mdx, mdx-4cv | DIA, FDB, GAS, SOL | [23,24,34,35] |
| Histone | Nucleus | mdx, mdx-4cv | DIA, EDL, GAS | [23,25,33,35] |
| Serum albumin | Extracellular space | mdx, grmd | DIA, EDL, FDB, GAS, SOL, VL | [23,32,36] |
| Tubulin | Microtubules | mdx, mdx-4cv, grmd | DIA, GAS, VL | [23,33,35,36] |
| 78 kDa glucose-regulated protein | Endoplasmic reticulum | mdx | DIA, GAS | [25,33,37] |
| Transferrin | Extracellular space | mdx, mdx-4cv | DIA, EDL, FDB, GAS, INT | [23,33,35,37,39] |
| Filamin A, C | Actin cytoskeleton | mdx, mdx-4cv | DIA, GAS | [33,35] |
| Proteoglycans (aporin, lumican, prolargin, biglycan, decorin) | mdx, mdx-4cv | DIA, INT | [35,37] | |
| Elongation factors | Cytoplasm/ribosome | mdx | DIA, EDL, GAS, INT, SOL | [23] |
| Heat shock protein Hsp90 | Cytoplasm | mdx, mdx-4cv | DIA, EDL, FDB, GAS, SOL | [23,35] |
| Heat shock protein Hsp70 | Cytoplasm | mdx | DIA, GAS | [23,37,39] |
| Small heat shock proteins (HspB1, HspB5, HspB7) | Cytoplasm | mdx | DIA, GAS, INT, SOL | [21,23,24] |
| Desmin | Intermediate filaments | mdx, mdx-4cv | DIA, EOM, GAS | [32,33,35] |
| Vinculin | Cytoskeleton | mdx, mdx-4cv | DIA, GAS | [33,35] |
| Anti-trypsin | Extracellular region | mdx | DIA, EDL, FDB, GAS, SOL | [33,38] |
Abbreviations used: DIA, diaphragm; EDL, extensor digitorum longus; EOM, extraocular muscle; FDB, flexor digitorum brevis; GAS, gastrocnemius; INT, interosseus; SOL, soleus; VL, vastus lateralis.
Table 7.
Overview of decreased biomarker candidates and correlation to previously published proteomic studies of secondary changes in animal models of dystrophinopathy.
| Protein Name | Tissue Localization | Animal Models | Skeletal Muscles | References |
|---|---|---|---|---|
| Parvalbumin | Cytoplasm | mdx, mdx-4cv | DIA, FDB, GAS, INT, SOL | [24,33,35,37] |
| Myoglobin | Cytoplasm | mdx | DIA, EDL, FDB, SOL | [24,37] |
| Fatty acid-binding protein FABP3 | Cytoplasm | mdx | DIA, SOL, GAS | [33,37] |
| Carbonic anhydrase CA3 | Cytoplasm | mdx | DIA | [21] |
| Perilipin | Cytoplasm | mdx, mdx-4cv | DIA, FDB | [35] |
| Myozenin | Actin cytoskeleton | mdx, mdx-4cv | DIA, EDL, FDB, GAS | [34,35] |
| Glycogen phosphorylase, muscle | Cytoplasm | mdx, mdx-4cv | DIA, EDL, GAS | [23,24,35] |
Abbreviations used: DIA, diaphragm; EDL, extensor digitorum longus; FDB, flexor digitorum brevis; GAS, gastrocnemius; INT, interosseus; SOL, soleus.
From the comparative analysis of the existing literature, in conjunction with the proteomic findings from this report, a variety of other promising biomarker candidates other than vimentin and parvalbumin have been identified. Increased proteins included various annexins, desmin, vinculin, tubulin, lamin, embryonic isoforms of myosin heavy chains, obscurin, hemopexin, various histones, albumin, transferrin, filamin, elongation factors, a variety of molecular chaperones and anti-trypsin, as well as collagen and associated proteoglycans (Table 6). Decreased proteins with a great potential to be useful for studying particular aspects of the molecular pathogenesis of dystrophinopathy or for the systematic monitoring of experimental therapies are myoglobin, the fatty acid-binding protein isoform FABP3, carbonic anhydrase CA3, perilipin, myozenin, and the muscle isoform of glycogen phosphorylase (Table 7). Since the identified proteins cover a wide range of cellular and structural activities in skeletal muscle tissues, their combined usage for the determination of changes in a biomarker signature would cover the maintenance of the cytoskeletal network, the extracellular matrix, energy metabolism, metabolite transportation, the cellular stress response, the excitation-contraction-relaxation cycle and other core activities.
2.9. Immunoblotting Survey of mdx Skeletal Muscles
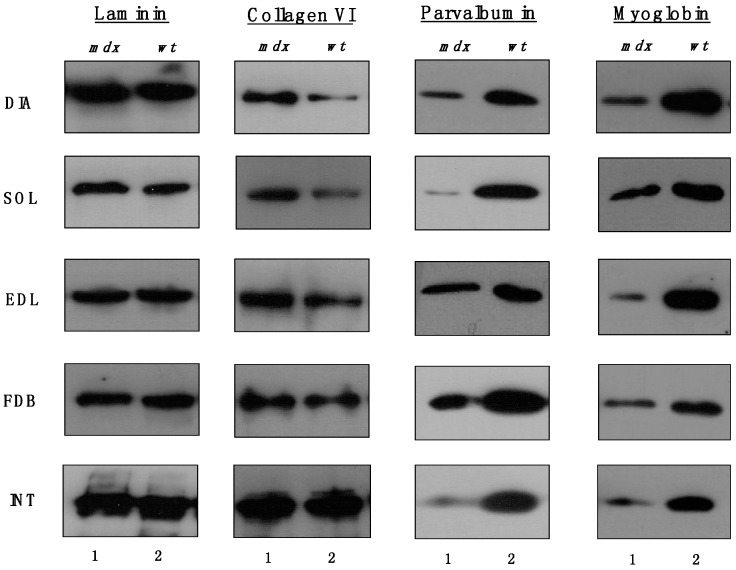
The immunoblots shown in Figure 5 confirmed the relatively unchanged concentration levels of laminin in dystrophin-deficient skeletal muscles and the tendency of increased collagen levels in all mdx preparations except INT muscle. All mdx muscles exhibited drastic decreases in myoglobin and parvalbumin, which agrees with previous findings [26,27,28].
Figure 5.
Comparative immunoblotting survey of dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx (lane 1) animal model of Duchenne muscular dystrophy vs. wild type (wt; lane 2) muscles. Antibody labelling was used to determine the concentration of laminin, collagen, parvalbumin and myoglobin.
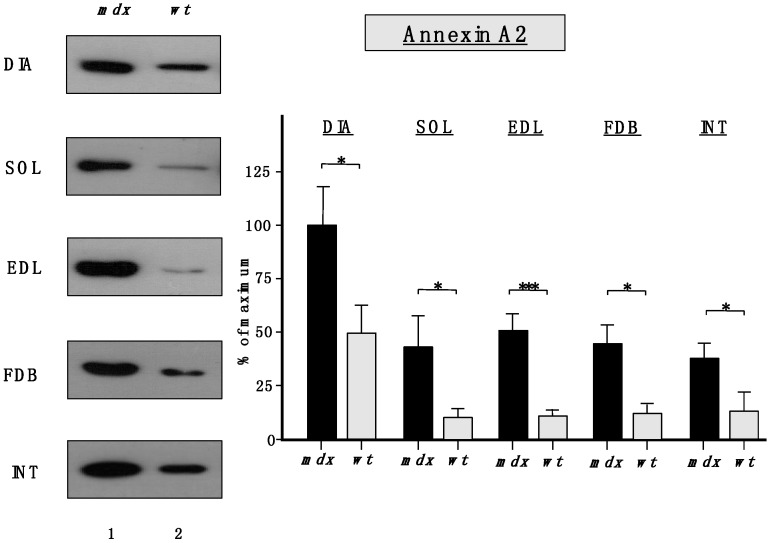
Importantly, Figure 6 clearly confirmed the findings from the proteomic survey of mdx muscles and demonstrated that annexins are significantly increased in the mdx muscles investigated in this study. The immunoblot analysis focused on annexin isoform A2.
Figure 6.
Comparative immunoblot analysis of dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx (lane 1) animal model of Duchenne muscular dystrophy vs. wild type (wt; lane 2) muscles. Antibody labeling was used to determine the concentration of annexin isoform A2 and is shown on the left side of this figure. The graphical representation of the immuno-decoration levels for annexin in mdx vs. wild type (wt) muscles is shown on the right side of this figure (Student’s t-test, unpaired; n = 4; * p < 0.05; *** p < 0.001).
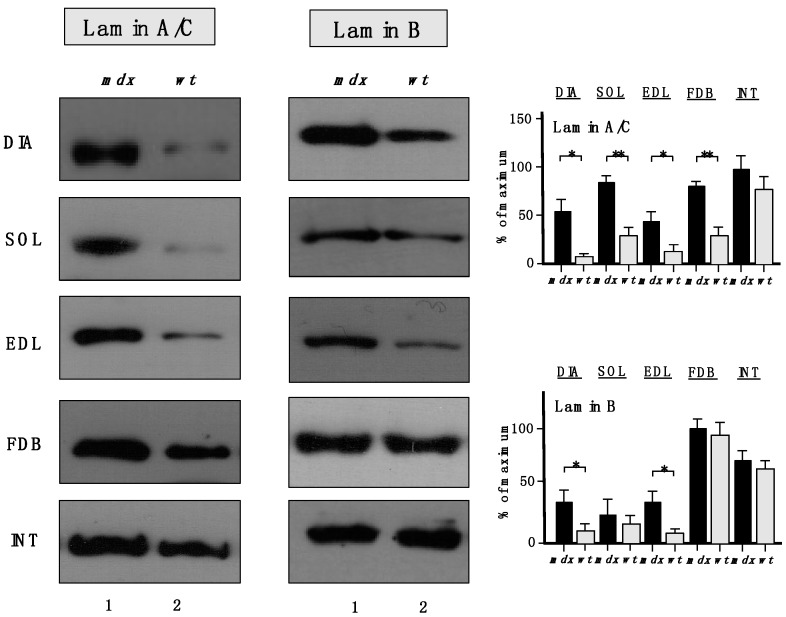
The increased levels of the nuclear envelope lamins, as suggested by proteomic analyses, were confirmed to be significant for lamin-A/C, with the exception of INT muscle (Figure 7). A drastic increase of lamin-B levels was also shown to occur in DIA and EDL muscles.
Figure 7.
Comparative immunoblot analysis of dystrophic diaphragm (DIA), soleus (SOL), extensor digitorum longus (EDL), flexor digitorum brevis (FDB), and interosseus (INT) muscles from the mdx (lane 1) animal model of Duchenne muscular dystrophy vs. wild type (wt; lane 2) muscles. Antibody labelling was used to determine the concentration of lamin isoform A/C and lamin isoform B, as shown on the left side of the figure. Graphical representation of the immuno-decoration levels for lamin A/C and lamin B in mdx vs. wild type (wt) muscles is shown on the right side of the figure (Student’s t-test, unpaired; n = 4; * p < 0.05; ** p < 0.01).
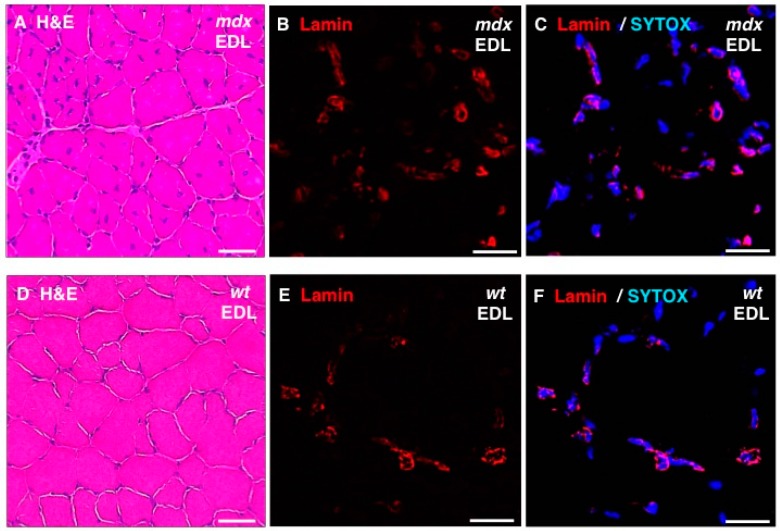
In addition, standard histochemical staining of transverse tissue sections with hematoxylin and eosin was used to estimate changes in the position and number of nuclei in dystrophic EDL muscles (Figure 8A,D). Immunofluorescence microscopy was employed to illustrate the localization and amounts of lamin in mdx fibres (Figure 8B,E) and these findings were correlated to the position of nuclei as judged by labelling of nucleic acids with SYTOX (Figure 8C,F). The histochemical analysis suggests that mdx fibres contain a considerably higher degree of central nucleation and an overall increased number of nuclei, which agrees with the previous histological profiling of the mdx mouse model of Duchenne muscular dystrophy [24,69]. Immunofluorescence microscopy revealed specific labelling of the nuclear envelope by antibodies to lamin A/C and the comparison between mdx and wild type EDL muscles suggests increased levels of lamin in dystrophin-deficient muscles.
Figure 8.
Histochemical analysis and immunofluorescence labelling of lamin in dystrophic mdx vs. wild type (wt) skeletal muscles. Shown are transverse sections of extensor digitorum longus (EDL) muscles from 100-day old mdx (A–C) and wild type (D–F) mice. Cryosections were stained with hematoxylin and eosin (H & E) (A,D) or analysed by immunofluorescence microscopy using antibodies to lamin A/C (B,C,E,F). In panels (C,F), merged images of SYTOX-based nucleic acid staining of nuclei (blue) and immunofluorescence labelling of lamin (red) are shown. In panels (A,D): bar equals 50μm; and in panels (B,C,E,F): bar equals 20 μm.
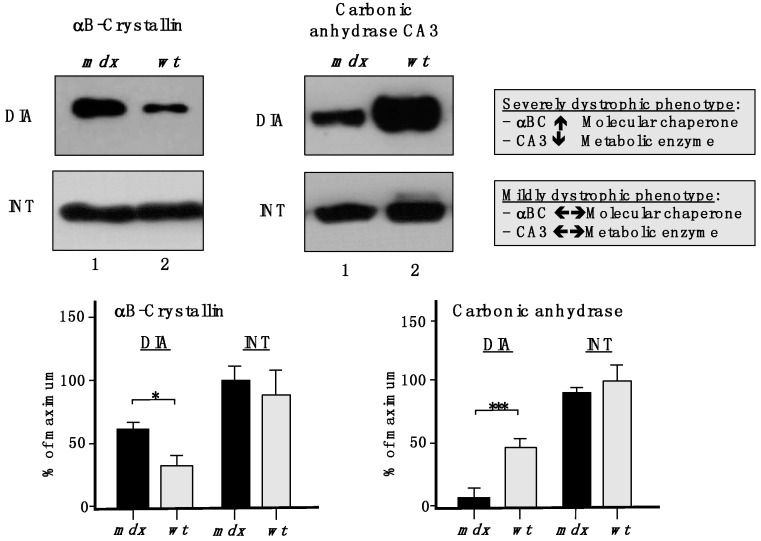
To show the drastic differences between a severely dystrophic phenotype vs. a mildly affected muscle, immunoblotting with two marker proteins is shown in Figure 9. In contrast to a drastic increase of the small heat shock protein αB-crystallin and a decrease in the CA3 isoform of carbonic anhydrase in mdx DIA muscle, both proteins exhibited comparable levels in mdx INT muscle vs. wild type INT muscle.
Figure 9.
Comparative immunoblotting to illustrate the drastic differences in biomarker concentrations between a severely dystrophic phenotype, the mdx diaphragm (DIA), vs. a mildly affected muscle, the mdx interosseus (INT) muscle. Lane 1 and 2 represent specimens from the mdx animal model of Duchenne muscular dystrophy vs. wild type (wt) specimens, respectively. Antibody labelling was used to determine the concentration of the small heat shock protein αB-crystallin and carbonic anhydrase isoform CA3, as shown in the upper part of the figure. The graphical representation of the immuno-decoration levels for αB-crystallin and carbonic anhydrase in mdx vs. wt muscles is shown in the lower part of the figure (Student’s t-test, unpaired; n = 4; * p < 0.05; *** p < 0.001).
The findings from the comparative proteomic profiling and the select immunoblotting survey of the mdx diaphragm, interosseus, flexor digitorum brevis, soleus, and extensor digitorum longus muscles clearly reflect differences in the number and extent of expression changes in proteins in severely vs. moderately or mildly dystrophic skeletal muscles. These pathobiochemical differences are probably due to variations in compensatory or adaptive mechanisms. The different subtypes of skeletal muscles studied in this report exhibit considerable dissimilarities in their contractile properties, their cellular size, motor unit organization, physiological adaptability, fibre type distribution, the extent of their calcium extrusion systems, vulnerability to proteolytic degradation, susceptibility to fibre degeneration, exposure to fatty tissue substitution, and predisposition to progressive myofibrosis. This would explain the differences in proteome-wide alterations related to proteins involved in the excitation-contraction-relaxation cycle, metabolite transportation, glycolysis, oxidative metabolism, the cytoskeletal network, the matrisome and the cellular stress response. Secondary changes in severely affected muscles that lack dystrophin are related to an (i) up-regulation of molecular chaperones, (ii) the compensatory increase in proteins of the intermediate filaments, (iii) the fibrosis-related increase in collagen levels, and (iv) the pathophysiological decrease in calcium binding proteins.
The new set of universal and muscle-associated biomarkers of progressive muscular dystrophy, such as annexin, lamin, and vimentin, can now be employed to establish improved predictive, diagnostic, prognostic and therapy-monitoring approaches focusing on murine models of dystrophinopathy [70]. Besides pharmacological treatments of general muscle wasting and cardio-respiratory complications, using glucocorticoids, diuretics and beta-blockers [71,72,73,74], new efforts to address the progressive nature of X-linked muscular dystrophy lie in cell-based, gene transfer, stop-codon read-through and exon-skipping procedures [75,76,77,78,79,80,81]. Proteomic markers can be extremely helpful in judging the overall effectiveness of these new therapeutic methods, since changes in these muscle proteins can give excellent indications of the reversal of specific damage pathways involved in progressive dystrophinopathies of model organisms [82].
3. Experimental Section
3.1. Materials
For the comparative proteomic analysis of mildly vs. severely affected mdx mouse skeletal muscles, materials and analytical grade chemicals were purchased from BioRad Laboratories (Hemel-Hempstead, Hertfordshire, UK) and Amersham Biosciences/GE Healthcare (Little Chalfont, Buckinghamshire, UK). Sequencing grade modified proteases (Lys-C and trypsin) were from Promega (Madison, WI, USA). Protease inhibitor cocktail tablets and chemiluminescence ECL kits were obtained from Roche (Mannheim, Germany). For immunoblotting, Whatman NC transfer membranes were purchased from Invitrogen (Carlsbad, CA, USA). Primary antibodies for immunoblotting were obtained from Santa Cruz Biotechnology, CA, USA (Sc-33701 to β-dystroglycan), Sigma Chemical Company, Dorset, UK (L-9393 to laminin), and Abcam, Cambridge, UK (ab6588 to collagen VI; ab85366 to carbonic anhydrase isoform CA3; ab11427 to parvalbumin; ab13496 to αB-crystallin; ab8984 to lamin-A/C; ab16048 to lamin B; ab41803 to annexin A2; and ab77232 to myoglobin). For immunofluorescence microscopy, a polyclonal rabbit antibody to lamin A/C (H-110) was purchased from Santa Cruz Biotechnology (Santa Cruz, TX, USA). Peroxidase-conjugated secondary antibodies were from Chemicon International (Temecula, CA, USA). The nucleic acid stain SYTOX Green and goat anti-rabbit IgG Alexa Fluor 647 conjugate was obtained from Invitrogen (Darmstadt, Germany). Tissue-Tek O.C.T. compound was from Sakura (Alphen aan de Rijn, Netherlands) and the mounting media Neo-Mount and Mowiol were purchased from Merck-Millipore (Schwalbach, Germany). All other chemicals were obtained from Sigma Chemical Company (Dorset, UK).
3.2. Preparation of Protein Extracts from the mdx Mouse Model of Duchenne Muscular Dystrophy
In order to compare the proteomic profile of dystrophin-deficient skeletal muscles with a highly degenerative vs. a mildly to moderately dystrophic phenotype, diaphragm, interosseus, flexor digitorum brevis, soleus, and extensor digitorum longus muscle were prepared from 100-day old male mdx mice, using a previously optimized method [24]. Wild type controls and mdx mice were obtained from the Animal Facility of the University Medicine Greifswald, Germany. Mice were kept under standard conditions and all procedures were carried out in accordance with German and Irish guidelines on the use of animals for scientific experiments; approved by the District Veterinary Office in Anklam, Germany. Animals were sacrificed by cervical dislocation after short ether anaesthesia and individual subtypes of skeletal muscles dissected and immediately quick-frozen in liquid nitrogen. For the comparative proteomic analysis of mdx tissues, muscle samples were transported to Maynooth University on dry ice and stored at −80 °C prior to usage. Tissue specimens were homogenised in a lysis buffer containing 7 M urea, 2 M thiourea, 65 mM CHAPS, 100 mM DTT; at a ratio of 1:10 (w/v). In order to prevent the potential proteolytic degradation of sensitive muscle proteins and aid the extraction process, the homogenization buffer was supplemented with a protease inhibitor cocktail and DNAase, respectively, as previously described in detail [83]. Muscle preparations were homogenised with a hand-held homogenizer model IKA T10 Basic Homogenizer from Fisher Scientific (Dublin, Ireland). Crude muscle extracts were then incubated for 2.5 h at 4 °C with gentle agitation using a Thermomixer from Eppendorf (Hamburg, Germany). Samples were centrifuged at 4 °C for 20 min at 14,000 g and the protein-containing supernatant fraction from both normal and mdx preparations were then carefully removed and used for the comparative proteomic analysis using label-free mass spectrometry. Protein concentrations were determined by the Bradford assay protocol [84].
3.3. Label-Free LC-MS/MS Analysis
Building on the previous gel-based analysis of various skeletal muscle subtypes [24], samples were separated here by a liquid chromatographic method. The proteomic profiling of 5 different subtypes of skeletal muscles from mutant mdx vs. wild type mice was carried out with a total of 40 different specimens. This included 4 biological repeats each of the 5 muscle subtypes (DIA, EDL, SOL, FDB, INT) from wild type mice and 4 biological repeats each of the 5 muscle subtypes (DIA, EDL, SOL, FDB, INT) from mdx mice. Preparation of individual protein fractions for label-free LC-MS/MS analysis was carried out in accordance with a previously optimised method [83]. Crude skeletal muscle protein samples were pre-treated with the ReadyPrep 2D clean up kit from BioRad Laboratories (Hemel-Hempstead, Hertfordshire, UK). The protein pellets created from the clean up kit were re-suspended in a label-free solubilisation buffer containing 6 M urea, 2 M thiourea and 10 mM Tris-Cl, pH 8.0 in LC-MS grade water. Re-suspended protein samples were then carefully vortexed, sonicated and centrifuged to ensure pellets were fully re-suspended. For label-free MS analysis, volumes were initially equalised with label-free solubilisation buffer and kept to a minimum. All samples were reduced for 30 min with 10 mM DTT and alkylated for 20 min in the dark with 25 mM iodoacetamide in 50 mM ammonium bicarbonate. Proteolytic digestion was initially carried out with Lys-C at a ratio of 1:100 (protease/protein) for 4 h at 37 °C. Samples were then diluted with 50 mM ammonium bicarbonate four times that of the initial volume. The second digestion step using trypsin was performed at a ration of 1:25 (protease/protein) overnight at 37 °C. Ahead of MS analysis the digested protein preparations were finally diluted at a ratio of 3:1 (v/v) with 2% TFA in 20% ACN, and then again vortexed and sonicated to ensure an even protein suspension.
An Ultimate 3000 nanoLC system (Dionex) coupled to a an LTQ Orbitrap XL mass spectrometer from Thermo Fisher Scientific (Dublin, Ireland) was used for the nano LC-MS/MS analysis of muscle proteins in the Proteomics Facility of the National Institute for Cellular Biotechnology, Dublin City University, as previously described [85,86]. Digested peptide mixtures (5 μL volume) were loaded onto a C18 trap column (C18 PepMap, 300 μm id × 5 mm, 5 μm particle size, 100 Å pore size; Dionex). Desalting was carried out at a flow rate of 25 μL/min in 0.1% TFA and 2% ACN for 5 min. The trap column was switched on-line with an analytical PepMap C18 column (75 μm id × 500 mm, 3 μm particle, and 100 Å pore size; Dionex). Peptides generated from skeletal muscle proteins were eluted with the following binary gradients: solvent A (2% ACN and 0.1% formic acid in LC-MS grade water) and 0%–25% solvent B (80% ACN and 0.08% formic acid in LC-MS grade water) for 240 min and 25%–50% solvent B for a further 60 min. The column flow rate was set to 350 nL/min. Data was acquired with Xcalibur software, version 2.0.7 (Thermo Fisher Scientific). The MS apparatus was operated in data-dependent mode and externally calibrated. Survey MS scans were acquired in the Orbitrap in the 400–1800 m/z range with the resolution set to a value of 30,000 at m/z 400 and lock mass set to 445.120025 u. CID fragmentation was carried out in the linear ion trap with up to three of the most intense ions (1+, 2+ and 3+) per scan [86]. Within 40 s, a dynamic exclusion window was applied. A normalised collision energy of 35%, an isolation window of 3 m/z, and one microscan were used to collect suitable tandem mass spectra [87].
3.4. Quantitative Profiling by Label-Free LC-MS/MS Analysis
Progenesis label-free LC-MS software version 3.1 from Non-Linear Dynamics (Newcastle upon Tyne, UK) was used to process the raw data generated from LC-MS/MS analysis. Data alignment was based on the LC retention time of each sample, allowing for any drift in retention time given and adjusted retention time for all runs in the analysis [86]. A reference run was established with the sample run that yielded most features (i.e., peptide ions). The retention times of all of the other runs were aligned to this reference run and peak intensities were then normalized. Prior to exporting the MS/MS output files to MASCOT (www.matrixscience.com) for protein identification, a number of criteria were employed to filter the data. This data included (i) peptide features with ANOVA < 0.05 between experimental groups, (ii) mass peaks (features) with charge states of +1, +2 and +3, and (iii) greater than one isotope per peptide [83]. A MASCOT generic file was generated from all exported MS/MS spectra from Progenesis software. The MASCOT generic file was used for peptide identification with MASCOT (version 2.2) and searched against the UniProtKB-SwissProt database (downloaded in January 2013) with 16,638 proteins (taxonomy: Mus musculus). The following search parameters were used for protein identification: (i) MS/MS mass tolerance set at 0.5 Da; (ii) peptide mass tolerance set to 20 ppm; (iii) carbamidomethylation set as a fixed modification; (iv) up to two missed cleavages were allowed; and (v) methionine oxidation set as a variable modification. On average, 3 out of 4 peptides were identified without a missed cleavage. For further consideration and re-importation back into Progenesis LC-MS software for further analysis, only peptides with ion scores of 40 and above were chosen. Importantly, the following criteria were applied to assign a muscle-associated protein as differentially expressed: (i) an ANOVA score between experimental groups of ≤ 0.05, (ii) proteins with ≥ 2 peptides matched, and (iii) a MASCOT score > 40 [85].
The bioinformatics analysis of potential protein interactions was carried out with standard software programmes and applied to catalogue the clustering of molecular functions and to identify potential protein interactions of the MS-identified muscle proteins with a changed concentration in the dystrophic mdx skeletal muscles [40]. Analyses were performed with the PANTHER (http://pantherdb.org; version 8.1) comprehensive database of protein families for the cataloguing of molecular functions [41].
3.5. Verification of Proteomic Findings Using Comparative Immunoblotting
In order to verify potential concentration changes of specific proteins in highly degenerative vs. mildly to moderately affected mdx muscles and confirm the proteomic data of this study, diaphragm, interosseus, flexor digitorum brevis, soleus, and extensor digitorum longus muscle were analysed by comparative immunoblotting surveys. Gel electrophoretic separation, membrane transfer and antibody incubation were performed by previously optimized methods [88]. The one-dimensional separation of muscle proteins from dystrophic vs. normal mice was performed using standard 10% PAGE gels. The electrophoretic transfer to Whatman Protan nitrocellulose sheets was carried out using a standardized wet transfer technique at 100 V for 70 min and 4 °C in a Transblot Cell from BioRad Laboratories (Hemel-Hempstead, Hertfordshire, UK). Membrane sheets were blocked for 1 h in a protein solution (5% (w/v) fat-free milk powder in phosphate-buffered saline) to prevent non-specific antibody binding. Nitrocellulose sheets were incubated in appropriately diluted primary antibody for a minimum of 3 h at 4 °C with gentle agitation. Membranes were thoroughly washed and incubated with peroxidase-conjugated secondary antibodies for 1 h at room temperature with gentle agitation. Membranes were washed again and antibody-labelled protein bands were visualised by the enhanced chemiluminescence method. Densitometric scanning and analysis of immunoblots was performed with a HP PSC-2355 scanner and ImageJ (NIH, USA) and Graph-Pad Prism software (San Diego, CA, USA).
3.6. Histochemical Analysis and Immunofluorescence Microscopy
Standard histochemical analysis and optimized immunofluorescence microscopy was carried out to characterize select mdx muscle specimens used in this study. Freshly dissected EDL muscles from 100-day old mdx and age-matched wild type mice were embedded in Tissue-Tek O.C.T. compound and frozen in −90 °C tempered petroleum ether [89]. Transverse sections of 10 µm thickness were cut at −25 °C using a Frigocut 2800n cryostat (Leica, Wetzlar, Germany) and air-dried overnight. For a histological overview, tissue sections were stained with hematoxylin and eosin (H & E) according to standard protocols [24], dehydrated in alcohol and mounted with Neo-Mount (Merck). For immunolabelling of lamin, acetone-fixed sections were initially blocked in a solution of phosphate-buffered saline (pH 7.4) containing 10% (v/v) normal goat serum and 0.5% (v/v) saponin and then incubated with a polyclonal rabbit antibody against lamin A/C. Primary antibodies were detected by a Alexa Fluor 647 conjugate and nuclei were counter-stained with the high-affinity nucleic acid stain SYTOX Green (Invitrogen). Samples were mounted with Mowiol (Merck-Millipore). All images were taken with an inverse fluorescence microscope model BZ9000 from Keyence Corporation (Mechelen, Belgium).
4. Conclusions
Label-free mass spectrometry, in combination with comparative immunoblotting, was successfully employed for the systematic identification of proteome-wide changes in mildly affected muscles vs. severely degenerated muscles from the mdx mouse model of dystrophinopathy. Striking intra-muscular increases in annexin, vimentin, and lamin were identified that might be exploitable for the future establishment of a comprehensive diagnostic biomarker signature in murine studies. Compensatory mechanisms and/or cellular variances may be responsible for differing secondary changes in individual mdx muscles. Differential markers were shown to be albumin, anti-trypsin, myoglobin, Hsp90, elongation factors, transferrin, collagens, parvalbumin, and myozenin. These proteins can now be further characterized to determine their potential usefulness as diagnostic, prognostic or therapy-monitoring markers in the field of animal model research and its biomedical applications.
Acknowledgments
This project was funded by the Higher Education Authority (HEA). The Programme for Research in Third Level Institutions PRTLI Cycle 5 is co-funded by the Irish Government and the European Union under Ireland’s EU Structural Funds Programme 2007–2013. Research was also supported by project grants from Muscular Dystrophy Ireland, the European Commission (FP7-REGPOT-2010, Grant No. 264143) and the Deutsche Duchenne Stiftung aktion benni & co e.V., as well as equipment grants from the Irish Health Research Board and the Higher Education Authority.
Author Contributions
Ashling Holland designed and performed the analytical experiments and the bioinformatics analysis. Kay Ohlendieck and Heinrich Brinkmeier conceived this collaborative study. Paula Meleady and Michael Henry are in charge of the Proteomics Suite of Dublin City University and have been involved in the mass spectrometric analysis. Claudia K. Winkler and Mirjam Krautwald were involved in the preparation of samples of dystrophic muscle specimens, the histochemical analysis and immunofluorescence microscopy. All authors were involved in interpreting of data, as well as the writing and correcting of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: Samples of the compounds are not available from the authors.
References
- 1.Hoffman E.P., Brown R.H., Jr., Kunkel L.M. Dystrophin: The protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Koenig M., Monaco A.P., Kunkel L.M. The complete sequence of dystrophin predicts a rod-shaped cytoskeletal protein. Cell. 1988;53:219–228. doi: 10.1016/0092-8674(88)90383-2. [DOI] [PubMed] [Google Scholar]
- 3.Muntoni F., Torelli S., Ferlini A. Dystrophin and mutations: One gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2:731–740. doi: 10.1016/S1474-4422(03)00585-4. [DOI] [PubMed] [Google Scholar]
- 4.Campbell K.P., Kahl S.D. Association of dystrophin and an integral membrane glycoprotein. Nature. 1989;338:259–262. doi: 10.1038/338259a0. [DOI] [PubMed] [Google Scholar]
- 5.Yoshida M., Ozawa E. Glycoprotein complex anchoring dystrophin to sarcolemma. J. Biochem. 1990;108:748–752. doi: 10.1093/oxfordjournals.jbchem.a123276. [DOI] [PubMed] [Google Scholar]
- 6.Ervasti J.M., Ohlendieck K., Kahl S.D., Gaver M.G., Campbell K.P. Deficiency of a glycoprotein component of the dystrophin complex in dystrophic muscle. Nature. 1990;345:315–319. doi: 10.1038/345315a0. [DOI] [PubMed] [Google Scholar]
- 7.Ibraghimov-Beskrovnaya O., Ervasti J.M., Leveille C.J., Slaughter C.A., Sernett S.W., Campbell K.P. Primary structure of dystrophin-associated glycoproteins linking dystrophin to the extracellular matrix. Nature. 1992;355:696–702. doi: 10.1038/355696a0. [DOI] [PubMed] [Google Scholar]
- 8.Rybakova I.N., Patel J.R., Ervasti J.M. The dystrophin complex forms a mechanically strong link between the sarcolemma and costameric actin. J. Cell Biol. 2000;150:1209–1214. doi: 10.1083/jcb.150.5.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ohlendieck K. Towards an understanding of the dystrophin-glycoprotein complex: Linkage between the extracellular matrix and the membrane cytoskeleton in muscle fibers. Eur. J. Cell Biol. 1996;69:1–10. [PubMed] [Google Scholar]
- 10.Ervasti J.M., Sonnemann K.J. Biology of the striated muscle dystrophin-glycoprotein complex. Int. Rev. Cytol. 2008;265:191–225. doi: 10.1016/S0074-7696(07)65005-0. [DOI] [PubMed] [Google Scholar]
- 11.Beggs A.H., Hoffman E.P., Snyder J.R., Arahata K., Specht L., Shapiro F., Angelini C., Sugita H., Kunkel L.M. Exploring the molecular basis for variability among patients with Becker muscular dystrophy: Dystrophin gene and protein studies. Am. J. Hum. Genet. 1991;49:54–67. [PMC free article] [PubMed] [Google Scholar]
- 12.Bushby K., Finkel R., Birnkrant D.J., Case L.E., Clemens P.R., Cripe L., Kaul A., Kinnett K., McDonald C., Pandya S., et al. Diagnosis and management of Duchenne muscular dystrophy, part 1: Diagnosis, and pharmacological and psychosocial management. Lancet Neurol. 2010;9:77–93. doi: 10.1016/S1474-4422(09)70271-6. [DOI] [PubMed] [Google Scholar]
- 13.Diegoli M., Grasso M., Favalli V., Serio A., Gambarin F.I., Klersy C., Pasotti M., Agozzino E., Scelsi L., Ferlini A., et al. Diagnostic work-up and risk stratification in X-linked dilated cardiomyopathies caused by dystrophin defects. J. Am. Coll. Cardiol. 2011;58:925–934. doi: 10.1016/j.jacc.2011.01.072. [DOI] [PubMed] [Google Scholar]
- 14.Shin J., Tajrishi M.M., Ogura Y., Kumar A. Wasting mechanisms in muscular dystrophy. Int. J. Biochem. Cell Biol. 2013;45:2266–2279. doi: 10.1016/j.biocel.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mosqueira M., Zeiger U., Förderer M., Brinkmeier H., Fink R.H. Cardiac and respiratory dysfunction in Duchenne muscular dystrophy and the role of second messengers. Med. Res. Rev. 2013;33:1174–1213. doi: 10.1002/med.21279. [DOI] [PubMed] [Google Scholar]
- 16.Vallejo-Illarramendi A., Toral-Ojeda I., Aldanondo G., López de Munain A. Dysregulation of calcium homeostasis in muscular dystrophies. Expert Rev. Mol. Med. 2014;16:e16. doi: 10.1017/erm.2014.17. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura A., Takeda S. Mammalian models of Duchenne Muscular Dystrophy: Pathological characteristics and therapeutic applications. J. Biomed. Biotechnol. 2011 doi: 10.1155/2011/184393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng R., Banks G.B., Hall J.K., Muir L.A., Ramos J.N., Wicki J., Odom G.L., Konieczny P., Seto J., Chamberlain J.R. Animal models of muscular dystrophy. Prog. Mol. Biol. Transl. Sci. 2012;105:83–111. doi: 10.1016/B978-0-12-394596-9.00004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Partridge T.A. The mdx mouse model as a surrogate for Duchenne muscular dystrophy. FEBS J. 2013;280:4177–4186. doi: 10.1111/febs.12267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Plantié E., Migocka-Patrzałek M., Daczewska M., Jagla K. Model Organisms in the Fight against Muscular Dystrophy: Lessons from Drosophila and Zebrafish. Molecules. 2015;20:6237–6253. doi: 10.3390/molecules20046237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doran P., Martin G., Dowling P., Jockusch H., Ohlendieck K. Proteome analysis of the dystrophin-deficient MDX diaphragm reveals a drastic increase in the heat shock protein cvHSP. Proteomics. 2006;6:4610–4621. doi: 10.1002/pmic.200600082. [DOI] [PubMed] [Google Scholar]
- 22.Lewis C., Ohlendieck K. Proteomic profiling of naturally protected extraocular muscles from the dystrophin-deficient mdx mouse. Biochem. Biophys. Res. Commun. 2010;396:1024–1029. doi: 10.1016/j.bbrc.2010.05.052. [DOI] [PubMed] [Google Scholar]
- 23.Gardan-Salmon D., Dixon J.M., Lonergan S.M., Selsby J.T. Proteomic assessment of the acute phase of dystrophin deficiency in mdx mice. Eur. J. Appl. Physiol. 2011;111:2763–2773. doi: 10.1007/s00421-011-1906-3. [DOI] [PubMed] [Google Scholar]
- 24.Carberry S., Brinkmeier H., Zhang Y., Winkler C.K., Ohlendieck K. Comparative proteomic profiling of soleus, extensor digitorum longus, flexor digitorum brevis and interosseus muscle from the mdx mouse model of Duchenne muscular dystrophy. Int. J. Mol. Med. 2013;32:544–556. doi: 10.3892/ijmm.2013.1429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Matsumura C.Y., de Oliveira B.M., Durbeej M., Marques M.J. Isobaric Tagging-Based Quantification for Proteomic Analysis: A Comparative Study of Spared and Affected Muscles from mdx Mice at the Early Phase of Dystrophy. PLoS ONE. 2013;8:e65831. doi: 10.1371/journal.pone.0065831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holland A., Carberry S., Ohlendieck K. Proteomics of the dystrophin-glycoprotein complex and dystrophinopathy. Curr. Protein Pep. Sci. 2013;14:680–697. doi: 10.2174/13892037113146660083. [DOI] [PubMed] [Google Scholar]
- 27.Brinkmeier H., Ohlendieck K. Chaperoning heat shock proteins: Proteomic analysis and relevance for normal and dystrophin-deficient muscle. Proteomics Clin. Appl. 2014;8:875–895. doi: 10.1002/prca.201400015. [DOI] [PubMed] [Google Scholar]
- 28.Dowling P., Holland A., Ohlendieck K. Mass spectrometry-based identification of muscle-associated and muscle-derived proteomic biomarkers of dystrophinopathies. J. Neuromusc. Dis. 2014;1:15–40. [PubMed] [Google Scholar]
- 29.Ohlendieck K., Campbell K.P. Dystrophin-associated proteins are greatly reduced in skeletal muscle from mdx mice. J. Cell Biol. 1991;115:1685–1694. doi: 10.1083/jcb.115.6.1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ohlendieck K., Matsumura K., Ionasescu V.V., Towbin J.A., Bosch E.P., Weinstein S.L., Sernett S.W., Campbell K.P. Duchenne muscular dystrophy: Deficiency of dystrophin-associated proteins in the sarcolemma. Neurology. 1993;43:795–800. doi: 10.1212/WNL.43.4.795. [DOI] [PubMed] [Google Scholar]
- 31.Drexler H.C., Ruhs A., Konzer A., Mendler L., Bruckskotten M., Looso M., Günther S., Boettger T., Krüger M., Braun T. On marathons and Sprints: An integrated quantitative proteomics and transcriptomics analysis of differences between slow and fast muscle fibers. Mol. Cell. Proteomics. 2012;11 doi: 10.1074/mcp.M111.010801. M111.010801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doran P., Dowling P., Donoghue P., Buffini M., Ohlendieck K. Reduced expression of regucalcin in young and aged mdx diaphragm indicates abnormal cytosolic calcium handling in dystrophin-deficient muscle. Biochim. Biophys. Acta. 2006;1764:773–785. doi: 10.1016/j.bbapap.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 33.Rayavarapu S., Coley W., Cakir E., Jahnke V., Takeda S., Aoki Y., Grodish-Dressman H., Jaiswal J.K., Hoffman E.P., Brown K.J., Hathout Y., Nagaraju K. Identification of disease specific pathways using in vivo SILAC proteomics in dystrophin deficient mdx mouse. Mol. Cell. Proteomics. 2013;12:1061–1073. doi: 10.1074/mcp.M112.023127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Carberry S., Zweyer M., Swandulla D., Ohlendieck K. Proteomics reveals drastic increase of extracellular matrix proteins collagen and dermatopontin in aged mdx diaphragm muscle. Int. J. Mol. Med. 2012;30:229–234. doi: 10.3892/ijmm.2012.1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Holland A., Dowling P., Meleady P., Henry M., Zweyer M., Mundegar R.R., Swandulla D., Ohlendieck K. Label-free mass spectrometric analysis of the mdx-4cv diaphragm identifies the matricellular protein periostin as a potential factor involved in dystrophinopathy-related fibrosis. Proteomics. 2015 doi: 10.1002/pmic.201400471. in press. [DOI] [PubMed] [Google Scholar]
- 36.Guevel L., Lavoie J.R., Perez-Iratxeta C., Rouger K., Dubreil L., Feron M., Talon S., Brand M., Megeney L.A. Quantitative proteomic analysis of dystrophic dog muscle. J. Proteome Res. 2011;10:2465–2478. doi: 10.1021/pr2001385. [DOI] [PubMed] [Google Scholar]
- 37.Carberry S., Zweyer M., Swandulla D., Ohlendieck K. Application of fluorescence two-dimensional difference in-gel electrophoresis as a proteomic biomarker discovery tool in muscular dystrophy research. Biology. 2013;2:1438–1464. doi: 10.3390/biology2041438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ge Y., Molloy M.P., Chamberlain J.S., Andrews P.C. Proteomic analysis of mdx skeletal muscle: Great reduction of adenylate kinase 1 expression and enzymatic activity. Proteomics. 2003;3:1895–1903. doi: 10.1002/pmic.200300561. [DOI] [PubMed] [Google Scholar]
- 39.Carberry S., Zweyer M., Swandulla D., Ohlendieck K. Comparative proteomic analysis of the contractile-protein-depleted fraction from normal versus dystrophic skeletal muscle. Anal. Biochem. 2014;446:108–115. doi: 10.1016/j.ab.2013.08.004. [DOI] [PubMed] [Google Scholar]
- 40.Mi H., Muruganujan A., Thomas P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–D386. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.PANTHER Gene List Analysis. [(accessed on 10 September 2014)]. Available online: http://pantherdb.org/
- 42.Hauerslev S., Sveen M.L., Duno M., Angelini C., Vissing J., Krag T.O. Calpain 3 is important for muscle regeneration: Evidence from patients with limb girdle muscular dystrophies. BMC Musculoskelet. Disord. 2012;13:43. doi: 10.1186/1471-2474-13-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bär H., Strelkov S.V., Sjöberg G., Aebi U., Herrmann H. The biology of desmin filaments: How do mutations affect their structure, assembly, and organisation? J. Struct. Biol. 2004;148:137–152. doi: 10.1016/j.jsb.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 44.Selbert S., Fischer P., Menke A., Jockusch H., Pongratz D., Noegel A.A. Annexin VII relocalization as a result of dystrophin deficiency. Exp. Cell Res. 1996;222:199–208. doi: 10.1006/excr.1996.0025. [DOI] [PubMed] [Google Scholar]
- 45.Bizzarro V., Petrella A., Parente L. Annexin A1: Novel roles in skeletal muscle biology. J. Cell. Physiol. 2012;227:3007–3015. doi: 10.1002/jcp.24032. [DOI] [PubMed] [Google Scholar]
- 46.Gruenbaum Y., Medalia O. Lamins: The structure and protein complexes. Curr. Opin. Cell Biol. 2015;32C:7–12. doi: 10.1016/j.ceb.2014.09.009. [DOI] [PubMed] [Google Scholar]
- 47.Amthor H., Egelhof T., McKinnell I., Ladd M.E., Janssen I., Weber J., Sinn H., Schrenk H.H., Forsting M., Voit T., et al. Albumin targeting of damaged muscle fibres in the mdx mouse can be monitored by MRI. Neuromuscul. Disord. 2004;14:791–796. doi: 10.1016/j.nmd.2004.08.004. [DOI] [PubMed] [Google Scholar]
- 48.Fumagalli L., Businaro R., Nori S.L., Toesca A., Pompili E., Evangelisti E., Giannetti S., Ippoliti F., de Renzis G. Protease inhibitors in mouse skeletal muscle: Tissue-associated components of serum inhibitors and calpastatin. Cell. Mol. Biol. 1996;42:535–546. [PubMed] [Google Scholar]
- 49.Miravitlles M. Alpha-1-antitrypsin and other proteinase inhibitors. Curr. Opin. Pharmacol. 2012;12:309–314. doi: 10.1016/j.coph.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 50.Jonigk D., Al-Omari M., Maegel L., Müller M., Izykowski N., Hong J., Hong K., Kim S.H., Dorsch M., Mahadeva R., et al. Anti-inflammatory and immunomodulatory properties of α1-antitrypsin without inhibition of elastase. Proc. Natl. Acad. Sci. USA. 2013;110:15007–15012. doi: 10.1073/pnas.1309648110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kanatous S.B., Mammen P.P. Regulation of myoglobin expression. J. Exp. Biol. 2010;213:2741–2747. doi: 10.1242/jeb.041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noble C.G., Song H. Structural studies of elongation and release factors. Cell. Mol. Life Sci. 2008;65:1335–1346. doi: 10.1007/s00018-008-7495-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang J., Pantopoulos K. Regulation of cellular iron metabolism. Biochem. J. 2011;434:365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Szőke D., Panteghini M. Diagnostic value of transferrin. Clin. Chim. Acta. 2012;413:1184–1189. doi: 10.1016/j.cca.2012.04.021. [DOI] [PubMed] [Google Scholar]
- 55.Desguerre I., Mayer M., Leturcq F., Barbet J.P., Gherardi R.K., Christov C. Endomysial fibrosis in Duchenne muscular dystrophy: a marker of poor outcome associated with macrophage alternative activation. J. Neuropathol. Exp. Neurol. 2009;68:762–773. doi: 10.1097/NEN.0b013e3181aa31c2. [DOI] [PubMed] [Google Scholar]
- 56.Klingler W., Jurkat-Rott K., Lehmann-Horn F., Schleip R. The role of fibrosis in Duchenne muscular dystrophy. Acta Myol. 2012;31:184–195. [PMC free article] [PubMed] [Google Scholar]
- 57.Kharraz Y., Guerra J., Pessina P., Serrano A.L., Muñoz-Cánoves P. Understanding the process of fibrosis in Duchenne muscular dystrophy. BioMed. Res. Int. 2014 doi: 10.1155/2014/965631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berchtold M.W., Brinkmeier H., Müntener M. Calcium ion in skeletal muscle: Its crucial role for muscle function, plasticity, and disease. Physiol. Rev. 2000;80:1215–1265. doi: 10.1152/physrev.2000.80.3.1215. [DOI] [PubMed] [Google Scholar]
- 59.Raymackers J.M., Debaix H., Colson-Van Schoor M., De Backer F., Tajeddine N., Schwaller B., Gailly P., Gillis J.M. Consequence of parvalbumin deficiency in the mdx mouse: Histological, biochemical and mechanical phenotype of a new double mutant. Neuromuscul. Disord. 2003;13:376–387. doi: 10.1016/S0960-8966(03)00031-2. [DOI] [PubMed] [Google Scholar]
- 60.Alderton J.M., Steinhardt R.A. Calcium influx through calcium leak channels is responsible for the elevated levels of calcium-dependent proteolysis in dystrophic myotubes. J. Biol. Chem. 2000;275:9452–9460. doi: 10.1074/jbc.275.13.9452. [DOI] [PubMed] [Google Scholar]
- 61.Mallouk N., Jacquemond V., Allard B. Elevated subsarcolemmal Ca2+ in mdx mouse skeletal muscle fibers detected with Ca2+-activated K+ channels. Proc. Natl. Acad. Sci. USA. 2000;97:4950–4955. doi: 10.1073/pnas.97.9.4950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dowling P., Doran P., Ohlendieck K. Drastic reduction of sarcalumenin in Dp427 (dystrophin of 427 kDa)-deficient fibres indicates that abnormal calcium handling plays a key role in muscular dystrophy. Biochem. J. 2004;379:479–488. doi: 10.1042/BJ20031311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Culligan K., Banville N., Dowling P., Ohlendieck K. Drastic reduction of calsequestrin-like proteins and impaired calcium binding in dystrophic mdx muscle. J. Appl. Physiol. 2002;92:435–445. doi: 10.1152/japplphysiol.00903.2001. [DOI] [PubMed] [Google Scholar]
- 64.Krüger J., Kunert-Keil C., Bisping F., Brinkmeier H. Transient receptor potential cation channels in normal and dystrophic mdx muscle. Neuromuscul. Disord. 2008;18:501–513. doi: 10.1016/j.nmd.2008.04.003. [DOI] [PubMed] [Google Scholar]
- 65.Nishimura R.N., Sharp F.R. Heat shock proteins and neuromuscular disease. Muscle Nerve. 2005;32:693–709. doi: 10.1002/mus.20373. [DOI] [PubMed] [Google Scholar]
- 66.Paepe B.D., Creus K.K., Weis J., Bleecker J.L. Heat shock protein families 70 and 90 in Duchenne muscular dystrophy and inflammatory myopathy: Balancing muscle protection and destruction. Neuromuscul. Disord. 2012;22:26–33. doi: 10.1016/j.nmd.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Takada F., Vander Woude D.L., Tong H.Q., Thompson T.G., Watkins S.C., Kunkel L.M., Beggs A.H. Myozenin: An alpha-actinin- and gamma-filamin-binding protein of skeletal muscle Z lines. Proc. Natl. Acad. Sci. USA. 2001;98:1595–1600. doi: 10.1073/pnas.041609698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Holland A., Ohlendieck K. Proteomic profiling of the contractile apparatus from skeletal muscle. Expert Rev. Proteomics. 2013;10:239–257. doi: 10.1586/epr.13.20. [DOI] [PubMed] [Google Scholar]
- 69.Duddy W., Duguez S., Johnston H., Cohen T.V., Phadke A., Gordish-Dressman H., Nagaraju K., Gnocchi V., Low S., Partridge T. Muscular dystrophy in the mdx mouse is a severe myopathy compounded by hypotrophy, hypertrophy and hyperplasia. Skelet. Muscle. 2015;5:16. doi: 10.1186/s13395-015-0041-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ohlendieck K. Proteomic identification of biomarkers of skeletal muscle disorders. Biomark. Med. 2013;7:169–186. doi: 10.2217/bmm.12.96. [DOI] [PubMed] [Google Scholar]
- 71.Arpan I., Willcocks R.J., Forbes S.C., Finkel R.S., Lott D.J., Rooney W.D., Triplett W.T., Senesac C.R., Daniels M.J., Byrne B.J., et al. Examination of effects of corticosteroids on skeletal muscles of boys with DMD using MRI and MRS. Neurology. 2014;83:974–980. doi: 10.1212/WNL.0000000000000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hor K.N., Mazur W., Taylor M.D., Al-Khalidi H.R., Cripe L.H., Jefferies J.L., Raman S.V., Chung E.S., Kinnett K.J., Williams K., et al. Effects of steroids and angiotensin converting enzyme inhibition on circumferential strain in boys with Duchenne muscular dystrophy: A cross-sectional and longitudinal study utilizing cardiovascular magnetic resonance. J. Cardiovasc. Magn. Reson. 2011;13:60. doi: 10.1186/1532-429X-13-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Viollet L., Thrush P.T., Flanigan K.M., Mendell J.R., Allen H.D. Effects of angiotensin-converting enzyme inhibitors and/or beta blockers on the cardiomyopathy in Duchenne muscular dystrophy. Am. J. Cardiol. 2012;110:98–102. doi: 10.1016/j.amjcard.2012.02.064. [DOI] [PubMed] [Google Scholar]
- 74.Goemans N., Buyse G. Current treatment and management of dystrophinopathies. Curr. Treat. Options Neurol. 2014;16:287. doi: 10.1007/s11940-014-0287-4. [DOI] [PubMed] [Google Scholar]
- 75.Partridge T.A. Impending therapies for Duchenne muscular dystrophy. Curr. Opin. Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 76.Malik V., Rodino-Klapac L.R., Viollet L., Wall C., King W., Al-Dahhak R., Lewis S., Shilling C.J., Kota J., Serrano-Munuera C., et al. Gentamicin-induced readthrough of stop codons in Duchenne muscular dystrophy. Ann. Neurol. 2010;67:771–780. doi: 10.1002/ana.22024. [DOI] [PubMed] [Google Scholar]
- 77.Popplewell L.J., Abu-Dayya A., Khana T., Flinterman M., Abdul Khalique N., Raju L., Øpstad C.L., Sliwka H.R., Partali V., Dickson G., et al. Novel cationic carotenoid lipids as delivery vectors of antisense oligonucleotides for exon skipping in Duchenne muscular dystrophy. Molecules. 2012;17:1138–1148. doi: 10.3390/molecules17021138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Koo T., Wood M.J. Clinical trials using antisense oligonucleotides in duchenne muscular dystrophy. Hum. Gene Ther. 2013;24:479–488. doi: 10.1089/hum.2012.234. [DOI] [PubMed] [Google Scholar]
- 79.Gintjee T.J., Magh A.S., Bertoni C. High throughput screening in duchenne muscular dystrophy: From drug discovery to functional genomics. Biology. 2014;3:752–780. doi: 10.3390/biology3040752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maffioletti S.M., Noviello M., English K., Tedesco F.S. Stem cell transplantation for muscular dystrophy: the challenge of immune response. BioMed. Res. Int. 2014 doi: 10.1155/2014/964010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Voit T., Topaloglu H., Straub V., Muntoni F., Deconinck N., Campion G., de Kimpe S.J., Eagle M., Guglieri M., Hood S., et al. Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): An exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol. 2014;13:987–996. doi: 10.1016/S1474-4422(14)70195-4. [DOI] [PubMed] [Google Scholar]
- 82.Doran P., Wilton S.D., Fletcher S., Ohlendieck K. Proteomic profiling of antisense-induced exon skipping reveals reversal of pathobiochemical abnormalities in dystrophic mdx diaphragm. Proteomics. 2009;9:671–685. doi: 10.1002/pmic.200800441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Holland A., Dowling P., Zweyer M., Swandulla D., Henry M., Clynes M., Ohlendieck K. Proteomic profiling of cardiomyopathic tissue from the aged mdx model of Duchenne muscular dystrophy reveals a drastic decrease in laminin, nidogen and annexin. Proteomics. 2013;13:2312–2323. doi: 10.1002/pmic.201200578. [DOI] [PubMed] [Google Scholar]
- 84.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 85.Holland A., Schmitt-John T., Dowling P., Meleady P., Henry M., Clynes M., Ohlendieck K. Intricate effects of primary motor neuronopathy on contractile proteins and metabolic muscle enzymes as revealed by label-free mass spectrometry. Biosci. Rep. 2014;34:331–343. doi: 10.1042/BSR20140029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Meleady P., Gallagher M., Clarke C., Henry M., Sanchez N., Barron N., Clynes M. Impact of miR-7 over-expression on the proteome of Chinese hamster ovary cells. J. Biotechnol. 2012;160:251–262. doi: 10.1016/j.jbiotec.2012.03.002. [DOI] [PubMed] [Google Scholar]
- 87.Meleady P., Hoffrogge R., Henry M., Rupp O., Bort J.H., Clarke C., Brinkrolf K., Kelly S., Müller B., Doolan P., et al. Utilization and evaluation of CHO-specific sequence databases for mass spectrometry based proteomics. Biotechnol. Bioeng. 2012;109:1386–1394. doi: 10.1002/bit.24476. [DOI] [PubMed] [Google Scholar]
- 88.Jockusch H., Holland A., Staunton L., Schmitt-John T., Heimann P., Dowling P., Ohlendieck K. Pathoproteomics of testicular tissue deficient in the GARP component VPS54: The wobbler mouse model of globozoospermia. Proteomics. 2014;14:839–852. doi: 10.1002/pmic.201300189. [DOI] [PubMed] [Google Scholar]
- 89.Steinberger M., Föller M., Vogelgesang S., Krautwald M., Landsberger M., Winkler C.K., Kasch J., Füchtbauer E.M., Kuhl D., Voelkl J., et al. Lack of the serum- and glucocorticoid-inducible kinase SGK1 improves muscle force characteristics and attenuates fibrosis in dystrophic mdx mouse muscle. Pflügers Arch. 2014 doi: 10.1007/s00424-014-1645-5. [DOI] [PubMed] [Google Scholar]