Abstract
Our recent study of the mechanism by which an epigenetic alteration, loss of imprinting (LOI) of Igf2, increases tumor risk, revealed a strong relationship between IGF2 dosage, the dynamics of signaling along the IGF2 axis, cell proliferation and tumor risk.1 Colon epithelia in a mouse model with LOI of Igf2 showed increased sensitivity to IGF1R blockade and abrogation of premalignant lesion development in LOI(+) mice. These results are consistent with the epigenetic progenitor model of cancer,2 in which epigenetic changes precede and heighten risk of cancer in response to oncogenic mutations. Thus, one can envision a highly targeted and focused chemoprevention strategy targeted to signaling pathways in nonmalignant cells that have undergone an epigenetic lesion, rather than a broad approach toward reversing epigenetic lesions that may have unintended consequences affecting the whole epigenome.
Keywords: epigenetics, epigenetic progenitor model, imprinting, colorectal cancer, signal transduction
Introduction
In our recent study, “Enhanced sensitivity to IGF-II signaling links loss of imprinting of Igf2 to increased cell proliferation and tumor risk”,1 we identified a novel type of connection between signal transduction, cell regulation, endocrinology and the epigenetic state of the cell, i.e., heritable changes other than DNA sequence that affect gene expression. Clearly, we have known for some time that many genes important in signal transduction are regulated epigenetically. Examples include the estrogen receptor,3 the Wnt pathway,4 the TNF-NFkB pathway,5 and even important oncogenic second messengers like PTEN.6 What was novel about the recent study is that it demonstrated that an epigenetic change, specifically loss of imprinting of Igf2 (LOI), changes the responsivity of the signaling pathway, thereby stably changing signaling dynamics itself.
How a cell interprets signals depends on the response machinery present in the cell. Cell A may respond to a given signal by expressing gene X, while cell B may instead respond to the same signal by expressing gene Y, or no gene at all. These ‘differences of opinion’ could be due to stochastic differences in the numbers of relevant intracellular molecules or, more interestingly, distinct histories of the cells reflected in the details of what signals the cells responded to previously and what proteomic makeup they had at the time of the arrival of the new signal. In multicellular organisms distinct cell histories can extend all the way to embryonic differentiation, which might endow the cells with the memory of belonging to a tissue, even if the cell is isolated from the tissue. This long-term memory, sometimes extending to the zygotic state, can be controlled by epigenetic modifications, reversibly silencing or activating certain genes or chromosomal loci through histone modification, siRNA expression, or DNA methylation. Thus the knowledge of the epigenetic state is critical to predicting cell responses to a particular combination of environmental and intracellular cues.
When the first draft sequence of the entire human genome emerged in early 2001,7 despite its enormous value for basic and applied genetics, it became quickly apparent that the job of understanding the relationship between the genetic code and cellular function was only half completed. For example, only 5% of the human genome is conserved, and of that, only 30% lies within the exons of protein-encoding genes.8 The rest lies in the so-called “dark matter” of the human genome—leading to efforts such as the ENCyclopedia Of DNA Elements (ENCODE)9 to identify regulatory components. Identifying genes and controlling regions, such as promoter, insulator and enhancer sites turns out to be a major undertaking in itself. However, once these sites are identified, there are still multiple regulatory levels related to various epigenetic marks modifying the accessibility of the DNA, such as histone acetylation, methylation and ubiquitination, and DNA methylation.
DNA methylation is a covalent modification of DNA in which a methyl group is transferred from S-adenosylmethionine to the C-5 position of cytosine by a family of DNA methyltransferases (DNMT’s). This usually occurs in the context of CpG dinucleotides in mammals.10 Histone modifications include acetylation, methylation and phosphorylation of dozens of residues on the amino-terminal tails as well as core regions of most histones, although primarily H3 and H4 have been studied.11 It has become increasingly apparent that epigenetic inheritance is a major component of the differences between one differentiated cell type and another, between somatic cells and stem cells, and between normal and diseased cells, including cancer. This view is supported by findings that although few genetic changes apparently account for the difference among differentiated cell types, the epigenetic pattern of the cell changes dramatically through differentiation and aging.2,12 The first known epigenetic alteration identified in cancer was altered DNA methylation, found initially in human colorectal cancer and premalignant adenomas.13,14
While epigenetic mechanisms are generally accepted as important surrogates for oncogene activation or tumor suppressor gene silencing in cancer initiation and progression, the idea that they might play a role in cancer predisposition is relatively new.1,15–17 Despite the fact that epigenetic alterations are found ubiquitously even in small benign neoplasms, relatively little attention has been given to the apparently normal tissues that ultimately become foci of cancer development, even though the cells in tissues can undergo repeated environmental, genetic and age dependent stress, toleration of which largely accounts for the long latency of cancer. Age-related association of DNA methylation has been observed, for example, in the estrogen receptor gene.18,19 Furthermore, we have recently found that DNA methylation patterns change significantly with age in the same individual, lending further credence to an epigenetic role in late-onset diseases.20 It is also important to bear in mind that while mutations have been found in many genes in human cancers, known germline mutations account for a very small fraction of cancer risk in the general population. For example, in colorectal cancer, the highly penetrant hereditary cancer syndromes account for <1% of cancer cases,21,22 even though 20% of CRC occurs in patients with two or more first or second degree relatives23 and there is a greater than two-fold risk for CRC among first degree relatives of patients with CRC.24
Colorectal Cancer
Carcinogenesis typically occurs through a series of genetic mutations, each giving the tumor cancer hallmarks such as self-sufficiency in growth signals, insensitivity to anti-growth signals, evasion of apoptosis, infinite replicative potential, the ability to induce angiogenesis, and finally the ability to invade tissue and metastasize.25 Having a predisposition for cancer can mean that one of the genetic mutations has already occurred in the germline, rather than somatically. A classic example of this is childhood retinoblastoma, which requires two “hits” to occur—if an individual has the germline mutation, he develops retinoblastoma bilaterally, and with a higher frequency, than if the mutation occurs sporadically—as the tumor only needs a single somatic mutation to occur, rather two sequential somatic mutations occurring in the same cell lineage.26 This paradigm is termed Knudson’s two-hit hypothesis.
Although there are many different paths for a cell to become cancerous, frequently such cells take advantage of a specific tissue- specific signaling pathway. For instance, in colorectal cancer, a possible weak link is the Wnt/β-catenin pathway. By an alteration or mutation of this pathway, cells in the colon frequently take the first step towards unrestricted growth. In fact, in sporadic colon carcinomas, over 90% carry an inactivated APC gene, whereas a minority have a mutant β-catenin or axin, part of the same complex27 (Fig. 1).
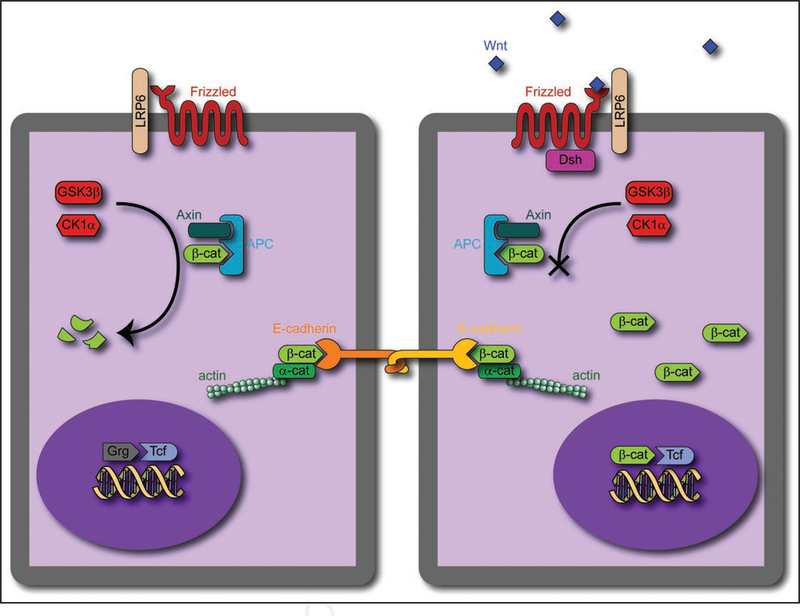
Figure 1.
Schematic of Wnt/β-catenin pathway. (Left) In a naiïve cell, p-catenin, when produced, binds to the APC/Axin complex, and is phosphorylated first by CK1 a, then by GSK3p. This phosphorylation leads to inactivation. The Tcf factor in the nucleus is bound by Groucho, a member of the Grg transcription factor family, which inhibits transcription. β-catenin also has a role in cell-cell adhesion, forming a complex with actin, β-catenin and E-cadherin to form a desmosome. (Right) In contrast, when Wnt is present, it binds to the Frizzled/LRP6 receptor complex, activating Disheveled (Dsh), which acts to inactivate β-catenin degradation, either through sequestering Axin or through inactivation of GSK3β. The accumulation of β-catenin in the cell allows for accumulation in the nucleus, allowing β-catenin to bind to Tcf and activate transcription.
The canonical Wnt/β-catenin signaling pathway ultimately controls the concentration of the free β-catenin and its localization within the cell (Fig. 1). Freshly produced β-catenin forms a complex with axin and adenomatous polyposis coli (APC). This allows the phosphorylation of β-catenin by glycogen synthase kinase-3 P (GSK3P) and casein kinase-1a (CK1a). The phospho- rylated β-catenin is then ubiquitinated by b-TrCP, and degraded.28 This proteosomic degradation endows β-catenin with a turnover time of around 20 minutes. The Wnt/β-catenin canonical pathway acts through the binding of the secreted morphogenic ligand Wnt to the complex of Frizzled and low density lipoprotein receptor-related protein (LRP) on the cell surface. This complex acts to inhibit the action of the β-catenin degradation complex in a poorly understood manner, though the recruitment of axin to the Wnt-activated receptor is a likely candidate.28 The inhibition of degradation causes β-catenin to accumulate first in the cytosol, then in the nucleus. Upon arrival in the nucleus, it binds to the Tcf and Lef families of transcription factors, activating a series of target genes. In intestinal epithelium, Tcf4 seems to be the primary factor driving intestinal epithelial stem cell self-renewal, and a Wnt signal gradient appears to control the differentiation of cells throughout the crypts, since a knockout of this factor denudes the intestine of its epithelial lining.29 The β-catenin/Tcf4 complex is likely the critical control point for determining the balance of differentiation and replication in intestinal crypts, likely through its control of c-MYC and p21CIP1/WAF1, regulators of the cell cycle.30
One hereditable perturbation to the Wnt signaling system occurs in familial adenomatous polyposis (FAP), in which the one of the germline APC alleles generally has a truncating mutation.27 If the allele from the unaffected parent undergoes a mutation or is lost (loss of heterogeneity), APC is inactivated, preventing the degradation of β-catenin. The probability of acquiring a single mutation (10−6 per cell division) which leads to the formation of adenomas is expected to be far higher than the probability acquiring two (10−12).26 Of course, this is not strictly accurate, as the first mutation might allow for larger zones of proliferative cells within the crypts, increasing the number of cells with a possibility of mutation. After identifying APC as a tumor suppressor gene from the study of FAP, later analysis showed that somatic mutations of APC occur in the majority of colorectal carcinomas and adenomas.27
Another hereditary colorectal cancer is hereditary nonpolyposis colorectal cancer (HNPCC), which acts through a deficiency in mismatch repair (MMR) genes. The deficiency in MMR genes leads to an increase in microsatellite instability, an increased possibility for DNA mutation, with mutation rates 2—3 times higher than normal.27 Nevertheless, initiation of adenomas occurs at the same rate as in the general population, much slower than in FAP, which may demonstrate adenomas as early as the preteen years.23,27 However, once an adenoma does develop, it rapidly progresses to malignancy, leading both FAP and HNPCC groups to develop carcinoma at approximately the same average age, 39 and 45 respectively.23
Signaling by Insulin-Like Growth Factor 2 (IGF2)
Genetic mutation is not the only change which may lead to a predisposition for cancer. Epigenetic marks such as histone modification and DNA methylation have also been linked to cancer.31 When epigenetic marks are used to silence or express genes in a parent-of- origin specific fashion, it is termed genomic imprinting. Genomic imprinting is a parent-of-origin-specific allele silencing, meaning that only the paternal or maternal copy of the gene is expressed, controlled in part by differentially methylated regions within or near the gene. However, if the imprinting marks are lost, the parent-specific pattern of allele expression is also lost, causing the imprinted gene to be either silenced or biallelically expressed.32
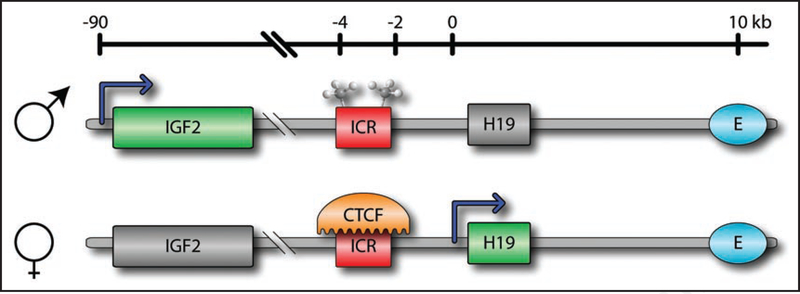
This epigenetic modification is exemplified by loss of imprinting (LOI) of the insulin-like-growth factor 2 gene (Igf2). Normally only the paternal copy of Igf2 is expressed, due to the methylation of the imprinting control region (ICR) (Fig. 2). Methylation of this region prevents binding of the insulating CTCF protein, allowing the enhancer region (E) to act in concert with the Igf2 promoter. In contrast, the maternal ICR is unmethylated, allowing CTCF binding and preventing the enhancer from controlling Igf2. When imprinting is lost, the maternal ICR becomes hypermethylated, causing the expression of its Igf2 copy as well. Recently, an alternative mechanism of LOI for Igf2 has been suggested, involving hypomethylation of the ICR as well as a region within the Igf2 gene, referred to as DMR0.33
Figure 2.
Normal imprinting of the Igf2/H19 locus. The imprinting control region (ICR) on the paternal chromosome is normally methylated, preventing the binding of the zinc-finger protein CTCF. This causes the upstream enhancer region (E) to act on the Igf2 gene, and silences H19. In contrast, the unmethylated ICR on the maternal chromosome allows CTCF binding, insulating the enhancer region from Igf2. This causes the expression of H19 and the silencing of Igf2.
Loss of imprinting leads to a 2—3 times higher dose of IGF2 in the individuals with this epigenetic alteration. Perhaps more importantly, LOI of human IGF2 has been associated with several types of cancer. Specifically, colorectal cancer is 5 times more prevalent in individuals who are LOI(+).34
IGF2 acts as a growth signal through the IGF family of receptors. There are three 3 receptors with significant affinity for IGF2, IGF1 receptor (IGF1R), IGF2 receptor (IGF2R) and the insulin receptor, which has two splicing variant isoforms (IRa and IRb). There are also IGF binding proteins 1–6 (IGFBPs) which are present in the extracellular space and act to stabilize and sequester IGFs.
IRa, IRb and IGF1R are members of the receptor tyrosine kinase family of receptors. These receptors are very similar to each other, e.g., IGF1R is 70% homologous to IR.35 These receptors are composed of two extracellular ligand binding 𝛼 subunits and two transmembrane p subunits with tyrosine kinase activity. Both IGF1R and IRa can effectively bind IGF2, having the highest specificity for this ligand. Between the insulin receptor splicing variants, IRa is dominant in fetal tissues, hematopoietic lineages, neural tissues and some malignant cancers, while IRb is dominant in most adult tissues.36,37 The relative expression of the different splicing variants may be altered through signaling, for example by applying a dose of insulin to the cells.38 This alternative splicing seems to be independent of mutation, suggesting that signaling changes the splicing, though transiently.39
The IGF2 receptor (IGF2R), one of two known mannose-6- phosphate receptors, is more commonly recognized for its role in protein trafficking. Upon binding IGF2, the receptor is internalized and trafficked to a lysosome, where dissociation and degradation of IGF2 takes place, with receptor then recycled back to the cell surface. This function is important for regulating the amount of IGF2 present in the extracellular space for signaling. This regulation was demonstrated in IGF2R knockout mice, which exhibit overgrowth and usually die perinatally, but are rescued by knocking down either Igflr or Igf2.40 The mechanism for rescue is through either removing the excess IGF2, with an Igf2 paternal knockout, or the removal of one of the major IGF2 receptors, by Igflr double knockout. As Igflr knockout by itself is normally lethal, it is thought that the IRa, in combination with the extra IGF2, compensates for the lack of the IGF1R pathway.
When either IGF1R or IRa binds IGF2, the tyrosine kinase domain of the receptor is autophosphorylated, activating its kinase activity (Fig. 3). The tyrosine kinase then phosphorylates an insulin receptor substrate (IRS), which subsequently recruits phosphatidylinositol 3-kinase (PI3K).41 PI3K then acts to convert the lipid phosphatidylinositol(4,5)P2 (PIP2) to phosphatidylinositol(3,4,5)P3 (PIP3), a process which is balanced by phosphatase and tensin homolog (PTEN) phosphatase activity, converting PIP3 back to PIP2.42 PIP3 then acts as a docking point for pleck- strin homology domain containing proteins, e.g., protein kinase B, also known as Akt. Once localized to the membrane by this binding, Akt is phosphorylated at Thr308 by 3-phosphoinositide- dependent protein kinase-1 (PDK1) leading to its partial activity, and at Ser473 by 3-phosphoinositide-dependent protein kinase-2 (PDK2) for full activity.42
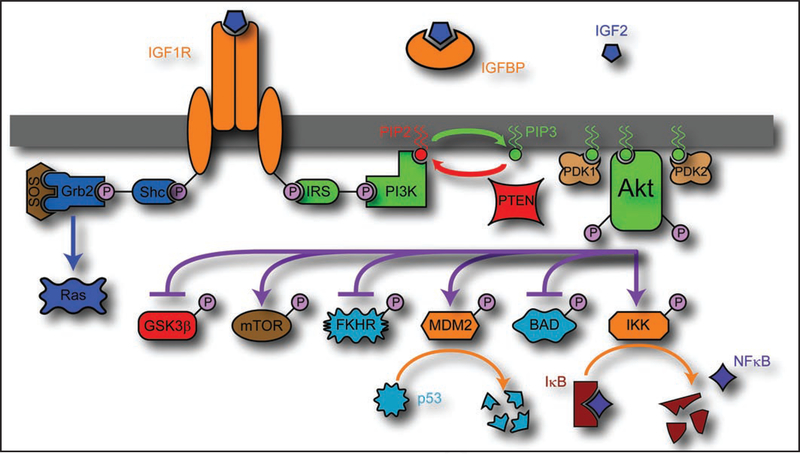
Figure 3.
Schematic of the IGF2 signaling pathway. IGF2 is present in the extracellular space, with free and IGFBP bound portions in equilibrium, acting to stabilize and control the concentration. IGF2 binds to IGF1R or INSR, which causes autophosphorylation of the receptor tyrosine kinase domain. The activated tyrosine kinase domain then acts to phosphorylate IRS, which then binds PI3K. PI3K acts to convert PIP2 to PIP3, which is balanced by the action of PTEN to convert PIP3 back to PIP2. PIP3 is a powerful second messenger, which, among other things, acts as a docking point for the pleckstrin homology domain of PKB/Akt. With Akt recruited to the membrane, it is then phosphorylated by PDK1 and PDK2, activating it. Akt then acts to phosphorylate a variety of factors, enhancing proliferation and preventing apoptosis. IGF1R may also activate other pathways, such as Shc/Grb2/SOS which activates the Ras pathway.
Akt is a major player in signal transduction, especially in ways which are relevant to cancer growth. It acts to promote cell survival, cell proliferation and to increase cell size. Two of Akt’s pro-apoptotic targets are BAD, a member of the BCL2 family, and FKHR, a member of the Forkhead family, both of which Akt inactivates through phosphorylation. It may also upregulate the NFkB pathway through phosphorylation and activation of IkB kinase (IKK), which then acts to degrade an NFkB sequestering protein, IkB. IkB usually binds to NFkB, keeping it in the cytoplasm, but upon IkB degradation, NFkB migrates to the nucleus, and activates a series of anti-apoptotic signals.43 Similarly, Akt participates in phosphorylation and activation of MDM2, a p53 binding protein. MDM2 acts by migrating to the nucleus after phosphorylation, and degrades p53, preventing the expression of apoptotic, cell cycle arresting or DNA repair genes.44 The mammalian target of rapamycin (mTOR) is also activated by Akt, either through direct phosphorylation45 or through inactivation of the tuberous sclerosis complex 2 through phosphorylation, which prevents it from interfering with Rheb, required for mTOR activation.46 The mTOR pathway is one of the major pathways activated by Akt, but also has a negative feedback role, only part of which is known, through downregulation of IRS.46 However, perhaps the most interesting target of Akt activation is GSK3P, mentioned previously in the context of Wnt signaling. Akt phosphorylates GSK3P, inactivating it, and preventing it from acting either in Wnt signaling through phosphorylation of β-catenin, or in cell cycle regulation, through the phosphorylation and subsequent degradation of cyclin D1.47 Consistent with the putative role of Akt in GSK3P inactivation, it has been shown that IGF1R activation, by itself, does not cause transcriptional activity of β-catenin through the Lef/Tcf complex, but rather acts as an enhancer to Wnt based signaling.48
The IR and IGF1R receptors also activate other pathways than the IRS/PI3K/Akt pathway. Shc is bound and phosphorylated by the tyrosine kinase receptor, allowing Shc to bind Grb2, which then binds Son Of Sevenless (Sos). Sos then interacts with Ras, causing it to release GDP and bind GTP, activating the Ras pathway, which involves the PI3K/Akt pathway, the Ral-GEF pathway, and the mitogen-activated protein kinase (MAPK) pathway.41
Also of potential importance is the direct interaction of IGF1R with P-catenin and E-cadherin. A major function of β-catenin in epithelial cells is its binding to the intracellular domain of cadherins, specifically to E-cadherin, a cell-cell adhesion protein. When cancer cells are transformed from epithelial-like cells to mesenchymal, highly malignant cells, it is often due to the loss of E-cadherin function.49 It has been shown that one of the possible causes of this loss of function, through this degradation of cell-cell adhesion, can be IGF signaling, with IGF1R inactivating E-cadherin through phosphorylation.48
Recent Results from Animal Models: IGF2 Addiction and Chemoprevention
As described above, LOI of IGF2 is a relatively common human epigenetic variant, important due to its association with Wilms tumor,50 Beckwith-Wiedemann syndrome,51 and risk of colorectal cancer.15,34 Our recently reported analysis1 showed that LOI of Igf2 increases expression of genes associated with proliferation in intestinal crypts and increases the chances for formation of aberrant crypt foci (ACFs). The most important point of this result was that the developing neoplastic growths have a stronger dependence on the IGF2 signaling pathway than those that form in wild-type samples. Blocking the kinase activity of IGF1R in vivo in our LOI mouse model substantially inhibited ACF formation in LOI mice, but had no effect on ACF inhibition in wild type mice. Thus, the colonic crypt cells in LOI mice were Igf2-addicted, i.e., dependent on continued IGF2 stimulation at least for neoplastic susceptibility if not proliferation per se. These cells were also likely IGF2-dependent for cell proliferation, as cell cycle genes showed increased expression in LOI mice, which was abated by IGF2 inhibition. This IGF2- addiction in epigenetically altered cells suggests a novel epigenetic chemoprevention strategy, in which cells with IGF2-addicted cells undergo reduced tumorigenesis in the colon upon IGF2 pathway blockade.
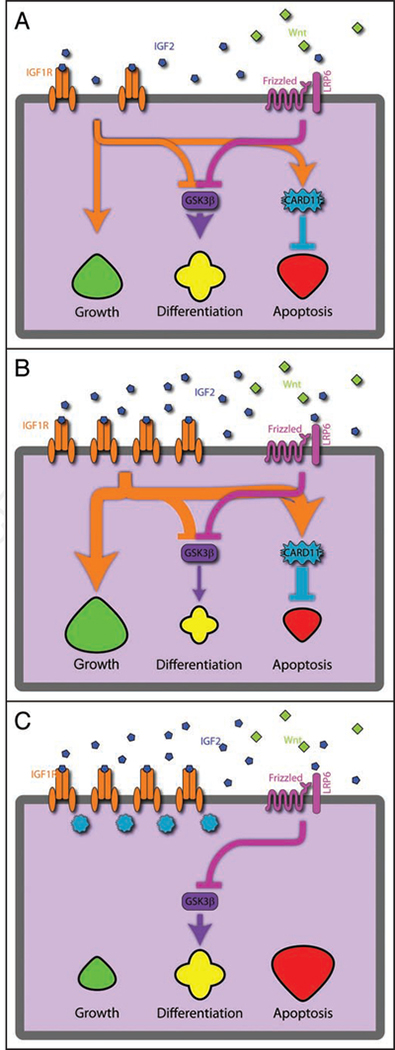
We propose a model linking epigenetic changes to the dynamics of signal transduction, illustrated in Figure 4. In the normal crypt cell, growth, differentiation and apoptosis are balanced, with the Wnt pathway thought to be the prime force behind controlling this equilibrium. When Igf2 is overexpressed epigenetically as in LOI, it leads to an increased signalling for a given amount of IGF2, at least in part mediated by overexpression of IGF1R, and subsequent increase in the downstream effects of the IGF2 axis. Cell growth is thus increased, differentiation is reduced or slowed, and apoptosis is inhibited. The IGF2 axis is linked to the primary differentiation pathway in the colon through inhibition of GSK3β, a major player in the Wnt pathway. In contrast, when treated with an IGF1R kinase inhibitor, cells regain equilibrium, and preneoplastic cells addicted to IGF2 by the epigenetic perturbation are now less prone to form tumors. A corollary of the model is that the genetic mutations in cells with epigenetic alterations that are most successful in leading to tumors are tailored to the IGF2-rich environment. When this strong signalling is abruptly removed by a drug, growth is decreased, and differentiation and apoptosis are increased, leading to a reduction in proliferative cells.
Figure 4.
Model of IGF2 effects on colonic epithelia. (A) With normal, monoallelic expression of IGF2, the IGF1R receptor acts to stimulate growth and inhibits apoptosis (through CARD11) and differentiation (through GSK3p). This is coupled to the Wnt pathway, as they both inhibit GSK3p. (B) In the case of LOI of Igf2, the biallelic expression of Igf2 leads to more IGF1R expression, as well as a reinforcement of the growth pathway while inhibiting the differentiation and apoptotic pathways. (C) However, when an inhibitor of IGF1R action is applied, these pathways are curtailed, leading to an increase in apoptotic pathways and reestablishment of the differentiation equilibrium.
LOI(+) has been shown to cause a marked expansion in the size of colonic crypts, as revealed by markers such as Musashi1, due to the mitogenic and proliferative effects of IGF2.17 Using RNA extracted from laser capture microdissected (LCM) crypts for microarray analysis, we found a marked difference between crypts extracted from LOI(+) and LOI(−) animals, accompanied by upregulation of proliferative genes, such as Cdc6, Mcm5, Mcm3, Chafla, Ligl and Ccnel, along with the expected doubling of Igf2 expression.1 This is likely due to the proliferative compartment of the crypts being expanded. Indeed, IGF2 has recently been shown to be one of the key ligands for stem-cell self-renewal and regulation.52
Considering that IGF2 is a soluble ligand, one must take into account how the local concentration of this ligand is regulated, as well has how the colon tissue is organized. IGF2 triggers a growth signal, but, due to a relatively low diffusivity and potentially high removal rates, it might only act only within a small spatial region, where its concentration is above the signaling threshold. The size of this region increases when the production of IGF2 doubles (through biallellic expression)—expanding the proliferative cell compartment which may enhance the probability of malignant growth when a mutation arises.53
To analyze the effect of this double dose of IGF2 and upregula- tion of proliferative genes, we treated our LOI(+) animal model with azoxymethane (AOM), a well established colon carcinogen.54 This is an especially realistic cancer model, as it allows the epigeneti- cally expanded crypt compartments to form before the exposure to carcinogen, similar to the gradual loss of imprinting in humans and the subsequent development of adenomas. AOM dosage in rodents usually seems to perturb the β-catenin signal pathway, through a mutation of either APC or β-catenin.55
After dosing the mice with carcinogen, we found that they developed ACFs, which have been identified as a precursor to colon cancer.56 Aberrant crypt foci are preneoplastic lesions often used by histologists as a measure of carcinogenesis. They are typically formed by either K-ras mutations or β-catenin/APC mutations.55 LOI(+) mice demonstrate a higher number of ACF after AOM treatment (1.6 fold higher), when compared to wild-type mice.
Receptors of mitogenic pathways, pathways frequently implicated in cancer formation, are often targeted by various therapeutic strategies, either through blocking of the receptor site or inhibition of the receptor tyrosine kinase domain.57,58 Two possible therapies exist for targeting IGF2 receptors—either a small molecule inhibitor targeting the tyrosine kinase domain,59 or a monoclonal antibody to directly block the ligand binding site.60 Using the former (NVP-AEW541; small molecule inhibitor), ACFs from these LOI(+) mice were found to be sensitive to perturbations of the IGF2 signaling pathway. The number of ACFs in LOI(+) mice dropped significantly when dosed (61% decrease in ACF, 56% decrease in ACF/cm2 colon), even below wild-type without the drug (37% ACF, 33% ACF/cm2). In contrast, LOI(−) mice show no significant effect by the drug, rather, they have slightly more ACFs after correcting for colon length. These data provide confirmation of the ability to exploit IGF2 addiction in LOI(+) cells as a strategy for cancer chemoprevention.
Signal Transduction in the Context of Loss of Imprinting
The increased dependence on IGF2 of developing neoplastic lesions in LOI(+) mice is an important result, with implications for tailored therapy, but the mechanics behind the dependence were somewhat opaque. In order to make a start at unravelling this mystery, we undertook a cellular level analysis. Using microfluidic devices, we tested the signal response of mouse embryonic fibroblasts (MEFs) derived from the LOI(+) mouse model.
In wild-type cells, IGF2 caused a transient phosphorylation of Akt, with a peak at 10—40 minutes and an eventual return to basal levels. However, LOI(+) cells showed a persistent phosphorylation of Akt at the lowest IGF2 dose (400 ng/mL). At the intermediate dose (800 ng/mL) of IGF2, the signal resembled a peak again, though delayed versus the wild-type cells. At the highest dose (1,600 ng/ mL) of IGF2, there was no observed peak at all. To check the cellular response to the kinase inhibitor, LOI(+) and wild-type MEFs were dosed with both a 400 ng/mL dose of IGF2 and a 3 μM dose of NVP-AEW541 for 60 minutes, and the Akt phosphorylation was measured. We found that the phosphorylation level was similar for wild-type cells with and without drug, but for LOI(+) cells, the phosphorylation was markedly weaker with the drug.
To help unravel this mystery, we decided to perform genetic analysis of these MEF cells. We used quantitative real-time PCR to determine the relative expression of IGF1R, IGF2R and IR in LOI(+) and wild-type MEF cells. We found that IGF1R and IR were significantly upregulated in LOI(+) MEFs as compared to WT MEFs. This indicated that the double-dose of IGF2 itself was able to cause an upregulation of the receptors which bind it. The underlying mechanism is not fully established, but this finding suggests the intriguing possibility of secondary changes in cell state caused by LOI(+). In fact, the epigenetic state of the cell may even have changed due to this sustained IGF2 double dose. The resulting increase in both IGF2 and IGF1R levels might spur tumor progression.61
It is likely that the colonic crypts, when developing the neoplastic lesions, are influenced by the upregulated IGF2 pathway. Developing ACFs usually require a mutation in the Wnt/p-catenin pathway, whether it be in P-catenin itself, APC, or a different gene.55 Though the majority of colorectal cancers seem to require a mutation first in the APC gene, allowing APC to claim the role of gatekeeper for CRC, these mutations are not all identical.62,63
Most APC mutations result in a truncated version of the protein which has no axin binding motifs, and only partial β-catenin downregulation function. Inactivation of both alleles of APC is usually required for most intestinal tumors, following Knudson’s two-hit hypothesis. However, the second mutation might be dependent on the nature of the first mutation.62 For example, in patients with FAP, the second genomic “hit” found in adenomas depends on the nature of the germ-line mutation. If the germ-line mutation still has partial function, the second hit must remove all function from the second allele. If, however, the germ-line mutation causes a complete loss of function, the second hit seems to retain partial function of APC. Thus it appears that a reduction but not complete elimination of signaling in the Wnt/β-catenin pathway is needed for cancer proliferation and growth. It will be important to test whether cells addicted to IGF2 from LOI have a different set point for Wnt/β-catenin pathway signaling. It is possible that alteration in IGF2 axis signaling perturbs the set point for carcinogenic β-catenin signaling, thereby altering the numbers of ACFs observed.
Implications for Diagnosis and Treatment
If these results are borne out on further study, and in particular in humans with LOI of IGF2 or other epigenetic changes that may confer risk of cancer, then they suggest a novel strategy of chemo- prevention for cancer. Currently, pharmaceutical research directed at epigenetic discoveries is targeted at reversing the epigenetic lesion, with drugs, for example, DNA demethylating drugs that have a broad effect across the genome, activating many genes that might not be desired.64 In contrast, if patients at risk of cancer have altered signaling dynamics specific to the at-risk cell population, then one might design targeted intervention with a high therapeutic ratio, i.e., benefit to side effect, and reduce risk of cancer before even small benign tumors develop. This would be a strategy akin to the use of lipid-lowering drugs to prevent heart disease. Obviously, much more research will be needed to connect these dots, but it is a hopeful and new line of thinking.
Acknowledgements
This work was supported by NIH Grant CA65145 (A.P.F.).
References
- 1.Kaneda A, Wang CJ, Cheong R, Timp W, Onyango P, Wen B, et al. Enhanced sensitivity to IGF-II signaling links loss of imprinting of IGF2 to increased cell proliferation and tumor risk. Proc Natl Acad Sci USA 2007; 104:20926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet 2006; 7:21–33. [DOI] [PubMed] [Google Scholar]
- 3.Ottaviano YL. Methylation of the estrogen receptor gene CpG island marks loss of estrogen receptor expression in human breast cancer cells. Cancer Res 1994; 54:2552–5. [PubMed] [Google Scholar]
- 4.Suzuki H, Watkins DN, Jair KW, Schuebel KE, Markowitz SD, Chen WD, et al. Epigenetic inactivation of SFRP genes allows constitutive WNT signaling in colorectal cancer. Nat Genet 2004; 36:417–22. [DOI] [PubMed] [Google Scholar]
- 5.Relaix F, Wei XJ, Wu X, Sassoon DA. Peg3/Pw1 is an imprinted gene involved in the TNF- NFkappaB signal transduction pathway. Nat Genet 1998; 18:287–91. [DOI] [PubMed] [Google Scholar]
- 6.Kang YH, Lee HS, Kim WH. Promoter methylation and silencing of PTEN in gastric carcinoma. Lab Invest 2002; 82:285–91. [DOI] [PubMed] [Google Scholar]
- 7.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature 2001; 409:860–921. [DOI] [PubMed] [Google Scholar]
- 8.Bejerano G, Haussler D, Blanchette M. Into the heart of darkness: large-scale clustering of human non-coding DNA. Bioinformatics 2004; 20:40–8. [DOI] [PubMed] [Google Scholar]
- 9.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 2007; 447:799–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet 2007; 39:457–66. [DOI] [PubMed] [Google Scholar]
- 11.Jenuwein T, Allis CD. Translating the histone code. Science 2001; 293:1074–80. [DOI] [PubMed] [Google Scholar]
- 12.Feinberg AP. The epigenetics of cancer etiology. Semin Cancer Biol 2004; 14:427–32. [DOI] [PubMed] [Google Scholar]
- 13.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature 1983; 301:89–92. [DOI] [PubMed] [Google Scholar]
- 14.Goelz SE, Vogelstein B, Hamilton SR, Feinberg AP. Hypomethylation of DNA from benign and malignant human colon neoplasms. Science 1985; 228:187–90. [DOI] [PubMed] [Google Scholar]
- 15.Cui H, Cruz-Correa M, Giardiello FM, Hutcheon DF, Kafonek DR, Brandenburg S, et al. Loss of IGF2 imprinting: a potential marker of colorectal cancer risk. Science 2003; 299:1753–5. [DOI] [PubMed] [Google Scholar]
- 16.Kaneda A, Feinberg AP. Loss of imprinting of IGF2: a common epigenetic modifier of intestinal tumor risk. Cancer Res 2005; 65:11236–40. [DOI] [PubMed] [Google Scholar]
- 17.Sakatani T, Kaneda A, Iacobuzio-Donahue CA, Carter MG, de Boom Witzel S, Okano H, et al. Loss of imprinting of Igf2 alters intestinal maturation and tumorigenesis in mice. Science 2005; 307:1976–8. [DOI] [PubMed] [Google Scholar]
- 18.Issa JP, Ottaviano YL, Celano P, Hamilton SR, Davidson NE, Baylin SB. Methylation of the oestrogen receptor CpG island links ageing and neoplasia in human colon. Nat Genet 1994; 7:536–40. [DOI] [PubMed] [Google Scholar]
- 19.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA 2005; 102:10604–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bjornsson HT, Sigurdsson MI, Fallin MD, Irizarry RA, Aspelund T, Cui H, et al. Intraindividual change over time in DNA methylation with familial clustering. JAMA 2008; 299:2877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samowitz WS, Curtin K, Lin HH, Robertson MA, Schaffer D, Nichols M, et al. The colon cancer burden of genetically defined hereditary nonpolyposis colon cancer. Gastroenterology 2001; 121:830–8. [DOI] [PubMed] [Google Scholar]
- 22.Percesepe A, Borghi F, Menigatti M, Losi L, Foroni M, Di Gregorio C, et al. Molecular screening for hereditary nonpolyposis colorectal cancer: a prospective, population-based study. J Clin Oncol 2001; 19:3944–50. [DOI] [PubMed] [Google Scholar]
- 23.Lynch HT, de la Chapelle A. Hereditary Colorectal Cancer. N Engl J Med 2003; 348:919–32. [DOI] [PubMed] [Google Scholar]
- 24.Johns LE, Houlston RS. A systematic review and meta-analysis of familial colorectal cancer risk. Am J Gastroenterol 2001; 96:2992–3003. [DOI] [PubMed] [Google Scholar]
- 25.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell 2000; 100:57–70. [DOI] [PubMed] [Google Scholar]
- 26.Knudson AG. Mutation and cancer: Statistical study of retinoblastoma. Proc Natl Acad Sci USA 1971; 68:820–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell 1996; 87:159–70. [DOI] [PubMed] [Google Scholar]
- 28.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 2004; 20:781–810. [DOI] [PubMed] [Google Scholar]
- 29.Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, et al. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet 1998; 19:379–83. [DOI] [PubMed] [Google Scholar]
- 30.van de Wetering M, Sancho E, Verweij C, de Lau W, Oving I, Hurlstone A, et al. The |3-Catenin/TCF-4 complex imposes a crypt progenitor phenotype on colorectal cancer cells. Cell 2002; 111:241–50. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer 2004; 4:143–53. [DOI] [PubMed] [Google Scholar]
- 32.Feinberg AP Genomic imprinting and gene activation in cancer. Nat Genet 1993; 4:110–3. [DOI] [PubMed] [Google Scholar]
- 33.Cui H, Onyango P, Brandenburg S, Wu Y, Hsieh CL, Feinberg AP Loss of imprinting in colorectal cancer linked to hypomethylation of H19 and IGF2. Cancer Res 2002; 62:6442–6. [PubMed] [Google Scholar]
- 34.Woodson K, Flood A, Green L, Tangrea JA, Hanson J, Cash B, et al. Loss of insulin-like growth factor-II imprinting and the presence of screen-detected colorectal adenomas in women. J Natl Cancer Inst 2004; 96:407–10. [DOI] [PubMed] [Google Scholar]
- 35.Ullrich A, Gray A, Tam AW, Yang-Feng T, Tsubokawa M, Collins C, et al. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J 1986; 5:2503–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vella V, Pandini G, Sciacca L, Mineo R, Vigneri R, Pezzino V, et al. A novel autocrine loop involving IGF-II and the insulin receptor isoform-A stimulates growth of thyroid cancer. Endocrine Soc 2002; 245–54. [DOI] [PubMed] [Google Scholar]
- 37.Frasca F, Pandini G, Scalia P, Sciacca L, Mineo R, Costantino A, et al. Insulin receptor isoform A, a newly recognized, high-affinity insulin-like growth factor II receptor in fetal and cancer cells. Mol Cell Biol 1999; 19:3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sell SM, Reese D, Ossowski VM. Insulin-inducible changes in insulin receptor mRNA splice variants. J Biol Chem 1994; 269:30769–72. [PubMed] [Google Scholar]
- 39.Norgren S, Zierath J, Wedell A, Wallberg-Henriksson H, Luthman H. Regulation of human insulin receptor RNA splicing in vivo. Proc Natl Acad Sci USA 1994; 91:1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ludwig T, Eggenschwiler J, Fisher P, D’Ercole AJ, Davenport ML, Efstratiadis A. Mouse mutants lacking the type 2 IGF receptor (IGF2R) are rescued from perinatal lethality inIgf2andIgf1rNull backgrounds. Dev Biol 1996; 177:517–35. [DOI] [PubMed] [Google Scholar]
- 41.Foulstone E, Prince S, Zaccheo O, Burns JL, Harper J, Jacobs C, et al. Insulin-like growth factor ligands, receptors and binding proteins in cancer. J Pathol 2005; 205:145–53. [DOI] [PubMed] [Google Scholar]
- 42.Vivanco I, Sawyers CL. The phosphatidylinositol 3-Kinase-AKT pathway in human cancer. Nat Rev Cancer 2002; 2:489–501. [DOI] [PubMed] [Google Scholar]
- 43.Romashkova JA, Makarov SS. NF[kappa]B is a target of AKT in anti-apoptotic PDGF signalling. Nature 1999; 401:86–90. [DOI] [PubMed] [Google Scholar]
- 44.Mayo LD, Donner DB. A phosphatidylinositol 3-kinase/Akt pathway promotes translocation of Mdm2 from the cytoplasm to the nucleus. Proc Natl Acad Sci USA 2001; 181–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nave BT, Ouwens M, Withers DJ, Alessi DR, Shepherd PR. Mammalian target of rapamycin is a direct target for protein kinase B: identification of a convergence point for opposing effects of insulin and amino-acid deficiency on protein translation. Biochem J 1999; 344:427–31. [PMC free article] [PubMed] [Google Scholar]
- 46.Hay N The Akt-mTOR tango and its relevance to cancer. Cancer Cell 2005; 8:179–83. [DOI] [PubMed] [Google Scholar]
- 47.Diehl JA, Cheng M, Roussel MF, Sherr CJ. Glycogen synthase kinase-3beta regulates cyclin D1 proteolysis and subcellular localization. Genes Dev 1998; 12:3499–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Playford MP, Bicknell D, Bodmer WF, Macaulay VM. Insulin-like growth factor 1 regulates the location, stability and transcriptional activity of beta-catenin. Proc Natl Acad Sci USA 2000; 210–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Christofori G, Semb H. The role of the cell-adhesion molecule E-cadherin as a tumour- suppressor gene. Trends Biochem Sci 1999; 24:73–6. [DOI] [PubMed] [Google Scholar]
- 50.DeBaun M, Niemitz E, McNeil D, Brandenburg S, Lee M, Feinberg A. Epigenetic alterations of H19 and LIT1 distinguish patients with Beckwith-Wiedemann syndrome with cancer and birth defects. Am J Hum Genet 2002; 70:604–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Feinberg AP, Oshimura M, Barrett JC. Epigenetic mechanisms in human disease. AACR 2002; 6784–7. [PubMed] [Google Scholar]
- 52.Bendall SC, Stewart MH, Menendez P, George D, Vijayaragavan K, Werbowetski-Ogilvie T, et al. IGF and FGF cooperatively establish the regulatory stem cell niche of pluripotent human cells in vitro. Nature 2007; 448:1015–21. [DOI] [PubMed] [Google Scholar]
- 53.Gatenby RA, Gillies RJ. A microenvironmental model of carcinogenesis. Nat Rev Cancer 2008; 8:56–61. [DOI] [PubMed] [Google Scholar]
- 54.Bissahoyo A, Pearsall RS, Hanlon K, Amann V, Hicks D, Godfrey VL, et al. Azoxymethane is a genetic background-dependent colorectal tumor initiator and promoter in mice: Effects of dose, route and diet. Toxicol Sci 2005; 88:340–5. [DOI] [PubMed] [Google Scholar]
- 55.Takahashi M, Wakabayashi K. Gene mutations and altered gene expression in azoxymethane- induced colon carcinogenesis in rodents. Cancer Sci 2004; 95:475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takayama T, Katsuki S, Takahashi Y, Ohi M, Nojiri S, Sakamaki S, et al. Aberrant crypt foci of the colon as precursors of adenoma and cancer. N Engl J Med 1998; 339:1277–84. [DOI] [PubMed] [Google Scholar]
- 57.Surmacz E Growth factor receptors as therapeutic targets: strategies to inhibit the insulinlike growth factor I receptor. Oncogene 2003; 22:6589–97. [DOI] [PubMed] [Google Scholar]
- 58.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Akiyama M, et al. Inhibition of the insulin-like growth factor receptor-1 tyrosine kinase activity as a therapeutic strategy for multiple myeloma, other hematologic malignancies, and solid tumors. Cancer Cell 2004; 5:221–30. [DOI] [PubMed] [Google Scholar]
- 59.Garcia-Echeverria C, Pearson MA, Marti A, Meyer T, Mestan J, Zimmermann J, et al. In vivo antitumor activity of NVP-AEW541-A novel, potent and selective inhibitor of the IGF-IR kinase. Cancer Cell 2004; 5:231–9. [DOI] [PubMed] [Google Scholar]
- 60.Roth RA, Cassell DJ, Wong KY, Maddux BA, Goldfine ID. Monoclonal antibodies to the human insulin receptor block insulin binding and inhibit insulin action. Proc Natl Acad Sci USA 1982; 79:7312–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lopez T, Hanahan D. Elevated levels of IGF-1 receptor convey invasive and metastatic capability in a mouse model of pancreatic islet tumorigenesis. Cancer Cell 2002; 1:339–53. [DOI] [PubMed] [Google Scholar]
- 62.Fodde R, Smits R, Clevers H. APC, Signal transduction and genetic instability in colorectal cancer. Nat Rev Cancer 2001; 1:55–67. [DOI] [PubMed] [Google Scholar]
- 63.Rowan AJ, Lamlum H, Ilyas M, Wheeler J, Straub J, Papadopoulou A, et al. APC mutations in sporadic colorectal tumors: A mutational “hotspot” and interdependence of the “two hits”. Proc Natl Acad Sci USA 2000; 97:3352–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 2004; 6:361–71. [DOI] [PubMed] [Google Scholar]