Significance
The tumor suppressor p53 plays a central role in mediating the cellular response to stress and DNA damage. p53 is a tetramer containing both structured and intrinsically disordered domains that function synergistically to regulate p53 activity. Using intein technology and NMR spectroscopy, we show that the disordered N-terminal transactivation domain of p53 makes intramolecular interactions with the structured DNA-binding domain. These interactions impair binding of p53 to nonspecific DNA sequences but not to p53-specific DNA sequences, providing a means whereby p53 can discriminate more effectively between cognate sites and the vastly more abundant noncognate sites in the genome.
Keywords: intrinsically disordered protein, intein, segmental isotope labeling, transcription factor, DNA recognition
Abstract
Atomic resolution characterization of the full-length p53 tetramer has been hampered by its size and the presence of extensive intrinsically disordered regions at both the N and C termini. As a consequence, the structural characteristics and dynamics of the disordered regions are poorly understood within the context of the intact p53 tetramer. Here we apply trans-intein splicing to generate segmentally 15N-labeled full-length p53 constructs in which only the resonances of the N-terminal transactivation domain (NTAD) are visible in NMR spectra, allowing us to observe this region of p53 with unprecedented detail within the tetramer. The N-terminal region is dynamically disordered in the full-length p53 tetramer, fluctuating between states in which it is free and fully exposed to solvent and states in which it makes transient contacts with the DNA-binding domain (DBD). Chemical-shift changes and paramagnetic spin-labeling experiments reveal that the amphipathic AD1 and AD2 motifs of the NTAD interact with the DNA-binding surface of the DBD through primarily electrostatic interactions. Importantly, this interaction inhibits binding of nonspecific DNA to the DBD while having no effect on binding to a specific p53 recognition element. We conclude that the NTAD:DBD interaction functions to enhance selectivity toward target genes by inhibiting binding to nonspecific sites in genomic DNA. This work provides some of the highest-resolution data on the disordered N terminus of the nearly 180-kDa full-length p53 tetramer and demonstrates a regulatory mechanism by which the N terminus of p53 transiently interacts with the DBD to enhance target site discrimination.
The tumor suppressor p53 plays a central role as a hub protein in numerous signaling pathways that regulate the cellular response to stress and damage. p53 comprises multiple domains including the intrinsically disordered N-terminal transactivation domain (NTAD) and proline-rich domain (PRD), the folded DNA-binding domain (DBD), a short, disordered region containing a nuclear localization signal (NLS), the folded tetramerization (TET) domain, and the disordered C-terminal regulatory domain (CTD) (Fig. 1A). The disordered NTAD contains two short amphipathic motifs, termed “AD1” and “AD2,” which transiently sample helical secondary structure and form binding sites for numerous regulatory proteins (1–5). These interactions are critical for the regulation of p53 function, with outcomes that include negative regulation, proteasomal degradation and stabilization, and activation of p53 response genes (6–12). The p53 NTAD is extensively and progressively phosphorylated in response to increasing levels of cellular stress (6, 13–18). The AD1 and AD2 motifs typically fold into ordered helical structure upon binding to regulatory partners (19–24).
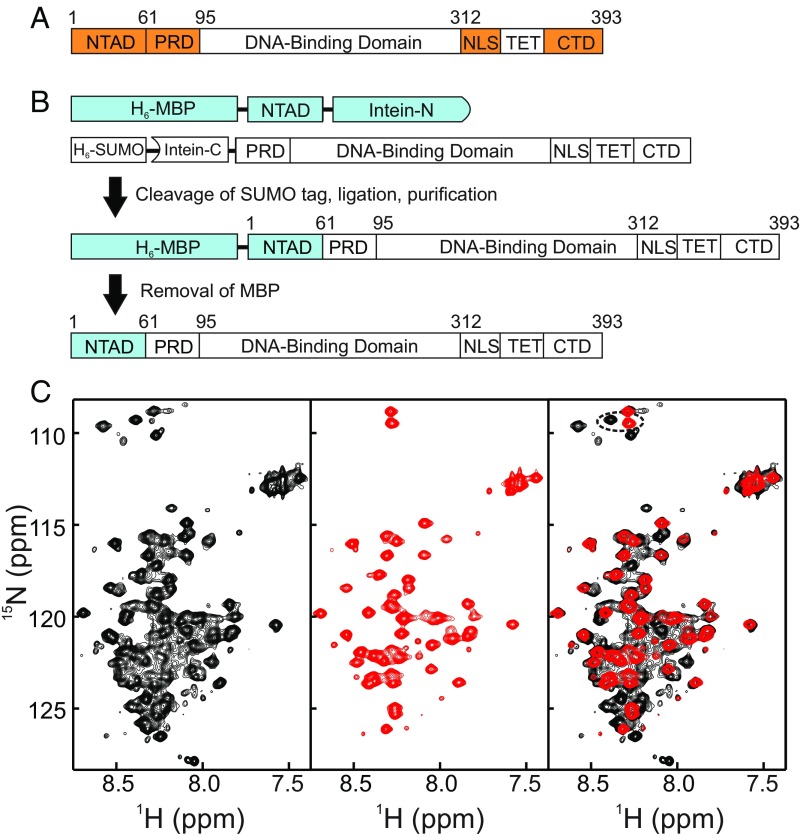
Fig. 1.
(A) Domain structure of p53. The domains colored orange are intrinsically disordered. (B) Strategy for intein-based segmental labeling of the p53 NTAD. (C) 1H-15N HSQC spectra of uniformly 15N-labeled full-length p53 (Left, black), 15NNTAD–p53 (Center, red), and overlay of the two spectra (Right). The circled resonance is perturbed by the intein splice site. Spectra in C were collected in NMR buffer at 25 °C.
While the importance of the NTAD in p53 function is clear, its structural characteristics and interactions within the full-length p53 tetramer are not. The predominant model of p53 structure proposes tetramerization through the TET domain, with the other domains extending from this core (25–27). While this model primarily depicts the NTAD as free and disordered (25, 26), later refinement using single-molecule FRET showed that the NTAD transiently samples conformations in which it is proximal to the DBD (28). However, given the resolution limit of the single-molecule FRET technique, these interactions could not be described in greater detail, and a functional role was not identified. Other studies have additionally implicated the NTAD in thermostability and in some aspects of DNA binding (29–33). These studies were notable for suggesting that the disordered N terminus may have a functional intramolecular role; however, the mechanisms are unknown, and molecular and structural characterization remains minimal. An alternative model of p53 structure, based on cryo-electron microscopy, proposes a role for the NTAD in tetramer formation through direct interactions with the C-terminal TET region (34). Thus, many questions remain about the structure and intramolecular interactions of the NTAD, and there is a need for higher-resolution experimental data to provide insights into its potential intramolecular role in the p53 tetramer.
Characterization of the disordered NTAD in the context of full-length p53 is challenging because of the high molecular weight and conformational heterogeneity of the tetramer (27, 35). The inherent flexibility of p53 hinders characterization by many structural techniques such as X-ray crystallography or high-resolution electron microscopy. In addition, the size of the assembled p53 tetramer—nearly 180 kDa—has limited solution studies such as NMR, as residues in the ordered domains display substantial resonance broadening, while those in the disordered domains experience resonance overlap and poor resolution (36, 37). Despite these limitations, several previous studies have sought to probe the full-length protein by solution methods such as fluorescence spectroscopy, cross-linking mass spectrometry, 13C-methyl NMR, and computational modeling (25, 28, 36, 38–42). While these studies have provided important details, they have generally relied on region-specific probes and therefore were resolution-limited.
This present work provides a more detailed characterization of the disordered NTAD in the context of full-length, tetrameric p53. Using segmental labeling, facilitated by trans-intein splicing, we have been able to observe nearly every residue of the NTAD within the p53 tetramer. Our work reveals a dynamic interaction between the NTAD and the folded DBD that modulates DNA binding. Paramagnetic relaxation enhancement (PRE) experiments show that the AD1 and AD2 motifs of the NTAD contact both the DNA-binding and distal surfaces of the DBD so that the NTAD competes directly with nonspecific DNA sequences for binding to the DBD.
Results
Segmental Labeling of Tetrameric p53 by trans-Intein Splicing.
Amide resonances of amino acids in the intrinsically disordered regions of p53 (the NTAD, PRD, NLS, and CTD domains) appear in the central, heavily overlapped region of the 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum of the uniformly labeled, full-length p53 tetramer, and individual cross-peaks are difficult to resolve (Fig. 1). To reduce spectral overlap and obtain high-quality NMR spectra, we utilized intein splicing to segmentally label the NTAD within tetrameric p53. Using the Npu DnaE trans-intein system (43, 44), we generated samples of tetrameric p53 with the NTAD (residues 1–61) labeled with an NMR-active isotope (15N), while residues 62–393 remained unlabeled and NMR invisible (Fig. 1 B and C). This entity is termed “15NNTAD–p53.” The intein splice site was selected as D61/E62, a site that is distant in the amino acid sequence from interaction sites (e.g., AD2 in the NTAD) or well-folded domains (e.g., the DBD). Careful selection of the splice site is important, since the Npu DnaE intein system inserts nonnative residues (GSCFNGT in the p53 constructs used here) at the splice site (43).
To segmentally label the p53 NTAD, residues 1–61 were fused to the N-terminal split intein fragment (IntN) and expressed in 15N-enriched medium. Residues 62–393 containing the PRD, DBD, NLS, TET, and CTD domains were fused to the C-terminal split intein (IntC) and expressed in unlabeled medium (e.g., natural-abundance 14N) as a fusion with an N-terminal H6SUMO tag [H6SUMO–IntC–p53(62–393)] (Fig. 1B). To improve expression levels, stabilizing mutations (M133L/V203A/N239Y/N268D) were introduced into the DBD (45, 46). The small ubiquitin-related modifier (SUMO) was cleaved from the expressed protein by incubation with the SUMO-specific protease ULP1, resulting in traceless removal of the expression tag (47). Nonnative residues N-terminal to the IntC fragment are inhibitory to the splicing reaction (48), and their removal using the SUMO/ULP1 system facilitated a nearly 90% splicing efficiency. Purification of the spliced, full-length p53 was initially problematic, as the final product oligomerizes with any remaining unspliced precursor components, resulting in a mixture of the desired segmentally labeled homotetramer and heterotetramers containing one or more precursor subunits that could not be separated. To overcome this problem, the IntN was expressed as a maltose-binding protein (MBP) fusion protein [H6MBP–p53(1–61)–IntN] (Fig. 1B), thereby increasing the charge and size differences between the ligated and unligated species and enabling purification of the fully ligated p53 homotetramer by ion-exchange chromatography.
The 1H-15N HSQC spectra of 15NNTAD–p53 show a dramatic improvement in resolution and quality compared with uniformly labeled full-length p53 (Fig. 1C). The cross-peaks in the spectrum of uniformly labeled p53 (black in Fig. 1C) are from residues in the disordered regions: The resonances of the folded DBD and TET domains are severely broadened in HSQC spectra of the 180-kDa tetramer, and their resonances are not visible. The HSQC cross-peaks of 15NNTAD–p53 (red in Fig. 1C) superimpose almost exactly with cross-peaks in the spectrum of uniformly labeled, full-length p53, demonstrating that segmental labeling does not perturb the chemical shifts of the observable resonances and hence that the intein ligation does not perturb the structure and intramolecular interactions of the NTAD (Fig. 1C). To confirm that intein splicing does not disrupt p53 oligomerization, we measured diffusion coefficients for 15NNTAD–p53 and natively expressed p53. The diffusion coefficients, 4.1 ± 0.3 and 4.3 ± 0.6 (×10−7⋅m2⋅s−1) for segmentally and uniformly labeled p53, respectively, are the same within experimental uncertainty (SI Appendix, Table S1), confirming that the intein splicing procedure does not result in protein that is substantially different in shape or size from the native protein.
The p53 NTAD Interacts with the DBD.
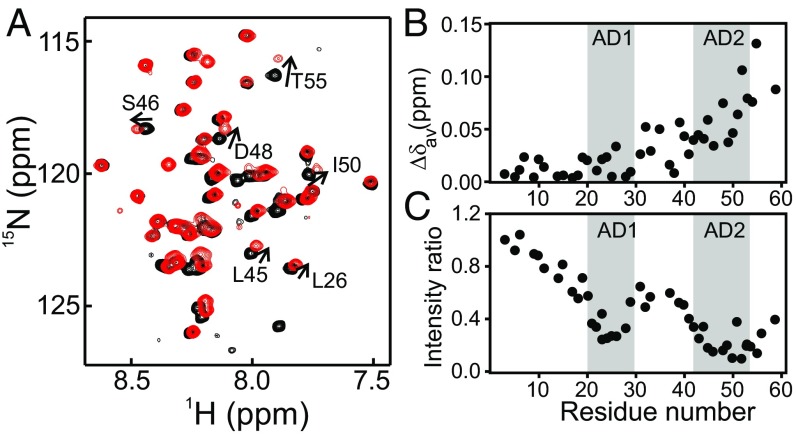
To determine the behavior of the NTAD within full-length p53, we collected a 1H-15N HSQC spectrum of 15NNTAD–p53 and compared it with the spectrum of the corresponding 15N-labeled NTAD peptide (Fig. 2A). The backbone 1H and 15N resonance assignments were made on the basis of published assignments for isolated NTAD peptides (49, 50). While the spectrum of the NTAD in 15NNTAD–p53 closely resembles that of the isolated domain, there are distinct differences that largely map to the amphipathic AD1 and AD2 interaction motifs (Fig. 2 B and C). Chemical shifts of residues in the AD2 region differ between the isolated NTAD and the segmentally labeled 15NNTAD–p53, whereas cross-peaks of residues within AD1 are only slightly shifted from their positions in the spectrum of the isolated peptide (Fig. 2B). We note that the chemical shift differences at the C-terminal end of the NTAD are not an artifact arising from comparisons with p53(1–61); except for residues 59 and 61 at the C terminus, the p53(1–61) chemical shifts are identical within experimental uncertainty to those of the longer p53(1–95) construct. In addition to the chemical shift changes, residues in both AD1 and AD2 have decreased cross-peak intensity in the p53 tetramer relative to the peptide (Fig. 2C). While the intensity loss is greater for AD2 than AD1, it is clear that intramolecular interactions within full-length p53 affect the conformational ensemble and dynamics of both motifs.
Fig. 2.
Comparison of NTAD NMR resonances in an isolated peptide and in the full-length p53 tetramer. (A) 1H-15N HSQC spectrum of an isolated 15N-labeled NTAD peptide [p53(1–61), black] and 15NNTAD–p53 tetramer (red) with selected residues labeled. The sample conditions were the same as in Fig. 1. (B) Differences in weighted averaged 1H and 15N NTAD chemical shifts between the p53(1–61) peptide and full-length p53. The amphipathic AD1 and AD2 interaction motifs are labeled. (C) Cross-peak intensities in 15NNTAD–p53 relative to p53(1–61) peptide. The intensity ratios shown are normalized to the resonance for Glu3. Weighted average chemical shift differences were calculated using the equation .
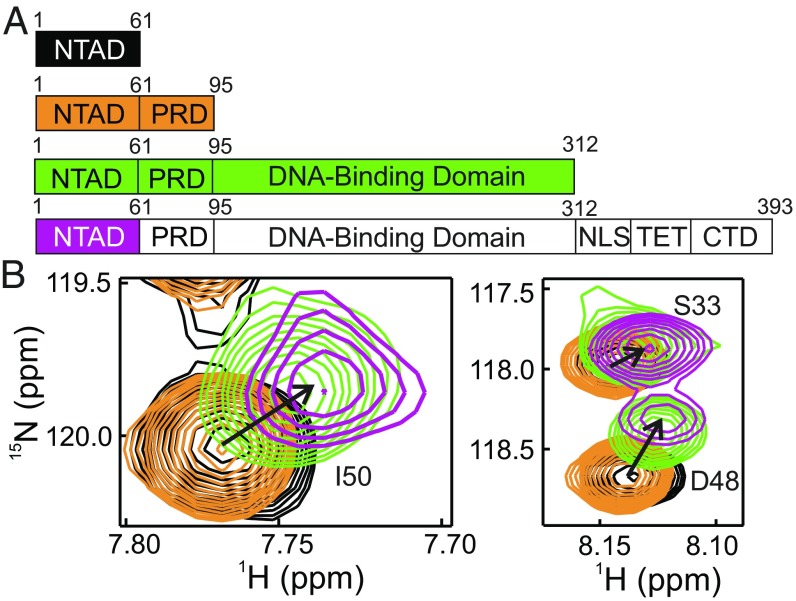
To determine the origin of the perturbations observed in the HSQC spectra of the NTAD in 15NNTAD–p53, truncation constructs were generated (Fig. 3A), and their 1H-15N HSQC spectra were acquired (Fig. 3B and SI Appendix, Fig. S1). Compared with the isolated NTAD [p53(1–61), black spectrum in Fig. 3B], the presence of residues 62–95 in the 15N-labeled p53(1–95) construct has little to no effect on the cross-peaks of residues 3–59 of the NTAD (orange spectrum in Fig. 3B), suggesting that the observed perturbations in spectra of full-length p53 do not result from interactions with the PRD. In contrast, a spectrum of 15N-labeled p53(1–312) displays substantial chemical shift changes relative to that of the isolated NTAD (green spectrum in Fig. 3B) and more closely resembles the HSQC spectrum of the 15NNTAD–p53 tetramer (purple spectrum in Fig. 3B). The resonances in the p53(1–312) construct that are shifted relative to the isolated p53(1–61) move toward, but do not reach the position of the cross-peaks in the full-length protein (Fig. 3B and SI Appendix, Fig. S1C). Cross-peaks in spectra of 15N-labeled p53(1–61), p53(1–312), and 15NNTAD–p53 fall along a line, suggesting that p53(1–312) and the segmentally labeled full-length p53 experience the same two-site exchange process between states in which the NTAD is free or bound to the DBD. The increased chemical shift perturbation in tetrameric p53 relative to the monomeric p53(1–312) construct indicates that the interactions between the NTAD and DBD are enhanced in the full-length protein, implying that the NTAD can interact with multiple DBD domains within the tetramer. Although intersubunit contacts cannot be recapitulated in the monomeric p53(1–312), the truncated construct is nevertheless a useful tool to study NTAD:DBD contacts. As p53(1–312) is unable to tetramerize, its mass remains relatively low (∼35 kDa), and cross-peaks of residues in the DBD are visible and well-resolved in standard 1H-15N HSQC spectra without 2H labeling; by using p53(1–312), the NTAD:DBD interaction can be probed by simultaneously observing resonances of the NTAD and DBD.
Fig. 3.
The p53 NTAD interacts with the DBD. (A) Truncation constructs used to identify intramolecular NTAD interactions. (B) Overlay of selected regions of the 1H-15N HSQC spectra of 15N-labeled NTAD peptide [p53(1–61), black], 15N-labeled NTAD-PRD peptide [p53(1–95), orange], the 15N-labeled NTAD–PRD–DBD construct [p53(1–312), green], and the 15NNTAD–p53 tetramer (purple). The sample conditions were the same as in Fig. 1. The full HSQC spectra are shown in SI Appendix, Fig. S1.
Electrostatics Contribute to the NTAD:DBD Interaction.
Upon increasing the NaCl concentration from 150 to 500 mM, the NTAD cross-peaks in the 1H-15N HSQC spectrum of p53(1–312) shift toward their position in spectra of the isolated NTAD, p53(1–61) (SI Appendix, Fig. S2A). The differences in chemical shift as a function of increasing salt concentration illustrate that, beyond 300 mM NaCl, the intramolecular interaction is greatly impaired, with chemical shift changes reduced to less than half their original values (SI Appendix, Fig. S2B). This trend of decreasing intramolecular interaction with increasing salt concentration reveals a critical electrostatic component in the NTAD:DBD interaction.
DNA Competes with the NTAD for Interaction with the DBD.
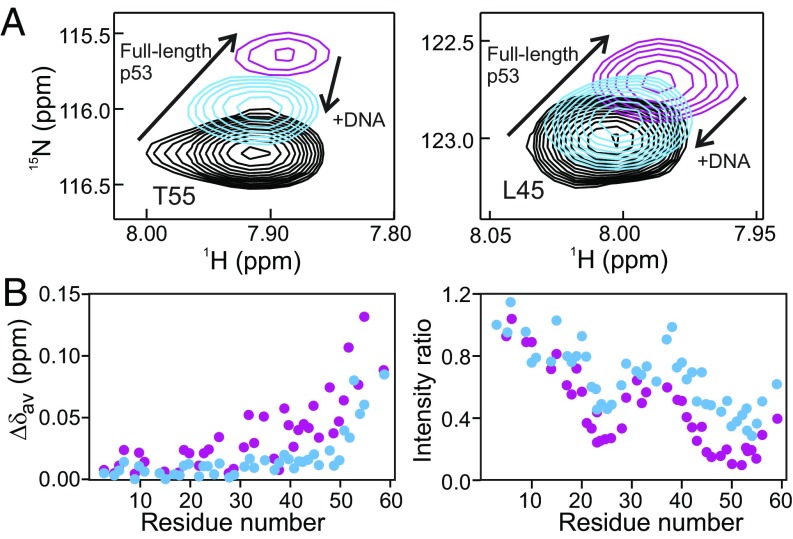
To determine how DNA binding affects the NTAD:DBD interaction, we added an oligonucleotide containing a 20-bp p53 consensus binding site (the sequence is shown in Materials and Methods) to 15NNTAD–p53. Upon the addition of DNA to 1:1 DNA:p53 tetramer stoichiometry, the cross-peaks of several NTAD residues shift toward their positions in spectra of the p53(1–61) construct, substantially decreasing the chemical shift and intensity differences between the isolated NTAD and the full-length p53 protein (Fig. 4). Cognate DNA binds very tightly to the p53 tetramer (with low-nanomolar affinity) (51), and the observation that DNA binding competes with intramolecular interactions between the DBD and the NTAD suggests that both DNA and the NTAD may bind to the same surface of the p53 DBD. It is notable, however, that after the addition of DNA, the chemical shifts of residues 51–55 still differ slightly from those of the isolated NTAD peptide, suggesting that the AD2 motif may make additional contacts with regions of the DBD other than the DNA-binding site. Similar changes were observed when the cognate DNA was added to the monomeric p53(1–312) construct, showing that the truncated monomer largely recapitulates the NTAD:DBD interactions observed in full-length p53. In addition to the changes in the NTAD resonances, the DBD cross-peaks, which can be observed in spectra of the monomeric p53(1–312) (black spectrum in Fig. 5 and SI Appendix, Fig. S3), are broadened beyond detection upon the addition of the cognate DNA (gold spectrum in Fig. 5A and SI Appendix, Fig. S3) due to the formation of a high molecular weight DNA complex.
Fig. 4.
Addition of a p53 cognate DNA sequence disrupts the NTAD:DBD interaction. (A) Selected cross-peaks from 1H-15N HSQC spectra of 15NNTAD–p53 before (purple) and after (blue) the addition of a 0.25× molar equivalent of a 20-bp cognate DNA sequence. The corresponding cross-peaks in the spectrum of the isolated 15N-labeled NTAD peptide [p53(1–61)] are shown in black. Since the p53 recognition element binds four DBDs, a molar ratio of 0.25:1.0 DNA:p53 monomer gives rise to a 1:1 complex. (B) Weighted average NTAD 1H and 15N chemical shift differences (Left) and ratio of cross-peak intensities (Right, normalized to the resonance for Glu3) between isolated 15N-labeled NTAD peptide and 15NNTAD–p53 before (purple circles) and after (blue circles) the addition of DNA. The sample conditions are as in Fig. 1.
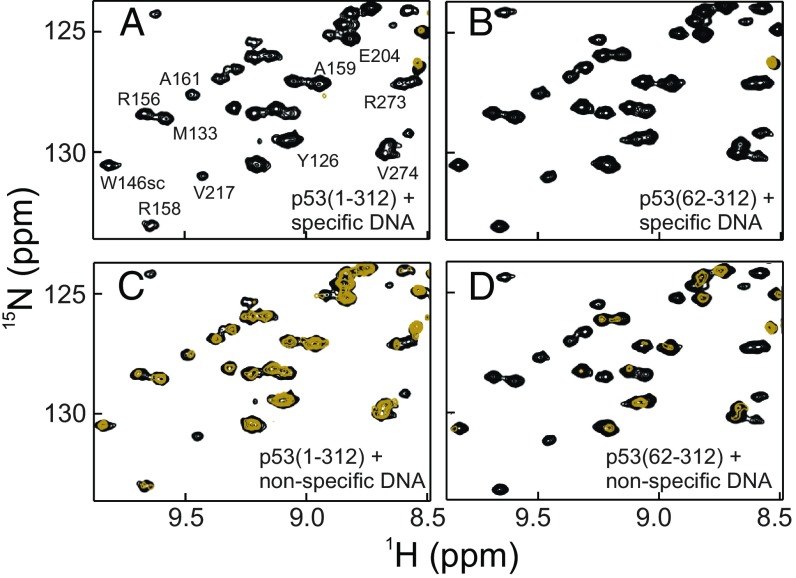
Fig. 5.
The specificity of DNA binding to monomeric p53(1–312) is affected by the NTAD. Regions of the 1H-15N HSQC spectra of 15N-labeled p53(1–312) (A and C) and 15N-labeled p53(62–312) (B and D) in the absence (black) and presence (gold) of a 0.25× equivalent of a 20-bp cognate DNA containing specific binding sites for four p53 DBDs (A and B) and of a 20-bp nonspecific DNA (C and D). For both constructs, the dispersed cross-peaks arising from residues in the DBD are broadened beyond detection upon formation of the cognate DNA complex. The dispersed DBD resonances of the nonspecific complex are broadened in the absence (D) but not in the presence (C) of the NTAD. Full 1H-15N HSQC spectra are shown in SI Appendix, Fig. S3. Sample conditions were the same as in Fig. 1.
The Disordered NTAD Regulates DNA Binding.
At salt concentrations lower than 175 mM, the isolated p53 DBD binds to both specific and nonspecific DNA oligomers with Kd values that differ by less than 10-fold (51, 52). Using constructs with and without the N-terminal 61 residues, we investigated whether the NTAD could regulate interactions with DNA. Upon the addition of a 20-bp DNA consensus sequence, all resonances associated with residues in the DBD are broadened beyond detection in the 1H-15N HSQC spectra of the monomeric p53(1–312) and p53(62–312) constructs (Fig. 5 A and B and SI Appendix, Fig. S3 A and B), suggesting that the NTAD has little to no effect on the binding of p53 to a target DNA. In contrast, there are large differences in cross-peak broadening upon the addition of a 20-bp nonspecific DNA to the p53(1–312) and p53(62–312) constructs (Fig. 5 C and D and SI Appendix, Fig. S3 C and D). DBD resonances of p53(62–312) are extensively broadened, losing nearly 80% of their intensity upon the addition of nonspecific DNA (Fig. 5D and SI Appendix, Fig. S3D). In contrast, the DBD cross-peaks in spectra of p53(1–312) decrease in intensity by less than 50% (Fig. 5C and SI Appendix, Fig. S3C), indicating that the NTAD competes successfully with the nonspecific DNA for binding to the DBD. These results demonstrate that, through its intramolecular interactions with the DBD, the NTAD brokers discrimination between specific and nonspecific DNA sequences.
While the p53(1–312) and p53(62–312) constructs clearly exhibit different DNA-binding behavior, these constructs lack the C-terminal domains required for oligomerization and high-affinity DNA binding (51, 53–55). To investigate the regulatory effect of the NTAD:DBD interactions in tetrameric full-length p53, we utilized fluorescence anisotropy to quantify the binding of full-length p53 and ∆NTAD p53 (residues 62–393, lacking the NTAD domain) to both a 20-bp nonspecific sequence and the p53 recognition element from the p21 gene (sequences are shown in Materials and Methods).
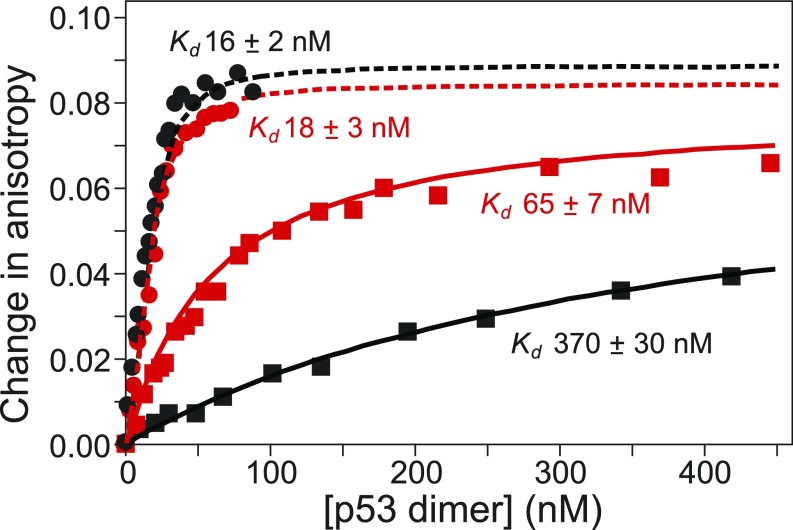
The fluorescence anisotropy measurements show that the full-length p53 tetramer and the truncated ∆NTAD p53 tetramer bind to the cognate p21 DNA sequence with dissociation constants Kd = 16 ± 2 nM and 18 ± 3 nM per dimer, respectively (Fig. 6). These experiments were repeated in triplicate and, since the measured dissociation constants are below the Kd for dissociation of tetramers into dimers (56), the data were fit to a 2:1 (p53 dimer-to-DNA) model (51, 57). The Kd values for p53 and ∆NTAD p53 are the same within experimental error and demonstrate that the NTAD has no effect on the interaction with cognate DNA, in agreement with the NMR data shown in Fig. 5 and SI Appendix, Fig. S3 and with ChIP assays (58). In contrast, a clear difference was observed between the two proteins for binding to the nonspecific DNA (Fig. 6). While the ΔNTAD p53 binds relatively tightly to nonspecific DNA, with a Kd of 65 ± 7 nM per dimer, the presence of the NTAD in full-length p53 reduces the affinity by over fivefold, to a Kd of 370 ± 30 nM per dimer. Since both dissociation constants are above the Kd for the p53 dimer–tetramer equilibrium (56), we infer that binding to DNA occurs primarily with p53 in a tetrameric form, and therefore both binding curves were fit to a 1:1 binding model (51, 57). The decreased affinity in the presence of the NTAD shows clearly that the N-terminal region substantially influences nonspecific DNA binding by the p53 tetramer. Taken together, the NMR and fluorescence data suggest a model in which the p53 NTAD can inhibit binding to nontarget DNA but does not affect binding to a cognate recognition element.
Fig. 6.
Measurement of DNA-binding affinity by fluorescence anisotropy. Plot of changes in fluorescence anisotropy upon the addition of increasing amounts of full-length p53 (residues 1–393) (black data points) and ∆NTAD p53 (residues 62–393) (red data points) to the 20-bp p53 recognition element from the p21 gene (circles) and to a 20-bp nonspecific DNA (squares). Fits are shown as solid lines for nonspecific DNA and as dashed lines for the cognate p21 DNA. All DNA samples were labeled with cyanine5. Kd values for the binding of full-length p53 and ∆NTAD p53 to the two DNA sequences are reported as a per-dimer Kd. Uncertainties are the SD of three independent measurements. The anisotropy data for binding to the specific DNA were fit to a 2:1 p53:DNA model, and binding to nonspecific DNA was fit to a 1:1 model.
NMR PRE Experiments Identify the NTAD Interaction Site on the DBD.
To identify the site of interaction between the NTAD and the DBD and to obtain insights into the mechanism for the observed NTAD regulatory effect, we utilized PRE experiments with a nitroxide spin label (TEMPO). A cysteine -knockout mutant of p53(1–312) was generated in which all exposed and semiexposed cysteine residues were mutated to serine, and nonnative cysteine residues were introduced by mutating either S15 or P58. The TEMPO spin label was attached to the introduced cysteine via maleimide coupling. S15C is located N-terminal to AD1, while P58C is C-terminal to AD2; these spin labels thus report independently on the interactions made by their respective subdomains. Coupling of TEMPO to S15C or P58C was accomplished with ∼80–90% efficiency, as measured by NMR chemical shift changes in surrounding residues. 1H-15N HSQC spectra were collected with both active (paramagnetic/oxidized, Ipara) and inactive (diamagnetic/reduced, Idia) spin labels. The peak heights for all nonoverlapping resonances were measured for both Ipara and for Idia spin labels, and the intensity ratio (Ipara/Idia) was used to identify the residues whose cross-peaks are broadened by proximity to the paramagnetic spin label (SI Appendix, Fig. S4). When the spin label is in the oxidized state, the 1H-15N HSQC spectra of both the S15C and P58C spin-labeled constructs exhibit substantial resonance broadening for residues close to the attachment site. With the spin label at S15C, cross-peaks of residues in both the AD1 and AD2 regions are broadened; similarly, attachment of the spin label at P58C broadens resonances associated with both the AD2 and AD1 motifs, confirming the presence of interactions between these motifs (SI Appendix, Fig. S4). Transient interactions between the AD1 and AD2 regions have been observed previously in isolated NTAD peptides (59). In addition, selective resonance broadening is observed for several DBD cross-peaks (SI Appendix, Fig. S4), indicating intramolecular interactions between the spin labeled NTAD and the DBD.
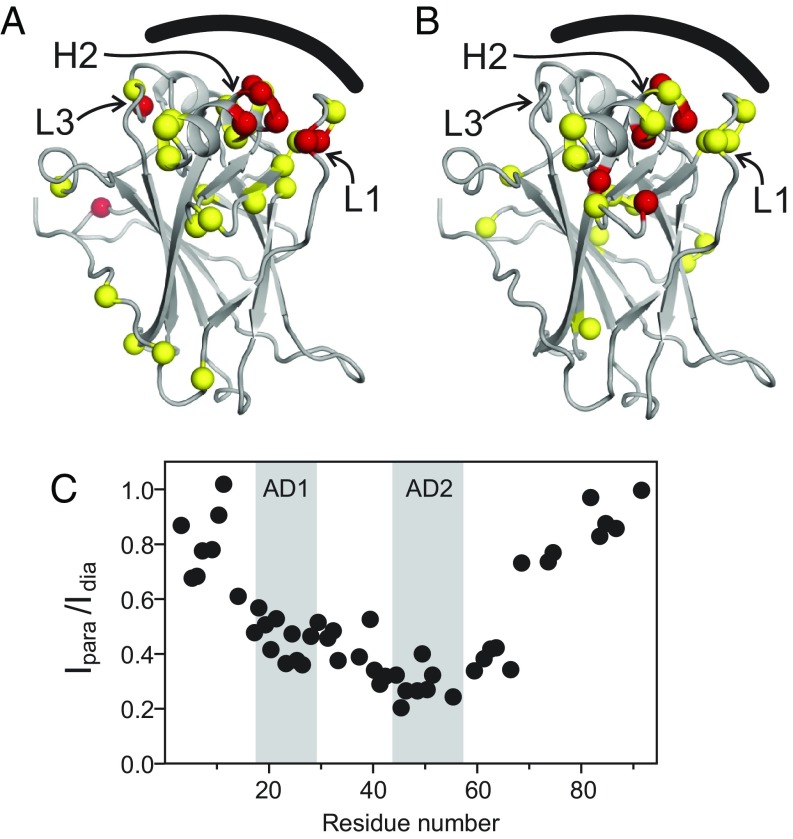
The residues in p53(1–312) whose amide cross-peaks are broadened by the paramagnetic spin labels at S15C and P58C are mapped to the structure of the DBD in Fig. 7. The PRE data show that both the AD1 and AD2 subdomains contact several regions on the surface of the DBD. With the spin label at P58C (AD2), the largest PRE is observed for residues located in the DNA-binding site; these include T118 and V122 in the L1 loop, G245 and to a lesser extent N247 in the L3 loop, A276, and G279, R280, and R283 in the C-terminal helix. The weak PRE observed for residues distant from the DNA-binding site (Fig. 7A) likely arises from transient interactions with the AD2 region. The contacts made by AD1 appear to be weaker and more widely distributed over the DBD surface, including the DNA-binding site (Fig. 7B).
Fig. 7.
NMR PRE experiments identify the NTAD:DBD interaction interface. (A and B) Structure of the p53 DBD (from Protein Data Bank ID code 3Q05) showing the location of residues whose HSQC cross-peaks are broadened by paramagnetic nitroxide spin labels at P58C (A) and S15C (B). The location of the DNA-binding surface is indicated by the black curved line. Spheres indicate residues that experience a >60% decrease in cross-peak intensity (red) or a 30–60% loss of intensity (yellow) in the presence of the paramagnetic spin label. (C) Magnitude of the PRE effects on NTAD resonances from a spin label attached at S121C on the DBD. The AD1 and AD2 regions are highlighted.
To confirm the interaction of AD1 and AD2 with the DNA-binding site, we repeated the PRE experiments with a spin label located on the DBD. The amide cross-peak of S121 is broadened by both NTAD-attached spin labels. This residue, which is situated on the L1 loop that forms part of the DNA-binding site, was mutated to a cysteine (S121C) to which the TEMPO spin label was attached. In the paramagnetic state, the spin label causes substantial broadening of HSQC cross-peaks of DBD residues that are in close spatial proximity to S121C (SI Appendix, Fig. S5), confirming spin-label attachment at the correct location. In addition to the PRE effects observed in the DBD, resonances associated with NTAD residues are also broadened (Fig. 7C). The NTAD PRE effects are localized to residues between the start of AD1 and the end of AD2, with residues N-terminal to AD1 and those within the PRD (residues 61–95) experiencing much less broadening. Residues in AD2 experience stronger PRE than those in AD1, with an average Ipara/Idia of 0.29 for residues 42–55 compared with an average of 0.45 for residues 18–26 (Fig. 7C). The stronger PRE effect observed in AD2 is consistent with the chemical shift and intensity data of Fig. 2. The S121C spin-label experiment confirms the results obtained using spin labels positioned near AD1 and AD2, demonstrating that both subdomains of the NTAD are involved in intramolecular interactions with the DNA-binding surface of the p53 DBD. The data show clearly, however, that AD2 interacts more intimately with the DNA-binding surface of the DBD and/or populates the DBD-bound state to a greater extent than does AD1. We note that the TEMPO spin label does not influence the interaction between the NTAD and the DBD; fully consistent PRE results are observed irrespective of the site of spin labeling, and attachment of the diamagnetic spin label at S121C causes no change in the 1H or 15N chemical shifts of NTAD backbone resonances.
Discussion
Trans-Intein Splicing Facilitates NMR Studies of Tetrameric p53.
Intein splicing has been applied successfully to several systems, including both ordered and disordered proteins (19, 60, 61). By combining a SUMO expression system (47) with intein splicing, we were able to achieve high reaction efficiency and obtain p53 segmentally labeled at the NTAD (15NNTAD–p53) in good yield. Additionally, by using an MBP expression and purification tag, we could successfully separate homotetrameric, ligated p53 from the unreacted precursors that readily formed heterotetramers with ligated monomers. The intein methodologies used in this work have broad applicability to proteins that are difficult to express (e.g., proteins requiring expression tags) or systems that form oligomers.
The 1H-15N HSQC spectra of 15NNTAD–p53 exhibit a dramatic increase in spectral resolution compared with the uniformly labeled protein (Fig. 1C). Resonances in the central region (8.0–8.5 ppm in the 1H dimension) of the uniformly 15N-labeled p53 spectrum, which arise from the disordered NTAD, PRD, NLS, and CTD domains, are heavily overlapped, making analysis difficult. However, for the NTAD segmentally 15N-labeled sample, distinct cross-peaks can be observed for most nonproline residues of the NTAD, allowing detailed investigation of its structure, dynamics, and interactions within the full-length p53 tetramer. This increased resolution opens the way for future studies of binding processes of intact p53, since the NTAD is extensively involved in interactions with regulatory proteins and undergoes substantial chemical shift changes upon complex formation (13, 19, 49, 62–64). These chemical shift perturbations could easily be obscured in spectra of uniformly labeled p53 due to the high degree of spectral overlap.
The NTAD Is Dynamically Disordered in Tetrameric p53.
When expressed as an isolated peptide, the NTAD of p53 is intrinsically disordered and fluctuates rapidly over an ensemble of conformational states (4, 59, 65). The HSQC spectrum of the 15NNTAD–p53 tetramer shows that the NTAD remains dynamically disordered in the full-length protein; the HSQC cross-peaks are sharp and, with the exception of certain AD2 residues, mostly have chemical shifts that are little changed from those in the free peptide (Fig. 2). In contrast to the intense cross-peaks of the disordered NTAD, PRD, NLS, and CTD residues, cross-peaks for the DBD and TET domain are broadened beyond detection in HSQC spectra of uniformly 15N-labeled p53 tetramer due to the slow tumbling of the globular core. Our NMR data are in full agreement with previous fluorescence studies that showed the NTAD to be dynamically disordered in full-length p53 (28). The observed dynamic disorder of the NTAD is consistent with the small-angle X-ray scattering model of p53 quaternary structure (25) but is inconsistent with a cryo-electron microscopy-based model in which oligomerization occurs through a three-helix bundle formed by two helices in the NTAD (in the AD1 and AD2 regions) and a C-terminal helix formed by the TET domain (34). Our observation that the NTAD residues are dynamically disordered in the 15NNTAD–p53 tetramer, with nearly random coil chemical shifts, is incompatible with this latter structural model: The NTAD resonances would be greatly broadened if it participated directly in oligomerization, and substantial changes in chemical shift associated with helix formation would also be expected.
The p53 NTAD Participates in a Transient Intramolecular Interaction with the DBD.
Although the NMR data overall provide convincing evidence that the N-terminal regions of the p53 tetramer are highly disordered, closer inspection reveals small differences in chemical shift and differential changes in cross-peak intensity relative to isolated p53(1–61) or p53(1–95) peptides that signify intramolecular interactions in the full-length protein (Fig. 2). The intensity changes show clearly that these interactions are mediated by both the AD1 and AD2 motifs, while chemical shift changes (Figs. 3 and 4) and paramagnetic spin labeling (Fig. 7 and SI Appendix, Figs. S4 and S5) reveal transient contacts with the DBD, in agreement with previous fluorescence and computational studies (28, 42). The interaction is strongly dependent upon electrostatics, and, since both subdomains contain numerous hydrophobic residues, there is also likely to be a substantial nonpolar contribution. The chemical shift changes (Fig. 2B), intensity losses (Fig. 2C), and PRE data (Fig. 7C) suggest that AD2 dominates the interactions with the DBD. AD2 appears to interact predominantly in the DNA-binding site (Fig. 7A), with weaker secondary interactions on a different face of the DBD. AD1 also interacts with the DNA-binding site, but the interactions appear to be weaker; additional contacts are also made with other regions of the DBD (Fig. 7B). The diminished interactions between AD1 and the DNA-binding surface could potentially result from competition between the AD1 and AD2 motifs. As AD2 appears to interact more tightly, it might be expected that AD2 would successfully outcompete AD1 for the preferred binding site on the DNA-binding surface, thereby forcing AD1 to bind elsewhere. Similar AD1/AD2 competition has been observed in interactions of the p53 NTAD with target proteins such as the TAZ2 domain of CBP (19, 64). However, as PRE measurements show that both the AD1 and AD2 subdomains contact the DNA-binding surface, any competition between them is likely to be dynamic, with rapid exchange between AD1 and AD2, as opposed to the formation of a stable AD2 complex that effectively blocks binding of AD1.
A previous single-molecule FRET analysis identified weak and transient interactions between the dynamically disordered NTAD and the DBD that are enhanced in the full-length p53 tetramer (28). Our NMR results are fully consistent with the FRET observations and have allowed us to probe the interaction at substantially higher resolution, showing that the dominant contacts are between the AD2 domain of the NTAD and the DNA-binding surface. Electrostatic interactions between the negatively charged NTAD, with seven aspartate/glutamate residues in AD2 and three in AD1, and the positively charged DNA-binding site clearly make a major contribution to binding, which is impaired but not completely abrogated at salt concentrations greater than 300 mM (SI Appendix, Fig. S2). The greater negative charge in AD2 likely explains its stronger interactions with the electropositive DNA-binding surface of the DBD (SI Appendix, Fig. S6).
Interactions Between the NTAD and the DBD Regulate DNA Binding.
Previous cellular and biochemical studies implicated the NTAD in modulating DNA binding and promoting dissociation from DNA (29, 31, 33). Our current studies provide insights into the mechanism by which the NTAD influences DNA binding and enhances target site selectivity. In the presence of the NTAD, the affinity for binding to a nonspecific DNA sequence is reduced by more than fivefold, while there is no observable effect on binding to a cognate p53 recognition element. This observation suggests an additional mechanism by which p53 resolves an apparent paradox—its ability to activate target genes despite binding affinities that would imply a lack of functional discrimination between specific and nonspecific sites (51, 52, 66). This lack of discrimination has been puzzling, given the overwhelming abundance of nonspecific genomic DNA sites relative to sequence-specific p53 target sites in vivo. In the absence of the N-terminal region, p53 constructs containing the DBD, the TET domain, and the CTD bind both specific and nonspecific DNA oligonucleotides with high affinity, with as little as a four- to sixfold difference in Kd (Fig. 6 and refs. 51, 52, and 67). The intrinsically disordered CTD functions as a nonspecific DBD that promotes rapid diffusion of p53 along genomic DNA to facilitate the search for specific target sites and enhance the ability of p53 to differentiate between cognate and nonspecific DNA sites (68–71). By inhibiting binding of the DBD to nonspecific DNA (Figs. 5 and 6), the NTAD is expected to function synergistically with the CTD to regulate DNA binding, coupling rapid diffusion on DNA with a high degree of binding-site discrimination.
Inhibition of nonspecific DNA binding occurs through competition between the NTAD and the DNA duplex for the binding surface of the p53 DBD. While specific DNA sequences bind tightly enough to fully displace the NTAD from the DNA-binding surface of the DBD, nonspecific DNA sequences bind more weakly and dissociate more rapidly (52) and cannot fully compete with the NTAD. It has been shown previously that the AD2 region of the p53 NTAD functions as a ssDNA mimic, binding to replication protein A (RPA) and positive cofactor 4 (PC4) in competition with ssDNA (18, 21). In the case of p53, it appears that the NTAD functions as an intramolecular DNA mimic, which competes effectively with binding of nonspecific DNA duplexes. It remains to be determined whether AD2 folds into a helical conformation on binding to the p53 DBD, as it does in its complexes with the DNA-binding surfaces of RPA and PC4, and whether helix formation is required in addition to electrostatic charge to appropriately mimic a DNA helix.
The observation that the NTAD interacts with the DBD to modulate DNA binding may also have implications for the primary protein-binding functions of the NTAD itself. This region mediates interactions between p53 and cellular regulatory proteins (5), including the transcriptional coactivators CBP and p300 that activate transcription of p53-regulated genes by acetylation of lysine residues in the CTD (72, 73). These apparently disparate functions—inhibition of DNA binding and recruitment of regulatory proteins—may synergize to afford an additional level of control over p53 activity. Binding to the DBD may serve to sequester the NTAD, especially the AD2 motif, from untimely interactions with other proteins, inhibiting recruitment of regulatory proteins that target the NTAD until p53 binds to a specific DNA recognition element and the NTAD is displaced from the DBD. Such a mechanism could ensure that the transcriptional coactivators CBP and p300, for example, do not bind to the p53 NTAD during the target-search process but are recruited to p53 only after it binds to a bona fide p53 target gene. This model is consistent with the observation that acetylation of p53 by CBP/p300 is DNA dependent (74); p300-mediated acetylation of p53 in vitro and in cells is greatly enhanced by binding to DNA containing a p53 consensus site, which would be expected to displace the NTAD from the DBD (Figs. 4 and 5) and thereby facilitate recruitment of CBP/p300. In contrast, binding to a nonspecific DNA sequence, which would not compete efficiently with NTAD binding (Fig. 5), is unable to stimulate p53 acetylation (74). Thus, the intramolecular interactions between the NTAD and DBD identified in the current work suggest another layer of regulatory control, one that influences both the DNA-binding activity of p53 and its interactions with the many cellular proteins that regulate p53 by binding to the NTAD.
Materials and Methods
Protein Expression and Purification.
Unless otherwise stated, all DBD-containing p53 constructs contained the quadruple M133L/V203A/N239Y/N268D superstabilizing mutations (45, 46). p53(1–393), the p53(88–312) cysteine-knockout mutant (C124S/C182S/C229S/C275S/C277S; C5×S), and all variations of p53(1–312) (e.g., wild type, M133L/V203A/N239Y/N268D, C5×S/S15C, C5×S/P58C, and C5×S/S121C) were expressed with N-terminal H6GB1 tags with a tobacco etch virus (TEV) protease cleavage site. The IntN for labeling the NTAD [p53(1–61)–IntN] was expressed as a fusion protein with an N-terminal H6MBP. The IntC [IntC–p53(62–393)] was expressed in fusion to an N-terminal H6SUMO tag. Proteins were expressed in LB or M9 medium using Escherichia coli BL21 (DE3) cells for H6SUMO–IntC–p53(62–393) and BL21 (DE3) DNAY cells for all other constructs. Cells were grown at 37 °C to an OD of ∼0.6–0.8, cooled to 16–18 °C, and induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside and, for constructs containing the p53 DBD, 150 µM ZnSO4. After growth for 16–20 h, cells were harvested by centrifugation and stored at −20 °C.
For all constructs, cells were lysed by sonication in 40–60 mL of lysis buffer [40 mM Tris (pH 8.0), 1 M NaCl, 5 mM DTT with one EDTA-free protease tablet (Roche)]. NaCl (1 M) was used to inhibit binding to endogenous DNA; success of the inhibition was monitored by A260/A280 measurements. All His-tagged proteins were initially purified by binding to His cOmplete Ni resin (Roche), washed in lysis buffer, and eluted using lysis buffer containing 250 mM imidazole.
With the exception of intein precursors, the proteins eluted from the Ni resin were dialyzed into buffer consisting of 20 mM Tris (pH 8.0), 200 mM NaCl, and 2 mM DTT, and the H6GB1 expression tag was cleaved by incubation with a 1:100–1:200 molar ratio of TEV protease. Dialysis and TEV cleavage were performed at 4 °C overnight. Following dialysis, p53(1–393), p53(88–312), and p53(1–312) constructs were further purified using a heparin HiTrap column (GE), eluting with a gradient of 40–500 mM NaCl. p53(1–393) was additionally purified by both HiTrap SP and HiTrap Q cation exchange using the same low- and high-salt buffers, while p53(1–312) and p53(88–312) constructs were purified by HiTrap Q. Following purification, these proteins were buffer exchanged into NMR buffer [20 mM Tris (pH 7.0), 150 mM NaCl, 2 mM DTT] and concentrated to 150–300 μM.
The intein splicing reaction was performed using a 1.2- to 1.4-fold excess of H6MBP–p53(1–61)–IntN precursor relative to the H6SUMO–IntC–p53(62–393) precursor containing the TET domain. Splicing reactions were typically carried out at protein concentrations of 20–30 µM. Following mixing, 1 mM Tris(2-carboxyethyl)phosphine (TCEP) and a 1:100–1:200 molar ratio of the SUMO protease ULP1 were added to the reaction mixture. Simultaneous SUMO tag cleavage and intein splicing occurred overnight at 4 °C during dialysis against buffer containing 20 mM Tris (pH 8.0), 200 mM NaCl, 2 mM DTT, and 0.5 mM TCEP. After splicing and cleavage, the ligation mixtures were purified using a HiTrap heparin column eluted with a salt gradient (40–700 mM NaCl) in buffer containing 20 mM Tris (pH 8.0) and 2 mM DTT. Separation of heterotetrameric p53 (full-length p53 tetramerized with unreacted precursors) from homotetramer was achieved using a Mono Q anion-exchange column. The separated MBP–p53 homotetramer was dialyzed against 20 mM Tris (pH 8.0), 200 mM NaCl, and 2 mM DTT with 1:100 molar equivalents of TEV protease to cleave the H6MBP fusion tag. Following cleavage, the NTAD-labeled full-length p53 (15NNTAD–p53) was purified from the H6MBP using a HiTrap SP cation-exchange column.
All purified proteins were either used immediately or stored at −80 °C after rapid freezing with liquid nitrogen. The purity of all proteins was assessed by SDS/PAGE (SI Appendix, Fig. S1), and the identities of the purified products were confirmed by LC-MS or MALDI MS. p53(1–61) and p53(1–95) were expressed and purified using the protocol for p53(13–61) described previously (49).
Validation of Superstable p53 for the Study of Intramolecular NTAD:DBD Interactions.
NMR and fluorescence experiments throughout this work were performed using a superstable variant of the p53 DBD containing four point mutations, M133L/V203A/N239Y/N268D, that substantially increase the stability of the DBD without compromising structure or function (45, 46). To verify that the observed NTAD:DBD interaction was not an artifact of working with a stabilized DBD, 1H-15N HSQC spectra were collected for wild-type p53(1–312). A comparison of the HSQC spectra of wild-type and superstable p53(1–312) reveals clear differences in the cross-peaks arising from the DBD due to the four point mutations (SI Appendix, Fig. S7A). However, there are only minor differences for cross-peaks corresponding to NTAD residues (SI Appendix, Fig. S7 B and C), with both differing substantially from the isolated NTAD (SI Appendix, Fig. S7D). These data demonstrate that the intramolecular NTAD:DBD interaction is largely unaffected by the change from the wild-type to the superstable p53 DBD, thus validating this construct for studies on the NTAD:DBD interaction.
Preparation of Duplex DNA.
For NMR studies, two DNA sequences were used, an engineered palindromic p53-consensus site DNA and a palindromic nonspecific DNA sequence. The sequence of the specific DNA was 5′-GAACATGTTCGAACATGTTC-3′ (previously used in ref. 36); the nonspecific sequence was 5′-ATGATGATGATCATCATCAT-3′. All DNA oligonucleotides were purchased from Integrated DNA Technologies. The DNA was dissolved in 20 mM Tris (pH 7.0) and 150 mM NaCl and was annealed by heating to 95 °C before cooling slowly over several hours to room temperature. The DNA duplexes were buffer exchanged into NMR buffer and concentrated before use.
Two 20-bp DNA oligomers were used for fluorescence anisotropy measurements. These included a specific DNA (the p53 recognition element in the p21 gene promoter) and a nonspecific DNA. These sequences were 5′-GAACATGTCCCAACATGTTG-3′ and 5′-GATGATGATGATGATGATGA-3′, respectively. Both oligonucleotides were labeled on the 5′ end with cyanine5 and were purchased from Integrated DNA Technologies. The fluorescently labeled DNA oligonucleotides were mixed 1:1 with their nonlabeled reverse complements and were annealed by the protocol described above.
Fluorescence Anisotropy Measurements.
Fluorescence anisotropy experiments were carried out using a Fluorolog-3 fluorimeter (Horiba). Excitation and emission wavelengths for the cyanine5 dye were set at 636 nm and 666 nm, respectively. The concentration of fluorescently labeled DNA was initially set at 5 nM, and p53 constructs were titrated in to concentrations of 0.1–10 μM depending on the binding affinity being measured. The concentration of labeled DNA was either held constant or allowed to decrease upon dilution (after the addition of p53). The two methods produced similar values and did not appear to affect the results (dilution was never more than a 50% decrease in DNA concentration). Experiments were carried out in NMR buffer at 25 °C with 2–5 min of incubation after each sequential addition of p53. Each experiment was performed in triplicate, and anisotropy curves were fit to equations described in ref. 51, using the 2:1 model for specific DNA and the 1:1 model for nonspecific DNA. Kd values are reported per dimer, and errors are derived from the SD of three independent measurements.
NMR Data Collection, Assignments, and Analysis.
NMR spectra were acquired on Bruker Avance 900 MHz, DRX 800 MHz, Avance 700 MHz, DRX 600 MHz, and Avance 600 MHz spectrometers. Unless otherwise stated, all spectra were recorded at 25 °C for samples in NMR buffer. Protein concentrations ranged from 30–200 μM depending on experiments. 1H-15N HSQC spectra were acquired using standard pulse sequences (75). Diffusion constants were measured using diffusion-ordered spectroscopy NMR experiments (76) with 150 µM protein.
Salt titrations for p53(1–312) and p53(1–61) were carried out with protein concentrations of 150 μM. The initial titration point had a NaCl concentration of 150 mM, and NaCl from a 5-M concentrated stock was added to this sample at 50-mM increments up to 500 mM NaCl. Nonspecific and specific DNA-binding NMR experiments were performed using 50-μM concentrations of p53(1–312) or p53(62–312). A 0.25× molar equivalent of DNA was used in NMR experiments; as four p53 monomers bind one DNA recognition element (51, 77), this ratio represents a 1:1 mixture.
Backbone resonance assignments for p53(1–61), p53(1–95), and NTAD-labeled full-length p53 were transferred from published data (19, 49, 50). Most NTAD cross-peaks in spectra of 15NNTAD–p53 are unshifted or are shifted only slightly from their chemical shifts in spectra of the isolated NTAD peptides, so transferring the assignments is straightforward. Assignments of AD2 cross-peaks, whose 15N chemical shifts differ by as much as 0.6 ppm in full-length p53 relative to the isolated peptides, were confirmed using truncation constructs with intermediate chemical shifts (see Fig. 3B for an example). The backbone chemical shifts of the DBD in the p53(1–312) superstable C5×S construct differed substantially from those of the previously assigned wild-type p53(94–312) construct (78). Backbone assignments for the superstable p53(88–312) C5×S construct were therefore made using standard 3D triple-resonance NMR spectroscopy (TROSY)-HNCACB and TROSY-HNCOCACB experiments (79); assignments of the p53(88–312) C5×S construct were transferable to the p53(1–312) C5×S/S15C, C5×S/P58C, and C5×S/S121C proteins, as the chemical shift of DBD cross-peaks between the two samples were similar.
Spin-Label Experiments.
4-Maleimido-TEMPO (Sigma) was used for PRE spin-label experiments with p53(1–312) superstable C5×S/S15C, C5×S/P58C, and C5×/S121C proteins. Maleimido-TEMPO was dissolved in 100% ethanol and was added in 2.5 molar excess relative to the p53 constructs. Reactions were typically carried out at protein concentrations of 80–120 μM in 20 mM Tris (pH 7.2), 150 mM NaCl, and 0.5 mM TCEP. The labeling reaction proceeded at room temperature for 2 h with light shaking, after which excess spin label was removed by buffer exchange on a gravity desalting column (NAP column; GE). 1H-15N HSQC spectra were acquired immediately after buffer exchange. Following acquisition of an HSQC spectrum with an active (paramagnetic) spin label, 2 mM DTT and 300 µM ascorbic acid were added to reduce and inactivate the TEMPO spin label. After incubation for 30 min at 25 °C, a second HSQC spectrum was collected with the spin label in the diamagnetic state. Concentrations for all spin-label NMR experiments were ∼50 μM of protein. The effect of the spin label was determined through intensity changes between the paramagnetic and diamagnetic states. The absolute cross-peak intensities in each state (Ipar and Idia) were derived from NMR spectra, and the Ipara/Idia ratio was used to identify resonances that are broadened by the paramagnetic TEMPO spin label.
Supplementary Material
Acknowledgments
We thank Gerard Kroon for expert assistance with NMR experiments, Maria Martinez-Yamout for assistance with intein ligation, and Rebecca Berlow and Shanshan Lang for suggestions on the manuscript. This work was supported by NIH Grants CA096865 and GM075995 and by the Skaggs Institute for Chemical Biology.
Footnotes
The authors declare no conflict of interest.
Data deposition: The NMR chemical shifts have been deposited in the BioMagResBank databank, www.bmrb.wisc.edu (accession no. 27599).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1814051115/-/DCSupplemental.
References
- 1.Ayed A, et al. Latent and active p53 are identical in conformation. Nat Struct Biol. 2001;8:756–760. doi: 10.1038/nsb0901-756. [DOI] [PubMed] [Google Scholar]
- 2.Dawson R, et al. The N-terminal domain of p53 is natively unfolded. J Mol Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 3.Candau R, et al. Two tandem and independent sub-activation domains in the amino terminus of p53 require the adaptor complex for activity. Oncogene. 1997;15:807–816. doi: 10.1038/sj.onc.1201244. [DOI] [PubMed] [Google Scholar]
- 4.Lee H, et al. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 5.Raj N, Attardi LD. The transactivation domains of the p53 protein. Cold Spring Harb Perspect Med. 2017;7:a026047. doi: 10.1101/cshperspect.a026047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schon O, Friedler A, Bycroft M, Freund SM, Fersht AR. Molecular mechanism of the interaction between MDM2 and p53. J Mol Biol. 2002;323:491–501. doi: 10.1016/s0022-2836(02)00852-5. [DOI] [PubMed] [Google Scholar]
- 7.Gu W, Shi XL, Roeder RG. Synergistic activation of transcription by CBP and p53. Nature. 1997;387:819–823. doi: 10.1038/42972. [DOI] [PubMed] [Google Scholar]
- 8.Lill NL, Grossman SR, Ginsberg D, DeCaprio J, Livingston DM. Binding and modulation of p53 by p300/CBP coactivators. Nature. 1997;387:823–827. doi: 10.1038/42981. [DOI] [PubMed] [Google Scholar]
- 9.Espinosa JM, Verdun RE, Emerson BM. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 10.Appella E, Anderson CW. Post-translational modifications and activation of p53 by genotoxic stresses. Eur J Biochem. 2001;268:2764–2772. doi: 10.1046/j.1432-1327.2001.02225.x. [DOI] [PubMed] [Google Scholar]
- 11.Bode AM, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 12.Xu Y. Regulation of p53 responses by post-translational modifications. Cell Death Differ. 2003;10:400–403. doi: 10.1038/sj.cdd.4401182. [DOI] [PubMed] [Google Scholar]
- 13.Lee CW, Ferreon JC, Ferreon AC, Arai M, Wright PE. Graded enhancement of p53 binding to CREB-binding protein (CBP) by multisite phosphorylation. Proc Natl Acad Sci USA. 2010;107:19290–19295. doi: 10.1073/pnas.1013078107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Teufel DP, Bycroft M, Fersht AR. Regulation by phosphorylation of the relative affinities of the N-terminal transactivation domains of p53 for p300 domains and Mdm2. Oncogene. 2009;28:2112–2118. doi: 10.1038/onc.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sakaguchi K, et al. Damage-mediated phosphorylation of human p53 threonine 18 through a cascade mediated by a casein 1-like kinase. Effect on Mdm2 binding. J Biol Chem. 2000;275:9278–9283. doi: 10.1074/jbc.275.13.9278. [DOI] [PubMed] [Google Scholar]
- 16.Dumaz N, Meek DW. Serine15 phosphorylation stimulates p53 transactivation but does not directly influence interaction with HDM2. EMBO J. 1999;18:7002–7010. doi: 10.1093/emboj/18.24.7002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lambert PF, Kashanchi F, Radonovich MF, Shiekhattar R, Brady JN. Phosphorylation of p53 serine 15 increases interaction with CBP. J Biol Chem. 1998;273:33048–33053. doi: 10.1074/jbc.273.49.33048. [DOI] [PubMed] [Google Scholar]
- 18.Rajagopalan S, Andreeva A, Teufel DP, Freund SM, Fersht AR. Interaction between the transactivation domain of p53 and PC4 exemplifies acidic activation domains as single-stranded DNA mimics. J Biol Chem. 2009;284:21728–21737. doi: 10.1074/jbc.M109.006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krois AS, Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein. Proc Natl Acad Sci USA. 2016;113:E1853–E1862. doi: 10.1073/pnas.1602487113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kussie PH, et al. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 21.Bochkareva E, et al. Single-stranded DNA mimicry in the p53 transactivation domain interaction with replication protein A. Proc Natl Acad Sci USA. 2005;102:15412–15417. doi: 10.1073/pnas.0504614102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Di Lello P, et al. Structure of the Tfb1/p53 complex: Insights into the interaction between the p62/Tfb1 subunit of TFIIH and the activation domain of p53. Mol Cell. 2006;22:731–740. doi: 10.1016/j.molcel.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 23.Feng H, et al. Structural basis for p300 Taz2-p53 TAD1 binding and modulation by phosphorylation. Structure. 2009;17:202–210. doi: 10.1016/j.str.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Miller Jenkins LM, et al. Characterization of the p300 Taz2-p53 TAD2 complex and comparison with the p300 Taz2-p53 TAD1 complex. Biochemistry. 2015;54:2001–2010. doi: 10.1021/acs.biochem.5b00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wells M, et al. Structure of tumor suppressor p53 and its intrinsically disordered N-terminal transactivation domain. Proc Natl Acad Sci USA. 2008;105:5762–5767. doi: 10.1073/pnas.0801353105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tidow H, et al. Quaternary structures of tumor suppressor p53 and a specific p53 DNA complex. Proc Natl Acad Sci USA. 2007;104:12324–12329. doi: 10.1073/pnas.0705069104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Joerger AC, Fersht AR. Structural biology of the tumor suppressor p53. Annu Rev Biochem. 2008;77:557–582. doi: 10.1146/annurev.biochem.77.060806.091238. [DOI] [PubMed] [Google Scholar]
- 28.Huang F, et al. Multiple conformations of full-length p53 detected with single-molecule fluorescence resonance energy transfer. Proc Natl Acad Sci USA. 2009;106:20758–20763. doi: 10.1073/pnas.0909644106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cain C, Miller S, Ahn J, Prives C. The N terminus of p53 regulates its dissociation from DNA. J Biol Chem. 2000;275:39944–39953. doi: 10.1074/jbc.M002509200. [DOI] [PubMed] [Google Scholar]
- 30.Hansen S, Lane DP, Midgley CA. The N terminus of the murine p53 tumour suppressor is an independent regulatory domain affecting activation and thermostability. J Mol Biol. 1998;275:575–588. doi: 10.1006/jmbi.1997.1507. [DOI] [PubMed] [Google Scholar]
- 31.Friedlander P, Legros Y, Soussi T, Prives C. Regulation of mutant p53 temperature-sensitive DNA binding. J Biol Chem. 1996;271:25468–25478. doi: 10.1074/jbc.271.41.25468. [DOI] [PubMed] [Google Scholar]
- 32.Hansen S, Hupp TR, Lane DP. CRC Cell Transformation Group Allosteric regulation of the thermostability and DNA binding activity of human p53 by specific interacting proteins. J Biol Chem. 1996;271:3917–3924. doi: 10.1074/jbc.271.7.3917. [DOI] [PubMed] [Google Scholar]
- 33.Wu Y, et al. Phosphorylation of p53 by TAF1 inactivates p53-dependent transcription in the DNA damage response. Mol Cell. 2014;53:63–74. doi: 10.1016/j.molcel.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okorokov AL, et al. The structure of p53 tumour suppressor protein reveals the basis for its functional plasticity. EMBO J. 2006;25:5191–5200. doi: 10.1038/sj.emboj.7601382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Joerger AC, Fersht AR. The tumor suppressor p53: From structures to drug discovery. Cold Spring Harb Perspect Biol. 2010;2:a000919. doi: 10.1101/cshperspect.a000919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bista M, Freund SM, Fersht AR. Domain-domain interactions in full-length p53 and a specific DNA complex probed by methyl NMR spectroscopy. Proc Natl Acad Sci USA. 2012;109:15752–15756. doi: 10.1073/pnas.1214176109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Veprintsev DB, et al. Core domain interactions in full-length p53 in solution. Proc Natl Acad Sci USA. 2006;103:2115–2119. doi: 10.1073/pnas.0511130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Arlt C, Ihling CH, Sinz A. Structure of full-length p53 tumor suppressor probed by chemical cross-linking and mass spectrometry. Proteomics. 2015;15:2746–2755. doi: 10.1002/pmic.201400549. [DOI] [PubMed] [Google Scholar]
- 39.Retzlaff M, et al. The regulatory domain stabilizes the p53 tetramer by intersubunit contacts with the DNA binding domain. J Mol Biol. 2013;425:144–155. doi: 10.1016/j.jmb.2012.10.015. [DOI] [PubMed] [Google Scholar]
- 40.D’Abramo M, et al. The p53 tetramer shows an induced-fit interaction of the C-terminal domain with the DNA-binding domain. Oncogene. 2016;35:3272–3281. doi: 10.1038/onc.2015.388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Demir Ö, Ieong PU, Amaro RE. Full-length p53 tetramer bound to DNA and its quaternary dynamics. Oncogene. 2017;36:1451–1460. doi: 10.1038/onc.2016.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chillemi G, et al. Molecular dynamics of the full-length p53 monomer. Cell Cycle. 2013;12:3098–3108. doi: 10.4161/cc.26162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iwai H, Züger S, Jin J, Tam PH. Highly efficient protein trans-splicing by a naturally split DnaE intein from Nostoc punctiforme. FEBS Lett. 2006;580:1853–1858. doi: 10.1016/j.febslet.2006.02.045. [DOI] [PubMed] [Google Scholar]
- 44.Aranko AS, Züger S, Buchinger E, Iwaï H. In vivo and in vitro protein ligation by naturally occurring and engineered split DnaE inteins. PLoS One. 2009;4:e5185. doi: 10.1371/journal.pone.0005185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nikolova PV, Henckel J, Lane DP, Fersht AR. Semirational design of active tumor suppressor p53 DNA binding domain with enhanced stability. Proc Natl Acad Sci USA. 1998;95:14675–14680. doi: 10.1073/pnas.95.25.14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Joerger AC, Allen MD, Fersht AR. Crystal structure of a superstable mutant of human p53 core domain. Insights into the mechanism of rescuing oncogenic mutations. J Biol Chem. 2004;279:1291–1296. doi: 10.1074/jbc.M309732200. [DOI] [PubMed] [Google Scholar]
- 47.Butt TR, Edavettal SC, Hall JP, Mattern MR. SUMO fusion technology for difficult-to-express proteins. Protein Expr Purif. 2005;43:1–9. doi: 10.1016/j.pep.2005.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zettler J, Schütz V, Mootz HD. The naturally split Npu DnaE intein exhibits an extraordinarily high rate in the protein trans-splicing reaction. FEBS Lett. 2009;583:909–914. doi: 10.1016/j.febslet.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 49.Ferreon JC, et al. Cooperative regulation of p53 by modulation of ternary complex formation with CBP/p300 and HDM2. Proc Natl Acad Sci USA. 2009;106:6591–6596. doi: 10.1073/pnas.0811023106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Teufel DP, Freund SM, Bycroft M, Fersht AR. Four domains of p300 each bind tightly to a sequence spanning both transactivation subdomains of p53. Proc Natl Acad Sci USA. 2007;104:7009–7014. doi: 10.1073/pnas.0702010104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weinberg RL, Veprintsev DB, Fersht AR. Cooperative binding of tetrameric p53 to DNA. J Mol Biol. 2004;341:1145–1159. doi: 10.1016/j.jmb.2004.06.071. [DOI] [PubMed] [Google Scholar]
- 52.Petty TJ, et al. An induced fit mechanism regulates p53 DNA binding kinetics to confer sequence specificity. EMBO J. 2011;30:2167–2176. doi: 10.1038/emboj.2011.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee W, et al. Solution structure of the tetrameric minimum transforming domain of p53. Nat Struct Biol. 1994;1:877–890. doi: 10.1038/nsb1294-877. [DOI] [PubMed] [Google Scholar]
- 54.Stürzbecher HW, et al. A C-terminal alpha-helix plus basic region motif is the major structural determinant of p53 tetramerization. Oncogene. 1992;7:1513–1523. [PubMed] [Google Scholar]
- 55.McKinney K, Prives C. Efficient specific DNA binding by p53 requires both its central and C-terminal domains as revealed by studies with high-mobility group 1 protein. Mol Cell Biol. 2002;22:6797–6808. doi: 10.1128/MCB.22.19.6797-6808.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rajagopalan S, Huang F, Fersht AR. Single-Molecule characterization of oligomerization kinetics and equilibria of the tumor suppressor p53. Nucleic Acids Res. 2011;39:2294–2303. doi: 10.1093/nar/gkq800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Arbely E, et al. Acetylation of lysine 120 of p53 endows DNA-binding specificity at effective physiological salt concentration. Proc Natl Acad Sci USA. 2011;108:8251–8256. doi: 10.1073/pnas.1105028108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu G, Xia T, Chen X. The activation domains, the proline-rich domain, and the C-terminal basic domain in p53 are necessary for acetylation of histones on the proximal p21 promoter and interaction with p300/CREB-binding protein. J Biol Chem. 2003;278:17557–17565. doi: 10.1074/jbc.M210696200. [DOI] [PubMed] [Google Scholar]
- 59.Lowry DF, Stancik A, Shrestha RM, Daughdrill GW. Modeling the accessible conformations of the intrinsically unstructured transactivation domain of p53. Proteins. 2008;71:587–598. doi: 10.1002/prot.21721. [DOI] [PubMed] [Google Scholar]
- 60.Minato Y, Ueda T, Machiyama A, Shimada I, Iwaï H. Segmental isotopic labeling of a 140 kDa dimeric multi-domain protein CheA from Escherichia coli by expressed protein ligation and protein trans-splicing. J Biomol NMR. 2012;53:191–207. doi: 10.1007/s10858-012-9628-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nabeshima Y, Mizuguchi M, Kajiyama A, Okazawa H. Segmental isotope-labeling of the intrinsically disordered protein PQBP1. FEBS Lett. 2014;588:4583–4589. doi: 10.1016/j.febslet.2014.10.028. [DOI] [PubMed] [Google Scholar]
- 62.Mujtaba S, et al. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 63.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 64.Arai M, Ferreon JC, Wright PE. Quantitative analysis of multisite protein-ligand interactions by NMR: Binding of intrinsically disordered p53 transactivation subdomains with the TAZ2 domain of CBP. J Am Chem Soc. 2012;134:3792–3803. doi: 10.1021/ja209936u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Borcherds W, et al. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- 66.von Hippel PH, Berg OG. On the specificity of DNA-protein interactions. Proc Natl Acad Sci USA. 1986;83:1608–1612. doi: 10.1073/pnas.83.6.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weinberg RL, Freund SM, Veprintsev DB, Bycroft M, Fersht AR. Regulation of DNA binding of p53 by its C-terminal domain. J Mol Biol. 2004;342:801–811. doi: 10.1016/j.jmb.2004.07.042. [DOI] [PubMed] [Google Scholar]
- 68.McKinney K, Mattia M, Gottifredi V, Prives C. p53 linear diffusion along DNA requires its C terminus. Mol Cell. 2004;16:413–424. doi: 10.1016/j.molcel.2004.09.032. [DOI] [PubMed] [Google Scholar]
- 69.Tafvizi A, Huang F, Fersht AR, Mirny LA, van Oijen AM. A single-molecule characterization of p53 search on DNA. Proc Natl Acad Sci USA. 2011;108:563–568. doi: 10.1073/pnas.1016020107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kim H, et al. p53 requires an intact C-terminal domain for DNA binding and transactivation. J Mol Biol. 2012;415:843–854. doi: 10.1016/j.jmb.2011.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Laptenko O, et al. The p53 C terminus controls site-specific DNA binding and promotes structural changes within the central DNA binding domain. Mol Cell. 2015;57:1034–1046. doi: 10.1016/j.molcel.2015.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gu W, Roeder RG. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell. 1997;90:595–606. doi: 10.1016/s0092-8674(00)80521-8. [DOI] [PubMed] [Google Scholar]
- 73.Luo J, et al. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci USA. 2004;101:2259–2264. doi: 10.1073/pnas.0308762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dornan D, Shimizu H, Perkins ND, Hupp TR. DNA-dependent acetylation of p53 by the transcription coactivator p300. J Biol Chem. 2003;278:13431–13441. doi: 10.1074/jbc.M211460200. [DOI] [PubMed] [Google Scholar]
- 75.Müller L. Sensitivity enhanced detection of weak nuclei using hetero-nuclear multiple quantum coherence. J Am Chem Soc. 1979;101:4481–4484. [Google Scholar]
- 76.Morris KF, Johnson CS. Diffusion-ordered two-dimensional nuclear magnetic resonance spectroscopy. J Am Chem Soc. 1992;114:3139–3141. [Google Scholar]
- 77.Wang Y, Schwedes JF, Parks D, Mann K, Tegtmeyer P. Interaction of p53 with its consensus DNA-binding site. Mol Cell Biol. 1995;15:2157–2165. doi: 10.1128/mcb.15.4.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Park SJ, Borin BN, Martinez-Yamout MA, Dyson HJ. The client protein p53 adopts a molten globule-like state in the presence of Hsp90. Nat Struct Mol Biol. 2011;18:537–541. doi: 10.1038/nsmb.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wittekind M, Mueller L. HNCACB, a high-sensitivity 3D NMR experiment to correlate amide-proton and nitrogen resonances with the alpha- and beta-carbon resonances in proteins. J Magn Reson. 1993;101:201–205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.