Significance
The presence of extracellular miRNAs in body fluids has been exploited as a brand-new source of biomarkers for different diseases. A fraction of those extracellular miRNAs, contained in extracellular vesicles and exosomes, are additionally being revealed as novel mediators of intercellular communication. Here, we show that systemic injection of exosomes transfected with synthetic miRNAs simulating those enriched in the plasma of obese mice robustly induces glucose intolerance, adipose inflammation, and hepatic steatosis in lean mice. These results support a role for exosomal miRNAs in the modulation of glucose and lipid metabolism in mice and may help us uncover thus far unexplored pathological mechanisms and provide us with novel therapeutic targets.
Keywords: miRNA, exosome, glucose intolerance, adiposity, dyslipidemia
Abstract
Obesity is frequently associated with metabolic disease. Here, we show that obesity changes the miRNA profile of plasma exosomes in mice, including increases in miR-122, miR-192, miR-27a-3p, and miR-27b-3p. Importantly, treatment of lean mice with exosomes isolated from obese mice induces glucose intolerance and insulin resistance. Moreover, administration of control exosomes transfected with obesity-associated miRNA mimics strongly induces glucose intolerance in lean mice and results in central obesity and hepatic steatosis. Expression of the candidate target gene Ppara is decreased in white adipose tissue but not in the liver of mimic-treated (MIMIC) mice, and this is accompanied by increased circulating free fatty acids and hypertriglyceridemia. Treatment with a specific siRNA targeting Ppara transfected into exosomes recapitulates the phenotype induced by obesity-associated miRNAs. Importantly, simultaneously reducing free fatty acid plasma levels in MIMIC mice with either the lipolysis inhibitor acipimox or the PPARα agonist fenofibrate partially protects against these metabolic alterations. Overall, our data highlight the central role of obesity-associated exosomal miRNAs in the etiopathogeny of glucose intolerance and dyslipidemia.
MicroRNAs (miRNAs) are small noncoding RNA molecules that function as negative regulators of translation (1), particularly during cellular transitions or in situations of stress, including maintenance of metabolic homeostasis (2). Importantly, miRNAs can be selectively secreted by most cell types (3). Changes in the profile of miRNAs circulating in blood can be detected in association with diverse pathological conditions, including metabolic disease (3). These extracellular miRNAs can be used as biomarkers to improve diagnosis and monitor response to therapy (4), and we have previously shown that the abundance of specific circulating miRNAs is altered in prediabetic subjects and glucose-intolerant mice alike (5). Circulating miRNAs can be found bound to protein complexes or associated with high-density lipoproteins (6, 7). They can also be located inside small vesicles called exosomes (8). Exosomes can be captured by acceptor cells, where the miRNAs they contain may induce transcriptomic changes, thus acting as a novel form of intercellular communication (8). Exosomal miRNAs have been shown to participate in tumor progression, angiogenesis, and metastasis (9). However, their role in metabolic diseases has been much less explored (10).
Type 2 diabetes is one of the most common metabolic diseases in the world, expected to affect almost a third of the population by 2050 (11). The ever-increasing rise in diabetes prevalence is closely associated with present-day obesity epidemics, as obesity is one of the major underlying causes of insulin resistance, which is a key component in the etiology of diabetes (12). Development of diabetes entails alterations in insulin-sensitive tissues such as the liver, the skeletal muscle, and the adipose depots, leading to a state of glucose intolerance or prediabetes that ultimately results in overt diabetes when pancreatic β-cells are unable to cope with the increased demand for insulin (13). Communication between different tissues is thus central to the maintenance of glucose homeostasis. Different circulating factors, including hormones, cytokines, and growth factors, are known to modulate interorgan cross-talk (14). In addition, a number of recent studies have evidenced that adipose-released exosomal miRNAs can regulate gene expression in other tissues (10, 15).
The study of exosomal miRNAs holds the promise of providing us with a better understanding of the etiology of metabolic disorders, which may then be exploited as a source of novel targets or as a new therapeutic strategy (16). Here, we focus on exploring the role of exosomal miRNAs during the development of obesity-associated glucose intolerance and dyslipidemia in mice.
Results
Diet-Induced Central Obesity Changes the miRNA Profile of Circulating Exosomes in Mice.
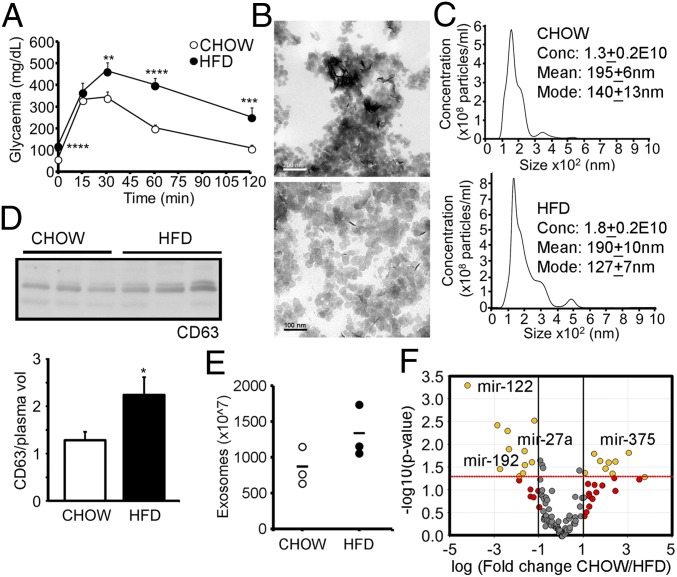
To explore the role of exosomes in the development of glucose intolerance, we took advantage of a mouse model that recapitulates many features of prediabetes (17). Male C57B6J mice were rendered glucose intolerant (Fig. 1A and SI Appendix, Fig. S1A and Table S1) by administration of a high-fat diet (HFD) for 15 wk. The HFD mice were hyperphagic (SI Appendix, Fig. S1B) and obese (SI Appendix, Fig. S1C). The adiposity index, a measure of central obesity calculated as the ratio of the weight of epididymal white adipose tissue (eWAT) and body weight, was significantly increased in the HFD mice (SI Appendix, Fig. S1D), which also displayed liver steatosis (SI Appendix, Fig. S1 E and F). We found a tight positive correlation between the glycemia area under the curve (AUC) obtained from the i.p. glucose tolerance test (IpGTT) and body weight (SI Appendix, Fig. S1G). Importantly, the correlation between the AUC and the adiposity index was also highly significant (SI Appendix, Fig. S1H). Hence, glucose tolerance is strongly associated with body weight and, particularly, with the percentage of fat. Obese mice were also dyslipidemic, presenting significantly increased plasma levels of triglycerides (TGs) (SI Appendix, Fig. S1I) and free fatty acids (FFAs) (SI Appendix, Fig. S1J).
Fig. 1.
Diet-induced central obesity changes the profile of circulating exosomal miRNAs. (A) IpGTT in C57B6J mice after 15 wk of HFD feeding. (B) Representative electron micrographs of exosomes isolated from mouse plasma. (C) Exosome size distribution determined by NTA in control and obese mice. (D) Western blot analysis of CD63 from equal volumes of plasma obtained from chow-fed and HFD mice and its quantification. (E) Number of exosomes from equal volumes of plasma estimated from esterase activity. (F) Volcano plot of real time RT-PCR profiling of the miRNA content of lean and obese plasma exosomes. Data are presented as mean ± SEM. n = 10 per group (A); n = 3 per group (B–E); n = 4 per group (F). *P < 0.05, **P < 0.01, ***P < 0.005, ****P < 0.001, Student’s t test.
Exosomes isolated from equal amounts of plasma from control and HFD mice were characterized by transmission electronic microscopy (Fig. 1B), nanoparticle tracking analysis (NTA) (Fig. 1C), and Western blot with exosomal marker CD63 (Fig. 1D), and by measuring exosome-associated esterase activity (Fig. 1E). The mean size of the particles obtained by NTA was larger than that evidenced by microscopy analysis, probably due to vesicle agglutination during the measurements. However, the values were not different between groups. All analyses showed a tendency for exosomes to be elevated in the HFD mice, which was only significant in the case of CD63 quantification (P = 0.07 for NTA; P = 0.1 for esterase activity). However, aside from their number, the composition of exosomes may also be modified by obesity. We focused on the miRNA content because exosomal miRNAs have been shown to participate in the establishment of metabolic phenotypes (10). Real-time RT-PCR miRNA profiling of exosomes isolated from plasma of the lean and HFD mice showed upregulation of 10 miRNAs in obese exosomes, whereas 9 were down-regulated (Fig. 1F and SI Appendix, Fig. S1K and Table S2). The miRNA pattern of obese exosomes was sufficiently different from that of lean exosomes to discriminate both populations (SI Appendix, Fig. S1L). Among the miRNAs increased in obesity, we identified miR-192 and miR-122, which have been previously shown by us and others to be increased in plasma of prediabetic or insulin-resistant subjects (5, 18). We performed a correlation analysis between each pair of miRNAs across all samples and selected those miRNAs with significant correlations to miR-122 (r ≥ 0.8, P < 0.01). Heat map representation of those miRNAs evidenced two populations (SI Appendix, Fig. S1M). We hypothesize that miRNAs showing a similar behavior might be under the same pattern of dynamic secretion. We also identified other miRNA families known to participate in the regulation of metabolism, including miR-27 and miR-30 (SI Appendix, Fig. S1M). Real-time RT-PCR analysis of the expression of miR-122, miR-192, and miR-27a-3p in control and HFD mice evidenced a decrease in all three miRNAs in the eWAT, whereas miR-122 and miR-192 were increased in the liver (SI Appendix, Fig. S1N). These data indicate that miRNA expression is regulated at the tissue level in obesity, and this is reflected in a change in the pattern of circulating exosomal miRNAs.
Exosomes from Obese Mice Induce Glucose Intolerance in Lean Mice.
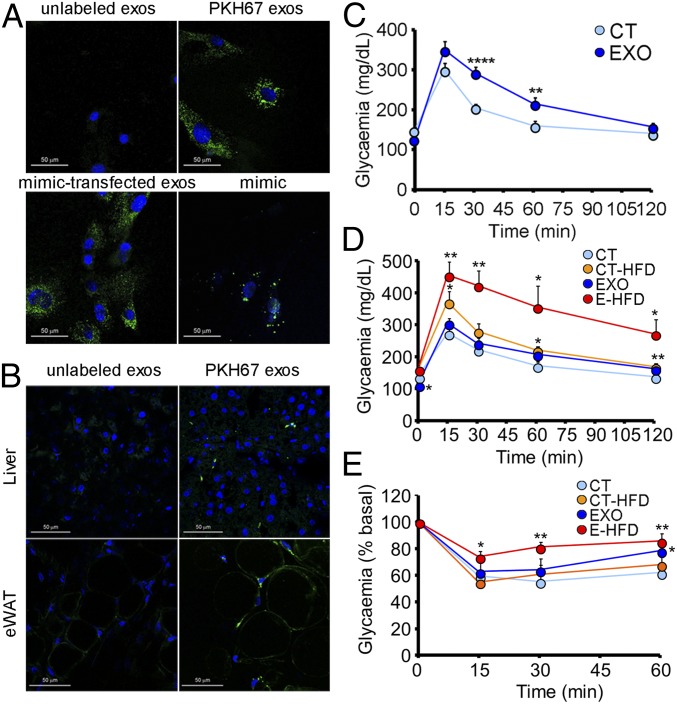
We first studied the ability of exosomes to be captured by acceptor cells. Fluorescently labeled exosomes added to 3T3-L1 cells for 24 h were easily detected by microscopy (Fig. 2A, upper). The presence of fluorescent signals was also evident in the liver (Fig. 2B, Upper) and the eWAT (Fig. 2B, Lower) of mice injected i.v. with PKH67-labeled exosomes, as has been demonstrated by others (10). To further corroborate that exosomes can reach those tissues, we injected mice with exosomes transfected with a nonmammalian miRNA, cel-miR-39-3p. Real-time RT-PCR analysis showed detectable levels of the exogenous miRNA in both the liver [cycle threshold (Ct) 25.6 ± 0.1] and the eWAT (Ct 32.8 ± 3).
Fig. 2.
Exosomes from obese mice induce glucose intolerance in lean mice. (A) 3T3-L1 cells after 24 h incubation with unlabeled exosomes (Upper left), exosomes labeled with fluorescent marker PKH67 (Upper right), unlabeled exosomes transfected with a fluorescent miRNA mimic (Lower left), and 3T3-L1 cells transfected with the same fluorescent mimic (Lower right). (B) Liver (Upper) and eWAT (Lower) sections of control mice after 6 h injection with PBS (Left) or exosomes labeled with PKH67 (Right). (C) IpGTT in chow-fed mice after 4 wk of biweekly injections of obese exosomes. (D and E) IpGTT (D) and insulin tolerance test (0.35 U/kg) (E) in C57B6J mice after 8 wk of biweekly systemic injections of obese exosomes, with HFD feeding during the last 4 wk. Data are presented as mean ± SEM. n = 3 per condition (A and B); n = 10 per group (C); n = 5 per group except n = 4 CT-HFD (D and E). *P < 0.05, **P < 0.01, ****P < 0.001 with respect to control (CT) group, Student’s t test.
Next, we injected control mice biweekly through the tail vein for 4 wk with exosomes isolated from the plasma of either lean or obese mice. Surprisingly, the mice treated with obese exosomes were rendered glucose intolerant (Fig. 2C) and insulin resistant (Fig. 2E and SI Appendix, Fig. S2 A and B). To test if obese exosomes predisposed the recipient mice to a worse metabolic outcome when subjected to an additional challenge, we administered HFD to half of the mice in each group during the next 4 wk. Mice fed HFD ingested more calories per day, independently of being treated with lean or obese exosomes (SI Appendix, Fig. S2C). Body weight increased in all experimental groups compared with the chow-fed control mice injected with lean exosomes (SI Appendix, Fig. S2D). However, significantly increased adiposity was only evident in the combined exosome-treated and HFD-fed (EXO-HFD) group (SI Appendix, Fig. S2E). Determination of the hepatic TG content showed a tendency for all experimental groups to be increased compared with the control, but the differences were not significant (SI Appendix, Fig. S2F). Superposition of a short-term HFD on the exosome-treated mice resulted in a dramatically exacerbated phenotype of glucose intolerance and insulin resistance (Fig. 2 D and E and SI Appendix, Fig. S2G and Table S1). Overall, these data show that modification of the population of circulating exosomes induces the development of metabolic alterations in mice.
Exosomes Transfected with Obesity-Associated miRNAs Induce Glucose Intolerance Dissociated from Obesity.
To determine if miRNAs are the ones influencing energetic metabolism, we isolated exosomes from lean mice and transfected them with a negative control or with a mixture of artificial miRNA mimics. We selected four of the miRNAs that we found to be more overexpressed in obese exosomes (miR-192, miR-122, miR-27a-3p, and miR-27b-3p). With this strategy, we aimed not to exactly reconstitute the obese exosomes but to isolate the effect of the miRNAs, discarding other sources of variation such as the protein content or lipid cover of the exosomes. As a control, we first transfected exosomes with a fluorescent miRNA mimic. Addition of these transfected exosomes to 3T3-L1 cells showed a pattern akin to that observed when adding exosomes labeled with PKH67 but clearly different from that observed when the cells were transfected with the same fluorescent miRNA mimic naked (Fig. 2A, Lower).
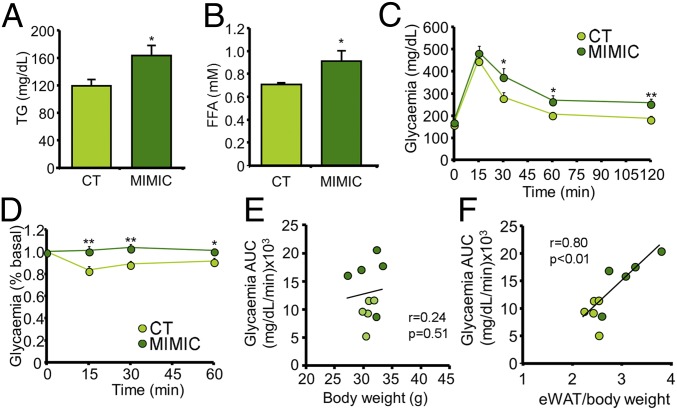
Lean mice fed standard chow were then injected through the tail vein biweekly for 4 wk with exosomes transfected with the selected mimics. After the treatment, body weight was no different between groups (SI Appendix, Fig. S3A) but the eWAT was significantly enlarged (SI Appendix, Fig. S3 B and C). Additionally, the mimic-treated (MIMIC) mice were dyslipidemic, with increased plasma levels of TGs (Fig. 3A) and FFAs (Fig. 3B). Importantly, as was the case with the mice injected with the obese exosomes (Fig. 2 C–E), the MIMIC mice were glucose intolerant (Fig. 3C and SI Appendix, Table S1) and insulin resistant (Fig. 3D and SI Appendix, Fig. S3D). Correlation between the glycemia AUC from the IpGTT and either body weight or percentage of eWAT indicated that glucose intolerance is significantly associated with an increase in eWAT (Fig. 3F) but not body weight (Fig. 3E). This is a different scenario from that observed in the HFD mice (SI Appendix, Fig. S1 G and H), where the AUC correlated even better with body weight than percentage of eWAT. Hence, the MIMIC mice replicated the phenotype of glucose intolerance and insulin resistance seen in the HFD mice but dissociated from obesity.
Fig. 3.
Exosomes transfected with obesity-associated miRNAs induce glucose intolerance dissociated from obesity. (A and B) Plasma TG (A) and FFA (B) concentrations from chow-fed mice after 4 wk of injections of exosomes loaded with mimics of four miRNAs enriched in obese exosomes. (C and D) IpGTT (C) and insulin tolerance test (0.175 U/kg) (D) in mice described in A and B. (E and F) Correlation between the glycemia AUC obtained from the IpGTT and either body weight (E) or percentage of eWAT (F). Data are presented as mean ± SEM. n = 5 per group (A–F). *P < 0.05, **P < 0.01, Student’s t test.
Mimic Treatment Induces eWAT Inflammation and Hepatic Steatosis.
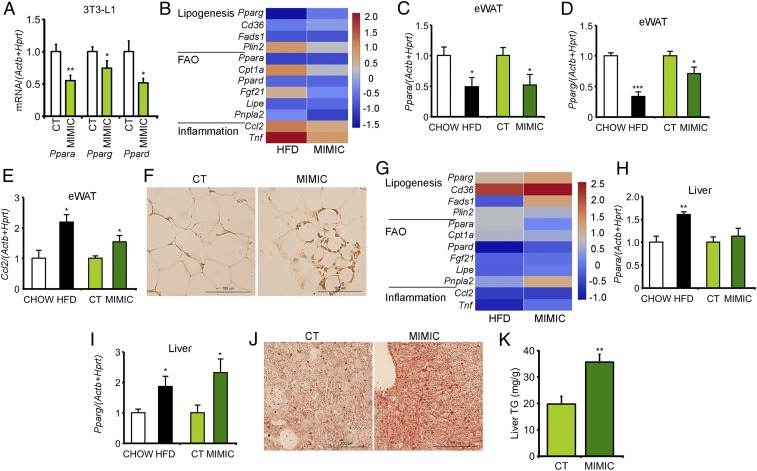
We used the Reactome Pathway Database to search for pathways affected by all of the miRNAs whose abundance was modified in obese exosomes. Analysis of the up-regulated miRNAs produced the most significant results, with the identification of pathways involved mostly in the regulation of transcription (SI Appendix, Table S3). Ingenuity Pathway Analysis (IPA) software (QIAGEN) was used to identify target genes specific to our four selected miRNAs, focusing on the liver and the eWAT. We selected these tissues because they are important insulin target organs and, in the case of the eWAT, we were surprised by the significant enlargement observed (SI Appendix, Fig. S3D). The analysis identified the Ppar family of transcription factors as one of the main canonical pathways affected and lipid metabolism as the main affected network (SI Appendix, Table S4). Accordingly, RT-PCR analysis of 3T3-L1 cells transfected with the four selected mimics evidenced that all three isoforms of the Ppar family were significantly down-regulated (Fig. 4A and SI Appendix, Fig. S4A).
Fig. 4.
Mimic treatment induces eWAT inflammation and hepatic steatosis. (A) mRNA expression level of Ppar family members in 3T3-L1 cells after transfection with the four selected obesity-associated miRNA mimics. (B and G) Heat maps showing differential mRNA expression of candidate target genes involved in lipogenesis, FAO, and inflammation between either HFD or MIMIC mice and respective controls in the eWAT (B) and the liver (G). (C–E) mRNA expression level of Ppara (C), Pparg (D), and Ccl2 (E) in the eWAT of HFD mice, MIMIC mice, and respective controls. (F) Representative H&E staining of eWAT sections from control and MIMIC mice. (H and I) mRNA expression level of Ppara (H) and Pparg (I) in the liver of HFD mice, MIMIC mice, and respective controls. (J and K) Representative Oil Red staining of liver sections (J) and TG quantification in the liver of control and MIMIC mice (K). Data are presented as mean ± SEM. At least n = 7 per condition (A); n = 4 per group (B–E, G–I, and K); n = 2 per group (F and J). *P < 0.05, **P < 0.01, ***P < 0.005 with respect to either control (CT) or CHOW groups, Student’s t test.
The eWAT of the MIMIC mice displayed decreased expression of adipogenic genes and fatty acid oxidation (FAO) pathways, with increased expression of inflammatory mediators. This is a pattern akin to that observed in the eWAT of the HFD mice (Fig. 4B). In particular, decreased expression of Ppara and Pparg (Fig. 4C and D) and increased expression of Ccl2 (Fig. 4E) were observed. Accordingly, the eWAT of the MIMIC mice also showed macrophage infiltration (Fig. 4F and SI Appendix, Fig. S4 B and C). In contrast, analysis of hepatic gene expression showed up-regulation of genes associated with de novo lipogenesis (DNL) in both the HFD and the MIMIC mice (Fig. 4G). Specifically, increased expression of Pparg (Fig. 4I) was observed, associated with a higher accumulation of hepatic lipid droplets (Fig. 4J) and increased TG content (Fig. 4K). On the other hand, Ppara expression remained unchanged in the MIMIC mice whereas it was increased in the HFD mice (Fig. 4H). Hence, mimic treatment induces robust gene expression changes associated with eWAT inflammation and hepatic steatosis, both of which are known components in the development of glucose intolerance and dyslipidemia (19).
Treatment with siPPARA-Transfected Exosomes Recapitulates the Central Obesity Phenotype of MIMIC Mice.
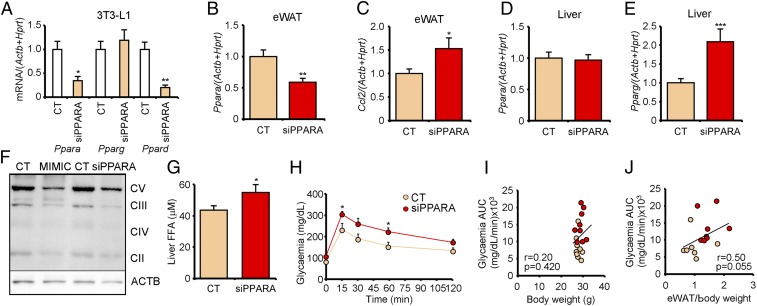
We hypothesized that decreased Ppara in the eWAT could explain the phenotype of the MIMIC mice. Decreased Ppara expression was also found in the eWAT of the HFD mice and is known to induce defective FAO and increased FFA delivery to the bloodstream (20), leading to hepatic steatosis. Accordingly, the MIMIC mice showed decreased mitochondrial content in the eWAT (Fig. 5F). Hence, we injected the mice with exosomes transfected with a siRNA targeting Ppara (siPPARA). First, transfection of 3T3-L1 cells showed that siPPARA effectively decreased Ppara mRNA levels (Fig. 5A). This was accompanied by a decrease in Ppard, the other catabolic isoform, but not the anabolic isoform Pparg.
Fig. 5.
Treatment with siPPARA-transfected exosomes recapitulates the central obesity phenotype of MIMIC mice. (A) mRNA expression level of Ppar family members in 3T3-L1 cells transfected with siPPARA siRNA. (B and C) mRNA expression of Ppara (B) and Ccl2 (C) in the eWAT of chow-fed mice after two injections of lean exosomes loaded with the siPPARA described in A. (D and E) mRNA expression of Ppara (D) and Pparg (E) in the liver of control and siPPARA mice. (F) Western blot analysis of mitochondrial complexes and housekeeping actin in the eWAT from MIMIC and siPPARA model mice. (G) FFA quantification in the liver of the mice described in B and C. (H) IpGTT in the mice described in B and C. (I and J) Correlation between the glycemia AUC obtained from the IpGTT and either body weight (I) or percentage of eWAT (J). Data are presented as mean ± SEM. n = 3 per condition (A); n = 4 per group (B–E); n = 2 per group (F); n = 8 per group (G); n = 5 per group (H); n = 9 per group (I); n = 8 control (CT) and 7 siPPARA (J). *P < 0.05, **P < 0.01, ***P < 0.005, Student’s t test.
Next, the control mice were treated for 1 wk with two systemic injections of siRNA-loaded exosome preparations. Remarkably, similar to the MIMIC mice, Ppara expression was decreased in the eWAT of the siPPARA mice (Fig. 5B) but not in the liver (Fig. 5D). This was accompanied by a phenotype of adipose inflammation (Fig. 5C) and activated hepatic DNL (Fig. 5E and SI Appendix, Fig. S5A). Interestingly, we observed a significant decrease in the mitochondrial content of the eWAT (Fig. 5F) and a concurrent increase in hepatic FFA (Fig. 5G) and TG content (SI Appendix, Fig. S5 B and C). It is surprising that Ppara was not decreased in the liver even though the exosomes were reaching the tissue (Fig. 2B). A possible explanation is that the obesity phenotype was opposing the effect of the exosomes in the liver. Accordingly, 48 h after a single injection of siPPARA-loaded exosomes, when the phenotype was not yet established, we observed comparable decreases in Ppara expression in both hepatocytes and adipocytes (SI Appendix, Fig. S5D). After treatment, the siPPARA mice were glucose intolerant (Fig. 5H and SI Appendix, Table S1). Correlation analysis again demonstrated that glucose intolerance in this model is associated with increased eWAT (Fig. 5J) but not body weight (Fig. 5I). Altogether, these data show that treatment with siPPARA-transfected exosomes decreased Ppara expression predominantly in the eWAT, and this was enough to recapitulate the central obesity phenotype of the MIMIC mice.
Decreasing FFA Plasma Levels Partially Revert the Pathologic Phenotype.
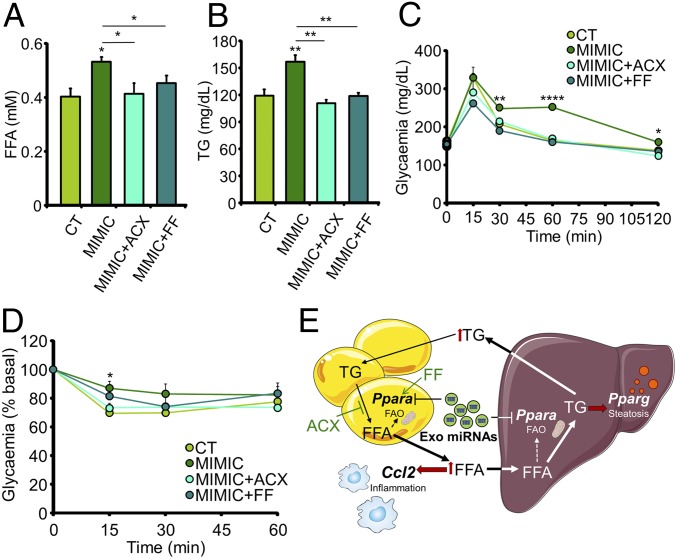
As the liver accumulates FFAs as a function of their circulating levels (21), hepatic steatosis may be secondary to adipose dysfunction. We expected that by decreasing FFA plasma levels we would be able to partially revert the pathological phenotype. Hence, we treated a new cohort of mice simultaneously with mimic-transfected exosomes and either the lipolysis inhibitor acipimox or the PPARα agonist fenofibrate, hence reducing plasma FFAs by enforcing their storage or favoring their oxidation, respectively. Circulating FFA and TG levels significantly increased in the MIMIC mice, but returned to control levels with administration of both drugs (Fig. 6 A and B). Importantly, in parallel with alleviation of dyslipidemia, both drugs mitigated the phenotype of glucose intolerance observed in the MIMIC mice (Fig. 6C and SI Appendix, Table S1) and normalized insulin resistance (Fig. 6D). A similar treatment with acipimox in the siPPARA-treated mice was also able to normalize dyslipidemia and glucose intolerance (SI Appendix, Fig. S6 A–D and Table S1). Overall, our data support the central role of obesity-associated exosomal miRNA-targeted adipose Ppara expression, reflected in increased FFA plasma levels, development of glucose intolerance, and dyslipidemia in mice.
Fig. 6.
Decreasing FFA plasma levels partially revert the pathologic phenotype. (A and B) Plasma FFA (A) and TG (B) concentrations from chow-fed mice after 4 wk of injections of exosomes loaded with mimics of four miRNAs enriched in obese exosomes and simultaneously administered acipimox (ACX) or fenofibrate (FF) orally. (C and D) IpGTT (C) and insulin tolerance test (0.5 U/kg) (D) in the mice described in A and B. (E) Proposed model: Injection of exosomes transfected with synthetic miRNAs simulating those enriched in obesity decreases Ppara expression and oxidative capacity in the eWAT. This is associated with increased FFA release to the bloodstream, which in turn induces adipose inflammation and hepatic steatosis. Treatment with the lipolysis inhibitor ACX or the PPARα agonist FF decreases plasma FFAs and partially reverts this phenotype. Data are presented as mean ± SEM. n = 5 per group (A–D). *P < 0.05, **P < 0.01, ****P < 0.001 with respect to the control (CT) group unless otherwise indicated, Student’s t test.
Discussion
Recent in vivo studies have demonstrated that exosomes can transfer mature miRNAs between organs, resulting in functional changes in the receiving cells (15) and affecting whole-body insulin sensitivity (10). We showed that obesity changes the profile of exosomal miRNAs in mice. We observed increases in exosomal miR-192 and miR-122 that were in accordance with previous evidence suggesting an important role for both miRNAs as novel circulating factors involved in insulin resistance (5, 18, 22). It was out of the scope of our study to identify the tissue that provides the main source of obesity exosomal miRNAs, but a number of reports have pointed to the eWAT (10, 15).
Importantly, we report that lean mice treated with exosomes from obese mice develop glucose intolerance and insulin resistance, which are further exacerbated by superposition of a short-term HFD. This effect, however, may be due to the induction of an inflammatory phenotype caused by the injection of contaminants present in the plasma of obese mice and coprecipitated with the vesicles. To avoid this unwanted effect and, at the same time, with the intention of focusing on the role of miRNAs and discarding other sources of variation such as the protein or lipid content of exosomes, we established an artificial model in which the only difference in the injected preparations was the miRNAs themselves. Remarkably, the mice injected with lean exosomes transfected with obesity-associated mimics replicated the phenotype of glucose intolerance seen in the HFD mice, hence pointing to a central role of exosomal miRNAs in orchestrating the phenotypic effects observed.
The MIMIC mice remained lean, but showed a significant enlargement of the epididymal adipose depot. Interestingly, we found a strong correlation between the percentage of eWAT and the glycemia AUC, suggesting that glucose tolerance in these mice was associated with central obesity rather than with total body weight. In this regard, the plasma abundance of miR-122 and miR-192 has been shown to correlate with waist circumference and visceral fat quantity in humans, as well as with an increased TG/HDL ratio (18). Inhibition of miR-122 reduces cholesterol and hepatic FA synthesis in mice and primates (23, 24). Similarly, miR-27a-3p and miR-27b-3p have been involved in the regulation of adipose function (25) and treatment of mice with miR-27 family mimics increases TG levels (26).
Our MIMIC mice displayed decreased expression of adipogenic genes and FAO pathways in the eWAT, associated with macrophage infiltration and enlargement of the tissue. This phenotype is also associated with increased FFA plasma levels, up-regulation of hepatic DNL genes, and accumulation of lipids. Importantly, both adipose inflammation and hepatic steatosis are known components in the early development of glucose intolerance and dyslipidemia. The decreased Ppara adipose expression found in the MIMIC mice might lead to defective FAO and increased FFA delivery to the periphery (20, 27, 28). To confirm that, we treated the mice with exosomes transfected with a siRNA targeting Ppara, thus decreasing its expression in the tissues targeted by the exosomes. We observed the main effect in the eWAT, where decreased Ppara expression was again accompanied by a phenotype of inflammation and tissue enlargement. In agreement with our results, the Ppara−/− mice displayed larger adipose stores with aging (28).
To provide further evidence of the role of the eWAT and the involvement of Ppara in the development of the phenotype, we used two alternative strategies, both aimed at decreasing FFA plasma levels. MIMIC mice were simultaneously administered either the lipolysis inhibitor acipimox, then decreasing FFA release to the bloodstream by enforcing their storage in the eWAT (29), or the PPARα agonist fenofibrate, then increasing oxidation. As expected, both strategies restored insulin sensitivity and significantly improved glucose tolerance in the MIMIC mice. Accordingly, fenofibrate has been shown to improve insulin sensitivity in patients with hypertriglyceridemia (30). PPARα activation was enough to revert the pathologic phenotype of the MIMIC mice, even though levels of the factor were decreased. These results, together with the data regarding the siPPARA-treated mice, support our notion that adipose Ppara is central to the phenotype observed (Fig. 6E).
Use of exosomes as vehicles for the transport of siRNAs may pave the way to strategies to specifically deliver bioactive cargo to target cells (16). Systemic injection of miRNA mimics/antagomiRs or siRNAs has been performed in mice (23, 24, 31). However, we want to stress two main differences between our experimental models and previous studies. On the one hand, in previous reports the siRNAs/miRNAs were administered either naked or coupled to an adjuvant such as cholesterol moieties or atelocollagen (23, 24, 31) whereas we packed them inside exosomes. Hence, we expected that we were delivering them to those tissues targeted by native exosomes. On the other hand, in contrast with the pharmacological doses frequently described of approximately 25 mg miRNA/kg mouse, we were injecting a 1,000-fold lower dose of 25 µg/kg, corresponding to 125 pmols miRNA.
Overall, we showed that obesity-associated exosomal miRNAs are active players in the first stages of the metabolic syndrome characterized by development of glucose intolerance, dyslipidemia, and central obesity in mice.
Materials and Methods
Experimental Animal Models.
HFD: mice were maintained in either a standard chow diet or an HFD for 15 wk. For the different treatments, mice were injected biweekly through the tail vein with native or transfected exosome preparations in PBS. EXO: mice were injected with 5 μg exosomes isolated from plasma of control or HFD mice for 4 wk. Half of the mice in each group were administered HFD during the next 4 wk while maintaining the injections. MIMIC: mice were injected with 25 μg exosomes transfected with a negative control or a mixture of miRNA mimics for 4 wk. A second cohort of mimic-injected mice was simultaneously daily administered either the lipolysis inhibitor acipimox or the PPARα agonist fenofibrate by oral gavage. siPPARA: mice were injected with 25 μg exosomes transfected with a nontargeting siRNA or a siRNA targeting Ppara for 1 wk. Glucose tolerance, insulin sensitivity, and circulating TG and FFA levels were determined after 6 h fasting (32). At killing, 1 mL blood was obtained for exosome isolation. Biodistribution studies: mice were injected with 50 μg exosomes transfected with a nonmammalian miRNA (cel-miR-39-3p) or PBS and killed 4 h afterward for RT-PCR analysis. A second cohort was injected with 100 μg PKH67-labeled exosomes or PBS 6 h before killing for immunohistochemistry analysis. Studies were approved by the Animal Research Committee of the University of Barcelona (register 404/13).
Exosome Characterization.
Exosomes isolated from mouse plasma by centrifugation were characterized by Western blot, NTA, transmission electron microscopy, and esterase ELISA. Real-time RT-PCR miRNA profiling was performed using predesigned panels with locked nucleic acid (LNA) primers (Exiqon). Differential expression was determined with GenEx software (Exiqon) by normalizing to the mean Ct of the plate. Exosomes were labeled with fluorescent dye PKH67 and transfected with 370 pmol fluorescent miRNA mimic, 125 pmol negative control (cel-miR-39-3p), artificial miRNA mimics (miR-192, miR-122, miR-27a-3p, and miR-27b-3p), or 190 pmol siRNA targeting Ppara or a negative control nontargeting siRNA using Exo-Fect (System Biosciences). For sequences and references, see SI Appendix, Table S5. 3T3-L1 cells were transfected with 8 pmol negative control or the four selected miRNA mimics using Metafectene Pro (Biontex) (33). For siRNA transfection, 60 nM siPPARA or a scrambled control was used (Applied Biosystems).
RNA/Protein Analyses and Immunohistochemistry.
For mRNA expression, 500 ng were retrotranscribed and analyzed by real-time RT-PCR. For tissue miRNA expression, 5 ng of total RNA was retrotranscribed and analyzed using commercial SYBRGreen primers (Exiqon). See SI Appendix, Table S6 for primer sequences and references. Protein extracted with radioimmunoprecipitation assay (RIPA) buffer was analyzed by Western blot with the Mitoprofiler (MitoSciences-Abcam) and antiactin (Sigma) antibodies. H&E and Oil Red staining was performed following the protocols at IHCWorld (www.ihcworld.com).
Statistical Analyses.
Differences between groups were determined by either t test analysis when only two groups were compared or by one-way ANOVA with t test analysis for posttest pairwise comparisons of three or more groups. Asterisks in figures indicate significance with respect to the control group unless otherwise specified. Correlation analyses were performed by Pearson regression.
Supplementary Material
Acknowledgments
We thank Dr. Hernando A. del Portillo from ISGlobal and Institut d’Investigació en Ciències de la Salut Germans Trias i Pujol (IGTP) for his help in the NTA analyses. We thank Anna Orduña for technical help and data discussion. We thank the members of the Electron Microscopy Unit from the CCiTUB for their help with exosome characterization. This work was supported by Grant EFSD/Lilly-2013 from the European Foundation for the Study of Diabetes (EFSD). It also received support from CIBERDEM and Project 2014_SGR_520 of the DURSI (Government of Catalonia).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1808855115/-/DCSupplemental.
References
- 1.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guay C, Regazzi R. Exosomes as new players in metabolic organ cross-talk. Diabetes Obes Metab. 2017;19:137–146. doi: 10.1111/dom.13027. [DOI] [PubMed] [Google Scholar]
- 3.Chen X, et al. Characterization of microRNAs in serum: A novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- 4.Párrizas M, Novials A. Circulating microRNAs as biomarkers for metabolic disease. Best Pract Res Clin Endocrinol Metab. 2016;30:591–601. doi: 10.1016/j.beem.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Párrizas M, et al. Circulating miR-192 and miR-193b are markers of prediabetes and are modulated by an exercise intervention. J Clin Endocrinol Metab. 2015;100:E407–E415. doi: 10.1210/jc.2014-2574. [DOI] [PubMed] [Google Scholar]
- 6.Arroyo JD, et al. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci USA. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13:423–435. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simons M, Raposo G. Exosomes–Vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–581. doi: 10.1016/j.ceb.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 9.Fong MY, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. 2015;17:183–194. doi: 10.1038/ncb3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ying W, et al. Adipose tissue macrophage-derived exosomal miRNAs can modulate in vivo and in vitro insulin sensitivity. Cell. 2017;171:372–384.e12. doi: 10.1016/j.cell.2017.08.035. [DOI] [PubMed] [Google Scholar]
- 11.Boyle JP, Thompson TJ, Gregg EW, Barker LE, Williamson DF. Projection of the year 2050 burden of diabetes in the US adult population: Dynamic modeling of incidence, mortality, and prediabetes prevalence. Popul Health Metr. 2010;8:29. doi: 10.1186/1478-7954-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson AMF, Olefsky JM. The origins and drivers of insulin resistance. Cell. 2013;152:673–684. doi: 10.1016/j.cell.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 13.Weir GC, Bonner-Weir S. Five stages of evolving beta-cell dysfunction during progression to diabetes. Diabetes. 2004;53(Suppl 3):S16–S21. doi: 10.2337/diabetes.53.suppl_3.s16. [DOI] [PubMed] [Google Scholar]
- 14.DeFronzo RA, et al. Type 2 diabetes mellitus. Nat Rev Dis Primers. 2015;1:15019. doi: 10.1038/nrdp.2015.19. [DOI] [PubMed] [Google Scholar]
- 15.Thomou T, et al. Adipose-derived circulating miRNAs regulate gene expression in other tissues. Nature. 2017;542:450–455. doi: 10.1038/nature21365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prattichizzo F, et al. Extracellular microRNAs and endothelial hyperglycaemic memory: A therapeutic opportunity? Diabetes Obes Metab. 2016;18:855–867. doi: 10.1111/dom.12688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kowalski GM, Bruce CR. The regulation of glucose metabolism: Implications and considerations for the assessment of glucose homeostasis in rodents. Am J Physiol Endocrinol Metab. 2014;307:E859–E871. doi: 10.1152/ajpendo.00165.2014. [DOI] [PubMed] [Google Scholar]
- 18.Shah R, et al. Extracellular RNAs are associated with insulin resistance and metabolic phenotypes. Diabetes Care. 2017;40:546–553. doi: 10.2337/dc16-1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Symonds ME, Sebert SP, Hyatt MA, Budge H. Nutritional programming of the metabolic syndrome. Nat Rev Endocrinol. 2009;5:604–610. doi: 10.1038/nrendo.2009.195. [DOI] [PubMed] [Google Scholar]
- 20.Li P, Zhu Z, Lu Y, Granneman JG. Metabolic and cellular plasticity in white adipose tissue II: Role of peroxisome proliferator-activated receptor-alpha. Am J Physiol Endocrinol Metab. 2005;289:E617–E626. doi: 10.1152/ajpendo.00010.2005. [DOI] [PubMed] [Google Scholar]
- 21.Liu J, Han L, Zhu L, Yu Y. Free fatty acids, not triglycerides, are associated with non-alcoholic liver injury progression in high fat diet induced obese rats. Lipids Health Dis. 2016;15:27. doi: 10.1186/s12944-016-0194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones A, et al. miRNA signatures of insulin resistance in obesity. Obesity (Silver Spring) 2017;25:1734–1744. doi: 10.1002/oby.21950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krützfeldt J, et al. Silencing of microRNAs in vivo with ‘antagomirs’. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 24.Esau C, et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006;3:87–98. doi: 10.1016/j.cmet.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 25.Sun L, Trajkovski M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism. 2014;63:272–282. doi: 10.1016/j.metabol.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 26.Xie W, et al. MiRNA-27 prevents atherosclerosis by suppressing lipoprotein lipase-induced lipid accumulation and inflammatory response in apolipoprotein E knockout mice. PLoS One. 2016;11:1–20. doi: 10.1371/journal.pone.0157085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Montagner A, et al. Liver PPARα is crucial for whole-body fatty acid homeostasis and is protective against NAFLD. Gut. 2016;65:1202–1214. doi: 10.1136/gutjnl-2015-310798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Costet P, et al. Peroxisome proliferator-activated receptor alpha-isoform deficiency leads to progressive dyslipidemia with sexually dimorphic obesity and steatosis. J Biol Chem. 1998;273:29577–29585. doi: 10.1074/jbc.273.45.29577. [DOI] [PubMed] [Google Scholar]
- 29.Blachère JC, Pérusse F, Bukowiecki LJ. Lowering plasma free fatty acids with Acipimox mimics the antidiabetic effects of the β 3-adrenergic agonist CL-316243 in obese Zucker diabetic fatty rats. Metabolism. 2001;50:945–951. doi: 10.1053/meta.2001.24868. [DOI] [PubMed] [Google Scholar]
- 30.Koh KK, Han SH, Quon MJ, Yeal Ahn J, Shin EK. Beneficial effects of fenofibrate to improve endothelial dysfunction and raise adiponectin levels in patients with primary hypertriglyceridemia. Diabetes Care. 2005;28:1419–1424. doi: 10.2337/diacare.28.6.1419. [DOI] [PubMed] [Google Scholar]
- 31.Tsukita S, et al. MicroRNAs 106b and 222 improve hyperglycemia in a mouse model of insulin-deficient diabetes via pancreatic β-cell proliferation. EBioMedicine. 2017;15:163–172. doi: 10.1016/j.ebiom.2016.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alcarraz-Vizan G, et al. BACE2 suppression promotes β-cell survival and function in a model of type 2 diabetes induced by human islet amyloid polypeptide overexpression. Cell Mol Life Sci. 2017;74:2827–2838. doi: 10.1007/s00018-017-2505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musri MM, et al. Histone demethylase LSD1 regulates adipogenesis. J Biol Chem. 2010;285:30034–30041. doi: 10.1074/jbc.M110.151209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.