Abstract
Chlamydia trachomatis is frequently detected in anorectal specimens from men and women. A recent hypothesis suggests that C. trachomatis is a natural commensal organism asymptomatically colonizing the gastrointestinal tract. In this study, we investigated the presence of chlamydial DNA and antigen in intestinal biopsy samples taken during colonoscopy. Cases (n = 32) were patients whose histopathology reports included the term ‘chlamydia’, suggesting a possible history of infection. Control patients (n = 234) did not have chlamydia mentioned in their histopathology report and all tested negative for Chlamydiaceae DNA by 23S ribosomal RNA-based real-time PCR. Amongst the cases, C. trachomatis DNA was detected in the appendix and colon of two female and one male patients. Chlamydia abortus DNA was present in the colon of a fourth female patient. Thus, chlamydial DNA could be demonstrated in intestinal biopsy samples proximal to the anorectal site and inclusions were identified in rectum or appendix of two of these patients by immunohistochemistry. However, the findings in two cases were compatible with sexually acquired C. trachomatis. The identification of C. trachomatis DNA/antigen does not prove the presence of active infection with replicating bacteria. Larger prospective studies on fresh tissue samples are required to confirm the data obtained in this study.
Keywords: Chlamydia trachomatis, rectal infection, colonoscopy, formalin-fixed and paraffin-embedded, biopsy specimens, gastrointestinal tract, reservoir
Chlamydia trachomatis DNA is present in intestinal biopsy samples of patients and might travel from the oral to the rectal site.
INTRODUCTION
Chlamydia trachomatis is the most commonly reported bacterial sexually transmitted infection (STI) worldwide, with an estimated 131 million new cases in 2012 (Newman et al.2015). Chlamydia trachomatis serovars D-K and L1-L3 primarily cause non-invasive genitourinary infections and invasive lymphogranuloma venereum, respectively. Genital C. trachomatis infection is found most often in 15 to 24-year-old women and 20 to 29-year-old men. Many patients are asymptomatic (women 70%–90%, men 30%–50%), thus most cases likely remain undiagnosed and unreported (Markle, Conti and Kad 2013). Factors, other than age, that have been associated with genital chlamydial infection include multiple sex partners, intermittent condom use, cervical ectopy, history of other STIs, and low educational status or socioeconomic position (Markle, Conti and Kad 2013; reviewed by Leonard and Borel 2014).
The role and pathogenicity of C. trachomatis in the human gastrointestinal (GI) tract are increasingly debated because of proposals to test and treat more women and men who have sex with men (MSM) for C. trachomatis detected in anorectal swabs using nucleic acid amplification tests (NAATs) (Dukers-Muijrers et al.2015). Genitourinary C. trachomatis primarily infects columnar mucosal epithelial cells in the endocervix in women, the urethra in women and men. It can also infect rectal columnar epithelium in both women and men. A systematic review found that almost 70% of women with genitourinary C. trachomatis infection were also positive for rectal C. trachomatis by NAAT, but reported anal intercourse was not associated with detection of anorectal C. trachomatis (Chandra et al.2018). Positive NAAT results for anorectal C. trachomatis could reflect the detection of non-viable chlamydial nucleic acid, rather than infection. The finding is also consistent with the hypothesis that auto-inoculation, transferring C. trachomatis between the genital and anorectal mucosa, can maintain infection in women in the absence of direct inoculation through anal intercourse (Rank and Yeruva 2014). Schachter et al. (1979, 1986) reported the presence of viable chlamydiae at different anatomical sites in a prospective study of 131 infants born to C. trachomatis-infected mothers. Chlamydiae were recovered by culture from the conjunctiva and nasopharynx within the first 3 weeks of life. Unexpectedly, several infants who had been previously repeatedly tested negative at the rectum became positive after 3–5 months after birth, suggesting that indirect routes of inoculation are possible.
Bavoil and colleagues have proposed that, as in animals, C. trachomatis is a natural commensal organism in the human GI tract, asymptomatically colonizing one or more sites (Bavoil et al.2017). Chlamydia muridarum infection studies in mice indicate that the GI tract, an immunologically protected site, may be a reservoir for genital re-infection (Perry and Hughes 1999; Igietseme, Portis and Perry 2001; Rank and Yeruva 2014), and long-term chlamydial colonization of the murine GI tract has been demonstrated (Yeruva et al.2013b; Zhang et al.2015). Moreover, eradication of chlamydial infection by azithromycin treatment in mice was less effective for GI than for genital infection (Yeruva et al. 2013a).
Although anorectal C. trachomatis is commonly diagnosed by NAATs in women and MSM (Van Liere et al. 2014, 2015; Dukers-Muijrers et al.2015; Chandra et al.2018), the presence of C. trachomatis DNA and/or antigen at more proximal GI sites such as ileum, cecum, appendix and colon have yet to be investigated. The objectives of this study were to examine intestinal biopsy samples taken during colonoscopy for the presence of chlamydial DNA and antigen in patients with and without a history of chlamydial infection and to examine factors associated with a history of chlamydial infection.
MATERIALS AND METHODS
We conducted this study using pathology records and histopathological specimens obtained during colonoscopy from patients with and without documentation of a history of chlamydial infection.
Definitions of cases and controls
We defined a case as a patient with a possible history of chlamydial infection investigated by colonoscopy at the cantonal hospital Winterthur. We searched for the term ‘chlamydia’ anywhere in the clinical history or in the comments section of histopathology reports, which are stored electronically. The time period analyzed ranged from 1999 to 2013. If a patient had more than one colonoscopy, we used the findings from only one examination. We chose only episodes in which the keyword ‘chlamydia’ appeared in the histopathology report and if it appeared more than once, we used the most recent information.
The control group consisted of 234 randomly but chronologically (according to numbers of pathology reports) selected patients at the cantonal hospital Winterthur whose histopathology record did not contain the word ‘chlamydia’. All these patients underwent colonoscopy in 2013.
We extracted the same information from the records of both cases and controls and coded each variable, as follows: group assignment (case or control), sex (female or male), clinical signs (none or any, such as abdominal pain, diarrhea etc.), biopsy site (ileum, colon, cecum, appendix, rectum) and results of NAATs done as part of this study to detect chlamydial species (positive or negative). Two pathologists reviewed all histopathology reports and categorized diagnoses as the following: no abnormality, polyp(s), neoplasia, infection/inflammation, ischemia/fibrosis, infection/inflammation AND polyp(s)/neoplasia. All polyps were of the hyperplastic type. Neoplasia included benign epithelial (adenoma) and malignant epithelial tumors (carcinoma). Information about prior or current antibiotic treatment was not available from the patient records.
Intestinal biopsy specimens
Formalin-fixed and paraffin-embedded (FFPE) tissue blocks from each patient were retrieved from the pathology archive of the cantonal hospital Winterthur. FFPE blocks containing intestinal anatomic localizations such as ileum, colon, cecum, appendix and rectum were chosen for the study. FFPE blocks containing biopsies taken from the esophagus, stomach and duodenum of the same patients were excluded. Pathology reports of hematoxylin and eosin (HE)-stained sections from all patients were retrieved from the database. The cantonal ethical committee of Zurich approved this project with the limitation that only samples dated prior to December 2013 were allowed to be included.
Statistical analysis
All data were entered in an Excel® spreadsheet and analyzed using Stata (version 15, Stata Corporation, College Station, Texas). We used descriptive statistics to compare demographic, clinical and histopathological characteristics of cases with a history of chlamydia and control patients. Because of the small number of biopsies for some anatomic sites, we considered all biopsies from each patient together and assigned a histopathological diagnosis for each patient. No patient had different abnormal findings at more than one site. We calculated odds ratios (OR, with 95% confidence intervals, CI) to examine the strength of association with a history of chlamydia. Owing to small numbers of observations for several strata, we did not conduct multivariable analysis.
DNA extraction
From each FFPE block, one 20-μm section was cut and deparaffinized in xylene by centrifugation at 13 800× g for 5 min. The supernatant containing the xylene was removed, and the sample washed twice in ethanol, centrifuged at 14 800× g for 5 min followed by removal of the supernatant. The remaining pellet was lysed with proteinase K (20 mg/ml, Roche Diagnostics, Mannheim, Germany) on a thermomixer at 56°C and 550 rpm overnight. DNA was extracted using the commercial DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. All extracts were examined with the Nanodrop® 1000 Version 3.7.1 (Thermo Fisher Scientific, Waltham, MA, USA) to determine DNA quantity and quality.
Real-time PCR for Chlamydiaceae
We examined all specimens for the presence of DNA by real-time PCR. Extracted DNA was analyzed on an Applied Biosystems 7500 Real-Time PCR System (Thermo Fisher Scientific) based on the 23S rRNA gene Chlamydiaceae family-specific real-time PCR as described by Ehricht et al. (2006), with primers Ch23SF (5΄CTGAAACCAGTAGCTTATAAGCGGT3΄), Ch23SR (5΄ACCTCGCCGTTTAACTTAACTCC3΄) and probe Ch23Sp (5΄FAMCTCATCATGCAAAAGGCACGCCGTAMRA3΄), modified according to Blumer et al. (2011) to include an internal positive control (enhanced green fluorescent protein, eGFP) (Hoffmann et al.2006), with primers EGFP1F (5΄GACCACTACCAGCAGAACAC3΄), EGFP10R (3΄CTTGTACAGCTCGTCCATGC5΄) and probe EGFP1HEX (5΄HEXAGCACCCAGTCCGCCCTGAGCABHQ13΄). The protocol by Blumer et al. (2011) was modified as follows: each 25 μl contained 12.5 μl Applied Biosystems TaqMan Fast Universal PCR MasterMix (2×) (Thermo Fisher Scientific), 500 nM of each primer Ch23SF and Ch23SR, 200 nM of probe Ch23Sp, 100 nM of each eGFP primer EGFP 1F and EGFP10R, 50 nM of eGFP probe EGFP1HEX, 0.25 μl eGFP template and 2.5 μl sample template (all primers and probes from Microsynth, Balgach, Switzerland). The products of the Chlamydiaceae and egfp PCRs consist of amplicons of 111 and 177 bp, respectively. The fast cycling profile of the instrument was used: initial denaturation at 95°C for 20 s, followed by 45 cycles of denaturation/elongation (95°C, 3 s and 60°C, 30 s). All samples were tested in duplicate and the cycle threshold was set at 0.1 for each run. A mean cycle threshold (Ct value) <38 was considered positive, and was used to calculate the corresponding chlamydial load as Chlamydiaceae 23S rRNA gene copy number per μl (Staub et al.2018). If several serial sections (see below) of the same FFPE block were positive, the sample with the lowest Ct value was used for the copy number calculation. If the amplification of internal control DNA was inhibited, the run was repeated following 1:10 dilution of the sample. A positive control containing a sevenfold dilution series of C. abortus DNA and a negative control of water instead of the template DNA were included in each run (Hoffmann et al.2015).
In PCR-positive cases after the initial screen, ten consecutive sections from each FFPE block were prepared alternating between repetition of the PCR analysis (n = 5) and chlamydial antigen detection by immunohistochemistry (IHC; n = 5) as described previously (Thierry et al.2016).
Chlamydial species identification by Arraymate Microarray
All samples positive by the Chlamydiaceae real-time PCR were further investigated using a species-specific 23S rRNA gene Arraymate microarray assay (Alere, Jena, Germany), as established by Borel et al. (2008). The current version carries 34 probes for 11 Chlamydiaceae species, three genus-specific probes, four family markers and 15 probes for Chlamydia-like organisms. Additionally, there are four internal control DNA probes and an internal staining control (biotin marker) (Hoffmann et al.2015). Each sample, including internal control DNA (Intype IC-DNA, Qiagen Labor, Leipzig, Germany), was amplified and biotin-labeled using a biotinylation PCR, as described by Borel et al. (2008), with 10 min of initialization (96°C) and 40 cycles of 94°C (denaturation), 50°C (annealing) and 72°C (elongation) for 30 s each. Four to 8 microliter of amplification product was loaded on the chip, which was processed according to manufacturer's instructions.
ompA PCR and sequencing
Samples identified as C. trachomatis by Arraymate Microarray were further characterized by conducting a PCR targeting a 219-bp sequence of the variable domain VD-1 region of the C. trachomatis ompA gene using the primer pair omp-V1-F (5΄-GACTTTGTTTTCGACCGTGTT-3΄) and omp-V1-R (5΄-ACAAATACATCAAACGATCCCA-3΄) as previously described (Jalal et al.2007; Kese et al.2011) using FastStart Taq DNA polymerase (Roche, Basel, Switzerland) under a temperature profile of 95°C for 5 min followed by 40 cycles of 95°C for 60 s, 65°C for 60 s and 72°C for 90 s. Amplicons were purified with the QIAquick PCR Purification Kit (Qiagen) and Sanger sequenced by Microsynth.
Chlamydial antigen detection by immunohistochemistry
The presence of chlamydial antigen in paraffin sections was examined on real-time PCR-positive cases using a Chlamydiaceae family-specific mouse monoclonal antibody targeting the chlamydial lipopolysaccharide (LPS, Clone ACI-P, Progen, Heidelberg, Germany) (Blumer et al.2011). A detection kit (Dako, Santa Clara, CA, USA) was used according to the manufacturer's instructions. After deparaffinization in xylene and rehydration through graded ethanol to water, the antigen retrieval was performed by a 10-min enzyme digestion (Proteinase K, Dako). The endogenous peroxidase activity was inhibited as described above, then slides were incubated for 60 min with the primary antibody diluted 1:200 in antibody diluent (Dako). The incubation with the link antibody and the substrate solution as well as the counterstaining with hematoxylin was performed as described above. For the positive control, intestinal tissue from gnotobiotic piglets experimentally infected with porcine Chlamydia suis strain S45 was used (Guscetti et al.2000).
RESULTS
We retrieved a total of 557 FFPE blocks from 266 patients. The majority of the FFPE control group blocks contained colon (n = 206) and, to a lesser extent, ileum (n = 99) and rectum (n = 25). Cecum and appendix were only available in seven and one patients, respectively. FFPE blocks of the case group contained colon (n = 26), ileum (n = 19), rectum (n = 15), appendix (n = 10) and cecum (n = 1). Patients in both groups underwent colonoscopy because of a risk of colon cancer (e.g. family history), for check-ups (e.g. after surgery), or to investigate clinical symptoms or signs (e.g. non-specific abdominal disorders, diarrhea etc.). Amongst 32 patients identified as cases, 27 underwent colonoscopy once, and five had between two to five examinations between 1999 and 2013. From 42 datasets of 32 patients, we examined 91 FFPE samples. In the control group, all patients (n = 234) underwent colonoscopy once in 2013. Compared with the control group, biopsies for cases were more often available from the appendix and rectum (Table 1).
Table 1.
Description of data, according to study group.
| Total | Case | Control | |||
|---|---|---|---|---|---|
| N = 266 | N = 32 | N = 234 | |||
| Variable | n (%)a | n (%)a | Odds ratio (95% CI) | P value | |
| Sex | 0.243 | ||||
| Female | 149 | 21 (65.6) | 128 (54.7) | 1 | |
| Male | 117 | 11 (34.4) | 106 (45.3) | 0.63 (0.29, 1.38) | |
| Age in years, mean (SD) | 58.8 (16.9) | 38.8 (22.3) | 61.6 (14.0) | <0.001 | |
| <60 years | 120 | 26 (81.3) | 94 (40.2) | 6.45 (2.47, 16.90) | |
| 60+ years | 146 | 6 (18.8) | 140 (59.8) | 1 | |
| Biopsy siteb,c | |||||
| Ileum | 115 | 16 (50.0) | 99 (42.3) | 1.36 (0.65, 2.86) | 0.410 |
| Colon | 225 | 19 (59.4) | 206 (88.0) | 0.20 (0.09, 0.46) | <0.001 |
| Cecum | 8 | 1 (3.1) | 7 (3.0) | 1.05 (0.12, 8.83) | 0.828 |
| Appendix | 11 | 10 (31.3) | 1 (0.4) | 105.9 (9.25, >1000) | <0.001 |
| Rectum | 40 | 15 (46.9) | 25 (10.7) | 7.38 (3.12, 17.4) | <0.001 |
| Histological diagnosis | <0.001 | ||||
| Infection/inflammation | 64 | 24 (75.0) | 40 (17.1) | 7.08 (2.49, 20.11) | |
| Infection+polyps/hyperplasia | 13 | 2 (6.3) | 11 (4.7) | 2.15 (0.37, 12.49) | |
| Polyps/hyperplasia | 35 | 0 (0.0) | 35 (15.0) | na | |
| Neoplasia | 85 | 0 (0.0) | 85 (36.3) | na | |
| Ischemia/fibrosis | 5 | 1 (3.1) | 4 (1.7) | 2.95 (0.27, 31.70) | |
| No abnormality | 64 | 5 (15.6) | 59 (25.2) | 1 | |
| Clinical signs | <0.001 | ||||
| Yes | 138 | 32 (100) | 106 (45.3) | na | |
| No | 128 | 0 (0.0) | 128 (54.7) | 1 | |
| Chlamydiaceae detected | <0.001 | ||||
| Yes | 4 | 4 (12.5) | 0 (0.0) | na | |
| No | 262 | 28 (87.5) | 234 (100) | 1 |
aNumbers presented are mean and standard deviation for age.
bTotal is greater than 276 because patients could have biopsy from more than one site.
cOdds ratio calculated separately for each site comparing cases and controls.
Abbreviations: na, not available (odds ratio could not be calculated because of zero count in one cell); SD, standard deviation.
Description of patients
Details of patient characteristics, presence of clinical signs and histopathological diagnosis are shown in Table 1.
There were more female patients amongst both cases (21/32) and controls (128/234). The age range of both groups was similar; year of birth ranged from 1924 to 1990 amongst cases and from 1926 to 1984 for controls, but the mean age of cases was lower than that of controls (38.8 versus 61.6 years).
Clinical symptoms or signs were present in all case patients (n = 32) and in 106 of 234 (45.3%) controls. The most frequent histopathological diagnoses, retrieved from the archived pathology reports, were infectious or inflammatory type lesions amongst cases (n = 24) and neoplasia (n = 85) or no abnormality (n = 59) amongst controls. Infection/inflammation was more common amongst cases than controls (OR 7.08, 95% CI 2.49–12.49) (Table 1).
Detection of Chlamydia
The real-time PCR for Chlamydiaceae was negative in all samples (n = 464) from 234 control patients.
Real-time PCR for Chlamydiaceae was positive in four tissues (appendix: n = 2; colon: n = 1; rectum: n = 1) from 4 of 32 (12.5%, 95% CI 3.5–29.0%) patients with a history of chlamydia (Table 2). Of these, C. trachomatis was identified in three patients and C. abortus in one patient by Arraymate microarray. Of the three C. trachomatis-positive patients, two were female and one was male.
Table 2.
Characteristics of patients with Chlamydiaceae detected.
| Biopsy available, PCR result (copy number per μl) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Case | Sex | Age, yr | Ileum | Colon | Cacum | Appendix | Rectum | Arraymate result | Immunhisto-chemistry |
| 1 | Female | 55 | No | No | No | Yes, +ve (7.8) | No | C. trachomatis | –ve |
| 2 | Male | 40 | Yes, –ve | Yes, –ve | No | No | Yes, +ve (48.2) | C. trachomatis | +ve, rectum |
| 3 | Female | 81 | Yes, –ve | Yes, +ve (349.1) | No | No | No | C. abortus | –ve |
| 4 | Female | 24 | No | No | No | Yes, +ve (186.2) | No | C. trachomatis | +ve, appendix |
yr, years; –ve, negative; +ve, positive.
The male C. trachomatis-positive patient (patient 2) was also HIV-positive, and the clinical record included positive test results for C. trachomatis and Neisseria gonorrhoeae. The histopathologic diagnosis was an ulcero-granulative colitis and active proctitis. From the tissues available (ileum, colon, rectum), only the rectum was positive by PCR (48.2 copies per μl) and Arraymate Microarray (positive for C. trachomatis). Sequencing of the PCR product in the VD-I region of ompA revealed 99% homology to the C. trachomatis LGV ompA-genotype L2. By IHC, three out of five serial sections of the rectum contained one or two chlamydial inclusions per section in the lamina propria (Fig. 1A). The ileum and colon were negative for Chlamydiaceae antigen detection by IHC (Table 2).
Figure 1.
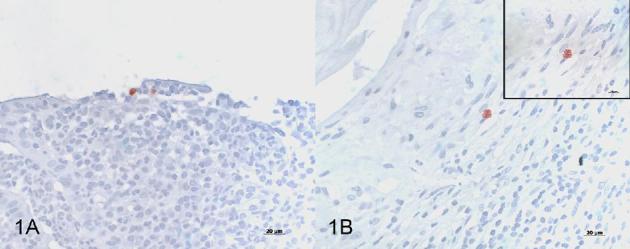
Immunohistochemistry using a Chlamydiaceae-family-specific mouse monoclonal antibody targeting the chlamydial lipopolysaccharide (LPS, Clone ACI-P, Progen), AEC/peroxidase method, hematoxylin counterstain. (A) Positive immunolabeling of 2 chlamydial inclusions in the epithelium of the rectal mucosa from patient 2 (magnification 400×). (B) Positive immunolabeling of a single chlamydial inclusion in the appendix tissue from patient 4 (magnification 400×). Inset: granular appearance of the chlamydial inclusion in B (magnification 1000×, oil immersion).
The younger female patient (24 years, patient 4) had a clinical history indicative of Chlamydia-associated adnexitis. The histological examination of the appendix revealed an acute partially fibrinous serositis. The appendix was positive for C. trachomatis by Arraymate, and by IHC four out of five serial sections contained 1 or 2 chlamydial inclusions in the lamina propria (Fig. 1B). The copy number in the PCR-positive appendix was 186.2 per μl and sequencing of the PCR product in the VD-I region of ompA revealed 99% homology to the C. trachomatis ompA-genotype F.
The older female patient (age 55 years, patient 1) suffered from unclear abdominal pain in the lower right quadrant, which was the reason for undergoing diagnostic laparoscopy and appendectomy. Histological investigation of the appendix revealed a mild, acute, granulocytic mucosal inflammation and local serositis. The available PCR-positive tissue (appendix) was positive for C. trachomatis by Arraymate but negative by IHC. The copy number in the PCR-positive appendix was 7.8 per μl and PCR targeting the VD-I region of ompA was not successful.
The C. abortus-positive patient (female, age 81 years, patient 3) underwent four colonoscopic examinations between 2011 and 2013. The Chlamydia-positive PCR result (349.1 copies per μl) was obtained from the colon sample in 2011. The clinical history at that time included bleeding from the anus and colitis ulcerosa as differential diagnosis. Histopathological examination revealed active colitis with cryptitis (inflammation of the colonic crypts) but the colon was negative for Chlamydiaceae by IHC. Follow-up examinations (n = 3) included the histopathological examination of the ileum and the colon which remained negative for Chlamydia by PCR. Diagnoses were colitis and polyp in 2012, ulcero-granulative colitis with epitheloid granulomas in 2012 and chronic-active colitis with cryptitis in 2013.
DISCUSSION
Chlamydia trachomatis is frequently detected in anorectal specimens from women and MSM. A recent hypothesis (Bavoil et al.2017) proposes that chlamydiae, taken up during oral sex practices, survive in and colonize the GI tract, reaching the rectum. Through fecal shedding, repeated contamination of the female lower genital tract might take place, with a possibility of reproductive sequelae in women if elementary bodies reach the endocervix (Bavoil et al.2017). Intestinal colonization by chlamydiae in animal hosts, e.g. birds, pigs and ruminants, resulting in fecal shedding with subsequent fecal-oral transmission, has been well-documented for decades (Bavoil et al.2017). However, anorectal PCR positivity for C. trachomatis in human patients is neither proof of active infection of the rectum, nor of colonization of the intestine. In this study, we were interested to assess whether chlamydiae can be detected in intestinal biopsy tissue samples.
Amongst a group of case patients, with pathology records indicating a documented or suspected chlamydial infection (n = 32), Chlamydia species were identified in only 12.5%; three patients were positive for C. trachomatis and one for C. abortus. Most patients in the case group did not have a current C. trachomatis infection detected by NAAT. They were identified using the search term ‘chlamydia’ in the clinical history or in the comments of the histopathology reports. It is probably rare that Chlamydia-positive patients undergo a colonoscopy. In contrast to the controls, the case group comprised a higher number of younger patients (mean 38.8) in which inflammatory lesions were diagnosed more frequently. The male C. trachomatis-positive patient had proctitis caused by an LGV genotype, which is known to result in colonoscopic investigation for suspected inflammatory bowel disease (de Vries 2014). The most frequent causes for infectious proctitis are STIs acquired through anal intercourse and include the pathogens N. gonorrhoeae, C. trachomatis (including LGV), herpes simplex virus and Treponema pallidum (de Vries 2014). There are ongoing epidemics of LGV amongst HIV-infected MSM in Switzerland (Kamarashev et al.2010) and elsewhere in Europe (Ward et al.2007; de Vrieze et al.2013; Peuchant et al.2016).
The two female C. trachomatis-positive patients had positive results in appendix tissue, the only submitted tissue from these patients. Spread of sexually transmitted C. trachomatis from the endocervix to the fallopian tube and then to the appendix and perihepatitis (Fitz-Hugh Curtis syndrome) are well-recognized complications of ascending chlamydial infection in women. In general, the number of appendix or cecum tissue specimens (11/32 cases and 8/234 controls) was too low to draw any conclusions. The cecum had the highest chlamydial load in an experimental study in ducks (Thierry et al.2016) and was the target tissue in the mouse GI tract following oral C. muridarum infection (Yeruva et al.2013b). Moreover, the cecum remained positive for up to 100 days post infection in the latter study (Yeruva et al.2013b).
The finding of C. abortus by Arraymate microarray in the colon of one female patient aged over 80 years is surprising. Chlamydia abortus is an animal pathogen causing abortion, stillbirth and neonatal losses in sheep and goats, and to a lesser extent, in cattle, but is also known for its zoonotic potential (Essig and Longbottom 2015). Pregnant women are at risk when they come into contact with the pathogen but intestinal carriage of C. abortus in humans has not been reported before. Data anonymization for ethical reasons made it impossible to determine whether the patient had any history of contact with ruminants. The patient suffered from colitis, but this finding seemed unlikely to be associated with an intestinal C. abortus infection, given that samples from follow-up investigations (n = 4) continuously revealed colitis while remaining Chlamydia-negative.
All Chlamydia-positive samples contained 23S rRNA gene copy numbers ranging from 7.8 to 349.1 copies per μl, and, where present, revealed only single IHC-positive cells per section in the lamina propria of the appendix or rectum. These findings are indicative of a low-level infection, which would fit with the colonization hypothesis (Bavoil et al.2017), or might be the result of contamination. Asymptomatic intestinal infection after experimental oral C. psittaci inoculation in ducks also resulted in the detection of few chlamydial inclusions by IHC (Thierry et al.2016). Comparable to our findings, single chlamydial inclusions were present in the epithelium of the avian cecum and its subepithelial tissue in the absence of histopathological lesions. In this avian study, chlamydial load measured by PCR was highest in the cecum which contrasted with the detection of very few inclusions by IHC. Sensitivity of IHC is much lower than PCR as only inclusions can be detected by IHC rather than individual elementary and reticulate bodies or DNA copies. Direct comparison between IHC and PCR might be difficult if sections have been taken from different levels of the FFPE blocks, although we tried to reduce this limitation in our study by taking alternating consecutive sections for each method. Moreover, PCR on retrospective FFPE material can be critical as preparation and storage of FFPE blocks leads to physical and chemical changes of DNA of the tissue reducing the length of amplifiable PCR fragments (Burach et al.2014). We tried to overcome this drawback by choosing PCR methods targeting DNA fragments smaller than 200 base pairs, and in case of a positive result, investigating serial sections of FFPE tissue. However, the limited length of the amplified 23S rRNA gene fragments will not allow comparative studies and phylogenetic analysis with corresponding C. trachomatis sequences on this material. Amplification of a VD-I region of ompA was only successful in two positive patients revealing homology to the LGV L2 ompA-genotype in the male and to ompA-genotype F in one female patient.
In the past, chlamydial infections have been linked to neoplastic diseases such as ocular MALT lymphoma (Ponzoni and Ferreri 2017). Moreover, chlamydial infections in animal hosts such as the pig can induce enteritis and diarrhea (Schautteet and Vanrompay 2011). Simkania negevensis infection rate in colonic biopsies was significantly higher in patients with Crohn's disease and ulcerative colitis compared to control patients undergoing colonoscopy suggesting that this Chlamydia-like bacterium might be involved in the inflammatory processes of chronic bowel diseases (Belluzzi et al.2017). The association of chlamydial infections and neoplastic or intestinal inflammatory processes prompted us to screen intestinal biopsy samples from randomly selected patients subjected to colonoscopy. In the control group (n = 234), we were not able to detect chlamydial DNA in 464 FFPE blocks containing ileum, colon, cecum, appendix or rectum. This could be explained in part by the limitations of this study. The control group were older patients undergoing colonoscopy and was therefore biased towards hyperplastic and neoplastic lesions, rather than younger adults who are at higher risk of sexually transmitted C. trachomatis infection. Also, inflammatory processes in the control group were less common. Due to these limitations, we think it would be premature to conclude that C. trachomatis in not present in the GI tract of patients without history of chlamydial infection. A larger sample size targeting the younger and sexually active population would be worth investigating in future studies.
Finally, we have to consider that the presence of C. trachomatis DNA/antigen in anorectal swabs or intestinal biopsy samples does not prove that a sustained infection with living/replicating bacteria has occurred. Such findings might represent a mere contamination with non-viable organisms, as NAATs cannot determine whether the organism is viable. Isolation studies or the use of other viability assays are impossible for FFPE material. Prospective studies on fresh tissue samples from the GI tract and with concurrent specimens from genitourinary, anorectal and oropharyngeal sites are required to confirm the findings obtained in this study.
Acknowledgements
We thank med. vet. Prisca Mattmann for assisting in PCR work. The authors wish to thank the laboratory team of the Institute of Veterinary Pathology, Vetsuisse Faculty, University of Zurich and the Cantonal Hospital Winterthur for technical help.
Part of this work was presented at the 14th International Symposium on Human Chlamydial Infections, Woudschoten, Zeist, The Netherlands, July 1–6, 2018 (N. Borel, A. Pospischil, H. Marti, T. Pesch, B. Prähauser, R. Güttinger, S. Wunderlin, H.M.B. Seth-Smith, R. Flury: Chlamydia in intestinal biopsy samples).
Conflict of interest. None declared.
REFERENCES
- Bavoil PM, Marques PX, Brotman R et al. . Does active oral sex contribute to female infertility? J Infect Dis 2017;216:932–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belluzzi A, Di Francesco A, Biondi R et al. . Simkania negevensis and Chlamydia pneumoniae in patients with inflammatory bowel diseases. In: 4th European Meeting on Animal Chlamydiosis, EMAC-4, Zagreb, Croatia. University of Zagreb, Croatia, 2017. p.18. [Google Scholar]
- Borel N, Kempf E, Hotzel H et al. . Direct identification of chlamydiae from clinical samples using a DNA microarray assay: a validation study. Mol Cell Probes 2008;22:55–64. [DOI] [PubMed] [Google Scholar]
- Blumer S, Greub G, Waldvogel A et al. . Waddlia, Parachlamydia and Chlamydiaceae in bovine abortion. Vet Microbiol 2011;152:385–93. [DOI] [PubMed] [Google Scholar]
- Burach F, Pospischil A, Hanger J et al. . Chlamydiaceae and Chlamydia-like organisms in the koala (Phascolarctos cinereus)–organ distribution and histopathological findings. Vet Microbiol 2014;172:230–40. [DOI] [PubMed] [Google Scholar]
- Chandra NL, Broad C, Folkard K et al. . Detection of Chlamydia trachomatis in rectal specimens in women and its association with anal intercourse: a systematic review and meta-analysis. Sex Transm Infect 2018;94:320–6. [DOI] [PubMed] [Google Scholar]
- de Vries HJ. Sexually transmitted infections in men who have sex with men. Clin Dermatol 2014;32:181–8. [DOI] [PubMed] [Google Scholar]
- de Vrieze NH, van Rooijen M, Schim van der Loeff MF et al. . Anorectal and inguinal lymphogranuloma venereum among men who have sex with men in Amsterdam, The Netherlands: trends over time, symptomatology and concurrent infections. Sex Transm Infect 2013;89:548–52. [DOI] [PubMed] [Google Scholar]
- Dukers-Muijrers NHTM, Schachter J, van Liere GAFS et al. . What is needed to guide testing for anorecteal and pharyngeal Chlamydia trachomatis and Neisseria gonorrhoeae in women and men? Evidence and opinion. BMC Infect Dis 2015;15:533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehricht R, Slickers P, Goellner S et al. . Optimized DNA microarray assay allows detection and genotyping of single PCR-amplifiable target copies. Mol Cell Probes 2006;20:60–63. [DOI] [PubMed] [Google Scholar]
- Essig A, Longbottom D. Chlamydia abortus: new aspects of infectious abortion in sheep and potential risk for pregnant women. Curr Clin Mico Rpt 2015;2:22–34. [Google Scholar]
- Guscetti F, Hoop R, Schiller I et al. . Experimental enteric infection of gnotobiotic piglets with Chlamydia psittaci strain of avian origin. J Vet Med B 2000;47:561–72. [DOI] [PubMed] [Google Scholar]
- Hoffmann B, Depner K, Schirrmeier H et al. . A universal heterologous internal control system for duplex real-time RT-PCR assays used in a detection system for pestiviruses. J Virol Methods 2006;136:200–9. [DOI] [PubMed] [Google Scholar]
- Hoffmann K, Schott F, Donati M et al. . Prevalence of chlamydial infections in fattening pigs and their influencing factors. PLoS One 2015;10:e0143576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igietseme JU, Portis JL, Perry LL. Inflammation and clearance of Chlamydia trachomatis in enteric and nonenteric mucosae. Infect Immun 2001;69:1832–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalal H, Stephen H, Alexander S et al. . Development of real-time PCR assays for genotyping of Chlamydia trachomatis. J Clin Microbiol 2007;45:2649–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamarashev J, Riess CE, Mosimann J et al. . Lymphogranuloma venereum in Zurich, Switzerland: Chlamydia trachomatis serovar L2 proctitis among men who have sex with men. Swiss Med Wkly 2010;140:209–12. [DOI] [PubMed] [Google Scholar]
- Kese D, Potocnik M, Maticic M et al. . Genotyping of Chlamydia trachomatis directly from urogenital and conjunctiva samples using an ompA gene pyrosequencing-based assay. FEMS Immunol Med Microbiol 2011;63:210–6. [DOI] [PubMed] [Google Scholar]
- Leonard CL, Borel N. Chronic chlamydial diseases: From atherosclerosis to urogenital infections. Curr Clin Micro Rpt 2014;1:61–72. [Google Scholar]
- Markle W, Conti T, Kad M. Sexually transmitted diseases. Prim Care 2013;40:557–87. [DOI] [PubMed] [Google Scholar]
- Newman L, Rowley J, Vander Hoorn S et al. . Global estimates of the prevalence and incidence of four curable sexually transmitted infections in 2012 based on systematic review and global reporting. PLoS One 2015;10:e0143304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry LL, Hughes S. Chlamydial colonization of multiple mucosae following infection by any mucosal route. Infect Immun 1999;67:3686–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peuchant O, Touati A, Sperandio C et al. . Changing pattern of Chlamydia trachomatis strains in Lymphogranuloma venereum outbreak, France, 2010–2015. Emerg Infect Dis 2016;22:1945–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponzoni M, Ferreri AJ. Bacteria associated with marginal zone lymphomas. Best Pract Res Clin Haematol 2017;30:32–40. [DOI] [PubMed] [Google Scholar]
- Rank RG, Yeruva L. Hidden in plain sight: chlamydial gastrointestinal infection and its relevance to persistence in human genital infection. Infect Immun 2014;82:1362–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachter J, Grossman M, Holt J et al. . Infection with Chlamydia trachomatis: involvement of multiple anatomic sites in neonates. J Infect Dis 1979;139:232–4. [DOI] [PubMed] [Google Scholar]
- Schachter J, Grossman M, Sweet RL et al. . Prospective study of perinatal transmission of Chlamydia trachomatis. J Am Med Assoc 1986;255:3374–7. [PubMed] [Google Scholar]
- Schautteet K, Vanrompay D. Chlamydiaceae infections in pigs. Vet Res 2011;42:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub E, Marti H, Biondi R et al. . Novel Chlamydia species isolated from snakes are temperature-sensitive and exhibit decreased susceptibility to azithromycin. Sci Rep 2018;8:5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thierry S, Vorimore F, Rossignol C et al. . Oral uptake of Chlamydia psittaci by ducklings results in systemic dissemination. PLoS One 2016;11:e0154860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Liere GAFS, Hoebe CJPA, Wolffs PFG et al. . High co-occurrence of anorectal chlamydia with urogenital chlamydia in women visiting an STI clinic revealed by routine universal testing in an observational study; a recommendation towards a better anorectal chlamydia control in women. BMC Infect Dis 2014;14:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Liere GAFS, van Rooijen MS, Hoebe CJ et al. . Prevalence of and factors associated with rectal-only Chlamydia and Gonorrhoea in women and in men who have sex with men. PLoS One 2015;10:e0140297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H, Martin I, Macdonald N et al. . Lymphogranuloma venereum in the United kingdom. Clin Infect Dis 2007;44:26–32. [DOI] [PubMed] [Google Scholar]
- Yeruva L, Melnyk S, Spencer N et al. . Differential susceptibilities to azithromycin treatment of chlamydial infection in the gastrointestinal tract and cervix. Antimicrob Agents Chemother 2013a;57:6290–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeruva L, Spencer N, Bowlin AK et al. . Chlamydial infection of the gastrointestinal tract: a reservoir for persistent infection. Pathog Dis 2013b;68:88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Huang Y, Gong S et al. . In vivo and ex vivo imaging reveals a long-lasting chlamydial Infection in the mouse gastrointestinal tract following genital tract inoculation. Infect Immun 2015;83:3568–77. [DOI] [PMC free article] [PubMed] [Google Scholar]


