Abstract
Chemically modified antisense oligonucleotides with phosphorothioate linkages (PS-ASOs) mediate site-specific cleavage of RNA by RNase H1 and are broadly used as research and therapeutic tools. PS-ASOs can enter cells via endocytic pathways and escape from membrane-enclosed endocytic organelles to reach target RNAs. We recently found that lysobisphosphatidic acid is required for release of PS-ASOs from late endosomes. Here, we evaluated the effects of other lipids on PS-ASO intracellular trafficking and activities. We show that free fatty acids, ceramide, and cholesterol increase PS-ASO activities. Free fatty acids induced formation of lipid droplets without changing the intracellular localization of PS-ASOs in early or late endosomes. Ceramide and cholesterol did not obviously induce the formation of lipid droplets, but cholesterol caused enlargement of endosome size and volume. Although none of those lipids appeared to influence PS-ASO internalization or intracellular trafficking processes, all led to an increase in leakiness of late endosomes. Thus, the membrane destabilization induced by these lipids likely contributes to PS-ASO release from late endosomes, which, in turn, increases PS-ASO activity.
Keywords: antisense oligonucleotides, free fatty acids, ceramide, cholesterol, intracellular trafficking, endosomal leakiness
Introduction
Antisense oligonucleotides with phosphorothioate backbones (PS-ASOs) are used as biological tools and therapeutic agents.1 PS-ASOs mediate sequence-specific RNA cleavage by RNase H1.1 To enhance pharmacological properties, PS-ASOs are usually designed as chimeric “gapmers” with deoxyribonucleotide in the middle flanked by five 2′-O-methoxyethyl (MOE) modified ribonucleotide residues on both sides.1 The PS backbone significantly increases protein binding compared to the phosphodiester (PO) backbone.1 Protein binding of PS-ASOs can mediate their cellular uptake by receptor or cell-surface proteins through endocytotic pathways.2, 3 This so-called “free uptake” route of cellular entry can result in RNase H1-mediated cleavage of target RNAs in both cytoplasm and nucleus.4 The mechanism of PS-ASO entry depends on the type of cell-surface protein bound and varies in different cell types3 and influences the pharmacological activity.5, 6
PS-ASOs are transported intracellularly mainly through endocytic pathways from early endosomes to late endosomes or lysosomes.7 Internalized PS-ASOs accumulate in late endosomes or lysosomes, and a small fraction is released to the cytosol and/or nucleus, contributing to target reduction.8 We and others have found that the pharmacological effects of PS-ASOs occur after they traffic to late endosomes and increase over time.9, 10 This observation suggests that a productive PS-ASO release stems from low levels of continuous leakage from late endosomes.9, 10
Different mechanisms have been proposed for PS-ASO escape from late endosomes.11, 12 Endosomal pH and mobility have significant impacts on endosomal escape.13 Changes in endosomal pH are believed to cause the swelling and rupture of the endosomal membranes to release endosomal contents into the cytoplasm.14 Endosomal mobility may generate shear stress on the vesicle membranes and result in a destabilization of the endosomal membranes. Greater endosomal mobility may even result in collisions with other cell organelles that result in endosomal bursting.11 In addition, membrane fusion events that constantly and actively drive vesicle budding and fusion during the trafficking processes could impact membrane leakiness.15 Finally, endosomal membrane lipid compositions directly affect membrane fusion properties and endosomal substance release.16 Lipid compositions influence the temporal stability of the membranes during the fusion events, and transient formation of non-bilayer lipid domains with increased permeability may allow PS-ASO release.17
We previously showed that a unique endosomal phospholipid, lysobisphosphatidic acid (LBPA), is required for PS-ASO release from late endosomes.9 Here, we evaluated the effects of other lipid species on PS-ASO intracellular trafficking and activities. We found that among tested lipid species, free fatty acids, ceramide, and cholesterol increased PS-ASO activities. These lipids did not alter PS-ASO internalization or intracellular trafficking processes significantly; however, all increased late endosome membrane leakiness. Thus, these exogenous lipids appeared to temporally or spatially destabilize the membranes, especially endosomal membranes, to potentiate the release process of PS-ASOs from late endosomes.
Results
Free Fatty Acids Increase PS-ASO Activities
Lipids are the building blocks of vesicles in which PS-ASOs are transported along endocytic pathways through free uptake.3, 12 PS-ASOs must traverse endosomal lipid bilayers before they enter the cytoplasm or nucleus to reach target RNAs.1 In an effort to better understand release pathways and to improve PS-ASO potency through free uptake, we evaluated the effects of different lipid species on PS-ASO activity. We hypothesized that changes in membrane dynamics resulting from alterations in cellular lipid composition would impact leakiness of the endosomal membranes, thus enhancing PS-ASO intracellular release.
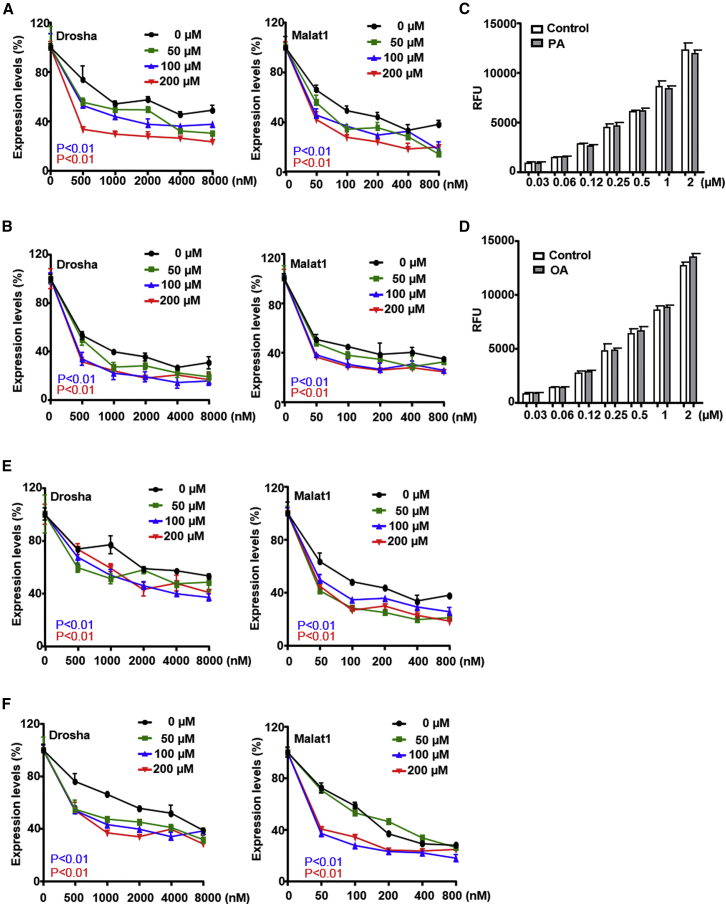
Free fatty acids were previously found to increase potency of fluoroarabino nucleic acid oligonucleotides.18, 19 To test how free fatty acids affect PS-ASO activities, A431 cells were pretreated with palmitic acid or oleic acid at concentrations up to 200 μM for 6 hr and then treated with either Drosha- or Malat1-specific PS-ASOs (IONIS IDs 25690 and 395254, respectively). No increase in cell death was observed due to free fatty acid treatment (data not shown). Both fatty acids modestly but statistically significantly increased activities of the PS-ASOs at all concentrations tested as determined by qRT-PCR analysis for target reduction (Figures 1A and 1B). To determine whether increased PS-ASO activities were due to elevated levels of uptake in cells treated with free fatty acids, we measured levels of uptake of Cy3-labeled PS-ASO (IONIS ID 446654) by flow cytometry. Cells were pretreated with free fatty acids for 6 hr and subsequently treated with Cy3-labeled PS-ASO at a range of concentrations. Uptake of PS-ASO increased in a concentration-dependent manner but pretreatment with free fatty acids did not significantly change levels of PS-ASO uptake (Figures 1C and 1D). These observations suggest that free fatty acids most likely affect PS-ASO intracellular trafficking and/or release, not cellular uptake.
Figure 1.
Free Fatty Acids Increase PS-ASO Activity
(A and B) A431 cells were pretreated with indicated concentrations of (A) palmitic acid or (B) oleic acid for 6 hr and were then treated with PS-ASOs targeting Drosha or Malat1 for 16 hr without the removal of fatty acids. The levels of Drosha and Malat1 were determined by qRT-PCR. Percent expression relative to non-PS-ASO-treated control is plotted. The error bars represent SDs from three independent experiments. p < 0.01 for 100 μM versus 0 μM (blue); p < 0.01 for 200 μM versus 0 μM (red). p values were computed by two-way ANOVA using Prism. (C and D) A431 cells were pretreated with (C) 200 μM palmitic acid (PA) or (D) 200 μM oleic acid (OA) for 6 hr or were not treated (control). Intracellular fluorescence of Cy3-PS-ASO (IONIS ID 446654) was quantified by flow cytometry at 2 hr. Relative fluorescence units (RFUs), indicative of uptake, are plotted versus PS-ASO concentration. (E and F) A431 cells were treated with indicated concentrations of PS-ASOs targeting Drosha or Malat1 for 4 hr, followed by replacement with medium without PS-ASOs but containing (E) 200 μM palmitic acid or (F) 200 μM oleic acid. After 20 hr, the levels of Drosha and Malat1 were determined by qRT-PCR. Percent expression relative to non-PS-ASO-treated control is plotted. The error bars represent SDs from three independent experiments. p < 0.01 for 100 μM versus 0 μM (blue); p < 0.01 for 200 μM versus 0 μM (red). p values were computed by two-way ANOVA using Prism.
To further confirm this observation, we tested effects of free fatty acids in an experimental setting in which PS-ASOs were incubated with cells and then were removed before fatty acids were applied to cells. Cells were pulsed with either Drosha- or Malat1-specific PS-ASOs for 4 hr followed by the replacement with fresh media with or without free fatty acids. Cells were collected for activity assays 20 hr after free fatty acid treatment. Both PS-ASO activities were higher with than without free fatty acid treatment (Figures 1E and 1F). Similar increase in PS-ASO activity was also observed in HeLa and HEK cells upon palmitic acid treatment (Figures S1A and S1B).
Fatty acids such as palmitoleic acid, stearic acid, heptadecylic acid, and nonadecylic acid also enhanced PS-ASO activities when cells were pretreated with PS-ASOs for 4 hr and then treated with fatty acids (Figure S1C). Palmitic acid treatment for 16 hr decreased uptake of PS-ASO (Figure S1D) but did not influence membrane fusion events measured by internalization rates of fluorescent lipid analog N-(lissamine rhodamine B sulfonyl)-phosphatidylethanolamine (N-Rh-PE) (Figure S1E).20 Together, these observations exclude the possibility that free fatty acids increase PS-ASO activities by increasing uptake of PS-ASOs and support our hypothesis that free fatty acids promote PS-ASO intracellular release from endosomes.
Free Fatty Acids Do Not Alter PS-ASO Intracellular Trafficking Processes
Free fatty acids are known to induce lipid-droplet formation.21 In A431 cells, we monitored lipid-droplet formation over time during treatment with free fatty acids. Lipid droplets, stained with Bodipy,22 began to accumulate as early as 4 hr after addition of fatty acids and increased significantly over time (Figure S2). The immediate formation of lipid droplets represents a rapid change in cellular lipid composition upon treatment of free fatty acids that could, in turn, promote the release of PS-ASO from endosomes in cells.
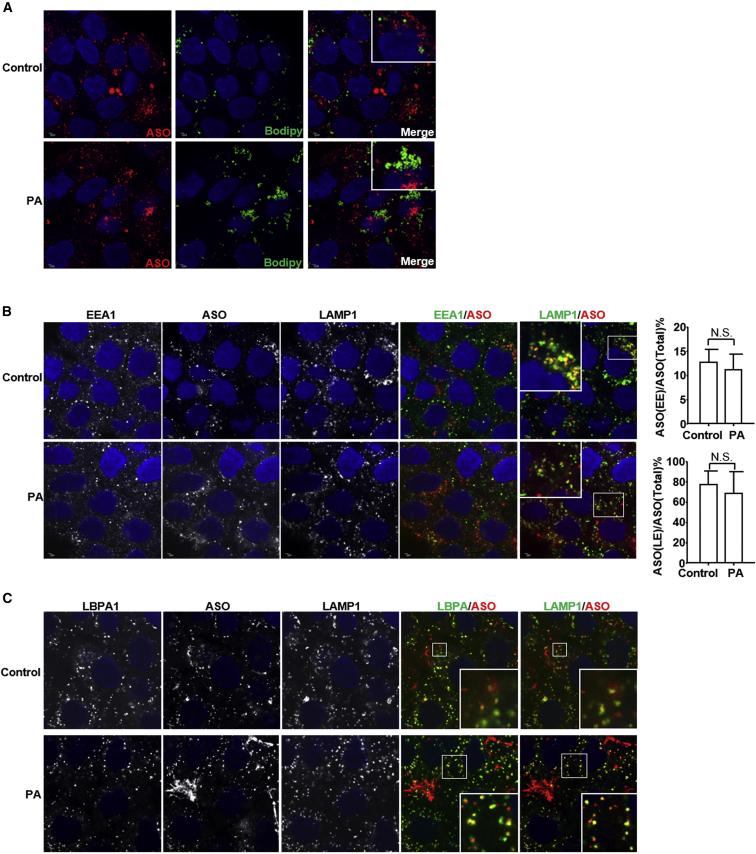
We therefore analyzed the localization of PS-ASOs in the presence of palmitic acid. A431 cells were pretreated with palmitic acid for 6 hr and then treated with Cy3-labeled PS-ASO (IONIS ID 446654) for 2 hr. PS-ASOs were detected in punctate structures, likely endosomes, and did not co-localize with lipid droplets (Figure 2A). This observation suggests that free fatty acids did not change the final destination of PS-ASOs in the endosomes upon induced formation of lipid droplets inside cells.
Figure 2.
Free Fatty Acids Do Not Affect PS-ASO Trafficking
(A) Representative immunofluorescence images of cells treated with 200 μM palmitic acid (PA) or not (control) for 6 hr prior to incubation with 2 μM Cy3-PS-ASO (IONIS ID 446654) for 2 hr. Cells were stained for lipid droplets (green) and PS-ASOs (red). Nuclei were stained with DAPI (blue). Merged images show possible co-localization sites. Scale bars, 2 μm. (B and C) Representative immunofluorescence images of cells pretreated with 200 μM palmitic acid for 6 hr and then incubated with 2 μM Cy3-PS-ASOs (IONIS ID 446654) for 2 hr before staining for (B) EEA1 (green) or (C) LBPA (green) and LAMP1 (green) and PS-ASO (red). Nuclei were stained with DAPI (blue). Merged images show possible co-localization sites. Scale bars, 2 μm. The PS-ASO-positive early endosomes or late endosomes were counted in 30 cells, and the percentage of the PS-ASO-positive early endosomes or late endosomes was calculated relative to the total numbers of the PS-ASO-positive organelles.
We previously showed that PS-ASOs are transported from early endosomes to late endosomes after internalization.23 We monitored PS-ASO trafficking in A431 cells pretreated with palmitic acid for 6 hr followed by incubation with Cy3-labeled PS-ASO (IONIS ID 446654) for another 2 hr. Cells were stained for EEA1, a marker of early endosomes, and LAMP1, a marker of late endosomes or lysosomes. At 2 hr, PS-ASOs exited from early endosomes and co-localized with LAMP1 (Figure 2B). Quantification of the co-localization between early endosomes and PS-ASOs or late endosomes and PS-ASOs showed that palmitic acid did not delay or accelerate this process (Figure 2B). This observation indicates that free fatty acids did not significantly alter PS-ASO intracellular trafficking processes.
As LBPA is important for endosomal membrane fusion events,9 we also tested whether free fatty acids changed LBPA abundance in late endosomes. Cells, pretreated with palmitic acid and incubated with Cy3-labeled PS-ASO (IONIS ID 446654) as above, were stained for LBPA and LAMP1. At 2 hr, PS-ASOs were co-localized with LBPA and LAMP1 in late endosomes. Palmitic acid changed neither the staining pattern of LBPA nor the co-localization pattern between PS-ASOs and LBPA in late endosomes (Figure 2C). Thus, free fatty acids increase PS-ASO activity without substantially altering endocytic pathways or PS-ASO intracellular trafficking.
Ceramide Increases PS-ASO Activity
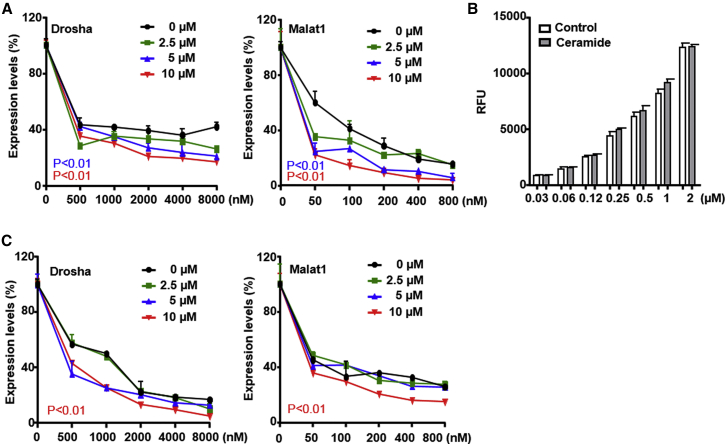
Free fatty acids contribute to ceramide synthesis.24 Ceramide is important in intra-endosomal membrane transport25 and intra-endosomal trafficking is important for PS-ASO release from late endosomes. The enhancement of PS-ASO activity in the presence of free fatty acids could be caused by changes in ceramide levels in late endosome membranes. We therefore tested whether ceramide increased PS-ASO activity. A431 cells were pretreated with C6 ceramide at concentrations up to 10 μM for 6 hr and subsequently treated with either Drosha- or Malat1-specific PS-ASOs. No increase in cell death was detected due to ceramide treatment (data not shown). Ceramide increased activities of both Drosha or Malat1 PS-ASOs with maximal effect at 10 μM (Figure 3A). Thus, our results show that ceramide increases PS-ASO activities, similar to the effects of free fatty acids.
Figure 3.
Ceramide Increases PS-ASO Activity
(A) A431 cells were pretreated with different concentrations of ceramide for 6 hr, followed by incubation with PS-ASOs targeting Drosha or Malat1 for 16 hr without the removal of ceramide. The levels of Drosha and Malat1 RNAs were determined by qRT-PCR. Percent expression relative to non-PS-ASO treated control is plotted. The error bars represent SDs from three independent experiments. p < 0.01 for 5 μM versus 0 μM (blue); p < 0.01 for 10 μM versus 0 μM (red). p values were computed by two-way ANOVA using Prism. (B) A431 cells were pretreated with 10 μM ceramide for 6 hr. Intracellular fluorescence of Cy3-PS-ASO (IONIS ID 446654) was quantified by flow cytometry at 2 hr. RFU, indicative of uptake, is plotted versus PS-ASO concentration. (C) A431 cells were treated with PS-ASOs targeting Drosha or Malat1 for 4 hr, and medium was replaced with medium without PS-ASOs but containing ceramide. After 20 hr, the levels of Drosha and Malat1 RNAs were determined by qRT-PCR. Percent expression relative to non-PS-ASO-treated control is plotted. The error bars represent SDs from three independent experiments. p < 0.01 for 10 μM versus 0 μM (red). p values were computed by two-way ANOVA using Prism.
We also measured Cy3-labeled PS-ASO uptake by flow cytometry in cells pretreated with ceramide for 6 hr. Pretreatment with ceramide did not significantly change levels of PS-ASO uptake (Figure 3B). Treatment with ceramide for 16 hr slightly decreased uptake of PS-ASOs (Figure S3A) and increased membrane fusion events as measured by analysis of N-Rh-PE uptake (Figure S3B). These observations suggest that ceramide treatment of cells does not increase PS-ASO cellular uptake but does increase membrane fusion rates.
We further tested the effects of ceramide on PS-ASO release from endosomes. Cells were incubated with Drosha- or Malat1-specific PS-ASOs for 4 hr. Medium was removed and replaced with medium with or without ceramide. PS-ASO activity was evaluated after 20 hr of ceramide treatment. Ceramide treatment enhanced PS-ASO activity with maximal effect at 10 μM (Figure 3C). Increase in PS-ASO activity was also observed in HeLa and HEK cells upon ceramide treatment (Figures S3C and S3D). These observations suggest that it promotes PS-ASO release from endosomes.
Ceramide Does Not Alter PS-ASO Intracellular Trafficking
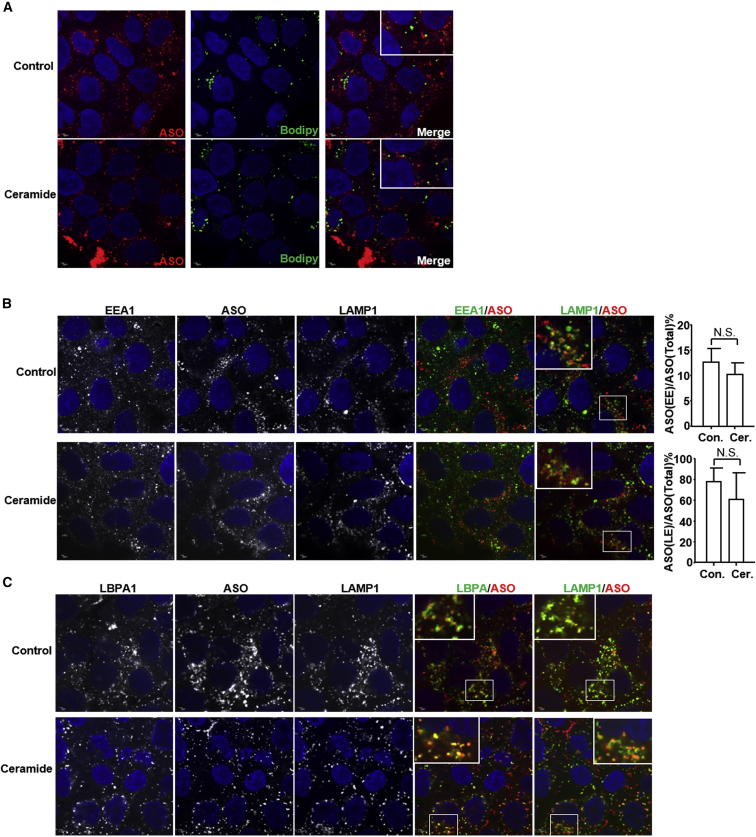
Unlike free fatty acids, ceramide did not stimulate lipid droplet formation significantly but did enhance PS-ASO activities (Figure 4A). We therefore examined the effects of ceramide on PS-ASO intracellular trafficking. A431 cells were pretreated with ceramide for 6 hr, incubated with Cy3-labeled PS-ASO (IONIS ID 446654), and stained with EEA1, LAMP1, and LBPA. After 2 hr, PS-ASOs were mainly co-localized with LAMP1, indicative of trafficking through early endosomes with and without ceramide pretreatment (Figure 4B). Quantification of the co-localization between early endosomes and PS-ASOs or late endosomes and PS-ASOs showed that ceramide did not alter the trafficking kinetics of PS-ASOs to late endosomes (Figure 4B). Moreover, ceramide did not change the staining pattern of LBPA in late endosomes (Figure 4C). Thus, like free fatty acids, ceramide increased PS-ASO activity without altering PS-ASO intracellular trafficking.
Figure 4.
Ceramide Does Not Affect PS-ASO Trafficking
(A) Representative immunofluorescence images of cells pretreated with 10 μM ceramide or untreated (control) for 6 hr, incubated with 2 μM Cy3-PS-ASOs (IONIS ID 446654) for 2 hr and stained for lipid droplets (green) and PS-ASOs (red). Nuclei were stained with DAPI (blue). Merged images are also shown. Scale bars, 2 μm. (B and C) Representative immunofluorescence images of cells pretreated with 10 μM ceramide or untreated (control) for 6 hr, incubated with 2 μM Cy3-PS-ASO for 2 hr, and stained for (B) EEA1 (green) or (C) LBPA (green) and LAMP1 (green) and PS-ASO (red). Nuclei were stained with DAPI (blue). Scale bars, 2 μm. The PS-ASO-positive early endosomes or late endosomes were counted in 30 cells, and the percentage of the PS-ASO-positive early endosomes or late endosomes was calculated relative to the total numbers of the PS-ASO-positive organelles.
Cholesterol Increases PS-ASO Activity
Another lipid species that can regulate vesicular trafficking and signaling is cholesterol.26 Cholesterol is enriched in the plasma membrane and abundant in endocytic systems.27, 28 Cholesterol binds other lipids, such as sphingolipids, to form dynamic platforms involved in membrane trafficking, vesicle endocytosis and/or exocytosis, and signal transduction.29
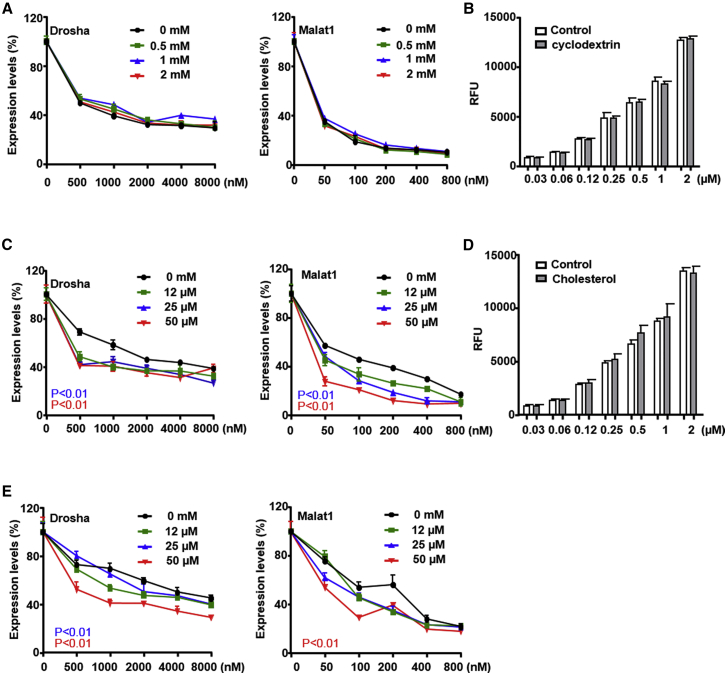
To test the effects of cholesterol on PS-ASO uptake, methyl-beta-cyclodextrin (MCD) was used to deplete plasma membranes cholesterol. A431 cells were pretreated with 2 mM MCD for 6 hr before PS-ASO treatment as previously described.30 Under these conditions, A431 cells survived and cholesterol levels were reduced to 60% of untreated cells (data not shown). After pretreatment of MCD, A431 cells were incubated with either Drosha- or Malat1-specific PS-ASOs. Treatment with MCD did not substantially change activities of either PS-ASO (Figure 5A). Surprisingly, MCD did not substantially alter Cy3-PS-ASO uptake (Figure 5B). Either the PS-ASO uptake pathways are cholesterol independent or the residual cholesterol is sufficient to maintain levels of PS-ASO uptake.
Figure 5.
Cholesterol Increases PS-ASO Activity
(A) A431 cells were pretreated with different concentrations of MCD for 6 hr, followed by incubation with PS-ASOs targeting Drosha or Malat1 for 16 hr without the removal of MCD. The levels of Drosha and Malat1 RNAs were determined by qRT-PCR. Percent expression relative to non-PS-ASO treated control is plotted. The error bars represent SDs from three independent experiments. (B) Intracellular fluorescence of Cy3-PS-ASO (IONIS ID 446654) was quantified at 2 hr by flow cytometry in A431 cells pretreated with 2 mM MCD for 6 hr. RFU is plotted versus PS-ASO concentration. (C) A431 cells were pretreated with different concentrations of MCD-complexed cholesterol in the presence of 1 μM Sandoz 58-035 for 6 hr, followed by incubation with PS-ASOs targeting Drosha or Malat1 RNA for 16 hr without the removal of MCD-complexed cholesterol. The levels of Drosha and Malat1 were determined by qRT-PCR. Percent expression relative to non-PS-ASO-treated control is plotted. The error bars represent SDs from three independent experiments; p < 0.01 for 25 μM versus 0 μM (blue); p < 0.01 for 50 μM versus 0 μM (red). p values were computed by two-way ANOVA using Prism. (D) Intracellular fluorescence of Cy3-PS-ASO (IONIS ID 446654) was quantified by flow cytometry at 2 hr to determine uptake (RFU) at indicated PS-ASO concentrations in A431 cells pretreated with 50 μM MCD-complexed cholesterol for 6 hr. (E) A431 cells were treated with PS-ASOs targeting Drosha or Malat1 RNA for 4 hr, followed by the replacement of fresh media without PS-ASOs but containing different concentrations of MCD-complexed cholesterol in the presence of 1 μM Sandoz 58-035 for another 20 hr. The levels of Drosha and Malat1 RNAs were determined by qRT-PCR. The error bars represent SDs from three independent experiments. p < 0.01 25 μM versus 0 μM (blue). p < 0.01 50 μM versus 0 μM (red) were computed by two-way ANOVA using Prism.
We then evaluated the effect of exogenous cholesterol on PS-ASO activities. A431 cells were pretreated with MCD-complexed cholesterol at different concentrations up to 50 μM in the presence of Sandoz 58-035 to inhibit Acyl-CoA:cholesterol acyltransferase (ACAT). After 6 hr, Drosha- or Malat1-specific PS-ASO was added. Interestingly, MCD-complexed cholesterol increased activities of both PS-ASOs (Figure 5C) at all tested concentrations. PS-ASO uptake, measured by flow cytometry, was not changed after cells were pretreated with exogenous MCD-complexed cholesterol for 6 hr (Figure 5D). However, treatment of cells for 16 hr with MCD-complexed cholesterol significantly decreased uptake of Cy3-PS-ASO (Figure S4A), but little change was observed in membrane fusion events (Figure S4B). These observations indicate that only prolonged exposure to free cholesterol is sufficient to interfere the uptake process of PS-ASOs. Nevertheless, as free cholesterol did not increase PS-ASO uptake in general, the increased activities of PS-ASOs were not caused by increased PS-ASO uptake upon pre-treatment of free cholesterol.
We also tested effects of cholesterol on PS-ASO release from endosomes. Cells were treated with either Drosha- or Malat1-specific PS-ASOs for 4 hr, and after the PS-ASO was removed, the cells were treated with MCD-complexed cholesterol. Activities of both PS-ASOs were increased upon cholesterol treatment with maximal effect at 50 μM (Figure 5E). Similar increase in PS-ASO activity was observed in HeLa and HEK cells upon MCD-complexed cholesterol treatment (Figures S4C and S4D). Thus, cholesterol also increases PS-ASO activity by promoting endosomal release of PS-ASOs.
Cholesterol Does Not Alter PS-ASO Intracellular Trafficking
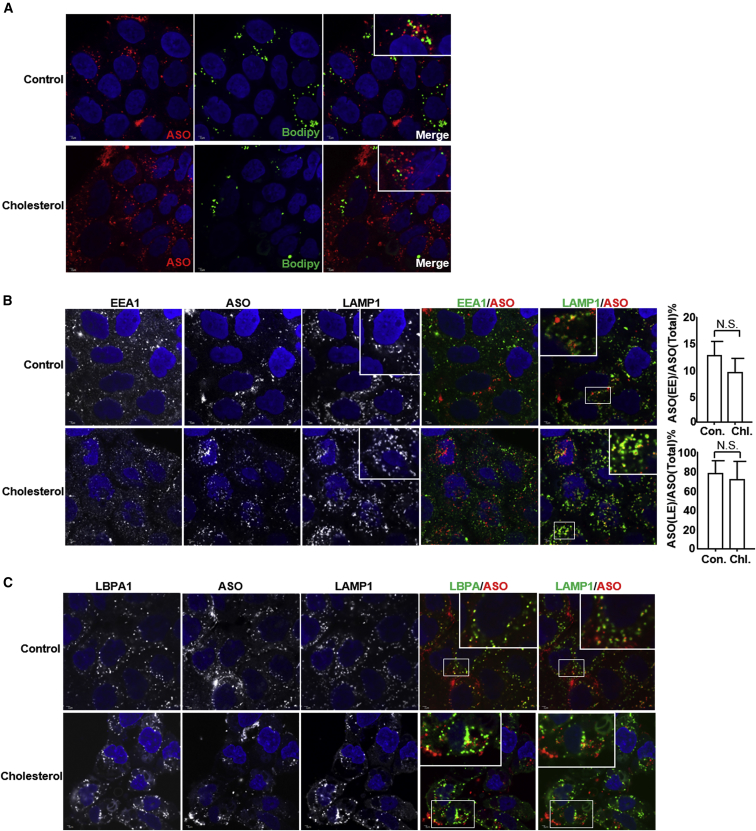
As cholesterol-enriched microdomains are important for vesicle endocytosis and exocytosis, we tested whether exogenous cholesterol altered PS-ASO trafficking. First, in the presence of Sandoz 58-035, which inhibits ACAT, exogenous cholesterol did not induce lipid-droplet formation (Figure 6A). Second, we stained EEA1 and LAMP1 in A431 cells pretreated with exogenous cholesterol for 6 hr and then incubated with Cy3-labeled PS-ASOs (IONIS ID 446654) for 2 hr (Figure 6B). Exogenous cholesterol did not change the morphology of early endosomes but increased the size of late endosomes. In cells treated with cholesterol, late endosomes had open interior spaces and LAMP1 was observed in outer membranes. Despite the morphologic changes in late endosomes in cells treated with cholesterol, PS-ASOs trafficked to late endosomes within 2 hr as usual (Figure 6B). Quantification of the co-localization between early endosomes and PS-ASOs or late endosomes and PS-ASOs showed that cholesterol did not significantly delay PS-ASO trafficking to late endosomes (Figure 6B). LBPA was co-localized with PS-ASOs inside late endosomes stained with LAMP1 in the cells treated with cholesterol (Figure 6C). These results are consistent with our previous observation that PS-ASOs and LBPA are co-localized inside late endosomes as punctate structures, likely intraluminal vesicles (ILVs).9 These results indicate that cholesterol did not interfere with sorting of PS-ASOs into ILVs after they traffic to late endosomes from early endosomes.
Figure 6.
Cholesterol Increases the Sizes of Late Endosomes but Does Not Affect PS-ASO Trafficking
(A) Representative immunofluorescence images of indicated cells stained for lipid droplets and PS-ASOs. Images of PS-ASOs (red) and lipid droplets (green) were merged to show the co-localization. Cells were pretreated with 50 μM MCD-complexed cholesterol in the presence of 1 μM Sandoz 58-035 for 6 hr before they were incubated with 2 μM Cy3-PS-ASO (IONIS ID 446654) for 2 hr prior to staining. Nuclei were stained with DAPI (blue). Scale bars, 2 μm. (B and C) Representative immunofluorescence images of indicated cells stained for EEA1, LBPA, LAMP1, and PS-ASOs. Images of PS-ASO (red) and EEA1 (green, B), LBPA (green, C), LAMP1 (green, B and C) were merged to show the co-localization. Cells were pretreated with 50 μM MCD-complexed cholesterol in the presence of 1 μM Sandoz 58-035 for 6 hr before they were incubated with 2 μM Cy3-PS-ASO (IONIS ID 446654) for 2 hr prior to staining. Nuclei were stained with DAPI (blue). Scale bars, 2 μm. The PS-ASO-positive early endosomes or late endosomes were counted in 30 cells, and the percentage of the PS-ASO-positive early endosomes or late endosomes was calculated relative to the total numbers of the PS-ASO-positive organelles.
Free Fatty Acids, Ceramide, and Cholesterol Increase the Leakiness of Endosomes
Lipids increased PS-ASO activities even when applied to cells after PS-ASO treatment. To analyze incorporation of those lipids into cellular organelles, we incubated cells with Bodipy-C16, NBD-cholesterol, or NBD-ceramide and evaluated fluorescence over time. We observed fluorescent signals in cells treated with all lipids as early as 4 hr after lipid addition (Figure S5A), indicating that the intracellular changes of membrane structures caused by these lipid species could occur before or during the PS-ASO release process to improve PS-ASO activities through free uptake.
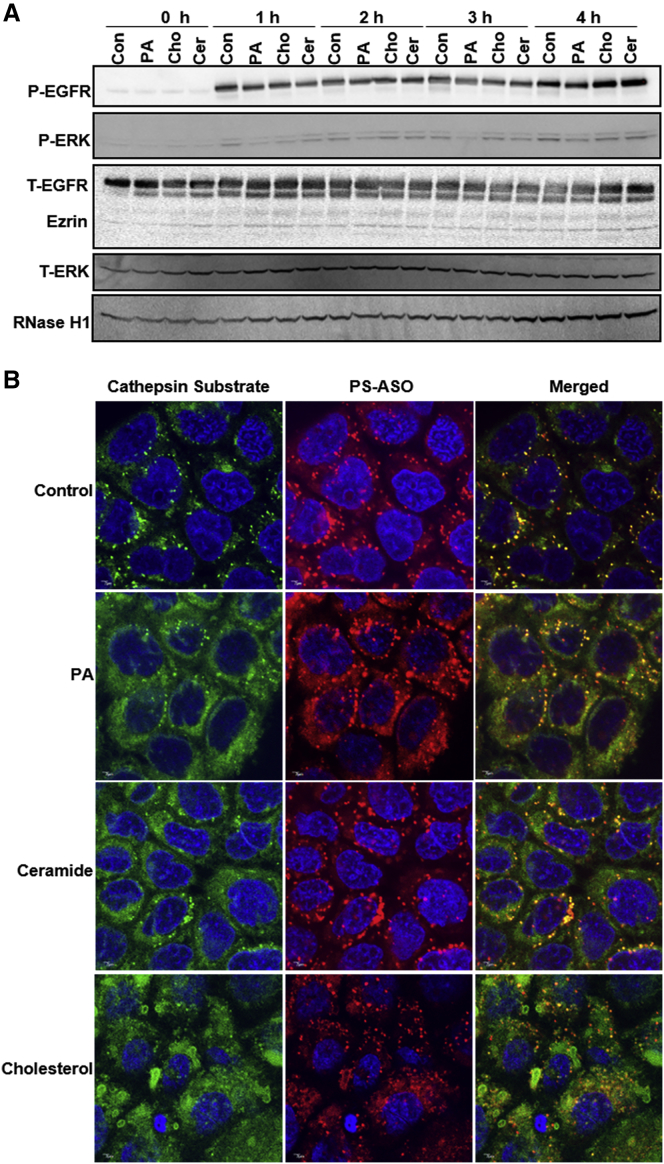
Free fatty acids, ceramide, and cholesterol can trigger signaling pathways and regulate different extracellular responses.15, 31 As epidermal growth factor receptor (EGFR) was shown to mediate productive uptake of PS-ASOs in A431 cells,6 we tested whether any of these lipid species enhanced EGFR signaling or internalization in A431 cells. A431 cells were pretreated with lipid for 6 hr and then were treated with epidermal growth factor (EGF). Downstream signaling of EGFR, manifested by phosphorylation of EGFR or extracellular signal-regulated kinase (ERK), was monitored by western blot. EGFR and ERK phosphorylation levels were modestly decreased in cells treated with free fatty acids, ceramide, and cholesterol (Figure 7A). This observation indicates that lipids did not increase PS-ASO activities by facilitating EGFR-mediated internalization. RNase H1 was also detected, and lipid treatment did not appear to affect RNase H1 levels, indicating that increased activities of PS-ASOs are not due to increased RNase H1 levels (Figure 7A).
Figure 7.
Lipids Do Not Significantly Change EGFR Cell Signaling Pathways but Increase the Leakiness of Endosomes
(A) Western analyses for phosphorylated EGFR (P-EGFR), phosphorylated ERK (P-ERK), total EGFR (T-EGFR), total ERK (T-ERK), and RNase H1 from lysates of cells that were pretreated with 200 μM palmitic acid (PA), 50 μM MCD-complexed cholesterol (Cho), or 10 μM ceramide (Cer) for 6 hr and then incubated with 100 nM EGF for the indicated time. Ezrin served as a loading control. (B) Representative immunofluorescent images of A431 cells treated with 2 μM Cy5-PS-ASO (IONIS ID 851810) and 200 μM palmitic acid (PA), 50 μM MCD-complexed cholesterol (Cho), or 10 μM ceramide (Cer) for 16 hr. Cells were stained for cathepsin substrate (green) and PS-ASOs (red). The nuclei were stained with DAPI (blue). Scale bars, 2 μm.
Endosomal leakiness may result in substance release from late endosomes.11 Since exogenous lipids enhance PS-ASO release from late endosomes, we assume that those lipids promote endosomal leakage. We tested this hypothesis using the fluorogenic Magic Red cathepsin L assay. Cathepsin L is present in late endosomes and lysosomes.32 When endosomes are leaky, cathepsin L activity can be detected in the cytosol, and fluorescent signal due to Magic Red is reduced in late endosomes.32 A431 cells were pretreated with lipid and Cy5-labeled PS-ASOs (IONIS ID 851810) for 16 hr and then stained with Magic Red. In cells not treated with lipid, Magic Red co-localized with Cy5-PS-ASOs in punctate structures that are late endosomes or lysosomes as showed in Figures 2B, 4B, and 6B. In lipid-treated cells, Magic Red signals were significantly reduced in late endosomes, and cytosols were diffusely fluorescent, although Cy5-PS-ASOs were still enriched in late endosomes (Figure 7B), suggesting that lipids did not cause general lysosomal lysis. These observations are also consistent with our observations that these lipid species were not toxic under the experimental conditions used. Thus, decreased Magic Red fluorescence in late endosomes coupled with increased fluorescence in the cytosol indicates an increase of endosomal leakiness in cells treated with lipids. The leakiness was most prominent in cells treated with cholesterol (Figure 7B). Taken together, increased endosomal leakiness could contribute to the increased intracellular release of PS-ASOs from cells treated with those lipids.
Lipid rafts exist as distinct ordered regions of the membranes. To examine whether the integrity of membranes was disturbed by lipid treatment, we stained lipid rafts in A431 cells treated or not with palmitic acid, ceramide, or cholesterol for 16 hr. The stained pattern was mainly enriched in plasma membranes regardless of whether cells were treated with the lipids or not (Figure S5B). This observation indicates that membrane integrity was not significantly affected by lipid treatment at the doses that increased PS-ASO activities. Thus, lipids may destabilize endosomal membranes temporally or spatially to increase PS-ASO release without perturbing the structural integrity of membranes in general.
Discussion
LBPA was previously identified as a lipid required for PS-ASO release from late endosomes.33 In this study, we demonstrated that other lipid species, such as free fatty acids, ceramide, and cholesterol, could also affect PS-ASO activities. PS-ASOs traffic from early endosomes to late endosomes immediately after their internalization.23 This kinetics of the endosomal trafficking was not significantly altered by those lipid species, suggesting that increased PS-ASO activity was not due to changed intracellular trafficking pathways. Instead, those lipid species all lead to an increase in membrane leakiness of late endosomes without disrupting membrane structural integrity. Thus, membrane destabilization, induced by exogenous lipids, could lead to increased endosomal leakage, resulting in PS-ASO release from late endosomes.
Interestingly, none of the lipids tested increased PS-ASO uptake, indicating that the increase in PS-ASO activities observed in the presence of exogenous lipids is directly linked to the enhanced release from late endosomes. This observation is consistent with our previous report that the amount of PS-ASO internalized is not directly correlated with PS-ASO activity and that the rate limiting step of PS-ASO-mediated RNA reduction is intracellular release.9 We previously observed that there is a significant delay from the time that PS-ASOs are internalized to the time that PS-ASOs mediate cleavage of their targets, suggesting that internalization and release of PS-ASOs are separable intracellular processes that can be manipulated individually.9 In support of this hypothesis, data reported here indicate that lipids influence the release of PS-ASOs from late endosomes.
Certain fatty acids (oleic acid and a variety of ω-6 polyunsaturated fatty acids but not the aliphatic palmitic acid) increase the potency of fluoroarabino nucleic acid oligonucleotides taken into cells through a free uptake mechanism.18 Interestingly, we found that both oleic acid and palmitic acid increased PS-ASO activities. The different effects of fatty acids could be attributed to differences in internalization processes and in release mechanisms between fluoroarabino nucleic acid oligonucleotides and PS-ASOs. The free uptake of oligonucleotides starts with protein binding at the cell surface.3 The differences in chemistries between fluoroarabino nucleic acid oligonucleotides and PS-ASOs could determine protein binding profiles, subsequent mechanism of internalization, and, ultimately, the intracellular release of PS-ASOs.34 Although internalization processes of PS-ASOs are still not fully understood, endosomal leakiness appears to affect the ultimate activity of PS-ASOs. We cannot exclude the possibility that certain lipids could alter internalization pathways.
Fatty acids may increase PS-ASO activity by promoting the production of ceramide, which acts to enhance membrane fusion and fission events.29 Endosomal membrane fusion is necessary for substance release from late endosomes.35 For example, the inner and outer membrane fusion events between ILVs and late endosomes, which are mediated by LBPA, significantly promote PS-ASO release from late endosomes.9 Although ceramide is known to trigger budding of exosome vesicles into late endosomes,25 it is still not clear how ceramide influences LBPA-mediated PS-ASO release from late endosomes.
Endogenous cholesterol increases lateral ordering of other lipids and decreases fluidity and permeability of membranes.26 It was a surprising observation that cholesterol treatment increased endosomal leakiness and PS-ASO activity. We speculate that exogenous cholesterol temporarily disorganizes or destabilizes existing membranes, resulting in increased leakiness of endosomal membranes that facilitates PS-ASO release and therefore activity.28 In addition, both free fatty acids and cholesterol promote autophagy,36 suggesting that these lipids can trigger membrane movement or re-organization to further influence cellular events such as PS-ASO release from late endosomes. Interestingly, a stronger leakiness of endosomes did not cause the strongest activity increase in cells treated with MCD-cholesterol. Release from late endosomes is a limiting factor for PS-ASO to gain activity through free uptake; however, that may not be the only determinant for PS-ASO activity mediated by RNase H1 cleavage. Although the stronger leakiness of endosomes was observed in cells treated with cholesterol, other determinants for PS-ASO activity may be altered differently in cells treated with different lipids.
PS-ASO release has been proposed to occur via a process mediated by lipid flip-flops in late endosome membranes.37, 38 It is also possible that the lipids recruit proteins that induce lipid segregation and membrane budding to increase PS-ASO release from late endosomes.39 One such protein could be ANXA2, which senses lipid packing defects or increased membrane curvature.23
The present work reiterates the importance of improving the potency of PS-ASOs through enhancing their productive intracellular release, which will help overcome the bottleneck of oligonucleotide therapeutics. An understanding on the mechanism of productive PS-ASO release will continue to guide the development of modifications or formulations for PS-ASOs that will result in better pharmacological effects. It is conceivable that ASO-lipid interactions can be modified to enhance PS-ASO release by inducing lipid membrane leakiness through engineering PS-ASOs using conjugates.
Materials and Methods
Chemicals, antibodies, siRNAs, ASOs, and qRT-PCR primer probe sets are listed in the Supplemental Information.
Cell Culture, Cell Treatment, and PS-ASO Activity
A431 cells were grown according to the protocols provided by the American Type Culture Collection.40 Cells were re-seeded at 50% confluency in 96-well plates for activity analysis. Cells were treated with free fatty acids (complexed with BSA), ceramide (dissolved in ethanol), or free cholesterol (complexed with beta-cyclodextrin) before or after they were incubated with PS-ASOs as described in figure legend. Cells were incubated with PS-ASOs for indicated times before they were collected for activity assay.
RNA Preparation and qRT-PCR
Total RNA was prepared using an RNeasy mini kit (QIAGEN, Valencia, CA, USA) from cells grown in 96-well plates (around 10,000 cells per well). qRT-PCR using TaqMan primer probe sets was performed essentially as described previously.33 In brief, approximately 50 ng total RNA in 5 μL water was mixed with 0.3 μL primer probe sets containing forward and reverse primers (10 μM of each) and fluorescently labeled probe (3 μM), 0.3 μL RT enzyme mix (QIAGEN), 4.4 μL RNase-free water, and 10 μL of 2× PCR reaction buffer in a 20 μL reaction. Reverse transcription was performed at 48°C for 10 min; 40 cycles of PCR were conducted at 94°C for 20 s and 60°C for 20 s using the StepOne Plus RT-PCR system (Applied Biosystems). Levels of mRNA were normalized to the amount of total RNA present in each reaction as determined for duplicate RNA samples using the RiboGreen assay (Life Technologies).
Immunofluorescence Staining
Cells were fixed with 4% paraformaldehyde for 30 min at room temperature and were permeabilized with 0.05% saponin (Sigma) in PBS for 5 min. Cells were treated with blocking buffer (1 mg/mL BSA in PBS) for 30 min and then incubated with primary antibodies (1:100 to 1:200 depending on antibody in blocking buffer) at room temperature for 2–4 hr, or at 4°C overnight. After three washes using 0.1% Triton in PBS, cells were incubated with fluorescently labeled secondary antibodies (1:200 in blocking buffer) at room temperature for 1–2 hr. After washing, slides were mounted with Prolong Gold anti-fade reagent with DAPI (Life Technologies) and imaged using a confocal microscope (Olympus FV-1000). Images were quantified using software of FV10-ASW 3.0 viewer.
Lipid Droplet, Magic Red MR-(RR)2, and Lipid Raft Staining
Bodipy 493/503 (Thermo Fisher Scientific) was used at 2 μg/mL to stain neutral lipids in fixed cells. The Vybrant Alexa Fluor 555 Lipid Raft Labeling Kit (Thermo Fisher Scientific) was used to stain lipid rafts in fixed cells according to the manufacturer’s protocol. Magic Red substrate 592/628 (Immunochemistry Technologies) was used to measure the leakiness of endosomes in live cells according to manufacturer’s protocol. Stained cells were imaged using confocal microscopy (Olympus FV-1000).
Protein Isolation and Western Blotting
Cells were lysed, and samples were incubated at 4°C for 30 min in RIPA buffer (50 mM Tris-HCl [pH 7.4], 1% Triton X-100, 150 mM NaCl, 0.5% sodium deoxycholate, and 0.5 mM EDTA). Proteins were separated by PAGE using 6% to 12% NuPAGE Bis-Tris gradient gels (Life Technologies) and electroblotted onto PVDF membranes using the iBLOT transfer system (Life Technologies). The membranes were blocked with 5% non-fat dry milk in PBS at 4°C for 30 min. Membranes were then incubated with primary antibodies (listed in the Supplemental Information) at room temperature for 3 hr. After three washes with PBS, the membranes were incubated with appropriate horseradish peroxidase (HRP)-conjugated secondary antibodies (1:2,000) at room temperature for 1 hr to develop the image using enhanced chemiluminescence (ECL) reagents (Abcam).
Flow Cytometry
Cy3-labeled PS-ASOs were added to A431 cells after treatment with different lipids. After 3 hr, cells were washed with PBS, trypsinized, and resuspended in PBS supplemented with 3% fetal bovine serum for analysis by flow cytometry using an Attune NxT Flow Cytometer (Thermo Fisher Scientific).
Author Contributions
S.W., X.-h.L., and S.T.C. conceived and designed the experiments; S.W. and N.A. performed the experiments; S.W., X.-h.L., and S.T.C. wrote the paper.
Conflicts of Interest
The authors declare no conflicts of interest.
Acknowledgments
We thank Dr. Wen Shen, Dr. Frank Bennett, and Dr. Joseph Ochaba for stimulating discussions. This work was supported by internal funding from Ionis Pharmaceuticals.
Footnotes
Supplemental Information includes Supplemental Materials and Methods five figures and can be found with this article online at https://doi.org/10.1016/j.omtn.2018.10.011.
Supplemental Information
References
- 1.Crooke S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017;27:70–77. doi: 10.1089/nat.2016.0656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Juliano R.L., Ming X., Nakagawa O. Cellular uptake and intracellular trafficking of antisense and siRNA oligonucleotides. Bioconjug. Chem. 2012;23:147–157. doi: 10.1021/bc200377d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Crooke S.T., Wang S., Vickers T.A., Shen W., Liang X.H. Cellular uptake and trafficking of antisense oligonucleotides. Nat. Biotechnol. 2017;35:230–237. doi: 10.1038/nbt.3779. [DOI] [PubMed] [Google Scholar]
- 4.Liang X.H., Sun H., Nichols J.G., Crooke S.T. RNase H1-Dependent Antisense Oligonucleotides Are Robustly Active in Directing RNA Cleavage in Both the Cytoplasm and the Nucleus. Mol. Ther. 2017;25:2075–2092. doi: 10.1016/j.ymthe.2017.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Prakash T.P., Graham M.J., Yu J., Carty R., Low A., Chappell A., Schmidt K., Zhao C., Aghajan M., Murray H.F. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014;42:8796–8807. doi: 10.1093/nar/gku531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang S., Allen N., Vickers T.A., Revenko A.S., Sun H., Liang X.H., Crooke S.T. Cellular uptake mediated by epidermal growth factor receptor facilitates the intracellular activity of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2018;46:3579–3594. doi: 10.1093/nar/gky145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Juliano R.L., Ming X., Carver K., Laing B. Cellular uptake and intracellular trafficking of oligonucleotides: implications for oligonucleotide pharmacology. Nucleic Acid Ther. 2014;24:101–113. doi: 10.1089/nat.2013.0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scott C.C., Vacca F., Gruenberg J. Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 2014;31:2–10. doi: 10.1016/j.semcdb.2014.03.034. [DOI] [PubMed] [Google Scholar]
- 9.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Intra-endosomal trafficking mediated by lysobisphosphatidic acid contributes to intracellular release of phosphorothioate-modified antisense oligonucleotides. Nucleic Acids Res. 2017;45:5309–5322. doi: 10.1093/nar/gkx231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Juliano R.L. Intracellular Trafficking and Endosomal Release of Oligonucleotides: What We Know and What We Don’t. Nucleic Acid Ther. 2018;28:166–177. doi: 10.1089/nat.2018.0727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varkouhi A.K., Scholte M., Storm G., Haisma H.J. Endosomal escape pathways for delivery of biologicals. J Control Release. 2011;151:220–228. doi: 10.1016/j.jconrel.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 12.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vermeulen L.M.P., Brans T., Samal S.K., Dubruel P., Demeester J., De Smedt S.C., Remaut K., Braeckmans K. Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano. 2018;12:2332–2345. doi: 10.1021/acsnano.7b07583. [DOI] [PubMed] [Google Scholar]
- 14.Lönn P., Kacsinta A.D., Cui X.S., Hamil A.S., Kaulich M., Gogoi K., Dowdy S.F. Enhancing Endosomal Escape for Intracellular Delivery of Macromolecular Biologic Therapeutics. Sci. Rep. 2016;6:32301. doi: 10.1038/srep32301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muallem S., Chung W.Y., Jha A., Ahuja M. Lipids at membrane contact sites: cell signaling and ion transport. EMBO Rep. 2017;18:1893–1904. doi: 10.15252/embr.201744331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bissig C., Gruenberg J. Lipid sorting and multivesicular endosome biogenesis. Cold Spring Harb. Perspect. Biol. 2013;5:a016816. doi: 10.1101/cshperspect.a016816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wickner W., Schekman R. Membrane fusion. Nat. Struct. Mol. Biol. 2008;15:658–664. doi: 10.1038/nsmb.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Souleimanian N., Deleavey G.F., Soifer H., Wang S., Tiemann K., Damha M.J., Stein C.A. Antisense 2′-Deoxy, 2′-Fluroarabino Nucleic Acids (2'F-ANAs) Oligonucleotides: In Vitro Gymnotic Silencers of Gene Expression Whose Potency Is Enhanced by Fatty Acids. Mol. Ther. Nucleic Acids. 2012;1:e43. doi: 10.1038/mtna.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khaled Z., Ho Y.Y., Benimetskaya L., Deckelbaum R.J., Stein C.A. Omega-6 polyunsaturated fatty acid-stimulated cellular internalization of phosphorothioate oligodeoxynucleotides: evidence for protein kinase C-zeta dependency. Biochem. Pharmacol. 1999;58:411–423. doi: 10.1016/s0006-2952(99)00126-4. [DOI] [PubMed] [Google Scholar]
- 20.Willem J., ter Beest M., Scherphof G., Hoekstra D. A non-exchangeable fluorescent phospholipid analog as a membrane traffic marker of the endocytic pathway. Eur. J. Cell Biol. 1990;53:173–184. [PubMed] [Google Scholar]
- 21.Xu S., Zhang X., Liu P. Lipid droplet proteins and metabolic diseases. Biochim Biophys Acta Mol Basis Dis. 2018;1864(5 Pt B):1968–1983. doi: 10.1016/j.bbadis.2017.07.019. [DOI] [PubMed] [Google Scholar]
- 22.Nevo-Yassaf I., Lovelle M., Nahmias Y., Hirschberg K., Sklan E.H. Live cell imaging and analysis of lipid droplets biogenesis in hepatatis C virus infected cells. Methods. 2017;127:30–36. doi: 10.1016/j.ymeth.2017.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Wang S., Sun H., Tanowitz M., Liang X.H., Crooke S.T. Annexin A2 facilitates endocytic trafficking of antisense oligonucleotides. Nucleic Acids Res. 2016;44:7314–7330. doi: 10.1093/nar/gkw595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ferreira N.S., Engelsby H., Neess D., Kelly S.L., Volpert G., Merrill A.H., Futerman A.H., Færgeman N.J. Regulation of very-long acyl chain ceramide synthesis by acyl-CoA-binding protein. J. Biol. Chem. 2017;292:7588–7597. doi: 10.1074/jbc.M117.785345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trajkovic K., Hsu C., Chiantia S., Rajendran L., Wenzel D., Wieland F., Schwille P., Brügger B., Simons M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science. 2008;319:1244–1247. doi: 10.1126/science.1153124. [DOI] [PubMed] [Google Scholar]
- 26.Ikonen E. Cellular cholesterol trafficking and compartmentalization. Nat. Rev. Mol. Cell Biol. 2008;9:125–138. doi: 10.1038/nrm2336. [DOI] [PubMed] [Google Scholar]
- 27.van Meer G., Voelker D.R., Feigenson G.W. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marrink S.J., de Vries A.H., Tieleman D.P. Lipids on the move: simulations of membrane pores, domains, stalks and curves. Biochim. Biophys. Acta. 2009;1788:149–168. doi: 10.1016/j.bbamem.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Lang T., Halemani N.D., Rammner B. Interplay between lipids and the proteinaceous membrane fusion machinery. Prog. Lipid Res. 2008;47:461–469. doi: 10.1016/j.plipres.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 30.Hissa B., Pontes B., Roma P.M., Alves A.P., Rocha C.D., Valverde T.M., Aguiar P.H., Almeida F.P., Guimarães A.J., Guatimosim C. Membrane cholesterol removal changes mechanical properties of cells and induces secretion of a specific pool of lysosomes. PLoS ONE. 2013;8:e82988. doi: 10.1371/journal.pone.0082988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fernandis A.Z., Wenk M.R. Membrane lipids as signaling molecules. Curr. Opin. Lipidol. 2007;18:121–128. doi: 10.1097/MOL.0b013e328082e4d5. [DOI] [PubMed] [Google Scholar]
- 32.Johnson D.E., Ostrowski P., Jaumouillé V., Grinstein S. The position of lysosomes within the cell determines their luminal pH. J. Cell Biol. 2016;212:677–692. doi: 10.1083/jcb.201507112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liang X.H., Sun H., Shen W., Crooke S.T. Identification and characterization of intracellular proteins that bind oligonucleotides with phosphorothioate linkages. Nucleic Acids Res. 2015;43:2927–2945. doi: 10.1093/nar/gkv143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller C.M., Donner A.J., Blank E.E., Egger A.W., Kellar B.M., Østergaard M.E., Seth P.P., Harris E.N. Stabilin-1 and Stabilin-2 are specific receptors for the cellular internalization of phosphorothioate-modified antisense oligonucleotides (ASOs) in the liver. Nucleic Acids Res. 2016;44:2782–2794. doi: 10.1093/nar/gkw112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bissig C., Gruenberg J. ALIX and the multivesicular endosome: ALIX in Wonderland. Trends Cell Biol. 2014;24:19–25. doi: 10.1016/j.tcb.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 36.Jaishy B., Abel E.D. Lipids, lysosomes, and autophagy. J. Lipid Res. 2016;57:1619–1635. doi: 10.1194/jlr.R067520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Juliano R.L., Carver K. Cellular uptake and intracellular trafficking of oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:35–45. doi: 10.1016/j.addr.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zelphati O., Szoka F.C., Jr. Mechanism of oligonucleotide release from cationic liposomes. Proc. Natl. Acad. Sci. USA. 1996;93:11493–11498. doi: 10.1073/pnas.93.21.11493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Drücker P., Pejic M., Galla H.J., Gerke V. Lipid segregation and membrane budding induced by the peripheral membrane binding protein annexin A2. J. Biol. Chem. 2013;288:24764–24776. doi: 10.1074/jbc.M113.474023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koller E., Vincent T.M., Chappell A., De S., Manoharan M., Bennett C.F. Mechanisms of single-stranded phosphorothioate modified antisense oligonucleotide accumulation in hepatocytes. Nucleic Acids Res. 2011;39:4795–4807. doi: 10.1093/nar/gkr089. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.