Abstract
Sirtuins are NAD+-dependent enzymes that govern cellular homeostasis by regulating the acylation status of their diverse target proteins. We recently demonstrated that both rod and cone photoreceptors rely on NAMPT-mediated NAD+ biosynthesis to meet their energetic requirements. Moreover, we found that this NAD+-dependent retinal homeostasis relies, in part, on maintenance of optimal activity of the mitochondrial sirtuins and of SIRT3 in particular. Nonetheless, it is unknown whether other sirtuin family members also play important roles in retinal homeostasis. Our results suggest that SIRT1, SIRT2, SIRT4, and SIRT6 are dispensable for retinal survival at baseline, as individual deletion of each of these sirtuins does not cause retinal degeneration by fundus biomicroscopy or retinal dysfunction by ERG. These findings have significant implications and inform future studies investigating the mechanisms underlying the central role of NAD+ biosyn-thesis in retinal survival and function.
Keywords: Sirtuins, NAD+, Retinal degeneration, Neurodegeneration, Photoreceptors, Retina
68.1. Introduction
Sirtuins are NAD+-dependent enzymes that are critically important for maintaining cellular homeostasis, especially under conditions of metabolic and nutritional stress. Mechanistically, sirtuins regulate cellular homeostasis by modifying the acylation status of their diverse target proteins. Since sirtuins require NAD+ as a cosubstrate, their function is exquisitely dependent on NAD+ availability (Imai et al. 2000; Verdin 2015). The sirtuin family includes numerous members, which localize to unique subcellular compartments: SIRT1 and SIRT2 are present in the nucleus and the cytoplasm (North and Verdin 2007; Tanno et al. 2007); SIRT6 is exclusively nuclear (Liszt et al. 2005); and SIRT3, SIRT4, and SIRT5 are present in the mitochondria (Schwer et al. 2002; Michishita et al. 2005). We recently demonstrated that NAMPT-mediated NAD+ biosynthesis is essential for photoreceptor survival and vision and that SIRT3 and SIRT5 play important roles in retinal homeostasis (Lin et al. 2016). However, whether the other mitochondrial sirtuin SIRT4 and the nuclear/cytoplasmic sirtuins, SIRT1, SIRT2, and SIRT6, play important roles in retinal function under baseline conditions has not been adequately explored. Therefore, we examined the effect of deleting these individual sirtuins on retinal survival and function.
68.2. Materials and Methods
68.2.1. Animals
All animal experiments were approved by the Animal Studies Committee and performed in accordance with the Washington University School of Medicine Animal Care and Use guidelines. We purchased floxed SIRT1 mice (Sirt1F/F), Sirt2−/− mice, and Sirt4−/− mice, along with strain-matched controls, from Jackson Laboratory (Bar Harbor, ME). We received floxed SIRT6 mice (Sirt6F/F) from Dr. Raul Mostoslavsky (Sebastian et al. 2012), rhodopsin-iCre75 transgenic mice from Dr. Ching-Kang Jason Chen (Li et al. 2005), and human red/green pigment-Cre (HRGP-Cre) transgenic mice from Dr. Yun Le (Le et al. 2004). We tested all mice by fundus biomicroscopy and electroretinography at roughly 2–3 months of age. Whenever possible, we compared mice to littermate controls. We confirmed that all mice were negative for the rd8 mutation of the Crb1 gene (Mattapallil et al. 2012) by PCR.
68.2.2. Assessment of Retinal Structure and Function
To assess retinal structure, we performed fundus biomicroscopy with the Micron III™ animal fundus camera (Phoenix Research Laboratories, Pleasanton, CA), as described previously (Lin et al. 2016). We examined multiple mice per genotype and present representative images. To assess retinal function, we performed electroretinography (ERG) with the UTAS-E3000 Visual Electrodiagnostic System running EM for Windows (LKC Technologies, Gaithersburg, MD), as described previously (Lin et al. 2016), testing at least three mice per genotype. For statistical analysis of the ERG data, we used GraphPad Prism version 5.0f (La Jolla, CA) to perform a mixed two-way ANOVA (within-subjects factor, flash intensity; between-subjects factor, genotype). We defined statistical significance as a p value <0.05.
68.3. Results
68.3.1. Deletion of Sirt1, Sirt2, Sirt4, or Sirt6 Does Not Cause Retinal Degeneration
To determine whether deletion of individual sirtuins causes retinal degeneration, we assessed the fundus appearance of mice lacking various individual sirtuins, including rod photoreceptor-specific SIRT1 conditional knockout (Sirt1−rod/−rod), cone photoreceptor-specific SIRT1 conditional knockout (Sirt1−cone/−cone), SIRT2 germline knockout (Sirt2−/−), SIRT4 germline knockout (Sirt4−/−), rod photoreceptor-specific SIRT6 conditional knockout (Sirt6−rod/−rod), and cone photoreceptor-specific SIRT6 conditional knockout (Sirt6−cone/−cone) mice. We tested photoreceptor-specific conditional knockouts for SIRT1 and SIRT6 because SIRT1 germline knockouts on our desired inbred background die shortly after birth (McBurney et al. 2003) and SIRT6 germline knockouts have a severe neurodegenerative phenotype, dying around 4 weeks of age (Mostoslavsky et al. 2006). At 2–3 months of age, none of the mice we tested had any evidence of retinal degeneration from their fundus appearances (Fig. 68.1a–d). These findings suggest that SIRT1, SIRT2, SIRT4, and SIRT6 are dispensable for retinal survival at baseline.
Fig. 68.1.
Sirt1−rod/−rod (a), Sirt1−cone/−cone (a), Sirt2−/− (b), Sirt4−/− (c), Sirt6−rod/−rod (d), and Sirt6−cone/−cone (d) mice all have normal-appearing fundi on retinal imaging at 2–3 months of age
68.3.2. Deletion of Sirt1, Sirt2, Sirt4, or Sirt6 Does Not Cause Retinal Dysfunction
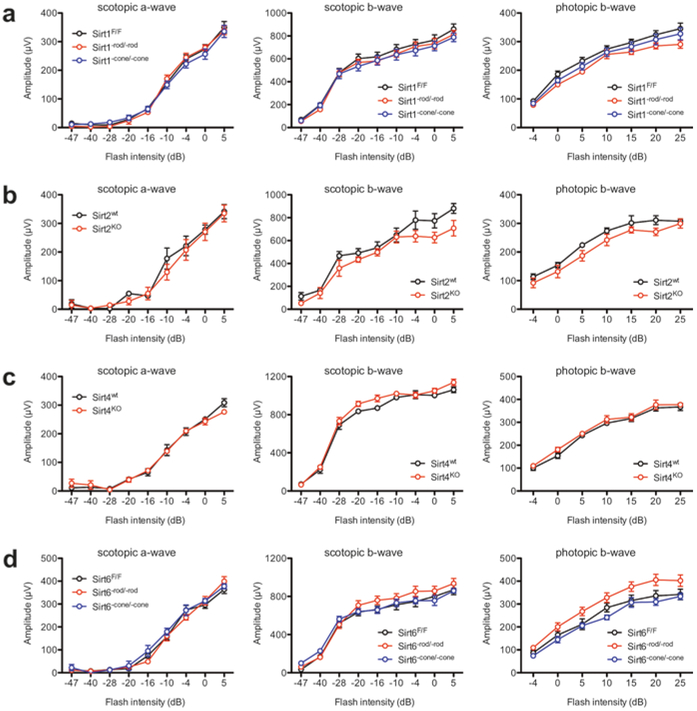
To determine whether deletion of individual sirtuins causes baseline retinal dysfunction, we also tested these mice with ERG. Consistent with the normal retinal appearances on fundus biomicroscopy, Sirt1−rod/−rod, Sirt1−cone/−cone, Sirt2−/−, Sirt4−/−, Sirt6−rod/−rod, and Sirt6−cone/−cone mice all had normal retinal function at baseline at 2–3 months of age (Fig. 68.2a–d). Once again, these findings suggest that these sirtuins are not essential for retinal function under basal conditions.
Fig. 68.2.
Sirt1−rod/−rod (a), Sirt1−cone/−cone (a), Sirt2−/− (b), Sirt4−/− (c), Sirt6−rod/−rod (d), and Sirt6−cone/−cone (d) mice all have normal scotopic and photopic retinal function by ERG at 2–3 months of age (N.S. by two-way mixed ANOVA). Graphs depict mean ± S.E.M (a–d)
68.4. Discussion
Sirtuins have been reported to play important functions in various mouse models of retinal disease (Shindler et al. 2007; Jaliffa et al. 2009; Chen et al. 2013; Zuo et al. 2013; Zeng and Yang 2015). However, we found that individual deletion of Sirt1, Sirt2, Sirt4, or Sirt6 did not cause retinal degeneration as assessed by fundus biomicroscopy nor did it cause retinal dysfunction by ERG. These findings suggest that these sirtuins are individually dispensable under basal conditions. These findings are important, as we previously demonstrated that NAMPT-mediated NAD+ biosyn-thesis is essential for retinal function and that SIRT3/SIRT5 may play important roles in NAD+-dependent retinal homeostasis under conditions of stress (Lin et al. 2016). These results expand on our previous findings and clarify the role of various sirtuin family members in retinal survival and function.
Further studies are necessary to determine whether these sirtuins become essential under conditions of stress, such as in light-induced degeneration or in other models of retinal dysfunction or retinal disease, since sirtuin-deficient mice tend not to show severe phenotypes until challenged by metabolic perturbations such as fasting (Hirschey et al. 2011). Moreover, our experiments in rod- and cone-specific conditional knockouts do not rule out the possibility that SIRT1 and SIRT6 play important roles in non-photoreceptor retinal cells. Indeed, SIRT6 germline knockout mice exhibit retinal dysfunction on ERG by 3 weeks of age, which has been hypothesized to be related to an important role for SIRT6 in inner retinal cells, such as bipolar cells, Müller cells, or perhaps both (Silberman et al. 2014). Cumulatively, our results suggest that SIRT1, SIRT2, SIRT4, and SIRT6 are individually dispensable for retinal survival and function under basal conditions, although further studies are necessary to explore how they interact with one another and whether they become essential under disease conditions.
Acknowledgments
This work was supported by NIH Grants R01 EY019287 (R.S.A.) and P30 EY02687 (Vision Core Grant); the C.M. and M.A. Reeves Foundation (R.S.A.); Research to Prevent Blindness (R.S.A.); the Hope Center (R.S.A.); the Lacy Foundation (S.K.); the Schulak Family Gift Fund for Retinal Research (R.S.A.); the Jeffrey Fort Innovation Fund (R.S.A.); and the Robert Machemer Foundation (S.K.). Additional funding comes from an unrestricted grant to the Department of Ophthalmology and Visual Sciences of Washington University School of Medicine from Research to Prevent Blindness. J.B.L. was supported by the Washington University in St. Louis Medical Scientist Training Program (NIH Grant T32 GM007200), the Washington University in St. Louis Institute of Clinical and Translational Sciences (NIH Grants UL1 TR000448, TL1 TR000449), the Washington University Diabetic Cardiovascular Disease Center, the American Federation for Aging Research, and the VitreoRetinal Surgery Foundation.
Contributor Information
J. B. Lin, Department of Ophthalmology & Visual Sciences, Washington University School of Medicine, St. Louis, MO, USA Neuroscience Graduate Program, Division of Biology and Biomedical Sciences, Washington University School of Medicine, St. Louis, MO, USA.
S. Kubota, Department of Ophthalmology & Visual Sciences, Washington University School of Medicine, St. Louis, MO, USA
R. Mostoslavsky, Massachusetts General Hospital Cancer Center, Harvard Medical School, Boston, MA, USA
R. S. Apte, Department of Ophthalmology & Visual Sciences, Washington University School of Medicine, St. Louis, MO, USA Department of Developmental Biology, Washington University School of Medicine, St. Louis, MO, USA; Department of Medicine, Washington University School of Medicine, St. Louis, MO, USA e-mail: apte@vision.wustl.edu.
References
- Chen J, Michan S, Juan AM et al. (2013) Neuronal sirtuin1 mediates retinal vascular regeneration in oxygen-induced ischemic retinopathy. Angiogenesis 16:985–992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Jing E et al. (2011) SIRT3 deficiency and mitochondrial protein hyper-acetylation accelerate the development of the metabolic syndrome. Mol Cell 44:177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M et al. (2000) Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 [DOI] [PubMed] [Google Scholar]
- Jaliffa C, Ameqrane I, Dansault A et al. (2009) Sirt1 involvement in rd10 mouse retinal degeneration. Invest Ophthalmol Vis Sci 50:3562–3572 [DOI] [PubMed] [Google Scholar]
- Le YZ, Ash JD, Al-Ubaidi MR et al. (2004) Targeted expression of Cre recombinase to cone photoreceptors in transgenic mice. Mol Vis 10:1011–1018 [PubMed] [Google Scholar]
- Li S, Chen D, Sauve Y et al. (2005) Rhodopsin-iCre transgenic mouse line for Cre-mediated rod-specific gene targeting. Genesis 41:73–80 [DOI] [PubMed] [Google Scholar]
- Lin JB, Kubota S, Ban N et al. (2016) NAMPT-Mediated NAD(+) biosynthesis is essential for vision in mice. Cell Rep 17:69–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M et al. (2005) Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280:21313–21320 [DOI] [PubMed] [Google Scholar]
- Mattapallil MJ, Wawrousek EF, Chan CC et al. (2012) The Rd8 mutation of the Crb1 gene is present in vendor lines of C57BL/6N mice and embryonic stem cells, and confounds ocular induced mutant phenotypes. Invest Ophthalmol Vis Sci 53:2921–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBurney MW, Yang X, Jardine K et al. (2003) The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol 23:38–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM et al. (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16:4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB et al. (2006) Genomic instability and aging-like pheno-type in the absence of mammalian SIRT6. Cell 124:315–329 [DOI] [PubMed] [Google Scholar]
- North BJ, Verdin E (2007) Interphase nucleo-cytoplasmic shuttling and localization of SIRT2 during mitosis. PLoS One 2:e784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA et al. (2002) The human silent information regulator (Sir)2 homo-logue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebastian C, Zwaans BM, Silberman DM et al. (2012) The histone deacetylase SIRT6 is a tumor suppressor that controls cancer metabolism. Cell 151:1185–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindler KS, Ventura E, Rex TS et al. (2007) SIRT1 activation confers neuroprotection in experimental optic neuritis. Invest Ophthalmol Vis Sci 48:3602–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silberman DM, Ross K, Sande PH et al. (2014) SIRT6 is required for normal retinal function. PLoS One 9:e98831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T et al. (2007) Nucleocytoplasmic shuttling of the NAD+−dependent histone deacetylase SIRT1. J Biol Chem 282:6823–6832 [DOI] [PubMed] [Google Scholar]
- Verdin E (2015) NAD(+) in aging, metabolism, and neurodegeneration. Science (New York, NY) 350:1208–1213 [DOI] [PubMed] [Google Scholar]
- Zeng Y, Yang K (2015) Sirtuin 1 participates in the process of age-related retinal degeneration. Biochem Biophys Res Commun 468:167–172 [DOI] [PubMed] [Google Scholar]
- Zuo L, Khan RS, Lee V et al. (2013) SIRT1 promotes RGC survival and delays loss of function following optic nerve crush. Invest Ophthalmol Vis Sci 54:5097–5102 [DOI] [PMC free article] [PubMed] [Google Scholar]