Abstract
Background
Diabetes mellitus (DM) poses a severe threat to global public health. Diabetic nephropathy (DN) is one of the most common complications of diabetes and the leading cause of end-stage renal disease (ESRD). Approximately 30–40% of DM patients in the world progress to ESRD, which emphasizes the effect of genetic factors on DN. Family clustering also supports the important role of hereditary factors in DN and ESRD. Therefore, a large number of genetic studies have been carried out to identify susceptibility genes in different diabetic cohorts. Extensive susceptibility genes of DN and ESRD have not been identified until recently.
Summary and Key Messages
Some of these associated genes function as pivotal regulators in the pathogenesis of DN, such as those related to glycometabolism and lipid metabolism. However, the functions of most of these genes remain unclear. In this article, we review several susceptibility genes according to their genetic functions to make it easier to determine their exact effect on DN and to provide a better understanding of the advancements from genetic studies. However, several challenges associated with investigating the genetic factors of DN still exist. For instance, it is difficult to determine whether these variants affect the expression of the protein they encode or other cytokines. More efforts should be made to determine how these genes influence the progression of DN. In addition, many results could not be replicated among races, suggesting that the association between genetic polymorphisms and DN is race-specific. Therefore, large, well-designed studies involving more relevant variables and ethnic groups and more relevant functional studies are urgently needed. These studies may be beneficial and retard the progression of DN by early intervention, especially for patients who carry certain risk alleles or genotypes.
Keywords: Susceptibility genes, Diabetic nephropathy, End-stage renal disease, Genetic studies, Gene polymorphism, Single nucleotide polymorphism
Introduction
Diabetes mellitus (DM) is a common, chronic, complex disorder of rapidly growing global importance accompanied by many complications, including retinopathy, neuropathy, and nephropathy. Until 2017, the International Diabetes Federation (IDF) reported that approximately 452 million adults suffer from DM worldwide, and the number may increase to 629 million by 2045 globally [1]. Diabetic nephropathy (DN) is one of the most conventional microvascular complications of DM and the leading cause of end-stage renal disease (ESRD), which results in high morbidity and mortality [2]. The main pathologic changes of DN are the accumulation of advanced glycation end products (AGEs), growth factors, and variations in hemodynamics and hormones, which result in proteinuria, hypertension, and constant decreased kidney function. Nevertheless, previous studies have shown that approximately 30–40% of DM patients progress to ESRD [3, 4], suggesting that genetic variations may have an impact on the initiation and development of DN and ESRD. It is well known that gene susceptibility to DN plays an important role in individuals, even with the same environmental exposure. Family clustering also supports the importance of hereditary factors in DN and ESRD [5]. Therefore, a myriad of genetic studies has been conducted to identify potential candidate genes in large diabetic cohorts [6], which may facilitate the exploration of the pathogenesis of DN.
With the advancements in genetic methods, including linkage and candidate gene studies and genome-wide association studies (GWAS), numerous candidate gene loci of DN have been identified. Considering the disparity of the study methods, study population, type of diabetes and phenotypes, it is not simple to understand the real effect of genetic variants.
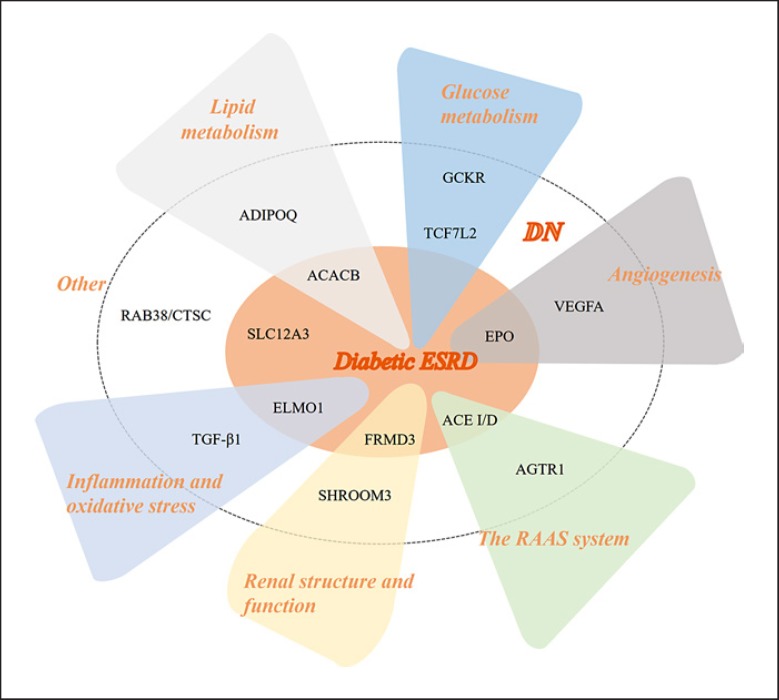
In this review, we summarize the current status, recent advancements and ongoing challenges of susceptibility genes in DN and ESRD according to their functions (Fig. 1). Detailed information on these genes is shown in Table 1. Herein, we classify and discuss these susceptibility genes in the development of DN according to their related functions.
Fig. 1.
The susceptibility genes in diabetic nephropathy. As shown in the figure, the susceptibility genes in diabetic nephropathy are divided into different categories according to their main functions.
Table 1.
The susceptibility genes in diabetic nephropathy
| Gene | SNP | MAF | Study type | Phenotype | DM type | Race | Sample size | OR/HR | p value | Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Lipid metabolism-related genes | ||||||||||
| ACACB | rs2268388 | A = 0.1631/817 | meta | T2DN | T2D | Japanese | 1,312 | 1.61 (additive) | 1.4×10–6 | [8] |
| meta | T2D-ESRD | T2D | European | 908 | 1.61 (additive) | 0.0006 | [8] | |||
| ADIPOQ | rs2241766 | G = 0.1514/758 | CC | T2DN | T2D | Korean | 708 | 1.96 (GG) | 0.049 | [19] |
| cohort | T2DN, male | T2D | Taiwanese | 556 | 1.81 (recessive TT vs. GT+GG, HR) | 0.019 | [18] | |||
| rs1063537 | T = 0.1442/722 | cohort | T2DN, male | T2D | Taiwanese | 556 | 1.89 (recessive CC vs. CT+TT, HR) | 0.013 | [18] | |
| Glucose metabolism-related genes | ||||||||||
| GCKR | rs1260326 | T = 0.2933/1469 | AA | estimated GFR | T2D | European | 2,097 | 1.12 (β) | 4.27×10–2 | [25] |
| TCF7L2 | rs7903146 | T = 0.2278/1141 | CC | T2DN | T2D | Caucasians | 1,355 | 1.97 (T) | <0.001 | [31] |
| rs11196218 | A = 0.2500/1252 | CC | T2DN | T2D | Chinese | 898 | 1.37 | 0.0051 | [30] | |
| Angiogenesis-related genes | ||||||||||
| EPO | rs1617640 | C = 0.3253/1629 | meta | DN | total | European | 2,572 | 1.47 | <0.05 | [42] |
| meta | PDR+ESRD | T1D | European | 7,007 | 1.31 | 2×10–9 | [39] | |||
| VEGFA | rs833061 | C = 0.3698/1852 | meta | T1DN | T1D | European | 543 | 0.67 | <0.00001 | [38] |
| Genes related to renal structure and function | ||||||||||
| FRMD3 | rs10868025 | G = 0.2800/1402 | meta | albuminuria with ESRD | T1D | 9 white | 1,705 | 1.45 (A [G]) | 5×10–7 | [81] |
| rs1888747 | C = 0.1979/991 | meta | albuminuria with ESRD | T1D | white | 1,705 | 1.45 (G [C]) | 6.3×10–7 | ||
| SHROOM3 | rs17319721 | A = 0.2238/1121 | meta | eGFR | T2D | Tayside, Scotland | 3,028 | 1.02 (A, HR) | 3.18×10–3 | [25] |
| Inflammation and oxidative stress-related genes | ||||||||||
| ELMO1 | rs10951509 | G = 0.4289/2148 | CC | T2D-ESRD | T2D | American Indians | 772 | 2.42 (A) | 0.002 | [78] |
| T2DN | T2D | Chinese | 200 | 1.76 (A) | 0.02 | [49] | ||||
| Intron 18+9170 A/G | ND | CC | T2DN | T2D | Japanese | 732 | 2.67 (GG vs. GA+AA) | 0.000008 | [46] | |
| rs1345365 | G = 0.4347/2177 | CC | T2D-ESRD | T2D | American Indians | 772 | 2.42 (A) | 0.001 | [78] | |
| CC | T2DN | T2D | Chinese | 200 | 3.27 (A) | 0.004 | [49] | |||
| TGF-β1 | rs1800470 | G = 0.4547/2277 | CC | T2D | T2D | Mexico inhabitants | 439 | 1.818 | 0.016 | [58] |
| 915 G>C | ND | CC | T2DN | T2D | Mexico inhabitants | 439 | 4.073 | 0.008 | [58] | |
| Genes related to the RAAS system | ||||||||||
| ACE I/D | rs1799752 | ND | CC | persistent microalbuminuria | T1D | Caucasians | 1,365 | 0.62 (DI/II) | 0.009 | [64] |
| CC | T2D-ESRD | T2D | Chinese Han | 432 | 2.23 (DD vs. II+ID) | 0.005 | [70] | |||
| AGTR1 | rs5186 (A1166C) | C = 0.1178/590 | CC | T1DN, male | T1D | Denmark, Finland, France, and Sweden | 3,561 | 1.27 (AA vs. AC+CC) | 0.03 | [73] |
| meta | DN | total | Caucasian, Asian | ND | 2.1 (CC vs. AA) | 0 | [75] | |||
| meta | DN | total | Caucasian, Asian | ND | 2.11 (dominant) | 0 | ||||
| Other susceptibility genes | ||||||||||
| SLC12A3 | rs11643718 | A = 0.0799/400 | CC | T2DN | T2D | Malaysian | 1,417 | 0.547 | 0.038 | [77] |
| CC | T2D-ESRD | T2D | Chinese | 372 | 2.2 (GA+AA) | 0.019 | [76] | |||
| CC | T2D-ESRD | T2D | Koreans | 358 | 2.295 | 0.003 | [79] | |||
| RAB38/CTSC | rs649529 | T = 0.4846/2427 | GWAS | UACR | T1D | European | 7,787 | 0.14 (β) | 5.8×10–7 | [82] |
MAF, global minor allele frequency; OR, odds ratio, the words in brackets mean contrast alleles or genotypes; HR, hazard ratio; β, β value; ND, no data; DN, diabetic nephropathy; T1D, type 1 diabetes; T2D, type 2 diabetes; ESRD, end-stage renal disease; T2D-ESRD, patients with T2D and end-stage renal disease; T1D-ESRD, patients with T1D and end-stage renal disease; T2DN, type 2 diabetes-related nephropathy; T1DN, type 1 diabetes-related nephropathy; PDR, proliferative diabetic retinopathy; eGFR, estimated glomerular filtration rate; UACR, urea albumin creatinine ratio; GWAS, genome-wide association study; CC, case-control study; cohort, prospective cohort; AA, association analysis; dominant, dominant model; recessive, recessive model; additive, additive model.
Lipid Metabolism-Related Genes
Previous studies have shown that an increase in renal lipid retention, which is related to the accumulation of biglycan, contributes to the development of DN. Dysregulation of genes related to lipid metabolism accounts for lipid deposition, resulting in the decline of glomerular filtration rate and inflammation. Variants in the acetyl-coenzyme A carboxylase beta (ACACB) and adiponectin (ADIPOQ) genes are likely involved in the development of DN.
ACACB
The ACACB gene is located on chromosome 12q24.1 and encodes acetyl-coenzyme A (CoA) carboxylase beta (ACC2/ACACB). ACC2 is a key rate-limiting enzyme for the β-oxidation of fatty acid. This gene catalyzes the transformation of acetyl-CoA into malonyl-CoA in the mitochondrial membrane and consequently inhibits carnitine palmitoyl transferase (CPT1) and reduces acyl-CoA transfer to the mitochondria in adipose and muscle tissues [7]. Although ACC2 is highly expressed in heart and skeletal muscles, the mRNA of ACC2 is expressed at a relatively low level in glomerular and tubular epithelial cells [8]. A single nucleotide polymorphism (SNP) in intron 18 of ACACB that was identified by a large-sample meta-analysis in Japanese individuals (rs2268388) showed a significant association with type 2 diabetes-related nephropathy (T2DN). Researchers have found the strongest associations in Asian [9], including Chinese [8], and Caucasian populations by case-control studies [8, 9]. In a recent meta-analysis, Li et al. [10] concluded that the T allele increases the susceptibility to T2DN. Functional in vitro studies have revealed that the activity of a DNA fragment containing rs2268388 was dramatically increased in proximal tubular epithelial cells (RPTECs), especially in T allele carriers [8]. Fatty acid oxidation persisted and 50% less fat accumulated in ACC2-deficient rats [11]. Overexpression of ACC2 leads to increased levels of proinflammatory cytokine, such as IL-6, in RPTECs, which could rely on activation of the p38-MAPK pathway [12]. In recent years, fatty acid toxicity was reported to play a critical role in T2D-associated renal injury. Moreover, a lack of ACC2 in high-glucose-cultured HK-2 cells resulted in a faster beta oxidation rate, less lipid deposition and malonyl-CoA content [13]. ACC2 can inhibit palmitic acid-induced autophagy, lower lipid accumulation, and restore cell viability [14], suggesting an important role for ACC2 in the development of DN. However, further studies are needed to clarify the concrete mechanisms of these polymorphisms in DN.
ADIPOQ
The ADIPOQ gene, which is located on chromosome 3q27, encodes adiponectin and has been identified as the susceptibility gene of cardiovascular disease, type 2 DM (T2D), obesity, and insulin resistance [15]. Adiponectin is mainly secreted by adipocytes and acts as a vital modulator in insulin resistance and lipid metabolism. It is commonly believed that adiponectin is insulin-sensitizing and facilitates β-cell oxidation [16], which also has anti-atherogenic and anti-inflammatory effects. The SNP rs17300539 (ADIPOQ_prom2GA) was initially found to be associated with DN in both Danish and French patients with T2D by linkage studies. Jorsal et al.[17] reported that the A allele may increase the risk of nephropathy in T1D. They found that carriers of the minor allele A in −11387 (rs17300539) and the non-A-allele in +2033 tended to have notably increased serum adiponectin levels in T1D, which predicted the progression of ESRD in a covariate-adjusted analysis. The polymorphisms of ADIPOQ also implicated the susceptibility of DN in T2D patients. The most popular SNP, rs266729 (+45T>G), showed an increased risk of DN in various populations with T2D, such as Taiwanese [18], Korean [19], and Egyptian [20]. Other SNPs, such as rs1063537, rs2241767, and rs2082940, are also related to DN [18]. Jorsal et al. [17] demonstrated that the level of adiponectin was increased in T1D patients with DN, which predicted the progression to ESRD. The authors also demonstrated links between ADIPOQ gene polymorphisms, including rs17300539, and the level of adiponectin. Furthermore, the deletion of ADIPOQ in diabetic mice increased the progression to kidney hypertrophy and glomerular enlargement. Albuminuria and oxidative stress were also increased in ADIPOQ-knockdown mice, while glycemia was invariable. In vitro, high-glucose-induced phosphorylation was decreased by adiponectin. In Akita/APN–/– (Ins2+/C96YAdipoq–/–) mice, the level of fibrosis and the inflammatory response were significantly increased. After high-glucose induction, adiponectin inhibited the transforming growth factor-β (TGF-β)/signal transduction molecule 2 (Smad2) pathway and the activation of nuclear factor-κB in mesangial cells [21].
Glucose Metabolism-Related Genes
There is no doubt that DN, as the main complication of diabetes, is associated with glucose metabolism. Polymorphisms of glucose metabolism-related genes, including glucokinase regulatory protein (GCKR) and transcription factor 7-like 2 (TCF7L2), are believed to be related to DN.
GCKR
Large-scale GWAS have illustrated that the GCKR gene is related to a reduction of renal function and chronic kidney disease (CKD) [22]. GCKR has been considered a susceptibility gene for diabetes [23], and many studies have been conducted to explore the association between GCKR and renal complications in T2D. GCKR, which is mapped to chromosome 2p23.3, encodes glucokinase regulatory protein (GKRP). GKRP is mainly produced in the liver and islet β-cells. By competitively combining with glucokinase (GCK) and forming a complex to inhibit the function of GCK, GKRP affects the affinity of GCK and fructose metabolites and consequently participates in the mediation of glucose metabolism [24]. The Genetics of Diabetes Audit and Research Tayside (GoDARTs) study evaluated the correlation between the estimated glomerular filtration rate (eGFR) and 16 candidate gene loci in 3,028 patients with T2D. They showed that the P446 L of rs1260326 in GCKR was associated with a higher baseline eGFR, especially in those with albuminuria [25], implying an association between GCKR variants and DN. Another SNP in GCKR, rs780094, is in intense linkage disequilibrium (LD) with rs1260326. Yan et al. [26] concluded that the A allele of rs780094 in GCKR was associated with diabetic kidney disease susceptibility in T2D patients, but the genotype was not significant. Overall, direct evidence is needed to confirm these associations.
TCF7L2
TCF7L2 is a susceptibility gene that is strongly associated with diabetes [27]. In recent years, studies have confirmed that the polymorphisms of TCF7L2 are correlated with T2DN. The TCF7L2 gene, which is also known as the T-cell transcription factor 4 (TGF4) gene, is located on 10q25.2-q25.3. TGF4 is a nuclear receptor gene that encodes TCF7L2, which contains the high migration rate group box, and plays a key role in the Wnt/β-catenin pathway. TCF7L2 is not only involved in the regulation of glucose homeostasis but also participates in regulating islet β-cell proliferation, differentiation, and insulin secretion [28]. Wu et al. [29] demonstrated in a Taiwan population that ADIPOQ, growth hormone secretagogue receptor (GHSR) and TCF7L2 may participate in the occurrence of T2DN in an interactive way. Subsequently, Fu et al. [30] illustrated that the genotype and allele frequency distribution of rs11196218 in TCF7L2 were significantly different compared to non-DN patients in a Chinese population, suggesting that TCF7L2 is related to the development of DN. Buraczynska et al.[31] found that rs7903146 in TCF7L2 was strongly correlated with DN in Caucasians, especially for the early onset of diabetes. Their study showed that in diabetic and nondiabetic ESRD patients, the T allele of rs7903146 increased the risk of developing DN. Hussain et al. [32] concluded that this association was not independent of DM. A recent systematic meta-analysis and review of previous studies deduced that TCF7L2 may affect the risk of DN, and subgroup analysis showed that this association was consistent in both Asians and Caucasians, but not in Africans [33]. Therefore, additional studies are needed to verify this conclusion. A previous functional study clarified that the effect of TCF7L2 on DN was related to the activin receptor-like kinase 1 (ALK1)/Smad1 pathway. Additionally, the AGEs increased the expression of TCF7L2 via TGF-β1, which helped transfer TCF7L2 from the cytoplasm to the nucleus. Subsequently, TCF7L2 combined with the promoter of ALK1 to increase its expression. ALK1 enhanced the effect of TGF-β and promoted the phosphorylation of cellular Smad1, resulting in glomerular sclerosis. These authors believed that the AGEs/TGF-β/TCF7L2/ALK1/Smad1 signaling pathway may play an important role in the development of DN [34].
Angiogenesis-Related Genes
Abnormal angiogenesis is a main characteristic of DN. Genes related to angiogenesis, such as the hormone erythropoietin (EPO) promoter gene and vascular endothelial growth factor A (VEGFA), are associated with DN.
EPO Promoter Gene
The EPO promoter gene is located on chromosome 7q22 and encodes EPO. EPO is a key factor involved in erythrocyte production and is widely used for the treatment of chronic renal failure and anemia after chemotherapy [35]. Circulating EPO is mainly produced by fibroblasts in the adult renal peritubular interstitial [36]. EPO is a powerful angiogenic factor in diabetic microvascular disease. A study of 19 SNPs was performed in 374 proliferative diabetic retinopathy (PDR)/ESRD and 239 T2D patients and suggested that only rs1617640 in EPO is associated with T2DM patients with PDR and ESRD. Subsequently, the authors verified the correlation in a GWAS of the Genetics of Kidneys in Diabetes (GoKinD) and Boston cohort of T1D. They simultaneously found that the correlation existed in patients with both PDR and DN, regardless of whether they progressed to ESRD [37]. A meta-analysis also showed that EPO was associated with DN and subgroup analysis showed a strong correlation in T1D [38]. However, in 2012, Williams et al. [39] proposed different results. The Genetics of Nephropa thy - an International Effort (GENIE) study, a larger and similar ancestry study, validated the correlation in European patients with T1D; however, they failed to repeat the results. Although the meta-analysis showed that the correlation reached genome-wide significance, they believed the reason for the difference lay in the requirement to establish a statistically lower threshold. Furthermore, it is not clear whether a DN susceptibility gene has the same effect on T1D and T2D. Additionally, the mechanism of EPO gene polymorphisms in DN has not yet been investigated.
VEGFA
The VEGFA gene is located on chromosome 6 (6p21.3). VEGFA is a cytokine that is highly correlated with diabetic microvascular diseases. It induces the proliferation of endothelial cells in the glomerulus and migrates and changes the permeability of various tissues. VEGFA is expressed in the kidney and is mainly distributed in vascular endothelial cells and podocytes. Therefore, VEGFA may be associated with diabetic microvascular complications. A previous study found that VEGFA expression was decreased in glomeruli cells in 7-week-old T1DM rats, but the occurrence of macroproteinuria, glomerular fibrosis, and apoptosis increased. In a T1D mouse model, a decrease in local VEGFA in the glomeruli promoted damage to endothelial cells, thereby aggravating glomerular injury [40]. VEGFA gene polymorphisms and the expression of the protein are closely related. Multiple loci are associated with DN, such as −2549 I/D/rs35569394, +405/rs2010963, and −1499C>T/rs833061 [41]. A meta-analysis of multiple loci in VEGFA found that rs833061 is the most significant SNP for DN, and a comprehensive analysis of 2 studies in Irishmen had a higher OR value of 2.08 [42].
Genes Related to Renal Structure and Function
Proteinuria and constant decreased kidney function in DN may be closely linked to the pathologic changes in renal structure and function. Glomerular podocyte dysfunction is extremely important for the initiation and progression of DN. Abnormalities in podocytes, such as podocyte hypertrophy or loss, are attributed to many factors. Some genes, including 4.1 protein ezrin, radixin, moesin (FERM) domain-containing 3 (FRMD3) (Table 1), and shroom3 (SHROOM3), which are related to renal structure and function, have been identified as susceptibility genes for DN.
SHROOM3
The SHROOM3 gene is located on 4q21.1 and encodes shroom3 protein, which is related to endothelial morphology. Shroom3 protein mainly regulates the morphogenesis of epithelial cells and tissues. In the rat kidney, the shroom3 protein is expressed in the condensing mesenchyme, Baumann's sac, and podocytes [43]. Knockout of this gene can lead to significant abnormalities in the rat glomeruli, which are characterized by the collapse and degeneration of glomerular cysts and remarkable damage to the arrangement and morphology of podocytes. In short, it maintains the normal structure and function of podocytes by adjusting the actin mesh [43]. A GWAS found that the rs17319721 SNP in the intronic region of the SHROOM3 gene was associated with CKD [44]. Due to its close correlation with renal structure function, some studies further verified that rs173197213 was related to the eGFR in T2D patients with proteinuria [25]. A recent study found that mutations in SHROOM3 disrupted its actin binding area, which could lead to the loss of podocytes and damage to the glomerular filtration barrier. However, the concrete correlation between T2DN and this gene remains unclear.
Inflammation and Oxidative Stress-Related Genes
Disorders of blood glucose and lipid metabolism are another main character of diabetes and DN, which promote inflammation and oxidative stress in patients with diabetes and DN. Several genes, such as engulfment and cell motility protein 1 (ELMO1), TGF-β, and nitric oxide synthase 3 (NOS3, eNOS), participate in the processes of inflammation and oxidative stress and are all involved in the pathogenesis of DN.
ELMO1
The ELMO1 gene is located on 7p14.2-p14.1 and encodes an evolutionarily conserved cytoplasmic protein with no obvious catalytic domains. ELMO1, a receptor that is located downstream of brain-specific angiogenesis inhibitor 1 (BAI1), forms a complex with Dock180, which functions as an unconventional guanine nucleotide exchange factor for the small GTPase Rac1, and hence regulates the actin cytoskeleton during phagocytosis and cell migration through the activation of Rac1 [45]. ELMO1 is a crucial factor for the pathogenesis of T2DN and certain nephropathy-associated variants differ across populations. Shimazaki et al. [46] identified ELMO1 as a susceptibility gene for DN by analyzing a large number of SNPs in Japanese populations; the strongest associated SNP is intron 18+9170. Another variation in intron 13 of the ELMO1 gene was also found to be related to DN in African-Americans [47]. Hanson et al. [48] performed a family study and found firm evidence for an association between two SNPs (rs1345365 and rs10951509) and DN, both of which are located in intron 13 and are in strong pairwise LD. In 2013, the relationship between ELMO1 gene polymorphisms and DN was first validated in a Chinese population; Wu et al. [49] confirmed that both variants, rs741301 and rs10951509, were associated with DN. Furthermore, the rs741301 polymorphism and duration of T2DM were identified as independent predictors of DN. Functional studies have found that a high level of Elmo1 expression aggravated the progression of DN and vice versa. The severity of renal fibrosis, the amount of urinary albumin excretion and changes in the ultrastructure of the glomerular basement membrane in Akita diabetic mice paralleled the genetic levels of ELMO1 [50]. Possible mechanisms by which ELMO1 is involved in the pathogenesis of DN include: (1) ELMO1 and oxidative stress: ELMO1 promotes the production of reactive oxygen species by activating Rac and increasing the expression of NAD(P)H oxidase, thereby increasing oxidative stress in the kidney, leading to renal oxidative damage [50]; (2) ELMO1 and renal fibrosis: increased expression of ELMO1 promotes the expression of fibrogenic genes (such as TGF-β1 and genes encoding type 1 collagen and fibronectin, etc.) and inhibits the expression of anti-fibrotic genes (such as matrix metalloproteinase genes), leading to excessive accumulation of extracellular matrix proteins and thickening of the glomerular basement membrane and consequently resulting in the initiation and progression of diabetic glomerulosclerosis [46]. ELMO1 also serves as a regulator of cyclooxygenase-2 (COX-2) activity, which aggravates glomerular injury and thus stimulates COX-2-mediated fibronectin accumulation in the development of glomerulosclerosis [51]. However, Sharma et al. [52] found that ELMO1 exerts a protective effect on the kidneys in zebrafish and human samples. Their results highlighted ELMO1 as an important factor for glomerular protection and renal cell survival by decreasing apoptosis, especially under diabetic conditions. Therefore, although some studies have proposed several plausible theories, the exact contribution of this gene to the development of DN is not yet clear.
TGF-β1
TGF-β1, one of three isoforms of the TGF-β family, is a multifunctional cytokine that modulates a myriad of cellular processes, including proliferation, differentiation, apoptosis, angiogenesis, extracellular matrix (ECM) formation, and immune processes [53]. It exerts its biological functions through a variety of signaling pathways, including the Smad and MAPK pathways [54]. In renal diseases, elevated expression of TGF-β1 can induce renal hypertrophy and promote excessive accumulation of ECM proteins, thus leading to renal fibrosis [55]. To be more specific, TGF-β1 is significantly enhanced in the renal tissues of patients with DN, especially in mesangial cells of diabetic glomeruli [56]. Therefore, it is likely that these pathological changes caused by TGF-β1 may contribute to the initiation and progression of DN.
The human TGF-β1 gene is located on chromosome 19q13.1–13.3 and includes 7 exons and 6 introns [57]. More than 10 SNPs in this gene are currently known. The hotspot mainly focuses on T869C (rs1800470; Leu10/Pro10; T29C, codon 10) in exon 1, which is located at position 10 in the signal peptide [55]. The T869C gene polymorphism has been linked with the risk of DN in different ethnic groups, such as Chinese [55], Egyptians [56], Mexicans [58], and Caucasians of Polish descent. The T869C gene polymorphism was notably different between diabetic patients with and without DN, and the DN group had a higher frequency of the CC/CT genotype [59]. C allele-containing genotypes may be susceptible, and the T allele/TT genotype may be a protective factor for DN [56, 58]. However, insignificant associations were also reported in Asians [60]. Zhou et al. [53] used a meta-analytic approach and found that the CC genotype may be considered a distinct genotype for DN, whereas the T allele is protective against DN in Asians but represents a risk factor for Caucasians. Notably, Jia et al. [59] conducted a meta-analysis and found that the T allele conferred a significantly reduced risk of DN compared with the C allele in the overall population. Obviously, these two studies contradict each other regarding the association between T alleles and DN risk in Caucasians. However, it is safe to conclude that the CC genotype may be a useful indicator to predict the risk of developing DN in Asians. The specific mechanism underlying the effect of the TGF-β1 T869C polymorphism on DN is not clear. Multiple studies conducted in various populations have suggested that the T869C polymorphism is associated with altered TGF-β1 protein expression and that the C allele is positively correlated with the serum TGF-β1 concentration [59]. It is believed that this effect may be attributed to substituting the C allele for T in T869C in which leucine is replaced by proline, thus accelerating the hydrolysis of the TGF-β1 precursor to a mature TGF-β1 protein [55]. In addition, the T869C SNP is associated with cholesterol and triglyceride plasma concentrations, and subjects with the CC+CT genotype showed higher plasma levels than those with the TT genotype, which may also be involved in the development of DN by altering the function and structure of podocytes and endothelial cells [58]. Another frequently studied SNP is the TGF-β1 G915C polymorphism (rs1800471; codon 25). The G allele has also been associated with increased TGF-β1 production [61]. Valladares-Salgado et al. [58] reported a positive association between the GC+CC genotypes of TGF-β1 G915C and DN in Mexicans, whereas El-Sherbini et al. [56] found no statistically significant association in Egyptians. Zhang et al. [62] also revealed that the G915C gene polymorphism was not associated with the risk of DN by conducting a meta-analysis that contained 7 studies. Overall, larger, well-designed studies that involve more relevant variables are warranted to elucidate the mechanism of the TGF-β1 gene polymorphism in DN susceptibility in the future.
Genes Related to the Renin-Angiotensin-Aldosterone System
The renin-angiotensin-aldosterone system (RAAS) regulates not merely blood pressure but also the internal pressure of the glomerulus, and hypertension is an independent risk factor of DN. Therefore, polymorphisms of RAAS-related genes, such as angiotensin-converting enzyme (ACE) and angiotensin II receptor type 1 (AGTR1), are closely related to the development of DN.
ACE I/D
The ACE gene, which is located on 17q23.3, contains 26 exons and 25 introns and mainly encodes ACE. ACE is one of the key enzymes of the RAAS and mainly transforms angiotensin I to angiotensin II, thus regulating the activity of angiotensin and bradykinin. The ACE gene is mainly expressed in the kidney, especially in the brush border of renal proximal tubules. It also exists in glomerular endothelial cells, mesangial cells, podocytes, and distal nephrons. Due to the crucial role of ACE in the RAAS, a large number of studies have linked its polymorphisms to the development of diabetic microvascular complications, such as DN [63]. ACE I/D is the most susceptible locus for DN. In the Diabetes Control and Complications Trial (DCCT) and following in the Epidemiology of Diabetes Interventions and Complications (EDIC) study, Boright et al. [64] found that the risk of developing refractory proteinuria and severe kidney diseases was lower in T1D patients with the II genotype than the I/D genotype. Therefore, they confirmed an association between the ACE I/D polymorphism and the development of DN in T1D. However, in another large-scale trial, T2D patients with the ID or DD genotype tended to have a lower incidence of end-stage renal failure, which contradicts the other 2 studies [65]. Ng [66] ascribed the different results to the haplotype diversity of the variants and underlined the necessity of an extensive haplotype analysis. The correlation between ACE I/D and DN remains controversial. However, a recent meta-analysis found that all genetic models of the ACE I/D polymorphism were significantly correlated with DN susceptibility in patients with T2DM [67]. Moreover, the average ACE activity of the DD carriers was twice as high as that of type II carriers, and the ACE I/D polymorphism affected approximately 47% of the ACE level. Similar results were obtained by subsequent studies [68], which validated that DD and ID+DD carriers had a higher ACE activity level. Another study showed that the eGFR decreased faster in patients with insulin-dependent diabetes (IDM) and those with DN who took captopril. These results indicated that the ACE I/D polymorphism affected the long-term benefits of patients with DN [65]. Follow-up observational studies suggested that the short- and long-term benefits of DD and II carriers with IDM and DN were similar to those with ARB [69]. This result might provide a new strategy for the treatment of DN. Recent studies have found that the frequency of the ID and DD genotype in diabetic ESRD patients with T2D was higher than that of non-DN patients, and DD carriers had higher HbA1c levels and diastolic blood pressure [70]. In summary, the ACE I/D polymorphism may participate in the development of DN and diabetic ESRD, but its specific mechanism requires further study.
AGTR1
The AGTR1 gene is located on 3q21–25 with a length of less than 55 kb, encoding the angiotensin II receptor type 1. It consists of five exons of which the first four are noncoding regions and the fifth is the coding region. The angiotensin II receptor mainly regulates the level of angiotensin II, a key enzyme involved in the RAAS. The RAAS regulates vasoconstriction, the reabsorption of sodium, and the inflammatory cascade, which is currently believed to exert a positive effect on the development of DN. Moreover, specific receptor blockers of AGTR1 exert a renal protective effect in DN which can lower blood pressure and reduce the occurrence of cardiovascular events [71]. Therefore, some studies have suggested that the polymorphism of AGTR1, especially the rs5186 (A1166C) mutation, is one of the potential candidate susceptibility genes for DN. The rs5186 variation is located in the 3′ end of the noncoding regions and may affect the stability and translation of the mRNA or play an important regulatory role in polymorphism LD. In a Swedish population, the A allele of rs5186 increased the risk of DN and may have a synergistic effect with smoking [72]. Subsequently, a study of 3,561 Caucasian patients with T1DM revealed that the risk of developing DN increased significantly in AA genotype carriers [73]. The correlation was verified in different ethnic groups, such as Asians and Caucasians [9, 72]. However, Currie et al. [74] found that the genotype and frequency of AGTR1 in 1467 Caucasian patients with T1DM were not related to DN. In 2012, a meta-analysis showed that the CC allele of the AGTR1 A1166C polymorphism was related to DN (compared with the AA genotype) [75]. Nevertheless, the correlation between AGTR1 polymorphism and DN requires studies with larger samples.
Other Susceptibility Genes
The functions of the following susceptibility genes involved in DN include various aspects and will be discussed separately. Some of these genes are listed in Table 1, such as solute carrier family 12 member 3 (SLC12A3), and RAB38/CTSC, and potassium voltage-gated channel subfamily J member 11 (KCNJ11).
SLC12A3
The SLC12A3 gene is located on 16q13 and encodes thiazide-sensitive Na-Cl cotransporter (NCC; TSC), a 12-transmembrane domain ion transporter protein that is preferentially expressed in renal distal convoluted tubules. It regulates the reabsorption of sodium ions and chloride ions and is a pivotal point of maintaining electrolyte homeostasis and regulating arterial blood pressure [76]. A loss of NCC function is responsible for Gitelman syndrome, an autosomal recessive disorder characterized by low blood pressure, hypocalciuria, hypokalemic metabolic alkalosis, and hypomagnesemia.
Functional studies using zebrafish and db/db mice as animal models have shown that SLC12A3 is of great importance for kidney cloacal development and the progression of DN [77]. The correlation between SLC12A3 gene polymorphism and DN has been demonstrated in different populations. The rs11643718 (Arg913Gln; G/A) variant in exon 23 is the most frequently studied SNP and is nonsynonymous. The protein structure of SLC12A3 was dramatically changed when the mutant allele 913Gln substituted the wild allele, Arg913 [77].
In a Japanese population, the Arg913Gln polymorphism was associated with urinary protein excretion in patients with T2D and reduced the risk of developing DN [78]. Similarly, in Malaysian populations, the Arg913Gln variant also exerted a protective effect on DN [77]. Additionally, in a Korean population, SNPs and haplotypes of the SLC12A3 gene, especially Arg913Gln, were significantly associated with ESRD caused by DN [79]. A recent case-control study suggested that Arg913Gln was associated with a high risk of DN-ESRD in Chinese T2DM patients undergoing hemodialysis and that the GA+AA genotype may be related to increased blood pressure and urinary albumin excretion rate [76]. However, genetic variation at the SLC12A3 locus does not explain the risk of advanced DN among Caucasians and North Indian populations with T2D [80].
Uncertainty still remains as to the precise mechanism of the SLC12A3 polymorphism in the development of DN. Inhibition of NCC by thiazide diuretics has been a cornerstone in the treatment of hypertension. Hypertension is an independent risk factor for the occurrence and progression of DN. Therefore, researchers have proposed that the Arg913Gln variant in the Chinese T2D population may promote the development of DN by increasing blood pressure [78], whereas some studies have found that the SLC12A3 polymorphism in the Japanese population has no significant effect on blood pressure [80]. Owing to the inconsistent results obtained from different ethnic groups, further studies are needed to clarify the exact mechanisms.
Conclusion
The pathogenesis of DN remains largely unknown and appears to be multifactorial, which may be owed to interactions between genetic and environmental factors. Numerous candidate gene loci have been found to be related to DN or diabetic ESRD until recently. Some of these genes function as pivotal regulators in the pathogenesis of DN, such as those related to glycometabolism and lipid metabolism. However, the functions of most genes remain unclear. In this article, we reviewed several susceptibility genes according to their genetic functions to make it easier to recognize their exact effect on DN and to provide a clearer understanding of the advancements in genetic studies. However, several challenges associated with investigating the genetic factors of DN still exist. For instance, it is difficult to determine whether these variants affect the expression of the protein they encode or other cytokines. More effort should be made to determine how the interactions between these genes influence the progression of DN. In addition, many results could not be replicated among races, suggesting that the association between genetic polymorphisms and DN is race-specific. Therefore, large, well-designed studies involving more relevant variables and ethnic groups and more relevant functional studies are urgently needed. These studies may be beneficial to retard the progression of DN by early intervention, especially for patients who carry certain risk alleles or genotypes.
Disclosure Statement
The authors declare no conflict of interest.
Acknowledgments
This work was supported by the National Key R&D Program of China (2016YFC1305501), the Key Program of the NSFC (81730018), the General Program of NSFC (81470960), and the Key Research & Development Plan of Hunan Province (2016JC2061).
References
- 1.Diabetes Atlas ID . IDF. ed 8. 2017. http://www.diabetesatlas.org/ [Google Scholar]
- 2.Groop PH, Thomas MC, Moran JL, Wadèn J, Thorn LM, Mäkinen VP, et al. FinnDiane Study Group The presence and severity of chronic kidney disease predicts all-cause mortality in type 1 diabetes. Diabetes. 2009 Jul;58((7)):1651–8. doi: 10.2337/db08-1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hill CJ, Cardwell CR, Patterson CC, Maxwell AP, Magee GM, Young RJ, et al. Chronic kidney disease and diabetes in the national health service: a cross-sectional survey of the U.K. national diabetes audit. Diabet Med. 2014 Apr;31((4)):448–54. doi: 10.1111/dme.12312. [DOI] [PubMed] [Google Scholar]
- 4.Marshall SM. Natural history and clinical characteristics of CKD in type 1 and type 2 diabetes mellitus. Adv Chronic Kidney Dis. 2014 May;21((3)):267–72. doi: 10.1053/j.ackd.2014.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Skrunes R, Svarstad E, Reisæter AV, Vikse BE. Familial clustering of ESRD in the Norwegian population. Clin J Am Soc Nephrol. 2014 Oct;9((10)):1692–700. doi: 10.2215/CJN.01680214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McDonough CW, Palmer ND, Hicks PJ, Roh BH, An SS, Cooke JN, et al. A genome-wide association study for diabetic nephropathy genes in African Americans. Kidney Int. 2011 Mar;79((5)):563–72. doi: 10.1038/ki.2010.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wakil SJ, Abu-Elheiga LA. Fatty acid metabolism: target for metabolic syndrome. J Lipid Res. 2009 Apr;50((5 Suppl)):S138–43. doi: 10.1194/jlr.R800079-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maeda S, Kobayashi MA, Araki S, Babazono T, Freedman BI, Bostrom MA, et al. A single nucleotide polymorphism within the acetyl-coenzyme A carboxylase beta gene is associated with proteinuria in patients with type 2 diabetes. PLoS Genet. 2010 Feb;6((2)):e1000842. doi: 10.1371/journal.pgen.1000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shah VN, Cheema BS, Sharma R, Khullar M, Kohli HS, Ahluwalia TS, et al. ACACβ gene (rs2268388) and AGTR1 gene (rs5186) polymorphism and the risk of nephropathy in Asian Indian patients with type 2 diabetes. Mol Cell Biochem. 2013 Jan;372((1–2)):191–8. doi: 10.1007/s11010-012-1460-2. [DOI] [PubMed] [Google Scholar]
- 10.Li T, Shi Y, Yin J, Qin Q, Wei S, Nie S, et al. The association between lipid metabolism gene polymorphisms and nephropathy in type 2 diabetes: a meta-analysis. Int Urol Nephrol. 2015 Jan;47((1)):117–30. doi: 10.1007/s11255-014-0843-6. [DOI] [PubMed] [Google Scholar]
- 11.Abu-Elheiga L, Matzuk MM, Abo-Hashema KA, Wakil SJ. Continuous fatty acid oxidation and reduced fat storage in mice lacking acetyl-CoA carboxylase 2. Science. 2001 Mar;291((5513)):2613–6. doi: 10.1126/science.1056843. [DOI] [PubMed] [Google Scholar]
- 12.Kobayashi MA, Watada H, Kawamori R, Maeda S. Overexpression of acetyl-coenzyme A carboxylase beta increases proinflammatory cytokines in cultured human renal proximal tubular epithelial cells. Clin Exp Nephrol. 2010 Aug;14((4)):315–24. doi: 10.1007/s10157-010-0296-x. [DOI] [PubMed] [Google Scholar]
- 13.Xu Y, Huang J, Xin W, Chen L, Zhao X, Lv Z, et al. Lipid accumulation is ahead of epithelial-to-mesenchymal transition and therapeutic intervention by acetyl-CoA carboxylase 2 silence in diabetic nephropathy. Metabolism. 2014 May;63((5)):716–26. doi: 10.1016/j.metabol.2014.02.010. [DOI] [PubMed] [Google Scholar]
- 14.Xin W, Zhao X, Liu L, Xu Y, Li Z, Chen L, et al. Acetyl-CoA carboxylase 2 suppression rescues human proximal tubular cells from palmitic acid induced lipotoxicity via autophagy. Biochem Biophys Res Commun. 2015 Jul;463((3)):364–9. doi: 10.1016/j.bbrc.2015.05.070. [DOI] [PubMed] [Google Scholar]
- 15.Fisman EZ, Tenenbaum A. Adiponectin: a manifold therapeutic target for metabolic syndrome, diabetes, and coronary disease? Cardiovasc Diabetol. 2014 Jun;13((1)):103. doi: 10.1186/1475-2840-13-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu M, Liu F. Regulation of adiponectin multimerization, signaling and function. Best Pract Res Clin Endocrinol Metab. 2014 Jan;28((1)):25–31. doi: 10.1016/j.beem.2013.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jorsal A, Tarnow L, Frystyk J, Lajer M, Flyvbjerg A, Parving HH, et al. Serum adiponectin predicts all-cause mortality and end stage renal disease in patients with type I diabetes and diabetic nephropathy. Kidney Int. 2008 Sep;74((5)):649–54. doi: 10.1038/ki.2008.201. [DOI] [PubMed] [Google Scholar]
- 18.Chung HF, Long KZ, Hsu CC, Mamun AA, Chiu YF, Tu HP, et al. Adiponectin gene (ADIPOQ) polymorphisms correlate with the progression of nephropathy in Taiwanese male patients with type 2 diabetes. Diabetes Res Clin Pract. 2014 Aug;105((2)):261–70. doi: 10.1016/j.diabres.2014.04.015. [DOI] [PubMed] [Google Scholar]
- 19.Choe EY, Wang HJ, Kwon O, Kim KJ, Kim BS, Lee BW, et al. Variants of the adiponectin gene and diabetic microvascular complications in patients with type 2 diabetes. Metabolism. 2013 May;62((5)):677–85. doi: 10.1016/j.metabol.2012.11.005. [DOI] [PubMed] [Google Scholar]
- 20.El-Shal AS, Zidan HE, Rashad NM. Adiponectin gene polymorphisms in Egyptian type 2 diabetes mellitus patients with and without diabetic nephropathy. Mol Biol Rep. 2014;41((4)):2287–98. doi: 10.1007/s11033-014-3082-0. [DOI] [PubMed] [Google Scholar]
- 21.Fang F, Bae EH, Hu A, Liu GC, Zhou X, Williams V, et al. Deletion of the gene for adiponectin accelerates diabetic nephropathy in the Ins2 (+/C96Y) mouse. Diabetologia. 2015 Jul;58((7)):1668–78. doi: 10.1007/s00125-015-3605-9. [DOI] [PubMed] [Google Scholar]
- 22.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010 May;42((5)):376–84. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Iwata M, Maeda S, Kamura Y, Takano A, Kato H, Murakami S, et al. Genetic risk score constructed using 14 susceptibility alleles for type 2 diabetes is associated with the early onset of diabetes and may predict the future requirement of insulin injections among Japanese individuals. Diabetes Care. 2012 Aug;35((8)):1763–70. doi: 10.2337/dc11-2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agius L. Hormonal and metabolite regulation of hepatic glucokinase. Annu Rev Nutr. 2016 Jul;36((1)):389–415. doi: 10.1146/annurev-nutr-071715-051145. [DOI] [PubMed] [Google Scholar]
- 25.Deshmukh HA, Palmer CN, Morris AD, Colhoun HM. Investigation of known estimated glomerular filtration rate loci in patients with type 2 diabetes. Diabet Med. 2013 Oct;30((10)):1230–5. doi: 10.1111/dme.12211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan D, Wang J, Jiang F, Zhang R, Sun X, Wang T, et al. Association between serum uric acid related genetic loci and diabetic kidney disease in the Chinese type 2 diabetes patients. J Diabetes Complications. 2016 Jul;30((5)):798–802. doi: 10.1016/j.jdiacomp.2016.02.018. [DOI] [PubMed] [Google Scholar]
- 27.Jainandunsing S, Koole HR, van Miert JN, Rietveld T, Wattimena JL, Sijbrands EJ, et al. Transcription factor 7-like 2 gene links increased in vivo insulin synthesis to type 2 diabetes. EBioMedicine. 2018 Apr;30:295–302. doi: 10.1016/j.ebiom.2018.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poy F, Lepourcelet M, Shivdasani RA, Eck MJ. Structure of a human Tcf4-beta-catenin complex. Nat Struct Biol. 2001 Dec;8((12)):1053–7. doi: 10.1038/nsb720. [DOI] [PubMed] [Google Scholar]
- 29.Wu LS, Hsieh CH, Pei D, Hung YJ, Kuo SW, Lin E. Association and interaction analyses of genetic variants in ADIPOQ, ENPP1, GHSR, PPARgamma and TCF7L2 genes for diabetic nephropathy in a Taiwanese population with type 2 diabetes. Nephrol Dial Transplant. 2009 Nov;24((11)):3360–6. doi: 10.1093/ndt/gfp271. [DOI] [PubMed] [Google Scholar]
- 30.Fu LL, Lin Y, Yang ZL, Yin YB. [Association analysis of genetic polymorphisms of TCF7L2, CDKAL1, SLC30A8, HHEX genes and microvascular complications of type 2 diabetes mellitus] Zhonghua Yi Xue Yi Chuan Xue Za Zhi. 2012 Apr;29((2)):194–9. doi: 10.3760/cma.j.issn.1003-9406.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 31.Buraczynska M, Zukowski P, Ksiazek P, Kuczmaszewska A, Janicka J, Zaluska W. Transcription factor 7-like 2 (TCF7L2) gene polymorphism and clinical phenotype in end-stage renal disease patients. Mol Biol Rep. 2014 Jun;41((6)):4063–8. doi: 10.1007/s11033-014-3275-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hussain H, Ramachandran V, Ravi S, Sajan T, Ehambaram K, Gurramkonda VB, et al. TCF7L2 rs7903146 polymorphism and diabetic nephropathy association is not independent of type 2 diabetes—a study in a south Indian population and meta-analysis. Endokrynol Pol. 2014;65((4)):298–305. doi: 10.5603/EP.2014.0041. [DOI] [PubMed] [Google Scholar]
- 33.Fan Z, Cai Q, Chen Y, Meng X, Cao F, Zheng S, et al. Association of the Transcription Factor 7 Like 2 (TCF7L2) Polymorphism With Diabetic Nephropathy Risk: A Meta-Analysis. Medicine (Baltimore) 2016 Mar;95((11)):e3087. doi: 10.1097/MD.0000000000003087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Araoka T, Abe H, Tominaga T, Mima A, Matsubara T, Murakami T, et al. Transcription factor 7-like 2 (TCF7L2) regulates activin receptor-like kinase 1 (ALK1)/Smad1 pathway for development of diabetic nephropathy. Mol Cells. 2010 Sep;30((3)):209–18. doi: 10.1007/s10059-010-0109-9. [DOI] [PubMed] [Google Scholar]
- 35.Coronado Daza J, Martí-Carvajal AJ, Ariza García A, Rodelo Ceballos J, Yomayusa González N, Páez-Canro C, et al. Early versus delayed erythropoietin for the anaemia of end-stage kidney disease. Cochrane Database Syst Rev. 2015 Dec;((12)):CD011122. doi: 10.1002/14651858.CD011122.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bunn HF. Erythropoietin. Cold Spring Harb Perspect Med. 2013 Mar;3((3)):a011619. doi: 10.1101/cshperspect.a011619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tong Z, Yang Z, Patel S, Chen H, Gibbs D, Yang X, et al. Genetics of Diabetes and Diabetic Complication Study Group Promoter polymorphism of the erythropoietin gene in severe diabetic eye and kidney complications. Proc Natl Acad Sci USA. 2008 May;105((19)):6998–7003. doi: 10.1073/pnas.0800454105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mooyaart AL, Valk EJ, van Es LA, Bruijn JA, de Heer E, Freedman BI, et al. Genetic associations in diabetic nephropathy: a meta-analysis. Diabetologia. 2011 Mar;54((3)):544–53. doi: 10.1007/s00125-010-1996-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams WW, Salem RM, McKnight AJ, Sandholm N, Forsblom C, Taylor A, et al. GENIE Consortium Association testing of previously reported variants in a large case-control meta-analysis of diabetic nephropathy. Diabetes. 2012 Aug;61((8)):2187–94. doi: 10.2337/db11-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sivaskandarajah GA, Jeansson M, Maezawa Y, Eremina V, Baelde HJ, Quaggin SE. Vegfa protects the glomerular microvasculature in diabetes. Diabetes. 2012 Nov;61((11)):2958–66. doi: 10.2337/db11-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang B, Cross DF, Ollerenshaw M, Millward BA, Demaine AG. Polymorphisms of the vascular endothelial growth factor and susceptibility to diabetic microvascular complications in patients with type 1 diabetes mellitus. J Diabetes Complications. 2003 Jan-Feb;17((1)):1–6. doi: 10.1016/s1056-8727(02)00181-2. [DOI] [PubMed] [Google Scholar]
- 42.Nazir N, Siddiqui K, Al-Qasim S, Al-Naqeb D. Meta-analysis of diabetic nephropathy associated genetic variants in inflammation and angiogenesis involved in different biochemical pathways. BMC Med Genet. 2014 Oct;15((1)):103. doi: 10.1186/s12881-014-0103-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Khalili H, Sull A, Sarin S, Boivin FJ, Halabi R, Svajger B, et al. Developmental origins for kidney disease due to shroom3 deficiency. J Am Soc Nephrol. 2016 Oct;27((10)):2965–73. doi: 10.1681/ASN.2015060621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, et al. Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet. 2009 Jun;41((6)):712–7. doi: 10.1038/ng.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park D, Tosello-Trampont AC, Elliott MR, Lu M, Haney LB, Ma Z, et al. BAI1 is an engulfment receptor for apoptotic cells upstream of the ELMO/Dock180/Rac module. Nature. 2007 Nov;450((7168)):430–4. doi: 10.1038/nature06329. [DOI] [PubMed] [Google Scholar]
- 46.Shimazaki A, Kawamura Y, Kanazawa A, Sekine A, Saito S, Tsunoda T, et al. Genetic variations in the gene encoding ELMO1 are associated with susceptibility to diabetic nephropathy. Diabetes. 2005 Apr;54((4)):1171–8. doi: 10.2337/diabetes.54.4.1171. [DOI] [PubMed] [Google Scholar]
- 47.Leak TS, Perlegas PS, Smith SG, Keene KL, Hicks PJ, Langefeld CD, et al. Variants in intron 13 of the ELMO1 gene are associated with diabetic nephropathy in African Americans. Ann Hum Genet. 2009 Mar;73((2)):152–9. doi: 10.1111/j.1469-1809.2008.00498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hanson RL, Millis MP, Young NJ, Kobes S, Nelson RG, Knowler WC, et al. ELMO1 variants and susceptibility to diabetic nephropathy in American Indians. Mol Genet Metab. 2010 Dec;101((4)):383–90. doi: 10.1016/j.ymgme.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu HY, Wang Y, Chen M, Zhang X, Wang D, Pan Y, et al. Association of ELMO1 gene polymorphisms with diabetic nephropathy in Chinese population. J Endocrinol Invest. 2013 May;36((5)):298–302. doi: 10.3275/8525. [DOI] [PubMed] [Google Scholar]
- 50.Hathaway CK, Chang AS, Grant R, Kim HS, Madden VJ, Bagnell CR, Jr, et al. High Elmo1 expression aggravates and low Elmo1 expression prevents diabetic nephropathy. Proc Natl Acad Sci USA. 2016 Feb;113((8)):2218–22. doi: 10.1073/pnas.1600511113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang C, Sorokin A. Upregulation of fibronectin expression by COX-2 is mediated by interaction with ELMO1. Cell Signal. 2011 Jan;23((1)):99–104. doi: 10.1016/j.cellsig.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharma KR, Heckler K, Stoll SJ, Hillebrands JL, Kynast K, Herpel E, et al. ELMO1 protects renal structure and ultrafiltration in kidney development and under diabetic conditions. Sci Rep. 2016 Nov;6((1)):37172. doi: 10.1038/srep37172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou TB, Jiang ZP, Qin YH, Drummen GP. Association of transforming growth factor-β1 T869C gene polymorphism with diabetic nephropathy risk. Nephrology (Carlton) 2014 Feb;19((2)):107–15. doi: 10.1111/nep.12176. [DOI] [PubMed] [Google Scholar]
- 54.Barnette DN, Hulin A, Ahmed AS, Colige AC, Azhar M, Lincoln J. Tgfβ-Smad and MAPK signaling mediate scleraxis and proteoglycan expression in heart valves. J Mol Cell Cardiol. 2013 Dec;65:137–46. doi: 10.1016/j.yjmcc.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mou X, Liu Y, Zhou D, Hu Y, Ma G, Shou C, et al. Different risk indictors of diabetic nephropathy in transforming growth factor-beta1 T869C CC/CT genotype and TT genotype. Iran J Public Health. 2016 Jun;45((6)):761–7. [PMC free article] [PubMed] [Google Scholar]
- 56.El-Sherbini SM, Shahen SM, Mosaad YM, Abdelgawad MS, Talaat RM. Gene polymorphism of transforming growth factor-β1 in Egyptian patients with type 2 diabetes and diabetic nephropathy. Acta Biochim Biophys Sin (Shanghai) 2013 Apr;45((4)):330–8. doi: 10.1093/abbs/gmt003. [DOI] [PubMed] [Google Scholar]
- 57.Fujii D, Brissenden JE, Derynck R, Francke U. Transforming growth factor beta gene maps to human chromosome 19 long arm and to mouse chromosome 7. Somat Cell Mol Genet. 1986 May;12((3)):281–8. doi: 10.1007/BF01570787. [DOI] [PubMed] [Google Scholar]
- 58.Valladares-Salgado A, Angeles-Martínez J, Rosas M, García-Mena J, Utrera-Barillas D, Gómez-Díaz R, et al. Association of polymorphisms within the transforming growth factor-β1 gene with diabetic nephropathy and serum cholesterol and triglyceride concentrations. Nephrology (Carlton) 2010 Sep;15((6)):644–8. doi: 10.1111/j.1440-1797.2010.01302.x. [DOI] [PubMed] [Google Scholar]
- 59.Jia H, Yu L, Gao B, Ji Q. Association between the T869C polymorphism of transforming growth factor-beta 1 and diabetic nephropathy: a meta-analysis. Endocrine. 2011 Dec;40((3)):372–8. doi: 10.1007/s12020-011-9503-0. [DOI] [PubMed] [Google Scholar]
- 60.Ahluwalia TS, Khullar M, Ahuja M, Kohli HS, Bhansali A, Mohan V, et al. Common variants of inflammatory cytokine genes are associated with risk of nephropathy in type 2 diabetes among Asian Indians. PLoS One. 2009;4((4)):e5168. doi: 10.1371/journal.pone.0005168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodrigues KF, Pietrani NT, Sandrim VC, Vieira CM, Fernandes AP, Bosco AA, et al. Association of a large panel of cytokine gene polymorphisms with complications and comorbidities in type 2 diabetes patients. J Diabetes Res. 2015;2015:605965. doi: 10.1155/2015/605965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang J, Guan YL, Xiao Y, Zhang XW. A meta-analysis of the association of G915C, G800A, C509T gene polymorphism of transforming growth factor-β1 with diabetic nephropathy risk. Ren Fail. 2014 Mar;36((2)):321–6. doi: 10.3109/0886022X.2013.832320. [DOI] [PubMed] [Google Scholar]
- 63.Wang Y, Peng W, Zhang X, Qiao H, Wang L, Xu Z, et al. The association of ACE gene polymorphism with diabetic kidney disease and renoprotective efficacy of valsartan. J Renin Angiotensin Aldosterone Syst. 2016 Sep;17((3)):17. doi: 10.1177/1470320316666749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boright AP, Paterson AD, Mirea L, Bull SB, Mowjoodi A, Scherer SW, et al. DCCT/EDIC Research Group Genetic variation at the ACE gene is associated with persistent microalbuminuria and severe nephropathy in type 1 diabetes: the DCCT/EDIC Genetics Study. Diabetes. 2005 Apr;54((4)):1238–44. doi: 10.2337/diabetes.54.4.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hadjadj S, Fumeron F, Roussel R, Saulnier PJ, Gallois Y, Ankotche A, et al. DIABHYCAR Study Group DIAB2NEPHROGENE Study Group SURDIAGENE Study Group Prognostic value of the insertion/deletion polymorphism of the ACE gene in type 2 diabetic subjects: results from the Non-insulin-dependent Diabetes, Hypertension, Microalbuminuria or Proteinuria, Cardiovascular Events, and Ramipril (DIABHYCAR), Diabete de type 2, Nephropathie et Genetique (DIAB2NEPHROGENE), and Survie, Diabete de type 2 et Genetique (SURDIAGENE) studies. Diabetes Care. 2008 Sep;31((9)):1847–52. doi: 10.2337/dc07-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ng, DP: Prognostic value of the insertion/deletion polymorphism of the ACE gene in type 2 diabetic subjects: Results from the Non-Insulin-Dependent Diabetes, Hypertension, Microalbuminuria or Proteinuria, Cardiovascular events, and Ramipril (DIABHYCAR), Diabete de type 2, Nephropathie et Genetique (DIAB2NEPHROGENE), and Survie, Diabete de type 2 et Genetique (SURDIAGENE) studies: Response to Hadjadj et al Diabetes Care. 2008;31:e78–e79. doi: 10.2337/dc08-1091. [DOI] [PubMed] [Google Scholar]
- 67.Ahmad N, Jamal R, Shah SA, Gafor AH, Murad NA. Renin-angiotensin-aldosterone system gene polymorphisms and type 2 diabetic nephropathy in asian populations: an updated meta-analysis. Curr Diabetes Rev. 2018 Jul;14 doi: 10.2174/1573399814666180709100411. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 68.Felehgari V, Rahimi Z, Mozafari H, Vaisi-Raygani A. ACE gene polymorphism and serum ACE activity in Iranians type II diabetic patients with macroalbuminuria. Mol Cell Biochem. 2011 Jan;346((1–2)):23–30. doi: 10.1007/s11010-010-0587-2. [DOI] [PubMed] [Google Scholar]
- 69.Andersen S, Tarnow L, Cambien F, Rossing P, Juhl TR, Deinum J, et al. Long-term renoprotective effects of losartan in diabetic nephropathy: interaction with ACE insertion/deletion genotype? Diabetes Care. 2003 May;26((5)):1501–6. doi: 10.2337/diacare.26.5.1501. [DOI] [PubMed] [Google Scholar]
- 70.Lu M, Zhang J, Li M, Ge X, Dai X, Zhao J, et al. The angiotensin-I converting enzyme gene I/D variation contributes to end-stage renal disease risk in Chinese patients with type 2 diabetes receiving hemodialysis. Mol Cell Biochem. 2016 Nov;422((1–2)):181–8. doi: 10.1007/s11010-016-2819-6. [DOI] [PubMed] [Google Scholar]
- 71.Brenner BM, Cooper ME, de Zeeuw D, Keane WF, Mitch WE, Parving HH, et al. RENAAL Study Investigators Effects of losartan on renal and cardiovascular outcomes in patients with type 2 diabetes and nephropathy. N Engl J Med. 2001 Sep;345((12)):861–9. doi: 10.1056/NEJMoa011161. [DOI] [PubMed] [Google Scholar]
- 72.Möllsten A, Kockum I, Svensson M, Rudberg S, Ugarph-Morawski A, Brismar K, et al. The effect of polymorphisms in the renin-angiotensin-aldosterone system on diabetic nephropathy risk. J Diabetes Complications. 2008 Nov-Dec;22((6)):377–83. doi: 10.1016/j.jdiacomp.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 73.Möllsten A, Vionnet N, Forsblom C, Parkkonen M, Tarnow L, Hadjadj S, et al. A polymorphism in the angiotensin II type 1 receptor gene has different effects on the risk of diabetic nephropathy in men and women. Mol Genet Metab. 2011 May;103((1)):66–70. doi: 10.1016/j.ymgme.2011.01.004. [DOI] [PubMed] [Google Scholar]
- 74.Currie D, McKnight AJ, Patterson CC, Sadlier DM, Maxwell AP, UK Warren 3/GoKinD Study Group Investigation of ACE, ACE2 and AGTR1 genes for association with nephropathy in Type 1 diabetes mellitus. Diabet Med. 2010 Oct;27((10)):1188–94. doi: 10.1111/j.1464-5491.2010.03097.x. [DOI] [PubMed] [Google Scholar]
- 75.Ding W, Wang F, Fang Q, Zhang M, Chen J, Gu Y. Association between two genetic polymorphisms of the renin-angiotensin-aldosterone system and diabetic nephropathy: a meta-analysis. Mol Biol Rep. 2012 Feb;39((2)):1293–303. doi: 10.1007/s11033-011-0862-7. [DOI] [PubMed] [Google Scholar]
- 76.Zhang R, Zhuang L, Li M, Zhang J, Zhao W, Ge X, et al. Arg913Gln of SLC12A3 gene promotes development and progression of end-stage renal disease in Chinese type 2 diabetes mellitus. Mol Cell Biochem. 2018 Jan;437((1–2)):203–10. doi: 10.1007/s11010-017-3120-z. [DOI] [PubMed] [Google Scholar]
- 77.Abu Seman N, He B, Ojala JR, Wan Mohamud WN, Östenson CG, Brismar K, et al. Genetic and biological effects of sodium-chloride cotransporter (SLC12A3) in diabetic nephropathy. Am J Nephrol. 2014;40((5)):408–16. doi: 10.1159/000368916. [DOI] [PubMed] [Google Scholar]
- 78.Nishiyama K, Tanaka Y, Nakajima K, Mokubo A, Atsumi Y, Matsuoka K, et al. Polymorphism of the solute carrier family 12 (sodium/chloride transporters) member 3, SLC12A3, gene at exon 23 (+78G/A: Arg913Gln) is associated with elevation of urinary albumin excretion in Japanese patients with type 2 diabetes: a 10-year longitudinal study. Diabetologia. 2005 Jul;48((7)):1335–8. doi: 10.1007/s00125-005-1785-4. [DOI] [PubMed] [Google Scholar]
- 79.Kim JH, Shin HD, Park BL, Moon MK, Cho YM, Hwang YH, et al. SLC12A3 (solute carrier family 12 member [sodium/chloride] 3) polymorphisms are associated with end-stage renal disease in diabetic nephropathy. Diabetes. 2006 Mar;55((3)):843–8. doi: 10.2337/diabetes.55.03.06.db05-1013. [DOI] [PubMed] [Google Scholar]
- 80.Ng DP, Nurbaya S, Choo S, Koh D, Chia KS, Krolewski AS. Genetic variation at the SLC12A3 locus is unlikely to explain risk for advanced diabetic nephropathy in Caucasians with type 2 diabetes. Nephrol Dial Transplant. 2008 Jul;23((7)):2260–4. doi: 10.1093/ndt/gfm946. [DOI] [PubMed] [Google Scholar]
- 81.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, et al. DCCT/EDIC Research Group Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes. 2009 Jun;58((6)):1403–10. doi: 10.2337/db08-1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, et al. DCCT/EDIC Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes. 2016 Mar;65((3)):803–17. doi: 10.2337/db15-1313. [DOI] [PMC free article] [PubMed] [Google Scholar]