Abstract
The Arabidopsis PIF4 and BES1/BZR1 transcription factors antagonize light signaling by facilitating co‐activated expression of a large number of cell wall‐loosening and auxin‐related genes. While PIF4 directly activates expression of these targets, BES1 and BZR1 activity switch from a repressive to an activator function, depending on interaction with TOPLESS and other families of regulators including PIFs. However, the complexity of this regulation and its role in diurnal control of plant growth and brassinosteroid (BR) levels is little understood. We show by using a protein array that BES1, PIF4, and the BES1‐PIF4 complex recognize different DNA elements, thus revealing a distinctive cis‐regulatory code beneath BES1‐repressive and PIF4 co‐activation function. BES1 homodimers bind to conserved BRRE‐ and G‐box elements in the BR biosynthetic promoters and inhibit their expression during the day, while elevated PIF4 competes for BES1 homodimer formation, resulting in de‐repressed BR biosynthesis at dawn and in response to warmth. Our findings demonstrate a central role of PIF4 in BR synthesis activation, increased BR levels being essential to thermomorphogenic hypocotyl growth.
Keywords: Arabidopsis, BES1, BR biosynthesis, PIF4, thermomorphogenesis
Subject Categories: Plant Biology, Signal Transduction, Transcription
Introduction
The plant steroid hormones brassinosteroids (BRs) act antagonistically to light in the regulation of hypocotyl elongation (Li et al, 1996; Szekeres et al, 1996). Defects in BR synthesis or BR signaling result in de‐etiolated growth in the dark, severe dwarfism, delayed senesce, and increased stress tolerance, highlighting a central role of these hormones in the control of plant growth and development (Chory et al, 1991; Clouse et al, 1996; Li et al, 1996; Szekeres et al, 1996). In a screen for mutants impaired in thermomorphogenic growth, independent mutations in BR biosynthetic genes have also been identified, indicating that enhanced hypocotyl growth in response to elevated temperatures relies on increased BR levels (Gray et al, 1998; Ibanez et al, 2018). The mechanism by which external light and temperature signals are integrated into BR signaling is, however, not fully understood. BRs are perceived by the transmembrane receptor kinase BRASSINOSTEROID INSENSITIVE 1 (BRI1), which binds these hormones via a conserved island region in the LRR extracellular domain (Kinoshita et al, 2005). Binding of BRs releases BRI1 from inhibition by BRI1 KINASE INHIBITOR 1 (BKI1) and enables BRI1 to associate with the BAK1 (BRI1 ASSOCIATED PROTEIN KINASE 1) co‐receptor (Li & Nam, 2002). Trans‐phosphorylation of these receptor kinases activates the BRI1 cytosolic kinase domain and triggers a phosphorylation cascade that leads to activation of the BRI1 SUPPRESSOR 1 (BSU1) phosphatase (Mora‐Garcia et al, 2004; Kim et al, 2011). Activated BSU1 is then responsible of dephosphorylating Y200 of BR‐INSENSITIVE 2 (BIN2) and inactivates this negative signaling GSK3 kinase (Kim et al, 2011), which allows nuclear accumulation of the non‐phosphorylated BES1 (BRI1‐EMS‐SUPPRESSOR 1) and BZR1 (BRASSINAZOLE RESISTANT 1) factors, acting as key regulators of BR‐regulated gene expression (He et al, 2005; Yin et al, 2005; Sun et al, 2010; Yu et al, 2011).
BES1 and BZR1 are transcriptional repressors, but they activate gene expression in concert with other transcriptional regulators (Yin et al, 2005; Sun et al, 2010). Genome‐wide analyses showed that BES1/BZR1 up‐ or down‐regulate thousands of BR‐responsive genes, although the exact mechanism for this dual regulatory function is not fully understood. In the context of down‐regulated genes (such as those for BR biosynthetic enzymes), it has been proposed that BES1 and BZR1 bind as monomers to conserved BRRE‐elements (Sun et al, 2010; Wang et al, 2012) and recruit the co‐repressor TPL to suppress their expression (Oh et al, 2014; Ryu et al, 2014; Espinosa‐Ruiz et al, 2017). In contrast, BZR1 and BES1 are thought to induce gene expression through their binding to G‐ and E‐box cis‐elements. Recognition of these elements is accepted to be mediated through heterodimerization with PIF4, to co‐activate together multiple elongation‐related targets (Sun et al, 2010; Oh et al, 2012). DNA binding specificity of the monomeric BES1 and BZR1 proteins, and the BES1‐PIF4 complex is however not well established, as it is largely unknown how interaction with PIF4 is dynamically regulated.
Here, we analyzed DNA recognition by BES1, PIF4, and the BES1‐PIF4 complex and show that these proteins bind different cis‐elements. We show that BES1 recognizes both BRRE‐ and G‐box elements as homodimer but not in a monomeric form. Moreover, interaction with PIF4 changes BES1 DNA binding specificity to recognition of a CATGTG element (PBE or PIF‐binding element), which is enriched in the PIF + BES‐UP target promoters. Notably, BR‐ biosynthesis genes and PIF + BES‐activated targets display similar peak expression levels at dawn, coinciding with maximal activity of the PIF4 factor (Nozue et al, 2007). This suggests a role of PIF4 in modulating BES1‐repressive activity, by competing for BES1 homodimer formation. Consistent with this function, we observed that expression of BR biosynthetic genes is strongly induced in PIF4 over‐expressor lines, while it is suppressed in pifq mutants. PIF4 and PIF5 accumulate under warm temperatures (Koini et al, 2009; Foreman et al, 2011), and we show that the DWF4, CPD, and BR6ox2 genes are up‐regulated at 28°C in wild‐type plants but not in pifq mutants. Together, our findings reveal a critical role of PIF4 in increasing BR levels in response to warm temperatures, and we demonstrate that this regulation is essential for thermomorphogenic growth.
Results
Association with PIF4 modifies BES1 DNA binding specificity
To investigate the molecular mechanisms that dictate the switch of BES1 activity from a repressive to co‐activator function, we performed protein binding microarray (PBM) studies (Godoy et al, 2011) to define the DNA recognition motifs of BES1, PIF4, and PIF4‐BES1 proteins. To this end, we expressed the PIF4‐His and BES1‐MBP‐tagged constructs in Escherichia coli and used the affinity‐purified proteins for PBM hybridization followed by detection with anti‐His or anti‐MBP antibodies. To obtain the PIF4‐BES1 complex, both PIF4‐His and BES1‐MBP proteins were co‐expressed from the same vector and purified through a His‐affinity matrix. This protein fraction was then used for PBM hybridization and detection with an anti‐MBP antibody, to secure that obtained signals did correspond to the BES1‐MBP/PIF4‐His heterodimer. BES1‐MBP was found in these studies to bind with high affinities a 5′‐CACGTG‐3′ (G‐box) DNA motif and the 5′‐CGTGTG‐3′ and 5′‐CGTGCG‐3′ (BRRE‐box) elements, whereas PIF4‐His recognized both a G‐box and 5′‐CATGTG‐3′ (PBE‐box) motif (Fig 1A and B). More interestingly, the PIF4‐BES1 complex did not recognize the BRRE‐ cis‐elements, but displayed preferential binding to the PBE‐box variant (Fig 1A and B), previously described as a PIF‐binding element (Zhang et al, 2013; Pfeiffer et al, 2014). These results highlight that interaction with PIF4 alters BES1 DNA recognition and prevents this factor from binding to the BRRE‐repressive motifs, while it recruits the BES1 protein to PBE‐box target elements.
Figure 1. Complex formation with PIF4 modifies BES1 DNA binding specificity and its transcriptional function.
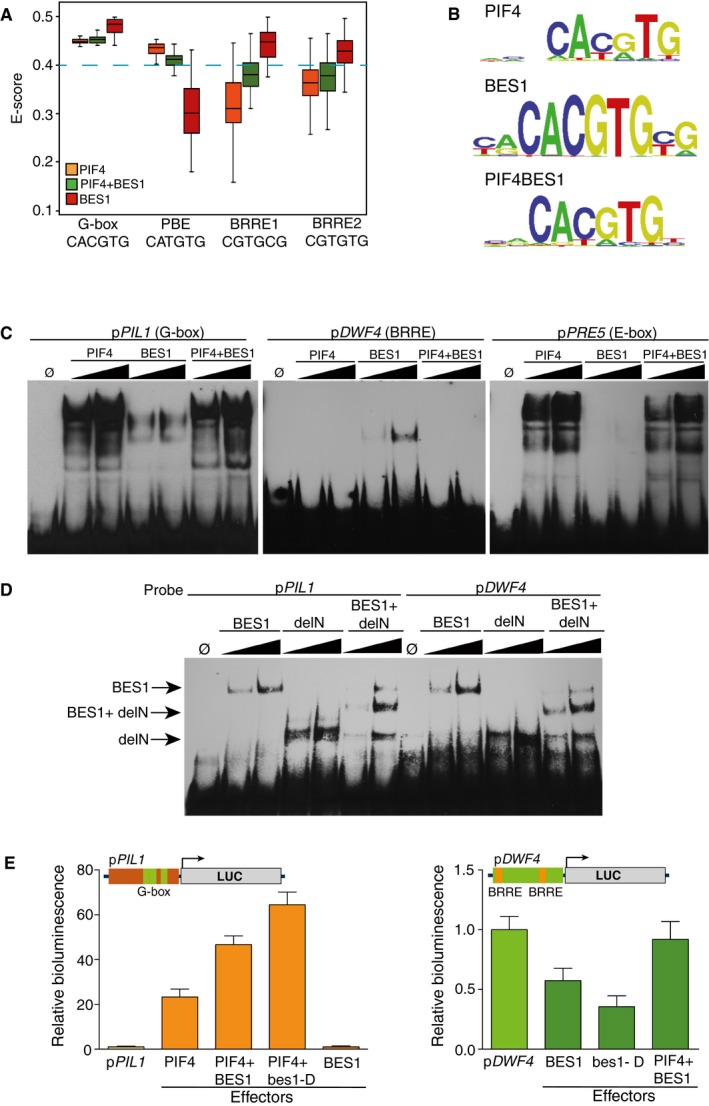
- Box plot showing the enrichment scores (E‐scores) of all possible 8‐mers containing the G‐box (CACGTG), PBE‐box (CATGTG), and BRRE‐elements (CGTGC/TG). Boxes represent the 25–75% quartiles and the black line the median of distribution. Bars indicate the 1–25% (above) and 75–100% (below) quartiles. E‐scores above 0.4 denote that binding of the proteins toward the indicated DNA element is statistically significant. Dashed blue line indicates the 0.4 threshold.
- Sequence logo representation of the top scoring 8‐mers obtained by hybridization with the PIF4, BES1, and PIF4‐BES1 proteins.
- Electrophoretic mobility shift assays (EMSA) showing interaction of the PIF4, BES1, and PIF4‐BES1 proteins with the conserved G‐box, PBE‐, and BRRE‐elements in the PIL1 (At2g46970), PRE5 (At3g28857), and DWF4 (At3g50660) promoters. Increasing amounts of protein were used for the assay.
- BES1 binds both BRRE‐ and G‐box elements as a homodimer. A deletion of BES1 (delN) fused to MBP (MBP‐delN) and the complete protein (MBP‐BES1) was co‐expressed in E. coli. Formation of intermediate mobility bands, indicative of DNA recognition by a dimeric form of the protein, was tested in EMSA assays. A signal corresponding to the dimeric full‐length MBP‐BES1 and MBP‐delN complex was detected with both DNA probes. Increasing amounts of the proteins were used in the assay.
- Co‐expression of the PIF4 and BES1 effector constructs leads to synergistic pPIL1 activation and reverses BES1‐dependent inhibition of the pDWF4 reporter. The pPIL1 and pDWF4 promoters including three G‐boxes (green boxes) and two BRRE‐elements (orange boxes) were fused to the firefly luciferase reporter gene (LUC) and co‐transfected with 35S::PIF4, 35S::BES1, and 35S::bes1‐D effector constructs into Nicotiana benthamiana leaves. Leaf disks were collected 48 h after infiltration, and luciferase activity was measured in a microplate luminometer. Error bars represent SD (n = 24).
To confirm these results, we performed electrophoretic mobility shift assays (EMSA) using DNA probes for the pPIL1, pPRE5, and pDWF4 promoters. These genes have been reported to be directly activated by PIF4 (PIL1) and the PIF4 + BES1 complex (PRE5) (Leivar & Quail, 2011; Hornitschek et al, 2012; Oh et al, 2012) or repressed by BES1/BZR1 (DWF4) (Kim et al, 2006) and show conserved G‐box (PIL1), PBE‐ (PRE5) and BRRE‐ (DWF4) motifs in their promoter regions (Fig 1C). Consistent with the PBM results, the PIF4, BES1, and PIF4 + BES1 proteins bound with comparable affinities to the G‐box element in the pPIL1 promoter. The PBE‐cis‐element in pPRE5 was bound by PIF4 and PIF4 + BES1, but it was not recognized by the BES1 factor. On the other side, the BRRE‐ binding site in the pDWF4 promoter was only recognized by BES1 (Fig 1C). Overall, these results confirm those obtained by PBM hybridization and demonstrate that DNA binding specificities of the PIF4 and BES1 factors differ from those of the PIF4 + BES1 complex, suggesting that they regulate a different set of genomic targets. Moreover, DNA affinity purification sequencing (DAP‐seq) studies with the BZR1 or BEH2‐BEH4 proteins identified an identical recognition motif as we report here for BES1 (O'Malley et al, 2016). Genomic regions bound by these factors are significantly enriched in the BRRE‐ and G‐box elements (Appendix Fig S1B and C), indicating that all these proteins recognize identical DNA motifs and likely have a redundant function. BES1 and BZR1 were observed in phylogenetic tree analyses to be more closely related than the rest of family members (Appendix Fig S1A). These factors share almost identical bHLH domains, and PIF4‐BZR1 is thereby predicted to bind the same PBE‐ motif as identified here for PIF4‐BES1.
BES1 and BZR1 were proposed to bind the BRRE‐binding motif as monomers, while recognizing the E‐box elements on heterodimerization with other TFs (Wang et al, 2012). To assess this possibility, a truncated form of BES1 (delN in Fig 1D) lacking the C‐terminal end was fused to the MBP‐tag and employed for EMSA studies. Both delN and full‐length BES1 proteins were co‐expressed in E. coli and subsequently analyzed with the pDWF4 (BRRE‐box) and pPIL1 (G‐box) probes for formation of intermediate mobility DNA complexes. As shown in Fig 1D, an intermediate band corresponding to the delN + BES1 dimeric protein was observed with both DNA probes. This indicates that BES1 binds both the BRRE‐ and G‐box as a dimer, instead of a monomeric form. This finding has important functional implications, as it suggests that complex formation with other factors will hamper BRRE‐ recognition, by interfering with BES1 homodimerization.
PIF4 interaction changes transcriptional activity of the BES1 factor
To examine how PIF4‐BES1 complex formation affects transcriptional outputs by these factors, the pPIL1::Luc and pDWF4::Luc reporter constructs were used in transient transactivation assays (Fig 1E). The two reporters were infiltrated in Nicotiana benthamiana leaves together with the 35S::PIF4, 35S::BES1, and the constitutively active 35S::bes1‐D (Yin et al, 2002) effector constructs, either singly or in combination. As expected, PIF4 activated the pPIL1::Luc reporter, and a synergistic activation of this construct was observed on co‐expression of PIF4 with the BES1 or bes1‐d proteins (Fig 1D). BES1 alone, however, did not activate this promoter, showing that BES1 is unable to up‐regulate the PIL1 gene by itself. The expression of the pDWF4::Luc reporter was suppressed by BES1, and more so by the constitutively active bes1‐d protein (Fig 1E). Remarkably, co‐expression of BES1 and PIF4 restored pDWF4::Luc expression, hence indicating that increased PIF4 levels reverse transcriptional repression by BES1. This suggests that PIF4 disrupts BES1 dimerization, thus competing for BES1 binding to its cognate BRRE‐ sites in the pDWF4 promoter.
Notably, in yeast two‐hybrid and bimolecular fluorescence complementation (BiFC) assays, we observed that interaction with PIF4 not only implicates the reported BES1 N‐terminal DNA‐binding domain (Oh et al, 2012), but also requires of the BES1 C‐terminal region (Appendix Fig S2A). This region includes the ETHYLENE RESPONSE FACTOR (ERF)‐associated amphiphilic repression (EAR) domain for TOPLESS (TPL) protein–protein interaction (Ryu et al, 2014) and a BIN2‐docking motif (Peng et al, 2010), with mutations within the EAR domain being established to inactivate BES1 function (Espinosa‐Ruiz et al, 2017). Thus, it is formally possible that mutations in the C‐terminal EAR lessen PIF4 binding affinity, while PIF4 complex formation hinders interaction with the co‐repressor TPL. As such, PIF4 physical contact with the BES1 C‐terminal region may be critical to reverse BES1‐repressive activity and to recruitment of this factor to the promoters of co‐activated loci.
The PBE‐cis‐element is overrepresented in the promoters of PIF4 and BES1 co‐activated targets
To further explore whether PBE‐elements are enriched in the promoters of BES1‐ and PIF4‐regulated loci, we conducted microarray studies on dark grown bes‐1D and bes1‐D;pif4pif5 plants. Differentially expressed genes (DEGs) obtained from these studies were combined with published datasets for BR‐regulated gene expression (Goda et al, 2004, 2008; Nemhauser et al, 2004, 2006; Vert et al, 2005) and with genes reported to be differentially expressed in bzr1‐1D and bri1‐116 mutants (Sun et al, 2010; Oh et al, 2012). This led to a total of 5,679 BR‐responsive genes equally distributed between UP‐ and DOWN‐regulated transcripts (Fig 2A; Appendix Table S1). A list of 5,324 PIF‐regulated genes was also obtained by merging our pif4pif5 data, with previous gene expression profiles of PIF4‐OX lines and pifq mutants (Leivar et al, 2009; Lorrain et al, 2009; Shin et al, 2009; Nozue et al, 2011; Hornitschek et al, 2012; Oh et al, 2012; Zhang et al, 2013; Appendix Table S2). As for BR‐responsive, these genes were evenly distributed among activated and repressed genes, and as expected, they showed a significant overlap with the BR‐ dataset, with 40% of the PIF‐activated genes being also induced by BRs, while 33% of the PIF‐repressed genes are BR‐repressed, versus to 9.4% or 8.9% expected randomly (Fig 2A).
Figure 2. The PBE‐cis‐element is overrepresented in the promoters of PIF + BES1 co‐activated targets.
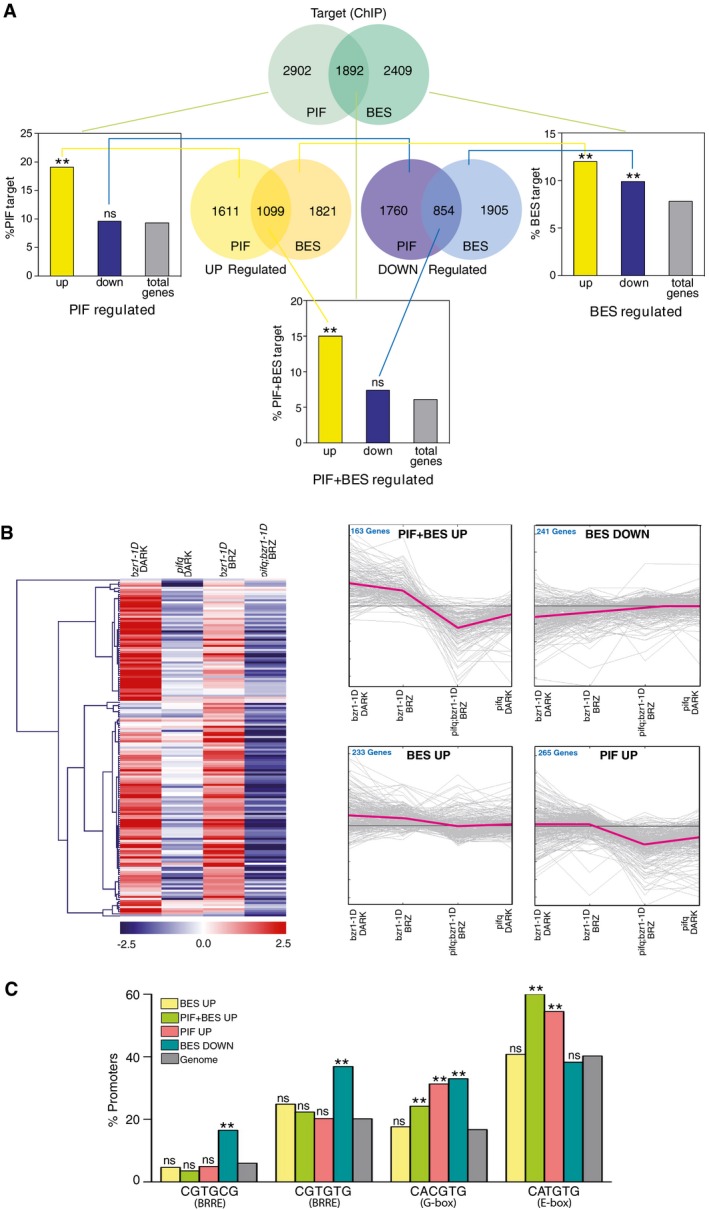
- Venn diagrams representing the genes up‐regulated (yellow) and down‐regulated (purple) by PIFs or BES1/BZR1 (BES), and the direct target loci (green) of these transcription factors. Gene sets derive from our bes1‐D and bes1‐D;pif4pif5 microarray analyses and from published RNA‐seq studies of pifq, bzr1‐1D, and bri1‐116 mutants (Appendix Tables S1 and S2). The direct target PIF and BES datasets were obtained from published ChIP‐chip or ChIP‐seq experiments with these factors (Appendix Tables S3 and S4). Histogram bars represent the percentage of PIF‐, BES‐, and PIF + BES‐regulated genes that are directly bound by these transcription factors, as compared to all the Arabidopsis genes (total genes, in gray) (chi‐squared test, **P‐value < 0.01, ns indicates no significant differences).
- Heat map and fold‐change graph representation of the genes identified in (A). Expression data were obtained from Genevestigator, and heat map and clustering analyses were performed using the MeV software. PIF + BES‐UP indicates genes up‐regulated by PIF and BES and identified as targets of both the PIF and BES factors. BES‐DOWN are BES‐repressed targets, while BES‐UP corresponds to BES‐induced targets. The PIF‐UP cluster includes PIF‐induced genes directly bound by the PIF factors.
- Percentages of occurrence of the indicated motifs in the PIF, BES, or PIF + BES target loci, as compared with all Arabidopsis genes (genome, in gray) (chi‐squared test, **P‐value < 0.01, ns indicates no significant differences).
We next gathered all genes that in ChIP‐chip or ChIP‐seq studies had been identified as directly targeted by PIFs or BES1/BZR1 (Sun et al, 2010; Yu et al, 2011; Hornitschek et al, 2012; Oh et al, 2012; Zhang et al, 2013), yielding a list of 4,301 BES1‐ and 4,794 PIF‐target loci (Appendix Tables S3 and S4). Overall, around 40% of the PIF‐ targets overlapped with genes directly regulated by BES1 or BZR1, in agreement with a coordinated function of these factors in gene regulation (Fig 2A). Moreover, by contrasting each the DEG and directly regulated datasets, we observed that PIF‐activated genes are significantly enriched among the PIF‐bound targets (19.1% of genes, versus 9.3% expected by chance, P‐value < 0.01), while a comparable enrichment was not observed for PIF‐repressed genes (9.6% of genes, in a similar range as expected randomly). This indicates that PIFs act mainly as transcriptional activators and that down‐regulated genes correspond to indirect targets of these factors (Fig 2A). As for BES1/BZR1 function, both induced and repressed genes were similarly overrepresented amid the BES1‐specific targets (12% of the induced and 9.9% of the repressed genes, versus 7.8% expected randomly), thus reflecting a dual role of these factors as transcriptional activators and repressors (Fig 2A). Finally, BES and PIF shared loci were exclusively enriched in genes with an up‐regulated pattern of expression (14.9% induced and 6% repressed genes, versus 5.8% randomly expected), consistent with the reported role of BES1/BZR1 in co‐activated expression of a subset of PIF targets (Oh et al, 2012).
These targets showed in a heat map to be differently misregulated in bzr1‐1d and pifq mutants (Fig 2B), hence identifying four main gene clusters that we designated as PIF + BES‐UP, PIF‐UP, BES‐UP, and BES‐DOWN. The PIF + BES‐UP cluster includes 163 genes, which are up‐regulated in bzr1‐1d seedlings, but whose expression is suppressed in bzr1‐1d;pifq and pifq mutants (Fig 2B). Moreover, bzr1‐1d activation of these genes is reduced by the BR biosynthesis inhibitor brassinazole (BRZ), which is shown to promote BIN2‐mediated phosphorylation and destabilization of the PIF4 factor (Bernardo‐Garcia et al, 2014). The PIF‐UP cluster includes 265 genes whose transcription is mostly induced by PIFs, since their expression is not affected in bzr1‐1D seedlings. Conversely, the BES‐UP and BES‐DOWN clusters contain 233 and 241 genes, which are respectively activated or repressed in bzr1‐1D plants, but whose expression is not affected in pifq mutants (Fig 2B).
We next interrogated the upstream regulatory regions of these target loci for the elements identified in the protein array. Remarkably, the 5′‐CGTGCG‐3′ and 5′‐CGTGTG‐3′ (BRRE) motifs were found to be overrepresented only in the promoters of genes in the BES‐DOWN cluster, consistent with the reported role of BES1 and BZR1 in negative BR feedback regulation, by binding to conserved BRRE‐cis‐elements in the BR biosynthetic promoters (Sun et al, 2010). The PBE 5′‐CATGTG‐3′ element was enriched in the PIF4‐UP and PIF + BES‐UP promoters, but not in the promoters of BES‐UP or BES‐DOWN genes, in agreement with our PBM and EMSA studies showing that BES1 does not recognize this cis‐element. Moreover, in line with the finding that PIF4, BES1, and PIF4‐BES1, bind all of them a 5′‐CACGTG‐3′ G‐box motif, this element was overrepresented in the promoters of PIF‐UP, PIF + BES‐UP, and BES‐DOWN genes (Fig 2C), while genes in the BES1‐UP cluster were not enriched in any of these elements. This suggests that BES1 activates these targets through physical interaction with other factors than PIFs and is recruited to a different DNA element.
Taken together, these data indicate that BES1/BZR1 repress BES‐DOWN targets by binding to G‐box and BRRE‐elements in their promoters, while PIF4 recruits these factors to a new cis‐element, the PBE (5′‐CATGTG‐3′) motif, to co‐activated expression of PIF + BES targets. Notably, PIF4 binds both G‐box and PBE‐elements in the absence of BES1, but interaction with BES1/BZR1 synergistically activates these genes. Also, PIF4 competes for BES1/BZR1 binding to the BRRE‐elements, which contributes to de‐repression of the BES‐DOWN targets, including BR biosynthetic genes. We propose that BES1/BZR1 inhibition of BR levels creates a negative feedback loop that restrains excessive PIFs activity. This way, dynamic changes in BES1 and PIF4 nuclear levels tightly control plant growth, combinatorial function of these factors providing a highly sensitive mechanism for rapid modulation of cell elongation in response to environmental cues.
Light stabilizes the BES1 protein
Constitutively active bes1‐d and bzr1‐1d proteins display significantly increased stability, which suggests that regulation of BES1 and BZR1 nuclear accumulation is critical to transcriptional control by these factors (Wang et al, 2002; Yin et al, 2002). The GSK3 kinase BRASSINOSTEROID INSENSITIVE 2 (BIN2) phosphorylates and inactivates BES1 and BZR1, in addition to promote proteasomal degradation of these factors (He et al, 2002; Vert & Chory, 2006; Wang et al, 2013a). Although it is largely accepted that BIN2‐dependent phosphorylation is the principal mechanism that controls BES1/BZR1 stability, other studies demonstrated that these factors are regulated in response to cues other than BR signaling (Wang et al, 2013b; Kim et al, 2014). Here, we used pBES1::BES1‐GFP lines to analyze the pattern of BES1 protein accumulation in short days (Fig 3A). Time course studies revealed that levels of the BES1 protein increase directly after dawn and remain high for the first 2–4 light hours, to be later reduced to lower levels as observed at night (Fig 3A). A band corresponding to the phosphorylated BES1 protein was visible across the whole 24‐h interval, whereas non‐phosphorylated BES1 was observed primarily after dawn (Fig 3A). To assess whether BES1 protein accumulation mirrored its expression profile, we performed qRT–PCR analyses of the BES1 and BZR1 transcripts in wild‐type plants. As seen in Fig 3B, elevated levels of these transcripts were detected at night, whereas their expression was reduced during the day, reaching basal levels by ZT6. BES1 was also expressed to considerably higher levels than BZR1, inconsistency between protein accumulation and transcript levels suggesting that BES1 is under post‐translational control (Fig 3A and B). To further test this hypothesis, we analyzed levels of the protein in 35S::BES1‐GFP lines. Notably, a higher accumulation of the BES1‐GFP protein is still observed in these plants during the day (Fig 3C), although transcript levels were slightly elevated at night (Appendix Fig S5D).
Figure 3. The BES1 protein is stabilized in the light.
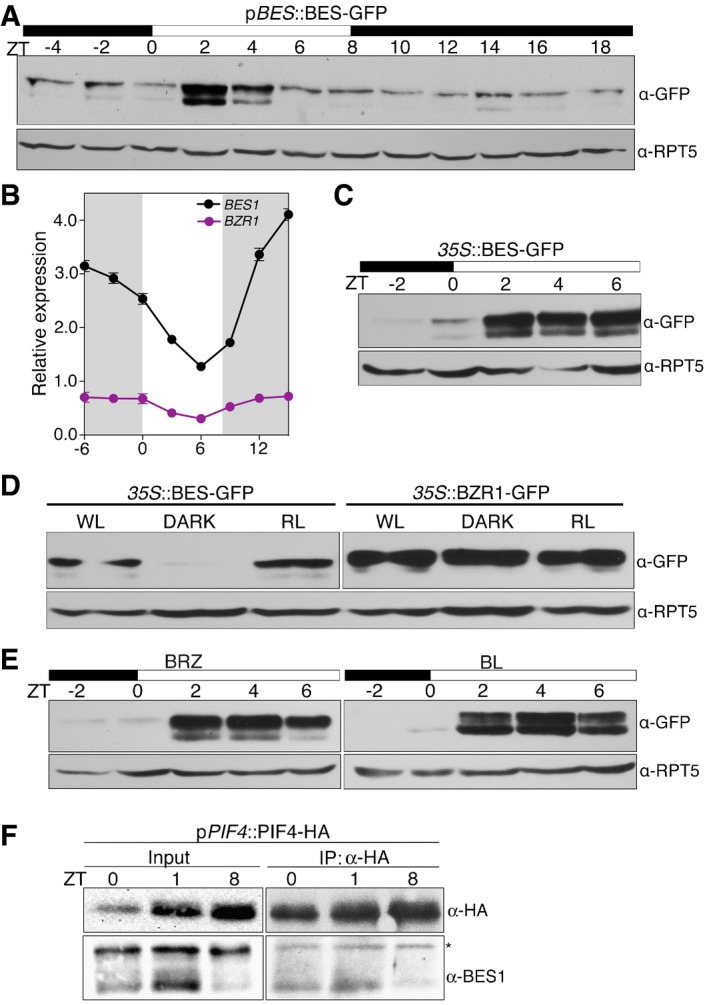
- Time course analysis of BES1 protein accumulation levels. pBES1::BES1‐GFP seedlings were grown under short days for 6 days, and samples were harvested each 2 h for a 24‐h interval. Total protein extracts were used for Western blot analysis with an anti‐GFP antibody to detect the BES1‐GFP protein. Hybridization with anti‐RPT5 antibody is included as a loading control. Dark and light periods are represented as black and white bars, respectively. The experiment was repeated twice with similar results.
- Diurnal oscillation of BES1 and BZR1 transcripts. Col‐0 seedlings were grown for 6 days under short days, and samples were harvested every 3 h. BES1 and BZR1 expression levels were determined by qRT–PCR analyses and normalized to PP2A. Relative levels are the mean of three technical replicates, and error bars represent standard deviation. The experiment was performed three times with similar results. Dark periods are indicated by shading.
- Western blot analyses of 35S::BES‐GFP seedlings grown for 6 days under short days. Samples were collected every 2 h, starting 2 h before lights on (ZT‐2) to ZT6. Western blots were hybridized with an anti‐GFP antibody to detect the BES1‐GFP protein and anti‐RPT5 as a loading control.
- BES1 but not BZR1 is stabilized in the light. 35S::BES1‐GFP and 35S::BZR1‐GFP seedlings were grown for 6 days in continuous white light (WL), red light (RL), and continuous dark (DARK), and the BES1 and BZR1 proteins analyzed by Western blot using an anti‐GFP antibody. Hybridization with an anti‐RPT5 antibody was used as a loading control.
- BES1 accumulation is independent of brassinosteroid levels. Six‐day‐old 35S::BES‐GFP seedlings were treated overnight with 100 nM epi‐brassinolide (BL) or 0.5 μM brassinazole (BRZ). Material was collected at the indicated time points and analyzed by Western blot hybridization with anti‐GFP and anti‐RPT5 antibodies.
- In vivo analysis of PIF4‐BES1 complex formation. Six‐day‐old pPIF4::PIF4‐HA seedlings grown under short‐day conditions were collected at ZT0, ZT1, and ZT8. PIF4‐HA was immunoprecipitated with an anti‐HA antibody and the pulled‐down BES1 protein immunodetected with an anti‐BES antibody. *indicates a non‐specific band by the primary antibody used to immunoprecipitation.
Source data are available online for this figure.
To further assess whether light affects BES1 protein stability, we analyzed protein levels in plants grown in continuous white light (WL), red light (RL), or in darkness (DARK). Remarkably, BES1 was detected in WL and RL, but not in darkness, in opposite to BZR1 which accumulated to constant levels (Fig 3D). Hence, in our conditions, light stabilized BES1, but did not affect the BZR1 protein. Lastly, we investigated whether light effects are mediated through changes in BR levels. As shown in Fig 3E, application of the BR biosynthesis inhibitor BRZ increased the ratio of phosphorylated to non‐phosphorylated BES1, while a preferential accumulation of the non‐phosphorylated protein was observed on treatment with epi‐brassinolide (eBL). However, neither BRZ nor eBL had an effect on BES1 levels at night, which demonstrates that BES1 stabilization during early morning responds to the presence of light.
PIF4 mediates de‐repressed expression of BR biosynthetic genes
PIF4 control of cell elongation is defined by a coincidence mechanism, whereby the circadian clock regulates PIF4 gene expression while light inactivates the PIF4 protein, via phyB‐dependent phosphorylation and proteasomal degradation (Nozue et al, 2007; Lorrain et al, 2008; Niwa et al, 2009). Therefore, to gain additional understanding on the functional significance of PIF4‐BES1 interaction in diurnal regulation of hypocotyl growth, we investigated expression levels of the XTR7, PRE5, and IAA19 genes in detail. Wild‐type, PIF4‐OX, pifq, and bes1‐d plants were grown in short‐day (SD) conditions, and levels of these transcripts analyzed over a 24‐h time course. These genes belong to the PIF + BES‐UP cluster, and so we reasoned that their peak expression levels should reflect PIF4 and BES1 temporal association. The BES‐DOWN CPD, DWF4, and BR6ox2 (CYP85A2) BR biosynthetic genes were similarly analyzed as readout of BES1 homodimer formation.
In wild‐type plants, XTR7, PRE5, and IAA19 displayed a narrow peak of expression at dawn (Fig 4), in accordance with their previously reported expression patterns (Nozue et al, 2007; Nomoto et al, 2012). This peak is however lost in pifq mutants (Fig 4), which corroborates an essential role of PIFs in transcriptional activation and recruitment of the BES1 factor to these promoters. In line with this function, transcripts of XTR7, PRE5, and IAA19 were strongly up‐regulated in PIF4‐OX lines at night, while their expression levels were reduced upon illumination (Fig 4), consistent with the light‐mediated inhibition of PIF4 by phyB (Lorrain et al, 2008). Furthermore, in constitutive bes1‐D and bzr1‐1D mutants, expression of these genes was elevated only at dawn, supporting that BES1/BZR1‐mediated co‐activation of these targets requires of transcriptionally active PIFs (Appendix Figs S3 and S4), while loss‐of‐function bes1 and bzr1 mutants showed normal levels of expression of these transcripts (Appendix Fig S5A), in support of a redundant function of BES1 and BZR1 in their regulation. These results suggest that PIF4 is limiting to PIF4‐BES1 complex formation and that interaction between these factors mostly takes place at dawn, when increased PIF4 transcription and the absence of light allow PIF4 accumulation in its active form. Western blot detection of BES1 in pPIF4::PIF4‐HA extracts immunoprecipitated with an anti‐HA antibody actually showed that endogenous BES1 is more efficiently pulled‐down at ZT0–ZT1 than ZT8, although levels of the PIF4‐HA protein are still elevated by the end of the day (Fig 3F). Also, the nuclear non‐phosphorylated form of BES1 is detected in these fractions, consistent with an interaction of these proteins in the nucleus. On the other side, an analogous dawn‐phased peak of expression as for PIF + BES‐UP targets was observed for the CPD, DWF4, and BR6ox2 genes (Fig 4). Transcript levels of these BES‐DOWN targets later decreased during the day, to slowly rise at night. More remarkably, their peak expression was suppressed in pifq mutants, whereas they were strongly up‐regulated at night in PIF4‐OX lines. Reduced levels of CPD, DWF4, and BR6ox2 transcripts were also detected in the constitutive bes1‐d and bzr1‐1D mutants (Appendix Figs S3 and S4), in agreement with a function of BES1 in negative regulation of these targets. Overall, these findings indicate that PIF4 de‐represses BR biosynthetic gene transcription, presumably by preventing dimeric BES1 accumulation, which negatively regulates these genes.
Figure 4. PIF4 mediates de‐repressed expression of BR biosynthetic genes.
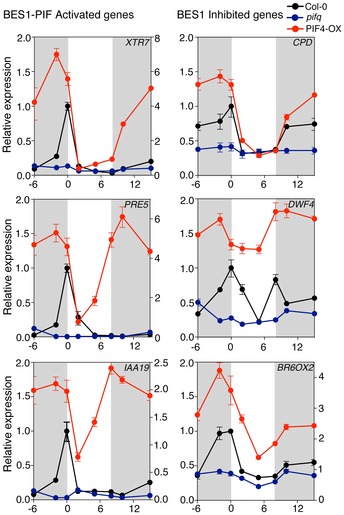
Col‐0 (black), PIF4‐OX (red), and pifq (blue) seedlings were grown for 6 days under short days, and samples were harvested at the indicated time points. Total RNA was used for qRT–PCR quantification of PIF + BES‐UP and the BR biosynthetic BES‐DOWN transcripts, using PP2A as endogenous control. Error bars represent SD of three technical replicates. Dark periods are shown in gray. Relative expression of the XTR7, PRE5, IAA19, and BR6ox2 genes in PIF4‐OX lines is referred to the right y axis of the graph. The experiment was repeated three times with similar results.
Notably, BES1 was transcribed in our growth conditions to higher levels at night and the protein showed a preferential stabilization in the light (Fig 3), while PIF4 transcriptional activity is highest at dawn, when rise in PIF4 transcription coincides with absence of light (Nozue et al, 2007; Niwa et al, 2009). This suggests that during late night, BES1 accumulates mainly in a complex with PIF4, which explains the observed rise in PIF + BES1‐UP transcript levels and up‐regulated expression of CPD, DWF4, and BR6ox2 genes. This temporal rise in BR levels may also play a critical role in mediating BIN2 inactivation during late night, thus favoring nuclear accumulation of both the transcriptionally active BES1 and PIF4 proteins. Immediately after dawn, however, opposite regulation of these factors by light is anticipated to modify their nuclear balance toward an excess of BES1. This allows BES1 dimerization, with BES1 repressive action then leading to BR biosynthetic gene inhibition as observed during the day. At the same time, a drop in BR levels is predicted to result in BES1 inactivation and thus would account for the gradual up‐regulation of BR biosynthetic genes observed during early night. Overall, these findings underscore that the opposite effects of light on PIF4 and BES1 protein stability have a relevant role in orchestrating combinatorial activity of these factors and in diurnal regulation of BR levels.
PIF4 promotes thermomorphogenic hypocotyl elongation by activating BR synthesis
PIF4, and to a lesser extent PIF5, are key activators of thermosensory growth (Koini et al, 2009; Stavang et al, 2009; Kumar et al, 2012). Warmer temperatures were shown to reduce binding of the evening complex (EC), ELF3, ELF4, and LUX proteins to its target promoters, which leads to increased PIF4 transcription during the night (Box et al, 2015; Raschke et al, 2015) and activated expression of auxin‐related genes, such as YUCCA8 and several SAURs (Stavang et al, 2009; Franklin et al, 2011). As we had found that PIF4 positively regulates CPD, DWF4, and BR6ox2 gene expression, we asked whether this regulation plays a relevant role in thermomorphogenesis. In wild‐type plants, we observed that CPD, DWF4, and BR6ox2 transcripts were elevated during the night in seedlings grown at 28°C compared to those grown at 22°C. The greatest temperature‐induced increase in these genes occurred in the late night, with temperature showing much smaller effects during the day (Fig 5A). A similar up‐regulation of these genes was not observed in pifq mutants, demonstrating that PIFs play an essential role in temperature‐dependent activation of BR synthesis (Fig 5A). Analyses of XTR7 transcript levels (used as a positive control) showed that this PIF4 target is strongly up‐regulated at 28°C during late night. By contrast, in the pifq mutant, it is expressed to basal levels and does not respond to temperature (Fig 5B).
Figure 5. PIF4 activates expression of BR‐synthesis genes in response to elevated temperatures.
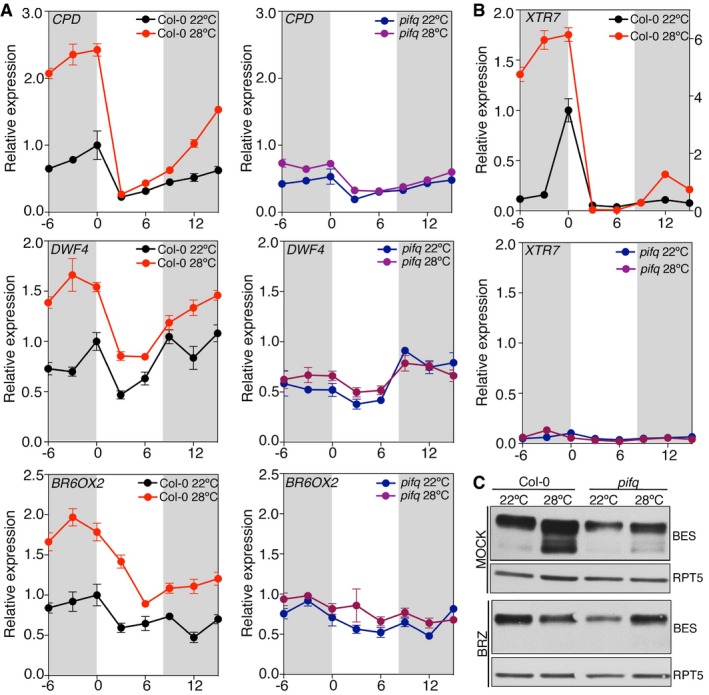
- Col‐0 and pifq seedlings were grown under short days at 22 and 28°C, and samples were collected at the indicated time points. Expression of BR biosynthetic genes was analyzed by qRT–PCR, using the constitutive PP2A gene as internal control. Error bars represent SD of three technical replicates. The experiment was repeated twice with similar results.
- Activation of the XTR7 PIF + BES‐UP target was studied as a positive control of the experiment. Error bars represent SD of three technical replicates. Dark periods are shown in gray. The experiment was performed three times with similar results.
- Increased levels of dephosphorylated BES1 at elevated temperatures depend on PIFs activation of BR synthesis. Col‐0 and pifq seedlings were grown for 6 days under short days and 22°C or 28°C, on half strength MS media (mock) or media supplemented with 0.5 μM BRZ. BES1 was analyzed by Western blot hybridization with an anti‐BES1 antibody. RPT5 was used as a loading control.
Source data are available online for this figure.
As previously reported, pif4pif5 mutants showed reduced temperature‐mediated hypocotyl elongation (Koini et al, 2009; Stavang et al, 2009) and we observed an even more temperature‐unresponsive phenotype in pifq mutants (Appendix Fig S6). Moreover, eBL restituted thermomorphogenic growth of both mutants, whereas the inhibitor BRZ compromised temperature‐induced growth of wild‐type plants, demonstrating that PIF‐mediated control of BR synthesis is essential to this response. Consistent with this regulation, an increased proportion of non‐phosphorylated BES1 was observed in wild‐type seedlings grown at 28°C (Fig 5C). By opposite, levels of non‐phosphorylated BES1 were significantly reduced in the pifq mutant, despite these plants accumulated comparable levels of phosphorylated BES1 when grown in the presence of BRZ (Fig 5C).
Collectively, these data illustrate that PIF4‐dependent control of BR synthesis not only contributes to coordinate hypocotyl elongation during late night, but is critical to the promotion of hypocotyl growth in warmth.
Discussion
In this study, we showed that BES1 binds to DNA as a dimeric and not as a monomeric protein and that complex formation with PIF4 modifies the DNA cis‐elements recognized by this factor (Fig 1). Dimeric BES1 acts as a transcriptional repressor by binding to G‐box and BRRE‐element which are found to be enriched in the promoters of BR biosynthetic genes and BES1‐DOWN targets, and recruits the co‐repressor TPL to these loci (Ryu et al, 2014; Espinosa‐Ruiz et al, 2017). Interaction with TPL relies on an EAR domain located at the C‐terminal end of the BES1 and BZR1 proteins. We observed that complex formation with PIF4 (Appendix Fig S2A and B) requires of the BES1 C‐terminal region, in addition to the N‐terminal DNA‐binding HLH domain (Appendix Fig S2A and C). Therefore, physical contact with PIF4 presumably hinders TPL interaction and mediates the switch of BES1‐repressive activity into a co‐activator function. Interestingly, mutations in the EAR domain result in BES1 inactivation (Ryu et al, 2014; Espinosa‐Ruiz et al, 2017), and it is possible that this effect is caused in part by weakened PIF4 interaction. Our findings showed that PIF4 modifies the DNA binding specificity of BES1, to recognition of a 5′‐CATGTG‐3′ motif, previously identified as a PIF‐binding element (PBE) (Zhang et al, 2013; Pfeiffer et al, 2014). Notably, this motif is overrepresented in the promoters of genes that are up‐regulated by PIF4 or the PIF4‐BES1 complex (Fig 2), underscoring that combinatorial regulation by PIF4 and BES1 is responsible of co‐activated expression of multiple target loci. Moreover, these elements are not enriched in genes activated by BES1 independent of PIFs, raising the possibility that BES1 binds other families of TFs to co‐activate these targets.
Our results reveal that PIF4‐BES1 interaction plays an important role in diurnal regulation of BR synthesis, most probably by competing for BES1 dimerization. In our growth conditions, BES1 transcript levels were elevated at night while decreased during the day, as opposed to the BES1 protein that is stabilized during early day (Fig 3). We showed that the pattern of BES1 protein accumulation is not regulated by BR signaling, but is dependent on light, as application of eBL or the inhibitor BRZ changed the ratio of phosphorylated to non‐phosphorylated protein, but did not increase BES1 protein levels during late night (Fig 3). Although COP1 has been reported to degrade phosphorylated BZR1 in darkness (Kim et al, 2014), and SINAT E3 ligases to degrade the dephosphorylated BES1 protein in the light (Nolan et al, 2017; Yang et al, 2017), a similar light‐dependent degradation of these proteins was not observed in our study, highlighting that growth conditions strongly affect BES1/BZR1 stability. These results underscore that regulation of BES1/BZR1 protein levels is far more complex than previously anticipated and that future studies will be required to understand light‐dependent control of BES1 accumulation.
The BES1 gene was shown in a recent report to encode two different isoforms, designated as BES1‐Long (BES1‐L) and BES1‐Short (BES1‐S), with BES1‐L having a stronger activity in promoting BR signaling (Jiang et al, 2015). All experiments in this work were carried out with the regular BES1‐S isoform as we observed that BES1‐L does not display the same light‐dependent stabilization as BES1‐S (Appendix Fig S5B). Transcript levels for the BES1‐L isoform were also less abundant than for the shorter protein (Appendix Fig S5C), which suggests that BES1‐S is the preponderant form in our growth conditions, and has a role in diurnal BR levels regulation as it is stabilized by light.
We could observe that transcription of BR biosynthetic genes was elevated at dawn, coinciding with the peak expression of PIF + BES‐UP targets (Fig 4). This expression profile suggests that increased levels of the PIF4 protein during late night de‐repress these BES‐DOWN targets. Light, on the other hand, inactivates PIF4 while stabilizes BES1, enabling BES1 dimerization to suppress BR biosynthetic gene expression during daytime. Consistent with this model of regulation, CPD, DWF4, and BR6ox2 were induced in PIF4‐OX lines, whereas their transcript levels were notably reduced in pifq and bes1‐d/bzr1‐1D mutants (Fig 4, Appendix Figs S3 and S4).
Whether PIF4 positively regulates BR biosynthetic genes by directly binding to these targets remains an open question. In fact, ChIP‐PCR studies showed that PIF4 binds the DWF4 and BR6ox2 promoters (Oh et al, 2012; Wei et al, 2017), and in our EMSA studies, both the G‐box and PBE‐elements were equally recognized by the PIF4 and PIF4‐BES1 proteins (Fig 1). Hence, it is possible that PIF4 and PIF4‐BES1 directly bind the conserved G‐box elements in the BR biosynthetic promoters. Otherwise, PIF4 may also facilitate BR biosynthetic gene expression by interfering with BES1 homodimer formation, hence precluding dimeric BES1 of binding to the G‐box and BRRE‐elements in these promoters, and inhibition of their expression through recruitment of the co‐repressor TPL (Oh et al, 2014; Ryu et al, 2014).
Furthermore, we provide evidence showing that this regulation is critical to thermomorphogenic hypocotyl elongation. We showed that warm temperatures cause up‐regulated expression of BR biosynthetic genes, and that this response is compromised in the pifq mutant, which fails to show temperature‐mediated hypocotyl elongation at 28°C (Fig 5 and Appendix Fig S6). Notably, BL restores thermomorphogenic growth of pif4pif5 and pifq mutants, while the inhibitor BRZ blocks enhanced elongation of wild‐type plants (Appendix Fig S6), hence demonstrating that the elongation defects of pifq mutants are caused by impaired thermo‐responsive activation of BR biosynthetic genes. In agreement with a prevalent role of this regulation in temperature‐induced hypocotyl growth, mutations affecting the DWF7 and ROT3 genes were recently identified in a screen for okapi mutants that reversed the hypersensitive thermo‐response of Arabidopsis Rrs‐7 accessions (Ibanez et al, 2018). Defects in these BR biosynthetic genes also block response to temperature in the Col‐0 background, whereas BL rescues thermomorphogenic defects of these mutants (Ibanez et al, 2018), evidencing that temperature‐induced hypocotyl elongation is strictly dependent on BRs.
We propose that increased PIF4 transcription as a result of elevated temperatures allows PIF4 protein accumulation during late night and alters PIF4/BES1 nuclear balance toward an overabundance of PIF4. Most BES1 is then bound to PIF4, and the PIF4‐BES1 complex is therefore dominant over the BES1 dimeric protein. This leads to activation of the auxin‐related and cell wall‐modifying PIF‐UP and PIF + BES‐UP targets supporting cell growth, in addition to de‐repressed transcription of BR biosynthetic genes. Increased BR levels in turn, positively feedback on this regulation, by signaling inactivation of BIN2, which phosphorylates and destabilizes the PIF3 and PIF4 factors (Bernardo‐Garcia et al, 2014; Ling et al, 2017), in addition to modulate BES1 and BZR1 subcellular localization by phosphorylation of these proteins (Ryu et al, 2010, 2007; Fig 6). Indeed, BZR1 was recently reported to translocate into the nucleus at elevated temperatures (Ibanez et al, 2018), and we showed that dephosphorylated BES1 levels increase in the wild type at 28°C (Fig 5). Hence, the switch in BES1 transcriptional activity from a repressive into a co‐activator function, coupled with recognition of distinct promoter cis‐elements by the dimeric BES1 and heterodimeric PIF4‐BES1 proteins, provides mechanistic detail on how concerted function of these factors coordinates thermomorphogenic growth.
Figure 6. Model for BR‐dependent diurnal and ambient temperature control of hypocotyl elongation.
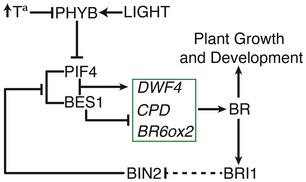
In this model, an increase in PIF4 protein levels as a consequence of elevated temperatures promotes formation of the PIF4‐BES1 complex, and activated expression of the DWF4, CPD, and BR6ox2 BR biosynthetic genes. Increased BR levels support plant growth by inactivating BIN2 and promoting accumulation of the active PIF4 and BES1 proteins. In response to light, phyB destabilizes PIF4, and the BES1 dimeric protein is dominant over the PIF4‐BES1 complex. Dimeric BES1 represses expression of BR biosynthetic genes and leads to decreased BRs levels. As a consequence, BIN2 is reactivated and phosphorylates PIF4 and BES1, inactivating these factors, which leads to increased transcription of BR biosynthetic genes during the night. Light and temperature cues thus control PIF4 and BES1 levels, the nuclear balance of both factors providing a highly sensitive mechanism for rapid modulation of plant growth and development in response to environmental cues.
Materials and Methods
Plant material and growth conditions
Arabidopsis thaliana Col‐0 wild‐type plants were used in this study. The bes1‐D (Yin et al, 2002; introgressed into Col‐0), pif4pif5 (de Lucas et al, 2008), and pifq (Shin et al, 2009) mutants, as well as pBES1‐BES1‐GFP (Yin et al, 2002) and 35S::BZR1‐GFP (Wang et al, 2002) lines were in the Col‐0 background. 35S::BES1‐GFP plants were generated by cloning the BES1 CDS (BES1topof‐BES1‐R) without stop codon into the pENTR™/D‐TOPO vector and subsequent mobilization by LR clonase (Invitrogen) reaction into the pGWB5 vector. This construct was introduced into Agrobacterium tumefaciens (GV3101) and used for Col‐0 plants transformation by floral dipping. The double‐mutant bes1‐D;pif4pif5 was generated by crossing pif4pif5 and bes1‐D mutants. Seedlings were grown for 6 days under short‐day conditions (8‐h light/16‐h dark) at 22°C, or under continuous white light, red light, or dark conditions. For thermomorphogenic studies, seedlings were grown under short‐day conditions at 22°C for 24 h and then transferred to 28°C or kept at 22°C for five additional days.
Seeds were surface‐sterilized for 15 min in 70% (v/v) ethanol and 0.01% (v/v) Triton X‐100, followed by two washes of 2 min in 96% (v/v) ethanol, and let dry in a sterile bench. Seeds were then sown on half strength Murashige and Skoog (MS) media with Gamborg's vitamins (Duchefa) and 1% sucrose (w/v), supplemented with 0.8% agar (w/v), and stratified at 4°C for 4 days in the dark. Germination was synchronized by 4 h illumination, before transferring plates to darkness for 20 additional hours. Brassinazole (TCI America) and epi‐brassinolide (SIGMA) treatments were performed at 0.5 and 0.1 μM, respectively. After germination, seedlings were moved to the plates containing the different treatments and grown in the conditions indicated. Treatments in liquid medium were performed by moving seedlings to multiwell plates containing 1.5 ml half strength MS liquid media supplemented with 0.5% Suc/well and the corresponding treatments and incubation for 16 h with continuous shaking.
For hypocotyl growth measurements, plants were grown for 6 days on vertical plates. Plates were photographed, and the length of the hypocotyl was determined with the ImageJ software.
Yeast two‐hybrid assay
Yeast two‐hybrid interaction assays of the PIF4 and BES1 proteins were performed with the GAL4 Two‐Hybrid System (Clontech). The complete CDS of PIF4 and BES1 was amplified with primers PIF4YFPf/PIF4YFPr and BES1topof/BES1‐R, cloned into the pENTRY™/D‐TOPO vector, and subsequently mobilized into the pGADT7 and pGBKT7 (Clontech) Gateway‐compatible vectors. PIF4 deletions were generated by using the following primer combinations: PIF4YFPf/APB_PIF4r for PIF4del1, PIF4sinAPBf/PIF4YFPr for PIF4del2, AtPIF4del3f/PIF4YFPr for PIF4del3, and AtPIF4del4f/PIF4YFPr for PIF4del4. For BES1 deletions, the following primer combinations were used: BES1del2f/BES1‐R for BES1del2, BES1topof/BES1del3r for BES1del3, and BES1del4f/BES1‐R for BES1del4. BES1del1 was obtained by HindIII/SacI digestion of the BES1del2 construct in the TOPO vector. Constructs were transformed into the AH109 yeast strain, by the lithium acetate method, and reporter gene activation was analyzed by selection on SD‐LW and SD‐LWHA plates.
Bimolecular fluorescence complementation assay
Full‐length coding regions for the Arabidopsis PIF4 and BES1 proteins and the BES1del2 deletion in the pENTR™/D‐TOPO vector (Invitrogen) were mobilized by LR‐reaction (Invitrogen) into the binary pBiFC vectors containing the N‐ and C‐terminal YFP fragments (YFPN43 and YFPC43). The BES1delN deletion was obtained by PCR amplification with primers BES1topof/BES1delNr. Generated constructs were transformed into the A. tumefaciens GV3101 strain and co‐infiltrated into N. benthamiana leaves, using the indicated combinations. The p19 protein was used to suppress gene silencing (Voinnet et al, 2003). Agrobacteria strains bearing the BiFC constructs and the p19 silencing suppressor were suspended in 10 mM MES pH 5.5, 10 mM MgSO4, and 150 μM acetosyringone at a OD600 ratio of 0.7:0.7:1.0, respectively, for infiltration. One day after infiltration, leaves were treated with 10 μM MG132 (VWR Calbiochem) and 24 h later they were observed under a Leica TCS SP5 laser scanning confocal microscope, with an excitation beam splitter TD 488/561/633 and an emission band width between 495 and 556 nm.
Electrophoretic mobility shift assay
Electrophoretic mobility shift assay assays were performed with biotin‐labeled probes. To this end, DNA fragments for the pPIL1, pPRE5, and pDWF4 promoter regions were amplified by using 5′‐biotinylated oligonucleotides (Appendix Table S5). Gels were transferred after electrophoretic separation to a nitrocellulose membrane and detected with the LightShift Chemiluminescent EMSA Kit (Pierce). The PIF4‐His and MBP‐BES1 recombinant proteins were incubated in a 20 μl reaction mixture containing 20 mM Hepes pH 7.9, 50 mM KCl, 2 mM MgCl2, 1 mM EDTA, 1 mM DTT, 50 ng/ml poly (dI‐dC), 10% glycerol, and 0.1% NP‐40 for 5 min at room temperature. The biotin‐labeled probes were then added to the mixture and incubated on ice for 20 more minutes, before separation on 4% native polyacrylamide gels in 0.5× TBE buffer. The labeled probes were detected with streptavidin, according to the instructions provided by the manufacturer (Pierce).
Protein binding microarray
For expression of the recombinant proteins, BES1 was cloned into the pMAL‐c2X vector (NEB), encoding the MBP (maltose binding protein) in front of the Gateway recombination cassette. This fusion was then inserted into the pCOLADuet‐1 (Novagen) vector by amplification with primers BES‐COLA‐2F (includes an RcaI restriction site, compatible with NcoI) and BES‐COLA‐2R, and PCR product digestion with the RcaI and AscI enzymes, before ligation into the NcoI/AscI sites (MCS1) of the pCOLADuet‐1 vector. The PIF4 CDS was cloned into the vector pET‐23a (+) (Novagen) by amplification with primers PIF4‐1F and PIF4‐1R, which respectively include the NdeI and EcoRI sites (Appendix Table S5), besides a linker of five glycines. The PIF4 His‐tag fusion was then amplified using primers PIF4‐COLA‐2F and PIF4‐COLA‐2R (includes an AvrII recognition site), and cut with NdeI and AvrII, for ligation into the pCOLADuet‐1 NdeI/AvrII sites (MCS2). Escherichia coli Rossetta cells were used to express the recombinant proteins. The cultures were grown until an OD600 of 0.6 and then induced with 1 mM isopropyl‐β‐D‐thiogalactopyranoside (IPTG) for 2 h at 37°C. The PIF4 6XHis protein was purified through Ni‐NTA agarose beads (Qiagen), and the MBP‐BES1 fusion through an amylose resin (NEB), following the specifications of the manufacturers. For the PBM, a 10 nucleotides design was selected. Generation of the double‐stranded microarray, protein incubation, and immunological detection of the protein‐DNA complexes was as described by Godoy et al (2011). Three independent hybridizations were performed with the purified protein fractions obtained from recombinant E. coli cultures expressing the PIF4‐His, MBP‐BES1, and PIF4‐His + MBP‐BES1 proteins.
RNA extraction and quantitative real‐time PCR analysis
Total RNA was isolated from whole seedlings using the High Pure Isolation Kit (Roche) and DNase‐treated when bound to the column, following the manufacturer instructions. One microgram of RNA was reverse‐transcribed into cDNA using the Transcriptor First‐Strand cDNA Synthesis Kit (Roche) and random hexamers. The cDNA reaction was diluted to 40 μl, and real‐time PCR amplification performed with 1 μl of cDNA, using the Power SYBR Green PCR Master Mix (Applied Biosystems), and a 7500 Real‐Time PCR System (Applied Biosystems). Expression levels were calculated relative to PP2A by means of the ΔΔCT threshold cycle method (Applied Biosystems). Primer sequences are provided in Appendix Table S5. Quantitative RT–PCR determinations were done in at least two independent biological replicates with similar results.
Protein extraction and Western blot analysis
Seedlings were homogenized in an extraction buffer containing (50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 2.5 mM EDTA, 10% glycerol, 25 mM β‐glycerophosphate, 0.1% NP‐40, 10 mM NaF, 2 mM Na3VO4, 100 μM PMSF, and 5 μM β‐mercaptoethanol and protease inhibitors). Extracts were centrifuged twice at 13,000 g for 10 min at 4°C, and protein concentration in the supernatant was determined by the Bradford assay (Bio‐Rad). Protein samples were boiled in TMx2 loading buffer for 5 min and 30 μg of protein separated on 8% SDS–PAGE gels. Gels were transferred to 0.2 μm nitrocellulose membranes (Amersham) and homogeneous protein transfer confirmed by Ponceau red staining. Membranes were incubated in blocking buffer (1× PBS buffer including 0.1% Tween 20 and 5% dried nonfat milk) for 1 h at room temperature, before overnight incubation at 4°C with anti‐GFP (Roche), anti‐BES1, or anti‐RPT5 (Kwok et al, 1999) antibodies. Finally, membranes were incubated for another hour with a peroxidase‐conjugated secondary antibody. The proteins were visualized with the Supersignal West Pico and Femto substrates (Pierce).
Luciferase assay
For the luciferase assay, N. benthamiana leaves were infiltrated Agrobacterium strains bearing the constructs expressing the PIL1 (pPIL1‐1F/pPIL1‐1R) or DWF4 (pDWF4‐F/pDWF4‐R) promoters fused to the Luciferase reporter gene (pLUC‐Trap3 vector), and the 35S::BES1‐GFP, 35S::PIF4‐HA, or 35S::bes1‐D‐GFP effector constructs, to test for activated expression of these genes. Two days after infiltration, 0.5‐cm‐diameter disks were collected from the leaves and transferred to 96‐well microtiter plates containing 165 μl 0.5× MS liquid media and 35 μl of 1× D‐Luciferin substrate (20 μg/ml; Promega). Luminescence was measured with a LB 960 Microplate Luminometer (Berthold). One disk was used per well and at least 12 disk replicates per sample, and the average Luminescence values represented.
In vivo co‐IP assay
Six‐day‐old pPIF4::PIF4‐HA seedlings grown under short‐day conditions were collected at ZT0, ZT1, and ZT8 and cross‐linked with 1% formaldehyde for 15 min under vacuum. The reaction was terminated by adding glycine to a final concentration of 125 mM and samples were frozen in liquid nitrogen, before homogenization in protein extraction buffer (25 mM Tris–HCl, pH 8.0, 75 mM NaCl, 0.2% Nonidet P‐40, 10 mM β‐mercaptoethanol, 1 mM PMSF, protease inhibitors (Roche), and 10 μM MG132). Extracts were centrifuged twice for 15 min at 13,000 g and 4°C, and the supernatants kept for co‐immunoprecipitation assays. 40 μl of anti‐HA sepharose beads was added to the tissue extracts, and the mixture was incubated for 3 h at 4°C. Beads were washed four times with 500 μl extraction buffer and the retained proteins eluted by incubation for 15 min at 65°C with 50 μl of pre‐heated 95°C 2× SDS protein loading buffer. 10 μl (anti‐HA) and 40 μl (anti‐BES1) of the Co‐IP samples were used for immunodetection as described previously.
Microarray analysis
Microarray was performed with bes1‐D, pif4pif5, bes1‐D;pif4pif5, and Col‐0 genotypes. 6‐day‐old seedlings grown in the dark were used for RNA extraction and Affymetrix microarray hybridization. Total RNA was extracted from whole seedlings with the High Pure RNA Isolation Kit (Roche). Three biological replicates were used for expression studies. Background correction and normalization were performed using the LIMMA package (Smyth & Speed, 2003). Genes were considered as differentially expressed if fold change was > 2 and the corrected P‐value (Rank Products) < 0.1. Enrichment in DNA elements in the 1‐kb upstream region of these genes was analyzed using the motif analysis tool from TAIR. Statistical analyses were performed using the SPSS statistical package. Hierarchical clustering of the directly regulated genes was obtained using the MEV TIGR free software (http://mev.tm4.org/).
Statistical analysis
For the analysis of the protein binding microarrays, raw signal intensities were normalized and adjusted with the Perl script “normalize_agilent_array.pl”, and enrichment scores (E‐Scores) for all the possible 8‐mers and position weight matrices for the best scoring 8‐mers were obtained using the Perl script “seed_and_wobble.pl”. These analyses were carried out with the PBM Analysis Suite (http://the_brain.bwh.harvard.edu/PBMAnalysisSuite/index.html). Box plots were obtained using the SPSS statistical program, and logos for the best motifs with enoLOGOS (http://www.benoslab.pitt.edu/cgi-bin/enologos/enologos.cgi).
To analyze the overlap of PIF and BES targets and PIF‐ and BES‐regulated genes, lists from published arrays, ChIP‐ChIP and ChIP‐seq data, were constructed (Appendix Tables S1–S4). Genes were categorized as direct targets or UP‐/DOWN‐regulated if present in at least one list, excluding those found to be simultaneously up‐ and down‐regulated in different datasets. Lists of the different PIF‐ and BES‐regulated and target genes were used to construct Venn diagrams using the Venny tool (http://bioinfogp.cnb.csic.es/tools/venny/). A chi‐squared test was performed to calculate gene enrichment of the overlapping datasets compared to total Arabidopsis genes (TAIR) using the SPSS statistical program. Gene enrichment was considered statistically significant with a P‐value < 0.01.
Data accessibility
Microarray results have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with the accession code GSE85211.
Author contributions
SP, AE‐R, and CM designed the experiments. MdL performed initial studies, and SB‐G carried out the yeast two‐hybrid assays. CM generated the purified proteins and carried out the BiFC BES1‐PIF4 interaction analyses. JMF‐Z hybridized the protein binding microarray and analyzed the corresponding DNA recognition motifs. CM performed the EMSA, Co‐IP, and Western blot studies. qRT–PCR analyses were carried out by AE‐R and CM, and the transactivation assays by SP. Microarray hybridization and promoter cis‐element studies were carried by AE‐R. CM and SP wrote the manuscript. All authors revised the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 5
Acknowledgements
We thank Prof. Yanhai Yin (Iowa State University, USA) for providing the pBES1::BES1‐GFP lines and the anti‐BES1 antibody, Prof. Zhi‐Yong Wang (Carnegie Institution, USA) for the 35S::BZR1‐GFP lines, and Prof. Xuelu Wang (Huazhong Agricultural University, China) for the 35S::BES1‐L and 35S::BES1‐S lines. We also acknowledge Dr Scott Hayes (Centro Nacional de Biotecnologia, Spain) for his comments on the manuscript. We are also thankful to Dr. Alberto Sanz (IE University, Spain) for help with statistical analysis. This work was supported by grants BIO2014‐60064‐R and BIO2017‐90056‐R from the Spanish Ministerio de Economía y Competitividad.
The EMBO Journal (2018) 37: e99552
References
- Bernardo‐Garcia S, de Lucas M, Martinez C, Espinosa‐Ruiz A, Daviere JM, Prat S (2014) BR‐dependent phosphorylation modulates PIF4 transcriptional activity and shapes diurnal hypocotyl growth. Genes Dev 28: 1681–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Box MS, Huang BE, Domijan M, Jaeger KE, Khattak AK, Yoo SJ, Sedivy EL, Jones DM, Hearn TJ, Webb AA, Grant A, Locke JC, Wigge PA (2015) ELF3 controls thermoresponsive growth in Arabidopsis . Curr Biol 25: 194–199 [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA (1991) Phenotypic and genetic analysis of det2, a new mutant that affects light‐regulated seedling development in Arabidopsis . Plant Cell 3: 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clouse SD, Langford M, McMorris TC (1996) A brassinosteroid‐insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol 111: 671–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinosa‐Ruiz A, Martinez C, de Lucas M, Fabregas N, Bosch N, Cano‐Delgado AI, Prat S (2017) TOPLESS mediates brassinosteroid control of shoot boundaries and root meristem development in Arabidopsis thaliana . Development 144: 1619–1628 [DOI] [PubMed] [Google Scholar]
- Foreman J, Johansson H, Hornitschek P, Josse EM, Fankhauser C, Halliday KJ (2011) Light receptor action is critical for maintaining plant biomass at warm ambient temperatures. Plant J 65: 441–452 [DOI] [PubMed] [Google Scholar]
- Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA, Gray WM (2011) Phytochrome‐interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108: 20231–20235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin‐regulated and brassinosteroid‐regulated genes in Arabidopsis . Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sasaki E, Akiyama K, Maruyama‐Nakashita A, Nakabayashi K, Li W, Ogawa M, Yamauchi Y, Preston J, Aoki K, Kiba T, Takatsuto S, Fujioka S, Asami T, Nakano T, Kato H, Mizuno T, Sakakibara H, Yamaguchi S, Nambara E et al (2008) The AtGenExpress hormone and chemical treatment data set: experimental design, data evaluation, model data analysis and data access. Plant J 55: 526–542 [DOI] [PubMed] [Google Scholar]
- Godoy M, Franco‐Zorrilla JM, Perez‐Perez J, Oliveros JC, Lorenzo O, Solano R (2011) Improved protein‐binding microarrays for the identification of DNA‐binding specificities of transcription factors. Plant J 66: 700–711 [DOI] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M (1998) High temperature promotes auxin‐mediated hypocotyl elongation in Arabidopsis . Proc Natl Acad Sci USA 95: 7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Yang Y, Li J, Wang ZY (2002) The GSK3‐like kinase BIN2 phosphorylates and destabilizes BZR1, a positive regulator of the brassinosteroid signaling pathway in Arabidopsis . Proc Natl Acad Sci USA 99: 10185–10190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307: 1634–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, Lopez‐Vidriero I, Franco‐Zorrilla JM, Solano R, Trevisan M, Pradervand S, Xenarios I, Fankhauser C (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71: 699–711 [DOI] [PubMed] [Google Scholar]
- Ibanez C, Delker C, Martinez C, Burstenbinder K, Janitza P, Lippmann R, Ludwig W, Sun H, James GV, Klecker M, Grossjohann A, Schneeberger K, Prat S, Quint M (2018) Brassinosteroids dominate hormonal regulation of plant thermomorphogenesis via BZR1. Curr Biol 28: 303–310 e303 [DOI] [PubMed] [Google Scholar]
- Jiang J, Zhang C, Wang X (2015) A recently evolved isoform of the transcription factor BES1 promotes brassinosteroid signaling and development in Arabidopsis thaliana . Plant Cell 27: 361–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HB, Kwon M, Ryu H, Fujioka S, Takatsuto S, Yoshida S, An CS, Lee I, Hwang I, Choe S (2006) The regulation of DWARF4 expression is likely a critical mechanism in maintaining the homeostasis of bioactive brassinosteroids in Arabidopsis . Plant Physiol 140: 548–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Guan S, Burlingame AL, Wang ZY (2011) The CDG1 kinase mediates brassinosteroid signal transduction from BRI1 receptor kinase to BSU1 phosphatase and GSK3‐like kinase BIN2. Mol Cell 43: 561–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B, Jeong YJ, Corvalan C, Fujioka S, Cho S, Park T, Choe S (2014) Darkness and gulliver2/phyB mutation decrease the abundance of phosphorylated BZR1 to activate brassinosteroid signaling in Arabidopsis . Plant J 77: 737–747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinoshita T, Cano‐Delgado A, Seto H, Hiranuma S, Fujioka S, Yoshida S, Chory J (2005) Binding of brassinosteroids to the extracellular domain of plant receptor kinase BRI1. Nature 433: 167–171 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature‐mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19: 408–413 [DOI] [PubMed] [Google Scholar]
- Kumar SV, Lucyshyn D, Jaeger KE, Alos E, Alvey E, Harberd NP, Wigge PA (2012) Transcription factor PIF4 controls the thermosensory activation of flowering. Nature 484: 242–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok SF, Staub JM, Deng XW (1999) Characterization of two subunits of Arabidopsis 19S proteasome regulatory complex and its possible interaction with the COP9 complex. J Mol Biol 285: 85–95 [DOI] [PubMed] [Google Scholar]
- Leivar P, Tepperman JM, Monte E, Calderon RH, Liu TL, Quail PH (2009) Definition of early transcriptional circuitry involved in light‐induced reversal of PIF‐imposed repression of photomorphogenesis in young Arabidopsis seedlings. Plant Cell 21: 3535–3553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leivar P, Quail PH (2011) PIFs: pivotal components in a cellular signaling hub. Trends Plant Sci 16: 19–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Nagpal P, Vitart V, McMorris TC, Chory J (1996) A role for brassinosteroids in light‐dependent development of Arabidopsis . Science 272: 398–401 [DOI] [PubMed] [Google Scholar]
- Li J, Nam KH (2002) Regulation of brassinosteroid signaling by a GSK3/SHAGGY‐like kinase. Science 295: 1299–1301 [DOI] [PubMed] [Google Scholar]
- Ling JJ, Li J, Zhu D, Deng XW (2017) Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2‐mediated PIF3 phosphorylation and degradation in darkness. Proc Natl Acad Sci USA 114: 3539–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome‐mediated inhibition of shade avoidance involves degradation of growth‐promoting bHLH transcription factors. Plant J 53: 312–323 [DOI] [PubMed] [Google Scholar]
- Lorrain S, Trevisan M, Pradervand S, Fankhauser C (2009) Phytochrome interacting factors 4 and 5 redundantly limit seedling de‐etiolation in continuous far‐red light. Plant J 60: 449–461 [DOI] [PubMed] [Google Scholar]
- de Lucas M, Daviere JM, Rodriguez‐Falcon M, Pontin M, Iglesias‐Pedraz JM, Lorrain S, Fankhauser C, Blazquez MA, Titarenko E, Prat S (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451: 480–484 [DOI] [PubMed] [Google Scholar]
- Mora‐Garcia S, Vert G, Yin Y, Cano‐Delgado A, Cheong H, Chory J (2004) Nuclear protein phosphatases with Kelch‐repeat domains modulate the response to brassinosteroids in Arabidopsis . Genes Dev 18: 448–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis . PLoS Biol 2: E258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475 [DOI] [PubMed] [Google Scholar]
- Niwa Y, Yamashino T, Mizuno T (2009) The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana . Plant Cell Physiol 50: 838–854 [DOI] [PubMed] [Google Scholar]
- Nolan TM, Brennan B, Yang M, Chen J, Zhang M, Li Z, Wang X, Bassham DC, Walley J, Yin Y (2017) Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev Cell 41: 33–46 e37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomoto Y, Kubozono S, Yamashino T, Nakamichi N, Mizuno T (2012) Circadian clock‐ and PIF4‐controlled plant growth: a coincidence mechanism directly integrates a hormone signaling network into the photoperiodic control of plant architectures in Arabidopsis thaliana . Plant Cell Physiol 53: 1950–1964 [DOI] [PubMed] [Google Scholar]
- Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448: 358–361 [DOI] [PubMed] [Google Scholar]
- Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock‐, light‐, and growth‐correlated genes reveals PHYTOCHROME‐INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis . Plant Physiol 156: 357–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Wang ZY (2012) Interaction between BZR1 and PIF4 integrates brassinosteroid and environmental responses. Nat Cell Biol 14: 802–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Zhu JY, Ryu H, Hwang I, Wang ZY (2014) TOPLESS mediates brassinosteroid‐induced transcriptional repression through interaction with BZR1. Nat Commun 5: 4140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley RC, Huang SC, Song L, Lewsey MG, Bartlett A, Nery JR, Galli M, Gallavotti A, Ecker JR (2016) Cistrome and epicistrome features shape the regulatory DNA landscape. Cell 165: 1280–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng P, Zhao J, Zhu Y, Asami T, Li J (2010) A direct docking mechanism for a plant GSK3‐like kinase to phosphorylate its substrates. J Biol Chem 285: 24646–24653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer A, Shi H, Tepperman JM, Zhang Y, Quail PH (2014) Combinatorial complexity in a transcriptionally centered signaling hub in Arabidopsis . Mol Plant 7: 1598–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raschke A, Ibanez C, Ullrich KK, Anwer MU, Becker S, Glockner A, Trenner J, Denk K, Saal B, Sun X, Ni M, Davis SJ, Delker C, Quint M (2015) Natural variants of ELF3 affect thermomorphogenesis by transcriptionally modulating PIF4‐dependent auxin response genes. BMC Plant Biol 15: 197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Park J, Choe S, Hwang I (2007) Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19: 2749–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu H, Kim K, Cho H, Hwang I (2010) Predominant actions of cytosolic BSU1 and nuclear BIN2 regulate subcellular localization of BES1 in brassinosteroid signaling. Mol Cells 29: 291–296 [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Bae W, Hwang I (2014) Control of early seedling development by BES1/TPL/HDA19‐mediated epigenetic regulation of ABI3. Nat Commun 5: 4138 [DOI] [PubMed] [Google Scholar]
- Shin J, Kim K, Kang H, Zulfugarov I, Bae G, Lee C‐H, Lee D, Choi G (2009) Phytochromes promote seedling light responses by inhibiting four negatively‐acting phytochrome‐interacting factors. Proc Natl Acad Sci USA 106: 7660–7665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smyth GK, Speed T (2003) Normalization of cDNA microarray data. Methods 31: 265–273 [DOI] [PubMed] [Google Scholar]
- Stavang JA, Gallego‐Bartolome J, Gomez MD, Yoshida S, Asami T, Olsen JE, Garcia‐Martinez JL, Alabadi D, Blazquez MA (2009) Hormonal regulation of temperature‐induced growth in Arabidopsis . Plant J 60: 589–601 [DOI] [PubMed] [Google Scholar]
- Sun Y, Fan X‐Y, Cao D‐M, Tang W, He K, Zhu J‐Y, He J‐X, Bai M‐Y, Zhu S, Oh E, Patil S, Kim T‐W, Ji H, Wong W, Rhee S, Wang Z‐Y (2010) Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis . Dev Cell 19: 765–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz‐Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de‐etiolation in Arabidopsis . Cell 85: 171–182 [DOI] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J (2005) Molecular mechanisms of steroid hormone signaling in plants. Annu Rev Cell Dev Biol 21: 177–201 [DOI] [PubMed] [Google Scholar]
- Vert G, Chory J (2006) Downstream nuclear events in brassinosteroid signalling. Nature 441: 96–100 [DOI] [PubMed] [Google Scholar]
- Voinnet O, Rivas S, Mestre P, Baulcombe D (2003) An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J 33: 949–956 [DOI] [PubMed] [Google Scholar]
- Wang ZY, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T, Chory J (2002) Nuclear‐localized BZR1 mediates brassinosteroid‐induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2: 505–513 [DOI] [PubMed] [Google Scholar]
- Wang Z‐Y, Bai M‐Y, Oh E, Zhu J‐Y (2012) Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet 46: 701–724 [DOI] [PubMed] [Google Scholar]
- Wang C, Shang JX, Chen QX, Oses‐Prieto JA, Bai MY, Yang Y, Yuan M, Zhang YL, Mu CC, Deng Z, Wei CQ, Burlingame AL, Wang ZY, Sun Y (2013a) Identification of BZR1‐interacting proteins as potential components of the brassinosteroid signaling pathway in Arabidopsis through tandem affinity purification. Mol Cell Proteomics 12: 3653–3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Sun S, Zhu W, Jia K, Yang H, Wang X (2013b) Strigolactone/MAX2‐induced degradation of brassinosteroid transcriptional effector BES1 regulates shoot branching. Dev Cell 27: 681–688 [DOI] [PubMed] [Google Scholar]
- Wei Z, Yuan T, Tarkowska D, Kim J, Nam HG, Novak O, He K, Gou X, Li J (2017) Brassinosteroid biosynthesis is modulated via a transcription Factor cascade of COG1, PIF4, and PIF5. Plant Physiol 174: 1260–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Li C, Cai Z, Hu Y, Nolan T, Yu F, Yin Y, Xie Q, Tang G, Wang X (2017) SINAT E3 ligases control the light‐mediated stability of the brassinosteroid‐activated transcription factor BES1 in Arabidopsis . Dev Cell 41: 47–58 e44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Z, Mora‐Garcia S, Li J, Yoshida S, Asami T, Chory J (2002) BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 109: 181–191 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J (2005) A new class of transcription factors mediates brassinosteroid‐regulated gene expression in Arabidopsis . Cell 120: 249–259 [DOI] [PubMed] [Google Scholar]
- Yu X, Li L, Zola J, Aluru M, Ye H, Foudree A, Guo H, Anderson S, Aluru S, Liu P, Rodermel S, Yin Y (2011) A brassinosteroid transcriptional network revealed by genome‐wide identification of BESI target genes in Arabidopsis thaliana . Plant J 65: 634–646 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Mayba O, Pfeiffer A, Shi H, Tepperman JM, Speed TP, Quail PH (2013) A quartet of PIF bHLH factors provides a transcriptionally centered signaling hub that regulates seedling morphogenesis through differential expression‐patterning of shared target genes in Arabidopsis . PLoS Genet 9: e1003244 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Review Process File
Source Data for Figure 1
Source Data for Figure 3
Source Data for Figure 5
Data Availability Statement
Microarray results have been deposited in the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) with the accession code GSE85211.


