Abstract
Increasing evidence indicates that tumor-initiating cells (TICs) are responsible for the occurrence, development, recurrence, and development of the drug resistance of cancer. MicroRNA (miRNA) plays a significant functional role by directly regulating targets of TIC-triggered non-small-cell lung cancer (NSCLC), but little is known about the function of the miR-30 family in TICs. In this study, we found the miR-30 family to be downregulated during the spheroid formation of NSCLC cells, and patients with lower miR-30a/c expression had shorter overall survival (OS) and progression-free survival (PFS). Moreover, transmembrane 4 super family member 1 (TM4SF1) was confirmed to be a direct target of miR-30a/c. Concomitant low expression of miR-30a/c and high expression of TM4SF1 correlated with a shorter median OS and PFS in NSCLC patients. miR-30a/c significantly inhibited stem-like characteristics in vitro and in vivo via suppression of its target gene TM4SF1, and then it inhibited the activity of the mTOR/AKT-signaling pathway. Thus, our data provide the first evidence that TM4SF1 is a direct target of miR-30a/c and miR-30a/c inhibits the stemness and proliferation of NSCLC cells by targeting TM4SF1, suggesting that miR-30a/c and TM4SF1 may be useful as tumor biomarkers for the diagnosis and treatment of NSCLC patients.
Keywords: miR-30 family, TM4SF1, NSCLC, biomarker, TICs
miRNAs play a significant functional role in TIC-triggered NSCLC. Herein, Ma et al. demonstrate that TM4SF1 is a direct target of miR-30a/c and miR-30a/c inhibits the stemness and proliferation of NSCLC cells by targeting TM4SF1, suggesting that miR-30a/c and TM4SF1 may be useful as tumor biomarkers for NSCLC.
Introduction
According to the newest statistics published by the American Cancer Society, lung cancer is the most lethal cancer in both males and females,1 of which non-small-cell lung cancer (NSCLC) accounts for about 85% of all lung cancers.2 Despite the great improvement in recent molecular-targeted therapy, 5-year survival rates remain poor due to the development of resistance and eventual relapse.3 For this reason, additional analysis of the mechanisms resulting in the occurrence and the development of NSCLC is required to improve the survival rate of lung cancer.
Data from leukemia, germ cell tumors, and a number of solid tumors support the notion that cancers are maintained by a subpopulation of self-renewing and evolving tumor-initiating cells (TICs),4, 5, 6, 7 also known as the cancer stem cell (CSC) model.8 Recent studies have suggested that TICs located within lung tumors, which display the capacity for renewal and reproduction of tumor heterogeneity, are responsible for NSCLC progression, resistance to therapy, and the hierarchical organization of tumor cells.9 Thus, identification of the factors critical to the various states of lung TICs (LTICs) may open new avenues in NSCLC therapy. Moreover, based on the CSC theory, the presence of LTICs may be of prognostic relevance to patients with NSCLC.10 However, the manner in which LTICs sustain their self-renewal remains largely unknown, and the clinical significance of LTICs for NSCLC has yet to be fully established.
MicroRNAs (miRNAs) are a class of conserved, small non-coding RNAs, approximately 20–22 nt, which could inhibit gene expression by binding to the 3′ UTR of their target mRNAs.11 Evidence has demonstrated that miRNAs play vital roles in regulating the self-renewal and tumorigenesis of TICs.12, 13 miR-30 has been described as a hub for the miRNA oncogenesis signal network in solid tumors,14 whose up- or downregulation has profound impacts on tumorigenesis.15 Bockhorn et al.16 has identified that miR-30c inhibits human breast tumor chemotherapy resistance by regulating Twinfilin-1 (TWF1) and interleukin 11 (IL-11). Intriguingly, miR-30 has been reported to inhibit the self-renewal and metastasis of breast TICs,17 repress the stem cell and cancer phenotypes via targeting LIN28,18 and play a regulatory role in self-renewal and neural differentiation in glioma cells.19 However, the exact role of miR-30 in regulating the biological behavior of LTICs remains unclear.
To better identify miRNAs involved in the LTICs self-renewing and differentiation processes, in this study, we analyzed genome-wide transcriptional profiling using an miRNA microarray in lung CSCs (LCSCs) at different stages. We found that expression levels of several tumor-related miRNAs were altered during spheroid formation of NSCLC cell lines. We further demonstrated that miR-30a/c inhibits self-renewal and the tumorigenesis of LTICs by targeting the oncogene transmembrane 4 super family member 1 (TM4SF1).
Results
Isolation and Identification of LCSCs
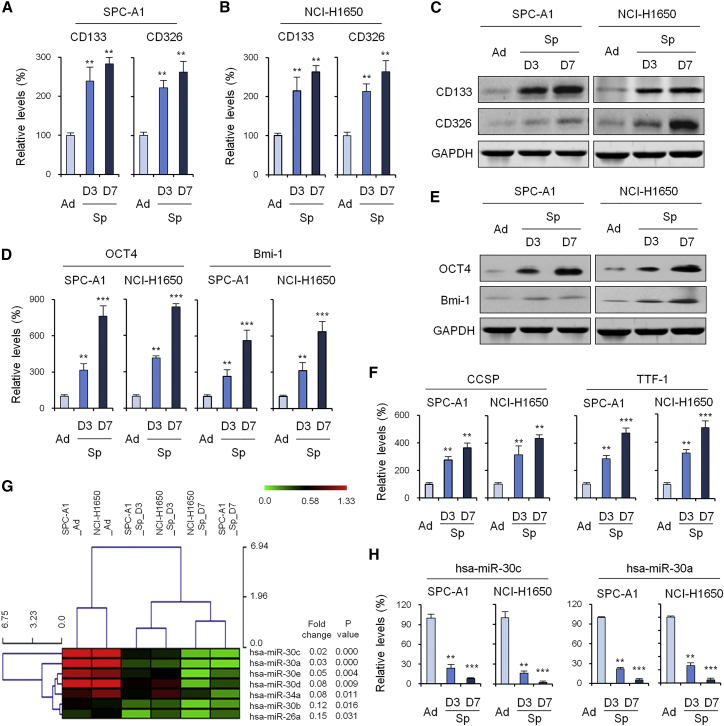
To obtain tumor spheres from NSCLC cell lines in vitro, SPC-A1 and NCI-H1650 cells were first cultured as adherent monolayers. Cells were then transferred to stem cell medium containing epidermal growth factor (EGF) and basic fibroblast growth factor (bFGF). After 3 days in culture, macroscopic and spherical or oval tumor spheres were formed. After another 7 days of continued proliferation, SPC-A1 and NCI-H1650 cells showed spherical suspension growth. Because CD133 and CD326 are well-known LCSC surface markers, we measured CD133 and CD326 mRNA expression and protein expression using qRT-PCR and western blot. The results showed that the cultured tumorigenic lung tumor spheres exhibited stem-like features, with gradual and significant increases in the expression of CD133 and CD326 at both the mRNA (Figures 1A and 1B) and protein (Figure 1C) levels. We detected the expression levels of embryonic stem cell markers Oct-4 and Bmi-1 using RNA extracted from adherent and spheroids of SPC-A1 and NCI-H1650 cells. The results showed that the mRNA and protein levels of Oct-4 and Bmi-1 were significantly increased in cells grown as spheres compared with those grown in adherence (Figures 1D and 1E). Moreover, tumor spheres from SPC-A1 and NCI-H1650 cells overexpressed lung stem cell markers, such as Clara cell secretory protein (CCSP) and thyroid transcription factor 1 (TTF-1) (Figure 1F).
Figure 1.
Isolation and Identification of Lung Cancer Stem Cells
(A and B) CD133 and CD326 mRNA expression during spheroid formation of SPC-A1 (A) and NCI-H1650 (B) cell lines by qRT-PCR. (C) CD133 and CD326 protein expression during spheroid formation of SPC-A1 and NCI-H1650 cell lines by western blot. (D and E) Embryonic stem cell markers (OCT 4 and Bmi-1) expression during spheroid formation of SPC-A1 and NCI-H1650 cell lines by qRT-PCR (D) and western blot (E). (F) Lung stem cell markers (CCSP and TTF-1) expression during spheroid formation of SPC-A1 and NCI-H1650 cell lines by qRT-PCR. (G) Unsupervised hierarchical clustering of the expression data during spheroid formation of SPC-A1 and NCI-H1650 cell lines. (H) Expression of miR-30c/a during spheroid formation of SPC-A1 and NCI-H1650 cell lines by qRT-PCR. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (**p < 0.01 and ***p < 0.001 vs. control).
Subsequently, miRNA expression of adherent and spheroids of SPC-A1 and NCI-H1650 cells was detected by microarray. We performed unsupervised hierarchical clustering of the expression data in order to evaluate the miRNA diversity among these cells. When comparing the expression levels of miRNAs from both sample groups, a handful of reported differential miRNAs, including miRNA-124a,3 miRNA-26a,5 and miRNA-34a,12 has been discovered by this method. Moreover, our results showed that the gene expression level of the miR-30 family was gradually and significantly decreased during the formation of tumor spheres from SPC-A1 and NCI-H1650 cells (Figure 1G).
To validate these differentially regulated miRNAs, we examined the miR-30 family at the level of transcription in RNA extracted from adherent and spheroids of SPC-A1 and NCI-H1650 cells. The amounts of the indicated transcripts, analyzed by real-time RT-PCR, were significantly lower in cells grown as spheres than in those grown in adherence. Among them, the levels of expression of miR-30c and -30a in tumor spheres from SPC-A1 and NCI-H1650 cells were extremely significantly lower than those of the control group (p < 0.001) (Figure 1H).
Differentiation and CSC Marker Expression in LCSCs
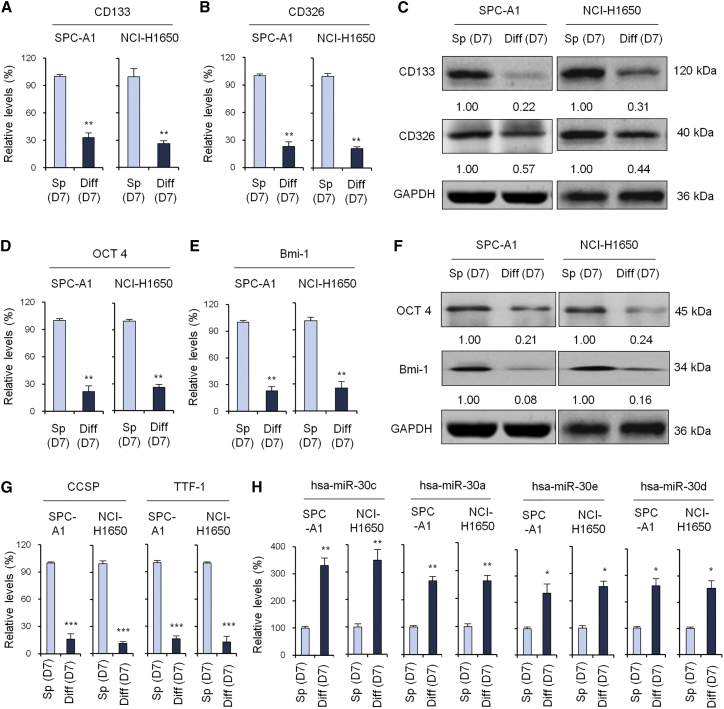
Tumor spheres cultured in suspension may re-differentiate into adherent cancer cells when added to routine medium containing serum. Next, we detected CSC marker expression in tumor spheres from SPC-A1 and NCI-H1650 cells cultured in suspension in serum-free chemically defined medium to maintain stemness or in monolayers after the addition of fetal bovine serum (FBS) to induce differentiation. The results showed that the mRNA expression (Figures 2A and 2B) and protein expression (Figure 2C) of stem cell-associated genes, including CD133 and CD326, in re-differentiated cells were significantly reduced after FBS treatment. The expression levels of stem cell markers, such as embryonic Oct-4 and Bmi-1 (Figures 2D–2F), and the lung stem cell markers CCSP and TTF-1 (Figure 2G) in re-differentiated cells were also significantly reduced following FBS treatment. Conversely, the expression of the miR-30 family, including miR-30c, -30a, -30e, and -30d, was significantly higher in adherent SPC-A1 and NCI-H1650 cells than in the tumor spheres cultured in suspension (Figure 2H).
Figure 2.
Cancer Stem Cell Marker Expression of Spheroid Cell and Differentiated Cells
(A) qRT-PCR to evaluate CD133 expression of tumor sphere cell and differentiated cell in SPC-A1 and NCI-H1650 cell lines. (B) qRT-PCR to evaluate CD326 expression of tumor sphere cell and differentiated cell in SPC-A1 and NCI-H1650 cell lines. (C) Western blot analysis to evaluate CD133 and CD326 expression of tumor sphere cell and differentiated cell in SPC-A1 and NCI-H1650 cell lines. (D–F) qRT-PCR (D and E) and western blot (F) to evaluate the expression of embryonic stem cell markers OCT 4 (D and F) and Bmi-1 (E and F). (G) qRT-PCR to evaluate the expression of lung stem cell markers CCSP and TTF-1. (H) qRT-PCR to evaluate the expression of miR-30c, -30a, -30e, and -30d in tumor spheres and differentiated cells. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (*p < 0.05, **p < 0.01, and ***p < 0.001 vs. control).
Knockdown and Overexpression Experiments and Spheroid Formation, Cell Growth, and Apoptosis Assays
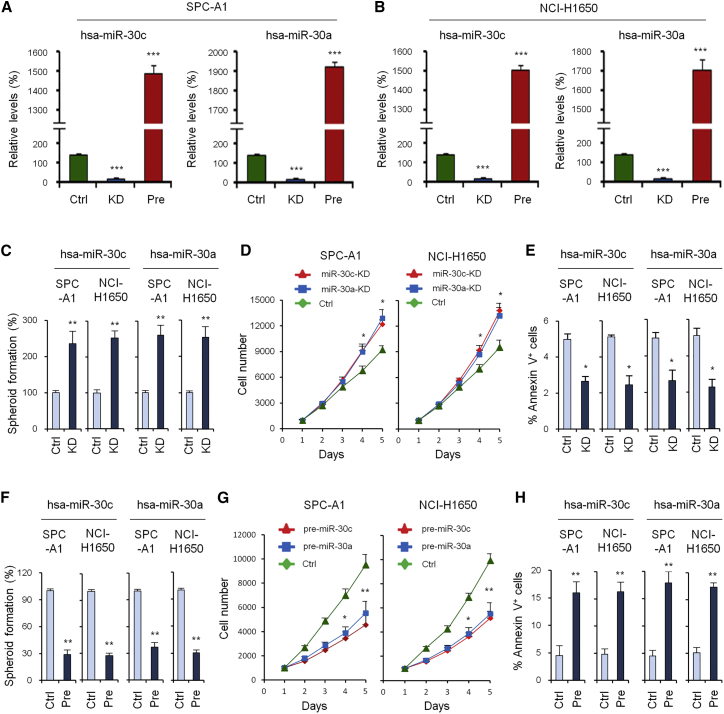
To further investigate the role of hsa-miR-30c/a in NSCLC, we constructed hsa-miR-30c/a overexpression and knockdown cells in SPC-A1 and NCI-H1650 cell lines, and we verified their expression by qRT-PCR (Figures 3A and 3B). The spheroid formation assay showed the hsa-miR-30c/a knockdown group had a stronger spheroid formation ability than the control group (Figure 3C), while the overexpression group showed inhibited spheroid formation (Figure 3F). Cell growth assays showed that, compared with the control group, the knockdown group was found to promote cell growth (Figure 3D), while the overexpression group could inhibit cell growth (Figure 3G). The apoptosis assay showed that, compared with the control group, the knockdown group could inhibit cell apoptosis (Figure 3E), while the overexpression group could promote cell apoptosis (Figure 3H).
Figure 3.
Knockdown and Overexpression Experiments, Spheroid Formation Assay, Cell Growth Assays, and Apoptosis Assay
(A) Overexpression and knockdown of miR-30c/a verified by qRT-PCR in the SPC-A1 cell line. (B) Overexpression and knockdown of miR-30c/a verified by qRT-PCR in the NCI-H1650 cell line. (C) Spheroid formation assay of the control group and knockdown groups of miR-30c/a at day 7. (D) Cell growth assays of the control group and knockdown groups of miR-30c/a. (E) Apoptosis assay of the control group and knockdown groups of miR-30c/a. (F) Spheroid formation assay of the control group and overexpression groups of miR-30c/a. (G) Cell growth assays of the control group and overexpression groups of miR-30c/a. (H) Apoptosis assay of the control group and overexpression groups of miR-30c/a. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (*p < 0.05, **p < 0.01, and ***p < 0.001 vs. control).
Prediction of Target Genes of hsa-miR-30c and hsa-miR-30a
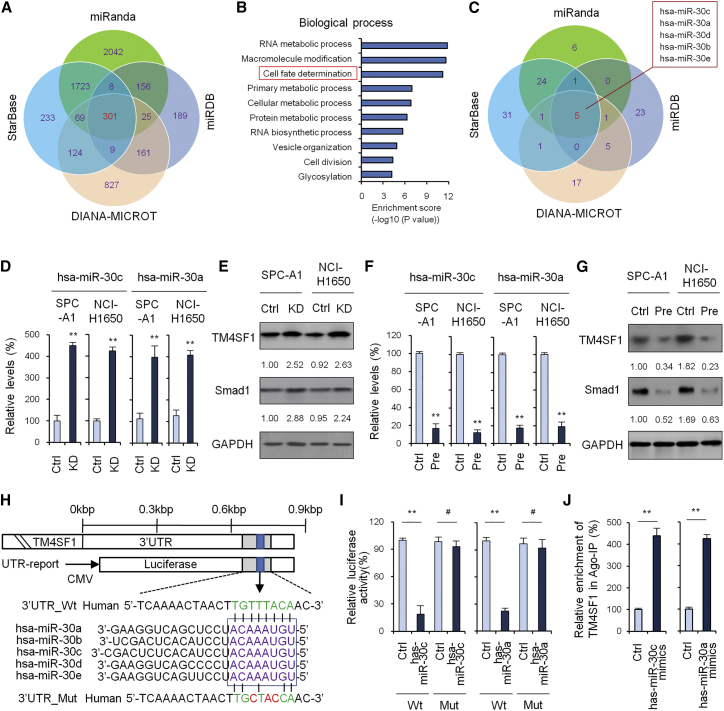
To predict target genes of hsa-miR-30c and hsa-miR-30a, we used 4 target gene prediction software packages: miRanda, miRDB, DIANA-MICROT, and StarBase. Based on this analysis, we found 301 common target genes (Figure 4A). Gene ontology (GO) analysis showed the top 3 GO terms in biological process to be RNA metabolic process, macromolecule modification and cell fate determination (Figure 4B). Of these, TM4SF1 was included as a cell fate determination term. Next, we used the 4 prediction software packages to explore potential target miRNAs of TM4SF1; the results showed there to be 5 intersections: hsa-miR-30c, hsa-miR-30a, hsa-miR-30d, hsa-miR-30b, and hsa-miR-30e (Figure 4C).
Figure 4.
Prediction of Target Genes of hsa-miR-30c and hsa-miR-30a
(A) Prediction of target genes of hsa-miR-30c and hsa-miR-30a using 4 target gene prediction software packages: miRanda, miRDB, DIANA-MICROT, and StarBase. 301 common target genes were found. (B) GO analysis of the 301 common target genes. (C) Prediction of potential target miRNA of TM4SF1 using 4 target gene prediction software. (D) TM4SF1 expression examined by qRT-PCR when hsa-miR-30c/a were knocked down. (E) TM4SF1 expression examined by western blot when hsa-miR-30c/a were knocked down. (F) TM4SF1 expression examined by qRT-PCR when hsa-miR-30c/a were overexpressed. (G) TM4SF1 expression examined by western blot when hsa-miR-30c/a were overexpressed. (H) The 3′ UTR-binding site and mutation site of miR-30 family of the TM4SF1 gene. (I) Luciferase reporter assay to verify whether hsa-miR-30c/a directly targeted TM4SF1. (J) RIP-IP assays were performed to co-immunoprecipitate the Ago2 complexes from SPC-A1 cells transfected with either hsa-miR-30c/a mimic or negative control mimic. Real-time PCR assays were performed on RNA samples isolated from the Ago2 coIP fractions to measure the relative enrichment of the TM4SF1 mRNA. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (**p < 0.01 and #p > 0.05 vs. control).
We then performed qRT-PCR and western blot analysis to verify whether TM4SF1 was a target gene of hsa-miR-30c and hsa-miR-30a. The results showed that TM4SF1 was obviously upregulated in the hsa-miR-30c/a knockdown group over control levels (Figures 4D and 4E), while TM4SF1 was visibly downregulated in the hsa-miR-30c/a overexpression group (Figures 4F and 4G). The 3′ UTR-binding site and mutation site of the miR-30 family of the TM4SF1 gene are shown in Figure 4H. Next, we performed luciferase reporter assay to further verify whether hsa-miR-30c/a directly targeted TM4SF1. As shown in Figure 4I, ectopic expression of hsa-miR-30c/a decreased the luciferase activity of the 3′ UTRs of TM4SF1. However, hsa-miR-30c/a did not have an inhibitory effect on luciferase activity of 3′ UTR_Mut of TM4SF1.
Mature miRNAs form stable complexes with Argonaute proteins (such as Ago2), which are the core of the RNAi-induced silencing complex (RISC). The miRNA then directs RISC binding to mRNA molecules containing specific targeting sequences, and this results in translation inhibition and/or enhanced mRNA degradation.20 To further confirm that hsa-miR-30c/a directly interacts with TM4SF1 mRNA, we tested hsa-miR-30c/a-mediated binding of RISC to TM4SF1 mRNA using the Ago2-based ribonucleoprotein-immunoprecipitation (IP) assay (RIP-IP). One irrelevant binding miRNA was used as a negative control. As shown in Figure 4J, the Ago2 co-immunoprecipitation (coIP) fraction from cells treated with the hsa-miR-30c/a mimic was significantly enriched for TM4SF1 mRNA compared to cells treated with control miRNA mimics.
TM4SF1 and miR-30c/a Expression in Different Differentiation Cells
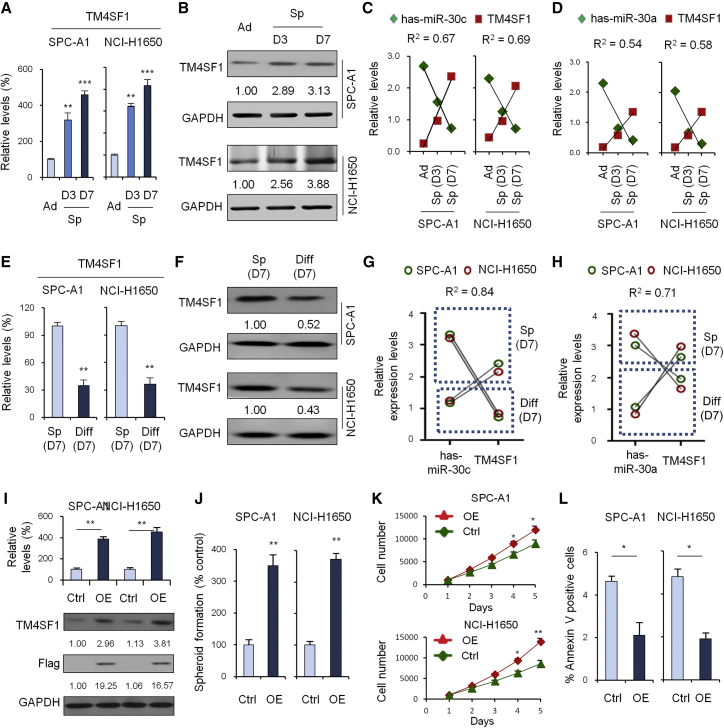
We performed qRT-PCR and western blot to investigate the expression of TM4SF1 in 0-, 3-, and 7-day SPC-A1 and NCI-H1650 cells of spheroid culture. As shown in Figures 5A and 5B, TM4SF1 expression increased over time and was expressed at high levels in spheroid (7-day) cells. Correction analysis showed there to be a significant negative correlation between has-miR-30c/a and TM4SF1 expression (Figures 5C and 5D). qRT-PCR and western blot analysis of spheroid cells (day 7) and differentiated cells showed differences from the differentiated cells: TM4SF1 was found to be significantly highly expressed in spheroid cells (Figures 5E and 5F). Correction analysis in Figures 5G and 5H showed a significant negative correlation, as above.
Figure 5.
TM4SF1 and miR-30c/a Expression in Different Differentiated Cells
(A) qRT-PCR to investigate the expression of TM4SF1 in 0-, 3-, and 7-day SPC-A1 and NCI-H1650 cells of spheroid culture. (B) Western blot to investigate the expression of TM4SF1 in 0-, 3-, and 7-day SPC-A1 and NCI-H1650 cells of spheroid culture. (C) Correction analysis of has-miR-30c and TM4SF1 expression. (D) Correction analysis of has-miR-30a and TM4SF1 expression. (E) qRT-PCR analysis of spheroid cells (day 7) and differentiated cells. (F) Blot analysis of spheroid cells (day 7) and differentiated cells. (G) Correction analysis of has-miR-30c and TM4SF1 expression. (H) Correction analysis of has-miR-30a and TM4SF1 expression. (I) Overexpression of TM4SF1 verified by qRT-PCR and western blot. (J) Spheroid formation assay of the control group and the TM4SF1 overexpression group at day 7. (K) Cell growth assays of the control group and the TM4SF1 overexpression group. (L) Apoptosis assay of the control group and the TM4SF1 overexpression group. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (*p < 0.05, **p < 0.01, and ***p < 0.001 vs. control).
Next, we wanted to investigate the influence of TM4SF1 overexpression on cell function. First, we verified overexpression of TM4SF1 by qRT-PCR and western blot (Figure 5I). A spheroid formation assay showed that, compared with the control group, the overexpression group could promote spheroid formation (Figure 5J). Cell growth assays showed that, compared with the control group, the overexpression group could promote cell growth (Figure 5K). An apoptosis assay showed that, compared with the control group, the overexpression group could inhibit cell apoptosis (Figure 5L).
miR-30c/a Regulates TM4SF1 Expression
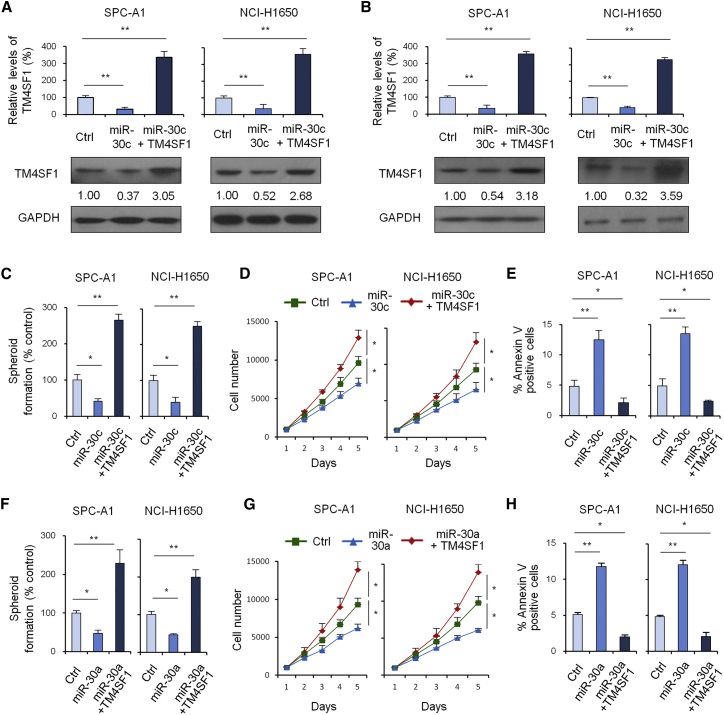
To further verify whether miR-30c/a regulates TM4SF1 expression, we performed qRT-PCR and western blot analysis of TM4SF1 expression in 3 groups of cells, including the control group, the miR-30c overexpression group, and the miR-30c and TM4SF1 overexpression group. The results showed miR-30c/a could downregulate TM4SF1 expression (Figures 6A and 6B). A spheroid formation assay showed that the miR-30c/a group could inhibit spheroid formation in a way not observed in the control group (Figures 6C and 6F). Cell growth assays showed that, compared with the control group, the miR-30c/a group could inhibit cell growth (Figures 6D and 6G). An apoptosis assay showed that, compared with the control group, the miR-30c/a group could promote cell apoptosis (Figures 6E and 6H). All these results suggested that miR-30c/a could directly inhibit TM4SF1 expression and function.
Figure 6.
miR-30a/c Regulate TM4SF1 Expression
(A) qRT-PCR and western blot analysis of TM4SF1 expression in 3 groups of cells: the control group, miR-30c group, and miR-30c + TM4SF1 group. (B) qRT-PCR and western blot analysis of TM4SF1 expression in 3 groups of cells: the control group, miR-30a group, and miR-30a + TM4SF1 group. (C) Spheroid formation assay of the control group, miR-30c group, and miR-30c + TM4SF1 group. (D) Cell growth assays of the control group, miR-30c group, and miR-30c + TM4SF1 group. (E) Apoptosis assay of the control group, miR-30c group, and miR-30c + TM4SF1 group. (F) Spheroid formation assay of the control group, miR-30a group, and miR-30a + TM4SF1 group. (G) Cell growth assays of the control group, miR-30a group, and miR-30a + TM4SF1 group. (H) Apoptosis assay of the control group, miR-30a group, and miR-30a + TM4SF1 group. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (*p < 0.05 and **p < 0.01 vs. control).
miR-30c/a Inhibits Tumor Growth In Vivo by Targeting TM4SF1
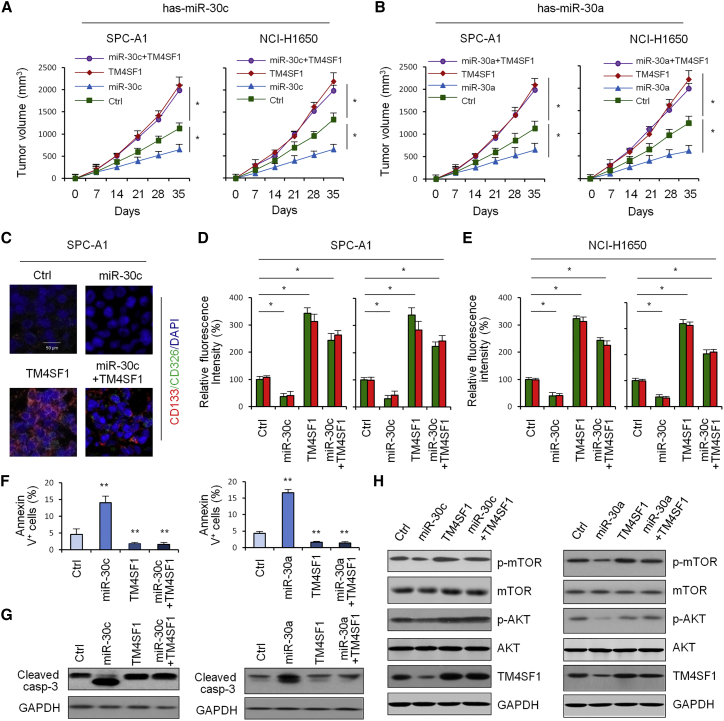
To further investigate whether and how miR-30c/a affects lung cancer formation in vivo, we overexpressed miR-30c/a and/or TM4SF1 in SPC-A1 and NCI-H1650 cell lines. Figures 7A and 7B show that TM4SF1 could promote NSCLC tumor growth, while miR-30c/a could inhibit tumor growth in vivo by targeting TM4SF1. Next, we observed staining of CSC surface markers CD326 and CD133 of 4 group tumor samples under a fluorescent microscope (Figure 7C). We quantitatively analyzed relative fluorescence intensity of the 4 samples. Figures 7D and 7E show that CD326 and CD133 expression was upregulated in NSCLC tissues compared with paracarcinoma tissues, TM4SF1 could promote CSC surface marker expression, and miR-30c could inhibit CSC surface marker expression by targeting TM4SF1. The apoptosis assay in Figure 7F showed that the rate of apoptosis was lower in NSCLC tissues compared with paracarcinoma tissues, TM4SF1 could inhibit cell apoptosis, and miR-30c could promote cell apoptosis by targeting TM4SF. We also investigated how TM4SF1 and miR-30c affect apoptotic signal molecules cleaved-caspase-3 by western blot. The results showed that miR-30c can promote cell apoptosis by targeting TM4SF (Figure 7G). Next, we performed western blot analysis to determine whether miR-30a/c and TM4SF1 affect the activity of the mTOR/AKT-signaling pathway. The results showed that miR-30a/c inhibited the activity of the mTOR/AKT-signaling pathway (Figure 7H).
Figure 7.
miR-30c/a Inhibit Tumor Growth In Vivo by Targeting TM4SF1
(A) Tumor volume curves of the control group, miR-30c group, TM4SF1 group, and miR-30c + TM4SF1 group. (B) Tumor volume curves of the control group, miR-30a group, TM4SF1 group, and miR-30a + TM4SF1 group. (C) Staining of cancer stem cell surface markers CD326 and CD133. (D) Quantitative analysis of relative fluorescence intensity of the para group, control group, miR-30c group, TM4SF1 group, and miR-30c + TM4SF1 group in SPC-A1 cells. Green, CD326; red, CD133. (E) Quantitative analysis of relative fluorescence intensity of the para group, control group, miR-30c group, TM4SF1 group, and miR-30c + TM4SF1 group in HCI-H1650 cells. Green, CD326; red, CD133. (F) Apoptosis assay of control group, miR-30c group, TM4SF1 group, and miR-30c + TM4SF1 group in HCI-H1650 cells. (G) Influence of miR-30c and TM4SF1 expression on apoptotic signal molecules cleaved-caspase-3 expression by western blot. (H) Influence of miR-30c and TM4SF1 expression on AKT/mTOR pathway-associated protein expressions by western blot. Data are shown as the means ± SDs of three independent experiments. Statistical analyses were performed with one-way ANOVA (*P<0.05 and **P<0.01 vs. control).
miR-30c/a and TM4SF1 Expression in NSCLC Tissue
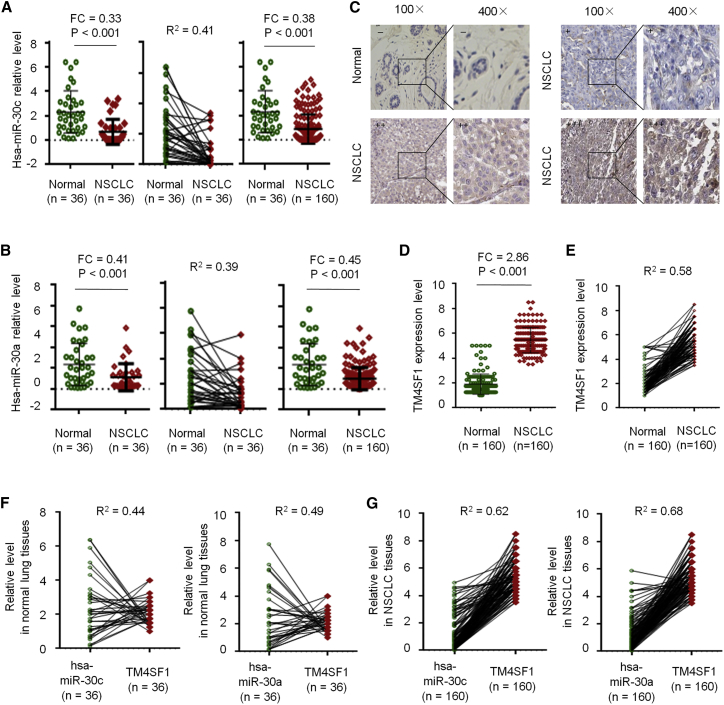
To further investigate miR-30c/a expression level in NSCLC tissue, we performed qRT-PCR in 36 paired NSCLC tissues and 124 normal lung tissues. The results showed that, when compared with normal tissues, miR-30c/a was significantly downregulated in NSCLC (Figures 8A and 8B). Immunohistochemistry (IHC) staining and qRT-PCR analysis showed that TM4SF1 was significantly upregulated in NSCLC (Figures 8C–8E). Next, correction analysis showed a significant negative correlation between miR-30c/a and TM4SF1 expression (Figures 8F and 8G).
Figure 8.
miR-30c/a and TM4SF1 Expression in NSCLC Tissue
(A) miR-30c expression by qRT-PCR in 36 paired NSCLC tissues and 124 normal lung tissues. (B) miR-30a expression by qRT-PCR in 36 paired NSCLC tissues and 124 normal lung tissues. (C) IHC staining of TM4SF1 expression in normal and NSCLC tissues. (D) qRT-PCR analysis of TM4SF1 expression in normal and NSCLC tissues. (E) Correlation analysis. (F) Correlation analysis of miR-30c and TM4SF1 expression. (G) Correlation analysis of miR-30a and TM4SF1 expression.
Clinical Significance of miR-30c/a and TM4SF1 in NSCLC
Kaplan-Meier survival curves were plotted and log rank analysis was performed to evaluate the prognostic value of miR-30c/a and TM4SF1 in NSCLC. The results indicated that miR-30c/a high expression was correlated with longer overall survival (OS) and progression-free survival (PFS) (Figures S1A and S1B) in NSCLC patients, while TM4SF1 high expression was correlated with shorter OS and PFS (Figure S1C) in NSCLC patients. Because the data suggested that TM4SF1 expression was negatively correlated with miR-30c/a expression and TM4SF1 was a direct target of miR-30c/a, we further examined the prognostic value of TM4SF1 expression together with miR-30c/a levels using multivariate analysis of OS and PFS by Kaplan-Meier survival analysis. The results showed that NSCLC patients with high miR-30c/a and low TM4SF1 expression levels had significantly greater levels of OS and PFS (Figures S1D and S1E), which suggested that miR-30c/a and TM4SF1 might have prognostic value and could be useful as tumor biomarkers for the diagnosis of NSCLC patients.
Discussion
Lung cancer is the most lethal cancer in the world. Its 5-year survival rates vary from 4%–17% depending on the stage and regional differences. Every year, 1.8 million people are diagnosed with lung cancer, and 1.6 million people die as a result of the disease.21 Despite advances in therapies, such as chemotherapy, radiotherapy, and targeted therapies, 5-year survival rates remain poor. A better understanding of the basis of lung cancer heterogeneity and drug resistance is vital to improving the outcome of lung cancer patients.
The CSC model proposes that tumors are hierarchically organized, and within the cancer population of the tumors, there are CSCs, also known as TICs, which are tumorigenic cells biologically distinct from other subpopulations.22 The CSC model has attracted a significant amount of attention in recent years as a viable explanation for the heterogeneity, drug resistance, dormancy, recurrence, and metastasis of various tumors.23 More and more evidence has shown that LCSCs are responsible for the occurrence, development, recurrence, and development of drug resistance.24 However, the manner in which LCSCs sustain their self-renewal remains largely unknown, and the clinical significance of LCSCs for NSCLC has yet to be fully established.
An miRNA is a small non-coding RNA molecule containing about 22 nt that functions in RNA silencing and post-transcriptional regulation of gene expression.25 miRNAs regulate many biological processes, including cell cycle regulation, cellular growth, proliferation, differentiation, apoptosis, metabolism, neuronal patterning, and aging, and dysregulation of miRNAs plays an important role in tumorigenesis, development, metastasis, and prognosis.26 The miR-30 family includes 5 members, hsa-miR-30c, hsa-miR-30a, hsa-miR-30d, hsa-miR-30b, and hsa-miR-30e, all of which share the same seed sequence and are encoded by 6 genes located on human chromosomes 1, 6, and 8.27 miR-30 was described as a hub for the miRNA oncogenesis signal network in solid tumors,28 whose up- and down-modulation has profound impacts on tumorigenesis.29
In a report by Fan et al.,30 miR-30 was found to suppress the lung cancer cell 95D epithelial-mesenchymal transition and invasion through targeted regulation of Snail. Chan et al.31 found that the miR-30 family may target PI3K-SIAH2 and that the associated predicted interaction network may serve as a novel putative theranostic panel in NSCLC. Yu et al.32 found that matrix metalloproteinase-19 promotes metastatic behavior in vitro and is associated with increased mortality in NSCLC. Zhong et al.33 found that miR-30b/c inhibits NSCLC cell proliferation by targeting Rab18. Low expression of miR-30c was reported to promote invasion by inducing an epithelial-mesenchymal transition in NSCLC.34 Moreover, miR-30 has been reported to play an important role in CSCs. miR-30 reduction maintains self-renewal and inhibits apoptosis in breast TICs,35 and the miR-30 family promotes migratory and invasive abilities in CD133+ pancreatic cancer stem-like cells.36 Che et al.37 found that miR-30 overexpression promotes glioma stem cells by regulating the Jak/STAT3-signaling pathway. Zhong et al.38 found that miR-30 regulated reprogramming factor LIN28 in embryonic stem cells and cancer cells. However, the exact role of miR-30 in regulating the biological behavior of LCSCs remains unclear.
In this study, through the genome-wide transcriptional profiling analysis with an miRNA microarray with different stages of LCSCs, a handful of reported differential miRNAs, including miRNA-124a,3 miRNA-26a,5 and miRNA-34a,12 has been discovered by this method. Tumor-suppressive microRNA-124a has been reported to inhibit stemness and enhance gefitinib sensitivity of NSCLC cells by targeting ubiquitin-specific protease 14.3 Lu et al.4 found that miR-26a inhibits CSC-like phenotype and tumor growth by targeting Jagged1, one of the Notch ligand, and that its tumor-suppressive effects are mediated through the inhibition of Jagged1/Notch signaling. Basak et al.12 showed that miR-34a is a negative regulator of the tumorigenic properties of NSCLC cells and CD44hi stem-like NSCLC cells, and they establish a strong rationale for developing miR-34a as a novel therapeutic agent against NSCLC. Moreover, we found that miR-30s were significantly altered during spheroid formation of NSCLC cell lines. qRT-PCR analysis verified that miR-30 expression was significantly downregulated during spheroid formation, and overexpression of miR-30a/c inhibited spheroid formation, inhibited cell growth, and promoted cell apoptosis.
To further explore the regulating mechanism, we performed bioinformatics analysis and confirmed TM4SF1 as a direct target of miR-30a/c by qRT-PCR, western blot, and luciferase reporter assays. TM4SF1, a small plasma membrane glycoprotein that regulates cell motility and proliferation, is highly expressed in different carcinomas, and it has been reported to be a new vascular therapeutic target in cancer.39 Previous studies have shown that TM4SF1 plays an important role in cell proliferation, invasion, metastasis, and radiotherapy resistance in different carcinomas. Upregulation of TM4SF1 predicts poor prognosis in invasive breast cancer,40 and TM4SF1 regulates apoptosis, cell cycle, and ROS metabolism via the PPARγ-SIRT1 feedback loop in human bladder cancer cells.41 Besides, Shih et al.42 found that TM4SF1 is critical to endothelial cell function and tumor angiogenesis. Cao et al.43 found that TM4SF1 regulates pancreatic cancer migration and invasion in vitro and in vivo. Sun et al.44 found that TM4SF1 regulates breast cancer cell migration and apoptosis through the PI3K/AKT/mTOR pathway. Xue et al.45 found that TM4SF1 promotes the self-renewal of esophageal cancer stem-like cells and is regulated by miR-141. Choi et al.46 found that TM4SF4 overexpression in radiation-resistant lung carcinoma cells activates IGF1R via the elevation of IGF1. However, the regular role of miR-30 on TM4SF1 in LCSCs remains unclear. In this study, our data showed that TM4SF1 was significantly upregulated during spheroid formation, and overexpression of TM4SF41 promoted spheroid formation, promoted cell growth, and inhibited cell apoptosis. Moreover, TM4SF1 was found to be a direct target of miR-30a/c, and miR-30a/c significantly inhibited stem-like characteristics in vitro and in vivo via the suppression of its target gene TM4SF1.
The PI3K/AKT/mTOR pathway is an intracellular signaling pathway important to regulating the cell cycle. In this way, it is directly related to cellular quiescence, proliferation, cancer, and longevity. Aziz et al.47 found the PI3K/AKT/mTOR pathway to be a central regulator of ovarian cancer. Raphael et al.48 found aberrations in the PI3K/AKT/mTOR pathway to be the most common genomic abnormalities in many kinds of breast cancer. It has been reported that TM4SF1 regulated breast cancer cell migration and apoptosis through the PI3K/AKT/mTOR pathway.49 Our data first showed that miR-30a/c significantly inhibited the activity of the mTOR/AKT-signaling pathway by targeting the oncogene TM4SF1.
In conclusion, our data confirmed that miR-30a/c significantly inhibited stem-like characteristics in vitro and in vivo via suppression of the target gene TM4SF1 and then inhibited the activity of the mTOR/AKT-signaling pathway. Low expression of miR-30a/c and high expression of TM4SF1 were found to be independent prognostic factors in NSCLC patients. Thus, our data provide the first evidence that TM4SF1 is a direct target of miR-30a/c and miR-30a/c inhibits stemness and proliferation of NSCLC cells by targeting TM4SF1, which suggests that miR-30a/c and TM4SF1 may be useful as tumor biomarkers for the diagnosis and treatment of NSCLC patients.
Materials and Methods
Tissue Sample and Ethics Statement
Fresh NSCLC samples were collected from patients undergoing surgical resection between 2008 and 2012 in the Department of Biobank, China-Japan Union Hospital and Shanghai Tenth People’s Hospital, which included paired tumor and adjacent non-cancerous tissues (n = 36) and a large cohort of individual NSCLC biopsies (n = 160), and they were classified according to the current WHO classification. Two experienced pathologists independently confirmed NSCLC diagnosis. The study was approved by the Ethical Committee of Shanghai Tenth People’s Hospital (SHSY-IEC-pap-15-18). Each subject provided written informed consent before participating in this study.
Cell Lines
Human NSCLC cell lines SPC-A1 and NCI-H1650 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China), and they were cultured in DMEM (Invitrogen, Carlsbad, CA, USA) and supplemented with 10% (v/v) FBS, 100 U/mL penicillin, and 100 mg/mL streptomycin. Cell lines were routinely tested for mycoplasma contamination, and they have been authenticated with short-tandem repeat analysis. Cell culture was conducted at 37°C in a humidified 5% CO2 incubator.
Tumor Sphere Formation Assays
The SPC-A1 and NCI-H1650 cell lines were cultured with ultra-low attachment surface plates (Nunc, Penfield, NY, USA) in serum-free DMEM (Gibco, Life Technologies), supplemented with 20 ng/mL human EGF (hEGF; R&D Systems, Minneapolis, MN, USA), 10 ng/mL hbFGF (R&D Systems), 4 mg/mL heparin sulfate (Sigma, St. Louis, MO), 0.15% BSA (Sigma), and 1% penicillin G-streptomycin.50 Sphere-forming capacity was determined by the ability of cells to form three-dimensional spheroids in culture in a period of 3–7 days. To assess long-term proliferation and fold expansion, spheres were disassociated and passaged every 7 days using 0.05% trypsin-EDTA (Invitrogen, Carlsbad, CA, USA). Cells were counted using a Countess (Invitrogen) automated cell counter. To induce tumor stem cell differentiation, the cultured tumor spheres of SPC-A1 and NCI-H1650 NSCLC cells were incubated with 10% (v/v) FBS for 7 days at 37°C in a humidified chamber supplemented with 5% CO2.
Plasmid Construction and Transfection
Overexpression of hsa-miR-30a/c and TM4SF1 was performed using the plasmid murine stem cell virus (pMSCV) retroviral plasmid. The hsa-miR-30a/c sponge plasmid was constructed by inserting eight tandemly arrayed hsa-miR-30a/c-binding sites into the 3′ UTR of Discosoma sp. red fluorescent protein (dsRed). All constructs were confirmed by sequencing. The plasmids were transiently transfected into target cells with Lipofectamin 2000 (Life Technologies, Gaithersburg, MD, USA). To generate stable cell lines with specific gene overexpression or knockdown, the plasmids were packaged into retroviruses with the amphotropic Phoenix packaging cell line and infected into target cells, followed by puromycin and hygromycin selection of infected cells.
RNA Extraction and qPCR
Total RNA, including miRNAs from normal lung tissue, NSCLC samples, and cell lines, was isolated using Trizol reagent (Life Technologies, Grand Island, NY, USA), according to the manufacturer’s instructions. RNA concentration was measured using a NanoDrop2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Electrophoresis on 1.5% denaturing agarose gels was performed to evaluate the quality of all RNA specimens. qPCR was conducted using the TaqMan Universal PCR Kit (Life Technologies, Carlsbad, CA, USA). The same amount of first-strand cDNA from each sample was used to detect the mRNA expression levels using specific primers. U6 and GAPDH were used as the endogenous control, and the 2−ΔΔCT method was used to analyze expression level.
Microarray Analysis
For microarray analysis of miRNA gene expression, Agilent Array platform was employed. The sample preparation and microarray hybridization were performed based on the manufacturer’s standard protocols. Briefly, 1 μg total RNA from each sample was amplified and transcribed into fluorescent cDNA using the manufacturer’s Agilent’s Quick Amp Labeling protocol (version 5.7, Agilent Technologies). Agilent Quick Amp Labeling Kit was used for sample labeling. Hybridization was performed in Agilent’s SureHyb Hybridization Chambers. The labeled cRNAs were hybridized onto the Whole Human Genome Oligo Microarray (4 × 44,000; Agilent Technologies). After having washed the slides, the arrays were scanned by the Agilent Scanner G2505B. Agilent Feature Extraction software (version 11.0.1.1) was used to analyze the acquired array images. Quantile normalization and subsequent data processing were performed using the GeneSpring GX version (v.)11.5.1 software package (Agilent Technologies).
Luciferase Reporter Assays
Human TM4SF1 3′ UTR oligonucleotides containing wild-type (WT) or mutant (Mut) hsa-miR-30a/c binding site were subcloned into the XhoI and NotI sites of the pGL3 luciferase reporter plasmid (Promega, Madison, WI, USA). For the luciferase assay, SPC-A1 and NCI-H1650 cells were seeded into 24-well plates and cultured for 24 hr, after which cells were co-transfected with either the WT or Mut reporter plasmid. Then 48 hr after transfection, the luciferase assay was performed using the Dual-Luciferase Kit (Promega, Madison, WI, USA).
Antibodies, Western Blot Analysis, and Immunofluorescence
Western blot analyses were performed by standard methods using the indicated antibodies. Densitometry was performed on scanned blots using NIH ImageJ software version 1.45 to quantify the average pixel density per band area after normalization to the pixel density of the corresponding loading controls. The rabbit monoclonal antibodies for CD133 (ab216323), CD326 (ab32392), TM4SF1 (ab113504), Smad1 (ab63356), Caspase-3 (ab13585), p-mTOR (ab131538), mTOR (ab2732), p-AKT (ab38449), and AKT (ab182729) and GAPDH antibody (ab181602) were purchased from Abcam (Cambridge, UK) and used for western blot analyses. The goat anti-rabbit immunoglobulin G (IgG) (Merck) and goat anti-mouse IgG (Merck) antibodies were used for western blot analyses. Antibody dilutions were 1:1,000 for primary antibodies and 1:5,000 for secondary antibodies in western blotting. Data are representative of 3–4 independent experiments.
Cell Viability Assays
For the cell viability assays, 3,000 cells were seeded into 96-well plates, transfected, and cultured overnight. Next, CCK8 (10 L) reagent was then added to each well, the absorbance (A) was measured at 450 nm after 1 hr, and the relative cell viability rate was calculated. All experiments were performed in triplicate.
Flow Cytometry Analysis
Cell apoptosis was determined by flow cytometry analysis. Cells were collected, washed with cold PBS, fixed in cold 70% ethanol, treated with DNase-free RNase (100 μg/mL; RB473, Sangon Biotech, Shanghai, China), and stained with 50 μg/mL propidium iodide (P1112, Sangon Biotech) and the Annexin V-APC/7-AAD Kit (KGA-1025, KeyGEN Biotech, Nanjing, China). The cells were analyzed using a Gallios flow cytometer (Beckman Coulter) to quantify the proportion of cells in apoptosis status.
Immunofluorescence and IHC
For immunofluorescence studies, 2 × 104 cells were plated onto a matrigel-coated 2-well LabTekII chamber slide. Cells were fixed the next day with 4% paraformaldehyde, permeabilized, and blocked with 0.3% Triton X-100, 3% BSA in PBS, followed by incubation with rabbit monoclonal antibody for CD133 and CD326 (1:1,000, Abcam, UK) overnight at 4°C. The primary antibody was detected by incubation with the secondary antibody (1:5,000, Merck) for 1 hr at room temperature. The slides were mounted with ProLong Antifade reagent with DAPI (Invitrogen, Life Technologies) and photographed. Standard IHC and H&E staining were used to evaluate TM4SF1 expression levels in NSCLC samples. Serial sections were stained in parallel with the primary antibody replaced by PBS as controls.
RNA Immunoprecipitation Assay
The EZ-Magna RIP Kit (Millipore, USA) was applied to conduct the RIP assay according to the product specification. First, cells were collected and lysed in complete RIP lysis buffer. Then, the cell extract was incubated with RIP buffer containing magnetic beads conjugated to a human anti-Ago2 antibody (Millipore, USA). Samples were incubated with proteinase K with shaking to digest proteins, and the immunoprecipitated RNA was isolated. Subsequently, the NanoDrop spectrophotometer was used to measure the concentration of RNA, and the purified RNA was subjected to real-time PCR analysis.51
Bioinformatics Analysis
GO classification (http://www.geneontology.org/) was used to evaluate the biological function of the predicted target genes in three aspects, including biological process, molecular function, and cellular components. We used four online target gene prediction software, DIANA-microT (http://diana.imis.athena-innovation.gr/), miRDB (http://mirdb.org/miRDB/), StarBase (http://www.starbasemn.org/), and miRanda,52 to forecast several potential target genes of hsa-miR-30.
In Vivo Tumor Studies
To evaluate the role of hsa-miR-30 and TM4SF1 on tumor growth, 2.5 × 106 cells overexpressing hsa-miR-30 and/or TM4SF1were injected subcutaneously into the flanks of the nude mice, and the same numbers of vector control cells were injected contralaterally. Tumor progression was monitored by palpation and tumor size measured by a digital caliper. At the end of the 4 weeks, tumor volume was calculated using the following formula: volume = length × width2 × 0.52.
Statistical Analysis
Measurement data were expressed as mean ± SD. Categorical data were reported as numbers and percentages. The independent t test was used to calculate the difference between two groups of data. The chi-square test was used to evaluate the difference among different groups. Univariate survival analysis of OS and disease-free survival (DFS) were carried out using the Kaplan-Meier method. Spearman’s correlation coefficient was used to test the relationship of two independent groups. All calculations were performed with the SPSS 20.0 software program (SPSS, Chicago, IL, USA). The level of significance was chosen as p < 0.05.
Author Contributions
Y.-S.M., X.-M.Z., and D.F. designed the study. Y.-S.M., X.-M.Z., G.-X.L., S.-B.X., and D.F. performed the cytological experiments. X.-L.C., F.Y., L.-L.C., L.-K.H., T.L., W.-T.X., L.-J.P., H.-D.L., W.Z., W.W., R.S., H.-Y.S., and C.-Y.W. were involved in patient recruitment. F.Y., C.-Y.W., and Z.-W.L. contributed to study materials and consumables. Y.-S.M., G.-X.L., and D.F. wrote the manuscript. All authors agreed with the results and conclusions.
Conflicts of Interest
The authors have no conflicts of interest.
Acknowledgments
This study was supported partly by grants from the National Natural Science Foundation of China (81472202, 81772932, 81201535, 81302065, 81702243, 81472209, and 81301993), The Fundamental Research Funds for the Central Universities (22120170212 and 22120170117), the Shanghai Natural Science Foundation (12ZR1436000 and 16ZR1428900), the Shanghai Municipal Commission of Health and Family Planning (201540228 and 201440398), and the Jilin Provincial Science and Technology Department (20140414061GH).
Footnotes
Supplemental Information includes one figure and can be found with this article online at https://doi.org/10.1016/j.ymthe.2018.09.006.
Contributor Information
Chun-Yan Wu, Email: chunyan_wu2015@126.com.
Da Fu, Email: fu800da900@126.com.
Supplemental Information
References
- 1.Ma Y., Hou L., Yu F., Lu G., Qin S., Xie R., Yang H., Wu T., Luo P., Chai L. Incidence and physiological mechanism of carboplatin-induced electrolyte abnormality among patients with non-small cell lung cancer. Oncotarget. 2017;8:18417–18423. doi: 10.18632/oncotarget.12813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo P., Yang Q., Cong L.L., Wang X.F., Li Y.S., Zhong X.M., Xie R.T., Jia C.Y., Yang H.Q., Li W.P. Identification of miR-124a as a novel diagnostic and prognostic biomarker in non-small cell lung cancer for chemotherapy. Mol. Med. Rep. 2017;16:238–246. doi: 10.3892/mmr.2017.6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yu F., Liu J.B., Wu Z.J., Xie W.T., Zhong X.J., Hou L.K., Wu W., Lu H.M., Jiang X.H., Jiang J.J. Tumor suppressive microRNA-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 14. Cancer Lett. 2018;427:74–84. doi: 10.1016/j.canlet.2018.04.022. [DOI] [PubMed] [Google Scholar]
- 4.Lu J., Song G., Tang Q., Yin J., Zou C., Zhao Z., Xie X., Xu H., Huang G., Wang J. MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1. Oncogene. 2017;36:231–241. doi: 10.1038/onc.2016.194. [DOI] [PubMed] [Google Scholar]
- 5.Wang Y., Ma Y., Fang Y., Wu S., Liu L., Fu D., Shen X. Regulatory T cell: a protection for tumour cells. J. Cell. Mol. Med. 2012;16:425–436. doi: 10.1111/j.1582-4934.2011.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu S.D., Ma Y.S., Fang Y., Liu L.L., Fu D., Shen X.Z. Role of the microenvironment in hepatocellular carcinoma development and progression. Cancer Treat. Rev. 2012;38:218–225. doi: 10.1016/j.ctrv.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 7.Liu L.L., Fu D., Ma Y., Shen X.Z. The power and the promise of liver cancer stem cell markers. Stem Cells Dev. 2011;20:2023–2030. doi: 10.1089/scd.2011.0012. [DOI] [PubMed] [Google Scholar]
- 8.Hou L.K., Ma Y.S., Han Y., Lu G.X., Luo P., Chang Z.Y., Xie R.T., Yang H.Q., Chai L., Cai M.X. Association of microRNA-33a molecular signature with non-small cell lung cancer diagnosis and prognosis after chemotherapy. PLoS ONE. 2017;12:e0170431. doi: 10.1371/journal.pone.0170431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang X., Zhu Y., Ma Y., Wang J., Zhang F., Xia Q., Fu D. The role of cancer stem cells in cancer metastasis: new perspective and progress. Cancer Epidemiol. 2013;37:60–63. doi: 10.1016/j.canep.2012.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Liang Z., Kong R., He Z., Lin L.Y., Qin S.S., Chen C.Y., Xie Z.Q., Yu F., Sun G.Q., Li C.G. High expression of miR-493-5p positively correlates with clinical prognosis of non small cell lung cancer by targeting oncogene ITGB1. Oncotarget. 2017;8:47389–47399. doi: 10.18632/oncotarget.17650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma Y.S., Wu T.M., Lv Z.W., Lu G.X., Cong X.L., Xie R.T., Yang H.Q., Chang Z.Y., Sun R., Chai L. High expression of miR-105-1 positively correlates with clinical prognosis of hepatocellular carcinoma by targeting oncogene NCOA1. Oncotarget. 2017;8:11896–11905. doi: 10.18632/oncotarget.14435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basak S.K., Veena M.S., Oh S., Lai C., Vangala S., Elashoff D., Fishbein M.C., Sharma S., Rao N.P., Rao D. The CD44high tumorigenic subsets in lung cancer biospecimens are enriched for low miR-34a expression. PLoS ONE. 2013;8:e73195. doi: 10.1371/journal.pone.0073195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y., Fu D., Shen X.Z. The potential role of ubiquitin c-terminal hydrolases in oncogenesis. Biochim. Biophys. Acta. 2010;1806:1–6. doi: 10.1016/j.bbcan.2010.03.001. [DOI] [PubMed] [Google Scholar]
- 14.Pashaei E., Pashaei E., Ahmady M., Ozen M., Aydin N. Meta-analysis of miRNA expression profiles for prostate cancer recurrence following radical prostatectomy. PLoS ONE. 2017;12:e0179543. doi: 10.1371/journal.pone.0179543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang C., Cai L., Liu J., Wang G., Li H., Wang X., Xu W., Ren M., Feng L., Liu P., Zhang C. MicroRNA-30a-5p inhibits the growth of renal cell carcinoma by modulating GRP78 expression. Cell. Physiol. Biochem. 2017;43:2405–2419. doi: 10.1159/000484394. [DOI] [PubMed] [Google Scholar]
- 16.Bockhorn J., Dalton R., Nwachukwu C., Huang S., Prat A., Yee K., Chang Y.F., Huo D., Wen Y., Swanson K.E. MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat. Commun. 2013;4:1393. doi: 10.1038/ncomms2393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu F., Deng H., Yao H., Liu Q., Su F., Song E. Mir-30 reduction maintains self-renewal and inhibits apoptosis in breast tumor-initiating cells. Oncogene. 2010;29:4194–4204. doi: 10.1038/onc.2010.167. [DOI] [PubMed] [Google Scholar]
- 18.Faria A.M., Sbiera S., Ribeiro T.C., Soares I.C., Mariani B.M., Freire D.S., de Sousa G.R., Lerario A.M., Ronchi C.L., Deutschbein T. Expression of LIN28 and its regulatory microRNAs in adult adrenocortical cancer. Clin. Endocrinol. (Oxf.) 2015;82:481–488. doi: 10.1111/cen.12607. [DOI] [PubMed] [Google Scholar]
- 19.Chao C.C., Kan D., Lu K.S., Chien C.L. The role of microRNA-30c in the self-renewal and differentiation of C6 glioma cells. Stem Cell Res. (Amst.) 2015;14:211–223. doi: 10.1016/j.scr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 20.Lv D., Ma Q.H., Duan J.J., Wu H.B., Zhao X.L., Yu S.C., Bian X.W. Optimized dissociation protocol for isolating human glioma stem cells from tumorspheres via fluorescence-activated cell sorting. Cancer Lett. 2016;377:105–115. doi: 10.1016/j.canlet.2016.04.022. [DOI] [PubMed] [Google Scholar]
- 21.Liu W.T., Wang Y., Zhang J., Ye F., Huang X.H., Li B., He Q.Y. A novel strategy of integrated microarray analysis identifies CENPA, CDK1 and CDC20 as a cluster of diagnostic biomarkers in lung adenocarcinoma. Cancer Lett. 2018;425:43–53. doi: 10.1016/j.canlet.2018.03.043. [DOI] [PubMed] [Google Scholar]
- 22.Schmitt M., Schewe M., Sacchetti A., Feijtel D., van de Geer W.S., Teeuwssen M., Sleddens H.F., Joosten R., van Royen M.E., van de Werken H.J.G. Paneth cells respond to inflammation and contribute to tissue regeneration by acquiring stem-like features through SCF/c-Kit signaling. Cell Rep. 2018;24:2312–2328.e7. doi: 10.1016/j.celrep.2018.07.085. [DOI] [PubMed] [Google Scholar]
- 23.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A-demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. USA. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu H., Wu X.Y., Fan R.T., Wang X., Guo Y.Z., Wang R. Side population cells from long-term passage non-small cell lung cancer cells display loss of cancer stem cell-like properties and chemoradioresistance. Oncol. Lett. 2016;12:2886–2893. doi: 10.3892/ol.2016.4934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q., Wan L., Xiao C., Hu H., Wang L., Zhao J., Lei Z., Zhang H.T. Inhibition of LHX2 by miR-124 suppresses cellular migration and invasion in non-small cell lung cancer. Oncol. Lett. 2017;14:3429–3436. doi: 10.3892/ol.2017.6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vázquez I., Maicas M., Marcotegui N., Conchillo A., Guruceaga E., Roman-Gomez J., Calasanz M.J., Agirre X., Prosper F., Odero M.D. Silencing of hsa-miR-124 by EVI1 in cell lines and patients with acute myeloid leukemia. Proc. Natl. Acad. Sci. USA. 2010;107:E167–E168. doi: 10.1073/pnas.1011540107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bridge G., Monteiro R., Henderson S., Emuss V., Lagos D., Georgopoulou D., Patient R., Boshoff C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood. 2012;120:5063–5072. doi: 10.1182/blood-2012-04-423004. [DOI] [PubMed] [Google Scholar]
- 28.Volinia S., Galasso M., Costinean S., Tagliavini L., Gamberoni G., Drusco A., Marchesini J., Mascellani N., Sana M.E., Abu Jarour R. Reprogramming of miRNA networks in cancer and leukemia. Genome Res. 2010;20:589–599. doi: 10.1101/gr.098046.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kao C.J., Martiniez A., Shi X.B., Yang J., Evans C.P., Dobi A., deVere White R.W., Kung H.J. miR-30 as a tumor suppressor connects EGF/Src signal to ERG and EMT. Oncogene. 2014;33:2495–2503. doi: 10.1038/onc.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fan M.J., Zhong Y.H., Shen W., Yuan K.F., Zhao G.H., Zhang Y., Wang S.K. MiR-30 suppresses lung cancer cell 95D epithelial mesenchymal transition and invasion through targeted regulating Snail. Eur. Rev. Med. Pharmacol. Sci. 2017;21:2642–2649. [PubMed] [Google Scholar]
- 31.Chan L.W., Wang F., Meng F., Wang L., Wong S.C., Au J.S., Yang S., Cho W.C. MiR-30 family potentially targeting PI3K-SIAH2 predicted interaction network represents a novel putative theranostic panel in non-small cell lung cancer. Front. Genet. 2017;8:8. doi: 10.3389/fgene.2017.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G., Herazo-Maya J.D., Nukui T., Romkes M., Parwani A., Juan-Guardela B.M., Robertson J., Gauldie J., Siegfried J.M., Kaminski N., Kass D.J. Matrix metalloproteinase-19 promotes metastatic behavior in vitro and is associated with increased mortality in non-small cell lung cancer. Am. J. Respir. Crit. Care Med. 2014;190:780–790. doi: 10.1164/rccm.201310-1903OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhong K., Chen K., Han L., Li B. MicroRNA-30b/c inhibits non-small cell lung cancer cell proliferation by targeting Rab18. BMC Cancer. 2014;14:703. doi: 10.1186/1471-2407-14-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhong Z., Xia Y., Wang P., Liu B., Chen Y. Low expression of microRNA-30c promotes invasion by inducing epithelial mesenchymal transition in non-small cell lung cancer. Mol. Med. Rep. 2014;10:2575–2579. doi: 10.3892/mmr.2014.2494. [DOI] [PubMed] [Google Scholar]
- 35.Wang T., Li F., Tang S. MiR-30a upregulates BCL2A1, IER3 and cyclin D2 expression by targeting FOXL2. Oncol. Lett. 2015;9:967–971. doi: 10.3892/ol.2014.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsukasa K., Ding Q., Miyazaki Y., Matsubara S., Natsugoe S., Takao S. miR-30 family promotes migratory and invasive abilities in CD133(+) pancreatic cancer stem-like cells. Hum. Cell. 2016;29:130–137. doi: 10.1007/s13577-016-0137-7. [DOI] [PubMed] [Google Scholar]
- 37.Che S., Sun T., Wang J., Jiao Y., Wang C., Meng Q., Qi W., Yan Z. miR-30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 2015;36:6805–6811. doi: 10.1007/s13277-015-3400-8. [DOI] [PubMed] [Google Scholar]
- 38.Zhong X., Li N., Liang S., Huang Q., Coukos G., Zhang L. Identification of microRNAs regulating reprogramming factor LIN28 in embryonic stem cells and cancer cells. J. Biol. Chem. 2010;285:41961–41971. doi: 10.1074/jbc.M110.169607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin C.I., Merley A., Sciuto T.E., Li D., Dvorak A.M., Melero-Martin J.M., Dvorak H.F., Jaminet S.C. TM4SF1: a new vascular therapeutic target in cancer. Angiogenesis. 2014;17:897–907. doi: 10.1007/s10456-014-9437-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xing P., Dong H., Liu Q., Zhao T., Yao F., Xu Y., Chen B., Zheng X., Wu Y., Jin F., Li J. Upregulation of transmembrane 4 L6 family member 1 predicts poor prognosis in invasive breast cancer: A STROBE-compliant article. Medicine (Baltimore) 2017;96:e9476. doi: 10.1097/MD.0000000000009476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao R., Wang G., Qian K., Chen L., Ju L., Qian G., Wu C.L., Dan H.C., Jiang W., Wu M. TM4SF1 regulates apoptosis, cell cycle and ROS metabolism via the PPARγ-SIRT1 feedback loop in human bladder cancer cells. Cancer Lett. 2018;414:278–293. doi: 10.1016/j.canlet.2017.11.015. [DOI] [PubMed] [Google Scholar]
- 42.Shih S.C., Zukauskas A., Li D., Liu G., Ang L.H., Nagy J.A., Brown L.F., Dvorak H.F. The L6 protein TM4SF1 is critical for endothelial cell function and tumor angiogenesis. Cancer Res. 2009;69:3272–3277. doi: 10.1158/0008-5472.CAN-08-4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao J., Yang J.C., Ramachandran V., Arumugam T., Deng D.F., Li Z.S., Xu L.M., Logsdon C.D. TM4SF1 regulates pancreatic cancer migration and invasion in vitro and in vivo. Cell. Physiol. Biochem. 2016;39:740–750. doi: 10.1159/000445664. [DOI] [PubMed] [Google Scholar]
- 44.Sun Y., Xu Y., Xu J., Lu D., Wang J. Role of TM4SF1 in regulating breast cancer cell migration and apoptosis through PI3K/AKT/mTOR pathway. Int. J. Clin. Exp. Pathol. 2015;8:9081–9088. [PMC free article] [PubMed] [Google Scholar]
- 45.Xue L., Yu X., Jiang X., Deng X., Mao L., Guo L., Fan J., Fan Q., Wang L., Lu S.H. TM4SF1 promotes the self-renewal of esophageal cancer stem-like cells and is regulated by miR-141. Oncotarget. 2017;8:19274–19284. doi: 10.18632/oncotarget.13866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choi S.I., Kim S.Y., Lee J., Cho E.W., Kim I.G. TM4SF4 overexpression in radiation-resistant lung carcinoma cells activates IGF1R via elevation of IGF1. Oncotarget. 2014;5:9823–9837. doi: 10.18632/oncotarget.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Aziz A.U.R., Farid S., Qin K., Wang H., Liu B. PIM kinases and their relevance to the PI3K/AKT/mTOR pathway in the regulation of ovarian cancer. Biomolecules. 2018;8:E7. doi: 10.3390/biom8010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raphael J., Desautels D., Pritchard K.I., Petkova E., Shah P.S. Phosphoinositide 3-kinase inhibitors in advanced breast cancer: A systematic review and meta-analysis. Eur. J. Cancer. 2018;91:38–46. doi: 10.1016/j.ejca.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 49.McFall T., Patki M., Rosati R., Ratnam M. Role of the short isoform of the progesterone receptor in breast cancer cell invasiveness at estrogen and progesterone levels in the pre- and post-menopausal ranges. Oncotarget. 2015;6:33146–33164. doi: 10.18632/oncotarget.5082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang Q., Zhang X.W., Ma Y.S., Lu G.X., Xie R.T., Yang H.Q., Lv Z.W., Zhong X.M., Liu T., Huang S.X. Up-regulated microRNA-299 corrected with poor prognosis of glioblastoma multiforme patients by targeting ELL2. Jpn. J. Clin. Oncol. 2017;47:590–596. doi: 10.1093/jjco/hyw188. [DOI] [PubMed] [Google Scholar]
- 51.Wang Y., Liu Z., Yao B., Li Q., Wang L., Wang C., Dou C., Xu M., Liu Q., Tu K. Long non-coding RNA CASC2 suppresses epithelial-mesenchymal transition of hepatocellular carcinoma cells through CASC2/miR-367/FBXW7 axis. Mol. Cancer. 2017;16:123. doi: 10.1186/s12943-017-0702-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Betel D., Wilson M., Gabow A., Marks D.S., Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36 doi: 10.1093/nar/gkm995. D149–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.