Abstract
Approximately 90% of patients diagnosed with non-small cell lung cancer (NSCLC) die due to distant metastases. However, the complicated molecular and cellular mechanisms involved in lung cancer metastasis remain poorly understood. Copine III (CPNE3), a member of a Ca2+-dependent phospholipid-binding protein family, was identified as a novel metastasis-associated protein in NSCLC in our previous study, however, its function in metastasis remains unclear. Here, we found that CPNE3 was expressed at high levels in NSCLC tissues and advanced TNM stages and was significantly associated with poor prognosis. In addition, CPNE3 interacted with phosphorylated erb-b2 receptor tyrosine kinase 2 (pErbB2) and receptor of activated protein C kinase 1 (RACK1) and activated the focal adhesion (FA) signaling pathway in NSCLC cells. Moreover, knockdown of RACK1 inhibited cell motility in the CPNE3-overexpressed NSCLC cells. These findings offer mechanistic insights into the oncogenic roles of CPNE3 and the pivotal effects of CPNE3 as a biomarker and therapeutic target for NSCLC metastasis.
Keywords: CPNE3, NSCLC, RACK1, FAK.
Introduction
Lung cancer remains the leading cause of cancer-related mortality worldwide for both men and women1. Non-small cell lung cancer (NSCLC) accounts for approximately 85-90% of lung cancer cases, and approximately 90% of NSCLC patients die due to distant metastases rather than the primary tumor2. Although NSCLC has been investigated in numerous molecular studies aimed at developing new treatment strategies, the 5-year overall survival rates remain at 4-17%, and 65% of NSCLC patients are classified at stages III and IV at diagnosis3-5. However, the exact underlying metastatic mechanisms and development progress of NSCLC remain complex and poorly understood. The need to identify potential therapeutic targets to improve NSCLC treatment is urgent.
In our previous study, we found that CPNE3 was a novel oncogene in NSCLC, and it promoted NSCLC metastasis in vitro and in vivo6. CPNE3 possess two C2Ds (C2D-A and C2D-B) and an A domain; C2Ds are responsible for Ca2+ -dependent membrane-binding properties and A domains bind proteins7, 8, it may regulate molecular events at the interface of the cell membrane and cytoplasm. CPNE3 was identified as a ligand of ErbB2 and plays a more general role in carcinogenesis9. In prostate cancer, CPNE3 was reported to be a susceptibility gene10, and miR-133b repressed CPNE3 expression11. Moreover, CPNE3 plays a pivotal role in other diseases. High expression of CPNE3 predicted an adverse prognosis in acute myeloid leukemia12, and it was identified as a novel candidate gene for schizophrenia13. However, in NSCLC, the underlying mechanism of CPNE3 remains unknown.
In this study, we demonstrated that CPNE3 was highly expressed in NSCLC patients and was associated with poor outcomes. RNA-seq was performed, and it revealed that CPNE3 participates in the focal adhesion kinase (FAK) signaling pathway. To further investigate the underlying mechanism, we made use of co-immunoprecipitation and found that CPNE3 interacts with RACK1 and pErbB2, and the mobility potential in CPNE3 overexpression cells was reduced after knockdown of RACK1.
Materials and methods
Cell lines and cell culture
Two human NSCLC cell lines (SPC-A-1 and H1299) and human embryonic kidney 293T cells were used. SPC-A-1 cells were obtained from the Cellular Institute of Chinese Academy of Science (Shanghai, China) in 2007. H1299 and 293T cells were obtained from the American Type Culture Collection (ATCC). These cells were incubated at 37°C in a humidified air atmosphere containing 5% carbon dioxide in Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS), 100 µ/ml penicillin (Sigma-Aldrich), and 100 µg/ml streptomycin (Sigma-Aldrich). These cell lines were mycoplasma-free and authenticated by quality examinations of morphology and growth profile.
Clinical NSCLC tissue samples
The 62 paired patient samples of primary NSCLC tissues and matched adjacent non-cancerous tissues were obtained from the Department of Cancer, Huashan Hospital affiliated with Shanghai Fudan University from 2014 to 2017. Tissue samples were acquired from the routine therapeutic surgery of patients who did not receive anti-tumor treatment. Upon resection, human surgical specimens were immediately frozen in liquid nitrogen and stored at -80°C. Informed consent was obtained from all patients, and the research was approved by the Ethics Committee of Shanghai Fudan University. We performed global mRNAs profiling with microarray analysis to screen metastasis-associated genes. Data are available via Gene Expression Omnibus (GEO). Online-available data sets were downloaded from NCBI. 117 lung adenocarcinoma patients were involved in the Tomida' Cohort (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13213).
RNA isolation and quantitative real-time PCR assays
Total RNA samples from the cell lines and tissue samples used in this study were extracted with TRIzol reagent (Invitrogen, CA) according to the manufacturer's protocol and were quantified with Nanodrop 2000 (Thermo, Japan). A PrimeScript RT Reagent Kit (TaKaRa, China) was used to synthesize first-strand cDNA, which was used as the template for real-time polymerase chain reaction (qPCR). qPCR was performed three times with SYBR Green Premix Ex Taq (TaKaRa, China). The specific primers for the target genes are listed in Supplementary Table S1. Relative gene expression was analyzed in accordance with 7500 Software version 2.0.6 of Applied Biosystems. Gene expression levels were quantified according to the comparative ΔΔCt method, and β-actin was used as the internal control.
Plasmid construction and transfection
For CPNE3 overexpression, full-length CPNE3 cDNA was obtained from Han's Lab (Xiamen, China) and was then cloned and inserted into the lentiviral expression vector pWPXL (Addgene). For CPNE3 knockdown, CPNE3-shRNA was obtained from Open Biosystems (Thermo, Japan). To produce the intended lentivirus, the constructed plasmids and lentiviral vector packaging system were transfected into HEK-293T cells using Lipofectamine 2000 Reagent (Invitrogen). After 48 hours, the supernatants from the HEK-293T cultures were collected to infect SPC-A-1/H1299 cells.
For RACK1 knockdown, RACK1-siRNAs were obtained from RiboBio (shanghai, China). Cells were transfected with the indicated siRNAs using Lipofectamine 2000 Reagent (Invitrogen). After transfection for 48 hours, the cells were used for migration, invasion, RNA extraction, and immunoblotting assays.
Cell migration and invasion assays
Cell migration and invasion assays were performed in a 24-well plate with 8 µm pore size chamber inserts (Corning). Per well, 5×104 cells (migration assays) and 1×105 cells (invasion assays) were inoculated into the upper chamber. For the invasion assays, the wells contained Matrigel-coated membrane, which was diluted with serum-free culture medium. In both assays, cells were suspended in 200 µl of DMEM without FBS. In the lower chamber, 800 µl of DMEM supplemented with 10% FBS was added. After the appropriate incubation period, the cells that remained in the upper chamber were removed with a cotton swab, and the cells that moved to the bottom surface of the membrane were fixed with 100% methanol and stained with 0.1% crystal violet for 20 min. Then, the cells were imaged and counted under a CKX41 inverted microscope (Olympus, Japan). Assays were conducted independently three times.
Western blot analysis
The lysates were extracted from cultured cells and were then separated using 8% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by transferring onto nitrocellulose membranes (Millipore). Next, we blocked the membranes in 5% milk with phosphate-buffered saline. After one hour of blocking, the membranes were incubated with primary antibodies (Supplementary Table S2) at 4 °C overnight. The next day, the membranes were incubated with HRP-conjugated anti-mouse IgG (Sigma-Aldrich) or HRP-conjugated anti-rabbit IgG (Sigma-Aldrich) secondary antibodies (1:5000). Subsequently, visualization was performed with SuperSignal West Femto Maximum Sensitivity Substrate (Thermo, USA).
Co-immunoprecipitation
Co-immunoprecipitation was performed with SPC-A-1 pWPXL-CPNE3 cells. Equal amounts of protein (3000 μg) were preprocessed by protein A/G magnetic beads (Thermo Scientific). After two hours, the lysates were incubated with antibodies at 4 °C overnight, followed by an overnight incubation with beads. The next day, the beads were gently washed with phosphate-buffered saline (PBS) containing 1% Triton X-100 five times, and then the beads were loaded with 2× protein loading buffer at 100 °C for 10 min. IgG-bound, CPNE3-bound or RACK1-bound proteins were separated using SDS-PAGE and stained with a Coomassie brilliant blue R250 staining kit (Beyotime Biotechnology).
KEGG pathway analysis
To better interpret the RNA seq dataset generated, KEGG pathway annotations of the 849 significantly differentially expressed genes were analyzed using DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/).
Statistical analysis
The quantitative variables are presented as the means and S.E.M unless otherwise noted and were analyzed by Student's t-test between the two groups (two-tailed; p<0.05 was considered statistically significant). The overall survival time was defined as the length of time between surgery and death. Figures were generated with GraphPad Prism 5 software. All analyses were performed with SPSS software (version 19.0, Armonk).
Results
CPNE3 is overexpressed and associated with poor outcomes in NSCLC patients
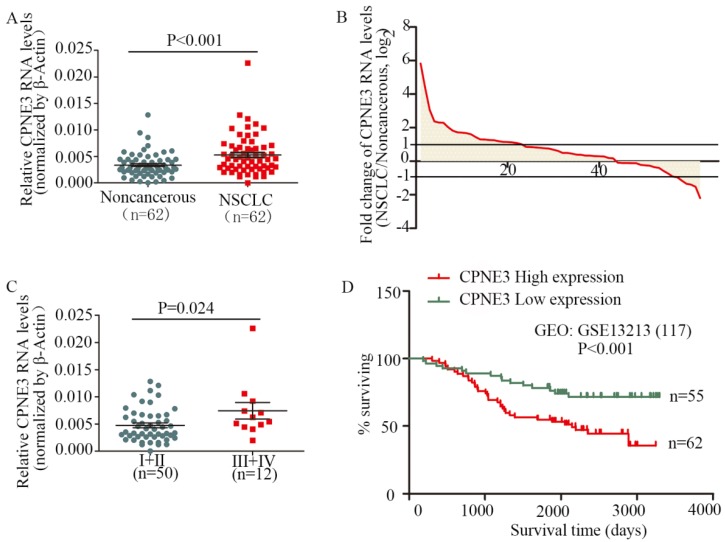
To assess the clinical significance of CPNE3, we analyzed CPNE3 RNA levels in 62 pairs of human NSCLC and their corresponding noncancerous lung tissues. Compared with corresponding nontumorous tissues, CPNE3 was significantly upregulated in NSCLC tissues (Fig. 1A). Moreover, the RNA expression levels of CPNE3 were upregulated in 69.4% (43/62) of NSCLC cases (Fig. 1B), and its expression was positively correlated with advanced TNM stages (p=0.024) (Fig. 1C) and tumor sizes (p=0.048), whereas CPNE3 levels were independent of age, gender, differentiation and metastasis (Supplementary Table S3). To better understand the impact of CPNE3 gene expression on the overall survival of lung cancer patients, we utilized NCBI Gene Expression Omnibus (GEO) datasets with accession number GSE13213 to assess the prognostic value of CPNE3. CPNE3 high/low mRNA expression was retrieved and identified according to the median value of gene expression. Importantly, high levels of CPNE3 RNA were remarkably associated with poor outcomes (p<0.001) in the 117-GEO-patient cohort (Fig. 1D). The baseline data of the 117 lung cancer patients are reported in Supplementary Table S4. Taken together, our data show that CPNE3 is highly expressed in lung cancer and is related to clinical severity and prognosis.
Figure 1.
CPNE3 is overexpressed and associated with poor outcomes in NSCLC patients. (A) CPNE3 RNA levels were quantified in 62 pairs of NSCLC tissues and adjacent normal tissues using qPCR. (B) Fold changes in CPNE3 expression in 62 paired tissues (NSCLC/noncancerous; upexpression, >1; no change, -1~1; downexpression, <-1.). (C) Clinical significance of CPNE3 in patients with NSCLC; high CPNE3 expression is positively correlated with TNM stage (III+IV). (D) Kaplan-Meier analyses of the correlation between CPNE3 RNA levels and overall survival in the 117-patient GEO cohort.
CPNE3 regulates the FAK signaling pathway in NSCLC cell lines
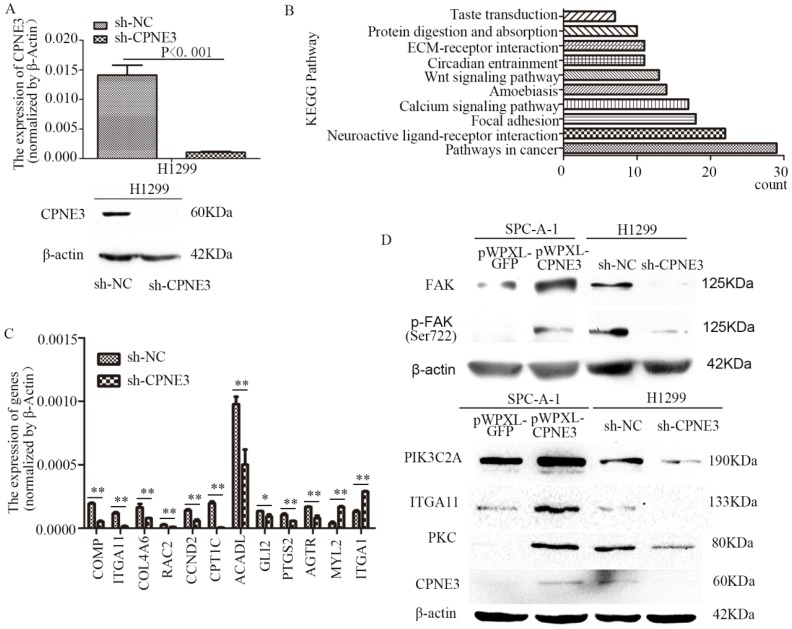
To seek the potential mechanism and identify changes in gene expression of CPNE3 in NSCLC cells, RNA-seq was performed in H1299 sh-NC/sh-CPNE3 cells (Fig. 2A) and 849 genes were found to be altered after sh-CPNE3 transfection (Supplementary Table S5). KEGG pathway annotations of the genes were analyzed using DAVID Bioinformatics Resources 6.8, in which the top three pathways (p<0.05) were Pathways in cancer, Neuroactive ligand-receptor interaction, and Focal adhesion (Figure 2B) (Supplementary Table S6). Then, 9 downregulated and 2 upregulated genes were selected for validation using qRT-PCR (Figure 2C), and we found that five genes, cartilage oligomeric matrix protein (COMP), integrin subunit alpha 11 (ITGA11), collagen type IV alpha 6 (COL4A6), Rac family small GTPase 2 (RAC2), and cyclin D2 (CCND2) were associated with the FAK signaling pathway. Therefore, we detected five important well-known signaling molecules (FAK, phosphorylated FAK [pFAK], phosphatidylinosito-4-phosphate 3-kinase catalytic subunit type 2 alpha [PIK3C2A], ITGA11, and protein kinase C [PKC]) in this pathway, and the results showed that overexpression of CPNE3 significantly increased the signaling molecules in SPC-A-1 cells, but knockdown of CPNE3 led to a significant decrease in H1299 cells (Fig. 2D). Consequently, these results supported the idea that CPNE3 was a crucial gene in the FAK signaling pathway in NSCLC cell metastasis.
Figure 2.
CPNE3 regulates the FAK signaling pathway in NSCLC cell lines. (A) Levels of mRNA and protein expression were validated after knockdown of CPNE3 in H1299 cells by qRT-PCR and western blotting. (B) The top ten KEGG pathways with p values <0.05. (C) mRNA expression levels of 9 downregulated and 2 upregulated genes selected for validation using qRT-PCR. (D) Protein expression levels of FAK, pFAK, PIK3C2A, ITGA11, and PKC were measured using western blot analysis in SPC-A-1 cells stably expressing pWPXL-CPNE3 and pWPXL-GFP and H1299 cells with stable knockdown of CPNE3 or a negative control. Data are representative of results from three independent experiments. *p< 0.05, **p<0.01 by Student's t-test.
CPNE3 interacts with pErbB2 and RACK1 in NSCLC cells
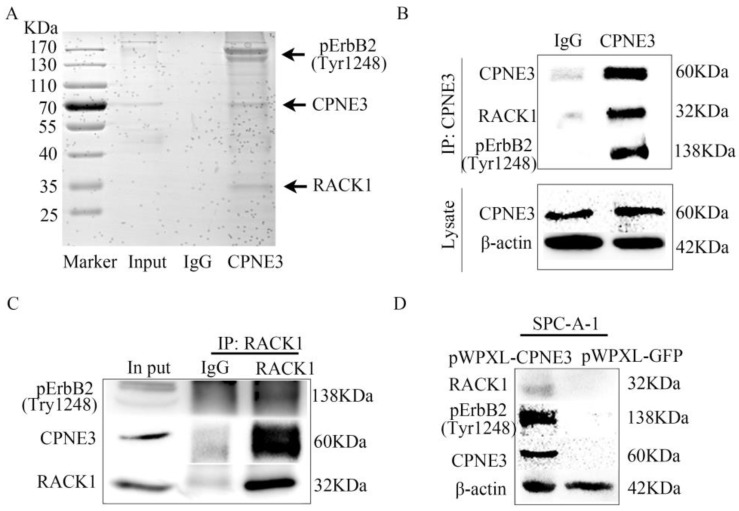
To further determine the underlying mechanism of CPNE3 in NSCLC, we next identified the protein partners of CPNE3 in SPC-A-1 pWPXL-CPNE3 cells through co-immunoprecipitation assays. First, the SDS-PAGE gel was stained by Coomassie brilliant blue R250 after IgG or CPNE3 pulldown. It showed that there were some proteins precipitated by CPNE3 in the positions 25 KDa-35 KDa and 130 KDa-170 KDa (Fig. 3A). Because CPNE3 has been reported to be a ligand of ErbB2 and bound to RACK1 in breast cancer, and because of the gel stained indication, the two proteins were selected for further investigation. We pulled down CPNE3 and probed for pErbB2 and RACK1, then found that CPNE3 specifically interacted with pErbB2 and RACK1, as shown in the immunoprecipitation assay (Fig. 3B). In addition, we pulled down RACK1 and probed for CPNE3 and pErbB2 and found that CPNE3 and pErbB2 were also precipitated by RACK1 in SPC-A-1 pWPXL-CPNE3 cells (Fig. 3C). Next, we examined whether the protein levels of RACK1 and pErbB2 were affected by CPNE3. This showed that overexpression of CPNE3 in SPC-A-1 cells led to a significant increase in RACK1 and pErbB2 protein expression (Fig. 3D). Consequently, these results support the conclusion that CPNE3 interacts with RACK1 and pErbB2 and functions as a positive regulator of RACK1 and pErbB2 protein expression in NSCLC cells.
Figure 3.
CPNE3 interacts with pErbB2 and RACK1 in NSCLC cells. (A) Coomassie brilliant blue R250 staining of an SDS-PAGE gel after IgG or CPNE3 pulldown. (B) Whole-cell lysates from SPC-A-1 pWPXL-CPNE3 cell lines were immunoprecipitated with an anti-CPNE3 antibody followed by IB with anti-pErbB2 and anti-RACK1 antibodies. IgG was used as a negative control. (C) Whole-cell lysates from SPC-A-1 pWPXL-CPNE3 cell lines were immunoprecipitated with an anti-RACK1 antibody followed by IB with anti-pErbB2 and anti-CPNE3 antibodies. IgG was used as a negative control. (D, E) mRNA expression levels of ErbB2 and RACK1 were measured using qRT-PCR in SPC-A-1 cells stably expressing pWPXL-CPNE3 and pWPXL-GFP and H1299 cells with stable knockdown of CPNE3 or the negative control. (F) Protein expression levels of CPNE3, pErbB2 and RACK1 were measured using western blot in SPC-A-1 cells stably expressing pWPXL-CPNE3 and pWPXL-GFP. Data are representative of results from three independent experiments. *p< 0.05 by Student's t-test.
Knockdown of RACK1 inhibits NSCLC cell migration and invasion
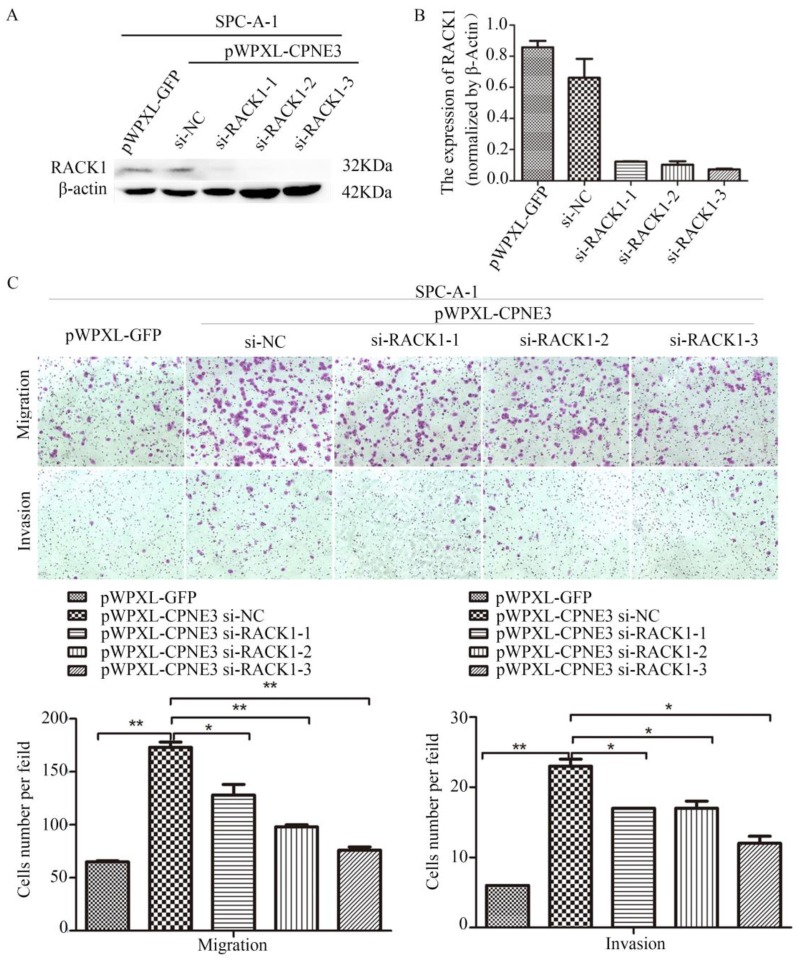
Our findings demonstrated that CPNE3 interacted with RACK1 in NSCLC cells; however, whether CPNE3 functions through RACK1 is largely unknown. Since CPNE3 interacted with RACK1 and affected its protein expression, it is plausible that CPNE3 functions through RACK1 and RACK1 has an important role in NSCLC cell metastasis14. To test this hypothesis, we knocked down RACK1 in CPNE3-overexpressed SPC-A-1 cells. Western blotting and qRT-PCR confirmed that RACK1 protein and mRNA levels were rescued (Fig. 4A and B). As hypothesized, RACK1 knockdown significantly reduced the CPNE3 overexpression-induced cell migration and invasion (Fig. 4C). The results revealed that CPNE3 might promote NSCLC cell migration and invasion via RACK1.
Figure 4.
Knockdown of RACK1 inhibits NSCLC cells migration and invasion. (A, B) Levels of protein and mRNA expression were validated after RACK1-specific siRNAs transfection in SPC-A-1 cells stably expressing pWPXL-CPNE3 and pWPXL-GFP by qRT-PCR and western blotting. (C) Knockdown of RACK1 significantly reduced the CPNE3 overexpression-induced cell migration and invasion capacities in SPC-A-1 cells. Data are representative of results from three independent experiments. *p< 0.05, **p<0.01 by Student's t-test.
Discussion
Currently, lung cancer has a 5-year survival rate that remains low, and metastatic spread is the leading cause of death of patients with NSCLC3. There is an urgent need for us to identify metastasis-related factors and to determine the underlying molecular mechanisms of NSCLC. In our previous study, we performed an iTRAQ proteomics approach on cell lines with different metastatic abilities and the same genomic background, which were derived from SPC-A-1 cells using a mouse-screening model15, and CPNE3 was shown to be an oncogenic protein, promoting NSCLC metastasis in vitro and vivo6. It was the first time that anyone had identified CPNE3 as a metastasis-associative protein in NSCLC, and our findings indicated that CPNE3 might play an important role in human carcinogenesis. Although several studies have investigated the role of CPNE3 in tumorigenesis, few researchers have elucidated the underlying mechanism in depth. There was only one report by C Heinrich, who suggested that CPNE3 interacts with ErbB2 and promotes tumor cell migration9, but he did not investigate NSCLC. CPNE3 is a functional protein, and it is also involved in other diseases16, 17. However, the exact underlying metastatic mechanisms in NSCLC remain poorly understood.
In the present study, CPNE3 was significantly highly expressed in NSCLC tissues compared with matched noncancerous lung tissues. Moreover, the RNA expression of CPNE3 was upregulated in the majority of NSCLC cases (69.4%). Importantly, high levels of CPNE3 in lung cancer tissues are correlated with advanced tumor TNM stage and large tumor size. Although it was not found to be significant in metastasis, we discussed that this may be because of tumor heterogeneity and the low numbers of the NSCLC sample. Consistently, high levels of CPNE3 RNA were remarkably associated with poor outcomes. Our results also strongly suggested that CPNE3 was related to the clinical severity of NSCLC patients, and it could be considered to be a clinical biomarker of prognosis in NSCLC.
Next, RNA-seq was used to compare the changes in gene expression after stable knockdown of CPNE3 in H1299 cells, and 849 genes were found to be altered, among which 494 genes were upregulated and 355 genes were downregulated. KEGG pathway analysis was performed on the dysregulated genes, revealing that these genes were involved in pathways involved in cancer, neuroactive ligand-receptor interaction, and focal adhesion. These results provide valuable information for further study of the molecular mechanisms in NSCLC metastasis. Furthermore, 9 downregulated and 2 upregulated genes were selected for validation using qRT-PCR, and they were all consistent with the findings of RNA-seq. Interestingly, we found that COMP, ITGA11, COL4A6, RAC2, and CCND2 were associated with the FAK signaling pathway18-20, which plays a fundamental role in cancer progression and metastasis21, 22. The focal adhesion proteins such as FAK, regulate cell spreading, adhesion, migration, proliferation and angiogenesis, and FAK expression correlates with tumor invasion, metastatic potential and poor disease outcome23. Consistently, our results suggested that overexpression of CPNE3 significantly increased the protein levels of FAK and pFAK and the key proteins (PIK3C2A, ITGA11, PKC) in the FAK signaling pathway. In contrast, knockdown of CPNE3 significantly reduced those proteins. The results indicated to us that CPNE3 might work as a pivotal protein in the FAK pathway in NSCLC metastasis, and further study is necessary to determine how CPNE3 affects the pathway.
In the present study, co-immunoprecipitation was performed to explore the protein partners of CPNE3 in NSCLC cells. According to the gel staining, ErbB2 and RACK1 were reported to interact with CPNE3 in breast cancer9, and so they were selected for further investigation. Consistently, our present evidence supported that CPNE3 interacted with pErbB2 and RACK1 in NSCLC cells. Furthermore, it was demonstrated that CPNE3 significantly increased pErbB2 and RACK1 protein levels. How does Copine-III interact with pErbB2 and RACK1? We had said that CPNE3 possessed two C2Ds, and C2Ds had been classified as Type I and Type II24 and the Type II C2D of PKC has been shown to directly bind specific pTyr-containing peptides25. According to our results, we consider it likely that Copine-III directly binds pErbB2 (pTyr1248). However, the manner by which the three proteins interact with each other requires further study.
ErbB2 has been shown to have a pivotal role in various types of human cancer, and the metastatic mechanism of ErbB2 is well known in NSCLC26, 27, so we focused our attention on RACK1. RACK1 is a member of the tryptophan-aspartate repeat (WD-repeat) family of proteins28. Many studies have reported on the role of RACK1 in tumorigenesis, and there are accumulating evidences now demonstrate multiple roles for RACK1 in regulating migration and invasion of tumour cells, including NSCLC 14. A study in non-small cell lung cancer, found that RACK1 mediates the assembly of a RACK1-PP2A-Akt signalling complex in response to EphB3 activation which results in reduced Akt phosphorylation and inhibition of cell migration 29. It suggests that RACK1 may act as a scaffolding protein, recruiting proteins to various transmembrane receptors and providing a platform for protein-protein interactions. RACK1 participates in numerous signaling events, and RACK1 was shown to associate with and be required for FAK activation 30, 31. There are evidences that Copine-III binds RACK1 and colocalizes with pFAK in the migrating front of breast cancer cells 9. Furthermore, Fei L et al. indicated that RACK1 promoted cellular proliferation by regulating G1/S progression in NSCLC cells32. Our previous findings demonstrated that overexpression of CPNE3 promoted NSCLC cell metastasis and proliferation. In the present study, we found that knockdown of RACK1 expression reduced cell migration and invasion ability in the CPNE3-overexpressed NSCLC cells, therefore demonstrating that CPNE3's effect on cell mobility and proliferation might largely dependent on RACK1, and then activate FAK pathway. Interestingly, C2 regulatory regions were found in a diverse range of proteins in addition to PKC and were the first protein domains identified capable of interacting with RACK1 33, which are conserved in Copine-III C2D-B. Therefore, it might be possible that RACK1, which is thought to direct PKCs to specific membrane locations 34, might also direct interact with Copine-III, and might be affected it expression at the transcription level. Taken together, we favor the hypothesis that CPNE3 promotes migration and invasion in non-small cell lung cancer by interacting with RACK1 via FAK signaling activation.
In summary, we identified a metastasis-related protein CPNE3 that is highly expressed in NSCLC tissues and is associated with poor patient survival. In addition, CPNE3 interacts with pErbB2 and RACK1 and activates the FAK signaling pathway in NSCLC cells. Moreover, knockdown of RACK1 inhibits cell migration and invasion ability in CPNE3-overexpressing NSCLC cells. Collectively, our study offers mechanistic insights into the oncogenic roles of CPNE3 and the pivotal effects of CPNE3 as a biomarker and therapeutic target for NSCLC metastasis.
Supplementary Material
Supplementary figures and tables.
Acknowledgments
This work was supported by grants from the National Natural Science Foundation of China (NO. 81772481, NO. 81502488) and the Natural Science Foundation of Shanghai (NO. 15ZR1439000). We are grateful for Dr. Didier Trono for sharing pWPXL lentivirus plasmids.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Flamini V, Jiang WG, Cui Y. Therapeutic role of MiR-140-5p for the treatment of non-small cell lung cancer. Anticancer Res. 2017;37(8):4319–4327. doi: 10.21873/anticanres.11825. [DOI] [PubMed] [Google Scholar]
- 3.Hirsch FR, Scagliotti GV, Mulshine JL. et al. Lung cancer: Current therapies and new targeted treatments. Lancet. 2017;389(10066):299–311. doi: 10.1016/S0140-6736(16)30958-8. [DOI] [PubMed] [Google Scholar]
- 4.Wood SL, Pernemalm M, Crosbie PA. et al. The role of the tumor-microenvironment in lung cancer-metastasis and its relationship to potential therapeutic targets. Cancer Treat Rev. 2014;40(4):558–566. doi: 10.1016/j.ctrv.2013.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury P, Sivajohanathan D, Chan A. et al. Postoperative adjuvant systemic therapy in completely resected Non-Small-Cell lung cancer: A systematic review. Clin Lung Cancer. 2017;18(3):259–273. doi: 10.1016/j.cllc.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 6.Lin HC, Zhang FL, Geng Q. et al. Quantitative proteomic analysis identifies CPNE3 as a novel metastasis-promoting gene in NSCLC. J Proteome Res. 2013;12(7):3423–3433. doi: 10.1021/pr400273z. [DOI] [PubMed] [Google Scholar]
- 7.Caudell EG, Caudell JJ, Tang CH. et al. Characterization of human copine III as a phosphoprotein with associated kinase activity. Biochemistry-Us. 2000;39(42):13034–13043. doi: 10.1021/bi001250v. [DOI] [PubMed] [Google Scholar]
- 8.Cowland JB, Carter D, Bjerregaard MD. et al. Tissue expression of copines and isolation of copines I and III from the cytosol of human neutrophils. J Leukoc Biol. 2003;74(3):379–388. doi: 10.1189/jlb.0203083. [DOI] [PubMed] [Google Scholar]
- 9.Heinrich C, Keller C, Boulay A. et al. Copine-III interacts with ErbB2 and promotes tumor cell migration. Oncogene. 2010;29(11):1598–1610. doi: 10.1038/onc.2009.456. [DOI] [PubMed] [Google Scholar]
- 10.Thomas G, Jacobs KB, Yeager M. et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40(3):310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 11.Mo W, Zhang J, Li X. et al. Identification of novel AR-targeted microRNAs mediating androgen signalling through critical pathways to regulate cell viability in prostate cancer. Plos One. 2013;8(2):e56592. doi: 10.1371/journal.pone.0056592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fu L, Fu H, Qiao J. et al. High expression of CPNE3 predicts adverse prognosis in acute myeloid leukemia. Cancer Sci. 2017;108(9):1850–1857. doi: 10.1111/cas.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cohen OS, Mccoy SY, Middleton FA. et al. Transcriptomic analysis of postmortem brain identifies dysregulated splicing events in novel candidate genes for schizophrenia. Schizophr Res. 2012;142(1-3):188–199. doi: 10.1016/j.schres.2012.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duff D, Long A. Roles for RACK1 in cancer cell migration and invasion. Cell Signal. 2017;35:250–255. doi: 10.1016/j.cellsig.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Jia D, Yan M, Wang X. et al. Development of a highly metastatic model that reveals a crucial role of fibronectin in lung cancer cell migration and invasion. Bmc Cancer. 2010;10:364–364. doi: 10.1186/1471-2407-10-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murata K, Yoshitomi H, Furu M. et al. MicroRNA-451 down-regulates neutrophil chemotaxis via p38 MAPK. Arthritis Rheumatol. 2014;66(3):549–559. doi: 10.1002/art.38269. [DOI] [PubMed] [Google Scholar]
- 17.Tan B, Liu L, Yang Y. et al. Low CPNE3 expression is associated with risk of acute myocardial infarction: A feasible genetic marker of acute myocardial infarction in patients with stable coronary artery disease. Cardiol J. 2018 doi: 10.5603/CJ.a2017.0155. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moon HE, Byun K, Park HW. et al. COMP-Ang1 potentiates EPC treatment of ischemic brain injury by enhancing angiogenesis through activating AKT-mTOR pathway and promoting vascular migration through activating Tie2-FAK pathway. Exp Neurobiol. 2015;24(1):55–70. doi: 10.5607/en.2015.24.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ratushnyy AY, Buravkova LB. Expression of focal adhesion genes in mesenchymal stem cells under simulated microgravity. Dokl Biochem Biophys. 2017;477(1):354–356. doi: 10.1134/S1607672917060035. [DOI] [PubMed] [Google Scholar]
- 20.Uehara K, Uehara A. Differentiated localizations of phosphorylated focal adhesion kinase in endothelial cells of rat splenic sinus. Cell Tissue Res. 2016;364(3):611–622. doi: 10.1007/s00441-015-2350-1. [DOI] [PubMed] [Google Scholar]
- 21.Sulzmaier FJ, Jean C, Schlaepfer DD. FAK in cancer: Mechanistic findings and clinical applications. Nat Rev Cancer. 2014;14(9):598–610. doi: 10.1038/nrc3792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benzina S, Harquail J, Guerrette R. et al. Breast cancer malignant processes are regulated by Pax-5 through the disruption of FAK signaling pathways. J Cancer. 2016;7(14):2035–2044. doi: 10.7150/jca.15200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ming Luo, Jun-Lin Guan. Focal Adhesion Kinase: a Prominent Determinant in Breast Cancer Initiation, Progression and Metastasis. Cancer Lett. 2010;289(2):127–139. doi: 10.1016/j.canlet.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nalefski EA, Falke JJ. The C2 domain calcium-binding motif: structural and functional diversity. Protein Sci. 1996;5(12):2375–2390. doi: 10.1002/pro.5560051201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Benes CH, Wu N, Elia AE. et al. The C2 domain of PKCdelta is a phosphotyrosine binding domain. Cell. 2005;121(2):271–280. doi: 10.1016/j.cell.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Lee JY, Hong M, Lee J. et al. An investigation of the role of gene copy number variations in sorafenib sensitivity in metastatic hepatocellular carcinoma patients. J Cancer. 2017;8(5):730–736. doi: 10.7150/jca.17887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang Z. ErbB receptors and cancer. Methods Mol Biol. 2017;1652:3–35. doi: 10.1007/978-1-4939-7219-7_1. [DOI] [PubMed] [Google Scholar]
- 28.Hu L, Lu F, Wang Y. et al. RACK1, a novel hPER1-interacting protein. J Mol Neurosci. 2006;29(1):55–65. doi: 10.1385/JMN:29:1:55. [DOI] [PubMed] [Google Scholar]
- 29.Li G, Ji XD, Gao H. et al. EphB3 suppresses non-small-cell lung cancer metastasis via a PP2A/RACK1/Akt signalling complex. Nat Commun. 2012;3:667–676. doi: 10.1038/ncomms1675. [DOI] [PubMed] [Google Scholar]
- 30.Kiely PA, Baillie GS, Barrett R. et al. Phosphorylation of RACK1 on tyrosine 52 by c-Abl is required for IGF-I-mediated regulation of focal adhesion kinase (FAK) J Biol Chem. 2009;284(30):20263–20274. doi: 10.1074/jbc.M109.017640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cox EA, Bennin D, Doan AT. et al. RACK1 regulates integrin-mediated adhesion, protrusion, and chemotactic cell migration via its Src-binding site. Mol Biol Cell. 2003;14(2):658–669. doi: 10.1091/mbc.E02-03-0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fei L, Ma Y, Zhang M. et al. RACK1 promotes lung cancer cell growth via an MCM7/RACK1/ Akt signaling complex. Oncotarget. 2017;8(25):40501–40513. doi: 10.18632/oncotarget.17120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCahill A, Warwicker J, Bolger GB. et al. The RACK1 scaffold protein: a dynamic cog in cell response mechanisms. Mol Pharmacol. 2002;62(6):1261–1273. doi: 10.1124/mol.62.6.1261. [DOI] [PubMed] [Google Scholar]
- 34.Kheifets V, Mochly-Rosen D. Insight into intra- and inter-molecular interactions of PKC: design of specific modulators of kinase function. Pharmacol Res. 2007;55(6):467–476. doi: 10.1016/j.phrs.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary figures and tables.