SUMMARY
Virus capsids are protein shells that protect the viral genome from environmental assaults, while maintaining the high internal pressure of the tightly packaged genome. To elucidate how capsids maintain stability under harsh conditions, we investigated the capsid components of the hyperthermophilic phage P74–26. We determined the structure of capsid protein gp87 and show that it has the same fold as decoration proteins in many other phages, despite lacking significant sequence homology. We also find that gp87 is significantly more stable than mesophilic homologs. Our analysis of the gp87 structure reveals that the core “β tulip” domain is conserved in trimeric capsid components across numerous double-stranded DNA viruses, including Herpesviruses. Moreover, this β barrel domain is found in anti-CRISPR protein AcrIIC1, suggesting a mechanism for the evolution of this Cas9 inhibitor. Our work illustrates the principles for increased stability of gp87, and extends the evolutionary reach of the β tulip domain.
INTRODUCTION
Tailed bacteriophages (also known as Caudoviruses) are the most common biological entities on Earth (Suttle, 2007; Wommack and Colwell, 2000). Caudoviruses assemble viral particles by actively packaging the viral genome inside a preformed, self-assembled capsid structure (Catalano, 2005). Because the DNA fills the capsid to nearly crystalline density, the pressure inside the capsid head is estimated to be extremely high (>6 MPa) (Evilevitch et al., 2004). Thus, the capsid structure must withstand high internal strain, in addition to the challenges imposed by the fluctuating external environment.
The capsids of Caudoviruses are primarily comprised of a single major capsid protein (MCP) of the HK97 fold, which assembles into an icosahedral shell (Aksyuk and Rossmann, 2011; Suhanovsky and Teschke, 2015). The MCP subunits interact with each other to build an interdigitated structure with high structural integrity. Proper construction of the capsid is directed by the portal ring that nucleates self-assembly of MCP, as well as a scaffolding protein that chaperones the MCP subunits to ensure the proper size and shape of the shell (Lurz et al., 2001; Prevelige and Fane, 2012; Prevelige et al., 1993). The MCP protein forms units called capsomers of five or six subunits, termed pentons or hexons, respectively. The capsid typically assembles with 11 pentons and a variable number of hexons, depending on the triangulation number of the virus shell. The capsid structure is altered during genome packaging, causing expansion of the shell, loss of the scaffolding protein, and a morphological change from nearly spherical to an icosahedral structure. The icosahedral shell is stable against the high internal pressure from the condensed DNA filling the capsid interior.
Many viruses have evolved mechanisms to stabilize the quasi and icosahedral 3-fold axes of the capsid, which are necessary for stabilization of the icosahedral shell. For example, the siphovirus HK97 uses covalent linkages at the 3-fold axes that form a “chain-mail” to stabilize the capsid structure (Wikoff et al., 2000). On the other hand, many viruses instead use auxiliary capsid proteins to add structural integrity, in particular the trimeric decoration proteins that bind at the 3-fold axes of the capsid icosahedron (Sternberg and Weisberg, 1977). Decoration proteins (also known as cementing proteins) play an important role in capsid stability and assembly. Cryoelectron microscopy (cryo-EM) structures reveal that decoration proteins specifically interact with the 3-fold axis by inserting their N-terminal region into a groove formed by neighboring capsomers (Lander et al., 2008). Decoration proteins only bind to the expanded, icosahedral capsid because the sites for binding are occluded in the smaller, spherical procapsids (Tang et al., 2006). Studies of the l phage decoration protein revealed that binding of decoration protein trimers strengthens the capsid assembly against both increased temperature and mechanical deformation (Gilcrease et al., 2005; Sae-Ueng et al., 2014; Sternberg and Weisberg, 1977). However, it is unclear how evolution modulates decoration protein-mediated stabilization. Moreover, it is unclear how decoration proteins are related to other proteins found throughout viral and cellular lineages.
Herpesviruses have a similar mechanism of capsid assembly as tailed bacteriophages, which implies that Herpesviruses evolved from Caudoviruses (Rixon and Schmid, 2014). Both classes of viruses use similar DNA-packaging machinery, with a terminase-class motor and portal complex that acts as both an entrance and exit for the viral genome (Bazinet and King, 1985; Hendrix, 1978; Hsiao and Black, 1978). Herpesvirus and Caudovirus capsids are primarily comprised of an MCP with the HK97 fold (Suhanovsky and Teschke, 2015) and direct self-assembly using a similar scaffolding protein (Fane and Prevelige, 2003). Herpesvirus particles have trimeric triplex proteins at the 3-fold axes of the capsid (Newcomb et al., 1993). Triplex is an attractive target for human cytomegalovirus (HCMV) vaccine development (Choi et al., 2016). The triplex complex is an integral part of both the immature procapsid and the final capsid shell, unlike decoration proteins of Caudoviruses (Heymann et al., 2003). Moreover, the triplex proteins are substantially larger and share no significant sequence similarity with decoration proteins. Thus, the shared mechanisms of particle assembly and evolutionary relationships of these capsid proteins remain unclear.
Phage structural proteins can be evolutionary sources of proteins that perform other functions, particularly phage-host interactions (Basler et al., 2012; Fraser et al., 2006; Veesler and Cambillau, 2011). A critical class of proteins that modulate phage-host interactions are the anti-CRISPR (Acr) proteins, which are encoded by phage and other mobile genetic elements to inhibit the CRISPR/Cas adaptive immune system of prokaryotes (Borges et al., 2017). Beyond their critical role in the molecular arms race between prokaryotes and viruses, Acr proteins are important for medicine and biotechnology. Acr proteins have been found in bacterial mobile elements, and have been proposed to increase the virulence of these bacterial strains (Maxwell, 2016). Acr proteins have been shown to improve CRISPR/Cas9-based genome editing by limiting off-target effects (Shin et al., 2017), as well as converting a CRISPR-associated nuclease into a transcriptional repressor (Bondy-Denomy et al., 2015). Despite their importance, the evolution of Acr proteins is still largely mysterious. As has been noted previously (Bondy-Denomy et al., 2013; Pawluk et al., 2016), Acr genes are often encoded near viral structural genes, which leads us to hypothesize that some Acr genes may have evolved from structural components of viruses.
Here we investigate phage decoration protein structure, function, and evolution using a hyperthermophilic phage. Phage P74–26 is found in hot springs and infects Thermus thermophilus bacteria (Yu et al., 2006). P74–26 has the longest tail of known viruses (~1 μm) and thrives at 70°C (Minakhin et al., 2008; Yu et al., 2006), a temperature at which related mesophilic phages are disabled in minutes (Bauer and Evilevitch, 2015). We identify the protein gp87 (P74p87) as the decoration protein of P74–26 phage. Our 1.7-Å resolution structure of gp87 reveals it has the same fold as the phage λ decoration protein, despite a lack of sequence similarity. We show that the hyperthermophilic protein is substantially more stable than its mesophilic homologs. Furthermore, we identify a conserved β barrel domain of the decoration protein that is found in Herpesvirus triplex proteins and the anti-CRISPR protein AcrIIC1, suggesting that these diverse proteins share a common evolutionary ancestor. Our work provides the groundwork for understanding the high stability of thermophilic viruses. Moreover, these studies illustrate a deep connection between the capsid machinery of tailed bacteriophage and Herpesviruses, and lead to a potential mechanism for the evolution of an Acr protein.
RESULTS
Major Components of P74–26 Virions
We purified P74–26 virions from infections of T. thermophilus strain HB8 using a combination of polyethylene glycol precipitation and gradient centrifugation. The P74–26 virions consist of an icosahedral capsid ~80 nm in diameter and long, non-contractile tails exceeding 800 nm in length (Figure 1A).
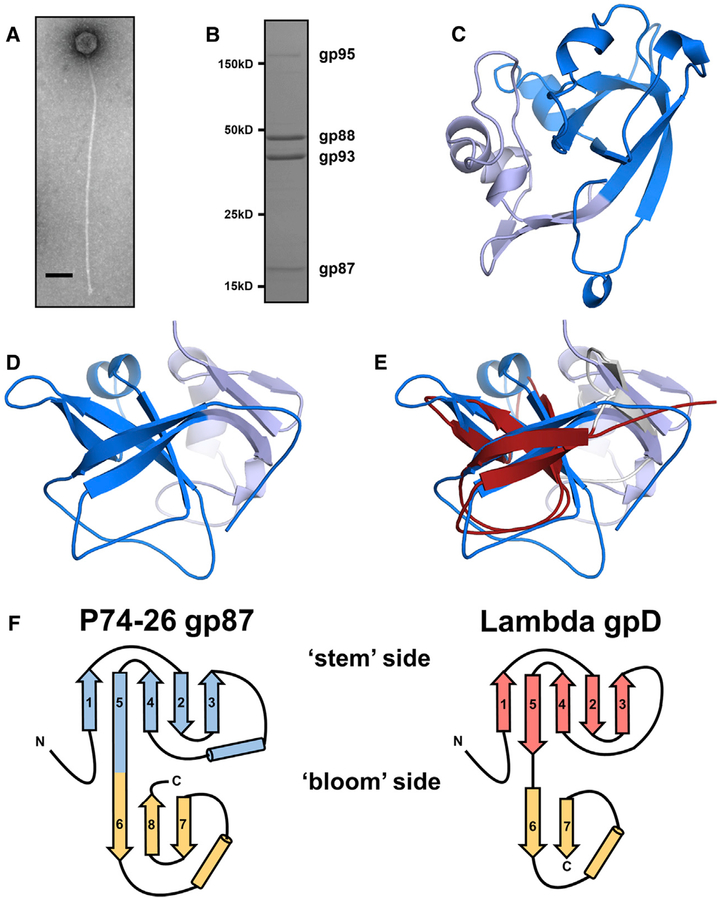
Figure 1. P74–26 gp87 Is a Thermophilic Capsid Decoration Protein.
(A) Negative-stain electron micrograph of purified P74–26 virion. Scale bar, 100 nm.
(B) SDS-PAGE analysis of P74–26 virions reveals major structural components including gp87, gp88 (MCP), gp93 (tail protein), and gp95 (tape measure protein).
(C and D) The 1.7-Å resolution structure of P74–26 gp87 with the five-stranded β tulip domain is highlighted in dark blue.
(E) Structure-based alignment of P74–26 gp87 (β tulip domain in blue) and λ decoration protein gpD (gray, β tulip domain in red, PDB: 1C5E) reveals significant structural homology despite high sequence variance.
(F) Topology diagrams of P74–26 gp87 and λ gpD reveal conserved architecture of β tulip domain flanked by a small mixed α/β domain.
To determine whether the P74–26 capsid is stabilized by covalent interactions (as in the phage HK97 [Duda, 1998; Wikoff et al., 2000]), we analyzed the proteome of P74–26 virions by SDS-PAGE. The MCP migrates on SDS-PAGE at its expected molecular weight (46.6 kDa), similar to results obtained previously (Figure 1B) (Minakhin et al., 2008). This result is in contrast to phage HK97, whose MCPs are linked covalently and run on SDS-PAGE as a smear at very high molecular weight (Duda, 1998). Thus, we conclude that the P74–26 capsid is stabilized by non-covalent interactions.
We hypothesized that P74–26 capsids are stabilized by decoration proteins. Our SDS-PAGE analysis reveals other major components of the virions: the tail tube protein (gp93;37.9 kDa), the tape measure protein (gp95; 550 kDa), and gp87 (16.3 kDa), whose function in the virion is unknown. Based on size, abundance, and gene location, we hypothesize that gp87 acts as a decoration protein. gp87 is similar in size to known decoration proteins gpD and SHP from phages λ and P21, respectively (~12 kDa for both). Moreover, the relative abundance of gp87 is close to that expected for a decoration protein (318 ± 6 copies per virion by gel band densitometry). Finally, the genes encoding decoration proteins and MCPs are usually proximal to each other (Casjens, 2005; Hatfull, 2008); the P74–26 MCP is encoded by gene 88, adjacent to gene 87.
P74–26 gp87 Structure Reveals High Similarity to Known Phage Decoration Proteins
To test whether gp87 is a decoration protein, we determined its atomic structure by X-ray crystallography. Purified recombinant P74–26 gp87 readily crystallized. We solved the structure to 2.3-Å resolution by single-wavelength anomalous dispersion using iodide ions soaked into the crystals, and phases subsequently were extended to a 1.7-Å resolution native dataset (Table 1; Figures 1C, 1D, and S1A). The structure reveals a core domain comprised of a five-stranded anti-parallel β barrel, followed by a mixed α/β subdomain. According to the nomenclature for β strand topology (Richardson, 1981), the barrel has a topology of (+3, +1, −2, −1) (Figure 1F). An α-helical linker connects strands 3 and 4. One end of the β barrel is flared open, while the other is capped with loops, such that the domain resembles a tulip. We refer to the domain as a β tulip due to this resemblance (Figure 2A). The smaller C-terminal domain consists of a three-stranded anti-parallel β sheet capped by an α helix and extended loop structure. The N-terminal 16 amino acids are not visible in the structure, most likely due to disorder. The crystal packing reveals a trimer of gp87 proteins arranged in a “head-to-tail” fashion (Figure 2B). The “bloom” end of the tulip is interacting with the mixed α/β domain of a neighbor, primarily through hydrophobic interactions. β tulip residues F27, L29, F31, I80, and L99 form an intermolecular hydrophobic cluster with the C-terminal domain residues I135 and F140 in the neighboring subunit (Figures S2A–S2C).
Table 1.
Data Collection and Refinement Statistics
| Data Collection | KI Derivative | Native (APS) |
|---|---|---|
| Space group | P 63 | P 63 |
| Wavelength | 1.54 (home source) | 0.979 (19-BM) |
| Resolution range | 29.38–2.34 | 25.84–1.69 |
| Unit cell dimensions | ||
| a, b, c (Å) | 89.74, 89.74, 31.36 | 89.52, 89.52, 31.39 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| No. of total reflections | 67,505 | 119,377 |
| No. of unique reflections | 6,255 | 16,355 |
| Multiplicity | 10.8(9.7) | 7.3 (7.2) |
| Completeness (%) | 98.9 (95.6) | 99.9(100) |
| Mean L/σI | 16.2 (3.2) | 14.3(2.3) |
| Wilson B factor | 32.14 | 26.76 |
| Rmerge | 0.125(0.728) | 0.057 (0.824) |
| Rmeas | 0.131 (0.768) | 0.062 (0.888) |
| Rpim | 0.039 (0.242) | 0.023 (0.328) |
| CC½ | 0.998 (0.847) | 0.999(0.810) |
| Refinement | ||
| Rwork (%) | 17.87 | |
| Rfree (%) | 21.26 | |
| RMSD | ||
| Bond lengths (Å) | 0.01 | |
| Bond angles (°) | 1.05 | |
| Ramachandran analysis (%) | ||
| Favored | 96.1 | |
| Rotamer outliers (%) | 0 | |
| Clashscore | 1.01 | |
| Average B factor | 36.8 | |
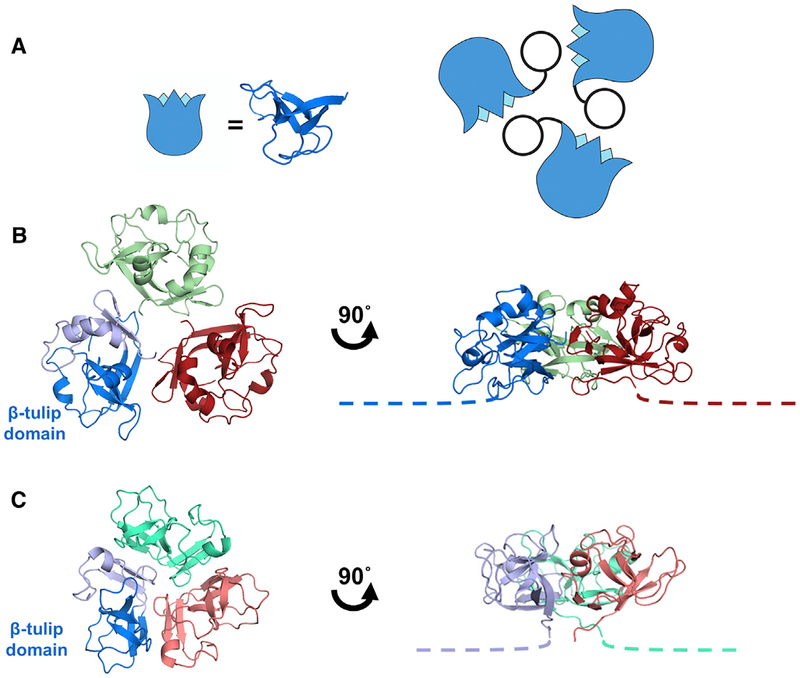
Figure 2. Interactions of the Decoration Protein for Capsid Stabilization.
(A) Model of decoration protein trimer highlighting positions of the β tulip domains (blue) within the assembly.
(B) P74–26 gp87 trimer highlights difference in trimer assembly characterized by a ~20° outward rotation of each of the gp87 trimer subunits (see also Video S2). The N-terminal capsid binding region of both crystal structures is disordered, and is drawn as proportional dotted lines in (B) and (C).
(C) Structure of the λ gpD trimer shows the orientation of gpD from the top of the capsid (left) and rotated 90° to the side (right).
The structure and assembly of P74–26 gp87 is very similar to that observed for other well-known decoration proteins. The gp87 protein exhibits the same fold as the gpD protein from phage λ and the SHP protein from lamboid phage P21 (Forrer et al., 2004; Yang et al., 2000) (Figures 1E, 1F, S1B, and S1C; Video S1). Both gpD and SHP contain an N-terminal β tulip domain and a C-terminal mixed α/β subdomain (Cα root-mean-square deviation [RMSD] compared with gp87 is 2.3 and 2.5Å, respectively). The similarity between the β tulip domains of gp87 and gpD is particularly high (Cα RMSD = 2.1Å; Table S1). Moreover, all three proteins have an N-terminal region that is disordered in the crystal structures (16, 14, and 11 residues for gp87, gpD, and SHP, respectively). The C-terminal domains have an overall similar fold (strand-loop-helix-strand). In gpD and SHP, the two β strands of the C-terminal subdomain form a parallel β sheet, whereas gp87 has an insertion of an extra β strand that results in the formation of an anti-parallel three-stranded β sheet (Figure S1D). Thus, the fold of gp87 is nearly identical to that of gpD and SHP.
The decoration proteins gpD and SHP also crystallized as trimers, with a similar (but not identical) arrangement around the 3-fold axis of the trimer as in gp87 (Figures 2B and 2C). The trimerization interface is comprised of the core β tulip domain of one subunit contacting the C-terminal domain of an adjacent subunit. This arrangement places the unstructured N-terminal region of each subunit at the “bottom” of the trimer such that it can attach to the capsid shell (Lander et al., 2008). Thus, the similarities in both tertiary and quaternary structure, and its high abundance in the virion lead us to conclude that gp87 is a decoration protein. The close structural and functional similarity is notable despite the low sequence homology between the thermophilic gp87 and its mesophilic cousins (~9% sequence identity) (Figure S1E).
Despite the lack of clear sequence similarity, we detect several residues that are conserved across decoration proteins. We built a structure-based sequence alignment of phage decoration proteins to identify any trends in conservation of residues throughout decoration proteins. We identify only seven residues that are largely conserved across known homologs (Figure S1E). We note two conserved residues (Asp 66 and Glu 83 in λ gpD) poised near the capsid binding surface of the decoration protein that may play a role in stabilizing the capsid when gpD is bound to the virion. Future studies will determine whether these putative interactions are important for stabilizing the capsid assembly.
P74–26 gp87 Is More Stable than Mesophilic Homologs
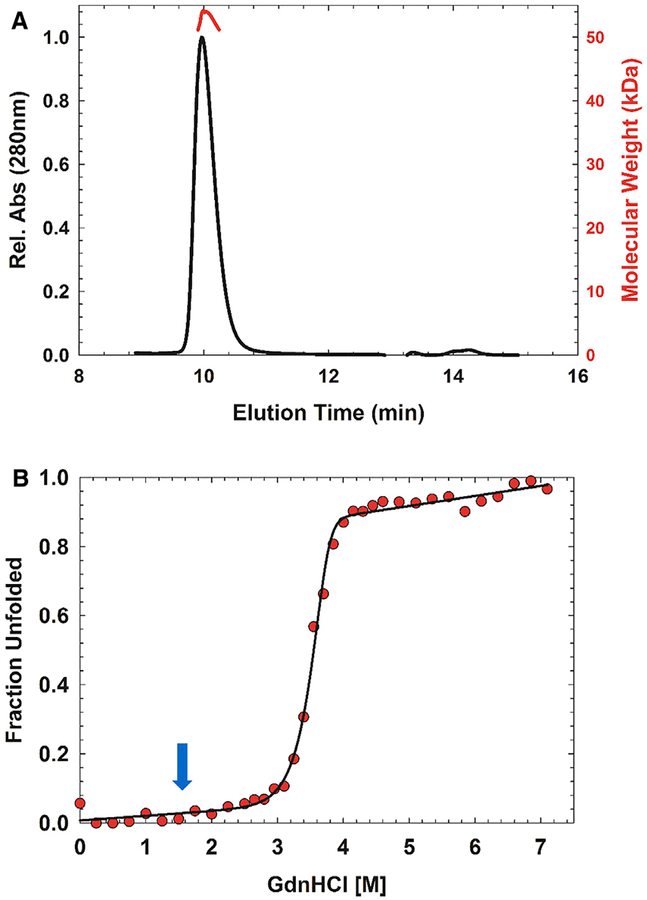
We investigated whether the thermophilic decoration protein is more stable than its mesophilic counterparts. We first investigated whether gp87 forms a stable quaternary assembly in solution using size-exclusion chromatography/multi-angle light scattering. We find that gp87 is a stable trimer in solution at ~20 mM, with a measured molecular mass of 53 kDa (the actual molecular mass of the trimer is 49 kDa) (Figure 3A). This result contrasts with λ gpD, which is monomeric in solution even up to millimolar concentrations under similar solvent conditions (Imber et al., 1980; Yang et al., 2000). However, gpD crystallized as a trimer. Thus, P74–26 gp87 forms a more stable trimer than λ gpD.
Figure 3. Thermophilic Decoration Protein Has Enhanced Stability Compared with Mesophilic Homologs.
(A) P74–26 gp87 forms a stable trimer in solution as determined by size-exclusion chromatography-multi-angle light scattering. Predicted molecular mass, 49 kDa; measured molecular mass, 52 kDa.
(B) Representative equilibrium fraction unfolding curve of P74–26 gp87 at 5 μM shows a steep unfolding transition from 3 to 4 M GdnHCl; excitation, 295 nm; emission, 325 nm. The solid line represents the global fit to a model of trimer to three unfolded monomers. Blue arrow indicates the comparable transition midpoint of λ gpD unfolding.
We investigated the stability of the trimer by guanidine hydrochloride (GdnHCl)-induced unfolding, using both circular dichroism (CD) and tryptophan fluorescence spectroscopies (Figures S2D–S2F). We measured unfolding across a range of protein concentrations (2.5–15 μM) to determine if trimerization is coupled with unfolding. Our CD and fluorescence data match each other quite well across all measured protein concentrations, with no hysteresis (Figures S2D–S2F). We observe a single, sharp, and reversible transition between 3 and 4 M GdnHCl with a midpoint of approximately 3.6 M GdnHCl (Figure 3B). In contrast, the midpoint of the λ gpD and P21 SHP unfolding curves are at 1.4 and 1.1 M GdnHCl, respectively (Forrer et al., 2004), indicating that gp87 trimer is substantially more stable than its mesophilic homologs. The data from the CD and fluorescence experiments were globally fit to a trimer to three unfolded monomers model (N3 ⇌ 3 U). Our attempts to fit the data to a three-state model failed to converge on a reasonable solution, indicating that trimer dissociation is coupled to unfolding. The estimated stability of the trimer was found to be 42.1 ±1.3 kcal (mol trimer)−1 with a denaturant dependence of 7.8 ±0.4 kcal (mol trimer)−1 M−1. In contrast, the stability estimated for the gpD monomer is ~5.2 kcal (mol)−1 (Forrer et al., 2004).
Similarities between Herpesvirus Triplex and Phage Decoration Proteins
Our finding that the decoration protein fold is specified with very low sequence conservation led us to investigate whether this fold is found in other proteins, but has been overlooked by classical comparative genomics approaches. We first investigated whether this ancient fold may be found in the Herpesviruses, because Herpesviruses are thought to be direct descendants of tailed bacteriophage (Baines and Weller, 2005; Chen et al., 2011; Duda et al., 2013; Rixon and Schmid, 2014; Selvarajan Sigamani et al., 2013). We focused on the triplex complex of Herpesviruses, because both triplex and phage decoration proteins are capsid components that bind and stabilize the 3-fold axes of the capsid (Lander et al., 2008; Yu et al., 2017). Thus, we hypothesized that the triplex proteins and phage decoration proteins may share an evolutionary origin.
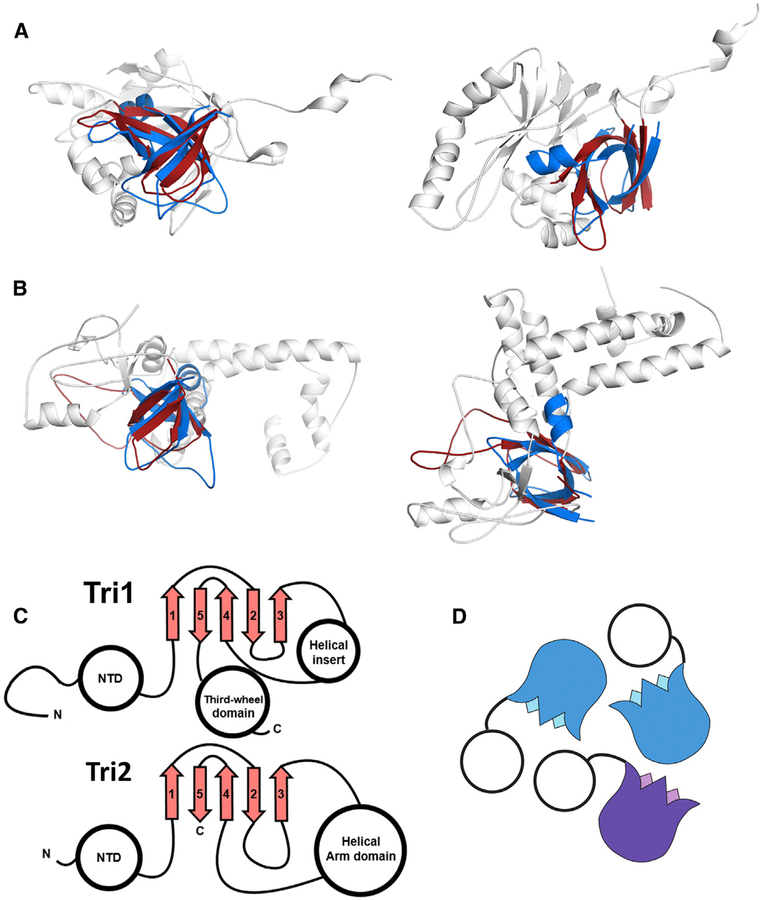
We find that core β tulip domain of phage decoration proteins is structurally similar to the core trimerization region of the triplex proteins (Figures 4A and 4B; Video S3). The triplex complex is a heterotrimer consisting of one copy of Tri1 and two copies of the Tri2 protein. Both Tri1 and Tri2 contain a cryptic β tulip domain centrally located in their structure. In both cases, the β tulip domains are discontinuous, with large insertions in loops between strands of the tulip (Figure 4C). The β tulip domains of Tri1 and Tri2 have the same basic size, shape, and topology as β tulip domains of phage decoration proteins (Cα RMSD = 2.4 and 2.2, respectively compared with gp87; Figures 4A–4C; Table S1). The β tulip region is the only region of obvious structural similarity between Tri1, Tri2, and decoration proteins. In all three subunits of the triplex trimer, the tulip domains participate in the primary trimerization interface (Figure 4D). Moreover, the triplex β tulip domains are arranged roughly parallel to the capsid shell, with the bloom end of the tulip as an interaction surface, similar to the phage decoration proteins. The N-terminal regions of triplex proteins are pointed toward the capsid surface and make key interactions with the HK97 fold of MCP (Yu et al., 2017), in an analogous manner as phage decoration proteins. Both Tri1 and Tri2 contain a helical region inserted in between strands 3 and 4 of the β tulip domain. This helical region is substantially larger in Tri2, which forms a “bracing arm” that tightly dimerizes the two Tri2 proteins. Insertion of helical regions is common between strands 3 and 4 of the β tulip domain, as seen in P74–26 gp87 and other β tulip proteins (see below). These similarities in tertiary and quaternary structure suggest a shared evolutionary history between the phage decoration proteins and the triplex proteins of Herpesviruses.
Figure 4. Structural Similarity of Phage Decoration Protein Trimers and the HCMV Triplex.
(A and B) The P74–26 gp87 β tulip domain (blue) is similar to central domain of HCMV Tri1 (A) and Tri2
(B) proteins (gray, β tulip domains in red; PDB: 5VKU).
(C) Central domain of HCMV Tri1 and Tri2 has conserved β tulip topology.
(D) HCMV triplex proteins form an asymmetric trimer consisting of two molecules of Tri2 (blue) and one molecule of Tri1 (purple).
β tulip Domains Are General Protein-Protein Interaction Motifs Enriched in Viruses
We sought to determine if other proteins contain β tulip domains. Unsurprisingly, sequence similarity search algorithms such as PSI-Blast (Altschul et al., 1997) found no significant homologs outside of decoration proteins or triplex proteins. Because the β tulip domain does not appear to be associated with a specific sequence motif, we searched for similar structures in the protein structure database using the programs DALI and VAST (Holm and Rosenström, 2010; Madej et al., 2014). Interestingly, β tulip domains were identified in three seemingly unrelated proteins: (1) AcrIIC1, (2) the tailspike protein gp12 of phage Φ29, and (3) the molybdenum metabolizing protein MoeA.
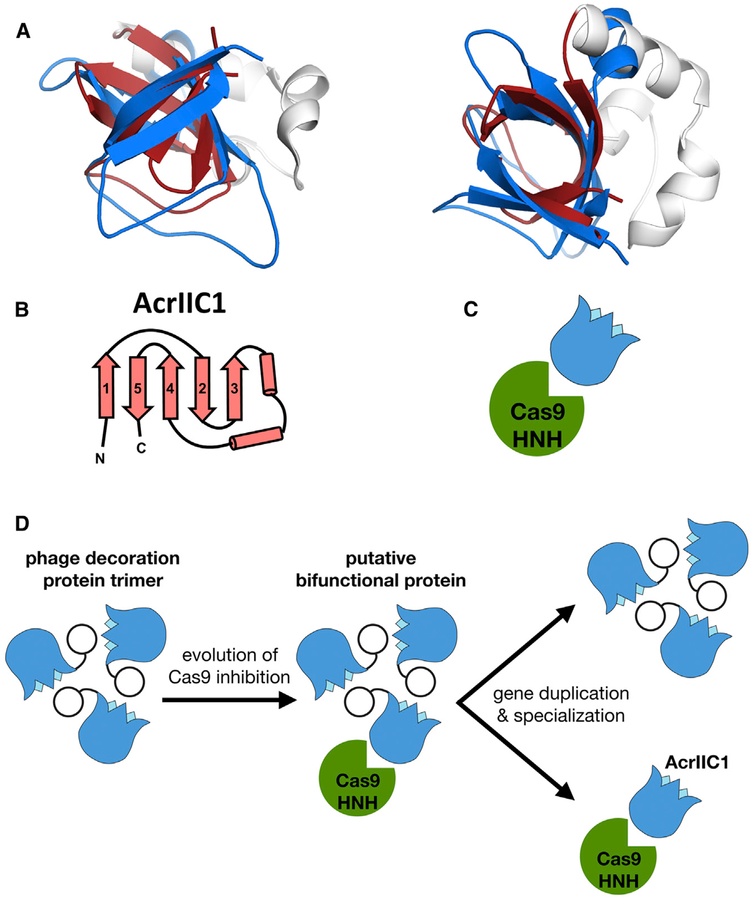
AcrIIC1 consists of a single β tulip with a small α-helical insertion (Figures 5A and 5B). This small protein is a broad spectrum Cas9 inhibitor and binds directly to the Cas9 HNH nuclease active site, preventing DNA cleavage (Harrington et al., 2017). Despite lacking any discernible sequence similarity, the β tulip of AcrIIC1 is structurally very similar to that of P74–26 gp87, with a Cα RMSD of 2.6Å (Figure 5A; Table S1). The β tulip domains of both AcrIIC1 and gp87 contain a helical linker that connects strands 3 and 4 of the tulip. Unlike other β tulip domains, AcrIIC1 is monomeric and interacts with its partner (Cas9) through the “stem” end of the β tulip (Figure 5C). Despite significant structure conservation between gp87 and AcrIIC1, gp87 does not inhibit NmeCas9 cleavage in vitro (Figure S3).
Figure 5. β tulip Domain Suggests Evolution of Acr Proteins from Phage Structural Proteins.
(A) Structural alignment of P74–26 gp87 β tulip domain (blue) with the anti-CRISPR protein AcrIIC1 (β tulip in red; PDB: 5VGB).
(B) Topology diagram of AcrIIC1 reveals conserved β tulip domain architecture.
(C) Cas9 binds stem side of the AcrIIC1 β tulip domain rather than the bloom side.
(D) Decoration protein trimers form interactions with neighboring subunits through the “bloom” end of the β tulip domain, leaving the “stem” end exposed. Anti-CRISPR AcrIIC1 binds to the Cas9 HNH domain through the stem end of the β tulip domain and may have evolved from a bifunctional phage decoration protein.
The Φ29 tailspike protein gp12 uses its tandem β tulip domains (termed the D4 region) to direct assembly of the tailspike trimer (Xiang et al., 2009). Recently, Rossmann and colleagues described structural homology between the C-terminal portion of gp12 and the decoration protein gp56 from the marine siphovirus TW1 (Wang et al., 2017). The TW1 decoration protein contains N- and C-terminal β tulip domains with significant structural similarity to other β tulip-containing proteins (Figures S4E–S4H; Table S1). Our structural comparison of gp12 with the decoration protein from P74–26 revealed that the D4 region of gp12 consists of two consecutive β tulip domains forming intramolecular contacts in a bloom-to-stem fashion (Figures S4A–S4D). This arrangement is echoed in the quaternary structure, as the bloom end of the C-terminal β tulip domain interacts with the stem end of the N-terminal tulip in the adjacent subunit of the trimer (Figure S4D). The N-terminal β tulip domain has a small b hairpin inserted between strands 4 and 5, which projects outward away from the ring of tulips. The molybdenum biosynthetic enzyme MoeA contains a C-terminal β tulip domain that aids in oligomerization of MoeA (Schrag et al., 2001). As often found in other β tulip domains, the MoeA tulip contains a short α-helical linker between strands 3 and 4 (Figures S5A and S5B).
DISCUSSION
Increased Stability of a Thermophilic Decoration Protein
P74–26 is a thermophilic virus found in hot springs between 60°C and 75°C. We sought to determine how the P74–26 virion maintains stability under these harsh conditions. We find that the P74–26 capsid is not stabilized by covalent crosslinks, unlike HK97 phage that stabilizes the capsid through “covalent chainmail” of the MCP (Wikoff et al., 2000). Instead, we hypothesize that gp87 acts as a decoration protein to stabilize the capsid. gp87 is present in the virion with a stoichiometry expected for a decoration protein, and adopts similar tertiary and quaternary structure as mesophilic decoration proteins. Despite the similar structure and function, gp87 is widely divergent at the sequence level from other known decoration proteins. The low sequence homology may preclude identification of decoration proteins using classical comparative genomics approaches; structural and/or biochemical analyses may be necessary for proper annotation.
Our structural and biochemical characterization of gp87 provides insight into the mechanisms of capsid stabilization. Phage P74–26 thrives at 70°C, a temperature at which phage λ disassembles readily (t1/2 = 5 min) (Bauer and Evilevitch, 2015). The P74–26 decoration protein is more stable than its mesophilic counterpart, gpD (Figure 3B) (Forrer et al., 2004). Furthermore, isolated gp87 is a stable trimer in solution with a stability of42.1 ± 1.3 kcal (mol trimer)−1, whereas gpD is monomeric in solution. Because our unfolding data show that gp87 unfolds in a single transition, we hypothesize that dissociation of the trimer and unfolding of monomers are tightly linked. Thus, we propose that gp87 has increased stability due to both tertiary and quaternary interactions.
Our analysis of the gp87 crystal structure reveals the likely mechanism for gp87 stabilization. Each subunit in the P74–26 decoration protein is larger, burying more hydrophobic surface within each protein’s core compared with the decoration proteins from phages λ and P21 (4,470, 3,382, and 3,828Å2 for gp87, gpD, and SHP, respectively) (Forrer et al., 2004). In particular, gp87 displays larger clusters of hydrophobic residues (I, L, V, and F) than gpD (Figures S2A–S2C). ILVF clusters have recently been hypothesized to be key determinants of folding and stability (Kathuria et al., 2016). The larger ILVF clusters in gp87 may increase the intrinsic stability of each subunit of the trimer. The ILVF clusters are more extensive in thermophilic gp87 than in mesophilic gpD. A gpD trimer contains six ILVF clusters, two per subunit. In contrast, a gp87 trimer has nine ILVF clusters, three of which span from the β tulip domain of one subunit to the C-terminal domain of the adjacent subunit (Figures S2A and S2B). Our structure also indicates that the interfacial interactions between decoration subunits increases stability of the complex. The interfaces of the trimeric assembly bury greater surface area in gp87 than SHP or gpD. The greater buried surface area is expected to afford greater stabilization of the trimer. Tighter interactions between subunits will link trimerization to the unfolding of decoration protein. Indeed, our unfolding data exhibit a single transition, supporting this model.
The gp87 structure also reveals insights into how the decoration protein interacts with the capsid to stabilize the overall assembly. The N-terminal region of decoration proteins is known to interact directly with MCP, forming an extended β sheet across the HK97 fold E loop (Lander et al., 2008). This structuring of the N-terminal region creates struts that stabilize the 3-fold axes in the icosahedral shell. The N-terminal region of gp87 is substantially longer than those of mesophilic gpD and SHP (27, 19, and 21 residues long, respectively). The longer arm could provide a stronger foothold for the trimer into the capsid structure, stabilizing the 3-fold axes. Moreover, the globular domains of gp87 are larger than those of its mesophilic cousins, which could allow more interaction surface between the core trimer and the MCP shell. Furthermore, we observe that the orientation of the gp87 subunit is different than observed for gpD and SHP, with each subunit rotated downward (toward the capsid shell) by ~20° (Video S2). The altered orientation could place the globular domains of gp87 in closer contact with the capsid shell to afford tighter contacts and greater stabilization. Finally, we identified several residues that are conserved across the known decoration proteins and we note that several of these residues appear to be poised to interact directly with the capsid shell. Interestingly, the two prime candidate residues for MCP interactions appear to interact directly with the capsid in cryo-EM structures of λ phage (Lander et al., 2008). Further structural and biochemical analyses will be necessary to uncover whether these interactions are important for stabilization of the capsid shell.
Evolutionary Relationship of Herpesvirus Triplex Proteins and Phage Decoration Proteins
Our analysis reveals that the β tulip domain of decoration proteins is structurally and functionally conserved in Herpesvirus triplex proteins. Although there is only 10% and 18% sequence identity between the β tulips of P74–26 gp87 with HCMV Tri1 and Tri2, their similarities deepen our understanding of the evolutionary linkage between these two classes of viruses. The structures of the β tulip domains are similar, with the same arrangement of β strands in the tulip. Both decoration protein and triplex β tulip domains participate in trimerization, in each case with the bloom side positioned at the interface. In addition, the β tulip domains arrange themselves in a similar manner. Both decoration proteins and triplex complexes position themselves at the 3-fold axes of the capsid icosahedron. In all cases, the C-terminal regions are pointed away from the capsid shell and the N-terminal region binds to the capsid surface, with direct interactions with the E loop of the MCP HK97 fold (Lander et al., 2008; Yu et al., 2017). Moreover, even the variable regions appear to have significant similarities. For example, gp87, Tri1, and Tri2 have a helical region inserted between strands 3 and 4 of the β tulip domain. In summary, the combined structural and organizational similarities illustrate the shared evolutionary history of Herpesvirus triplex and phage decoration proteins.
For the past decade or more, evidence of the shared evolutionary history of Herpesviruses and Caudoviruses has accumulated (Rixon and Schmid, 2014). In particular, the MCPs share the HK97 fold, although those of Herpesviruses have complicated elaborations (Suhanovsky and Teschke, 2015; Wikoff et al., 2000). Both virus families also direct the assembly of the capsid using a scaffolding protein that is part of procapsid but not found in the infectious particle (Fane and Prevelige, 2003; Johnson, 2010). Furthermore, the genome-packaging machinery and mechanism is largely conserved between the two classes of viruses (Bazinet and King, 1985; Hendrix, 1978; Hsiao and Black, 1978). Here we add to the similarities of these different classes of viruses, showing that the decoration proteins share an evolutionary history as well. Thus, the major genes involved in virus structure and assembly are conserved throughout Caudoviruses and Herpesviruses.
While a shared evolutionary history may imply mechanistic similarities between decoration and triplex proteins, there are substantial differences. First, the triplex proteins are not 3-fold symmetric as in phage decoration proteins (Figures 2A and 4D); the two Tri2 subunits grip each other in a tight embrace, while the Tri1 protein makes less-extensive contact (Yu et al., 2017). The heterotrimeric nature of the triplex complex has been hypothesized to allow each subunit in the complex to adopt a different role in capsid assembly (Saad et al., 1999). Decoration proteins attach to the capsid late in the assembly process after capsid expansion (Casjens and Hendrix, 1974; Dokland and Murialdo, 1993), while the triplex proteins bind the procapsid early in assembly before DNA is packaged and stay bound throughout all stages of virus particle assembly (Trus et al., 1996). This difference in assembly may reflect the fact that the triplex proteins embed between MCP capsomers and are an integral part of the capsid surface (Yu et al., 2017). This difference could be explained by the fact that the decoration proteins’ interactions with capsid components are less extensive and the decoration protein binding site is only accessible after capsid expansion (Lander et al., 2008; Tang et al., 2006).
The shared evolutionary history suggests that triplex proteins evolved from phage decoration proteins and, over the course of time, acquired a more integral role in capsid structure. As the triplex formed the basis for the 3-fold axes in Herpesvirus procapsids and capsids, additional domains were inserted into loops between strands of the β tulip fold. We note that MCP also underwent domain insertions within loops of the HK97 fold during the course of evolution, suggesting a common mechanism for elaboration of the basic folds found in viral capsids. We also note that the triplex’s role as a necessary component of the procapsid required evolution of a separate mechanism for strengthening the capsid against the internal pressure of packaged DNA. In Herpes Simplex Virus 1, this is achieved by the protein UL25 (Sae-Ueng et al., 2014; Snijder et al., 2017), which binds the penton vertices of the capsid. In the herpesvirus KSHV, the SCP protein crosslinks MCP subunits within hexons (Dai et al., 2015).
Evolutionary Origin of an Acr Protein
AcrIIC1 is a broad-spectrum inhibitor of Cas9 nuclease activity. AcrIIC1 binds directly to the HNH nuclease domain of Cas9, preventing target cleavage (Harrington et al., 2017; Pawluk et al., 2016). Acr inhibitors of Cas9 have found use in biotechnology applications to decrease off-target DNA cleavage (Shin et al., 2017). Despite the importance of Acr proteins, their evolution has remained mysterious (Pawluk et al., 2017).
Our discovery of the structural similarity of AcrIIC1 and phage decoration proteins suggests a shared evolutionary ancestor. AcrIIC1 consists primarily of a β tulip fold with a strong structural similarity to the β tulip domain of gp87 (Cα RMSD = 2.6Å, Figures 5A and 5B; Table S1). Much like other β tulips, AcrIIC1 contains a small helical insert between strands 3 and 4. However, we detect no significant sequence homology between AcrIIC1 and gp87 (6% sequence identity). This is not unexpected because the decoration proteins of λ and P74–26 phage share very little conservation, despite conserved function. Given the low sequence homology between β tulip-containing proteins, structure homology is much more useful than sequence similarity in identifying protein ancestry.
Our findings suggest shared ancestry between AcrIIC1 and phage decoration proteins, but do not clarify which preceded the other. Although it is formally possible that AcrIIC1 is the evolutionary origin for phage decoration proteins, we find it more likely that phage decoration proteins predated the evolution of AcrIIC1. AcrIIC1 uses the stem side of its β tulip domain to bind the Cas9 HNH domain (Harrington et al., 2017), in contrast to other β tulip proteins that interact primarily through the bloom side (Figure 5C). Thus, interaction with the Cas9 HNH nuclease could have evolved in a decoration protein without disrupting its central trimerization role (see Video S3). Gene duplication would then allow this putative bifunctional protein to specialize for each individual function (Figure 5D). One hurdle for this evolutionary pathway is that Acr proteins must act quickly upon phage infection to prevent CRISPR-mediated genome cleavage. However, decoration proteins are typically expressed late in infection (White et al., 2012) and typically remain outside the cell upon infection. How a decoration protein could be present during early stages of phage infection is not clear, although these proteins could be packaged with the genome and injected into the host cell, as is found for many phage proteins (Falco et al., 1977; Mullaney and Black, 2014; Strauss and King, 1984; Zhao et al., 2016). Alternatively, we propose that AcrIIC1 could have evolved in the context of a prophage, where the Cas9 inhibitory effect could have evolved for gene regulation or other functions (Bondy-Denomy et al., 2015).
The evolution of Acr proteins has been mysterious. There are currently over 20 unique families of Acr proteins that have been reported (Pawluk et al., 2017). These proteins are often small (50–150 residues) and have no clear sequence or structural features in common. Moreover, the mechanisms of Acr inhibition are diverse (Bondy-Denomy et al., 2015). Because of their remarkable diversity and small size, Acr proteins have been proposed to evolve de novo from small open reading frames (Pawluk et al., 2017). Here we show that the AcrIIC1 protein most likely evolved from the phage decoration protein fold. Future experiments (outside the scope of this paper) could investigate the evolutionary path between decoration and Acr proteins, perhaps by utilizing decoration proteins as phage display platforms (Yang et al., 2000).
Because β tulip domains have extremely low or no sequence homology, we could only identify this relation by structural similarity rather than sequence motifs. We propose that some Acr proteins may have evolved from other phage proteins, but their structural homology has been masked by the rapid evolution and insertion of new structural elements. Regardless, de novo evolution remains a likely explanation for many Acr proteins. Recent work from the Cordes group has shown that proteins from young, “non-coding” genes can have rudimentary ability to fold into compact, native-like states, suggesting a mechanism for evolution of new folds (Bungard et al., 2017).
The β tulip Fold as a Widespread ProteinInteraction Motif
Although this structural motif is quite small, we could only identify β tulip domains in a handful of proteins. There are other examples of five-stranded b barrels (e.g., HIV gp120 [Pan et al., 2015]), but these domains use a different topology than found in β tulip domains, suggesting that they evolved independently. The β tulip fold is structurally conserved despite lacking strong sequence conservation. This is reminiscent of the HK97 fold (Suhanovsky and Teschke, 2015), as well as the well-studied triosephosphate isomerase-barrel fold (Nagano et al., 2002).
The β tulip fold is enriched in viral proteins. Of the five classes of β tulip proteins that we identified, four of them are viral in origin: phage decoration proteins, Herpesvirus triplex proteins, phage tailspike proteins, and phage Acr proteins. The only outlier is MoeA, a molybdenum cofactor biosynthesis enzyme that is found throughout life. Whether the MoeA β tulip evolved through convergent or divergent evolution is still unknown. Regardless, we propose that the viral β tulip proteins share a common ancestor.
Recently, Rossmann and colleagues have elucidated the structure of the capsid-stabilizing protein gp56 from the marine siphovirus TW1, which shares structural homology with phage λ gpD and the tailspike protein from phage Φ29 (Wang et al., 2017). TW1 gp56 consists of two β tulip domains arranged in bloom to stem orientation around the central axis of the trimer (Figure S4H). The N-terminal β tulip features a short α-helical linker between strands 4 and 5 (Figure S4G). This contrasts with the other β tulip containing proteins, as α-helical linkers appear to be most common between strands 3 and 4. Given the similarity between gp56 and Φ29 tailspike, Rossmann and colleagues noted that in general the “gp56-like fold” is capable of forming stable trimers in capsid stabilizing proteins, but also in proteins with differing functions (Wang et al., 2017).
One of the viral β tulip proteins is the Φ29 phage tailspike protein, which forms a homotrimeric enzyme complex used for recognition and digestion of host cell wall structure (Moreno and Bluzat-Moreno, 1978). The D4 region of the tailspike protein contains a pair of β tulip domains arrayed in a head-to-tail fashion around the trimerization interface. Both β tulip domains have significant structure homology to P74–26 gp87 (Cα RMSD = 1.7 and 1.5Å for N- and C-terminal β tulip domains of D4, respectively; Figures S4A–S4D; Table S1). The tandem β tulip domains in D4 act as an “autochaperone” to allow trimerization of the D1, D2, and D3 regions. Removal of D4 results in non-productive assembly of the tailspike trimer (Xiang et al., 2009), most likely due to kinetic traps in the folding of the highly interdigitated β helix motifs found in the D1, D2, and D3 domains (Betts and King, 1999). (We note that an adenovirus uses the tailspike β helix fold as a decoration protein to stabilize the capsid 3-fold axes [Menéndez-Conejero et al., 2017]. Thus, both the β tulip and β helix regions of the tailspike have evolved to stabilize the capsid.) Interestingly, Rossmann and colleagues have noted that the D4 region of gp12 shows significant sequence homology to the minor capsid protein VP260 of Paramecium bursaria chlorella virus 1 (PBCV1) and some bacterial adhesion proteins (Xiang et al., 2009). The MCP of PBCV1 exhibits a jelly roll fold instead of the HK97 fold found in Caudo-viruses and Herpesviruses (Nandhagopal et al., 2002). Therefore, we propose the β tulip domain plays a role in capsid stabilization and molecular recognition across a broad swath of viral lineages, not just those of the HK97 fold viral lineage.
Our analysis reveals some general features of the β tulip domain as a protein interaction module. The β tulip domain tends to function in the context of a multimer, particularly trimers (gpD, gp87, triplex, gp12, and MoeA), although AcrIIC1 is an exception. β tulip domains act as protein-protein interaction modules, with the primary interaction surface mediated by the bloom side of the tulip, in particular strand 1. Again, AcrIIC1 is an exception. Finally, the β tulip motif is quite malleable in terms of sequence as well as inserted structural elements. Insertions of auxiliary elements, in particular in between strands 3 and 4 of the tulip, are common. Insertion of extra domains into the fundamental fold appears to be a common feature of capsid coat proteins; for example, HCMV MCP has multiple insertions in loops of its core HK97 fold (Yu et al., 2017). The alterations in sequence and structure of the β tulip domain shown here provides an example of the flexibility of the fold, as well as highlights the challenges in uncovering this fold in other proteins using comparative genomics.
STAR★METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Thermus thermophilus: strain HB8 | Laboratory of Dr. K. Severinov | ATCC: 27634 |
| Thermus bacteriophage P74–26 | Laboratory of Dr. K. Severinov | TAX: 466052 |
| Deposited Data | ||
| P74–26 Head Decoration Protein structure | This Paper | PDB: 6BL5 |
| Oligonucleotides | ||
| P74–26 gp87 forward
primer: GATCGGATCCATGGATAAAATTCAACTG |
This Paper | N/A |
| P74–26 gp87 reverse
primer: GATCCTCGAGTCAG CGCGTGTAGTCAAAGAAATAG |
This Paper | N/A |
| Recombinant DNA | ||
| pSMT3 vector | Yunus and Lima, 2009 | N/A |
| Software and Algorithms | ||
| Phenix | Adams et al., 2010 | http://www.phenix-online.org/ |
| HKL 3000 | Otwinowski and Minor, 1997 | http://www.hkl-xray.com/hkl-3000 |
| COOT | Emsley and Cowtan, 2004 | http://www2.mrc-lmb.cam.ac.uk/personal/pemsley/coot/ |
| ImageJ | Schneider et al., 2012 | http://imagej.nih.gov/ij/ |
| DALI server | Holm and Rosenstrom, 2010 | http://ekhidna.biocenter.helsinki.fi/dali_server/ |
| NCBI VAST | Madej et al., 2014 | http://www.ncbi.nlm.nih.gov/Structure/VAST/vast.shtml |
| PDBeFold | Krissinel and Henrick, 2004 | www.ebi.ac.uk/msd-srv/ssm/ |
| Clustal Omega | Sievers et al., 2011 | www.ebi.ac.uk/Tools/msa/clustalo/ |
| InterProSurf | Negi et al., 2007 | http://curie.utmb.edu/prosurf |
| BASiC Networks | Kathuria et al., 2016 | http://biotools.umassmed.edu/ccss/ccssv2/basic.cgi |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Brian Kelch (brian.kelch@umassmed.edu).
EXPERIMENTAL MODEL DETAILS
In this study, Thermus thermophilus strain HB8 was used to propagate phage P74–26. Expression of recombinant gp87 for structural characterization and in vitro studies was performed in E. coli strain BLR (DE3). T. thermophilus was cultured by shaking at 65°C in Thermus medium (0.8% (w/v) Tryptone, 0.4% (w/v) Yeast Extract, 0.3% (w/v) NaCl, 1 mM MgCl2, 0.5 M CaCl2). E.coli was cultured by shaking at 37°C in Terrific Broth (2.4% w/v Yeast Extract, 2% w/v Tryptone, 0.4% v/v glycerol, 0.017 M KH2PO4, 0.072 M K2HPO4).
METHOD DETAILS
Growth and Purification of P74–26 Virions
Phage stocks were prepared using fresh overnight cultures of Thermus thermophilus strain HB8 grown at 65°C in Thermus Growth Medium (0.8% (w/v) Tryptone, 0.4% (w/v) Yeast Extract, 0.3% (w/v) NaCl, 1 mM MgCl2, 0.5 M CaCl2). For preparation of P74–26 phage stock, 6 mL of fresh T. thermophilus (OD600 = 1.0) was inoculated with 4 mL of purified phage stock at 1×106 Plaque Forming Units per mL (PFU/mL) for adsorption. Adsorption reaction mixture was incubated for 10 minutes at 65°C, then inoculated into 1 L Thermus Growth Medium. The culture was then incubated at 65°C, shaking for 4–5 hours, yielding a high-titer phage lysate (>1×109 PFU/mL). Lysates were spun at 4,000 × g for 20 minutes at 4°C to remove cell debris, then the supernatant was treated with DNase I and RNase A to a final concentration of 2 Units/mL and 1 μg/mL, respectively and incubated at 30°C for one hour. Solid NaCl was added to the phage stock to a final concentration of 1 M while stirring, then culture was incubated on ice for one hour and spun at 11,000 × g for 20 minutes at 4°C. To precipitate virions, solid PEG-8,000 was added to a final concentration of 10% (w/v) while stirring and phage stock was incubated on ice overnight.
To pellet virions, precipitated phage stock was spun at 11,000 × g for 20 minutes at 4°C. The phage pellet was then resuspended in 2 mL of resuspension buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM MgS04). 0.4 g solid CsCl was added to resuspension. Phage resuspension was then added to CsCl step gradients (five steps: 1.2 g/mL (2 mL), 1.3 g/mL (2 mL), 1.4 g/mL (2 mL), 1.5 g/mL (2 mL), and1.7 g/mL (1 mL); made in 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM MgS04) prepared in 12 mL ultracentrifuge tubes (Seton Scientific). Gradients were spun in Beckman SW-40Ti swinging bucket rotor at 38,000 RPM for 18 hours at 4°C. P74–26 virions, which sediment at ~1.5 g/mL CsCl, were isolated and dialyzed twice overnight at 4°C into 2 L of 50 mM Tris-HCl pH 8.0, 10 mM NaCl, 10 mM MgCl2.
SDS-PAGE Analysis
30uL samples of virions (~1×1011 PFU/mL) were run on 12% SDS-PAGE gels. Samples were incubated in SDS loading buffer for five minutes at 95°C and run on gels at 180 V for 45 minutes. Gels were fixed with 50% (v/v) ethanol, 10% (v/v) acetic acid and then stained with Coomassie Blue in 5% (v/v) ethanol, 7.5% (v/v) acetic acid. Gels were imaged on an Amersham Imager 600 (GE Healthcare). Densitometry was performed using ImageJ (Schneider et al., 2012).
Electron Microscopy
CsCl gradient purified virions (~1×1011 PFU/mL) were applied to 400-mesh copper grids (Electron Microscopy Sciences) coated in carbon. 3.5 μL samples were applied to carbon surface of grid and incubated for 30 seconds, then excess sample was removed from grids. Following sample application grids were stained with 1% Uranyl Acetate (pH 4.5) and visualized in a Philips CM120 electron microscope (120 kV) fitted with Gatan Orius SC1000 camera. Micrographs were collected at a magnification of 19,500X.
Cloning, Expression, and Purification of gp87
P74–26 gp87 was synthesized by the Genscript Corporation and subcloned into BamHI and XhoI sites of the pSMT3 vector with a cleavable N-terminal His6-SUMO tag (Yunus and Lima, 2009). Restriction enzymes were purchased from New England BioLabs, and oligonucleotide primers were obtained from Integrated DNA Technologies. P74–26 gp87 forward primer: GATCGGATCCATGGA TAAAATTCAACTG; P74–26 gp87 reverse primer: GATCCTCGAGTCAGCGCGTGTAGTCAAAGAAATAG.
P74–26 gp87 was expressed in E. coli BLR-DE3 cells containing the pSMT3-gp87 plasmid (Yunus and Lima, 2009). Cultures were grown in Terrific Broth supplemented with 30 mg/mL kanamycin at 37°C to an OD600 of 0.7. Cultures were then incubated at 4°C for 20 minutes, then overnight expression at 18°C was induced with a final concentration of 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Cells were then pelleted and resuspended in buffer A (50 mM Tris-HCl pH 7.5, 300 mM KCl, 20 mM Imidazole, 5 mM 2-mercaptoethanol (βME), 10% (v/v) Glycerol). All subsequent purification steps were performed at room temperature. Cells were lysed in a cell disruptor and pelleted. Cleared lysate was then applied to 2×5 mL His-Trap columns (GE Healthcare) pre-equilibrated in buffer A. P74–26 gp87 was then eluted with buffer B (50 mM Tris-HCl pH 7.5, 300 mM KCl, 500 mM Imidazole, 5 mM βME, 10% (v/v) Glycerol). Eluate was dialyzed into 50 mM Tris-HCl pH 7.5, 150 mM KCl, 2 mM DTT and the His-SUMO tag was cleaved with Ubiquitin-like Specific Protease 1 (ULP1) overnight. The dialyzed protein was passed over a 5 mL His-Trap to remove cleaved His-SUMO tag. Cleaved eluate was then concentrated to 60 μM, aliquotted and flash frozen in liquid nitrogen, then stored at −80°C.
Crystallization, Data Collection, and Structure Determination
P74–26 gp87 native crystals were formed by hanging-drop vapor diffusion at 25°C. 1 mg/mL protein was mixed 1:1 with well solution containing 100 mM Tris-HCl pH 7.0, 19.5% (w/v) PEG 3350. Crystals were soaked in cryoprotectant (100 mM Tris-HCl pH 7.0, 21% (w/v) PEG 3350) and flash frozen in liquid nitrogen prior to data collection. Initial native dataset was collected to 1.9Å using a MicroMax007-HF/Rigaku Saturn 944 CCD detector x-ray diffraction system at wavelength 1.54Å (home source). Following native dataset collection, the native crystal was thawed, soaked in well solution supplemented with 1 M KI for 60 seconds, and flash frozen again. A KI derivative dataset was then immediately collected to 2.3Å, with anomalous signal extending to 2.8Å according to PHENIX xtriage. Using a separate crystal formed under identical crystallization conditions, an additional native dataset was later collected to1.7Å at Advanced Photon Source beamline 19-BM at wavelength 0.979Å. Diffraction datasets were processed using HKL3000 (Otwinowski and Minor, 1997). P74–26 gp87 structure was solved by SAD phasing of the 2.3Å derivative dataset using the iodide anomalous signal with PHENIX autosol (Dauter et al., 2000; Zwart et al., 2008). 11 iodides were found per monomeric subunit. PHENIX auto build was used to generate the initial model of gp87. The final model was obtained through iterative rounds of structure refinement in PHENIX refine (Adams et al., 2010) and manual model building in COOT (Emsley and Cowtan, 2004). Water molecules were placed using the Find Waters tool in COOT. Final model of gp87 consists of residues 17–145, where the N-terminal 16 residues are unresolved in the crystal structure. The final model was validated using Mol Probity and PDB validation tools [Chen et al., 2010]. Data collection and refinement statistics are provided in Table 1. Structure was deposited in the Protein Data Bank (www.rcsb.org), PDB code: 6BL5.
Folding and Refolding Analysis
P74–26 gp87 stocks at final concentrations of 2.5, 5, 10, or 15 μM were prepared in Tris-Buffered Saline, pH 7.5 (Boston BioProducts)and either 7.5M guanidine hydrochloride (GdnHCl) or no GdnHCl. These stocks were mixed using a Hamilton Microlab 500 series titrator to yield final samples with the desired concentration of GdnHCl. GdnHCl concentration of stocks was determined using an ABBE Mark II refractometer (Reichert). Fluorescence emission spectra for GdnHCl titrations at 2.5, 5, and 10 μM concentrations of gp87 were collected at 25°C using a Fluoromax 4 Spectrofluorometer (Horiba Scientific). P74–26 gp87 was excited at 295 nm and emission spectra were collected from 310 to 400 nm for each sample. To check for hysteresis, we measured fluorescence after 24, 48, and 120 hours of incubation at 25°C. No significant difference in fluorescence was seen between 24 hours and 120 hours, indicating that folding/unfolding reached equilibrium. Circular dichroism (CD) spectra of titrations of gp87 at 5 μM and 15 μM final concentrations were collected at 25°C using a Jasco-810 spectropolarimeter (Jasco, Inc) equipped with a temperature control system. Equilibrium unfolding was monitored from 215–260 nm. The Trp fluorescence and CD data were globally fit using single-value decomposition analysis (Simler et al., 2004) to a trimer unfolding to three monomers model (N3 ⇌ 3 U) across all wavelengths for all spectra collected. Data was fit using an in-house least squares fitting program Savuka (Gualfetti et al., 1999).
SEC-MALS
P74–26 gp87 was run on tandem size exclusion chromatography – multi-angle light scattering (SEC-MALS) by injecting a 100 μL sample at a concentration of 1 mg/mL (~60 μM) in 50 mM Tris-HCl pH 7.5, 150 mM KCl, 2 mM DTT. Protein was filtered through a 0.2 μM syringe filter. Elution was monitored using a Dawn Heleos-II MALS detector and Optilab T-rex differential refractive index detector (Wyatt Technology). Elution peaks were selected and molar mass was determined using the ASTRA6 analysis program (Wyatt Technology). The peak for the gp87 trimer eluted after 10 minutes at a concentration of ~20 μM as measured by refractive index at 658 nm and absorbance at 280 nm.
NmeCas9 Inhibition Assay
Purification of NmeCas9 and anti-CRISPR proteins is as described in Pawluk et al. (Pawluk et al., 2016). NmeCas9 sgRNA targeting N-TS4B (Amrani et al., 2017, bioRxiv) was generated by in vitro T7 transcription (Epicentre). In vitro cleavage assay was performed as previously described (Pawluk et al., 2016). Briefly, NmeCas9 (50 nM) was pre-incubated with anti-CRISPR or gp87 protein in reaction buffer (20 mM HEPES-KOH pH 7.5, 150 mM KCl, 1 mM DTT, and 5 mM MgCl2) for 10 minutes at room temperature. Next, in vitro T7 transcribed sgRNA (75 nM) was added to the mixture and incubated for another 20 minutes. pUC19 plasmid containing the target site N-TS4B was linearized by Scal digestion. The linearized plasmid was added at ~1 nM final concentration. The reactions were incubated at 37°C for 1 hour, treated with proteinase K at 50°C for 10 minutes, and visualized after electrophoresis in a 1% agarose/TAE gel.
QUANTIFICATION AND STATISTICAL ANALYSIS
Homology searches using both full-length P74–26 gp87 and the β tulip domain alone were performed using NCBI BLAST, DALI protein structure comparison server, and NCBI Vector Alignment Search Tool (VAST) (Holm and Rosenström, 2010; Madej et al., 2014). Structure-based sequence alignment between P74–26 gp87 and λ gpD was performed using the PDBeFold program (EMBL-EBI - www.ebi.ac.uk/msd-srv/ssm/) (Krissinel and Henrick, 2004). Multiple sequence alignments with homologous proteins were performed using the Clustal Omega program (EMBL-EBI - www.ebi.ac.uk/Tools/msa/clustalo/) (Sievers et al., 2011). Cα RMSD and Z-score calculations for full-length protein and β tulip domain alignments were determined using PDBeFold. β tulip domains for HCMV triplex proteins were defined as follows: HCMV Tri1 – residues 31–83, 130–149; HCMV Tri2 – residues 91–111, 135–139, 283–306. Surface area calculations for gp87, gpD, and SHP were performed using the program InterProSurf (Negi et al., 2007). ILVF cluster analysis of hydrophobic networks gp87 and gpD was performed using the program BASiC Networks (Kathuria et al., 2016).
DATA AND SOFTWARE AVAILABILITY
The structure of the Head Decoration Protein gp87 from Thermus phage P74–26 is deposited in the Protein Data Bank (rcsb.org) under the PDB ID code 6BL5.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank J. Hayes, and Drs. C. Gaubitz, O. Bilsel, K. Maxwell, and members of the Schiffer and Royer labs for helpful comments and discussions. The authors thank Drs. C. Gaubitz and W. Harshbarger for assistance with X-ray data collection. Drs. K. Severinov and L. Minakhin kindly provided strains of Thermus thermophilus and phage P74–26 stocks. The authors also thank A. Mir for help with the in vitro NmeCas9 cleavage assay. Plasmid pSMT3 was obtained from Dr. C. Lima. The work was supported by The Pew Charitable Trusts (to B.A.K.) and by the National Institute of General Medical Sciences (R01-GM125797 to E.J.S.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures, one table, and three videos can be found with this article online at https://doi.org/10.1016/j.str.2018.04.008.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adams PD, Afonine PV, Bunkóczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung L-W, Kapral GJ, Grosse-Kunstleve RW, et al. (2010). PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr 66, 213–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aksyuk AA, and Rossmann MG (2011). Bacteriophage assembly. Viruses 3, 172–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, and Lipman DJ (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrani N, Gao XD, Liu P, Gupta A, Edraki A, Ibraheim R, Sasaki KE, Zhu LJ, Wolfe SA, and Sontheimer EJ (2017). NmeCas9 is an intrinsically high-fidelity genome editing platform. bioRxiv https://doi.org/10.1101/172650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JD, and Weller SK (2005). Cleavage and packaging of Herpes Simplex Virus 1 DNA In Viral Genome Packaging Machines: Genetics, Structure, and Mechanism, Catalano C, ed. (Landes Bioscience; ), pp. 135–150. [Google Scholar]
- Basler M, Pilhofer M, Henderson GP, Jensen GJ, and Mekalanos JJ (2012). Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer DW, and Evilevitch A (2015). Influence of internal DNA pressure on stability and infectivity of phage λ. J. Mol. Biol 427, 3189–3200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazinet C, and King J (1985). The DNA translocating vertex of dsDNA bacteriophage. Annu. Rev. Microbiol 39, 109–129. [DOI] [PubMed] [Google Scholar]
- Betts S, and King J (1999). There’s a right way and a wrong way: in vivo and in vitro folding, misfolding and subunit assembly of the P22 tailspike. Structure 7, 131–139. [DOI] [PubMed] [Google Scholar]
- Bondy-Denomy J, Pawluk A, Maxwell KL, and Davidson AR (2013). Bacteriophage genes that inactivate the CRISPR/Cas bacterial immune system. Nature 493, 429–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bondy-Denomy J, Garcia B, Strum S, Du M, Rollins MF, Hidalgo-Reyes Y, Wiedenheft B, Maxwell KL, and Davidson AR (2015). Multiple mechanisms for CRISPR-Cas inhibition by anti-CRISPR proteins. Nature 526, 136–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges AL, Davidson AR, and Bondy-Denomy J (2017). The discovery, mechanisms, and evolutionary impact of anti-CRISPRs. Annu. Rev. Virol 4, 37–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bungard D, Copple JS, Yan J, Chhun JJ, Kumirov VK, Foy SG, Masel J, Wysocki VH, and Cordes MHJ (2017). Foldability of a natural de novo evolved protein. Structure 25, 1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casjens SR (2005). Comparative genomics and evolution of the tailed-bacteriophages. Curr. Opin. Microbiol 8, 451–458. [DOI] [PubMed] [Google Scholar]
- Casjens SR, and Hendrix RW (1974). Locations and amounts of major structural proteins in bacteriophage lambda. J. Mol. Biol 88, 535–545. [DOI] [PubMed] [Google Scholar]
- Catalano CE (2005). Viral Genome Packaging: Genetics, Structure, and Mechanism (Landes Bioscience; ). [Google Scholar]
- Chen DH, Baker ML, Hryc CF, Dimaio F, Jakana J, Wu W, Dougherty M, Haase-Pettingell C, Schmid MF, Jiang W, et al. (2011). Structural basis for scaffolding-mediated assembly and maturation of a dsDNA virus. Proc. Natl. Acad. Sci. USA 108, 1355–1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, and Richardson DC (2010). MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr 66, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi KY, Root M, and McGregor A (2016). A novel non-replication-competent cytomegalovirus capsid mutant vaccine strategy is effective in reducing congenital infection. J. Virol 90, 7902–7919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Gong D, Xiao Y, Wu TT, Sun R, and Zhou ZH (2015). CryoEM and mutagenesis reveal that the smallest capsid protein cements and stabilizes Kaposi’s sarcoma-associated herpesvirus capsid. Proc. Natl. Acad. Sci. USA 112, 649–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dauter Z, Dauter M, and Rajashankar KR (2000). Novel approach to phasing proteins: derivatization by short cryo-soaking with halides. Acta Crystallogr. D Biol. Crystallogr 56, 232–237. [DOI] [PubMed] [Google Scholar]
- Dokland T, and Murialdo H (1993). Structural transitions during maturation of bacteriophage lambda capsids. J. Mol. Biol 233, 682–694. [DOI] [PubMed] [Google Scholar]
- Duda RL (1998). Protein chainmail: catenated protein in viral capsids. Cell 94, 55–60. [DOI] [PubMed] [Google Scholar]
- Duda RL, Oh B, and Hendrix RW (2013). Functional domains of the HK97 capsid maturation protease and the mechanisms of protein encapsidation. J. Mol. Biol 425, 2765–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, and Cowtan K (2004). Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- Evilevitch A, Castelnovo M, Knobler CM, and Gelbart WM (2004). Measuring the force ejecting DNA from phage. J. Phys. Chem 108, 6838–6843. [Google Scholar]
- Falco SC, Laan KV, and Rothman-Denes LB (1977). Virion-associated RNA polymerase required for bacteriophage N4 development. Proc. Natl. Acad. Sci. USA 74, 520–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fane BA, and Prevelige PE (2003). Mechanism of scaffolding-assisted viral assembly. Adv. Protein Chem 64, 259–299. [DOI] [PubMed] [Google Scholar]
- Forrer P, Chang C, Ott D, Wlodawer A, and Plückthun A (2004). Kinetic stability and crystal structure of the viral capsid protein SHP. J. Mol. Biol 344, 179–193. [DOI] [PubMed] [Google Scholar]
- Fraser JS, Yu Z, Maxwell KL, and Davidson AR (2006). Ig-like domains on bacteriophages: a tale of promiscuity and deceit. J. Mol. Biol 359, 496–507. [DOI] [PubMed] [Google Scholar]
- Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, and Casjens SR (2005). Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol 187, 2050–2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualfetti PJ, Bilsel O, and Matthews CR (1999). The progressive development of structure and stability during the equilibrium folding of the alpha subunit of tryptophan synthase from Escherichia coli. Protein Sci 8, 1623–1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington LB, Doxzen KW, Ma E, Liu JJ, Knott GJ, Edraki A, Garcia B, Amrani N, Chen JS, Cofsky JC, et al. (2017). A broad-spectrum inhibitor of CRISPR-Cas9. Cell 170, 1224–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatfull GF (2008). Bacteriophage genomics. Curr. Opin. Microbiol 11, 447–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrix RW (1978). Symmetry mismatch and DNA packaging in large bacteriophages. Proc. Natl. Acad. Sci. USA 75, 4779–4783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heymann JB, Cheng N, Newcomb WW, Trus BL, Brown JC, and Steven AC (2003). Dynamics of herpes simplex virus capsid maturation visualized by time-lapse cryo-electron microscopy. Nat. Struct. Biol 10, 334–341. [DOI] [PubMed] [Google Scholar]
- Holm L, and Rosenström P (2010). Dali server: conservation mapping in 3D. Nucleic Acids Res 38, 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CL, and Black LW (1978). Head morphogenesis of bacteriophage T4. III. The role of gene 20 in DNA packaging. Virology 91, 26–38. [DOI] [PubMed] [Google Scholar]
- Imber R, Tsugita A, Wurtz M, and Hohn T (1980). Outer surface protein of bacteriophage lambda. J. Mol. Biol 139, 277–295. [DOI] [PubMed] [Google Scholar]
- Johnson JE (2010). Virus particle maturation: insights into elegantly programmed nanomachines. Curr. Opin. Struct. Biol 20, 210–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathuria SV, Chan YH, Nobrega RP, Ozen A, and Matthews CR (2016). Clusters of isoleucine, leucine, and valine side chains define cores of stability in high-energy states of globular proteins: sequence determinants of structure and stability. Protein Sci 25, 662–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, and Henrick K (2004). Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. D Biol. Crystallogr 60, 2256–2268. [DOI] [PubMed] [Google Scholar]
- Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, and Johnson JE (2008). Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure 16, 1399–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurz R, Orlova EV, Günther D, Dube P, Dröge A, Weise F, van Heel M, and Tavares P (2001). Structural organisation of the head-to-tail interface of a bacterial virus. J. Mol. Biol 310, 1027–1037. [DOI] [PubMed] [Google Scholar]
- Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, and Bryant SH (2014). MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res 42, 297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KL (2016). Phages fight back: inactivation of the CRISPR-Cas bacterial immune system by anti-CRISPR proteins. PLoS Pathog 12, e1005282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menéndez-Conejero R, Nguyen TH, Singh AK, Condezo GN, Marschang RE, van Raaij MJ, and San Martín C (2017). Structure of a reptilian adenovirus reveals a phage tailspike fold stabilizing a vertebrate virus capsid. Structure 25, 1562–1573. [DOI] [PubMed] [Google Scholar]
- Minakhin L, Goel M, Berdygulova Z, Ramanculov E, Florens L, Glazko G, Karamychev VN, Slesarev AI, Kozyavkin SA, Khromov I, et al. (2008). Genome comparison and proteomic characterization of Thermus thermophilus bacteriophages P23–45 and P74–26: siphoviruses with triplex-forming sequences and the longest known tails. J. Mol. Biol 378, 468–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno F, and Bluzat-Moreno FG (1978). Evidence that the neck appendages are adsorption organelles in Bacillus subtilis bacteriophage phi29. J. Virol 27, 831–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullaney JM, and Black LW (2014). Bacteriophage T4 capsid packaging and unpackaging of DNA and proteins. Methods Mol. Biol 1108, 69–85. [DOI] [PubMed] [Google Scholar]
- Nagano N, Orengo CA, and Thornton JM (2002). One fold with many functions: the evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol 321, 741–765. [DOI] [PubMed] [Google Scholar]
- Nandhagopal N, Simpson AA, Gurnon JR, Yan X, Baker TS, Graves MV, Van Etten JL, and Rossmann MG (2002). The structure and evolution of the major capsid protein of a large, lipid-containing DNA virus. Proc. Natl. Acad. Sci. USA 99, 14758–14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negi SS, Schein CH, Oezguen N, Power TD, and Braun W (2007). InterProSurf: a web server for predicting interacting sites on protein surfaces. Bioinformatics 23, 3397–3399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newcomb WW, Trus BL, Booy FP, Steven AC, Wall JS, and Brown JC (1993). Structure of the herpes simplex virus capsid. Molecular composition of the pentons and the triplexes. J. Mol. Biol 232, 499–511. [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, and Minor W (1997). Processing of X-ray diffraction data. Methods Enzymol 276, 307–326. [DOI] [PubMed] [Google Scholar]
- Pan R, Gorny MK, Zolla-Pazner S, and Kong XP (2015). The V1V2 region of HIV-1 gp120 forms a five-stranded beta barrel. J. Virol 89, 8003–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Amrani N, Zhang Y, Garcia B, Hidalgo-Reyes Y, Lee J, Edraki A, Shah M, Sontheimer EJ, Maxwell KL, and Davidson AR (2016). Naturally occurring off-switches for CRISPR-Cas9. Cell 167, 1829–1838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluk A, Davidson AR, and Maxwell KL (2017). Anti-CRISPR: discovery, mechanism and function. Nat. Rev. Microbiol 16, 12–17. [DOI] [PubMed] [Google Scholar]
- Prevelige PE, and Fane BA (2012). Building the machines: scaffolding protein functions during bacteriophage morphogenesis. Adv. Exp. Med. Biol 726, 325–350. [DOI] [PubMed] [Google Scholar]
- Prevelige PE Jr., Thomas D, and King J (1993). Nucleation and growth phases in the polymerization of coat and scaffolding subunits into icosahedral procapsid shells. Biophys. J 64, 824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson JS (1981). The anatomy and taxonomy of protein structure. Adv. Protein Chem 34, 167–339. [DOI] [PubMed] [Google Scholar]
- Rixon FJ, and Schmid MF (2014). Structural similarities in DNA packaging and delivery apparatuses in Herpesvirus and dsDNA bacteriophages. Curr. Opin. Virol 5, 105–110. [DOI] [PubMed] [Google Scholar]
- Saad A, Zhou ZH, Jakana J, Chiu W, and Rixon FJ (1999). Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol 73, 6821–6830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sae-Ueng U, Liu T, Catalano CE, Huffman JB, Homa FL, and Evilevitch A (2014). Major capsid reinforcement by a minor protein in herpes-viruses and phage. Nucleic Acids Res 42, 9096–9107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, and Eliceiri KW (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrag JD, Huang W, Sivaraman J, Smith C, Plamondon J, Larocque R, Matte A, and Cygler M (2001). The crystal structure of Escherichia coli MoeA, a protein from the molybdopterin synthesis pathway. J. Mol. Biol 310, 419–431. [DOI] [PubMed] [Google Scholar]
- Selvarajan Sigamani S, Zhao H, Kamau YN, Baines JD, and Tang L (2013). The structure of the Herpes simplex virus DNA-packaging terminase pUL15 nuclease domain suggests an evolutionary lineage among eukaryotic and prokaryotic viruses. J. Virol 87, 7140–7148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Jiang F, Liu J-J, Bray NL, Rauch BJ, Baik SH, Nogales E, Bondy-Denomy J, Corn JE, and Doudna JA (2017). Disabling Cas9 by an anti-CRISPR DNA mimic. Sci. Adv 3, e1701620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol 7, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simler BR, Doyle BL, and Matthews CR (2004). Zinc binding drives the folding and association of the homo-trimeric gamma-carbonic anhydrase from Methanosarcina thermophila. Protein Eng. Des. Sel 17, 285–291. [DOI] [PubMed] [Google Scholar]
- Snijder J, Radtke K, Anderson F, Scholtes L, Corradini E, Baines J, Heck AJR, Wuite GJL, Sodeik B, and Roos WH (2017). The vertex specific proteins pUL17 and pUL25 mechanically reinforce Herpes Simplex Virus capsids. J. Virol 91, https://doi.org/10.1128/JVI.00123-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sternberg N, and Weisberg R (1977). Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol 117, 733–759. [DOI] [PubMed] [Google Scholar]
- Strauss H, and King J (1984). Steps in the stabilization of newly packaged DNA during phage P22 morphogenesis. J. Mol. Biol 172, 523–543. [DOI] [PubMed] [Google Scholar]
- Suhanovsky MM, and Teschke CM (2015). Nature’s favorite building block: deciphering folding and capsid assembly of proteins with the HK97-fold. Virology 479–480, 487–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suttle CA (2007). Marine viruses – major players in the global ecosystem. Nat. Rev. Microbiol 5, 801–812. [DOI] [PubMed] [Google Scholar]
- Tang L, Gilcrease EB, Casjens SR, and Johnson JE (2006). Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure 14, 837–845. [DOI] [PubMed] [Google Scholar]
- Trus BL, Booy FP, Newcomb WW, Brown JC, Homa FL, Thomsen DR, and Steven AC (1996). The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly. J. Mol. Biol 263, 447–462. [DOI] [PubMed] [Google Scholar]
- Veesler D, and Cambillau C (2011). A common evolutionary origin for tailed-bacteriophage functional modules and bacterial machineries. Microbiol. Mol. Biol. Rev 75, 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Hardies SC, Fokine A, Klose T, Jiang W, Cho BC, and Rossmann MG (2017). Structure of the marine siphovirus TW1: evolution of capsid-stabilizing proteins and tail spikes. Structure 26, 238–248. [DOI] [PubMed] [Google Scholar]
- White HE, Sherman MB, Brasilès S, Jacquet E, Seavers P, Tavares P, and Orlova EV (2012). Capsid structure and its stability at the late stages of bacteriophage SPP1 assembly. J. Virol 86, 6768–6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, and Johnson JE (2000). Topologically linked protein rings in the bacteriophage HK97 capsid. Science 289, 2129–2133. [DOI] [PubMed] [Google Scholar]
- Wommack KE, and Colwell RR (2000). Virioplankton: viruses in aquatic ecosystems. Microbiol. Mol. Biol. Rev 64, 69–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Leiman PG, Li L, Grimes S, Anderson DL, and Rossmann MG (2009). Crystallographic insights into the autocatalytic assembly mechanism of a bacteriophage tail spike. Mol. Cell 34, 375–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Forrer P, Dauter Z, and Conway JF (2000). Novel fold and capsid-binding properties of the lambda-phage display platform protein gpD. Nat. Struct. Biol 7, 230–237. [DOI] [PubMed] [Google Scholar]
- Yu MX, Slater MR, and Ackermann H-W (2006). Isolation and characterization of Thermus bacteriophages. Arch. Virol 151, 663–679. [DOI] [PubMed] [Google Scholar]
- Yu X, Jih J, Jiang J, and Zhou ZH (2017). Atomic structure of the human cytomegalovirus capsid with its securing tegument layer of pp150. Science 356, https://doi.org/10.1126/science.aam6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yunus AA, and Lima CD (2009). Purification of SUMO conjugating enzymes and kinetic analysis of substrate conjugation. Methods Mol. Biol 497, 167–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Speir JA, Matsui T, Lin Z, Liang L, Lynn AY, Varnado B, Weiss TM, and Tang L (2016). Structure of a bacterial virus DNA-injection protein complex reveals a decameric assembly with a constricted molecular channel. PLoS One 11, e0149337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwart PH, Afonine PV, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, McKee E, Moriarty NW, Read RJ, Sacchettini JC, et al. (2008). Automated structure solution with the PHENIX suite. Methods Mol. Biol 426, 419–435. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.