Abstract
Variation at the FADS1/FADS2 gene cluster is functionally associated with differences in lipid metabolism and is often hypothesized to reflect adaptation to an agricultural diet. Here, we test the evidence for this relationship using both modern and ancient DNA data. We show that almost all the inhabitants of Europe carried the ancestral allele until the derived allele was introduced ∼8,500 years ago by Early Neolithic farming populations. However, we also show that it was not under strong selection in these populations. We find that this allele, and other proposed agricultural adaptations at LCT/MCM6 and SLC22A4, were not strongly selected until much later, perhaps as late as the Bronze Age. Similarly, increased copy number variation at the salivary amylase gene AMY1 is not linked to the development of agriculture although, in this case, the putative adaptation precedes the agricultural transition. Our analysis shows that selection at the FADS locus was not tightly linked to the initial introduction of agriculture and the Neolithic transition. Further, it suggests that the strongest signals of recent human adaptation in Europe did not coincide with the Neolithic transition but with more recent changes in environment, diet, or efficiency of selection due to increases in effective population size.
Keywords: Human evolution, selection, ancient DNA, agriculture, diet
Introduction
Human history has seen a number of major transitions in diet (Luca et al. 2010). The most recent was the transition to a modern “industrialized” diet based on intensive farming and highly processed food. Before that, many parts of the world saw a dramatic transition from a diet based on hunting and gathering to a diet heavily based on the products of agriculture. The transition occurred independently several times in different parts of the world, but the earliest known example is the Fertile Crescent, at least 10,500 years ago. From there agriculture spread gradually northwest through Anatolia into Europe, and eastwards to the Indian subcontinent (Bellwood 2004). Even outside these periods of transition, differences in diet based on both cultural preferences and food source availability would have been a major aspect of environmental differences between human populations—differences that would likely lead to genetic adaptation. Thus, by identifying and studying the evolution of genetic adaptations to diet, we learn not only about historical changes in diet, but also about the genetic basis of diet-related phenotypic differences among present-day human populations.
One previously identified adaptation involves the fatty acid desaturase genes FADS1 and FADS2. These genes encode proteins which catalyze key steps in the ω-3 and ω-6 lipid biosynthesis pathways (Nakamura and Nara 2004). These pathways synthesize long-chain (LC) polyunsaturated fatty acids (PUFA) necessary for cell- and, particularly, neuronal-membrane development from short-chain (SC) PUFA (Darios and Davletov 2006). The evolutionary interaction with diet stems from the fact that different diets contain different ratios of SC- and LC-PUFA. Specifically, diets that are high in meat or marine products contain relatively high LC-PUFA levels, and thus may require lower levels of FADS1 and FADS2 activity compared with diets that are high in plant-based fats (Ameur et al. 2012; Mathias et al. 2012; Fumagalli et al. 2015; Kothapalli et al. 2016; Buckley et al. 2017; Ye et al. 2017). As a result of this environmental interaction, these genes have been repeatedly targeted by natural selection.
Most dramatically, a derived haplotype (“haplotype D”, following Ameur et al. 2012) containing the 3′ end of FADS1 is at very high frequency in present-day African populations and intermediate to high frequency in present-day Eurasians (Ameur et al. 2012; Mathias et al. 2012), and experienced ancient positive selection in Africa (Ameur et al. 2012; Mathias et al. 2012). The derived haplotype increases expression of FADS1 (Ameur et al. 2012) and likely represents an adaptation to a high ratio of SC- to LC-PUFA, that is, to a plant-based diet. In Europe, direct evidence from ancient DNA has shown that haplotype D was rare around 10,000 years before present (BP) but has increased in frequency since then, likely due to selection, to its present-day frequency of ∼60% in Europe (Mathieson et al. 2015). This increase was plausibly associated with the adoption—starting around 8,500 BP in Southeastern Europe before spreading North and West—of an agricultural lifestyle and diet that would have a higher SC- to LC-PUFA ratio than the earlier hunter–gatherer diet (Mathieson et al. 2015; Buckley et al. 2017; Ye et al. 2017). Interestingly, the Altai Neanderthal genome shares at least a partial version of haplotype D (Buckley et al. 2017; Harris et al. 2017), suggesting that the functional variation at this locus may predate the split of Neanderthals and modern humans.
Other haplotypes at the locus have been shown to be under selection in different populations in more recent history. In particular, another haplotype that is common in, but largely restricted to, the Greenlandic Inuit population reduces the activity of FADS1, is associated with PUFA levels, and is likely an adaptation to a diet that is extremely high in LC-PUFA from marine sources (Fumagalli et al. 2015). Conversely, a variant that increases expression of FADS2 has been selected in South Asian populations—and may be a specific adaptation to a vegetarian diet (Kothapalli et al. 2016). Reflecting its important role in lipid metabolism, variation at the FADS locus also contributes significantly to variation in lipid levels in present-day populations. As well as directly contributing to variation in PUFA levels, SNPs in haplotype D are among the strongest genome-wide association study (GWAS) signals for triglyceride and cholesterol levels (Teslovich et al. 2010). Thus, the complex evolutionary history of the region is not only informative about ancient human diets, but also potentially relevant for understanding the distribution of lipid-related disease risk both within and between populations. We therefore aimed to characterize the history of the region, by combining inference from ancient and modern DNA data, in order to understand the evolutionary basis of this important functional variation and its relationship with changes in diet. We also compared the evolutionary history of the FADS locus with the histories of other loci involved in dietary adaptation, to see whether we could detect shared patterns of adaptation.
Results
Haplotype Structure at FADS1
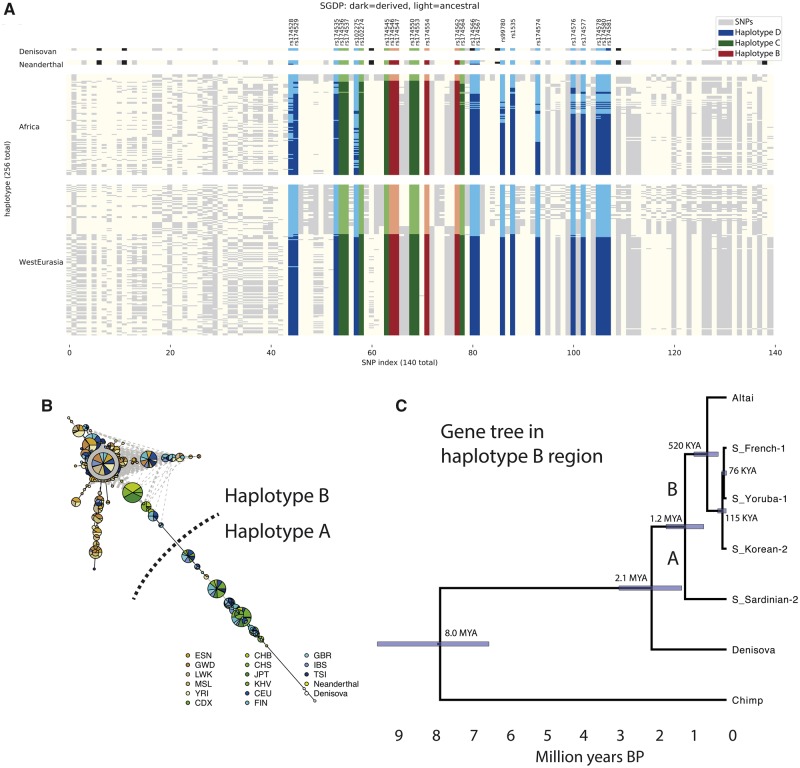
We began by investigating 300 high-coverage whole-genome sequences from the Simons Genome Diversity Project (SGDP; Mallick et al. 2016), as well as three archaic human genomes (Meyer et al. 2012; Prufer et al. 2014, 2017), to investigate the fine-scale haplotype structure in the region (fig. 1, supplementary fig. 1, Supplementary Material online). By clustering haplotypes using a graph-adjacency algorithm (Materials and Methods), we defined three nested “derived” haplotypes (fig. 1A, table 1, supplementary table 1, (fig. 1, Supplementary Material online). Haplotype D extends over 63 kb, is largely restricted to Eurasia, and is comparable to the haplotype D defined by Ameur et al. (2012). Haplotype C is a 36 kb portion of haplotype D that is shared between African and Eurasian populations. Finally, haplotype B is a 15 kb region that represents the portion of haplotype D that is shared between modern humans and both the Altai (Prufer et al. 2014) and Vindija (Prufer et al. 2017) Neanderthals. We denote the modern human haplotype carrying ancestral alleles at haplotype-defining SNPs as haplotype A (the “ancestral” haplotype). This analysis also allows us to prioritize likely causal SNPs, which are currently unknown (Buckley et al. 2017; Ye et al. 2017). If the derived SNP that was selected in modern humans was also selected in Neanderthals, then it must lie in haplotype B, which is defined by just four SNPs (rs174546, rs174547, rs174554, and rs174562).
Table 2.
Summary of ABC Results for Three Possible Episodes of Selection.
| P (New mut.) | Start (yrs BP) | Start CrI (yrs BP) | Sel. Coeff. | |
|---|---|---|---|---|
| 1. Selection for the derived allele in the ancestors of present-day Africans | 0.74 | 310,548 | 202,409–492,231 | 0.001–0.002 |
| 2. Selection for the ancestral allele in the ancestors of present-day Europeans | 0.00 | 389,661 | 131,876–1,108,594 | 0.000–0.002 |
| 3. Selection for the derived allele in Europe | 0.00 | 2,538 | 1,695–4,048 | 0.034–0.069 |
We inferred the probability of selection on a new mutation (as opposed to standing variation), estimated selection-onset time, 95% credible intervals for this time, and 95% credible intervals for the selection coefficient. See supplementary tables 2 and 3, Supplementary Material online for more detailed results.
Fig. 1.
Haplotype structure at the FADS1 region. (A) Haplotypes from the SGDP African and West Eurasian populations (Mallick et al. 2016) and the Neanderthal ( Prufer et al. 2014, 2017 ) and Denisovan (Meyer et al. 2012) genomes. Each column represents a SNP and each row a phased haplotype. Dark and light colors represent derived and ancestral alleles at each SNP and blue, green, and red colors indicate SNPs that are part of haplotypes D, C, and B, respectively. (B) Haplotype network for region B constructed from 1000 Genomes (1000 Genomes Project Consortium 2015) and archaic samples. Green, blue and brown indicate East Asian, European and African populations respectively. (C) Gene tree for the haplotype B region inferred for representative haplotypes.
Table 1.
Haplotypes Described in the Text.
| Haplotype | Length (kb) | State | Shared |
|---|---|---|---|
| A | – | Ancestral | – |
| B | 15 | Derived | Neanderthal and present-day humans |
| C | 36 | Derived | Present-day humans |
| D | 63 | Derived | Present-day Eurasians |
Note.—Summary of the four haplotypes discussed. See supplementary table 1, Supplementary Material online for defining SNPs.
Using data from the 1000 Genomes project (1000 Genomes Project Consortium 2015), we replicate the observation (Mathias et al. 2012; Buckley et al. 2017; Harris et al. 2017) that haplotypes in the haplotype B region fall into two clusters (fig. 1B, supplementary fig. S2, Supplementary Material online). One cluster contains Eurasian-ancestry individuals that carry haplotype A (and all individuals with Native American ancestry). The other cluster contains Neanderthals, Eurasians that carry haplotype D, and almost all present-day African-ancestry individuals. A small number of intermediate haplotypes are either recombinant or the result of phasing errors. We inferred the phylogeny of the haplotype B region using BEAST2 (fig. 1C;Bouckaert et al. 2014). The region is small and has a very low recombination rate (average 0.03 cM/mb according to the International HapMap Consortium 2007 combined recombination map), so we ignored possible recombination events. Assuming that the common ancestor of human and chimp haplotypes was 6.6–10.0 Ma—corresponding to a genome-wide mutation rate of ∼4–6 × 10−10 per-base per-year (Amster and Sella 2016; Scally 2016)—we infer that the most recent common ancestor (MRCA) of the present-day African (C) and European (D) haplotypes was around 76,000 BP, of haplotypes B and C around 520,000 BP, and that of haplotypes A and B around 1.2 million BP. These dates are consistent with the estimates of Harris et al. (2017) and suggest that the European-specific haplotype D diverged around the time of the out-of-Africa bottleneck and the diversification of non-African lineages (Mallick et al. 2016). They also suggest that modern human ancestral and derived haplotypes coexisted in the Denisovan-Neanderthal-modern human ancestral population, with the Neanderthal haplotype splitting off around the time of mean human-Neanderthal divergence 550–765,000 BP (Prufer et al. 2014).
ABC Analysis of Ancient Selection in Africa and Recent Selection in Eurasia
Having established the structure and phylogeny of the haplotypes in the region, we used approximate Bayesian computation ( ABC; Wegmann et al. 2010; Peter et al. 2012 ) to infer the strength and timing of selection on the derived haplotype in Africa (table 2, supplementary table 2, Supplementary Material online), first described by Mathias et al. (2012). Using data from the 1000 Genomes project, treating the derived allele of rs174546 as the selected SNP, and fixing the mutation rate to 1.25 × 10−8 per-base per-generation, we infer that selection most likely but not definitively acted on a new mutation (posterior probability of new mutation 0.74, range 0.13–0.99, see Peter et al. 2012 for details of the inference approach) and estimate that selection began 202,000–492,000 BP (supplementary table 2, Supplementary Material online). This estimate is uncertain though, with 95% credible intervals (CrIs) in different populations ranging from 85,000 to 1,426,000 BP. Mathias et al. (2012) estimated 85,000 ± 84,000 years for the onset of selection in Africa. However, that analysis assumed a Human–Chimpanzee split of 6.5 Ma, now thought to be an underestimate and, if appropriately rescaled, would overlap the low end of our estimate. Overall, the evidence suggests that selection began around or before the time of the deepest splits among human populations (Mallick et al. 2016; Schlebusch et al. 2017) 200,000–250,000 BP. This is consistent with the observation that the derived allele is shared among all present-day African populations including those, such as Khoe-San and Mbuti, which were substantially diverged from the ancestors of other present-day African populations at least 100,000 BP (Mallick et al. 2016; Schlebusch et al. 2017).
In contrast, when we analyze present-day European genomes, we find that strong (selection coefficient s = 3.4–6.9%) selection for the derived allele acted on standing variation (posterior probability = 1.0) much more recently—starting between 1700 and 4000 BP (95% CrIs in different populations range from 1100 to 12,000 BP, supplementary table 2, Supplementary Material online). This recent selection is hard to reconcile with the suggestion (Mathieson et al. 2015; Ye et al. 2017) that it is closely linked to the development of agriculture, at least 10,500 BP (Bellwood 2004). It is also unclear why the derived haplotype was at such low frequency in pre-agricultural Europeans (Mathieson et al. 2015; Ye et al. 2017), when ABC results and derived allele sharing between African populations would imply that the derived allele would have been at high frequency before the split of African and non-African ancestral populations. To resolve these questions, we turned to ancient DNA data.
Low Frequency of the Derived FADS1 Allele in Upper Palaeolithic Eurasia
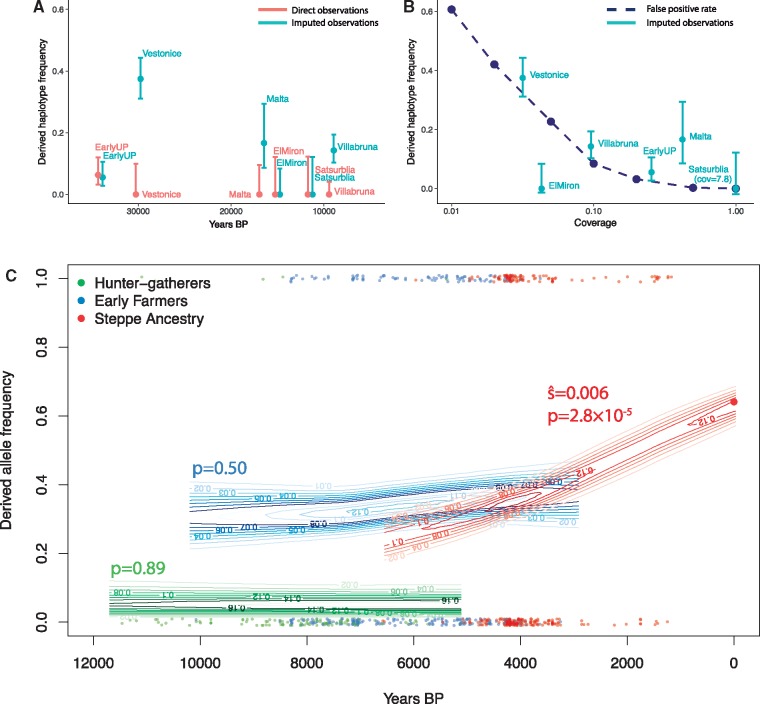
We first investigated why the derived haplotype was at such low frequency in pre-agricultural Europeans, by analyzing data from 52 Paleolithic and Mesolithic individuals dated between 45,000 and 8,000 BP (Fu et al. 2014; Raghavan et al. 2014; Jones et al. 2015; Fu et al. 2016; Sikora et al. 2017). We infer the presence of haplotype D based on five SNPs that were typed on the “1240k” capture array used for most of these samples (Fu et al. 2016). The derived haplotype is rare in all populations (fig. 2A, “Direct observations”). When we tried to use imputation to increase sample size, the results were inconsistent. Specifically, imputed data suggested a much higher frequency in most of the ancient population groupings, for example around 40% frequency in the Věstonice population. This higher frequency agrees with a previous analysis of this data by Ye et al. (2017). However, we find that imputation is unreliable for these data because many of the Upper Paleolithic samples have extremely low coverage. This leads to a high rate of false positive inference of the derived allele because it is at relatively high frequency in the present-day populations used as a reference panel. Older samples tend to have lower coverage and thus higher false positive rates, leading to a spurious inference of a decline in frequency over time. We estimated the false positive rate as a function of coverage by simulating low coverage data from present-day samples that carry the ancestral allele. Nonzero derived allele frequency estimates in the imputed data are consistent with the estimated false positive rate (Materials and Methods, fig. 2B). In fact, we find reliable evidence of the derived allele in only one individual—from the 34,000 BP site of Sunghir (Sikora et al. 2017). Therefore, it seems likely that the derived allele was rare among early “out-of-Africa” populations, at least in Western Eurasia.
Fig. 2.
Direct ancient DNA evidence for the history of FADS1. (A) Derived haplotype frequency estimated from direct observation of SNPs on the haplotype (red) and imputed data (blue) in Upper Palaeolithic individuals (45,000–10,000 BP; Fu et al. 2016). (B) Estimated imputation false positive rate as a function of coverage (dashed line). Imputed allele frequencies in Upper Palaeolithic populations plotted for comparison at the median coverage in that population. (C) Allele frequencies at rs174546 over the past 12,000 years estimated from 1,055 (669 with coverage) ancient and 99 modern individuals. Each point is an ancient pseudo-haploid individual call, at the bottom of the plot if it is ancestral and the top if it derived. Contours indicate the posterior probability of allele frequencies in the ancient populations and P-values for nonzero selection coefficients are indicated.
This lack of the derived allele in early non-Africans is surprising because our ABC analysis suggested that selection within the ancestral population began at least 200,000 years ago, and the derived allele would therefore be expected to have been at high frequency when African and non-African ancestors diverged. Further, the derived allele is shared between present-day African populations that were genetically isolated before the split of present-day African and non-African ancestors (Mallick et al. 2016; Schlebusch et al. 2017). Ye et al. (2017) determined that the ancestral allele must have been selected in the ancestors of present-day Europeans, but located this selection in Europe, after the out of Africa bottleneck. Our analysis suggests that in fact, this selection was much older, occurring either before or during the bottleneck. To test this, we used the same ABC approach we used to investigate the derived allele to infer selection on the ancestral allele (Materials and Methods, table 1, supplementary table 2, Supplementary Material online). We infer that the ancestral allele was selected from standing variation in the ancestors of present-day Europeans (95% CrI = 0.0–0.2%), starting 132,000–1,109,000 BP (95% CrI). This large CrI overlaps the CrIs for selection on the derived allele in African ancestors and means that we do not have power to resolve the order of these selective events. We also estimated the timing of selection using an independent approach based on haplotype decay (Smith et al. 2018), qualitatively supporting ancient selection for the derived allele (supplementary table 4, Supplementary Material online). These results support the observation that if there was selection on the ancestral allele in the ancestors of present-day Europeans, it occurred mostly before the date of the earliest ancient samples with genetic data.
Selection for the Derived FADS1 Allele in Europe Was Not Closely Linked to Early Agriculture
The derived haplotype was almost absent in Upper Palaeolithic and Mesolithic Europe (before ∼8,500 BP), but is today at a frequency of ∼60%. Previous analysis of both ancient and modern DNA has inferred strong selection for the allele over the past 10,000 years (Mathieson et al. 2015; Field et al. 2016; Buckley et al. 2017; Ye et al. 2017). This has been interpreted to mean that selection for the derived allele was driven by the development of agriculture, around 10,500 BP—a reasonable interpretation since the derived allele is plausibly an adaption to a diet high in plant fats and low in animal fats. However, our ABC analysis suggests that selection in Europe might actually be restricted to the past few thousand years and thus postdate the development of agriculture by many millennia. To test this directly, we analyzed data from 1,055 ancient Europeans who lived between 12,000 and 1,000 BP (Keller et al. 2012; Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Allentoft et al. 2015; Gunther et al. 2015; Haak et al. 2015; Jones et al. 2015; Mathieson et al. 2015; Cassidy et al. 2016; Hofmanová et al. 2016; Martiniano et al. 2016; Schiffels et al. 2016; Jones et al. 2017; Lazaridis et al. 2017; Lipson et al. 2017; Mathieson et al. 2018; Olalde et al. 2018). We divided these individuals into three populations, based on their genome-wide ancestry (Materials and Methods). First, individuals with “hunter–gatherer ancestry” were the Mesolithic inhabitants of Europe or their descendants. Second, “Early Farmers” were people from Neolithic Northwest Anatolia or their descendants, possibly admixed with hunter–gatherers, who migrated throughout Europe. Finally, people with “Steppe ancestry” had ancestry that was originally derived from Bronze Age steppe populations like the Yamnaya (Lazaridis et al. 2014; Allentoft et al. 2015; Haak et al. 2015). Because transitions between these populations involved dramatic genetic discontinuity, with 75–100% of ancestry replaced (Allentoft et al. 2015; Haak et al. 2015), we analyzed each of them separately.
In each of these three populations, we estimated the frequency and selection coefficient of the derived allele by fitting a hidden Markov Model (HMM) to the time series of observations (Materials and Methods, fig. 2C;Mathieson and McVean 2013). We estimate that the selection coefficient in both hunter–gatherer and Early Farmer populations was not significantly different from zero (i.e., no evidence that the allele was under selection), and a selection coefficient of 0.6% (95% CI 0.4–1.5%) in Steppe-ancestry populations. This selection coefficient is lower than that estimated using ABC. One possible explanation is that the HMM forces the selection coefficient to be constant in each population, so if selection operated for only part of the time represented, or in a subset of the population, its strength might be underestimated. However, both the ancient DNA and ABC analyses are consistent with a relatively recent onset of selection and show that, although the derived allele was present in early farming populations, it was not strongly selected. Plausibly, the derived allele was introduced to the ancestors of the early farmers through admixture with a population that carried “basal Eurasian” ancestry (Lazaridis et al. 2016) not found in Palaeolithic or Mesolithic Europe. Alternatively, the allele may have been retained in some, unsampled, Upper Paleolithic Eurasian population, or driven to higher frequency somewhere by one or more additional ancient episodes of selection.
Because agriculture was introduced to different parts of Europe at different times, spreading broadly from south to north, we split the ancient DNA data into Northern and Southern European groups (Materials and Methods) and reran the analysis, obtaining similar results in both groups (supplementary fig. 3, Supplementary Material online). To confirm the key observation that the derived allele was not under selection in Early Farmers, we restricted to this group and fitted a logistic regression to the observations with date, hunter–gatherer ancestry, Steppe ancestry and latitude as covariates. None of these four coefficients were significantly different from zero (P = 0.19, 0.89, 0.41, and 0.78 respectively), confirming that selection for the allele is not being masked by variation in ancestry (for example by increasing hunter–gatherer ancestry over time), or in the geographic position of sampled individuals (for example because agriculture began at different dates at different latitudes). Finally, we split the Early Farmer and Steppe ancestry populations into 4000 year chunks and analyzed them separately (supplementary fig. 4, Supplementary Material online). The frequency of the derived allele is higher in later Early Farmers (after 6,000 BP), compared with early Early Farmers (40% vs. 29%, Fisher’s exact test P = 0.026), perhaps reflecting a shift in ancestry. But we find no evidence of selection within either early or later Early Farmers, or in early Steppe ancestry populations (before 4,000 BP).
Patterns of Population Differentiation at Other Lipid-Associated Alleles
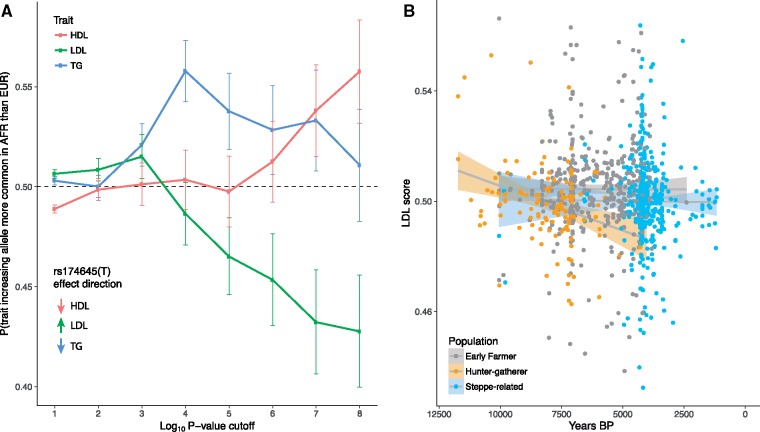
SNPs that tag the derived haplotype are among the strongest genome-wide associations with lipid levels (Teslovich et al. 2010). To test whether the selective events we observe were consequences of more general selection on lipid levels, we investigated patterns of African-European population differentiation between variants associated with three blood lipid traits–triglycerides (TG), high-density cholesterol (HDL) and low-density cholesterol (LDL; Teslovich et al. 2010). We find that, for HDL and TG, trait-increasing alleles tend to be more common in African than European populations, whereas for LDL, the trait increasing allele is more common in European populations (fig. 3A, Materials and Methods). These effects are in the opposite direction to those of the FADS1 haplotype. The derived allele, which is more common in African than European populations, tends to decrease HDL and TG and increase LDL. We conclude that selection on FADS1 was not driven by its effect on overall blood lipid levels (which, if anything, moved in the opposite direction), but by its specific effect on PUFA synthesis. We further find that there is no significant difference in the frequency of LDL-increasing alleles when comparing the three ancient populations (fig. 3B). Since they do differ in the frequency of the FADS1 allele, this suggests that recent selection on FADS1 was also not driven by selection more generally on lipid levels.
Fig. 3.
Trends in lipid-associated allele frequencies. (A) Probability that the trait-increasing allele is more common in African than European populations in the 1000 Genomes Project data (1000 Genomes Project Consortium 2015) for lipid-related traits. We show results for each trait with different P-value cutoffs. Vertical bars represent 95% confidence intervals. Inset shows the direction of effect for the derived FADS1 haplotype, using rs174546 as the tag SNP. (B) LDL score for 1,003 ancient individuals, classified according to ancestry, as a function of their age. LDL score is the proportion of significant LDL-associated variants at which the ancient individual carries the trait-increasing allele (Materials and Methods).
Patterns of Population Differentiation at Other Diet-Associated Variants
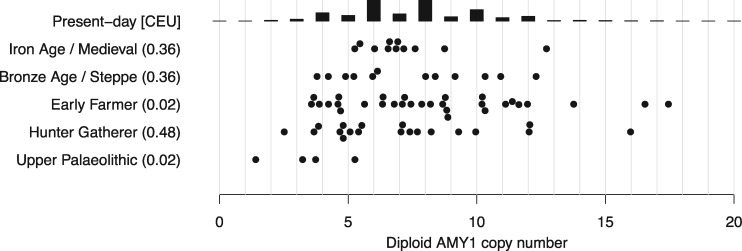
We investigated whether other variants that have been suggested to be associated with the adoption of agriculture showed similar temporal patterns of selection to FADS1. The salivary amylase gene AMY1 is highly copy-number variable among present-day populations, ranging from a diploid copy number of 2 (the ancestral state) to 17 (Groot et al. 1991; Perry et al. 2007; Usher et al. 2015), with a mean of 6.7 copies in present-day Europeans (Usher et al. 2015). It has been suggested that increased copy number improves the digestion of starchy food and is therefore an adaptation to an agricultural diet that is relatively rich in starch (Perry et al. 2007). To date, one Early Farmer has been reported to have a relatively high AMY1 copy number of 16 (Lazaridis et al. 2014), whereas six hunter–gatherers had 6–12 copies (Lazaridis et al. 2014; Olalde et al. 2014; Günther et al. 2018). Inchley et al. (2016) showed that the initial expansion of AMY1 copy number predated the out-of-Africa bottleneck, and found no evidence of recent selection. To investigate this directly, we called AMY1 copy number in 76 ancient West Eurasian individuals with published shotgun sequence data (Keller et al. 2012; Meyer et al. 2012; Skoglund et al. 2012; Fu et al. 2014; Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Prufer et al. 2014; Raghavan et al. 2014; Skoglund et al. 2014; Fu et al. 2015; Gunther et al. 2015; Cassidy et al. 2016; Kılınç et al. 2016; Omrak et al. 2016; Schiffels et al. 2016; Prufer et al. 2017; Saag et al. 2017; Günther et al. 2018). We counted the number of reads that mapped to regions around each of the three AMY1 copies in the human reference genome (Usher et al. 2015), and compared with bootstrap estimates of mean read depth with a linear correction for GC content (Materials and Methods, fig. 4, supplementary table 5, Supplementary Material online). We find no significant difference between AMY1 copy number in present day Europeans and ancient Europeans from the Iron Age/Medieval, Bronze Age, Neolithic, or Mesolithic periods. In particular, Mesolithic pre-agricultural hunter–gatherers have a mean copy number of 7.2—not statistically different from present-day Europeans. Neolithic Early Farmers have a mean copy number of 8.4—slightly higher, but not significantly different (at a Bonferroni-corrected significance level of 0.01) from present-day populations (P = 0.02). We do find that four Upper Palaeolithic individuals dating between ∼45,000–20,000 BP have lower AMY1 copy number than present-day Europeans although, with a small sample size, this is also not statistically significant (mean 3.4, P = 0.02). These results therefore suggest an expansion in copy number sometime earlier than ∼10,000 BP and thus predating the development of agriculture.
Fig. 4.
AMY1 copy number. Inferred in ancient samples, arranged by time and subsistence strategy, compared with the distribution in a present-day population (CEU) with Northern European ancestry (Usher et al. 2015). Parentheses: t-test P-values for difference between CEU and ancient populations.
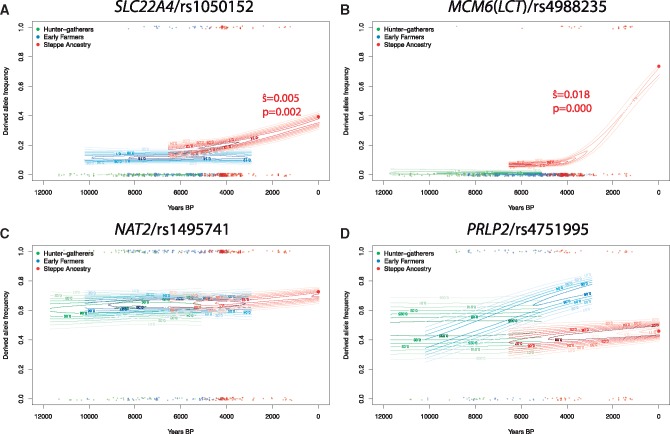
Finally, we also investigated the history of other mutations that have been suggested to be involved in adaptation to agricultural subsistence. It has been proposed, based on both ancient and modern DNA, that the ergothioneine transporter gene SLC22A4—and in particular the nonsynonymous variant 503 F (rs1050152)—was targeted by selection in Early Neolithic farming populations (Huff et al. 2012; Mathieson et al. 2015). However, analysis of our larger ancient DNA data set reveals a more complicated story, with an allele frequency trajectory similar to that of the FADS1 derived allele (fig. 5A). Specifically, the derived allele of rs1050152 is absent in hunter–gatherers, and present at low frequency in Early Farmers and Bronze Age populations. But, similar to the derived FADS1 allele, it does not increase in frequency—and therefore does not appear to be under selection—in Early Farming populations. Strong selection on this allele likely only operated in the past few thousand years. This timescale is similar to the timescale over which the European lactase persistence variant (Enattah et al. 2002) became common in Europe ( fig. 5B;Burger et al. 2007; Allentoft et al. 2015; Mathieson et al. 2015 ). The “slow acetylator” variant of the NAT2 gene has been hypothesized to be advantageous in agricultural populations (Luca et al. 2008; Magalon et al. 2008; Sabbagh et al. 2011). However, we find that a SNP (rs1495741) that tags the “fast acetylator” phenotype (Garcia-Closas et al. 2011) shows no change in frequency over the past 10,000 years (fig. 5C). Finally, we find that a variant (rs4751995) in the gene PLRP2 that is relatively common in present-day populations with a cereal-based diet (Hancock et al. 2010) is also more common in Early Farmers than hunter–gatherers, although it shows no evidence of selection in any of these populations (fig. 5D).
Fig. 5.
Allele frequency trajectories for other putative agricultural adaptation variants. As in figure 2C, estimated allele frequency trajectories and selection coefficients in different ancient European populations. Significant selection coefficients are labelled.
Discussion
The combination of high-quality genome sequence data from present-day people, and large ancient DNA data sets, provides new opportunities to investigate and understand the process of human adaptation. In particular, the temporal aspect of ancient DNA data adds another dimension to our analysis, allowing us to make precise inference of the timing of selection. We show that the derived allele was very rare or absent in all European populations, until it was introduced around 8,500 BP by migration from early farmers carrying basal Eurasian ancestry. Although the derived allele is plausibly advantageous in populations that consume a plant-based diet, we find no evidence that it was actually selected in early European Farmers. Conversely, the low frequency of the derived allele in Upper Palaeolithic Europe is consistent with isotopic evidence that protein intake was dominated by animal protein at this time (Richards 2009). Finally, it is not known whether the Neanderthal allele would have had the same function as the derived modern human allele. But if it did, it would not support the claim that the derived allele is associated with a plant-based diet since Neanderthals, like Upper Palaeolithic modern humans, are thought to have consumed a largely meat-based diet (Bocherens 2009). Therefore, the derived/ancestral state at the major FADS haplotype may not be a simple marker of plant- versus meat-based diet or more recent hunter–gatherer versus farmer subsistence, but could reflect a more complex pattern of interaction based on unknown dietary and genetic factors.
Our results support and extend several recent results about the evolutionary history of the FADS1 allele. It has previously been shown that the derived allele was anciently selected in Africa (Mathias et al. 2012). We find that the ancestral allele was rare in early non-African populations, suggesting that there may have been selection for the ancestral allele as proposed by Ye et al. (2017), but that it must have been before or during—rather than after—the out-of-Africa bottleneck. The fixation of the ancestral allele in present-day Native Americans has been interpreted as evidence for selection in their Siberian or Beringian ancestors (Amorim et al. 2017; Harris et al. 2017; Hlusko et al. 2018). However, our analysis shows that this may not be the case, because the derived allele was likely very rare in Eurasia at the time of the Native American-Eurasian split 20–25,000 BP (Raghavan et al. 2015). Both Ye et al. (2017) and Buckley et al. (2017) argue that recent selection in Europe for the derived allele was driven by changes in diet. Our results support this view, with the caveat that the relevant changes were not simply those associated with the Neolithic transition. Finally, Buckley et al. (2017) found evidence for differential selective pressures across Europe and propose the marker rs174594 as the target of selection. ABC analysis is consistent with selection on rs174594 across Europe (supplementary table 6, Supplementary Material online) but it is in strong LD with rs174546 and we were unable to test the model of Buckley et al. (2017) further because rs174594 is not on the capture array that was used to generate most of the ancient DNA data we analyzed.
Similarly, due to limited ancient DNA data, we were not able to resolve the history of the FADS1 allele in East Asia. ABC analysis was very sensitive to the exact demography that we assumed. When we capped recent Ne at 45,000, we found that the ancestral allele was selected in the ancestors of present-day East Asians, although with a large CrI (72,000–1,221,000 BP). The 39,000-year-old Tianyuan individual did not carry the derived haplotype, further suggesting that it was absent in the Upper Paleolithic ancestors of East Asians as well as Europeans. We estimated that the derived allele was selected more anciently than in Europe (i.e., ∼62,600 BP with 95% CrI 27,300–158,000 BP). The most common East Asian derived haplotype is also an outgroup to the common European and African haplotypes (supplementary fig. 2, Supplementary Material online), which would be consistent with deriving from a separate, older, event. More ancient DNA from East Asia will help resolve this question, although we note that agriculture developed later in East Asia than in Western Eurasia, so it is likely that selection on the derived FADS1 allele was also unassociated with the development of agriculture.
In the case of FADS1 and all the other examples we investigated, the proposed agricultural adaption was either not temporally linked with the initial development of agriculture or showed no evidence of selection in Early Farmer populations. Instead, the three variants with any evidence of selection were strongly selected at some point between the Bronze Age and the present day, that is, in the past ∼4000 years. This time period is one in which there is relatively limited ancient DNA data, and so we are unable to determine the timing of selection any more accurately. Future research should address the question of why this recent time period saw the most rapid changes in apparently diet-associated genes. One plausible hypothesis is that the change in environment or diet at this time was actually more dramatic than the earlier change associated with the initial development agriculture. For example, Early Neolithic populations may have retained some proportion of their diet from hunter–gatherer strategies, and only later transitioned to a completely agricultural diet. Other environmental factors like pathogen load or climate might also affect the selective pressures. Another hypothesis is that effective population sizes were so small, or populations so structured, before the Bronze Age that selection did not operate efficiently on variants with small selection coefficients. For example, analysis of present-day genomes from the United Kingdom suggests that effective population size increased by a factor of 100–1,000 in the past 4500 years (Browning and Browning 2015). Larger ancient DNA data sets from the past 4000 years will likely resolve this question.
Materials and Methods
Identifying and Analyzing FADS1 Haplotypes
We defined derived haplotypes using the following procedure. Within the region ±50 kb from rs174546 (hg19 chr11: 61519830–61619830) there are 140 common (MAF >0.05) SNPs when considering the SGDP (600 haplotypes) and archaic (6 haplotypes) samples, and restricting to sites where the ancestral allele can be determined based on the chimpanzee genome (PanTro2 Chimpanzee Sequencing Analysis Consortium 2005). For each pair of SNPs within these 140, we compute the number of “mismatches” between ancestral and derived states. For example, if SNP 1 at haplotype H is in the ancestral state, but SNP 2 at H is in the derived state, then this counts as one mismatch. 1000 Genomes data were used to set the ancestral/derived state for each SNP (we used the chimpanzee allele if the allele was not present in 1000 Genomes). Counting up these mismatches for all pairs of SNPs, we obtain a 140 × 140 symmetric “mismatch” matrix M. We transform this matrix into an “adjacency” matrix A by setting each entry to 1 if the number of mismatches is below some threshold t, and 0 otherwise. In other words, if M[i, j] ≤ t, A[i, j] = 1, otherwise A[i, j] = 0. This adjacency matrix can then be interpreted as a graph, with SNPs as the nodes and edges between SNPs if they are connected (i.e., have a low number of mismatches).
From this graph, we find the largest clique (connected component where every pair of nodes is connected). This procedure can be interpreted as a way to find a subset of SNPs that are all in high LD with each other. The problem of finding the largest clique in a graph is NP-hard, but we use the Bron–Kerbosch algorithm which is more efficient in practice than brute force (Bron and Kerbosch 1973).
We use the procedure above to define three nested haplotypes: One considering all modern non-African populations (haplotype D), one considering all modern human populations (haplotype C) and one considering all modern and Neanderthal samples (haplotype B). For the mismatch thresholds, we use 12, 12, and 3 respectively (this last lower threshold accommodates the small sample size of archaic populations). Haplotype D contains 25 SNPs, haplotype C contains 11 SNPs, and haplotype B contains 4 SNPs. When there is more than one maximal clique of SNPs to choose from, we select one that is a subset of a larger core. This means that haplotype B is a subset of haplotype C and haplotype C is a subset of haplotype D. Note that our haplotype D differs slightly from the derived haplotype defined by Ameur et al. (2012) which was 28 SNPs long.
We constructed a haplotype network for the haplotype B region from 1000 Genomes European, East Asian and African haplotypes, using the R package “pegas” (Paradis 2010). We inferred the phylogenetic relationship between the haplotypes by picking a single individual from the SGDP that was homozygous for each of the representative haplotypes and inferring the tree relating the haplotypes using BEAST2 (Bouckaert et al. 2014). We rooted the tree with chimpanzee and used a uniform [6.6–10.0] Ma prior for human–chimp divergence. This corresponds to a genome-wide mutation rate of ∼4–6 × 10−10 per-base per-year.
ABC and Startmrca Analysis
To quantify the strength and timing of selection in different populations, we used ABC, implemented in the ABCtoolbox package (Wegmann et al. 2010). We use the pipeline implemented by Peter et al. (2012), which uses a model selection approach to distinguish selection on a de novo mutation (SDN) from selection on standing variation (SSV). Intuitively, this approach simulates data under both the SDN and SSV models, for a range of parameters, and then selects the simulations that best match the observed data, in the sense of being close in the space of a set of predefined summary statistics. The parameters of the selected simulations are then used to estimate a posterior distribution for the parameters of interest. In addition to SDN/SSV model selection, we estimated two continuous parameters: The selection-onset time and the selection coefficient. To perform the simulations for ABC we used mbs (Teshima and Innan 2009), which creates a selected allele frequency trajectory backward in time from a specified present-day frequency. This implicitly creates a range of selection-onset times in the past, which we used as a prior. For the selection strength, we used a uniform prior of [–4,–1] on log10 of the selection coefficient. For the SSV model, we used a uniform prior of [0–0.2] for the allele frequency at the selection start time.
We chose the derived allele (C) of rs174546 as the putatively selected allele, and analyzed 50 kb on either side for a total region length of L = 100 kb. We fixed the mutation rate at 1.25 × 10−8 per base per generation, and used the recombination rate from the combined HapMap 2 map (International HapMap Consortium 2007). Since the recombination map may have changed over time, we also repeated the analysis using a constant recombination rate of 1.1785 × 10−8 per base per generation (the average rate in this region). We also set the current effective population size Ne to 10,000, and the heterozygote advantage at 0.5. This mutation rate is at the low end of human mutation rate estimates (Scally 2016), but a higher mutation rate would only lead to more recent estimates for the onset of selection. For each region (AFR, EAS, and EUR) and each model (SDN and SSV), we simulated 1 million data sets. Each data set had a sample size of n = 170 haplotypes and a length of L = 100 kb to match the 1000 Genomes data. Within each region we chose five representative populations for further analysis: ESN, GWD, LWK, MSL, and YRI for AFR; CDX, CHB, CHS, JPT, and KHV for EAS; and CEU, FIN, GBR, IBS, and TSI for EUR. On the basis of the current frequency of the selected allele in the subpopulations for each region, we computed simulation priors: [0.984, 0.992] for AFR, [0.304, 0.670] for EAS, and [0.575, 0.699] for EUR. We also used population-specific demographies from PSMC, based on Yoruba (AFR), Han (EAS), and French (EUR) individuals from the SGDP (Mallick et al. 2016).
For each simulated data set we computed 27 summary statistics (as described in Peter et al. 2012). During the ABC estimation phase, we retained the top 500 simulated data sets with statistics closest to each real data set (15 in total, one for each subpopulation), and computed posterior distributions for the selection-onset time and selection coefficient. We computed combined estimates (supplementary tables 2 and 3, Supplementary Material online) by assuming a lognormal distribution for the posteriors and averaging over the five population-specific estimates. Finally, we averaged over the two (SDN/SSV) models tested.
For EAS and EUR, we also wanted to test for ancient selection on the ancestral allele (T) of rs174546. To this end, we merged the subpopulations for both EAS and EUR, and selected 295 haplotypes with the ancestral allele for each. Then we ran our ABC procedure in the same way as before, except with a “current” allele frequency of 0.999999 (mbs does not allow 1 for technical reasons). The EAS results were very sensitive to demography—particularly to the value of present-day Ne—so we restricted maximum Ne to the value of 45,000 inferred by Gravel et al. (2011).
The main limitations of the ABC approach are that it does not model complex modes of selection and relies on simulating under an appropriate range of parameters. We checked that ancient selection would not obscure more recent signals of selection by simulating data under a realistic model of alternating selection coefficients using SLiM2 (Haller and Messer 2017), and rerunning the ABC analysis (supplementary fig. 5, Supplementary Material online). Similarly, the results are conditional on simulating under the correct demography and with an appropriate range of parameters. For our key results, we checked that the distributions of simulated summary statistics were broadly consistent with the observed values, indicating that our simulations are reasonable (supplementary figs. 6–8, Supplementary Material online). When we modeled selection on the ancestral allele, we noticed that the observed values of the singleton count statistics were often outside the simulated distribution (supplementary fig. 7, Supplementary Material online)—most likely because we merged five populations leading to an increase in the rate of singletons—so we excluded these four statistics from the ABC estimation for selection on the ancestral allele.
Startmrca (Smith et al. 2018) is a method for estimating the selection-onset time of a beneficial allele. It uses an HMM (in the “copying” style of Li and Stephens 2003) to model present-day haplotypes as imperfect mosaics of the selected haplotype and a reference panel of nonselected haplotypes. We used this method to estimate the selection-onset time for the ancestral allele of rs174546 in the EAS and EUR superpopulations described above. We used YRI individuals that are homozygous for the derived allele as the reference panel. Following Smith et al. (2018), we used a 1 Mb region surrounding rs174546, the HapMap combined recombination map in this region (International HapMap Consortium 2007), 100 haplotypes in the selected population, and 20 haplotypes in the reference population. To obtain selection-onset time estimates for each population, we ran five independent MCMC chains, each with 10,000 iterations. We postprocessed the results by discarding the first 6,000 iterations (burn-in), and retaining the remaining successful iterations over all five chains. To obtain CrIs, we took the 2.5 and 97.5 quantiles of each resulting distribution (supplementary table 3.2, Supplementary Material online).
We also analyzed selection for the derived allele of rs174546 in the EAS and EUR superpopulations. In this case, there were not enough AFR individuals homozygous for the ancestral allele to use as a reference population. So instead we used a “local” reference population, that is, EAS and EUR individuals homozygous for the ancestral allele. We also use a lower prior for the derived allele ([0–4,000] generations versus [0–20,000] for the ancestral). We did not analyze AFR because the selection is likely too old for the method, and there is no suitable reference population. We note that startmrca tends to underestimate the time of very ancient selective events (the “star” genealogy assumption becomes less appropriate). It also does not account for SSV, which would result in an overestimation of the selection-onset time, so our startmrca quantitative results are likely unreliable.
Ancient DNA Analysis
We first analyzed 52 Palaeolithic and Mesolithic samples (Fu et al. 2014; Jones et al. 2015; Fu et al. 2016; Sikora et al. 2017) for the presence of the derived FADS1 allele. These samples were typed on a capture array (“1240k capture”) that contains 5 of the 25 SNPs that define haplotype D. We divided the samples into population groups as defined by Fu et al. (2016) and inferred the allele frequency in each of these populations by maximizing the following likelihood function:
where rij and aij are the number of reference and alternative reads from individual i at SNP j, N is the number of individuals in the population. B(x, n, p) is the binomial probability of seeing x successes out of n trials with probability p, and ɛ, δ, γ are small error probabilities, which we set to 0.1, 0, 0.1 for transversions, 0.15, 0.05, 0 for C > T or G > A transitions, and 0, 0.05, 0.15 for T > C or A > G transitions. This implies a conservative 10% rate of contamination or error, and a 5% deamination rate. We computed binomial confidence intervals assuming an effective sample size of .
To impute the FADS1 haplotype in the Upper Palaeolithic and Mesolithic samples, we computed genotype likelihoods at each typed SNP in the 5 Mb region around rs174546, for each individual, assuming a binomial distribution of reference and alternative allele counts and a 5% deamination rate. We then imputed using BEAGLE 4.1 (Browning and Browning 2016), and the 1000 Genomes reference panel downloaded from bochet.gcc.biostat.washington.edu/beagle/1000_Genomes_phase3_v5a. We filtered out imputed SNPs with a genotype probability of <0.8 and used the remaining SNPs to determine the presence of the allele. In order to estimate the false positive rate, we simulated read data at different coverages for 50 individuals from the 1000 Genomes EAS (East Asian) super-population who are homozygous for the ancestral allele. We computed genotype likelihoods and imputed as for the ancient data, having first removed the 50 test individuals from the reference panel. We performed 10 simulations for each coverage level and used the frequency at which the derived allele was imputed as an estimate of the false positive rate.
To analyze the Holocene history of FADS1 and other alleles, we assembled a data set of 1078 published ancient samples, most of which were typed on the “1240k” capture array which targets ∼1.24 million SNPs. We used the pseudohaploid version of these data, where each individual has a single allele at each SNP, from a randomly selected read. We classified these individuals into “hunter–gatherer,” “Early Farmer,” and “Steppe ancestry” populations as follows. First, we ran supervised ADMIXTURE (Alexander et al. 2009) with K = 4, and the four populations: WHG, EHG, Anatolia_Neolithic, and Yamnaya_Samara fixed to have cluster membership 1, as previously described (Mathieson et al. 2018). We then classified individuals based on their inferred ancestry. If they had >25% ancestry from the Yamnaya_Samara cluster and dated later than 6,000 BP, we classified them as “Steppe ancestry”. If they had <25% ancestry from the Yamnaya_Samara cluster, >50% from the Anatolia_Neolithic cluster and dated earlier than 2,000 BP, we classified them as “Early Farmer”. Finally, if they had <25% ancestry from the Yamnaya_Samara cluster and <50% ancestry from the Anatolia_Neolithic cluster and were dated earlier than 5,100 BP, we classified them as “hunter–gatherer”. We excluded 23 samples that did not fit into any of these classifications, leaving an analysis data set of 1,055 samples of which 669 had coverage at rs174546. These classifications are informed by previous analysis that combined genetic, chronological and archaeological information, and largely correspond to classifications that would be derived from archaeological context alone. We estimated allele frequency trajectories and selection coefficients separately for each population using a method that fits a HMM to the observed frequencies (Mathieson and McVean 2013), assuming an effective population size (Ne) of 10,000 in each population. We also analyzed separately the ancient individuals from Northern and Southern Europe (supplementary fig. 3, Supplementary Material online). For this analysis, we defined “Northern Europe” to be the region east of 13°E and north of 45°N, or west of 13°E and north of 49°N, with “Southern Europe” as the complement. This corresponds approximately to the region reached by the Neolithic transition by 7,000 BP (Fort 2015).
To call AMY1 copy number we assembled a set of 76 ancient genomes with shotgun sequence data that had nonzero mapped coverage at the locus. The majority of published ancient shotgun genomes have zero coverage, presumably because the copy number variable region was masked during alignment. We counted the number of reads that mapped to the any of the three AMY1 duplicate regions in the human reference genome (Usher et al. 2015) and compared the total to the average read depth in 1000 random regions of chromosome 1, of the same size as the AMY1 duplicate regions. We fitted a linear model of coverage as a function of GC content to these 1000 regions, for each individual, and used this to correct our estimates for GC bias.
Analysis of Lipid GWAS Hits
We tested the directionality of lipid-associated alleles using genome-wide association meta-analysis results for LDL, HDL, and TG (Teslovich et al. 2010). Specifically, we constructed a list of SNPs with P-values below a given cutoff by iteratively selecting the SNP with the lowest P-value and then removing all SNPs within 500 kb. For each of these SNPs we extracted allele frequencies in the EUR and AFR super-populations and then tested whether trait increasing alleles were more common in AFR than EUR (fig. 3A).
To analyze the LDL hits in ancient samples, we first identified all SNPs with an association P-value <10−6 that were on the capture array used to genotype the majority of the ancient samples. We iteratively removed SNPs within 250 kb of the most-associated SNPs to produce an independent set of associated SNPs. For each (pseudohaploid) individual, we constructed the “LDL score” by counting the proportion of these SNPs at which that individual carried the trait-increasing allele (fig. 3B). We found no significant differences with respect to ancestry when we fitted a binomial generalized linear model with ancestry as a covariate. We also fitted a model including date as a covariate to test for significant differences over time, also with a nonsignificant result.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank Rasmus Nielsen, Joshua Schraiber, and two anonymous reviewers for helpful comments on an earlier draft, and Benjamin Peter for help running the ABC methods.
References
- Alexander DH, Novembre J, Lange K.. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19(9):1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allentoft ME, Sikora M, Sjogren KG, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlstrom T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522(7555):167–172. [DOI] [PubMed] [Google Scholar]
- Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, et al. 2012. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am J Hum Genet. 90(5):809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amorim CE, Nunes K, Meyer D, Comas D, Bortolini MC, Salzano FM, Hunemeier T.. 2017. Genetic signature of natural selection in first Americans. Proc Natl Acad Sci U S A. 114(9):2195–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amster G, Sella G.. 2016. Life history effects on the molecular clock of autosomes and sex chromosomes. Proc Natl Acad Sci U S A. 113(6):1588–1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellwood P. 2004. First farmers: the origins of agricultural societies. Malden, MA: Wiley-Blackwell. [Google Scholar]
- Bocherens H. 2009. Neanderthal Dietary Habits: review of the Isotopic Evidence In: Hublin J-J, Richards MP, editors. The evolution of hominin diets. New York, NY: Springer; p. 241–250. [Google Scholar]
- Bouckaert R, Heled J, Kuhnert D, Vaughan T, Wu CH, Xie D, Suchard MA, Rambaut A, Drummond AJ.. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol. 10(4):e1003537.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning BL, Browning SR.. 2016. Genotype imputation with millions of reference samples. Am J Hum Genet. 98(1):116–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL.. 2015. Accurate non-parametric estimation of recent effective population size from segments of identity by descent. Am J Hum Genet. 97(3):404–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bron C, Kerbosch J.. 1973. Finding all cliques of an undirected graph [H]. Commun ACM. 16(9):575–577. [Google Scholar]
- Buckley MT, Racimo F, Allentoft ME, Jensen MK, Jonsson A, Huang H, Hormozdiari F, Sikora M, Marnetto D, Eskin E, et al. 2017. Selection in europeans on fatty acid desaturases associated with dietary changes. Mol Biol Evol. 34(6):1307–1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger J, Kirchner M, Bramanti B, Haak W, Thomas MG.. 2007. Absence of the lactase-persistence-associated allele in early Neolithic Europeans. Proc Natl Acad Sci U S A. 104(10):3736–3741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy LM, Martiniano R, Murphy EM, Teasdale MD, Mallory J, Hartwell B, Bradley DG.. 2016. Neolithic and Bronze Age migration to Ireland and establishment of the insular Atlantic genome. Proc Natl Acad Sci U S A. 113(2):368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chimpanzee Sequencing Analysis Consortium 2005. Initial sequence of the chimpanzee genome and comparison with the human genome. Nature 437:69–87. [DOI] [PubMed] [Google Scholar]
- Darios F, Davletov B.. 2006. Omega-3 and omega-6 fatty acids stimulate cell membrane expansion by acting on syntaxin 3. Nature 440(7085):813–817. [DOI] [PubMed] [Google Scholar]
- Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I.. 2002. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 30(2):233–237. [DOI] [PubMed] [Google Scholar]
- Field Y, Boyle EA, Telis N, Gao Z, Gaulton KJ, Golan D, Yengo L, Rocheleau G, Froguel P, McCarthy MI, et al. 2016. Detection of human adaptation during the past 2000 years. Science 354(6313):760–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort J. 2015. Demic and cultural diffusion propagated the Neolithic transition across different regions of Europe. J R Soc Interface. 12:20150166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Hajdinjak M, Moldovan OT, Constantin S, Mallick S, Skoglund P, Patterson N, Rohland N, Lazaridis I, Nickel B, et al. 2015. An early modern human from Romania with a recent Neanderthal ancestor. Nature 524(7564):216–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prufer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514(7523):445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Posth C, Hajdinjak M, Petr M, Mallick S, Fernandes D, Furtwangler A, Haak W, Meyer M, Mittnik A, et al. 2016. The genetic history of Ice Age Europe. Nature 534(7606):200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A, et al. 2015. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science 349(6254):1343–1347. [DOI] [PubMed] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 5:5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Closas M, Hein DW, Silverman D, Malats N, Yeager M, Jacobs K, Doll MA, Figueroa JD, Baris D, Schwenn M, et al. 2011. A single nucleotide polymorphism tags variation in the arylamine N-acetyltransferase 2 phenotype in populations of European background. Pharmacogenet Genomics. 21(4):231–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Genomes P, Bustamante CD.. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci U S A. 108:11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groot PC, Mager WH, Frants RR.. 1991. Interpretation of polymorphic DNA patterns in the human alpha-amylase multigene family. Genomics 10(3):779–785. [DOI] [PubMed] [Google Scholar]
- Günther T, Malmström H, Svensson EM, Omrak A, Sánchez-Quinto F, Kılınç GM, Krzewińska M, Eriksson G, Fraser M, Edlund H, et al. 2018. Population genomics of Mesolithic Scandinavia: investigating early postglacial migration routes and high-latitude adaptation. PLoS Biol. 16(1):e2003703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther T, Valdiosera C, Malmstrom H, Urena I, Rodriguez-Varela R, Sverrisdottir OO, Daskalaki EA, Skoglund P, Naidoo T, Svensson EM, et al. 2015. Ancient genomes link early farmers from Atapuerca in Spain to modern-day Basques. Proc Natl Acad Sci U S A. 112(38):11917–11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522(7555):207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW.. 2017. SLiM 2: flexible, interactive forward genetic simulations. Mol Biol Evol. 34(1):230–240. [DOI] [PubMed] [Google Scholar]
- Hancock AM, Witonsky DB, Ehler E, Alkorta-Aranburu G, Beall C, Gebremedhin A, Sukernik R, Utermann G, Pritchard J, Coop G, et al. 2010. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci U S A. 107(Suppl 2):8924–8930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris DN, Ruczinski I, Yanek LR, Becker LC, Becker DM, Guio H, Cui T, Chilton FH, Mathias RA, O'Connor T.. 2017. Evolution of hominin polyunsaturated fatty acid metabolism: from africa to the new world. Biorxiv doi:10.1101/175067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlusko LJ, Carlson JP, Chaplin G, Elias SA, Hoffecker JF, Huffman M, Jablonski NG, Monson TA, O'Rourke DH, Pilloud MA, et al. 2018. Environmental selection during the last ice age on the mother-to-infant transmission of vitamin D and fatty acids through breast milk. Proc Natl Acad Sci U S A. doi:10.1073/pnas.1711788115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmanová Z, Kreutzer S, Hellenthal G, Sell C, Diekmann Y, Díez-del-Molino D, van Dorp L, López S, Kousathanas A, Link V, et al. 2016. Early farmers from across Europe directly descended from Neolithic Aegeans. Proc Natl Acad Sci U S A. 113(25):6886–6891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff CD, Witherspoon DJ, Zhang Y, Gatenbee C, Denson LA, Kugathasan S, Hakonarson H, Whiting A, Davis CT, Wu W, et al. 2012. Crohn's disease and genetic hitchhiking at IBD5. Mol Biol Evol. 29(1):101–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inchley CE, Larbey CD, Shwan NA, Pagani L, Saag L, Antao T, Jacobs G, Hudjashov G, Metspalu E, Mitt M, et al. 2016. Selective sweep on human amylase genes postdates the split with Neanderthals. Sci Rep. 6:37198.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium. 2007. A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ER, Gonzalez-Fortes G, Connell S, Siska V, Eriksson A, Martiniano R, McLaughlin RL, Llorente MG, Cassidy LM, Gamba C.. 2015. Upper Palaeolithic genomes reveal deep roots of modern Eurasians. Nature Commun. 6:8912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones ER, Zarina G, Moiseyev V, Lightfoot E, Nigst PR, Manica A, Pinhasi R, Bradley DG.. 2017. The neolithic transition in the baltic was not driven by admixture with early European farmers. Curr Biol. 27(4):576–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 1000 Genomes Project Consortium. 2015. A global reference for human genetic variation. Nature 526(7571):68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, Leidinger P, Backes C, Khairat R, Forster M, et al. 2012. New insights into the Tyrolean Iceman's origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 3:698.. [DOI] [PubMed] [Google Scholar]
- Kılınç GM, Omrak A, Özer F, Günther T, Büyükkarakaya AM, Bıçakçı E, Baird D, Dönertaş HM, Ghalichi A, Yaka R, et al. 2016. The demographic development of the first farmers in anatolia. Curr Biol. 26(19):2659–2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kothapalli KSD, Ye K, Gadgil MS, Carlson SE, O’Brien KO, Zhang JY, Park HG, Ojukwu K, Zou J, Hyon SS, et al. 2016. Positive selection on a regulatory insertion–deletion polymorphism in FADS2 influences apparent endogenous synthesis of arachidonic acid. Mol Biol Evol. 33(7):1726–1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Mittnik A, Patterson N, Mallick S, Rohland N, Pfrengle S, Furtwängler A, Peltzer A, Posth C, Vasilakis A, et al. 2017. Genetic origins of the Minoans and Mycenaeans. Nature 548(7666):214–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Nadel D, Rollefson G, Merrett DC, Rohland N, Mallick S, Fernandes D, Novak M, Gamarra B, Sirak K, et al. 2016. Genomic insights into the origin of farming in the ancient Near East. Nature 536(7617):419–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513(7518):409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Stephens M.. 2003. Modeling linkage disequilibrium and identifying recombination hotspots using single-nucleotide polymorphism data. Genetics 165(4):2213–2233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipson M, Szecsenyi-Nagy A, Mallick S, Posa A, Stegmar B, Keerl V, Rohland N, Stewardson K, Ferry M, Michel M, et al. 2017. Parallel palaeogenomic transects reveal complex genetic history of early European farmers. Nature 551(7680):368–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F, Bubba G, Basile M, Brdicka R, Michalodimitrakis E, Rickards O, Vershubsky G, Quintana-Murci L, Kozlov AI, Novelletto A.. 2008. Multiple advantageous amino acid variants in the NAT2 gene in human populations. PLoS ONE. 3(9):e3136.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luca F, Perry GH, Di Rienzo A.. 2010. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 30:291–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalon H, Patin E, Austerlitz F, Hegay T, Aldashev A, Quintana-Murci L, Heyer E.. 2008. Population genetic diversity of the NAT2 gene supports a role of acetylation in human adaptation to farming in Central Asia. Eur J Hum Genet. 16(2):243–251. [DOI] [PubMed] [Google Scholar]
- Mallick S, Li H, Lipson M, Mathieson I, Gymrek M, Racimo F, Zhao M, Chennagiri N, Nordenfelt S, Tandon A, et al. 2016. The Simons Genome Diversity Project: 300 genomes from 142 diverse populations. Nature 538(7624):201–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martiniano R, Caffell A, Holst M, Hunter-Mann K, Montgomery J, Müldner G, McLaughlin RL, Teasdale MD, van Rheenen W, Veldink JH, et al. 2016. Genomic signals of migration and continuity in Britain before the Anglo-Saxons. Nat Commun. 7:10326.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias RA, Fu W, Akey JM, Ainsworth HC, Torgerson DG, Ruczinski I, Sergeant S, Barnes KC, Chilton FH.. 2012. Adaptive evolution of the FADS gene cluster within Africa. PLoS ONE. 7(9):e44926.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Alpaslan-Roodenberg S, Posth C, Szecsenyi-Nagy A, Rohland N, Mallick S, Olalde I, Broomandkhoshbacht N, Candilio F, Cheronet O, et al. 2018. The genomic history of southeastern Europe. Nature 555:197–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, Lazaridis I, Rohland N, Mallick S, Patterson N, Roodenberg SA, Harney E, Stewardson K, Fernandes D, Novak M, et al. 2015. Genome-wide patterns of selection in 230 ancient Eurasians. Nature 528(7583):499–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathieson I, McVean G.. 2013. Estimating selection coefficients in spatially structured populations from time series data of allele frequencies. Genetics 193(3):973–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge MT, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prufer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science 338(6104):222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura MT, Nara TY.. 2004. Structure, function, and dietary regulation of delta6, delta5, and delta9 desaturases. Annu Rev Nutr. 24:345–376. [DOI] [PubMed] [Google Scholar]
- Olalde I, Allentoft ME, Sanchez-Quinto F, Santpere G, Chiang CW, DeGiorgio M, Prado-Martinez J, Rodriguez JA, Rasmussen S, Quilez J, et al. 2014. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507(7491):225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I, Brace S, Allentoft ME, Armit I, Kristiansen K, Booth T, Rohland N, Mallick S, Szecsenyi-Nagy A, Mittnik A, et al. 2018. The Beaker phenomenon and the genomic transformation of northwest Europe. Nature 555:190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omrak A, Gunther T, Valdiosera C, Svensson EM, Malmstrom H, Kiesewetter H, Aylward W, Stora J, Jakobsson M, Gotherstrom A.. 2016. Genomic evidence establishes anatolia as the source of the European neolithic gene pool. Curr Biol. 26(2):270–275. [DOI] [PubMed] [Google Scholar]
- Paradis E. 2010. pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26(3):419–420. [DOI] [PubMed] [Google Scholar]
- Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. 2007. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 39(10):1256–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R.. 2012. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8(10):e1003011.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, de Filippo C, Grote S, Mafessoni F, Korlevic P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyregne S, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science 358(6363):655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prufer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2014. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 505(7481):43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW Jr, Orlando L, Metspalu E, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505(7481):87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Steinrucken M, Harris K, Schiffels S, Rasmussen S, DeGiorgio M, Albrechtsen A, Valdiosera C, Avila-Arcos MC, Malaspinas AS, et al. 2015. Genomic evidence for the Pleistocene and recent population history of Native Americans. Science 349(6250):aab3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards MP. 2009. Stable isotope evidence for European upper paleolithic human diets In: Hublin J-J, Richards MP, editors. The evolution of hominin diets. New York, NY: Springer; p. 251–257. [Google Scholar]
- Saag L, Varul L, Scheib CL, Stenderup J, Allentoft ME, Saag L, Pagani L, Reidla M, Tambets K, Metspalu E, et al. 2017. Extensive farming in estonia started through a sex-biased migration from the steppe. Curr Biol. 27(14):2185–2193.e2186. [DOI] [PubMed] [Google Scholar]
- Sabbagh A, Darlu P, Crouau-Roy B, Poloni ES.. 2011. Arylamine N-acetyltransferase 2 (NAT2) genetic diversity and traditional subsistence: a worldwide population survey. PLoS ONE. 6(4):e18507.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scally A. 2016. The mutation rate in human evolution and demographic inference. Curr Opin Genet Dev. 41:36–43. [DOI] [PubMed] [Google Scholar]
- Schiffels S, Haak W, Paajanen P, Llamas B, Popescu E, Loe L, Clarke R, Lyons A, Mortimer R, Sayer D, et al. 2016. Iron age and Anglo-Saxon genomes from East England reveal British migration history. Nature Commun. 7:10408.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlebusch CM, Malmstrom H, Gunther T, Sjodin P, Coutinho A, Edlund H, Munters AR, Vicente M, Steyn M, Soodyall H, et al. 2017. Southern African ancient genomes estimate modern human divergence to 350,000 to 260,000 years ago. Science 358(6363):652–655. [DOI] [PubMed] [Google Scholar]
- Sikora M, Seguin-Orlando A, Sousa VC, Albrechtsen A, Korneliussen T, Ko A, Rasmussen S, Dupanloup I, Nigst PR, Bosch MD, et al. 2017. Ancient genomes show social and reproductive behavior of early Upper Paleolithic foragers. Science 358(6363):659–662. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Malmstrom H, Omrak A, Raghavan M, Valdiosera C, Gunther T, Hall P, Tambets K, Parik J, Sjogren KG, et al. 2014. Genomic diversity and admixture differs for Stone-Age Scandinavian foragers and farmers. Science 344(6185):747–750. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Malmstrom H, Raghavan M, Stora J, Hall P, Willerslev E, Gilbert MT, Gotherstrom A, Jakobsson M.. 2012. Origins and genetic legacy of Neolithic farmers and hunter–gatherers in Europe. Science 336(6080):466–469. [DOI] [PubMed] [Google Scholar]
- Smith J, Coop G, Stephens M, Novembre J.. 2018. Estimating time to the common ancestor for a beneficial allele. Mol Biol Evol. 35(4):1003–1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teshima KM, Innan H.. 2009. mbs: modifying Hudson's ms software to generate samples of DNA sequences with a biallelic site under selection. BMC Bioinformatics. 10:166.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. 2010. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466(7307):707–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usher CL, Handsaker RE, Esko T, Tuke MA, Weedon MN, Hastie AR, Cao H, Moon JE, Kashin S, Fuchsberger C, et al. 2015. Structural forms of the human amylase locus and their relationships to SNPs, haplotypes and obesity. Nat Genet. 47(8):921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wegmann D, Leuenberger C, Neuenschwander S, Excoffier L.. 2010. ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics. 11:116.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye K, Gao F, Wang D, Bar-Yosef O, Keinan A.. 2017. Dietary adaptation of FADS genes in Europe varied across time and geography. Nat Ecol Evol. 1:167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.