Abstract
Background and Aims
Recent meta-analysis of genome-wide association studies have identified over 241 inflammatory bowel disease susceptibility loci. However, the known variants only account for a fraction of inflammatory bowel disease heritability. To identify additional susceptibility loci, we performed a trans-ethnic meta-analysis as well as an Asian-specific meta-analysis, using all published Immunochip association results of inflammatory bowel disease.
Methods
An inverse-variance fixed-effects meta-analysis was carried out across Korean and East Asian Immunochip datasets of 4156 cases and 4904 controls [Asian ancestry]. A trans-ethnic meta-analysis of inflammatory bowel disease was performed together with the European datasets of 38 155 cases and 48 485 controls genotyped on the immunochip using a Bayesian approach, Meta-Analysis of Trans-ethnic Association studies [MANTRA].
Results
We identified seven novel associations, including three novel susceptibility loci at MYO10-BASP1, PPP2R3C/KIAA0391/PSMA6/NFKB1A and LRRK1 as well as four novel secondary associations within previously known loci at NCF4, TSPAN32, CIITA and VANGL2. The new loci further implicate alterations in B cell biology in Crohn’s disease pathogenesis. The effects of five loci were universal across European and Asian ancestries, whereas the NCF4 and CIITA loci showed significant heterogeneity between European and East Asian populations. In addition, 103 previously known IBD loci showed supporting evidence of association with nominal significance [p < 0.05] in Asians.
Conclusions
Our findings of new loci not previously associated with IBD support the importance of studying inflammatory bowel disease genetics in diverse populations.
Keywords: Inflammatory bowel disease, meta-analysis, trans-ethnic
1. Introduction
Inflammatory bowel disease [IBD] is a chronic inflammatory disorder of the gastrointestinal tract. Crohn’s disease [CD] and ulcerative colitis [UC] are the two major subtypes of IBD. IBD is believed to develop due to dysregulated mucosal immune responses to gut flora in genetically susceptible individuals.1
Although the incidence of IBD is lower in Asian populations than in Western populations, the incidence of IBD is rapidly increasing throughout Asia due to environmental changes.2–5 Epidemiological and clinical studies indicate that the phenotype and clinical course of IBD differs between Asians and Europeans.2–7 First, there is a male predominance of CD in Asia, with male to female ratios ranging from 1.67:1 to 2.9:1. Second, ileocolonic disease is predominant in Asian cases, comprising about two-thirds of CD cases, while colonic disease is only noted in ~10% of cases. It is unclear whether these Asian-specific clinical characteristics of IBD are solely due to the difference in the environments between Asia and the West, which underscores the necessity of genetic studies of IBD in Asian populations.
Recent large-scale studies on populations of European ancestry have markedly advanced our understanding of IBD-related genetics. A meta-analysis by the International IBD Genetics Consortium [IIBDGC] of genome-wide association studies [GWASs] and Immunochip data from 96 486 individuals with multiple ancestries [including Asian samples] identified >200 susceptibility loci for IBD, and reported an overlap in the directionality of the odds ratios [ORs] between the cohorts with European and Asian ancestry.8 The latest genome-wide meta-analysis performed on populations with European ancestry reported 241 susceptibility loci for IBD.9 However, it is clear that additional studies are needed to expand our understanding of the genetic architecture of IBD. Despite the differences in the clinical characteristics of IBD between different ethnicities, the number of studies in non-European populations is limited. Several GWASs on CD or UC have been performed in Asian populations.10–15 These studies provided insights into the genetic architecture of each disease, but not a broad view of the genetic basis of IBD in Asians. In the recent largest-to-date Asian-specific GWAS of IBD, we identified two new susceptibility loci to IBD and confirmed associations with 28 established IBD loci in Koreans.16 In the present study, we aimed to identify [i] novel genetic variants associated with IBD in Asians through a meta-analysis of Korean and East Asian studies and [ii] additional IBD susceptibility loci through a trans-ethnic meta-analysis of Korean, East Asian, and European studies. Here we present the findings of the meta-analyses, including seven novel associations identified at the level of genome-wide significance.
2. Methods
2.1. Study population
The samples of the three Immunochip datasets are described in Supplementary Table 1. Samples in the Korean dataset were used in our previous published Immunochip studies of CD and UC,12,13 respectively. The summary statistics of Korean IBD data can be downloaded from a webpage [https://drive.google.com/drive/folders/1L1Zu4G0yzVuB0Ea11HkF-XQqr9pX15-0]. Their clinical characteristics are described in Supplementary Table 2. Previously published summary statistics of the East Asian and European IBD Immunochip datasets available at the IIBDGC [https://www.ibdgenetics.org/downloads.html] were used. The East Asian study comprised 2054 cases from Japan, 453 cases from Korea, 317 cases from China, and 3719 controls. The European dataset comprised 38 155 cases and 48 485 controls.
2.2. Quality control
The quality control [QC] procedures of the Korean, East Asian and European Immunochip datasets are described in previous publications.8,12,13 As Korean Immunochip studies were performed on CD and UC separately, standard QC procedures were applied using the PLINK v1.9 software [https://www.cog-genomics.org/plink2] as described previously for the Korean IBD dataset.12,13 Briefly, single nucleotide polymorphisms [SNPs] with missingness rates of >2%, a minor allele frequency [MAF] of <1%, or failing the Hardy–Weinberg equilibrium [p < 1 × 10–5] test were excluded. Samples with a high proportion of missing genotypes [>4%] and identity by descent [PI_HAT >0.2] were removed. QC was conducted on each dataset separately and the combined set of samples using a common approach. To control for population stratification, genotypes that passed QC filters were merged with HapMap Phase III data [194 individuals] from three populations: European [CEU], Asian [CHB + JPT], and African [YRI]. A principal-components analysis was subsequently performed on the merged dataset, and population outliers were excluded [Supplementary Figure 1]. For the meta-analysis, 87 overlapping cases [79 CD and 8 UC cases that were present in both Korean and East Asian datasets] were excluded from the Korean dataset. Finally, the Korean dataset consisted of 1332 cases and 1185 controls, with 89 051 SNPs [Supplementary Table 3].
2.3. Statistical analysis
Association tests were performed on the Korean dataset of IBD, CD and UC using a logistic regression analysis in PLINK v1.9. A quantile–quantile [Q–Q] plot was generated using R [3.2.0] [http://www.r-project.org/] to evaluate the overall significance of the genome-wide associations and the potential impact of population stratification. The impact of population stratification was also evaluated by calculating the genomic control inflation factor [λGC] and the genomic inflation factor for 1000 cases and 1000 controls [λGC1000] using a set of 3120 “null” SNPs that are not associated with autoimmune diseases [Supplementary Table 1]. After QC, 2117 SNPs were used as null markers to generate the Q–Q plot shown in Supplementary Figure 2 [λGC = 0.98]. Both the Q–Q plots and genomic inflation factor [λGC] of Immunochip test statistics showed that the three Immunochip association analyses had negligible inflation due to population stratification. A Manhattan plot was generated with −log10p by using R [3.2.0]. For regional plots of novel associations that were identified in the Asian meta-analysis, imputation was performed using ImpG version 1.0 [https://github.com/huwenboshi/ImpG]17 and the Asian reference data [JPT + CHB] from the 1000 Genomes Project [February 2012 release] [http://www.1000genomes.org/]. To assess whether candidate novel signals were due to long-range linkage disequilibrium [LD] with variants in previously reported loci, a conditional analysis in the regions identified in trans-ethnic meta-analysis was performed. For all variants in candidate loci that were <3 Mb away from a known locus, conditional analysis was performed on each of the three datasets separately (East Asian/European dataset, GCTA [cnsgenomics.com/software/gcta/]; Korean dataset, PLINK v1.9) followed by a meta-analysis, or on the combined dataset. Secondary SNPs with conditional p < 5 × 10–8 or Log10BF > 6 were assumed to be independent from the reported lead SNP in the region.
2.3.1. Asian-specific meta-analysis
For the Asian-specific meta-analyses, Korean and East Asian association results were combined using the inverse-variance method under the assumption of a fixed effect as implemented in METAL.18 Between-study heterogeneity was quantified using the I2 heterogeneity score, and statistical significance was assessed using the Q test statistic. For the fixed-effects model, significance was defined as pmeta < 5 × 10–8.
2.3.2. Trans-ethnic meta-analysis
For the trans-ethnic meta-analyses, the three independent datasets were combined using two approaches. First, a meta-analysis was performed using the METAL software assuming fixed effects across studies and using inverse-variance weighting. Second, the ethnic-specific Immunochip summary statistics were combine using the MANTRA [Meta-Analysis of Trans-ethnic Association Studies] package, a meta-analysis software tool allowing for heterogeneity in allelic effects caused by differences in LD structure in diverse populations.19 MANTRA results are reported as Log10 Bayes’ factors [Log10BF]. A Log10BF > 6 was considered to be a genome-wide significant threshold value, and SNPs with posterior probability of heterogeneity [phet] > 0.5 were interpreted as having significant heterogeneity. Both fixed effects results and MANTRA results are reported.
2.4. eQTL and bioinformatics analysis
To gain more insight into the potential functional roles of the novel IBD loci, a cis-eQTL analysis was performed by searching publicly available expression data generated from eQTL Blood Browser,20 the Genotype-Tissue Expression [GTEx] database21 and Geuvadis/1000 Genomes resources.22 Whole blood, small intestine, transverse colon, and sigmoid colon were selected in the GTEx browser because they are the most important tissues in mucosal immunity. To explore epigenetic profiles of genomic locations associated with IBD, ENCODE histone modification data, HaploReg and Regulome DB were used to examine whether any of the SNPs or their proxies [r2 ≥ 0.8 in the 1000 genomes of JPT + CHB reference panel] were annotated as transcription factor binding or enhancer elements. Evidence of prior association signals with autoimmune diseases or other immune-related phenotypes was searched for in the Ensembl, UCSC Genome Bioinformatics, and GeneCards databases. When the SNP was not directly typed, a proxy SNP was used [r2 ≥ 0.8].
3. Results
3.1. Asian-specific meta-analysis
To identify additional IBD risk loci in Asians, we first performed a meta-analysis using the Korean IBD Immunochip dataset and the summary statistics from the East Asian Immunochip dataset.8,12,13 The combined dataset consisted of 4156 IBD cases and 4904 controls, with 1332 cases and 1185 controls from the Korean population and 2824 cases and 3719 controls from the East Asian population. Association analysis of the Korean dataset alone did not show any novel IBD loci with genome-wide significance [Supplementary Figure 3]. In a meta-analysis of the combined dataset, 12 of the previously reported regions exceeded statistical significance [pmeta < 5 × 10–8], including two novel associations in Asians at rs2072711 in NCF4-CSF2RB at 22q13 [OR = 1.21; 95% confidence interval [CI], 1.13 – 1.29; pAsian-meta = 5.07 × 10–9] and at rs12928665 in CIITA at 16p13 [OR = 1.19; 95% CI, 1.13 – 1.25; pAsian-meta = 3.93 × 10–9] [Supplementary Table 4, Supplementary Figure 4]. We also examined the 245 previously established European IBD-associated loci [276 independent SNPs] in the Asian meta-analysis.8,9 Of 193 SNPs in 174 loci [including 36 proxy SNPs with r2 ≥ 0.8] available, 111 SNPs from 102 loci were replicated at nominal p < 0.05 in association analyses for IBD, CD or UC [excluding rs864745 and rs10995235 with effects in opposite directions; Supplementary Table 5]. An additional locus [JAK2], not in LD with the European SNP, reached a genome-wide significant association in the Asian meta-analysis [Supplementary Table 4], resulting 112 SNPs being replicated at 103 loci in Asians.
3.2. Trans-ethnic meta-analysis
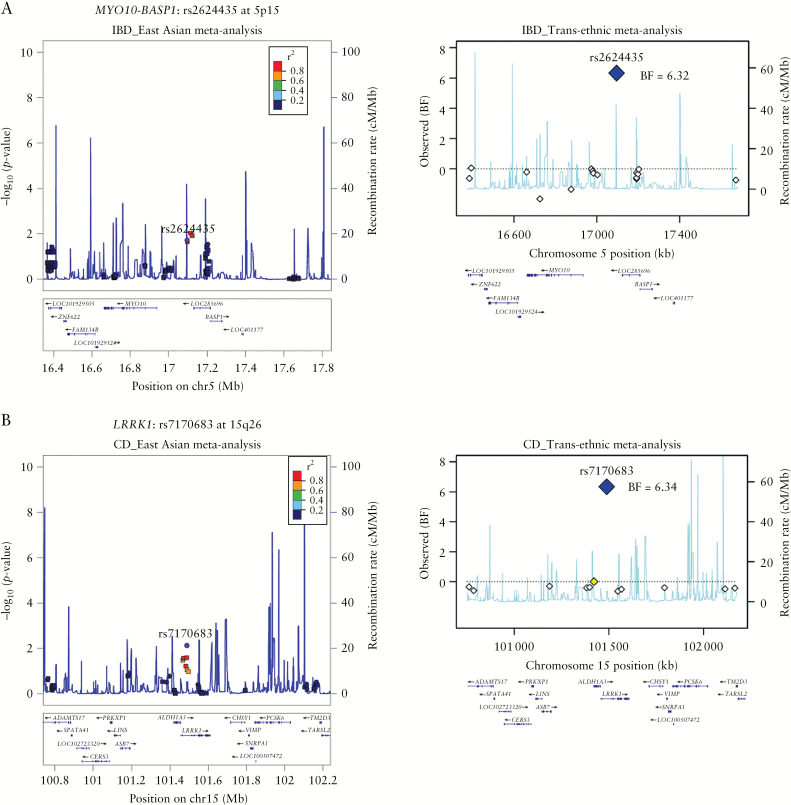
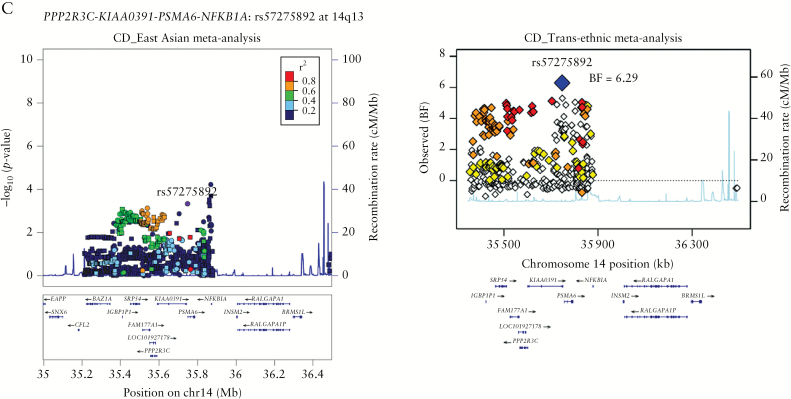
We performed a trans-ethnic meta-analysis of IBD using three independent Immunochip datasets. The combined dataset consisted of 42 311 IBD cases and 53 389 controls, with 1332 cases and 1185 controls from Korean population, 2824 cases and 3719 controls from East Asian population, and 38 155 cases and 48 485 controls from the European population. After applying stringent quality controls, we tested 89 051, 106 681, 126 098 and 80 291 SNPs for association in the Korean, East Asian, European and combined cohort, respectively [Supplementary Table 1]. We performed a meta-analysis using [i] inverse-variance based meta-analysis in METAL software and [ii] the MANTRA package [Supplementary Figure 5]. Following meta-analyses, the SNPs within 241 known loci9 in the MHC region [25−34 Mb, hg19] and known genes were removed.9,23 Twenty-two loci showed significant associations with IBD in the MANTRA analyses [Log10BF > 6], of which 19 loci also showed significant associations in the fixed-effects trans-ethnic METAL analyses [p < 5 × 10–8, Supplementary Table 6]. As these loci were <3 Mb away from known loci except for rs2624435 near MYO10-BASP1, we performed a conditional analysis for 21 variants [Supplementary Table 7]. Only three loci [CIITA, TSPAN32 and NCF4] showed genome-wide significant associations following conditional analyses on the reported European lead SNPs, suggesting the presence of three novel secondary associations within previously reported loci [Table 1]. The MANTRA analyses of CD showed eight loci with Log10BF > 6 [Supplementary Table 6]. Only two of these loci, rs57275892 [PSMA6-NFKBIA] and rs7170683 [LRRK1], were novel [Table 1]. Of the three loci with Log10BF > 6 in the MANTRA analyses of UC [Supplementary Table 6], rs17371986 [VANGL2] showed an independent association within a previously known locus following conditional analysis [Supplementary Table 7]. Of seven novel associations, only rs12928665 [CIITA] and rs2072711 [NCF4] showed evidence of heterogeneity between Asian and European populations [phet ≥ 0.91; Table 1, Supplementary Table 6].
Table 1.
Seven novel associations identified through Asian and trans-ethnic meta-analysis on inflammatory bowel disease.
| Chr | SNP | Position | Effect allele | Study | Effect allele frequency | Inflammatory bowel disease | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| [hg19] | Controls | Cases | OR | p | p random | p Q | |||||
| [N] | [N] | [95% CI] | |||||||||
| 22 | rs2072711 | 37 268 555 | A | Korean | 0.32 | 1 185 | 1 332 | 1.21 | 0.002 | ||
| [1.07–1.36] | |||||||||||
| [NCF4, CSF2RB] | East Asian | 0.26 | 3 719 | 2 824 | 1.21 | 6.55 × 10–7 | |||||
| [1.13–1.28] | |||||||||||
| European | 0.17 | 48 485 | 38 155 | 1.00 | 0.954 | ||||||
| [0.97–1.03] | |||||||||||
| Combined1 † | 4 904 | 4 156 | 1.21 | 5.07 × 10 –9** | 5.07 × 10 –9** | 0.97 | |||||
| [1.13–1.29] | |||||||||||
| Combined2‡ | 53 389 | 42 311 | 1.03 | 0.027 | 0.121 | 4.42 × 10–7 | |||||
| [1.00–1.05] | |||||||||||
| 16 | rs12928665 | 10 971 474 | A | Korean | 0.43 | 1 185 | 1 332 | 0.86 | 0.008 | ||
| [CIITA] | [0.75–0.97] | ||||||||||
| East Asian | 0.46 | 3 719 | 2 824 | 0.83 | 1.29 × 10–7 | ||||||
| [0.76–0.90] | |||||||||||
| European | 0.76 | 48 485 | 38 155 | 0.94 | 1.29 × 10–8 | ||||||
| [0.91–0.96] | |||||||||||
| Combined1 † | 4 904 | 4 156 | 0.84 | 3.93 × 10 –9** | 3.93 × 10 –9** | 0.61 | |||||
| [0.79–0.89] | |||||||||||
| Combined2 ‡ | 53 389 | 42 311 | 0.92 | 1.23 × 10 –13** | 0.006 | 0.002 | |||||
| [0.90–0.94] | |||||||||||
| 11 | rs2074023 | 2 325 581 | A | Korean | 0.20 | 1 185 | 1 332 | 0.88 | 0.061 | ||
| [TSPAN32] | [0.74–1.01] | ||||||||||
| East Asian | 0.25 | 3 719 | 2 824 | 0.87 | 8.45 × 10–4 | ||||||
| [0.79–0.95] | |||||||||||
| European | 0.40 | 48 485 | 38 155 | 0.95 | 1.71 × 10–7 | ||||||
| [0.93–0.97] | |||||||||||
| Combined1† | 4 904 | 4 156 | 0.87 | 1.29 × 10–4 | 1.29 × 10–4 | 0.92 | |||||
| [0.81–0.94] | |||||||||||
| Combined2 ‡ | 53 389 | 42 311 | 0.94 | 1.05 × 10 –9** | 0.005 | 0.09 | |||||
| [0.92–0.96] | |||||||||||
| 5 | rs2624435 | 17 095 269 | A | Korean | 0.14 | 1 185 | 1 332 | 0.92 | 0.287 | ||
| [MYO10, BASP1] | [0.76–1.08] | ||||||||||
| East Asian | 0.14 | 3 719 | 2 824 | 0.89 | 0.035 | ||||||
| [0.79–1.00] | |||||||||||
| European | 0.23 | 48 485 | 38 155 | 0.94 | 1.93 × 10–7 | ||||||
| [0.92–0.96] | |||||||||||
| Combined1† | 4 904 | 4 156 | 0.90 | 0.019 | 0.019 | 0.79 | |||||
| [0.83–0.98] | |||||||||||
| Combined2 ‡ | 53 389 | 42 311 | 0.94 | 1.80 × 10 –8** | 1.80 × 10 –8** | 0.62 | |||||
| [0.92–0.96] | |||||||||||
| 15 | rs7170683 | 101 488 582 | A | Korean | 0.48 | 1 185 | 1 332 | 0.98 | 0.748 | ||
| [LRRK1] | [0.87–1.09] | ||||||||||
| East Asian | 0.47 | 3 719 | 2 824 | 0.98 | 0.673 | ||||||
| [0.91–1.06] | |||||||||||
| European | 0.52 | 48 485 | 38 155 | 0.96 | 1.98 × 10–4 | ||||||
| [0.94–0.98] | |||||||||||
| Combined1† | 4 904 | 4 156 | 0.98 | 0.597 | 0.597 | 0.97 | |||||
| [0.93–1.05] | |||||||||||
| Combined2 ‡ | 53 389 | 42 311 | 0.97 | 2.11 × 10–4 | 2.11 × 10–4 | 0.81 | |||||
| [0.95–0.98] | |||||||||||
| 14 | rs57275892 | 35 747 271 | A | Korean | 0.35 | 1 185 | 1 332 | 0.80 | 1.98 × 10–4 | ||
| [PPP2R3C, KIAA0391, PSMA6, NFKBIA] | [0.69–0.92] | ||||||||||
| East Asian | 0.33 | 3 719 | 2 824 | 0.92 | 0.025 | ||||||
| [0.84–0.99] | |||||||||||
| European | 0.17 | 48 485 | 38 155 | 0.95 | 1.50 × 10–4 | ||||||
| [0.92–0.98] | |||||||||||
| Combined1† | 4 904 | 4 156 | 0.88 | 9.25 × 10–5 | 0.029 | 0.06 | |||||
| [0.83–0.94] | |||||||||||
| Combined2 ‡ | 53 389 | 42 311 | 0.94 | 5.55 × 10–7 | 0.013 | 0.02 | |||||
| [0.92–0.96] | |||||||||||
| 1 | rs17371986 | 160 402 259 | A | Korean | 0.83 | 1 185 | 1 332 | 0.88 | |||
| [VANGL2] | [0.73–1.02] | 0.080 | |||||||||
| East Asian | 0.79 | 3 719 | 2 824 | 0.95 | 0.205 | ||||||
| [0.86–1.03] | |||||||||||
| European | 0.79 | 48 485 | 38 155 | 0.95 | 5.18 × 10–5 | ||||||
| [0.93–0.98] | |||||||||||
| Combined1† | 4 904 | 4 156 | 0.93 | 0.047 | 0.047 | 0.39 | |||||
| [0.86–1.00] | |||||||||||
| Combined2 ‡ | 53 ,389 | 42 311 | 0.95 | 8.27 × 10–6 | 8.27 × 10–6 | 0.55 | |||||
| [0.93–0.97] | |||||||||||
| Chr | Crohn's disease | Ulcerative colitis | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cases | OR | p | prandom | pQ | Cases | OR | p | prandom | pQ | |
| (N) | (95% CI) | [N] | [95% CI] | |||||||
| 22 | 637 | 1.29 | 6.95 × 10–4 | 695 | 1.14 | 0.082 | ||||
| [NCF4, CSF2RB] | [1.11–1.49] | [0.98–1.31] | ||||||||
| 1 690 | 1.26 | 2.17 × 10–7 | 1 134 | 1.15 | 0.009 | |||||
| [1.17–1.35] | [1.04–1.26] | |||||||||
| 20 550 | 0.96 | 0.008 | 17 647 | 1.05 | 0.008 | |||||
| [0.92–0.99] | [1.01–1.08] | |||||||||
| 2 327 | 1.27 | 6.01 × 10 -10** | 6.01 × 10 -10** | 0.83 | 1 829 | 1.15 | 0.002 | 0.002 | 0.89 | |
| [1.18–1.37] | [1.05–1.25] | |||||||||
| 22 877 | 1.00 | 0.986 | 0.218 | 1.31 × 10-10 | 19 476 | 1.06 | 3.20 × 10–4 | 0.017 | 0.15 | |
| [0.97–1.03] | [1.03–1.09] | |||||||||
| 16 | 637 | 0.90 | 0.140 | 695 | 0.82 | 0.004 | ||||
| [CIITA] | [0.76–1.04] | [0.69–0.96] | ||||||||
| 1 690 | 0.81 | 1.07 × 10–6 | 1 134 | 0.85 | 8.92 × 10–4 | |||||
| [0.73–0.90] | [0.76–0.95] | |||||||||
| 20 550 | 0.94 | 6.61 × 10–6 | 17 647 | 0.94 | 2.36 × 10–5 | |||||
| [0.91–0.97] | [0.91–0.97] | |||||||||
| 2 327 | 0.84 | 7.53 × 10–7 | 3.48 × 10–4 | 0.22 | 1 829 | 0.84 | 1.38 × 10–5 | 1.38 × 10–5 | 0.64 | |
| [0.78–0.90] | [0.78–0.91] | |||||||||
| 22 877 | 0.93 | 2.22 × 10 –9** | 0.020 | 0.005 | 19 476 | 0.93 | 4.50 × 10 –8** | 0.007 | 0.03 | |
| [0.90–0.95] | [0.90–0.95] | |||||||||
| 11 | 637 | 0.82 | 0.026 | 695 | 0.93 | 0.361 | ||||
| [TSPAN32] | [0.65–0.99] | [0.77–1.09] | ||||||||
| 1 690 | 0.87 | 0.004 | 1 134 | 0.88 | 0.017 | |||||
| [0.77–0.96] | [0.77–0.98] | |||||||||
| 20 550 | 0.95 | 1.04 × 10–5 | 17 647 | 0.94 | 1.73 × 10–5 | |||||
| [0.92–0.97] | [0.92–0.97] | |||||||||
| 2 327 | 0.86 | 2.94 × 10–4 | 2.94 × 10–4 | 0.58 | 1 829 | 0.89 | 0.013 | 0.013 | 0.56 | |
| [0.79–0.93] | [0.81–0.98] | |||||||||
| 22 877 | 0.94 | 4.18 × 10–7 | 0.012 | 0.07 | 19 476 | 0.94 | 1.44 × 10–6 | 1.44 × 10–6 | 0.41 | |
| [0.92–0.96] | [0.92–0.96] | |||||||||
| 5 | 637 | 1.05 | 0.628 | 695 | 0.80 | 0.028 | ||||
| [MYO10, BASP1] | [0.86–1.24] | [0.61–1.00] | ||||||||
| 1 690 | 0.90 | 0.099 | 1 134 | 0.89 | 0.093 | |||||
| [0.78–1.02] | [0.74–1.03] | |||||||||
| 20 550 | 0.96 | 0.003 | 17 647 | 0.93 | 3.64 × 10–6 | |||||
| [0.93–0.99] | [0.91–0.96] | |||||||||
| 2 327 | 0.94 | 0.262 | 0.525 | 0.19 | 1 829 | 0.86 | 0.008 | 0.008 | 0.42 | |
| [0.85–1.04] | [0.76–0.96] | |||||||||
| 22 877 | 0.96 | 0.001 | 0.001 | 0.41 | 19 476 | 0.93 | 2.80 × 10–7 | 0.005 | 0.26 | |
| [0.93–0.98] | [0.90–0.96] | |||||||||
| 15 | 637 | 0.86 | 0.027 | 695 | 1.11 | 0.117 | ||||
| [LRRK1] | [0.72–0.99] | [0.98–1.25] | ||||||||
| 1 690 | 0.93 | 0.073 | 1 134 | 1.08 | 0.097 | |||||
| [0.84–1.01] | [0.99–1.18] | |||||||||
| 20 550 | 0.94 | 4.97 × 10–7 | 17 647 | 0.98 | 0.212 | |||||
| [0.92–0.96] | [0.96–1.01] | |||||||||
| 2 327 | 0.91 | 0.007 | 0.007 | 0.34 | 1 829 | 1.09 | 0.024 | 0.024 | 0.76 | |
| [0.84–0.97] | [1.01–1.18] | |||||||||
| 22 877 | 0.94 | 1.99 × 10 –8** | 1.99 × 10 –8** | 0.39 | 19 476 | 0.99 | 0.608 | 0.345 | 0.04 | |
| [0.92–0.96] | [0.97–1.02] | |||||||||
| 14 | 637 | 0.82 | 0.006 | 695 | 0.79 | 7.59 × 10–4 | ||||
| [PPP2R3C, KIAA0391, PSMA6, NFKBIA] | [0.67–0.96] | [0.65–0.93] | ||||||||
| 1 690 | 0.89 | 0.014 | 1 134 | 0.96 | 0.399 | |||||
| [0.81–0.98] | [0.86–1.06] | |||||||||
| 20 550 | 0.93 | 4.89 × 10–6 | 17 647 | 0.98 | 0.355 | |||||
| [0.90–0.96] | [0.95–1.02] | |||||||||
| 2 327 | 0.87 | 4.34 × 10–4 | 7.98 × 10–4 | 0.30 | 1 829 | 0.90 | 0.008 | 0.167 | 0.03 | |
| [0.81–0.94] | [0.82–0.97] | |||||||||
| 22 877 | 0.92 | 2.38 × 10 –8** | 3.91 × 10 –4 | 0.20 | 19 476 | 0.97 | 0.064 | 0.145 | 0.01 | |
| [0.89–0.95] | [0.94–1.00] | |||||||||
| 1 | 637 | 0.83 | 0.046 | 695 | 0.92 | 0.341 | ||||
| [VANGL2] | [0.66–1.01] | [0.74–1.10] | ||||||||
| 1 690 | 0.99 | 0.913 | 1 134 | 0.88 | 0.037 | |||||
| [0.89–1.10] | [0.77–1.00] | |||||||||
| 20 550 | 0.99 | 0.576 | 17 647 | 0.92 | 1.95 × 10–8 | |||||
| [0.96–1.02] | [0.89–0.95] | |||||||||
| 2 327 | 0.95 | 0.276 | 0.368 | 0.09 | 1 829 | 0.89 | 0.023 | 0.023 | 0.74 | |
| [0.87–1.04] | [0.81–0.98] | |||||||||
| 22 877 | 0.99 | 0.384 | 0.414 | 0.17 | 19 476 | 0.92 | 1.58 × 10 –9** | 1.58 × 10 –9** | 0.83 | |
| [0.96–1.02] | [0.89–0.94] | |||||||||
Chr, chromosome; CI, confidence interval; OR, odds ratio; Position, chromosome position. Genome-wide significant results from the combined analysis are indicated in bold.
**Association results at the genome-wide significance level [p < 5.0 × 10–8].
†Combined1: Korean+East Asian dataset.
‡Combined2: Korean+East Asian+European dataset.
3.3. Novel loci
All three newly discovered SNPs are non-coding variants, rs2624435 in MYO10-BASP1 at 5p15 for IBD, rs57275892 in PPP2R3C/KIAA0391/PSMA6/NFKB1A at 14q13, and rs7170683 in LRRK1 at 15q26 for CD. The SNP densities across the MYO10-BASP1 and LRRK1 loci were sparse [Figure 1A, 1B]; however, upon examining the eQTL database, rs2624435 showed an association with BASP1 expression levels in blood, and rs7170683 showed an association with both LRRK1 and ALDH1A3 expression levels in lymphoblastoid cell lines of European ancestry [Supplementary Table 8]. A novel IBD susceptibility locus at rs2624435 is located between MYO10 and BASP1. The biological function of the Brain Acid Soluble Protein 1 [BASP1] gene is not well known, except for its involvement in nephrogenesis as a transcriptional co-suppressor,24 whereas an unconventional myosin, Myosin X [MYO10], is implicated in phagocytosis via its role in filopodia induction. Macrophages lacking Myo10 showed markedly reduced filopodia formation.25 A novel CD susceptibility locus at rs7170683 is located in the Leucine-rich repeat kinase 1 [LRRK1]. LRRK1 belongs to a member of the ROCO family of proteins with multiple functional domains including ankyrin-like repeats, leucine-rich repeats [LRRs], a Ras-like GTPase domain [ROC] and an adjacent C-terminal domain [COR], and a serine–threonine kinase domain. Its homolog LRRK2 was shown to be associated with CD23 and Parkinson’s disease.26 LRRK1 is involved in a variety of functions including autophagy and osteoclast differentiation. A recent knock-out mice study reported that LRRK1 plays a critical role in B cell development and antibody production by regulating NF-kB signaling.27 The other novel CD susceptibility SNP [rs57275892] at 14q13 is located ~500 bp upstream of PSMA6 [Figure 1C] in a LD region of 645 kb [35.212−35.857 Mb] that includes BAZ1A, SRP54, FAM177A1, PPP2R3C, KIAA0391, PSMA6 and NFKB1A. Although rs57275892 is located ~500 bp upstream of PSMA6, eQTL analyses showed that it was associated most significantly with the expression of a nearby gene, protein phosphatase 2 regulatory subunit B″ gamma [PPP2R3C] [p = 2.00 × 10–22] in whole blood [Supplementary Table 8]. PPP2R3C is a regulatory subunit of a serine/threonine phosphatase, protein phosphatase 2. Knock-out and transgenic mice studies show that PPP2R3C is involved in both the survival of germinal center B cells and the differentiation of peritoneal B cells into autoantibody-producing plasma cells, suggesting possible roles in inflammatory diseases.28
Figure 1.
Signal plots for the three novel IBD loci: [A] rs2624435 at 5p15, [B] rs7170683 at 15q26 and [C] rs57275892 at 14q13. The left panels represent signal plots from the fixed-effects meta-analyses of Korean and East Asian–ancestry individuals. SNPs are plotted according to their chromosomal positions [NCBI Build 37] with –log10p values from the Asian meta-analysis in the region flanking 750 kb on either side of the marker SNP. The circles indicate the genotyped SNPs, and the squares indicate the imputed SNPs. The most strongly associated SNP in the discovery stage is shown as a small purple circle. Linkage disequilibrium [LD; r2 values] between the lead SNP and the other SNPs are indicated using colours. The relative location of the annotated genes and the direction of transcription are shown in the lower portion of the figure. The estimated recombination rates of the Asian samples from the 1000 Genomes Project [Nov 2014] are plotted to reflect the local LD structure. Plots are generated using LocusZoom. The right panel presents signal plots for the MANTRA association signal after trans-ethnic meta-analysis of Korean, East-Asian, and European Immunochip data. Each point represents an SNP that passed the QC in the MANTRA analysis, plotted with their BF [on a log10 scale] as a function of genomic position [NCBI Build 37]. Plots were generated using R.
3.4. Novel secondary associations within previously known loci
The association signal [rs2074023] at chromosome 11p15 is located within intron 1 of Tetraspanin32 [TSPAN32, Log10BF = 7.49, pmeta = 1.05 × 10–9] in a LD region of 117.8 kb that includes C11orf21, TSPAN32, CD81 and TSSC4 [Table 1, Supplementary Figure 6]. rs2074023 is located ~452 kb away from the previously reported rs9076118,9 [r2 < 0.2]; however, conditional logistic regression analysis on rs907611 supported the independent effects of the two SNPs [Supplementary Table 7]. Examination of the eQTL database for rs2074023 showed that it is associated with the expression levels of both C11orf21 and TSPAN32 in lymphoblastoid cell lines and small intestine, whereas previously reported rs907611 is associated with the expression levels of CTSD in blood [Supplementary Table 8], supporting their independent effects. A search of HaploReg v4 for rs2074023 showed a Regulome DB score 1b, indicating that it is likely to affect binding and to be linked to expression of a gene target [Supplementary Table 9]. TSPAN32 is a member of the tetraspanins of integral membrane proteins with functional roles in cell motility, membrane fusion, proliferation and immunity. Tetraspanins, of which over 30 have been identified in humans, can associate with one another and with other molecules such as integrins or proteins of the immunoglobulin superfamily to form a network on the surfaces of many different cell types.29 Knock-out mice experiments have shown that TSPAN32 may play a role in the negative regulation of peripheral T-lymphocyte proliferation,30 suggesting that it may have roles in inflammatory diseases. The biological function of the other gene, C11orf21, is unknown; however, rs7944004, located in 5.7 kb 3' of C11orf21 [14.4 kb away from rs2074023 with r2 = 0.75], is associated with chronic lymphocytic leukemia.31 The association signal [rs17371986] at chromosome 1q23 is located at 3.8 kb from the 3' end of VANGL planar cell polarity protein 2 gene [VANGL2] in a LD region of 110.7 kb that includes NCSTN, NHLH1 and VANGL2 [Table 1, Supplementary Figure 6]. The previously reported rs4656958 [~2 kb 5' of ITLN1] is located ~454 kb away from rs17371986 [r2 < 0.2]; however, conditional logistic regression analysis of rs4656958 supported the independent effects of the two SNPs [Supplementary Table 7]. Examination of the eQTL database for rs17371986 showed that it is associated with VANGL2 expression levels in the sigmoid colon in the GTEx database [Supplementary Table 8]. As VANGL2 is involved in the regulation of planar cell polarity, its involvement in UC susceptibility is not obvious.
3.5. Loci showing heterogeneity between Asian and European populations
Of the two novel IBD associations that showed significant heterogeneity between Asian and European populations, association effects of NCF4 were mapped to two independent SNPs in two populations [phet = 1.0, Supplementary Table 6 and Supplementary Figure 6]. The top SNP rs2072711 identified in the Asian-specific meta-analyses showed much stronger association with CD than with UC, whereas the recently reported European top SNP rs4821544 [10 kb away from rs2072711, r2 < 0.2 in ASN] showed a significant association with CD [OR = 1.07, p = 6.75 × 10–8] only [Supplementary Table 10]. rs2072711 showed weak associations with CD and UC in the European dataset with different effect directions, but rs4821544 failed to show associations with IBD in the Asian samples. The conditional analysis on rs4821544 did not abolish the association at rs2072711 in East Asian [p = 6.55 × 10–7, pcondition = 6.15 × 10–7] and Korean samples [p = 2.16 × 10–3, pcondition = 1.95 × 10–3], suggesting two independent associations in the NCF4 locus. rs2072711 is located on chromosome 22q12 in a LD region of ~60.3 kb, which includes two genes, NCF4 [neutrophil cytosolic factor 4] and CSF2RB [colony stimulating factor 2 receptor beta common subunit]. Examination of eQTL databases for rs2072711 showed that it had a much stronger association with the mRNA expression levels of NCF4 [p = 6.20 × 10–15 in lymphoblastoid, p = 4.35 × 10–55 in blood] than with those of CSF2RB [p = 1.10 × 10–5 in lymphoblastoid] [Supplementary Table 8].20,32,33NCF4 encodes a cytosolic regulatory component of superoxide-producing phagocyte NADPH oxidase, a multicomponent enzyme system important for host defense. The risk alleles for both rs4821544 and rs2072711 were associated with decreased expression of NCF4 in the blood eQTL database, p = 3.24 × 10–28 and p = 4.35 × 10–55, respectively.20
rs12928665 in intron 1 of the MHC class II transactivator gene [CIITA] at chromosome 16p13 showed significant heterogeneity between Asian and European populations [Log10BF = 7.76, phet = 1.00; ptrans-ethnic meta = 1.23 × 10–13pQ = 2.14 × 10–3] [Supplementary Table 6 and Supplementary Figure 6]. Of note is the significant difference in frequency of rs12928665 between Asian and European populations [Supplementary Table 11]. Previous studies have reported two independent association signals, rs529866 in the SOCS1 locus8.9 [401.8 kb away from rs12928665] for CD and rs11641184 in the LITAF locus8 [733.1 kb from rs12928665] for IBD. The conditional analysis on rs529866, rs11641184 and both did not abolish the association at rs12928665, suggesting three independent associations in the CIITA-SOCS1-LITAF locus [Supplementary Table 11]. Previously, rs4781011 of CIITA [intron 2, 3838 bp away from rs12928665] was reported to be associated with UC in Europeans, but not to be present at 200 IBD susceptibility loci or present at the level of genome-wide significance.34 LD between rs12928665 and rs4781011 in Asians [JPT + CHB] was low [r2 < 0.2], whereas it was almost complete in Europeans [r2 = 0.95]. Indeed, association of rs12928665 with IBD [p = 1.29 × 10–8] was stronger than that of rs4781011 in the European dataset [p = 4.21 × 10–8], suggesting that the association previously reported might have been due to an indirect association caused by the LD between the two SNPs. Examination of the eQTL database for the three SNPs showed that they were associated with the expression levels of different genes in blood, indicating that their effects are independent [Supplementary Table 8]. CIITA is an important transcription factor for the expression of HLA class II molecules and is involved in the expression of HLA class I molecules.35,36
4. Discussion
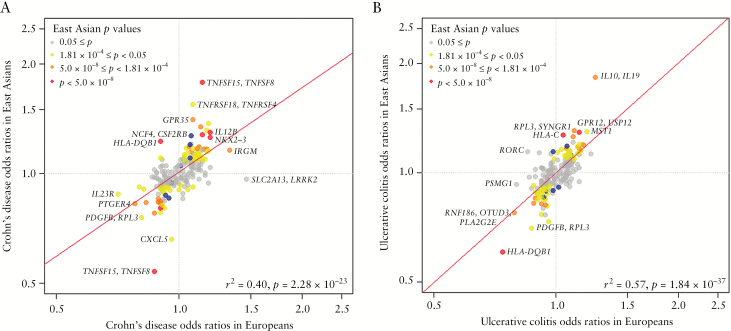
Here we present the largest Immunochip meta-analysis of IBD, using all published Immunochip association results of IBD. Despite the limitation of using only 80 291 common SNPs for the analyses, we identified seven novel associations, including three novel susceptibility loci at MYO10-BASP1 for IBD, PPP2R3C/KIAA0391/PSMA6/ NFKB1A and LRRK1 for CD, and four novel secondary associations within previously known loci at NCF4, TSPAN32, CIITA and VANGL2. In addition, we were able to replicate 103 loci of known IBD susceptibility loci in Asian samples [Supplementary Table 5]. The comparison of the effect sizes for all 193 SNPs from known loci and seven novel associations identified in the present study between Asians and Europeans revealed a positive correlation in the direction of effects for both CD and UC [Figure 2] [r2 = 0.40 and p = 1.54 × 10–23 for CD; r2 = 0.57 and p = 1.84 × 10–37 for UC]. This observation is consistent with a previous large-scale study that reported substantial genetic overlap between Europeans and Asians.8
Figure 2.
Comparison of odds ratios [ORs] for the 193 reported and 7 newly identified Crohn’s disease and ulcerative colitis risk variants in Europeans and East Asians [based on the minor allele in Europeans]. The 193 reported SNPs are listed in Supplementary Table 5 and 7 newly identified SNPs in Table 1. Each dot represents the ORs [on a log scale] for each SNP in [A] Crohn’s disease and [B] ulcerative colitis. Colour denotes the range of association p values for Crohn’s disease or ulcerative colitis in the discovery dataset. The red line refers to linear regression, which was weighted by the inverse of the variance of the log[ORs] in the discovery dataset. The correlation coefficients and p values of the linear model are shown at the bottom-right corner.
Of the two novel IBD associations showing significant heterogeneity between Asian and European populations, the association effects of NCF4 were mapped to two independent SNPs in two populations. The fact that the risk alleles for both SNPs were associated with decreased expression of NCF4 in blood eQTL database suggests that the heterogeneity of the most significant signals might be due to the LD difference between Asians and Europeans [Supplementary Figure 7]. The NCF4-CSF2RB region showed genome-wide significant associations with both IBD and CD in Asians, but with CD only in Europeans. Previous studies have shown association of rs4821544 with ileal CD in Europeans37–39 and its effects on reactive oxygen species production following stimulation with GM-CSF.40 Stronger association of NCF4 in Asians could be due to the fact that ileocolonic disease is the most common type of CD in Asian populations, whereas ileal, colonic and ileocolonic disease occur in equal proportions in the CD of western populations.
In this study, we conducted the largest Immunochip trans-ethnic meta-analysis of IBD and discovered seven novel associations, including three novel susceptibility loci and four novel independent associations within previously known loci. The new loci suggest that additional factors are involved in the T cell biology and B cell immunity of IBD. In conclusion, this trans-ethnic study advances our understanding of the genetic architecture of IBD susceptibility by discovering novel associations and revealing allelic heterogeneity between Asian and European populations.
Web Resources
The URLs for data presented herein are as follows:
METAL, http://csg.sph.umich.edu/abecasis/metal/
The 1000 Genome Project, http://www.1000genomes.org/
UCSC Genome Browser, http://genome.ucsc.edu/
GCTA, http://cnsgenomics.com/software/gcta/
IIBDGC, https://www.ibdgenetics.org/
RegulomeDB v2, http://www.broadinstitute.org/mammals/haploreg/haploreg.php
Genotype–Tissue Expression [GTEx] project, http://www.gtexportal.org/home
eQTL Blood Browser, http://www.genenetwork.nl/bloodeqtlbrowser/
Geuvadis/1000 Genomes resources, http://www.ebi.ac.uk/Tools/geuvadis-das/
Funding
This work was supported by a Korean Health Technology R&D Project grant through the Korea Health Industry Development Institute to S-KY [A120176], funded by the Ministry of Health & Welfare, as well as a Mid-career Researcher Program grant through the National Research Foundation of Korea to KS [2014R1A2A1A09005824, 2017R1A2A1A05001119], funded by the Ministry of Science, Information & Communication Technology and Future Planning, the Republic of Korea. MH was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education [NRF-2016R1A6A3A01013069].
Conflict of Interest
None declared.
Supplementary Data
Supplementary data are available at ECCO-JCC online.
Author Contributions
KS and S-KY obtained financial support. KS conceived and designed the study and supervised the data analysis and interpretation. BDY and BH participated in the study design and supervised the data analysis and interpretation. S-KY, BDY, SHP and HSL recruited subjects and participated in the diagnostic evaluation. TH, KDT, JIR, JL and DPBM, participated in genotyping and supervised the data analysis. SYB and T-HK provided additional control data. MH, SJ, JH, JB, WL, YL, BMK and SBL performed data analyses. KS and MH drafted the manuscript. KS revised the manuscript.
Supplementary Material
Acknowledgments
We would like to thank all the participating patients and healthy donors who provided the DNA and clinical information for this study. This work was supported by the PLSI supercomputing resources of the Korea Institute of Science and Technology Information. Ethics approval was given by the Institutional Review Board of the Asan Medical Center.
Glossary
Abbreviations:
- CD
Crohn’s disease
- CI
confidence interval
- GWAS
genome-wide association study
- IBD
inflammatory bowel disease
- LD
linkage disequilibrium
- MAF
minor-allele frequency
- OR
odds ratio
- SNP
single nucleotide polymorphism
- UC
ulcerative colitis
References
- 1. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011;474:307–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thia KT, Loftus EV Jr, Sandborn WJ, Yang SK. An update on the epidemiology of inflammatory bowel disease in Asia. Am J Gastroenterol 2008;103:3167–82. [DOI] [PubMed] [Google Scholar]
- 3. Yang SK, Yun S, Kim JH et al. . Epidemiology of inflammatory bowel disease in the Songpa–Kangdong district, Seoul, Korea, 1986–2005: a KASID study. Inflamm Bowel Dis 2008;14:542–9. [DOI] [PubMed] [Google Scholar]
- 4. Ng SC, Tang W, Ching JY et al. . Incidence and phenotype of inflammatory bowel disease based on results from the Asia–Pacific Crohn’s and colitis epidemiology study. Gastroenterology 2013;145:158–62.e2. [DOI] [PubMed] [Google Scholar]
- 5. Park SJ, Kim WH, Cheon JH. Clinical characteristics and treatment of inflammatory bowel disease: a comparison of Eastern and Western perspectives. World J Gastroenterol 2014;20:11525–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Burisch J, Pedersen N, Čuković-Čavka S et al. ; EpiCom-group. East–West gradient in the incidence of inflammatory bowel disease in Europe: the ECCO-EpiCom inception cohort. Gut 2014;63:588–97. [DOI] [PubMed] [Google Scholar]
- 7. Cosnes J, Gower-Rousseau C, Seksik P, Cortot A. Epidemiology and natural history of inflammatory bowel diseases. Gastroenterology 2011;140:1785–94. [DOI] [PubMed] [Google Scholar]
- 8. Liu JZ, van Sommeren S, Huang H et al. ; International Multiple Sclerosis Genetics Consortium; International IBD Genetics Consortium. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet 2015;47:979–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. de Lange KM, Moutsianas L, Lee JC et al. . Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet 2017;49:256–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang SK, Hong M, Zhao W et al. . Genome-wide association study of ulcerative colitis in Koreans suggests extensive overlapping of genetic susceptibility with Caucasians. Inflamm Bowel Dis 2013;19:954–66. [DOI] [PubMed] [Google Scholar]
- 11. Yang SK, Hong M, Zhao W et al. . Genome-wide association study of Crohn’s disease in Koreans revealed three new susceptibility loci and common attributes of genetic susceptibility across ethnic populations. Gut 2014;63:80–7. [DOI] [PubMed] [Google Scholar]
- 12. Yang SK, Hong M, Choi H et al. . Immunochip analysis identification of 6 additional susceptibility loci for Crohn’s disease in Koreans. Inflamm Bowel Dis 2015;21:1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ye BD, Choi H, Hong M et al. . Identification of ten additional susceptibility loci for ulcerative colitis through immunochip analysis in Koreans. Inflamm Bowel Dis 2016;22:13–9. [DOI] [PubMed] [Google Scholar]
- 14. Asano K, Matsushita T, Umeno J et al. . A genome-wide association study identifies three new susceptibility loci for ulcerative colitis in the Japanese population. Nat Genet 2009;41:1325–9. [DOI] [PubMed] [Google Scholar]
- 15. Yamazaki K, Umeno J, Takahashi A et al. . A genome-wide association study identifies 2 susceptibility loci for Crohn’s disease in a Japanese population. Gastroenterology 2013;144:781–8. [DOI] [PubMed] [Google Scholar]
- 16. Yang SK, Hong M, Oh H et al. . Identification of loci at 1q21 and 16q23 that affect susceptibility to inflammatory bowel disease in Koreans. Gastroenterology 2016;151:1096–9.e4. [DOI] [PubMed] [Google Scholar]
- 17. Pasaniuc B, Zaitlen N, Shi H et al. . Fast and accurate imputation of summary statistics enhances evidence of functional enrichment. Bioinformatics 2014;30:2906–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Morris AP. Transethnic meta-analysis of genomewide association studies. Genet Epidemiol 2011;35:809–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Westra HJ, Peters MJ, Esko T et al. . Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet 2013;45:1238–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. GTEx Consortium. Human Genomics. The Genotype–Tissue Expression [GTEx] pilot analysis: multitissue gene regulation in humans. Science 2015;348; 648–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lappalainen T, Sammeth M, Friedländer MR et al. ; Geuvadis Consortium. Transcriptome and genome sequencing uncovers functional variation in humans. Nature 2013;501:506–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jostins L, Ripke S, Weersma RK et al. ; International IBD Genetics Consortium [IIBDGC]. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012;491:119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Green LM, Wagner KJ, Campbell HA, Addison K, Roberts SG. Dynamic interaction between WT1 and BASP1 in transcriptional regulation during differentiation. Nucleic Acids Res 2009;37:431–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horsthemke M, Bachg AC, Groll K et al. . Multiple roles of filopodial dynamics in particle capture and phagocytosis and phenotypes of Cdc42 and Myo10 deletion. J Biol Chem 2017;292:7258–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zimprich A, Biskup S, Leitner P et al. . Mutations in LRRK2 cause autosomal-dominant Parkinsonism with pleomorphic pathology. Neuron 2004;44:601–7. [DOI] [PubMed] [Google Scholar]
- 27. Morimoto K, Baba Y, Shinohara H et al. . LRRK1 is critical in the regulation of B-cell responses and CARMA1-dependent NF-κB activation. Sci Rep 2016;6:25738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kitabatake M, Soma M, Zhang T et al. . JNK regulatory molecule G5PR induces IgG autoantibody–producing plasmablasts from peritoneal B1a cells. J Immunol 2015;194:1480–8. [DOI] [PubMed] [Google Scholar]
- 29. Jones EL, Demaria MC, Wright MD. Tetraspanins in cellular immunity. Biochem Soc Trans 2011;39:506–11. [DOI] [PubMed] [Google Scholar]
- 30. Gartlan KH, Belz GT, Tarrant JM et al. . A complementary role for the tetraspanins CD37 and Tssc6 in cellular immunity. J Immunol 2010;185:3158–66. [DOI] [PubMed] [Google Scholar]
- 31. Berndt SI, Skibola CF, Joseph V et al. . Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nat Genet 2013;45:868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dimas AS, Deutsch S, Stranger BE et al. . Common regulatory variation impacts gene expression in a cell type-dependent manner. Science 2009;325:1246–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fehrmann RS, Jansen RC, Veldink JH et al. . Trans-eQTLs reveal that independent genetic variants associated with a complex phenotype converge on intermediate genes, with a major role for the HLA. PLoS Genet 2011;7:e1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McGovern DP, Gardet A, Törkvist L et al. ; NIDDK IBD Genetics Consortium. Genome-wide association identifies multiple ulcerative colitis susceptibility loci. Nat Genet 2010;42:332–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chang CH, Flavell RA. Class II transactivator regulates the expression of multiple genes involved in antigen presentation. J Exp Med 1995;181:765–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Steimle V, Otten LA, Zufferey M, Mach B. Complementation cloning of an MHC class II transactivator mutated in hereditary MHC class II deficiency [or bare lymphocyte syndrome]. Cell 1993;75:135–46. [PubMed] [Google Scholar]
- 37. Rioux JD, Xavier RJ, Taylor KD et al. . Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet 2007;39:596–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Roberts RL, Hollis-Moffatt JE, Gearry RB, Kennedy MA, Barclay ML, Merriman TR. Confirmation of association of IRGM and NCF4 with ileal Crohn’s disease in a population-based cohort. Genes Immun 2008;9:561–5. [DOI] [PubMed] [Google Scholar]
- 39. Muise AM, Xu W, Guo CH et al. ; NEOPICS. NADPH oxidase complex and IBD candidate gene studies: identification of a rare variant in NCF2 that results in reduced binding to RAC2. Gut 2012;61:1028–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Somasundaram R, Deuring JJ, van der Woude CJ, Peppelenbosch MP, Fuhler GM. Linking risk conferring mutations in NCF4 to functional consequences in Crohn’s disease. Gut 2012;61:1097; author reply 1097–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.