Abstract
Following proliferation of oogonia in mammals, great numbers of germ cells are discarded, primarily by apoptosis, while the remainder form primordial follicles (the ovarian reserve) that determine fertility and reproductive lifespan. More massive, rapid, and essentially total loss of oocytes, however, occurs when the transcription factor Lhx8 is ablated—though the cause and mechanism of germ cell loss from the Lhx8-/- ovaries has been unknown. We found that Lhx8−/− ovaries maintain the same number of germ cells throughout embryonic development; rapid decrease in the pool of oocytes starts shortly before birth. The loss results from activation of autophagy, which becomes overwhelming within the first postnatal week, with extracellular matrix proteins filling the space previously occupied by follicles to produce a fibrotic ovary. Associated with this process, as early as a few days before birth, Lhx8-/- oocytes failed to repair DNA damage—which normally occurs when meiosis is initiated during embryonic development; and DNA damage repair genes were downregulated throughout the oocyte short lifespan. Based on gene expression analyses and morphological changes, we propose a model in which lineage-restricted failure of DNA repair triggers germ cell autophagy, causing premature depletion of the ovarian reserve in Lhx8-/- mice.
Keywords: ovarian reserve, Lhx8, autophagy, apoptosis, reproduction, DNA damage, oocyte
Summary Sentence
Ablation of Lhx8 causes premature loss of germ cells by autophagy associated with impairment of DNA damage repair during meiosis.
Introduction
During the process of mammalian primordial follicle formation (follicular assembly), most oocytes are discarded by attrition [1] while the remainder are largely incorporated into primordial follicles soon after birth [2,3]. Good-quality oocytes are prerequisite to the formation of a competent pool of ovarian follicles (the ovarian reserve), which is complete within several days after birth in mice [4]. Any disruption of this embryonic process can result in a smaller ovarian reserve and consequent reduction in fertility and reproductive lifespan.
Once formed, primordial follicles can remain quiescent up to 2 years in mice and several decades in women—until menstrual/estrus cycles cease, if they are not selected for ovulation. However, following the establishment of the reserve, some primordial follicles undergo progressive and irreversible recruitment throughout the female reproductive lifespan [5]. The first step in recruitment is independent of the release of ovarian gonadotropins, and is characterized by the transition from primordial to growing (primary and preantral) follicles. Subsequent steps, leading to the eventual release of oocytes for fertilization, are instead controlled by gonadotropin-regulated cycle events [6]. Strict regulation of these processes is paramount to avoid premature exhaustion of the primordial follicle reservoir [5].
Lhx8 is a transcription factor involved in the morphogenesis of several organs. In the central nervous system, Lhx8 is necessary for the correct proliferation and development of cholinergic neurons [7]. Lhx8 also plays a critical role in regulating the epithelia-mesenchyme interactions necessary for palatal shelve fusion [8], and tooth morphogenesis [9]. In the ovary, Lhx8 is expressed in oocytes and indispensable for both the survival and the maturation of ovarian follicles [10]. When Lxh8 is ablated in mice, primordial follicles still form but do not progress beyond the primordial stage and soon disappear from the ovary, leaving it empty before puberty [10].
Recent in vitro analyses have identified TGATTG as the core of a putative LHX8 DNA-binding sequence and showed that LHX8 directly regulates the transcription of several genes including germ cell-specific Nobox [11]. The mechanism of action of Lhx8 and the cause of oocyte death in its absence have, however, been unresolved. Here, we present evidence that oocyte loss occurs primarily via autophagy when Lhx8 is ablated. We infer that autophagy is likely activated by failure of repair of meiosis-associated DNA damage during embryonic development, resulting in an end-stage fibrotic ovary. Our study shows evidence that in Lhx8-/- ovaries, mechanisms other than apoptosis, prominently including autophagy, are responsible for the decrease in germ cell numbers around and after birth, leading to the premature loss of the ovarian reserve.
Materials and methods
Animals
Lhx8 mutant mice were obtained from Dr Heiner Westphal at the National Institute of Child Health and Human Development (Bethesda, MD) [8], and were maintained on a mixed 129, C57BL/6, SF1 genetic background. Mice were euthanized ethically according to Animal Care and Use Committee (ACUC)-approved NIA Standard Operation Procedures.
RNA extraction and real-time PCR
Gonads were collected from pups at 0 and 7 days postnatum (P0 and P7). For each genotype, ovaries from three animals were processed, providing biological replicates. Total RNA was extracted from ovaries using a Precellys 24 homogenizer (Bertin Technologies, France), followed by an RNeasy isolation kit protocol (Qiagen). RNA concentration and purity were determined using Nanodrop ND-1000 spectrophotometer, and confirming ratios A260:230 and A260:280 greater than 1.8. RNA integrity was assessed using an Agilent BioAnalyzer. Complementary DNA was generated employing either the RT2 Nano PreAMP cDNA synthesis kit (SA Biosciences) or the Ovation Pico WTA Sytem V2 (Nugen) followed by affinity purification (Zymo Research). Real-time PCR was performed using an ABI 7300 real time PCR system (Applied Biosystems). RT2 Profiler PCR arrays were used for autophagy, apoptosis, DNA damage signaling pathway, extracellular matrix and adhesion molecules (Qiagen, Supplementary Table 1–4). Normalization of PCR arrays was done by scaling the average expression of oocyte-specific genes Vasa, Kit, and Sohlh1 (Qiagen PPM25712A, PPM05195A, and PPM39140A, respectively) to adjust for changes in germ cell number. The following Taqman probes (Applied Biosystems) were used: Spo11 (Mm0048876_m1), and Msx1 (Mm00440330_m1). Normalization of these real-time PCRs was done by scaling the expression of Sdha (Mm01352366_m1).
Western blotting
Ovaries were homogenized (Precellys 24, Bertin, France) and protein extracted in RIPA buffer supplemented with proteinase inhibitor cocktail tablets (Roche Diagnostic). Proteins were analyzed using the following antibodies: LC3B (Abcam, ab48394, dil 1:2000), β-actin (Bethyl, A300-491A, dil 1:2000). Band intensity analysis was performed using Molecular Imaging Software (ver. 4.0.5; Kodak).
Microarray expression profiling
For each genotype, gonads from three pups were separately processed. Total RNA was obtained, amplified, purified (as described above), and labeled for MouseWG-6 v2 Expression BeadChip arrays (Illumina).
Microarray data were analyzed using DIANE 6.0, a spreadsheet-based microarray analysis program based on SAS JMP7.0.
Raw microarray data were subjected to filtering by detection of P value and Z normalization; the data were further tested for significant changes as previously described [12]. Sample quality was assessed by analysis of scatter plots, principal component analysis, and gene sample z-score-based hierarchical clustering to exclude possible outliners. The ANOVA test was used to eliminate genes with larger variances (P ≥ 0.05) within each comparing group. Genes were determined to be differentially expressed after calculating the Z ratio, which indicates the fold difference between experimental groups, and false discovery rate (fdr), which controls for the expected proportion of false-rejected hypotheses. Individual genes with P value ≤ 0.05, absolute value of Z ratio≥ 1.5, and fdr≤ 0.3 were considered significantly changed. Hierarchical clustering/K-means clustering and principal components analysis were performed to identify clustering within groups. Array data for each experimental animal was also hierarchically clustered with Ilumina Bead Studio version 2.0. The Parameterized Analysis of Gene Enrichment (PAGE) algorithm was employed for gene set enrichment analysis using data for all of the genes in each sample as input against the data set supplied by the Gene Ontology (GO) Institute. For each relevant comparison, the lists of differentially expressed genes and Z ratios were entered into the PAGE Pathway Analysis software to organize them according to known biological pathways. The Enrichment z-scores for each functional grouping were calculated based on mRNA abundance changes (z-ratio) predicting these interactions and networks by z-test. The P value was calculated by comparing the number of user-specified genes of interest participating in a given function or pathway relative to the total number of occurrences of these genes in all functional/pathway annotations stored in the knowledge base. All of the pathways must have at least three genes found in the microarray gene set. The P value ≤ 0.05 and fdr ≤ 0.3 were the cutoff criteria for the significant pathway/GO selection. Significance of functions and pathways was calculated using the right-tailed Fisher exact test.
Histological preparations
Ovaries were placed in a solution of 4% paraformaldehyde in PBS, and fixed at 4°C for 1 h to overnight, depending on the developmental stage. After fixation, gonads were embedded in paraffin and sectioned at a thickness of 5 μm.
For immunofluorescence on ovary sections, we used the protocol previously described [13]. Briefly, following deparaffinization and rehydration of ovary sections, heat-mediated antigen retrieval was performed in a EMS-820 precision pulsed laboratory microwave oven (Electron Microscopy Sciences) by heating the slides in 10 mM citrate buffer, pH 6.0 at 90°C for 5 min. Sections were incubated with primary antibody overnight at 4°C. Incubation with Alexa Fluor secondary antibodies (Invitrogen) was of 30 min. The slides were finally incubated with DAPI (Dojindo Molecular Technologies) at room temperature for 15 min before mounting. Antibodies against LC3B (ab48394, dil 1:200), BECN1 (ab55878, dil 1:100), ATG7 (ab53255, dil 1:200), SYCP3 (ab15093, dil 1:400), CHEK1 (ab47574, dil 1:200), and phospho-γH2A (ab2893, dil 1:400) were from Abcam; LAMA1 (LS-C25114, dil 1:100) was from LifeSpan Biosciences; VASA (560189, dil 1:100) was from BD Pharmingen. Images visualized in a Deltavision fluorescence microscope (Applied Precision) and processed using Adobe Photoshop CS.
To identify apoptotic cells, we used the DeadEnd Fluorimetric TUNEL system (Promega) in combination with VASA staining to visualize germ cells. For each stage, ovaries from three gonads from different animals per genotype were analyzed for germ cell number. For each gonad, VASA-positive germ cells were counted on every fifth section and added to obtain an index of the total number of germ cells per gonad. The total numbers were then divided by the number of gonads at each stage to get an average value [14].
For connective tissue staining, we performed a Heidenhain's AZAN modification of Mallory's triple stain [15]. Briefly, after deparaffinization and rehydration, sections were incubated in azocarmine G solution for 20 min at 56°C. Differentiation was performed in aniline-ethanol, and sections were then treated with phosphotungstic acid solution for 1 h. Final staining with aniline blue-orange G solution was for 20–45 min before clearing and mounting.
Ovaries for electron microscopy were fixed for 2 h at room temperature in 4% formaldehyde+2% glutaraldehyde in 0.1 Cacodylate buffer, pH 7.2. The samples were then processed, embedded in plastic, and analyzed at the Optical Microscopy and Analysis Laboratory of the National Cancer Institute (Frederick MD).
Quantification of fluorescence intensity for CHEK1was done using the software ImageJ (imagej.nih.gov). The Corrected Total Cell Fluorescence (CTCF) was calculated by selecting the cell area and applying the following formula per developers’ recommendation: CTCF = integrated density – (area of selected cell × mean fluorescence of background readings).
Statistical analyses
For germ cell number and gene expression using real-time PCR, statistical analysis was performed by the unpaired t-test (GraphPad).
Results
Oocyte loss due to autophagy in Lhx8-/- mice
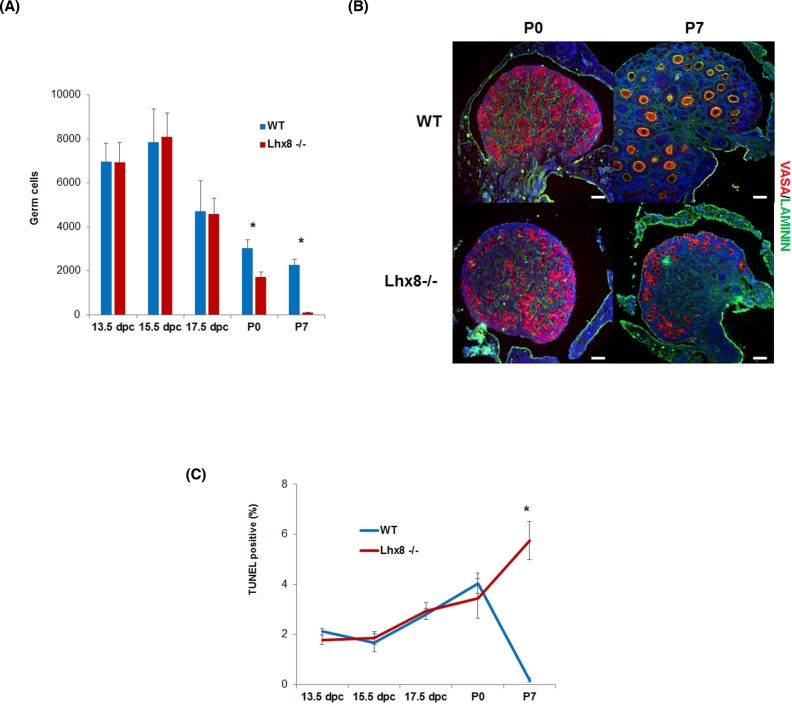
We established Lhx8 homozygous mutant offspring by crossing heterozygous mice (i.e., Lhx8+/– × Lhx8+/–). During embryo development, we saw no difference in survival between 13.5, 15.5, and 17.5 dpc. However, we confirmed the previous report that Lhx8 mutation was detrimental for viability after birth [8], and extended the analysis of survival to 21 days after birth (P21). Although Lhx8-/- mice were born alive at the expected Mendelian ratio, only 37%, 18%, and 6% of the Lhx8-/- mice that were born reached P7, P14, and P21, respectively (Supplementary Figure 1). Germ cells were not lost during embryonic development, and their number, estimated by quantitative morphometry by immunofluorescence staining of VASA, remained virtually the same in Lhx8-/- and wild-type embryos from 13.5 dpc (days postcoitum) through 17.5 dpc (Figure 1A). A sharp drop in germ cell number was evident; however, starting by P0, when ∼40% of germ cells had disappeared from Lhx8-/- ovaries compared to wild-type (Figure 1A). At P7, only sparse germ cells remained, mostly localized in the cortical region of the ovary (Figure 1B).
Figure 1.
Germ cell dynamics and survival in Lhx8-/- ovaries. (A) Germ cell numbers at 13.5, 15.5, 17.5 dpc, P0, and P7 in wild-type (blue) and Lhx8-/- (red) ovaries (n = 3 per genotype). Throughout embryonic development, there was no difference between wild-type and Lhx8-/- ovaries. However, the number of germ cells dropped dramatically in Lhx8-/- ovaries at birth. (B) Immunostaining for germ cell-specific VASA (red), and laminin (green) in wild-type and Lhx8-/- ovaries at P0, and P7, showing progressive loss of germ cells (the middle section of each ovary is displayed). Nuclei are stained blue with DAPI. Bar = 40 μm. (C) Percentage of TUNEL-positive germ cells in ovaries at 13.5, 15.5, 17.5, P0, and P7. Wild-type and Lhx8-/- ovaries showed no significant difference (n = 3 per genotype) from 13.5 dpc to P0. At P7, a larger but still limited number of positive germ cells were found in Lhx8-/- ovaries. Data in panels (A and C) are represented as mean values ± SEM. (*) P < 0.05, unpaired t-test. dpc = days post coitum; P0 = 0 dpn; P7 = 7 dpn; P21 = 21 dpn.
To better understand the involvement of Lhx8 ablation in oocyte death, we carried out immunofluorescence with Vasa antibody as a marker of germ cells and TUNEL assays to detect DNA fragmentation, a feature commonly associated with apoptosis. From 13.5 dpc to P0, time point at which a large fraction of oocytes had already been lost, Lhx8-/- ovaries showed no greater TUNEL positivity than the low level observed in wild-type controls (Figure 1C). By P7, the number of TUNEL-positive germ cells was appreciably higher in Lhx8-/- ovaries, though still relatively low (Figure 1C).
The results at P7 were consistent with previous findings suggesting that apoptosis was not the only process involved in the perinatal death of the germ cells [16]. To try to identify the mechanism responsible for oocyte loss by P7, we analyzed gene expression profiles of ovaries at P0 and P7. At P0, the vast majority of genes that were upregulated in Lhx8-/- ovaries were in the autophagy pathway (Table 1). These comprised genes involved in autophagosome formation, including Atg4c, Atg9b, and Atg12; genes involved in protein transport, such as Atg3 and Atg7; and regulatory genes such as Ctsb and Rb1. When we analyzed ovaries at P7, again many genes involved in autophagy, including Atg4a, Atg4c, Atg7, Ulk1, Ulk2, and Wipi1, were upregulated, concomitant with the dramatic reduction of ovarian reserve (Table 1). Although genes in the apoptosis pathway were not differentially expressed at P0, several were upregulated by P7, the stage of rapid oocyte demise. Both positive and negative regulators were augmented (Table 1 and Discussion). Positive regulators of apoptosis included Apaf1, Cidea, and Dfbb; negative regulators, Aft5 and Prdx2. In addition—and in line with the observation of TUNEL-positive cells at this developmental stage—a number of caspases were also upregulated, most significantly Casp9 and Casp14.
Table 1.
Genes involved in apoptosis and autophagy. Ratio of expression of genes involved in apoptosis and autophagy in P0 and P7 Lhx8-/- compared to wild-type ovaries (n = 3 per genotype). At P0, apoptotic genes did not show changes in expression.
| P0 | P7 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Apoptosis | Autophagy | Apoptosis | Autophagy | ||||||||
| Gene | Fold change | P value | Gene | Fold change | P value | Gene | Fold change | P value | Gene | Fold change | P value |
| Atg12 | 2.25 | 0.003 | Apaf1 | 10.46 | 0.002 | Atg12 | 6.32 | 0.007 | |||
| Atg3 | 1.95 | 0.003 | Aft5 | 13.83 | 0.006 | Atg4a | 11.62 | 0.005 | |||
| Atg4c | 1.80 | 0.002 | Bcl2l11 | 17.71 | 0.009 | Atg4c | 20.25 | 0.007 | |||
| Atg7 | 1.67 | 0.007 | Casp1 | 12.55 | 0.009 | Atg4d | 11.84 | <0.001 | |||
| Atg9b | 1.84 | 0.007 | Casp14 | 9.27 | 0.002 | Atg7 | 8.18 | 0.002 | |||
| Ctsb | 2.01 | 0.001 | Casp7 | 12.25 | 0.009 | Cdkn1b | 25.35 | 0.007 | |||
| Prkaa1 | 2.21 | 0.001 | Casp9 | 10.12 | 0.002 | Cln3 | 22.70 | 0.007 | |||
| Rb1 | 2.18 | 0.001 | Cidea | 6.11 | 0.005 | Ctsd | 14.25 | 0.005 | |||
| Tmem74 | 1.97 | 0.004 | Cideb | 17.99 | 0.005 | Cxcr4 | 19.71 | 0.000 | |||
| Dapk1 | 30.90 | 0.006 | Dapk1 | 54.52 | 0.006 | ||||||
| Dffb | 10.29 | 0.006 | Eif4g1 | 17.11 | 0.004 | ||||||
| Prdx2 | 10.73 | 0.006 | Hdac6 | 13.46 | 0.010 | ||||||
| Hspa8 | 22.60 | 0.007 | |||||||||
| Htt | 13.99 | 0.004 | |||||||||
| Irgm1 | 15.90 | 0.008 | |||||||||
| Pik3c3 | 13.21 | 0.009 | |||||||||
| Pik3cg | 9.51 | 0.004 | |||||||||
| Pik3r4 | 6.82 | 0.002 | |||||||||
| Prkaa1 | 16.96 | 0.007 | |||||||||
| Rgs19 | 15.91 | 0.008 | |||||||||
| Ulk1 | 19.85 | 0.006 | |||||||||
| Ulk2 | 22.44 | 0.005 | |||||||||
| Wipi1 | 18.67 | 0.006 | |||||||||
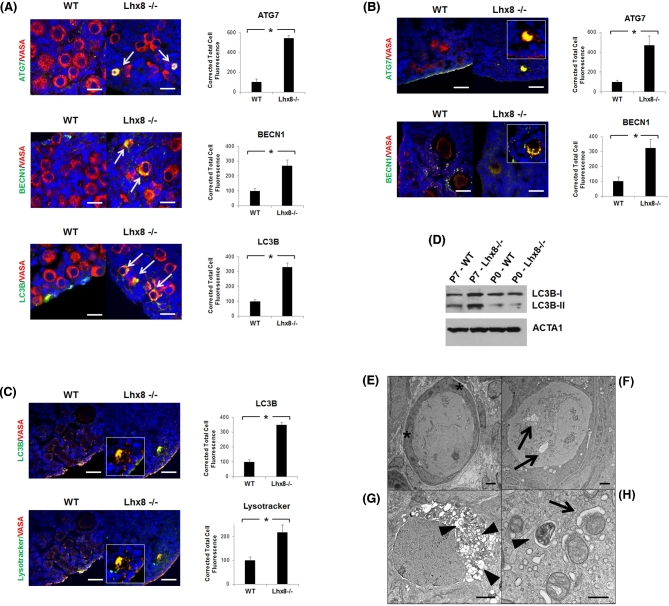
Immunofluorescence at P2 showed some Lhx8-/- oocytes positive for autophagic markers ATG7, BECN1, and LC3B (Figure 2A). At P7, when the majority of germ cells have been lost in Lhx8-/- ovaries, immunofluorescence confirmed that virtually all the remaining germ cells stained for the same autophagic markers. (Figure 2B–C). By contrast, wild-type ovaries were negative for such immunostaining. Notably, colocalization of lysosome-specific LysoTracker with LC3B, which is recruited to the autophagosome during autophagy, identified autolysosomes that result from the fusion of lysosomes and autophagic vesicles, suggesting that autophagy was ongoing. During autophagy, the soluble form of LC3B (i.e., LC3B-I) is converted into the membrane-bound LC3B-II, participating in the initiation of autophagosome formation [17]. Immunoblot analysis showed higher conversion of LC3B from LC3B-I into LC3B-II in P7 Lhx8-/- ovaries; the densitometric ratio of LC3B-I and LC3B-II in Lhx8-/- compared to wild-type ovaries was 2.21 and 2.66 respectively, consistent with the occurrence of autophagy and formation of autophagosomes when Lhx8 is ablated (Figure 2D).
Figure 2.
Germ cell loss results from autophagy and results in fibrotic ovaries. (A) Immunohistochemistry of P2 wild-type and Lhx8-/- ovaries for autophagy markers ATG7, BECN1, and LC3B showing first signs of autophagy activation (arrows). Cytoplasmic colocalization with VASA (red) results in yellow color, which is more evident at higher magnification. Nuclei are stained blue with DAPI. Bar = 20 μm. Quantification of fluorescence intensity for each autophagy marker is shown next to their images. Data are represented as mean values ± SEM. (*) P < 0.05. (B and C) Immunohistochemistry at P7 showed that the few remaining oocytes in Lhx8-/- ovaries were positive for the markers of autophagy. Quantification of fluorescence intensity for each autophagy marker is shown next to their images. Data are represented as mean values ± SEM. (*) P < 0.05. (C) Lysotracker, a stain for lysosomes, colocalized with LC3B and identified autolysosomes. VASA was used as an oocyte marker. Nuclei are stained blue with DAPI. Bar = 40 μm. Quantification of fluorescence intensity for LC3B and Lysotracker is shown next to their images. Data are represented as mean values ± SEM. (*) P < 0.05. (D) Western blot confirming an increased conversion of LC3B into the shorter LC3B-II form, associated with autophagy, in P7 Lhx8-/- ovaries compared to wild type. Actin was used as loading control. (E) Electron microscopy of P7 wild-type ovary shown as reference. Asterisks mark two of the flat granulosa cells surrounding the oocyte. (F-H) Electron microscopy of P7 Lhx8-/- ovary sections. Oocytes presenting several degrees of autophagy, ranging from a few autophagic vacuoles (F, arrows) to cytoplasm completely filled with autolysosomes (G, arrowheads). Bar = 2 μm. (H) Autophagy involving mitochondria, showing a mitochondrion in the early phase of engulfment by a nascent autophagosome (arrow), and another being digested in an autolysosome (arrowhead). Bar = 500 nm.
The morphology of dying oocytes in Lhx8-/- ovaries at P7 was further validated and characterized at higher resolution by electron microscopy. Imaging of ultrastructure demonstrated the presence of oocytes in various stages of autophagy, and again with no signs of apoptosis (Figure 2F–H). Interestingly, immunostaining of LC3B confirmed the appearance of the first autophagic oocytes at 17.5 dpc Lhx8-/- (Supplementary Figure 2). Although only a few oocytes were affected in each ovary at this stage, none were observed in wild-type ovaries. The results are thus consistent with the initiation of subsequent accelerated decline in oocyte cell count by P0. Taken together, these results suggested an initial activation of autophagy around the time of birth (evidenced by upregulation of autophagic factors), which then became more dramatic and morphologically evident at P7.
By P7, virtually all oocytes were undergoing autophagic degeneration, ranging from a few autophagic vacuoles (Figure 2F) to more severe instances in which the cytoplasm was filled with autophagosomes engulfing all intracellular structures (Figure 2G). In addition, activation of the autophagic pathway eventually led to mitophagy (i.e., elimination of mitochondria by autophagy), as evidenced by morphological ultrastructure in electron micrographs (Figure 2H).
Pathways altered during germ cell loss
Expression profiling of ovaries suggested possible mechanisms that might account for the induction of cell death and autophagy. Microarray analysis at P0 and P7 further confirmed that the most downregulated genes were oocyte-specific. These included Nrlp14, Zp3, Oosp1, Bmp15, Fbxw15 [18], H1foo [19], Zp1, and Gdf9 (Supplementary Table 5). As expected, GO classification scored “Reproduction” and “Reproductive Process” as highly downregulated biological processes in Lhx8-/- ovaries at P0 and P7, with a combined fraction of downregulated pathway genes of 71% and 76%, respectively (Supplementary Table 6). Only a small number of genes in these GO classes were upregulated, and those have also been implicated in other processes observed in Lhx8-/- ovaries. They included genes involved in extracellular matrix remodeling (Adamts2); autophagy (S100a11) [20]; mitochondrial fragmentation (Ggnbp1) [21]; and response to stress (regulator of G-protein signaling 2 [Rgs2]) [22].
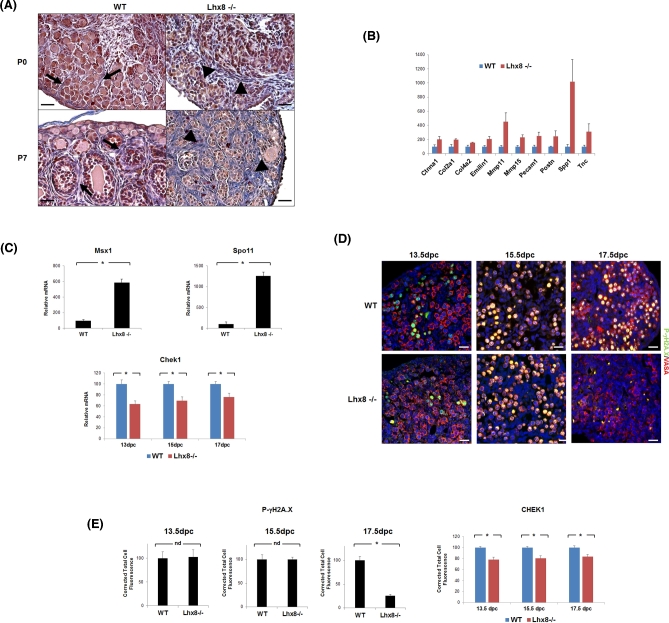
Although GO classes that were affected in Lhx8-/- ovaries were largely downregulated, a few GO classes were upregulated, especially including “Extracellular Matrix” and “Extracellular Matrix Part.” The genes in these two classes jointly accounted for 65 upregulated and only 13 downregulated genes at P7 (Supplementary Table 6). Consistent with remodeling and severe fibrosis accompanying reduction of follicle numbers and autophagy, the upregulated genes included numerous collagen and matrix metallopeptidases genes involved in fibrosis—among them Col1a1, Col3a1, Col4a1, Col4a4, Col4a5, Col5a1, Col6a1, Col6a2, Col6a3, Col15a1, Col8a1, Col16a1, Col23a1, Mmp2, Mmp11, Mmp14, Mmp15, and Mmp23 [23]. Direct gene expression analyses and ovary section staining further validated these results. When Heidenhain's AZAN modification of Mallory's triple stain was used to visualize the connective component of the gonad (Figure 3A), the connective tissue in wild-type newborn ovaries stained faintly (blue stain), revealing an organized structure of thin collagen fibers surrounding clusters of oocytes and primordial follicles. At P7, collagen was more evident and surrounded developing follicles. By contrast, collagen fibers were dramatically overexpressed at P0 in Lhx8-/- ovaries, especially closer to the medulla, and were even more pervasive and intensely stained by P7, when the number of germ cells had greatly decreased and the remaining follicular structure was completely disorganized. These results thus showed connective tissue replacing oocytes and follicles in Lhx8-/- mice, leading to fibrotic ovaries. Additionally, real-time PCR of extracellular membrane (ECM) genes in newborn ovaries confirmed the upregulation of tested genes, including Col2a1, Col4a2, Mmp11, Mmp15, Postn, and Tnc—all of which are involved in ECM remodeling and fibrinogenesis (Figure 3B) [24,25].
Figure 3.
Fibrosis and impaired DNA damage repair in Lhx8-/- ovaries. (A) Visualization of fibrosis by Heidenhain's AZAN modification of Mallory's triple stain. Cells are stained red and collagen fibers blue. Basement membrane in wild-type ovaries at P0 and P7 comprises thin layers surrounding clusters of germ cells (at P0, arrows) and follicles (at P7, arrows). Lhx8-/- ovaries at both P0 and increasingly at P7 showed signs of fibrosis as germ cells were replaced by thick sheets of collagen (arrowheads). Bar = 30 μm. (B) Expression of marker genes involved in ECM remodeling and fibrinogenesis in P0 wild-type and Lhx8-/- ovaries (n = 3 per genotype). Data for the extracellular matrix and adhesion molecules RT2 PCR array are represented as mean values ± SEM. P < 0.05 for all genes. (C) Expression of meiosis marker genes Msx1 and Spo11 in P0 wild-type and Lhx8-/- ovaries, and DNA damage repair marker gene Chek1 at 13.5, 15.5, and 17.5 dpc (n = 3 per genotype). Data are represented as mean values ± SEM. (*) P < 0.05. (D) Immunohistochemistry of wild-type and Lhx8-/- ovaries at 13.5, 15.5, and 17.5 dpc for phospho-γH2A.X (green) and VASA (red). Stages 13.5 and 15.5 dpc show no difference between wild-type and Lhx8-/- ovaries. However, phospho-γH2A.X is selectively not expressed in Lhx8-/- ovaries at 17.5 dpc. Nuclei are stained blue with DAPI. Bar = 20 μm. (E) CHEK1 and phospho-γH2A.X expression in wild-type and Lhx8-/- ovaries at 13.5, 15.5, and 17.5 dpc. Fluorescence intensity measurements showed a downregulation of CHEK1 in Lhx8-/- ovaries throughout all assayed embryonic stages, whereas phospho-γH2A.X is downregulated starting from 17.5 dpc. Data are represented as mean values ± SEM. (*) P < 0.05.
Possibly, more proximal to the direct fate of oocytes, genes in DNA damage repair pathways were also significantly downregulated by P0. The genes included Atr and Rad1, which detect DNA insults [26–28]; Chek1 and Brca2, downstream targets involved in mediating and transducing damage signals [29,30]; and Rad18, Xrcc6, and Pold3, required for repair of several types of DNA damage [31–33]. Real-time PCR assays confirmed the downregulation of DNA damage repair markers and extended results to additional genes including Fancc, H2afx, Mlh3, Rad51c, and Xrcc1 (Table 2) [34–38]. At P0 and P7, of 84 genes tested belonging to the DNA damage repair pathway, a total of 34 (40%) and 39 (46%) genes respectively were found significantly dysregulated (Table 2). Indeed, two genes involved in meiosis initiation and induction of DNA breaks during recombination, Msx1 and Spo11, were upregulated in Lhx8-/- ovaries at P0 (Figure 3C).
Table 2.
Genes involved in DNA damage repair. Ratio of expression of genes involved in DNA damage repair pathway in P0 and P7 Lhx8-/- compared to wild-type ovaries (n = 3 per genotype).
| P0 | P7 | ||||
|---|---|---|---|---|---|
| Gene | Fold change | P value | Gene | Fold change | P value |
| Brca2 | –1.77 | 0.021 | Apex1 | –2.16 | 0.031 |
| Chek1 | –2.37 | 0.003 | Brca1 | –1.82 | 0.030 |
| Ercc1 | –2.58 | 0.001 | Brca2 | –2.89 | 0.013 |
| Fancc | –2.74 | 0.018 | Chaf1a | –3.03 | 0.016 |
| Gadd45a | –1.70 | 0.007 | Chek1 | –5.75 | 0.032 |
| Gtf2h2 | –2.00 | 0.011 | Dclre1a | –2.02 | 0.001 |
| H2afx | –1.51 | 0.037 | Ercc1 | –1.41 | 0.096 |
| Lig1 | –1.67 | 0.032 | Exo1 | –3.55 | 0.012 |
| Mgmt | –3.60 | 0.007 | Fancc | –1.78 | 0.001 |
| Mlh3 | –2.44 | 0.015 | Gtf2h1 | –1.59 | 0.021 |
| Msh2 | –2.25 | 0.001 | Gtf2h2 | –2.09 | 0.007 |
| Msh3 | –1.94 | 0.014 | Hus1 | –2.10 | 0.021 |
| Ogg1 | –2.26 | <0.001 | Lig1 | –1.65 | 0.027 |
| Parp1 | –2.03 | 0.000 | Mbd4 | –1.43 | 0.024 |
| Parp2 | –2.25 | 0.005 | Mgmt | –3.38 | 0.021 |
| Pinx1 | –3.38 | 0.006 | Mif | –2.33 | 0.020 |
| Pold1 | –1.72 | 0.016 | Msh3 | –1.40 | 0.024 |
| Pold3 | –2.29 | 0.026 | Mutyh | –2.33 | 0.005 |
| Rad1 | –2.19 | 0.011 | Nthl1 | –2.34 | 0.001 |
| Rad18 | –1.78 | 0.009 | Pinx1 | –3.98 | <0.001 |
| Rad21 | –1.75 | 0.026 | Pms1 | –1.63 | 0.015 |
| Rad23a | –1.94 | 0.001 | Pold1 | –2.58 | 0.006 |
| Rad51c | –2.00 | 0.015 | Pold3 | –2.80 | 0.004 |
| Rad9 | –2.10 | 0.012 | Pole | –2.59 | 0.019 |
| Rad9b | –1.58 | 0.047 | Polk | –1.64 | 0.011 |
| Rbbp4 | –1.76 | 0.001 | Rad1 | –2.58 | 0.011 |
| Smc1a | –1.34 | 0.029 | Rad17 | –1.75 | 0.019 |
| Smc3 | –1.55 | 0.021 | Rad18 | –2.07 | 0.002 |
| Terf1 | –2.09 | 0.027 | Rad51l1 | –1.67 | 0.004 |
| Trp53 | –1.93 | <0.001 | Srd5a2 | 22.60 | 0.001 |
| Xrcc1 | –3.08 | <0.001 | Tdg | –1.19 | 0.024 |
| Xrcc3 | –1.45 | 0.034 | Terf1 | –2.00 | 0.050 |
| Xrcc6 | –5.94 | 0.001 | Tnp1 | 5.90 | 0.011 |
| Xrn2 | –1.50 | 0.047 | Wrnip1 | –1.63 | 0.017 |
| Xpa | –1.60 | 0.024 | |||
| Xpc | –2.09 | 0.038 | |||
| Xrcc1 | –2.95 | 0.026 | |||
| Xrcc3 | –1.45 | 0.011 | |||
| Xrcc6 | –2.61 | 0.032 | |||
To test whether the changes in repair-related genes were truly associated with an impairment of DNA damage repair, we analyzed embryonic ovaries at the time of meiosis and performed immunohistochemistry staining for the phosphorylated form of γH2A.X, which is generated in response to DNA damage and is required for subsequent efficient repair. In wild-type ovaries at 13.5 (when meiosis starts), 15.5, and 17.5 dpc, the majority of oocytes were positive for γH2A.X as expected (Figure 3D). In Lhx8-/- samples, however, γH2A.X positivity dramatically decreased by 17.5 dpc (Figure 3D–E), consistent with reduced or defective DNA damage recognition and repair.
To exclude the alternative possibility that the different pattern of γH2A.X expression was due to a delay or a different rate of meiotic division in Lhx8-/- mice, we assessed progression through meiosis by immunostaining the synaptonemal complexes with anti-SCP3 antibody. Comparison of wild-type and Lhx8-/- ovaries showed a similar proportion of SCP3-positive oocytes between 13.5 and 17.5 dpc (Supplementary Figure 3). However, gene expression analysis by microarray and real-time PCR showed a downregulation of Chek1, which was confirmed by quantification of immunofluorescence staining (Figure 3E). CHEK1—a member of the DNA damage checkpoint pathway—is involved in the initiation of DNA damage response, and is indispensable for the regulation of meiosis. These results were consistent with a normal rate of progression through meiosis, but with deficient DNA damage repair as an early, and possibly critical, insult in Lhx8-/- oocytes.
Discussion
Activation of autophagy in Lhx8-/- follicles
A previous study reported germ cell death in Lhx8-/- mice that seemed independent of apoptosis [10], but did not address other possible pathways. We now attribute the inexorable loss of germ cells and failure of ovarian follicle formation in the absence of Lhx8 to autophagy rather than apoptosis, starting around the time of birth. Some apoptosis was seen in wild-type controls with rates and trend comparable to those previously reported [39], and was similar in Lhx8-/- ovaries up to the time of birth. As for activation of autophagy, it has been previously noted as an additional mechanism affecting the establishment of the ovarian reserve in wild-type mice, especially when placental nutrient levels decrease following birth [16]. In a comparable way, in absence of Lhx8, severe reduction in germ cell numbers could be accounted for by autophagic processes. Activation of autophagy started around P0, initially by upregulation of genes involved in the formation and function of autophagic vacuoles. This was soon followed by the execution of the autophagic program, as shown by increased levels of autophagic proteins and morphological appearance of germ cells positive for autophagic markers such as LC3B, BECN1, and ATG7. Moreover, electron microscopy confirmed that the few oocytes remaining in the ovary at P7 were undergoing autophagy, ranging from a few autophagic vacuoles to autophagosomes completely filling the cytoplasm (Figure 2B–D). Notably, no sign of autophagy was seen in somatic cells either by immunofluorescence staining of autophagic markers or by ultrastructural examination using electron microscopy. This specificity of cell death to the oocytes is consistent with the specific expression of Lhx8 in the germ cell compartment of the ovary. By P7, the majority of remaining oocytes were dramatically filled with autophagosomes, which could account for the greater conversion of LC3B-I into LC3B-II even in the presence of a smaller remaining pool of germ cells.
The precise event that determines whether apoptosis or autophagy will initiate first or at all remains unknown. However, recent studies have shown that although factors belonging to apoptosis and autophagy pathways can interact in cross talk that decides the fate of the cell [40,41], autophagy can precede or induce caspase cascade and apoptosis [42–44]. Our findings in Lhx8-/- ovaries are consonant, with genes of the autophagy pathway upregulated before any in the apoptosis pathway (Table 1). Thus, we saw only small numbers of TUNEL-positive oocytes until P0, suggesting that apoptosis at the time of birth is limited compared to the autophagy that was demonstrably dominant at P7.
The striking upregulation of ECM genes and deposition of fibrous tissue could then be a consequence of germ cell loss, with connective tissue expanding to fill the space previously occupied by follicles. However, ECM factors have also been directly associated with the regulation of induction of autophagy [45 and reviewed by 46], suggesting the possibility of a more active role in the overall process in Lhx8-/- ovaries.
DNA damage and autophagy
With a possible link to initiation of autophagy, oocytes sustain double-strand breaks during meiotic recombination, beginning around 13.5 dpc [47], when Lxh8 expression has been detected in oocytes [48]. Consistent with its function in meiosis, in Lhx8-/- ovaries we observed dysregulation of Msx1, a gene involved in promoting meiosis [49] and identified as a putative target of LHX8 [11] (Figure 2G), and another initiator of meiosis, Stra8, was earlier reported to be upregulated [10]. Furthermore, SPO11, directly involved in the induction of DNA breaks for recombination during meiosis [50], was also dysregulated at P0 in Lhx8-/- ovaries (Figure 2G); thus, significant DNA damage was still ongoing at that stage.
As further evidence of relevant changes in DNA damage dynamics in the Lhx8-/- ovaries, of 84 genes belonging to this pathway that were tested by real-time PCR, 40% and 46% were dysregulated at P0 and P7, respectively. Finally, immunostaining of phospho-γH2A.X showed a consistent pattern. There was no significant difference in expression between wild-type and Lhx8-/- ovaries during early stages of development (Figure 3C), in accord with the finding that germ cell numbers were virtually the same in these ovaries (Figure 1A). But in contrast to persistent significant staining of wild-type at 17.5 dpc, phospho-γH2A.X, which is required for the assembly of DNA repair factors at the damaged sites, was undetectable in Lhx8-/- ovaries (Figure 3C). Additionally, despite normal progression of meiosis (as evidenced by SYCP3 staining), reduced production of CHEK1 added to the downregulation of genes of the DNA damage signaling pathway, suggesting an impairment in activation of the DNA checkpoint and damage repair. The activation of CHEK1 initiates a cascade of events responsible for cell cycle arrest and DNA repair [51]; and in cell culture experiments, CHEK1 has been found actively participating in the survival mechanism triggered by autophagy in response to DNA damage [52]. Taken together, these results suggest that endogenous double-strand breaks occur normally in Lhx8-/- oocytes, as in their wild-type counterparts, starting from the initiation of meiotic recombination at 13.5 dpc. However, in the Lhx8-/- oocytes, these DNA insults are not properly resolved later on during embryonic development.
Disruption of the DNA damage response has also been shown to induce degradation of repair proteins by autophagy [53]. We infer that when LHX8-mediated transcription fails in meiotic oocytes, accumulated DNA damage becomes a plausible candidate for the decisive defect initiating autophagy.
The amount of DNA damage is affected by the continuing entrance of oocytes into meiosis in successive waves during embryonic and perinatal development until they arrest at the prophase of meiosis I (dictyate stage) by P5 [54]. The DNA repair capacity of oocytes is correspondingly elevated starting at P0 during the dictyate stage [55], but DNA damage resistance decreases in the dictyate state [56], so that oocytes in primordial follicles become very sensitive to DNA damage and highly prone to cell elimination if DNA damage is not repaired [57, 58]. This is indeed consistent with what we observed in the present study, i.e., early loss of oocytes at the primordial follicle stage when Lhx8 is ablated. Interestingly, a recent study showed that conditional ablation of Lhx8 at the primordial follicle stage led to oocyte loss in primordial follicles, whereas knockout of Lhx8 in oocytes of primary follicles resulted in the death of primary follicles [59], suggesting that oocyte loss is likely to occur at the follicular stage when Lhx8 is knocked out. This is also consistent with our hypothesis that LHX8 might be required to maintain the integrity of DNA in the oocytes, because repair of DNA breaks induced during prophase I continues until meiosis I has been completed—a process that begins with the resumption of meiosis at puberty [47]. Therefore, deleting Lhx8 at different stages of folliculogenesis may result in oocyte death from the point at which such ablation occurs.
The hypothesis of DNA damage/autophagy/death and its further testing
A growing number of observations to date are consistent with the hypothesis that impairment of the DNA repair machinery in Lhx8-/- oocytes would increase autophagy, leading to the degradation of DNA damage repair enzymes that include CHEK1. In the absence of Lhx8, the inadequate repair of DNA damage would occur during and following meiosis—the time when normally Lhx8 begins to be expressed—a process that precedes primordial follicle formation and correlates with the substantial loss of germ cells that would become significant by P0 (Figure 1A). In other words, we propose that when Lhx8 is ablated, repair mechanisms are disarmed and unrepaired DNA damage activates massive autophagy, leading to sterile fibrotic ovaries.
It is relevant that autophagy, a major adaptive responses to stress, in fact promotes survival in response to DNA damage [60–65]. Thus, autophagy might be initiated not as programmed cell death but as a survival mechanism. It could possibly prolong the time necessary to check and eventually repair the DNA damage necessary for meiosis completion. Instead, if DNA damage persists and autophagy becomes pervasive and/or protracted, the oocytes in primordial follicles would die; and resultant loss of follicles would be followed by the observed progressive fibrosis.
To test this hypothesis, further work will be required to characterize better the pathways involving LHX8 in germ cell and ovary development. It could be informative to determine how Lhx8 itself is regulated. In other systems, FGF8 and WNT/b-catenin signaling were shown to regulate LHX8 [66], but this has not been investigated in the ovary. It would also be of interest to determine the extent to which genes of the autophagy or DNA damage repair pathways are directly regulated by LHX8.
Relevance to human reproduction of the process observed in mice also remains to be determined. Although the time scale is obviously different, and some events do not occur during the same developmental stage (for example, primordial follicle formation occurs in utero in humans) or are conceptually distinct (mice do not undergo proper menopause), the dynamics of follicle development in mice and humans show many similarities. For example, germ cell cyst breakdown, primordial follicle assembly, and follicle maturation are superficially comparable and many—though not all—genes regulating these processes are orthologous. Studies conducted on Caucasian and Korean populations did not find LHX8 variants associated with premature ovarian insufficiency, in which menopause occurs before age 40 [67, 68], but the contribution of LHX8 to fertility and premature menopause beyond 40 years of age has yet to be assessed.
Supplementary data
Supplementary data are available at BIOLRE online.
Supplementary Figure 1. Survival of Lhx8-/- mice. Newborn mice from each litter were counted and their survival followed up to P21. Data show the total number of pups and the number of Lhx8-/- mice per litter (n = 25). At P0, Lhx8-/- mice are born alive and at the expected Mendelian ratio of 25%. However, their number decrease over time and only 6% reach P21.
Supplementary Figure 2. First occurrence of autophagy in Lhx8-/- ovaries at 17.5 dpc. Immunohistochemistry of wild-type and Lhx8-/- ovaries at 17.5 dpc for autophagy marker LC3B (green) with VASA (red) as an oocyte marker. Although rare at this stage of development, a few oocytes begin to show positive immunostaining for LC3B (arrows, superimposition with VASA results in yellow color). Conversely, wild-type ovaries are negative. Nuclei are stained blue with DAPI. Bar = 20 μm.
Supplementary Figure 3. SYCP3 expression through embryonic development of wild-type and Lhx8-/- oocytes. Immunohistochemistry of wild-type and Lhx8-/- ovaries at 13.5, 15.5, and 17.5 dpc for SYCP3 (green) and VASA (red). SYCP3 staining shows oocytes at similar stages of meiosis in both wild-type and Lhx8-/- ovaries. Nuclei are stained blue with DAPI. Bar = 10 μm.
Supplementary Table 1. Assessed genes involved in autophagy pathway. List of 84 genes in autophagy pathway RT2 Profiler PCR array.
Supplementary Table 2. Assessed genes involved in apoptosis pathway. List of 84 genes in apoptosis pathway RT2 Profiler PCR array.
Supplementary Table 3. Assessed genes involved in DNA damage signaling pathway. List of 84 genes in DNA damage signaling pathway RT2 Profiler PCR array.
Supplementary Table 4. Assessed ECM and adhesion molecules genes. List of 84 genes belonging to extracellular matrix and adhesion molecules RT2 Profiler PCR array.
Supplementary Table 5. Top 20 downregulated genes in Lhx8-/- ovaries. Most of the top 20 downregulated genes in P7 Lhx8-/- ovaries by microarray analysis are oocyte-specific (asterisks).
Supplementary Table 6. Gene Ontology (GO) of genes dysregulated in Lhx8-/- ovaries. GO classes of Lhx8-/- ovaries by microarray analysis at P7, showing the number of total genes (Genes), upregulated (Genes Up) and downregulated (Genes Down) genes, and the zscore (ClassScore). Classes involving reproduction (i.e., “reproduction,” “reproductive process”) are among the top ones downregulated, whereas those involving the extracellular matrix (including “extracellular matrix,” “extracellular matrix part,” “extracellular region”) are among the top ones upregulated.
Acknowledgments
We thank the NIA Comparative Medicine Section for service with breeding, genotyping, and handling of the mice analyzed here. We are grateful to H. Westphal for the provision of the Lhx8-/- mice and to E. Lehrmann and Y. Zhang for technical assistance with the microarray experiments.
Footnotes
Grant Support: This work was supported entirely by the Intramural Research Program of the NIH, National Institute on Aging.
References
- 1. Reynaud K, Driancourt MA. Oocyte attrition. Mol Cell Endocrinol 2000; 163:101–108. [DOI] [PubMed] [Google Scholar]
- 2. Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol 2001; 234:339–351. [DOI] [PubMed] [Google Scholar]
- 3. Pepling ME. Follicular assembly: mechanisms of action. Reproduction 2012; 143:139–149. [DOI] [PubMed] [Google Scholar]
- 4. Gallardo TD, John GB, Shirley L, Contreras CM, Akbay EA, Haynie JM, Ward SE, Shidler MJ, Castrillon DH. Genomewide discovery and classification of candidate ovarian fertility genes in the mouse. Genetics 2007; 177:179–194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Elvin JA, Matzuk MM. Mouse models of ovarian failure. Rev Reprod 1998; 3:183–195. [DOI] [PubMed] [Google Scholar]
- 6. McGee EA, Hsueh AJ. Initial and cyclic recruitment of ovarian follicles. Endocr Rev 2000; 21:200–214. [DOI] [PubMed] [Google Scholar]
- 7. Zhao Y, Marin O, Hermesz E, Powell A, Flames N, Palkovits M, Rubenstein JL, Westphal H. The LIM-homeobox gene Lhx8 is required for the development of many cholinergic neurons in the mouse forebrain. Proc Natl Acad Sci USA 2003; 100:9005–9010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhao Y, Guo YJ, Tomac AC, Taylor NR, Grinberg A, Lee EJ, Huang S, Westphal H. Isolated cleft palate in mice with a targeted mutation of the LIM homeobox gene lhx8. Proc Natl Acad Sci USA 1999; 96:15002–15006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grigoriu M, Tucker AS, Sharpe PT, Pachnis V. Expression and regulation of Lhx6 and Lhx7, a novel subfamily of LIM homeodomain encoding genes, suggests a role in mammalian head development. Development 1998; 125:2063–2074. [DOI] [PubMed] [Google Scholar]
- 10. Choi Y, Ballow DJ, Xin Y, Rajkovic A. Lim Homeobox Gene, Lhx8, Is Essential for Mouse Oocyte Differentiation and Survival1. Biol Reprod 2008; 79:442–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Park M, Jeon S, Jeong JH, Park M, Lee DR, Yoon TK, Choi DH, Choi Y. Identification and characterization of lhx8 DNA binding elements. Dev Reprod 2012; 16:379–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cheadle C, Vawter MP, Freed WJ, Becker KG. Analysis of microarray data using Z score transformation. J Mol Diagn 2003; 5:73–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pelosi E, Omari S, Michel M, Ding J, Amano T, Forabosco A, Schlessinger D, Ottolenghi C. Constitutively active Foxo3 in oocytes preserves ovarian reserve in mice. Nat Comms 2013; 4:1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Tilly JL. Ovarian follicle counts–not as simple as 1, 2, 3. Reprod Biol Endocrinol 2003; 1:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Brown GG. An Introduction to Histotechnology. New York: Appleton-Century-Crofts; 1978:39–51. [Google Scholar]
- 16. Rodrigues P, Limback D, McGinnis LK, Plancha CE, Albertini DF. Multiple mechanisms of germ cell loss in the perinatal mouse ovary. Reproduction 2009; 137:709–720. [DOI] [PubMed] [Google Scholar]
- 17. Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol 2008; 445:77–88. [DOI] [PubMed] [Google Scholar]
- 18. De La Chesnaye E Kerr B, Paredes A, Merchant-Larios H, Méndez JP, Ojeda SR. Fbxw15/Fbxo12J is an F-box protein-encoding gene selectively expressed in oocytes of the mouse ovary. Biol Reprod 2008; 78:714–725. [DOI] [PubMed] [Google Scholar]
- 19. Hayakawa K, Ohgane J, Tanaka S, Yagi S, Shiota K. Oocyte-specific linker histone H1foo is an epigenomic modulator that decondenses chromatin and impairs pluripotency. Epigenetics 2012; 7:1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ghislat G, Knecht E. New Ca2+-dependent regulators of autophagosome maturation. Commun Integr Biol 2012; 5:308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Aihara T, Nakamura N, Honda S, Hirose S. A novel potential role for gametogenetin-binding protein 1 (GGNBP1) in mitochondrial morphogenesis during spermatogenesis in mice. Biol Reprod 2009; 80:762–770. [DOI] [PubMed] [Google Scholar]
- 22. Nguyen CH, Zhao P, Sobiesiak AJ, Chidiac P. RGS2 is a component of the cellular stress response. Biochem Biophys Res Commun 2012; 426:129–134. [DOI] [PubMed] [Google Scholar]
- 23. Pardo A, Selman M. Matrix metalloproteases in aberrant fibrotic tissue remodeling. Proc Am Thorac Soc 2006; 3:383–388. [DOI] [PubMed] [Google Scholar]
- 24. Naik PK, Bozyk PD, Bentley JK, Popova AP, Birch CM, Wilke CA, Fry CD, White ES, Sisson TH, Tayob N, Carnemolla B, Orecchia P et al. . Periostin promotes fibrosis and predicts progression in patients with idiopathic pulmonary fibrosis. Am J Physiol Lung Cell Mol Physiol 2012; 303:L1046–L1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. El-Karef A, Yoshida T, Gabazza EC, Nishioka T, Inada H, Sakakura T, Imanaka-Yoshida K. Deficiency of tenascin-C attenuates liver fibrosis in immune-mediated chronic hepatitis in mice. J Pathol 2007; 211:86–94. [DOI] [PubMed] [Google Scholar]
- 26. Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev 2000; 14:397–402. [PMC free article] [PubMed] [Google Scholar]
- 27. Lehmann AR, Hoeijmakers JH. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol 2000; 10:479–482. [DOI] [PubMed] [Google Scholar]
- 28. Lindsey-Boltz LA, Bermudez VP, Hurwitz J, Sancar A. Purification and characterization of human DNA damage checkpoint Rad complexes. Proc Natl Acad Sci USA 2001; 98:11236–11241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saintigny Y, Delacôte F, Varès G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J 2001; 20:3861–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moynahan ME, Pierce AJ, Jasin M. BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 2001; 7:263–272. [DOI] [PubMed] [Google Scholar]
- 31. Watanabe K, Iwabuchi K, Sun J, Tsuji Y, Tani T, Tokunaga K, Date T, Hashimoto M, Yamaizumi M, Tateishi S. RAD18 promotes DNA double-strand break repair during G1 phase through chromatin retention of 53BP1. Nucleic Acids Res 2009; 37:2176–2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Goedecke W, Eijpe M, Offenberg HH, van Aalderen M, Heyting C. Mre11 and Ku70 interact in somatic cells, but are differentially expressed in early meiosis. Nat Genet 1999; 23:194–198. [DOI] [PubMed] [Google Scholar]
- 33. Longley MJ, Pierce AJ, Modrich P. DNA polymerase delta is required for human mismatch repair in vitro. J Biol Chem 1997; 272:10917–10921. [DOI] [PubMed] [Google Scholar]
- 34. Niedzwiedz W, Mosedale G, Johnson M, Ong CY, Pace P, Patel KJ. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell 2004; 15:607–620. [DOI] [PubMed] [Google Scholar]
- 35. Paull TT, Rogakou EP, Yamazaki V, Kirchgessner CU, Gellert M, Bonner WM. A critical role for histone H2AX in recruitment of repair factors to nuclear foci after DNA damage. Curr Biol 2000; 10:886–895. [DOI] [PubMed] [Google Scholar]
- 36. Lipkin SM, Wang V, Jacoby R, Banerjee-Basu S, Baxevanis AD, Lynch HT, Elliott RM, Collins FS. MLH3: a DNA mismatch repair gene associated with mammalian microsatellite instability. Nat Genet 2000; 24:27–35. [DOI] [PubMed] [Google Scholar]
- 37. Sigurdsson S, Van Komen, S, Bussen, W, Schild D, Albala JS, Sung P. Mediator function of the human Rad51B-Rad51C complex in Rad51/RPA-catalyzed DNA strand exchange. Genes Dev 2001; 15:3308–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Brem R, Hall J. XRCC1 is required for DNA single-strand break repair in human cells. Nucleic Acids Res 2005; 33:2512–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pepling ME, Spradling AC. Mouse ovarian germ cell cysts undergo programmed breakdown to form primordial follicles. Dev Biol 2001; 234:339–351. [DOI] [PubMed] [Google Scholar]
- 40. Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8:741–752. [DOI] [PubMed] [Google Scholar]
- 41. Djavaheri-Mergny M, Maiuri MC, Kroemer G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010; 29:1717–1719. [DOI] [PubMed] [Google Scholar]
- 42. González-Polo RA, Boya P, Pauleau AL, Jalil A, Larochette N, Souquère S, Eskelinen EL, Pierron G, Saftig P, Kroemer G. The apoptosis/autophagy paradox: autophagic vacuolization before apoptotic death. J Cell Sci 2005; 118:3091–3102. [DOI] [PubMed] [Google Scholar]
- 43. Espert L, Denizot M, Grimaldi M, Robert-Hebmann V, Gay B, Varbanov M, Codogno P, Biard-Piechaczyk M. Autophagy is involved in T cell death after binding of HIV-1 envelope proteins to CXCR4. J Clin Invest 2006; 116:2161–2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scott RC, Juhász G, Neufeld TP. Direct induction of autophagy by Atg1 inhibits cell growth and induces apoptotic cell death. Curr Biol 2007; 17:1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pratt J, Roy R, Annabi B. Concanavalin-A-induced autophagy biomarkers requires membrane type-1 matrix metalloproteinase intracellular signaling in glioblastoma cells. Glycobiology 2012; 22:1245–1255. [DOI] [PubMed] [Google Scholar]
- 46. Neill T, Schaefer L, Iozzo RV. Instructive roles of extracellular matrix on autophagy. Am J Pathol 2014; 184:2146–2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Cohen PE, Pollack SE, Pollard JW. Genetic analysis of chromosome pairing, recombination, and cell cycle control during first meiotic prophase in mammals. Endocr Rev 2006; 27:398–426. [DOI] [PubMed] [Google Scholar]
- 48. Zhang LJ, Pan B, Chen B, Zhang XF, Liang GJ, Feng YN, Wang LQ, Ma JM, Li L, Shen W. Expression and epigenetic dynamics of transcription regulator Lhx8 during mouse oogenesis. Gene 2012; 506:1–9. [DOI] [PubMed] [Google Scholar]
- 49. Le Bouffant R, Souquet B, Duval N, Duquenne C, Hervé R, Frydman N, Robert B, Habert R, Livera G. Msx1 and Msx2 promote meiosis initiation. Development 2011; 138:5393–5402. [DOI] [PubMed] [Google Scholar]
- 50. Mahadevaiah SK, Turner JM, Baudat F, Rogakou EP, de Boer P, Blanco-Rodríguez J, Jasin M, Keeney S, Bonner WM, Burgoyne PS. Recombinational DNA double-strand breaks in mice precede synapsis. Nat Genet 2001; 27:271–276. [DOI] [PubMed] [Google Scholar]
- 51. Harper JW, Elledge SJ. The DNA damage response: ten years after. Mol Cell 2007; 28:739–745. [DOI] [PubMed] [Google Scholar]
- 52. Liou JS, Wu YC, Yen WY, Tang YS, Kakadiya RB, Su TL, Yih LH. Inhibition of autophagy enhances DNA damage-induced apoptosis by disrupting CHK1-dependent S phase arrest. Toxicol Appl Pharmacol 2014; 278:249–258. [DOI] [PubMed] [Google Scholar]
- 53. Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, Botrugno OA, Parazzoli D, Oldani A, Minucci S, Foiani M. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature 2011; 471:74–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hogan B, Beddington R, Constantini F, Lacey E. Manipulating the Mouse Embryo: a Laboratory Manual, 2nd ed New York: Cold Spring Harbor Laboratory Press; 1994:33–38. [Google Scholar]
- 55. Guli CL, Smyth DR. UV-induced DNA repair is not detectable in pre-dictyate oocytes of the mouse. Mutat Res 1988; 208:115–119. [DOI] [PubMed] [Google Scholar]
- 56. Baker TG. Comparative aspects of the effects of radiation during oogenesis. Mutat Res 1971; 11:9–22. [DOI] [PubMed] [Google Scholar]
- 57. Russell WL. Effect of the interval between irradiation and conception on mutation frequency in female mice. Proc Natl Acad Sci USA 1965; 54:1552–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Livera G, Petre-Lazar B, Guerquin MJ, Trautmann E, Coffigny H, Habert R. p63 null mutation protects mouse oocytes from radio-induced apoptosis. Reproduction 2008; 135:3–12. [DOI] [PubMed] [Google Scholar]
- 59. Ren Y, Suzuki H, Jagarlamudi K, Golnoski K, McGuire M, Lopes R, Pachnis V, Rajkovic A. Lhx8 regulates primordial follicle activation and postnatal folliculogenesis. BMC Biol 2015; 13:39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ 2007; 14:548–558. [DOI] [PubMed] [Google Scholar]
- 61. Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 2007; 3:500–510. [DOI] [PubMed] [Google Scholar]
- 62. Polager S, Ofir M, Ginsberg D. E2F1 regulates autophagy and the transcription of autophagy genes. Oncogene 2008; 27:4860–4864. [DOI] [PubMed] [Google Scholar]
- 63. Elliott A, Reiners JJ Jr. Suppression of autophagy enhances the cytotoxicity of the DNA-damaging aromatic amine p-anilinoaniline. Toxicol Appl Pharmacol 2008; 232:169–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kang KB, Zhu C, Yong SK, Gao Q, Wong MC. Enhanced sensitivity of celecoxib in human glioblastoma cells: Induction of DNA damage leading to p53-dependent G1 cell cycle arrest and autophagy. Mol Cancer 2009; 8:66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ 2007; 14:500–510. [DOI] [PubMed] [Google Scholar]
- 66. Landin Malt A, Cesario JM, Tang Z, Brown S, Jeong J. Identification of a face enhancer reveals direct regulation of LIM homeobox (Lhx8) by wingless-Int(WNT)/b-catenin signaling. J Biol Chem 2014; 289:30289–30301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Qin Y, Zhap H, Kovanci E, Simpson JL, Chen KZ, Rajkovic A. Analysis of LHX8 mutation in premature ovarian failure. Fertil Steril 2008; 89:1012–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Jeon S, Won HJ, Kim YS, Lyn SW, Seok HH, Kim NK, Lee WS, Shim SH, Yoon TK, Choi Y. Novel single-nucleotide polymorphisms of LHX8 gene in Korean women with premature ovarian insufficiency. Genes Genom 2010; 32:397–400. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.