Novel diagnostic approaches for Zika and dengue are on the rise but may not make it to the market because of bottlenecks in access to samples for validation. An international reference laboratory response is needed to address these challenges, which include networks of in-country laboratories, with well-characterized samples to facilitate assay validation and ensure quality control.
Keywords: Dengue, Zika, flavivirus, arbovirus, laboratory, diagnostics, serology, surveillance
Abstract
Epidemics of dengue, Zika, and other arboviral diseases are increasing in frequency and severity. Current efforts to rapidly identify and manage these epidemics are limited by the short diagnostic window in acute infection, the extensive serologic cross-reactivity among flaviviruses, and the lack of point-of-care diagnostic tools to detect these viral species in primary care settings. The Partnership for Dengue Control organized a workshop to review the current landscape of Flavivirus diagnostic tools, identified current gaps, and developed strategies to accelerate the adoption of promising novel technologies into national programs. The rate-limiting step to bringing new diagnostic tools to the market is access to reference materials and well-characterized clinical samples to facilitate performance evaluation. We suggest the creation of an international laboratory-response consortium for flaviviruses with a decentralized biobank of well-characterized samples to facilitate assay validation. Access to proficiency panels are needed to ensure quality control, in additional to in-country capacity building.
Zika virus (ZIKV) and the dengue viruses (DENVs) are arthropod-borne viruses (arboviruses) of the Flaviviridae family that cocirculate in tropics and subtropics along with other arboviruses that share the same Aedes species mosquito vectors [1]. Several factors, including viral evolution, redistribution of vectors, ineffective vector control strategies, population growth, urbanization, and globalization have contributed to the global spread of DENV, ZIKV, and other arboviruses [2].
Up to 400 million DENV infections are estimated to occur every year [3], and infection with any of the 4 DENV serotypes (DENV1–4) can cause severe and sometimes fatal disease. The geographical expansion of dengue is increasingly associated with more-severe disease outcomes [2, 4]. ZIKV is following the global spread of DENV [2]. ZIKV infections were first thought to only cause sporadic and mild disease in parts of Africa and Asia [5]. A major Zika outbreak with a high attack rate occurred for the first time in 2007. During a subsequent outbreak in the Pacific (French Polynesia) in 2013, ZIKV was linked to severe neurological disease in humans [6]. The recent explosive outbreak in the Americas unmasked the association between prenatal ZIKV infections and severe birth defects [2, 6].
No specific therapeutic options exist for DENV or ZIKV infections. For DENV, a vaccine was recently licensed but has not yet been implemented widely in any of the affected countries [7]; for ZIKV, at least 45 vaccine candidates are now in development, but a licensed vaccine will not be available for years to come [8]. There is an urgent need for highly specific diagnostic assays that can identify and discriminate between cocirculating DENV and ZIKV for efficient case management, surveillance, control, and vaccine trials. In May 2017, the Partnership for Dengue Control (PDC) [9] organized a workshop with approximately 80 key stakeholders and thought leaders to address critical issues related to the diagnosis and surveillance of ZIKV and DENV. The workshop was organized around 3 questions: What is the status of Zika and dengue diagnostic tools? What technological innovations might be available in the near, intermediate, and long-term future? and What is needed to make these technologies readily available where they are most needed? The following is a summary of key outcomes that emerged from the meeting.
WHAT IS THE STATUS OF ZIKA AND DENGUE DIAGNOSTIC TOOLS?
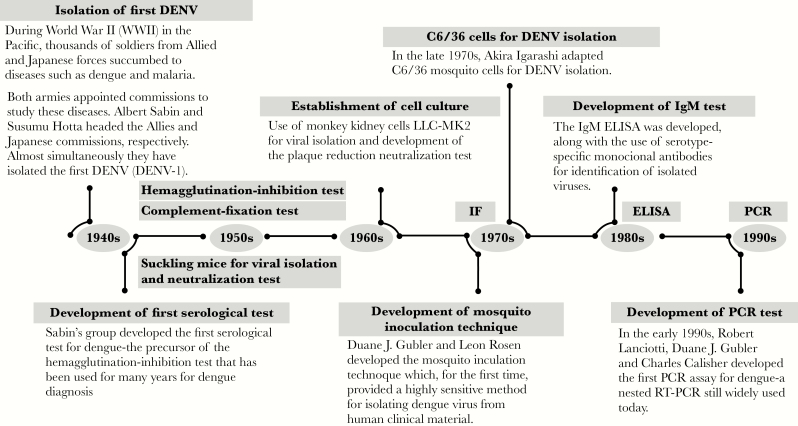
Individuals infected with DENV and ZIKV may be asymptomatic or display a similar constellation of initial clinical symptoms [10]. Hence, virus-specific assays are required for accurate diagnosis. Since the first isolation of DENV during World War II [11, 12], a number of diagnostic methods commonly used for viral detection, such as viral isolation, plaque reduction neutralization test (PRNT), the immunoglobulin M (IgM) enzyme-linked immunosorbent assay (ELISA), and, in the 1990s, reverse transcription–polymerase chain reaction (RT-PCR) [13] were developed for DENV (Figure 1) and other medically relevant flaviviruses.
Figure 1.
Historical time line of the development of dengue diagnostic tools, 1940s–1990s. Dengue virus (DENV) was first isolated in the early 1940s by groups led by Albert Sabin and Susumu Hotta. A number of viral isolation, serological, and molecular methods have since been developed. IF, immunofluorescence; ELISA, enzyme-linked immunosorbent assay; IgM, immunoglobulin M; RT-PCR, reverse transcription–polymerase chain reaction.
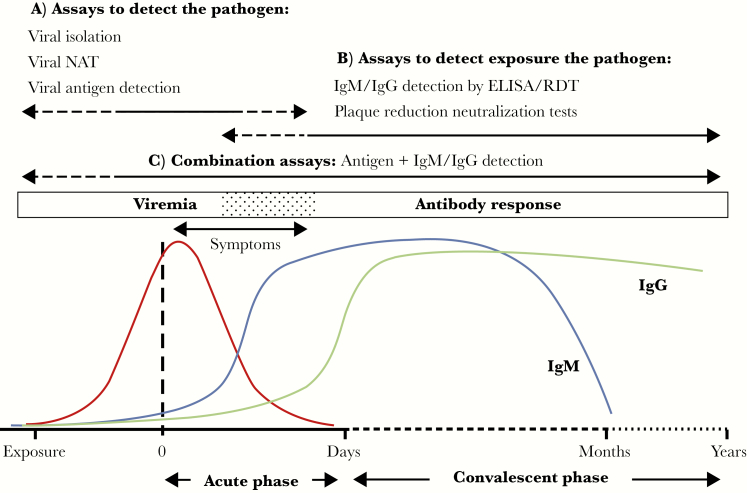
Assays to detect DENV and ZIKV can be divided into 2 main categories: (1) assays to detect the pathogen (viral isolation, viral nucleic acid testing, or viral antigen detection); and (2) assays to detect exposure to the pathogen (detection of virus-specific antibodies such as IgM, immunoglobulin G, and immunoglobulin A). Assay selection depends both on the timing of sample collection and the purpose of testing (Figure 2). The viremic period of flaviviral infections is transient and short-lived; the duration of viral shedding and the presence of ZIKV RNA can be variable across sample types (eg, serum, whole blood, urine, saliva, and amniotic fluid) [6, 14] and different hosts (eg, pregnant women and other adults) [15]. A negative viral isolation and/or nucleic acid test result does not exclude the presence of a current infection.
Figure 2.
Schematic representation of the typical kinetics of flaviviral infections (adapted from Peeling et al 2010 with permission). As for most arboviral infections, viremia (red line) normally precedes the onset of clinical symptoms (0) and lasts for a few days after symptom onset. A, During the acute phase, flaviviral infections are best detected by viral isolation, nucleic acid amplification tests (NATs), and antigen detection assays (eg, the dengue virus NS1 assay). B, During the course of infection, viral-specific immunoglobulin M (IgM; blue line) and immunoglobulin G (IgG; green line) antibodies are produced and can be used to detect exposure to the pathogen. Current infections can be diagnosed by the detection of IgM and IgG antibodies in paired acute and convalescent phase samples (ie, detection of seroconversion of a ≥4-fold rise in the IgG or total antibody titer). IgM antibodies can persist for several months, and IgG antibodies are known to persist for several years. C, Combination assays that detect both antigen and antibodies are applicable throughout the entire spectrum of disease. ELISA, enzyme-linked immunosorbent assay; RDT, rapid diagnostic test.
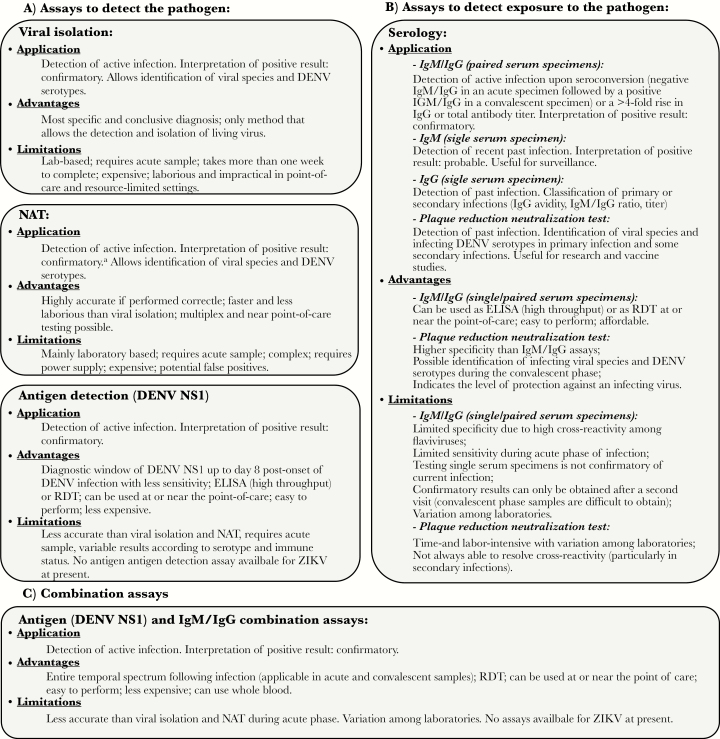
In the convalescent phase of infection, serologic methods are preferred, although paired acute and convalescent phase samples are required to distinguish current from past infections [16]. The major challenge of ZIKV and DENV diagnosis by serologic analysis is the extensive cross-reactivity of antibody responses resulting from prior flaviviral infections and/or vaccination [17–19]. Figure 3 details the applications, advantages, and limitations of the different types of assays available for the detection of DENV and ZIKV infections.
Figure 3.
Applications, advantages, and limitations of the main diagnostic tests in use for the detection of flaviviral infections. A, Assays to detect the pathogen directly: viral isolation, nucleic acid amplification tests (NATs), and antigen detection assays. B, Assays to detect exposure to infection. C, Combination assays to detect both the pathogen and exposure to infection. aRNA copy number is not an accurate measure of infectious virus (viral RNA can persist for longer periods than infectious virus). DENV, dengue virus; ELISA, enzyme-linked immunosorbent assay; IgG, immunoglobulin G; IgM, immunoglobulin M; RDT, rapid diagnostic test; ZIKV, Zika virus.
LANDSCAPE OVERVIEW AND EXISTING GAPS
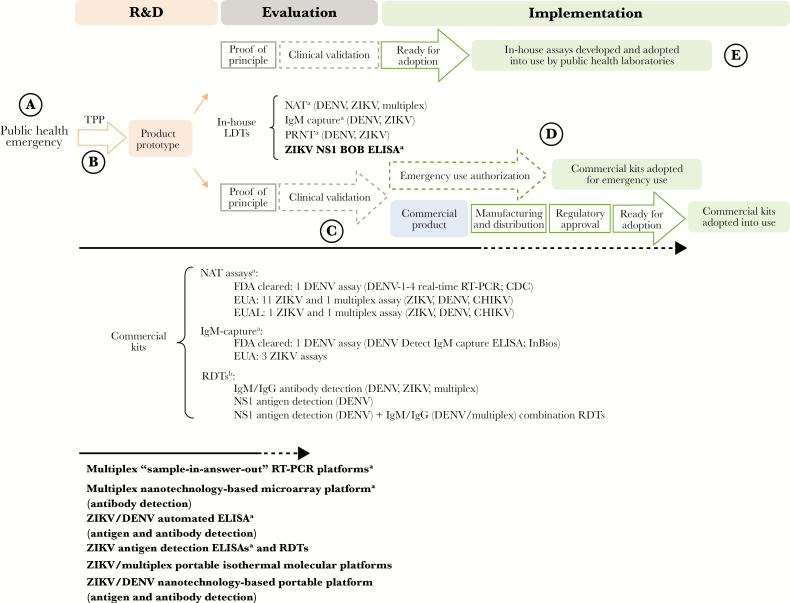
Both in-house laboratory-developed tests (LDTs) and commercial kits are available to detect ZIKV and DENV infections (Figure 4). Most of the available technologies require laboratory facilities with appropriate diagnostic competence (Figure 4); point-of-care assays remain limited. Zika commercial kits include nucleic acid tests and serologic assays. The current ZIKV nucleic acid tests have not yet gone through much rigorous evaluations [20], and the evidence is even scarcer for serologic assays. Antigen detection assays for the diagnosis of ZIKV infections are currently not available on the market.
Figure 4.
Diagnostic landscape to detect Zika virus (ZIKV) and dengue virus (DENV) infections and the pathway to adoption. The Zika public health emergency triggered significant efforts toward the development of new diagnostic assays. Commercial kits and in-house laboratory-developed tests (LDTs) are available to detect ZIKV and DENV infections. Multiplex assays that simultaneously detect ZIKV and other arboviral infections (DENV, CHIKV and others) are also available. Target product profiles (TPP) are used to define the desired technical and operational characteristics of a test. Quality-assured clinical laboratories can develop, validate, and then implement their in-house LDTs. Commercial kits require clinical validation, scale production, distribution, and regulatory approval to be adopted for wide use. Two DENV commercial kits (1 nucleic acid amplification test [NAT] and 1 immunoglobulin M [IgM]–capture enzyme-linked immunosorbent assay [ELISA]) have been cleared by the Food and Drug Administration (FDA). Emergency use authorization mechanisms by the FDA (the Emergency Use Authorization [EUA] authority) and the World Health Organization (the Emergency Use Assessment and Listing [EUAL] procedure) were put into place to accelerate adoption of ZIKV commercial kits in response to the Zika public health emergency. In bold are the types of assays that are currently in the pipeline (most assays are stuck at the evaluation stage because of the lack of access to well-characterized clinical specimens). BOB, blockade of binding; CDC, Centers for Disease Control and Prevention; IgG, immunoglobulin G; PRNT, plaque-reduction neutralization test; RT-PCR, reverse transcription–polymerase chain reaction; R&D, research and development. aAssays that require laboratory infrastructure. bNo rapid diagnostic test (RDT) has been cleared by the FDA nor received EUA/EUAL approval to date.
Performance of commercial dengue diagnostic tools has improved over the last decade. These include 2 Food and Drug Administration (FDA)–approved assays (1 RT-PCR and 1 IgM-capture ELISA). Additionally, there are several rapid lateral flow assays (RDTs) for the detection of DENV NS1 antigen, DENV-specific IgM antibodies, or both (Figure 4), none of which are FDA approved. RDTs hold promise as future point-of-care applications; however, the clinical performance of these assays has been highly variable [21].
While dengue serologic assays have been clinically validated, their specificity has decreased by cross-reactivity in the context of the recent cocirculation of ZIKV [22]. To date, very few dengue and Zika diagnostic assays have been adequately and independently evaluated using clinical specimens from both ZIKV-infected and DENV-infected populations. Diagnostic assays that can accurately detect and distinguish cocirculating flaviviral infections and predict severe disease outcomes at or near the point-of-care are urgently needed.
WHAT TECHNOLOGICAL INNOVATIONS MIGHT BE AVAILABLE IN THE NEAR, INTERMEDIATE, AND LONG-TERM FUTURE?
Different companies and research groups were invited to present technologies to detect DENV and ZIKV. In the following, we discuss the technologies in the pipeline (Figure 4) and their potential to change the paradigm of flaviviral diagnosis.
Pipeline Assays to Detect the Pathogen
Nucleic-Acid Testing
Simpler and faster alternatives to traditional RT-PCR methods have the potential to be used at or near the point-of-care. These include cartridge-based sample-in-answer-out multiplex real-time RT-PCR assays that can simultaneously detect ZIKV, DENV1–4, other arboviruses (eg, chikungunya virus, an alphavirus), and other viruses (3-plex to 6-plex combinations) from a single specimen in <2 hours. Arboviral assays are being developed for use on existing industry platforms that were previously validated and implemented for other molecular tests. This strategy illustrates the usefulness of open-platform systems that can easily incorporate newly developed molecular amplification methods to suit an emergent medical need, such as the ZIKV epidemic. Another advantage of these systems is the ability to transmit data wirelessly and monitor the results remotely. The disadvantages are that these platforms are costly and that some require technical expertise and laboratory infrastructure that are not widely available.
The development of more-portable molecular platforms linked to faster isothermal amplification methods independent of thermal cycling, such as recombinase polymerase amplification [23] and loop-mediated isothermal amplification, are also underway for singleplex and multiplex detection of ZIKV and other arboviruses. In prototype formats, results can be achieved in <1 hour, and assays can potentially be applied to settings without electricity or highly trained users. Proof-of-principle studies exist [23–30], but further simplification of sample preparation and more-robust clinical performance evaluation will be required. Innovative technologies, such as clustered regularly interspaced short palindromic repeats (CRISPR)–based RNA sensing, robotics, microfluidics, smartphones, and 3D printers, are being used to develop these assays. Other nucleic acid testing innovations include the use of paper-based strips for multiplex detection of ZIKV, DENV, and chikungunya virus end point RT-PCR products [31].
Antigen Detection Assays
High-affinity monoclonal antibodies that recognize specific epitopes on ZIKV antigens are required to develop antigen detection assays and are either in development or were developed for NS1 [32, 33], including the development of RDTs [34].
Pipeline Assays to Detect Past Exposure
Given our understanding of the cross-reactivity of current antibody detection methods for flaviviruses, there is a lack of reliable reference diagnostic tools against which to compare newly developed specific serologic assays. Detection of virus-specific neutralizing antibodies by PRNT can be useful to discriminate viral species and serotypes in primary infections. However, the specificity of PRNT in sequential DENV infections or sequential DENV and ZIKV infections and at early time points after infection is limited [32, 35, 36]. Interestingly, little cross-neutralization is detected in samples collected during the late convalescent phase (ie, >2 months) after DENV and ZIKV infection [37]. These observations highlight the importance of the timing of sample collection and the history of exposure to past infections to inform the serodiagnosis of flaviviral infections. It is critical to evaluate multiple flaviviruses simultaneously in neutralization assays to interpret the results appropriately.
Strongly neutralizing human monoclonal antibodies target quaternary structure epitopes that typically bind across envelope proteins displayed on the surface of the viral particles [38–40]. Epitopes with high sequence homology among serotypes and viral species can trigger cross-reactive antibody responses, whereas unique epitopes can lead to type-specific antibody responses. This information is being used to rationally design ZIKV and DENV envelope and NS1 recombinant antigens for specific serologic assays.
Isolated human ZIKV type-specific anti-NS1 monoclonal antibodies were used to identify type-specific recognition sites on ZIKV NS1 protein by antibody competition assays [32]. One of these antibodies was adapted to a competition-based ELISA, in which serum antibodies are measured for their ability to block the binding of a ZIKV NS1-specific monoclonal antibody to solid-phase ZIKV NS1 [41]. This approach, named “ZIKV NS1 blockade-of-binding (BOB) ELISA,” was shown to be more specific than traditional ELISAs. Clinical validation in large multicenter cohorts of patients stratified by exposure to DENV and ZIKV infection, immune status, and timing of sample collection confirmed the high specificity and sensitivity of the assay [41]. The ZIKV NS1 blockade-of-binding ELISA has been implemented in laboratories of 6 different countries (Brazil, Italy, Nicaragua, Switzerland, United Kingdom, and United States).
Nanotechnology assays have also been developed, including simple-to-use readout platforms with data connectivity that use disposable microfluidic cartridges for rapid detection of ZIKV and DENV antibodies/antigen, and a multiplex serologic assay that uses near‐infrared fluorescence enhanced imaging on a nanoscale plasmonic gold microarray antigen platform (12-plex) for antibody detection on 2 different channels [42]. The latter was shown to detect and distinguish IgG antibodies from ZIKV- and DENV-infected patients, as well as determine the timing of exposure to infection by measuring IgG avidity.
WHAT IS NEEDED TO MAKE THESE TECHNOLOGIES AVAILABLE IN THE FIELD?
For the last 25 years, routine diagnostic approaches have mainly included laboratory-based RT-PCR, IgM detection, and PRNT. Recognition of Zika as a public health emergency of international concern has galvanized the development of new diagnostic assays to detect flaviviral infections. While these efforts must be encouraged, it is equally important to look downstream and identify the issues around translating research into a product that is available for use in the field, robust, easy to use, reasonably inexpensive, and accurate and has demonstrable clinical impact. Previous research and development experience has shown that the path from diagnostic development to adoption is long and fragmented [16]. There is a massive loss between the number of tests that undergo initial development and the ones that are taken into adoption for routine use.
HOW CAN WE ACCELERATE TRAVEL ON THE PATHWAY FROM DISCOVERY TO ADOPTION?
The 5 major steps identified for optimizing the time to adoption (Figure 4A–E) are discussed below and summarized in Table 1.
Table 1.
Summary Table of the Challenges and Drivers Along the Pathway to Adoption
| Stepa | Challenge(s) | Drivers |
|---|---|---|
| A | Market failure due to uncertainty and lack of demand of public health emergencies | Research and development models for diagnostic preparedness, including product development; product development partnerships, such as CEPI.dx; other innovative financing models |
| B | TPP: the performance characteristics that are set in the TPP are aspirational in nature; TPPs are often deemed to be too stringent | Risk and benefit models to set accuracy targets may help inform use of diagnostic tests when they do not meet the minimum or ideal characteristics set in the TPP |
| C | Lack of clinical samples and resources for clinical validations | Development of international reference standard for assay comparability; improved access to qualified field laboratory networks; access to proficiency panels; development of standardized protocols |
| D | Regulatory approval that is region specific, nontransparent, complex, slow, and costly | Establishment of regulatory networks, common strategies, information sharing, and early partnerships |
| E | Limited in-country capacity for wide adoption. | Mechanisms for appropriate transfer of technology in a more streamlined fashion; regulation of quality of local laboratories and in-house assays for national scale-up and sustainable implementation |
Abbreviation: TPP, target product profile.
aSteps along the pathway to adoption that require optimization (Figure 4A–E).
Market Uncertainty
For diagnostic countermeasures to be readily available, research and development must happen before rather than in response to an outbreak [43]. The unpredictable and episodic nature of outbreaks brings uncertainty to the market, and diagnostic companies are left unable to adequately forecast demand and establish business models that allow a return on investment. Even when a product is brought into the market, there is no guarantee that it will be adopted by national health authorities. Once a public health emergency of international concern has ended, sustained manufacturing support of the product may be at stake. Sustainable markets are required to ensure that validated, approved, high-quality diagnostic tools remain available for use in the next outbreak event. As such, innovative financial incentives are needed to achieve sustainable emergency preparedness for diagnostic tools. From investments in product development to the establishment of partnerships and the creation of models to support scalable adoption into national programs, a variety of mechanisms have been proposed or established to overcome some of the challenges.
The WHO R&D Blueprint for Actions to Prevent Epidemics has initiated a call for open-platform technologies to improve research and development preparedness against global health emergencies, so that in the event of an epidemic diagnostic kits can be made available in a short time frame [44]. Furthermore, there was a call for a coalition between diagnostic preparedness efforts and programs that finance and manage the development of vaccines [43]. As a result, CEPI∙dx, a new partnering model between the Coalition for Epidemic Preparedness Innovations, the Foundation for Innovative New Diagnostics, and other diagnostic partners, has been created.
Target Product Profiles (TPPs)
TPPs are used to define the desired minimum technical and operational characteristics of diagnostic tests, to ensure the development of the most-impactful products. TPPs are aspirational in nature; however, excessively stringent requirements may deter industry partners from developing new products and lead to a lack of diagnostic tests meeting those requirements. Strategies on how to best define the desired characteristics of TPPs and/or inform the use of diagnostic tests when those requirements are not met have been proposed. For example, a slightly less accurate test might provide a higher public health impact in terms of increased access to testing, compared with a more accurate but expensive or complex test (this was the approach used to approve the use of HIV self-testing and malaria RDTs in the past). As such, it is important to consider how the assay will be used, in which setting, and for what purpose (eg, surveillance, early warning, or clinical management at the point-of-care), as different applications will have different technical and operational requirements. A weighted risk and benefits approach within different use scenarios may be more appropriate not only to define but also to guide regulatory approval and adoption.
Assay Optimization and Clinical Validation
Internationally accepted reference preparations to compare and potentially standardize the different assays are crucially important [45]. The WHO has established numerous reference preparations, most of them as WHO international standards. For ZIKV RNA, the biological standard for molecular tests was characterized for the majority of nucleic acid tests available [46], and the complete sequence of the ZIKV of this reference preparation was published [47] and established as a WHO international standard. Lack of access to biobanks of well-characterized clinical specimens delays the process of test optimization, clinical validation, and product adoption. This lack was identified as the most significant bottleneck along the pathway from development to adoption.
Of note, the pathway to adoption of in-house assays and of commercial kits differs substantially. Quality-assured clinical laboratories can develop, validate, and then implement their in-house assays. In contrast, commercial diagnostic kits go through regulatory approval processes that may require large clinical validation studies, manufacturing under a quality management system, and some level of distribution capacity. The different streams of test development make it challenging to determine relative comparability of the accuracy of the different tests because very few of them share the same calibration controls (ie, internal positive controls used for measuring the reactivity of a diagnostic test) or screening panels (ie, a small set of coded samples that include high-positive, low-positive, cross-reactive, and negative samples, to measure the specificity and sensitivity of a diagnostic test). Obtaining irreplaceable clinical specimens is costly; the same test materials cannot be used throughout the development process. Access to clinical samples becomes even more challenging during an outbreak, with multiple demands to prioritize assay validation in a short time frame and the inability to do head-to-head comparisons.
A coordinated network of quality-assured laboratories with staff that are well trained in assay validation and performance evaluation could be leveraged a priori. Such an approach would alleviate pressure on the countries involved in outbreak response, yet provide access to clinical samples and data in a way that may be acceptable to the different parties. Of note, local restrictions on the export of clinical samples (as witnessed during the ZIKV outbreak) limits sample sharing for product validation outside the affected countries [43]. The involvement of a network of capable local laboratories would have the advantage to overcome the need for out-of-country sample transfer and facilitate country involvement and capacity development at an early stage of product development. A transparent and fair process of engagement needs to be put into place to minimize distrust and ensure access and equitable sharing of specimens and data. The creation of a governance system to provide access to reference panels and protocols for test validation has been proposed.
Regulatory Approval
Regulation is essential to ensure the safety, quality, and effectiveness of diagnostic tests, yet >50% of countries do not independently regulate in vitro diagnostic tools [48]. The regulatory landscape for in vitro diagnostic tools is highly variable, and regulatory approval mechanisms vary from country to country. This makes assay uptake processes slow, costly, and nontransparent. Regulatory harmonization between international and national regulatory agencies, coupled with coordinated information sharing among the different interest groups (industry, regulators, researchers, laboratories, health systems, and patients), is required.
During outbreak events, emergency use authorizations are generally used to provide regulatory oversight for diagnostic tools that have not previously been evaluated and yet are urgently needed for global response. Both the FDA and the WHO have implemented programs (the Emergency Use Authorization [EUA] authority and the Emergency Use Assessment and Listing [EUAL] procedure, respectively) to address the evaluation of new diagnostic tools in an emergency setting. It is important to note that in both cases, EUA and/or EUAL approval does not extend approval for use outside of an emergency setting. Once an emergency has ended, industry will need to seek approval for regular use of their products in the intended settings, using either FDA 510(k) or WHO prequalification procedures. The data obtained during EUA and/or EUAL approval may be included in the application package for regular approval; however, it is likely that additional data will be required for full approval. In these instances, it can be challenging for industry to provide sufficient data, as limited access to well-characterized samples can prevent evaluation of the products to the extent required for FDA and WHO approval. As of May 2017, several ZIKV diagnostic assays have received approval through the EUA (15 assays) and/or EUAL (2 assays) [45]; however, no single ZIKV assay has been cleared by the FDA to date (Figure 4).
Sustainable In-Country Capacity
Sustainable in-country capacity is needed for diagnostic tools to respond to the intermediate and long-term infectious diseases threats. Higher-cost commercial kits are unlikely to solve this issue at a national level in many resource-constrained countries. Therefore, key reagents, protocols, and quality control standards (eg, proficiency panels) must be made available to national reference laboratories and other such public-sector entities to ensure wide and sustainable adoption.
CONCLUSIONS
Promising technologies for detection of ZIKV and DENV infections are currently in the pipeline. These technologies have the potential to address many of the current challenges of epidemic flaviviral diseases. The rate-limiting bottleneck is early access to calibration controls and screening panels, as well as access to well-characterized samples for development, validation, and comparison of the performance of different assays. Proficiency testing for both serologic and molecular diagnostic tools should be developed for all regions of endemicity, paired with capacity building. We suggest that an international reference laboratory response for flaviviruses is needed, which would include networks of in-country laboratories and preparation of protocols for evaluation studies. This could be achieved through initiatives such as the Global Dengue and Aedes Transmitted Disease Consortium, the European Virus Archive, the future EVD-LabNet by the European Centre for Disease Prevention and Control, or the Zika research consortia funded by the European Commission [49]. The knowledge obtained should be put into the public domain. Researchers and policymakers alike need to ensure mechanisms for greater reagent availability and sharing of standard reagents, such as reference materials, antigens, monoclonal antibodies, cell lines, control sera, and standardized protocols. While this workshop focused on challenges for arbovirus diagnostic development, the key outcomes highlighted here translate to all pathogens of epidemic potential.
Notes
Acknowledgments. We thank the Fondation Merieux, for supporting the meeting; Richard Vaux, Benedicte Pansier, and Severine Comignani, for their assistance with logistics; and Raman Preet, for assistance for all ZikaPLAN consortium–related matters during this meeting.
Financial support. This work was supported by the Gates Foundation; the European Union’s 2020 Research and Innovation Programme, through the ZikaPLAN consortium (grant 734584); the EPSRC IRC in Early-Warning Sensing Systems for Infectious Diseases (EP/K031953/1); bioMérieux (unrestricted grant); Sanofi Pasteur (unrestricted grant); Takeda (unrestricted grant); the Foundation for Innovative and New Diagnostics (in-kind support); the World Health Organization (WHO) Special Program for Research and Training in Tropical Diseases (in-kind support); and the WHO R&D Blueprint (in-kind support).
Potential conflicts of interest. All authors: No reported conflicts of interest. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1. Musso D, Gubler DJ. Zika virus. Clin Microbiol Rev 2016; 29:487–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Wilder-Smith A, Gubler DJ, Weaver SC, Monath TP, Heymann DL, Scott TW. Epidemic arboviral diseases: priorities for research and public health. Lancet Infect Dis 2017; 17:e101–6. [DOI] [PubMed] [Google Scholar]
- 3. Bhatt S, Gething PW, Brady OJ et al. The global distribution and burden of dengue. Nature 2013; 496:504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guzman MG, Gubler DJ, Izquierdo A, Martinez E, Halstead SB. Dengue infection. Nat Rev Dis Primers 2016; 2:16055. [DOI] [PubMed] [Google Scholar]
- 5. Faye O, Freire CC, Iamarino A et al. Molecular evolution of Zika virus during its emergence in the 20(th) century. PLoS Negl Trop Dis 2014; 8:e2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Baud D, Gubler DJ, Schaub B, Lanteri MC, Musso D. An update on Zika virus infection. Lancet 2017; 390:2099–109. [DOI] [PubMed] [Google Scholar]
- 7. Wilder-Smith A, Vannice KS, Hombach J, Farrar J, Nolan T. Population perspectives and World Health Organization recommendations for CYD-TDV Dengue vaccine. J Infect Dis 2016; 214:1796–9. [DOI] [PubMed] [Google Scholar]
- 8. Durbin A, Wilder-Smith A. An update on Zika vaccine developments. Expert Rev Vaccines 2017; 16:781–7. [DOI] [PubMed] [Google Scholar]
- 9. Gubler DJ. The partnership for dengue control—a new global alliance for the prevention and control of dengue. Vaccine 2015; 33:1233. [DOI] [PubMed] [Google Scholar]
- 10. Duffy MR, Chen TH, Hancock WT et al. Zika virus outbreak on Yap Island, Federated States of Micronesia. N Engl J Med 2009; 360:2536–43. [DOI] [PubMed] [Google Scholar]
- 11. HOTTA S. Experimental studies on dengue. I. Isolation, identification and modification of the virus. J Infect Dis 1952; 90:1–9. [DOI] [PubMed] [Google Scholar]
- 12. SABIN AB. Research on dengue during World War II. Am J Trop Med Hyg 1952; 1:30–50. [DOI] [PubMed] [Google Scholar]
- 13. Lanciotti RS, Calisher CH, Gubler DJ, Chang GJ, Vorndam AV. Rapid detection and typing of dengue viruses from clinical samples by using reverse transcriptase-polymerase chain reaction. J Clin Microbiol 1992; 30:545–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schaub B, Vouga M, Najioullah F et al. Analysis of blood from Zika virus-infected fetuses: a prospective case series. Lancet Infect Dis 2017; 17:520–7. [DOI] [PubMed] [Google Scholar]
- 15. Suy A, Sulleiro E, Rodó C et al. Prolonged Zika virus viremia during pregnancy. N Engl J Med 2016; 375:2611–3. [DOI] [PubMed] [Google Scholar]
- 16. Peeling RW, Artsob H, Pelegrino JL et al. Evaluation of diagnostic tests: dengue. Nat Rev Microbiol 2010; 8:S30–8. [DOI] [PubMed] [Google Scholar]
- 17. Priyamvada L, Hudson W, Ahmed R, Wrammert J. Humoral cross-reactivity between Zika and dengue viruses: implications for protection and pathology. Emerg Microbes Infect 2017; 6:e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Keasey SL, Pugh CL, Jensen SM et al. Antibody responses to Zika virus infections in environments of flavivirus endemicity. Clin Vaccine Immunol 2017; 24:e00036–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Andrade DV, Harris E. Recent advances in understanding the adaptive immune response to Zika virus and the effect of previous flavivirus exposure. Virus Res 2017; doi: 10.1016/j.virusres.2017.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Waggoner JJ, Pinsky BA. Zika virus: diagnostics for an emerging pandemic threat. J Clin Microbiol 2016; 54:860–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blacksell SD. Commercial dengue rapid diagnostic tests for point-of-care application: recent evaluations and future needs?J Biomed Biotechnol 2012; 2012:151967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. van Meer MPA, Mögling R, Klaasse J et al. Re-evaluation of routine dengue virus serology in travelers in the era of Zika virus emergence. J Clin Virol 2017; 92:25–31. [DOI] [PubMed] [Google Scholar]
- 23. Teoh BT, Sam SS, Tan KK et al. Early detection of dengue virus by use of reverse transcription-recombinase polymerase amplification. J Clin Microbiol 2015; 53:830–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gootenberg JS, Abudayyeh OO, Lee JW et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science 2017; 356:438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pardee K, Green AA, Takahashi MK et al. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016; 165:1255–66. [DOI] [PubMed] [Google Scholar]
- 26. Yaren O, Alto BW, Gangodkar PV et al. Point of sampling detection of Zika virus within a multiplexed kit capable of detecting dengue and chikungunya. BMC Infect Dis 2017; 17:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee D, Shin Y, Chung S, Hwang KS, Yoon DS, Lee JH. Simple and highly sensitive molecular diagnosis of Zika virus by lateral flow assays. Anal Chem 2016; 88:12272–8. [DOI] [PubMed] [Google Scholar]
- 28. Chan K, Weaver SC, Wong PY et al. Rapid, affordable and portable medium-throughput molecular device for Zika virus. Sci Rep 2016; 6:38223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abd El Wahed A, Sanabani SS, Faye O et al. Rapid molecular detection of Zika virus in acute-phase urine samples using the recombinase polymerase amplification assay. PLoS Curr 2017; 9:doi: 10.1371/currents.outbreaks.a7f1db2c7d66c3fc0ea0a774305d319e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Priye A, Bird SW, Light YK, Ball CS, Negrete OA, Meagher RJ. A smartphone-based diagnostic platform for rapid detection of Zika, chikungunya, and dengue viruses. Sci Rep 2017; 7:44778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Niwa K, Oribe A, Okumura H et al. Tag/hybridization-based sensitive detection of polymerase chain reaction products. Anal Biochem 2014; 464:12–6. [DOI] [PubMed] [Google Scholar]
- 32. Stettler K, Beltramello M, Espinosa DA et al. Specificity, cross-reactivity, and function of antibodies elicited by Zika virus infection. Science 2016; 353:823–6. [DOI] [PubMed] [Google Scholar]
- 33. Rose N, Pinho-Nascimento CA, Ruggieri A et al. Generation of monoclonal antibodies against native viral proteins using antigen-expressing mammalian cells for mouse immunization. BMC Biotechnol 2016; 16:83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bedin F, Boulet L, Voilin E, Theillet G, Rubens A, Rozand C. Paper-based point-of-care testing for cost-effective diagnosis of acute flavivirus infections. J Med Virol 2017; 89:1520–7. [DOI] [PubMed] [Google Scholar]
- 35. Patel B, Longo P, Miley MJ, Montoya M, Harris E, de Silva AM. Dissecting the human serum antibody response to secondary dengue virus infections. PLoS Negl Trop Dis 2017; 11:e0005554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Beltramello M, Williams KL, Simmons CP et al. The human immune response to Dengue virus is dominated by highly cross-reactive antibodies endowed with neutralizing and enhancing activity. Cell Host Microbe 2010; 8:271–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collins MH, McGowan E, Jadi R et al. Lack of durable cross-neutralizing antibodies against Zika virus from Dengue virus infection. Emerg Infect Dis 2017; 23:773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rouvinski A, Guardado-Calvo P, Barba-Spaeth G et al. Recognition determinants of broadly neutralizing human antibodies against dengue viruses. Nature 2015; 520:109–13. [DOI] [PubMed] [Google Scholar]
- 39. Dejnirattisai W, Wongwiwat W, Supasa S et al. A new class of highly potent, broadly neutralizing antibodies isolated from viremic patients infected with dengue virus. Nat Immunol 2015; 16:170–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Teoh EP, Kukkaro P, Teo EW et al. The structural basis for serotype-specific neutralization of dengue virus by a human antibody. Sci Transl Med 2012; 4:139ra83. [DOI] [PubMed] [Google Scholar]
- 41. Balmaseda A, Stettler K, Medialdea-Carrera R et al. Antibody-based assay discriminates Zika virus infection from other flaviviruses. Proc Natl Acad Sci U S A 2017; 114:8384–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang B, Pinsky BA, Ananta JS et al. Diagnosis of Zika virus infection on a nanotechnology platform. Nat Med 2017; 23:548–50. [DOI] [PubMed] [Google Scholar]
- 43. Perkins MD, Dye C, Balasegaram M et al. Diagnostic preparedness for infectious disease outbreaks. Lancet 2017; 390:2211–4. [DOI] [PubMed] [Google Scholar]
- 44. World Health Organization. About the R&D Blueprint http://www.who.int/blueprint/about/en/. Accessed 10 January 2018.
- 45. Chua A, Prat I, Nuebling CM, Wood D, Moussy F. Update on Zika diagnostic tests and WHO’s related activities. PLoS Negl Trop Dis 2017; 11:e0005269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Baylis SA, Hanschmann KO, Schnierle BS, Trösemeier JH, Blümel J; Zika Virus Collaborative Study Group Harmonization of nucleic acid testing for Zika virus: development of the 1st World Health Organization International Standard. Transfusion 2017; 57:748–61. [DOI] [PubMed] [Google Scholar]
- 47. Trosemeier JH, Musso D, Blumel J, Theze J, Pybus OG, Baylis SA. Genome sequence of a candidate World Health Organization reference strain of Zika virus for nucleic acid testing. Genome Announc 2016; 4:doi: 10.1128/genomeA.00917-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Special Program for Research and Training in Tropical Diseases. Diagnostics for tuberculosis: global demand and market potential Geneva: World Health Organization, 2006. http://www.who.int/tdr/publications/documents/tbdi.pdf. Accessed 10 January 2018.
- 49. Wilder-Smith A, Preet R, Renhorn KE et al. ZikaPLAN: Zika preparedness Latin American Network. Glob Health Action 2017; 10:1398485. [DOI] [PMC free article] [PubMed] [Google Scholar]