Abstract
Here we present a Caenorhabditis elegans N-glycan shotgun array. This nematode serves as a model organism for many areas of biology including but not limited to tissue development, host–pathogen interactions, innate immunity, and genetics. Caenorhabditis elegans N-glycans contain structural motifs that are also found in other nematodes as well as trematodes and lepidopteran species. Glycan binding toxins that interact with C. elegans glycoconjugates also do so with some agriculturally relevant species, such as Haemonchus contortus, Ascaris suum, Oesophagostomum dentatum and Trichoplusia ni. This situation implies that protein–carbohydrate interactions seen with C. elegans glycans may also occur in other species with related glycan structures. Therefore, this array may be useful to study these relationships in other nematodes as well as trematode and insect species. The array contains 134 distinct glycomers spanning a wide range of C. elegans N-glycans including the subclasses high mannose, pauci mannose, high fucose, mammalian-like complex and phosphorylcholine substituted forms. The glycans presented on the array have been characterized by two-dimensional separation, ion trap mass spectrometry, and lectin affinity. High fucose glycans were well represented and contain many novel core structures found in C. elegans as well as other species. This array should serve as an investigative platform for carbohydrate binding proteins that interact with N-glycans of C. elegans and over a range of organisms that contain glycan motifs conserved with this nematode.
Keywords: fucosyl, glycoform, mass spectrometry, MSALAD, pauci mannose
Introduction
Glycan arrays first appeared in the scientific literature in 2002 (Wang et al. 2002). First arrays included those produced with bacterial polysaccharides, glycoproteins, glycolipids and proteoglycans. Sources ranged from natural to those produced by chemical synthesis (Fukui et al. 2002). Over the past 15 years a wealth of information has been gleaned from glycan array analytical platforms. Protein–carbohydrate recognition is a common event in biology and these array platforms allow screening of many glycoconjugates against potential carbohydrate binding proteins (CBPs), intact viruses or microbes, and other carbohydrate binding entities. They have been used in a wide range of applications to interrogate carbohydrate recognition including but not limited to enzyme recognition (Vidal-Melgosa et al. 2015), innate immune function, bacterial (Byres et al. 2008) and viral adhesin (Stevens et al. 2006) as well as carbohydrate recognition of antibodies in cancer research and vaccine development (Pedersen et al. 2011; Muthana and Gildersleeve 2014). There are essentially three sources of glycans used for array purposes. These are synthetically derived ones which can include chemo-enzymatic methods, those produced from glycan libraries composed of both synthetic and natural compounds, and those derived exclusively from a natural source such as a whole organism or tissue. The present work describes our Caenorhabditis elegans N-glycan array and lays the background for possible uses based on the current understanding of this organism and those that are related to it through shared glycoconjugate structure. To date, it is the most complete array produced representing this nematode.
The free-living nematode C. elegans is a simple animal consisting of 959 somatic cells. All its somatic cells are mapped throughout development. It possesses a complex life cycle consisting of four larval stages, L1–L4, an alternative L3 stage, Dauer and an adult stage. The genome has been sequenced in its entirety. Caenorhabditis elegans has served as a model for the study of tissue development, innate immunity and host–pathogen interactions among other scientific areas (Girard et al. 2007). Glycosylation processes in this organism have been investigated over the past two decades. While glycosylation processes in this organism have not been fully defined, a strong base of knowledge has accumulated in the literature showing structural cross-over between its glycome and other organisms. A range of glycan-dependent interactions have been established in C. elegans and this is especially the case for its interactions with pathogens. Glycosyltransferases are well represented in the C. elegans genome (Warren et al. 2002) and in some cases, such as GNT-I, there is redundancy hinting at breadth of the glycosyltransferase repertoire in this simple organism (Chen et al. 2003). The developmental stages have specific patterns of glycan expression (Cipollo et al. 2005; Geyer et al. 2012) reflecting stage specific functions for the glycan subclasses. The C. elegans glycome is complex with nearly 200 glycoforms reported in the N-glycans alone (Cipollo et al. 2002, 2004b, 2005; Haslam et al. 2002; Hanneman et al. 2006; Palaima et al. 2010; Paschinger et al. 2008; Parsons et al. 2014; Yan et al. 2015a, 2015b).
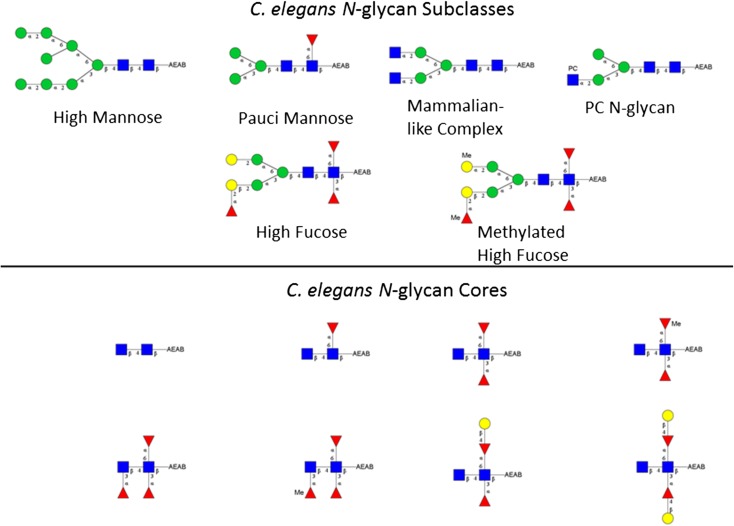
The C. elegans N-glycome can be divided into five subclasses including high mannose, pauci mannose, mammalian-like abbreviated complex, phosphocholine substituted, and high fucose. Biosynthesis of C. elegans N-glycans has been partially described (Schachter 2004; Paschinger et al. 2008) (see Figure 3 for structural orientation). The high mannose glycans are the archetypal high mannose glycans found in nearly all eukaryotes consisting of Man5-9GlcNAc2. Pauci mannose glycans arise through ER and Golgi trimming of the high mannose glycans to the compositions Man3-4GlcNAc2. They are formed subsequent to transient addition of GlcNAc to the Manα1,3-linked arm of the core. They may also contain Fucα1,6- at the aglycone-most GlcNAc of the core. Caenorhabditis elegans makes complex glycans that are similar to higher eukaryotes with extension of the canonical Man3GlcNAc2 (±core Fuc) with antennary GlcNAc. Up to five added GlcNAc residues have been reported (Haslam et al. 2002; Cipollo et al. 2005). LacdiNAc (GalNAcβ1-4GlcNAc) extensions have also been reported (Haslam et al. 2002) but are likely low in abundance although the β1,4-N-acetylgalactosaminyltransferase has been cloned and characterized (Kawar et al. 2005). Complex glycans can be modified with the addition of phosphorylcholine. This modification has been reported in the N-glycans in a range of other nematodes and trematodes (Sambasivam et al. 1993; Haslam et al. 1997; Morelle et al. 2000) and recently in lepidopteran species (Stanton et al. 2017). In pathogenic nematode and trematode species these glycans are thought to promote B- and T-cell anergy or tolerance, contributing to immune avoidance of the parasite (Harnett et al. 1999). The high fucose N-glycans are the most complex subclass by far. The aglycone-most GlcNAc can be both Fucα1,3- and Fucα1,6- substituted and both can be extended by the addition of Galβ1,4-. The penultimate GlcNAc can also be Fucα1,3- substituted and the core Manβ1,4- residue may be extended with Fucα1,2Gal-. The high fucose N-glycans can also contain Fucα1,2Galβ1,2- antennae that extend from the core Manα1,3- and Manα1,6- arms (Haslam et al. 2002). The high fucose subclass contains glycans with as many as four Fuc residues. Additionally, both Gal and Fuc can be methylated possibly serving as a stop signal for further extension. Importantly, seminal work was done using array technology to reveal the enzymatic basis for the complex core structure of C. elegans. Yan et al. (2013) produced a limited glycan array to interrogate C. elegans fucosyltransferase activity. Using core end extended glycoforms the authors investigated C. elegans fucosyltransferases in both solution and on array to reveal key enzymatic steps of the complex cores. This work helped to clarify controversy about the core structures by providing an enzymatic basis for the glycan structures reported (Yan et al. 2013).
Fig. 3.
Representative C. elegans AEAB labeled N-glycans. Subclasses are shown. The upper panel shows high mannose, pauci mannose, PC N-glycan, high fucose and methylated high fucose glycan representatives. The lower panel shows C. elegans core diversity as detected in this study. Designated linkages shown are inferred from the literature and our data.
A number of interactions that involve, or are likely to involve, CBP interactions with C. elegans glycoconjugates have been studied. Most of these involve recognition of C. elegans glycoconjugates by bacterial and fungal pathogens. Others involve C. elegans lectin interactions with glycoconjugates that could be self or non-self entities. Undoubtedly, many CBP interactions occur in the C. elegans niche. Below are several examples where pathogen or pathogenic factor recognition of C. elegans glycoconjugates have been studied.
Many glycosylation deficient C. elegans mutants have altered susceptibilities to microbial pathogens. Some C. elegans glycosylation patterns are required for pathogenic interactions when the nematode is co-cultured with pathogenic bacteria, or when exposed to virulence factors. The bus mutants bus-2 and bus-4, deficient in Ce core I O-glycans (Palaima et al. 2010; Parsons et al. 2014), are deficient in O-glycan in the rectum, anus and cuticle leading to an inability of Microbacterium nematophilum to infect these tissues (Gravato-Nobre et al. 2005). Yersinia species (Y. pestis and Y. pseudotuberculosis), that bind to the head region of wild-type C. elegans, causing an inability to feed, are unable to colonize the head region of these bus mutants (Sun et al. 2012). Thus, the bus mutants avoid starvation. Likely O-glycans are required for these pathogenic interactions. Mutations in the bus mutants may have an indirect effect on the innate immune system as well. The bus-2 mutants have altered fucosyl glycan staining in the hind gut, a primary site of innate immune function (Palaima et al. 2010) and our unpublished work suggests that the innate immune system in bus-2 and other bus mutants is altered.
The C. elegans bre mutants are deficient in production of glycolipids, and are resistant to Bacillus thuringiensis pore-forming toxins (Griffitts et al. 2003). The bre-5 gene encodes UDP-GlcNAc:Man GlcNAc transferase required in glycolipid synthesis. The bre-2, -3 and -4 are also active in this pathway. The bre-1 gene encodes a protein with significant homology to a GDP-mannose 4,6-dehydratase, a protein that catalyzes the first committed step in the biosynthesis of GDP-fucose from GDP-mannose. The above bre mutants are deficient in glycolipid and all have diminished susceptibility to the B. thuringiensis CRY toxin CRY-5A (Barrows et al. 2007). Glycolipids derived from C. elegans have been shown to be receptors for CRY-5B (Griffitts et al. 2005). The loss in bre mutant susceptibility to CRY toxins provides a specific example of glycoconjugate recognition by pathogenic factors of C. elegans glycoconjugates. That CRY toxins are active on other nematodes and insects (de Maagd et al. 2001) demonstrates that C. elegans can be a good model for investigation of protein–carbohydrate interactions for other nematode species as well as insects. The CRY toxins are used transgenically in agriculture as alternatives to more toxic chemical pesticides in defense of crops (Jouzani et al. 2017). Study of these toxins in the context of C. elegans bre mutants greatly aided in understanding of the Cry5B target across species and the toxin has been crystalized with its deminimus glycoconjugate ligand (Hui et al. 2012). There are over 200 Cry protein homologs many of which are active against nematodes and insect (de Maagd et al. 2001). A better understanding of their specificities could be beneficial to agricultural defense through the use of these proteins as they are less toxic than conventional pesticides.
The Coprinopsis cinerea mushroom lectin CCL2 is toxic in wild-type C. elegans. Its carbohydrate ligand is the trisaccharide GlcNAc(Fucα1,3)β1,4GlcNAc, a component of a significant percentage of C. elegans N-glycan cores (Cipollo et al. 2004b, Hanneman et al. 2006; Poltl et al. 2007). The CGL2 lectin of the same fungus recognizes the Galβ1,4Fucα1,6GlcNAc trisaccharide, a more novel moiety of C. elegans N-glycan core (Hanneman et al. 2006; Butschi et al. 2010). Recently both proteins were found to be toxic in H. contortus, inhibiting larval development (Heim et al. 2015). This helminth is the cause of significant economic loss in sheep and goat production. Ascaris suum and Oesophagostomum dentatum, which also produce the Gal β 1,4Fucα1,6GlcNAc N-glycan cores are also susceptible to CGL-2 lectin toxicity (Butschi et al. 2010).
The C. elegans genome encodes 278C-lectin homologs many of which have been linked to innate immune defense from pathogenic microbes. Their expression patterns are partially pathogen specific with overlapping specificities. Most of them contain a signal sequence and are likely secreted (Schulenburg et al. 2008). The specificities of the lectins are almost completely unknown. Only CLEC-79 has been investigated in this regard. Glycan array analysis revealed its ligand to be Galβ1-3GalNAc, which is the T-antigen, a common disaccharide in O-glycans of higher eukaryotes. It also forms the core of C. elegans Core-I O-glycans (Guerardel et al. 2001). Whether CLEC-79 recognizes self or other glycoconjugates is unknown. In the case of C. elegans Core-I O-glycans, the core T-antigen (Galβ1-3GalNAc) is highly substituted with beta linked Glc and GlcA with little overall T-antigen disaccharide detected as a percentage of overall O-glycan (Cipollo et al. 2004b; Palaima et al. 2010; Parsons et al. 2014). However, the possibility remains that this or other C. elegans C-type lectin may recognize self-ligand in the innate immunity cascade or other biological processes.
While it is not the purpose of this report to exhaustively review all carbohydrate protein interactions seen in C. elegans, the above examples demonstrate that pathogens of C. elegans target its glycoconjugates and that there is structural cross-over between the glycans of this nematode and those of other nematodes, trematodes and insect species. Similar structural motifs are recognized by pathogens in other species. This situation strongly suggests that, at least in some instances, C. elegans can serve as a model for these glycan-dependent pathogenic interactions and these have biomedical implications both in humans and in other mammals. Additionally, C. elegans has a vast repertoire of lectins whose specificities have remained largely unexplored, which identified them as fertile ground for exploration with current array technologies.
In this work we report a C. elegans N-glycan array containing 134 distinct glycoforms. These glycans were released from C. elegans glycoproteins with the N-glycanase PNGase A. Due to the broad specificity of this enzyme all five N-glycan subclasses are well represented and include a wide range of the high fucose N-glycans. This array should have broad applications to investigate protein–carbohydrate interactions that occur in the C. elegans model and those organisms that contain homologous N-glycan structures.
Results
Workflow
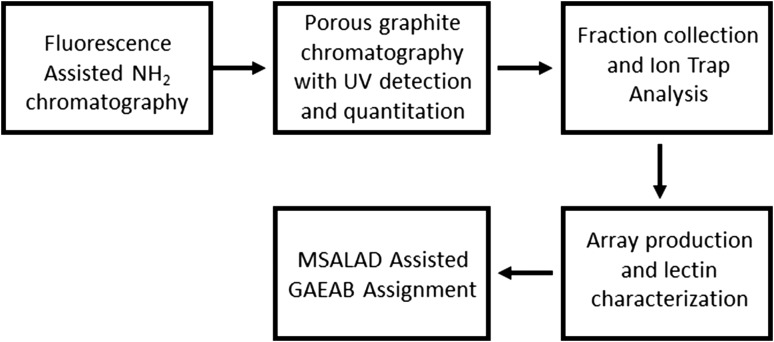
The workflow is shown in Figure 1. The two-dimensional HPLC strategy is based on that used in our previous publication (Song et al. 2009). 2-Amino-N-(2-amino-ethyl)-benzamide (AEAB) glycan derivatives (GAEABs) were produced as described in Materials and methods. Yield was approximated at greater than 90% as only trace amounts of label-less glycans were detected by mass spectrometry in the second dimension. Ten grams wet weight of mixed stages nematodes were used as glycan source. The first-dimension separation was performed by NH2 chromatography of the AEAB derivatized N-glycans released from C. elegans mixed stage, whole nematode extracts using PNGase A. Fluorescence detection was used for quantitation of saccharide contained in each fraction. Eleven fractions were collected (labeled Fractions 15–25 in the metadata file included in supplemental materials). The Chromatogram is shown in Supplementary data, Figure S1. Approximately 320 nmols of GAEABs were yielded based on integrated fluorescence.
Fig. 1.
Workflow diagram. Caenorhabditis elegans N-glycans were first separated using a low resolution NH2 column chromatography. Chromatography and quantitation of saccharides was assisted with fluorescence detection. Resultant fractions were individually separated using high resolution porous graphite chromatography in line with ion trap mass spectrometry. Infusion into the mass spectrometer as well as fraction collection was facilitated using a nanomate Triversa instrument. Glycan fractions were quantitated based on photodiode array detected UV absorbance and arrayed accordingly. An array of lectins was used to characterize arrayed GAEABs. Assignment of absorbance, lectin signal and mass spectrometry data were aided with the use of MSALAD and GlycoWorkBench.
Between 20 and 50 nmols of GAEABs from each NH2 fraction was introduced to a second-dimension separation using a Porous Graphite HPLC/MS ion trap system. The HPLC/MS system was equipped with a Nanomate Triversa source and fraction collector implemented such to split the eluent for approximately 500 nL to the ion trap mass spectrometer and the remainder was collected into 200 μL fractions. An in-line UV detector was used to allow quantitation of the AEAB saccharides to facilitate accurate sample amounts to be printed to array. Porous graphite HPLC is a high resolution system capable of separating glycan stereoisomers (Lipniunas et al. 1996). In this work over two hundred peaks were detected. Extracted ion chromatogram detection revealed up to eight isoforms per glycan composition based on peak separation at ½ height, molecular ion mass, elusion position differences greater than 0.5 min (fraction to fraction), and MS2 profile characteristics.
Assisted glycome characterization
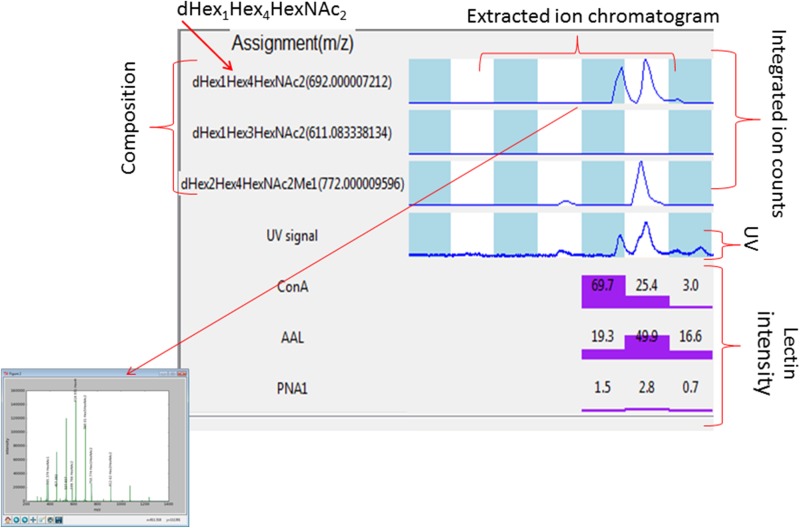
We used an in-house bioinformatics software program, MSALAD, to assist in visualization and analysis of the datasets. The program integrates UV, extracted ion chromatograms, mass spectrometric (MS and MS2) and lectin affinity (or other CBP) data in an interactive visualization platform. Compositional assignments are automated as are total glycan abundances per fraction. MS2 assignment is partially automated and served as a first pass for structural interrogation. See Figures 2 and 4 for example displays and further detail. MSALAD displays for all detected glycomers can be found in Supplementary data, Figures S2–S12. Data can be arranged in a range of formats for analysis via spreadsheet. Glycomers were defined using analytical parameters. The term “glycomer” is defined in the next paragraphs. The MS data can be searched glycome-wide for composition as well as fragment mass and intensities. Using this approach, we could identify key fragment ions and use them to assign glycomer distribution. This approach was particularly useful for core structure assignment but also useful for other structural components such as Pc substitution, antennary Fuc, HexNAc substitutions and others. Subsequent multiple passes through the data were performed manually.
Fig. 2.
Example MSALAD display and assignment aid. Glycan compositions are assigned using MSALAD monomer database. The software searches datasets for glycans by iteratively matching deconvoluted masses to a structural database. The database iteratively builds glycans based on monomer and substituent sets. Extracted ion chromatographs of assigned glycan compositions are displayed within each LC/MS run. Integrated counts for each composition are shown. Compositions are displayed along with ion m/z from which they were derived. The UV absorbance trace is shown. Point and click displays assigned MS2 mass spectrum. Normalized lectin intensities for Con A, HPA, AAL, PNA and SNA are shown. MSALAD was used as a first pass aid in assignment.
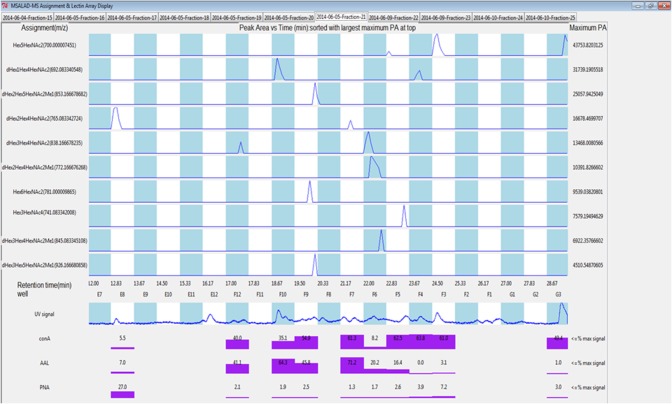
Fig. 4.
MSALAD display of Fraction 21 data. Separation of isomers can be seen for Hex5HexNAc2, dHex1Hex4HexNAc2, dHex2Hex4HexNAc2 and dHex3Hex4HexNAc2. Strong binding of PNA lectin (Gal) is seen for glycomer 28 in well E8. This glycomer has terminally linked Gal on the core.
Estimate of glycoforms present
Glycomers identified in this study are shown in Supplementary data, Table S1. The following steps were used to estimate the number of glycoforms: (1) Detected glycans were assigned compositions according to m/z and sorted. Metadata remain associated to indicate elution time, parent ion m/z and associate deconvoluted neutral mass, LC dimension 1 (NH2 chromatography) fraction, dimension 2 (PGC chromatography) elution time, and associated lectin data. (2) Glycans with the same composition that eluted in the second dimension within a user defined time increment are candidates to be the same glycomer. The first dimension is consulted for fractional overlap. (3) MS2 data are inspected to verify glycomer assignments. Since the mass spectrometer conditions are essentially identical for each spectrum collected, and daughter ion m/z and ion intensity are reflective of their origin, spectra of like glycans will have virtually identical spectra whereas those that are different will not. (4) Lectin data were used to infer composition. The lectins Con A, AAL and PNA lectins were useful for ascribing Man, Fuc and Gal containing compositions. To summarize, since it is possible that neighboring fractions taken from the first dimension can have the same GAEABs we consulted first and second dimension elution times, MS1 and MS2 data, and lectin intensities to judge which peaks were likely to represent the same species. When fractions coincided in both dimensions, mass matched and MS2 fragmentation patterns were essentially identical, those GAEABs within the fractional overlap regions were determined to be the same glycans. Using this approach, we identified 146 putative glycoforms referred to herein as glycomers. All of these were printed except for 1, 49, 60, 82, 87, 100, 108, 112, 125, 142, 145 and 146 due to low abundance. The metadata can be seen in the supplemental materials metadata_FNL file. The reader should note that we use the term “glycomer” here rather than a more definitive term such as isomer when referring to these 146 carbohydrate entities. While they are highly characterized information is not complete enough to specifically define all linkages, branch structures and anomeric configurations. Therefore, we use the term glycomer.
Determination of subclass members
MSALAD derived extracted ion intensities were used to group glycans by composition and calculate the % total ion intensity for each glycan subclass. In the case of the C. elegans N-glycome the following compositions were used to define each noted subclass: Hex5-9HexNAc2 are high mannose glycans; Fuc0-1Hex1-3HexNAc2 are pauci mannose glycans; Me0-2dHex2-4Hex3-5HexNAc2 are high fucose glycans (plus dHex1Hex3-4HexNAc2 with antennary Fuc; these were excluded from Pauci); Me0-2dHex0-1Hex3-6HexNAc3-4 are complex and; Pc1-3Hex3HexNAc3 are phosphocholinyl glycans. In this study we found that the majority of glycans were either high mannose (17%) or high fucose (48.8%) glycans. Pauci mannose glycans were 15.6%. Complex glycans were 2.1% and phosphocholinyl glycans were 1.3% based on ion intensity. About 11.5% of the fucosyl glycans had at least one methyl substitution. The distribution of compositions is shown in Supplementary data, Table S1. While the ionization potentials across the range of glycans detected are not likely to be the same, this analysis can serve as a first approximation of glycan composition distributions. The C. elegans relative distributions reported here are similar compared to published data that used semi-quantitative permethylation analysis of PNGase A released glycans (Cipollo et al. 2004b). The MS data used in this study was generated in positive polarity mode. Ions were predominantly of the [M+nH]n+ form. While many structural attributes could be assigned, these ions generally do not produce significant cross ring fragments, limiting the ability to assign linkage information. However, much of the structure can be inferred based on known linkages and branch configurations present in the C. elegans N-glycome. See Figure 3 for examples of structural detail inferred based on the current understanding of C. elegans glycosylation and our data. Discussion on structural features in the paragraphs that follow will be discussed in these terms.
Structural features of C. elegans N-glycan array glycomers
Pauci Mannose and High Fucose Glycans
It is known that Fuc can rearrange under mass spectrometry conditions in native and reducing end glycans derivatives (Harvey et al. 2002; Wuhrer et al. 2006). We did observe low abundance fragment ion intensities that were consistent with rearrangement. As part of the screening process we chose a cutoff of 10% base peak height to discount fragment signals where Fuc was present to limit the use of Fuc transfer information in structural assignments. This resulted in structural assignment trends that are expected based on known C. elegans glycan configurations and biosynthetic pathways. However, we cannot rule out the possibility that some Fuc assignments may be in error.
The pauci mannose glycoforms (dHex0-1Hex1-4HexNAc2) included glycomers 1–2, 4, 6–7, 11–14, 16, 18, 20, 24 and 100–103 (Supplementary data, Table S1). These glycans are reported to rise through transient addition of GlcNAcβ1,2 to the core followed by addition of Fucα1,3- to the aglycone GlcNAc. After this action a Golgi N-acetyl glucosidase removes the added GlcNAc forcing the formation of either pauci mannose or possibly high fucose glycans (Schachter 2004). The tendency within this group was for strong Con A and AAL signal as can be expected since their compositions contain primarily Man and Fuc. See Supplementary data, Figures 2–12 and supplemental Table 2 metadata_FNL for details.
Glycomers 3, 5, 15, 17, 19, 21–23, 25–99 (Supplementary data, Table S1) were of the high fucose subtype (dHex1-4Hex3-5HexNAc2). The core chitobiose could be substituted with up to three Fuc residues. The reducing end GlcNAc can contain both Fucα1,6- and Fucα1,3- substitutions whereas the penultimate GlcNAc can be substituted with Fucα1,3-. See Figure 3 for structural examples. There was evidence in the data that all three fucose residues can be substituted with a Methyl group. A Galβ1,4 extension of the core Fucα1,6 was observed in some glycomers (glycomers 47–48, 50, 63–64). Substitution of both Fucα1,6- and Fucα1,3- with Galβ1,4- was also observed (i.e., glycomer 65), however, these compounds were low in intensity in these PNGase A-released glycans. Some fucosyl glycans were not arrayed including 49, 60, 82, and 87 due to low overall abundance of glycans in the corresponding PGC fraction. Fucα1,2Gal- antennary substitutions were also observed (glycomers 68, 72, 74–75). Some of these Fuc were also methylated (i.e., 63, 79, 88). Patterns consistent with antennary Gal methyl substitution were also observed (i.e., glycomers 80–81, 95). Con A and AAL bound strongly to this glycan class. It is notable that glycomers with methyl Fuc substitution tended to produce less AAL lectin signal intensity. Those with core or antennary Gal tended to bind PNA lectin such as 5 and 85.
A likely useful aspect of this array is that it is comprised of at least one representative of nearly every high fucose glycan biosynthetic intermediate known. This should allow some level of dissection of binding preferences for proteins analyzed on the array. For instance, glycomers 1–6 represent a range of different single Fuc substitutions of the core or antennae. There are representatives of penultimate GlcNAc substitution, aglycone GlcNAc substitution, single antennary Fuc substitution and Gal capping of the aglycone Fucα1,6GlcNAc. This set may be informative to dissect preferences of proteins that bind fucosylated cores or prefer Fuc located in the antennae for example. To inspect the array for structures of interest it is useful to compare those in Figure 3 with those on the array seen in Supplementary data, Table S1. Cross reference with metadata_FNL file will reveal the positions of each GAEAB glycomer on the array. Although fucosyl forms 1, 49, 60, 82 and 87 were not printed due to low abundance, all possible fucosyl substructures are still represented.
Complex and PC substituted glycans
Mammalian-like complex glycans were of the form dHex0-1Hex3HexNAc3-4 (glycomers 8–10, 104–106, 110–111, see Supplementary data, Table S1). Larger forms were of very low abundance and therefore not arrayed. Fucα1,6- can substitute the core aglycone GlcNAc. These glycans can contain GlcNAcβ1,2- extensions of the core Man arms and can also contain core bisecting GlcNAcβ1,4-. PC substituted glycoforms on the array were limited. Three Pc1Hex3HexNAc3 glycomers were arrayed (glycomers 107–109). All three glycoforms eluted well apart thus representing three isomers.
High mannose and intermediate glycans
High mannose glycans are of the form Man5-8GlcNAc2 (glycomers 117–120, 122–125, 127–134, 135–137, 141–146, see Supplementary data, Table S1). There were an estimated 27 glycomers in this group. As expected, Con A signal was high for most glycans in this class and signal intensity tended to increase with the ion intensity detected for these glycomers. Our assignments gave eight glycomers for Man5GlcNAc2, 10 glycomers for Man6GlcNAc2, five glycomers for Man7GlcNAc2 and four glycomers for Man8GlcNAc2. Some of these signals were very low and could have been representatives of degradation intermediates or rarer synthetic intermediates. It is possible that some of these were also representative of Golgi processing intermediates that contain Gal but were not abundant enough to provide adequate PNA signal. The Gal substitution of high mannose glycans has been previously reported and these are likely to be intermediates in the formation of high fucose glycans (Cipollo et al. 2004b). Possibly, low abundance assigned Man5GlcNAc2 and Man6GlcNAc2 glycomers (where some Man may be Gal) may contain Gal as these would be biosynthetically more likely to contain this substitution. Also, conforming to the compositions Hex5-7HexNAc2 were glycomers 121, 126, 139–140 and 138. These had Gal indicating PNA lectin signal and were likely intermediates in the formation of high fucose glycans.
Discussion
We have produced a C. elegans N-glycan array. The array contains 146 distinct glycomers characterized by elution positions in two-dimensional chromatography, MS1 and MS2 datasets, and lectin characterization data. An in-house informatics software MSALAD was used to aid in integration of datasets and facilitate structural assignment. Based on the data and structural inferences derived from published work concerning C. elegans N-glycans much of the structural detail of the arrayed glycans can be inferred.
The high mannose glycans present include structural isomers of the compositions Man5-8GlcNAc2. Man9GlcNAc2 was not present in amounts required for inclusion in the array (approximately 0.5 nmol for 300 slides). Interestingly a Man5Gal1GlcNAc2 structure was present (glycomer 126) which we have previously reported (Cipollo et al. 2004b). This glycan has a Gal extension from core Man and likely represents an intermediate in the synthesis of the high fucose C. elegans N-glycans.
Seemingly, the least interesting subclass present on the array is the pauci mannose oligosaccharides. These are composed of Fuc0-2Man1-4GlcANc2. The most abundant among these in the glycome are the canonical Man3GlcNAc2 and its fucosylated form Fuc1Man3GlcNAc2. However, these are biosynthetically interesting in that likely their synthesis requires first the addition of Fucα1,6- to the core aglycone GlcNAc followed by addition of GlcNAcβ1,2- to the core Manα1,3- arm, followed by removal of Man residues from the upper and central arm (Schachter 2004). The fate of the saccharide is then determined by the action of Golgi N-acetylglucosaminidase, which can remove the GlcNAcβ1,2- leaving a dead-end product. If the saccharide avoids the enzyme’s activity, presumably, the fate is towards mammalian like complex and Pc substituted forms. It is intriguing that the Mana1,3 arm appears to be substituted with either GlcNAcβ1,2- or Galβ1,2-. Whether these reactions compete is not known. If they do, then these two steps are important in determination of the end fate to be either high fucose or mammalian-like complex and Pc substituted. Essentially, all intermediates for these two processes are present on the array.
The mammalian-like complex glycans present on the array include Man3-5GlcNAc3-4. Larger members of this class have been reported by us and others (Haslam et al. 2002; Zhu et al. 2004; Cipollo et al. 2005; Paschinger et al. 2008). However, the larger forms are of very low abundance, in our experience, and were not detected in our preparations. Seven glycomers representative of this class are present on the array (glycomers 8–10, 104–106 and 110).
Three phosphocholine N-glycans are present on the array. All contain a single phosphocholine (PC) substitution. Low abundance forms with up to 4 PC substitutions have been reported by us and others (Cipollo et al. 2002, 2005; Haslam et al. 2002; Yan et al. 2015c). The biosynthesis of these glycans has been characterized (Houston and Harnett 2004; Cipollo et al. 2004a). The C. elegans PC N-glycans are structurally similar or identical to those reported in other nematode, trematode and lepidopteran species (Haslam et al. 1997; Paschinger and Wilson 2015; Poltl et al. 2007). Harnett et al. have reported that these glycans are involved in processes that render the host immune system suppressed through interactions with both B- and T-cells (Wilson et al. 2003) and may modulate complement activation (Ahmed et al. 2016). Although limited in number, the presence of a subset of these glycans on this array may serve for further investigation of relevant interactions of immune system components or other proteins with this class of glycans.
The array’s most numerous and abundant glycan subclass was high fucose. Eighty one of these are present on the array (Glycomers 15, 17, 19, 21–24, 26–99). The complexity of this subclass strongly suggests that they play many roles in C. elegans biology. A range of core structures are present with up to three Fuc present on the chitobiose core. Methyl group substitution is seen on all three of these Fuc residues. In no case was Gal substitution of Fuc indicated when the Fuc was methylated (MeFuc). Both core and antennary MeFuc was observed. Methylated Gal (MeGal) was also present but primarily in the antennae. As described previously, a subset of these glycans have been reported to act as receptors for the Coprinopsis cinerea mushroom toxic lectins CCL2 and CGL2, which bind the core trisaccharides GlcNAc(Fucα1,3)β1,4GlcNAc, and Gal β 1,4Fucα1,6GlcNAc, respectively. These core sequences are present or likely to be present in other trematodes and nematodes such as Haemonchus contortus (Haslam et al. 1996, 1998; Paschinger and Wilson 2015) and Pristionchus pacificus (Yan et al. 2015c). This subclass of glycans are likely to be targets for many CBPs encountered in the natural C. elegans ecological niche and those of other nematodes, trematodes and lepidopteran species that also contain these glycan structures or close structural homologs. A simple blast search of CCL2 reveals a range of homologs in various fungal species such as Pleurotus ostreatus, a known predator of nematodes (Satou et al. 2008). Similarly the fungus Laccaria bicolor, also with anti-nematode properties (Wohlschlager et al. 2014), bears a galectin homolog of CGL-2 (Martin et al. 2008). The array presented here may serve as an analytical platform to investigate other microbial lectins and other toxins with regard to nematodes, trematodes and insect species where structural homologies exist between their glycome and that of C. elegans.
Caenorhabditis elegans possesses a large repertoire of lectins. Those best described are the galectins and C-type lectins. Caenorhabditis elegans galectins LEC-6 and LEC-10 have been reported to recognize glycan targets in the nematode intestine, pharynx and rectal valve and fut-8, bre-1 and galt-1 mutants do not bind these lectins in these tissues (Maduzia et al. 2011). All three of the encoded enzymes are required for the synthesis of the Galβ1,4Fuc N-glycan core modification. LEC-1 through -4 also show affinity for β-galactosides (Nemoto-Sasaki et al. 2008). However, their preferences were investigated by frontal affinity using largely mammalian type oligosaccharides and their substructures. The authors concluded that some lectin specialization, in terms of C. elegans niche specific glycoconjugate interactions, likely exists in these lectins (Nemoto-Sasaki et al. 2008). Clearly, the array presented here could prove useful for further investigation of C. elegans galectin preferences. As described previously, there are 278 C. elegans C-type lectins. Other than CLEC-79, which recognizes Galβ1,3GalNAc as discussed earlier, to our knowledge, no other ligand preference has been described. These lectins are widely expected to be active as part of C. elegans adaptive immunity (Pees et al. 2016). However, these proteins are largely unexplored in terms of ligand preferences, another area where this array could prove useful.
Here we have presented a highly characterized C. elegans N-glycan array. The high mannose, pauci mannose, mammalian like complex, PC substituted and high fucose glycoforms are represented. In particular, the high fucose glycoforms, with 83 glycomers, represents more than half of the array. Most conceivable intermediates are likely to be present which promises a high degree of interrogation capability for CBPs that target this class. The major glycoforms in the other four classes are also present with three PC glycomers, eight mammalian-like complex, 36 high mannose including some Gal substituted intermediates, and 16 pauci mannose glycomers. This array should prove useful for screening of binding preferences for a wide range of CBPs including those in the nematode, trematode and insect niches and others where structural conservation with the arrayed glycans is contained.
Materials and methods
The array was constructed using the guidelines provided by the Minimum Information for a Glycomics Experiment Project (MIRAGE) as reported here (Liu et al. 2016).
Materials
Maltooligosaccharide, bovine pancreas ribonuclease B and sodium cyanoborohydride were purchased from Sigma Aldrich. The 2-amino-N-(2-aminoethyl)-benzamide (AEAB) used as linker was synthesized by condensation of methyl anthranilate with ethylenediamine as previously described to yield the AEAB-HCl salt (Song et al. 2009). Lectins used were purchased from Vector Laboratories and included Griffonia Simplicifolia Lectin I (GSL-1), (Con A), PHA-L, Griffonia Simplicifolia Lectin II (GSL-II), Ricinus Communis Agglutinin I (RCA-I), Helix pomatia lectin(HPA), Sambucus Nigra (SNA), Aleuria Aurantia Lectin (AAL), Maackia Amurensis Lectin (MAL), Wheat Germ Agglutinin (WGA) and Peanut Agglutinin (PNA). Biotinylated lectins were from Vector Labs (Burlingame, CA). All chemicals used were ACS grade or higher.
Preparation of C. elegans N-glycans
Caenorhabditis elegans N2 Bristol strain mixed stages nematodes were used as the N-glycan source. Nematodes were grown in large scale culture and harvested as previously described (Parsons et al. 2014). Approximately 10 g (wet weight) of flash frozen nematodes were processed to yield glycoprotein rich extracts and glycans were released from protein using PNGase A as previously described (Palaima et al. 2010).
Porous graphite solid phase extraction and quality assessment
The glycans were purified by porous graphite (Hypersep Hypercarb 100 mg/1 mL, Thermoscientific; 60,106-302) solid phase extraction. Briefly, columns were equilibrated using sequential 1 mL additions of 100%, 60% and 30% acetonitrile/0.1% TFA followed by 0.1% TFA. Glycans diluted in 1% butanol were then applied and washed with 3 mL of 0.1% TFA in water. Eluted fractions from 30% acetonitrile/0.1% TFA and 60% acetonitrile/0.1% TFA were collected and rotary evaporated for further analysis.
An aliquot of approximately 5 μg of N-glycans was permethylated (Ciucanu and Kerek 1984) and their quality assessed by MALDI-TOF MS on a Perseptive Biosystems Voyager DE RF MALDI-TOF mass spectrometer. Data were processed using DataExplorer (Perseptive Biosystems). The permethylated glycans were suspended in 50% acetonitrile/water, spotted onto a MALDI plate and mixed 1:1 with 2,5-DHB suspended in 1 mM sodium acetate, 50% acetonitrile and analyzed in positive polarity reflectron mode.
Preparation of AEAB oligosaccharide derivatives
Equal volumes of 0.5 M 2-amino-N-(2-amino-ethyl)-benzamide (AEAB) in DMSO and 1 M cyanoborohydride in 7:3 (v/v) DMSO/acetic acid were added to dried glycan samples (10–1000 mg). The solution was vortexed for 2 min and incubated at 65°C in a heating block for 2 h. The samples were allowed to cool at room temperature and the glycans precipitated by the addition of 10 volumes of acetonitrile followed by incubation at –20°C for 30 min. The precipitate was collected by centrifugation and dried by rotary evaporation. The AEAB glycans (GAEAB) were reconstituted in distilled water.
First dimension separation: NH2 chromatography
Approximately 250 μg of GAEAB were analyzed. The aminated saccharides were separated by NH2 chromatography on an Agilent 1200 HPLC system equipped with a G1311A quaternary pump, G1322A degasser, G1364C fraction collector and a G1321A fluorescence detector. An Agilent Zorbax NH2 250 × 4.6 mm column was used. Particle size was 5 μm. A three solvent system was used where solvent A = 100% acetonitrile, solvent B = 250 mM ammonium acetate, and solvent C = HPLC grade water. Starting conditions were solvent A = 80%, solvent B = 4% and solvent C = 16%. A linear gradient was applied over 90 min ending at solvent A = 10%, solvent B = 50% and solvent C = 40%. Fluorescence detection was accomplished with excitation performed at 330 nm and emission at 420 nm. Quantitation was accomplished with establishment of a linear curve using lactose-AEAB saccharide derivative. Twenty-five fractions were collected and their glycan amounts estimated in preparation for second dimension separation.
Second dimension separation: porous graphite with ion trap LC/MS
A porous graphite (PGC) column (Thermo Scientific, 4.6 mm × 150 mm) was used with the following binary solvent system. Solvent A: Acetonitrile with 0.1% formic acid and solvent B: water with 0.1%formic acid. A biphasic gradient was used. Phase 1 was 5% acetonitrile for 10 min followed by phase 2 where acetonitrile was increased linearly from 5% to 50% over 63 min. An Advion Triversa Nanomate automated nanospray chip-based fraction collector (ANCSF) was used as the ion trap source and fraction collector. A 1:100 split was employed with one part injected into the ion trap and the remaining sample collected in 96 well plates for further processing and analysis. Infusion into the ion trap was approximately 500 nL/min through 5 mm nozzles.
The Thermo LTQ XL ion trap was programmed for a six stage MS experiment. The first segment performed a scan of 150–2000 m/z. The remaining scans were designed to perform MSn scans of up to the five most abundant ions in the targeted scan using the following positive mode settings: source voltage 5 kV, capillary temperature 170°C, and normalized collision energy 35%. Normalized collision energy is calculated by the instrument’s Xcalibur® software, which applies a linear function that is dependent upon precursor ion mass and the amplitude of the resonance excitation radio frequency value. A photodiode array was used to detect UV absorbance at 330 nm. Quantitation of eluted saccharide was accomplished using a linear curve produced using lactose-AEAB.
Printing, binding assay and scanning
NHS-activated slides were purchased from Schott. Non-contact printing was performed using a Piezorray Printer (Perkin Elmer) as previously described (Yu et al. 2014). Biotinylated lectins were used in the binding assay and the bound lectins were detected by a secondary incubation with Cy5-SA. The slides were scanned with a Perkin Elmer ProScanArray microarray scanner equipped with four lasers covering an excitation range from 488 to 637 nm. The Lectin binding assays, image scanning methods using the ScanArray Express software, and methods details are described in Yu et al. (2012, 2014). The specific layout of the glycans on the array are provided in supplemental materials file metadata_FNL. Relative fluorescence units for all lectins was expressed as percent for display in MSALAD.
Supplementary Material
Abbreviations
AEAB, 2-amino-N-(2-aminoethyl)-benzamide; AAL, Aleuria Aurantia Lectin; Con A, concanavalin A lectin; CPB, carbohydrate binding protein; GAEAB, 2-amino-N-(2-aminoethyl)-benzamide labeled glycan; GSL-1, Griffonia Simplicifolia Lectin I; GSL-II, Griffonia Simplicifolia Lectin II (GSL-II); HPA, helix pomatia lectin; MAL, Maackia Amurensis Lectin; PHA-L, Phaseolus Vulgaris Leucoagglutinin; PNA, Peanut Agglutinin; RCA-I, Ricinus Communis Agglutinin I; SNA, Sambucus Nigra; WGA, Wheat Germ Agglutinin
Supplementary data
Funding
This work was supported by grants 5P41GM103694-06 to R.C., R01 GM085448 to D.S., by Oak Ridge Institute for Science and Education and Program Grant Z01 BJ 02044-09 to J.F.C.
Conflict of interest statement
None declared.
References
- Ahmed UK, Maller NC, Iqbal AJ, Al-Riyami L, Harnett W, Raynes JG. 2016. The carbohydrate-linked phosphorylcholine of the parasitic nematode product ES-62 modulates complement activation. J Biol Chem. 291:11939–11953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrows BD, Haslam SM, Bischof LJ, Morris HR, Dell A, Aroian RV. 2007. Resistance to Bacillus thuringiensis toxin in Caenorhabditis elegans from loss of fucose. J Biol Chem. 282:3302–3311. [DOI] [PubMed] [Google Scholar]
- Butschi A, Titz A, Walti MA, Olieric V, Paschinger K, Nobauer K, Guo X, Seeberger PH, Wilson IB, Aebi M et al. . 2010. Caenorhabditis elegans N-glycan core beta-galactoside confers sensitivity towards nematotoxic fungal galectin CGL2. PLoS Pathog. 6:e1000717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byres E, Paton AW, Paton JC, Lofling JC, Smith DF, Wilce MC, Talbot UM, Chong DC, Yu H, Huang S et al. . 2008. Incorporation of a non-human glycan mediates human susceptibility to a bacterial toxin. Nature. 456:648–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Spence AM, Schachter H. 2003. Isolation of null alleles of the Caenorhabditis elegans gly-12, gly-13 and gly-14 genes, all of which encode UDP-GlcNAc: Alpha-3-D-mannoside beta1,2-N-acetylglucosaminyltransferase I activity. Biochimie. 85:391–401. [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad AM, Costello CE, Hirschberg CB. 2004. b. srf-3, a mutant of Caenorhabditis elegans, resistant to bacterial infection and to biofilm binding, is deficient in glycoconjugates. J Biol Chem. 279:52893–52903. [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad AM, Costello CE, Hirschberg CB. 2005. N-glycans of Caenorhabditis elegans are specific to developmental stages. J Biol Chem. 280:26063–26072. [DOI] [PubMed] [Google Scholar]
- Cipollo JF, Awad A, Costello CE, Robbins PW, Hirschberg CB. 2004. a. Biosynthesis in vitro of Caenorhabditis elegans phosphorylcholine oligosaccharides. Proc Natl Acad Sci U S A. 101:3404–3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollo JF, Costello CE, Hirschberg CB. 2002. The fine structure of Caenorhabditis elegans N-glycans. J Biol Chem. 277:49143–49157. [DOI] [PubMed] [Google Scholar]
- Ciucanu I, Kerek F. 1984. A simple and rapid method for the permethylation of carbohydrates. Carbohydr Res. 131:209–217. [Google Scholar]
- de Maagd RA, Bravo A, Crickmore N. 2001. How Bacillus thuringiensis has evolved specific toxins to colonize the insect world. Trends Genet. 17:193–199. [DOI] [PubMed] [Google Scholar]
- Fukui S, Feizi T, Galustian C, Lawson AM, Chai W. 2002. Oligosaccharide microarrays for high-throughput detection and specificity assignments of carbohydrate-protein interactions. Nat Biotechnol. 20:1011–1017. [DOI] [PubMed] [Google Scholar]
- Geyer H, Schmidt M, Muller M, Schnabel R, Geyer R. 2012. Mass spectrometric comparison of N-glycan profiles from Caenorhabditis elegans mutant embryos. Glycoconj J. 29:135–145. [DOI] [PubMed] [Google Scholar]
- Girard LR, Fiedler TJ, Harris TW, Carvalho F, Antoshechkin I, Han M, Sternberg PW, Stein LD, Chalfie M. 2007. WormBook: The online review of Caenorhabditis elegans biology. Nucleic Acids Res. 35:D472–D475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravato-Nobre MJ, Nicholas HR, Nijland R, O’Rourke D, Whittington DE, Yook KJ, Hodgkin J. 2005. Multiple genes affect sensitivity of Caenorhabditis elegans to the bacterial pathogen Microbacterium nematophilum. Genetics. 171:1033–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffitts JS, Haslam SM, Yang T, Garczynski SF, Mulloy B, Morris H, Cremer PS, Dell A, Adang MJ, Aroian RV. 2005. Glycolipids as receptors for Bacillus thuringiensis crystal toxin. Science. 307:922–925. [DOI] [PubMed] [Google Scholar]
- Griffitts JS, Huffman DL, Whitacre JL, Barrows BD, Marroquin LD, Muller R, Brown JR, Hennet T, Esko JD, Aroian RV. 2003. Resistance to a bacterial toxin is mediated by removal of a conserved glycosylation pathway required for toxin-host interactions. J Biol Chem. 278:45594–45602. [DOI] [PubMed] [Google Scholar]
- Guerardel Y, Balanzino L, Maes E, Leroy Y, Coddeville B, Oriol R, Strecker G. 2001. The nematode Caenorhabditis elegans synthesizes unusual O-linked glycans: Identification of glucose-substituted mucin-type O-glycans and short chondroitin-like oligosaccharides. Biochem J. 357:167–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanneman AJ, Rosa JC, Ashline D, Reinhold VN. 2006. Isomer and glycomer complexities of core GlcNAcs in Caenorhabditis elegans. Glycobiology. 16:874–890. [DOI] [PubMed] [Google Scholar]
- Harnett W, Deehan MR, Houston KM, Harnett MM. 1999. Immunomodulatory properties of a phosphorylcholine-containing secreted filarial glycoprotein. Parasite Immunol. 21:601–608. [DOI] [PubMed] [Google Scholar]
- Harvey DJ, Mattu TS, Wormald MR, Royle L, Dwek RA, Rudd PM. 2002. “Internal residue loss”: Rearrangements occurring during the fragmentation of carbohydrates derivatized at the reducing terminus. Anal Chem. 74:734–740. [DOI] [PubMed] [Google Scholar]
- Haslam SM, Coles GC, Munn EA, Smith TS, Smith HF, Morris HR, Dell A. 1996. Haemonchus contortus glycoproteins contain N-linked oligosaccharides with novel highly fucosylated core structures. J Biol Chem. 271:30561–30570. [DOI] [PubMed] [Google Scholar]
- Haslam SM, Coles GC, Reason AJ, Morris HR, Dell A. 1998. The novel core fucosylation of Haemonchus contortus N-glycans is stage specific. Mol Biochem Parasitol. 93:143–147. [DOI] [PubMed] [Google Scholar]
- Haslam SM, Gems D, Morris HR, Dell A. 2002. The glycomes of Caenorhabditis elegans and other model organisms. Biochem Soc Symp. 69:117–134. [PubMed] [Google Scholar]
- Haslam SM, Khoo KH, Houston KM, Harnett W, Morris HR, Dell A. 1997. Characterisation of the phosphorylcholine-containing N-linked oligosaccharides in the excretory-secretory 62 kDa glycoprotein of Acanthocheilonema viteae. Mol Biochem Parasitol. 85:53–66. [DOI] [PubMed] [Google Scholar]
- Heim C, Hertzberg H, Butschi A, Bleuler-Martinez S, Aebi M, Deplazes P, Kunzler M, Stefanic S. 2015. Inhibition of Haemonchus contortus larval development by fungal lectins. Parasit Vectors. 8:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston KM, Harnett W. 2004. Structure and synthesis of nematode phosphorylcholine-containing glycoconjugates. Parasitology. 129:655–661. [DOI] [PubMed] [Google Scholar]
- Hui F, Scheib U, Hu Y, Sommer RJ, Aroian RV, Ghosh P. 2012. Structure and glycolipid binding properties of the nematicidal protein Cry5B. Biochemistry. 51:9911–9921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouzani GS, Valijanian E, Sharafi R. 2017. Bacillus thuringiensis: A successful insecticide with new environmental features and tidings. Appl Microbiol Biotechnol. 101:2691–2711. [DOI] [PubMed] [Google Scholar]
- Kawar ZS, Haslam SM, Morris HR, Dell A, Cummings RD. 2005. Novel poly-GalNAcbeta1-4GlcNAc (LacdiNAc) and fucosylated poly-LacdiNAc N-glycans from mammalian cells expressing beta1,4-N-acetylgalactosaminyltransferase and alpha1,3-fucosyltransferase. J Biol Chem. 280:12810–12819. [DOI] [PubMed] [Google Scholar]
- Lipniunas PH, Neville DC, Trimble RB, Townsend RR. 1996. Separation of biosynthetic oligosaccharide branch isomers using high-performance liquid chromatography on a porous two-dimensional graphite stationary phase. Anal Biochem. 243:203–209. [DOI] [PubMed] [Google Scholar]
- Liu Y, McBride R, Stoll M, Palma AS, Silva L, Agravat S, Aoki-Kinoshita KF, Campbell MP, Costello CE, Dell A et al. . 2016. The minimum information required for a glycomics experiment (MIRAGE) project: Improving the standards for reporting glycan microarray-based data. Glycobiology. 27:280–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduzia LL, Yu E, Zhang Y. 2011. Caenorhabditis elegans galectins LEC-6 and LEC-10 interact with similar glycoconjugates in the intestine. J Biol Chem. 286:4371–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Aerts A, Ahren D, Brun A, Danchin EG, Duchaussoy F, Gibon J, Kohler A, Lindquist E, Pereda V et al. . 2008. The genome of Laccaria bicolor provides insights into mycorrhizal symbiosis. Nature. 452:88–92. [DOI] [PubMed] [Google Scholar]
- Morelle W, Haslam SM, Olivier V, Appleton JA, Morris HR, Dell A. 2000. Phosphorylcholine-containing N-glycans of Trichinella spiralis: Identification of multiantennary lacdiNAc structures. Glycobiology. 10:941–950. [DOI] [PubMed] [Google Scholar]
- Muthana SM, Gildersleeve JC. 2014. Glycan microarrays: Powerful tools for biomarker discovery. Cancer Biomark. 14:29–41. [DOI] [PubMed] [Google Scholar]
- Nemoto-Sasaki Y, Hayama K, Ohya H, Arata Y, Kaneko MK, Saitou N, Hirabayashi J, Kasai K. 2008. Caenorhabditis elegans galectins LEC-1-LEC-11: Structural features and sugar-binding properties. Biochim Biophys Acta. 1780:1131–1142. [DOI] [PubMed] [Google Scholar]
- Palaima E, Leymarie N, Stroud D, Mizanur RM, Hodgkin J, Gravato-Nobre MJ, Costello CE, Cipollo JF. 2010. The Caenorhabditis elegans bus-2 mutant reveals a new class of O-glycans affecting bacterial resistance. J Biol Chem. 285:17662–17672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons LM, Mizanur RM, Jankowska E, Hodgkin J, O Rourke D, Stroud D, Ghosh S, Cipollo JF. 2014. Caenorhabditis elegans bacterial pathogen resistant bus-4 mutants produce altered mucins. PLoS One. 9:0107250, doi:10.1371/journal.pone. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschinger K, Gutternigg M, Rendic D, Wilson IB. 2008. The N-glycosylation pattern of Caenorhabditis elegans. Carbohydr Res. 343:2041–2049. [DOI] [PubMed] [Google Scholar]
- Paschinger K, Wilson IB. 2015. Two types of galactosylated fucose motifs are present on N-glycans of Haemonchus contortus. Glycobiology. 25:585–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen JW, Blixt O, Bennett EP, Tarp MA, Dar I, Mandel U, Poulsen SS, Pedersen AE, Rasmussen S, Jess P et al. . 2011. Seromic profiling of colorectal cancer patients with novel glycopeptide microarray. Int J Cancer. 128:1860–1871. [DOI] [PubMed] [Google Scholar]
- Pees B, Yang W, Zarate-Potes A, Schulenburg H, Dierking K. 2016. High Innate immune specificity through diversified C-type lectin-like domain proteins in invertebrates. J Innate Immun. 8:129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltl G, Kerner D, Paschinger K, Wilson IB. 2007. N-glycans of the porcine nematode parasite Ascaris suum are modified with phosphorylcholine and core fucose residues. FEBS J. 274:714–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambasivam H, Rassouli M, Murray RK, Nagpurkar A, Mookerjea S, Azadi P, Dell A, Morris HR. 1993. Studies on the carbohydrate moiety and on the biosynthesis of rat C-reactive protein. J Biol Chem. 268:10007–10016. [PubMed] [Google Scholar]
- Satou T, Kaneko K, Li W, Koike K. 2008. The toxin produced by pleurotus ostreatus reduces the head size of nematodes. Biol Pharm Bull. 31:574–576. [DOI] [PubMed] [Google Scholar]
- Schachter H. 2004. Protein glycosylation lessons from Caenorhabditis elegans. Curr Opin Struct Biol. 14:607–616. [DOI] [PubMed] [Google Scholar]
- Schulenburg H, Hoeppner MP, Weiner J 3rd, Bornberg-Bauer E. 2008. Specificity of the innate immune system and diversity of C-type lectin domain (CTLD) proteins in the nematode Caenorhabditis elegans. Immunobiology. 213:237–250. [DOI] [PubMed] [Google Scholar]
- Song X, Xia B, Stowell SR, Lasanajak Y, Smith DF, Cummings RD. 2009. Novel fluorescent glycan microarray strategy reveals ligands for galectins. Chem Biol. 16:36–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton R, Hykollari A, Eckmair B, Malzl D, Dragosits M, Palmberger D, Wang P, Wilson IB, Paschinger K. 2017. The underestimated N-glycomes of lepidopteran species. Biochim Biophys Acta. 1861:699–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 355:1143–1155. [DOI] [PubMed] [Google Scholar]
- Sun YC, Guo XP, Hinnebusch BJ, Darby C. 2012. The Yersinia pestis Rcs phosphorelay inhibits biofilm formation by repressing transcription of the diguanylate cyclase gene hmsT. J Bacteriol. 194:2020–2026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal-Melgosa S, Pedersen HL, Schuckel J, Arnal G, Dumon C, Amby DB, Monrad RN, Westereng B, Willats WG. 2015. A new versatile microarray-based method for high throughput screening of carbohydrate-active enzymes. J Biol Chem. 290:9020–9036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Liu S, Trummer BJ, Deng C, Wang A. 2002. Carbohydrate microarrays for the recognition of cross-reactive molecular markers of microbes and host cells. Nat Biotechnol. 20:275–281. [DOI] [PubMed] [Google Scholar]
- Warren CE, Krizus A, Roy PJ, Culotti JG, Dennis JW. 2002. The Caenorhabditis elegans gene, gly-2, can rescue the N-acetylglucosaminyltransferase V mutation of Lec4 cells. J Biol Chem. 277:22829–22838. [DOI] [PubMed] [Google Scholar]
- Wilson EH, Deehan MR, Katz E, Brown KS, Houston KM, O’Grady J, Harnett MM, Harnett W. 2003. Hyporesponsiveness of murine B lymphocytes exposed to the filarial nematode secreted product ES-62 in vivo. Immunology. 109:238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wohlschlager T, Butschi A, Grassi P, Sutov G, Gauss R, Hauck D, Schmieder SS, Knobel M, Titz A, Dell A et al. . 2014. Methylated glycans as conserved targets of animal and fungal innate defense. Proc Natl Acad Sci U S A. 111:E2787–E2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuhrer M, Koeleman CA, Hokke CH, Deelder AM. 2006. Mass spectrometry of proton adducts of fucosylated N-glycans: Fucose transfer between antennae gives rise to misleading fragments. Rapid Commun Mass Spectrom. 20:1747–1754. [DOI] [PubMed] [Google Scholar]
- Yan S, Brecker L, Jin C, Titz A, Dragosits M, Karlsson NG, Jantsch V, Wilson IB, Paschinger K. 2015. a. Bisecting galactose as a feature of N-glycans of wild-type and mutant Caenorhabditis elegans. Mol Cell Proteomics. 14:2111–2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Jin C, Wilson IB, Paschinger K. 2015. b. Comparisons of Caenorhabditis fucosyltransferase mutants reveal a multiplicity of isomeric N-glycan structures. J Proteome Res. 14:5291–5305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Serna S, Reichardt NC, Paschinger K, Wilson IB. 2013. Array-assisted characterization of a fucosyltransferase required for the biosynthesis of complex core modifications of nematode N-glycans. J Biol Chem. 288:21015–21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Wilson IB, Paschinger K. 2015. c. Comparison of RP-HPLC modes to analyse the N-glycome of the free-living nematode Pristionchus pacificus. Electrophoresis. 36:1314–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Lasanajak Y, Song X, Hu L, Ramani S, Mickum ML, Ashline DJ, Prasad BV, Estes MK, Reinhold VN et al. . 2014. Human milk contains novel glycans that are potential decoy receptors for neonatal rotaviruses. Mol Cell Proteomics. 13:2944–2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Mishra S, Song X, Lasanajak Y, Bradley KC, Tappert MM, Air GM, Steinhauer DA, Halder S, Cotmore S et al. . 2012. Functional glycomic analysis of human milk glycans reveals the presence of virus receptors and embryonic stem cell biomarkers. J Biol Chem. 287:44784–44799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu S, Hanneman A, Reinhold VN, Spence AM, Schachter H. 2004. Caenorhabditis elegans triple null mutant lacking UDP-N-acetyl-D-glucosamine:alpha-3-D-mannoside beta1,2-N-acetylglucosaminyltransferase I. Biochem J. 382:995–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.