Abstract
Calorie restriction confers health benefits distinct from energy deficit by exercise. We characterized the adipose-transcriptome to investigate the molecular basis of the differential phenotypic responses. Abdominal subcutaneous fat was collected from 24 overweight participants randomized in three groups (N = 8/group): weight maintenance (control), 25% energy deficit by calorie restriction alone (CR), and 25% energy deficit by calorie restriction with structured exercise (CREX). Within each group, gene expression was compared between 6 months and baseline with cutoffs at nominal p ≤ .01 and absolute fold-change ≥ 1.5. Gene-set enrichment analysis (false discovery rate < 5%) was used to identify significantly regulated biological pathways. CR and CREX elicited similar overall clinical response to energy deficit and a comparable reduction in gene transcription specific to oxidative phosphorylation and proteasome function. CR vastly outweighed CREX in the number of differentially regulated genes (88 vs 39) and pathways (28 vs 6). CR specifically downregulated the chemokine signaling-related pathways. Among the CR-regulated genes, 27 functioned as transcription/translation regulators (eg, mRNA processing or transcription/translation initiation), whereas CREX regulated only one gene in this category. Our data suggest that CR has a broader effect on the transcriptome compared with CREX which may mediate its specific impact on delaying primary aging.
Keywords: Caloric Restriction, Transcriptional regulation
Calorie restriction and exercise are arguably the two most successful lifestyle interventions that have consistently been shown to confer health benefits. Although both treatments are effective in achieving energy deficit (even if exercise is more difficult), and thus induce clinically relevant weight loss and improvements in metabolic health including lipid profile and glucose homeostasis (1,2), ample literature suggests that calorie restriction alone elicits favorable health outcomes beyond its effect on energy metabolism. Notably, there is evidence for calorie restriction as the only treatment to delay the decline of biological functions due to advancing age (primary aging) and to the onset of chronic diseases triggered by environmental factors (secondary aging) (3). Calorie restriction has also been shown to extend maximal life span in many species ranging from yeast to rodents and perhaps also in nonhuman primates (4–6). SIRT1 activation has been proposed as one of the key molecular mediators of the effects of calorie restriction. Although not entirely conclusive, the protective effects of calorie restriction against cancers, neurodegenerative and vascular diseases, as well as its effects on metabolic alterations in white adipose tissue, liver, and skeletal muscle, have all been reported to be associated with SIRT1 activation (7–9). Such an extensive effect on multiple tissues, together with the ability of SIRT1 to modify histone and thus induce epigenetic modifications (10), implicates that calorie restriction is likely to impact upstream pathways on global transcriptional and/or translational levels.
In the current study, we focused on the white adipose tissue to understand how calorie restriction may differ from exercise in regulating the transcription landscape and molecular pathways operative in this tissue. White adipose tissue is one of the first responders to energy deficit by mobilizing lipid stores to maintain energy substrate supply (11). Driven by a rapid reduction in circulating insulin, FoxO1 and SREBP-1c (the predominant isoform of the Forkhead box O family and sterol regulatory element binding protein, respectively, in adipose tissue) trigger a complex transcriptional cascade that involves SIRT1 and PPARγ, and collectively inhibits adipogenesis and promotes lipolysis (12,13). A current study showed that adipose tissue–specific ablation of SIRT1 activity alone was sufficient to induce systemic metabolic dysfunctions in mice, an effect completely abolished by calorie restriction (14). By prescribing calorie restriction alone or in combination with exercise with a similar energy deficit, we aimed to distinguish the effect of calorie restriction from energy deficit per se on transcriptomic response in subcutaneous adipose tissue in humans. We hypothesized that calorie restriction would (i) induce shifts in gene expression that are related to energy metabolism as a direct response to energy deficit and (ii) elicit distinct transcriptional changes that are beyond the effect of energy deficit per se and are indicative of calorie restriction–specific benefits on aging.
Research Design and Methods
Participants and Interventions
Overweight (25 ≤ body mass index ≤ 30kg/m2) but otherwise healthy men and women were recruited for the Comprehensive Assessment of the Long-term Effects of Reducing Intake of Energy (CALERIE) trial Phase 1 (ClinicalTrials.gov Identifier: NCT00099151). Details of the study were described elsewhere (15). Briefly, participants were randomized into one of the following 6-month interventions: (i) weight maintenance (Control); (ii) 25% calorie restriction of baseline energy requirements (CR); (iii) 12.5% calorie restriction plus 12.5% increase in energy expenditure by structured exercise (CREX); and (iv) very low calorie diet to achieve 15% weight loss followed by weight maintenance. Outcome measures were taken at baseline and at 6 months. Data from fully adherent participants of the Control, CR, and CREX groups were included in the current analysis (N = 8 per group, 4 men and 4 women; Table 1). The study was approved by the CALERIE Data Safety Monitoring Board and the Institutional Review Board of the Pennington Biomedical Research Center. Written informed consent was obtained from all participants.
Table 1.
Effects of Calorie Restriction (CR) and Calorie Restriction Plus Exercise (CREX) on Metabolic Parameters (N = 8/group)
| Control | CR | CREX | ||||
|---|---|---|---|---|---|---|
| Baseline | 6 Months | Baseline | 6 Months | Baseline | 6 Months | |
| Sex (M/F) | 4/4 | 4/4 | 4/4 | |||
| Age (y) | 35.9±2.7 | 39.0±2.1 | 37.9±1.8 | |||
| Weight (kg) | 81.1±2.0 | 82.3±2.1 | 82.6±4.4 | 73.2±4.1* | 85.0±3.9 | 75.7±3.7* |
| BMI (kg/m2) | 27.3±0.7 | 27.7±0.8 | 27.7±0.5 | 24.5±0.5*,§ | 27.9±0.6 | 24.8±0.6*,‖ |
| Body composition | ||||||
| Body fat (%) | 30.7±2.3 | 30.8±2.8 | 31.9±3.0 | 26.9±3.4* | 30.9±2.5 | 25.1±2.6* |
| Fat-free mass (kg) | 56.4±3.0 | 57.1±3.2 | 56.7±4.6 | 53.8±4.5† | 59.1±4.4 | 57.1±4.3† |
| Fat mass (kg) | 24.7±1.6 | 25.2±2.2 | 25.9±2.3 | 19.4±2.4* | 25.9±1.7 | 18.6±1.7* |
| Serum profile | ||||||
| Glucose (mg/dL) | 90.1±1.5 | 92.2±2.6 | 89.6±2.2 | 87.5±3.1 | 90.9±2.2 | 91.8±2.6 |
| Insulin (µU/mL) | 13.5±1.1 | 13.9±2.1 | 9.4±1.8 | 7.1±1.4‡,‖ | 9.7±1.2 | 7.8±0.8‖ |
| Free fatty acids (mM) | 0.42±0.09 | 0.46±0.10 | 0.46±0.06 | 0.64±0.12 | 0.40±0.02 | 0.55±0.12 |
| Triglycerides (mg/dL) | 136±21 | 158±20 | 135±37 | 90±18‖ | 105±21 | 85±13‖ |
| TNF-α (pg/mL) | 7.1±1.9 | 11.3±3.4‡ | 10.3±2.8 | 10.9±4.1 | 5.8±2.0 | 7.4±2.3 |
| IL-6 (pg/mL) | 159±81 | 151±83 | 54±16 | 124±68 | 106±45 | 135±60 |
| CRP (mg/dL) | 0.32±0.10 | 0.23±0.07 | 0.29±0.11 | 0.23±0.08 | 0.15±0.04 | 0.11±0.04 |
| Leptin (ng/mL) | 17.5±4.6 | 19.2±5.0 | 18.5±4.7 | 11.0±3.3† | 14.3±3.1 | 7.8±2.1† |
| Adiponectin (µg/mL) | 2.6±0.2 | 2.6±0.2 | 3.3±0.4 | 3.7±0.4‡ | 3.6±0.4 | 3.8±0.4 |
| Energy metabolism | ||||||
| 24-h energy expenditure (kcal/d) | 2148±115 | 2127±104 | 2071±132 | 1871±126† | 2198±141 | 2004±116‡ |
| Sleeping metabolic rate (kcal/d) | 1662±71 | 1629±89 | 1594±108 | 1463±99‡ | 1700±106 | 1565±83‡ |
| 24-h respiratory quotient | 0.90±0.01 | 0.90±0.02 | 0.91±0.01 | 0.88±0.01 | 0.88±0.01 | 0.86±0.01 |
| Sleep respiratory quotient | 0.90±0.01 | 0.87±0.02 | 0.89±0.01 | 0.85±0.01‡ | 0.87±0.02 | 0.82±0.02‡ |
| Carbohydrate oxidation (g/d) | 323±13 | 311±26 | 327±27 | 255±28‡ | 277±25 | 240±29 |
| Protein oxidation (g/d) | 80.3±7.3 | 82.1±9.2 | 68.5±7.2 | 61.0±5.3 | 97.8±9.0 | 73.8±5.4‡ |
| Fat oxidation (g/d) | 47.3±7.2 | 49.3±14.1 | 42.6±7.8 | 56.4±11.3 | 64.5±13.2 | 71.5±10.2 |
| SI (10–4 µU.mL−1.min−1) | 2.5±0.4 | 2.6±0.5 | 3.4±0.6 | 4.8±1.6 | 3.6±0.5 | 5.8±0.9† |
| AIRg (µU.mL−1.min−1) | 852±170 | 732±118 | 666±152 | 504±82 | 616±236 | 345±109 |
Note: BMI = body mass index; CRP = C-reactive protein; IL = interleukin; TNF = tumor necrosis factor.
*p < .001, †p < .01, and ‡p < .05 vs baseline (paired-samples t test).
§ p < .01 and ‖p < .05 vs control at 6 months (one-way analysis of variance).
Clinical Assessments
Whole-body composition was assessed using dual-energy x-ray absorptiometry (QDA 4500A, Hologic, Bedford, MA). Sedentary 24-hour energy expenditure was measured in a whole-room respiratory chamber (15). Whole-body insulin sensitivity (SI) was determined by the insulin-modified frequently sampled intravenous glucose tolerance test as described previously (16). Fasting serum concentrations of lipids (free fatty acids, triglycerides, and cholesterols), inflammatory markers (highly sensitive C-reactive protein, tumor necrosis factor-α, and interleukin-6), and adipokines (leptin and adiponectin) were measured using standard procedures.
Adipose Tissue Collection and Sample Processing
Abdominal subcutaneous adipose tissue was collected in fasting conditions by needle biopsies. Approximately 50mg of the tissue was immediately fixed in 2% osmium tetrachloride (w/v) / 0.05M collidine-HCl, and the remaining sample was snap frozen in liquid nitrogen. Total RNA was extracted from the frozen tissue (> 1.5g wet weight) using the RNeasy Mini Kit (Qiagen, Valencia, CA). The yield and purity of RNA (optical density ratio 260/280) were determined by spectrophotometry (NanoDrop ND-1000, NanoDrop Technologies, Wilmington, DE). RNA was resuspended and its integrity was determined using the RNA 6000 Nano Assay Chip Kit on the Bioanalyzer 2100 and the 2100 Expert software (Agilent Technologies, Santa Clara, CA).
Microarray Hybridization and Data Processing
RNA was amplified and purified using the MessageAmp II aRNA Amplification Kit (Life Technologies, Carlsbad, CA). Gene expression profiling was performed using the Sentrix Array Matrix (Illumina Inc, San Diego, CA) that contained 47,312 probes. Transcripts with a detection p value more than .05 across all samples were considered unexpressed and removed from further analysis. Signals from the remaining 30,330 transcripts were log-transformed to base 2, quantile-normalized, and adjusted for sex, race, and age via the Partek Genomics Suite software (version 6.6; Partek, St Louis, MS). Two samples (one in Control and one in CR) were identified as outliers by principal components analysis and thus removed. Probes were annotated using the Illumina microarray annotation package (Illumina Inc). Gene expression was compared between 6 months and baseline within each treatment group. To reduce the noise effect from some genes (high within-group variation of expression), we imposed a “consistency” filter by which genes showing directionally consistent changes in expression (between 6 months and baseline) in the majority of the subjects in each group (5/7 for Control and 6/8 in CR and CREX) were retained for analysis. This resulted in the retainment of 13,482, 17,156, and 10,174 probes for the Control, CR, and CREX groups, respectively (Supplementary Table 1).
Quantitative Real-time PCR (RT-PCR) for Gene Validation
RNA from subcutaneous fat (200ng for each sample) was reverse transcribed to cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA). Based on their predicted physiological relevance in adipogenesis, lipid metabolism, and epigenetic regulation, CEBPZ, CGI-85, SNCG, and ABCA1 were selected to validate the microarray data. Their mRNA expression was quantified using the ABI PRISM 7900HT Sequence Detection System with Custom TaqMan Array Micro Fluidic Cards (Life Technologies). mRNA expression was normalized to cyclophilin B and reported as arbitrary units.
Adipocyte Sizing and Counting
The procedures for sizing and counting adipocytes were adapted from Hirsch and Gillian (17) and Pasarica and colleagues (18). Briefly, the adipose tissue sample was fixed in osmium/collidine-HCl followed by repeated rinsing with 0.154M NaCl and filtering to remove tissue debris. The sample was then digested with 8M urea in 0.154M NaCl to yield a suspension of fixed free cells in 0.01% Triton X-100 solution (v/v) ready for analysis. The number of adipocytes (cells per mg wet weight of tissue) and the average cell size (volume) were measured using a Multisizer 3 Coulter Counter (Beckman Coulter, Fullerton, CA).
Statistical Analysis
For clinical outcomes, values are expressed as means ± SEMs. Paired-samples t test was used to compare within-group treatment effect. One-way or two-way analysis of variance with Bonferroni post hoc tests were used to compare treatment differences at the same time point. Statistical analyses were performed by using the IBM SPSS Statistics (version 21; IBM Corporation) and the GraphPad Prism Program (version 5.04, GraphPad Software Inc., San Diego, CA). Significance was accepted at p value less than .05.
For microarray analysis, genes with statistically significant differences in expression within each treatment group were identified via a regularized paired t test (CyberT; http://cybert.ics.uci.edu). Genes with a nominal p value ≤ .01 and absolute 6-months-to-baseline fold-change ≥ 1.5 were considered as significantly differentially expressed. Biological pathways with enrichment for differentially expressed genes were identified via gene-set enrichment analysis (19) using the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway repository (20). Pathways were considered significant at a false discovery rate less than 5%.
Results
Metabolic Outcomes
Baseline metabolic parameters were not different across the groups (Table 1). Six-month interventions with either CR or CREX induced similar weight loss of 11% (−9.4±0.7kg for CR and −9.3±1.0kg for CREX respectively vs baseline; p < .001), whereas the Control group remained weight stable. Both CR and CREX led to significant reduction in fat mass (−6.5±0.6kg for CR and −7.2±0.7kg for CREX respectively vs baseline; p < .001), and it contributed to 70% and 80% of total weight loss in CR and CREX, respectively. Consistent with reductions in whole body fat mass, plasma leptin was lower after CR (−7.5±1.9ng/mL vs baseline; p < .01) and CREX (−6.5±1.5ng/mL vs baseline; p < .01), but none of the adipose-derived inflammatory markers were altered in the circulation (Table 1). At the end of the intervention, circulating levels of insulin in both CR (7.1±1.4 µU/mL) and CREX (7.8±0.8 µU/mL) were significantly lower than that of the Control (13.9±2.1 µU/mL; p < .05).
Compared with baseline, both intervention groups induced ~10% reduction in both sedentary 24-hour energy expenditure (−200±35 kcal/d for CR, p < .01; −194±59 kcal/d for CREX, p < .05) and sleeping metabolic rate (−131±39 kcal/d for CR and −135±47 kcal/d for CREX; p < .05). We found evidence for a shift toward fat utilization in both CR and CREX with significant decrease in sleep respiratory quotient (−0.03±0.01 for CR and −0.05±0.02 for CREX respectively vs baseline; p < .05). Circulating triglyceride levels also trended lower after CR (−45.1±20.2mg/dL vs baseline; p = .06) and CREX (−19.9±11.2mg/dL vs baseline; p = .12) interventions.
Overall Transcriptional Response in Adipose Tissue
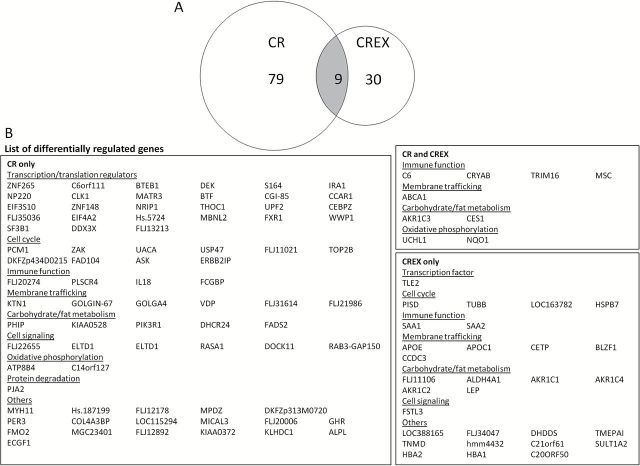
The adipose tissue transcriptome was not different across the groups at baseline. Using a nominal p value ≤ .01 and absolute 6-months-to-baseline fold-change ≥ 1.5, no genes were found to be differentially regulated in the Control group, whereas 88 and 39 genes were differentially regulated by CR and CREX, respectively, with only 9 genes overlapping between the two groups (Figure 1). Based on the GeneCards database (http://www.genecards.org), genes regulated by CR or CREX were broadly classified into similar functional categories including regulation of transcription/translation, immune function, membrane trafficking, energy metabolism, cell cycle, and signaling. Interestingly, 27 out of the 88 CR-regulated genes functioned in various aspects of transcription or translation regulation (eg, RNA splicing, mRNA export, histone methylation, or transcription/translation initiation), whereas the evidence for differential expression of these genes was substantially weaker in the CREX samples (Supplementary Table 2). We used RT-PCR to specifically determine the effect of CR on the mRNA expression of two of these genes: CEBPZ (a negative transcriptional regulator of adipogenesis (21)) and CGI-85 (a regulator of epigenetic histone modification (22)). Consistent with the microarray data, CR induced a 2.3-fold (p < .05) and 2.1-fold (p < .05) increase in the mRNA expression of CEBPZ and CGI-85, respectively, whereas CREX and Control were without effect (Supplementary Figure 1).
Figure 1.
Effects of calorie restriction (CR) and calorie restriction plus exercise (CREX) on the transcriptome in subcutaneous adipose tissue (N = 7–8/group). The number of differentially regulated genes by CR and CREX (overlapping genes in the gray area) were indicated in a Venn diagram (A) and listed in (B). Gene expression was profiled using the Sentrix Array Matrix and compared between 6 months and baseline within each group.
Pathway Analysis
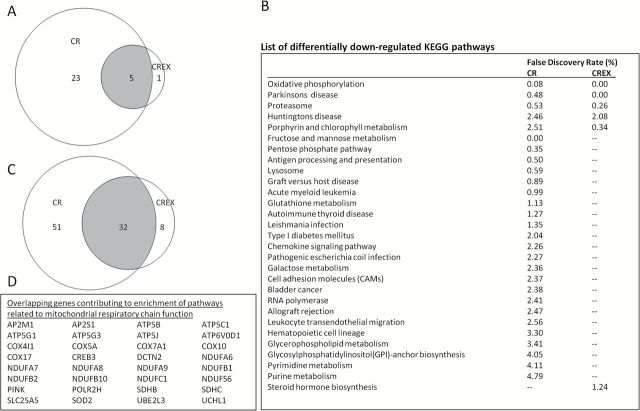
Gene-set enrichment analysis was performed to identify pathways that were enriched for genes displaying consistent patterns of over- or underexpression between 6 months and baseline samples, even if the magnitude of the changes were small and did not satisfy the criterion for a priori defined statistical significance. With a threshold for false discovery rate < 5%, none of the treatments resulted in any significantly upregulated pathways after 6 months of intervention. CR and CREX downregulated 28 and 6 KEGG pathways, respectively, and 5 pathways were commonly regulated by both treatments (Figures 2A and B). Several of the overlapping pathways were enriched by common genes that were involved in mitochondrial respiratory chain function. Specifically, genes encoding subunits of ATP synthase, cytochrome c oxidase, NADH dehydrogenase, and succinate dehydrogenase were largely responsible for the enrichment of the “oxidative phosphorylation,” “Parkinsons disease,” and the “Huntingtons disease” pathways that were downregulated by both CR and CREX (Figures 2C and D and Supplementary Figure 2). Additionally, we observed a distinct effect of CR on downregulating the chemokine signaling-related pathways. A comparison based on the shared genes that contributed to significant enrichment of these pathways is shown in Supplementary Table 3 and Supplementary Figure 3.
Figure 2.
Pathways downregulated by calorie restriction (CR) and calorie restriction plus exercise (CREX) in subcutaneous adipose tissue (N = 7–8/group). The number of differentially regulated pathways by CR and CREX (overlapping pathways in the gray area) were indicated in a Venn diagram (A) and listed in (B). (C) Venn diagram showing the number of genes that contributed to the enrichment of the “oxidative phosphorylation,” “Parkinsons disease,” and “Huntingtons disease” KEGG pathways in CR and CREX (overlapping genes in the gray area and listed in (D)). Biological pathways with enrichment for differentially regulated genes were identified via gene-set enrichment analysis using the KEGG pathway repository. False discovery rate values were only reported in significantly regulated pathways (< 5%).
Adipose Tissue Characteristics
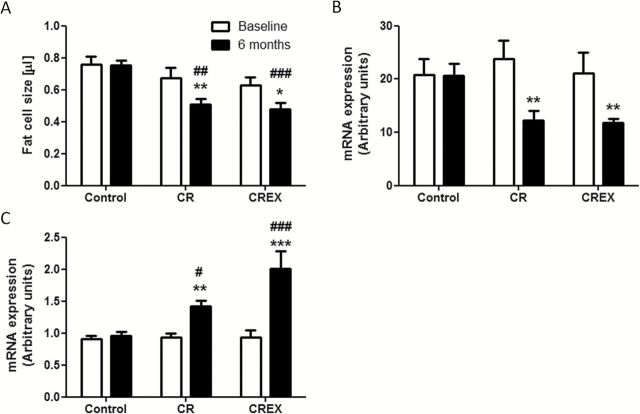
Both CR and CREX reduced the mean fat cell size by 22% as compared with baseline (p < .05; Figure 3A). We used RT-PCR to specifically validate changes in the expression of genes that play a role in cellular lipid transport. CR and CREX reduced the mRNA expression of SNCG, a negative regulator of adipose triglyceride lipase-mediated lipolysis (23), by 48% and 44%, respectively (p < .01; Figure 3B), and increased the mRNA level of ABCA1 that is involved in lipid efflux pathways (24) (1.5-fold for CR and 2-fold for CREX; p < .01; Figure 3C). Collectively these data suggest that CR and CREX had similar impact on genetic regulation to promote lipid removal.
Figure 3.
Effect of calorie restriction (CR) and calorie restriction plus exercise (CREX) on fat cell size (A) and the mRNA expression of SNCG (B) and ABCA1 (C) in adipose tissue (N = 7–8/group). Subcutaneous fat was fixed in osmium/collidine-HCl and digested in urea to yield a suspension of fixed free cells, which was then counted and sized using a Multisizer 3 Coulter Counter. mRNA expression was quantified using real-time PCR and reported in arbitrary units after normalization to cyclophilin B. ***p < .001, **p < .01, and *p < .05 compared with baseline within the group. ###p < .001, ##p < .01, and #p < .05 compared with Control at the same time point.
Discussion
Calorie restriction has long been used to achieve energy deficit as part of weight management regimen. It is now clear that the health benefits of calorie restriction exceed those directly associated with weight loss, but whether these are merely a consequence of energy deficit or are specific to calorie restriction are largely unknown. An important feature of the current study was to compare the changes in transcriptome in response to similar energy deficit induced by calorie restriction alone (CR) or by a combination of CR and increased levels of structured exercise (CREX). Such design allowed us to tease out the effects of CR from that of weight loss because as expected both restricted groups achieved similar weight loss. Focusing on the global transcriptional changes, we showed that both CR and CREX were equally effective in modulating gene expression in the white adipose tissue to adapt to energy deficit, but overall CR had a substantially more diverse impact on genes and pathways than CREX. A further finding was that about one third of the CR-regulated genes are transcription/translation regulators. Our data strongly suggest that regulating the transcriptome is one of the key molecular mechanisms by which CR confers favorable health outcomes.
To date there are only a few studies that investigated the metabolic effects of matched energy deficit, either by calorie restriction and/or by exercise, in humans. By design and consistent with other studies (25,26), CR and CREX induced similar reduction in body weight and total fat mass. Further, we showed that CR and CREX had comparable effects on decreasing energy expenditure and shifting whole-body energy substrate utilization toward fat oxidation (as evidenced by the reduction in sleep respiratory quotient). Together our data suggest that at the same level of energy deficit irrespective of how it might be achieved, CR and CREX elicit equally effective clinical response to restore energy balance.
We then asked whether the effects of CR and CREX on energy metabolism were driven by similar molecular mechanisms. Here we provide evidence for global transcriptional changes in the white adipose tissue for critical metabolic adaptations to reduced energy availability, that is, to mobilize energy storage and to reduce energy consumption. First, both CR and CREX upregulated genes that were involved in membrane trafficking, an essential step in lipolysis and lipid export to other tissues (27). The downregulation of the proteasome pathway has also been implicated in reducing lipid content and adipocyte differentiation (28,29). Physiological relevance of these transcriptional changes is supported by the reduction in fat cell size and the trends of increasing circulating free fatty acids and whole-body fat oxidation in both groups. Second, our pathway analysis indicated that both CR and CREX downregulated key genes in oxidative phosphorylation, for example, genes encoding subunits of ATP synthase, cytochrome c oxidase, and NADH dehydrogenase in the mitochondria. Similar changes in the expression of these genes have also been reported in the subcutaneous fat of participants following calorie restriction protocols (30,31). Since lipogenesis and adipogenesis are both energy-consuming processes, such transcriptional changes could be interpreted as an adaptive response to reduce lipid deposition in the face of negative energy balance. It is also possible that these changes indicate reprogramming of mitochondrial functions in the adipose tissue. Seminal work from de Cabo and colleagues (32,33) suggest that changes in mitochondria during calorie restriction may be an adaptive mechanism to increase bioenergetic efficiency in the face of energy deficit. Along this notion, improvements in mitochondrial function have been extensively reported in key energy-metabolizing tissues of calorie-restricted rodents (34) as well as in the skeletal muscle of our CALERIE participants who underwent 6 months of CR (35). This, together with the downregulation of the proteasome pathway, implicates a role of calorie restriction in reducing oxidative protein damage and the need for protein degradation and clearance as observed in rodent skeletal muscle (36,37). Our results suggest that a similar effect of CR (and CREX) is also extended to the adipose tissue, which possibly helps preserve functional capacity.
Despite comparable transcriptional and clinical response in energy metabolism, we showed that CR vastly outweighed CREX in the total number of differentially regulated genes (88 vs 39) and pathways (28 vs 6). This suggests that calorie restriction is probably eliciting molecular changes beyond adaptations to energy deficit per se. Wheatley and colleagues (38) reported that calorie restriction uniquely altered the expression of 496 transcripts in the visceral fat of diet-induced obese mice, as compared with only 20 transcripts specifically regulated by exercise. Such a broad influence of calorie restriction on the transcriptome is unlikely to be a direct impact on individual genes but rather a more coordinated effect upstream at the level of global transcription regulation. Among the 88 genes that are differentially regulated by CR, 23 are known to be directly involved in the synthesis, processing, and transport of mRNA, or the modulation of the transcription process. Further, the upregulation of CGI-85 (methylates histone) and FLJ35036 (interacts with methylated DNA) provides evidence for CR to impact on transcription at the level of epigenomic changes. Along this notion, it has been shown that individuals who respond to calorie restriction (as defined by significant weight loss) exhibit specific DNA methylation in regions of chromosomes that contained genes related to weight control and insulin secretion (39). Epigenetic histone modifications (40) and chromatin remodeling (41) have been proposed as key molecular mechanisms underlying the benefits of calorie restriction. Together, our data suggest that CR regulates the overall transcriptional function, and this does not appear to be a primary response to energy deficit per se but rather a distinct effect of calorie restriction. Genomic effects may also be the key regulator of the aging process. Pioneering work from the laboratories of Weindruch and Spindler showed that most differential gene expression induced by aging in rodents was at least partly or completely reversed by calorie restriction (42,43). The Spindler group further showed that shifting mice from long-term calorie-restricted to control diet reversed 90% of the transcriptional changes induced by calorie restriction and returned the animals to an aging rate similar to the controls (44), implicating a causal relationship between calorie restriction, gene expression, and aging. Consistent with this notion, we reported earlier that in our CALERIE study CR improved biomarkers of longevity in humans, as evidenced by reductions in both fasting insulin level and core body temperature (15). In a recent cohort of nonobese individuals who underwent a 2-year calorie restriction protocol, we also showed a decrease in 24-hour core temperature at 12 and 24 months compared with baseline (45).
An important question then is why calorie restriction elicits such a unique transcriptional response. Although both CR and CREX participants achieved the same level of energy deficit, CR solely did so by limiting energy intake and thus it is logical to hypothesize that the nutrient-responsive pathways are the key mechanistic links. In multicellular organisms, nutrient sensing involves a complex network of pathways with insulin/insulin-like growth factor (IGF)-1 and the mammalian target of rapamycin (mTOR) axes as two of the major players (46). Calorie restriction downregulated the IGF-1–mediated signaling cascade in mice (47), and this has been causally linked to its protective effect against various forms of cancer (48–50). A recent study provided the first evidence for long-term calorie restriction to downregulate the insulin/IGF pathway at both the transcriptional and activity levels in human skeletal muscle (51). Low glucose availability causes a rapid drop in circulating insulin, which in turn drives changes in the insulin/IGF-1 pathway, but whether calorie restriction also modifies the amino acid–sensing mTOR signaling remains controversial (52–54). Differences in the duration and level of protein restriction may explain the variable effects of calorie restriction on mTOR. It should also be noted that the nutrient-sensing pathways are likely to be the first responder to calorie restriction. The subsequent decrease in energy level, that is, a reduction in the NADH/NAD+ and ATP/ADP ratios as in any events of energy deficit, then activates sirtuins and AMP-dependent kinase (41) which may then feedback to the nutrient-sensing and other energy metabolizing pathways for a coordinated effort to restore energy balance. This may also explain the largely comparable effects of CR and CREX on the transcriptome to mobilize fat storage and improving mitochondrial efficiency.
It should be noted that we used increased levels of exercise to distinguish calorie restriction from energy deficit per se, but our data did not allow direct comparison between the effects of calorie restriction and exercise training alone. Investigating calorie restriction and exercise in the context of matched energy deficit is technically challenging and thus has only been attempted in a limited number of studies (25,26). Also, even in studies with great precision in controlling energy in and energy out, different regimes of calorie restriction and exercise training elicit vastly different metabolic responses and that limit the relevance of efficacy comparisons. Available literature to date largely agrees that calorie restriction and exercise training overlap in a wide range of health benefits from weight loss to protection against some age-related diseases (55). Extension of maximal life span, however, remains as a unique feature of calorie restriction that so far cannot be replicated by any form of exercise training (56,57).
The current study used a genomic approach to explore the molecular mechanisms by which calorie restriction may confer benefits to both life span and health span in humans. An important extension of this line of work would be to select candidate target genes and pursue functional analysis at the cellular level in human samples, as the relevance of animal findings to humans is not entirely clear due to the differences in metabolism and mortality risks (58). Finally, given the enormous challenge (and an almost impossible task) of maintaining drastic lifestyle changes such as life-long calorie restriction, identifying specific molecular targets will be critical for the development of calorie restriction mimetics (59). Data from both animals (60) and humans (61) suggest that these agents mimic some effects of calorie restriction, at least in the short term, but whether one can achieve healthy life-span extension in humans is yet to be determined.
Conclusion
At the same level of energy deficit, calorie restriction alone or in combination with increased levels of exercise elicit similar metabolic adaptations in overweight but otherwise healthy individuals. Calorie restriction broadly impacts on the transcriptome, and this is clearly beyond the sole effect of energy deficit. Specifically, calorie restriction differentially regulates genes that modulate the overall transcriptional function, which may be one of the key molecular mechanisms by which calorie restriction confers systemic benefits and delay primary aging.
Funding
This work was supported by research grant U01 AG20478 and partially supported by a NORC Center Grant P30DK072476.
Supplementary Material
References
- 1. Fontana L, Klein S, Holloszy JO. Effects of long-term calorie restriction and endurance exercise on glucose tolerance, insulin action, and adipokine production. Age (Dordr). 2010;32:97–108. doi:10.1007/s11357-009-9118-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Williams PT, Krauss RM, Vranizan KM, Wood PD. Changes in lipoprotein subfractions during diet-induced and exercise-induced weight loss in moderately overweight men. Circulation. 1990;81:1293–1304. [DOI] [PubMed] [Google Scholar]
- 3. Holloszy JO, Fontana L. Caloric restriction in humans. Exp Gerontol. 2007;42(8):709–712. doi:10.1016/j.exger.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Speakman JR, Hambly C. Starving for life: what animal studies can and cannot tell us about the use of caloric restriction to prolong human lifespan. J Nutr. 2007;137:1078–1086. [DOI] [PubMed] [Google Scholar]
- 5. Colman RJ, Anderson RM, Johnson SC, et al. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–204. doi:10.1126/science.1173635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattison JA, Roth GS, Beasley TM, et al. Impact of caloric restriction on health and survival in rhesus monkeys from the NIA study. Nature. 2012;489:318–321. doi:10.1038/nature11432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kume S, Uzu T, Kashiwagi A, Koya D. SIRT1, a calorie restriction mimetic, in a new therapeutic approach for type 2 diabetes mellitus and diabetic vascular complications. Endocr Metab Immune Disord Drug Targets. 2010;10:16–24. [DOI] [PubMed] [Google Scholar]
- 8. Pani G. Neuroprotective effects of dietary restriction: Evidence and mechanisms. Semin Cell Dev Biol. 2015;40:106–114. doi:10.1016/j.semcdb.2015.03.004 [DOI] [PubMed] [Google Scholar]
- 9. Bordone L, Guarente L. Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol. 2005;6:298–305. doi:10.1038/nrm1616 [DOI] [PubMed] [Google Scholar]
- 10. Zhang T, Kraus WL. SIRT1-dependent regulation of chromatin and transcription: linking NAD(+) metabolism and signaling to the control of cellular functions. Biochim Biophys Acta. 2010;1804:1666–1675. doi:10.1016/j.bbapap.2009.10.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ida M, Hirata M, Odori S, et al. Early changes of abdominal adiposity detected with weekly dual bioelectrical impedance analysis during calorie restriction. Obesity (Silver Spring). 2013;21:E350–E353. doi:10.1002/oby.20300 [DOI] [PubMed] [Google Scholar]
- 12. White UA, Stephens JM. Transcriptional factors that promote formation of white adipose tissue. Mol Cell Endocrinol. 2010;318(1–2):10–14. doi:10.1016/j.mce.2009.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kousteni S. FoxO1, the transcriptional chief of staff of energy metabolism. Bone. 2012;50:437–443. doi:10.1016/j.bone.2011.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Xu C, Cai Y, Fan P, et al. Calorie restriction prevents metabolic aging caused by abnormal SIRT1 function in adipose tissues. Diabetes. 2015;64:1576–1590. doi:10.2337/db14-1180 [DOI] [PubMed] [Google Scholar]
- 15. Heilbronn LK, de Jonge L, Frisard MI, et al. Effect of 6-month calorie restriction on biomarkers of longevity, metabolic adaptation, and oxidative stress in overweight individuals: a randomized controlled trial. JAMA. 2006;295(13):1539–1548. doi:10.1001/jama.295.13.1539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Larson-Meyer DE, Heilbronn LK, Redman LM, et al. Effect of calorie restriction with or without exercise on insulin sensitivity, beta-cell function, fat cell size, and ectopic lipid in overweight subjects. Diabetes Care. 2006;29:1337–1344. doi:10.2337/dc05-2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hirsch J, Gallian E. Methods for the determination of adipose cell size in man and animals. J Lipid Res. 1968;9:110–119. [PubMed] [Google Scholar]
- 18. Pasarica M, Xie H, Hymel D, et al. Lower total adipocyte number but no evidence for small adipocyte depletion in patients with type 2 diabetes. Diabetes Care. 2009;32:900–902. doi:10.2337/dc08-2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi:10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: kyoto encyclopedia of genes and genomes. Nucl Acids Res. 1999;27(1):29–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tang QQ, Lane MD. Role of C/EBP homologous protein (CHOP-10) in the programmed activation of CCAAT/enhancer-binding protein-beta during adipogenesis. Proc Natl Acad Sci U S A. 2000;97:12446–12450. doi:10.1073/pnas.220425597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schotta G, Sengupta R, Kubicek S, et al. A chromatin-wide transition to H4K20 monomethylation impairs genome integrity and programmed DNA rearrangements in the mouse. Genes Dev. 2008;22(15):2048–2061. doi:10.1101/gad.476008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Millership S, Ninkina N, Rochford JJ, Buchman VL. γ-synuclein is a novel player in the control of body lipid metabolism. Adipocyte. 2013;2:276–280. doi:10.4161/adip.25162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Schmitz G, Langmann T. Structure, function and regulation of the ABC1 gene product. Curr Opin Lipidol. 2001;12:129–140. [DOI] [PubMed] [Google Scholar]
- 25. Weiss EP, Racette SB, Villareal DT, et al. Improvements in glucose tolerance and insulin action induced by increasing energy expenditure or decreasing energy intake: a randomized controlled trial. Am J Clin Nutr. 2006;84(5):1033–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Weiss EP, Albert SG, Reeds DN, et al. Calorie restriction and matched weight loss from exercise: independent and additive effects on glucoregulation and the incretin system in overweight women and men. Diabetes Care. 2015;38:1253–1262. doi:10.2337/dc14-2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thompson BR, Lobo S, Bernlohr DA. Fatty acid flux in adipocytes: the in’s and out’s of fat cell lipid trafficking. Mol Cell Endocrinol. 2010;318(1–2):24–33. doi:10.1016/j.mce.2009.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Dasuri K, Zhang L, Ebenezer P, et al. Proteasome alterations during adipose differentiation and aging: links to impaired adipocyte differentiation and development of oxidative stress. Free Radic Biol Med. 2011;51:1727–1735. doi:10.1016/j.freeradbiomed.2011.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Prince AM, May JS, Burton GR, Lyle RE, McGehee RE., Jr Proteasomal degradation of retinoblastoma-related p130 during adipocyte differentiation. Biochem Biophys Res Commun. 2002;290:1066–1071. doi:10.1006/bbrc.2001.6291 [DOI] [PubMed] [Google Scholar]
- 30. Mutch DM, Pers TH, Temanni MR, et al. A distinct adipose tissue gene expression response to caloric restriction predicts 6-mo weight maintenance in obese subjects. Am J Clin Nutr. 2011;94(6):1399–1409. doi:10.3945/ajcn.110.006858 [DOI] [PubMed] [Google Scholar]
- 31. Capel F, Klimcáková E, Viguerie N, et al. Macrophages and adipocytes in human obesity: adipose tissue gene expression and insulin sensitivity during calorie restriction and weight stabilization. Diabetes. 2009;58:1558–1567. doi:10.2337/db09-0033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martin-Montalvo A, de Cabo R. Mitochondrial metabolic reprogramming induced by calorie restriction. Antioxid Redox Signal. 2013;19:310–320. doi:10.1089/ars.2012.4866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. López-Lluch G, Hunt N, Jones B, et al. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–1773. doi:10.1073/pnas.0510452103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barazzoni R, Zanetti M, Bosutti A, et al. Moderate caloric restriction, but not physiological hyperleptinemia per se, enhances mitochondrial oxidative capacity in rat liver and skeletal muscle–tissue-specific impact on tissue triglyceride content and AKT activation. Endocrinology. 2005;146:2098–2106. doi:10.1210/en.2004-1396 [DOI] [PubMed] [Google Scholar]
- 35. Civitarese AE, Carling S, Heilbronn LK, et al. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4(3):e76. doi:10.1371/journal.pmed.0040076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hepple RT, Qin M, Nakamoto H, Goto S. Caloric restriction optimizes the proteasome pathway with aging in rat plantaris muscle: implications for sarcopenia. Am J Physiol Regul Integr Comp Physiol. 2008;295:R1231–R1237. doi:10.1152/ajpregu.90478.2008 [DOI] [PubMed] [Google Scholar]
- 37. Lass A, Sohal BH, Weindruch R, Forster MJ, Sohal RS. Caloric restriction prevents age-associated accrual of oxidative damage to mouse skeletal muscle mitochondria. Free Radic Biol Med. 1998;25:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wheatley KE, Nogueira LM, Perkins SN, Hursting SD. Differential effects of calorie restriction and exercise on the adipose transcriptome in diet-induced obese mice. J Obes. 2011;2011:265417. doi:10.1155/2011/265417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bouchard L, Rabasa-Lhoret R, Faraj M, et al. Differential epigenomic and transcriptomic responses in subcutaneous adipose tissue between low and high responders to caloric restriction. Am J Clin Nutr. 2010;91:309–320. doi:10.3945/ajcn.2009.28085 [DOI] [PubMed] [Google Scholar]
- 40. Chung KW, Kim DH, Park MH, et al. Recent advances in calorie restriction research on aging. Exp Gerontol. 2013;48(10):1049–1053. doi:10.1016/j.exger.2012.11.007 [DOI] [PubMed] [Google Scholar]
- 41. Vaquero A, Reinberg D. Calorie restriction and the exercise of chromatin. Genes Dev. 2009;23(16):1849–1869. doi:10.1101/gad.1807009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee CK, Klopp RG, Weindruch R, Prolla TA. Gene expression profile of aging and its retardation by caloric restriction. Science. 1999;285:1390–1393. [DOI] [PubMed] [Google Scholar]
- 43. Cao SX, Dhahbi JM, Mote PL, Spindler SR. Genomic profiling of short- and long-term caloric restriction effects in the liver of aging mice. Proc Natl Acad Sci U S A. 2001;98:10630–10635. doi:10.1073/pnas.191313598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dhahbi JM, Kim HJ, Mote PL, Beaver RJ, Spindler SR. Temporal linkage between the phenotypic and genomic responses to caloric restriction. Proc Natl Acad Sci U S A. 2004;101:5524–5529. doi:10.1073/pnas.0305300101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ravussin E, Redman LM, Rochon J, et al. A 2-year randomized controlled trial of human caloric restriction: feasibility and effects on predictors of health span and longevity. J Gerontol A Biol Sci Med Sci. 2015;70(9):1097–1104. doi:10.1093/gerona/glv057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dilova I, Easlon E, Lin SJ. Calorie restriction and the nutrient sensing signaling pathways. Cell Mol Life Sci. 2007;64:752–767. doi:10.1007/s00018-007-6381-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Thompson AC, Bruss MD, Nag N, Kharitonenkov A, Adams AC, Hellerstein MK. Fibroblast growth factor 21 is not required for the reductions in circulating insulin-like growth factor-1 or global cell proliferation rates in response to moderate calorie restriction in adult mice. PLoS One. 2014;9:e111418. doi:10.1371/journal.pone.0111418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Harvey AE, Lashinger LM, Hays D, et al. Calorie restriction decreases murine and human pancreatic tumor cell growth, nuclear factor-κB activation, and inflammation-related gene expression in an insulin-like growth factor-1-dependent manner. PLoS One. 2014;9:e94151. doi:10.1371/journal.pone.0094151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Galet C, Gray A, Said JW, et al. Effects of calorie restriction and IGF-1 receptor blockade on the progression of 22Rv1 prostate cancer xenografts. Int J Mol Sci. 2013;14:13782–13795. doi:10.3390/ijms140713782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nogueira LM, Lavigne JA, Chandramouli GV, Lui H, Barrett JC, Hursting SD. Dose-dependent effects of calorie restriction on gene expression, metabolism, and tumor progression are partially mediated by insulin-like growth factor-1. Cancer Med. 2012;1:275–288. doi:10.1002/cam4.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mercken EM, Crosby SD, Lamming DW, et al. Calorie restriction in humans inhibits the PI3K/AKT pathway and induces a younger transcription profile. Aging Cell. 2013;12:645–651. doi:10.1111/acel.12088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bonkowski MS, Dominici FP, Arum O, et al. Disruption of growth hormone receptor prevents calorie restriction from improving insulin action and longevity. PLoS One. 2009;4:e4567. doi:10.1371/journal.pone.0004567 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 53. Sharma N, Castorena CM, Cartee GD. Tissue-specific responses of IGF-1/insulin and mTOR signaling in calorie restricted rats. PLoS One. 2012;7:e38835. doi:10.1371/journal.pone.0038835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Dogan S, Johannsen AC, Grande JP, Cleary MP. Effects of intermittent and chronic calorie restriction on mammalian target of rapamycin (mTOR) and IGF-I signaling pathways in mammary fat pad tissues and mammary tumors. Nutr Cancer. 2011;63:389–401. doi:10.1080/01635581.2011.535968 [DOI] [PubMed] [Google Scholar]
- 55. Mercken EM, Carboneau BA, Krzysik-Walker SM, de Cabo R. Of mice and men: the benefits of caloric restriction, exercise, and mimetics. Ageing Res Rev. 2012;11:390–398. doi:10.1016/j.arr.2011.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Holloszy JO. Exercise and food restriction in rats. J Nutr. 1992;122(3 suppl):774–777. [DOI] [PubMed] [Google Scholar]
- 57. Weindruch R, Walford RL, Fligiel S, Guthrie D. The retardation of aging in mice by dietary restriction: longevity, cancer, immunity and lifetime energy intake. J Nutr. 1986;116:641–654. [DOI] [PubMed] [Google Scholar]
- 58. Barzilai N, Bartke A. Biological approaches to mechanistically understand the healthy life span extension achieved by calorie restriction and modulation of hormones. J Gerontol A Biol Sci Med Sci. 2009;64(2):187–191. doi:10.1093/gerona/gln061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Minor RK, Allard JS, Younts CM, Ward TM, de Cabo R. Dietary interventions to extend life span and health span based on calorie restriction. J Gerontol A Biol Sci Med Sci. 2010;65(7):695–703. doi:10.1093/gerona/glq042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fok WC, Zhang Y, Salmon AB, et al. Short-term treatment with rapamycin and dietary restriction have overlapping and distinctive effects in young mice. J Gerontol A Biol Sci Med Sci. 2013;68(2):108–116. doi:10.1093/gerona/gls127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Timmers S, Konings E, Bilet L, et al. Calorie restriction-like effects of 30 days of resveratrol supplementation on energy metabolism and metabolic profile in obese humans. Cell Metab. 2011;14:612–622. doi:10.1016/j.cmet.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.