SUMMARY
Microbial patterns are recognized by cell surface receptors to initiate pattern-triggered immunity (PTI) in plants. Receptor-like cytoplasmic kinases (RLCK), such as BIK1, and calcium-dependent protein kinases (CPK) are engaged during PTI to activate the NADPH oxidase RBOHD for reactive oxygen species (ROS) production. It is unknown if protein kinases besides CPKs and RLCKs participate in RBOHD regulation. We screened mutants in all 10 Arabidopsis MAP4 kinases (MAP4Ks) and identified the conserved MAP4K SIK1 as a positive regulator of PTI. sik1 mutants were compromised in their ability to elicit the ROS burst in response to microbial features and exhibited compromised PTI to bacterial infection. SIK1 directly interacts with, phosphorylates and stabilizes BIK1 in a kinase activity-dependent manner. Furthermore, SIK1 directly interacts with and phosphorylates RBOHD upon flagellin perception. Thus, SIK1 positively regulates immunity by stabilizing BIK1 and activating RBOHD to promote the extracellular ROS burst.
Keywords: SIK1, MAP4K, PTI, ROS, BIK1, RBOHD, phosphorylation, autoimmunity
In Brief
Zhang et al. identify the conserved MAP4 kinase SIK1 as required for pattern-triggered immunity in plants. SIK1 associates with, phosphorylates and stabilizes the central immune regulator BIK1. Upon perception of pathogens, SIK1 and activated BIK1 phosphorylate the NADPH oxidase RBOHD to enhance ROS production and promote defense.
Graphical Abstract
INTRODUCTION
Plants rely on their innate immune system to actively recognize and respond to pathogenic organisms. Immune receptors with extracellular domains act to perceive conserved microbial patterns, resulting in pattern-triggered immunity (PTI). These microbial features are referred to as pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs). Well-characterized PAMPs include bacterial flagellin and fungal chitin (Thomma et al., 2011). PTI responses include the generation of reactive oxygen species (ROS), a transient influx of calcium from the apoplast, activation of mitogen-activated protein kinases (MAPKs) and calcium-dependent protein kinases (CPKs), as well as transcriptional reprogramming of immune-related genes (Couto and Zipfel, 2016).
The Arabidopsis receptor FLS2 is a well-characterized pattern recognition receptor (PRR) that perceives a conserved 22 amino acid epitope from bacterial flagellin (flg22) (Chinchilla et al., 2006). The kinase BIK1, a member of the receptor-like cytoplasmic kinase (RLCK) family, is important for downstream immune signaling (Tang et al., 2017). BIK1 associates with the FLS2 complex and is rapidly phosphorylated upon flg22 perception. Activated BIK1 is released from the FLS2 complex and directly phosphorylates the NADPH oxidase (NOX) RBOHD to activate extracellular ROS production (Kadota et al., 2014; Li et al., 2014). The extracellular ROS burst is significantly compromised in the bik1 mutant, but MAPK activation is not affected (Feng et al., 2012; Zhang et al., 2010), indicating that BIK1’s primary role is to enhance the extracellular ROS burst. BIK1 also positively regulates immune responses triggered by other PAMPs, suggesting that BIK1 serves as a central and convergent regulator of distinct PRR-dependent pathways (Couto and Zipfel, 2016). For example, BIK1 associates with the CERK1 PRR and is required for chitin-induced immune responses (Zhang et al., 2010). Given the importance of BIK1 in immune signaling, its’ protein stability is tightly regulated during PTI (Liang et al., 2016; Monaghan et al., 2014; Wang et al., 2018).
The extracellular ROS burst can act as an antimicrobial molecule, strengthen the plant cell wall through oxidative cross-linking, and act as a local and systemic messenger to induce downstream immune responses (Kadota et al., 2015; Wang et al., 2014). Plasma-membrane localized NOXs produce O2- in the apoplast which can then be rapidly converted to H2O2 and can diffuse into the cytosol (Kadota et al., 2015; Tian et al., 2016). In plants, NOXs belong to the respiratory burst oxidase homolog (RBOH) family. The Arabidopsis RBOH family contains 10 members, and RBOHD and RBOHF are important for pathogen-induced ROS burst and ETI-induced cell death (Nühse et al., 2007; Torres et al., 2002; Torres et al., 2005). Positive regulation of plant NOX activity primarily occurs through post-translational modifications, including phosphorylation of RBOHD’s N-terminus at activating sites as well as conformational changes induced by Ca2+ binding to EF-hand motifs (Kadota et al., 2015). BIK1 and CPKs have been shown to be important for activation of RBOHD by phosphorylating its N-terminal regions (Dubiella et al., 2013; Kadota et al., 2014; Li et al., 2014). However, it is unknown if other protein kinases besides CPKs and RLCKs participate in the regulation of RBOHD to ensure its robust activation upon pathogen or PAMP perception.
The mitogen-activated protein kinase kinase kinase kinase (MAP4K) family is evolutionarily conserved and related to yeast STE20 (Dan et al., 2001). In yeast and humans, MAP4Ks can directly activate MAPK cascades, but can also phosphorylate diverse substrates (Brenner et al., 2009; Leberer et al., 1997). In humans, MAP4Ks are also involved in immunity and activate NF-κB immune signaling (Brenner et al., 2009; Jiao et al., 2015). In Arabidopsis, the MAP4K family contains 10 members. The SIK1 MAP4K regulates cell proliferation and cell expansion (Xiong et al., 2016). The Arabidopsis BLUS1 MAP4K is phosphorylated by phototropins and serves as a primary regulator of stomatal opening (Takemiya et al., 2013). However, the functions and mechanisms of MAP4Ks in plant immunity remain unexplored.
Here, we show that sik1 mutants were compromised in the ROS burst upon PAMP perception and displayed compromised immunity to infection with the bacterial pathogen Pseudomonas syringae, indicating that MAP4K family is involved in plant innate immunity. In addition, we show that SIK1 phosphorylates BIK1 and stabilizes it in a kinase activity-dependent manner. Interestingly, SIK1 also associates with RBOHD in Arabidopsis and phosphorylates RBOHD’s N-terminus. Together, these results indicate SIK1 positively regulates PTI by stabilizing BIK1 and activating RBOHD to promote the extracellular ROS burst.
RESULTS
sik1 mutants are compromised in flg22-induced ROS production
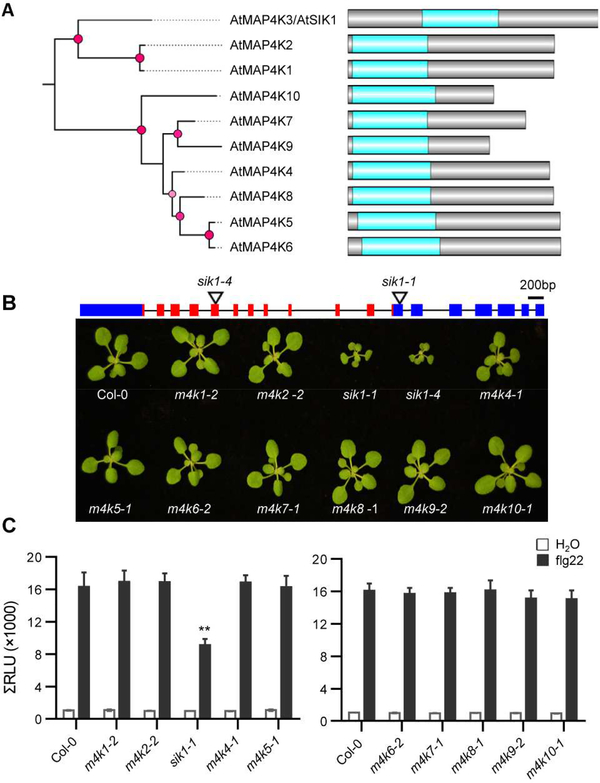
In order to define the role of plant MAP4Ks, we first identified homozygous T-DNA insertion mutants in the 10 Arabidopsis MAP4K members. Sequence analyses and PCR confirmed the T-DNA insertions and RT-PCR analyses verified the absence of full-length transcripts in T-DNA lines, indicating that these lines are null mutants (Figure S1). We were unable to amplify MAP4K8 and MAP4K9 transcripts in wild-type Col-0 (Figure S1B), likely due to the low expression levels of these genes in leaf tissue. Unlike the other Arabidopsis MAP4K members, SIK1 (MAP4K3, At1g69220) exhibits unique domain architecture with a central kinase domain and additional N- and C-terminal regions (Figure 1A). Interestingly, the sik1 mutants displayed a dwarf phenotype, which is consistent with previous findings (Xiong et al., 2016), whereas other mutant lines did not show obvious differences in plant growth compared to wild-type Col-0 (Figure 1B).
Figure 1. The SIK1 MAP4K is involved in flg22-induced ROS production.
(A) Domain architecture and phylogeny of the Arabidopsis MAP4K family. Red circles represent nodes with posterior probability greater than 0.5. MAP4K kinase domains are highlighted in blue. (B) Top: SIK1 gene structure and T-DNA insertion sites. Black lines indicate introns. Boxes indicate exons. Red boxes represent exons in the kinase domain. Bottom: growth phenotypes of three-week-old map4k T-DNA insertion mutants. (C) The ROS burst in T-DNA mutants of the MAP4K family after treatment with 100 nM flg22 or water. Total relative luminescent units (RLU) were detected over a 35-min period. Values are means ± SEM of RLU (n=12). Asterisks indicate significant differences (Dunnett’s test, p < 0.01). Experiments were repeated three times with similar results. See also Figure S1 and S2.
To analyze the role of MAP4Ks in plant immunity, we phenotyped each mutant for alterations in the flg22-induced ROS burst. ROS production was detected using a luminol-based assay with water used as a negative control. The ROS burst was significantly reduced in the sik1–1 mutant upon flg22 treatment compared to wild-type Col-0 (Figure 1C and S2). No alterations in the flg22-induced ROS burst were observed in the other nine T-DNA mutants (Figure 1C and S2). These results indicate that SIK1 is involved in flg22-induced ROS production.
SIK1 is a functional and conserved kinase in land plants
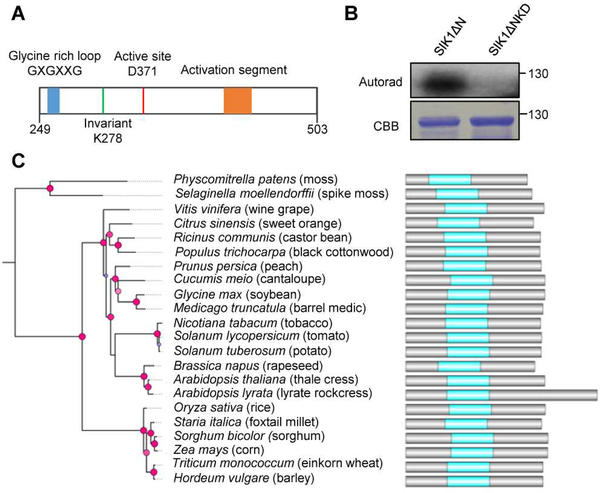
SIK1’s kinase domain exhibits conserved features of active protein kinases (Figure S3). It contains a characteristic glycine-rich loop with the consensus GXGXXG motif (Figure 2A), which is a structural hallmark of protein kinases and nucleotide binding proteins (Taylor and Kornev, 2011). It also possesses an invariant lysine residue at position 278 and a conserved aspartate (D371) (Figure 2A), which are generally required for kinase activity. A conserved activation segment was identified in the SIK1 kinase domain (Figure 2A), and the DFG motif in this segment has been shown to be important for function in other kinases by coordinating Mg2+ at the active site (Taylor and Kornev, 2011). These features indicate that SIK1 is an active kinase.
Figure 2. SIK1 is an active protein kinase and is ubiquitous in land plants.
(A) Diagram of conserved motifs in SIK1’s kinase domain. (B) SIK1 kinase activity. Recombinant MBP-SIK1ΔN and the kinase dead variant MBP-SIK1ΔNKD were purified from E. coli. In vitro kinase assays were initiated by adding γ−32P-ATP to the indicated recombinant proteins. Phosphorylated proteins were visualized by autoradiography (upper panel). SDS-PAGE gel stained with Coomassie Brilliant Blue (bottom panel). Similar results were obtained in two independent experiments. (C) SIK1 is conserved in land plants. Phylogeny of SIK1 homologs in selected plant species and their corresponding domain architecture. Phylogeny was determined by maximum likelihood in PhyML. The SH-like support values of the branches were mapped to circles from blue to red, representing the lower and higher support values. The red circles represent nodes with posterior probability greater than 0.5. See also Figure S3.
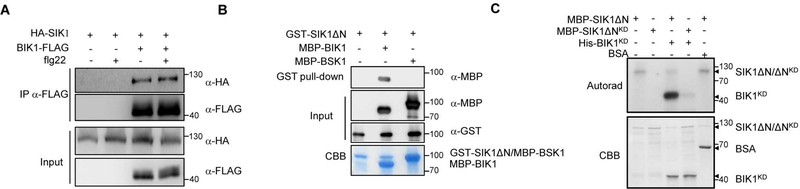
We next experimentally examined the kinase activity of SIK1 using an in vitro phosphorylation assay. Since full-length SIK1 is toxic to Escherichia coli (Xiong et al., 2016), truncated SIK1 protein lacking its N-terminal region was utilized (SIK1ΔN, amino acids 249–836). We purified recombinant SIK1ΔN and its kinase-dead variant K278E (SIK1ΔNKD) from E. coli, and evaluated their kinase activities by analyzing autophosphorylation using a radiolabeled assay. The SIK1ΔN protein can autophosphorylate itself, but the kinase-dead variant K278 cannot (Figure 2B), demonstrating that SIK1 is an active kinase.
To determine if SIK1 is conserved in plants, we searched for homologs in other plant species by BLAST analyses. SIK1 homologs are ubiquitous in land plants and they all possess conserved domain architecture with a central kinase domain (Figure 2C). The kinase domains of these homologs are highly conserved (92–99% amino acid identity) and also share conserved features with SIK1 (Figure S3).
sik1 mutants exhibit enhanced resistance to Pseudomonas syringae due to high levels of SA
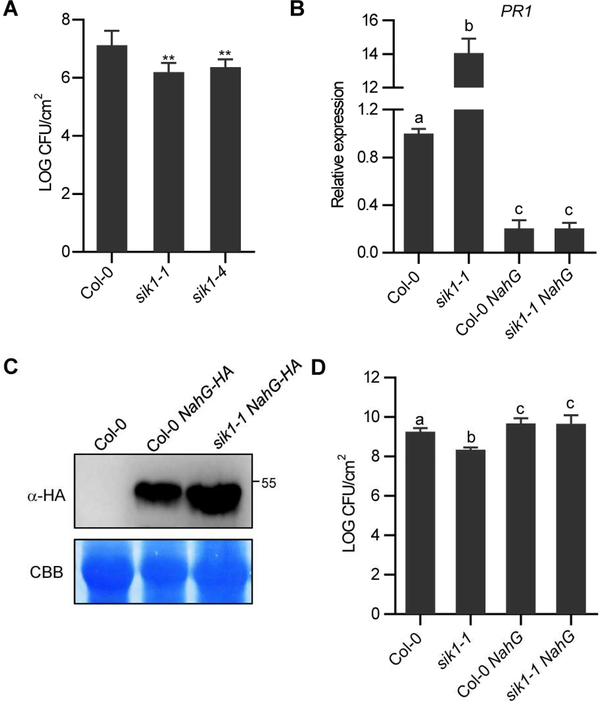
The compromised ROS production in sik1–1 led us to propose that SIK1 may positively regulate PTI. We inoculated two sik1 knockouts (sik1–1 and sik1–4) with the virulent bacterial pathogen Pseudomonas syringae pv. tomato DC3000 (Pst). Unexpectedly, the sik1 mutants displayed significantly lower bacterial titers than Col-0 (Figure 3A). Since activation of plant immunity can restrict the delivery of type III effectors (Crabill et al., 2010), we evaluated the delivery of the Pst AvrRpm1 effector in sik1–1 using the adenylate cyclase assay. Pst is still able to deliver AvrRpm1 into sik1–1 (Figure S4A), indicating that effector delivery is not grossly altered in sik1–1. The sik1 mutants also exhibit a lesion-mimic phenotype under long-day conditions (Figure S4B). The presence of dwarf and lesion mimic phenotypes are frequently associated with higher levels of salicylic acid (SA) and autoimmune responses (Rodriguez et al., 2015). The sik1 mutants exhibited significantly higher levels of basal SA (Figure S4C), constitutive expression of the SA-response defense marker gene PR1 (Figure S4D), and higher PR1 protein accumulation compared to wild-type Col-0 (Figure S4E). Jasmonic acid (JA) levels were decreased in the sik1 mutants (Figure S4F), consistent with the antagonism between SA and JA pathways (Fu and Dong, 2013). No significant alterations in the abundance of abscisic acid (ABA) and cis-(+)-12-oxo-phytodienoic acid (OPDA) were detected in sik1 mutants (Figure S4F).
Figure 3. sik1 mutants display resistance to Pst because of high SA accumulation.
(A) Titers of P. syringae DC3000 (Pst) three days post-infiltration. The data are shown as means ± SEM (n ≥ 4). Asterisks indicate significant differences (Dunnett’s test, p < 0.01). Similar results were obtained in three independent experiments. (B) Relative expression of the PR1 gene in Col-0 and sik1 mutants using qPCR. The data are shown as means ± SEM (n≥3) Different letters indicate significant differences (Fisher’s LSD, p < 0.01). (C) Immunoblot analyses with anti-HA demonstrating NahG expression in NahG-HA transgenic lines. (D) Titers of P. syringae DC3000 in NahG-expressing plants three days-post infiltration. The data are shown as means ± SEM (n≥3) and analyzed as described in (B). Experiments were repeated three times with similar results. See also Figure S4.
To test if high levels of SA leads to enhanced resistance to Pst in sik1, we generated transgenic plants constitutively expressing the salicylate hydroxylase NahG, which degrades SA (Gaffney et al., 1993), in both Col-0 and sik1–1 backgrounds. Both Col-0 NahG and sik1–1 NahG transgenic lines exhibited comparable levels of PR1 gene expression (Figure 3B) and NahG protein (Figure 3C), indicating similar SA levels. Therefore, we should be able to uncouple pleiotropic effects from high SA levels by using the sik1–1 NahG transgenic line. NahG expression did not completely restore the dwarf phenotype in sik1 mutant (Figure S4B). However, NahG expression suppressed the sik1–1 resistance phenotype to virulent Pst (Figure 3D). Together, these results demonstrate that the enhanced resistance in the sik1 mutants is caused by high SA accumulation.
sik1 NahG plants are compromised in flg22-triggered immunity
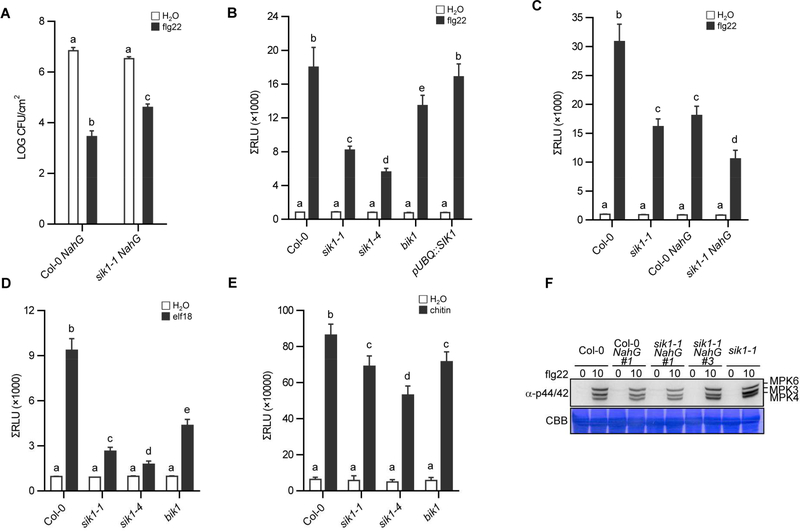
To investigate if SIK1 is required for PAMP-induced resistance to Pst, a flg22-protection assay was carried out. Pre-treatment with PAMPs, such as flg22, results in enhanced resistance to subsequent inoculation with virulent bacteria (Zipfel et al., 2004). Col-0 NahG and sik1 NahG leaves were infiltrated with water or flg22 one day prior to inoculation of the same leaf with virulent Pst. After flg22 pretreatment, sik1 NahG carried approximately 14-fold more bacteria than Col-0 NahG (Figure 4A), indicating that sik1 NahG is substantially inhibited in flg22 responsiveness. Taken together, these results suggest that SIK1 positively regulates flg22-triggered immunity to Pst.
Figure 4. sik1 mutants exhibit compromised PAMP-triggered immunity and ROS burst.
(A) Susceptibility of the sik1–1 NahG line #1 to Pst. Leaves were pretreated with either water or flg22 (1 μM) 24 h before inoculation with Pst. Statistically significant differences (Fisher’s LSD; p < 0.01) between the Col-0 NahG and the sik1–1 NahG lines are indicated. Similar results were obtained in three independent experiments. (B and C) The ROS burst in the indicated lines after treatment with 100 nM flg22 or water. Total relative luminescent units (RLU) were detected over a 40-min period. (D) The ROS burst in the indicated lines after treatment with 100 nM elf18 or water. Total relative luminescent units (RLU) were detected over a 40-min period. (E) The ROS burst in the indicated lines after treatment with 10 μM chitin or water. For chitin induced ROS burst, the more sensitive probe L012 was used. Total relative luminescent units (RLU) were detected over a 40-min period. Values in (B-E) are means ± SEM of RLU (n≥12). Different letters indicate significant differences (Fisher’s LSD, p < 0.01). Similar results were obtained in three independent experiments. (F) MAPK activation was detected after flg22 treatment by an immunological assay. The expected identities of the respective bands are marked on the right. This experiment was repeated twice with similar results. See also Figure S5.
sik1 mutants are compromised in different PAMP-induced ROS burst but not in MAPK activation
To confirm the role of SIK1 in ROS regulation, we analyzed a second independent homozygous T-DNA mutant line (sik1–4) as well as a UBQ::SIK1-GFP complementation line (Xiong et al., 2016). Flg22-induced ROS production was significantly reduced in both sik1–1 and sik1–4 compared to wild-type Col-0 (Figure 4B and S5A). ROS production in the SIK1 complementation line was not significantly different from wild-type Col-0, further confirming the role of SIK1 in flg22-induced extracellular ROS burst (Figure 4B). The knockout mutant of the PTI regulator BIK1 also displays high basal levels of SA accumulation and compromised extracellular ROS production (Veronese et al., 2006; Zhang et al., 2010). The bik1 mutant, which has been shown to be compromised in both elf18- and chitin-induced ROS burst, was used as a control (Zhang et al., 2010). Interestingly, the ROS production in sik1 mutants was even lower than that in bik1 after flg22 treatment (Figure 4B), highlighting the importance of SIK1 in ROS production. PBL1 is a close homolog of BIK1, and is also required for PTI signaling. Consistent with previous results (Zhang, 2010), ROS production was further reduced in the bik1 pbl1 double mutant compared to the bik1 single mutant after flg22 treatment (Figure S5B and S5C). ROS production in the sik1 mutants was still lower than that in the bik1 pbl1 mutant (Figure S5B and S5C). We also observed a reduced ROS burst in sik1 NahG compared to Col-0 NahG (Figure 4C and S5D), indicating that the compromised ROS production in sik1 mutants is not due to high SA accumulation. Interestingly, NahG expression significantly reduced ROS production after flg22 treatment in Col-0 and sik1 (Figure 4C), demonstrating that SA is involved in regulation of apoplastic ROS production. Chitin- and elf18-induced ROS production was also reduced in sik1 mutants compared to wild-type Col-0 (Figure 4D, 4E, S5E and S5F), indicating that SIK1 regulates responsiveness to a broad range of PAMPs. In parallel to ROS production, PTI also induces MAPK activation (Couto and Zipfel, 2016; Yu et al., 2017). We did not observe compromised PAMP-triggered MAPK activation in sik1 and sik1 NahG compared to wild-type Col-0 and Col-0 NahG (Figure 4F). MAPK activation was slightly higher in the sik1 mutant, but not in sik1 NahG lines (Figure 4F). Together, these results indicate that SIK1 primarily regulates PAMP-induced ROS production.
SIK1 associates with and phosphorylates BIK1
To investigate how SIK1 modulates ROS production, we first analyzed the abundance of the FLS2 immune receptor and the central immune regulator BIK1 in sik1 mutant lines. The compromised flg22-induced ROS burst in the sik1 mutant is not caused by reduced levels of the FLS2 immune receptor (Figure S6A) or reduced transcriptional expression of BIK1 (Figure S6B). The sik1 mutants phenocopied the bik1 mutant with respect to decreased ROS production after PAMP treatment and high basal levels of SA. Therefore, we then investigated if SIK1 associates with BIK1. To test this hypothesis, we expressed BIK1-FLAG and HA-SIK1 in Arabidopsis, and performed co-immunoprecipitation (co-IP). A strong association between SIK1 and BIK1 was observed in the presence and absence of flg22 (Figure 5A), supporting the hypothesis that SIK1 resides within the same protein complex as BIK1.
Figure 5. SIK1 directly interacts with and phosphorylates BIK1.
(A) Co-immunoprecipitation of HA-SIK1 and BIK1-FLAG after co-expression in Arabidopsis. The indicated constructs were transformed into Col-0 protoplasts. Protoplasts were treated with 1 μM flg22 or water, and then total proteins were subjected to anti-FLAG immunoprecipitations. (B) SIK1 interacts with BIK1 in vitro. GST-tagged SIK1ΔN and MBP-tagged BIK1 were purified from E. coli, and protein-protein interaction was examined by a GST pull-down assay. MBP-BSK1 was used as a control. CBB, Coomassie Brilliant Blue staining. (C) SIK1 phosphorylates BIK1. In vitro kinase assays were initiated by adding γ−32P-ATP to the indicated recombinant proteins. Phosphorylated proteins were visualized by autoradiography (top panel). SDS-PAGE gel stained with Coomassie Brilliant Blue (bottom panel).
To test if SIK1 directly interacts with BIK1, we purified GST-SIK1ΔN, MBP-BIK1 and MBP-BSK1 recombinant proteins from E. coli. BSK1 is an Arabidopsis RLCK that is involved in both brassinosteriod perception and innate immunity (Shi et al., 2013). The GST pull-down assay showed that SIK1ΔN specifically interacts with BIK1 compared to BSK1 (Figure 5B). We next investigated SIK1’s ability to phosphorylate BIK1 in vitro. SIK1ΔN phosphorylated a kinase dead variant BIK1KD (BIK1K105A/K106A), but the SIK1ΔNKD did not (Figure 5C), indicating that trans-phosphorylation of BIK1 was due to SIK1ΔN phosphorylation. To identify BIK1 sites phosphorylated by SIK1, we co-expressed BIK1KD with SIK1ΔN or SIK1ΔNKD in E. coli and then purified BIK1KD recombinant protein for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analyses. Twelve serine (S) residues, twelve threonine (T) residues and one tyrosine (Y) residue were identified as potential BIK1 phosphorylated sites (Table S1) and only detected after co-expression with SIK1ΔN. Among these 25 residues, 10 (S26, S32, S33, S34, T35, T42, T50, S54, T56 and T64) are in the N-terminus, 10 (S71, T120, Y168, S193, S206, S219, S233, S236, T314 and T341) are in the kinase domain, and five (S360, T362, T368, T375 and T377) are in the C-terminus. The total protein sequence coverage was similar for BIK1KD after co-expression with SIK1ΔN and SIK1ΔNKD (89%). All corresponding peptides were also identified in SIK1ΔN KD samples, but were not phosphorylated. These results further demonstrate that SIK1 is an active kinase and can phosphorylate BIK1 at multiple sites in vitro. Among these residues, phosphorylation of S236 has been shown to be important for BIK1 stability (Wang et al., 2018).
SIK1 stabilizes the central immune regulator BIK1
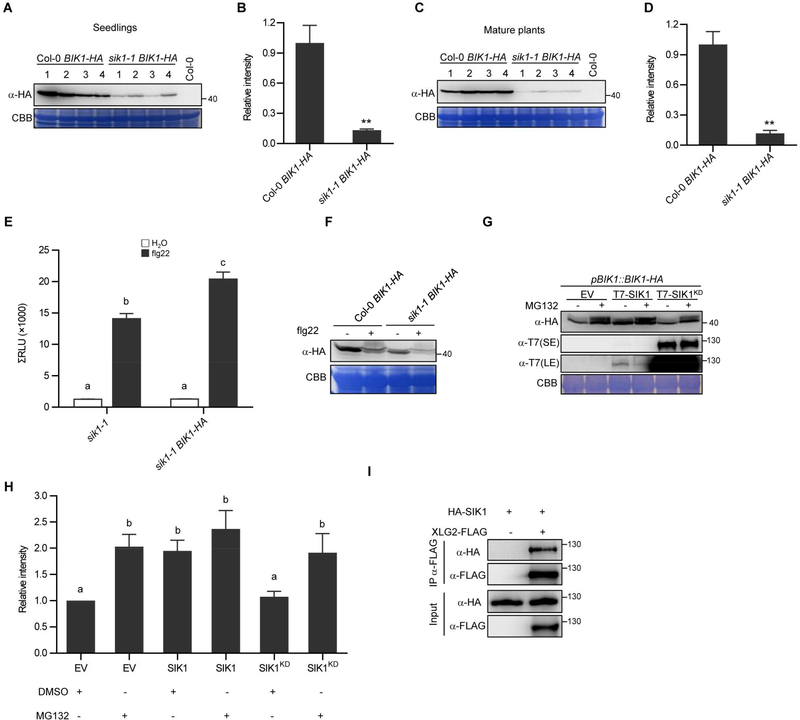
BIK1 is a central component of PRR-mediated immune responses and the accumulation of BIK1 is tightly controlled (Liang et al., 2016; Monaghan et al., 2014). Because the sik1 and bik1 mutants share several phenotypes, we investigated if the stability of BIK1 is regulated by SIK1. We generated pBIK1::BIK1-HA transgenic lines in both Col-0 and sik1–1 genetic backgrounds, and detected the BIK1-HA protein accumulation by western blot in eight independent transgenic lines from each genetic background. The BIK1 protein levels were dramatically lower in seedlings and mature plants of the sik1–1 BIK1-HA transgenic lines compared to that in the Col-0 BIK1-HA lines (Figure 6A-D). To investigate if the reduced BIK1 protein accumulation was caused by differential transcription of BIK1 in sik1–1, we analyzed the BIK1-HA transcripts in these transgenic lines. No obvious differences were observed for BIK1-HA transcripts between independent Col-0 and sik1–1 transgenic lines (Figure S6C), indicating that the reduced BIK1 protein accumulation in the sik1–1 genetic background is modulated at the protein level.
Figure 6. SIK1 regulates BIK1 stability.
(A) BIK1-HA accumulation in seedlings of the independent transgenic lines. (B) Quantification of band intensity based on eight independent transgenic lines per genotype, including those shown in (A). (C) BIK1-HA protein levels in mature plants of the independent transgenic lines. (D) Quantification of band intensity based on eight transgenic lines per genotype, including those shown in (C). Asterisks indicate significant differences (Student’s t test, p<0.01) for (B and D). (E) flg22-induced ROS burst in sik1–1 and sik1–1 BIK1-HA transgenic lines. Total relative luminescent units (RLU) were detected over a 40-min period. Similar results were obtained in two independent experiments. (F) Detection of BIK1-HA in transgenic lines after flg22 treatment. Protoplasts were isolated from Col-0 BIK1-HA and sik1–1 BIK1-HA transgenic lines, and treated with water or flg22 for 10 min. CBB, Coomassie Brilliant Blue staining. (G) Enhanced BIK1 stability by SIK1 expression. The indicated constructs were expressed in pBIK1::BIK1-HA protoplasts. The accumulation of BIK1 and SIK1 was determined by immunoblot analyses after treatment for 6h with either 0.2% DMSO (−) or 50 μM MG132 (+). KD = kinase dead variant. SE = short exposure; LE = long exposure. This experiment was repeated three times with similar results. (H) Quantification of BIK1-HA band intensity based on three replicates. Different letters indicate significant differences (Fisher’s LSD, p < 0.01) for (E and H). (I) Co-immunoprecipitation of HA-SIK1 and XLG2-FLAG after co-expression in Arabidopsis. See also Figure S6.
Transgenic overexpression of BIK1 can increase BIK1 protein accumulation and increase PAMP-induced ROS production (Liang et al., 2016). To further investigate if the reduced BIK1 accumulation is responsible for the compromised ROS production in sik1 mutants, we measured the flg22-induced ROS burst in the sik1–1 mutant and sik1–1 BIK1-HA transgenic lines. Expression of BIK1-HA can significantly enhance ROS production in the sik1 mutant (Figure 6E and S6D). These results indicate that SIK1 regulates extracellular ROS production through BIK1 stabilization.
Next, we investigated at what point SIK1 functions to stabilize BIK1. In the absence of PAMP perception, BIK1 exhibits reduced accumulation in the sik1–1 mutant (Figure 6A-D). Treatment with flg22 did not rescue BIK1-HA accumulation in the sik1 mutant compared to that in Col-0 (Figure 6F), indicating that SIK1 mainly stabilizes BIK1 at a resting state. After flg22 treatment, we detected a partial shift of BIK1 protein by western blot, which has been previously demonstrated to be phosphorylated BIK1 (Lu et al., 2010). We detected a similar pattern of BIK1 mobility in Col-0 and the sik1–1 mutant after flg22 perception, indicating that sik1–1 is not compromised in BIK1 hyperphosphorylation (Figure 6F). We next generated protoplasts from the bik1 complementation line pBIK1::BIK1-HA. Expression of T7-SIK1 in pBIK1::BIK1-HA enhanced BIK1 accumulation (Figure 6G and 6H). However, expression of a T7-SIK1 kinase-dead variant (SIK1KD) did not enhance BIK1 accumulation (Figure 6G and 6H), indicating that SIK1 kinase activity is required for BIK1 stability. Protein accumulation of PBL1, a close homolog of BIK1, was not affected in sik1–1 (Figure S6E). Interestingly, wild-type T7-SIK1 accumulated at a low level, whereas T7-SIK1KD robustly accumulated (Figure 6G), suggesting that SIK1 protein accumulation is also tightly regulated in a kinase activity-dependent manner. These results strongly support that SIK1 regulates extracellular ROS burst through BIK1 stabilization.
Similar to BIK1, SIK1 associates with the PRR FLS2 at a resting state (Figure S6F). It was reported recently that heterotrimeric G proteins are directly coupled with the FLS2-BIK1 immune complex to positively regulate FLS2-mediated immune responses by stabilizing BIK1 at a resting state (Liang et al., 2016; Liang et al., 2018). These previous findings prompted us to investigate the link between SIK1 and heterotrimeric G proteins. We expressed the extra-large G protein subunit XLG2 and SIK1 in Arabidopsis, and performed co-IP. Interestingly, XLG2 associates with SIK1 in planta (Figure 6I), indicating that SIK1 enhances BIK1 stability at a resting state by coupling with heterotrimeric G proteins.
SIK1 associates with and phosphorylates RBOHD
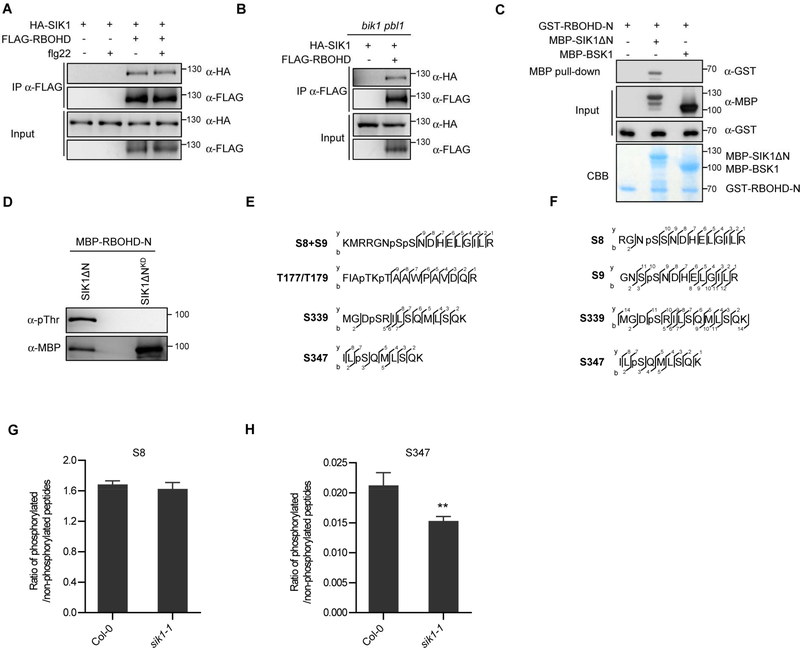
PAMP-induced ROS production was lower in the sik1 mutants than bik1 pbl1 after flg22 perception (Figure S5B and S5C), indicating that SIK1 may also regulate the extracellular ROS independently of RLCKs. In order to investigate if SIK1 directly regulates RBOHD, we first expressed SIK1 and RBOHD in Arabidopsis and performed co-IP to test their interaction. A specific signal for HA-SIK1 was clearly observed in the FLAG-RBOHD immunoprecipitate (Figure 7A), indicating that SIK1 can associate with RBOHD. SIK1 remains physically associated with RBOHD after flg22 treatment (Figure 7A), indicating that the SIK1-RBOHD association is not dynamic.
Figure 7. SIK1 directly interacts with RBOHD and phosphorylates its N-terminus.
(A) SIK1 associates with RBOHD in Col-0. HA-SIK1 and FLAG-RBOHD were co-expressed in Col-0 protoplasts, and co-immunoprecipitation was performed. Protoplasts were treated with 1 μM flg22 or water, and then total proteins were subjected to antiFLAG immunoprecipitations. (B) SIK1 associates with RBOHD in bik1 pbl1. The indicated constructs were co-expressed and subjected to anti-FLAG immunoprecipitations. (C) MBP pull-down assay demonstrates direct interaction between SIK1 and RBOHD N-terminus in vitro. GST-RBOHD-N, MBP-SIK1ΔN, and MBP-BSK1 recombinant proteins were affinity purified from E. coli, and pull-down assays were performed using Amylose beads. (D) SIK1 phosphorylates RBOHD’s N-terminus. MBP-RBOHD-N was co-expressed with His-SIK1ΔN or His-SIK1ΔNKD in E.coli, and then MBP-RBOHD-N was purified. Phosphorylation was detected by anti-pThr immunoblot (top panel). Total MBP-RBOHD-N was detected by anti-MBP immunoblot (bottom panel). (E) RBOHD phosphorylation sites identified in vitro by LC-MS/MS. The observed y and b ions are numbered. (F) RBOHD phosphosites identified in vivo upon flg22 treatment. The observed y and b ions are numbered. (G and H) Quantification of phosphorylation of S8 and S347 residues in vivo. FLAG-RBOHD was transiently expressed in Col-0 and sik1–1 protoplasts, and treated with 1 μM flg22 for 10 min. Total proteins were subjected to immunoprecipitation with anti-FLAG beads followed by on bead digestion. Phosphorylated peptides were quantified by PRM. Data are means ±SE of four biological replicates. Asterisks indicate significant differences (Student’s t test, p<0.05). Similar results were obtained in two independent experiments. See also Figure S7.
To investigate if SIK1 associates with RBOHD through BIK1 or PBL1 RLCK proteins, we examined the SIK1-RBOHD association in the bik1 pbl1 genetic background. Interestingly, SIK1 still associates with RBOHD in the bik1 pbl1 double mutant (Figure 7B), indicating that SIK1 may directly interact with RBOHD. We purified GST-RBOHD-N, MBP-SIK1ΔN and MBP-BSK1 recombinant proteins from E. coli and conducted an MBP pull-down assay. The result clearly showed that SIK1ΔN specifically interacts with the N-terminus of RBOHD (Figure 7C).
Positive regulation of RBOHD activity is mainly through phosphorylation of its N-terminus (Kadota et al., 2015). Therefore, to test if SIK1 can directly phosphorylate the N-terminus of RBOHD, we co-expressed MBP-RBOHD-N with SIK1ΔN or SIK1ΔNKD in E.coli, and purified MBP-RBOHD-N for detection of phosphorylation and LC-MS/MS analyses. SIK1ΔN phosphorylated the RBOHD N-terminus (MBP-RBOHD-N) but the SIK1ΔNKD did not (Figure 7D), indicating that trans-phosphorylation of MBP-RBOHD-N was due to SIK1ΔN. LC-MS/MS identified six potential phosphorylation residues (S8, S9, T177, T179, S339 and S347) in RBOHD’s N-terminus only after co-expression with SIK1ΔN (Figure 7E, and S7). Among these residues, S339 and S347 are essential for RBOHD activity and are also phosphorylated by BIK1 or CPKs upon PAMP perception (Kadota et al., 2014; Li et al., 2014). Phosphorylation of S8, S9, T177 and T179 residues have not been previously reported. To test if these phosphorylation sites occur after PAMP perception, we expressed FLAG-RBOHD in Arabidopsis protoplasts, and enriched FLAG-RBOHD protein using anti-FLAG agarose beads for mapping phosphorylation sites in vivo by mass spectrometry. The spectra demonstrated phosphorylation of S8, S9, S339 and S347 residues after flg22 treatment in vivo (Figure 7F). To further determine if ratio of phosphorylation of these residues is altered in the sik1–1 mutant, parallel reaction monitoring (PRM) was conducted. Unmodified peptides for the S339 site were undetectable by mass spectrometry and S9 phosphorylation was not detected by PRM, so we focused on the S8 and S347 residues. We did not observe a significant difference for phosphorylation of residue S8 between Col-0 and the sik1–1 mutant (Figure 7G). However, phosphorylation of S347 was significantly reduced in sik1–1 (Figure 7H), suggesting that SIK1 contributes to S347 phosphorylation in vivo. These results suggest that SIK1 positively regulates ROS by directly phosphorylating RBOHD at promoting sites in addition to enhancing BIK1 stability.
DISCUSSION
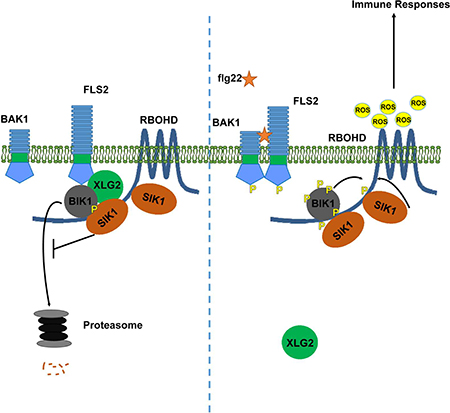
Multiple MAP4Ks have been identified across eukaryotes based on their homology to the yeast kinase STE20 (Dan et al., 2001). Here, we characterized the Arabidopsis MAP4K SIK1 as an important component of plant immune responses that positively regulates extracellular ROS production and promotes PTI against the bacterial pathogen Pst. SIK1 associates with and stabilizes the central immune regulator BIK1 as well as directly phosphorylates RBOHD to promote the ROS burst. Importantly, we further demonstrated that, similar to yeast STE20, the Arabidopsis MAP4K SIK1 also associates with heterotrimeric G proteins. These results indicate that the STE20 MAP4K family exerts similar function to transduce signals from surface-localized receptors to downstream responses via heterotrimeric G proteins. Our results not only reveal regulatory mechanisms that ensure robust immune responses, but also reveal the importance and functional conservation of the STE20 family across eukaryotes.
The post-translational modification and stability of PRRs and their associated members are tightly regulated in order to ensure appropriate and robust immune activation. Overexpression of BIK1 results in enhanced defense responses, indicating that BIK1 contributes to immune signaling in a dose dependent manner (Monaghan et al., 2014). SIK1-mediated stabilization of BIK1 likely contributes to appropriate and robust immune signaling. Expression of BIK1-HA significantly enhances ROS production in the sik1–1 mutant, further highlighting the importance of BIK1 accumulation for mediating PTI responses. Heterotrimeric G proteins also maintain BIK1 stability by attenuating the proteasome-dependent degradation of BIK1 (Liang et al., 2016; Wang et al., 2018). Interestingly, the yeast STE20 MAP4K associates with the βγ-subunits of heterotrimeric G proteins to activate downstream signaling upon perception of pheromone by the G protein-coupled receptor (Leberer et al., 1997). In this study, we demonstrated that SIK1 stabilizes BIK1 at a resting state and associates with XLG2, indicating that SIK1 may work together with the heterotrimeric G proteins to regulate BIK1 protein accumulation.
SIK1 promotes BIK1 stability in a kinase dependent manner, indicating that phosphorylation by SIK1 may play a positive role in BIK1 protein stability. Previously, the CPK28 kinase was demonstrated to phosphorylate BIK1 and enhance BIK1 turnover by the proteasome (Monaghan et al., 2014). Protein turnover can also be inhibited by phosphorylation. For example, to ensure activation of defense-related genes, the transcriptional factor ERF6 is stabilized by MPK3/MPK6-mediated phosphorylation (Meng et al., 2013). Phosphorylation of BIK1 S236 and T237 residues stabilizes BIK1 (Wang et al., 2018), and we show here that S236 is phosphorylated by SIK1 in vitro, indicating that SIK1 modulates BIK1 accumulation by direct phosphorylation. SIK1 associates with XLG2 to enhance BIK1 stability at a resting state, thus it is likely that BIK1 S236 phosphorylation occurs in the absence of pathogen perception. Given the opposing phenotypes of the cpk28 and sik1 knockouts, CPK28 likely phosphorylates different BIK1 residues, resulting in an opposite effect on BIK1 protein turnover. BIK1 activity and abundance is regulated by multiple mechanisms (Couto et al., 2016; Liang et al., 2016; Monaghan et al., 2014), further highlighting its importance in plant immune signaling.
The NADPH oxidase RBOHD is required for the extracellular ROS burst in response to PAMP perception (Kadota et al., 2015). This ROS burst is critical for mediating many PTI responses, including stomatal closure, callose deposition, systemic signaling, and inhibition of bacterial growth (Kadota et al., 2015). RBOHD is positively regulated by N-terminal phosphorylation mediated by multiple kinases to ensure robust immune responses (Kadota et al., 2015). BIK1 is able to phosphorylate RBOHD at four critical residues: S39, S339, S343 and S347 whose phosphorylation levels rapidly increase upon PAMP perception (Kadota et al., 2014; Li et al., 2014). RBOHD S347 was also shown to be phosphorylated by CPKs (Kadota et al., 2014). We show here that SIK1 directly interacts with and phosphorylates RBOHD’s N-terminus. Among these residues, S8, S9, S339 and S347 are phosphorylated in vivo after flg22 treatment. Interestingly, phosphorylation of S347 is significantly reduced in sik1–1, further confirming that SIK1 affects RBOHD’s phosphorylation status. S347 phosphorylation has been shown to be required for RBOHD activation (Kadota et al., 2014; Li et al., 2014; Nühse et al., 2007). These findings indicate that, in addition to regulating BIK1 protein accumulation, SIK1 directly interacts with and phosphorylates RBOHD to ensure a robust extracellular ROS burst.
Although sik1 and bik1 knockouts are clearly compromised in PTI responses, they also exhibit autoimmune phenotypes including high levels of basal SA and enhanced resistance to virulent Pst. Phosphorylated BIK1 was recently demonstrated to be directly involved in regulating levels of the defense hormones JA and SA (Lal et al., 2018). When autoimmune effects are removed, the corresponding genes of some mutants, such as saul1, have been shown to facilitate immune signaling (Tong et al., 2017). Similarly, when the high level of basal SA was removed by the expression of the SA dehydrogenase NahG, the sik1 knockout was compromised in flg22-induced bacterial growth inhibition and no longer exhibited enhanced resistance to virulent Pst. These results highlight the importance of careful phenotyping of autoimmune mutants.
SIK1 homologs are present across land plants and display similar domain architecture, with a long N-terminal region and central kinase domain. The conservation of SIK1 in diverse plants indicates that homologs may use similar mechanisms to regulate RLCK stability and RBOHD phosphorylation. Thus, modification of SIK1 expression or stability could be a promising strategy to enhance crop disease resistance. Our study has uncovered a role of a conserved MAP4K family member in plant innate immunity, which motivates us to investigate the functions of other MAP4K members in the future.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to will be fulfilled by the Lead Contact, Gitta Coaker (glcoaker@ucdavis.edu).
EXPERIMENTAL MODELS AND SUBJECT DETAILS
Arabidopsis thaliana
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the wild-type control. Arabidopsis T-DNA insertion lines for MAP4K members (Table S2) were obtained from the Arabidopsis Biological Resource Center (ABRC) and are in the Col-0 genetic background. T-DNA insertion mutants were genotyped by PCR using T-DNA specific and the gene-specific primers as listed in Table S3. Gene-specific primers (Table S3) were then used for RT-PCR to detect full-length transcripts. Seeds of bik1 (Lu et al., 2010), bik1 pbl1 (Zhang et al., 2010), the bik1 complementation line pBIK1::BIK1-HA (Lin et al., 2013), fls2 (SALK_062054) (Ranf et al., 2012), sik1–4 (Xiong et al., 2016) and the sik1–4 complementation line pUBQ::SIK1 (Xiong et al., 2016) used in this study were described previously.
Arabidopsis seeds were stratified for 3 days in the dark at 4°C then sown on soil. To detect lesion mimic phenotypes, Arabidopsis plants were grown at 22°C, with a 16-h-light/8-h-dark photoperiod (100 μmol m−2 s−1). For all other experiments, Arabidopsis plants were grown at 23°C and 70% relative humidity with a 10-h-light/14-h-dark photoperiod (100 μmol m−2 s−1).
Nicotiana benthamiana
N. benthamiana was grown in growth chamber at 22°C under a long-day photoperiod (16 h light and 8 h dark).
Bacterial Strains
P. syringae pv. tomato (Pst) DC3000 was grown at 28°C on nutrient yeast-glycerol agar (NYGA) with appropriate antibiotics.
METHOD DETAILs
Phylogenetic analyses and prediction of protein domain architecture
SIK1 orthologs were determined by bi-directional BLAST. Multiple sequence alignments were generated by T-Coffee (Tommaso et al., 2011). Trees were built using PhyML with default settings (Guindon et al., 2010), and displayed with FigTree (http://tree.bio.ed.ac.uk/software/figtree) and iTOL (http://itol.embl.de). The kinase domains in SIK1, SIK1 orthologs, and MAP4K family members were predicted using Pfam (Finn et al., 2016), and domain architecture was illustrated by DOG (Ren et al., 2009).
PAMP-triggered ROS Burst
Leaf disks of four-week-old Arabidopsis plants were harvested with a cork borer (4.76 mm) and ROS burst was detected after addition of 100 nM flg22 or elf18 peptide as previously described (Lin et al., 2015). For the chitoheptaose-induced ROS assay, the L-012 chemiluminescent probe was used because it is more sensitive than luminol (Liang et al., 2013). For FLS2 protein detection, total protein extract from the indicated lines were separated on SDS-PAGE and immunoblot was performed using anti-FLS2 (1:5,000) (Agrisera) primary antibody followed by anti-rabbit-HRP (1:2,000) (BioRad) secondary antibody.
Phytohormone measurement
Arabidopsis rosette leaves from five-week-old plants were harvested and ground in liquid nitrogen. Extraction and quantification of SA, JA, ABA, and OPDA were carried out by gas chromatography-mass spectrometry (GC-MS) as previously described (Chehab et al., 2008; Engelberth and Engelberth, 2009) using dihydro-JA, deuterated SA, and ABA as internal standards.
Quantitative RT-PCR
Total RNA from Arabidopsis leaves was extracted via the Trizol method. One microgram of total RNA was reverse transcribed with an oligo(dT) primer using M-MLV Reverse Transcriptase (Promega). The relative expression levels of the tested genes were normalized to the AtEF1α in Arabidopsis. PCR primers are provided in Table S3. Quantitative RT-PCR was performed on a CFX96 real-time System (Bio-Rad) under the following conditions: 95°C for 30 s, then 40 cycles at 95°C for 5 s and 60°C for 15 s, followed by a melting curve analysis to validate specificity. The relative expression level of each gene was determined using the delta-delta Ct method (Livak and Schmittgen, 2001).
Transgenic Lines
The NahG gene was PCR amplified from pCIB200-NahG (Gaffney et al., 1993), cloned into pENTR/D-TOPO (Thermo Fisher Scientific), and recombined into pGWB14 (Nakagawa et al., 2007) by LR reaction (Thermo Fisher Scientific). pBIK1::BIK1-HA construct was described previously (Zhang et al., 2010). Binary vectors were then transformed into Agrobacterium tumefaciens strain C58C1, and transgenic lines were generated by floral dip (Clough and Bent, 1998). Transformants were selected for one-half-strength Murashige and Skoog medium supplemented with 15 μg ml−1 Hygromycin B. For NahG-trangenic lines, expression of NahG-3×HA was confirmed by immunoblot in T3 homozygous lines. Horseradish peroxidase (HRP)-conjugated anti-HA antibody (Sigma) was used at a 1:1,000 concentration in conjunction with SuperSignal West Pico Chemiluminescent Substrate (Pierce) for detection. PR1 protein expression in NahG-transgenic lines was detected using anti-PR1 antibody (Wang et al., 2005). The PR1 mRNA abundance was measured by qPCR. PR1 protein expression, ROS, and bacterial growth assays were performed on sik1 NahG-HA line #1. MAPK activation was performed on sik1 NahG-HA lines #1 and #3. For BIK1-HA transgenic lines, expression of BIK1-HA protein was confirmed by western blot using an anti-HA antibody in independent T1 transgenic lines.
Bacterial inoculation
P. syringae pv. tomato (Pst) DC3000 was grown at 28°C for 2 days on NYGA containing 100 mg ml−1 rifampicin and 25 mg ml−1 kanamycin. Bacterial cells were collected and diluted to the appropriate concentration with 5 mM MgCl2. To measure bacterial growth, Arabidopsis leaves were infiltrated with the Pst DC3000 at a concentration of 1 × 105 colony-forming units ml−1 (CFU ml−1). Bacterial titers were detected 3 days post-inoculation (dpi) as previously described (Liu et al., 2009). For the flg22 protection assay, five-week-old plants were first infiltrated with 1 μM flg22 or H2O 24 h before infiltration with 1 × 105 CFU ml-1. Bacterial titers were determined 2 dpi.
Recombinant protein purification and kinase assays
SIK1 variants were cloned as maltose-binding protein (MBP) fusions for recombinant protein expression in E. coli. SIK1ΔN refers to amino acids 249–836. SIK1 cDNA fragments were PCR amplified and cloned into pENTR/D-TOPO (Invitrogen). SIK1ΔN KD (K278E) variants were generated by PCR-based site-directed mutagenesis. The resulting pENTR SIK1 constructs were recombined into a modified pMAL-C4X vector (New England Biolabs) containing a Gateway cassette. The RBOHD N-terminus was cloned into the pMAL-C4X vector by LR reaction. Primers used are listed in Table S3.
His-BIK1KD was expressed and purified as previously described (Lal et al., 2016). The remaining expression constructs were transformed into E. coli BL21(DE3) and protein expression was induced by addition of 0.3 mM IPTG to a 250 ml culture at OD600 = 0.4, followed by incubation at 28°C for 4 h. Cells were harvested and resuspended in column buffer (20 mM Tris-HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT, and 0.1% Triton X-100) for MBP-tagged proteins, and buffer A (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 20 mM imidazole, and 0.1% Triton X-100) for His-tagged proteins, followed by lysis using a microfluidizer. Cell lysates were centrifuged at 20,000g to separate insoluble cell debris, and soluble supernatant was applied to MBPTrap HP (GE Healthcare) or HisTrap HP (GE Healthcare) 5 ml column on an AKTA FPLC (GE Healthcare). MBP fusion proteins were eluted with 10 mM maltose in column buffer; His-BIK1KD was eluted with a gradient of 20–500 mM imidazole in buffer A.
In vitro kinase assays were performed in kinase buffer (20 mM Tris-HCl pH 7.5, 10 mM MgCl2, 1 mM CaCl2, 100 μM ATP, 1 mM DTT). 0.5 μg of SIK1ΔN or SIK1ΔNKD and 3–5 μg of substrate were used per reaction. Reactions were initiated by incubating at 30°C for 30 min and stopped with the addition of 3 × Laemmli sample buffer. Samples were subjected to immunoblot with anti-pThreonine antibody (Cell Signaling) at a 1:2000 concentration, followed by anti-rabbit-HRP secondary antibody (BioRad). Blots were visualized with SuperSignal West Pico Chemiluminescent Substrate (Pierce) for detection. For radiolabeled kinase assays, 10 μCi of γ−32P-ATP was added to each reaction and performed as described above. The proteins were separated on a 12% SDS-PAGE gel and phosphorylated proteins were visualized by X-ray film exposure.
In vitro pull-down assays
The coding region of BIK1, BSK1, RBOHD-N, and SIK1 was amplified using PCR. For GST-tagged proteins, GST vector (Addgene #29707) linearized with SspI was used. MBP-tagged proteins were cloned into the MBP vector (Addgene #29708) linearized with SspI. Cloning was performed using Gibson assembly (Gibson et al., 2009). Sequenced plasmids were transformed into the BL21 expression strain of E. coli.
Recombinant proteins were purified separately, and in vitro GST pull-downs were performed by incubating 1 μg of GST-SIK1ΔN with 4 μg of MBP-BIK1, or MBP-BSK1 with GST beads calibrated with pull-down buffer (30 mM HEPES, 300 mM NaCl, 0.2% triton X-100, pH 7.5). Proteins were incubated with moderate shaking at 4°C for 2 h. GST beads were washed 3 times with 100 column volumes of wash buffer (30 mM HEPES, 350 mM NaCl, 0.2% triton X-100, pH 7.5) for 2 h each. Protein bound to GST resin was eluted by boiling the resin in SDS sample buffer and analyzed by western blot. Similar protocol was followed for GST-RBOHD-N and MBP-SIK1ΔN pull-down assay.
Phosphorylation site identification
To identify BIK1 phosphorylation sites in vitro, the kinase inactive His-BIK1 recombinant protein was co-expressed with SIK1ΔN or SIK1ΔNKD in E. coli BL21(DE3) and purified using a HisTrap HP column. The purified recombinant protein was then digested with trypsin (Minkoff et al., 2014). Briefly, the recombinant protein was reduced with 5 mM DTT, alkylated with 15 mM iodoacetamide, and digested overnight with trypsin (Promega) at 37°C. To identify RBOHD phosphorylatio n sites, the RBOHD N terminus was co-expressed with SIK1ΔN or SIK1ΔNKD in Rosetta(DE3), and purified using a MBPTrap HP column. The RBOHD-N recombinant protein was separated on a 10% precast protein gel (BioRad), and stained with Coomassie brilliant blue. The band for MBP-RBOHD-N was excised and subjected to tryptic in-gel digestion (Elmore et al., 2012). Tryptic peptides were analyzed by LC-MS/MS using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). The Peptides were identified using X!Tandem (Craig and Beavis, 2004). Parameters were set for 20 ppm peptide tolerance. Phosphorylation of Ser, Tyr or Thr residues were allowed as variable modifications. X!Tandem results were combined in Scaffold version 4 (Searle, 2010) and exported to Excel (Microsoft Office).
To quantify phosphosites in vivo, FLAG-RBOHD (Li et al., 2014) was transiently expressed in Col-0 and sik1–1 protoplasts, and after 12 h incubation protoplasts were treated with 1 μM flg22 for 10 min. Then total proteins were subjected to immunoprecipitation using anti-FLAG beads and followed by on bead digestion with trypsin. Isolation list for phosphopeptide and control peptides, as shown in Table S4, were acquired on the Orbitrap mass spectrometer using Skyline software (MacLean et al., 2010). The peptides were analyzed using an Orbitrap Fusion Lumos mass spectrometer (Thermo Fisher Scientific). The resulted raw data were imported into Skyline and the peak areas were then exported into Excel (Microsoft) for further analysis.
MAPK activity assay
Four-week-old Arabidopsis plants were spray with 10 μM flg22 or water containing 0.025% Silwet L-77 for 10 min and frozen in liquid nitrogen. Tissue samples were ground in liquid nitrogen and resuspended in 50 mM HEPES (pH 7.5), 50 mM NaCl, 10 mM EDTA, 0.2% Triton X-100, 1 × complete protease inhibitors (Thermo Fisher Scientific) and 1 × phosphatase inhibitors (Thermo Fisher Scientific). The protein concentration was determined using the Pierce 660 nm protein assay supplemented with the Ionic Detergent Compatibility Reagent (Thermo Fisher Scientific). Equal amounts of total protein were loaded on a 12% SDS-PAGE gel, and an antiphospho-p44/42 (Erk1/2) (Cell Signaling) monoclonal antibody was used to determine the phosphorylation state of MPK3, MPK4 and MPK6.
RLCK stability in Arabidopsis protoplasts
To generate gateway compatible vectors for transient expression in protoplasts, the 35S-T7-R1-CmR-ccdB-R2 fragment from pGWB27 (Nakagawa et al., 2007) was PCR amplified and cloned into pUC19 vector (New England Biolabs). pENTR clone of SIK1 was recombined into the modified vector mentioned above by LR reaction. BAK1 cDNA fragment was cloned as an XhoI and BstBI fragment into pUC19–35S-FLAG-RBS (Li et al., 2005). PBL1 was cloned into pUC19–35S-FLAG-RBS using KpnI and BstBI restriction sites. All constructs were transiently expressed in Arabidopsis protoplasts as previously described (Yoo et al., 2007). Protein accumulation was analyzed by immunoblot with anti-HA-HRP (1:1,000) (Roche), anti-T7-HRP (1:3,000) (Millipore), and anti-FLAG-HRP (1:2,000) (Sigma) antibodies.
Co-immunoprecipitation
To test the association between SIK1 and BIK1, RBOHD or XLG2, the Arabidopsis protoplasts were transfected with the indicated constructs (Liang et al., 2016; Li et al., 2014). After 12 h of incubation, protoplasts were treated with 1 μM flg22 for 10 min, and total protein was isolated with an extraction buffer (50 mM Tris-HCl pH7.5, 50 mM NaCl, 10% glycerol, 5 mM DTT, 1mM Na2MoO4.2H2O, 1% IGEPAL, 1 × complete protease inhibitor and 1 × phosphatase inhibitors). The extract was pre-cleared by centrifugation at 14,000 rpm for 10 min. Total protein was incubated with an agarose-conjugated anti-Flag antibody (Sigma) for 2 h, and then washed four times with a buffer containing 50 mM Tris-HCl pH7.5, 50 mM NaCl, 5 mM DTT, 1mM Na2MoO4.2H2O and 1% IGEPAL. Immunoprecipitates were separated on a 10% SDS-PAGE gel and detected by anti-HA or anti-Flag immunoblot.
To test association between SIK1 ΔN and FLS2 in N. benthamiana, Agrobacterium suspensions containing p1776-SIK1ΔN (35S: T7-SIK1ΔN) or pEarlygate103-FLS2 (35S: FLS2-GFP-His) were co-infiltrated into N. benthamiana leaves. One gram of leaf tissue was collected at 48 hpi, and homogenized in IP buffer containing 50 mM Tris-HCl pH7.5, 150 mM NaCl, 10% glycerol, 10 mM DTT, 1mM Na2MoO4.2H2O, 2% IGEPAL (Sigma), 1 × complete protease inhibitor (Thermo Fisher Scientific) and 1 × phosphatase inhibitors (Thermo Fisher Scientific). The homogenate was pre-cleared by centrifugation at 14,000 rpm for 20 min and further filtered using two layers of cheesecloth. Subsequently, 30 μl of GFP-Trap (Chromotek) was added to the homogenate and incubated at 4°C for 2 h. Beads were washed 3 times with IP buffer and re-suspended in 3 × Laemmli buffer. Samples were separated on SDS-PAGE and immunoblot was performed using anti-T7-HRP (1: 3,000) (Millipore) antibody, or anti-GFP (1: 500) (Abcam) primary antibody followed by anti-rabbit-HRP (1: 2,000) (BioRad) secondary antibody.
Effector translocation assays
The adenylate cyclase (CyaA) delivery assay was performed as previously described, with slight modifications (Crabill et al., 2010). Five-week-old Col-0 and sik1–1 leaves were infiltrated with Pst DC3000 containing avrRpm1-CyaA (Casper-Lindley et al., 2002; Mudgett and Staskawicz, 1999) at 3 × 107 CFU ml-1. Eight hours post-infiltration, 0.56 cm2 leaf discs from six individual plants were harvested as a biological replicate. Samples were flash frozen in liquid nitrogen and stored at −80°C. To extract cAMP, leaf discs were grounded to fine powder and re-suspended in 200 μl of 0.1 M HCl. Samples were diluted 300-fold with 0.1 M HCl. cAMP levels were quantified by using a direct cAMP ELISA kit (Enzo). Three biological replicates were performed.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistical analyses were performed using Prism 7 software (GraphPad). The data are presented as mean ± SEM. For luminal-based ROS burst and bacterial growth curve assays, n represents the number of individual plants. For quantification of BIK1-HA band intensity, PRM, and hormone measurement, n represents the number of experimental replicates. Student’s t test was used to compare means for two groups. One-way ANOVA with Dunnett’s multiple-comparison test was performed to compare means from several groups against a control group mean. Fisher’s LSD test was used to compare means between groups. Statistical analyses and the exact value of n are described in detail in the figures and figure legends.
Supplementary Material
Highlights.
The conserved MAP4K SIK1 is required for PAMP-induced ROS production in Arabidopsis
SIK1 binds, phosphorylates and stabilizes the central immune regulator BIK1
sik1 mutants display resistance to P. syringae due to high salicylic acid accumulation
SIK1 binds to and phosphorylates an NADPH oxidase to enhance ROS production for defense
ACKNOWLEDGMENTS
We thank Q. Gong for the sik1–4 complementation seeds, P. He for the bik1 complementation seeds and Q. Wang for assistance in phylogenetic analyses. This work was supported by grants from NIH (R01-GM092772) and NSF (MCB-1054298) awarded to G.C.; the National Natural Science Foundation of China (31672008, 31301613) to M.Z.; NSF (IOS-1146128) to B.D.; NSF (NSF-IOS-1354434; NSF-DBI-0723722, NSF-DBI-1042344) to S.P.D.-K.; NIH (R01-GM107311) and NSF (IOS-1036491, IOS-1352478) grants to K.D.; M.Z.’s visiting was partially supported by the National Science Fund for Distinguished Young Scholars (31625023), the Priority Academic Program Development of Jiangsu Higher Education Institutions and the Outstanding Youth Fund of Jiangsu Province (BK20160016).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Ashby AM, Watson MD, Loake GJ, Shaw CH (1988). Ti plasmid-specified chemotaxis of Agrobacterium tumefaciens C58C1 toward vir-inducing phenolic compounds and soluble factors from monocotyledonous and dicotyledonous plants. J. Bacteriol 170, 4181–4187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D, Brechmann M, Rohling S, Tapernoux M, Mock T, Winter D, Lehmann WD, Kiefer F, Thome M, Krammer PH, et al. (2009). Phosphorylation of CARMA1 by HPK1 is critical for NF-κB activation in T cells. Proc. Natl. Acad. Sci. USA 106, 14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper-Lindley C, Dahlbeck D, Clark ET, and Staskawicz BJ (2002). Direct biochemical evidence for type III secretion-dependent translocation of the AvrBs2 effector protein into plant cells. Proc. Natl. Acad. Sci. USA 99, 8336–8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chehab EW, Kaspi R, Savchenko T, Rowe H, Negre-Zakharov F, Kliebenstein D, and Dehesh K (2008). Distinct roles of jasmonates and aldehydes in plant-defense responses. PLoS ONE. 3, e1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Bauer Z, Regenass M, Boller T, and Felix G (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell. 18, 465–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, Kemmerling B, Nurnberger T, Jones JDG, Felix G, and Boller T (2007). A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 448, 497–500. [DOI] [PubMed] [Google Scholar]
- Clough SJ, and Bent AF (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Couto D, Niebergall R, Liang X, Bucherl CA, Sklenar J, Macho AP, Ntoukakis V, Derbyshire P, Altenbach D, Maclean D, et al. (2016). The Arabidopsis protein phosphatase PP2C38 negatively regulates the central immune kinase BIK1. PLoS Pathog. 12, e1005811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couto D, and Zipfel C (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immunol 16, 537–552. [DOI] [PubMed] [Google Scholar]
- Crabill E, Joe A, Block A, van Rooyen JM, and Alfano JR (2010). Plant immunity directly or indirectly restricts the injection of type III effectors by the Pseudomonas syringae type III secretion system. Plant Physiol. 154, 233–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig R, and Beavis RC (2004). TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 20, 1466–1467. [DOI] [PubMed] [Google Scholar]
- Dan I, Watanabe NM, and Kusumi A (2001). The Ste20 group kinases as regulators of MAP kinase cascades. Trends Cell Biol. 11, 220–230. [DOI] [PubMed] [Google Scholar]
- Dubiella U, Seybold H, Durian G, Komander E, Lassig R, Witte CP, Schulze WX, and Romeis T (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA 110, 8744–8749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley KW, Haag JR, Pontes O, Opper K, Juehne T, Song K, and Pikaard CS (2006). Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Elmore JM, Liu J, Smith B, Phinney B, and Coaker G (2012). Quantitative proteomics reveals dynamic changes in the plasma membrane during Arabidopsis immune signaling. Mol. Cell. Proteomics 11, M111.014555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberth MJ, and Engelberth J (2009). Monitoring plant hormones during stress responses. J. Vis. Exp e1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng F, Yang F, Rong W, Wu X, Zhang J, Chen S, He C, and Zhou JM (2012). A Xanthomonas uridine 5’-monophosphate transferase inhibits plant immune kinases. Nature. 485, 114–118. [DOI] [PubMed] [Google Scholar]
- Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu ZQ, and Dong X (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol 64, 839–863. [DOI] [PubMed] [Google Scholar]
- Gaffney T, Friedrich L, Vernooij B, Negrotto D, Nye G, Uknes S, Ward E, Kessmann H, and Ryals J (1993). Requirement of salicylic-acid for the induction of systemic acquired-resistance. Science. 261, 754–756. [DOI] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang RY, Venter JC, Hutchison CA 3rd, and Smith HO (2009). Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, and Gascuel O (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol 59, 307–321. [DOI] [PubMed] [Google Scholar]
- Jiao S, Zhang Z, Li C, Huang M, Shi Z, Wang Y, Song X, Liu H, Li C, Chen M, et al. (2015). The kinase MST4 limits inflammatory responses through direct phosphorylation of the adaptor TRAF6. Nat. Immunol 16, 246–257. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Shirasu K, and Zipfel C (2015). Regulation of the NADPH oxidase RBOHD during plant immunity. Plant Cell Physiol. 56, 1472–1480. [DOI] [PubMed] [Google Scholar]
- Kadota Y, Sklenar J, Derbyshire P, Stransfeld L, Asai S, Ntoukakis V, Jones JD, Shirasu K, Menke F, Jones A, et al. (2014). Direct regulation of the NADPH oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell 54, 43–55. [DOI] [PubMed] [Google Scholar]
- Lal NK, Fisher AJ, and Dinesh-Kumar SP (2016). Arabidopsis receptor-like cytoplasmic kinase BIK1: purification, crystallization and X-ray diffraction analysis. Acta Crystallogr. Sect. F 72, 738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal NK, Nagalakshmi U, Hurlburt NK, Flores R, Bak A, Sone P, Ma X, Song G, Walley J, Shan L, et al. (2018). The receptor-like cytoplasmic kinase BIK1 localizes to the nucleus and regulates defense hormone expression during plant innate immunity. Cell Host Microbe. 23, 485–497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laluk K, Luo H, Chai M, Dhawan R, Lai Z, and Mengiste T (2011). Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 23, 2831–2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leberer E, Thomas DY, and Whiteway M (1997). Pheromone signalling and polarized morphogenesis in yeast. Curr. Opin. Genet. Dev 7, 59–66. [DOI] [PubMed] [Google Scholar]
- Li J, Henty-Ridilla JL, Staiger BH, Day B, and Staiger CJ (2015).Capping protein integrates multiple MAMP signalling pathways to modulate actin dynamics during plant innate immunity. Nat Commun. 6, 7206. doi: 10.1038/ncomms8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Li M, Yu L, Zhou Z, Liang X, Liu Z, Cai G, Gao L, Zhang X, Wang Y, et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe. 15, 329–338. [DOI] [PubMed] [Google Scholar]
- Li X, Lin H, Zhang W, Zou Y, Zhang J, Tang X, and Zhou JM (2005). Flagellin induces innate immunity in nonhost interactions that is suppressed by Pseudomonas syringae effectors. Proc. Natl. Acad. Sci. USA 102, 12990–12995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ding P, Lian K, Wang J, Ma M, Li L, Li L, Li M, Zhang X, Chen S, et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife. 5, e13568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Ma M, Zhou Z, Wang J, Yang X, Rao S, Bi G, Li L, Zhang X, Chai J, et al. (2018). Ligand-triggered de-repression of Arabidopsis heterotrimeric G proteins coupled to immune receptor kinases. Cell Res. 28, 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Cao Y, Tanaka K, Thibivilliers S, Wan J, Choi J, Kang CH, Qiu J, and Stacey G (2013). Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 341, 1384–1387. [DOI] [PubMed] [Google Scholar]
- Lin W, Li B, Lu D, Chen S, Zhu N, He P, and Shan L (2014). Tyrosine phosphorylation of protein kinase complex BAK1/BIK1 mediates Arabidopsis innate immunity. Proc. Natl. Acad. Sci. USA 111, 3632–3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W, Lu D, Gao X, Jiang S, Ma X, Wang Z, Mengiste T, He P, and Shan L (2013). Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc. Natl. Acad. Sci. USA 110, 12114–12119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin ZJ, Liebrand TW, Yadeta KA, and Coaker G (2015). PBL13 Is a serine/threonine protein kinase that negatively regulates Arabidopsis immune responses. Plant Physiol. 169, 2950–2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, and Coaker G (2009). RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol. 7, e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, and Schmittgen TD (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lu D, Wu S, Gao X, Zhang Y, Shan L, and He P (2010). A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Pro. Natl. Acad. Sci. USA 107, 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean B, Tomazela DM, Shulman N, Chambers M, Finney GL, Frewen B, Kern R, Tabb DL, Liebler DC, MacCoss MJ (2010). Skyline: an open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 26, 966–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Xu J, He Y, Yang K, Mordorski B, Liu Y, and Zhang S (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell. 25, 1126–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkoff BB, Burch HL, and Sussman MR (2014). A pipeline for 15N metabolic labeling and phosphoproteome analysis in Arabidopsis thaliana. Methods Mol. Biol 1062, 353–379. [DOI] [PubMed] [Google Scholar]
- Monaghan J, Matschi S, Shorinola O, Rovenich H, Matei A, Segonzac C, Malinovsky FG, Rathjen JP, MacLean D, Romeis T, et al. (2014). The calcium-dependent protein kinase CPK28 buffers plant immunity and regulates BIK1 turnover. Cell Host Microbe. 16, 605–615. [DOI] [PubMed] [Google Scholar]
- Mudgett MB, and Staskawicz BJ (1999). Characterization of the Pseudomonas syringae pv. tomato AvrRpt2 protein: demonstration of secretion and processing during bacterial pathogenesis. Mol. Microbiol 32, 927–941. [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, and Kimura T (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng 104, 34–41. [DOI] [PubMed] [Google Scholar]
- Nühse TS, Bottrill AR, Jones AME, and Peck SC (2007). Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranf S, Grimmer J, Poschl Y, Pecher P, Chinchilla D, Scheel D, and Lee J (2012). Defense-related calcium signaling mutants uncovered via a quantitative high-throughput screen in Arabidopsis thaliana. Mol. Plant 5, 115–130. [DOI] [PubMed] [Google Scholar]
- Ren J, Wen L, Gao X, Jin C, Xue Y, and Yao X (2009). DOG 1.0: illustrator of protein domain structures. Cell Res. 19, 271–273. [DOI] [PubMed] [Google Scholar]
- Rodriguez E, El Ghoul H, Mundy J, and Petersen M (2016). Making sense of plant autoimmunity and “negative regulators”. FEBS J. 283, 1385–1391. [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011). Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 7, e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Searle BC (2010). Scaffold: A bioinformatic tool for validating MS/MS-based proteomic studies. Proteomics. 10, 1265–1269. [DOI] [PubMed] [Google Scholar]
- Shi H, Yan H, Li J, Tang D (2013). BSK1, a receptor-like cytoplasmic kinase, involved in both BR signaling and innate immunity in Arabidopsis. Plant Signal. Behav 8, e24996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemiya A, Sugiyama N, Fujimoto H, Tsutsumi T, Yamauchi S, Hiyama A, Tada Y, Christie JM, and Shimazaki KI (2013). Phosphorylation of BLUS1 kinase by phototropins is a primary step in stomatal opening. Nat. Commun 4, 2094. [DOI] [PubMed] [Google Scholar]
- Tang D, Wang G, and Zhou JM (2017). Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 29, 618–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, and Kornev AP (2011). Protein kinases: evolution of dynamic regulatory proteins. Trends Biochem. Sci 36, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma BPHJ, Nurnberger T, and Joosten MHAJ (2011). Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell. 23, 4–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian S, Wang X, Li P, Wang H, Ji H, Xie J, Qiu Q, Shen D, and Dong H (2016). Plant aquaporin AtPIP1;4 links apoplastic H2O2 induction to disease immunity pathways. Plant Physiol. 171, 1635–1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, and Notredame C (2011). T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 39, W13–W17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong M, Kotur T, Liang W, Vogelmann K, Kleine T, Leister D, Brieske C, Yang S, Ludke D, Wiermer M, et al. (2017). E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 215, 1516–1532. [DOI] [PubMed] [Google Scholar]
- Torres MA, Dangl JL, and Jones JD (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. USA 99, 517–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres MA, Jones JD, and Dangl JL (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet 37, 1130–1134. [DOI] [PubMed] [Google Scholar]
- van Wersch R, Li X, and Zhang Y (2016). Mighty dwarfs: Arabidopsis autoimmune mutants and their usages in genetic dissection of plant immunity. Front. Plant Sci 7, 1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veronese P, Nakagami H, Bluhm B, AbuQamar S, Chen X, Salmeron J, Dietrich RA, Hirt H, and Mengiste T (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 18, 257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, El-Shetehy M, Shine MB, Yu K, Navarre D, Wendehenne D, Kachroo A, and Kachroo P (2014). Free radicals mediate systemic acquired resistance. Cell Rep. 7, 348–355. [DOI] [PubMed] [Google Scholar]
- Wang D, Weaver ND, Kesarwani M, and Dong X (2005). Induction of protein secretory pathway is required for systemic acquired resistance. Science. 308, 1036–1040. [DOI] [PubMed] [Google Scholar]
- Wang J, Grubb L, Wang J, Liang X, Li L, Gao C, Ma M, Feng F, Li L, Zhang X, et al. (2018). A regulatory module controlling homeostasis of a plant immune kinase. Mol. Cell 69, 493–504. [DOI] [PubMed] [Google Scholar]
- Xiong J, Cui X, Yuan X, Yu X, Sun J, and Gong Q (2016). The Hippo/STE20 homolog SIK1 interacts with MOB1 to regulate cell proliferation and cell expansion in Arabidopsis. J. Exp. Bot 67, 1461–1475. [DOI] [PubMed] [Google Scholar]
- Xu J, Wei X, Yan L, Liu D, Ma Y, Guo Y, Peng C, Zhou H, Yang C, Lou Z, et al. (2013). Identification and functional analysis of phosphorylation residues of the Arabidopsis BOTRYTIS-INDUCED KINASE1. Protein Cell. 4, 771–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, and Sheen J (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc 2, 1565–1572. [DOI] [PubMed] [Google Scholar]
- Yu X, Feng B, He P, and Shan L (2017). From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Li W, Xiang T, Liu Z, Laluk K, Ding X, Zou Y, Gao M, Zhang X, Chen S, et al. (2010). Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 7, 290–301. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JDG, Felix G, and Boller T (2004). Bacterial disease resistance in Arabidopsis through flagellin perception. Nature. 428, 764–767. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.