Abstract
To investigate genetic predispositions for MYCN-amplified neuroblastoma, we performed a meta-analysis of three genome-wide association studies totaling 615 MYCN-amplified high-risk neuroblastoma cases and 1869 MYCN-nonamplified non-high-risk neuroblastoma cases as controls using a fixed-effects model with inverse variance weighting. All statistical tests were two-sided. We identified a novel locus at 3p21.31 indexed by the single nucleotide polymorphism (SNP) rs80059929 (odds ratio [OR] = 2.95, 95% confidence interval [CI] = 2.17 to 4.02, Pmeta = 6.47 × 10-12) associated with MYCN-amplified neuroblastoma, which was replicated in 127 MYCN-amplified cases and 254 non-high-risk controls (OR = 2.30, 95% CI = 1.12 to 4.69, Preplication = .02). To confirm this signal is exclusive to MYCN-amplified tumors, we performed a second meta-analysis comparing 728 MYCN-nonamplified high-risk patients to identical controls. rs80059929 was not statistically significant in MYCN-nonamplified high-risk patients (OR = 1.24, 95% CI = 0.90 to 1.71, Pmeta = .19). SNP rs80059929 is within intron 16 in the KIF15 gene. Additionally, the previously reported LMO1 neuroblastoma risk locus was statistically significant only in patients with MYCN-nonamplified high-risk tumors (OR = 0.63, 95% CI = 0.53 to 0.75, Pmeta = 1.51 × 10-8; Pmeta = .95). Our results indicate that common genetic variation predisposes to different neuroblastoma genotypes, including the likelihood of somatic MYCN-amplification.
Neuroblastoma is characterized by a broad spectrum of clinical behavior (1). Children with high-risk disease are treated with high-dose chemotherapy, surgery, autologous stem cell transplant, radiation, and immunotherapy (2). A core high-risk criterion is MYCN oncogene amplification (2), which is present in approximately 20% of all neuroblastoma tumors. Among high-risk patients, half have MYCN-amplification. Multiple studies have demonstrated that the biology of MYCN-amplified and MYCN-nonamplified high-risk neuroblastoma is disparate (3,4). However, the genetic events predisposing patients to MYCN-amplified neuroblastoma remain unclear.
Genome-wide association studies (GWAS) comparing neuroblastoma patients to healthy controls have identified several susceptibility loci associated with clinical risk group (5–12). Many of these susceptibility loci modify oncogenic drivers or have tumor suppressor activity (13–15). Here, we performed a meta-analysis of three GWAS to test the hypothesis that in addition to phenotypic risk group, specific germline susceptibility loci are associated with the development of somatic MYCN-amplification. We then performed a second meta-analysis of MYCN-nonamplified high-risk cases to identify associations differentially associated with MYCN-amplified or MYCN-nonamplified high-risk neuroblastoma.
Patients were divided into three sets based on genotype array platform, public availability of data, and use in published studies. Set 1 comprised the 1662 patients from dbGaP project phs000124.v2.p1 (8). Set 2 comprised data from 2242 patients for whom data is not yet publically available, but who were analyzed previously (9). Set 3 was the only set genotyped on the Illumina OmniExpress 770 array and comprised the remaining 1245 patients neither previously published nor publically available. Only patients of self-reported European ancestry were included. All patients had informed consent for the Children’s Oncology Group ANBL00B1 (NCT00904241) after institutional review board approval.
Prior to association testing, we performed genome-wide quality control (QC), principal component analysis (PCA) to confirm ancestry, and imputation using single nucleotide polymorphisms (SNPs) common for cases and controls in each set as previously described (16). We removed nine samples and 86 913 SNPs from Set 1, 63 samples and 60 979 SNPs from Set 2, and 45 samples and 94 851 SNPs from Set 3. Following imputation, we performed GWAS for each individual set separately for MYCN-amplified and MYCN-nonamplified high-risk cases compared with non-high-risk controls in SNPTEST (17) using a frequentist additive score test. The analysis workflow is detailed in Supplementary Figure 1 (available online).
Two meta-analyses were performed on these GWAS results. The first meta-analysis combined association evidence from the three MYCN-amplified high-risk case sets (n = 615) and 1869 non-high-risk neuroblastoma controls. The second meta-analysis combined association evidence from the three MYCN-nonamplified high-risk GWAS (n = 728) and the same 1869 non-high-risk controls. The meta-analyses used a fixed-effects model with inverse variance weighting implemented in METAL (18). The METAL output included tests of heterogeneity, the Cochran’s test (Phet), and I2 of effect sizes within each meta-analysis for each SNP (19). SNPs were excluded based on absence in any of the three input data sets, not being an SNP, or I2 being greater than 75%.
The replication set comprised 127 MYCN-amplified and 140 MYCN-nonamplified high-risk cases, and 254 non-high-risk neuroblastoma patients as controls, all of European ancestry, genotyped on the OmniExpress platform. Genotyped SNPs were QC-filtered using PLINK (20) for minor allele frequency greater than 0.01, an Hardy–Weinberg equilibrium P value of less than 1 × 10-4, a missingness rate of less than 0.1, and missing SNP rate of 0.01. Ancestry was inferred by PCA incorporating the 1000 Genomes Project population v. 3 reference panel (21) and stratified using multidimensional scaling analysis in PLINK. Prephasing and imputation were performed as described above. Imputed SNPs with an info score of less than 0.7 were excluded. Association testing was performed in SNPTEST using a frequentist additive score test, with a P value of less than .05 considered statistically significant. All statistical tests were two-sided.
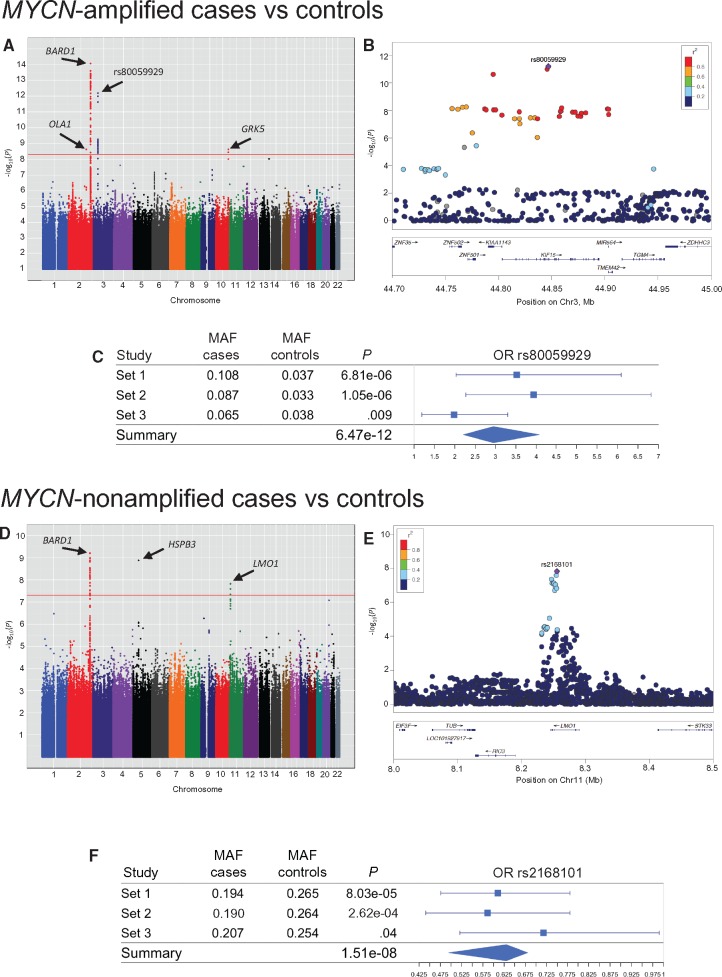
We identified a novel association that surpassed genome-wide statistical significance between MYCN-amplified neuroblastoma and a locus at 3p21.31, indexed by the SNP rs80059929 (OR = 2.95, 95% CI = 2.17 to 4.02, Pmeta = 6.47 × 10-12) (Figure 1A) located in intron 16 within the KIF15 gene locus with a minor allele frequency of 8.6% in cases compared with 3.6% in controls. This discovery was confirmed in an independent replication set (rs80059929 OR = 2.30, 95% CI = 1.12 to 4.69, Preplication = .02). The locus containing this SNP spans nearly 150 kb (defined by SNPs with Pmeta < 1×10-6 and r2 ≥ 0.6 with rs80059929) (Figure 1B;Supplementary Table 1, available online). All three discovery studies and the replication study had the same risk allele for rs80059929 with a P value of less than .05 (Figure 1C). SNPs reaching genome-wide statistical significance were also found in or near BARD1, OLA1, and GRK5 (Table 1). BARD1 was previously identified as a high-risk neuroblastoma susceptibility locus using high-risk cases from Set 1 (6). The associations in OLA1 and GRK5 loci were not statistically significant in the replication set, although the association in OLA1 was suggestive (Preplication = .11).
Figure 1.
Specific genomic loci associated with the development of MYCN-amplified high-risk neuroblastoma. A) Manhattan plot showing single nucleotide polymorphisms (SNPs) statistically significantly associated with MYCN-amplified neuroblastoma. B) LocusZoom (22) plot of novel loci identified at 3p21.31 shows that the most strongly associated SNP is located in intron 16 within the KIF15 gene locus. C) Forest plot shows the P value and odds ratio with 95% confidence interval for rs80059929 from each set and meta-analysis. Error bars indicate 95% confidence intervals. All statistical tests were two-sided. D) Manhattan plot showing SNPs statistically significantly associated with MYCN-nonamplified high-risk neuroblastoma. E) LocusZoom of previously identified loci on chromosome 11 shows the SNPs in LMO1 that were not associated with the development of MYCN-amplified disease. F) Forest plot shows the P value and odds ratio with 95% confidence interval for rs2168101 from each set and meta-analysis. Error bars indicate 95% confidence intervals. All statistical tests were two-sided. MAF = minor allele frequency; OR = odds ratio.
Table 1.
Index SNPs in loci statistically significantly associated with the development of MYCN-amplified or MYCN-nonamplified high-risk neuroblastoma
| Gene | SNP | CHR | bp | Minor allele frequencies |
MYCN cases, non-HR controls |
MYCN-nonamp HR cases, non-HR controls |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Controls, % | MYCN cases, % | MYCN-nonamp HR cases, % | OR (95% CI) | SE | Pmeta* | I2 | OR (95% CI) | SE | Pmeta* | I2 | ||||||
| BARD1 (5') | rs777318† | 2 | 215736845 | 31.0 | 42.7 | 38.6 | 1.67 (1.46 to 1.91) | 0.11 | 8.51E-14 | 32.2 | 1.41 (1.24 to 1.60) | 0.09 | 1.69E-07 | 64.8 | ||
| KIF15 | rs80059929† | 3 | 44846722 | 3.6 | 8.6 | 4.6 | 2.95 (2.17 to 4.02) | 0.47 | 6.47E-12 | 47.0 | 1.24 (0.90 to 1.71) | 0.2 | .19 | 31.9 | ||
| OLA1 | rs189431406† | 2 | 175113136 | 1.1 | 3.1 | 1.5 | 5.66 (3.08 to 10.4) | 1.76 | 2.36E-08 | 21.2 | 1.41 (0.75 to 2.65) | 0.45 | .29 | 0 | ||
| GRK5 | rs2168222† | 10 | 121055101 | 1.7 | 4.0 | 2.4 | 4.38 (2.61 to 7.36) | 1.16 | 2.37E-08 | 0 | 1.77 (1.03 to 3.04) | 0.49 | .04 | 0 | ||
| BARD1 (5') | rs36086595‡ | 2 | 215680293 | 27.1 | 38.2 | 35.9 | 1.68 (1.46 to 1.93) | 0.12 | 3.67E-13 | 55.0 | 1.51 (1.33 to 1.73) | 0.1 | 6.18E-10 | 7.0 | ||
| HSPB3 (5') | rs78132525‡ | 5 | 53717252 | 3.1 | 3.7 | 6.5 | 1.27 (0.84 to 1.93) | 0.27 | .25 | 30.6 | 2.98 (2.09 to 4.24) | 0.54 | 1.31E-09 | 0 | ||
| LMO1 | rs2168101‡ | 11 | 8255408 | 26.2 | 26.3 | 19.6 | 1.01 (0.85 to 1.19) | 0.09 | .95 | 0 | 0.63 (0.53 to 0.75) | 0.13 | 1.51E-08 | 0 | ||
Fixed-effects model with inverse variance weighting of P values obtained from frequentist additive score test were used to calculate Pmeta. All statistical tests were two-sided. bp = base pair; CHR = chromosome; CI = confidence interval; HR = high-risk; OR = odds ratio; SE = standard error; SNP = single nucleotide polymorphism.
Index SNPs in loci statistically significantly associated with the development of MYCN-amplified high-risk neuroblastoma.
Index SNPs in loci statistically significantly associated with the development of MYCN-nonamplified high-risk neuroblastoma.
A second meta-analysis evaluated the association of 3p21.31 and MYCN-nonamplified high-risk disease. Here, we found no association in the MYCN-nonamplified high-risk cases compared with non-high-risk controls (rs80059929 OR = 1.24, 95% CI = 0.90 to 1.71, Pmeta = .19) (Figure 1D). There was statistically significant heterogeneity between the meta-analyses (Phet = 1.4 × 10-4, I2 = 93.1%). In contrast, we found that the previously identified high-risk neuroblastoma susceptibility locus in LMO1 was statistically significantly associated with MYCN-nonamplified high-risk neuroblastoma (rs2168101 OR = 0.63, 95% CI = 0.53 to 0.75, Pmeta = 1.51 × 10-8), but not with MYCN-amplified disease (rs2168101 OR = 1.01, 95% CI = 0.85 to 1.19, Pmeta = .95; Phet = 7.0 × 10-5, I2 = 93.7%) (Figure 1, D–F, and Table 1; Supplementary Table 2, available online). This association was replicated in 140 MYCN-nonamplified cases and 254 non-high-risk controls (rs2168101 OR = 0.61, 95% CI = 0.42 to 0.88, Preplication = 2.9 × 10-3). SNPs reaching genome-wide statistical significance were again found in or near BARD1.
Thus, we found that MYCN-amplified high-risk neuroblastoma and MYCN-nonamplified high-risk neuroblastoma have both shared and unique germline genetic architecture. We identified a novel susceptibility locus at 3p21.31 uniquely associated with MYCN-amplified disease. Additionally, neuroblastoma-associated variants previously described in LMO1 were statistically significant only in patients with MYCN-nonamplified high-risk disease.
The causal DNA variant at the 3p21.31 locus was not identified and expression quantitative loci (eQTLs) were not found, which are limitations of our study. Thus, the mechanisms leading to somatic MYCN-amplification remain unclear. Fine-mapping and additional functional studies will be needed to determine how germline DNA variants at 3p21.3 increase predisposition to MYCN-amplified neuroblastoma tumors. Ultimately, these studies may lead to the discovery of new genomic biomarkers and provide insight to the development of novel treatment strategies for this aggressive pediatric disease.
Funding
This work was supported in part by the Neuroblastoma Children’s Cancer Society (SLC); the Children’s Neuroblastoma Cancer Foundation (SLC); the Matthew Bittker Foundation (SLC); the Elise Anderson Neuroblastoma Research Fund (SLC); the Cancer Research Foundation (MAA); and the Conquer Cancer Foundation of the American Society of Clinical Oncology (MAA). Also, it was supported by the National Institutes of Health (R01CA124709, R00CA151869 to SJD, and K12CA139160 and T32GM007019 to MAA).
Notes
The opinions, results, and conclusions reported in this paper are those of the authors and are independent from the funding sources. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health. The funders had no role the design of the study; the collection, analysis, or interpretation of the data; the writing of the manuscript; or the decision to submit the manuscript for publication.
We thank the University of Chicago Computational Research Institute for use of the Tarbell High-Performance Computing cluster and the University of Chicago Center for Data Intensive Science and the Open Commons Consortium for use of the Bionimbus Protected Data Cloud.
Supplementary Material
References
- 1. Pinto NR, Applebaum MA, Volchenboum SL, et al. Advances in risk classification and treatment strategies for neuroblastoma. J Clin Oncol. 2015;3327:3008–3017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cohn SL, Pearson AD, London WB, et al. The International Neuroblastoma Risk Group (INRG) classification system: An INRG Task Force report. J Clin Oncol. 2009;272:289–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Shohet JM, Ghosh R, Coarfa C, et al. A genome-wide search for promoters that respond to increased MYCN reveals both new oncogenic and tumor suppressor microRNAs associated with aggressive neuroblastoma. Cancer Res. 2011;7111:3841–3851. [DOI] [PubMed] [Google Scholar]
- 4. Valentijn LJ, Koster J, Haneveld F, et al. Functional MYCN signature predicts outcome of neuroblastoma irrespective of MYCN amplification. Proc Natl Acad Sci U S A. 2012;10947:19190–19195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nguyen le B, Diskin SJ, Capasso M, et al. Phenotype restricted genome-wide association study using a gene-centric approach identifies three low-risk neuroblastoma susceptibility Loci. PLoS Genet. 2011;73:e1002026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Capasso M, Devoto M, Hou C, et al. Common variations in BARD1 influence susceptibility to high-risk neuroblastoma. Nat Genet. 2009;416:718–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Wang K, Diskin SJ, Zhang H, et al. Integrative genomics identifies LMO1 as a neuroblastoma oncogene. Nature. 2011;4697329:216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Maris JM, Mosse YP, Bradfield JP, et al. Chromosome 6p22 locus associated with clinically aggressive neuroblastoma. N Engl J Med. 2008;35824:2585–2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Diskin SJ, Capasso M, Schnepp RW, et al. Common variation at 6q16 within HACE1 and LIN28B influences susceptibility to neuroblastoma. Nat Genet. 2012;4410:1126–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Diskin SJ, Capasso M, Diamond M, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;1064:dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Diskin SJ, Hou C, Glessner JT, et al. Copy number variation at 1q21.1 associated with neuroblastoma. Nature. 2009;4597249:987–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gamazon ER, Pinto N, Konkashbaev A, et al. Trans-population analysis of genetic mechanisms of ethnic disparities in neuroblastoma survival. J Natl Cancer Inst. 2013;1054:302–309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oldridge DA, Wood AC, Weichert-Leahey N, et al. Genetic predisposition to neuroblastoma mediated by a LMO1 super-enhancer polymorphism. Nature. 2015;5287582:418–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Russell MR, Penikis A, Oldridge DA, et al. CASC15-S is a tumor suppressor lncRNA at the 6p22 neuroblastoma susceptibility locus. Cancer Res. 2015;7515:3155–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bosse KR, Diskin SJ, Cole KA, et al. Common variation at BARD1 results in the expression of an oncogenic isoform that influences neuroblastoma susceptibility and oncogenicity. Cancer Res. 2012;728:2068–2078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hungate EA, Vora SR, Gamazon ER, et al. A variant at 9p21.3 functionally implicates CDKN2B in paediatric B-cell precursor acute lymphoblastic leukaemia aetiology. Nat Commun. 2016;7:10635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Marchini J, Howie B, Myers S, et al. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;397:906–913. [DOI] [PubMed] [Google Scholar]
- 18. Willer CJ, Li Y, Abecasis GR.. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;2617:2190–2191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Higgins JP, Thompson SG, Deeks JJ, et al. Measuring inconsistency in meta-analyses. BMJ. 2003;3277414:557–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Purcell S, Neale B, Todd-Brown K, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;813:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Genomes Project C, Auton A, Brooks LD, et al. A global reference for human genetic variation. Nature. 2015;5267571:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: Regional visualization of genome-wide association scan results. Bioinformatics. 2010;2618:2336–2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.