McNally and Roll-Mecak review the molecular mechanism of microtubule-severing enzymes and their diverse roles in processes ranging from cell division to ciliogensis and morphogenesis.
Abstract
Microtubule-severing enzymes generate internal breaks in microtubules. They are conserved in eukaryotes from ciliates to mammals, and their function is important in diverse cellular processes ranging from cilia biogenesis to cell division, phototropism, and neurogenesis. Their mutation leads to neurodegenerative and neurodevelopmental disorders in humans. All three known microtubule-severing enzymes, katanin, spastin, and fidgetin, are members of the meiotic subfamily of AAA ATPases that also includes VPS4, which disassembles ESCRTIII polymers. Despite their conservation and importance to cell physiology, the cellular and molecular mechanisms of action of microtubule-severing enzymes are not well understood. Here we review a subset of cellular processes that require microtubule-severing enzymes as well as recent advances in understanding their structure, biophysical mechanism, and regulation.
Introduction
Microtubules are central to the life of all eukaryotic cells. They form a dynamic scaffold for the cell, provide tracks for intracellular trafficking, and mediate chromosome segregation. These essential functions are dependent on their dynamic properties and organization. Therefore, cells dedicate a large number of factors to regulate the dynamics and morphology of microtubule arrays. Microtubule networks are sculpted by the action of polymerases, depolymerases, and severing enzymes (Roll-Mecak and McNally, 2010; Akhmanova and Steinmetz, 2015). While polymerases and depolymerases act at microtubule ends to catalyze the addition or removal of tubulin subunits, severing enzymes break microtubules along their lengths. If the newly generated microtubules are stable and regrow, then severing enzymes can act as microtubule amplifiers; if the newly severed microtubules are unstable, severing enzymes can act as depolymerases (Ribbeck and Mitchison, 2006; Roll-Mecak and Vale, 2006). Therefore, severing enzymes have the potential to be both positive and negative regulators of microtubule mass, and thus are implicated in regulating microtubule dynamics in a wide range of basic cellular processes ranging from ciliogenesis to cell division, neurogenesis, and phototropism.
Microtubule severing was first reported in Xenopus laevis egg extracts (Vale, 1991). The first microtubule-severing enzyme was purified from sea urchin eggs (McNally and Vale, 1993) and named katanin after a Japanese samurai sword. Katanin belongs to the large family of ATPases associated with diverse cellular activities (AAA ATPases). Members of this family are represented in all major cellular pathways and have one common property: they use the energy of ATP hydrolysis to disassemble or remodel protein complexes, nucleic acid complexes, and aggregated or misfolded proteins (Olivares et al., 2015). Katanin is composed of an AAA ATPase catalytic subunit (p60) and a regulatory subunit (p80; Hartman et al., 1998; Fig. 1) that form a stable complex in vivo (McNally and Vale, 1993; Wang et al., 2017). Katanin p60 and p80 mutants have the same phenotype in several species, indicating the essential nature of the complex (Mains et al., 1990; Sharma et al., 2007; Wang et al., 2017). To date, katanin (McNally and Vale, 1993), spastin (Evans et al., 2005; Roll-Mecak and Vale, 2005), and fidgetin (Mukherjee et al., 2012) have been shown to have microtubule-severing activity in vitro (reviewed in Roll-Mecak and McNally, 2010), while katanin-like 1 (KATNL1) has been shown to destabilize microtubules in cells when overexpressed (Sonbuchner et al., 2010). All three enzymes have a similar domain architecture that consists of an AAA ATPase C-terminal core and an N-terminal microtubule interacting and trafficking domain (MIT; Fig. 1). Katanin-like 2 (KATNL2), the one katanin homologue that is predicted to contain a lis homology domain at the N terminus, does not sever microtubules when overexpressed in cells (Cheung et al., 2016) and has not been shown to possess microtubule-severing activity in vitro. Together with vacuolar protein 4 (VPS4), the severing enzymes form the meiotic clade of AAA ATPases. All members of this AAA subfamily disassemble large noncovalent protein polymers, the microtubule for severing enzymes or ESCRTIII polymers in the case of VPS4 (Monroe and Hill, 2016). The phylogenetic relationship between microtubule-severing enzymes and their close relatives is summarized in Fig. 1.
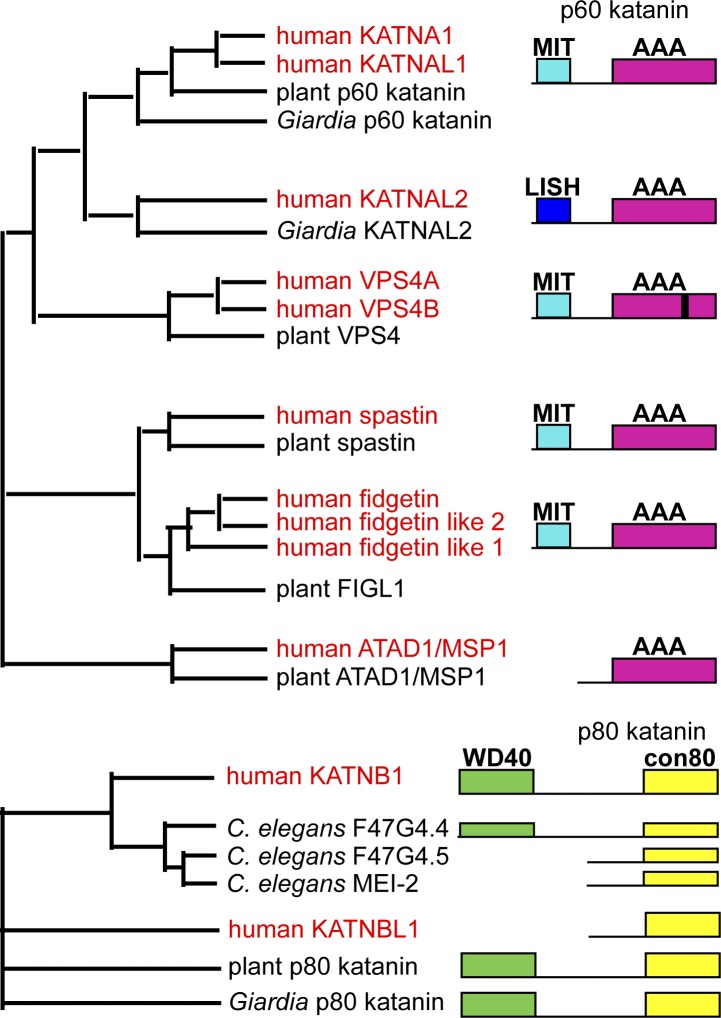
Figure 1.
Phylogeny of microtubule-severing enzymes. Top: Evolutionary relationship among microtubule-severing AAA ATPases, katanin p60, spastin, and fidgetin, as well as the closely related VPS4 and MSP1 proteins that act on nonmicrotubule substrates. Homologues from animals, plants, and basal eukaryotes were identified with BLASTp and aligned with M-COFFEE (Moretti et al., 2007). Phylogenetic trees were computed with RAxML (Stamatakis, 2006). The black line in the VPS4 AAA domain indicates a β-domain insertion. VPS4 has clear homologues in archaea while microtubule-severing enzymes do not. Bottom: Evolutionary relationship among p80 katanin homologues. Most clades of eukaryotes have a katanin p80 with an N-terminal WD40 domain and a C-terminal con80 domain (conserved p80 domain). WD40-less versions appear to have independently evolved in nematodes and chordates. All human homologues are shown in red.
An extensive body of research over the last 20 years has elucidated how microtubule plus end dynamics is regulated by a large array of molecular players (Akhmanova and Steinmetz, 2010). While the dynamic behavior of microtubule plus ends is easily imaged at the cell periphery, severing events that happen deeper in the cell, in denser microtubule networks, are not easily visible by light microscopy. As a result, severing has been visualized directly only in a few systems that have larger spacing between microtubules, most notably in plant cells (Nakamura et al., 2010; Lindeboom et al., 2013; Zhang et al., 2013). Thus, despite the importance of microtubule-severing enzymes to normal physiology, their cellular mechanism has proven a challenge to dissect. We start by providing examples of in vivo systems where the connection between microtubule severing and cellular phenotype is best understood, highlight cellular processes where microtubule-severing enzymes are important, and end with recent in vitro reconstitution and structural studies that shed light on the mechanism of microtubule severing.
Cellular functions of microtubule-severing enzymes
The most insight into the multifaceted functions of microtubule severing in cells has been obtained from the cortex of plant cells, where enzyme mediated microtubule severing can be clearly observed in vivo by time-lapse imaging (Lindeboom et al., 2013; Zhang et al., 2013; Wang et al., 2017). Severing events have also been observed in the ultra-thin lamellipodium of cultured animal cells (Zhang et al., 2011) and linked to katanin function; however, in this case, it was not possible to correlate the severing event with the presence of an enzyme at the severing site as in plants. Loss of microtubule-severing enzymes also generates strong phenotypes in spindles, neurons, and cilia. However, the high density of microtubules in these systems has so far precluded direct observation of severing events.
Control of plant cell shape: microtubule array reorientation and amplification
The clearest direct observations of microtubule severing in vivo were reported in the cortex of plant cells (Wightman and Turner, 2007; Nakamura et al., 2010; Lindeboom et al., 2013; Zhang et al., 2013). These observations illustrate the two opposing paradigms for microtubule severing in vivo: to either prune or amplify microtubule arrays. Specific types of plant cells have highly parallel arrays of cortical microtubules tightly associated with the plasma membrane along their lengths (Barton et al., 2008). New microtubules are often nucleated from the wall of preexisting microtubules at discrete 40° angles by a complex of γ-TuRC (Murata et al., 2005; Nakamura et al., 2010) and augmin (Liu et al., 2014). The resulting oblique microtubules that deviate from the parallel arrangement of microtubules in the array frequently encounter microtubules in the parallel array at a steep angle (>45°; Fig. 2 A). A minority of these steep-angled collisions result in a catastrophe (Chi and Ambrose, 2016), whereas a majority result in a crossover (Wightman and Turner, 2007), where the new oblique microtubule polymerizes over the preexisting microtubule (Barton et al., 2008). Most of the time, the new microtubule is severed at the crossover (Wightman and Turner, 2007), and 85% of severing events generate a new plus end that depolymerizes (Zhang et al., 2013). In contrast, the newly generated minus ends are stable (Fig. 2 A). As a result, the free microtubule fragment polymerizes along a curved arc (Wightman and Turner, 2007), “moves” by treadmilling (Shaw et al., 2003), and, when it encounters another microtubule at a shallow angle, is “zippered” into a bundle with the preexisting microtubule.
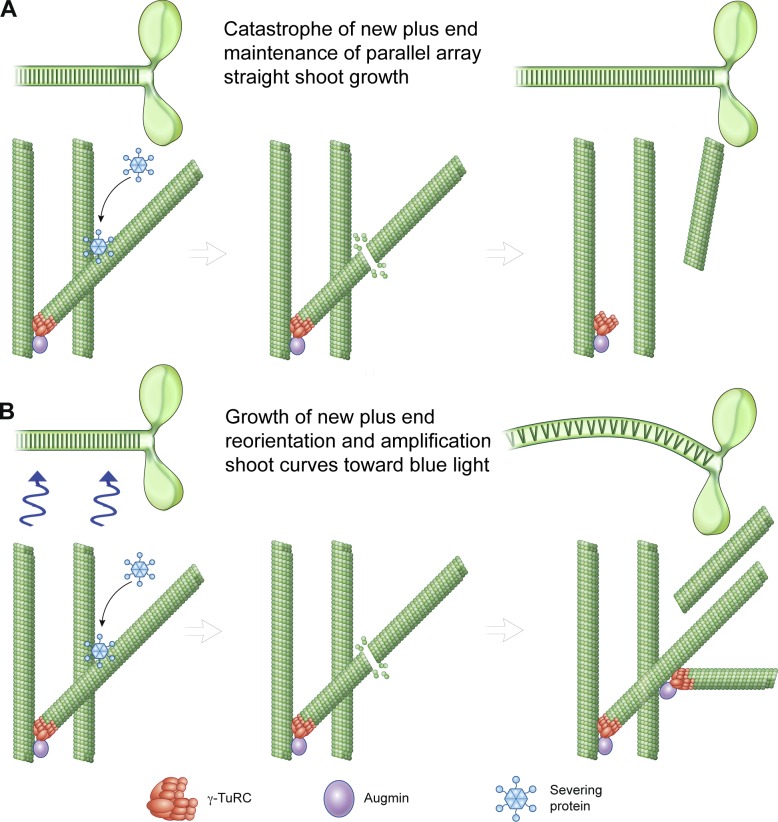
Figure 2.
Regulation of microtubule array geometry by severing enzymes. (A) Maintenance of a parallel array of cortical microtubules in Arabidopsis by katanin-dependent severing at microtubule crossovers, followed by catastrophe of the new plus end and treadmilling (not shown) and reorientation of the free microtubule. (B) During reorientation of Arabidopsis shoots in response to light, severing at crossovers followed by continued polymerization of new plus ends, and further branched nucleation, result in reorientation of the cortical microtubule array.
Katanin localizes at crossover sites, and severing events at crossovers are not observed in a p60 katanin mutant (Lindeboom et al., 2013; Zhang et al., 2013; Wang et al., 2017). The molecular mechanism for targeting katanin to crossovers is not well understood, but it does require the p80 regulatory subunit, which is sufficient for this localization in vivo (Wang et al., 2017). Moreover, purified Caenorhabditis elegans katanin p60:p80 shows a preference for microtubule crossovers in vitro (McNally et al., 2014), suggesting that the ability to recognize this microtubule geometry lies solely within the katanin molecule. Thus, katanin-dependent severing results in depolymerization of one microtubule that is not parallel to the preexisting microtubule array and generates a second microtubule that can be more readily aligned with the parallel array. The resulting parallel microtubule bundles help orient cellulose fibers that control cell shape and growth polarity. Consistent with this, katanin mutants have orthogonally overlapping cortical microtubules (Burk et al., 2001), disordered cellulose fibers (Burk and Ye, 2002), and abnormal cell shape (Bichet et al., 2001; Burk et al., 2001).
Katanin-dependent severing of microtubules at crossovers was also directly observed in Arabidopsis thaliana hypocotyl cells (cells in the stem of the germinating seedling) during phototropism, a process that involves the reorientation of microtubules and cellulose fibers to facilitate the movement of the plant toward light. However, in this context, severing is used as a microtubule amplifier (Lindeboom et al., 2013). Elegant time-lapse imaging showed that the new plus ends generated by katanin severing at crossovers between an oblique microtubule and one from the existing parallel microtubule array are stable and regrow at high frequency (Fig. 2 B). Since the new microtubule generated through severing at crossovers continues to grow at a shallow angle with respect to its parent oblique microtubule, it will continue to cross over parallel microtubules from the old array and thus get severed, leading to a rapid amplification in microtubule numbers and effectively creating a new microtubule array at 90° to the original one (Fig. 2 B). This reorientation of the microtubule array is essential for blue light–stimulated shoot growth (Lindeboom et al., 2013). Thus, severing at crossovers can stabilize a preexisting parallel microtubule array if the newly generated plus ends catastrophe, but promotes the generation of a novel, perpendicular array if the new plus ends are stable and regrow.
Microtubules grow through the addition of GTP-tubulin dimers at their ends, which hydrolyze the nucleotide once they incorporate into the lattice. The delay between the rate of GTP hydrolysis and tubulin dimer addition to the ends leads to the formation of a “GTP cap,” a layer of GTP-tubulin at the end of the microtubule that protects the microtubule from depolymerization (Carlier and Pantaloni, 1981, 1982; Mitchison and Kirschner, 1984; Carlier et al., 1987). Thus, in vitro and in cells, new plus ends generated through laser or mechanical ablation quickly depolymerize (Nicklas et al., 1989; Walker et al., 1989; Tran et al., 1997; Tirnauer et al., 2004) as a result of the exposure of the GDP-tubulin lattice, which is unstable in the absence of a stabilizing GTP cap at the new end. This raises the interesting question of how the plus ends generated by katanin are stabilized, because severing would also expose the unstable GDP-microtubule lattice. This fundamental question was recently answered through in vitro reconstitution studies that revealed that katanin as well as spastin extract GDP-tubulin dimers out of the microtubule in an ATP hydrolysis–dependent manner, creating nanodamage sites that are then repaired by the spontaneous incorporation of GTP-tubulin (Vemu et al., 2018). As a result, the newly severed microtubule ends emerge with a high density of GTP-tubulin, which stabilizes the polymer against spontaneous depolymerization, leading to amplification of microtubule mass and number (Vemu et al., 2018). The mechanism for microtubule amplification through severing and elongation has analogies to that in the actin system, where severing by cofilin followed by growth of the severed fragment at the fast-growing barbed end can be used to generate new filaments at the leading edge of motile cells (DesMarais et al., 2005; Ribbeck and Mitchison, 2006).
Cell migration: Severing as a potential leading edge catastrophe mechanism
Animal cell migration is driven by localized actin polymerization, which pushes a thin leading edge of the cell forward, but microtubules are important in regulating the direction of these protrusions and thus the direction of cell migration. Time-lapse imaging of individual microtubules in Drosophila melanogaster S2 cells showed a higher catastrophe frequency within 3 µm of the leading edge, with 30% of the depolymerizing events preceded by a microtubule breakage event close to the plus end and in the vicinity of the cortex (Zhang et al., 2011). These catastrophes generate microtubules that are nearly perpendicular to the leading edge. Katanin localizes at the cortex in these cells. Unlike in plants, it was not possible to directly visualize the enzyme at severing sites. The severing events observed in S2 cells are reminiscent of the ones initially observed in newt lung cells, where they coincided with the pronounced local buckling of the microtubule (Waterman-Storer and Salmon, 1997). Katanin depletion by RNAi in S2 cells results in microtubules that continue to polymerize and thus bend along the cortex (Zhang et al., 2011). The katanin-depleted cells exhibit increased rates of actin-driven protrusions. Although S2 cells do not exhibit directed cell migration, this result suggests that localized cortical katanin could contribute to directed migration by locally generating microtubules perpendicular to the leading edge, thereby locally inhibiting actin-driven protrusions. One of the two human katanin catalytic subunits (Zhang et al., 2011) and fidgetin-like 2 (Charafeddine et al., 2015) concentrate at the leading edge of migratory cells, and their depletion results in increased cell migration and wound healing. Depletion of fidgetin in cultured neurons also increases axon elongation (Leo et al., 2015). Thus, the role of leading edge microtubule severing might be of broader significance; however, severing has not been directly observed in these latter cases, and fidgetin depletion has an opposite effect in mammalian astrocytes, where it inhibits migration (Hu et al., 2017).
Debranching, release of minus ends from centrosomes, and poleward flux
New microtubules are often nucleated from γ-TuRC at centrosomes in animal cells or γ-TuRC associated with the wall of a preexisting microtubule by the augmin complex in both plant and animal cells. Release of microtubule minus ends from their initial nucleation site has been directly observed or inferred in several different cell types. “Poleward flux” or movement of tubulin subunits toward mitotic spindle poles is an example of inferred release of minus ends to allow net depolymerization of minus ends that would otherwise be capped by γ-TuRC. Katanin-mediated release of microtubule minus ends from γ-TuRC–associated branched nucleation sites has been directly observed in plants (Nakamura et al., 2010), but the molecular mechanism is unclear. This release could be mediated by severing so close to the minus end that the γ-TuRC–proximal microtubule fragment is not observed, or at the junction between γ-tubulin and α-tubulin at the microtubule minus end. Differentiating between these two possible mechanisms will require reconstitution with purified components and ultrastructural analyses using superresolution fluorescence and EM. Release of microtubule minus ends from γ-TuRC–associated nucleation sites in centrosomes has been directly observed by time-lapse imaging in animal cells (Keating et al., 1997), and katanin has been implicated in this release in interphase cells (Dong et al., 2017) and neurons (Ahmad et al., 1999) by indirect assays. Katanin binds to the microtubule minus end binding proteins CAMSAP/patronin (Jiang et al., 2014) and ASPM (Jiang et al., 2017) and is most dramatically concentrated near microtubule minus ends at spindle poles in vertebrate cells (McNally and Thomas, 1998). p80 katanin knockout human cells have markedly reduced rates of poleward flux in their mitotic spindles (Jiang et al., 2017) that could be driven by severing near minus ends, depolymerization of minus ends, or uncapping from γ-TuRCs. Strikingly, RNAi-mediated depletion of spastin or fidgetin, but not katanin, slows poleward flux in the spindles of cultured Drosophila cells (Zhang et al., 2007), suggesting that different severing proteins can exchange roles over evolutionary time. Because kinesin-13s are also involved in poleward flux in both vertebrate (Ganem et al., 2005) and Drosophila cells (Rogers et al., 2004), and because individual microtubules cannot be imaged at mitotic spindle poles, the detailed mechanistic role of severing in poleward flux is not clear.
Both p80 katanin and ASPM knockout cells have a reduced mass of astral microtubules and are defective in spindle orientation (Jiang et al., 2017). Mutations in p80 katanin cause microcephaly in humans, mice, zebrafish, and Drosophila (Hu et al., 2014; Mishra-Gorur et al., 2014) with specific loss of neurons derived from asymmetric cell divisions documented in Drosophila (Mishra-Gorur et al., 2014) and mouse (Hu et al., 2014). These results suggest that severing at spindle poles is especially important for generating functional astral microtubule arrays that are used for spindle orientation during asymmetric divisions in the developing brain. The severing-dependent microtubule amplification observed in blue light–stimulated plant shoots (Lindeboom et al., 2013) and recently demonstrated in vitro (Vemu et al., 2018) suggests how severing might increase the number of astral microtubules in neuronal precursors.
C. elegans oocyte meiosis
In C. elegans, complete loss-of-function katanin mutants assemble acentriolar oocyte meiotic spindles that are composed of oblique overlapping microtubules (Mains et al., 1990) with no apparent clustering of microtubule minus ends (McNally and McNally, 2011), unlike wild-type spindles that are composed of highly bundled parallel/anti-parallel microtubule arrays with two discrete spindle poles. Electron tomography revealed that the microtubules in the meiotic spindles of a katanin-null mutant are much longer than in the wild type (Srayko et al., 2006). No coordinated separation of chromosomes into two distinct masses occurs in these spindles (Yang et al., 2003), and embryos produced from these defective meioses are haploid, pentaploid, or aneuploid and never advance to hatching, even if fertilized by wild-type sperm (Mains et al., 1990; Yang et al., 2003; Hara and Kimura, 2013). Three different p60 katanin point mutants with no detectable in vitro severing activity that are expressed in oocytes have this katanin null phenotype (McNally et al., 2014; Joly et al., 2016), indicating that severing is required to convert oblique overlapping microtubules into an array of parallel/anti-parallel bundled microtubules. An appealing idea is that severing at crossovers culls orthogonal microtubules, thus promoting parallel/anti-parallel bundling as observed in plants. Interestingly, meiotic spindles in katanin-null mutants have reduced microtubule mass and number (McNally et al., 2006; Srayko et al., 2006), indicating that severing functions as a microtubule amplifier in this system as it does during phototropism in Arabidopsis. Partial loss-of-function katanin mutants assemble unusually long meiotic spindles with a linear relationship between in vitro severing activity and in vivo meiotic spindle length (McNally et al., 2014). Katanin activity contributes to meiotic spindle scaling also in Xenopus, where the higher activity of Xenopus tropicalis katanin compared with Xenopus laevis is responsible for the difference in spindle size between the two species. This difference in activity is due to an inhibitory phosphorylation site on katanin p60 (Loughlin et al., 2011; Whitehead et al., 2013). The role of microtubule-severing proteins in oocyte meiotic spindles of other species is an area awaiting exploration.
Cytokinesis, abscission, and nuclear envelope resealing
Katanin is required for completion of cytokinesis in the ciliate Tetrahymena thermophila (Sharma et al., 2007) and the African trypanosome parasite Trypanosoma brucei (Benz et al., 2012), where it is thought to sever stable subcortical microtubule bundles that would otherwise prevent division. During abscission, the final stage of animal cytokinesis, a stable microtubule bundle called the midbody presents a barrier to division. Katanin localizes at the midbody, where it is required for abscission (Matsuo et al., 2013), as does the short isoform of spastin (Connell et al., 2009; Guizetti et al., 2011). Abscission requires membrane remodeling through the action of VPS4, an AAA ATPase closely related to spastin and katanin that remodels ESCRTIII filaments (Lata et al., 2008). Spastin, ESCRTIII, and VPS4 are linked by a complex protein interaction network that is thought to coordinate severing of the midbody with membrane remodeling (Yang et al., 2008; Allison et al., 2013). Finally, microtubule bundles on either side of the abscission site are in a position to interfere with nuclear envelope reassembly at the end of mitosis. A network of spastin, ESCRTIII, and VPS4 coordinates microtubule severing and membrane remodeling to allow the final resealing of the two daughter nuclear envelopes at the site where midzone microtubules are in the way (Vietri et al., 2015). Microtubules at the midbody are glutamylated (Yu et al., 2015), and spastin has been shown to be potently regulated by glutamylation (Lacroix et al., 2010; Valenstein and Roll-Mecak, 2016), suggesting a mechanism for selective severing of microtubule bundles at this structure.
Ciliogenesis
Cilia are complex sensory and motile organelles found in almost all cells. Ciliary assembly involves multiple steps: the centrioles form basal bodies, dock onto the cortex, and start growing the cilium. This growth requires continuous trafficking into the growing structure, and impairment of this process negatively affects cilia length. Katanin mutants in Tetrahymena (Sharma et al., 2007) and Chlamydomonas (Dymek et al., 2004; Dymek and Smith, 2012) have paralyzed flagella shorter than wild type and are missing the central pair microtubules of the axoneme. p80 katanin knockout mouse fibroblasts have supernumerary primary cilia and centrioles (Hu et al., 2014). Defective cilia beating and length heterogeneity has been reported in ependymal cells of the brain in mice with a mutant form of KATNAL1 (Banks et al., 2018). These results indicate that katanin has a deeply conserved role in ciliogenesis, but its mechanism of action in cilia remains to be elucidated.
Severing enzymes in neurons
Spastin mutations cause hereditary spastic paraplegia (HSP), a disorder characterized by the slow degeneration of axons (Hazan et al., 1999). Disease mutants have impaired microtubule-severing activity (Evans et al., 2005; Roll-Mecak and Vale, 2005). Spastin mutant mice exhibit defects in fast axonal transport and accumulate axonal swellings composed of stalled cargos (Kasher et al., 2009). Axonal swellings are also observed in human HSP patients (Kasher et al., 2009) and neurons differentiated from induced pluripotent stem cells derived from HSP patients (Denton et al., 2014). In the latter, axonal swellings were reduced after treatment with a microtubule-targeting drug (vinblastine), suggesting that spastin-mediated severing is necessary to maintain optimal microtubule dynamics for fast axonal transport. Administration of vinblastine also ameliorates the spasticity observed in spastin mutant flies (Trotta et al., 2004). Interestingly, spastin-null flies have fewer microtubules at the neuromuscular junction, especially in the more distant boutons (Sherwood et al., 2004), consistent with a role of severing as a microtubule number amplifier.
In Drosophila, a p60 katanin homologue is required for programmed dendrite pruning (Lee et al., 2009), and fidgetin is required for injury-induced dendrite pruning (Tao et al., 2016). Regulated microtubule disassembly is an intermediate in each of these pruning processes. Intriguingly, the same p60 katanin gene is required for elaboration of dendritic arbors, and the mutant dendrites have fewer growing microtubule plus ends (Stewart et al., 2012). This would suggest that in the same cell type, severing can promote net microtubule disassembly or amplify microtubule number and mass. Adding to this complexity, tissue-specific depletion of the canonical Drosophila p60 katanin results in increased elaboration of dendritic arbors (Mao et al., 2014). Severing is also implicated in regulation of synapse number, as supernumerary but defective synaptic boutons are observed in both spastin (Sherwood et al., 2004) and p60 katanin (Mao et al., 2014) mutant flies. Finally, spastin is required for efficient axon, but not dendrite regeneration (Stone et al., 2012; Rao et al., 2016). We propose that the wide range of neuronal functions for severing proteins is due to their ability to promote culling or amplification of microtubules depending on the dynamics of the newly severed ends. In vitro reconstitution experiments at physiological severing enzyme concentrations with dynamic microtubules (Vemu et al., 2018) and in the presence of microtubule-associated proteins (MAPs) abundant in neurons will be key to understanding how these various facets of microtubule severing can be regulated. Moreover, both spastin and katanin are regulated by glutamylation in cells (Sharma et al., 2007; Lacroix et al., 2010). Using microtubules with quantitatively defined levels of tubulin glutamylation, it was shown that spastin displays a graded response to the number of posttranslationally added glutamates on the tubulin tails (Valenstein and Roll-Mecak, 2016), indicating that the enzyme has different severing activities in the axon or dendrite, which contain microtubules with different degrees of glutamylation (Park and Roll-Mecak, 2018). Regulation of katanin by glutamylation has yet to be demonstrated in an in vitro reconstituted system. The diverse consequences of microtubule severing in vivo are likely due to the structure and dynamics of the microtubule ends generated by severing and the differential targeting of microtubule-severing enzymes to different microtubule geometries or to microtubules decorated with different MAPs and tubulin posttranslational modifications. We focus the remainder of this review on recent exciting developments on the mechanism of microtubule severing and the structure of severing enzymes.
Molecular mechanism of microtubule severing
Severing enzymes have the fascinating capacity to disassemble a polymer of considerable girth and stiffness. Microtubules are 25 nm in width and have a persistent length on the orders of hundreds of micrometers to millimeters (Hawkins et al., 2010). They are formed through the lateral association of 13 protofilaments (this number can vary depending on organism and cell type), linear polymers of head-to-tail tubulin heterodimers. Thus, in order to sever a microtubule, 13 longitudinal (between tubulin dimers within a protofilament) and lateral polymerization interfaces (between dimers in adjacent protofilaments) need to be disrupted. In contrast, actin-severing proteins like cofilin or gelsolin have to disrupt only two polymerization interfaces (Dominguez and Holmes, 2011). Microtubule-severing enzymes accomplish this remarkable task by using the energy of ATP hydrolysis. How severing enzymes are able to harness the energy from ATP hydrolysis to disrupt the microtubule lattice is still not well understood, but structural and in vitro reconstitution studies in the last year have brought us one step closer to unraveling their mechanism.
Spiral and rings: Insights into the power stroke for microtubule severing from katanin structures
Katanin p60, spastin, and fidgetin have a similar domain architecture that consists of an evolutionary conserved AAA catalytic core required for severing that is linked to an N-terminal MIT domain through a poorly conserved linker (Fig. 1; Roll-Mecak and McNally, 2010; Sharp and Ross, 2012). The spastin or katanin AAA domain and part of the linker are sufficient for severing activity (White et al., 2007; Roll-Mecak and Vale, 2008; Grode and Rogers, 2015). The minimal katanin AAA domain has no detectable severing activity (Grode and Rogers, 2015), while the AAA domain of spastin and a short part of the linker have dramatically impaired microtubule-severing activity (Roll-Mecak and Vale, 2008). Katanin and spastin assemble into hexamers via their AAA domains in an ATP-dependent manner (Hartman and Vale, 1999; Roll-Mecak and Vale, 2008; Zehr et al., 2017), and single-molecule fluorescence experiments have shown that microtubule-bound active katanin is a hexamer (Díaz-Valencia et al., 2011). Both enzymes exist as monomers at physiological concentrations even when bound to ATP (Hartman and Vale, 1999; Roll-Mecak and Vale, 2008; Nithianantham et al., 2018). Oligomerization is stimulated by the microtubule (Hartman and Vale, 1999; Eckert et al., 2012; Wen and Wang, 2013), presumably ensuring that the enzymes are activated only on the relevant substrate. Oligomerization is required for severing activity, and mutations at hexamerization interfaces inactivate both spastin and katanin (Roll-Mecak and Vale, 2008; Zehr et al., 2017). Several spastin HSP mutations are at hexamerization interfaces (Roll-Mecak and Vale, 2008).
Structural and functional studies of spastin catalyzed allowed the proposal of a mechanism for microtubule severing (White et al., 2007; Roll-Mecak and Vale, 2008). In the presence of ATP, spastin hexamerizes into a ring structure with a central, positively charged pore. Loops lining this pore engage the negatively charged intrinsically disordered C-terminal tails of tubulin that decorate the microtubule surface and are required for severing by both spastin and katanin (Roll-Mecak and Vale, 2005; Johjima et al., 2015; Valenstein and Roll-Mecak, 2016). Mutations of conserved residues in the pore loops inactivate the enzyme while still preserving ATPase (White et al., 2007; Roll-Mecak and Vale, 2008; Johjima et al., 2015). In light of the structural similarity of the spastin AAA ring to bacterial unfoldases such as those of the ClpX family (Olivares et al., 2016), it was proposed that severing enzymes break the microtubule by engaging the C-terminal tails of tubulin through this central pore and pulling on them with their conserved pore loops to dislodge the tubulin dimer out of the microtubule lattice. This repeated action would gradually unravel microtubule lattice interactions and generate internal breaks in the microtubule (Roll-Mecak and Vale, 2008). Direct demonstration for tubulin extraction out of the microtubule by these enzymes has only been recently obtained using negative stain electron microscopic analyses of severing reaction intermediates. These revealed that both spastin and katanin progressively remove tubulin subunits out of the microtubule lattice, generating nanodamage sites along the microtubule, and that these nanodamage sites progress to mesoscale severing events (Vemu et al., 2018).
While the common theme of AAA ATPases is that they form hexameric assemblies, structures in this state have been few and far between until the recent technical advances in cryo-EM. Severing enzymes were no exception as it was difficult to capture their physiological oligomeric assemblies in diffracting crystals. X-ray crystal structures of monomeric AAA domains for spastin (Roll-Mecak and Vale, 2008; Taylor et al., 2012), katanin (Zehr et al., 2017; Nithianantham et al., 2018), and fidgetin-1 (Peng et al., 2013) are known and show a high degree of structural similarity consistent with their primary sequence conservation (Fig. 3 A). They consist of a nucleotide-binding domain (NBD) and a helix-bundle domain (HBD) arranged as a crescent with the nucleotide-binding site at the hinge between these domains (Fig. 3 A). Likewise, an isolated structure of the MIT domain from katanin is known (Fig. 3 B; Iwaya et al., 2010). The MIT domain for both spastin and katanin binds to regulatory proteins, including the p80 subunit for katanin and an ESCRTIII component for spastin (Fig. 3, C and D; Yang et al., 2008; Rezabkova et al., 2017).
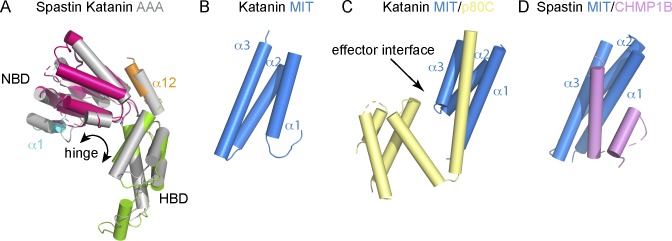
Figure 3.
Structural domains in microtubule-severing enzymes. (A) Superposition of the x-ray structure of the AAA domains of spastin (PDB ID 3B9P; Roll-Mecak and Vale, 2008) and katanin (PDB ID 5WC1; Zehr et al., 2017). Spastin AAA domain, gray. Katanin NBD, hot pink; HBD, light green; the N-terminal helix α1 is part of the fishhook and colored cyan; the C-terminal helix α12 specific to AAA ATPases of the meiotic clade is colored orange. (B) Structure of the katanin MIT domain (PDB ID 2RPA; Iwaya et al., 2010). (C) Structure of the complex between the katanin p60 MIT domain and the p80 conserved C-terminal domain (PDB ID 5NBT; Rezabkova et al., 2017); MIT domain, blue; p80 C-terminal domain, yellow. (D) Structure of the spastin MIT domain in complex with CHMP1B (PDB ID 3EAB; Yang et al., 2008). MIT domain, blue; CHMP1B, lavender. For reference, the domain organization in spastin and katanin can be seen in Fig. 1.
The first atomistic model for a microtubule-severing enzyme in its assembled hexameric state was reported recently using cryo-EM (Zehr et al., 2017). It revealed an asymmetric hexamer with the AAA modules bound to ATP and arranged in a right-handed spiral that creates an ∼40-Å-wide gate between the bottom and top protomers (Fig. 4). Comparison of the apo and nucleotide-bound structures from the crystal and cryo-EM structures revealed that flexing of the NBD and HDB domains around the nucleotide-binding site facilitates the ATP-driven assembly of the hexamer (Zehr et al., 2017; Nithianantham et al., 2018). Two conserved substrate binding loops (loops 1 and 2) line the central channel in a staircase arrangement optimized for substrate transfer among protomers (Fig. 5). The structure also revealed that the C-terminal part of the linker that connects the MIT and AAA domains is in fact not disordered, but forms a fishhook-shaped structure crowning the AAA ring close to the central channel (Fig. 5 A). On the opposite face, the structure revealed a belt of interconnected helices (α12 in Fig. 3 A) that link one protomer to the other and are critical for hexamerization (Fig. 5 B). These C-terminal helices are conserved in all members of the meiotic clade of AAA ATPases (Monroe and Hill, 2016). Notably, early screens for defects in C. elegans meiosis isolated katanin mutations (Mains et al., 1990) at the interface between these helices. In vitro assays demonstrated that mutations in these helices inactivate both katanin- and spastin- severing activity (Roll-Mecak and Vale, 2008; Zehr et al., 2017).
Figure 4.
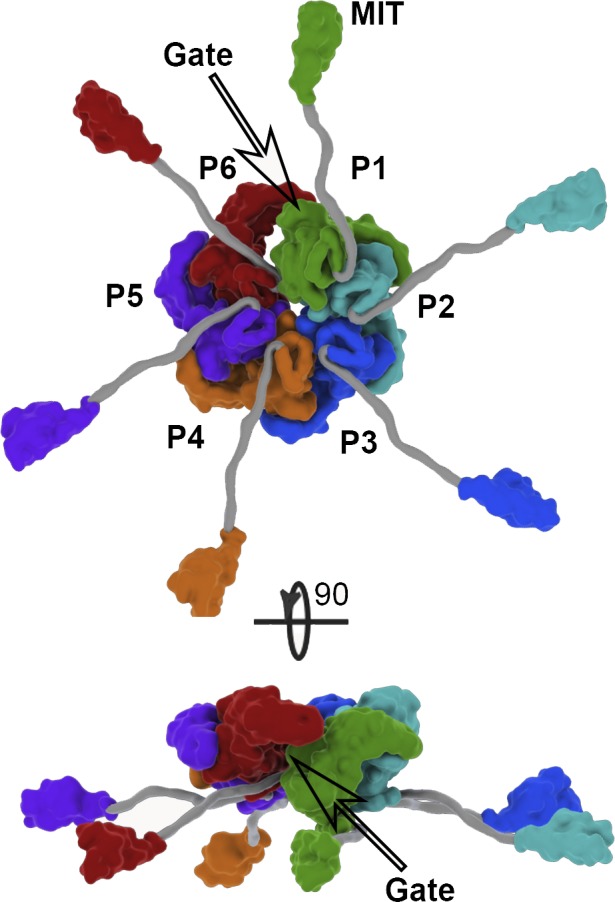
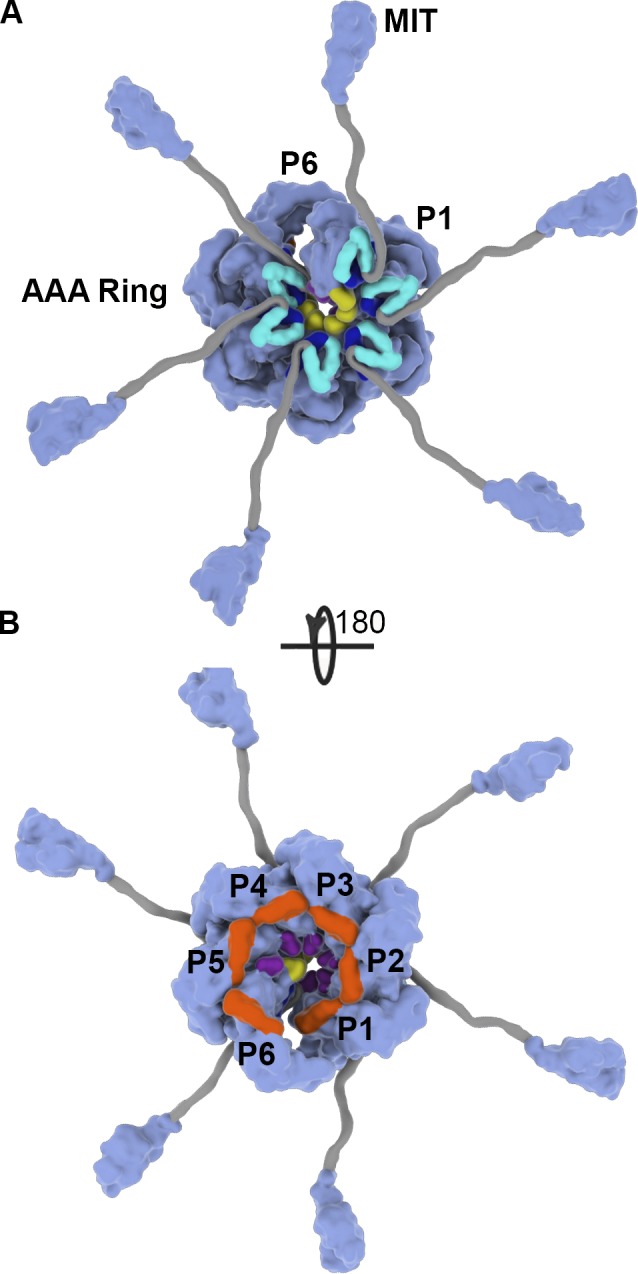
Katanin forms an asymmetric hexamer with a spiral arrangement of the AAA domains. Orthogonal views of the molecular surface of the assembled katanin hexamer in the spiral conformation showing the central AAA ring with radiating arms composed of a poorly conserved flexible linker and an N-terminal MIT domain. The AAA ring structure was determined by single-particle cryo-EM to 4.4 Å (PDB ID 5WC0; Zehr et al., 2017); the MIT domain structure was determined by NMR (PDB ID 2RPA; Iwaya et al., 2010); the structure of the flexible linker (gray) has not been experimentally determined and was modeled to have an approximate span as the one reported from small angle x-ray scattering (Zehr et al., 2017). Protomer P1, green; P2, cyan; P3, blue; P4, orange; P5, purple; and P6, red.
Figure 5.
Structural elements unique to katanin augment the AAA ring and participate in hexamerization. (A and B) Views of the molecular surface of the assembled katanin hexamer in the spiral conformation highlighting important functional elements. Fishhook, cyan; pore loop 1, yellow; pore loop 2, magenta; C-terminal helices, brick red.
The katanin cryo-EM structures also shed light on how hexamerization stimulates ATPase activity as the arginine fingers that are required to stabilize the ATP hydrolysis transition state are contributed in trans by the neighboring protomer. Their mutation, or mutation of the equivalent Arg residues in spastin, inactivates ATPase and severing activity (Roll-Mecak and Vale, 2008; Zehr et al., 2017). The asymmetric architecture of the hexamer excludes a concerted ATPase mechanism whereby all AAA engines would fire at the same time since the boundary P6 protomer does not have an Arg finger contributed in trans (Zehr et al., 2017). Classification of the katanin hexamers on cryo-EM grids revealed a second conformation that resembles a closed ring in which the ATP is missing from the boundary P1 protomer (Zehr et al., 2017). The loss of nucleotide leads to a rotation of the NBD in this protomer to close the 40-Å gate between the boundary protomers P1 and P6 by making contacts with the HBD in P6 (Fig. 6 A). This movement results in an ∼20-Å displacement of the substrate binding loops in protomer P1 (Fig. 6 B) and led to the proposal that the sequential ATP hydrolysis in protomers around the AAA ring results in the processive pulling of the tubulin substrate through the central pore with ∼20-Å steps (Fig. 6). An analogous mechanism was recently visualized for other AAA ATPases such as Hsp104 (Gates et al., 2017), VPS4 (Han et al., 2017), and Yme1 (Puchades et al., 2017). A sequential ATP hydrolysis cascade around the ring is an attractive way to ensure processive pulling of the tubulin tail through the pore. However, it is also possible that when working against the microtubule load, there is slippage, and the enzyme switches to a stochastic mechanism for ATP hydrolysis and translocation where protomers fire randomly around the ring, as has been shown for ClpB (Doyle et al., 2012) or Hsp104 with certain substrates (DeSantis et al., 2012). Last, it is also possible that the mechanical work is performed repeatedly by the boundary protomers while the remaining four protomers acts as a static scaffold. We note that the mechanism of ATP hydrolysis in the spastin or katanin ring has yet to be experimentally addressed through kinetic studies. It is also not yet clear at what point in the cycle the enzyme disengages from the tail and whether it leaves the microtubule with the dislodged tubulin subunit. Spastin is less efficient at severing hyperglutamylated microtubules despite retaining robust ATPase (Valenstein and Roll-Mecak, 2016), suggesting that the coupling between the chemical and mechanical cycles is substrate-dependent, as observed with other AAA family unfoldases.
Figure 6.
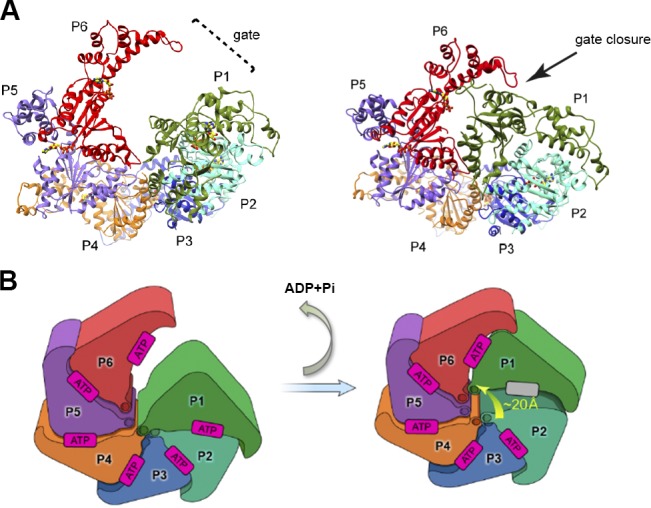
Proposed mechanism for tubulin tail translocation through the katanin central channel. (A) Left: Spiral conformation (PDB ID 5WC0; Zehr et al., 2017) of the hexamer with ATP bound in all six protomers. Right: ATP hydrolysis and release in the boundary protomer P1 transitions the hexamer to the ring conformation (PDB ID 5WCB; Zehr et al., 2017) that closes the gate between P1 and P6, and translocates the pore loops. (B) Schematic of conformational changes triggered by hydrolysis and release of ATP in the boundary protomer P1 that result in closure of the P1–P6 gate and translocation of the pore loops together with the tubulin substrate by ∼20 Å. Adapted from Zehr et al. (2017). Figure is reprinted with permission from Nature Structural and Molecular Biology.
Microtubule recognition by severing enzymes
The mechanism of substrate engagement by severing enzymes is still unknown, as there are currently no structures available of microtubule complexes or complexes of the enzymes with the isolated tubulin tails that are required for severing. It is still unclear how the pore loops coordinate ATP hydrolysis with substrate binding, hexamerization, and translocation. Moreover, in addition to the contact between the microtubule and the AAA ring, the MIT domains and part of the linker also bind the microtubule (Hartman and Vale, 1999; Roll-Mecak and Vale, 2005). NMR and x-ray studies have shown that the MIT domains of spastin and katanin are composed of a three-helix bundle (Fig. 3, B and D; Yang et al., 2008; Iwaya et al., 2010). How the MIT domain and linker recognize the microtubule is not known. Small angle x-ray scattering studies of the assembled katanin hexamer demonstrated that the MIT domains reside on average at the tip of flexible arms that emanate from the AAA ATPase ring (Fig. 4; Zehr et al., 2017), an architecture similar to that initially visualized for spastin (Roll-Mecak and Vale, 2008). Thus, spastin and katanin likely use multivalent interactions mediated by the flexible arms to anchor the AAA engine to the microtubule lattice and start pulling on the tubulin substrate through the ATP hydrolysis-driven remodeling of the AAA ring. This multivalent engagement of the microtubule ensures a slow off-rate of the enzyme from the microtubule, possibly contributing to high processivity. It is not clear what the valency of this interaction is. Given the polar nature of the microtubule polymer, it is not possible for all six arms to be engaged simultaneously unless the hinge points with the microtubule-binding domain are flexible.
The MIT domain of katanin binds with high affinity to the C-terminal domain of the regulatory p80 subunit, and this interaction is required for p60:p80 complex formation (Hartman et al., 1998; McNally and McNally, 2011; Rezabkova et al., 2017). p80 katanin also has a WD40 domain at the N terminus that is required for targeting to centrosomes (Hartman et al., 1998). Not all katanin p80 isoforms have the WD40 domain, likely reflecting different localizations of the enzyme (Fig. 1). While the p80 subunit is not required for severing (Hartman et al., 1998; Loughlin et al., 2011), it enhances it by increasing the affinity for the microtubule (McNally and McNally, 2011; Joly et al., 2016). The p80 C-terminal domain forms an extended helix-turn-helix fold with its N-terminal helix binding in a groove formed by two helices of the p60 MIT domain (Fig. 3 C; Rezabkova et al., 2017). The analogous interface is used by spastin to bind CHMP1B (Fig. 3 D; Yang et al., 2008). The interface formed by the MIT and p80 C-terminal domain is used as a binding platform for three known katanin regulators that target the enzyme to the growing minus ends of microtubules: CAMSAP2, CAMSAP3, and ASPM (Jiang et al., 2017, 2018). Mutations that destabilize the p80 C-terminal domain cause microcephaly (Mishra-Gorur et al., 2014).
Open questions and new directions
The recent katanin structures gave us a first glimpse into the mechanochemical cycle of these enzymes. Structures in different nucleotide states and with the tubulin substrate will be needed to fully understand the force generating conformational changes during severing. A fundamental unanswered question is how tubulin dimers are removed from the lattice. While the proposed mechanism of tubulin tail translocation through the pore is attractive, it has been proposed based on analogy with AAA ATPases such as Hsp104 and ClpX and still awaits direct experimental proof. Is tubulin extracted without unfolding? Such a mechanism would be analogous to that used by NSF to disassemble SNARE complexes (Zhao and Brunger, 2016). Or is it partially unfolded from the C terminus until it is distorted enough to allow dissociation of the bound GTP and loss of the tubulin subunit from the lattice? Such a mechanism is more analogous to that found for Rubisco activase, which is thought to remodel a short peptide segment of Rubisco to induce a conformational change that leads to the release of the inhibitory sugar phosphate (Portis et al., 2008). It is also possible that tubulin is completely unfolded by threading it through the central pore. Hydrogen deuterium exchange experiments demonstrated that VPS4 completely unfolds the ESCRTIII subunits, leading to the unraveling of the ESCRT polymer (Yang et al., 2015). Such a mechanism would have profound physiological implications for severing enzymes since it would generate denatured tubulin that needs to be cleared. Tubulin does not spontaneously fold and needs an elaborate chaperone system for folding and heterodimer assembly (Lundin et al., 2010).
Another open question is how many tubulin subunits need to be removed from the microtubule until the rest of the subunits spontaneously or cooperatively dissociate. Recent studies have shown that microtubules nanodamaged by spastin or katanin through tubulin dimer removal are long-lived enough to have a chance to repair, indicating that stability is not catastrophically compromised with the removal of the first tubulin dimers (Vemu et al., 2018). In the case of VPS4, ∼50% of the ESCRT III subunits need to be actively removed in order for the lattice to disintegrate completely (Yang et al., 2015). Studies have shown that AAA ATPases are capable of producing large forces. ClpX, a bacterial AAA ATPase that unfolds polypeptides, exerts upwards of 20 pN of force (Aubin-Tam et al., 2011; Maillard et al., 2011). The energy required to remove a tubulin subunit from the microtubule lattice has not been experimentally determined, and neither has the force generated by a severing enzyme during the ATPase cycle, in large part due to difficulties in engineering handles on the tubulin substrate for force measurements. Advances in the production of recombinant engineered mammalian tubulin (Minoura et al., 2013; Ti et al., 2016; Vemu et al., 2016) make it now possible to overcome this hurdle. What is also unknown is how many rounds of ATP hydrolysis the enzyme undergoes before disengaging from the tubulin tail and the microtubule. The two do not necessarily need to be coordinated as there are separate points of contact with the AAA central channel and the MIT/linkers.
From a cell biological perspective, a major challenge in the following years will be to find ways to better visualize dense microtubule arrays where severing is important. Neurons pose particular challenges for this, but this is one area where recent advances in cryo-EM tomography hold great promise to probe these microtubule arrays at higher resolution (Lučič et al., 2013). Regulation and targeting of microtubule severing will also continue to be an important topic since unchecked severing is likely highly deleterious to the cell. The precise graded regulation of spastin by the number of glutamates on the tubulin tails (Valenstein and Roll-Mecak, 2016) underscores the importance of knowing not only where the microtubule-severing enzymes are localized, but also the posttranslational status of the microtubules they interact with. Interestingly, katanin shows localization at plus and minus ends in mammalian cells (Jiang et al., 2017) and microtubule crossovers in plant cortical arrays (Lindeboom et al., 2013; Wang et al., 2017) that is dependent on the p80 subunit. C. elegans katanin also prefers microtubule crossovers in in vitro reconstituted assays (McNally et al., 2014). As cells have multiple p80 isoforms, it will be important to establish how these contribute to the differential localization of katanin. Another fertile area of future exploration will be to understand how microtubule-severing enzymes are regulated by MAPs, especially since the tubulin C-terminal tails that are essential for severing are also part of the binding sites for many MAPs. For example, cellular studies have shown that katanin is inhibited by tau (Qiang et al., 2006), while experiments with Xenopus extracts revealed inhibition by the MAP4 homologue XMAP230 (McNally et al., 2002). Proteomic studies have now identified many interacting partners for severing enzymes (Cheung et al., 2016); however, most of these remain to be validated and studied in greater depth to understand their mechanism of action. The interaction between spastin and the ESCRTIII component CHMP1B (Reid et al., 2005; Yang et al., 2008) raises the interesting question of whether CHMP1B regulates spastin microtubule-severing activity to facilitate coordination between the microtubule cytoskeleton and membrane remodeling during abscission.
Last, but arguably most important for a microtubule regulator, we are just now starting to understand the direct effects severing enzymes have on microtubule structure and dynamics. Recent electron microscopic studies and in vitro reconstituted quantitative microtubule dynamics assays with purified severing enzymes at physiological concentrations revealed that both spastin and katanin create nanodamage sites along the microtubule shaft that are spontaneously healed with GTP-tubulin subunits from the soluble pool (Vemu et al., 2018). This ATP hydrolysis-dependent process produces GTP-tubulin islands in microtubules that can act as rescue sites and generates newly severed ends enriched in GTP-tubulin that stabilizes them against spontaneous depolymerization. These GTP-enriched islands serve as a platform for the recruitment of proteins that recognize the GTP (or GDP-Pi)-bound state of tubulin such as end-binding protein 1 (Vemu et al., 2018). Thus, severing enzymes do not simply disassemble microtubules, they also remodel the microtubule lattice. An important next step is to demonstrate this mechanism of tubulin exchange in cells and understand how it is regulated. In vivo, these newly severed GTP-enriched plus ends likely serve as a platform for the many microtubule dynamics regulators that are recruited to the plus ends in an end-binding protein 1–dependent manner (Akhmanova and Steinmetz, 2015) and thus can either stabilize or destabilize the newly severed plus ends. Almost 30 years after the discovery of the first microtubule-severing enzyme, many fundamental questions about the molecular and cellular mechanism of this family of proteins still remain unanswered, while recent studies have opened exciting new avenues for the exploration of their in vivo functions and biochemical mechanism. New genomic editing tools coupled with advances in high-resolution imaging and in vitro reconstitution assays with purified enzymes, engineered recombinant tubulin, and diverse microtubule array geometries obtained through micropatterning will shed light on the multifaceted roles of severing enzymes in regulating microtubule array architecture and dynamics. Given the involvement of severing enzymes in human disease, pursuit of these basic questions also holds promise for improved diagnostics and therapeutic interventions.
Acknowledgments
We thank Bara Krautz (Science Animated, Zurich, Switzerland) for help with katanin model illustrations and Ethan Tyler (Medical Research Arts, National Institutes of Health, Bethesda, MD) for the severing panels.
F.J. McNally is supported by National Institute of General Medical Sciences grant 1R01GM-079421 and U.S. Department of Agriculture National Institute of Food Agriculture Hatch project 1009162. A. Roll-Mecak is supported by the intramural programs of the National Institute of Neurological Disorders and Stroke and the National Heart, Lung, and Blood Institute.
The authors declare no competing financial interests.
Author contributions: F.J. McNally and A. Roll-Mecak wrote and edited the manuscript.
References
- Ahmad F.J., Yu W., McNally F.J., and Baas P.W.. 1999. An essential role for katanin in severing microtubules in the neuron. J. Cell Biol. 145:305–315. 10.1083/jcb.145.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2010. Microtubule +TIPs at a glance. J. Cell Sci. 123:3415–3419. 10.1242/jcs.062414 [DOI] [PubMed] [Google Scholar]
- Akhmanova A., and Steinmetz M.O.. 2015. Control of microtubule organization and dynamics: two ends in the limelight. Nat. Rev. Mol. Cell Biol. 16:711–726. 10.1038/nrm4084 [DOI] [PubMed] [Google Scholar]
- Allison R., Lumb J.H., Fassier C., Connell J.W., Ten Martin D., Seaman M.N., Hazan J., and Reid E.. 2013. An ESCRT-spastin interaction promotes fission of recycling tubules from the endosome. J. Cell Biol. 202:527–543. 10.1083/jcb.201211045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubin-Tam M.E., Olivares A.O., Sauer R.T., Baker T.A., and Lang M.J.. 2011. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 145:257–267. 10.1016/j.cell.2011.03.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banks G., Lassi G., Hoerder-Suabedissen A., Tinarelli F., Simon M.M., Wilcox A., Lau P., Lawson T.N., Johnson S., Rutman A., et al. . 2018. A missense mutation in Katnal1 underlies behavioural, neurological and ciliary anomalies. Mol. Psychiatry. 23:713–722. 10.1038/mp.2017.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton D.A., Vantard M., and Overall R.L.. 2008. Analysis of cortical arrays from Tradescantia virginiana at high resolution reveals discrete microtubule subpopulations and demonstrates that confocal images of arrays can be misleading. Plant Cell. 20:982–994. 10.1105/tpc.108.058503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benz C., Clucas C., Mottram J.C., and Hammarton T.C.. 2012. Cytokinesis in bloodstream stage Trypanosoma brucei requires a family of katanins and spastin. PLoS One. 7:e30367 10.1371/journal.pone.0030367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bichet A., Desnos T., Turner S., Grandjean O., and Höfte H.. 2001. BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J. 25:137–148. 10.1046/j.1365-313x.2001.00946.x [DOI] [PubMed] [Google Scholar]
- Burk D.H., and Ye Z.-H.. 2002. Alteration of oriented deposition of cellulose microfibrils by mutation of a katanin-like microtubule-severing protein. Plant Cell. 14:2145–2160. 10.1105/tpc.003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk D.H., Liu B., Zhong R., Morrison W.H., and Ye Z.-H.. 2001. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 13:807–827. 10.1105/tpc.13.4.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier M.F., and Pantaloni D.. 1981. Kinetic analysis of guanosine 5′-triphosphate hydrolysis associated with tubulin polymerization. Biochemistry. 20:1918–1924. 10.1021/bi00510a030 [DOI] [PubMed] [Google Scholar]
- Carlier M.F., and Pantaloni D.. 1982. Assembly of microtubule protein: role of guanosine di- and triphosphate nucleotides. Biochemistry. 21:1215–1224. 10.1021/bi00535a017 [DOI] [PubMed] [Google Scholar]
- Carlier M.F., Didry D., and Pantaloni D.. 1987. Microtubule elongation and guanosine 5′-triphosphate hydrolysis. Role of guanine nucleotides in microtubule dynamics. Biochemistry. 26:4428–4437. 10.1021/bi00388a036 [DOI] [PubMed] [Google Scholar]
- Charafeddine R.A., Makdisi J., Schairer D., O’Rourke B.P., Diaz-Valencia J.D., Chouake J., Kutner A., Krausz A., Adler B., Nacharaju P., et al. . 2015. Fidgetin-Like 2: a microtubule-based regulator of wound healing. J. Invest. Dermatol. 135:2309–2318. 10.1038/jid.2015.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung K., Senese S., Kuang J., Bui N., Ongpipattanakul C., Gholkar A., Cohn W., Capri J., Whitelegge J.P., and Torres J.Z.. 2016. Proteomic Analysis of the Mammalian Katanin Family of Microtubule-severing Enzymes Defines KATNBL1 as a Regulator of Mammalian Katanin Microtubule-severing. Mol. Cell. Proteomics. 15:1658–1669. 10.1074/mcp.M115.056465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi Z., and Ambrose C.. 2016. Microtubule encounter-based catastrophe in Arabidopsis cortical microtubule arrays. BMC Plant Biol. 16:18 10.1186/s12870-016-0703-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connell J.W., Lindon C., Luzio J.P., and Reid E.. 2009. Spastin couples microtubule severing to membrane traffic in completion of cytokinesis and secretion. Traffic. 10:42–56. 10.1111/j.1600-0854.2008.00847.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton K.R., Lei L., Grenier J., Rodionov V., Blackstone C., and Li X.J.. 2014. Loss of spastin function results in disease-specific axonal defects in human pluripotent stem cell-based models of hereditary spastic paraplegia. Stem Cells. 32:414–423. 10.1002/stem.1569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeSantis M.E., Leung E.H., Sweeny E.A., Jackrel M.E., Cushman-Nick M., Neuhaus-Follini A., Vashist S., Sochor M.A., Knight M.N., and Shorter J.. 2012. Operational plasticity enables hsp104 to disaggregate diverse amyloid and nonamyloid clients. Cell. 151:778–793. 10.1016/j.cell.2012.09.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DesMarais V., Ghosh M., Eddy R., and Condeelis J.. 2005. Cofilin takes the lead. J. Cell Sci. 118:19–26. 10.1242/jcs.01631 [DOI] [PubMed] [Google Scholar]
- Díaz-Valencia J.D., Morelli M.M., Bailey M., Zhang D., Sharp D.J., and Ross J.L.. 2011. Drosophila katanin-60 depolymerizes and severs at microtubule defects. Biophys. J. 100:2440–2449. 10.1016/j.bpj.2011.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez R., and Holmes K.C.. 2011. Actin structure and function. Annu. Rev. Biophys. 40:169–186. 10.1146/annurev-biophys-042910-155359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong C., Xu H., Zhang R., Tanaka N., Takeichi M., and Meng W.. 2017. CAMSAP3 accumulates in the pericentrosomal area and accompanies microtubule release from the centrosome via katanin. J. Cell Sci. 130:1709–1715. 10.1242/jcs.198010 [DOI] [PubMed] [Google Scholar]
- Doyle S.M., Hoskins J.R., and Wickner S.. 2012. DnaK-dependent disaggregation by Caseinolytic Peptidase B, ClpB, mutants reveals functional overlap in the N-terminal domain and nucleotide-binding domain-1 pore tyrosine. J. Biol. Chem. 287:28470–28479. 10.1074/jbc.M112.383091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., and Smith E.F.. 2012. PF19 encodes the p60 catalytic subunit of katanin and is required for assembly of the flagellar central apparatus in Chlamydomonas. J. Cell Sci. 125:3357–3366. 10.1242/jcs.096941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymek E.E., Lefebvre P.A., and Smith E.F.. 2004. PF15p is the chlamydomonas homologue of the Katanin p80 subunit and is required for assembly of flagellar central microtubules. Eukaryot. Cell. 3:870–879. 10.1128/EC.3.4.870-879.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckert T., Link S., Le D.T., Sobczak J.-P., Gieseke A., Richter K., and Woehlke G.. 2012. Subunit Interactions and cooperativity in the microtubule-severing AAA ATPase spastin. J. Biol. Chem. 287:26278–26290. 10.1074/jbc.M111.291898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans K.J., Gomes E.R., Reisenweber S.M., Gundersen G.G., and Lauring B.P.. 2005. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 168:599–606. 10.1083/jcb.200409058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Upton K., and Compton D.A.. 2005. Efficient mitosis in human cells lacking poleward microtubule flux. Curr. Biol. 15:1827–1832. 10.1016/j.cub.2005.08.065 [DOI] [PubMed] [Google Scholar]
- Gates S.N., Yokom A.L., Lin J., Jackrel M.E., Rizo A.N., Kendsersky N.M., Buell C.E., Sweeny E.A., Mack K.L., Chuang E., et al. . 2017. Ratchet-like polypeptide translocation mechanism of the AAA+ disaggregase Hsp104. Science. 357:273–279. 10.1126/science.aan1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grode K.D., and Rogers S.L.. 2015. The non-catalytic domains of Drosophila katanin regulate its abundance and microtubule-disassembly activity. PLoS One. 10:e0123912 10.1371/journal.pone.0123912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guizetti J., Schermelleh L., Mäntler J., Maar S., Poser I., Leonhardt H., Müller-Reichert T., and Gerlich D.W.. 2011. Cortical constriction during abscission involves helices of ESCRT-III-dependent filaments. Science. 331:1616–1620. 10.1126/science.1201847 [DOI] [PubMed] [Google Scholar]
- Han H., Monroe N., Sundquist W.I., Shen P.S., and Hill C.P.. 2017. The AAA ATPase Vps4 binds ESCRT-III substrates through a repeating array of dipeptide-binding pockets. eLife. 6:e31324 10.7554/eLife.31324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara Y., and Kimura A.. 2013. An allometric relationship between mitotic spindle width, spindle length, and ploidy in Caenorhabditis elegans embryos. Mol. Biol. Cell. 24:1411–1419. 10.1091/mbc.e12-07-0528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartman J.J., and Vale R.D.. 1999. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 286:782–785. 10.1126/science.286.5440.782 [DOI] [PubMed] [Google Scholar]
- Hartman J.J., Mahr J., McNally K., Okawa K., Iwamatsu A., Thomas S., Cheesman S., Heuser J., Vale R.D., and McNally F.J.. 1998. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 93:277–287. 10.1016/S0092-8674(00)81578-0 [DOI] [PubMed] [Google Scholar]
- Hawkins T., Mirigian M., Selcuk Yasar M., and Ross J.L.. 2010. Mechanics of microtubules. J. Biomech. 43:23–30. 10.1016/j.jbiomech.2009.09.005 [DOI] [PubMed] [Google Scholar]
- Hazan J., Fonknechten N., Mavel D., Paternotte C., Samson D., Artiguenave F., Davoine C.-S., Cruaud C., Dürr A., Wincker P., et al. . 1999. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nat. Genet. 23:296–303. 10.1038/15472 [DOI] [PubMed] [Google Scholar]
- Hu W.F., Pomp O., Ben-Omran T., Kodani A., Henke K., Mochida G.H., Yu T.W., Woodworth M.B., Bonnard C., Raj G.S., et al. . 2014. Katanin p80 regulates human cortical development by limiting centriole and cilia number. Neuron. 84:1240–1257. 10.1016/j.neuron.2014.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z., Feng J., Bo W., Wu R., Dong Z., Liu Y., Qiang L., and Liu M.. 2017. Fidgetin regulates cultured astrocyte migration by severing tyrosinated microtubules at the leading edge. Mol. Biol. Cell. 28:545–553. 10.1091/mbc.e16-09-0628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwaya N., Kuwahara Y., Fujiwara Y., Goda N., Tenno T., Akiyama K., Mase S., Tochio H., Ikegami T., Shirakawa M., and Hiroaki H.. 2010. A common substrate recognition mode conserved between katanin p60 and VPS4 governs microtubule severing and membrane skeleton reorganization. J. Biol. Chem. 285:16822–16829. 10.1074/jbc.M110.108365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Hua S., Mohan R., Grigoriev I., Yau K.W., Liu Q., Katrukha E.A., Altelaar A.F., Heck A.J., Hoogenraad C.C., and Akhmanova A.. 2014. Microtubule minus-end stabilization by polymerization-driven CAMSAP deposition. Dev. Cell. 28:295–309. 10.1016/j.devcel.2014.01.001 [DOI] [PubMed] [Google Scholar]
- Jiang K., Rezabkova L., Hua S., Liu Q., Capitani G., Altelaar A.F.M., Heck A.J.R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2017. Microtubule minus-end regulation at spindle poles by an ASPM-katanin complex. Nat. Cell Biol. 19:480–492. 10.1038/ncb3511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang K., Faltova L., Hua S., Capitani G., Prota A.E., Landgraf C., Volkmer R., Kammerer R.A., Steinmetz M.O., and Akhmanova A.. 2018. Structural Basis of Formation of the Microtubule Minus-End-Regulating CAMSAP-Katanin Complex. Structure. 26:375–382.e4. 10.1016/j.str.2017.12.017 [DOI] [PubMed] [Google Scholar]
- Johjima A., Noi K., Nishikori S., Ogi H., Esaki M., and Ogura T.. 2015. Microtubule severing by katanin p60 AAA+ ATPase requires the C-terminal acidic tails of both α- and β-tubulins and basic amino acid residues in the AAA+ ring pore. J. Biol. Chem. 290:11762–11770. 10.1074/jbc.M114.614768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joly N., Martino L., Gigant E., Dumont J., and Pintard L.. 2016. Microtubule-severing activity of the AAA+ ATPase Katanin is essential for female meiotic spindle assembly. Development. 143:3604–3614. 10.1242/dev.140830 [DOI] [PubMed] [Google Scholar]
- Kasher P.R., De Vos K.J., Wharton S.B., Manser C., Bennett E.J., Bingley M., Wood J.D., Milner R., McDermott C.J., Miller C.C., et al. . 2009. Direct evidence for axonal transport defects in a novel mouse model of mutant spastin-induced hereditary spastic paraplegia (HSP) and human HSP patients. J. Neurochem. 110:34–44. 10.1111/j.1471-4159.2009.06104.x [DOI] [PubMed] [Google Scholar]
- Keating T.J., Peloquin J.G., Rodionov V.I., Momcilovic D., and Borisy G.G.. 1997. Microtubule release from the centrosome. Proc. Natl. Acad. Sci. USA. 94:5078–5083. 10.1073/pnas.94.10.5078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacroix B., van Dijk J., Gold N.D., Guizetti J., Aldrian-Herrada G., Rogowski K., Gerlich D.W., and Janke C.. 2010. Tubulin polyglutamylation stimulates spastin-mediated microtubule severing. J. Cell Biol. 189:945–954. 10.1083/jcb.201001024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lata S., Schoehn G., Jain A., Pires R., Piehler J., Gőttlinger H.G., and Weissenhorn W.. 2008. Helical structures of ESCRT-III are disassembled by VPS4. Science. 321:1354–1357. 10.1126/science.1161070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H.-H., Jan L.Y., and Jan Y.-N.. 2009. Drosophila IKK-related kinase Ik2 and Katanin p60-like 1 regulate dendrite pruning of sensory neuron during metamorphosis. Proc. Natl. Acad. Sci. USA. 106:6363–6368. 10.1073/pnas.0902051106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leo L., Yu W., D’Rozario M., Waddell E.A., Marenda D.R., Baird M.A., Davidson M.W., Zhou B., Wu B., Baker L., et al. . 2015. Vertebrate Fidgetin Restrains Axonal Growth by Severing Labile Domains of Microtubules. Cell Reports. 12:1723–1730. 10.1016/j.celrep.2015.08.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindeboom J.J., Nakamura M., Hibbel A., Shundyak K., Gutierrez R., Ketelaar T., Emons A.M.C., Mulder B.M., Kirik V., and Ehrhardt D.W.. 2013. A mechanism for reorientation of cortical microtubule arrays driven by microtubule severing. Science. 342:1245533 10.1126/science.1245533 [DOI] [PubMed] [Google Scholar]
- Liu T., Tian J., Wang G., Yu Y., Wang C., Ma Y., Zhang X., Xia G., Liu B., and Kong Z.. 2014. Augmin triggers microtubule-dependent microtubule nucleation in interphase plant cells. Curr. Biol. 24:2708–2713. 10.1016/j.cub.2014.09.053 [DOI] [PubMed] [Google Scholar]
- Loughlin R., Wilbur J.D., McNally F.J., Nédélec F.J., and Heald R.. 2011. Katanin contributes to interspecies spindle length scaling in Xenopus. Cell. 147:1397–1407. 10.1016/j.cell.2011.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučič V., Rigort A., and Baumeister W.. 2013. Cryo-electron tomography: the challenge of doing structural biology in situ. J. Cell Biol. 202:407–419. 10.1083/jcb.201304193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundin V.F., Leroux M.R., and Stirling P.C.. 2010. Quality control of cytoskeletal proteins and human disease. Trends Biochem. Sci. 35:288–297. 10.1016/j.tibs.2009.12.007 [DOI] [PubMed] [Google Scholar]
- Maillard R.A., Chistol G., Sen M., Righini M., Tan J., Kaiser C.M., Hodges C., Martin A., and Bustamante C.. 2011. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 145:459–469. 10.1016/j.cell.2011.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mains P.E., Kemphues K.J., Sprunger S.A., Sulston I.A., and Wood W.B.. 1990. Mutations affecting the meiotic and mitotic divisions of the early Caenorhabditis elegans embryo. Genetics. 126:593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao C.-X., Xiong Y., Xiong Z., Wang Q., Zhang Y.Q., and Jin S.. 2014. Microtubule-severing protein Katanin regulates neuromuscular junction development and dendritic elaboration in Drosophila. Development. 141:1064–1074. 10.1242/dev.097774 [DOI] [PubMed] [Google Scholar]
- Matsuo M., Shimodaira T., Kasama T., Hata Y., Echigo A., Okabe M., Arai K., Makino Y., Niwa S., Saya H., and Kishimoto T.. 2013. Katanin p60 contributes to microtubule instability around the midbody and facilitates cytokinesis in rat cells. PLoS One. 8:e80392 10.1371/journal.pone.0080392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., and Thomas S.. 1998. Katanin is responsible for the M-phase microtubule-severing activity in Xenopus eggs. Mol. Biol. Cell. 9:1847–1861. 10.1091/mbc.9.7.1847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally F.J., and Vale R.D.. 1993. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 75:419–429. 10.1016/0092-8674(93)90377-3 [DOI] [PubMed] [Google Scholar]
- McNally K.P., and McNally F.J.. 2011. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol. Biol. Cell. 22:1550–1560. 10.1091/mbc.e10-12-0951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K.P., Buster D., and McNally F.J.. 2002. Katanin-mediated microtubule severing can be regulated by multiple mechanisms. Cell Motil. Cytoskeleton. 53:337–349. 10.1002/cm.10080 [DOI] [PubMed] [Google Scholar]
- McNally K., Audhya A., Oegema K., and McNally F.J.. 2006. Katanin controls mitotic and meiotic spindle length. J. Cell Biol. 175:881–891. 10.1083/jcb.200608117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally K., Berg E., Cortes D.B., Hernandez V., Mains P.E., and McNally F.J.. 2014. Katanin maintains meiotic metaphase chromosome alignment and spindle structure in vivo and has multiple effects on microtubules in vitro. Mol. Biol. Cell. 25:1037–1049. 10.1091/mbc.e13-12-0764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minoura I., Hachikubo Y., Yamakita Y., Takazaki H., Ayukawa R., Uchimura S., and Muto E.. 2013. Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett. 587:3450–3455. 10.1016/j.febslet.2013.08.032 [DOI] [PubMed] [Google Scholar]
- Mishra-Gorur K., Çağlayan A.O., Schaffer A.E., Chabu C., Henegariu O., Vonhoff F., Akgümüş G.T., Nishimura S., Han W., Tu S., et al. . 2014. Mutations in KATNB1 cause complex cerebral malformations by disrupting asymmetrically dividing neural progenitors. Neuron. 84:1226–1239. 10.1016/j.neuron.2014.12.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., and Kirschner M.. 1984. Dynamic instability of microtubule growth. Nature. 312:237–242. 10.1038/312237a0 [DOI] [PubMed] [Google Scholar]
- Monroe N., and Hill C.P.. 2016. Meiotic Clade AAA ATPases: Protein Polymer Disassembly Machines. J. Mol. Biol. 428:1897–1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti S., Armougom F., Wallace I.M., Higgins D.G., Jongeneel C.V., and Notredame C.. 2007. The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res. 35:W645–W648. 10.1093/nar/gkm333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S., Diaz Valencia J.D., Stewman S., Metz J., Monnier S., Rath U., Asenjo A.B., Charafeddine R.A., Sosa H.J., Ross J.L., et al. . 2012. Human Fidgetin is a microtubule severing the enzyme and minus-end depolymerase that regulates mitosis. Cell Cycle. 11:2359–2366. 10.4161/cc.20849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Sonobe S., Baskin T.I., Hyodo S., Hasezawa S., Nagata T., Horio T., and Hasebe M.. 2005. Microtubule-dependent microtubule nucleation based on recruitment of γ-tubulin in higher plants. Nat. Cell Biol. 7:961–968. 10.1038/ncb1306 [DOI] [PubMed] [Google Scholar]
- Nakamura M., Ehrhardt D.W., and Hashimoto T.. 2010. Microtubule and katanin-dependent dynamics of microtubule nucleation complexes in the acentrosomal Arabidopsis cortical array. Nat. Cell Biol. 12:1064–1070. 10.1038/ncb2110 [DOI] [PubMed] [Google Scholar]
- Nicklas R.B., Lee G.M., Rieder C.L., and Rupp G.. 1989. Mechanically cut mitotic spindles: clean cuts and stable microtubules. J. Cell Sci. 94:415–423. [DOI] [PubMed] [Google Scholar]
- Nithianantham S., McNally F.J., and Al-Bassam J.. 2018. Structural basis for disassembly of katanin heterododecamers. J. Biol. Chem. 293:10590–10605. 10.1074/jbc.RA117.001215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivares A.O., Baker T.A., and Sauer R.T.. 2016. Mechanistic insights into bacterial AAA+ proteases and protein-remodelling machines. Nat. Rev. Microbiol. 14:33–44. 10.1038/nrmicro.2015.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., and Roll-Mecak A.. 2018. The tubulin code in neuronal polarity. Curr. Opin. Neurobiol. 51:95–102. 10.1016/j.conb.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W., Lin Z., Li W., Lu J., Shen Y., and Wang C.. 2013. Structural insights into unusually strong ATPase activity of the AAA domain of Caenorhabditis elegans fidgetin like-1 (FIGL-1) protein. J. Biol. Chem. 288:29305–29312. 10.1074/jbc.M113.502559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portis A.R. Jr., Li C., Wang D., and Salvucci M.E.. 2008. Regulation of Rubisco activase and its interaction with Rubisco. J. Exp. Bot. 59:1597–1604. 10.1093/jxb/erm240 [DOI] [PubMed] [Google Scholar]
- Puchades C., Rampello A.J., Shin M., Giuliano C.J., Wiseman R.L., Glynn S.E., and Lander G.C.. 2017. Structure of the mitochondrial inner membrane AAA+ protease YME1 gives insight into substrate processing. Science. 358:eaao0464 10.1126/science.aao0464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiang L., Yu W., Andreadis A., Luo M., and Baas P.W.. 2006. Tau protects microtubules in the axon from severing by katanin. J. Neurosci. 26:3120–3129. 10.1523/JNEUROSCI.5392-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao K., Stone M.C., Weiner A.T., Gheres K.W., Zhou C., Deitcher D.L., Levitan E.S., and Rolls M.M.. 2016. Spastin, atlastin, and ER relocalization are involved in axon but not dendrite regeneration. Mol. Biol. Cell. 27:3245–3256. 10.1091/mbc.e16-05-0287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid E., Connell J., Edwards T.L., Duley S., Brown S.E., and Sanderson C.M.. 2005. The hereditary spastic paraplegia protein spastin interacts with the ESCRT-III complex-associated endosomal protein CHMP1B. Hum. Mol. Genet. 14:19–38. 10.1093/hmg/ddi003 [DOI] [PubMed] [Google Scholar]
- Rezabkova L., Jiang K., Capitani G., Prota A.E., Akhmanova A., Steinmetz M.O., and Kammerer R.A.. 2017. Structural basis of katanin p60:p80 complex formation. Sci. Rep. 7:14893 10.1038/s41598-017-14194-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K., and Mitchison T.J.. 2006. Meiotic spindle: sculpted by severing. Curr. Biol. 16:R923–R925. 10.1016/j.cub.2006.09.048 [DOI] [PubMed] [Google Scholar]
- Rogers G.C., Rogers S.L., Schwimmer T.A., Ems-McClung S.C., Walczak C.E., Vale R.D., Scholey J.M., and Sharp D.J.. 2004. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 427:364–370. 10.1038/nature02256 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., and McNally F.J.. 2010. Microtubule-severing enzymes. Curr. Opin. Cell Biol. 22:96–103. 10.1016/j.ceb.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2005. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 15:650–655. 10.1016/j.cub.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2006. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 175:849–851. 10.1083/jcb.200611149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roll-Mecak A., and Vale R.D.. 2008. Structural basis of microtubule severing by the hereditary spastic paraplegia protein spastin. Nature. 451:363–367. 10.1038/nature06482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma N., Bryant J., Wloga D., Donaldson R., Davis R.C., Jerka-Dziadosz M., and Gaertig J.. 2007. Katanin regulates dynamics of microtubules and biogenesis of motile cilia. J. Cell Biol. 178:1065–1079. 10.1083/jcb.200704021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp D.J., and Ross J.L.. 2012. Microtubule-severing enzymes at the cutting edge. J. Cell Sci. 125:2561–2569. 10.1242/jcs.101139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S.L., Kamyar R., and Ehrhardt D.W.. 2003. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 300:1715–1718. 10.1126/science.1083529 [DOI] [PubMed] [Google Scholar]
- Sherwood N.T., Sun Q., Xue M., Zhang B., and Zinn K.. 2004. Drosophila spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2:e429 10.1371/journal.pbio.0020429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonbuchner T.M., Rath U., and Sharp D.J.. 2010. KL1 is a novel microtubule severing enzyme that regulates mitotic spindle architecture. Cell Cycle. 9:2403–2411. 10.4161/cc.9.12.11916 [DOI] [PubMed] [Google Scholar]
- Srayko M., O’toole E.T., Hyman A.A., and Müller-Reichert T.. 2006. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr. Biol. 16:1944–1949. 10.1016/j.cub.2006.08.029 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 22:2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Stewart A., Tsubouchi A., Rolls M.M., Tracey W.D., and Sherwood N.T.. 2012. Katanin p60-like1 promotes microtubule growth and terminal dendrite stability in the larval class IV sensory neurons of Drosophila. J. Neurosci. 32:11631–11642. 10.1523/JNEUROSCI.0729-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M.C., Rao K., Gheres K.W., Kim S., Tao J., La Rochelle C., Folker C.T., Sherwood N.T., and Rolls M.M.. 2012. Normal spastin gene dosage is specifically required for axon regeneration. Cell Reports. 2:1340–1350. 10.1016/j.celrep.2012.09.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Feng C., and Rolls M.M.. 2016. The microtubule-severing protein fidgetin acts after dendrite injury to promote degeneration. J. Cell Sci. 129:3274–3281. 10.1242/jcs.188540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.L., White S.R., Lauring B., and Kull F.J.. 2012. Crystal structure of the human spastin AAA domain. J. Struct. Biol. 179:133–137. 10.1016/j.jsb.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ti S.C., Pamula M.C., Howes S.C., Duellberg C., Cade N.I., Kleiner R.E., Forth S., Surrey T., Nogales E., and Kapoor T.M.. 2016. Mutations in Human Tubulin Proximal to the Kinesin-Binding Site Alter Dynamic Instability at Microtubule Plus- and Minus-Ends. Dev. Cell. 37:72–84. 10.1016/j.devcel.2016.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirnauer J.S., Salmon E.D., and Mitchison T.J.. 2004. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol. Biol. Cell. 15:1776–1784. 10.1091/mbc.e03-11-0824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran P.T., Walker R.A., and Salmon E.D.. 1997. A metastable intermediate state of microtubule dynamic instability that differs significantly between plus and minus ends. J. Cell Biol. 138:105–117. 10.1083/jcb.138.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trotta N., Orso G., Rossetto M.G., Daga A., and Broadie K.. 2004. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr. Biol. 14:1135–1147. 10.1016/j.cub.2004.06.058 [DOI] [PubMed] [Google Scholar]
- Vale R.D. 1991. Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell. 64:827–839. 10.1016/0092-8674(91)90511-V [DOI] [PubMed] [Google Scholar]
- Valenstein M.L., and Roll-Mecak A.. 2016. Graded control of microtubule severing by tubulin glutamylation. Cell. 164:911–921. 10.1016/j.cell.2016.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A., Atherton J., Spector J.O., Szyk A., Moores C.A., and Roll-Mecak A.. 2016. Structure and dynamics of single-isoform recombinant Neuronal Human Tubulin. J. Biol. Chem. 291:12907–12915. 10.1074/jbc.C116.731133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vemu A., Szczesna E., Zehr E.A., Spector J.O., Grigorieff N., Deaconescu A.M., and Roll-Mecak A.. 2018. Severing enzymes amplify microtubule arrays through lattice GTP-tubulin incorporation. Science. 361:eaau1504 10.1126/science.aau1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vietri M., Schink K.O., Campsteijn C., Wegner C.S., Schultz S.W., Christ L., Thoresen S.B., Brech A., Raiborg C., and Stenmark H.. 2015. Spastin and ESCRT-III coordinate mitotic spindle disassembly and nuclear envelope sealing. Nature. 522:231–235. 10.1038/nature14408 [DOI] [PubMed] [Google Scholar]
- Walker R.A., Inoué S., and Salmon E.D.. 1989. Asymmetric behavior of severed microtubule ends after ultraviolet-microbeam irradiation of individual microtubules in vitro. J. Cell Biol. 108:931–937. 10.1083/jcb.108.3.931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu W., Wang G., Li J., Dong L., Han L., Wang Q., Tian J., Yu Y., Gao C., and Kong Z.. 2017. KTN80 confers precision to microtubule severing by specific targeting of katanin complexes in plant cells. EMBO J. 36:3435–3447. 10.15252/embj.201796823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterman-Storer C.M., and Salmon E.D.. 1997. Actomyosin-based retrograde flow of microtubules in the lamella of migrating epithelial cells influences microtubule dynamic instability and turnover and is associated with microtubule breakage and treadmilling. J. Cell Biol. 139:417–434. 10.1083/jcb.139.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen M., and Wang C.. 2013. The nucleotide cycle of spastin correlates with its microtubule-binding properties. FEBS J. 280:3868–3877. 10.1111/febs.12385 [DOI] [PubMed] [Google Scholar]
- White S.R., Evans K.J., Lary J., Cole J.L., and Lauring B.. 2007. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 176:995–1005. 10.1083/jcb.200610072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead E., Heald R., and Wilbur J.D.. 2013. N-terminal phosphorylation of p60 katanin directly regulates microtubule severing. J. Mol. Biol. 425:214–221. 10.1016/j.jmb.2012.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wightman R., and Turner S.R.. 2007. Severing at sites of microtubule crossover contributes to microtubule alignment in cortical arrays. Plant J. 52:742–751. 10.1111/j.1365-313X.2007.03271.x [DOI] [PubMed] [Google Scholar]
- Yang B., Stjepanovic G., Shen Q., Martin A., and Hurley J.H.. 2015. Vps4 disassembles an ESCRT-III filament by global unfolding and processive translocation. Nat. Struct. Mol. Biol. 22:492–498. 10.1038/nsmb.3015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Rismanchi N., Renvoisé B., Lippincott-Schwartz J., Blackstone C., and Hurley J.H.. 2008. Structural basis for midbody targeting of spastin by the ESCRT-III protein CHMP1B. Nat. Struct. Mol. Biol. 15:1278–1286. 10.1038/nsmb.1512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.Y., McNally K., and McNally F.J.. 2003. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev. Biol. 260:245–259. 10.1016/S0012-1606(03)00216-1 [DOI] [PubMed] [Google Scholar]
- Yu I., Garnham C.P., and Roll-Mecak A.. 2015. Writing and reading the tubulin code. J. Biol. Chem. 290:17163–17172. 10.1074/jbc.R115.637447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehr E., Szyk A., Piszczek G., Szczesna E., Zuo X., and Roll-Mecak A.. 2017. Katanin spiral and ring structures shed light on power stroke for microtubule severing. Nat. Struct. Mol. Biol. 24:717–725. 10.1038/nsmb.3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Rogers G.C., Buster D.W., and Sharp D.J.. 2007. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 177:231–242. 10.1083/jcb.200612011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D., Grode K.D., Stewman S.F., Diaz-Valencia J.D., Liebling E., Rath U., Riera T., Currie J.D., Buster D.W., Asenjo A.B., et al. . 2011. Drosophila katanin is a microtubule depolymerase that regulates cortical-microtubule plus-end interactions and cell migration. Nat. Cell Biol. 13:361–370. 10.1038/ncb2206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Fishel E., Bertroche T., and Dixit R.. 2013. Microtubule severing at crossover sites by katanin generates ordered cortical microtubule arrays in Arabidopsis. Curr. Biol. 23:2191–2195. 10.1016/j.cub.2013.09.018 [DOI] [PubMed] [Google Scholar]
- Zhao M., and Brunger A.T.. 2016. Recent advances in deciphering the structure and molecular mechanism of the AAA+ ATPase N-ethylmaleimide-sensitive factor (NSF). J. Mol. Biol. 428:1912–1926. 10.1016/j.jmb.2015.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]