Mitochondrial apoptotic priming predicts response to cancer chemotherapy, but the mechanisms underlying variability in this mitochondrial phenotype among closely related tumors are poorly understood. Ariës et al. show that PRC2 loss-of-function mutations induce resistance to mitochondrial apoptosis in T-ALL.
Abstract
The tendency of mitochondria to undergo or resist BCL2-controlled apoptosis (so-called mitochondrial priming) is a powerful predictor of response to cytotoxic chemotherapy. Fully exploiting this finding will require unraveling the molecular genetics underlying phenotypic variability in mitochondrial priming. Here, we report that mitochondrial apoptosis resistance in T cell acute lymphoblastic leukemia (T-ALL) is mediated by inactivation of polycomb repressive complex 2 (PRC2). In T-ALL clinical specimens, loss-of-function mutations of PRC2 core components (EZH2, EED, or SUZ12) were associated with mitochondrial apoptosis resistance. In T-ALL cells, PRC2 depletion induced resistance to apoptosis induction by multiple chemotherapeutics with distinct mechanisms of action. PRC2 loss induced apoptosis resistance via transcriptional up-regulation of the LIM domain transcription factor CRIP2 and downstream up-regulation of the mitochondrial chaperone TRAP1. These findings demonstrate the importance of mitochondrial apoptotic priming as a prognostic factor in T-ALL and implicate mitochondrial chaperone function as a molecular determinant of chemotherapy response.
Introduction
Conventional combination chemotherapy remains the primary treatment modality with curative potential for most patients with metastatic cancer. Effective chemotherapy regimens typically combine drugs with distinct mechanisms of action to minimize emergence of resistance. Nevertheless, treatment responses are often highly variable among patients with clinically indistinguishable tumors. For example, while intensive combination chemotherapy cures a substantial fraction of T cell acute lymphoblastic leukemia (T-ALL), front-line therapy fails in 15–25% of children and 40–50% of adults with the disease, and these patients have a very poor prognosis (Goldberg et al., 2003; Marks et al., 2009). Strikingly, chemotherapy resistance in T-ALL often manifests as induction failure (Goldberg et al., 2003; Oudot et al., 2008), in which the first cycle of intensive combination chemotherapy fails to induce disease remission. Despite considerable insights into the molecular mechanisms underlying cellular transformation, the pathobiology of resistance to conventional chemotherapy remains poorly understood.
Decreased proliferation rate provides one mechanism for chemotherapy resistance in some contexts. For example, culture of leukemic cells in nonproliferative conditions induces resistance to antimetabolites and vinca alkaloids (Schabel et al., 1965), and recent work has revealed that relapses can arise from rare hypoproliferative cells in ALL (Ebinger et al., 2016). However, several drugs with a central role in contemporary T-ALL therapy, such as asparaginase, corticosteroids, and DNA-damaging agents, remain highly active against hypoproliferative leukemic cells (Schabel et al., 1965; Mauer, 1975). Additionally, some studies of proliferation rate in pretreatment ALL clinical specimens have suggested that chemotherapy resistance is associated with increased, rather than decreased, proliferation (Scarffe et al., 1980; Dow et al., 1982). Thus, proliferation rate cannot fully explain primary chemotherapy resistance. Loss-of-function mutations of TP53 or CDNK2A have been shown to mediate chemotherapy resistance in model systems (Lowe et al., 1994; Schmitt et al., 1999). However, TP53 mutations are very rare at T-ALL diagnosis (Hsiao et al., 1994), whereas CDKN2A deletions are common but lack a consistent association with treatment failure (Rubnitz et al., 1997; Ramakers-van Woerden et al., 2001; Gutierrez et al., 2010). Drug resistance mutations are identified more commonly at relapse, including TP53 mutations and activating mutations of the NT5C2 nucleotidase that induce resistance to 6-mercaptopurine (Hsiao et al., 1994; Meyer et al., 2013; Tzoneva et al., 2013), but these are very rare in treatment-naive patients, indicating selection under evolutionary pressure. Thus, the molecular genetics underlying primary chemotherapy resistance in T-ALL remain poorly understood.
Pretreatment resistance to mitochondrial apoptosis is a cellular phenotype that predicts resistance to cytotoxic chemotherapy in a range of human cancers (Ni Chonghaile et al., 2011; Vo et al., 2012; Bhola et al., 2016), findings that we extend here to T-ALL. However, the molecular mechanisms underlying the striking phenotypic variability in chemotherapy response among patients with seemingly identical tumors remain poorly understood. Here, we show that loss-of-function mutations in any of three core components of polycomb repressive complex 2 (PRC2; EZH2, EED, or SUZ12) are associated with resistance to mitochondrial apoptosis. PRC2 depletion in human T-ALL cells induced resistance to mitochondrial apoptosis induction by multiple antileukemic drugs with distinct mechanisms of action, including dexamethasone, doxorubicin, and vincristine. PRC2 inactivation led to transcriptional up-regulation of the LIM domain transcription factor CRIP2 and downstream up-regulation of the TRAP1 gene, which encodes a mitochondrial chaperone protein of the HSP90 family (Felts et al., 2000; Kang et al., 2007). Importantly, we found that TRAP1 overexpression was necessary for induction of chemotherapy resistance downstream of PRC2 inactivation, and pharmacologic inhibition of TRAP1 synergized with dexamethasone and doxorubicin. These findings demonstrate the prognostic importance of mitochondrial apoptotic priming in T-ALL and implicate mitochondrial chaperone function as a key determinant of chemotherapy response.
Results
Mitochondrial apoptosis resistance predicts primary chemotherapy resistance in T-ALL
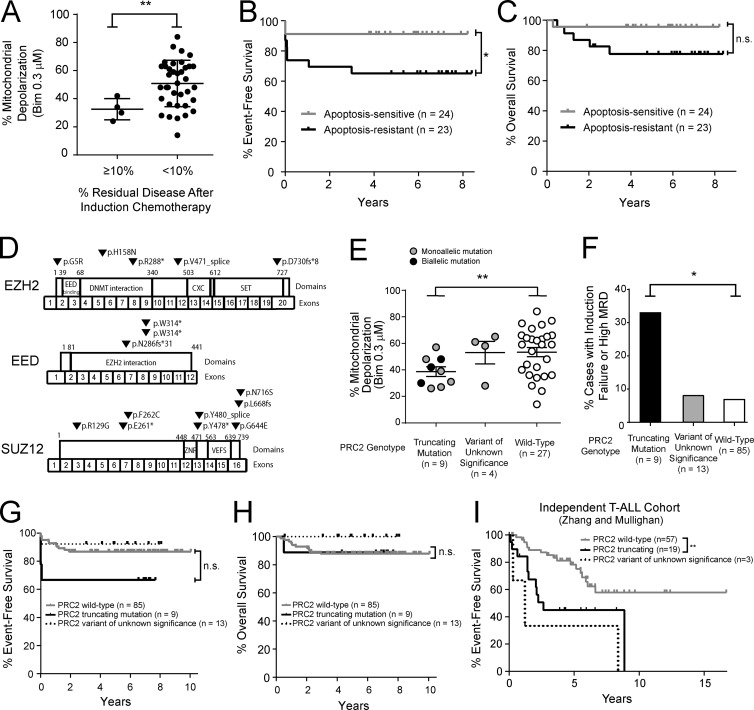
To investigate mechanisms underlying phenotypic variability in chemotherapy response, we focused on childhood T-ALL because combination chemotherapy is often curative, but treatment resistance commonly presents as failure of induction chemotherapy (Goldberg et al., 2003; Oudot et al., 2008). Induction failure, in which the first cycle of intensive combination chemotherapy fails to induce disease remission, strongly suggests primary or preexisting chemotherapy resistance. To test whether mitochondrial apoptosis resistance predicts T-ALL treatment failure, we analyzed a cohort of T-ALL specimens collected before the initiation of therapy in children treated on contemporary clinical trials (Table S1). BH3 profiling was performed to assess mitochondrial apoptotic priming, based on the ability of a fixed dose of pro-apoptotic peptide encoding the active site of BIM (also known as BCL2L11) to trigger loss of mitochondrial membrane potential (Ni Chonghaile et al., 2011). Resistance to mitochondrial apoptosis was associated with high levels of residual leukemia in the bone marrow at the end of this initial phase of chemotherapy (Fig. 1 A), based on the 10% cutoff that most robustly predicts outcome in a large cohort of childhood T-ALL (Wood et al., 2014). To assess whether mitochondrial apoptosis resistance predicts survival, we classified patients into apoptosis-sensitive or apoptosis-resistant groups based on whether they were above or below the median mitochondrial depolarization by BH3 profiling. Mitochondrial apoptosis resistance predicted significantly inferior event-free survival (65% versus 91% at 5 yr; P = 0.0376; Fig. 1 B), as well as a trend toward inferior overall survival that did not reach statistical significance (78% versus 96% at 5 yr; P = 0.091; Fig. 1 C). No other clinical features were significant predictors of mitochondrial apoptosis resistance in this cohort (Table S2).
Figure 1.
PRC2 mutations are associated with resistance to mitochondrial apoptosis in human T-ALL. (A) T-ALL blasts were collected before the initiation of chemotherapy from children treated on DFCI 05001 or COG AALL0434 clinical trials, and BH3 profiling analysis was performed to assess mitochondrial apoptotic priming, based on the degree of mitochondrial depolarization in response to 0.3 µM BIM peptide. Results were compared with the degree of residual leukemia in the bone marrow following the initial induction phase of combination chemotherapy. P = 0.008 by Welch t test. Number of samples per group: MRD ≥ 10%, n = 4; MRD < 10%, n = 37. Each data point represents percent mitochondrial depolarization in an independent patient sample. (B and C) Comparison of event-free survival (P = 0.0376 by log-rank test; B) and overall survival (P = 0.091 by log-rank test; C) among T-ALL cases classified as apoptosis sensitive or resistant based on whether mitochondrial depolarization was above or below the mean. Number of samples per group: apoptosis sensitive, n = 24; apoptosis resistant, n = 23. (D) Targeted exome sequencing and array CGH revealed mutations or deletions of EZH2, EED, or SUZ12 in 13 of 40 T-ALL cases analyzed by both BH3 profiling and sequencing analysis. (E) Association of PRC2 genotype with percent mitochondrial depolarization by BH3 profiling in primary T-ALL patient samples. Truncating mutations were defined as stop or frameshift mutations predicted to result in premature termination of translation or deletions identified by array CGH analysis. Variants of unknown significance are missense substitutions or splice region variants of unknown functional consequence. Each data point represents percent mitochondrial depolarization in an independent patient sample. Color of each circle reflects the allelic nature of the mutation in each sample (blank circle, WT); cases with two distinct heterozygous mutations of the same gene were presumed to have biallelic mutations. P = 0.007 by Welch t test for truncating mutations versus WT cases. The significance of other comparisons was not assessed. Number of samples per group: truncating mutation, n = 9; variant of unknown significance, n = 4; WT, n = 27. (F) Association of PRC2 mutation type with resistance to induction chemotherapy. P = 0.040 by Welch t test for truncating mutations versus WT cases. The significance of other comparisons was not assessed. Number of samples per group: truncating mutation, n = 9; variant of unknown significance, n = 13; WT, n = 85. (G and H) Comparison of event-free survival (P = 0.09 by log-rank test; G) and overall survival (P = 0.87 by log-rank test; H) among T-ALL cases by PRC2 mutation type. Note that the data shown in F–H include T-ALL cases analyzed by BH3 profiling and genomic analyses shown in A–E, as well as an additional cohort of T-ALL cases subjected to sequencing analysis only, on which BH3 profiling data were not available (see Table S1). Number of samples per group: WT, n = 85; truncating mutations, n = 9; variant of unknown significance, n = 13. (I) Validation of the association of PRC2 mutations with outcome in an independent cohort of T-ALL cases treated on St. Jude or Associazione Italiana di Ematologia e Oncologia Pediatrica clinical trials and subjected to sequencing and copy number analyses, as described (Zhang et al., 2012). P = 0.002 by log-rank test. Number of samples per group: WT, n = 57; truncating mutations, n = 19; variant of unknown significance, n = 3. *, P ≤ 0.05; **, P ≤ 0.01; n.s., P > 0.05.
PRC2 mutations are associated with resistance to mitochondrial apoptosis in T-ALL
Our findings further strengthen the link between priming of the mitochondrial apoptotic machinery and chemotherapy response in diverse human cancers (Ni Chonghaile et al., 2011; Vo et al., 2012), but the molecular genetics underlying phenotypic variability in mitochondrial priming remain poorly understood. To investigate underlying mechanisms, we began by applying targeted exome sequencing, array comparative genomic hybridization (CGH), and RNA sequencing analysis (RNA-seq) to all cases with sufficient available material (Tables S1 and S3–S6) The best-known regulators of mitochondrial apoptosis are genes of the BCL2 family (Davids and Letai, 2012; Czabotar et al., 2014), but RNA-seq revealed no difference in expression of any of the known pro- or antiapoptotic BCL2 family genes between apoptosis-sensitive versus resistant primary patient samples (Fig. S1 A). Prior work has implicated mutations of TP53, CDKN2A, PTEN, NOTCH1, or FBW7 in chemotherapy response (Lowe et al., 1994; Schmitt et al., 1999; Gutierrez et al., 2009; Mansour et al., 2009), but none of these were associated with mitochondrial apoptotic priming in this cohort (Fig. S1 B).
Resistance to mitochondrial apoptosis was associated with deletions or mutations of EZH2, EED, or SUZ12, which encode core components of PRC2. In the 40 T-ALL samples analyzed by both BH3 profiling and exon sequencing analysis, 13 (33%) harbored mutations or deletions of EZH2, EED, or SUZ12 (Fig. 1 D; Tables S1, S4, and S5). Nine of the PRC2 mutant cases harbored truncating mutations, which included seven cases with frameshift or stop mutations predicted to induce premature termination of translation and two cases with heterozygous deletions involving EED. An additional four cases harbored missense or splice region variants of unknown significance. PRC2 truncating mutations were associated with resistance to mitochondrial apoptosis, an effect that was similar in cases with monoallelic or biallelic PRC2 mutations (Fig. 1 E).
We then asked whether PRC2 mutations predict treatment response in T-ALL. For this, we included all T-ALL cases analyzed by sequencing analysis, which included 67 cases in addition to those analyzed by BH3 profiling (Table S1). In this larger cohort, 22 of 107 T-ALL cases (20.5%) harbored PRC2 mutations. This revealed that truncating PRC2 mutations were associated with a poor response to induction chemotherapy, with a trend toward inferior event-free survival that did not reach statistical significance (Fig. 1, F and G), but there was no difference in overall survival (Fig. 1 H). PRC2 mutations have been reported to be particularly common among T-ALL cases with the so-called early T cell precursor (ETP) immunophenotype (Zhang et al., 2012), which is linked to poor clinical outcome in some cohorts (Coustan-Smith et al., 2009; Gutierrez et al., 2010), but PRC2 mutations were more strongly linked to apoptosis resistance than ETP status (Table S2). To validate the association of PRC2 mutations with inferior clinical outcome in an independent cohort, we leveraged a separate cohort of T-ALL diagnostic clinical specimens previously subjected to genomic analysis (Zhang et al., 2012), which revealed that truncating PRC2 mutations were associated with inferior survival in this independent cohort (Fig. 1 I). The PRC2 complex has been implicated as a tumor suppressor in T-ALL (Ntziachristos et al., 2012; Simon et al., 2012; Zhang et al., 2012; Danis et al., 2016), but whether PRC2 regulates mitochondrial apoptosis is unknown.
PRC2 depletion induces resistance to mitochondrial apoptosis in T-ALL
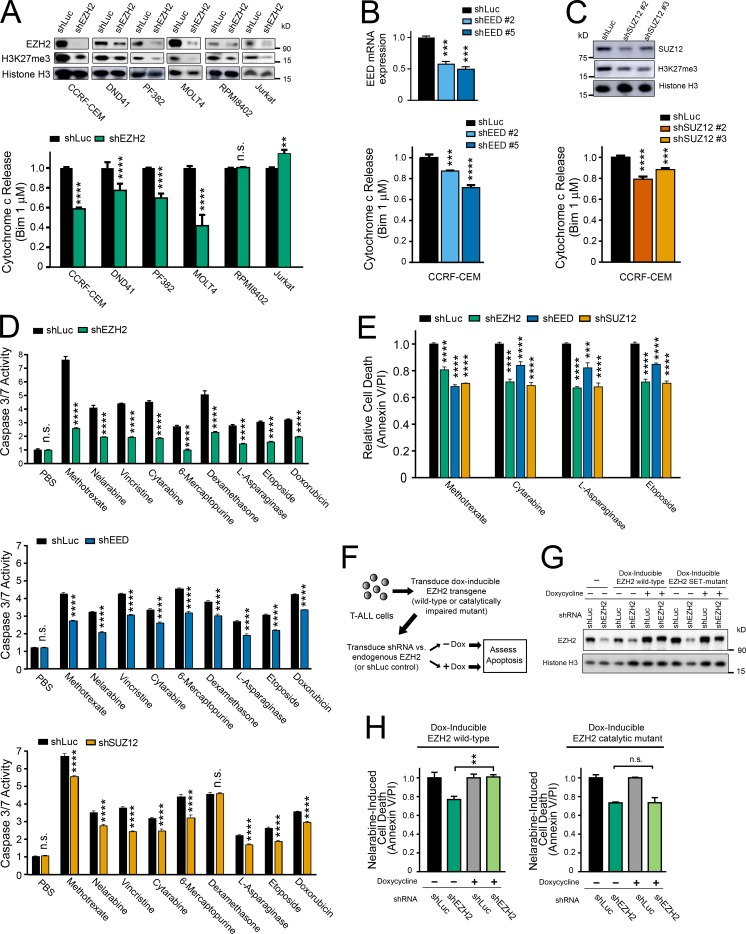
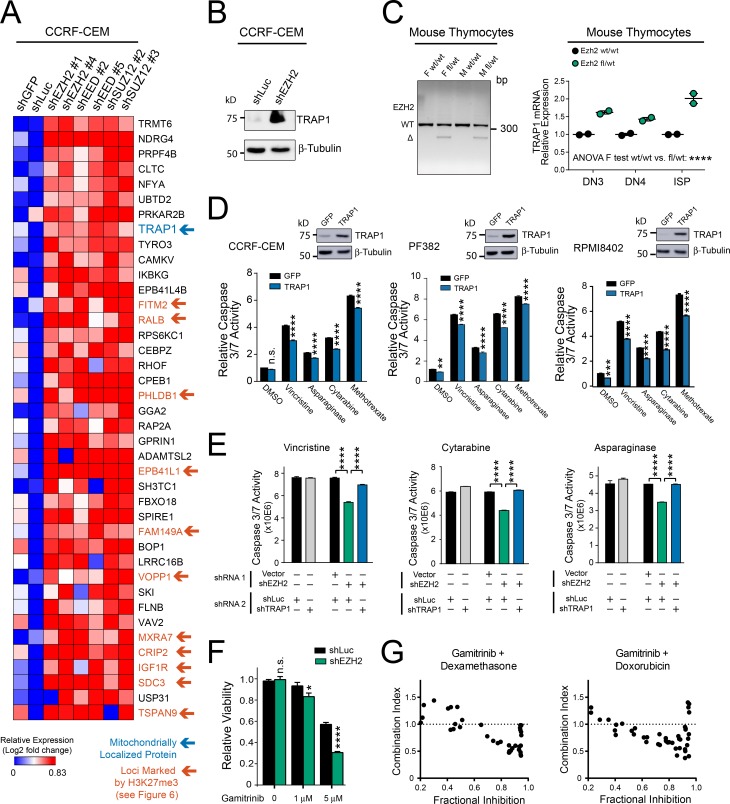
To test whether PRC2 regulates mitochondrial apoptosis in T-ALL, we began by depleting EZH2, a catalytic PRC2 subunit, from a panel of human T-ALL cell lines using RNA interference. BH3 profiling was then performed to assess the ability of BIM peptide to induce cytochrome c release, indicating mitochondrial outer membrane permeabilization. EZH2 depletion induced mitochondrial apoptosis resistance in most of the cell lines tested (Fig. 2 A). To confirm that this phenotype was PRC2-dependent, we depleted EED or SUZ12, which encode core PRC2 components and found that this phenocopied the effect of EZH2 depletion on mitochondrial priming (Fig. 2, B and C). To test whether PRC2 also regulates chemotherapy-induced apoptosis, we treated CCRF-CEM T-ALL cells depleted of PRC2 core components with a panel of chemotherapeutics that form the backbone of contemporary T-ALL therapy. PRC2 depletion induced resistance to apoptosis induction, as assessed by caspase 3/7 activity or by annexin V/propidium iodide staining, by a broad range of chemotherapeutics with distinct mechanisms of action (Fig. 2, D and E). Of note, EZH2 depletion had no significant effect on cell cycle distribution of these cells (data not shown) and induced resistance to drugs such as dexamethasone and asparaginase that are cytotoxic to both proliferating and nonproliferating leukemic cells (Mauer, 1975).
Figure 2.
PRC2 depletion induces resistance to chemotherapy-induced apoptosis in human T-ALL. (A) The indicated human T-ALL cell lines were transduced with shRNA targeting EZH2 or Luciferase control, and knockdown efficacy was assessed using Western blot analysis (top). FACS-based BH3 profiling was then performed to assess the degree of cytochrome c release following treatment with 1 µM BIM peptide for 30 min (bottom). BH3 profiling results were normalized to shLuc control for each cell line. P < 0.0001 for CCRF-CEM, P < 0.0001 for DND41, P < 0.0001 for PF382, P < 0.001 for MOLT4, P = 0.99 for RPMI8402, and P = 0.003 for Jurkat, as assessed using two-way ANOVA with a Sidak adjustment for multiple comparisons between shLuc and shEZH2 only for each cell line. (B and C) CCRF-CEM cells were transduced with shRNAs targeting EED, SUZ12, or Luciferase control, and knockdown efficacy was assessed using quantitative reverse transcription PCR (B, top) or Western blot analysis (C, top). BH3 profiling was then performed as in (A, bottom). Significance was assessed by one-way ANOVA with Tukey adjustment for multiple comparisons. P values for shLuc versus shEED no. 2 = 0.0012 and shLuc versus shEED no. 5 = 0.0004 in B; P values for shLuc versus shSUZ12 no. 2 < 0.0001; shLuc versus shSUZ12 no. 3 = 0.0009 in C. (D) CCRF-CEM cells were transduced with shRNAs targeting EZH2, EED, SUZ12, or Luciferase control, treated with vehicle (PBS) or each of the indicated chemotherapeutic agents, and apoptosis induction was assessed by caspase 3/7 activity assay. Results were normalized to vehicle-treated shLuc cells. Significance was assessed by two-way ANOVA with a Sidak adjustment for multiple comparisons between control or PRC2-targeting shRNA only for each drug. For shLuc versus shEZH2, P = 1.00 for PBS and < 0.0001 for all other drugs; for shLuc versus shEED, P = 0.99 for PBS and < 0.0001 for all other drugs; for shLuc versus shSUZ12, P = 0.98 for PBS, P = 0.54 for dexamethasone, and P < 0.0001 for all other drugs. (E) CCRF-CEM cells were transduced with the indicated shRNAs, treated with the indicated chemotherapeutic drugs, and cell death was assessed using annexin V and propidium iodide staining. Significance was assessed by two-way ANOVA with a Sidak adjustment for multiple comparison between shRNAs only for each drug. For all comparisons, P < 0.0001 except P = 0.0007 for shLuc versus shEED in asparaginase-treated conditions. (F) Schema of experimental design to rescue shEZH2-induced resistance to chemotherapy-induced apoptosis using doxycycline-inducible transgenes encoding WT EZH2, or a catalytically defective triple mutant of EZH2. The EZH2 shRNA used targets the 3′UTR of the endogenous gene, which is not present in the EZH2 transgenes used for rescue. (G) CCRF-CEM cells manipulated as shown in F were subjected to Western blot analysis for the indicated proteins. (H) CCRF-CEM cells experimentally manipulated as shown in F were treated with nelarabine for 48 h, and induction of apoptosis was assessed by caspase 3/7 activity assay. Significance was assessed by one-way ANOVA with Tukey adjustment for multiple comparisons. P = 0.01 for shEZH2 –dox versus shEZH2 +dox in cells transduced with doxycycline-inducible WT EZH2; P = 1.00 for shEZH2 −dox versus shEZH2 +dox in cells transduced with doxycycline-inducible mutant EZH2. All bar charts represent the mean ± SEM of at least n = 3 biological replicates from one representative experiment, and each experiment was repeated independently at least twice. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., P > 0.05.
To test whether regulation of mitochondrial apoptosis is dependent on the catalytic activity of PRC2, we tested whether shRNA depletion of endogenous EZH2 could be rescued by restoring expression of WT EZH2 or of an EZH2 mutant with impaired methyltransferase catalytic activity (Kim et al., 2015). Apoptosis resistance upon EZH2 depletion was rescued by expression of a WT EZH2 transgene that escapes shRNA targeting, but not by the methyltransferase-defective mutant (Fig. 2, F–H). We then asked whether apoptosis induction following EZH2 depletion might be mediated by EZH1, based on recent findings that transcriptional activation after EZH2 depletion may be mediated by noncanonical EZH1-containing PRC2 complexes (Xu et al., 2015). However, combined knockdown of both EZH1 and EZH2 revealed that EZH1 is not required for induction of apoptosis resistance following EZH2 inactivation (Fig. S2).
Depletion of the catalytic PRC2 subunit EZH2 induces chemotherapy resistance in T-ALL
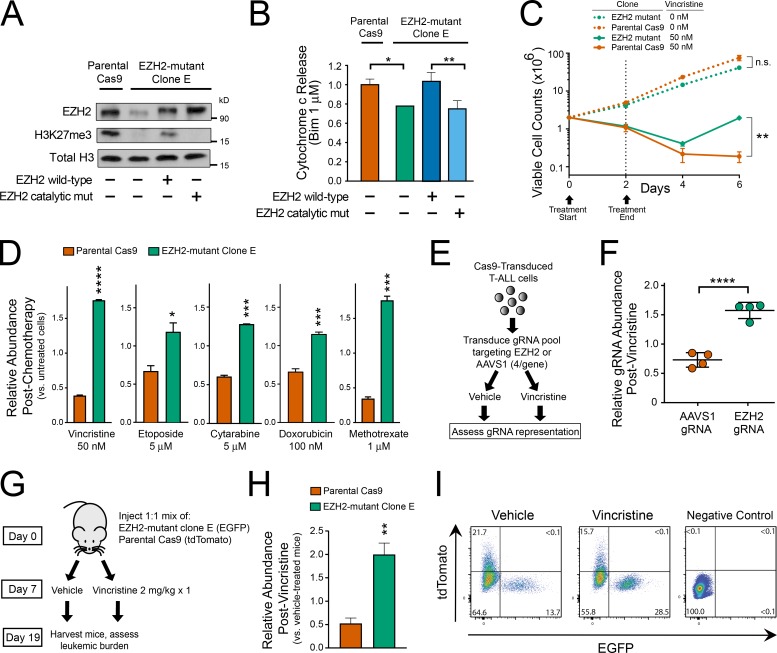
To test whether EZH2 depletion improves leukemic cell fitness upon chemotherapy treatment and to definitively rule out off-target shRNA effects, we then generated single-cell cloned isogenic EZH2-haploinsufficient or control T-ALL cells. Parental CCRF-CEM cells were first transduced with Cas9 and a guide RNA (gRNA) targeting exon 5 of EZH2, and single-cell cloning yielded an EZH2 mutant clone harboring mutations in three of the four alleles in this tetraploid cell line (Table S7), which we termed clone E. This EZH2-deficient clone exhibited low protein levels of EZH2 and of trimethylated histone H3 lysine 27 (the histone mark placed by EZH2) by Western blot analysis (Fig. 3 A), and resistance to mitochondrial apoptosis by BH3 profiling (Fig. 3 B), when compared with Cas9-transduced parental controls. Importantly, both of these effects were rescued by the transduction of WT EZH2, but not by a catalytically impaired EZH2 mutant (Fig. 3, A and B), indicating that they are caused by loss of EZH2 enzymatic activity. We then treated control (Cas9 parental) or EZH2 mutant clone E cells with a 2-d pulse of vincristine, followed by a subsequent release from chemotherapy. EZH2 mutant cells remained sensitive to vincristine-induced growth suppression, but had a significant survival advantage upon chemotherapy treatment, as indicated by their improved ability to grow out after release from vincristine (Fig. 3 C).
Figure 3.
EZH2 depletion induces chemotherapy resistance in T-ALL. (A) CCRF-CEM cells were transduced with Cas9 and EZH2 targeting gRNA, single-cell cloned, and next-generation sequencing was used to identify a clone with EZH2 haploinsufficiency (EZH2 mutant clone E). Western blot analysis was performed to assess expression of the indicated proteins in basal conditions or following transduction with doxycycline-induced constructs encoding WT EZH2 or a catalytically defective triple mutant. One representative experiment is shown, which was repeated independently. (B) BH3 profiling was performed on the cells shown in A. Results shown are the mean ± SEM of n = 3 biological replicates. Significance was assessed by one-way ANOVA with Tukey adjustment for multiple comparisons. P = 0.017 for parental Cas9 versus EZH2 mutant clone; P = 0.004 for clone E transduced with EZH2 WT versus EZH2 catalytic mutant. Bar charts represent the mean of three biological replicates, and the experiment was repeated independently. (C) EZH2 mutant clone E cells or their parental Cas9 controls were treated with the indicated doses of vincristine for 48 h and then released from chemotherapy. Viable cell counts were obtained by trypan blue exclusion at the indicated time points. Significance assessed by Welch t test (P = 0.07 for untreated EZH2 mutant versus untreated parental Cas9; P = 0.006 for vincristine-treated EZH2 mutant versus vincristine-treated parental Cas9). Results shown are the mean ± SEM of n = 3 biological replicates. Representative data of at least two independent experiments shown. (D) EGFP-transduced EZH2 mutant clone E cells were mixed at 1:1 ratio with tdTomato-transduced parental Cas9 controls. The resultant pool of cells was treated with vehicle control or the indicated chemotherapeutics for 48 h and subsequently released from chemotherapy. Relative abundance of each clone was assessed by flow cytometry analysis 4 d after chemotherapy release. Results are normalized to the abundance of each clone in nonchemotherapy-treated controls. Significance assessed by Welch t test, with P < 0.0001 for vincristine, P = 0.0281 for etoposide, P = 0.0003 for cytarabine, P = 0.0008 for doxorubicin, and P = 0.0004 for methotrexate. Results shown are the mean ± SEM of n = 3 biological replicates, and the experiment was repeated independently. (E) Experimental design to assess relative fitness of CCRF-CEM cells transduced with gRNAs targeting the catalytic domain of EZH2, or the AAVS1 safe-harbor genomic locus, in control or chemotherapy-treated conditions. (F) CCRF-CEM cells manipulated as shown in E were treated with 30 nM vincristine or vehicle for 48 h, released from vincristine for 12 d, and gRNA representation was assessed by next-generation sequencing. Relative abundance of each gRNA was normalized to its abundance in vehicle-treated controls. Significance assessed by Welch t test; P = 0.0001. (G) Experimental design to assess in vivo chemosensitivity of EZH2 mutant clone E cells or their parental Cas9 controls in NRG-immunodeficient mice. (H) FACS analysis of splenic cells harvested from mice after treatment as indicated in G. Each bar is the mean of five independent mice. Significance was assessed by Welch t test; P = 0.0026. (I) Representative FACS plots from mice analyzed in H. Control mice without fluorescent leukemia are shown as the negative control for setting FACS gates. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., P > 0.05.
We then asked whether the fitness advantage of EZH2 mutant cells remains in co-culture experiments, an experimental design that ensures identical experimental conditions. For this, we first transduced EZH2 mutant clone E cells with EGFP, and Cas9-transduced parental controls with tdTomato, sorted cells with successful expression of the fluorescent marker, and mixed these in a 1:1 ratio. These cells were then subjected to 48-h pulses of treatment with various chemotherapeutics, followed by a 4-d release, and relative abundance of each clone was assessed by flow cytometry. This revealed that EZH2 mutant clone E cells had significantly improved fitness following treatment with not only vincristine, but also with a broad range of chemotherapeutic drugs with distinct mechanisms of action (Fig. 3 D). To rule out the possibility that this effect was unique to this EZH2 mutant clone, we used an alternative experimental approach that avoided single-cell cloning. We first identified CRISPR/Cas9 gRNAs (gRNAs) targeting the catalytic SET domain of EZH2 or the AAVS1 safe-harbor locus (Sadelain et al., 2012; n = 4 gRNAs for each locus) and ensured that each of these gRNAs efficiently cut their target locus (data not shown). A pool of CCRF-CEM cells were then transduced with this pool of gRNAs and subsequently treated with a 48-h pulse of vincristine (or vehicle control), followed by a release from drug treatment (Fig. 3 E). Analysis of gRNA representation 14 d after the start of vincristine treatment revealed a significant enrichment of all of the EZH2-targeting gRNAs following vincristine treatment (Fig. 3 F).
To test the in vivo relevance of these findings, we injected mice with equal amounts of EZH2 mutant clone E cells labeled with EGFP, and Cas9-transduced control cells labeled with tdTomato. 7 d after injection, mice were treated with vehicle or vincristine (n = 5 mice per group; Fig. 3 G). Mice were harvested on day 19 after injection, and the relative abundance of EZH2 mutant versus parental clones was assessed by flow cytometry. This revealed that EZH2 mutant T-ALL cells had a significant in vivo fitness advantage following treatment with chemotherapy (Fig. 3, H and I). Collectively, these findings indicate that EZH2 inactivation induces chemotherapy resistance in T-ALL.
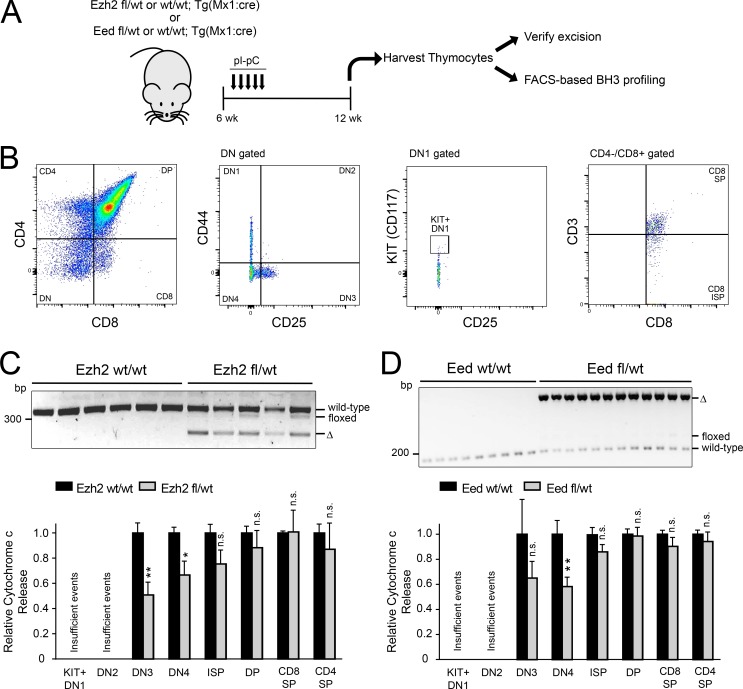
Haploinsufficiency for the core PRC2 components Ezh2 or Eed is sufficient to induce mitochondrial apoptosis resistance in mouse thymocytes
We then asked whether PRC2 regulates mitochondrial apoptosis during normal T cell development. Mx1:Cre transgenic mice, which were either WT, heterozygous, or homozygous for floxed Ezh2 or Eed alleles ((Shen et al., 2008; Neff et al., 2012), were treated with polyinosine-polycytosine acid (pIpC) to induce Cre activity in hematopoietic cells. Subsequently, thymocytes were harvested and stained for markers of T cell developmental stage, and mitochondrial apoptotic priming was assessed using BH3 profiling (Fig. 4, A and B). Haploinsufficiency for either Ezh2 or Eed was sufficient to induce apoptosis resistance in late double-negative thymocytes, but not at more mature stages of T cell development (Fig. 4, C and D). Ezh2 or Eed haploinsufficiency did not significantly alter thymocyte numbers at the stages of thymocyte development assessed (data not shown), but we were unable to perform BH3 profiling in Ezh2 or Eed homozygous knockout thymocytes because the thymus of these mice was markedly hypocellular and populated by thymocytes that escaped biallelic gene deletion (data not shown), suggesting that the previously described requirement for Ezh2 in normal T cell development is PRC2-dependent (Su et al., 2005; Simon et al., 2012).
Figure 4.
Haploinsufficiency for the core PRC2 components Ezh2 or Eed is sufficient to induce mitochondrial apoptosis resistance in mouse thymocytes. (A) Schema of experimental design. (B) FACS gating and sorting strategy to isolate thymocytes at the indicated developmental stages. Negative sorting was also performed to exclude cells expressing B cell, NK cell, red blood cell, or granulocytic markers. (C and D) Mx1:Cre transgenic, Ezh2 fl/WT (n = 5) or WT/WT (n = 6) siblings were treated with pIpC, and thymocytes were harvested as indicated in A and stained with a panel of antibodies to determine T cell developmental stage (B). PCR was performed on genomic DNA to verify appropriate deletion of Ezh2 (C, top). Stained thymocytes were subjected to BH3 profiling analysis using 1 µM BIM peptide for 25 min, and cytochrome c release was assessed by stage of T cell development using FACS analysis (C, bottom). Results are shown for all stages of T cell development that consistently yielded >250 FACS events for analysis. Significance was assessed by two-way ANOVA with Sidak adjustment for multiple comparisons between WT/WT versus fl/WT only for each stage of T cell development. P = 0.0066 for DN3, 0.049 for DN4, 0.44 for immature single positive (ISP), 0.96 for double positive (DP), 1.0 for CD8 SP, and 0.94 for CD4 SP. (D) Mx1:Cre transgenic, Eed fl/WT (n = 12) or WT/WT (n = 9) siblings were treated with pIpC, and thymocytes were harvested as indicated in A. PCR was performed to verify Eed deletion (D, top), and staining for T cell developmental markers and BH3 profiling analysis were performed as in D (bottom). Results are shown for all stages of T cell development that consistently yielded >250 FACS events for analysis. Significance was assessed by two-way ANOVA with Sidak adjustment for multiple comparisons between WT/WT versus fl/WT only for each stage of T cell development. P = 0.37 for DN3, 0.0017 for DN4, 0.76 for ISP, 1.0 for DP, 0.96 for CD8 SP, and 1.0 for CD4 single positive (SP). *, P ≤ 0.05; **, P ≤ 0.01; n.s., P > 0.05.
TRAP1 overexpression is necessary for induction of mitochondrial apoptosis resistance downstream of PRC2 inactivation
We then turned our attention to the mechanism linking PRC2 to the mitochondrial apoptotic machinery. PRC2 is best known as a chromatin-modifying complex whose activity is associated with transcriptional repression (Margueron and Reinberg, 2011). Thus, we began by investigating genes whose transcription is up-regulated upon depletion of PRC2, using RNA-seq of T-ALL cells depleted of PRC2 core components (Table S8). A recent study has shown that EZH2 loss induces chemotherapy resistance in acute myeloid leukemia in association with HOXB7 and HOXA9 up-regulation and undefined downstream consequences (Göllner et al., 2017), but PRC2 knockdown did not up-regulate expression of any HOXA or HOXB cluster gene in these cells (data not shown). PRC2 depletion had no consistent effects on transcription of any of the known pro- or antiapoptotic BCL2 family genes (Fig. S3), suggesting that PRC2 regulates mitochondrial priming via alternative apoptotic regulators.
PRC2 depletion induced apoptosis resistance as assessed by BH3 profiling, an assay performed after cytoplasmic membrane permeabilization and release of at least some soluble cytoplasmic proteins. Thus, we reasoned that PRC2 was likely to regulate mitochondrial apoptosis via factors intrinsic to, or stably associated with, the mitochondria. Analysis of the genes most significantly up-regulated following PRC2 depletion revealed a single transcript known to encode a mitochondrial protein, TRAP1 (Fig. 5 A). TRAP1 is a nuclearly encoded, mitochondrially localized chaperone of the HSP90 family (Felts et al., 2000; Kang et al., 2007). Using Western blot analysis, we confirmed that PRC2 depletion up-regulates TRAP1 protein expression (Fig. 5 B). Analysis of Trap1 mRNA expression in mouse Ezh2-haploinsufficient versus control thymocytes revealed increased Trap1 mRNA expression in Ezh2-haploinsufficient cells (Fig. 5 C), indicating that TRAP1 regulation by EZH2 is evolutionarily conserved.
Figure 5.
TRAP1 overexpression is necessary for induction of mitochondrial apoptosis resistance downstream of PRC2 inactivation. (A) CCRF-CEM human T-ALL cells were transduced with the indicated shRNAs, and RNA-seq was performed. Heat map depicts the genes most highly up-regulated following PRC2 depletion, based on DESEQ2 analysis for differentially expressed genes. Blue arrow denotes mitochondrial localized proteins; orange arrows denote genes whose loci are marked by H3K27me3 on ChIP-seq analysis (as investigated further in Fig. 6). (B) Western blot analysis for the indicated proteins in CCRF-CEM cells transduced with the indicated shRNA. The experiment was repeated twice, and a representative blot is shown. (C) Mx1:Cre transgenic, Ezh2 fl/WT or WT/WT siblings were treated with pIpC 4 wk after birth, and thymocytes were harvested 12 wk after birth. PCR was performed to verify excision of Ezh2 (left). Mouse thymocytes of the indicated developmental stages were FACS sorted and subjected to qRT-PCR analysis for Gapdh and Trap1 (right). Data points represent the mean expression of individual mice, with two mice analyzed per genotype. Significance was assessed by two-way ANOVA including main effect terms (using an F test) for Ezh2 genotype and T cell developmental stage (P = 0.0001). No interaction term was included in the model, so pairwise comparisons were not assessed. (D) The indicated human T-ALL cells were transduced with doxycycline-inducible, neomycin-resistant constructs encoding GFP or TRAP1. Western blot analysis was performed to assess expression of the indicated proteins (top) 72 h after the start of doxycycline treatment. At that time point, cells were treated with the indicated chemotherapeutics for 48 h, and apoptosis induction was assessed using a caspase 3/7 activity assay. Significance was assessed by two-way ANOVA with a Sidak adjustment for multiple comparisons between GFP and TRAP1 only for each treatment. For CCRF-CEM cells, P = 0.25 for DMSO and P < 0.0001 for all drugs. For PF382, P = 0.0048 for DMSO and P < 0.0001 for all other drugs. For RPMI 8402, P = 0.0002 for DMSO and P < 0.0001 for all other drugs. Results shown are the mean ± SEM of n = 3 biological replicates of a representative experiment, all of which were repeated at least twice. (E) CCRF-CEM cells were transduced with a neomycin-resistant “shRNA 1” targeting EZH2 or empty vector control and a puromycin-resistant “shRNA 2” targeting either TRAP1 or Luciferase control. Following antibiotic selection, cells were treated with the indicated chemotherapeutics for 48 h, and apoptosis induction was assessed using a caspase 3/7 activity assay. Differences were assessed by two-way ANOVA analysis, with Tukey adjustment for multiple comparisons. In all three drug conditions, P < 0.0001 for the comparison of shVector-shLuc versus shEZH2-shLuc–transduced cells, and P < 0.0001 for shEZH2-shLuc versus shEZH2-shTRAP1–transduced cells. Results shown are the mean ± SEM of n = 3 biological replicates from a representative experiment, all of which were repeated at least twice. (F) CCRF-CEM cells were transduced with the indicated shRNAs, treated with gamitrinib at the indicated doses, and viability was assessed using Cell TiterGlo. Differences were assessed by two-way ANOVA analysis with Sidak adjustment for multiple comparisons between shLuc and shGFP only for each gamitrinib dose. P = 0.97 for DMSO, P = 0.023 at 1 µM, and P < 0.0001 at 5 µM. Results shown are the mean ± SEM of n = 3 biological replicates from a representative experiment, which was repeated independently. (G) CCRF-CEM cells were transduced with shEZH2 no. 1, treated with gamitrinib in combination with either dexamethasone or doxorubicin at a range of doses, and viability was assessed using Cell TiterGlo. Synergy was assessed using combination index analysis. Data are shown for dose combinations with fractional inhibition of 0.2–0.95. Note synergistic interactions (combination index < 0.7) at dose combinations approaching maximal efficacy, as assessed by fractional inhibition of cell viability. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., P > 0.05.
We then asked whether TRAP1 overexpression is sufficient to induce resistance to chemotherapy-induced apoptosis in cells with physiological PRC2 activity. Transduction of TRAP1 into human T-ALL cell lines induced partial resistance to chemotherapy-induced apoptosis (Fig. 5 D). To test whether TRAP1 overexpression is required for mitochondrial apoptosis resistance following PRC2 inactivation, we used RNA interference to deplete either EZH2 or control, in combination with depletion of TRAP1 or shLuciferase control. Depletion of TRAP1 completely blocked the emergence of apoptosis resistance following EZH2 knockdown (Fig. 5 E and Fig. S4). We also took advantage of gamitrinib, an ATP-competitive small molecule inhibitor of HSP90-family chaperones that inhibits mitochondrial TRAP1 and HSP90 function by virtue of its specific mitochondrial localization (Kang et al., 2009). While gamitrinib had some toxicity to control T-ALL cells, EZH2-depleted cells demonstrated increased sensitivity to this drug (Fig. 5 F), indicating that PRC2 deficiency imparts an increased dependence on mitochondrial chaperone function. Furthermore, gamitrinib had synergistic antileukemic activity in combination with dexamethasone or doxorubicin at highly effective dose combinations (Fig. 5 G).
PRC2 induces TRAP1 expression and mitochondrial apoptosis resistance via CRIP2
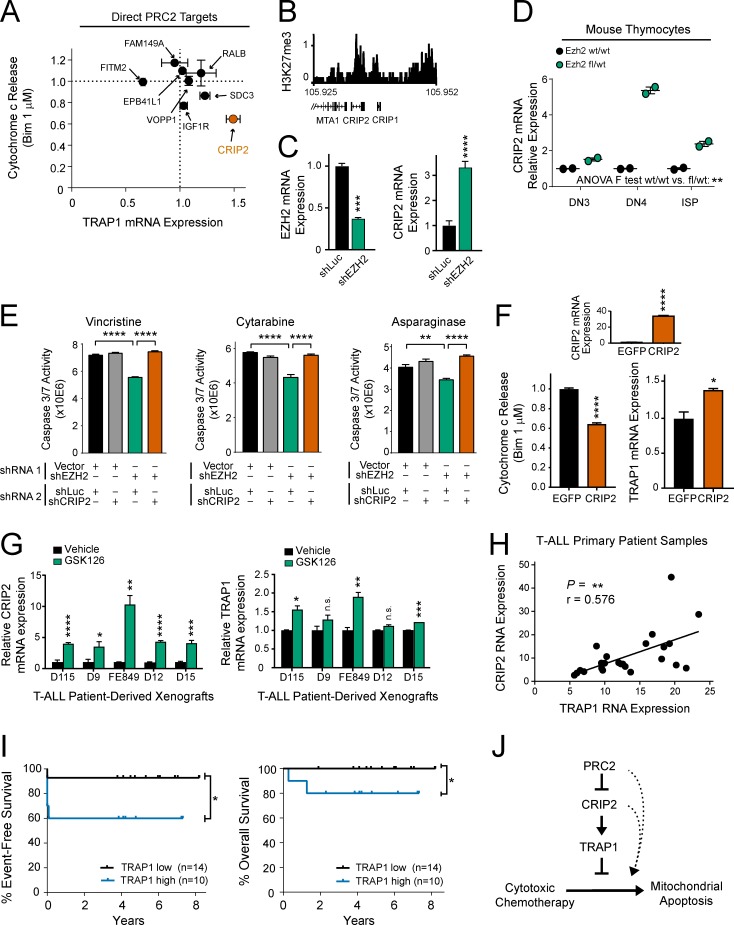
To define how PRC2 regulates TRAP1 expression, we performed chromatin immunoprecipitation-sequencing (ChIP-seq) analysis for the histone mark placed by PRC2, trimethylated histone H3 lysine 27 (H3K27me3), in CCRF-CEM cells (Table S9). We noted that there were no significant H3K27me3 peaks detected in CCRF-CEM cells following EZH2 knockdown (data not shown), indicating that EZH2 is the primary H3K27me3 methyltransferase in these cells. Analysis of ChIP-seq in PRC2 proficient CCRF-CEM cells revealed no detectable H3K27me3 at the TRAP1 locus (data not shown), indicating that regulation of TRAP1 by PRC2 is indirect. To identify the relevant direct target(s) of PRC2 that regulate TRAP1 expression and apoptosis resistance, we first combined ChIP-seq and RNA-seq analysis to define the direct canonical transcriptional targets of PRC2, defined as genes whose loci were marked by H3K27me3 at baseline and whose RNA expression increased following PRC2 depletion (Fig. 5 A). We then transduced each of these genes individually into T-ALL cells, to define those that regulate TRAP1 expression and mitochondrial apoptosis resistance using quantitative reverse transcription PCR (qRT-PCR) and BH3 profiling analysis, respectively. We found that CRIP2 was the direct PRC2 target whose up-regulation most strongly induced apoptosis resistance and TRAP1 expression (Fig. 6, A and B).
Figure 6.
PRC2 regulates TRAP1 overexpression via CRIP2. (A) Direct canonical transcriptional targets of PRC2, defined as genes whose loci are marked by H3K27me3 at baseline and whose transcription is activated following PRC2 depletion, were transduced individually into human CCRF-CEM T-ALL cells. BH3 profiling (1 µM BIM peptide for 25 min) and qRT-PCR analysis were used to assess induction of apoptosis resistance and TRAP1 mRNA expression, respectively, and each is shown normalized to levels in EGFP-transduced control cells. Note that overexpression of the direct PRC2 targets PHLDB1, MXRA7, and TSPAN9 was toxic to CCRF-CEM cells; thus, these targets were not investigated further. Results shown are the mean ± SEM of n = 3 biological replicates. (B) ChIP-seq for H3K27me3 at the CRIP2 locus in PRC2-proficient CCRF-CEM cells. (C) CCRF-CEM cells were transduced with the indicated shRNAs and subjected to qRT-PCR analysis for expression of the indicated genes. P values calculated by Welch t test (EZH2 mRNA, P = 0.0004; CRIP2 mRNA, P < 0.0001). Results shown are the mean ± SEM of n = 3 biological replicates from one representative experiment, which was repeated twice. (D) Mx1:Cre transgenic, Ezh2 WT/WT or fl/WT mice were treated with pIpC 4 wk after birth, and thymocytes were harvested 12 wk after birth. Thymocytes of the indicated developmental stages were FACS sorted and subjected to qRT-PCR analysis for Gapdh and Crip2 RNA expression. Results are shown from individual mice from n = 2 mice per group. Significance was assessed by two-way ANOVA including main effect terms (using an F test) for Ezh2 genotype and T cell developmental stage (P = 0.008). No interaction term was included in the model, so pairwise comparisons were not assessed. (E) CCRF-CEM cells were transduced with a neomycin-resistant “shRNA 1” targeting EZH2 or empty vector control and a puromycin-resistant “shRNA 2” targeting either CRIP2 or Luciferase control. Cells were then treated with the indicated chemotherapeutic drugs, and effects on apoptosis were assessed by caspase 3/7 activity. Differences were assessed by two-way ANOVA analysis, with Tukey adjustment for multiple comparisons. In vincristine- and cytarabine-treated cells, P < 0.0001 for shVector-shLuc versus shEZH2-shLuc and shEZH2-shLuc versus shEZH2-shCRIP2–transduced cells. In asparaginase-treated cells, P < 0.0036 for shVector-shLuc versus shEZH2-shLuc, and P < 0.0001 shEZH2-shLuc versus shEZH2-shCRIP2–transduced cells. Results shown are the mean ± SEM of n = 3 biological replicates from one representative experiment, which was repeated independently. (F) CCRF-CEM cells were transduced with CRIP2 or EGFP. qRT-PCR analysis was performed to assess expression of CRIP2 mRNA (top, P < 0.0001 by Welch t test) or TRAP1 mRNA (bottom right, P = 0.027 by Welch t test). Effects on mitochondrial apoptosis were assessed by BH3 profiling using 1 µM BIM peptide for 25 min (bottom, P < 0.0001 by Welch t test). Results shown are the mean ± SEM of n = 3 biological replicates from one representative experiment, which was repeated independently. (G) T-ALL cells from primary patients were expanded in immunodeficient mice, harvested, treated in short-term culture with vehicle or 1 µM GSK126 for 6 d, and qRT-PCR analysis was performed for the indicated genes. P values calculated by Welch t test. For CRIP2, P = 0.0001 for D115, P = 0.031 for D9, P = 0.0015 for FE849, P < 0.0001 for D12, and P = 0.0007 for D15. For TRAP1, P = 0.033 for D115, P = 0.16 for D9, P = 0.006 for FE849, P = 0.08 for D12, and P = 0.0001 for D15. Results shown are the mean ± SEM of n = 3 biological replicates from one representative experiment, which was repeated independently. (H) Correlation of CRIP2 and TRAP1 mRNA expression in RNA-seq data from primary T-ALL patient samples. Pearson correlation coefficient (r = 0.576) reported P value (P = 0.003) indicating a significant positive correlation between CRIP2 and TRAP1 RNA expression. (I) Comparison of event-free survival and overall survival among T-ALL cases on DFCI 05001 or COG AALL0434 clinical trials classified as TRAP1 high or low expressing based on whether RNA-seq TRAP1 expression was above or below the median. A one-sided log-rank test was performed due to the hypothesized direction of event-free survival (P = 0.028) and overall survival (P = 0.043) differences. Number of samples per group: TRAP1 low, n = 14; TRAP1 high, n = 10. (J) Proposed model to explain regulation of chemotherapy-induced apoptosis by PRC2. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., P > 0.05.
CRIP2 encodes a LIM (named after the LIN-11, ISL-1, and MEC-3 proteins in Caenorhabditis elegans)-only domain protein that has been implicated as a transcriptional regulator of NF-κB signaling in nasopharyngeal carcinoma (Cheung et al., 2011). We confirmed that EZH2 depletion induced expression of CRIP2 in T-ALL cells (Fig. 6 C) and that Ezh2 haploinsufficiency induced Crip2 expression in nontransformed mouse T cell progenitors (Fig. 6 D). To test whether Crip2 up-regulation is necessary for induction of apoptosis resistance following Ezh2 inactivation, we combined transduction of one shRNA, targeting either EZH2 or empty vector control, with a second shRNA, targeting either CRIP2 or shLuciferase control. CRIP2 knockdown completely prevented the ability of EZH2 depletion to induce resistance to several chemotherapeutics with distinct mechanisms of action (Fig. 6 E and Fig. S5). BH3 profiling and qRT-PCR analysis of T-ALL cells transduced with EGFP or CRIP2 revealed that CRIP2 overexpression is sufficient to induce TRAP1 mRNA up-regulation and resistance to mitochondrial apoptosis (Fig. 6 F). In a panel of primary T-ALL cells that have been expanded in immunocompromised mice, treatment with the EZH2 inhibitor GSK126 induced transcriptional regulation of both CRIP2 and TRAP1 (Fig. 6 G). Moreover, RNA-seq analysis of primary T-ALL patient samples revealed that expression of CRIP2 and TRAP1 were strongly positively correlated (Fig. 6 H). We then asked whether TRAP1 overexpression predicts chemotherapy failure in childhood T-ALL. We classified patients into TRAP1 high or low groups based on mean TRAP1 expression in pretreatment T-ALL lymphoblast specimens and compared survival among these groups. TRAP1 up-regulation predicted inferior event-free and overall survival in children with T-ALL treated on recent clinical trials (Fig. 6 I). Collectively, these findings support a model in which loss of PRC2 induces transcriptional up-regulation of its direct target CRIP2, which subsequently activates expression of the mitochondrial chaperone TRAP1, leading to resistance to chemotherapy-induced mitochondrial apoptosis (Fig. 6 J).
Discussion
The studies described here demonstrate that PRC2 genotype provides one molecular explanation for phenotypic variability in mitochondrial apoptotic priming between chemotherapy-sensitive versus resistant T-ALL cell populations. Thus, PRC2 function is a key molecular determinant of treatment response in T-ALL. PRC2 haploinsufficiency was sufficient to induce apoptosis resistance in nontransformed T cell progenitors, suggesting that chemotherapy resistance can evolve before oncogenic transformation. These findings thus provide a model in which the mutational path to T cell transformation, which evolves in the absence of chemotherapy-induced selective pressure, can nevertheless result in the formation of a highly chemoresistant leukemic clone if the genetic lesions acquired result in high-level resistance to mitochondrial apoptosis.
We found that PRC2 mutations predict early treatment failure in independent cohorts of childhood T-ALL, in line with the previously reported association of EZH2 inactivation with treatment failure in T-ALL (Zhang et al., 2012) and in acute myeloid leukemia (Göllner et al., 2017). Of note, this association was not observed in a recent whole genome–sequencing study of T-ALL (Liu et al., 2017), but cases of primary chemotherapy resistance appear to have been excluded from this study because their post-chemotherapy specimens harbored leukemia, and there was no alternative source of germline DNA available for the whole genome–sequencing approach applied. It will be of considerable interest to prospectively validate the ability of PRC2 mutations to predict primary chemotherapy resistance in future clinical trials.
Deletion of PRC2 genes induced apoptosis resistance at late double-negative stages of T cell development, but not in more mature T cell progenitors. PRC2 function can be highly context dependent for reasons that are not well understood, even in seemingly related cell types. For example, while EZH2 is a tumor suppressor in immature lymphocytes of the T cell lineage (Simon et al., 2012; Danis et al., 2016), it functions as a driver oncogene in B-lineage lymphocytes (Souroullas et al., 2016). In T cell progenitors, we note that the switch from PRC2-dependent to PRC2-independent regulation of mitochondrial apoptosis occurs in association with profound changes in regulation of apoptosis by TCR signaling. Thus, one speculative possibility is that loss of PRC2-dependent transcriptional repression may potentiate preTCR-driven survival signaling in double-negative T cell progenitors, where this triggers antiapoptotic signals. As these cells mature, PRC2 loss may cease to be antiapoptotic as part of the switch to negative selection, where TCR signaling becomes pro-apoptotic (Klein et al., 2014).
Our data do not rule out a role for differential induction of BCL2 family proteins and mitochondrial apoptotic priming by chemotherapeutics as an additional determinant of treatment response. However, our findings implicate TRAP1 up-regulation as one important mechanism linking PRC2 loss to induction of mitochondrial apoptosis resistance in T-ALL. Indeed, TRAP1 was required for the induction of resistance to chemotherapy-induced apoptosis upon PRC2 depletion, and pharmacologic TRAP1 inhibition synergized with dexamethasone and doxorubicin, antileukemic drugs with distinct mechanisms of action. TRAP1 is an HSP90 family chaperone that localizes to the mitochondrial inner membrane and inter-membrane space (Pridgeon et al., 2007). TRAP1 inhibits a nonapoptotic, caspase-independent form of cell death termed mitochondrial permeability transition-driven necrosis (Kang et al., 2007) and has also been shown to inhibit some features of mitochondrial apoptosis (Pridgeon et al., 2007; Costantino et al., 2009). While mitochondrial permeability transition-driven necrosis and BCL2-regulated mitochondrial apoptosis are distinct cell death pathways (Baines et al., 2005; Nakagawa et al., 2005; Galluzzi et al., 2015), these do share some cellular machinery (Karch et al., 2013), and it will be of interest to define precisely how TRAP1 regulates mitochondrial apoptosis.
The broad range of cytotoxic chemotherapeutics used for T-ALL therapy are used in combination because their distinct mechanisms of action impair emergence of resistance. However, our findings suggest that their antileukemic activity nevertheless converges on a common downstream pathway, mitochondrial apoptosis, whose impairment is sufficient to simultaneously induce resistance to all chemotherapeutics that we tested. Our data implicate up-regulation of the LIM domain transcription factor CRIP2, and downstream activation of TRAP1 chaperone functions as a key mediator of apoptosis resistance and clinical response to cytotoxic chemotherapy. These findings suggest that the clinical development of TRAP1 inhibitors may provide a therapeutic strategy to specifically reverse apoptosis resistance and improve clinical outcomes for patients with high-risk subsets of T-ALL.
Materials and methods
Patient samples
113 diagnostic T-ALL samples were obtained from the bone marrow or peripheral blood of children before the initiation of chemotherapy from children with newly diagnosed T-ALL enrolled on Dana-Farber Cancer Institute (DFCI) Study 05-001 (Place et al., 2015) or Children’s Oncology Group (COG) Study AALL0434. Of these, 47 had a suitable viably frozen specimen for BH3 profiling, and the remainder were included in the sequencing analyses only (Table S1). Samples were purified using Ficoll-Paque reagent before viable freezing or genomic DNA extraction. Viably frozen samples were stored in liquid nitrogen. All samples were collected with informed consent and Institutional Review Board (IRB) approval of the respective institutions, in accordance with the Declaration of Helsinki. End-induction minimal residual disease analysis was assessed by flow cytometry (COG AALL0434) or PCR analysis for T cell receptor gene rearrangements (DFCI 05001) per the clinical standard on each protocol. The percent bone marrow blast cutoff used to define induction failure was different between these two clinical trials (≥5% on DFCI 05-001 or ≥ 25% in COG AALL0434), therefore protocol-defined induction failure or ≥5% bone marrow blasts were considered equivalent for the purposes of assessing the effect of PRC2 genotype on response to induction chemotherapy.
BH3 profiling of primary T-ALL patient samples
The sequence of the peptide mimetics of the BIM BH3 domain and inert PUMA2A negative control, as well as methods of synthesis, were as previously described (Ni Chonghaile et al., 2011). BH3 profiling of 47 primary patient samples was performed using the plate-based fluorimetry method to assess effects of BIM BH3 peptide or PUMA2A negative control on mitochondrial membrane potential, as previously described (Chonghaile et al., 2014). In brief, BH3 peptides at 70 μM/liter were plated in triplicate on a black 384-well plate. Cells were gently permeabilized with 0.005% digitonin and loaded with the fluorescent mitochondrial dye JC-1. The cells were plated on top of the peptides at 2.5 × 10E4 cells per well. The loss of mitochondrial potential was measured on the Tecan Saffire2 at an excitation of 545 nm and an emission of 590 nm over 3 h. The percentage of mitochondrial depolarization for the peptides was calculated by normalization to the solvent-only control DMSO (0%) and the positive control carbonyl cyanide-4-(trifluoromethoxy)phenyl hydrazone (FCCP; 100%) at 60 min.
Targeted exome sequencing of T-ALL patient samples
T-ALL diagnostic specimens were purified using Ficoll-Paque reagent and viably frozen. Genomic DNA (gDNA) was extracted using the AllPrep DNA/RNA mini kit (OPv1 cohort; Qiagen) or the DNeasy kit (OPv3mod cohort; Qiagen) according to the manufacturer’s instructions. Targeted exon sequencing was performed at the Center for Cancer Genome Discovery at the DFCI using an Illumina sequencing platform for all protein-coding exons of the genes shown in Table S3. In brief, 200 ng of double-stranded gDNA was fragmented to an average of 150 bp or 250 bp using Covaris ultrasonication (LE220 Focused-ultrasonicator; Covaris). Fragmented DNA was purified using Agencourt AMPure XP beads (Beckman Coulter). Size-selected DNA was then ligated to sequencing adaptors using sample-specific barcodes, and libraries were constructed (SPRIworks HT; Beckman Coulter) and quantified using qPCR (Kapa Biosystems) or MiSeq (Illumina, Inc.). For targeted exon enrichment, libraries were pooled in equal mass to a total of 500 ng, and regions of interest were captured using custom-designed baits (SureSelect Target Enrichment system; Agilent Technologies). All captures were sequenced on the HiSeq 2500 platform (Illumina, Inc.) in Rapid Run Mode.
Pooled sample reads were deconvoluted and sorted using Picard tools. Reads were aligned to the reference sequence b37 edition from the Human Genome Reference Consortium using Burrows-Wheeler Aligner using parameters “-q, 5; -l, 32; -k, 2; -o, 1.” Duplicate reads were removed using the Picard tools (Li and Durbin, 2009). The alignments were further refined using the Genome Analysis Toolkit for localized realignment around indel sites. Recalibration of the quality scores was also performed using Genome Analysis Toolkit tools (McKenna et al., 2010; DePristo et al., 2011).
Mutation analysis for single-nucleotide variants was performed using MuTect v1.1.4 (Cibulskis et al., 2013) and annotated by Oncotator or Variant Effect Predictor (McLaren et al., 2010). MuTect was run in single or paired mode using internal control Centre d’Etude du Polymorphisme Humain as the “matched” normal in paired mode. We used the SomaticIndelDetector tool that is part of the Genome Analysis Toolkit for indel calling, as described previously (Abedalthagafi et al., 2014). Mutation calls were made for those variants predicted to result in a nonsynonymous amino acid alteration, frameshift mutation, stop codon, or to alter a splice site and for variants with at least 10 reads of the mutant allele.
Germline variant filters were applied. Variants were filtered against the 6,500-exome release of the Exome Sequencing Project database and the Genome Aggregation Database (gnomAD v.2.0). Variants represented in either database at >0.1% frequency were excluded from further analysis. Catalogue of Somatic Mutations in Cancer (COSMIC v.80) annotations were added to the variant calls, if available. Sequencing was performed in two cohorts (OPv1 and OPv3mod) for the genes indicated in Table S3. In the OPv1 cohort, EED (NM_003797.3) was sequenced by Sanger sequencing at Genewiz, because this gene was not included in the OPv1 sequencing platform used. These data are from primary patient samples collected before the 2015 update of the National Institutes of Health (NIH) Genomic Data Sharing policy, and the informed consent obtained from these patients only allows their data to be deposited as “Controlled Access” data behind a dbGap firewall. Users with appropriate IRB and institutional protections can access the data. Data from targeted exon sequencing from primary T-ALL patient samples is available in the dbGap controlled-access database, study ID phs001513.
RNA-seq of T-ALL patient samples
RNA was extracted from T-ALL patient samples using the AllPrep DNA/RNA Mini kit (Qiagen) according to the manufacturer’s instructions. RNA samples were then treated with Ambion Turbo DNase (Thermo Fisher Scientific), and DNA contamination was confirmed to be <10% for all samples. RNA quantity was determined using the Qubit RNA Assay kit (Thermo Fisher Scientific), and RNA quality was determined on an Agilent Bioanalyzer using the RNA Pico kit (Agilent). Using the NEBNext Ultra RNA Library Prep kit for Illumina (New England Biolabs), 50–100 ng of total RNA was converted into a DNA library following the manufacturer’s protocol. Following library construction, DNA libraries were then put through quality control. Library quantity was determined using the Qubit High Sensitivity DNA kit (Thermo Fisher Scientific), and library size was determined using the Bioanalyzer High Sensitivity Chip kit (Agilent). Finally, libraries were put through qRT-PCR using the Universal Library Quantification kit for Illumina (Kapa Biosystems) and run on the 7900HT Fast Real-Time PCR machine (Applied Biosystems). Libraries passing quality control were diluted to 2 nM using sterile water and then sequenced on the HiSeq 2000 (Illumina) at a final concentration of 12 pM on a single-read flowcell with 50 sequencing cycles, following all manufacturer protocols. Alignments were performed with STAR aligner (version 2.3.1z4) against the hg19 w ERCC92 genome (ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/dna/). Cufflinks package was used to generate fragments per kilobase of transcript per million mapped reads values. These data are from primary patient samples collected before the 2015 update of the NIH Genomic Data Sharing policy, and the informed consent obtained from these patients only allows their data to be deposited as “Controlled Access” data behind a dbGap firewall. Users with appropriate IRB and institutional protections can access the data. Data from RNA-seq from primary T-ALL patient samples is available in the dbGap controlled-access database, study ID phs001513.
Microarray-based CGH analysis of T-ALL patient samples
T-ALL patient samples were profiled for DNA copy number analysis on SurePrint G3 Human 4×180K CGH Microarrays. Patient and control gDNAs were labeled with Cy3 and Cy5 dyes (PerkinElmer), and hybridization was performed according to the manufacturer’s instructions (Agilent Technologies), followed by data analysis using the arrayCGHbase tool (Menten et al., 2005). Segmentation was performed with the BioConductor DNAcopy package, as previously described. Log2 copy number ratio for heterozygous deletion was defined as −0.5 to −1.5 (corresponding to 35–70% of normal copy number), and log2 copy number ratio for homozygous deletion was defined as less than −1.5 (corresponding to <35% of normal copy number). Array CGH data are available in the NCBI Gene Expression Omnibus as GSE96624.
Cell lines and cell culture
Cell lines were obtained from ATCC or DSMZ, as follows: CCRF-CEM, ATCC cat no. CCL-119; DND41, DSMZ cat no. ACC-525; PF382, DSMZ cat no. ACC-38; MOLT4, ATCC cat no. CRL-1582; RPMI 8402, DSMZ cat no. ACC-290; Jurkat, ATCC cat no. TIB-152; and 293T, ATCC cat no. CRL-11268. Cell lines were cultured in DMEM or RPMI 1640 (Thermo Fisher Scientific) with 10% FBS (Sigma-Aldrich) or tetracycline system–approved FBS (Clontech) and 1% penicillin/streptomycin (Thermo Fisher Scientific) at 37°C and 5% CO2. Cell line identities were validated using short tandem repeats profiling at the DFCI Institute Molecular Diagnostics Laboratory, and mycoplasma contamination was excluded using the MycoAlert Mycoplasma Detection kit (Lonza), most recently in April 2018.
RPMI-8402 is listed in the database of commonly misidentified cell lines reported by the international cell line authentication committee. However, short tandem repeat testing verified RPMI-8402, and this cell line harbors a deletion immediately upstream of TAL1 resulting in its aberrant overexpression, and a NOTCH1 mutation that is unique among the T-ALL cell lines tested (O’Neil et al., 2007), mutations that are pathognomonic of human T-ALL. Thus, we have high confidence that RPMI-8402 is a faithful cell line model of human T-ALL.
shRNA and expression plasmids
The following shRNA vectors in pLKO.1 with puromycin resistance were obtained from the RNAi consortium of the Broad Institute: shGFP (TRCN000007218); shLuciferase (TRCN0000072243); shEZH2 no. 1 (TRCN0000040073); shEZH2 no. 4 (TRCN0000040076); shEED no. 2 (TRCN0000021205); shEED no. 5 (TRCN0000021208); shSUZ12 no. 2 (TRCN0000038725); shSUZ12 no. 3 (TRCN0000038727); shEZH1 no. 3 (TRCN0000002441); shTRAP1 no. 4 (TRCN0000244240); and shCRIP2 no.3 (TRCN0000243399). For experiments combining shEZH2 knockdown with knockdown of a second gene (EZH1, CRIP2, or TRAP1), shEZH2 no. 1 was also cloned into a neomycin-resistant pLKO.1 vector generated by the S. Stewart laboratory (Washington University, St. Louis, MO) and obtained from Addgene (13425).
pInducer20 expression constructs encoding WT or a catalytically impaired triple mutant of EZH2 (F672I/H694A/R732K) were previously described (Kim et al., 2015). TRAP1 and EGFP protein-coding sequences were amplified in an attB-flanked PCR product from plasmid EX-Z0100-Lv105 or EX-LV105-EGFP, respectively (Genecopoeia), cloned using the Gateway clonase BP into pDONR221, and subsequently cloned into pInducer20 (44012; Addgene) using the Gateway clonase LR recombination (Thermo Fisher Scientific). EPB41L1 was obtained from pDONR221 EPB41L1 (DNASU clone ID 43369), and Gateway-cloned into destination vector pLenti CMV Puro DEST (w118-1; 17452; Addgene). AttB-flanked PCR products of RALB, IGF-1R and dTomato were amplified from Addgene vectors nos. 50989, 11212, and 48687 and subsequently Gateway-cloned into pLenti CMV Puro DEST. FAM149A, CRIP2, VOPP1, FITM2, TSPAN9, SDC3, and MXRA7 coding sequences were synthesized as attB-flanked oligonucleotides by Eurofins Genomics, and Gateway-cloned into pLenti CMV Puro DEST. PHLDB1 was produced by Eurofins as multiple GeneStrands with flanking attL sides (5 × 500–1,000 bp) and assembled with the Gibson Assembly Cloning kit according to protocol; afterward, LR recombination was used to transfer PHLDB1 into the pLenti CMV Puro DEST destination vector. All destination plasmids were deposited into the Addgene repository (nos. 107499–107510).
Lentiviral production, infection, and selection
Lentiviruses were generated by cotransfecting pLKO.1, pInducer20, or pLenti CMV Puro DEST plasmids of interest together with packaging vectors psPAX2 and pMD2.G using Fugene (Promega). Lentiviral infection was performed by spinoculating T-ALL cell lines with virus-containing media (1,500 g × 90 min) in the presence of 8 µg/ml polybrene (Merck Millipore). Selection with antibiotics was begun 24 h after infection with neomycin (700 µg/ml for a minimum of 5 d; Thermo Fisher Scientific), puromycin (1 µg/ml for a minimum of 48 h; Thermo Fisher Scientific), or blasticidin (15 µg/ml for a minimum of 7 d; Invivogen). After 7 d of selection, shLuc, shEZH2, shEED, or shSUZ12 cells were used for BH3 profiling or chemotherapy exposure experiments. Transgene expression in pInducer20-based constructs was induced by treatment with 750 ng/µl doxycycline (Sigma-Aldrich) for at least 72 h before beginning BH3 profiling or chemotherapy exposure experiments. pLenti CMV Puro DEST cells were selected for 10 d before BH3 profiling and qRT-PCR analysis. CCRF-CEM clone E (EZH2 mutant) and parental Cas9 control cells were infected with pLenti CMV Puro DEST EGFP or tdTomato, respectively, and EGFP or tdTomato positive cells were sorted using a Facsaria II (BD Biosciences). Sorting efficiency (>99.5%) was confirmed by FACS analysis 48 h after sorting before starting experiments.
qRT-PCR and primers
RNA was isolated using RNeasy kit (Qiagen), and cDNA was made using SuperScript III first-strand cDNA synthesis kit (Thermo Fisher Scientific). qRT-PCR was performed using Power SYBR green PCR Master Mix (Thermo Fisher Scientific) and 7500 real-time PCR system (Applied Biosystems). Primers used were as follows (all 5′–3′): Human Bactin F: CTGGCACCCAGCACAATG; Human Bactin R: GCCGATCCACACGGAGTACT; Human EZH2 F: TGGAAAGAACGGAAATCTTAAACC; Human EZH2 R: GGATGACTTGTGTTGGAAAATCC; Human EZH1 F: GAGTTGGTCGATGCCCTGAAT; Human EZH1 R: AGCATGTCGCTTTCTCTTTCTT; Human EED F: GTGACGAGAACAGCAATCCAG; Human EED R: TATCAGGGCGTTCAGTGTTTG; Human SUZ12 F: AGGCTGACCACGAGCTTTTC; Human SUZ12 R: GGTGCTATGAGATTCCGAGTTC; Human TRAP1 F: TTGCAGGCAGGACGACTGT; Human TRAP1 R: GCTTGGCCGTCAGACACC; Human CRIP2 F: CCAAATGCCCCAAGTGCGACAAG; Human CRIP2 R: CGGGCGTCAGCGTCTTGCT; Human BAD F: CCCAGAGTTTGAGCCGAGTG; Human BAD R: CCCATCCCTTCGTCGTCCT; Human BAK1 F: GTTTTCCGCAGCTACGTTTTT; Human BAK1 R: GCAGAGGTAAGGTGACCATCTC; Human BAX F: CCCGAGAGGTCTTTTTCCGAG; Human BAX R: CCAGCCCATGATGGTTCTGAT; Human BCL-2 F: GGTGGGGTCATGTGTGTGG; Human BLC-2 R: CGGTTCAGGTACTACGTCATCC; Human BCL-B F: GCCAGGTTACGGCAGATTCA; Human BCL-B R: GAAGGTCACGAGCGTCACC; Human BID F: ATGGACCGTAGCATCCCTCC; Human BID R: GTAGGTGCGTAGGTTCTGGT; Human BIM F: TAAGTTCTGAGTGTGACCGAGA; Human BIM R: GCTCTGTCTGTAGGGAGGTAGG; Human BMF F: GAGCCATCTCAGTGTTGGAG; Human BMF R: GCCAGCATTGCCATAAAAGAGTC; Human BNIP3 F: CAGGGCTCCTGGGTAGAACT; Human BNIP3 R: CTACTCCGTCCAGACTCATGC; Human BOK F: GTCTTCGCTGCGGAGATCAT; Human BOK R: CATTCCGATATACGCTGGGAC; Human BCL-W F: CGTCCCCGTATAGAGCTGTG; Human BCL-W R: GGACAAGTGCAGGAGTGGAT; Human BCL-XL F: GACTGAATCGGAGATGGAGACC; Human BCL-XL R: GCAGTTCAAACTCGTCGCCT; Human BFL1 F: AGTGCTACAAAATGTTGCGTTC; Human BFL1 R: GGCAATTTGCTGTCGTAGAAGTT; Human BIK F: GGAGCTGCTCATACAGGAGG; Human BIK R: GTGGCTTACAGACGCTGCC; Human HRK F: CCTTTCAAGCTCTGGGCTC; Human HRK R: CAGGCGGAACTTGTAGGAAC; Human MCL1 F: GTGCCTTTGTGGCTAAACACT; Human MCL1 R: AGTCCCGTTTTGTCCTTACGA; Human NOXA F: AAGTTTCTGCCGGAAGTTCA; Human NOXA R: GCAAGAACGCTCAACCGAG; Human PUMA F: GACCTCAACGCACAGTACGAG; Human PUMA R: AGGAGTCCCATGATGAGATTGT; Mouse GAPDH F: CGTGGAGTCTACTGGTGTCTTCAC; Mouse GAPDH R: TTGGCTCCACCCTTCAAGTG; Mouse TRAP1 F: AGCCGAGGACAAGGAGGAG; Mouse TRAP1 R: GCATCCGTCTGAAGGTGAATC; Mouse CRIP2 F: CCCCATCGAGGTCCCTGTGGT; Mouse CRIP2 R: ACTAGAGGCTTTGCTGGGACCCT; Mouse Bactin F: GGCTGTATTCCCCTCCATCG; and Mouse Bactin R: CCAGTTGGTAACAATGCCATGT.
Western blot and antibodies
Cells were lysed in radioimmunoprecipitation assay buffer (Merck Millipore) supplemented with cOmplete protease inhibitor (Roche) and PhosSTOP phosphatase inhibitor (Roche). 20 µg of protein lysate was mixed with Laemmli sample buffer (Bio-Rad) and β-mercaptoethanol (Sigma-Aldrich) before being run on a 4–12% Novex Bis-Tris polyacrylamide gel (Thermo Fisher Scientific). Blots were transferred to polyvinylidene fluoride membrane (Thermo Fisher Scientific) and blocked with 5% milk (Thermo Fisher Scientific) or 5% BSA (New England Biolabs) in phosphate-buffered saline with 0.1% Tween (Boston Bioproducts) and probed with the following antibodies: Total histone H3 (1:1,000, no. 4499; Cell Signaling Technology), Tri-methyl-histone H3 Lys 27 (1:1,000; no. 07-449; Merck Millipore), EZH2 (1:1,000; no. 5246; Cell Signaling Technology), SUZ12 (1:1,000; no. 3737; Cell Signaling Technology), TRAP1 (1:1,000; no. 612344; BD Biosciences), β-tubulin (1:1,000; no. 2128; Cell Signaling Technology). Secondary detection of horseradish peroxidase–linked antibodies (Cell Signaling Technology) with horseradish peroxidase substrate (Thermo Fisher Scientific) was visualized using Amersham Imager 600 (GE Healthcare Life Sciences). All antibodies were validated using the shRNA knockdown or overexpression experiments shown in this manuscript.
BH3 profiling of T-ALL cell lines
BH3 profiling of T-ALL cell lines was performed by flow cytometry to assess effects of BIM BH3 peptide or PUMA2A negative control on cytochrome c release, as previously described (Bhola et al., 2016). 100,000 T-ALL cells were incubated with 1 µM BIM or inactive PUMA2A peptide (Tufts University Core Facility, Boston, MA) in DTEB buffer (135 mM trehalose, 50 mM KCl, 20 µM EDTA, 20 µM EGTA, 5 mM succinate, 0.1% BSA, and 10 mM Hepes-KOH; final pH, 7.5) containing 0.002% (wt/vol) digitonin (Sigma-Aldrich) for 25 or 30 min, as indicated. Cells were then fixed in 2% paraformaldehyde (Alfa Aesar). Cytochrome c was then stained using 1:40 anti–cytochrome c antibody conjugated to either Alexa Fluor 488 or 647 (nos. 560263 and 558709; BD Biosciences) in cytochrome c–staining buffer (20% FBS, 10% BSA, 1% Saponin, and 3 mM Sodium Azide in PBS) overnight at 4°C and analyzed on an LSR-II (BD Biosciences).
Assessment of chemotherapy-induced apoptosis
T-ALL cells (10,000 or 40,000 per well) were seeded in 96-well plates and incubated with chemotherapeutic agents at the doses indicated below for 48 h. Chemotherapy doses were as follows unless otherwise indicated: asparaginase, 10 international units/ml (Sigma-Aldrich); dexamethasone, 10 µM (Sigma-Aldrich); vincristine, 1 µM (Selleckchem); doxorubicin, 1 µM (Sigma-Aldrich); etoposide, 10 µM (Sigma-Aldrich); cytarabine, 10 µM (Selleckchem); nelarabine, 10 µM (Sigma-Aldrich); 6-mercaptopurine, 10 µM (Abcam); and methotrexate, 10 µM (Selleckchem). Annexin V and propidium iodide staining were assessed using the Apoptosis Detection kit II (BD Biosciences), and caspase 3/7 activity was assessed using the Caspase Glo 3/7 Assay (Promega) according to the manufacturer’s instructions.
Cell cycle
Vybrant DyeCycle violet stain was used to determine cell cycle according to manufacturer’s protocol (Life Technologies and Thermo Fisher Scientific). 500,000 cells were incubated with 500 µl RPMI + 1 µl Dyecycle for 90 min at 37°C, and fluorescence was measured using a LSR-II Flow Cytometer (BD Biosciences).
Mice
Ezh2 and Eed conditional knockout mice were previously described (Shen et al., 2008; Neff et al., 2012), and crossed to Mx1:Cre transgenic mice (stock no. 003556; The Jackson Laboratory), all maintained in the C57BL/6 background (The Jackson Laboratory). All mice used for experiments in this study were generated from crosses of siblings that were Ezh2 or Eed fl/wt, crossed to animals that were either fl/wt or wt/wt. In all experiments, only one of the parents also carried a single copy of the Mx1:Cre transgene.
Ezh2 and Eed genotyping and assessment for Cre-induced deletion were performed with 3-primer PCR using the following primers (all 5′–3′): Ezh2-F: CCCATGTTTAAGGGCATAGTGACATG; Ezh2-R: ATGTGCAGGTCAGTCAGCAACTTCAG: Ezh2-onelox: TCGAGGGACCTAATAACTTCGTATAGCA; Eed-F: CTACGGGCAGGAGGAAGAG; Eed-R: GGGGGAGAGGGAGTTGTC; Eed-onelox: CCACATAGGCTCATAGAATTG; MXCre-F: GCGGTCTGGCAGTAAAAACTATC; MXCre-R: GTGAAACAGCATTGCTGTCACTT.
No statistical methods were used to predetermine sample size. 6 Ezh2 wt/wt (5 males and 1 female) mice were compared with 5 Ezh2 fl/wt (1 male and 4 females) siblings and 9 Eed wt/wt mice (3 males and 6 females) to 12 Eed wt/fl (7 male and 5 female) siblings. Animals were excluded from analysis only if PCR analysis revealed ineffective deletion of Ezh2 or Eed by pIpC treatment, which was a preestablished criterion, and when this occurred, the entire cohort of mice from that experimental batch was excluded from analysis. No randomization nor blinding was done. Mice were handled in strict accordance with Good Animal Practice as defined by the Office of Laboratory Animal Welfare. All animal work was done with Boston Children’s Hospital Institutional Animal Care and Use Committee approval (protocol no. 15-10-3058R).
Engraftment of human cell lines for in vivo drug treatment studies were performed into NRG NOD.Cg-Rag1tm1MomIl2rgtm1Wjl/SzJ immunodeficient mice (stock no. 007799; Jackson Laboratory).
Staining, sorting, BH3 profiling, and qRT-PCR of mouse thymocytes
Ezh2 or Eed fl/wt or wt/wt mice that also expressed the Mx1:Cre transgene were treated beginning at 6 wk of age with five doses of 15 µg/g pIpC (Sigma-Aldrich) given every other day to excise Ezh2 or Eed. Mice were sacrificed 4 wk after the last pIpC dose and the thymus was isolated. Mouse thymocytes were dissociated through a 40-µm mesh filter, and red blood cells were lysed using the BD Biosciences Red Blood Cell Lysis reagent. Isolated thymocytes were stained for the following markers of T cell development using the following antibodies from BD Biosciences: CD3e-PerCP (1:100; no. 553067), CD4-BV650 (1:400; no. 563747), CD117-PE-Cy7 (1:200, no. 558163), CD44-APC (1:200; no. 559250), CD8a-APC-Cy7 (1:400; no. 557654), CD25-BV421 (1:200; no. 562606), and CD28-PE (1:200; no. 553297). Cells were also stained for CD19-Biotin (1:400; no. 553784), Ter-119-Biotin (1:400; no. 553672), NK1.1-Biotin (1:400; no. 553163), and Gr1-Biotin (1:400, no. 553125; all from BD Biosciences) for negative selection using a Streptavidin-V500 secondary antibody (1:100; no. 561419; BD).
BH3 profiling was then performed on these stained cells based on flow cytometry for cytochrome c release, as previously described (Bhola et al., 2016). In brief, thymocytes were incubated with 1 µM active hBIM peptide or PUMA2A inactive control for 30 min in DTEB buffer (135 mM trehalose, 50 mM KCl, 20 µM EDTA, 20 µM EGTA, 5 mM succinate, 0.1% BSA, and 10 mM Hepes-KOH; final pH, 7.5) containing 0.002% (wt/vol) digitonin (Sigma-Aldrich) and fixed in 2% paraformaldehyde (Alfa Aesar). Cytochrome c was then stained using anti–cytochrome c antibody Alexa Fluor 488 (1:40; no. 560263; BD Falcon) in cytochrome c staining buffer (20% FBS, 10% BSA, 1% Saponin, and 3 mM Sodium Azide in PBS) overnight at 4°C. Samples were analyzed on an LSR-II instrument (BD Biosciences). Results are reported only for thymocyte subsets in which >250 events were consistently captured.
For qRT-PCR purposes, stained thymocytes were sorted for the indicated populations using a Facsaria II (BD Biosciences). RNA isolation, cDNA synthesis and qRT-PCR analysis was performed as described in quantitative reverse transciptase PCR and primers section.
Generation of EZH2 mutant CCRF-CEM clones using CRISPR-Cas9
A gRNA targeting exon 5 of EZH2 (NM_004456) was generated by annealing the oligonucleotides (F: 5′-CACCGTTATGATGGGAAAGTACACG-3′; R: 5′-CAATACTACCCTTTCATGTGCCAAA-3′) and cloning into pHK09-combomod-Puro (provided by D. Bauer Lab, Boston, USA), which is a modified version of LentiGuide-Puro (52963; Addgene) harboring sequence modifications to optimize gRNA stability. In brief, 1 µl of 100 µm forward and reverse oligo was mixed with 1 µl 10× T4 DNA Ligation Buffer (New England Biolabs), 6.5 µl ddH2O, 0.5 µl T4 PNK (New England Biolabs), and annealed for 30 min at 37°C and 5 min 95°C. 1 µl phosphoannealed oligo 1:500 was then ligated into 1 µl of BsmBI-digested lentiviral pHK09-puro plasmid using 1 µl Quick Ligase (New England Biolabs), 5 µl 2× Quick Ligase buffer (New England Biolabs) and 2 µl ddH2O for 5 min at room temperature.
Lentivirus of pHK09-combomod-puro and Lenti-Cas9-Blast (52962; Addgene) was produced as described in the Lentiviral production section below. CCRF-CEM cells were infected with Lenti-Cas9-Blast and selected in blasticidin for 7 d, whereafter cells were infected with pHK09-puro and selected with puromycin for at least 48 h. Single-cell clones were made and gRNA cutting frequency was determined by PCR amplification of the gRNA target locus (F: 5′-TTAGGTGGAAGATGAAACTGTTTTA-3′; R: 5′-TCATGCCCTATATGCTTCATAAA-3′), followed by next-generation sequencing at the MGH CCIB DNA Core facility. Cutting efficiency was assessed using CrispRVariantsLite v1.1 (Lindsay et al., 2016). The EZH2 mutant clone described was then infected with pInducer20 EZH2 WT or EZH2 mutant, selected in neomycin for 5 d, and used for BH3-profiling and Western blot.
Pooled CRISPR/Cas9 gRNA transduction and chemotherapy treatment of CCRF-CEM cells
A pooled gRNA library with four unique gRNAs targeting the catalytic SET domain of EZH2, and four unique gRNAs targeting the safe-harbor AAVS1 locus located in intron 1 of the PPP1R12C gene, were designed as described (Sanjana et al., 2014), and cloned into the pHKO9-combomod-puro lentiviral vector as described above. gRNA target sequences were as follows: EZH2-SET_a, 5′-ATTGCTGGCACCATCTGACG-3′; EZH2-SET_b, 5′-AAGATGAAGCTGACAGAAGA-3′; EZH2-SET_c, 5′-GTGGTGGATGCAACCCGCAA-3′; EZH2-SET_d, 5′-AAAACAGCTCTTCGCCAGTC-3′; AAVS1_a, 5′-AGCGGCTCCAATTCGGAAGT-3′; AAVS1_b, 5′-GCTCAAAGTGGTCCGGACTC-3′; AAVS1_c, 5′-GAGAGGTGACCCGAATCCAC-3′; and AAVS1_d, 5′-AGTTCTTAGGGTACCCCACG-3′.
Lentivirus was produced by cotransfecting equal amounts of each of these gRNAs together with the psPAX2 and pMD2.G vectors as described below. Virus was concentrated using the Beckmann XL-90 ultracentrifuge (Beckman Coulter) at 100,000 g (24,000 rpm) for 2 h at 4°C. Viral titers were determined using AlamarBlue staining, as described (https://portals.broadinstitute.org/gpp/public/resources/protocols). CCRF-CEM cells expressing Cas9 protein were plated in 96-well format (40,000 cells per well in 100 l RPMI medium), and infected with 12,000 infectious units of lentivirus encoding this pool of gRNAs, for a multiplicity of infection of 0.3. Infected cells were selected 48 h post-infection with puromycin at 1 µg/ml for 7 d. Infected cells were treated with vehicle (RPMI medium) or 30 nM of vincristine for 48 h in 24-well format (400,000 cell per well in 1 ml RPMI medium), and then transferred into fresh media without vincristine. Cells were harvested 14 d after start of vincristine treatment, and genomic DNA was extracted using DNeasy Blood and Tissue kit (Qiagen). gRNA sequences were PCR-amplified using pHKO9 sequencing primers (pHKO9: F, 5′-TCTTGTGGAAAGGACGAAACACCG-3′; pHKO9, R, 5′-TCTACTATTCTTTCCCCTGCACTGT-3′), PCR-purified using the QIAquick PCR purification kit (Qiagen), sequenced using CRISPR sequencing at the MGH CCIB DNA Core facility described above, and gRNA cutting efficiency was calculated using an in-house python script.
Chemotherapy treatment of CCRF-CEM EZH2 mutant clone E and parental Cas9 controls
Clone E (EZH2 mutant) and the parental Cas9 control were plated and split every other day at a density of 0.5 million/ml. Cells were treated with vincristine (50 nM) or vehicle (DMSO) for 48 h; afterward, they were released into nonchemotherapy-containing growth media (RPMI 1640 with 10% FBS), and viable cell counts were assessed by trypan blue exclusion using a Countess II Automated Cell Counter (Thermo Fisher Scientific) according to the manufacturer’s instructions.
To assess relative fitness of EZH2 mutant clone E cells versus parental Cas9 controls upon chemotherapy treatment in co-culture experiments, EGFP-transduced EZH2 mutant clone E cells, and tdTomato-transduced Parental Cas9 cells were mixed at a 1:1 ratio and treated with vincristine (50 nM), etoposide (5 µM), cytarabine (10 µM), doxorubicin (100 nM), and methotrexate (1 µM) for 48 h, and then released into standard growth media for 4 d. The percent EGFP and dTomato cells was assessed with FACS analysis using the LSR II flow cytometer (BD Biosciences), and data were analyzed using FlowJo version 10.4.2.
To assess relative fitness of EZH2 mutant clone E cells versus parental Cas9 controls upon in vivo chemotherapy treatment, EGFP-transduced EZH2 mutant clone E cells and tdTomato-transduced Parental Cas9 cells were mixed at a 1:1 ratio and injected by tail vein injection (0.5 million cells/mouse) into 8-wk-old NRG mice (stock no. 007799; The Jackson Laboratory). Mice were treated with irradiation (450 cGy) 3 h before transplantation. Mice (n = 5 per group) were treated with either vehicle (0.9% sodium chloride solution) or vincristine 2 mg/kg via intraperitoneal injection. Mice were monitored daily and supportive care (hydrogel, bacon bites, and 300 µl normal saline solution via subcutaneous injection) were administered daily for the duration of the experiment. Mice were sacrificed at day 19 after leukemia injection, the spleen was isolated and dissociated through a 40-µM mesh filter, and red blood cells were lysed using Red Blood Cell Lysis reagent (BD Biosciences). Mice spleen cells were analyzed by FACS analysis as described above.
RNA-seq of T-ALL cell lines after PRC2 depletion
RNA was isolated from an equal number of CCRF-CEM cells 8 d after infection with shGFP, shLuciferase, shEZH2 no. 1, shEZH2 no. 4, shEED no. 2, shEED no. 5, shSUZ12 no. 2, or shSUZ12 no. 3. RNA was extracted using RNeasy kit (Qiagen). Samples were treated with Ambion Turbo DNase (Thermo Fisher Scientific) to yield DNA contamination below 10%. 3 μl of 1:10 ERCC spike-in controls (Ambion; Thermo Fisher Scientific) were added to 5,000 ng of RNA. Quality of RNA was assessed using the Agilent 2100 Bioanalyzer. Library preparation and sequencing were performed as for the T-ALL patient samples, as described above.
RNA sequences were aligned with STAR aligner (version 2.3.1z4) against the hg19 w ERCC92 genome (ftp://ftp.ensembl.org/pub/release-75/fasta/homo_sapiens/dna/). The RUV-Seq package was used to normalize to spike-in controls. FeatureCounts software was used to produce integer counts (raw read counts) for all genes in the supplied genome annotation file, which were then normalized using DESeq package, and these normalized read counts are shown in Table S7. Two control shRNAs (shLuciferase; shGFP) were compared with six PRC2 shRNA (shEZH2 no. 1, shEZH2 no. 4, shEED no. 2, shEED no. 5, shSUZ12 no. 2, and shSUZ12 no. 3), using DESeq2 analysis. Genes that were significantly up-regulated following PRC2 depletion were defined based on log2 fold-change >0.45 and nominal P value <0.02, as calculated by DESeq2 analysis, and the heat map was generated using GENE-E. RNA-seq of T-ALL cells following depletion of PRC2 genes is available as NCBI GEO entry GSE95648.
ChIP-seq
Chromatin immunoprecipitation-sequencing was performed as described (Knoechel et al., 2014). In brief, CCRF-CEM cells transduced with shRNA-targeting control (Luciferase) or EZH2 were cross-linked using formaldehyde, and sonication was performed to fragment chromatin to 200–700-bp size range using a Covaris E220 Focused-ultrasonicator. Immunoprecipitation was then performed on five million cells with anti-H3K4me3 antibody (no. 39155; Active Motif), which was validated as described (Knoechel et al., 2014). Protein A/G dynabeads were used to pull down antibody-bound chromatin, followed by washing and elution. Cross-linking was reversed with proteinase K treatment, and DNA was cleaned up using Solid Phase Reversible Immobilization beads (Beckman Coulter). ChIP DNA was quantified using the Qubit system (Invitrogen). Sequencing libraries for ChIP-seq were prepared according to Illumina’s instructions. ChIP DNA and input were sequenced with the Illumina NextSeq 500 platfom. ChIP-seq reads were aligned to the reference genome (hg19) using BWA (version 0.7.13; Li and Durbin, 2009) with default parameters and duplicated reads were filtered out with Picard. Reads with <20 bases of mapping quality were removed from the alignment. ChIP peaks for H3K27me3 were called by Homer (Heinz et al., 2010) using default parameters requiring putative peak to have threefold more normalized tags in the ChIP experiment than the input and a cumulative poisson P value threshold of 0.001. Putative peaks that were identified in regions of genomic duplication or repeat elements were filtered out. Note that there were no significant peaks identified in CCRF-CEM cells transduced with shEZH2, indicating that EZH2 is the primary H3K27me3 “writer” in these cells. ChIP-seq peaks were called significant if the maximum peak intensity was > 0.7, a cutoff verified by manual examination of peaks using Integrative Genomics Viewer version 2.3.93. ChIP-seq of T-ALL cells following depletion of EZH2 is available as NCBI GEO entry GSE112147.
Gamitrinib single agent and synergy testing
CCRF-CEM cells were infected with the indicated shRNAs, and puromycin selection was begun 24 h after infection. After 8 d of puromycin selection, treatment with vehicle (DMSO) or the indicated doses of gamitrinib was performed for 48 h, after which cell viability was assessed using CellTiter-Glo (Promega). Synergy of gamitrinib in combination with dexamethasone or doxorubicin was tested in CCRF-CEM cells infected with shEZH2 no. 1 selected for 8 d in puromycin. In brief, 5,000 cells were plated in 384-well format using a Biotek MicroFlo Select Dispenser. Drug concentration combinations, across a serial twofold dilution range around previously determined IC50 concentrations, were plated in quadruplicate onto the cells using a Bravo Automated Liquid Handling Platform (Agilent). Viability was then assessed after 48 h of treatment using CellTiter-Glo (Promega). The Chou-Talalay method was used to calculate combination indexes of the fractional inhibition using CompuSyn software. Fractional inhibition was defined as 1, relative cell viability.
GSK126 treatment of patient-derived xenografts
Human T-ALL patient-derived xenografts were engrafted into immunodeficient mice as described (Townsend et al., 2016). Patient-derived cells were cultured in the presence of recombinant human cytokines stem cell factor (50 ng/ml), Flt3-liter (20 ng/ml) and IL-7 (10 ng/ml; R&D Systems) at 37° under 5% CO2, as described (Reynolds et al., 2014; Burns et al., 2018). Cells were treated with vehicle or 1 µM GSK126 (Cayman Chemical) for 6 d, and qRT-PCR was performed as described above.
Statistical analyses
Overall survival was calculated from the time of registration to the time of death censored at the last known follow up. Event-free survival was calculated from the time of registration to relapse or death and was censored at the last known follow up. Induction failures were included as events at time zero. The log-rank test was used to test for differences in survival between groups, and the method of Kaplan and Meier was used to construct survival curves.
Significance of RNA-seq read counts, which are not normally distributed, was performed using DESeq analysis, which accounts for the negative binomial distribution of RNA-seq data, as described above. For two-group comparisons of continuous measures, a two-tailed Welch unequal variances t test was used. For three-group comparisons of continuous measures, ANOVA was performed and a Tukey adjustment for multiple comparisons was used. For analysis of two effects, a two-way ANOVA model was constructed and, unless indicated, included an interaction term between the two effects. Post hoc adjustment for multiple comparisons for two-way ANOVA included either a Tukey or Sidak adjustment depending on the pairwise comparisons of interest.
Data shown as bar graphs represent the mean and standard error of the mean of a minimum of three biological replicates. For experiments performed in duplicate, results from each replicate are shown as individual data points. All P values reported are two-sided except for the one-sided analysis of the association of TRAP1 overexpression with inferior survival in Fig. 6 I, as indicated, due to the hypothesized direction of the difference before testing. P values were considered significant if <0.05.
Online supplemental material
Fig. S1 shows the lack of association of mitochondrial apoptosis resistance in primary T-ALL patient samples with RNA expression of BCL2 family genes or with oncogenic mutations previously associated with treatment response. Fig. S2 shows that shRNA depletion of EZH1 does not reverse chemotherapy resistance induction by shRNA knockdown of EZH2. Fig. S3 shows that shRNA depletion of PRC2 subunits has no consistent effect on mRNA expression of any known pro- or antiapoptotic BCL2 family genes. Fig. S4 shows effects of TRAP1- and EZH2-targeting shRNAs on expression of their target mRNAs and that shRNA depletion of TRAP1 reverses the ability of EZH2 depletion to induce chemotherapy resistance. Fig. S5 shows effects of CRIP2 and EZH2-targeting shRNAs on expression of their target mRNAs, and that shRNA depletion of CRIP1 reverses the ability of EZH2 depletion to induce chemotherapy resistance. Table S1 shows a summary of clinical features and genotyping of the primary T-ALL patient samples analyzed. Table S2 shows association of clinical features with mitochondrial apoptotic priming. Table S3 shows the genes whose protein-coding exons were sequenced by targeted exome sequencing. Table S4 shows results of targeted exome sequencing. Table S5 shows resuls of array CGH analysis. Table S6 shows results of RNA-seq of primary T-ALL patient samples. Table S7 shows the genotype of EZH2-mutant CCRF-CEM clone E cells. Table S8 shows RNA-seq of CCRF-CEM cells transduced with control or PRC2-targeting shRNAs. Table S9 shows result of H3K27me3 ChIP-seq in CCRF-CEM cells.
Supplementary Material
Acknowledgments
We thank Alex Kentsis, Marc Mansour, and Daniel Bauer for critical discussions, and Jeremy Ryan, Taras Kreslavsky, Ronald Mathieu, Erin Powers, Christine Reynolds, Mina Jacob, Yana Pikman, Paolo Cifani, Meaghan McGuinness, Connor McGuckin, and Divya Vinjamur for experimental advice and assistance. We thank Aaron Thorner, Paul Van Hummelen, and Matthew Ducar for assistance with targeted exome sequencing analysis. We thank Jian Xu for SUZ12 shRNA hairpins, Tobias Neff and Scott Armstrong for Ezh2- and Eed-floxed mice, Dario Altieri for gamitrinib, and Andrew Weng and David Weinstock for some of the patient-derived xenograft specimens utilized. We thank John Gilbert for editorial assistance. We are grateful to the patients and families who provided samples for these studies.
This work was supported by National Institutes of Health (NIH) grants R01 CA193651, R21 CA167124, the Boston Children’s Hospital Translational Research Program, and the V Foundation for Cancer Research. The Children’s Oncology Group (COG) work was supported by NIH grants U10 CA98543 (COG Chair’s grant), U10 CA98413 (COG Statistical Center), U24 CA114766 (COG Specimen Banking), U10 CA 180886 (COG Operations Center), and U10 CA 180899 (COG Statistics and Data Center). I.M. Ariës is a recipient of a Ter Meulen grant from the Royal Dutch Academy. L. Hinze was supported by the German National Academic Foundation and the Biomedical Education Program. S. Balbach was supported by German Cancer Aid. S.P. Hunger is the Jeffrey E. Perelman Distinguished Chair in the Department of Pediatrics, Children's Hospital of Philadelphia. P. Van Vlierberghe and S. Peirs are supported by the Fund for Scientific Research Flanders, Children Cancer Fund Ghent, Belgian Foundation Against Cancer, Belgian Stand Up To Cancer Foundation, and a Starting Grant from the European Research Council. A. Gutierrez is a Clinical Investigator of the Damon Runyon Cancer Research Foundation and supported by an Investigatorship at Boston Children’s Hospital.
The authors declare no competing financial interests.
Author contributions: conceptualization, A. Gutierrez; methodology, I.M. Ariës, K. Bodaar, S.A. Karim, T. Ni Chonghaile, M.A. Burns, M. Pfirrmann, S. Peirs, B. Menten, P. Van Vlierberghe, B. Knoechel, A.G. Letai, and A. Gutierrez; investigation, I.M. Ariës, K. Bodaar, S.A. Karim, T. Ni Chonghaile, L. Hinze, M. Burns, M. Pfirrmann, J. Degar, J.T. Landrigan, S. Balbach, S. Peirs, B. Menten, R. Isenhart, M.L. Loh, S.P. Hunger, D.T. Teachey, K.R. Rabin, S.S. Winter, K.P. Dunsmore, B.L. Wood, L.B. Silverman, P. Van Vlierberghe, B. Knoechel, A.G. Letai, and A. Gutierrez; formal analysis, I.M. Ariës, K. Bodaar, S.A. Karim, T. Ni Chonghaile, M. Burns, S. Peirs, B. Menten, R. Isenhart, K.E. Stevenson, D.S. Neuberg, M. Devidas, M.L. Loh, S.P. Hunger, D.T. Teachey, K.R. Rabin, S.S. Winter, S.E. Sallan, P. Van Vlierberghe, S.H. Orkin, B. Knoechel, A.G. Letai, and A. Gutierrez; resources, M.L. Loh, S.P. Hunger, D.T. Teachey, K.R. Rabin, S.S. Winter, P. Van Vlierberghe, B. Knoechel, A.G. Letai, and A. Gutierrez; supervision, A. Gutierrez; writing, I.M. Ariës, K. Bodaar, and A. Gutierrez.
References
- Abedalthagafi M.S., Merrill P.H., Bi W.L., Jones R.T., Listewnik M.L., Ramkissoon S.H., Thorner A.R., Dunn I.F., Beroukhim R., Alexander B.M., et al. . 2014. Angiomatous meningiomas have a distinct genetic profile with multiple chromosomal polysomies including polysomy of chromosome 5. Oncotarget. 5:10596–10606. 10.18632/oncotarget.2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines C.P., Kaiser R.A., Purcell N.H., Blair N.S., Osinska H., Hambleton M.A., Brunskill E.W., Sayen M.R., Gottlieb R.A., Dorn G.W., et al. . 2005. Loss of cyclophilin D reveals a critical role for mitochondrial permeability transition in cell death. Nature. 434:658–662. 10.1038/nature03434 [DOI] [PubMed] [Google Scholar]
- Bhola P.D., Mar B.G., Lindsley R.C., Ryan J.A., Hogdal L.J., Vo T.T., DeAngelo D.J., Galinsky I., Ebert B.L., and Letai A.. 2016. Functionally identifiable apoptosis-insensitive subpopulations determine chemoresistance in acute myeloid leukemia. J. Clin. Invest. 126:3827–3836. 10.1172/JCI82908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns M.A., Liao Z.W., Yamagata N., Pouliot G.P., Stevenson K.E., Neuberg D.S., Thorner A.R., Ducar M., Silverman E.A., Hunger S.P., et al. . 2018. Hedgehog pathway mutations drive oncogenic transformation in high-risk T-cell acute lymphoblastic leukemia. Leukemia. 32:2126–2137. 10.1038/s41375-018-0097-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung A.K., Ko J.M., Lung H.L., Chan K.W., Stanbridge E.J., Zabarovsky E., Tokino T., Kashima L., Suzuki T., Kwong D.L., et al. . 2011. Cysteine-rich intestinal protein 2 (CRIP2) acts as a repressor of NF-kappaB-mediated proangiogenic cytokine transcription to suppress tumorigenesis and angiogenesis. Proc. Natl. Acad. Sci. USA. 108:8390–8395. 10.1073/pnas.1101747108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chonghaile T.N., Roderick J.E., Glenfield C., Ryan J., Sallan S.E., Silverman L.B., Loh M.L., Hunger S.P., Wood B., DeAngelo D.J., et al. . 2014. Maturation stage of T-cell acute lymphoblastic leukemia determines BCL-2 versus BCL-XL dependence and sensitivity to ABT-199. Cancer Discov. 4:1074–1087. 10.1158/2159-8290.CD-14-0353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K., Lawrence M.S., Carter S.L., Sivachenko A., Jaffe D., Sougnez C., Gabriel S., Meyerson M., Lander E.S., and Getz G.. 2013. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat. Biotechnol. 31:213–219. 10.1038/nbt.2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantino E., Maddalena F., Calise S., Piscazzi A., Tirino V., Fersini A., Ambrosi A., Neri V., Esposito F., and Landriscina M.. 2009. TRAP1, a novel mitochondrial chaperone responsible for multi-drug resistance and protection from apoptotis in human colorectal carcinoma cells. Cancer Lett. 279:39–46. 10.1016/j.canlet.2009.01.018 [DOI] [PubMed] [Google Scholar]
- Coustan-Smith E., Mullighan C.G., Onciu M., Behm F.G., Raimondi S.C., Pei D., Cheng C., Su X., Rubnitz J.E., Basso G., et al. . 2009. Early T-cell precursor leukaemia: a subtype of very high-risk acute lymphoblastic leukaemia. Lancet Oncol. 10:147–156. 10.1016/S1470-2045(08)70314-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czabotar P.E., Lessene G., Strasser A., and Adams J.M.. 2014. Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat. Rev. Mol. Cell Biol. 15:49–63. 10.1038/nrm3722 [DOI] [PubMed] [Google Scholar]
- Danis E., Yamauchi T., Echanique K., Zhang X., Haladyna J.N., Riedel S.S., Zhu N., Xie H., Orkin S.H., Armstrong S.A., et al. . 2016. Ezh2 Controls an Early Hematopoietic Program and Growth and Survival Signaling in Early T Cell Precursor Acute Lymphoblastic Leukemia. Cell Reports. 14:1953–1965. 10.1016/j.celrep.2016.01.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davids M.S., and Letai A.. 2012. Targeting the B-cell lymphoma/leukemia 2 family in cancer. J. Clin. Oncol. 30:3127–3135. 10.1200/JCO.2011.37.0981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M., et al. . 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43:491–498. 10.1038/ng.806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dow L.W., Chang L.J., Tsiatis A.A., Melvin S.L., and Bowman W.P.. 1982. Relationship of pretreatment lymphoblast proliferative activity and prognosis in 97 children with acute lymphoblastic leukemia. Blood. 59:1197–1202. [PubMed] [Google Scholar]
- Ebinger S., Özdemir E.Z., Ziegenhain C., Tiedt S., Castro Alves C., Grunert M., Dworzak M., Lutz C., Turati V.A., Enver T., et al. . 2016. Characterization of Rare, Dormant, and Therapy-Resistant Cells in Acute Lymphoblastic Leukemia. Cancer Cell. 30:849–862. 10.1016/j.ccell.2016.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felts S.J., Owen B.A., Nguyen P., Trepel J., Donner D.B., and Toft D.O.. 2000. The hsp90-related protein TRAP1 is a mitochondrial protein with distinct functional properties. J. Biol. Chem. 275:3305–3312. 10.1074/jbc.275.5.3305 [DOI] [PubMed] [Google Scholar]
- Galluzzi L., Bravo-San Pedro J.M., Vitale I., Aaronson S.A., Abrams J.M., Adam D., Alnemri E.S., Altucci L., Andrews D., Annicchiarico-Petruzzelli M., et al. . 2015. Essential versus accessory aspects of cell death: recommendations of the NCCD 2015. Cell Death Differ. 22:58–73. 10.1038/cdd.2014.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J.M., Silverman L.B., Levy D.E., Dalton V.K., Gelber R.D., Lehmann L., Cohen H.J., Sallan S.E., and Asselin B.L.. 2003. Childhood T-cell acute lymphoblastic leukemia: the Dana-Farber Cancer Institute acute lymphoblastic leukemia consortium experience. J. Clin. Oncol. 21:3616–3622. 10.1200/JCO.2003.10.116 [DOI] [PubMed] [Google Scholar]
- Göllner S., Oellerich T., Agrawal-Singh S., Schenk T., Klein H.U., Rohde C., Pabst C., Sauer T., Lerdrup M., Tavor S., et al. . 2017. Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat. Med. 23:69–78. 10.1038/nm.4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Sanda T., Grebliunaite R., Carracedo A., Salmena L., Ahn Y., Dahlberg S., Neuberg D., Moreau L.A., Winter S.S., et al. . 2009. High frequency of PTEN, PI3K, and AKT abnormalities in T-cell acute lymphoblastic leukemia. Blood. 114:647–650. 10.1182/blood-2009-02-206722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez A., Dahlberg S.E., Neuberg D.S., Zhang J., Grebliunaite R., Sanda T., Protopopov A., Tosello V., Kutok J., Larson R.S., et al. . 2010. Absence of biallelic TCRgamma deletion predicts early treatment failure in pediatric T-cell acute lymphoblastic leukemia. J. Clin. Oncol. 28:3816–3823. 10.1200/JCO.2010.28.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao M.H., Yu A.L., Yeargin J., Ku D., and Haas M.. 1994. Nonhereditary p53 mutations in T-cell acute lymphoblastic leukemia are associated with the relapse phase. Blood. 83:2922–2930. [PubMed] [Google Scholar]
- Kang B.H., Plescia J., Dohi T., Rosa J., Doxsey S.J., and Altieri D.C.. 2007. Regulation of tumor cell mitochondrial homeostasis by an organelle-specific Hsp90 chaperone network. Cell. 131:257–270. 10.1016/j.cell.2007.08.028 [DOI] [PubMed] [Google Scholar]
- Kang B.H., Plescia J., Song H.Y., Meli M., Colombo G., Beebe K., Scroggins B., Neckers L., and Altieri D.C.. 2009. Combinatorial drug design targeting multiple cancer signaling networks controlled by mitochondrial Hsp90. J. Clin. Invest. 119:454–464. 10.1172/JCI37613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karch J., Kwong J.Q., Burr A.R., Sargent M.A., Elrod J.W., Peixoto P.M., Martinez-Caballero S., Osinska H., Cheng E.H., Robbins J., et al. . 2013. Bax and Bak function as the outer membrane component of the mitochondrial permeability pore in regulating necrotic cell death in mice. eLife. 2:e00772 10.7554/eLife.00772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Kim W., Howard T.P., Vazquez F., Tsherniak A., Wu J.N., Wang W., Haswell J.R., Walensky L.D., Hahn W.C., et al. . 2015. SWI/SNF-mutant cancers depend on catalytic and non-catalytic activity of EZH2. Nat. Med. 21:1491–1496. 10.1038/nm.3968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein L., Kyewski B., Allen P.M., and Hogquist K.A.. 2014. Positive and negative selection of the T cell repertoire: what thymocytes see (and don’t see). Nat. Rev. Immunol. 14:377–391. 10.1038/nri3667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoechel B., Roderick J.E., Williamson K.E., Zhu J., Lohr J.G., Cotton M.J., Gillespie S.M., Fernandez D., Ku M., Wang H., et al. . 2014. An epigenetic mechanism of resistance to targeted therapy in T cell acute lymphoblastic leukemia. Nat. Genet. 46:364–370. 10.1038/ng.2913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., and Durbin R.. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 25:1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay H., Burger A., Biyong B., Felker A., Hess C., Zaugg J., Chiavacci E., Anders C., Jinek M., Mosimann C., and Robinson M.D.. 2016. CrispRVariants charts the mutation spectrum of genome engineering experiments. Nat. Biotechnol. 34:701–702. 10.1038/nbt.3628 [DOI] [PubMed] [Google Scholar]
- Liu Y., Easton J., Shao Y., Maciaszek J., Wang Z., Wilkinson M.R., McCastlain K., Edmonson M., Pounds S.B., Shi L., et al. . 2017. The genomic landscape of pediatric and young adult T-lineage acute lymphoblastic leukemia. Nat. Genet. 49:1211–1218. 10.1038/ng.3909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe S.W., Bodis S., McClatchey A., Remington L., Ruley H.E., Fisher D.E., Housman D.E., and Jacks T.. 1994. p53 status and the efficacy of cancer therapy in vivo. Science. 266:807–810. 10.1126/science.7973635 [DOI] [PubMed] [Google Scholar]
- Mansour M.R., Sulis M.L., Duke V., Foroni L., Jenkinson S., Koo K., Allen C.G., Gale R.E., Buck G., Richards S., et al. . 2009. Prognostic implications of NOTCH1 and FBXW7 mutations in adults with T-cell acute lymphoblastic leukemia treated on the MRC UKALLXII/ECOG E2993 protocol. J. Clin. Oncol. 27:4352–4356. 10.1200/JCO.2009.22.0996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R., and Reinberg D.. 2011. The Polycomb complex PRC2 and its mark in life. Nature. 469:343–349. 10.1038/nature09784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marks D.I., Paietta E.M., Moorman A.V., Richards S.M., Buck G., DeWald G., Ferrando A., Fielding A.K., Goldstone A.H., Ketterling R.P., et al. . 2009. T-cell acute lymphoblastic leukemia in adults: clinical features, immunophenotype, cytogenetics, and outcome from the large randomized prospective trial (UKALL XII/ECOG 2993). Blood. 114:5136–5145. 10.1182/blood-2009-08-231217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer M.D. 1975. Cell kinetics and practical consequences for therapy of acute leukemia. N. Engl. J. Med. 293:389–393. 10.1056/NEJM197508212930807 [DOI] [PubMed] [Google Scholar]
- McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., and DePristo M.A.. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. 10.1101/gr.107524.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren W., Pritchard B., Rios D., Chen Y., Flicek P., and Cunningham F.. 2010. Deriving the consequences of genomic variants with the Ensembl API and SNP Effect Predictor. Bioinformatics. 26:2069–2070. 10.1093/bioinformatics/btq330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menten B., Pattyn F., De Preter K., Robbrecht P., Michels E., Buysse K., Mortier G., De Paepe A., van Vooren S., Vermeesch J., et al. . 2005. arrayCGHbase: an analysis platform for comparative genomic hybridization microarrays. BMC Bioinformatics. 6:124 10.1186/1471-2105-6-124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J.A., Wang J., Hogan L.E., Yang J.J., Dandekar S., Patel J.P., Tang Z., Zumbo P., Li S., Zavadil J., et al. . 2013. Relapse-specific mutations in NT5C2 in childhood acute lymphoblastic leukemia. Nat. Genet. 45:290–294. 10.1038/ng.2558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T., Shimizu S., Watanabe T., Yamaguchi O., Otsu K., Yamagata H., Inohara H., Kubo T., and Tsujimoto Y.. 2005. Cyclophilin D-dependent mitochondrial permeability transition regulates some necrotic but not apoptotic cell death. Nature. 434:652–658. 10.1038/nature03317 [DOI] [PubMed] [Google Scholar]
- Neff T., Sinha A.U., Kluk M.J., Zhu N., Khattab M.H., Stein L., Xie H., Orkin S.H., and Armstrong S.A.. 2012. Polycomb repressive complex 2 is required for MLL-AF9 leukemia. Proc. Natl. Acad. Sci. USA. 109:5028–5033. 10.1073/pnas.1202258109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Chonghaile T., Sarosiek K.A., Vo T.T., Ryan J.A., Tammareddi A., Moore V.G., Deng J., Anderson K.C., Richardson P., Tai Y.T., et al. . 2011. Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science. 334:1129–1133. 10.1126/science.1206727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ntziachristos P., Tsirigos A., Van Vlierberghe P., Nedjic J., Trimarchi T., Flaherty M.S., Ferres-Marco D., da Ros V., Tang Z., Siegle J., et al. . 2012. Genetic inactivation of the polycomb repressive complex 2 in T cell acute lymphoblastic leukemia. Nat. Med. 18:298–301. 10.1038/nm.2651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neil J., Grim J., Strack P., Rao S., Tibbitts D., Winter C., Hardwick J., Welcker M., Meijerink J.P., Pieters R., et al. . 2007. FBW7 mutations in leukemic cells mediate NOTCH pathway activation and resistance to gamma-secretase inhibitors. J. Exp. Med. 204:1813–1824. 10.1084/jem.20070876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudot C., Auclerc M.F., Levy V., Porcher R., Piguet C., Perel Y., Gandemer V., Debre M., Vermylen C., Pautard B., et al. . 2008. Prognostic factors for leukemic induction failure in children with acute lymphoblastic leukemia and outcome after salvage therapy: the FRALLE 93 study. J. Clin. Oncol. 26:1496–1503. 10.1200/JCO.2007.12.2820 [DOI] [PubMed] [Google Scholar]
- Place A.E., Stevenson K.E., Vrooman L.M., Harris M.H., Hunt S.K., O’Brien J.E., Supko J.G., Asselin B.L., Athale U.H., Clavell L.A., et al. . 2015. Intravenous pegylated asparaginase versus intramuscular native Escherichia coli L-asparaginase in newly diagnosed childhood acute lymphoblastic leukaemia (DFCI 05-001): a randomised, open-label phase 3 trial. Lancet Oncol. 16:1677–1690. 10.1016/S1470-2045(15)00363-0 [DOI] [PubMed] [Google Scholar]
- Pridgeon J.W., Olzmann J.A., Chin L.S., and Li L.. 2007. PINK1 protects against oxidative stress by phosphorylating mitochondrial chaperone TRAP1. PLoS Biol. 5:e172 10.1371/journal.pbio.0050172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakers-van Woerden N.L., Pieters R., Slater R.M., Loonen A.H., Beverloo H.B., van Drunen E., Heyman M., Moreno T.C., Rots M.G., van Wering E.R., et al. . 2001. In vitro drug resistance and prognostic impact of p16INK4A/P15INK4B deletions in childhood T-cell acute lymphoblastic leukaemia. Br. J. Haematol. 112:680–690. 10.1046/j.1365-2141.2001.02586.x [DOI] [PubMed] [Google Scholar]
- Reynolds C., Roderick J.E., LaBelle J.L., Bird G., Mathieu R., Bodaar K., Colon D., Pyati U., Stevenson K.E., Qi J., et al. . 2014. Repression of BIM mediates survival signaling by MYC and AKT in high-risk T-cell acute lymphoblastic leukemia. Leukemia. 28:1819–1827. 10.1038/leu.2014.78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubnitz J.E., Behm F.G., Pui C.H., Evans W.E., Relling M.V., Raimondi S.C., Harrison P.L., Sandlund J.T., Ribeiro R.C., Grosveld G., and Downing J.R.. 1997. Genetic studies of childhood acute lymphoblastic leukemia with emphasis on p16, MLL, and ETV6 gene abnormalities: results of St Jude Total Therapy Study XII. Leukemia. 11:1201–1206. 10.1038/sj.leu.2400779 [DOI] [PubMed] [Google Scholar]
- Sadelain M., Papapetrou E.P., and Bushman F.D.. 2012. Safe harbours for the integration of new DNA in the human genome. Nat. Rev. Cancer. 12:51–58. 10.1038/nrc3179 [DOI] [PubMed] [Google Scholar]
- Sanjana N.E., Shalem O., and Zhang F.. 2014. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 11:783–784. 10.1038/nmeth.3047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarffe J.H., Hann I.M., Evans D.I., Morris Jones P., Palmer M.K., Lilleyman J.S., and Crowther D.. 1980. Relationship between the pretreatment proliferative activity of marrow blast cells and prognosis of acute lymphoblastic leukaemia of childhood. Br. J. Cancer. 41:764–771. 10.1038/bjc.1980.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schabel F.M. Jr., Skipper H.E., Trader M.W., and Wilcox W.S.. 1965. Experimental evaluation of potential anticancer agents. XIX. Sensitivity of nondividing leukemic cell populations to certain classes of drugs in vivo. Cancer Chemother. Rep. 48:17–30. [PubMed] [Google Scholar]
- Schmitt C.A., McCurrach M.E., de Stanchina E., Wallace-Brodeur R.R., and Lowe S.W.. 1999. INK4a/ARF mutations accelerate lymphomagenesis and promote chemoresistance by disabling p53. Genes Dev. 13:2670–2677. 10.1101/gad.13.20.2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X., Liu Y., Hsu Y.J., Fujiwara Y., Kim J., Mao X., Yuan G.C., and Orkin S.H.. 2008. EZH1 mediates methylation on histone H3 lysine 27 and complements EZH2 in maintaining stem cell identity and executing pluripotency. Mol. Cell. 32:491–502. 10.1016/j.molcel.2008.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon C., Chagraoui J., Krosl J., Gendron P., Wilhelm B., Lemieux S., Boucher G., Chagnon P., Drouin S., Lambert R., et al. . 2012. A key role for EZH2 and associated genes in mouse and human adult T-cell acute leukemia. Genes Dev. 26:651–656. 10.1101/gad.186411.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souroullas G.P., Jeck W.R., Parker J.S., Simon J.M., Liu J.Y., Paulk J., Xiong J., Clark K.S., Fedoriw Y., Qi J., et al. . 2016. An oncogenic Ezh2 mutation induces tumors through global redistribution of histone 3 lysine 27 trimethylation. Nat. Med. 22:632–640. 10.1038/nm.4092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su I.H., Dobenecker M.W., Dickinson E., Oser M., Basavaraj A., Marqueron R., Viale A., Reinberg D., Wülfing C., and Tarakhovsky A.. 2005. Polycomb group protein ezh2 controls actin polymerization and cell signaling. Cell. 121:425–436. 10.1016/j.cell.2005.02.029 [DOI] [PubMed] [Google Scholar]
- Townsend E.C., Murakami M.A., Christodoulou A., Christie A.L., Köster J., DeSouza T.A., Morgan E.A., Kallgren S.P., Liu H., Wu S.C., et al. . 2016. The Public Repository of Xenografts Enables Discovery and Randomized Phase II-like Trials in Mice. Cancer Cell. 29:574–586. 10.1016/j.ccell.2016.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzoneva G., Perez-Garcia A., Carpenter Z., Khiabanian H., Tosello V., Allegretta M., Paietta E., Racevskis J., Rowe J.M., Tallman M.S., et al. . 2013. Activating mutations in the NT5C2 nucleotidase gene drive chemotherapy resistance in relapsed ALL. Nat. Med. 19:368–371. 10.1038/nm.3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vo T.T., Ryan J., Carrasco R., Neuberg D., Rossi D.J., Stone R.M., Deangelo D.J., Frattini M.G., and Letai A.. 2012. Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell. 151:344–355. 10.1016/j.cell.2012.08.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood B.L., Winter S., Dunsmore K.P., Devidas M., Chen S., Asselin B.L., Esiashvili N., Loh M., Winick N.J., Carroll W.L., et al. . 2014. T-Lymphoblastic Leukemia (T-ALL) Shows Excellent Outcome, Lack of Significance of the Early Thymic Precursor (ETP) Immunophenotype, and Validation of the Prognostic Value of End-Induction Minimal Residual Disease (MRD) in Children’s Oncology Group (COG) study AALL0434. Blood (ASH annual meeting abstracts) 124:001. [Google Scholar]
- Xu J., Shao Z., Li D., Xie H., Kim W., Huang J., Taylor J.E., Pinello L., Glass K., Jaffe J.D., et al. . 2015. Developmental control of polycomb subunit composition by GATA factors mediates a switch to non-canonical functions. Mol. Cell. 57:304–316. 10.1016/j.molcel.2014.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Ding L., Holmfeldt L., Wu G., Heatley S.L., Payne-Turner D., Easton J., Chen X., Wang J., Rusch M., et al. . 2012. The genetic basis of early T-cell precursor acute lymphoblastic leukaemia. Nature. 481:157–163. 10.1038/nature10725 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.