Abstract
TET2 somatic mutations occur in ~10% of DLBCLs but are of unknown significance. Herein we show that TET2 is required for the humoral immune response and is a DLBCL tumor suppressor. TET2 loss of function disrupts transit of B-cells through germinal centers (GC), causing GC hyperplasia, impaired class switch recombination, blockade of plasma cell differentiation and a pre-neoplastic phenotype. TET2 loss was linked to focal loss of enhancer hydroxymethylation and transcriptional repression of genes that mediate GC exit such as PRDM1. Notably, these enhancers and genes are also repressed in CREBBP-mutant DLBCLs. Accordingly, TET2 mutation in patients yields a CREBBP-mutant gene expression signature, CREBBP and TET2 mutations are generally mutually exclusive, and hydroxymethylation loss caused by TET2 deficiency impairs enhancer H3K27 acetylation. Hence TET2 plays a critical role in the GC reaction and its loss of function results in lymphomagenesis through failure to activate genes linked to GC exit signals.
Keywords: DLBCL, GC B-cells, PC differentiation, epigenetics, DNA methylation, TET proteins
INTRODUCTION
Diffuse large B-cell lymphomas (DLBCLs) are aggressive tumors and the most common form of non-Hodgkin lymphoma (1). These tumors arise from fully mature and differentiated B-cells transiting the germinal center (GC) reaction. GCs form transiently when resting B-cells are activated by T-cells reacting to an antigenic challenge. GC B-cells replicate at an accelerated rate and diversify their immunoglobulin (Ig) genes through somatic hypermutation mediated by the enzyme activation induced cytosine deaminase (AICDA) (2). This clonal expansion and mutagenesis enables the emergence of B-cells with de novo high affinity immunoglobulins (Ig) against specific antigens. Only those GC B-cells expressing high affinity B-cell receptors (i.e. Ig) for specific antigen are selected by T-cells to survive and exit the GC reaction, after which they subsequently differentiate into either memory or long-lived antibody secreting PC (3). This proliferative and mutagenic phenotype makes GC B-cells especially prone to undergo malignant transformation (4).
Epigenetic mechanisms play a central role in the entry and exit of B-cells to the GC reaction, and are commonly disrupted in DLBCL (5). Indeed somatic mutations of histone modifier genes have emerged as a genetic hallmark of this disease (6). Recent studies have shed light into the mechanism though which the commonly mutated histone modifiers KMT2D, EZH2, CREBBP and EP300 contribute to pathogenesis. In the case of EZH2, gain of function mutations result in increased H3K27me3 mediated silencing of gene promoters for cell checkpoint and PC differentiation genes (7,8). KMT2D loss of function mutation results in focal reduction of H3K4me1 and hence in impaired activation of gene enhancers that normally respond to CD40 signaling and control expression of tumor suppressor genes such as TNFAIP3 (9,10). CREBBP loss of function mutation causes focal loss of H3K27 acetylation of enhancers and hence transcriptional repression of genes involved in GC exit signals including the BCR, cytokine, CD40 and antigen presentation pathways (11,12). Hence disruption of histone modifying complexes is well documented as a driving force in malignant transformation of GC B-cells.
In contrast, not much is known in DLBCL regarding the contribution and relevance of the other major arm of epigenetic modifiers, in particular the enzymes that mediate covalent modifications of DNA. It is known that GC B-cells acquire a cytosine hypomethylation signature that is highly dependent on the actions of AICDA, possibly due to replacement of methylated cytosines with unmethylated nucleotides during somatic hypermutation (13). Cytosine methylation profiling studies have shown that DLBCLs feature aberrant cytosine methylation signatures and increased inter- and intra-tumor epigenetic DNA methylation heterogeneity, which is also directly linked to the presence of AICDA (14–17). Virtually nothing is known about cytosine hydroxymethylation in the humoral immune response and DLBCL.
Intriguingly, genome sequencing studies show that ~10% of DLBCLs display somatic mutations of TET2 (6,18), a dioxygenase that converts 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) (19). TET2 is well known to be frequently mutated in myeloid disorders including acute myeloid leukemia and myelodysplasia (20–22). Somatic mutations of TET2 in HSCs are also linked to the development of angioimmunoblastic T-cell lymphoma (23). 5hmC has been described as a necessary intermediate step in DNA demethylation in cooperation with the base excision repair machinery, but is also an independent epigenetic mark linked to transcriptional activation (24–26). Previous studies in hematopoietic stem/progenitor cells (HSPC) and embryonic stem cells have shown that 5hmC is enriched in gene bodies and enhancers and can modulate enhancer activity, affecting gene expression (26–30).
Notably, it has been shown that DLBCL patients with TET2 mutations harbor the same mutation in their HSPCs (31). Since TET2 mutations are typically linked to HSCs and may be present in individuals with healthy hematopoiesis it is not clear whether the presence of TET2 mutations in DLBCL is a passenger event or can be specifically shown to function as a tumor suppressor in this disease. Here, we investigate whether and how TET2 loss of function is linked to malignant transformation of GC B-cells by performing functional experiments in mouse models and linking these effects to observations from human patients with TET2 mutant DLBCLs.
RESULTS
Tet2 deletion in hematopoietic cells induces GC hyperplasia
Since DLBCLs arise from GC B-cells, we studied whether TET2 loss affected GC B-cell formation. We first examined Tet2 expression in the humoral immune response based on published RNA-seq profiles (32) and observed that Tet2/TET2 was expressed in both naïve and GC B-cells in mouse and human cells, with reduced levels in further differentiated cells (Supplementary Fig. S1A). Expression was validated using qPCR in mouse GC B cells (Supplementary Fig. S1B). To reflect the situation in human DLCBL patients where TET2 mutations are observed in HSCs, we performed conditional deletion of Tet2 in HSCs in mice under the control of Vav-Cre to delete this gene in the HSC compartment. To determine if HSC knockout of Tet2 could manifest effects in mature B-cells undergoing the GC reaction, we induced T cell-dependent antigen response with sheep red blood cell injections (SRBC) in WT (Vav-Cre/Tet2+/+) and Tet2-deficient (Vav-Cre/Tet2−/−) mice. Animals were sacrificed ten days later and spleens examined by flow cytometry and immunohistochemistry. We observed a significant increase in the percentage of GC B-cells (B220+GL7+CD95+) from Vav-Cre/Tet2−/− mice compared to Vav-Cre/Tet2+/+ mice (p<0.001, Fig. 1A). Tet2 deletion in Vav-Cre/Tet2−/− GC B cells was confirmed by qPCR (Supplementary Fig. S1B). In contrast, there was no difference in the percentage of total B cells (B220+ cells) or the other B cell subsets in the spleen between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice including mature B cells (B220+IgD+IgM+), transitional B cells (B220+IgDintIgM+), follicular B cells (B220+CD23+CD21+), marginal zone B cells (B220+CD23lowCD21+) and plasmablasts/PC (CD138+) (Fig. 1B and Supplementary Fig. S1C). Immunohistochemical analysis of the spleens using PNA and Ki-67 revealed an increase in the size of the GCs of Vav-Cre/Tet2−/− compared to Vav-Cre/Tet2+/+ mice (Fig. 1C and D), but not in the number of GCs between Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ animals (Fig. 1C and E).
Figure 1. Tet2 deletion in hematopoietic cells induces GC hyperplasia.
A, Representative flow cytometry plot and quantification of (B220+CD95+GL7+) GC B-cells from Vav-Cre/Tet2+/+ (n=5) and Vav-Cre/Tet2−/− (n=5) mice at day 10 after immunization with SRBC. B, Quantification of flow cytometry data corresponding to total B cells (B220+), mature B cells (B220+IgD+IgM+), transitional B cells (B220+IgDintIgM+), follicular B cells (B220+CD23+CD21+), marginal zone B cells (B220+CD23lowCD21+) and plasmablasts/PC (CD138+) in the spleens of Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice. C, Representative histologic sections of formalin-fixed, paraffin-embedded spleens from Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice. Sections were stained with H&E and antibodies specific for B220, PNA and Ki-67. D, E, Quantification of GC area (D) and number of GCs (E) in the spleens of Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice. two-tailed t test, ***p<0.001 ****p<0.0001.
GC hyperplasia induced by Tet2 loss of function is B-cell autonomous.
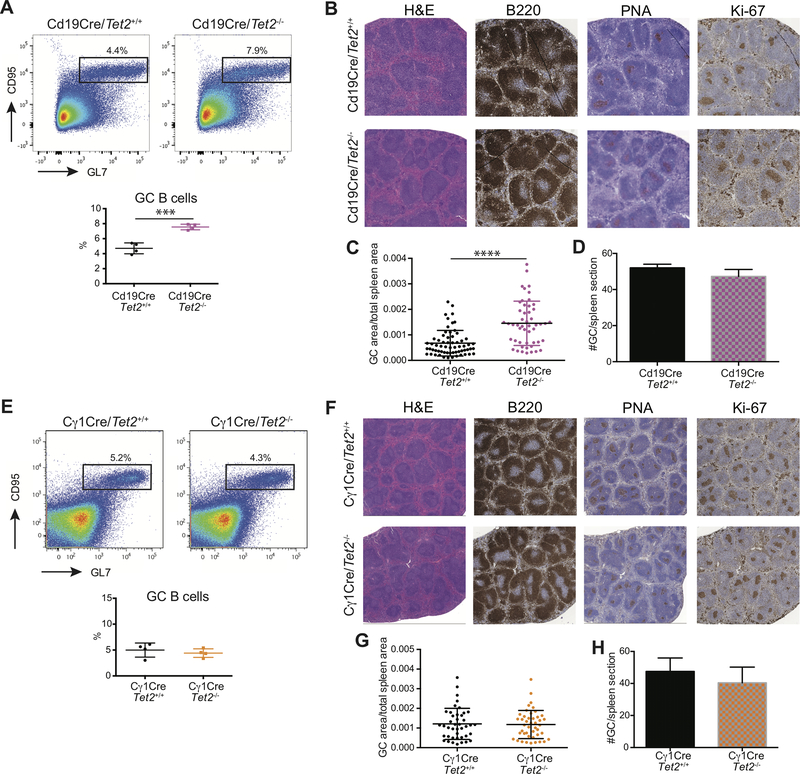
Because the Vav-Cre allele knocks out Tet2 in all hematopoietic lineages, we cannot exclude that this GC is caused by Tet2 loss of function in some other cell type. Therefore, we generated Cd19-Cre/Tet2−/− mice, in which Tet2 loss of function is specifically induced during B-cell development. These mice were immunized with SRBC as described above to examine GC formation. Similar to Vav-Cre/Tet2−/− mice, Cd19-Cre/Tet2−/− animals also displayed a significant increase in the proportion of GC B-cells after immunization (Fig. 2A), whereas other mature B-cell subpopulations were not affected (Supplementary Fig. S2A). As in the Vav-Cre model, we also observed GC hyperplasia as shown by immunohistochemistry with staining for PNA and Ki67 (Fig 2B). This was due to expansion in the size, not the number, of GCs in Cd19-cre/Tet2−/− animals compared to Cd19-cre/Tet2+/+ mice (Fig. 2C and D). This result indicates that GC hyperplasia is B-cell autonomous and not dependent on TET2 loss of function in other cell lineages.
Figure 2. GC hyperplasia induced by Tet2 loss of function is B-cell autonomous.
A, Representative flow cytometry plot and quantification of (B220+CD95+GL7+) GC B-cells from Cd19-Cre/Tet2+/+ (n=4) and Cd19-Cre/Tet2−/− (n=4) mice at day 10 after immunization with SRBC. B, Representative histologic sections of formalin-fixed, paraffin-embedded spleens from Cd19-Cre/Tet2+/+ and Cd19-Cre/Tet2−/− mice. Sections were stained with H&E and antibodies specific for B220, PNA and Ki-67. C, D, Quantification of GC area (C) and number of GCs (D) in the spleens of Cd19-Cre/Tet2+/+ and Cd19-Cre/Tet2−/− mice. E, Representative flow cytometry plot and quantification of (B220+CD95+GL7+) GC B-cells from Cγ1-Cre/Tet2+/+ (n=5) and Cγ1-Cre/Tet2−/− (n=5) mice at day 10 after immunization with SRBC. F, Representative histologic sections of formalin-fixed, paraffin-embedded spleens from Cγ1-Cre/Tet2+/+ and Cγ1-Cre/Tet2−/− mice. Sections were stained with H&E and antibodies specific for B220, PNA and Ki-67. G, H, Quantification of GC area (G) and number of GCs (H) in the spleens of Cγ1-Cre/Tet2+/+ and Cγ1-Cre/Tet2−/− mice. two-tailed t test, ***p<0.001 ****p<0.0001.
We next induced deletion of TET2 specifically in already formed GC B-cells crossing Tet2f/f mice with those bearing the Cγ1-Cre (GC specific) allele. In contrast to the Vav-Cre and CD19-Cre models, the immunized Cγ1-Cre/Tet2−/− mice showed no differences in the percentage of GC B-cells between TET2-deficient (Cγ1-Cre/Tet2−/−) and WT (Cγ1-Cre/Tet2+/+) mice (Fig. 2E). Likewise, the percentage of total B cells and other B cell subpopulations was similar in Cγ1-Cre /Tet2−/− and Cγ1-Cre /Tet2+/+ mice (Supplementary Fig. S2B). We also found that the number and size of GCs was comparable in Cγ1-Cre /Tet2−/− and Cγ1-Cre /Tet2+/+ mice (Fig. 2F-H). We confirmed efficient Tet2 deletion in both CD19-Cre/Tet2−/− and Cγ1-Cre/Tet2−/− GC B cells (Supplementary Fig. S2C and D), excluding the possibility that the different phenotypes reflected differences in excision efficiency by the two Cre lines. The differential effect in the Cd19-cre vs. Cγ1-Cre is consistent with what has been observed with other DLBCL epigenetic tumor suppressor models (10,12).
Tet2 loss of function impairs affinity maturation and PC differentiation in a B-cell autonomous manner
The aberrant GC phenotype induced by Tet2 loss of function prompted us to also determine whether it is required for immunoglobulin affinity maturation. Therefore we analyzed the ability of Vav-Cre/Tet2−/− mice to generate specific antibodies against the T cell-dependent antigen NP-Chicken Gamma Globulin (NP-CGG). Mice were immunized with NP-CGG and then received a booster shot at day 21, after which serum was collected at day 35 to detect Ig and bone marrow at day 60 to identify high affinity PC (Fig. 3A). ELISA analysis of serum revealed a significant reduction in the titers of both low (NP26) and high (NP7) affinity IgG1 antibodies in Vav-Cre/Tet2−/− mice compared to Vav-Cre/Tet2+/+ mice (p<0.001 Fig. 3B). The defect disproportionately affected the high affinity species, consistent with a perturbation in AICDA driven immunoglobulin affinity maturation during the GC response (p<0.01 Fig. 3C). We also tested lambda light chain maturation (Igλ) and observed the same pattern of reduced production of both low and high affinity Igλ in Vav-Cre/Tet2−/− mice (NP26 p<0.0001; NP4 p<0.001; Fig. 3B), again with disproportionate effect on the high affinity species (ratio NP7/NP26 p<0.01 Fig. 3C). Notably, we found a similar reduction in high and low affinity IgG1 and Igλ, predominantly in high affinity antibodies, in Cγ1-Cre/Tet2−/− mice compared to Cγ1-Cre /Tet2+/+ mice (Supplementary Fig. S3A and B), indicating that TET2 deletion at the GC stage also impairs affinity maturation. In addition, we performed ELISPOT assays in bone marrow at day 60 to measure the abundance of NP-specific antibody secreting cells. This analysis yielded a marked reduction in cells that secrete high and low affinity IgG1 NP antibodies in the bone marrow of Vav-Cre/Tet2−/− mice (p<0.01 Fig. 3D. In contrast the abundance of non-class switched anti-NP IgM secreting cells was not significantly reduced compared to controls (Fig. 3E). Importantly, this effect was observed regardless of whether TET2 loss of function was induced in HSCs (Vav-Cre/Tet2−/−), early B-cells (Cd19-Cre/Tet2−/−) or in GCs (Cγ1-Cre/Tet2−/−) (Fig. 3D and Supplementary Fig. S3C-F). These findings are suggestive of an impairment in GC exit functions and induction of the PC differentiation program. Accordingly, we observed a reduction in the percentage of PC in the spleen of SRBC immunized constitutive Tet2-deficient mice (Tet2−/−) (n=5) as compared to controls Tet2+/+ (n=4) (p<0.01 Fig. 3F). This GC exit/PC differentiation block is consistent with the mechanism of malignant transformation induced by loss of function of epigenetic regulators in DLBCL (9–12). Importantly this effect is B-cell autonomous and observed regardless of whether Tet2 deletion was induced in HSCs, Pre-B-cells or GC B-cells.
Figure 3. Tet2 loss of function impairs affinity maturation and PC differentiation in a B-cell autonomous manner.
A, Schematic diagram of the protocol of primary and secondary immunizations. B, Thirty five days after immunization (fourteen days after boost), NP-specific antibodies (IgG1 and Igλ) were measured in the sera of Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice by ELISA. C, NP7/NP26 ratio of NP-specific antibodies detected in B. D, E, Sixty days after immunization (forty days after boost), NP-specific IgG1 (D) and IgM (E) secreting cells from the bone marrow of Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice were quantified by ELISPOT. two-tailed t test, *p<0.05 **p<0.01 ***p<0.001. F, Splenic PC formation after SRBC immunization of Tet2+/+ and Tet2−/− mice; shown as FACS profiles (left) and quantitation (right) on day 32; each symbol represents one mouse. G, Schematic illustration of the in vitro GC B-cells (iGCB) and PB (iPB) culture system. H, Number of live Tet2+/+ and Tet2−/− B cells cultured on 40LB with IL4 on D4 and IL21 on D8. I, J, Flow cytometry plots of iGCB cells on D4 (I) and D8 (J); the gated area shows the live iGCB cells (CD19+GL7+FAS+). Histograms represent the percentage (upper) and the cell number (lower) of iGCB cells. K, L, Flow cytometry plots of iPB cells on D4 (K) and D8 (L); the gated area shows the live iPB cells (CD19+GL7+FAS+). Histograms represent the percentage (upper) and the cell number (lower) of iPB cells. For D4 symbols represent triplicate of 5 independent experiments. For D8 symbols represent triplicate of 4 independent experiments. All p values were calculated using unpaired Student’s t-test, *p < 0.05, **p < 0.005, ***p < 0.0005 in all experiments.
To further establish the cell autonomous nature of this phenotype, we sorted naïve B-cells from Tet2−/− animals and compared their behavior in vitro under culture conditions that mimic the GC reaction (33) (Fig. 3G). This method involves the plating of naïve B-cells together with 40LB cells and sequential exposure to IL-4 for four days followed by IL-21 for another four days (33). We plated equal numbers of Tet2−/− and Tet2+/+ cells and observed significant increase in the amount of B-cells both at the four-day (D4) and eight-day (D8) timepoints derived from the Tet2−/− mice (p<0.0005 Fig. 3H). Furthermore, flow cytometry analysis showed significantly increased numbers of induced GC B-cells (iGC; CD19+GL7+CD95+) derived from Tet2−/− vs Tet2+/+ mice at D4 and D8 (Fig. 3I and J). In marked contrast, the induced plasmablast (iPB) population (CD19+CD138+) was significantly lower in Tet2−/− compared to Tet2+/+ cells at both timepoints (Fig. 3K and L).
These in vitro data mirror the in vivo results and indicate that Tet2 loss in B cells induces expansion of GC B-cells with corresponding blockade of PC differentiation in a B-cell autonomous fashion. Along these lines analysis of CD19+CD138+ Tet2−/− iPB showed significantly higher expression of the GC B cell–associated marker GL7, lower expression of CD138 and higher expression of CD19 compared to Tet2+/+ cells (Supplementary Fig. S3G). Analysis of IgG1 and IgM expression on D4 iGC B cells showed that the percentage of IgG1-switched Tet2−/− iGC B cells was significantly lower compared to Tet2+/+ cells, suggesting a defect in class switch recombination, similar to what observed in vivo (Supplementary Fig. S3H).
Tet2-deficient GC B cells exhibit repression of genes involved in GC exit.
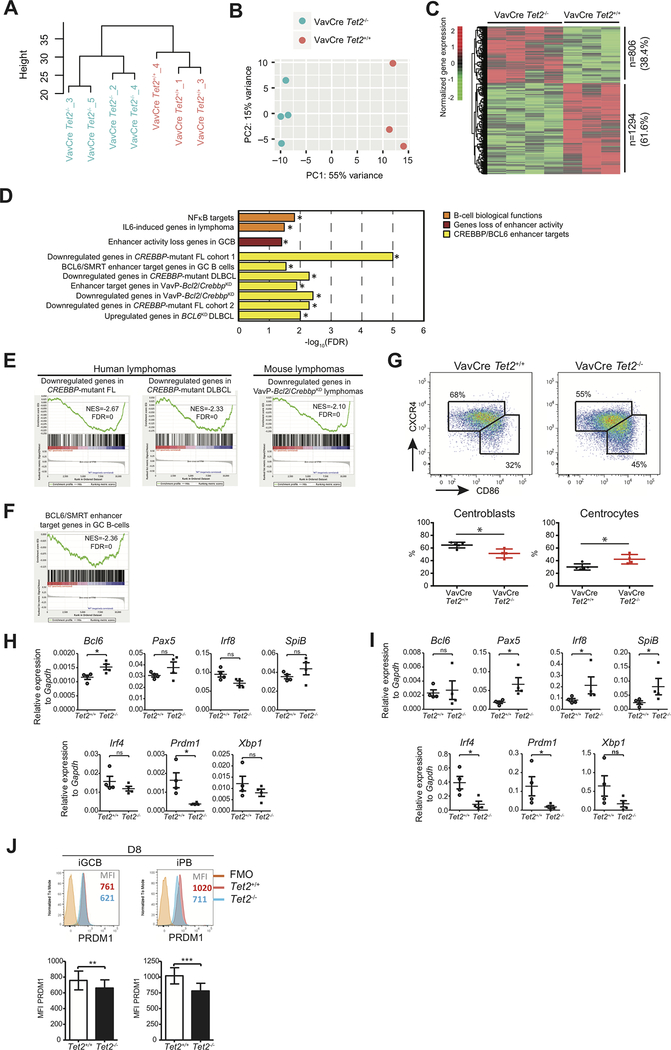
In order to identify putative mechanisms responsible for the GC disruption and putative tumor suppressor function of TET2, we analyzed the impact of Tet2 loss of function on the transcriptional signatures of GC B cells by RNAseq. Unsupervised hierarchical clustering and principal component analysis showed a clear difference in transcriptional profiles between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− sorted GC B-cells (Fig. 4A and B). We then performed a supervised analysis and identified 2100 differentially expressed genes (DEGs) between Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ GC B cells (FC>1.2, p<0.05; Fig. 4C). Notably, the majority of these were skewed towards being more repressed in Tet2-deficient GC B-cells (n=1294, 61.6%), suggesting that TET2 is mainly a transcriptional activator in GC B-cells. Gene set enrichment analysis (GSEA) was performed against an extensive database of GC and DLBCL relevant gene sets. This analysis indicated significant enrichment for downregulation of genes involved in the exit of GC B-cells from the GC reaction such as NFκB and IL6 induced genes (Fig. 4D and Supplementary Table 1). Enrichment of those gene sets among downregulated genes in Vav-Cre/Tet2−/− was independently confirmed using a hypergeometric test (Supplementary Fig. S4A).
Figure 4. Tet2-deficient GC B cells exhibit repression of genes involved in GC exit.
A, B, Hierarchical clustering (A) and principal component analysis (B) of RNAseq data of Vav-Cre/Tet2+/+ (n=3) and Vav-Cre/Tet2−/− (n=4) GC B cells. C, Normalized gene expression levels of 2100 differentially expressed genes between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− GC B cells. Expression levels were scaled by row. D, Minus log 10 of the FDR scores for the enrichment of 10 gene sets involved in the exit of GC B-cells from the GC. FDR scores were obtained using Gene Set Enrichment Analysis (GSEA) as described in the methods section. E, F GSEA enrichment plots showing correlation of different genesets with ranked expression change between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− GC B cells. NES, normalized enrichment score; FDR, false discovery rate. G, Representative flow cytometry plot and quantification of centroblasts (CXCR4highCD86high) and centrocytes (CXCR4lowCD86low) in the GC of Vav-Cre/Tet2+/+ (n=5) and Vav-Cre/Tet2−/− (n=5) mice at day 10 after immunization with SRBC. H, Expression of Bcl6, Pax5, Irf8, SpiB, Irf4, Prdm1 and Xbp1 by iGCB cells on D8, as measured by quantitative RT-PCR (n=4). I, Expression of Bcl6, Pax5, Irf8, SpiB, Irf4, Prdm1 and Xbp1 by iPB on D8, as measured by quantitative RT-PCR (n=4). J, Representative PRDM1 intracellular staining profile of iGCB (CD19+CD138−) and iPB (CD19+CD138+) at D8. Numbers indicate the median fluorescence intensity (MFI) of PRDM1. All p values were calculated using unpaired Student’s t test, *p < 0.05 and ns: not significant, in all experiments.
We also noted enrichment for repression of genes whose enhancers are normally repressed during the GC reaction but are induced upon GC exit (Fig. 4D). The genes controlled by these enhancers are repressed in GC B-cells by the BCL6 transcriptional repressor with its corepressor SMRT, and reactivated upon GC exit by the CREBBP histone acetyltransferase (11,34). These genes are accordingly aberrantly repressed in DLBCL and FL patients with CREBBP somatic mutations (11,12). Notably, genes repressed in TET2 deficient GC B-cells were significantly enriched for genes that are also downregulated in murine and primary human CREBBP deficient lymphomas (Fig. 4D and E and Supplementary Table 1). Likewise there was significant enrichment for BCL6/SMRT regulated genes as well as genes that are upregulated upon BCL6 knockdown (Fig. 4D and F and Supplementary Table 1). Hence TET2 function is associated with regulation of genes whose enhancers are switched off in the GC reaction by BCL6 and activated upon GC exit, through the actions of CREBBP.
These data prompted us to examine the impact of TET2 deficiency on B-cells exiting the GC reaction. GCs are divided into two regions. The GC dark zone is composed of proliferating and mutating GC B-cells called centroblasts, whereas the GC light zone contains non-cycling GC B-cells that have migrated from the dark zone, which are called centrocytes. Centrocytes can be induced to exit the GC reaction by T follicular helper (TFH) cells (3). Vav-Cre/Tet2−/− mice manifested disruption and skewing in the polarity of the GC reaction, with a relative and significant increase in the proportion of centrocytes (Fig. 4G) as compared to control mice. We observed similar skewed proportion towards centrocytes in Cd19-Cre/Tet2−/− (Supplementary Fig. S4B) but not in Cγ1-Cre/Tet2−/− mice (Supplementary Fig. S4C). These results suggest that Tet2 deficiency impairs the ability of GC B-cells to exit the GC reaction, leading to their accumulation as centrocytes. To confirm that the changes in gene expression observed in total GC B-cells from Vav-Cre/Tet2−/− mice were not a consequence of the skewed GC B-cell populations in these animals, we performed RNAseq in GC B-cells from Cγ1-Cre/Tet2−/− and Cγ1-Cre/Tet2+/+ mice, which do not exhibit such skewing. This analysis again showed clear separation between TET2-deficient and WT GC B-cells (Supplementary Fig. S5A and B), with predominant downregulation of gene expression after TET2 deletion in Cγ1-Cre/Tet2−/− GC B-cells compared to Cγ1-Cre/Tet2+/+ cells. More specifically, 1768 genes were down-regulated (59.5%) while 1204 were upregulated (40.5%) as a result of TET2 loss-of-function in Cγ1Cre/Tet2−/− GC B-cells (FC>1.2 FDR<0.05; Supplementary Fig. S5C). Similar to the case of Vav-Cre/Tet2−/− GC B-cells, genes downregulated in Cγ1Cre/Tet2−/− GC B-cells featured enrichment for genes downregulated in human FL and DLBCL with CREBBP mutations and mouse Crebbp knockdown cells, and NFκB targets (Supplementary Fig. S5D and E).
To further explore the centrocyte defect in the absence of Tet2 we used qPCR to examine the expression of several key transcription factors (TF) that must be switched off (Pax5, Irf8, SpiB, Bcl6) for exit from the GC, or switched on (Irf4, Xbp1 and Prdm1) for GC exit and the transition to the PC phenotype, using our in vitro GC culture system. This analysis yielded progressively more significant aberrant expression of these genes as these iGC cells transited the ex vivo GC reaction and attempted to differentiate to the PC state. Thus, we observed a significant increase of the GC TFs Bcl6 and the corresponding reduction in the GC exit and PC TF Prdm1 at D8 in Tet2−/− iGCB compared to Tet2+/+ cells (p<0.05, Fig. 4H). Similar results were obtained for iPB at D8, with higher expression of Pax5, Irf8 and SpiB and lower expression of Irf4 and Prdm1 in Tet2−/− than Tet2+/+ cells (p<0.05, Fig. 4I). Flow cytometry for the PC master regulator PRDM1 indicated a reduction in protein expression at D8 in Tet2−/− iGCB (p<0.01) and iPB (p<0.001) compared to Tet2+/+ cells (Fig. 4J). Together these results show that Tet2 loss causes failure to induce genes repressed in the GC reaction and which drive terminal differentiation of GC B-cells.
TET2 deficiency results in reduction of enhancer cytosine hydroxymethylation and histone acetylation
To gain insight into the mechanism through which Tet2 loss of function perturbs gene expression we next examined the genomic distribution of 5hmC using hMeDIP-Seq in purified GC B-cells from Vav-Cre/Tet2−/− vs Vav-Cre/Tet2+/+ mice. 5hmC peaks were called using MACS2 and were found to be distributed predominantly in intergenic areas and introns in both WT and Tet2-deficient GC B-cells (Fig. 5A). This result is in line with previous publications showing that TET2 loss of function largely affects DNA hydroxymethylation at enhancers (29,35–40). We previously mapped the location of gene enhancers in murine GC-derived B-cells based on histone modifications (11). When these peaks were overlapped with the 5hmC peaks in Vav-Cre/Tet2+/+ GC B-cells we observed that at least 70% of enhancers were marked by 5hmC (Fig. 5B). Next, we identified the differentially hydroxymethylated regions (DHMR) between Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ GC B cells. This analysis yielded 24,645 DHMR lost in Vav-Cre/Tet2−/− GC B cells (representing 5.5% of the total 5hmC peaks) and only 51 DHMR gained in Vav-Cre/Tet2−/− cells (Fig. 5C). This focal loss of peaks was predominantly located in intergenic and intronic areas in accordance with the general distribution of 5hmC (Fig. 5D). Plotting the read density for these distal and intronic enhancer DHMRs illustrated the reduction in 5hmC reads at these sites (Fig. 5E and Supplementary Fig. S6A).
Figure 5. TET2 deficiency results in reduction of enhancer cytosine hydroxymethylation and histone acetylation.
A, Genomic distribution of 5hmC peaks in GC B-cells from Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− mice. B, Overlap of 5hmC peaks with enhancers in Vav-Cre/Tet2+/+ GC B-cells. C, D, Number (C) and genomic distribution (D) of DHMR between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− GC B-cells. E, Heatmap of the 5hmC enrichment in all DHMR overlapping with distal enhancers in Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− GC B-cells. Region spans 1 kbp up and down from the DHMR’ peak center. Heatmap was generated using deeptools (73). F, Minus log 10 of the FDR scores for the enrichment of 11 gene sets involved in the exit of GC B-cells from the GC. FDR scores were obtained using hypergeometric test for a total of 1825 genes with enhancers overlapping with DHMR. G, Correlation of differential hydroxymethylation in distal enhancers and differential gene expression between Vav-Cre/Tet2+/+ and Vav-Cre/Tet2−/− GC B-cells. H, GSEA enrichment plots showing correlation of genes associated with peaks losing 5hmC and H3K27ac signals. I, Normalized gene expression level of 253 leading edge genes associated with peaks losing 5hmC and H3K27ac signals in Vav-Cre/Tet2−/− and VavP-CrebbpKD B cells, respectively. J, Aggregation plots of 5hmC and H3K27ac enrichment in peaks losing both signals, associated with 253 genes shown in figure 5I.
To determine the function of genes affected by reduction of 5hmC we performed GSEA against our database of gene sets. Similar to what we observed by gene expression profiling, there was significant enrichment for gene sets involved in GC exit and PC differentiation including NFκB, IL6, CD40 and antigen presentation (Fig. 5F). From the mechanistic perspective hypo DHMRs were significantly associated with enhancers that are repressed in GC B-cells and induced during GC exit (Fig. 5F). Enhancers with 5hmC loss in Vav-Cre/Tet2−/− cells were enriched for genes that are repressed in CREBBP-deficient lymphomas in mice, as well as CREBBP mutant human patients with FL and DLBCL (Fig. 5F). There was also the reciprocal significant enrichment for BCL6-SMRT target enhancers and genes induced upon BCL6 knockdown (Fig. 5F). Similar results were obtained using a hypergeometric test (Supplementary Fig. S6B). These data suggest that genes downregulated in Vav-Cre/Tet2−/− GC B-cells are also affected by loss of enhancer 5hmC. To determine how robust this association was we performed an integrative analysis for 5hmC fold change and gene expression log 2 transformed fold change between Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ GC B-cells. This analysis revealed significant association of distal enhancer (up to 100 Kb) or intronic reduction in 5hmC with transcriptional repression (Fig. 5G: p=0.016 and Supplementary Fig. S6C: p=0.009, chi-squared test). Thus TET2 loss of function results in focal reduction of enhancer hydroxymethylation, which is linked to transcriptional repression of the corresponding genes.
The data also suggest that TET2 and CREBBP either cooperate or are mutually dependent on each other to activate gene enhancers that regulate exit from the GC reaction. To address this point, we first analyzed the genes with distal enhancers containing DHMR shown in Fig. 5E. We found that 60% of those genes (426 out of 703) were also associated with H3K27Ac signal loss in CREBBP-deficient B cells (p=5×10−31, hypergeometric test), indicating that there was a significant correlation between genes containing DHMR due to TET2 deficiency and those losing H3K27Ac in the absence of CREBBP. To investigate this putative association further, we identified the peaks that lose both 5hmC and H3K27Ac in Vav-Cre/Tet2−/− and Crebbp knockdown murine B-cells (11), respectively. We found 2619 peaks with at least 35% reduction of both signals. Only 0.43% of all promoters overlapped with both 5hmC and H3K27ac peaks losing signal, while 6.1% of all enhancers overlapped with both 5hmC and H3K27ac signal loss (p=0, chi-square test, Supplementary Fig. S6D). We then performed GSEA for the genes overlapping those peaks and found that they were significantly skewed towards repression in Vav-Cre/Tet2−/− and Crebbp knockdown murine B-cells (FDR<0.01 and p=2.8×10−04, Fig. 5H). Analysis of the leading edge of those gene sets identified 253 genes downregulated in both TET2- and CREBBP-deficient B cells, including Prdm1, NF-κB related genes and genes involved in GC B-cell migration and differentiation such as Il-9r, Ccr1, Cxcr5 and Tlr6 (Fig. 5I and Supplementary Table S2). Analysis of 5hmC and H3K27Ac distribution in murine TET2 or CREBBP loss of function GC B-cells confirmed the significant reduction of both signals in these 253 genes (Fig. 5J).
Aberrant repression of Prdm1 contributes to the phenotype of TET2-deficient B cells.
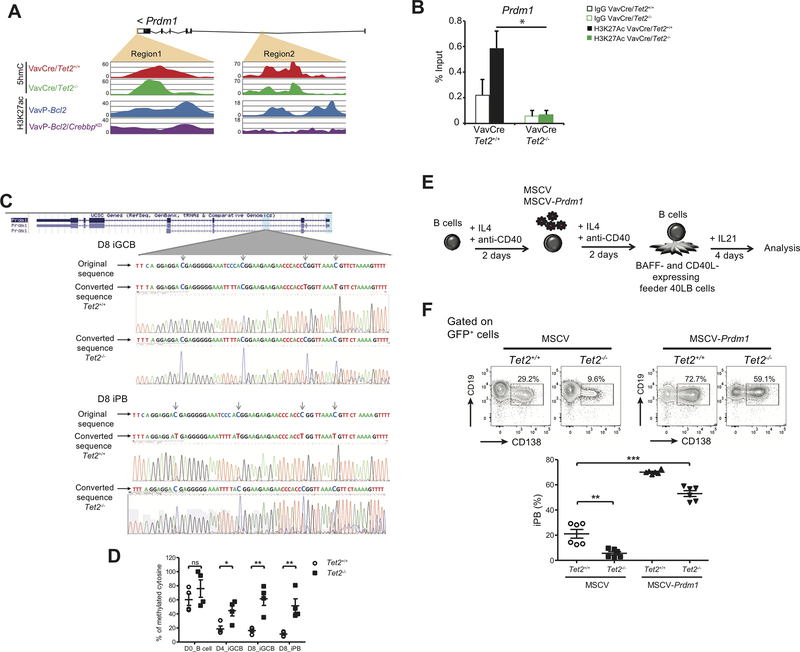
Given the defect in PC differentiation manifested by Tet2-deficient GC B-cells we next focused on the link between this phenotype and aberrant silencing of the Prdm1 gene. This locus featured two regions with concordant reduction in overlapping 5hmC and H3K27Ac peaks in the absence of TET2 and CREBBP respectively (Fig. 6A; orange areas). With the selected cutoff of at least 35% loss of both 5hmC and H3K27ac signals, we did not identify additional regions in the proximity of Prdm1 gene affected in this manner (data not shown). To confirm the link between 5hmC and H3K27Ac we performed H3K27Ac QChIP and observed a significant reduction in this histone mark in Vav-Cre/Tet2−/− compared to Vav-Cre/Tet2+/+ GC B cells (Fig. 6B). As a second validation we examined H3K27Ac by QCHIP at the Il9r locus where we had observed overlapping 5hmC and H3K27Ac (Supplementary Fig. S6E and F). TET2 loss of function has also been associated with DNA hypermethylation. We therefore performed bisulfite sequencing of a region within intron 2 described by Barwick et al. (41), as becoming hypomethylated in PB as compared to mature B-cells. This analysis detected increased methylation in Tet2−/− D8 iGC and D8 iPB compared to Tet2+/+ cells (Fig. 6C). DNA methylation in intron2 was then quantified using methylation-sensitive restriction enzyme qPCR. Data is represented as the percentage of methylated cytosine in Fig. 6D. No significant differences in Prdm1 methylation between Tet2−/− and Tet2+/+ naïve (D0) B cells were observed, whereas significantly higher percentage of methylated cytosine was observed in sorted Tet2−/− D4 iGCB (p<0;05), D8 iGCB (p<0.01) and D8 iPB (p<0.01) cells compared to Tet2+/+ cells (Fig. 6D). In Tet2+/+ cells, DNA methylation of this region decreased dramatically between naïve (D0) and D4 iGCB and further decreased in D8 iGCB and iPB. In contrast, DNA methylation in Tet2−/− cells decreased slightly between naïve (D0) and D4 iGCB and then remained at equivalent levels in D8 iGCB and iPB. These data suggest that reduction in expression of Prdm1 might also be linked to DNA hypermethylation. Finally, to determine whether aberrant repression of PRDM1 contributes to the Tet2-deficient phenotype, we transduced naive B cells with retrovirus control (MSCV) or retrovirus expressing Prdm1 (MSCV-Prdm1) (Fig. 6E). We analyzed GFP+ cells four days after culture in the presence of IL21 and observed that ectopic Prdm1 expression rescued the differentiation of TET2-deficient B cells (Fig. 6F).
Figure 6. Aberrant repression of Prdm1 contributes to the phenotype of TET2-deficient B cells.
A, Read-density tracks of H3K27ac histone modification as well as 5hmC levels in Prdm1 locus in Vav-Cre/Tet2−/−, Vav-Cre/Tet2+/+, VavP-Bcl2 and VavP-Bcl2/CrebbpKD B-cells. Areas marked in orange indicate regions losing 5hmC and H3K27ac signals. B, H3K27Ac enrichment in region 1 shown in A. C, (Top) Prdm1 DNA methylation status in iGCB cells on D8. Shown are DNA original sequence (upper line), representative bisulfite-converted DNA sequence of Tet2+/+ iGCB cells (middle line), and representative bisulfite-converted DNA sequence of Tet2−/− iGCB cells (lower line). The experiment was performed with 3 biological replicates. (Bottom) Prdm1 DNA methylation status in iPB on D8. Shown are DNA original sequence (upper line), representative bisulfite-converted DNA sequence of Tet2+/+ iPB (middle line), and representative bisulfite-converted DNA sequence of Tet2−/− iPB (lower line). The experiment was performed with 2 biological replicates. D, Quantification of DNA methylation by Methylation-sensitive restriction enzyme qPCR. Differentially methylated region represented as the percentage of methylated cytosine in Prdm1 gene were validated in Day0 (D0, naïve) B cells, day4 (D4) iGCB cells, day8 (D8) iGCB cells and day8 (D8) iPB cells (n=4). E, Schematic illustration of the ectopic expression of PRDM1 in B cells. Briefly, IL4 and anti-CD40 treated naïve B cells for 2 days and were infected with a retrovirus expressing GFP or PRDM1-GFP. After 2 more days of culture with IL4 and anti-CD40, cells were harvested and cultured in the presence of 40LB feeder cells and IL21 for 4 days. F, GFP-positive cells were analyzed for CD19 and CD138 expression. Representative plots of FACS analysis of the in vitro induced CD19+CD138+ population. Histograms represent the percentage of iPB (CD19+CD138+); symbols represent triplicate of 2 independent experiments.
TET2 mutations in human DLBCLs manifest the Tet2-deficient GC gene signature and similarities to CREBBP mutant cases.
Differentiation blockade such as that caused by Tet2 deficiency in GC B-cells is a canonical hallmark of DLBCL and hence we predicted that over time loss of TET2 would lead to progression of GCs towards a more frankly pre-neoplastic phenotype. To ascertain whether this was the case we observed Vav-Cre/Tet2−/− and Vav-Cre/Tet2−+/+ control mice for a longer period of time, during which they were immunized on a monthly basis to induce GC formation repeatedly. Animals were sacrificed at ten months, which is the time point at which Vav-Cre/Tet2−/− animals were beginning to manifest signs of morbidity. We observed splenomegaly in Vav-Cre/Tet2−/− mice compared to Vav-Cre/Tet2−+/+ mice based on the spleen to body weight ratio (Supplementary Fig. S7A).
We performed a detailed histological analysis using B220, the GC-specific marker PNA and the proliferation marker Ki-67. This analysis showed disruption of splenic architecture and nodular expansion of abnormal PNA+ and Ki-67 positive GC-derived B-cells in the spleens of Vav-Cre/Tet2−/− as compared to Vav-Cre/Tet2−+/+ mice (Supplementary Fig. S7B). Vav-Cre/Tet2−/− animals also displayed B cells outside of the spleen with involvement of lungs and liver (Supplementary Fig. S7C). In all cases T-cells (CD3+) were intermingled with the B-cells, suggesting that full transformation might not have been achieved and/or B-cells require T-cells signals/support. We further validated these findings by flow cytometry quantifying the proportion of CD95+GL7+ GC B-cells among the total population of B220+ cells. We observed a significant increase in GC B-cells in the Vav-Cre/Tet2−/− mice as compared to Vav-Cre/Tet2+/+ mice (p<0.05, Supplementary Fig. S7D). Neoplastic B-cells in mice manifest reduced staining for IgD, IgM, CD21 and CD23 (42). Along these lines, Vav-Cre/Tet2−/− mice displayed significant reduction in B220+IgD+IgM+ cells as compared to Vav-Cre/Tet2−+/+ mice (p<0.01, Supplementary Fig. S7E) and greater proportion of B220+IgDlowIgMlow/int B cells (p<0.05, Supplementary Fig. S7E). Likewise, Vav-Cre/Tet2−/− mice featured significant expansion of abnormal B220+CD23lowCD21low B cells (p<0.01, Supplementary Fig. S7F).
To investigate whether Tet2 loss of function manifested a classical tumor suppressor phenotype for B-cell lymphomas, we next explored whether Tet2 loss of function would cooperate with a canonical DLBCL oncogene. To avoid the confounding effect of Tet2 loss in other cell lineages, we crossed the GC-specific Cγ1-Cre/Tet2−/− mice with the IμBcl6 mouse model, that directs constitutive expression of the Bcl6 proto-oncogene in GC B-cells, and develop DLBCL after long latency (42). We compared and contrasted the phenotype of Cγ1-Cre/Tet2−/−;IμBcl6 to Cγ1-Cre/Tet2−/− or IμBcl6 only. All mice were immunized on a monthly basis to induce GC formation and elicit the DLBCL phenotype and were sacrificed at seven months, the timepoint at which Cγ1-Cre/Tet2−/−;IμBcl6 animals showed signs of morbidity but prior to the known timing for development of overt lymphoma in IμBcl6 mice.
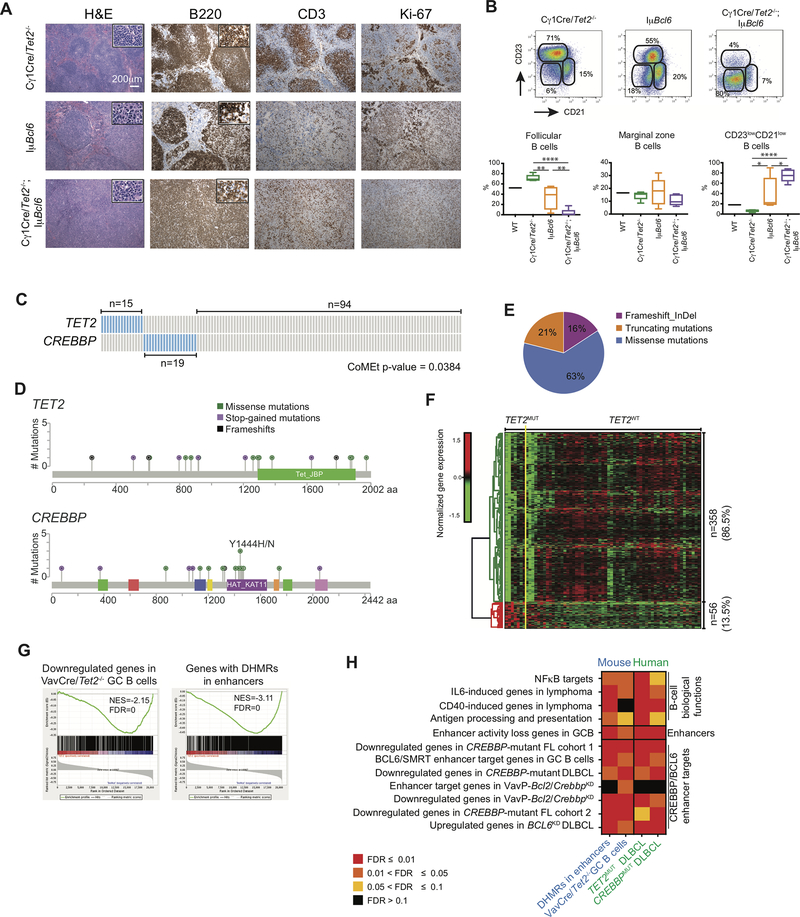
Anatomic analysis of these animals revealed significant and much more severe splenomegaly in Cγ1-Cre/Tet2−/−;IμBcl6 mice as compared to the Cγ1-Cre/Tet2−/− or IμBcl6 cohorts (Supplementary Fig. S7G and H). We performed a detailed histological analysis of the spleens of these mice using H&E staining, as well as immunohistochemistry for B220, CD3 and the proliferation marker Ki-67. Strikingly, 100% of Cγ1-Cre/Tet2−/−;IμBcl6 mice (n=12) displayed either follicular or diffuse lymphoma, which frequently led to the total effacement of splenic architecture. In these mice, B220+cells displayed a high proliferation index (>50% of cells Ki67 positive) and CD3+ T cells were frequently mixed among neoplastic cells (Fig. 7A and Supplementary Fig. S7I). In marked contrast, only 50% of IμBcl6 mice (n=6) displayed enlarged spleens, with less severe architectural effacement due to enlarged follicles and/or diffuse lymphoid infiltrates of intermediate/large B220+ elements. Ki-67+ cells ranged from 20–40% of the total cells (Fig. 7A and Supplementary Fig. S7I). Age-matched Cγ1-Cre/Tet2−/− mice (n=7) displayed no evidence of lymphomas and had normal splenic architecture, with well-defined primary and secondary follicles (Fig. 7A and Supplementary Fig. S7I). These results were confirmed using flow cytometry. B cells in the spleen of all Cγ1-Cre/Tet2−/−;IμBcl6 mice were mostly neoplastic (B220+CD23lowCD21low), compared to IμBcl6 (p<0.05, Fig. 7B) or Cγ1-Cre/Tet2−/− mice (p<0.0001, Fig. 7B). This result correlated with a decrease in the proportion of normal splenic follicular B-cells in Cγ1-Cre/Tet2−/−;IμBcl6 mice compared to IμBcl6 (p<0.01, Fig. 7B) and Cγ1-Cre/Tet2−/− mice (p<0.0001, Fig. 7B). All together, these results demonstrate that Tet2 loss of function results in accelerated formation of GC-derived lymphomas.
Figure 7. TET2 mutations in human DLBCLs manifest the Tet2-deficient GC gene signature, and similarities to CREBBP mutant cases.
A, Representative histologic sections of formalin-fixed, paraffin-embedded spleens from Cγ1Cre/Tet2−/−;IμBcl6, Cγ1Cre/Tet2−/− and IμBcl6 mice. Sections were stained with H&E and antibodies specific for B220, CD3 and Ki-67. B, Representative flow cytometry plot and quantification of CD23+CD21+ (follicular B cells), CD23−CD21+ (marginal zone B cells) and CD23−CD21− B cells in the spleens of Cγ1Cre/Tet2−/−;IμBcl6, Cγ1Cre/Tet2−/− and IμBcl6 mice; two-tailed t test, *p < 0.05 **p < 0.01 ****p < 0.0001. C, Mutation matrix for TET2 and CREBBP mutations in a cohort of 128 DLBCL. Blue color indicates any type of mutation while grey indicate no known mutations of the gene. Each of the 128 columns indicates one patient sample. Mutual exclusivity on TET2 and CREBBP mutations was computed using CoMEt. D, Lolliplot of the TET2 and CREBBP mutations, plotted using MutationMapper (74,75). E, Distribution of insertions/deletions and mutations among TET2 mutant DLBCL. F, Normalized gene expression levels of 414 differentially expressed genes between TET2 mutant (TET2MUT) and non-mutant (TET2WT) DLBCL. TET2MUT indicate 7 samples with either stop-codon of frameshift mutation, while TET2WT indicate 60 samples without any kind of mutation in TET2 or CREBBP genes. Expression levels were scaled by row. G, GSEA enrichment plots showing correlation of different genesets with ranked expression change between TET2MUT and TET2WT DLBCL. NES, normalized enrichment score; FDR, false discovery rate. H, Heatmap of the FDR scores for the enrichment of 12 gene sets involved in the exit of GC B-cells from the GC. FDR scores were obtained using hypergeometric test for mouse genes with enhancers overlapping DHMR, described as “DHMR in enhancers” and shown previously Fig. 6F. Column described as “Vav-Cre/Tet2−/− GC B cells” presents the FDR scores obtained using GSEA, also shown on Fig. 5D. Columns “TET2MUT DLBCL” and “CREBBPMUT DLBCL” indicate GSEA-based FDR scores of the enrichment of the gene sets in DLBCL.
A second link to human lymphomas lies in the gene expression profile of Tet2-deficient GC B-cells, which is highly similar to the signature induced by the well-established lymphoma epigenetic tumor suppressor CREBBP. This scenario suggesting that these two tumor suppressors have overlapping effects, in which case they would most likely not occur together. To address this point we performed targeted resequencing of TET2 and CREBBP in a cohort of 128 DLBCL patients, for whom gene expression microarrays were also available (43). TET2 mutations were detected in 12% of the cases (n=15), consisting of frameshift, indels and missense mutations (Fig. 7C-E and Supplementary Table S3), which is in line with previous studies (6,18). In addition, 15% of these DLBCLs (n=19) had somatic missense of mutations of CREBBP, eleven of them affecting the HAT domain (Fig. 7C and D and Supplementary Table S3). We focused on HAT mutations since these have been reported to induce the most severe phenotype (44). Notably, we observed that TET2 mutations were significantly mutually exclusive with CREBBP mutations (Fig. 7C; p=0.0384). There was also a trend towards mutual exclusivity in the patient cohort reported by Reddy et al. (6), although it was not statistically significant (p=0.16). In addition, we found that TET2 mutated cases were predominantly classified as GCB-DLBCL subtype (11/15), similarly to what has been reported for CREBBP mutated DLBCL (11).
Since gene expression arrays were available for these patients we performed a supervised analysis to determine genes differentially expressed between TET2 mutant cases vs non-TET2 and non-CREBBP mutant DLBCLs. Even though DLBCLs are extremely heterogeneous we were still able to identify a set of 358 genes (86.5%) differentially downregulated and 56 genes (13.5%) differentially upregulated in TET2 mutant DLBCLs (FC>1.5, FDR<0.05, Fig. 7F). This skewing towards transcriptional repression is similar to what was observed in Vav-Cre/Tet2−/− GC B cells (Fig. 5C). To determine whether the human TET2 mutant signature was similar to the Vav-Cre/Tet2−/− signature from murine GC B-cells we performed GSEA and observed significant enrichment for genes downregulated in Vav-Cre/Tet2−/− GC B cells (FDR=0, Fig. 7G). Genes affected by DHMRs in mice were also significantly enriched among genes downregulated in human TET2 mutant cases (FDR=0, Fig. 7G).
In addition, we compared and contrasted the relative enrichment of gene sets linked to TET2 loss of function in the gene expression profiles of Vav-Cre/Tet2−/− GC B-cells, DHMR profiles of Vav-Cre/Tet2−/− GC B-cells, human TET2 mutant DLBCLs and in human CREBBP mutant lymphomas from our new cohort. These included the GC exit gene sets linked to NFκB, IL6, CD40 and antigen presentation, the enhancers normally repressed in human GC B-cells, the various murine and human CREBBP mutant signatures, and the BCL6-SMRT repressed enhancers and genes induced upon BCL6 knockdown. Overall this analysis yielded significant enrichment of almost all of these gene sets across the board in both the murine and human gene expression profiles (Fig. 7H). Collectively these data indicate the relevance of the murine Tet2 knockout experiments to the human disease. These results also underline the link between TET2 and CREBBP in coordinating the GC exit transcriptional program through activation of gene enhancers and suggest that TET2 and CREBBP mutations induce malignant transformation through at least partially overlapping mechanisms of action. Finally, our findings suggest that lymphomas with TET2 loss of function might become addicted to HDAC3, as we previously demonstrated for CREBBP-deficient lymphoma cells (11). To further investigate this hypothesis, we transduced TET2 WT DLBCL cell lines (OCI-Ly18 and MD901) with two different lentiviral shRNA for TET2 or a control shRNA and confirmed TET2 knockdown by qPCR (Supplementary Fig. S8A and B). We then treated the cells with our HDAC3 selective inhibitor (45) or vehicle for 4 days and quantified the number of live cells (Annexin V− DAPI−) in each condition. We observed that TET2 loss of function made the cells more sensitive to HDAC3 inhibition, as reflected by the increase in growth inhibition after HDAC3i treatment in shTET2 compared to shScr (Supplementary Fig. S8C and D). Whether patients with TET2 mutations could benefit from treatment with HDAC3 inhibitors requires further investigation.
DISCUSSION
Herein we report that TET2 is a tumor suppressor in DLBCL and not merely a passenger mutation that might be inherited from aberrant HSCs. This work expands on the known role of TET2 in myeloid malignancies and T-cell malignancies (23,31,46). Hence the transforming effect of TET2 deficiency goes beyond previous results showing B1 transformation in non-immunologically challenged mice (47) and oncogenic potential in fully mature B2-cells participating in the GC reaction. Tet2 deficiency resulted in impairment of B-cells to undergo terminal differentiation in a B-cell autonomous manner as manifested by the deficient formation of GC-derived antibody secreting cells and reduced titers of high affinity antibodies. This phenotype is indicative of a defect in the ability of B-cells to exit the GC reaction, which is a canonical feature of DLBCL lymphomagenesis (48). Indeed, Tet2 loss of function in GC B-cells results in accelerated formation of DLBCLs in cooperation with constitutive expression of BCL6, a major oncogene in DLBCL, highlighting the significance of TET2 mutations in primary DLBCL patients. The GC reaction is a cyclical process, whereby proliferative centroblasts from the dark zone migrate to the light zone as centrocytes, that interact with TFH cells and follicular dendritic cells (3). Centrocytes that encode for higher affinity B-cell receptors are selected to return to the dark zone as centroblasts for further proliferation and mutagenesis. Repeated rounds of somatic hypermutation lead to enrichment and selection of B-cells with the highest affinity BCR, which then undergo class switch recombination and differentiate to PC that confer long term immunity by secreting high affinity antibodies (3). Tet2 loss of function impairs this process as manifested by failure of GC B-cells to undergo these final steps, which indicates that TET2 is not only a tumor suppressor but is also critically required for B-cells to successfully undergo the high-affinity humoral immune response.
Exit from the GC reaction requires the engagement of epigenetic mechanisms that activate enhancers for genes that must be induced to resolve the GC phenotype and mediate the PC phenotype (9,11,34). Some of the key signaling pathways driving this process are CD40, NFκB (downstream of BCR signaling), antigen presentation genes such as MHC class II, and cytokines such as IL-6 and others (4,49). Our data suggest that the defect in class switch recombination and PC differentiation evident in Tet2-deficient GCs is linked to aberrant repression of enhancers that regulate these genes. These enhancers accordingly manifested a reduction in the 5hmC mark, suggesting that TET2 is required to induce enhancer activation and concordant transcriptional activation of genes in these pathways.
Indeed we find that TET2 is part of a more complex enhancer switching mechanism controlling egress of B-cells from the GC reaction. Specifically, GC exit enhancers require histone acetyl-transferase CREBBP, which is disrupted by somatic loss of function mutations in DLBCL and is a bona fide DLBCL tumor suppressor (11,12). During the GC reaction CREBBP target enhancers are repressed and H3K27-deacetylated by the transcriptional repressor BCL6 through its recruitment of SMRT/HDAC3 complexes (11,34). BCL6/SMRT complexes are then suppressed in the light zone downstream of BCR and CD40 signaling, enabling CREBBP to induce H3K27 acetylation and activation of GC exit enhancers and genes (11,34,50–53). CREBBP mutations disrupt this balance resulting in sustained repression of these genes, and a corresponding defect in GC exit (11,12). Along these lines we show that there is striking similarity in the sets of genes aberrantly repressed in Tet2- and Crebbp-deficient mouse GC-derived B-cells and most importantly, in primary human DLBCL patients. We find that loss of enhancer 5hmC is associated with loss of H3K27 acetylation, suggesting that CREBBP requires the presence of TET2 and the 5hmC mark to mediate H3K27 acetylation. A functional link between TET2 and CREBBP is supported by other studies, which show that CREBBP and p300 can acetylate TET2, increasing its enzymatic activity and protein stability (40), and that 5hmC co-localizes with active enhancers bound by p300 (54). The fact that TET2 mutant DLBCL patients manifest similar gene expression profiles as B-cells in the Tet2-deficient mouse models support the relevance of our experimental systems to the human disease. Our data suggest that TET2 loss of function most likely mediates malignant transformation by suppressing/impairing CREBBP mediated enhancer acetylation, disrupting GC exit transcriptional programming and causing accumulation of B-cells in the light zone, which manifests as GC hyperplasia.
CREBBP mutations are generally described as early lesions in DLBCL and induce a similar phenotype as Tet2 deficiency with defective PC formation (12,55). Along these lines it is notable that TET2 somatic mutations have been demonstrated to be present in the HSCs of TET2 mutant DLBCL patients and are present at high variant allele frequencies in DLBCL. Hence TET2 loss of function mutations are the only known lymphoma driving mutations shown to occur at the HSC level. Even though Tet2 loss of function causes B-cell autonomous effects, our mouse models that disrupts Tet2 expression in HSCs is likely the most physiologically relevant. CREBBP mutations likely occur at a later stage than TET2 mutations during early B-cell development. Regardless, both TET2 and CREBBP are early hits during lymphomagenesis, which represents an additional similarity between these lesions. The fact that CREBBP and TET2 mutations tend to be mutually exclusive in DLBCL patients lends further support to this model, consistent with these being pathway mutations with a shared mechanism of action. TET2 loss of function can also lead to gain of cytosine methylation (18), although this effect is less pronounced than loss of 5hmC (26,56,57). Although the loss of 5hmC seems to adequately explain the aberrant transcriptional profiles of TET2 deficient lymphomas, the potential contribution of 5mC should not be excluded and is worth exploring in future studies, as suggested by our analysis of the Prdm1 locus.
In conclusion, we propose that TET2 be considered a bona fide tumor suppressor in DLBCL and included in targeted resequencing panels used to characterize mature B-cell lymphomas. Perturbation of chromatin mechanisms is a hallmark of DLBCL, with >50% of the recently identified 150 recurrent somatic mutations affecting either transcription factors or epigenetic modifiers (6). TET2 thus joins the set of alleles in DLBCL that disrupt the GC reaction through perturbations in chromatin modifications, a finding with implications for design and interpretation of studies testing the impact of epigenetic modifier drugs in DLBCL patients. In this regard, our results showing sensitivity to HDAC3 inhibition of TET2-deficient lymphomas suggest that this new therapeutic approach should be tested in patients with TET2 mutations.
METHODS
Animal models
Vav-Cre/Tet2f/f and Tet2f/f mice were a generous gift from R. Levine, Memorial Sloan Kettering Cancer Center (46). Cγ1Cre mice (B6.129P2(Cg)-Ighg1tm1(cre)Cgn/J; stock number 010611) and Cd19-Cre mice (B6.129P2(C)-Cd19tm1(cre)Cgn/J; stock number 006785) were obtained from The Jackson Laboratory. IμBcl6 mice for the lymphoma studies were obtained from R. Dalla-Favera (42), Columbia University. All mice were followed until any one of several criteria for euthanizing were met, including severe lethargy and more than 10% body weight loss in accordance with our Weill Cornell Medicine Institutional Animal Care and Use Committee–approved animal protocol (protocol #: 2011–0031). All animals were maintained according to the guidelines of the Research Animal Resource Center of Weill Cornell Medicine.
Mice carrying constitutive inactivated Tet2 alleles (Tet2−/−) and CD19-Cre/Tet2−/− mice have been previously described (31,47). These mice, including wild type C57BL/6 mice, were maintained in the breeding facility of SCEA, Gustave Roussy, Villejuif, France) and were sacrificed at the indicated times. Experiments with Tet2−/− mice were conducted according to the Gustave Roussy Institutional guidelines and authorized by the Direction Départementale des Services Vétérinaires du Val de Marne.
DLBCL Patient Samples
A cohort of 128 tumors with a pathological diagnosis of DLBCL were interrogated by targeted sequencing, as previously described (43). Informed written consent was obtained from all the subjects. In brief, 500ng-1μg of tumor DNA was sheared by sonication using a Covaris S2 instrument, and end repaired, A-tailed and ligated with Illumina TruSeq adaptors (Bio-O Scientific) using Kapa HyperPrep Kits (Kapa Biosystems). Adapter ligated products were enriched using 6 cycles of PCR, and 12-plexed for hybrid capture using a custom hybrid capture reagent (Nimblegen) that included all coding exons for CREBBP and TET2. Each pool was sequenced with 100bp paired end reads on a single lane of a HiSeq 2500 Instrument in high output mode at the Hudson Alpha Institute for Biotechnology. Raw sequencing reads were aligned to the human genome (hg19) using a BWA-Mem (58), realigned around InDels using GATK, sorted and deduplicated using Piccard tools, and variants were called according to a consensus between VarScan2 (59) and GATK Unified Genotyper (60). This approach has been validated to have a specificity of 92.9% and a sensitivity of 86.7% (44). Average on-target rate for this dataset was 88% and average depth of coverage 641X (min = 263X, max = 1396X). Affymetrix U133 plus 2 gene expression microarrays were performed on 84 matched DLBCL tumors previously (61) (GEO Accession, GSE10846). All human subjects research was approved by the respective institutional review board.
Mouse B-cell isolation
Spleens of IμBCL6 and IμBCL6+shTet2 mice were dissected and cell suspensions were prepared. B cells were positively selected using anti-B220 magnetic microbeads (Miltenyi Biotech). For the isolation of GC B-cells, Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ mice were immunized intraperitoneally with SRBCs (1 × 108 cells per mouse) or NP-CGG ratio 20–25 (Biosearch Technologies) in alum (1:1) to induce GC formation. Mice were sacrificed 10 days after immunization, spleens were dissected and mononuclear cells were purified using Histopaque gradient centrifugation. Cell suspensions were enriched in B cells by positive selection with anti-B220 magnetic microbeads and GC B-cells (B220+GL7+FAS+DAPI−) were sorted using a BD FACSAria II or a BD Influx sorter (BD Biosciences).
In vitro generation of iGCB and iPB
Splenic murine B cells were isolated from 2- to 3-month-old mice using the mouse untouched B-cell isolation kit (CD43 (Ly-48) MicroBeads, Miltenyi Biotec) according to the manufacturer’s instructions (yield > 90% CD19+ B220+). 40LB cells expressing CD40L and produce BAFF, are a kind gift of D. Kitamura (33) and cultured in DMEM media with 10% FBS and Penicillin G/Streptomycin.
For B cells culture RPMI-1640 medium (ThermoFisher Scientific) was supplemented with 10% FCS, 0,055 mM 2-ME, 10 mM HEPES, 1 mM sodium pyruvate, 100 units/ml penicillin, and 100 μg/ml streptomycin (GIBCO). Purified B cells (110 × 103 cells per well) were cultured in a 6-well plate in the presence of 40LB cells (0,5 × 106 cells per well) that had been irradiated with 80 Gy of γ-ray. rIL-4 (1 ng/ml; Peprotech ) was added to the primary culture for 4 days, and on day 4, the cells were replated onto a new irradiated feeder layer and cultured with rIL-21 (10 ng/ml; Peprotech). Cells were harvested at days 4 (D4) or 8 (D8) and processed for flow cytometric analysis and RNA or protein isolation.
For viral transduction naïve B-cells were first stimulated by anti-CD40 antibody (3μg/ml; COGER) and rIL4 (25 ng/ml; Peprotech), transduced after 48 hours and plated on 40LB cells 48 hours after.
A PRDM1 (human) cDNA fragment was obtained from Genescript, and was subcloned in an MSCV-IRES-GFP retrovirus backbone. Viral particles were generated as described (62).
DLBCL cell cultures
Human DLBCL cell lines were grown in medium containing RPMI-1640, 10% FBS, 1% HEPES, 1% glutamine, and 1% penicillin/streptomycin. OCI-Ly18 was obtained from Ontario Cancer Institute in June 2011 and MD901 was provided by Jose Angel Martinez-Climent (CIMA, Pamplona, Spain) in June 2011. Cell line authentication testing was performed at IDEXX BioResearch (http://www.idexxbioresearch.com/cellcheck), using methods recommended by the American National Standards Institute (ANSI ASN-0002–2011). The cell lines were confirmed to be of human origin and tested for evidence of cross-species contamination (mouse, rat, Chinese hamster and African Green monkey). Short tandem repeat (STR) testing was performed and the genetic profile obtained was compared to the established cell line profile to confirm the cell lines are consistent with the established profile. These cell lines have also been routinely tested for Mycoplasma contamination in the laboratory. OCI-Ly18 and MD901 cells were transduced with shRNAs against TET2, or Scr as control, for 4 days and then treated with a selective HDAC3 inhibitor (20 μmol/L) or vehicle (DMSO) for 96 hours. The shRNA sequences to knockdown human TET2 were shTET2#1: CCTCAAGCATAACCCACCAAT; shTET2#2: CCTTATAGTCAGACCATGAAA.
ELISA and ELISPOT
For analysis of T cell–dependent antibody production, mice were immunized intraperitoneally with 100 μg NP-CGG ratio 20–25 in alum (1:1). On day 35 after immunization, serum was collected and titers of isotype-specific antibodies to NP were measured in plates coated with NP26-BSA or NP7-BSA with the SBA Clonotyping System, according to the manufacturer’s protocol (5300–05; Southern Biotechnology). Results were assessed by spectrophotometric measurement of absorbance at 405nm using a Biotek Synergy Neo Alpha Plate Reader (BioTek). Background readings of absorbance in negative control wells were <0.050. For enzyme-linked immunospot (ELISPOT) assay, we collected bone marrow cells on day 60 after immunization. Cells were incubated for 20 h at 37°C on NP26-BSA-coated or NP4-BSA-coated 96-well MultiScreen-HA filter plates (Millipore). NP-specific spots were visualized with goat antibody to mouse IgG1 (1034–05) or IgM (1021–05) conjugated to horseradish peroxidase (Southern Biotechnology), and color was visualized by the addition of 3,3′,5,5′-tetramethylbenzidine (Southern Biotechnology). Plates were evaluated using an automated Zeiss Elispot reader system (ZellNet Consulting, Inc).
Flow cytometry analysis
Flow cytometry analysis of spleen cell suspensions was performed using the following fluorescent-labeled anti–mouse antibodies: APC-conjugated anti-B220 (BD Biosciences #553092), anti-CD138 (BD Biosciences #558626) and anti-IgM (eBioscience #17–5790-82); PE-conjugated anti-IgD (BD Biosciences #558597), anti-CD23 (eBioscience #5010271) and anti-CD95 (BD Biosciences #554258); PECy7-conjugated anti-CD21 (BioLegend #123420), anti-CD95 (BD Biosciences #557653), anti-GL7 (BD Biosciences # 561530) and anti-CD86 (BD Biosciences #560582); PEVio770-conjugated anti-B220 (Miltenyi Biotec #130–102-308); FITC-conjugated anti-CXCR4 (BD Biosciences #551967) and anti-IgG1 (ebioscience #11–4011-85); Brilliant Violet 421-conjugated anti-CD138 (BioLegend #142507); PerCP-Cy5.5-conjugated anti-GL7 (BioLegend #144610), anti-CD138 (BioLegend #142510) and anti-IgM (BD Biosciences #550881), APC-Cy7-conjugated anti-CD19 (ebioscience #47–0193-82), AlexaFluor647-conjugated anti-BLIMP1 (BD Biosciences #563643). Sytox blue (ThermoFisher Scientific) or DAPI was used for the exclusion of dead cells. Data was acquired on a BD FACSCanto II (BD Biosciences) and analyzed using FlowJo v10.1 software (TreeStar).
Histology and immunohistochemistry
Mice organs were fixed in 4% formaldehyde and embedded in paraffin. 4 micron-sections were deparaffinized and heat antigen-retrieved in citrate buffer pH 6.4 and endogenous peroxidase (HRP) activity was blocked by treating the sections with 3% hydrogen peroxide in methanol. Indirect immunohistochemistry was performed with antispecies-specific biotinylated secondary antibodies followed by avidin–horseradish peroxidase or avidin-AP, and developed by Vector Blue or DAB color substrates (Vector Laboratories). Sections were counterstained with hematoxylin if necessary. The following primary antibodies were used: biotin-conjugated anti-B220 (BD Biosciences 550286), anti-CD3 (Abcam ab16669), anti-PNA (Vector Laboratories B1075) and anti-Ki67 (Abcam ab16667). Photomicrographs were scanned using Aperio eSlide Manager (Leica Biosystems).
Genomic DNA and RNA extraction
Murine gDNA was extracted using the Puregene Gentra cell kit (Qiagen) and eluted in TE. The quality of purified DNA was checked using an Agilent 2100 Bioanalyzer (Agilent Technologies). Total RNA was extracted from murine GC B cells or tumors using Trizol (LifeTechnologies) and RNeasy Mini Kit (Qiagen) with DNase treatment. RNA concentration was determined using Qubit (LifeTechnologies) and integrity was verified using Agilent 2100 Bioanalyzer.
Targeted Bisulfite DNA sequencing and quantification of methylation.
Genomic DNA (200 ng) was modified by bisulfite treatment according to the manufacturer’s instructions (MethylDetector, Active Motif). DNA fragments were PCR-amplified and direct sequencing reaction was performed using standard conditions (Applied Biosystems). Primers were: 5’- GTTGATAGTAGAAAGTTGTTTTGG and 5’- CTCTAACTCAAAATCATTCTATTCT.
The CpG methylation-sensitive digestion of genomic DNA was carried out with the EpiJET DNA Methylation Analysis Kit based on MspI/HpaII digestion (Thermo Scientific) following manufacturer’s instructions using 100 ng purified genomic DNA. qPCR was performed using SYBR Green Select Master Mix (Thermo Scientific) following manufacturer’s instructions. The primers for common PCR were as follows: forward: 5’- CTCTGACTCTGGTCTGAAGT-3’; reverse: 5’- GTCTCCTGCTTCGTGTTATC-3’. The methylation level was quantified using the 2−ΔCt method and presented as the percent cytosine methylation at the target-gene site.
Chromatin immunoprecipitation (ChIP)
Cells were crosslinked with 1% formaldehyde and neutralized with 0.125 M glycine. Cell lysates were sonicated to a length of 300–500 base pairs with a Covaris E-220 Focused-ultrasonicator (Covaris), and proteins were immunoprecipitated with antibody to H3K27Ac (Active Motif 39133) or IgG (ab-37415; abcam) as control. After complete washing, immunoprecipitated DNA was eluted in elution buffer and reverse-crosslinked overnight at 65 °C. DNA was purified and quantified by real-time PCR. Primer sequences are shown in Supplementary Table S4. Enrichment was calculated relative to input.
Quantitative real-time PCR
cDNA synthesis from RNA was performed using the Verso cDNA Synthesis kit (Thermo Scientific). Tet2 expression was detected using the Green FastMix kit (Applied Biosystems) on a QuantStudio6 Flex Real-Time PCR System (Applied Biosystems) or using TaqMan Universal PCR Master Mix (Applied Biosystems) on a ABI PRISM 7500 (Applied Biosystems). Gene expression was normalized to Rpl13 or Hprt using the ΔC(t) method and results were represented as mRNA expression. Primer sequences are shown in Supplementary Table S4. All mouse TaqMan probes were purchased from Applied Biosystems: Pax5 (Mm00435501), Irf4 (Mm00516431), Irf8 (Mm00492567), Xbp1 (Mm00457357), SpiB (Mm03048233) Bcl6 (Mm00477633) and Prdm1 (Mm00476128). Polr2a (Mm00839493) and Gapdh (Mm99999915) expression level was used for normalization expressed values relative to control using the ΔΔCT method.
Hydroxymethylated DNA immunoprecipitation sequencing (hMeDIP-seq)
A total of 1,5 ug of genomic was sonicated with a Covaris S220. End Repair, A-Tailing, and ligation of the pre-annealed adapters was performed before immunoprecipitation with the 5hmC antibody (1 ug/mL, Active Motif 39791). A 10% of the volume was set aside before immunoprecipitation as input. Spike-ins (Diagenode AF-107–0040) were used in order to control the efficiency of the 5hmC pull down. Amplification of the libraries was performed using Kapa HiFi Hot Start Ready Mix (Kapa Biosystems, KK2601) for 12 cycles. Concentrations and quality of the final DNA libraries was measured by Qubit (LifeTechnologies) and Agilent 2100 Bioanalyzer respectively.
Sequencing of the libraries was performed in an Illumina HiSeq2500 multiplexing a single pull down and a single input on each lane. Reads were trimmed using cutadapt 1.9.1 (CITATION DOI: http://dx.doi.org/10.14806/ej.17.1.200) and aligned by bowtie 2.0.5. Unique reads were extracted using an in-house pipeline. MACS2 (63) peak caller “callpeak” was used to detect broad peaks in 5hmC Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ datasets after applying “--nomodel --extsize 200 -B --SPMR --broad --bw 200 --broad-cutoff 0.1” parameters. Subsequently, “bdgdiff” was used from MACS2 to detect differential regions between Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ samples. For UCSC visualization purposes, --SPMR normalization was complementary invoked. For genomic annotation of DHMR regions, we used mouse GRC38 genome from gencode as a reference genome sequence. DHMR regions were annotated for introns, exons, promoters and distal enhancers. Promoter regions were defined as 2kb upstream or downstream of transcription start site (TSS) of the gene. For distal enhancers, we first added 2kb upstream promoter region to the gene boundary and then defined distal enhancer as 100kb upstream or downstream region around it. For genome annotation of 5hmC peaks, they were overlapped with distal enhancers, introns and promoter regions. Fold enrichment was averaged for genes containing multiple 5hmC peaks. Separately, transcriptome datasets from Vav-Cre/Tet2−/− and Vav-Cre/Tet2+/+ cells were compared against each other to obtain a list of differential genes at fold-change cutoff of 1.2 and p-value less than 0.05.
Identification of peaks losing 5hmC and H3K27ac signals
First, the BAM files with the mapped reads for hMeDIP-Seq (5hmC signal) and ChIP-Seq (H3K27ac signal, (11)) were partitioned into 50 bp long bins and normalized using Ashoor et al. approach (64), to allow the cross comparison between replicates. The mean read depth coverage from each knockout or knockdown condition was then compared with the mean signal strength in the corresponding bins in WT condition. Next, all bins fulfilling the following criteria were connected into temporary peaks: (1) minimal number of 30 reads in WT condition, that is the minimal sum of the reads in all WT replicates; (2) at least 35% signal loss in knockout or knockdown condition; (3) the maximum distance between the peaks losing signal cannot exceed 51 bp and the minimal temporary peak length needs to be composed of at least 4 bins. The above analysis was conducted separately for hMeDIP-Seq and ChIP-Seq data using “merge” and “intersect” functions from bedtools package (65) in combination with in-house scripts. Next, only the temporary peaks losing 5hmC or H3K27ac signals, which were located not further than 300 bp from each other, were retained and overlapped with the H3K27ac and 5hmC peaks from the corresponding mice models in WT condition. Finally, the collection of peaks losing both 5hmC and H3K27ac signals was assigned to genes, if located in the region up to 100 kb upstream or downstream around them.
RNA sequencing (RNA-seq)
Libraries were generated using the TruSeq-stranded-mRNA sample kit (Illumina) following the Illumina single-end library preparation protocol. Single-end sequencing (SE50) was performed on Illumina HiSeq2500. RNA sequencing results were aligned to mm10 or hg19, respectively, using STAR (66) and annotated to RefSeq using the Rsubread package (67). Differentially expressed genes were detected using DESeq2 (68) with fold-change cutoff of 1.2 and p-value less than 0.05.
Microarray data analysis
Raw microarray readings were processed using limma (version 3.30.13) package (69), including extraction from CEL files and normalization with RMA function. For the purpose of the GSEA analysis, normalized expression levels were subsequently exported as GCT files, and postprocessed as described in “Gene Set Enrichment Analysis” paragraph. For the purpose of the differential gene expression analysis, microarray probes expression levels were transformed to log2 values and condensed using avereps function. Differentially expressed genes (DEGs) were called using lmFit and eBayes functions. False discovery rate values lower than or equal to 0.05 were considered as significant, considering recomputed FDR scores for DEGs with a minimum of 1.5 fold change.
Gene set enrichment analysis
All GSEA analyses were conducted using the same settings as in case of the previously published hematological cancer studies (70), with an exception of the permute parameter which value was set to “gene_set” as a consequence of dealing with sample sizes smaller then 7. The input files are all the genes’ normalized expression levels from each of the datasets:
-
(1)
TET2mut DLBCL: 7 TET2 mutated samples compared against 60 samples with none of these genes mutated.
-
(2)
CREBBPmut DLBCL: 7 CREBBP mutated samples compared against 60 samples with none of these genes mutated
-
(3)
Vav-Cre/Tet2−/−: 4 Vav-Cre/Tet2−/− GC B-cells were compared against 3 Vav-Cre/Tet2+/+ GC B-cells.
All gene sets used in GSEA study were based on the previous publications in Melnick or Straudt (9,11,34,71) laboratories. In addition, the enrichment of these gene sets among the 1825 genes which enhancers overlap with differentially hydroxymethylated regions in mice, or 1294 genes which were downregulated in Vav-Cre/Tet2−/− mouse, was conducted using hypergenometric test, followed by Benjamini–Hochberg correction for multiple testing. Gene sets were also tested for the enrichment in the shuffled gene lists as an additional background control.
Statistical analysis
Mutual exclusivity of CREBBP and TET2 mutations in human DLBCL patients cohort was computed using CoMEt (72).
Data deposition
Raw data from RNA sequencing and hMeDIPseq in murine GC B-cells were deposited in GEO under accession number GSE111700.
Supplementary Material
Statement of significance.
We show that TET2 is required for exit of the GC, B cell differentiation and is a tumor suppressor for mature B-cells. Loss of TET2 phenocopies CREBBP somatic mutation. These results advocate for sequencing TET2 in lymphoma patients and for the testing of epigenetic therapies to treat these tumors.
ACKNOWLEDGEMENTS
We thank people from the Epigenomics Core WCM, Genomics Core WCM, Flow Cytometry Core WCM, Laboratory of Comparative Pathology WCM and MSKCC, Edward Holson and Kdac Therapeutics Inc. for the HDAC3 inhibitor, the Gustave Roussy Platforms, including Patrick Gonin, Yann Lecluse, Philippe Rameau and Olivia Bawa; and Dr. Daisuke Kitamura (Tokyo University of Science, Japan) for the kind gift of the 40LB cell line.
Research reported in this publication was supported by the Office of the Director of the National Institutes of Health under Award Number S10OD019986 to Hospital for Special Surgery. P.M.D. is supported by a Lymphoma Research Foundation Post-doctoral Fellowship. H.G. is supported by a grant from Fondation de France (No 00067113). O.E. is supported by NSF CAREER, LLS SCOR, Hirschl Trust Award, Starr Cancer Consortium I6-A618, NIH 1R01CA19454. M. R. G. is supported by R01 CA201380. O.A.B. is supported by the Institut National du Cancer (2013-PLBIO-09, 2016-PLBIO-068 and INCa-DGOS-INSERM 12551), Association Laurette Fugain and équipe labelisée Ligue Nationale Contre le Cancer. S.A. is supported by Comité Val-d’Oise de la Ligue contre le cancer. S. L. is supported by Leukemia Research Foundation Research Grant, the Jackson Laboratory Cancer Center New Investigator Award and the Jackson Laboratory Director’s Innovation Fund. Research reported in this publication was partially supported by the National Cancer Institute of the National Institutes of Health under Award Number P30CA034196. A. M. is supported by the Chemotherapy Foundation, The Follicular Lymphoma Research Consortium, Leukemia and Lymphoma Society SCOR Grant #7012–16 and the Starr Cancer Consortium.
Abbreviations:
- (GC)
germinal centers
- (DLBCL)
diffuse large B-cell lymphoma
- (AICDA)
activation-induced cytidine deaminase
- (Ig)
immunoglobulin
- (HSPC)
hematopoietic stem/progenitor cells
- (NP-CGG)
NP-Chicken Gamma Globulin
- (5mC)
5-methylcytosine
- (5hmC)
5-hydroxymethylcytosine
- (DHMR)
differentially hydroxymethylated regions
Footnotes
Disclosure of Potential Conflict of Interest
A. Melnick reports receiving research support from Janssen. R. Levine is in the supervisory board of Qiagen, SAB for Loxo, C4 therapeutics and Isoplexis; and he is a consultant and receives research support from Roche and Celgene. No potential conflicts of interest were disclosed by the other authors.
REFERENCES
- 1.Coiffier B, Thieblemont C, Van Den Neste E, Lepeu G, Plantier I, Castaigne S, et al. Long-term outcome of patients in the LNH-98.5 trial, the first randomized study comparing rituximab-CHOP to standard CHOP chemotherapy in DLBCL patients: a study by the Groupe d’Etudes des Lymphomes de l’Adulte. Blood 2010;116(12):2040–5 doi 10.1182/blood-2010-03-276246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Silva NS, Klein U. Dynamics of B cells in germinal centres. Nat Rev Immunol 2015;15(3):137–48 doi 10.1038/nri3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mesin L, Ersching J, Victora GD. Germinal Center B Cell Dynamics. Immunity 2016;45(3):471–82 doi 10.1016/j.immuni.2016.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klein U, Dalla-Favera R. Germinal centres: role in B-cell physiology and malignancy. Nat Rev Immunol 2008;8(1):22–33 doi 10.1038/nri2217. [DOI] [PubMed] [Google Scholar]
- 5.Klose RJ, Bird AP. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 2006. [DOI] [PubMed] [Google Scholar]
- 6.Reddy A, Zhang J, Davis NS, Moffitt AB, Love CL, Waldrop A, et al. Genetic and Functional Drivers of Diffuse Large B Cell Lymphoma. Cell 2017;171(2):481–94 e15 doi 10.1016/j.cell.2017.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beguelin W, Popovic R, Teater M, Jiang Y, Bunting KL, Rosen M, et al. EZH2 is required for germinal center formation and somatic EZH2 mutations promote lymphoid transformation. Cancer Cell 2013;23(5):677–92 doi 10.1016/j.ccr.2013.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caganova M, Carrisi C, Varano G, Mainoldi F, Zanardi F, Germain PL, et al. Germinal center dysregulation by histone methyltransferase EZH2 promotes lymphomagenesis. J Clin Invest 2013;123(12):5009–22 doi 10.1172/JCI70626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ortega-Molina A, Boss IW, Canela A, Pan H, Jiang Y, Zhao C, et al. The histone lysine methyltransferase KMT2D sustains a gene expression program that represses B cell lymphoma development. Nat Med 2015;21(10):1199–208 doi 10.1038/nm.3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Dominguez-Sola D, Hussein S, Lee JE, Holmes AB, Bansal M, et al. Disruption of KMT2D perturbs germinal center B cell development and promotes lymphomagenesis. Nat Med 2015;21(10):1190–8 doi 10.1038/nm.3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jiang Y, Ortega-Molina A, Geng H, Ying HY, Hatzi K, Parsa S, et al. CREBBP Inactivation Promotes the Development of HDAC3-Dependent Lymphomas. Cancer Discov 2017;7(1):38–53 doi 10.1158/2159-8290.CD-16-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang J, Vlasevska S, Wells VA, Nataraj S, Holmes AB, Duval R, et al. The CREBBP Acetyltransferase Is a Haploinsufficient Tumor Suppressor in B-cell Lymphoma. Cancer Discov 2017;7(3):322–37 doi 10.1158/2159-8290.CD-16-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dominguez PM, Teater M, Chambwe N, Kormaksson M, Redmond D, Ishii J, et al. DNA Methylation Dynamics of Germinal Center B Cells Are Mediated by AID. Cell Rep 2015;12(12):2086–98 doi 10.1016/j.celrep.2015.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaknovich R, Cerchietti L, Tsikitas L, Kormaksson M, De S, Figueroa ME, et al. DNA methyltransferase 1 and DNA methylation patterning contribute to germinal center B-cell differentiation. Blood 2011;118(13):3559–69 doi 10.1182/blood-2011-06-357996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De S, Shaknovich R, Riester M, Elemento O, Geng H, Kormaksson M, et al. Aberration in DNA methylation in B-cell lymphomas has a complex origin and increases with disease severity. PLoS Genet 2013;9(1):e1003137 doi 10.1371/journal.pgen.1003137 PGENETICS-D-12-01703 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan H, Jiang Y, Boi M, Tabbo F, Redmond D, Nie K, et al. Epigenomic evolution in diffuse large B-cell lymphomas. Nat Commun 2015;6:6921 doi 10.1038/ncomms7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teater M, Dominguez PM, Redmond D, Chen Z, Ennishi D, Scott DW, et al. AICDA drives epigenetic heterogeneity and accelerates germinal center-derived lymphomagenesis. Nat Commun 2018;9(1):222 doi 10.1038/s41467-017-02595-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Asmar F, Punj V, Christensen J, Pedersen MT, Pedersen A, Nielsen AB, et al. Genome-wide profiling identifies a DNA methylation signature that associates with TET2 mutations in diffuse large B-cell lymphoma. Haematologica 2013;98(12):1912–20 doi 10.3324/haematol.2013.088740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ito S, Shen L, Dai Q, Wu SC, Collins LB, Swenberg JA, et al. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011;333(6047):1300–3 doi 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abdel-Wahab O, Mullally A, Hedvat C, Garcia-Manero G, Patel J, Wadleigh M, et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 2009;114(1):144–7 doi 10.1182/blood-2009-03-210039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Delhommeau F, Dupont S, Della Valle V, James C, Trannoy S, Masse A, et al. Mutation in TET2 in myeloid cancers. N Engl J Med 2009;360(22):2289–301 doi 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 22.Smith AE, Mohamedali AM, Kulasekararaj A, Lim Z, Gaken J, Lea NC, et al. Next-generation sequencing of the TET2 gene in 355 MDS and CMML patients reveals low-abundance mutant clones with early origins, but indicates no definite prognostic value. Blood 2010;116(19):3923–32 doi 10.1182/blood-2010-03-274704. [DOI] [PubMed] [Google Scholar]
- 23.Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, et al. Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 2012;120(7):1466–9 doi 10.1182/blood-2012-02-408542. [DOI] [PubMed] [Google Scholar]
- 24.Wu H, D’Alessio AC, Ito S, Wang Z, Cui K, Zhao K, et al. Genome-wide analysis of 5-hydroxymethylcytosine distribution reveals its dual function in transcriptional regulation in mouse embryonic stem cells. Genes Dev 2011;25(7):679–84 doi 25/7/679 [pii] 10.1101/gad.2036011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cimmino L, Abdel-Wahab O, Levine RL, Aifantis I. TET family proteins and their role in stem cell differentiation and transformation. Cell Stem Cell 2011;9(3):193–204 doi 10.1016/j.stem.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rampal R, Alkalin A, Madzo J, Vasanthakumar A, Pronier E, Patel J, et al. DNA hydroxymethylation profiling reveals that WT1 mutations result in loss of TET2 function in acute myeloid leukemia. Cell Rep 2014;9(5):1841–55 doi 10.1016/j.celrep.2014.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stroud H, Feng S, Morey Kinney S, Pradhan S, Jacobsen SE. 5-Hydroxymethylcytosine is associated with enhancers and gene bodies in human embryonic stem cells. Genome Biol 2011;12(6):R54 doi 10.1186/gb-2011-12-6-r54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 2014;56(2):286–97 doi 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rasmussen KD, Jia G, Johansen JV, Pedersen MT, Rapin N, Bagger FO, et al. Loss of TET2 in hematopoietic cells leads to DNA hypermethylation of active enhancers and induction of leukemogenesis. Genes Dev 2015;29(9):910–22 doi 10.1101/gad.260174.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cimmino L, Dolgalev I, Wang Y, Yoshimi A, Martin GH, Wang J, et al. Restoration of TET2 Function Blocks Aberrant Self-Renewal and Leukemia Progression. Cell 2017;170(6):1079–95 e20 doi 10.1016/j.cell.2017.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Quivoron C, Couronne L, Della Valle V, Lopez CK, Plo I, Wagner-Ballon O, et al. TET2 inactivation results in pleiotropic hematopoietic abnormalities in mouse and is a recurrent event during human lymphomagenesis. Cancer Cell 2011;20(1):25–38 doi 10.1016/j.ccr.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 32.Bunting KL, Soong TD, Singh R, Jiang Y, Beguelin W, Poloway DW, et al. Multi-tiered Reorganization of the Genome during B Cell Affinity Maturation Anchored by a Germinal Center-Specific Locus Control Region. Immunity 2016;45(3):497–512 doi 10.1016/j.immuni.2016.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nojima T, Haniuda K, Moutai T, Matsudaira M, Mizokawa S, Shiratori I, et al. In-vitro derived germinal centre B cells differentially generate memory B or plasma cells in vivo. Nat Commun 2011;2:465 doi 10.1038/ncomms1475. [DOI] [PubMed] [Google Scholar]
- 34.Hatzi K, Jiang Y, Huang C, Garrett-Bakelman F, Gearhart MD, Giannopoulou EG, et al. A hybrid mechanism of action for BCL6 in B cells defined by formation of functionally distinct complexes at enhancers and promoters. Cell Rep 2013;4(3):578–88 doi 10.1016/j.celrep.2013.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Figueroa ME, Lugthart S, Li Y, Erpelinck-Verschueren C, Deng X, Christos PJ, et al. DNA methylation signatures identify biologically distinct subtypes in acute myeloid leukemia. Cancer Cell 2010;17(1):13–27 doi S1535–6108(09)00420–6 [pii] 10.1016/j.ccr.2009.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogdanovic O, Smits AH, de la Calle Mustienes E, Tena JJ, Ford E, Williams R, et al. Active DNA demethylation at enhancers during the vertebrate phylotypic period. Nat Genet 2016;48(4):417–26 doi 10.1038/ng.3522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hon GC, Song CX, Du T, Jin F, Selvaraj S, Lee AY, et al. 5mC oxidation by Tet2 modulates enhancer activity and timing of transcriptome reprogramming during differentiation. Mol Cell 2014;56(2):286–97 doi 10.1016/j.molcel.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahe EA, Madigou T, Serandour AA, Bizot M, Avner S, Chalmel F, et al. Cytosine modifications modulate the chromatin architecture of transcriptional enhancers. Genome Res 2017;27(6):947–58 doi 10.1101/gr.211466.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamazaki J, Jelinek J, Lu Y, Cesaroni M, Madzo J, Neumann F, et al. TET2 Mutations Affect Non-CpG Island DNA Methylation at Enhancers and Transcription Factor-Binding Sites in Chronic Myelomonocytic Leukemia. Cancer Res 2015;75(14):2833–43 doi 10.1158/0008-5472.CAN-14-0739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang YW, Wang Z, Xie W, Cai Y, Xia L, Easwaran H, et al. Acetylation Enhances TET2 Function in Protecting against Abnormal DNA Methylation during Oxidative Stress. Mol Cell 2017;65(2):323–35 doi 10.1016/j.molcel.2016.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barwick BG, Scharer CD, Bally APR, Boss JM. Plasma cell differentiation is coupled to division-dependent DNA hypomethylation and gene regulation. Nat Immunol 2016;17(10):1216–25 doi 10.1038/ni.3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cattoretti G, Pasqualucci L, Ballon G, Tam W, Nandula SV, Shen Q, et al. Deregulated BCL6 expression recapitulates the pathogenesis of human diffuse large B cell lymphomas in mice. Cancer Cell 2005;7(5):445–55 doi 10.1016/j.ccr.2005.03.037. [DOI] [PubMed] [Google Scholar]
- 43.Garcia-Ramirez I, Tadros S, Gonzalez-Herrero I, Martin-Lorenzo A, Rodriguez-Hernandez G, Moore D, et al. Crebbp loss cooperates with Bcl2 overexpression to promote lymphoma in mice. Blood 2017;129(19):2645–56 doi 10.1182/blood-2016-08-733469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Green MR, Kihira S, Liu CL, Nair RV, Salari R, Gentles AJ, et al. Mutations in early follicular lymphoma progenitors are associated with suppressed antigen presentation. Proc Natl Acad Sci U S A 2015;112(10):E1116–25 doi 10.1073/pnas.1501199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner FF, Lundh M, Kaya T, McCarren P, Zhang YL, Chattopadhyay S, et al. An Isochemogenic Set of Inhibitors To Define the Therapeutic Potential of Histone Deacetylases in beta-Cell Protection. ACS Chem Biol 2016;11(2):363–74 doi 10.1021/acschembio.5b00640. [DOI] [PubMed] [Google Scholar]
- 46.Moran-Crusio K, Reavie L, Shih A, Abdel-Wahab O, Ndiaye-Lobry D, Lobry C, et al. Tet2 loss leads to increased hematopoietic stem cell self-renewal and myeloid transformation. Cancer Cell 2011;20(1):11–24 doi 10.1016/j.ccr.2011.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mouly E, Ghamlouch H, Della-Valle V, Scourzic L, Quivoron C, Roos-Weil D, et al. B-cell tumor development in Tet2-deficient mice. Blood Adv 2018;2(6):703–14 doi 10.1182/bloodadvances.2017014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatzi K, Melnick A. Breaking bad in the germinal center: how deregulation of BCL6 contributes to lymphomagenesis. Trends Mol Med 2014;20(6):343–52 doi 10.1016/j.molmed.2014.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Victora GD, Nussenzweig MC. Germinal centers. Annu Rev Immunol 2012;30:429–57 doi 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 50.Niu H, Ye BH, Dalla-Favera R. Antigen receptor signaling induces MAP kinase-mediated phosphorylation and degradation of the BCL-6 transcription factor. Genes Dev 1998;12(13):1953–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polo JM, Juszczynski P, Monti S, Cerchietti L, Ye K, Greally JM, et al. Transcriptional signature with differential expression of BCL6 target genes accurately identifies BCL6-dependent diffuse large B cell lymphomas. Proc Natl Acad Sci U S A 2007;104(9):3207–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ranuncolo SM, Polo JM, Dierov J, Singer M, Kuo T, Greally J, et al. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat Immunol 2007;8(7):705–14 doi 10.1038/ni1478. [DOI] [PubMed] [Google Scholar]
- 53.Saito M, Gao J, Basso K, Kitagawa Y, Smith PM, Bhagat G, et al. A signaling pathway mediating downregulation of BCL6 in germinal center B cells is blocked by BCL6 gene alterations in B cell lymphoma. Cancer Cell 2007;12(3):280–92 doi 10.1016/j.ccr.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 54.Ichiyama K, Chen T, Wang X, Yan X, Kim BS, Tanaka S, et al. The methylcytosine dioxygenase Tet2 promotes DNA demethylation and activation of cytokine gene expression in T cells. Immunity 2015;42(4):613–26 doi 10.1016/j.immuni.2015.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Horton SJ, Giotopoulos G, Yun H, Vohra S, Sheppard O, Bashford-Rogers R, et al. Early loss of Crebbp confers malignant stem cell properties on lymphoid progenitors. Nat Cell Biol 2017;19(9):1093–104 doi 10.1038/ncb3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko M, Huang Y, Jankowska AM, Pape UJ, Tahiliani M, Bandukwala HS, et al. Impaired hydroxylation of 5-methylcytosine in myeloid cancers with mutant TET2. Nature 2010;468(7325):839–43 doi 10.1038/nature09586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shih AH, Jiang Y, Meydan C, Shank K, Pandey S, Barreyro L, et al. Mutational cooperativity linked to combinatorial epigenetic gain of function in acute myeloid leukemia. Cancer Cell 2015;27(4):502–15 doi 10.1016/j.ccell.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li H Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. 2013;1303.
- 59.Koboldt DC, Zhang Q, Larson DE, Shen D, McLellan MD, Lin L, et al. VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res 2012;22(3):568–76 doi 10.1101/gr.129684.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 2010;20(9):1297–303 doi 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 2008;359(22):2313–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Damm F, Mylonas E, Cosson A, Yoshida K, Della Valle V, Mouly E, et al. Acquired initiating mutations in early hematopoietic cells of CLL patients. Cancer Discov 2014;4(9):1088–101 doi 10.1158/2159-8290.CD-14-0104. [DOI] [PubMed] [Google Scholar]
- 63.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008;9(9):R137 doi 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashoor H, Louis-Brennetot C, Janoueix-Lerosey I, Bajic VB, Boeva V. HMCan-diff: a method to detect changes in histone modifications in cells with different genetic characteristics. Nucleic Acids Res 2017;45(8):e58 doi 10.1093/nar/gkw1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 2010;26(6):841–2 doi 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 2013;29(1): 15–21 doi 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liao Y, Smyth GK, Shi W. The Subread aligner: fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res 2013; 41(10):e108 doi 10.1093/nar/gkt214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12): 550 doi 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7): e47 doi 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, et al. MLL translocations specify a distinct gene expression profile that distinguishes a unique leukemia. Nat Genet 2002; 30 (1) 41–7 doi 10.1038/ng765. [DOI] [PubMed] [Google Scholar]
- 71.Shaker S, Bernstein M, Momparler RL. Antineoplastic action of 5-aza-2’-deoxycytidine (Dacogen) and depsipeptide on Raji lymphoma cells. Oncol Rep 2004;11 (6): 1253–6. [PubMed] [Google Scholar]
- 72.Leiserson MD, Wu HT, Vandin F, Raphael BJ. CoMEt: a statistical approach to identify combinations of mutually exclusive alterations in cancer. Genome Biol 2015; 16:160 doi 10.1186/s13059-015-0700-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 2016;44 (W1):W160–5 doi 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013;6(269):pl1 doi 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012;2(5):401–4 doi 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.