Abstract
Purpose:
Pediatric DMGs are highly malignant tumors with poor clinical outcomes. Over 70% of DMG patients harbor the histone 3 p.K27M (H3K27M) mutation, which correlates with a poorer clinical outcome, and is also used as a criterion for enrollment in clinical trials. Because complete surgical resection of DMG is not an option, biopsy at presentation is feasible, but re-biopsy at time of progression is rare. While imaging and clinical-based disease monitoring is the standard of care, molecular-based longitudinal characterization of these tumors is almost non-existent. To overcome these hurdles we examined if liquid biopsy allows measurement of disease response to precision therapy.
Experimental Design:
We established a sensitive and specific methodology which detects major driver mutations associated with pediatric DMGs using droplet digital PCR (n=48 subjects, n=110 specimens). Quantification of ctDNA for H3K27M was used for longitudinal assessment of disease response compared to centrally reviewed MRI data.
Results:
H3K27M was identified in cerebrospinal fluid (CSF) and plasma in 88% of patients with DMG, with CSF being the most enriched for ctDNA. We demonstrated the feasibility of multiplexing for detection of H3K27M, and additional driver mutations in patient’s tumor and matched CSF, maximizing the utility of a single source of liquid biome. A significant decrease in H3K27M plasma ctDNA agreed with MRI assessment of tumor response to radiotherapy in 83% (10/12) of patients.
Conclusions:
Our liquid biopsy approach provides a molecularly-based tool for tumor characterization, and is the first to indicate clinical utility of ctDNA for longitudinal surveillance of DMGs.
Keywords: circulating tumor DNA, diffuse midline gliomas, liquid biopsy, digital droplet PCR, histone 3 K27M
Introduction
High grade gliomas in children are rare, accounting for about 20% of all pediatric central nervous system (CNS) cancers. Nevertheless, pediatric brain cancers are the leading cause of cancer related mortality in children under 14 years of age (1). Among these, malignant tumors arising in midline brain structures (e.g. pons, thalamus, spinal cord) have one of the worst outcomes (2,3). Children with diffuse midline gliomas (DMGs) – including those with diffuse intrinsic pontine glioma (DIPG) - have a median survival of only 12 months. DMGs often harbor mutations in genes encoding canonical histones H3.1 (HIST1H3B/C), H3.2 (HIST2H3C), and non-canonical histone H3.3 (H3F3A) variants (2–5). The presence of these mutations often confers a poorer outcome as compared to tumors without these alterations (2). On the basis of distinct genetic features and clinical behavior, these tumors were recently re-classified by the World Health Organization (WHO) as “Diffuse midline glioma, H3 K27M-mutant” (6).
In recent years, the recognition that surgical biopsy of brainstem tumors can be performed safely has led to a far more detailed understanding of the biology of DMGs, and has prompted the development of therapies directed against tumor-specific alterations (7,8). However, the sensitive anatomic location of these tumors and the need for specialized surgical expertise has limited the broader adoption of surgical biopsy, especially at time of recurrence. Hence, MR imaging and clinical examination are currently widely used to diagnose, and assess response to therapy, although both of these methods are limited in sensitivity and specificity (9–11). The inability to accurately assess disease response and treatment-related molecular changes confer significant challenges, particularly for emerging biologically targeted strategies such as immunotherapy (12,13). Appropriate selection of tumor-specific treatment plans will require the ability to frequently monitor both the burden of disease and the ongoing genetic changes that clearly occur in these tumors over time.
The use of ctDNA to monitor disease progression is a non/minimally-invasive method that has been increasingly used for disease monitoring in adult cancers including glioblastoma multiforme (GBM), melanoma, lung, breast, and colon cancers (14–18), however such studies have not been applied to the pediatric population. Thus, there is an urgent need for the development of ctDNA assays for clinical applications in pediatric CNS patients.
Digital droplet PCR (ddPCR) is one of the most sensitive methods for detection and quantification of ctDNA when compared to Sanger sequencing, quantitative PCR, and next generation sequencing platforms (19). The high sensitivity of ddPCR is attributed to the fact that PCR reactions are performed in millions of isolated droplets, allowing for detection of rare, low mutation allelic frequencies (19,20). While Sanger sequencing of H3F3A p.K27M mutation in CSF of DIPG patients was shown to be feasible (21), such an approach does not allow for calculating mutation allelic frequencies, and lacks sensitivity for clinical applications. As the histone 3 K27M mutation is present at low mutant allelic frequencies, where one mutant allele exists among 30 alleles encoding isoforms of H3, a more sensitive approach is needed for detection of driver mutations in these tumors (22,23). Therefore, the goal of this study was to assess the feasibility of detecting histone 3 K27M (H3K27M) and other driver mutations in CSF and plasma, as well as to investigate the clinical utility of tracking H3K27M in plasma ctDNA for disease monitoring. By using plasma collected longitudinally through an ongoing clinical trial (PNOC003; NCT02274987) conducted by the Pacific Pediatric Neuro-Oncology Consortium (PNOC), we were also able to correlate ctDNA levels with treatment response, and progression.
Materials and methods
Biological specimens
All patient samples were collected after written informed consent was obtained by each patient or each patient’s guardian for participation in a clinical trial, or biorepository as approved by the respective Institutional Review Boards of the University of California San Francisco (IRB #14–13895), University of California San Diego (IRB #150450), and Children’s National Health System (IRB #1339, #747). All studies involving human subjects were conducted in accordance with the U.S. Common Rule. CSF and cyst fluid were collected from patients with DMG consented to the Children’s National (CN) brain tumor biorepository, while plasma was collected from patients with DIPG enrolled in the PNOC003 (NCT02274987) precision therapy clinical trial. This prospective I/II multi-institutional clinical trial (PNOC003, NCT02274987) led by PNOC enrolled newly diagnosed patients with DIPG to evaluate the feasibility of tumor genomic profiling of biopsies, and treated patients with precision therapy of up to four FDA-approved therapeutic agents (24). Blood was collected from enrolled patients for ctDNA analysis at the time of initial diagnosis, as well as with each subsequent MRI.
Whole blood was collected in a purple top potassium EDTA tube for isolation of plasma. The tube was then inverted to mix with the anticoagulant and spun at 2,000Xg for 15 minutes at 4°C. Blood was separated into three layers: plasma in the supernatant, white blood cell layer located between the plasma and red blood cell pellet. Plasma was aliquoted and stored at −80°C for future analyses.
Cerebrospinal fluid (CSF) was collected from subjects and processed according to the Children’s National (CN) brain tumor biorepository standardized protocol. Briefly, CSF was spun at 10,000Xg for 10 minutes to eliminate cellular debris and supernatants were collected, aliquoted and frozen at −80°C for future analyses. Data for CSF and plasma samples collected from healthy pediatric, and non-CNS malignant control pediatric subjects is shown in Supplementary Table 1. Time points for all CSF and plasma samples collected from subjects in this study is indicated in Supplementary Table 2.
DNA extractions
Genomic DNA was extracted from frozen tumor tissue using the Gentra Puregene kit (Qiagen, Valencia, CA). ctDNA was extracted from 1ml of frozen plasma, 500μl of frozen CSF, and 200μl of frozen cyst fluid using the QIAamp circulating nucleic acids kit (Qiagen, Valencia, CA) according to the manufacturer’s instructions. The ctDNA was subjected to pre-amplification prior to ddPCR using Q5 hot start high-fidelity master mix (New England Biolabs, Ipswich, MA), and 50 nM each of forward and reverse primers for each gene as shown in Supplementary Table 3. Pre-amplification was performed in an ABI 2720 thermocycler at 98°C for 3 minutes, and nine cycles of 98°C for 10 seconds, at annealing temperature for 3 minutes, 72°C for 30 seconds, and an extension of 72°C for 2 minutes. The pre-amplified product was diluted 1:5 with TE buffer, pH 8.0.
Digital Droplet PCR
All ddPCR reactions were conducted on the RainDrop ddPCR system according to the manufacturer’s instructions (Billerica, MA). The ddPCR reaction was conducted with 1X TaqMan genotyping mastermix (Life technologies, Carlsbad, CA), 1X RainDance droplet stabilizer, 12ul of the diluted pre-amplified PCR product, 900nM of the forward and reverse primers, and 200nM of the mutant and wild type probes (Supplementary Table 3). The prepared PCR reactions were added to the 8 wells in a Source microfluidics chip, which is loaded along with an empty strip of PCR tubes onto the RainDrop Source machine. The RainDrop Source generates millions of droplets by using the components of the PCR reaction and surfactant containing fluorocarbon oil. The dropletized samples are dispensed into the PCR strip and used for the PCR reaction in the second step. PCR reactions were performed in an ABI 2720 thermocycler using the optimized conditions for each primer and probe sets. The amplified products in each droplet were detected using the RainDrop Sense machine and the Sense microfluidics chip based on the fluorescence signal emitted by the wild type and mutant probes. The raw spectral data were analyzed using the RainDrop Analyst software to plot the signal for mutant and wild type alleles. In the analysis software, the calculated matrix function was used to apply spectral compensation on intact droplets. Negative, wild type, and mutant gates were applied based on the no template negative, and genomic DNA positive controls. All results were manually reviewed for false positives of the mutant alleles and background noise based on the no template negative control. Mutant allele frequency (MAF) was calculated by the percentage of mutant droplets divided by the sum of mutant and wild type droplets.
Magnetic Resonance (MR) Imaging
MR imaging of the brain was performed for all the patients according to the clinical protocol on either 1.5T or 3T scanners. Contrast-enhanced, FLAIR (fluid-attenuated inversion recovery) and T2-weighted images were used to measure tumor size. Anterior-posterior, transverse, and cranio-caudal dimensions were measured in mm on either PACS (Agfa Healthcare) or Horos (Horos Version 3.0) software packages. Tumor growth on MRI was defined as >25% increase in tumor volume-related FLAIR signal abnormality and/or development of new enhancing or nonenhancing lesions by an experienced neuro-oncologist (S.M.) and neuroradiology fellow (M.S.A.) with guidance from a board certified neuroradiologist (S.C.).
Statistical analysis
Plasma ctDNA serial assessments and central MRI tumor measurement analyses were conducted independently in a blinded manner. Temporal plasma ctDNA MAFs were associated to clinical outcomes, and MRI tumor volume measurements at the end of the study. None of the MAF values collected were normally distributed; therefore all analyses used nonparametric tests. All data points for plasma and CSF represent technical triplicates and duplicates, respectively. Error bars in all figures represent standard error of mean. Paired samples were analyzed by a Wilcoxon signed rank test, and unpaired values were analyzed by a Mann Whitney test to obtain p-values. A Kappa coefficient test was used to assess for agreement between plasma ctDNA and MRI evaluation of tumor response to radiation therapy. We assessed for at least a 50% reduction of H3K27M MAF in plasma ctDNA, in agreement with at least a 10% decrease in MRI tumor volume measurements at post radiation compared to baseline (n=12). For all analyses, a significance threshold of 0.05 was used to define statistical significance.
Results
A sensitive and specific platform for discriminating rare, tumor-associated circulating DNA
Clinicopathologic and genomic characteristics of the tumors of our pediatric DMG patient cohort are shown in Fig. 1a. As expected, patients with DMGs harbored a predictable combination of mutations: 94% harbored histone 3 mutations (79% with H3.3K27M, 15% with H3.1K27M), and 6% were H3 wild type. Our initial goal was to develop and validate a clinically relevant and minimally invasive liquid biopsy platform suitable for detection and quantification of somatic mutations associated with pediatric DMGs. To accomplish this goal, we validated our ddPCR probes for assessing mutation burden by using tumor genomic DNA from DMG patients harboring oncohistone H3F3A (p.K27M), HIST1H3B (p.K27M) and partner mutants in genes ACVR1 (p.G328V, p.R206H), PPM1D (p.E525X), PIK3R1 (p.K567E), and BRAF (p.V600E) (Supplementary Fig. 1a). Additionally, no mutant clusters were observed where control brain genomic DNA was used as template (Supplementary Fig. 1a). To assess the sensitivity, we used a range (2.5ng, 250pg, 25pg and 2.5pg) of genomic DNA as template for ddPCR and detected a corresponding linear decrease (10 fold) in MAF, indicating a low (2.5pg) amount of DNA to be suitable for detection by ddPCR (Supplementary Fig. 1b).
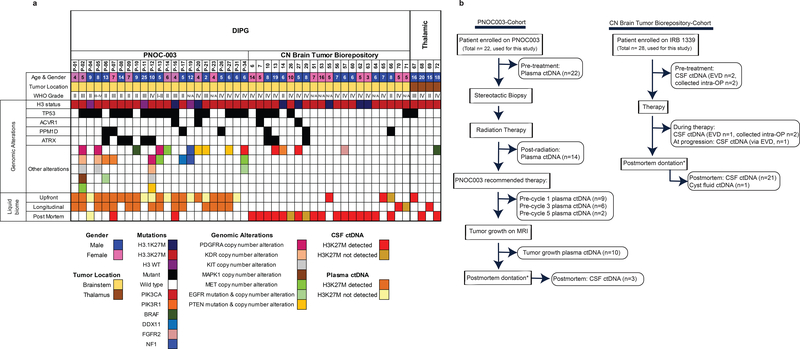
Figure 1. Overview of biofluid specimens utilized in this study for detection of ctDNA in diffuse midline glioma (DMG) patients.
(a) Demographic, clinical, and molecular information for all patients analyzed in this study. Genomic alterations were obtained by tumor sequencing data generated for PNOC003 trial, biorepository sequencing project, or other clinical studies. (b) Flow chart portraying biofluid samples analyzed in this study. Patients were either enrolled in PNOC003 trial (DIPG) or Children’s National (CN) brain tumor biorepository (DMG); *patients enrolled on IRB 1339 to donate specimens at postmortem.
To assess the specificity of our platform, we used a large cohort of control pediatric non-CNS malignant specimens (CSF = 16; plasma = 20) and probed for wild type and mutant histone ctDNA. Our data demonstrated that any allelic frequency detected as equal or below 0.001% to be false positive in the case of histone p.K27M analysis (Supplementary Table 1). We also analyzed genomic DNA, CSF, and plasma obtained from three DIPG patients who did not harbor histone 3 p.K27M mutations. As expected, histone mutant copies were not detected in any of the specimen tested indicating the specificity of our platform (Supplementary Fig. 1c).
CSF and plasma harbor ctDNA indicative of driver mutations associated with pediatric DMGs
Liquid biome specimens were analyzed from 84 subjects (48 DMG patients, and 36 non-CNS diseased controls), who were either consented for the Children’s National (CN) brain tumor biorepository, or enrolled on an ongoing clinical trial PNOC003 (NCT02274987) (Fig. 1b). A cohort of 110 biofluid samples (30 CSF, 79 plasma, and 1 cyst fluid) representing 48 DMG patients was processed for liquid profiling. CSF samples were collected at a single time point through the CN biorepository either at pre-treatment, during therapy, or at postmortem from 27 DMG patients, while serial sampling at pre-treatment and postmortem was available for one patient with DIPG. Plasma specimens were obtained starting at diagnosis, and longitudinally throughout treatment from patients with DIPG enrolled in PNOC003 (NCT02274987) (at initial biopsy n=22, during treatment n=15) (Fig. 1b, Supplementary Table 2). Three of the DIPG patients enrolled in PNOC003 also consented for CSF collection at postmortem via the CN biorepository, among which, two patients harbored H3 K27M and one was H3 wild type (Fig. 1).
A total of 28 CSF samples were analyzed, where we successfully detected histone 3 mutant and wild type alleles in 75% of CSF specimens collected at diagnosis, 67% of those collected during treatment, and 90% of those collected at postmortem (Fig. 2a). Tumor genome data was available for 24 of these subjects identifying one as histone 3 wild type, and 23 as H3 K27M mutants. The histone 3 mutation status of four tumor samples was not known. Our analysis of CSF specimens validated the histone 3 wild type specimen, as well as 20 of the 23 (87%) as H3 K27M mutant subjects. The four samples for which tumor genome data was not available, were shown to be H3K27M mutant by ddPCR analysis of CSF.
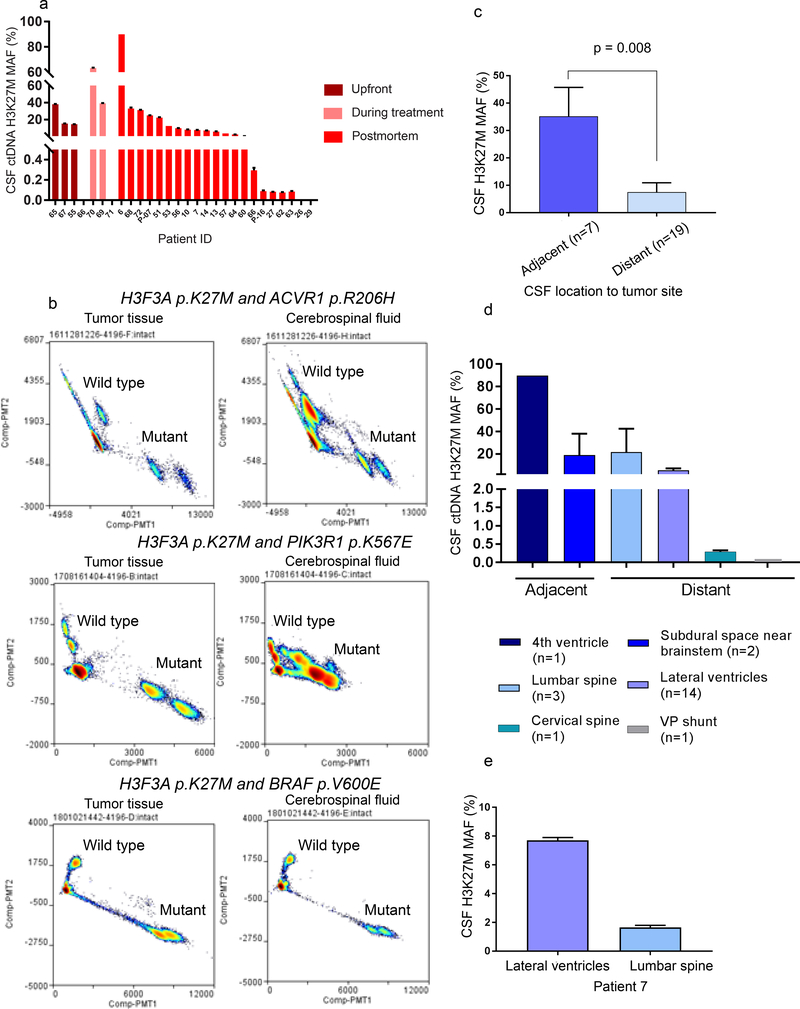
Figure 2. Detection of histone 3 p.K27M mutant (H3K27M) ctDNA in CSF of patients diagnosed with DMG.
(a) Detection of H3K27M ctDNA in CSF collected throughout disease. (b) Multiplexed detection of major driver mutations in tumor and matched CSF. (c) CSF collected from adjacent neuroanatomical locations yield a significantly (Mann Whitney U test) higher H3K27M mutation allelic frequency in DMG patients. (d) Cohort based and (e) matched intrapatient analysis indicating higher H3K27M MAF levels in CSF from adjacent locations in DIPG patients.
We have previously shown that in addition to oncohistone mutations, DMGs harbor obligate partner mutations in genes such as ACVR1, TP53, PPM1D, and PIK3R1 (25). We thus sought to determine whether biofluids represent a source for detection of multiple mutations for a particular patient. We first used tumor DNA known to harbor oncohistone and partner mutations (ACVR1 p.R206H, PIK3R1 p.K567E, or BRAF p.V600E) to optimize the experimental and analytical approach for multiplexed assays (Fig. 2b). We tested CSF obtained from these patients and showed the feasibility of detecting mutant and wild type alleles, for oncohistone and obligate partners in ACVR1, PIK3R1 or BRAF (Fig. 2b).
We then assessed the effect of CSF location on ctDNA abundance. CSF collected from lateral ventricles, lumbar spine, cervical spine, subdural (at biopsy), ventriculoperitoneal (VP) shunt, and the fourth ventricle (cisternae magna tap) was analyzed by ddPCR. CSF collected from the lateral ventricles was considered adjacent for thalamic tumors, while CSF collected from the fourth ventricle, and subdural space near brainstem during biopsy were considered adjacent locations for DIPGs. We detected a significantly higher MAF in CSF collected from adjacent locations compared to distant in DMG patients (Fig. 2c). This was confirmed by our cohort based, and matched analysis of CSF collected from patients diagnosed with DIPG (Fig. 2d-e). Nonetheless, we report successful detection of H3 K27M ctDNA in all locations tested.
We previously reported the suitability of cyst fluid for identification of tumor associated proteins (26). Here, we analyzed tumor cyst fluid, CSF, and tumor genomic DNA obtained at postmortem from a patient diagnosed with DIPG. We showed that the cyst fluid is highly enriched for tumor DNA as suggested by the high number of histone mutant DNA copies (Supplementary Fig. 2).
To assess the enrichment of ctDNA in CSF versus plasma, we tested these two biofluids and found a significantly higher allelic frequency of ctDNA in CSF (n=27 patients, 29 specimens) compared to plasma (n=20 subjects, 77 specimens) (p < 0.0001) (Fig. 3a). We used matched CSF-plasma specimens (n=2 subjects) and found an enrichment of ctDNA in CSF compared to plasma within the same patient (Fig. 3b). We successfully detected H3K27M ctDNA in 87% (20/23 subjects) of CSF samples and 90% (18/20 subjects) of plasma samples analyzed from patients known to harbor oncohistones in their primary tumors. Although detectable, the levels of ctDNA in plasma were low, possibly due to challenges of overcoming the blood brain barrier. Our data indicate that both CSF and plasma are a suitable source for detection of ctDNA, and identification of tumor-associated somatic mutations. These results emphasize the potential value of liquid biopsy for complementing standard surgical biopsies, and the need for a highly sensitive platform for the detection of ctDNA in biofluids from patients with DMG.
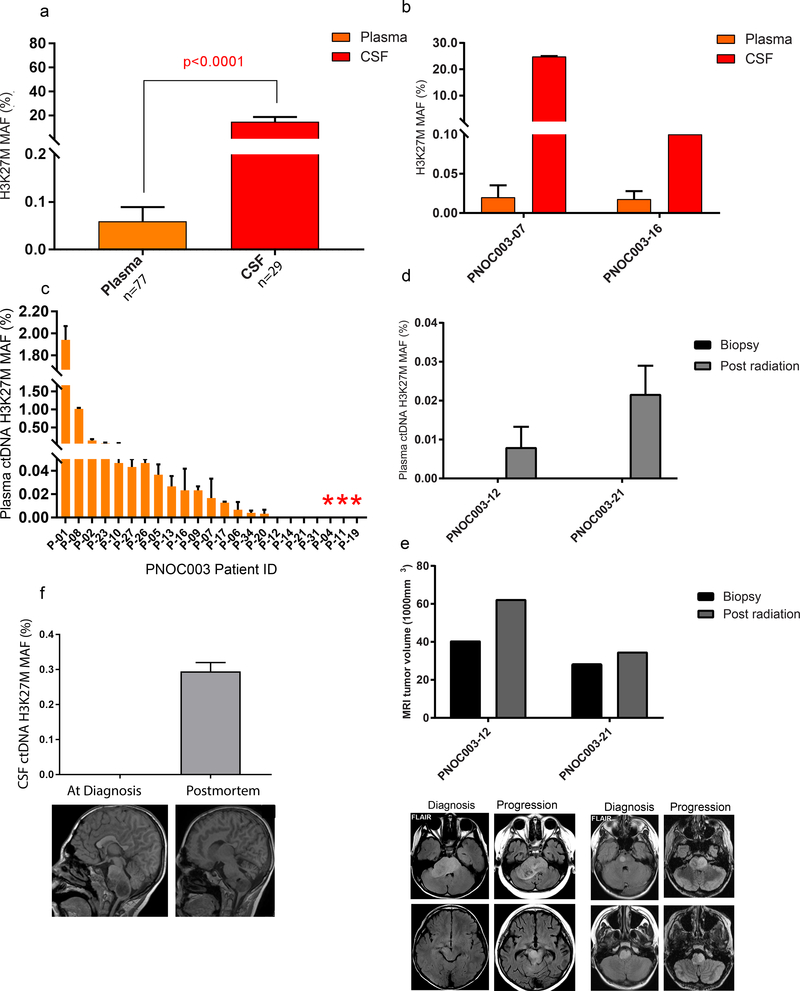
Figure 3. Abundance of histone 3 p.K27M mutation (H3K27M) ctDNA in biofluids of patients diagnosed with DMG.
(a) Significantly higher ctDNA MAFs for H3K27M were detected in CSF compared to plasma (Mann-Whitney U test). (b) Higher H3K27M MAFs were present in CSF compared to plasma in the same patient. (c) Plasma ctDNA collected at diagnosis in DIPG patients enrolled in the PNOC003 clinical trial analyzed for H3K27M. Red asterisk denotes histone 3 wild type DIPGs. (d) Detection of H3K27M plasma ctDNA at post radiation in two patients that lacked detection at diagnosis. (e) Increase in tumor size by MRI at post radiation compared to diagnosis in patients shown in (d). (f) Detection of H3K27M ctDNA in CSF collected at postmortem that lacked detection at diagnosis, in a patient known to harbor H3.3 K27M in tumor.
Treatment response assessment using ctDNA quantification
To investigate the clinical translation of our approach, we studied plasma obtained from subjects diagnosed with DIPG who enrolled in the ongoing clinical trial (PNOC003). Analysis of upfront and serial plasma samples probing for histone mutations (H3.3 and H3.1) indicated the following: i) histone mutations were successfully detected at diagnosis in 80% (n=16 of 20) of subjects harboring known histone mutations determined by genomic analysis of pretreatment biopsies (Fig. 3c); ii) histone mutation was not detected at diagnosis in 20% of subjects with histone mutations (n=4 of 20), iii) however, among these four subjects, histone mutation was subsequently detected following radiation therapy in two subjects where serial samples were available (Fig. 3d). MR image analysis of the two subjects for whom H3K27M ctDNA was absent at diagnosis, but present following radiation therapy indicated an increase in tumor volume following radiation (patient 12: 40.6mm3 to 62.4mm3, and patient 21: 28.6mm3 to 34.8mm3) (Fig. 3e) suggesting that changes in tumor architecture in response to radiation, or temporary disruption of the blood brain barrier (BBB) may contribute to the availability of ctDNA in plasma at different time points. Similarly, our analysis of serially collected CSF from a subject with H3 K27M DIPG at pre-treatment and at postmortem, revealed an increase in ctDNA abundance at the later stage of disease (Fig. 3f).
To assess if ctDNA can be developed as a surrogate biomarker of treatment response, we assessed the longitudinal plasma samples obtained from enrolled subjects (PNOC003). Overall we found that subjects with DIPG had a decrease in ctDNA following radiotherapy. These findings were comparable to changes in MRI tumor volume measurements, where a significant decrease was observed in tumor burden following radiation therapy as determined by a paired Wilcoxon signed rank analysis (n=14) (Fig. 4a). We report an agreement of 75% between ctDNA response (at least 50% decrease) and MRI tumor volume measurements (at least 10% decrease) at post radiation compared to baseline using the Kappa coefficient analysis (n = 12 subjects, kappa = 0.4, p = 0.078).
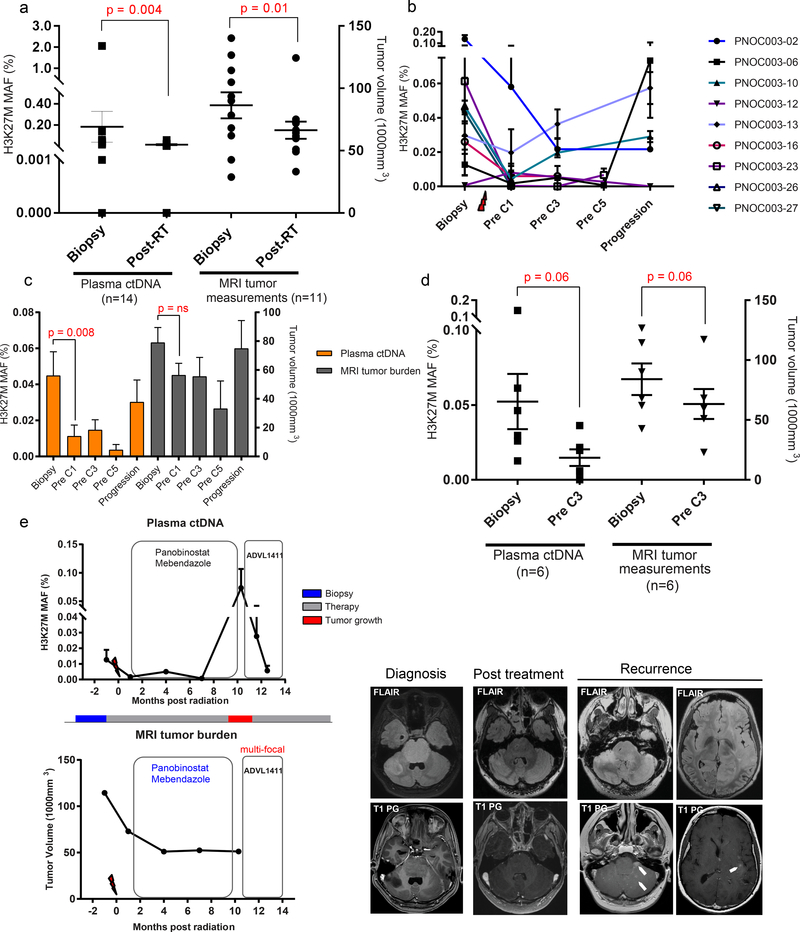
Figure 4. Temporal analysis of plasma ctDNA agrees with response to therapy in DIPG patients.
(a) Significant decrease in plasma ctDNA and MRI tumor volumes in response to radiation therapy. (b) Serial plasma ctDNA analysis for changes in H3K27M throughout course of treatment in patients that followed PNOC003 recommended therapy. (c) Fluctuations in plasma ctDNA and MRI tumor volumes in an average of nine patients (Pre C1) reflect similar trends post therapy, and at progression. (d) Decrease in plasma ctDNA and MRI tumor burden from diagnosis to pre-cycle 3 of PNOC003 recommended therapy. (e) Dynamic changes in plasma ctDNA and MRI tumor measurements in patient P-06 in response to therapy. FLAIR and T1-weighted post-gadolinium MR images prior to treatment demonstrating prominent expansile pontine mass and an additional focus within right cerebellar hemisphere that demonstrates evidence of enhancement. After treatment, the pontine and right cerebellar hemisphere lesions decreased in size. Paired Wilcoxon signed rank tests were used to derive all p-values.
We subsequently analyzed only the subjects who followed the recommended precision therapy (n=9), and found a decrease in plasma ctDNA from biopsy to early cycles of precision therapy (pre C1). Moreover, 60% (3/5) of patients showed an increase in plasma ctDNA at progression, suggesting that plasma ctDNA may agree with detecting tumor growth (Fig. 4b). The longitudinal changes in H3 K27M from biopsy to post therapy closely mirrored the trend of MRI-based clinical response in these subjects, characterized by a steady decrease following radiation and chemotherapy, and an increase at progression (Fig. 4c). Among subjects that followed PNOC003 recommended therapy, pre C3 was the time point with the highest sample availability for ctDNA analysis and radiographic evaluation. We found a decrease in plasma ctDNA congruent with the diminished tumor burden measured by MRI (Fig. 4d). Nonetheless, these findings infer the clinical utility of plasma ctDNA for monitoring response, comparable to MRI evaluation. These findings were further evaluated in one patient with DIPG who followed recommended therapy, where our analysis displayed that changes in ctDNA MAFs during the course of disease precisely reflected tumor response. Following enrollment in a different clinical trial, we detected a decrease in plasma ctDNA, indicative of a response to therapy (Fig. 4e). Notably, patient-specific temporal analysis of changes in MAF of histone 3 mutation indicated a close agreement of ctDNA level with both clinical course of the disease, and tumor response, as revealed by MR imaging volumetric analysis (P-02, P-10, P-13, P-27). In some patients (P-01, P-23), ctDNA levels showed fluctuations when compared to MRI assessment. This may be due to the limited sensitivity of MRI in detecting small variations in tumor size which needs further validation in a larger cohort (Supplementary Fig. 3).
We then assessed whether an increase in H3 K27M ctDNA accompanied tumor spread as determined by MRI, molecular, and/or histologic studies. Tumor extension was determined by MRI obtained during the course of the treatment, or molecular and histopathologic review of autopsied brain specimens. Tumor involvement beyond pons for patients enrolled in the clinical trial (PNOC003) was determined by central MRI review at diagnosis, during therapy, or at tumor growth. We found that increased levels of CSF ctDNA corresponded to tumor spread; however, the small sample size for the availability of CSF for DIPGs with tumor dissemination precluded statistical significance. On the contrary, plasma collected at various times throughout disease (diagnosis, during treatment, or at tumor growth) was not predictive or indicative of tumor spread beyond pons (Supplementary Fig. 4).
Discussion
Outcomes for children diagnosed with DMG have not changed despite decades of clinical research. Recent advancements in genomic, epigenomic, and proteomic profiling provide new opportunities for accelerating development of novel therapeutic interventions for patients with these tumors. Collection of postmortem specimens led to the identification of oncohistones as driver mutations as well as other partner mutations, and newer clinical trials are building upon this knowledge to identify rational, biologically targeted treatment approaches (4,5,25). However, the difficulty of performing repeated surgical biopsies in the pons is a barrier to the accurate assessment of responses to therapy.
Liquid biopsy has emerged as a useful tool for diagnosis, and measurement of efficacy in the treatment of adult cancers. ctDNA has been used to determine patient’s mutation profiles, as a biomarker for molecular-based disease monitoring, and recurrence in adult chronic lymphocytic leukemia, breast, and colon cancer (14,18,27). The only reported liquid biopsy approach for pediatric DMGs was established using Sanger sequencing that does not allow for quantification of ctDNA (21). Here we report the establishment of a sensitive, specific, rapid, and minimally-invasive methodology for tumor surveillance of pediatric DMGs by monitoring the liquid biome. Specifically, we show the detection of mutations associated with pediatric DMGs such as DIPG and thalamic tumors, using patients’ CSF and plasma.
Given the paucity of tumor DNA in these rare cancers, and the invasive nature of surgical biopsy, the multiplexed mutation analysis we used allows for maximized clinical utility of a single accessible biofluid specimen. Necrotic biopsy tumor tissue may sometimes result in degraded nucleic acid content unsuitable for subsequent sequencing analyses; in such cases, if CSF or cyst fluid is available, these may serve as more adequate materials for sequencing. CSF collected for clinical purposes may serve as a surrogate for detecting tumor driver mutations. Indeed, in many cases an extra-ventricular drain is placed to relieve pressure prior to any surgical interventions. In such cases, our platform will allow for detecting tumor-associated mutations in the advance of any surgical interventions.
We have shown that tumor-associated ctDNA can be quantified using ddPCR, which will allow for a rapid and more sensitive method for surveying tumor mutations. This represents a key advance particularly for tumors with limited tissue acquisition, or prohibitive sampling at multiple time points. Many of the ongoing trials have exclusion and inclusion criteria that are based on patient’s cancer mutation profile. For example newly launched clinical trials, including an immunotherapy trial will enroll only pediatric glioma patients who harbor the H3K27M mutation (NCT03416530, NCT02960230) and therefore establishing non-invasive tools for the detection of mutations, such a ctDNA, is critical.
Using plasma collected through the ongoing clinical trial (PNOC003), we show that MAF is readily obtained from ctDNA present in liquid biome, and that MAF might correlate with treatment response. Our report is the first to provide an indication of the potential utility of ctDNA for tumor profiling, and assessment of tumor response to therapy in pediatric patients with DMGs. These results indicate the feasibility of incorporating liquid biopsy as a sensitive and minimally invasive tool to inform clinical management for children with DMGs.
While we report the detection of ctDNA in both CSF and plasma, the exact mechanism of tumor DNA release into the biofluid is not well understood. A survey of published manuscripts indicated that ctDNA is released from multiple sources, including living cells, apoptotic and/or necrotic tumor cells (19,28–30). We also found that CSF yielded a significantly higher amount of ctDNA compared to plasma. This is most likely due to the location of biofluids in relation to site of tumor, and challenges of overcoming the BBB for release of ctDNA into plasma.
Our lack of detection of ctDNA in plasma obtained at biopsy from four subjects (H3 mutant by biopsy) may suggest high BBB integrity and thus lack of ctDNA. Subsequent analyses detected ctDNA in plasma drawn for two of these four subjects following radiation therapy, indicating the potential role of radiation therapy for temporary disruption of the BBB. Moreover, as the cellular turnover in a growing tumor increases, apoptotic and necrotic tumor cells are hypothesized to contribute to the increased release of ctDNA into biofluids (29). As such, we expected to observe an initial rise in ctDNA as tumor cells die and release DNA into biofluids. However, our tested time points fell after the completion of RT when the tumor mass was reduced in most patients as assessed by MRI, corresponding to the expectant reduced number of ctDNA. An issue to be determined is whether inclusion of earlier collection time points during the course of RT, would allow us to detect an initial spike in ctDNA immediately after early radiation treatments, followed by stabilization of ctDNA at maximal tumor response. Our study suggests that collection of multiple plasma draws at diagnosis and during treatment will be more informative as ctDNA may not be detectable in initial blood draws.
We have recently shown that DIPG tumor cells disseminate throughout the brain during the course of disease (25,31). Our ctDNA analysis in DIPG patients was indicative of tumor expansion beyond pons, where an increased amount of ctDNA in CSF was observed in patients who exhibited tumor spread; however, these findings were not replicated using ctDNA assessment in plasma. Studies of a larger cohort in clinical settings are required to assess the statistical significance of our finding.
More importantly, our results indicate the unique strength of liquid biopsy for assessing the molecular landscape of DMGs non-invasively, and potential for longitudinal assessment of tumor response to therapy, which is a new tool that is complementary to MR imaging. Similar to the clinical utility of ctDNA for monitoring response to therapy with respect to MRIs in adult GBMs (17), despite a small sample size, we found a significant reduction in ctDNA following RT. Similar patterns in temporal changes of ctDNA and MRI assessments of tumor response indicated that our ddPCR analysis of plasma is highly translational, and offers a novel platform for assessing tumor response, regression and/or progression in DMGs. Our liquid biopsy approach may also be expanded for facilitating diagnosis, longitudinal monitoring, and assessing recurrence in other childhood CNS cancers.
In summary, we have shown that CSF and plasma ctDNA analysis of children with DMG is feasible, shows promise for detecting mutational load, provides an additional means for molecular disease characterization, and most importantly is a clinically relevant and might serve as an additional method for assessing tumor response to treatment.
Supplementary Material
Translational Relevance.
Performing stereotactic biopsies in patients with diffuse midline glioma (DMG) including diffuse intrinsic pontine glioma (DIPG) has become more widely accepted. However, due to risks, costs and clinical regulations associated with the procedure, re-biopsy is rarely performed at disease progression. Inability to accurately assess disease response and molecular changes are major hurdles for developing successful therapeutic interventions. Liquid biopsy is a minimally invasive method for detecting tumor-associated circulating DNA (ctDNA). We investigated the feasibility of using liquid biopsy for detecting primary driver mutations in patients with DMG. We established a molecular-based tool to detect rare mutant alleles, with the objective of complementing diagnosis, and clinical monitoring. Each patient’s response to precision therapies was successfully monitored in real-time using ctDNA as a surrogate biomarker of tumor response. This is the first evidence of clinical translational utility to measure ctDNA for longitudinal tumor surveillance in this devastating pediatric disease.
Acknowledgements
The authors would like to acknowledge the generosity of all patients’ and their families. We would also like to thank all staff members of Pediatric Pacific Neuro-Oncology Consortium (PNOC) for their support. Human tissue and CSF was obtained from University of Maryland Brain and Tissue Bank, which is a brain and tissue repository of the NIH NeuroBioBank. E.P. is a pre-doctoral student in the Molecular Medicine Program of the Institute for Biomedical Sciences at The George Washington University. This work is from a dissertation to be presented to the above program in partial fulfillment of the requirements for the Ph.D. degree.
Financial support: This work was supported by funding from the V foundation (Atlanta, GA), Goldwin Foundation (St. Lincoln, NE), Pediatric Brain Tumor Foundation (Asheville, NC), Smashing Walnuts Foundation (Middleburg, VA), The Gabriella Miller Kids First Data Resource Center, Zickler Family Foundation (Chevy Chase, MD), Piedmont Community Foundation (Middleburg, VA), Musella Foundation (Hewlett, NY), Mathew Larson Foundation (Franklin Lake, NJ), Childhood Brain Tumor Foundation (Germantown, MD). M.S.A was supported by NIH T32 grant (Bethesda, MD), and American Society of Neuroradiology (Oak Brook, IL). National Center for Research Resources UL1TR001876.
Footnotes
Conflict of interest statement: the authors declare no potential conflicts of interest.
References
- 1.Ostrom QT, Gittleman H, de Blank PM, Finlay JL, Gurney JG, McKean-Cowdin R, et al. American Brain Tumor Association Adolescent and Young Adult Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2008–2012. Neuro-oncology 2016;18 Suppl 1:i1–i50 doi 10.1093/neuonc/nov297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Castel D, Philippe C, Calmon R, Le Dret L, Truffaux N, Boddaert N, et al. Histone H3F3A and HIST1H3B K27M mutations define two subgroups of diffuse intrinsic pontine gliomas with different prognosis and phenotypes. Acta neuropathologica 2015;130(6):815–27 doi 10.1007/s00401-015-1478-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mackay A, Burford A, Carvalho D, Izquierdo E, Fazal-Salom J, Taylor KR, et al. Integrated Molecular Meta-Analysis of 1,000 Pediatric High-Grade and Diffuse Intrinsic Pontine Glioma. Cancer cell 2017;32(4):520–37 e5 doi 10.1016/j.ccell.2017.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012;482(7384):226–31 doi 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 5.Wu G, Broniscer A, McEachron TA, Lu C, Paugh BS, Becksfort J, et al. Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nature genetics 2012;44(3):251–3 doi 10.1038/ng.1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Louis DN, Perry A, Reifenberger G, von Deimling A, Figarella-Branger D, Cavenee WK, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta neuropathologica 2016;131(6):803–20 doi 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 7.Plessier A, Le Dret L, Varlet P, Beccaria K, Lacombe J, Meriaux S, et al. New in vivo avatars of diffuse intrinsic pontine gliomas (DIPG) from stereotactic biopsies performed at diagnosis. Oncotarget 2017;8(32):52543–59 doi 10.18632/oncotarget.15002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carai A, Mastronuzzi A, De Benedictis A, Messina R, Cacchione A, Miele E, et al. Robot-Assisted Stereotactic Biopsy of Diffuse Intrinsic Pontine Glioma: A Single-Center Experience. World neurosurgery 2017;101:584–8 doi 10.1016/j.wneu.2017.02.088. [DOI] [PubMed] [Google Scholar]
- 9.Hargrave D, Chuang N, Bouffet E. Conventional MRI cannot predict survival in childhood diffuse intrinsic pontine glioma. Journal of neuro-oncology 2008;86(3):313–9 doi 10.1007/s11060-007-9473-5. [DOI] [PubMed] [Google Scholar]
- 10.Laprie A, Pirzkall A, Haas-Kogan DA, Cha S, Banerjee A, Le TP, et al. Longitudinal multivoxel MR spectroscopy study of pediatric diffuse brainstem gliomas treated with radiotherapy. International journal of radiation oncology, biology, physics 2005;62(1):20–31 doi 10.1016/j.ijrobp.2004.09.027. [DOI] [PubMed] [Google Scholar]
- 11.Riley GT, Armitage PA, Batty R, Griffiths PD, Lee V, McMullan J, et al. Diffuse intrinsic pontine glioma: is MRI surveillance improved by region of interest volumetry? Pediatric radiology 2015;45(2):203–10 doi 10.1007/s00247-014-3134-6. [DOI] [PubMed] [Google Scholar]
- 12.Chheda ZS, Kohanbash G, Okada K, Jahan N, Sidney J, Pecoraro M, et al. Novel and shared neoantigen derived from histone 3 variant H3.3K27M mutation for glioma T cell therapy. The Journal of experimental medicine 2018;215(1):141–57 doi 10.1084/jem.20171046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ochs K, Ott M, Bunse T, Sahm F, Bunse L, Deumelandt K, et al. K27M-mutant histone-3 as a novel target for glioma immunotherapy. Oncoimmunology 2017;6(7):e1328340 doi 10.1080/2162402X.2017.1328340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Science translational medicine 2016;8(346):346ra92 doi 10.1126/scitranslmed.aaf6219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsao SC, Weiss J, Hudson C, Christophi C, Cebon J, Behren A, et al. Monitoring response to therapy in melanoma by quantifying circulating tumour DNA with droplet digital PCR for BRAF and NRAS mutations. Scientific reports 2015;5:11198 doi 10.1038/srep11198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Science translational medicine 2015;7(302):302ra133 doi 10.1126/scitranslmed.aab0021. [DOI] [PubMed] [Google Scholar]
- 17.De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martinez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nature communications 2015;6:8839 doi 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schiavon G, Hrebien S, Garcia-Murillas I, Cutts RJ, Pearson A, Tarazona N, et al. Analysis of ESR1 mutation in circulating tumor DNA demonstrates evolution during therapy for metastatic breast cancer. Science translational medicine 2015;7(313):313ra182 doi 10.1126/scitranslmed.aac7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Diaz LA Jr., Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 2014;32(6):579–86 doi 10.1200/JCO.2012.45.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang W, Song Z, Zhang Y. A Comparison of ddPCR and ARMS for detecting EGFR T790M status in ctDNA from advanced NSCLC patients with acquired EGFR-TKI resistance. Cancer medicine 2017;6(1):154–62 doi 10.1002/cam4.978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang TY, Piunti A, Lulla RR, Qi J, Horbinski CM, Tomita T, et al. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta neuropathologica communications 2017;5(1):28 doi 10.1186/s40478-017-0436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bender S, Tang Y, Lindroth AM, Hovestadt V, Jones DT, Kool M, et al. Reduced H3K27me3 and DNA hypomethylation are major drivers of gene expression in K27M mutant pediatric high-grade gliomas. Cancer cell 2013;24(5):660–72 doi 10.1016/j.ccr.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Lewis PW, Muller MM, Koletsky MS, Cordero F, Lin S, Banaszynski LA, et al. Inhibition of PRC2 activity by a gain-of-function H3 mutation found in pediatric glioblastoma. Science 2013;340(6134):857–61 doi 10.1126/science.1232245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mueller S, Liang W, Gupta N, Magge S, Kilburn L, Crawford J, et al. DIPG-40. PNOC-003: PRECISION MEDICINE TRIAL FOR CHILDREN WITH DIFFUSE INTRINSIC PONTINE GLIOMA. Neuro-oncology 2017;19(suppl_4):iv14–iv doi 10.1093/neuonc/nox083.055. [Google Scholar]
- 25.Nikbakht H, Panditharatna E, Mikael LG, Li R, Gayden T, Osmond M, et al. Spatial and temporal homogeneity of driver mutations in diffuse intrinsic pontine glioma. Nature communications 2016;7:11185 doi 10.1038/ncomms11185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saratsis AM, Yadavilli S, Magge S, Rood BR, Perez J, Hill DA, et al. Insights into pediatric diffuse intrinsic pontine glioma through proteomic analysis of cerebrospinal fluid. Neuro-oncology 2012;14(5):547–60 doi 10.1093/neuonc/nos067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yeh P, Hunter T, Sinha D, Ftouni S, Wallach E, Jiang D, et al. Circulating tumour DNA reflects treatment response and clonal evolution in chronic lymphocytic leukaemia. Nature communications 2017;8:14756 doi 10.1038/ncomms14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Vaart M, Pretorius PJ. Circulating DNA. Its origin and fluctuation. Annals of the New York Academy of Sciences 2008;1137:18–26 doi 10.1196/annals.1448.022. [DOI] [PubMed] [Google Scholar]
- 29.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer research 2001;61(4):1659–65. [PubMed] [Google Scholar]
- 30.Stroun M, Lyautey J, Lederrey C, Olson-Sand A, Anker P. About the possible origin and mechanism of circulating DNA apoptosis and active DNA release. Clinica chimica acta; international journal of clinical chemistry 2001;313(1–2):139–42. [DOI] [PubMed] [Google Scholar]
- 31.Kambhampati M, Perez JP, Yadavilli S, Saratsis AM, Hill AD, Ho CY, et al. A standardized autopsy procurement allows for the comprehensive study of DIPG biology. Oncotarget 2015;6(14):12740–7 doi 10.18632/oncotarget.3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.