Abstract
Cycles of Cdc53/Cullin1 rubylation (a.k.a NEDDylation) protect ubiquitin-E3 SCF (Skp1-Cullin1-F-box protein) complexes from self-destruction and play an important role in mediating the ubiquitination of key protein substrates involved in cell cycle progression, development, and survival. Cul1 rubylation is balanced by the COP9 signalosome (CSN), a multi-subunit derubylase that shows 1:1 paralogy to the 26S proteasome lid. The turnover of SCF substrates and their relevance to various diseases is well studied, yet, the extent by which environmental perturbations influence Cul1 rubylation/derubylation cycles per se is still unclear. In this study, we show that the level of cellular oxidation serves as a molecular switch, determining Cullin1 rubylation/derubylation ratio. We describe a mutant of the proteasome lid subunit, Rpn11 that exhibits accumulated levels of Cullin1-Rub1 conjugates, a characteristic phenotype of csn mutants. By dissecting between distinct phenotypes of rpn11 mutants, proteasome and mitochondria dysfunction, we were able to recognize the high reactive oxygen species (ROS) production during the transition of cells into mitochondrial respiration, as a checkpoint of Cullin1 rubylation in a reversible manner. Thus, the study adds the rubylation cascade to the list of cellular pathways regulated by redox homeostasis.
Abbreviations: CRL, Cullin-RING ubiquitin ligase; CSN, COP9 signalosome; DTT, Dithiothreitol;; Grx1, Glutaredoxin; LC-MS, Liquid chromatography–mass spectrometry;; MPN, Mpr-1-PAD1-N terminal; NAC, N-acetyl-L-cysteine; NEDD8, Neural Precursor Cell Expressed, Developmentally Down-Regulated 8; PCI, proteasome/COP9/Initiation factor; ROS, Reactive Oxygen Species; RPN, regulatory particle non-ATPase; Rub1, Related Ubiquitin1; SCF, Skp, Cullin, F-box containing complex; TCA cycle, tricarboxylic acid cycle; TMT, Tandem mass tagging; Ub, Ubiquitin; Ubl, Ubiquitin-like protein; WT, Wild type
Keywords: 26S proteasome, Mitochondria, NEDD8/Rub1, Rpn11, Thiol switch, Ubiquitin
Graphical abstract
Highlights
-
•
Cullin rubylation status inversely correlates with the cellular redox state.
-
•
The yeast diauxic shift blocks cullin rubylation in a reversible manner.
-
•
Cullin rubylation status is determined by the oxidation state of individual cells.
-
•
The activity of rubylation cascade enzymes is perturbed by high ROS.
-
•
The loss of cullin rubylation correlates with the appearance of free Rub1 molecules.
1. Introduction
Functioning as reversible molecular switches, cysteine residues are inherent cellular sensors of reactive oxygen species (ROS), sensing and translating the redox status into specific cellular responses [25], [67], [78]. Indeed, cysteine thiol groups switch between reduced and oxidized forms, and are involved in various cellular pathways, including signaling phosphatases [35], [36], [41], [53], transcriptional regulation [4], [65], and control of DNA damage repair [60], as well as in the regulation of thiol-based enzymes [30]. A fundamental cellular mechanism driven by thiol-based enzymes is the post-translational modification of proteins by ubiquitin (Ub) and ubiquitin-like (Ubl) proteins [32], [74], [85]. The most studied outcome of Ub conjugation is proteolysis of the Ub-modified target by the 26S proteasome machine [23], [27]. Attachment of Ub and Ubls (such as SUMO, URM1, and NEDD8/Rub1) to target proteins requires an analogous cascade of enzymes initiated by ATP-dependent generation of a signature high-energy thioester intermediate [12]. Each of these cascades begins with activation of the Ubl by an ATP-consuming E1 (activation) enzyme, forming Ubl~E1 thioester bond on an acceptor cysteine. In a second step, the Ubl is transthiolized from the E1 enzyme to a catalytic cysteine on an E2 (conjugation) enzyme, forming, once again, a Ubl~E2 thioester bond [34]. Finally, the activated Ubl is transferred to amino groups on target proteins in a step mediated by a substrate-specific E3 ligase [32], [79]. Despite a presence of thiol-based enzymes in Ubl conjugation pathways, only a few alterations by redox have been reported [84]. For instance, elevated concentrations of hydrogen peroxide (H2O2) inhibit human SUMOylation in a reversible manner by the formation of a disulfide bridge between both catalytic cysteine of SUMO-E1 and SUMO-E2 enzymes [9]. This switch is required for cell survival when ROS is accompanied by DNA damage [83]. For Ub, the exposure to ROS leads to oxidation of active cysteines of the E1 and E2 enzymes in mammalian retina cells until the recovery by the antioxidant glutathione, eventually results in hyper-activation of the E1 and increased ubiquitination [56], [77]. Additionally, E2 enzymes display differential sensitivities to oxidizing chemicals, suggesting redox as a mechanism to divert Ub between different E2s for precise substrate ubiquitination [19]. A unique example is an allosteric hyper-activation of Ub-E2 through a thiol oxidation of its Ub E2 variant binding partner [98]. Oxidation as an activation signal was also documented for Urm1, which progresses via a thiocarboxilate intermediate at its carboxyl terminus [89].
Rub1 (Related to Ubiquitin-1, A.K.A. the yeast homolog of NEDD8) is the closest paralog of ubiquitin. Orthologues of NEDD8/Rub1 are required for vitality in all studied organisms with the notable exception of budding yeast [38], [50], [93]. Limited evidence suggests a sensitivity of the rubylation pathway to ROS [20], [37], although the molecular mechanism has not been characterized. Rub1 is activated by the heterodimeric E1 enzyme Ula1/Uba3 (a.k.a. NAE), and transferred through canonical trans-thiolation to its cognate E2 enzyme, forming a thioester Ubc12~Rub1 bonds [34]. Ubc12~Rub1 transiently interacts with Rbx1, a RING subunit of a modular class of multi-subunit cullin-RING ubiquitin E3 ligases (CRLs), triggering the transfer of Rub1 to a specific lysine residue on the cullin scaffold subunit, and leading to their activation [11]. Human CRL complexes mediate ~ 20% of protein ubiquitination, including key cellular factors participating in cell cycle progression, DNA damage repair, and survival [82], [91]. Cullin rubylation is strictly conserved in all eukaryotes [47], and is also important in cancer biology. Indeed, the development of a specific inhibitor for its E1, MLN4924, has promising therapeutic potential [99].
Cullins undergo cycles of Rub1 conjugation/deconjugation during their catalytic cycle. The hydrolysis of Rub1 (derubylation) from cullins is performed by the COP9 signalosome (CSN), an evolutionarily conserved multi-subunit complex [92]. In addition to enzymatic cullin-derubylation activity, the CSN also hinders both substrate recognition and ubiquitination by CRLs [13], [21], [24], [42], [55]. An interesting property of the CSN is its close homology to the proteasome lid, the peripheral subcomplex of the 26S proteasome [59]. Both CSN and the proteasome lid belong to the PCI (proteasome, CSN, and eIF3) family of complexes. In this family, six subunits in each complex bear the hallmark PCI domain and two subunits carry an MPN (Mpr1–PAD1–N-terminal) domain [28]. One of the MPN subunits is a catalytically active MPN+/JAMM metalloprotease: Csn5/Jab1 in the CSN [3], [16] and Rpn11 in the proteasome lid [52], [90], [94]. In accordance, the substrates of these two enzymes are also close paralogues: Cullin-Rub1 for Csn5, and poly-Ub marked proteins for Rpn11 [46].
In S. cerevisiae, neither CSN nor Rub1 are essential for viability. Nevertheless, the CSN complex drives canonical cullin derubylation, yet it is unclear for what mechanistic purpose [16], [51], [97]. This yeast CSN contains only six subunits: four PCI subunits (one of which, Rpn5, is shared with the proteasome), an endemic subunit harboring the S6CD domain [15], [58], [76], [95], and a sole MPN subunit, the catalytic Csn5. Mutants in any yeast CSN subunit share a characteristic biochemical phenotype of accumulation of Rub1-modified cullins [92], a potent tool to study how cullins rubylation/derubylation cycles link with physiology.
In previous research, we demonstrated that not only do CSN mutants influence cullins, mutations in all PCI subunits of the proteasome lid also alter ratios of rubylated to unrubylated Cdc53/cullin1 (yCul1) [95]. yCul1 is a highly conserved module that serves as the functional core of SCF (Skp1- Cdc53/cullin1-F-box protein) E3 ligases, the largest class of CRLs that regulates diverse processes including cell cycle and development [57], [75]. Here, we extend the CSN/lid comparison to MPN subunits and describe a mutant of Rpn11 that exclusively exhibit a high ratio of modified to unmodified yCul1. Further characterization of rpn11 phenotypes points to a redox-dependent checkpoint of cullin rubylation, balancing cullin derubylation by the CSN.
2. Results
2.1. yCul1-rubylation inversely correlates with mitochondrial respiration
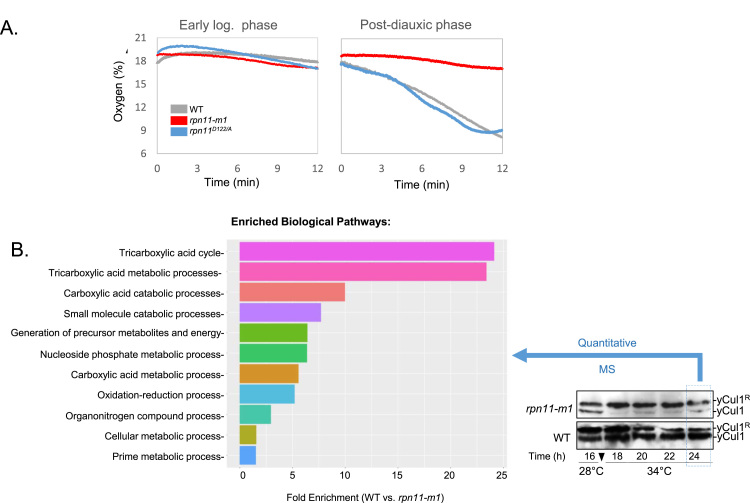
Accumulation of rubylated yCul1 (yCul1R) in mutants of the proteasome lid PCI subunits, suggests either direct or indirect functional link between proteasome and cullins [95]. To examine this hypothesis, we expanded our assessment of cullins to mutants of Rpn8 and Rpn11, the MPN-containing subunits of the proteasome lid. For Rpn11, two distinct mutants have been studied: the catalytic dead mutant, rpn11D122/A [52] and the carboxyl-terminal truncated mutant, rpn11-m1 [70]. Both mutants are temperature sensitive, and they share physiological and biochemical phenotypes such as cell cycle defects and an accumulation of ubiquitin chains [52], [69]. However, the loss of Rpn11 C- terminus in rpn11-m1 leads to an additional phenotype of mitochondrial dysfunction at temperatures higher than 32 °C [68], [71]. We tested these mutants for accumulation of yCul1R and observed that rpn11-m1 uniquely exhibited high accumulation of yCul1R, regardless of the growth phase, at a semi-restrictive temperature of 34 °C, for which, both mutants of Rpn11 are sensitive (Fig. 1a-d). We also examined the yCul1R accumulation in rpn81–307 (Fig. 1a), which lacks the Rpn8's carboxyl terminus and shows decreased quantity of fully assembled 26S proteasome [96]. In this case, the yCul1R/yCul1 ratio was unaffected relative to WT strains, suggesting that proteasome levels do not necessarily directly influence cullin rubylation. Neither was this phenotype observed in deletion mutants of other yeast deubiquitinating enzymes (data not shown), pointing to a unique feature of rpn11-m1 that is correlated with mitochondrial deficiencies at a semi-restrictive temperature [68], [71].
Fig. 1.
Rubylation status of yCul1/Cdc53 upon proteasome malfunction. (A) wild type (WT) and mutants of the proteasome lid MPN-containing subunits Rpn11 (rpn11-m1 and rpn11D122/A) and Rpn8 (rpn81–307), were grown at the semi-restrictive temperature of 34 °C for 6 h. yCul1 rubylation status was examined in whole cell extract by immunoblotting for yCul1/Cdc53. (B) WT and rpn11-m1 mutant cells were grown in YPD for 16 h at the permissive temperature of 28 °C. Cells were diluted to 0.5 OD600 and shifted to 34 °C for prolonged growth. yCul1 rubylation status was examined by immunoblotting at indicated time points. (C) WT, rpn11-m1 and rpn11D122/A, as well as the CSN mutant Δcsn5, were grown at 28 °C to log phase. yCul1 rubylation status was examined in whole cell extract by immunoblotting. Percentage of yCul1R/yCul1 of 3 experiments was quantified by ImageJ software (A, B, C bottom). (D) Schematic representation of phenotypes relating to yCul1 rubylation in Rpn11 mutants at 34 °C, summarized from A-C. (E) Proteasome lid (rpn11-m1, rpn11D122/A), base (rpt2RF), and catalytic core (α7ΔN) mutant strains were grown at 34 °C for 10 h to reach the diauxic shift. yCul1 rubylation status, stability of the proteasome substrate Sic1 and accumulation of ubiquitin chains were examined by immunoblotting for yCul1, Sic1 and Ub respectively. (F) Logarithmic culture of the Δpdr5 mutant strains with increased drug permeability was cultured in YPD at 34 °C and incubated with 10, 20 or 50 µM of MG132 for 3 h to inhibit proteasome 20S proteolytic activity. Drug permeability was assessed by examination of the stability of the short lived protein Cdc4 and accumulation of ubiquitin chains.
To further emphasize that an increase in yCul1R is not due to the proteasome dysfunction, we evaluated proteasome mutants for yCul1R/yCul1 ratio at a phase in which the budding yeast mitochondria apparatus becomes vital, i.e., following glucose exhaustion, towards the end of the fermentative phase. At this stage, cells switch to oxygenic respiration (i.e., “diauxic shift”), which allows catabolizing ethanol and other fermentation byproducts in the mitochondria via the tricarboxylic acid (TCA) cycle and oxidative phosphorylation. We assessed yCul1R/yCul1 ratio at 34 °C during the diauxic shift in several proteasome mutants, allocated in various proteasome sub-complexes: rpn11D122/A (proteasome lid catalytic dead mutant), rpt2RF (proteasome-base mutant), and α7ΔN (a 20S core particle mutant). Both rpn11D122/A- and rpt2RF-containing proteasomes form stable 26S complexes, however, both have been characterized as sensitive to general stress and shown impaired proteolytic activity [43], [52], [73], whereas α7ΔN displays an opposite phenotype of enhanced proteolytic activity [5]. Indeed, as expected, both, rpt2RF and rpn11D122/A displayed accumulation of ubiquitin chains and impaired proteasome substrate turnover, yet these proteasome mutants did not deviate from wild-type (WT) or from α7ΔN in their yCul1R/yCul1 ratio (Fig. 1e). Similarly, treatment of yeast cells with the proteasome inhibitor MG132 exhibited accumulation of ubiquitin conjugates, but did not show high yCul1R status (Fig. 1f). Taken together, these results reinforce mitochondrial defects rather than proteasome defects as a key factor in determining yCul1R/yCul1 ratio.
2.2. The accumulation of yCul1R in rpn11-m1 correlates with low ROS production
The well-documented mitochondria defects in rpn11-m1 are demonstrated in this study by the lack of a pronounced diauxic shift in this mutant (Fig. S1; [68], [69], [71]) as well as by the lack of oxygen consumption (i.e. respiration) at the post-diauxic phase (Fig. 2a). Hence, we hypothesized that rpn11-m1 is deviated from WT by not adjusting to metabolic alterations requiring de-novo transcription of genes attributed directly to aerobic metabolism [10], [48], [80]. We further posited that proteome modulation during a respiratory phase might underlie the alterations in yCul1 rubylation. Therefore, we applied a quantitative proteomic comparison at growth conditions when WT and rpn11-m1 are diverged in their yCul1 populations (Fig. 2b, Fig. S1). We used Amine Reactive Tandem Mass Tagging (TMT) isobaric labeling followed by a comprehensive liquid chromatography–mass spectrometry (LC-MS)/MS analysis to analyze the difference in protein expression between the WT and rpn11-m1 strains. Functional annotation analysis revealed that proteins participating in multiple metabolic pathways were significantly enriched in the WT proteome relative to rpn11-m1 (Table S3, data sheet). Specifically, enzymes catalyzing several key steps in the TCA cycle were under-represented in the proteome of rpn11-m1 (Fig. 2b, Table S3 gProfiler sheet), suggesting that this mutant inefficiently utilizes the TCA cycle for respiration. Interestingly, the proteomics data suggest a bottleneck in rpn11-m1 proteome, leading to the accumulation of alpha-ketoglutarate. These proteomics data include under-representation of Kgd1 and Kgd2 subunits of the mitochondrial alpha-ketoglutarate dehydrogenase complex and enrichment of Gdh2 that produces additional alpha-ketoglutarate from glutamate. Notably, the accumulation of alpha-ketoglutarate increases resistance of S. cerevisiae cells to H2O2 and other stresses [6].
Fig. 2.
Accumulation of yCul1Rinrpn11-m1correlates with mitochondria dysfunction. (A) WT (grey), rpn11D122/A (blue) and rpn11-m1 (red) yeast strains were grown in YPD at 34 °C for 4 or 16 h. For each strain, respiration rate of 5 × 107 cells was expressed by measuring dissolved oxygen concentrations over time. (B) Samples of rpn11-m1 and wild type were grown at a semi-restrictive temperature of 34 °C to identify the most pronounced yCul1 rubylation phenotype of the mutant (24 h in dashed square). Proteomes of both samples were compared by quantitative Mass Spectrometry (bottom). Biological pathways significantly enriched for protein abundance in the wild type samples relative to rpn11-m1 (log2FC (WT/rpn11-m1) > 2) were categorized using gProfileR [66] and manually curated to remove redundancy. Fold Enrichment values larger than 1 indicate that the category is overrepresented in the selected set of differentially expressed proteins detected using Mass Spectrometry (see also Table S3).
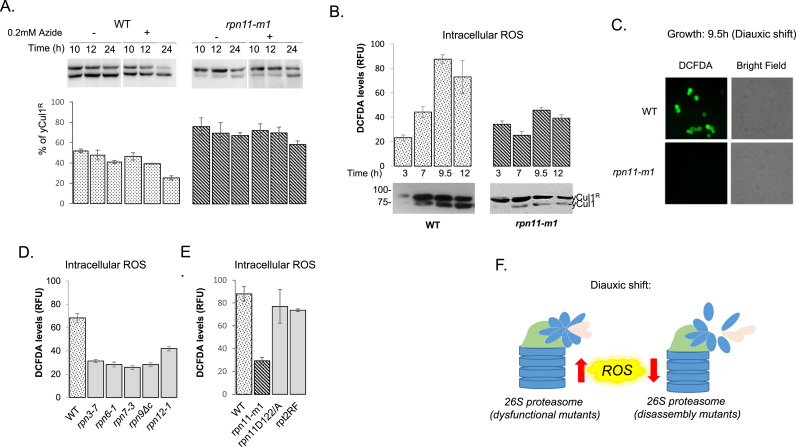
The TCA cycle precedes the electron transport chain, which creates the chemical gradient powering oxidative phosphorylation. In respiring cells, a leakage of the electron transport chain causes a build-up of reduced electron carriers in the mitochondria leading to elevated ROS [8], [29], [40], [64], [88]. This explains why the loss of TCA cycle enzymes (preceding the electron transport chains) was found to be associated with decreased ROS production [6], [40]. Since a loss of yCul1 rubylation was observed in an oxidative environment, we expected that the blockage of mitochondrial electron transport at the late exponential phase (i.e., start of oxygen consumption) would affect yCul1 rubylation status in respiring WT cells. Indeed, the inhibition of the respiratory electron transport chains by sodium azide (complex IV inhibitor) led to decreased yCul1R/yCul1 ratio in WT cells during the respiratory phase (Fig. 3a, left; S4a, b). Yet, similar treatment of rpn11-m1 cells did not affect yCul1 rubylation status even at a 75-fold higher concentration of azide (Fig. 3a, right; S4a, b). The insensitivity of rpn11-m1 to azide supports TCA cycle deficiency suggested by MS (Fig. 2b) as well as lack of oxygen uptake (Fig. 2a). Accordingly, the transition from primarily yCul1R to yCul1 in WT occurred precisely at the diauxic shift characterized by a peak in ROS levels (Fig. 3b, c). No such peak in ROS was observed for rpn11-m1 at 34 °C due to its attenuated respiration at the post-diauxic phase (Fig. 2a) and accumulation of antioxidants [45], perhaps explaining why yCul1 in this mutant was rubylated even during prolonged growth (Figs. 1b, 2b, 3b). In addition to rpn11-m1, the increased yCul1R/yCul1 ratio was also observed in mutants of the proteasome lid PCI subunits [95]. We showed that similar to rpn11-m1, the high yCul1R status in these mutants correlated with mitochondrial respiratory defects when grown on a non-fermentable carbon source, and with decreased ROS production when glucose in the environment was exhausted (Fig. 3d, S2a). Interestingly, the PCI mutants and rpn11-m1 share a structural feature of critical disassembly of proteasome lids onto 26S holoenzymes [26], [96], in contrast to the rpt2RF and rpn11D122/A mutants which form stable 26S holocomplexes [43], [52]. Consistent with these results, rpt2RF and rpn11D122/A (as well as WT cells) are also distinguished from the lid structural mutants by the accumulation of ROS at the late log phase (Fig. 3e, S2b). Thus, the data suggest that intact proteasome lid structure links the increased ROS during mitochondrial respiration with decreased yCul1R levels (Fig. 3f).
Fig. 3.
yCul1 rubylation status responds to oxidative environment. (A) WT and rpn11-m1 mutant cells were diluted to 0.5 OD600 in YPD and incubated at 34 °C with indicated amounts of azide. yCul1 Rubylation status was tested by immunoblotting at the indicated times. See Fig. S4a, b for additional information. (B) WT and rpn11-m1 strains were grown in YPD at 34 °C for the indicated time. Endogenous ROS was measured by fluorescence following treatment with 2′ 7′-dichlorofluorescein diacetate (DCFDA) for one hour at each time point. Representative immunoblots of yCul1 rubylation status are shown for comparison (bottom) (C) ROS in intact cells was visualized by fluorescence microscopy (100 ×, Nikon Eclipse E600 microscope). (D, E) proteasome lid disassembly mutants (rpn3–7, rpn6-1, rpn7-3, rpn9ΔC and rpn12-1) (D); and proteasome dysfunctional mutants, (rpt2RT and rpn11D122/A) (E), were grown in YPD at 37 °C for 9.5 h and oxidative stress was assessed similar to B. Results of 3 independent experiments were quantified by IMAGEJ. (F) Schematic presentation summarizing effect of proteasome mutants on ROS at the diauxic shift (9.5 h of growth). Note that rpt2RT and rpn11D122/A behave similar to WT in this regard, in contrast to proteasome disassembly mutants (rpn3–7, rpn6-1, rpn7-3, rpn9ΔC and rpn12-1).
2.3. yCul1 rubylation is sensitive to ROS
To test the direct effect of an oxidative environment on yCul1R/yCul1 ratio, H2O2 was added to cells at the early log phase. Treatment with H2O2 led to an accumulation of unmodified yCul1 in both WT and rpn11 mutant cells (Fig. 4a). Antioxidants, such as thioredoxin, glutathione synthase, and glutathione reductase, support the maintenance of an intracellular reducing environment during the transition to respiration. Deleting these genes led to lower levels of cellular antioxidant capacity, resulting in a decreased yCul1R/yCul1 ratio at the late log phase (Fig. 4b, S3a, 6 h). Conversely, treating cells at the diauxic shift with the reducing reagents dithiothreitol (DTT), or NAC (N-acetyl-L-cysteine) delayed the appearance of unrubylated yCul1 (Fig. 4c; S4d).
Fig. 4.
Decreased yCul1 rubylation linked to intra-cellular ROS. (A) WT, rpn11-m1 and rpn11D122/A mutants in logarithmic phase growth were treated with 4.4 mM of H2O2 for 2 h and yCul1 rubylation status was determined by immunoblotting. (B) Mutants of ROS scavengers, Δtrx2, Δgsh1 and Δglr1 were grown in YPD at 28 °C, and yCul1 rubylation status evaluated in whole cell extract by immunoblotting (see also Fig. S3) (C) Wild type cells growth for 16 h were diluted to 0.5OD600 in YPD and incubated for 6 h at 28 °C before addition of 2 mM DTT to growth media. Rubylation status of yCul1 was examined by immunoblotting at 6 h, and at 8, 10 h. yCul1 modification ratio was quantified (A, B, C bottom) (D) Cells were sorted by FACS according to their oxidation status into oxidized, reduced, or “mixed” (i.e., unsorted) populations (as assessed by the Grx1-roGFP2 sensor). (E) 106 WT cells were collected during sorting for each repeat of the different subpopulations (n = 3 repeats), followed by examination of the yCul1 Rubylation status by immunoblotting and quantification (see also Fig. S4). (F) schematic representation of the correlation between the oxidation state and yCul1 rubylation status in individual cells.
In order to directly link the intracellular redox state the yCul1 rubylation status, we applied a flow cytometry methodology to cells expressing a redox-sensitive Grx1-roGFP2 probe, which allows measurement of the redox state of individual cells within a heterogeneous population and sorting them according to their oxidative status, resulting in “oxidized” and “reduced” subpopulations [62]. The sorted populations (oxidized, reduced or “mixed” populations) were tested by immunoblotting for yCul1R/yCul1 ratio (Fig. 4e). The “mixed” population contains a similar amount of cells, taken before applying the sorting mode. By comparing cullin rubylation in the sorted populations, we observed a clear correlation between the oxidation status and yCul1 rubylation intensity in cells at the diauxic phase (between 6 and 24 h of growth) (Fig. 4e, left, S4c). Indeed, comparing the oxidized and reduced populations during prolonged growth of 48 h (during which the cullin modification status is very low, but far more cells are in the oxidized population) revealed a clear difference as well (Fig. 4e, right, S4b). A similar correlation was found when oxidized cells of the ∆glr1 strain, lacking the glutathione reductase gene and showing a constant mitochondrial oxidation [63], were sorted and subjected to immunoblotting (data not shown). Taken together, it appears that changes in yCul1R/yCul1 ratio observed in the heterogeneous population by immunoblotting overtime is a consequence of perturbation in cellular oxidative stress per se (Fig. 4f).
2.4. The rubylation enzymes Uba3 E1 and Ubc12 E2 are sensitive to ROS
The fast response of yCul1R to the redox status suggests a possible regulatory role for ROS in either stimulation of CSN derubylation activity or inhibition of the thiol -dependent rubylation enzymes. To verify whether rubylation cascade enzymes are sensitive to ROS, we examined the production of thioester-linked Ubc12~Rub1 during physiological oxidation (prolonged growth phase and the lack of fermentable carbon source) or by inducing oxidation by H2O2 treatment (Fig. 5). Subsequently, total protein samples were separated by protein gels under non-reducing conditions to preserve the thioester conjugates. Immunoblotting for Ubc12 showed that 1 mM of H2O2 promoted a significant reduction in Ubc12~Rub1 thioester forms (Fig. 5a, left). Note, that intracellular ROS measured under these conditions was comparable to endogenous ROS levels measured at the diauxic shift (Fig. S3b), and to concentrations for which, both the SUMO and the ubiquitin cascades are sensitive [9], [19]. Indeed, a lower ratio of Ubc12~Rub1 thioester was observed following the diauxic shift (Fig. 5a, middle) or by switching to the non-fermentable carbon source, glycerol, which forces the utilization of mitochondrial respiration (Fig. S5b). Inversely, switching back to glycolysis by the addition of glucose to starved cells in the stationary phase, suppressed intracellular ROS (Fig. 5a, right; S5a, b) and resulted in a reoccurrence of Ubc12~Rub1 thioester along with the appearance of yCul1R conjugates. Surprisingly, in contrast to the observations in WT cells, the Ubc12~Rub1 thioester forms were very highly maintained in the rpn11-m1 mutant cells at all growth times with a slight reduction, detected mainly by the emergence of free Ubc12 (Fig. 5b 8–24 h). Finally, the loss of thiol-sensitive Uba3 conjugates in cells treated with 1.1 mM H2O2 confirmed that the rubylation cascade responds to ROS from the E1 stage (Fig. 5c, right, 35–50 kD). Ultimately, yCul1 rubylation status depends also on CSN activity. In the future, it will be interesting to decipher whether CSN derubylase activity is stimulated by high ROS, similarly to other UBL-specific proteases such as SENP3, a deSUMOylase that is stabilized by high ROS levels and functions to deSUMOylate the p300 protein [33].
Fig. 5.
Respiration attenuates enzymes in the rubylation cascade. (A) WT cells expressing endogenous Ubc12-HA were grown at 34 °C and treated as detailed: YP medium was complemented by 4% of glucose (left and middle panels). Indicated concentrations of H2O2 were added for 2 h (left); or cells were grown untreated for indicated times (middle); or grown for 6 h with 0–2% of glucose in the medium (right). Both Ubc12~Rub1 thioester forms and yCul1 rubylation status were examined by immunoblotting in reduced (+ DTT) and non-reduced (-DTT) conditions. (B) rpn11-m1-HA mutant cells expressing endogenous levels of Ubc12-HA diluted to OD600 = 0.5 in YPD and re-incubated at 34 °C for indicated times. Δrub1 served as a control for a strain deficient in Rub1 (left). Ubc12~Rub1 thioester formation were examined over time. (C) WT cells expressing 6His-Flag-Uba3 diluted to OD600 = 0.5 at logarithmic phase were incubated for 2 h with indicated H2O2 concentrations. Uba3 and yCul1 expression and modification was assessed through immunoblotting in reduced (+ DTT) and non-reduced (-DTT) conditions. Asterisk for nonspecific bands, observed in control ∆uba3 and ∆rub1 [6His-Flag-Uba3] samples. (D) WT and mutant strains of the CSN/Rub1 axis (∆yuh1, ∆csn5, ∆dcn1, ∆rub1, ycul1K760/R) were grown to log phase in YPD. yCul1 rubylation status and the performance of free Rub1 were assessed by immunoblotting. (E) Cells at the pre-diauxic (OD600 =0.8) or post-diauxic phase (OD600 = 42) phases were examined for yCul1 rubylation status and the performance of free Rub1 by immunoblotting. Each experiment was repeated 3 times and a representative immunoblot is shown.
Finally, the loss of modified yCul1 is correlated with the performance of free Rub1 molecules and vice versa (Fig. 5d, e). For instance, at the early log phase, Rub1 is almost fully conjugated to cullins (Fig. 5e, OD600 0.8), a trademark of ∆csn5 mutant cells lacking the catalytic subunit of the CSN complex (Fig. 5d). Conversely, the free form of Rub1 is accumulated at the post-diauxic phase (Fig. 5e, OD600 42) in a similar manner as y-cul1K760/R, a mutant of yCul1 that cannot undergo rubylation, as well as to ∆yuh1, a deletion mutant of a hydrolytic enzyme processing Rub1 from its precursor. (Fig. 5d). Taken together, in this study, a mutant of Rpn11 previously characterized as displaying mitochondrial defects [45], [68], [69], [96] was used as a platform to identify the inhibitory effect of accumulated ROS during the diauxic shift on the rubylation cascade of enzymes. The inhibition of Rub1 cascade resulted in the loss of yCul1 rubylation and the appearance of free molecules of Rub1, presumably responding to the oxidative environment.
3. Discussion
Apart of being the catalytic subunit of the proteasome lid, the carboxyl terminal of Rpn11 plays additional roles in stabilizing the proteasome [96] and in mitochondrial integrity [68], [69], [71]. By dissecting the mitochondrial and proteasomal capacities of rpn11-m1 and pointing to a TCA cycle dysfunction in this mutant and in other proteasome disassembly mutants, we identified that redox homeostasis is fundamental for yCul1 rubylation/derubylation cycling. Since the link between 26S proteasome disassembly and malfunction of mitochondria was demonstrated also in higher organisms such as C. elegans [45], we believe that this indirect link between proteasome structure and cullin rubylation status is conserved across phyla.
Redox homeostasis breaks down naturally during the yeast diauxic shift, a potent experimental model to study rapid metabolic response to the production of endogenous ROS. We show that the dramatic loss of yCul1 rubylation status in WT at the diauxic shift, previously described as a carbon source dependence [97], is a physiological transition from primarily anaerobic glycolysis to mitochondrial respiration accompanied by ROS production. Indeed, we observed a loss of Ubc12~Rub1 thioester formation upon induction of ROS with a reversible attenuation. Our results are also supported by a high-throughput thioredoxin trapping screen performed by Ben-Lulu and colleagues in 2014, which suggested a reversible S-nitrosylation of human Ubc12 cysteines [7]. The sensitivity of Rub1 enzymatic cascade to oxidation is initiated by the E1 Uba3/Ula1. Future studies will focus on identifying specific residues in both enzymes, involved in the regulation of rubylation cycles by ROS. For instance, oxidation of the catalytic cysteine thiol group of either Uba3 or Ubc12 could lead to a reversible sulfenic acid (R-SOH) form. Another option could be the formation of a reversible disulfide bridge between cysteine of E1~E2 enzymes, similar to the SUMOylation enzymes [9]. However, the formation of this disulfide bridge requires a specific insertion of two amino acids in the SUMO E2, Ubc9, that is absent in the Rub1 E2, Ubc12 [83].
The turnover of SCF substrates has a direct relevance to various diseases [39], [82], [99]; however, not much is known about how fine-tuning of SCF properties through rubylation cycling affects disease progression. Modification of CRLs by Rub1 is critical for cell cycle progression and survival, and required for ubiquitination and degradation of many key cellular regulators, including cyclin E, p27, the DNA replication licensing factor Cdt-1, and the transcription factor inhibitor pIκBα. Unregulated turnover of these CRL-substrates is most relevant to promoting protein toxicity leading to pathologies such as cancer. Vice versa, inhibition of their degradation is more toxic to cancer cells than to normal cells, and was suggested as a treatment for various cancers [18]. Indeed, the rubylation cascade inhibitor MLN4924, leads to proteotoxicity and its clinical investigation as a treatment in several malignancies is ongoing [81], [99]. Similarly, we showed here that high endogenous ROS also inhibits the rubylation cascade and could thus provide another explanation to the effect of increased oxidative stress through “ROS induction therapy” on selectively killing cancer cells without affecting normal cells [87]. Our study suggests that reinforcing redox homeostasis in cells increases the capacity of proteolytic systems not only by proteasomal maintenance [45], but also through persisting SCF activity.
Yet, in budding yeast, the turnover of SCF/CRL substrates is barely regulated by cullin-rubylation. Why does this mechanism of cullin rubylation cycles exist in budding yeast if it is not necessary for the turnover of substrates? We hypothesize that reinforcement of health through the loss of cullin rubylation might lead to additional biological function beyond CRLs E3-activity. For instance, a side effect of the loss of cullin rubylation is the increased level of free Rub1 (Fig. 5d, e). The liberated Rub1 possibly performs additional, yet undefined, activities during high ROS. Indeed, a handful of reports point to a secondary, CRL-independent, function of Rub1 [1], [31], [54]. Such studies recognized and examined various Rub1 targets linked to natural cellular stress responses. Rub1 targets which are not cullins were also found when Rub1 was overexpressed or upon an increase in the Rub1 to ubiquitin ratio [1], [22], [31]. Under these conditions, atypical rubylation patterns were observed, and found to be triggered by the ubiquitination cascade of enzymes [1], [22], [31], [54].
In conclusion, our results indicate that the silencing of cullin rubylation is a natural mechanism occurring in a state of high respiratory stress. We suggest that this silencing is a cellular defense mechanism rather than a defect in proteostasis control. We further propose that the rubylation cascade enzymes serve as redox sensors allowing the liberation of Rub1 upon high ROS, which subsequently functions as a game changer, responding to oxidative environment and maintaining cellular needs.
4. Materials and methods
4.1. Yeast strains
This study includes widely used laboratory strains of Saccharomyces cerevisiae (SUB62, W303, BY4741). The experiments obtained a similar pattern of results for all studied strains. Genotypes and mating type of haploid progeny were determined by phenotype, auxotrophic analysis, PCR and immunoblotting. Double mutants or single mutants bearing genomic tags, were generated by mating of haploid strains, sporulation, and dissection of spores. Genotypes of haploid progeny were determined by a mating with haploid reference strains, PCR and immunoblotting.
4.2. Yeast media and growth conditions
Unless otherwise is specified, Saccharomyces cerevisiae cells were grown under standard growth conditions at a semi-restrictive temperature of 34 °C. All yeast strains were grown in glucose rich YPD medium (1% bactopeptone, 1% yeast extract, and 2% glucose) complemented by adenine hemisulfate. For all treatments: Unless stated otherwise, overnight grown starter cultures were diluted to OD600 = 0.5 and incubated for indicated times, temperatures or treatments, as described in figure legends. Growth phases were determined according to the growth response graph of the W303 wild type strain (Fig. S1) and related literature [10]. Maintenance of plasmids was achieved by culturing plasmid-containing strains in a selective synthetic complete (SC) media. For proteasome inhibition, MG132 (in DMSO) was added for 2 h. For mitochondria inhibition, sodium-azide (in DMSO) were directly added to yeast cultures. For exogenous induction of ROS, overnight cultures were diluted to OD600 = 0.5 and cultured for 3 h (logarithmic phase) before the addition of indicated concentrations of H2O2 directly to the growth medium for 2 h. Yeast strains and plasmids used in this study are listed in Supplemental Tables 1 and 2.
4.3. Endogenous oxidative stress measurements
2′ 7′-dichlorofluorescein diacetate (DCFDA) (Sigma) was directly added to YPD grown yeast cultures one hour prior to harvest, in a final concentration of 10 µg per 107 Cells, as was previously described by Allen et al. [2]. Cells were washed twice with a phosphate buffered saline (PBS) pH 7.2, and 2 × 107 cells were placed in a 96 wells plate for florescence measurement (Excitation = 485/20; Emission = 528/20). Cells were either visualized by FITC fluorescence filter (460–500 nm) using a Nikon Eclipse E600 fluorescent light microscope, or quantified through a BioTek, Synergy HT plate reader.
4.4. Statistical analysis
The values of Relative Fluorescence Units (RFU) of at least three independent experiments were analyzed by One-Way ANOVA followed by Turkey HSD test for pairwise comparison.
4.5. Antibodies
The following antibodies were used: anti-CDC53, anti-Sic1, anti-CDC4, anti-Actin, anti-HA (Santa Cruz); anti-Flag (Sigma); anti-ubiquitin (Dako), anti-NEDD8 (Abcam), anti-Rpn11 [95], anti-Rpn8 [96], anti-Rpn2 [72].
4.6. Immunoblotting
For denatured cells extracts, cells were harvested in trichloroacetic acid (TCA) as we previously described at Yu et al. [95]. Samples were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for immunoblotting. Analysis was done by X-ray film or ECL fusion FX 6 EDGE system. Experiments were repeated 3 times and a representative result is shown.
4.7. Quantification of immunoblots
Quantitative analysis of immunoblots was performed using ImageJ v1.40f software (http://imagej.net/). The yCul1 rubylation ratio was calculated by comparing the relative abundance of modified yCul1 out of the total amount of yCul1 (i.e. free and modified). The average of 3 experiments was calculated.
4.8. Sample preparation for mass spectrometry analysis
Yeast cultures were washed twice with DDW and once with chilled buffer A (25 mM Tris [pH 7.4], 10 mM MgCl2, 1 mM ATP). Pellet was resuspended in two volumes of buffer A and lysed using glass beads at 4 °C. Native lysates were clarified by centrifugation at 16,000×g for 15 min and digested with trypsin [44]. Peptides derived from four strains (1) rpn11-m1/Δcsn5, (2) rpn11-m1, (3) wild type (w303), and (4) Δcsn5 were labeled with TMT126, TMT127, TMT128, and TMT129 isobaric labeling reagents (Thermo Scientific), respectively, according to manufacturer's instructions. The labeled peptides were then combined and fractionated into 4 fractions using the SCX STAGE tip method [17]. SCX fractions were desalted with C18 STAGE tips and evaporated to dryness before being resuspended in 0.1% formic acid for mass spectrometry analysis. Data of rpn11-m1 and wild type (W303) had been analyzed for this study.
4.9. Mass spectrometry analysis
Samples were analyzed in technical duplicate by LC-MS/MS on a Thermo Scientific LTQ Orbitrap Elite mass spectrometry system equipped with a Proxeon Easy nLC 1000 ultra-high-pressure liquid chromatography and autosampler system. Samples were injected onto a C18 column (25 cm × 75 µm I.D. packed with ReproSil Pur C18 AQ 1.9 µm particles) in 0.1% formic acid and then separated with a two-hour gradient from 5% to 30% ACN in 0.1% formic acid at a flow rate of 300 ml/min. The mass spectrometer collected data using a data-dependent MS2/MS3 acquisition method [86]. A full scan was collected in the Orbitrap at 60,000 resolution (FWHM) with an AGC target of 1000,000 followed by 10 collision-induced dissociation (CID) MS/MS scans of the 10 most intense ions in the ion trap with an AGC target of 30,000, minimum signal of 500, isolation width of 2.0 m/z, normalized collision energy of 35.0, activation Q of 0.25, and activation time of 20.0 ms. The most intense ion in each MS/MS scan was further selection for high energy collision-induced dissociation (HCD) with a minimum signal of 500, isolation width of 4.0 m/z, normalized collision energy of 60.0, and an activation time of 50.0 ms. HCD scans were collected in the Orbitrap at 15,000 resolution (FWHM) and with an AGC target of 100,000. Charge state screening was enabled to reject unassigned or single charge states. Dynamic exclusion was enabled to exclude ions within + /− 10 p.p.m. with a repeat count of 1, a repeat duration of 20.0 s, an exclusion duration of 20.0 s, and for a maximum exclusion list size of 500.
Raw mass spectrometry data were analyzed by the Protein Prospector suite [49]. Data were searched against a database containing SwissProt S. cerevisiae protein sequences, concatenated to a decoy database where each sequence was randomized in order to estimate the false positive rate. The searches considered a precursor mass tolerance of + /− 20 p.p.m. and fragment ion tolerances of 0.8 da, and considered variable modifications for protein N-terminal acetylation, protein N-terminal acetylation and oxidation, glutamine to pyroglutamate conversion for peptide N-terminal glutamine residues, protein N-terminal methionine loss, protein N-terminal acetylation and methionine loss, and methionine oxidation, and constant modifications for carbamidomethyl cysteine and TMT 6-plex labeling modifications on lysine residues and the amino terminus of each peptide. Prospector data was filtered using a maximum protein expectation value of 0.01 and a maximum peptide expectation value of 0.05. TMT reporter ion intensities were extracted for each identification using an in-house Perl script. Missing values were imputed to the minimum intensity value. The data was normalized to equalize the median and log2FC values calculated.
4.10. Cell sorting using Grx1-roGFP2
Strains were transformed with the Grx1-roGFP2 plasmid using a modified One Step transformation protocol [14] with cells scraped from plate. Cells were then grown at 30 °C on 2–5 ml minimal medium supplemented with casein amino acid and -URA selection under aerobic conditions, diluted to approximately 0.25 OD600, and harvested at relevant time points following dilution. Sorting was conducted using predefined gates as described in [62], with Grx1-roGFP2 expression under oxidized and reduced conditions determining subpopulations. These gates were adjusted according to probe expression in each of the strains. 2 × 106 cells were taken per sample for continued analysis, with 3 repeats per subpopulation from separate initial colonies.
4.11. Respiration measurement
5 ml of cells in the studied growth phases in a concentration of 1 OD600/1 ml were transferred to a 50 ml falcon tube with a stir bar on a magnetic stirrer and placed in a controlled-temperature cabinet at 34 °C. For respiration rate measurements a 1 ml aliquot was transferred to a microfuge tube and placed in a dry block pre-heated to 34 °C. The medium was immediately covered with paraffin oil to prevent gas exchange between the medium and room air during the measurement. Oxygen consumption rate was measured by inserting to the medium, using a micromanipulator, a needle-type oxygen optode (NTH-PSt-1) connected to a Micro-Fiber Optic Oxygen Transmitter (Microx TX3; PreSens GmbH, Regensburg, Germany). Digital output was recorded and stored on a computer. The oxygen optode was pre-calibrated using oxygen-free and air saturated solutions according to the manufacturer's recommended procedures (https://www.presens.de/support-services/faqs/question/how-can-i-prepare-the-calibration-solutions-cal0-and-cal100-for-oxygen-sensors-35.html).
4.12. Ubc12~Rub1 thioester forms
Overnight cultures were diluted in SC medium complemented by either 4% glucose, 3% glycerol or as described at the figure legends. For each condition, 20OD600 of cells were collected after 4 h of growth, washed twice with DDW, and pellets were split for reduced and non-reduced protein extraction. Extraction and detection of thioester forms, was performed according to Rabut et al. [61].
Acknowledgements
We would like to thank Maya Schuldiner, Mordechai Choder, Agnès Delahodde and Gwenael Rabut for sharing yeast strains with us. We would like also to thank Moran Benhar for fruitful discussions.
Acknowledgments
Funding
This work was supported by the Israel Ministry of Science and Technology (MOST) – Italy Ministry of Foreign Affairs (MAE) [grant number 3-9022 to EP and TR]; Israel Science Foundation [grants no. 162/17 for EP, no. 1765/13 for DR and no. 909/14 for MHG]; Human Frontier Science Program [grant number CDA00064/2014 for DR]; University of Michigan – Israel Partnership for Research [grant number 2020978 to MHG]. LB and AS fellowships were supported by the PBC Fellowship Program of the Israeli Council for Higher Education.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.redox.2018.11.010
Appendix A. Supplementary material
Supplementary material
Supplementary material
References
- 1.Abidi N., Xirodimas D.P. Regulation of cancer-related pathways by protein NEDDylation and strategies for the use of NEDD8 inhibitors in the clinic. Endocr. Relat. Cancer. 2015;22:T55–T70. doi: 10.1530/ERC-14-0315. [DOI] [PubMed] [Google Scholar]
- 2.Allen S.A., Clark W., McCaffery J.M., Cai Z., Lanctot A., Slininger P.J., Liu Z.L., Gorsich S.W. Furfural induces reactive oxygen species accumulation and cellular damage in Saccharomyces cerevisiae. Biotechnol. Biofuels. 2010;3:2. doi: 10.1186/1754-6834-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ambroggio X.I., Rees D.C., Deshaies R.J. JAMM: a metalloprotease-Like zinc site in the proteasome and signalosome. PLoS Biol. 2004;2:e2. doi: 10.1371/journal.pbio.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antelmann H., Helmann J.D. Thiol-based redox switches and gene regulation. Antioxid. Redox Signal. 2011;14:1049–1063. doi: 10.1089/ars.2010.3400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bajorek M., Finley D., Glickman M.H. Proteasome disassembly and downregulation is correlated with viability during stationary phase. Curr. Biol. 2003;13:1140–1144. doi: 10.1016/s0960-9822(03)00417-2. [DOI] [PubMed] [Google Scholar]
- 6.Bayliak M., Burdyliuk N., Lushchak V. Growth on alpha-ketoglutarate increases oxidative stress resistance in the yeast saccharomyces cerevisiae. Int. J. Microbiol. 2017;2017:5792192. doi: 10.1155/2017/5792192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ben-Lulu S., Ziv T., Admon A., Weisman-Shomer P., Benhar M. A substrate trapping approach identifies proteins regulated by reversible S-nitrosylation. Mol. Cell Proteom. 2014;13:2573–2583. doi: 10.1074/mcp.M114.038166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bonawitz N.D., Rodeheffer M.S., Shadel G.S. Defective mitochondrial gene expression results in reactive oxygen species-mediated inhibition of respiration and reduction of yeast life span. Mol. Cell Biol. 2006;26:4818–4829. doi: 10.1128/MCB.02360-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bossis G., Melchior F. Regulation of SUMOylation by reversible oxidation of SUMO conjugating enzymes. Mol. Cell. 2006;21:349–357. doi: 10.1016/j.molcel.2005.12.019. [DOI] [PubMed] [Google Scholar]
- 10.Brauer M.J., Saldanha A.J., Dolinski K., Botstein D. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell. 2005;16:2503–2517. doi: 10.1091/mbc.E04-11-0968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Calabrese M.F., Scott D.C., Duda D.M., Grace C.R., Kurinov I., Kriwacki R.W., Schulman B.A. A RING E3-substrate complex poised for ubiquitin-like protein transfer: structural insights into cullin-RING ligases. Nat. Struct. Mol. Biol. 2011;18:947–949. doi: 10.1038/nsmb.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cappadocia L., Lima C.D. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem. Rev. 2017 doi: 10.1021/acs.chemrev.6b00737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cavadini S., Fischer E.S., Bunker R.D., Potenza A., Lingaraju G.M., Goldie K.N., Mohamed W.I., Faty M., Petzold G., Beckwith R.E., Tichkule R.B., Hassiepen U., Abdulrahman W., Pantelic R.S., Matsumoto S., Sugasawa K., Stahlberg H., Thoma N.H. Cullin-RING ubiquitin E3 ligase regulation by the COP9 signalosome. Nature. 2016;531:598–603. doi: 10.1038/nature17416. [DOI] [PubMed] [Google Scholar]
- 14.Chen D.C., Yang B.C., Kuo T.T. One-step transformation of yeast in stationary phase. Curr. Genet. 1992;21:83–84. doi: 10.1007/BF00318659. [DOI] [PubMed] [Google Scholar]
- 15.Cirigliano A., Stirpe A., Menta S., Mori M., Dell'Edera D., Pick E., Negri R., Botta B., Rinaldi T. Yeast as a tool to select inhibitors of the cullin deneddylating enzyme Csn5. J. Enzym. Inhib. Med. Chem. 2016;31:1632–1637. doi: 10.3109/14756366.2016.1160901. [DOI] [PubMed] [Google Scholar]
- 16.Cope G.A., Suh G.S., Aravind L., Schwarz S.E., Zipursky S.L., Koonin E.V., Deshaies R.J. Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of NEDD8 from CUL1. Science. 2002;298:608–611. doi: 10.1126/science.1075901. [DOI] [PubMed] [Google Scholar]
- 17.Dephoure N., Gygi S.P. A solid phase extraction-based platform for rapid phosphoproteomic analysis. Methods. 2011;54:379–386. doi: 10.1016/j.ymeth.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deshaies R.J. Proteotoxic crisis, the ubiquitin-proteasome system, and cancer therapy. BMC Biol. 2014;12:94. doi: 10.1186/s12915-014-0094-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doris K.S., Rumsby E.L., Morgan B.A. Oxidative stress responses involve oxidation of a conserved ubiquitin pathway enzyme. Mol. Cell Biol. 2012;32:4472–4481. doi: 10.1128/MCB.00559-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Downs C.A., Kumar A., Kreiner L.H., Johnson N.M., Helms M.N. H2O2 regulates lung epithelial sodium channel (ENaC) via ubiquitin-like protein Nedd8. J. Biol. Chem. 2013;288:8136–8145. doi: 10.1074/jbc.M112.389536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dubiel D., Rockel B., Naumann M., Dubiel W. Diversity of COP9 signalosome structures and functional consequences. FEBS Lett. 2015;589:2507–2513. doi: 10.1016/j.febslet.2015.06.007. [DOI] [PubMed] [Google Scholar]
- 22.Enchev R.I., Schulman B.A., Peter M. Protein neddylation: beyond cullin-RING ligases. Nat. Rev. Mol. Cell Biol. 2015;16:30–44. doi: 10.1038/nrm3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fischer E.S., Scrima A., Bohm K., Matsumoto S., Lingaraju G.M., Faty M., Yasuda T., Cavadini S., Wakasugi M., Hanaoka F., Iwai S., Gut H., Sugasawa K., Thoma N.H. The molecular basis of CRL4DDB2/CSA ubiquitin ligase architecture, targeting, and activation. Cell. 2011;147:1024–1039. doi: 10.1016/j.cell.2011.10.035. [DOI] [PubMed] [Google Scholar]
- 25.Fra A., Yoboue E.D., Sitia R. Cysteines as redox molecular switches and targets of disease. Front. Mol. Neurosci. 2017;10:167. doi: 10.3389/fnmol.2017.00167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.K. Fukunaga, T. Kudo, E.A. Toh, K. Tanaka, Y. Saeki Dissection of the assembly pathway of the proteasome lid in Saccharomyces cerevisiae, Biochem. Biophys. Res. Commun. [DOI] [PubMed]
- 27.Glickman M.H., Ciechanover A. The Ubiquitin-proteasome Proteolytic Pathway: destruction for the sake of construction. Physiol. Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 28.Glickman M.H., Rubin D.M., Coux O., Wefes I., Pfeifer G., Cjeka Z., Baumeister W., Fried V.A., Finley D. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 29.Grant C.M., MacIver F.H., Dawes I.W. Mitochondrial function is required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. FEBS Lett. 1997;410:219–222. doi: 10.1016/s0014-5793(97)00592-9. [DOI] [PubMed] [Google Scholar]
- 30.Groitl B., Jakob U. Thiol-based redox switches. Biochim. Biophys. Acta. 2014;1844:1335–1343. doi: 10.1016/j.bbapap.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hjerpe R., Thomas Y., Chen J., Zemla A., Curran S., Shpiro N., Dick L.R., Kurz T. Changes in the ratio of free NEDD8 to ubiquitin triggers NEDDylation by ubiquitin enzymes. Biochem. J. 2012;441:927–936. doi: 10.1042/BJ20111671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hochstrasser M. Origin and function of ubiquitin-like proteins. Nature. 2009;458:422–429. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang C., Han Y., Wang Y., Sun X., Yan S., Yeh E.T., Chen Y., Cang H., Li H., Shi G., Cheng J., Tang X., Yi J. SENP3 is responsible for HIF-1 transactivation under mild oxidative stress via p300 de-SUMOylation. EMBO J. 2009;28:2748–2762. doi: 10.1038/emboj.2009.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang D.T., Miller D.W., Mathew R., Cassell R., Holton J.M., Roussel M.F., Schulman B.A. A unique E1-E2 interaction required for optimal conjugation of the ubiquitin-like protein NEDD8. Nat. Struct. Mol. Biol. 2004;11:927–935. doi: 10.1038/nsmb826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. Reactive oxygen species promote TNFalpha-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell. 2005;120:649–661. doi: 10.1016/j.cell.2004.12.041. [DOI] [PubMed] [Google Scholar]
- 36.Kwon J., Lee S.R., Yang K.S., Ahn Y., Kim Y.J., Stadtman E.R., Rhee S.G. Reversible oxidation and inactivation of the tumor suppressor PTEN in cells stimulated with peptide growth factors. Proc. Natl. Acad. Sci. USA. 2004;101:16419–16424. doi: 10.1073/pnas.0407396101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.L Y.K., McIntosh C.J., Biasio W., Liu Y., Ke Y., Olson D.R., Miller J.H., Page R., Snyder P.M., McDonald F.J. Regulation of the delta and alpha epithelial sodium channel (ENaC) by ubiquitination and Nedd8. J. Cell Physiol. 2013;228:2190–2201. doi: 10.1002/jcp.24390. [DOI] [PubMed] [Google Scholar]
- 38.Lammer D., Mathias N., Laplaza J.M., Jiang W., Liu Y., Callis J., Goebl M., Estelle M. Modification of yeast Cdc53p by the ubiquitin-related protein Rub1p affects function of the SCFCdc4 complex. Genes Dev. 1998;12:914–926. doi: 10.1101/gad.12.7.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee M.H., Zhao R., Phan L., Yeung S.C. Roles of COP9 signalosome in cancer. Cell Cycle. 2011;10:3057–3066. doi: 10.4161/cc.10.18.17320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee Y.J., Burlet E., Galiano F., Circu M.L., Aw T.Y., Williams B.J., Witt S.N. Phosphate and succinate use different mechanisms to inhibit sugar-induced cell death in yeast: insight into the Crabtree effect. J. Biol. Chem. 2011;286:20267–20274. doi: 10.1074/jbc.M110.209379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leslie N.R., Bennett D., Lindsay Y.E., Stewart H., Gray A., Downes C.P. Redox regulation of PI 3-kinase signalling via inactivation of PTEN. EMBO J. 2003;22:5501–5510. doi: 10.1093/emboj/cdg513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lingaraju G.M., Bunker R.D., Cavadini S., Hess D., Hassiepen U., Renatus M., Fischer E.S., Thoma N.H. Crystal structure of the human COP9 signalosome. Nature. 2014;512:161–165. doi: 10.1038/nature13566. [DOI] [PubMed] [Google Scholar]
- 43.Lipson C., Alalouf G., Bajorek M., Rabinovich E., Atir-Lande A., Glickman M., Bar-Nun S. A proteasomal ATPase contributes to dislocation of ERAD substrates. J. Biol. Chem. 2008;283:7166–7175. doi: 10.1074/jbc.M705893200. [DOI] [PubMed] [Google Scholar]
- 44.Liu S., Zhao T., Song B., Zhou J., Wang T.T. Comparative proteomics reveals important viral-host interactions in HCV-infected human liver cells. PLoS One. 2016;11:e0147991. doi: 10.1371/journal.pone.0147991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Livnat-Levanon N., Kevei E., Kleifeld O., Krutauz D., Segref A., Rinaldi T., Erpapazoglou Z., Cohen M., Reis N., Hoppe T., Glickman M.H. Reversible 26S proteasome disassembly upon mitochondrial stress. Cell Rep. 2014;7:1371–1380. doi: 10.1016/j.celrep.2014.04.030. [DOI] [PubMed] [Google Scholar]
- 46.Lyapina S., Cope G., Shevchenko A., Serino G., Tsuge T., Zhou C., Wolf D.A., Wei N., Shevchenko A., Deshaies R.J. Promotion of NEDD-CUL1conjugate cleavage by COP9 signalosome. Science. 2001;292:1382–1385. doi: 10.1126/science.1059780. [DOI] [PubMed] [Google Scholar]
- 47.Lydeard J.R., Schulman B.A., Harper J.W. Building and remodelling Cullin-RING E3 ubiquitin ligases. EMBO Rep. 2013;14:1050–1061. doi: 10.1038/embor.2013.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Maris A.F., Assumpcao A.L., Bonatto D., Brendel M., Henriques J.A. Diauxic shift-induced stress resistance against hydroperoxides in Saccharomyces cerevisiae is not an adaptive stress response and does not depend on functional mitochondria. Curr. Genet. 2001;39:137–149. doi: 10.1007/s002940100194. [DOI] [PubMed] [Google Scholar]
- 49.Matsui N.M., Smith D.M., Clauser K.R., Fichmann J., Andrews L.E., Sullivan C.M., Burlingame A.L., Epstein L.B. Immobilized pH gradient two-dimensional gel electrophoresis and mass spectrometric identification of cytokine-regulated proteins in ME-180 cervical carcinoma cells. Electrophoresis. 1997;18:409–417. doi: 10.1002/elps.1150180315. [DOI] [PubMed] [Google Scholar]
- 50.Maytal-Kivity V., Pick E., Piran R., Hofmann K., Glickman M.H. The COP9 signalosome-like complex in S. cerevisiae and links to other PCI complexes. Int. J. Biochem. Cell Biol. 2003;35:706–715. doi: 10.1016/s1357-2725(02)00378-3. [DOI] [PubMed] [Google Scholar]
- 51.Maytal-Kivity V., Piran R., Pick E., Hofmann K., Glickman M.H. COP9 signalosome components play a role in the mating pheromone response of S. cerevisiae. EMBO Rep. 2002;12:1215–1221. doi: 10.1093/embo-reports/kvf235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Maytal-Kivity V., Reis N., Hofmann K., Glickman M.H. MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem. 2002;3:28. doi: 10.1186/1471-2091-3-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng T.C., Fukada T., Tonks N.K. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Mol. Cell. 2002;9:387–399. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 54.Mergner J., Kuster B., Schwechheimer C. DENEDDYLASE1 protein counters automodification of neddylating enzymes to maintain NEDD8 protein homeostasis in arabidopsis. J. Biol. Chem. 2017;292:3854–3865. doi: 10.1074/jbc.M116.767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mosadeghi R., Reichermeier K.M., Winkler M., Schreiber A., Reitsma J.M., Zhang Y., Stengel F., Cao J., Kim M., Sweredoski M.J., Hess S., Leitner A., Aebersold R., Peter M., Deshaies R.J., Enchev R.I. Structural and kinetic analysis of the COP9-Signalosome activation and the cullin-RING ubiquitin ligase deneddylation cycle. Elife. 2016;5 doi: 10.7554/eLife.12102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Obin M., Shang F., Gong X., Handelman G., Blumberg J., Taylor A. Redox regulation of ubiquitin-conjugating enzymes: mechanistic insights using the thiol-specific oxidant diamide. FASEB J. 1998;12:561–569. doi: 10.1096/fasebj.12.7.561. [DOI] [PubMed] [Google Scholar]
- 57.Patton E.E., Willems A.R., Sa D., Kuras L., Thomas D., Craig K.L., Tyers M. Cdc53 is a scaffold protein for multiple Cdc34/Skp1/F-box proteincomplexes that regulate cell division and methionine biosynthesis in yeast. Genes Dev. 1998;12:692–705. doi: 10.1101/gad.12.5.692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Pick E., Golan A., Zimbler J.Z., Guo L., Sharaby Y., Tsuge T., Hofmann K., Wei N. The minimal deneddylase core of the COP9 signalosome excludes the Csn6 MPN(-) domain. PLoS One. 2012;7:e43980. doi: 10.1371/journal.pone.0043980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pick E., Hofmann K., Glickman M.H. PCI complexes: beyond the proteasome, CSN, and eIF3 Troika. Mol. Cell. 2009;35:260–264. doi: 10.1016/j.molcel.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 60.Posey K.L., Gimble F.S. Insertion of a reversible redox switch into a rare-cutting DNA endonuclease. Biochemistry. 2002;41:2184–2190. doi: 10.1021/bi015944v. [DOI] [PubMed] [Google Scholar]
- 61.Rabut G., Le Dez G., Verma R., Makhnevych T., Knebel A., Kurz T., Boone C., Deshaies R.J., Peter M. The TFIIH subunit Tfb3 regulates cullin neddylation. Mol. Cell. 2011;43:488–495. doi: 10.1016/j.molcel.2011.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Radzinski M., Fassler R., Yogev O., Breuer W., Shai N., Gutin J., Ilyas S., Geffen Y., Tsytkin-Kirschenzweig S., Nahmias Y., Ravid T., Friedman N., Schuldiner M., Reichmann D. Temporal profiling of redox-dependent heterogeneity in single cells. Elife. 2018;7 doi: 10.7554/eLife.37623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Radzinski M., Reichmann D. Variety is the spice of life: how to explore a redox-dependent heterogeneity in genomically identical cellular populations. Curr. Genet. 2018 doi: 10.1007/s00294-018-0878-9. [DOI] [PubMed] [Google Scholar]
- 64.Rasmussen A.K., Chatterjee A., Rasmussen L.J., Singh K.K. Mitochondria-mediated nuclear mutator phenotype in Saccharomyces cerevisiae. Nucleic Acids Res. 2003;31:3909–3917. doi: 10.1093/nar/gkg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Reichmann D., Voth W., Jakob U. Maintaining a healthy proteome during oxidative stress. Mol. Cell. 2018;69:203–213. doi: 10.1016/j.molcel.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Reimand J., Arak T., Adler P., Kolberg L., Reisberg S., Peterson H., Vilo J. g: profiler-a web server for functional interpretation of gene lists (2016 update) Nucleic Acids Res. 2016;44:W83–W89. doi: 10.1093/nar/gkw199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rhee S.G., Woo H.A., Kang D. The role of peroxiredoxins in the transduction of H2O2 signals. Antioxid. Redox Signal. 2018;28:537–557. doi: 10.1089/ars.2017.7167. [DOI] [PubMed] [Google Scholar]
- 68.Rinaldi T., Hofmann L., Gambadoro A., Cossard R., Livnat-Levanon N., Glickman M.H., Frontali L., Delahodde A. Dissection of the carboxyl-terminal domain of the proteasomal subunit Rpn11 in maintenance of mitochondrial structure and function. Mol. Biol. Cell. 2008;19:1022–1031. doi: 10.1091/mbc.E07-07-0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rinaldi T., Pick E., Gambadoro A., Zilli S., Maytal-Kivity V., Frontali L., Glickman M.H. Participation of the proteasomal lid subunit Rpn11 in mitochondrial morphology and function is mapped to a distinct C-terminal domain. Biochem. J. 2004;381:275–285. doi: 10.1042/BJ20040008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rinaldi T., Ricci C., Porro D., Bolotin-Fukuhara M., Frontali L. A mutation in a novel yeast proteasomal gene produces a cell cycle arrest, overreplication of nuclear and mitochondrial DNA and an altered mitocondrial morphology. Mol. Biol. Cell. 1998;9:2917–2931. doi: 10.1091/mbc.9.10.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rinaldi T., Ricordy R., Bolotin-Fukuhara M., Frontali L. Mitochondrial effects of the pleiotropic proteasomal mutation mpr1/rpn11: uncoupling from cell cycle defects in extragenic revertants. Gene. 2002;286:43–51. doi: 10.1016/s0378-1119(01)00799-5. [DOI] [PubMed] [Google Scholar]
- 72.Rosenzweig R., Osmulski P.A., Gaczynska M., Glickman M.H. The central unit within the 19S regulatory particle of the proteasome. Nat. Struct. Mol. Biol. 2008;15:573–580. doi: 10.1038/nsmb.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rubin D.M., Glickman M.H., Larsen C.N., Dhruvakumar S., Finley D. Active site mutants in the six regulatory particle ATPases reveal multiple roles for ATP in the proteasome. EMBO J. 1998;17:4909–4919. doi: 10.1093/emboj/17.17.4909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Schulman B.A., Harper J.W. Ubiquitin-like protein activation by E1 enzymes: the apex for downstream signalling pathways. Nat. Rev. Mol. Cell Biol. 2009;10:319–331. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Seol J.H., Feldman R.M., Zachariae W., Shevchenko A., Correll C.C., Lyapina S., Chi Y., Galova M., Claypool J., Sandmeyer S., Nasmyth K., Deshaies R.J., Shevchenko A., Deshaies R.J. Cdc53/cullin and the essential Hrt1 RING-H2 subunit of SCF define a ubiquitin ligase module that activates the E2 enzyme Cdc34. Genes Dev. 1999;13:1614–1626. doi: 10.1101/gad.13.12.1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Serino G., Pick E. Duplication and familial promiscuity within the proteasome lid and COP9 signalosome kin complexes. Plant Sci. 2013;203–204:89–97. doi: 10.1016/j.plantsci.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 77.Shang F., Gong X., Taylor A. Activity of ubiquitin-dependent pathway in response to oxidative stress. J. Biol. Chem. 1997;272:23086–23093. doi: 10.1074/jbc.272.37.23086. [DOI] [PubMed] [Google Scholar]
- 78.Sies H. Hydrogen peroxide as a central redox signaling molecule in physiological oxidative stress: oxidative eustress. Redox Biol. 2017;11:613–619. doi: 10.1016/j.redox.2016.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smalle J., Vierstra R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu Rev. Plant Biol. 2004;55:555–590. doi: 10.1146/annurev.arplant.55.031903.141801. [DOI] [PubMed] [Google Scholar]
- 80.Soontorngun N. Reprogramming of nonfermentative metabolism by stress-responsive transcription factors in the yeast Saccharomyces cerevisiae. Curr. Genet. 2016 doi: 10.1007/s00294-016-0609-z. [DOI] [PubMed] [Google Scholar]
- 81.Soucy T.A., Smith P.G., Milhollen M.A., Berger A.J., Gavin J.M., Adhikari S., Brownell J.E., Burke K.E., Cardin D.P., Critchley S., Cullis C.A., Doucette A., Garnsey J.J., Gaulin J.L., Gershman R.E., Lublinsky A.R., McDonald A., Mizutani H., Narayanan U., Olhava E.J., Peluso S., Rezaei M., Sintchak M.D., Talreja T., Thomas M.P., Traore T., Vyskocil S., Weatherhead G.S., Yu J., Zhang J., Dick L.R., Claiborne C.F., Rolfe M., Bolen J.B., Langston S.P. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature. 2009;458:732–736. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 82.Soucy T.A., Smith P.G., Rolfe M. Targeting NEDD8-activated cullin-RING ligases for the treatment of cancer. Clin. Cancer Res. 2009;15:3912–3916. doi: 10.1158/1078-0432.CCR-09-0343. [DOI] [PubMed] [Google Scholar]
- 83.Stankovic-Valentin N., Drzewicka K., Konig C., Schiebel E., Melchior F. Redox regulation of SUMO enzymes is required for ATM activity and survival in oxidative stress. EMBO J. 2016;35:1312–1329. doi: 10.15252/embj.201593404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Stankovic-Valentin N., Melchior F. Control of SUMO and Ubiquitin by ROS: signaling and disease implications. Mol. Asp. Med. 2018 doi: 10.1016/j.mam.2018.07.002. [DOI] [PubMed] [Google Scholar]
- 85.Tanaka K., Suzuki T., Chiba T. The ligation systems for ubiquitin and ubiquitin-like proteins. Mol. Cells. 1998;8:503–512. [PubMed] [Google Scholar]
- 86.Ting L., Rad R., Gygi S.P., Haas W. MS3 eliminates ratio distortion in isobaric multiplexed quantitative proteomics. Nat. Methods. 2011;8:937–940. doi: 10.1038/nmeth.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trachootham D., Alexandre J., Huang P. Targeting cancer cells by ROS-mediated mechanisms: a radical therapeutic approach? Nat. Rev. Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 88.Trancikova A., Weisova P., Kissova I., Zeman I., Kolarov J. Production of reactive oxygen species and loss of viability in yeast mitochondrial mutants: protective effect of Bcl-xL. FEMS Yeast Res. 2004;5:149–156. doi: 10.1016/j.femsyr.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 89.Van der Veen A.G., Schorpp K., Schlieker C., Buti L., Damon J.R., Spooner E., Ploegh H.L., Jentsch S. Role of the ubiquitin-like protein Urm1 as a noncanonical lysine-directed protein modifier. Proc. Natl. Acad. Sci. USA. 2011;108:1763–1770. doi: 10.1073/pnas.1014402108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Verma R., Aravind L., Oania R., McDonald W.H., Yates J.R., III, Koonin E.V., Deshaies R.J. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 91.Wee S., Geyer R.K., Toda T., Wolf D.A. CSN facilitates Cullin-RING ubiquitin ligase function by counteracting autocatalytic adapter instability. Nat. Cell Biol. 2005;7:387–391. doi: 10.1038/ncb1241. [DOI] [PubMed] [Google Scholar]
- 92.Wei N., Serino G., Deng X.W. The COP9 signalosome: more than a protease. Trends Biochem. Sci. 2008;33:592–600. doi: 10.1016/j.tibs.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 93.Whitby F.G., Xia G., Pickart C.M., Hill C.P. Crystal structure of the human ubiquitin-like protein NEDD8 and interactions with ubiquitin pathway enzymes. J. Biol. Chem. 1998;273:34983–34991. doi: 10.1074/jbc.273.52.34983. [DOI] [PubMed] [Google Scholar]
- 94.Yao T., Cohen R.E. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 95.Yu Z., Kleifeld O., Lande-Atir A., Bsoul M., Kleiman M., Krutauz D., Book A., Vierstra R.D., Hofmann K., Reis N., Glickman M.H., Pick E. Dual function of Rpn5 in two PCI complexes, the 26S proteasome and COP9 signalosome. Mol. Biol. Cell. 2011:911–920. doi: 10.1091/mbc.E10-08-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yu Z., Livnat-Levanon N., Kleifeld O., Mansour W., Nakasone M.A., Castaneda C.A., Dixon E.K., Fushman D., Reis N., Pick E., Glickman M.H. Base-CP proteasome can serve as a platform for stepwise lid formation. Biosci. Rep. 2015;35 doi: 10.1042/BSR20140173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zemla A., Thomas Y., Kedziora S., Knebel A., Wood N.T., Rabut G., Kurz T. CSN- and CAND1-dependent remodelling of the budding yeast SCF complex. Nat. Commun. 2013;4:1641. doi: 10.1038/ncomms2628. [DOI] [PubMed] [Google Scholar]
- 98.Zhao Y., Long M.J.C., Wang Y., Zhang S., Aye Y. Ube2V2 is a rosetta stone bridging redox and ubiquitin codes, coordinating DNA damage responses. ACS Cent. Sci. 2018;4:246–259. doi: 10.1021/acscentsci.7b00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhao Y., Sun Y. Cullin-RING Ligases as attractive anti-cancer targets. Curr. Pharm. Des. 2013;19:3215–3225. doi: 10.2174/13816128113199990300. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material