Abstract
Despite claims that the mammalian Y Chromosome is on a path to extinction, comparative sequence analysis of primate Y Chromosomes has shown the decay of the ancestral single-copy genes has all but ceased in this eutherian lineage. The suite of single-copy Y-linked genes is highly conserved among the majority of eutherian Y Chromosomes due to strong purifying selection to retain dosage-sensitive genes. In contrast, the ampliconic regions of the Y Chromosome, which contain testis-specific genes that encode the majority of the transcripts on eutherian Y Chromosomes, are rapidly evolving and are thought to undergo species-specific turnover. However, ampliconic genes are known from only a handful of species, limiting insights into their long-term evolutionary dynamics. We used a clone-based sequencing approach employing both long- and short-read sequencing technologies to assemble ∼2.4 Mb of representative ampliconic sequence dispersed across the domestic cat Y Chromosome, and identified the major ampliconic gene families and repeat units. We analyzed fluorescence in situ hybridization, qPCR, and whole-genome sequence data from 20 cat species and revealed that ampliconic gene families are conserved across the cat family Felidae but show high transcript diversity, copy number variation, and structural rearrangement. Our analysis of ampliconic gene evolution unveils a complex pattern of long-term gene content stability despite extensive structural variation on a nonrecombining background.
Advances in DNA sequencing technologies, alongside the increasing affordability of large-scale sequencing projects, have dramatically expanded opportunities for interspecific comparative genomics (Lindblad-Toh et al. 2011; Zhang et al. 2014; Lamichhaney et al. 2015; Hu et al. 2017; Meadows and Lindblad-Toh 2017). However, standard short-read sequencing approaches still fail to accurately assemble repetitive portions of eukaryotic genomes (Eichler et al. 2004; Alkan et al. 2011; Huddleston et al. 2014; Chaisson et al. 2015; Khost et al. 2017). Ampliconic regions of the male-specific region of the Y Chromosome (MSY) are notably enriched with large, high-identity segmental duplications that are prone to complex structural arrangements (Skaletsky et al. 2003). These characteristics have prevented the construction of accurate assemblies of MSY amplicons using standard NGS approaches (Carvalho and Clark 2013; Skinner et al. 2016; Tomaszkiewicz et al. 2016), leading to their absence in all but a small number of genome assemblies (Skaletsky et al. 2003; Hughes et al. 2010, 2012; Bellott et al. 2014; Soh et al. 2014). As a result, most interspecific comparative analyses have been restricted to the ancestral single-copy region of the MSY (Li et al. 2013; Bellott et al. 2014; but see Murphy et al. 2006; Cortez et al. 2014).
The few studies investigating ampliconic regions of the MSY have found that these regions can differ drastically in both gene content and copy number between closely related species (Hughes et al. 2010; Cortez et al. 2014; Soh et al. 2014). Ampliconic regions typically harbor massively amplified gene families that exhibit testis-specific or testis-biased expression, suggesting a primary function in male gametogenesis (Lahn and Page 1997; Murphy et al. 2006; Paria et al. 2011; Soh et al. 2014; Ghenu et al. 2016; Oetjens et al. 2016). Accordingly, Y-linked mutations can have drastic effects on male reproductive fitness, producing phenotypes such as spermatogenic failure (Kuroda-Kawaguchi et al. 2001; Repping et al. 2002), abnormal sperm morphology (Touré et al. 2004, 2005; Cocquet et al. 2012; Campbell and Nachman 2014), and sex ratio distortion (Cocquet et al. 2012; Helleu et al. 2014). Thus, given the Y Chromosome's proclivity for accruing structural rearrangements and given the manifestation of male-biased sterility in interspecific mammalian hybrids (Haldane's rule), it seems likely that ampliconic gene divergence has played a critical role in the speciation process (Meiklejohn and Tao 2010; Good 2012; Soh et al. 2014). To investigate this possibility, however, a better understanding of ampliconic MSY interspecific variation and its evolutionary patterns is needed.
The cat family, Felidae, is an ideal system to study MSY evolution and its biological consequences across a broad range of divergence times (Murphy et al. 2006; Pearks Wilkerson et al. 2008; Li et al. 2013; Davis et al. 2015). This family contains 41 species divided among eight lineages that emerged ∼12 million years ago (mya) (Johnson et al. 2006; Li et al. 2016; Kitchener et al. 2017). Each of the eight species complexes diverged within the past 3–6 million years, presenting a diverse and parallel sample of radiations with which to examine a variety of evolutionary processes, notably interspecific hybridization (Li et al. 2016). There are numerous examples of anthropogenically derived felid hybrids, including several derived from crosses to the domestic cat, which are actively propagated as popular cat breeds (Gray 1972; Gershony et al. 2014; Davis et al. 2015). Some hybrid crosses exhibit sex-biased litters when F1s are backcrossed to nonhybrid parental species (Supplemental Table S1), and F1 and early generation backcross males exhibit hybrid male sterility (Davis et al. 2015). Thus, the felid radiation is a powerful model system to investigate the underlying genetic basis for phenotypes like sperm morphology and hybrid male sterility, including potential contributions from Y-linked components.
Murphy et al. (2006) used direct cDNA selection and fluorescence in situ hybridization (FISH) to identify four novel gene families within the ampliconic region of the domestic cat MSY, all of which exhibited testis-specific expression. These gene families included two that appeared to be unique to the felid lineage: TETY1 and CCDC71LY (formerly FLJ36031Y). Both genes derive from autosomal homologs from Chromosomes A3 and A2, respectively. In addition, Murphy et al. (2006) discovered an ampliconic expansion of an ancestral Y-linked gene, CUL4BY, which has since been shown to be multicopy within the dog and the horse (Paria et al. 2011). Another ancestral Y-linked gene, CLDN34Y (formerly TETY2), appears to be multicopy exclusively within Carnivora (Li et al. 2013). A fifth gene, TSPY1, is multicopy in nearly all mammalian Y Chromosomes examined (Bellott et al. 2014; Cortez et al. 2014) and is highly amplified across the domestic cat Y.
Although the transcriptional content of the ampliconic region of the domestic cat MSY has been previously explored, the actual DNA sequence, structure, and regulatory architecture have not been determined. Sequencing and assembling these regions are important to unveil undiscovered gene repertoires and enable structural variant analysis between closely related species which may contribute to reproductive isolation. We used a multipronged approach that combined BAC clone-based finishing, qPCR, FISH, and whole-genome sequence analyses to reveal the evolutionary timescale of ancestral and lineage-specific Y Chromosome gene loss/gain events and structural rearrangements within the cat family.
Results
Ampliconic BAC clone sequencing and assembly
We sequenced 48 randomly selected BAC clones from a candidate pool of over 500 positive clones identified in a screen of half of the RPCI-86 domestic cat clone filters, using overgo probes designed from five domestic cat ampliconic MSY gene transcripts: CCDC71LY, CLND34Y, CUL4BY, TETY1, and TSPY1 (Murphy et al. 2006). FISH analyses from Murphy et al. (2006) revealed that the majority of ampliconic genes were dispersed along the entire length of Chr Yq, suggesting it is composed of an array of similar repeat units. Clones were randomly selected from a suite of different autorad signal intensities. Six clones were sequenced using both the Pacific Biosciences (PacBio) RSII and Illumina MiSeq platforms, while the remaining 42 clones were sequenced only on the Illumina MiSeq. Each clone that was sequenced with PacBio assembled into a circular contig. The average length of assembled clone inserts was 113,475 bp (SD 17,779), which falls well within the range of expected insert lengths. De novo assembly of the 48 Illumina libraries resulted in 20 clones that passed our established quality control (QC) metrics, including five of the six clones sequenced on both the MiSeq and PacBio platforms. The clones that did not pass QC failed at one or more stages of the assembly process (Supplemental Figs. S1A–C).
Twenty-one clones were accurately assembled with these approaches (19 clones via Illumina sequencing and six clones using PacBio sequencing with five clones being assembled by both methods), typically into single contigs or scaffolds (Supplemental Table S2). The sequences represent ∼2.4Mb (∼10%) of the domestic cat ampliconic MSY. When we compared the MiSeq assemblies that passed QC to their PacBio counterparts, we found that there were no major disparities between the two sequencing approaches (99.9% average pairwise identity) (Supplemental Figs. S2–S6; Supplemental Table S3). It should be noted that we did not correct the PacBio assemblies with Illumina reads as we wanted to compare the two approaches, but this step has been shown to improve assembly accuracy (Walker et al. 2014).
Ampliconic sequence content
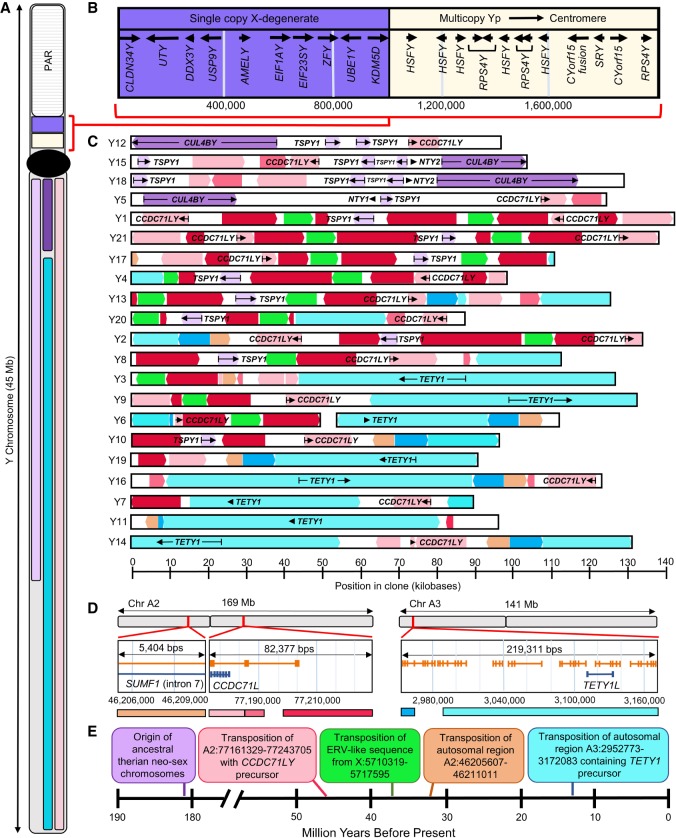
Analysis of ∼10% of the estimated ampliconic sequence on the domestic cat MSY revealed that it is composed of a mixture of sequence classes derived from several independent autosomal transposition events, as well as retention and expansion of ancestral X–Y orthologs (Fig. 1A–C). We identified three separate autosomal transpositions that occurred at different periods in the evolutionary history of the felid Y Chromosome. We dated the timing of these transpositions using relaxed molecular clock approaches (see Methods). The oldest transposition is estimated to have occurred ∼46 mya (24–66 mya 95% CI) (Fig. 1D,E; Supplemental Figs. S7–S10), >30 Myr prior to the most recent common ancestor (MRCA) of all extant felids, and near the estimated time of the feliform radiation (Eizirik et al. 2010). This event copied ∼83 kb from Chr A2 onto the ancestral feliform MSY, including the gene CCDC71L. The second autosomal transposition occurred 32 mya (18–53 mya 95% CI) (Fig. 1D,E; Supplemental Fig. S11), moving a 5.4-kb segment that also originated from Chr A2. The last autosomal transposition occurred ∼15 mya (11–23 mya 95% CI), which corresponds to the estimated age of the felid MRCA (Supplemental Fig. S12). This ∼100-kb Y-linked segment originated from a ∼219-kb segment from the short arm of Chr A3 (Fig. 1D,E) and includes the previously described ampliconic gene TETY1.
Figure 1.
Domestic cat Y Chromosome sequence content. (A) Ideogram of the domestic cat Y Chromosome. The purple and beige regions of the short arm represent the single and multicopy region of the Y Chromosome sequenced by Li et al. (2013). In the long arm, the colored blocks indicate the distribution of known ampliconic genes based on FISH analyses, with the corresponding genes being indicated in C. It is important to note that the resolution of the FISH analysis did not allow us to determine the extent to which the ampliconic region extends across the centromere. (B) Annotation of the Y Chromosome region as presented in Li et al. (2013). (C) Sequence content of 21 assembled BAC clones annotated with both transcripts (black arrows, text) and sequence origin (colors). Clones are placed in order by sequence content similarity, and this order is not reflective of their chromosomal position. (D) Annotated view of autosomal progenitor sequences with the representative color for each transposed region displayed below. Annotated loci are depicted in blue, and in silico–predicted loci are shown in orange. (E) Timeline of notable events in felid Y Chromosome evolution.
Three ampliconic MSY genes arose from ancestral X–Y homologs: CUL4BY, CLDN34Y, and TSPY1. No single scaffold contained the entire coding sequence of CUL4BY. Three scaffolds—Y12, Y15, and Y18—had sequences that corresponded to the first five to nine exons of the published sequence of this gene, while scaffold Y5 contained sequence derived from the last four exons of the published sequence of CUL4BY. While these sequences could be pseudogenized copies, they were typically located at the ends of sequenced clones oriented in such a way that they may represent fragments of a complete gene sequence. Numerous intact copies of TSPY1 (present in ∼52% of sequenced clones) were often found within a single clone. We were unable to identify any additional copies of CLDN34Y within any of the assembled ampliconic scaffolds (Li et al. 2013).
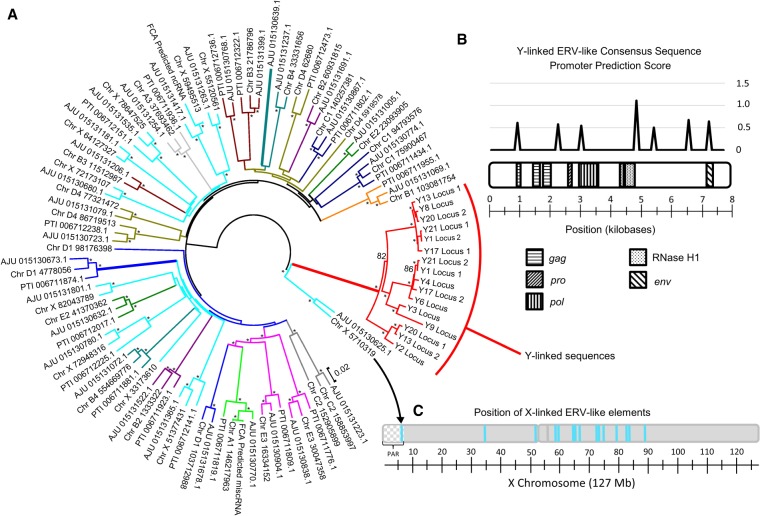
We identified a modified endogenous retrovirus (ERV) derived from an X-linked sequence that has spread along the long arm of the Y Chromosome (Fig. 1) after its initial retrotransposition ∼20 mya (14–27 mya 95% CI) (Fig. 1D,E; Supplemental Fig. S13). This ERV sequence is found on several autosomes but is enriched on the X and Y Chromosomes (χ2 = 283, d.f. = 19, P < 0.001) (Fig. 2A,C; Supplemental Table S4). Furthermore, the Y-linked copies of this sequence are highly diverged from their non–Chr Y counterparts (Fig. 2A), yet retain fragments of the genes encoded by ERVs (e.g., gag, pol, pro, env). Multiple sequences within the ERV consensus sequence could possibly act as promoters for nearby genes (Fig. 2B).
Figure 2.
Paralogous endogenous retrovirus (ERV)-like elements are enriched on the X and Y Chromosomes. (A) Phylogenetic reconstruction of relationships between ERV-like elements identified within the domestic cat genome (labeled by chromosome and coordinates), cheetah (AJU), and tiger (PTI). The branches are color-coded based on their predicted chromosome of origin. (B) Ideogram of consensus of Y-linked ERV-like sequences annotated with vestigial ERV genes (below) and the promoter prediction score (above) by position within the sequence. Scores below 0.5 can be ignored, while those ranging from 0.5 to 0.8 are considered marginal and between 0.8 and 1.0 as moderately likely; any score above 1.0 is considered as highly likely. (C) Position of ERV-like sequences along the domestic cat X Chromosome.
Ampliconic Y transcripts
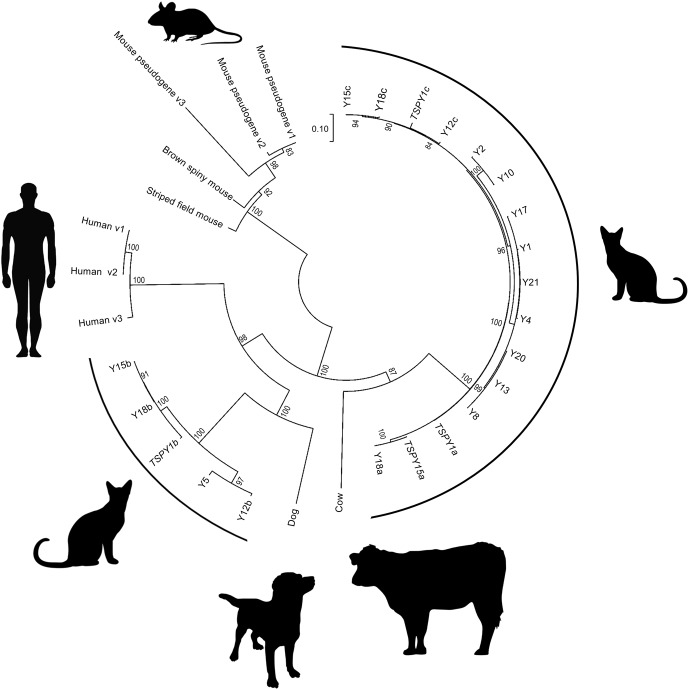
Murphy et al. (2006) reported three subfamilies of TSPY1: TSPY1a and TSPY1c sharing ∼98% pairwise identity, and TSPY1b only shares ∼64% identity with both TSPY1a and TSPY1c. We identified a large number of additional TSPY1 variants from testis RNAseq data, all of which fall into one of the two subgroups (Fig. 3). The TSPY1b subgroup clusters with TSPY1 genes found in dog and human, while the subgroup containing TSPY1a and TSPY1c are more similar to the cow gene. This suggests that TSPY1 was multicopy in the eutherian ancestor but that specific subfamilies were independently lost in several ordinal lineages.
Figure 3.
Phylogenetic reconstruction of the TSPY1 variants published for the domestic cat, dog, human, cow, and several rodent species, as well as TSPY1 transcript variants identified in this study (all clones starting with an uppercase “Y”).
Skaletsky et al. (2003) reported an antisense open reading frame (ORF), CYorf16, on the human MSY that was transcribed from each TSPY1 locus. A cat ortholog for CYorf16 was not found, although we did identify five novel TSPY1-like ORFs within a substantial number of the ampliconic scaffolds, ranging in length from 100 to 220 amino acids and including both sense and antisense orientations (Supplemental Fig. S14). Whether these candidate ORFs undergo active transcription and translation remains to be determined.
The CCDC71LY ampliconic gene family contains a large number of sense and antisense ORFs (99–322 amino acids long) (Supplemental Fig. S15). An array of frameshift and frameshift-recovering mutations has led to a diversity of putative peptide sequences composed of different combinations of shared sequence motifs (Supplemental Figs. S16–S18). Finally, two previously undocumented, testis-specific Y-linked transcripts were discovered in the ampliconic sequences, NTY1 and NTY2 (Novel Transcript on the Y), that show modest sequence similarity (66%). NTY1 was found on only one Y scaffold (Y5) and contains a single exon with an ORF of 183 amino acids. NTY2 was found in two scaffolds and contains a single exon with an ORF of 163 amino acids. These transcripts shared no significant similarity to any sequence in the NCBI nr database, and both were transcribed at levels comparable to other ampliconic genes (Supplemental Fig. S19).
Ampliconic structural evolution
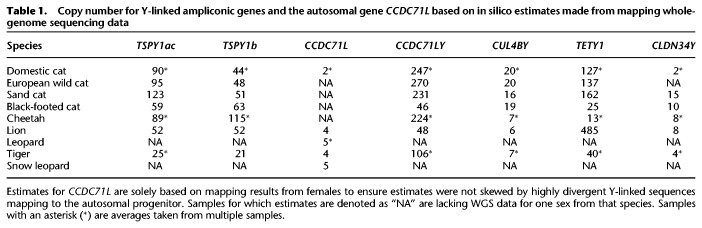
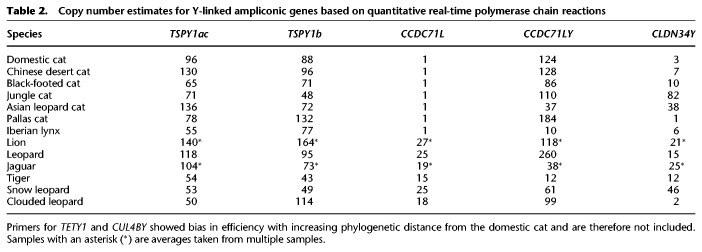
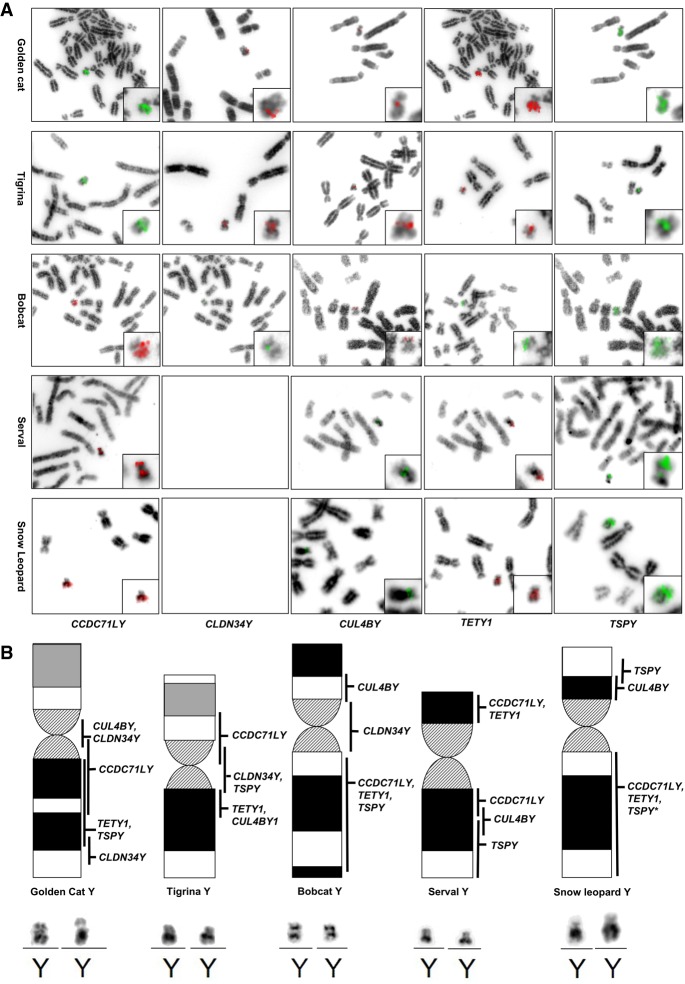
Both qPCR and in silico–based estimates of ampliconic gene copy number variation demonstrated a large degree of gene family size variation within and across species (Tables 1, 2; Supplemental Tables S5, S6). We also determined that the autosomal progenitor of CCDC71LY, CCDC71L, has undergone expansion within the Pantherinae, including the leopard, lion, tiger, jaguar, clouded leopard, and snow leopard (Tables 1, 2). To visually validate the amplification and chromosomal distribution of ampliconic genes, we conducted FISH with domestic cat cDNA probes on the chromosomes of five species from different felid clades: the Asian golden cat (Catopuma temminckii; Asian golden cat clade), tigrina (Leopardus tigrinus; Ocelot clade), bobcat (Lynx rufus; Lynx clade), serval (Leptailurus serval; Caracal clade), and the snow leopard (Panthera uncia; Panthera clade). These results confirmed a high degree of conservation of gene family content but drastic differences in structure (Fig. 4A,B; Supplemental Figs. S20–S24). For example, TSPY1 appears to be expanded along a majority of Yq in both the Asian golden cat and bobcat, but it is restricted to the proximal region of tigrina Yq. Similarly, CCDC71LY is massively amplified along tigrina Yp, but was found along Yq of all other felid Y Chromosomes that we analyzed (Li et al. 2013). While the FISH results support the conservation of ampliconic genes across Felidae, it should be noted that some lineages may have independently acquired novel Y-linked genes that would go undiscovered using this approach. Specifically, the cDNA hybridization signals were restricted to the distal and proximal portions of tigrina Yp and Yq, respectively, and the putative gene content of the remainder of the long arm is unknown.
Table 1.
Copy number for Y-linked ampliconic genes and the autosomal gene CCDC71L based on in silico estimates made from mapping whole-genome sequencing data
Table 2.
Copy number estimates for Y-linked ampliconic genes based on quantitative real-time polymerase chain reactions
Figure 4.
Distribution of ampliconic loci along the Y Chromosomes of five divergent felids. (A) FISH with probes designed for known ampliconic genes in metaphase preparations of several felid species. (B) Ideograms for each species with the predicted distribution of each ampliconic Y-linked gene indicated. Note the long arm of the Tigrina Y contains a substantial amount of sequence in which no known ampliconic genes were identified. (*) TSPY1 appears to be present along the long arm of the snow leopard Y, but it should be noted that the signal intensity of this probe was not as strong as other ampliconic genes expanded throughout this region.
Discussion
Assembly strategy and results
We generated ∼2.4 Mb (∼10%) of ampliconic sequence from the domestic cat Y Chromosome using two distinct sequencing platforms: Illumina MiSeq and PacBio RSII. While we were unable to confidently assemble every BAC clone sequence, we were able to accurately reconstruct many clones for a fraction of the cost that would have been required to conduct PacBio sequencing of every clone. Regardless of the approach taken, our results affirm previous studies that concluded that accurate assembly of Y Chromosome amplicons requires targeted sequencing approaches given their highly repetitive and structurally complex architecture (Hughes and Page 2015). The increasing availability of Y-linked sequence information for a variety of species has beneficial implications for many facets of biology. These resources can aid in elucidating historical and current population structures (Karmin et al. 2015; Hallast et al. 2016; Poznik et al. 2016; Jobling and Tyler-Smith 2017), allow for a more comprehensive understanding of male reproductive biology (Kuroda-Kawaguchi et al. 2001; Touré et al. 2004, 2005; Campbell et al. 2012), and provide a more complete substrate on which to investigate genome evolution (Murphy et al. 2006; Li et al. 2013).
Remarkable ampliconic protein coding diversity
During spermatogenesis, transcription across most of the genome is prolific, presumably due to permissive epigenetic regulation rather than playing any functional role (Soumillon et al. 2013), although it could be argued that spermatogenesis is a complex process requiring a large suite of disparate genes. We identified more than 50 testis-specific ampliconic gene family members, as well as two previously unidentified novel loci (NTY1 and NTY2), and validated their specificity through analysis of RNA-seq data from a suite of domestic cat tissues (Supplemental Fig. S19; Supplemental Table S7). Further examination of this diverse array of variants revealed a remarkable amount of putative protein coding ability. Specifically, within the largest two gene families, TSPY1 and CCDC71LY, we observed 66 and 83 ORFs, respectively, including both sense and antisense orientations (Supplemental Figs. S14–S18). While these ORFs appeared to be relatively conserved across the TSPY1 family, numerous small indels occurring between CCDC71LY variants have resulted in more variable peptide combinations. The presence of such diversity suggests that the unique evolutionary processes that shape the Y (e.g., lack of recombination, higher mutation rate, smaller effective population size, drift, and male-limited transmission) provide (or in the least permit) a great deal of potentially functional variation.
Co-opting ERVs
Within the Y scaffolds we identified an expansion of a novel MSY sequence derived from an ERV. While this particular class of ERV is found elsewhere throughout the genome, it is notably enriched along the sex chromosomes (Fig. 2). Furthermore, this sequence does not appear to be randomly distributed throughout the assembled Y scaffolds but was only found adjacent to a region that was transposed from Chr A2 ∼46 mya, suggesting the segments have been amplified as a single unit. The immediate ancestor of the Y-linked ERV-like elements is an ERV located within the pseudoautosomal region, suggesting a probable recombination-mediated mechanism by which the elements were acquired on the Y Chromosome from its X Chromosome predecessor.
Reduced recombination could account for the increased frequency of these potentially deleterious ERV-like insertions on the sex chromosomes, but the domestication of selfish genetic elements by the host to utilize their protein coding potential or to regulate transcriptional activity has been documented by several studies (e.g., Werren 2011; McLaughlin and Malik 2017). Recently, Davis et al. (2017) demonstrated that LTRs from some mouse ERVs regulate the expression of many lncRNAs in both spermatocytes and spermatids. These findings suggest that the acquisition, retention, and amplification of this Y-linked feline ERV could serve to regulate CCDC71LY and possibly other ampliconic Y-linked genes during spermatogenesis.
Extensive copy number variation across Felidae
We observed substantial intra- and interspecific copy number variation for all ampliconic gene families across Felidae, which is supported by differences in Y Chromosome length observed in FISH analyses. The discrepancies between qPCR and in silico approaches highlight the difficulties encountered in studying complex ampliconic regions of mammalian genomes. The differences we observed are likely both biological and methodological in nature. First, variation in Y Chromosome length has been observed within species, and the individuals used for qPCR and the in silico–based approaches were different. This highlights the volatile course of Y Chromosome evolution and suggests strong selective pressure to retain these genes throughout the felid radiation.
However, there are methodological sources for the observed disparities. While the qPCR primers were designed from multispecies alignments of ampliconic loci, the extensive gene family variation observed within the domestic cat alone suggests we were unable to capture additional interspecific variation within our primer design. For example, qPCR estimates for both TSPY1ac and TSPY1b were higher than those observed in the in silico–based approach, but the opposite pattern was observed in estimates of CCDC71LY copy number. This would be expected given the higher sequence similarity within TSPY1ac (98%) and TSPY1b (98.5%) than within CCDC71LY (∼93%). Mapping short-read data is likely more permissive of sequence divergence than qPCR, as just a few mismatches between primer and target sequence would lead to inefficiencies in PCR amplification.
Another issue we encountered when estimating copy number in silico was mismapping of reads, specifically when mapping more distantly related species to the domestic cat reference genome. This problem can arise when divergence between species causes sequence diversity between orthologs and paralogs to overlap. We were able to mitigate these issues by utilizing both male and female samples to estimate copy number, allowing us to better ensure Y-linked reads were not mapping to paralogous autosomal genes and skewing results.
Coexpansion of CCDC71L and CCDC71LY in Panthera
The autosomal progenitor of CCDC71LY, CCDC71L, has undergone amplification within Panthera (Tables 1, 2; Supplemental Tables S5, S6), perhaps in response to the amplification of CCDC71LY on Chr Y. This may suggest the presence of an evolutionary arms race, in which the autosomal copy was amplified in order to suppress biased transmission of the Y. In support of this hypothesis, when female hybrid F1 ligers (offspring of a male lion × female tiger cross) are backcrossed to the lions, the litters display skewed sex ratios (Supplemental Table S1). A similar process has been documented in Mus, in which mice deficient in copy number for the ampliconic Y-linked gene Sly produce female-biased litters and those with ampliconic X-linked Slx-deficient genotypes produce male-biased litters (Cocquet et al. 2012; Good 2012). In Panthera, however, the repressor of the Y-linked CCDC71LY would be autosomal and not sex linked and, depending on the molecular mechanisms behind repression, could conceivably lead to male-biased litters produced by CCDC71L-deficient males. A similar drive system has been found in Drosophila, but this involves the suppression of an X-linked sex-ratio distorter by an autosomal locus dubbed nmy (Tao et al. 2007). This suppressor evolved through a retrotransposition of the distorter itself, and repression appears to be facilitated through sequence homology: a plausible mechanism in Panthera if CCDC71LY and CCDC71L do in fact constitute such a system.
Another intriguing possibility is that CCDC71LY and CCDC71L do not share an antagonistic relationship but rather work cooperatively to regulate some particular function during reproduction. In humans, CCDC71L appears to be most highly expressed within the fallopian tube (Supplemental Fig. S25; DeLuca et al. 2012; The GTEx Consortium 2013; Carithers et al. 2015; Melé et al. 2015), while in the cat, its expression appears to be highest in both the fallopian tube and testes (Supplemental Fig. S26). These observations, taken together with the testis-specific expression of CCDC71LY, suggests the possibility of a functional relationship between the two gene families that is worthy of future investigation.
Y Chromosome stability
Despite increasing evidence in support of the longevity and stability of ancestral single-copy Y-linked genes throughout mammalian evolution, papers that espouse the Y Chromosome's inevitable demise are still published (Graves 2016). Although the evolutionary forces that shape the mammalian Y are in some ways more drastic than other regions of the genome, our study, along with several others before, indicates that portions of the Y Chromosome have not only remained stable, but most mammalian MSYs have accrued and amplified new genomic content (Murphy et al. 2006; Hughes et al. 2010, 2012; Bellott et al. 2014; Soh et al. 2014). While the ancestral therian Y Chromosomes did undergo an initial period of rapid degradation, the current model of gene loss on the Y predicts a nonlinear loss of loci that has all but ceased in many species (Bachtrog 2008, 2013; Hughes et al. 2012). Hughes et al. (2012), for example, demonstrated that the rhesus macaque MSY has not undergone any gene loss in the last 25 Myr, and gene loss along the human MSY has been restricted to the most recent evolutionary strata. Furthermore, Bellott et al. (2014) and Cortez et al. (2014) demonstrated that ancestral Y-linked genes that were retained on the eutherian Y were not random but were selectively retained due to dosage constraints and frequently maintain their ancestral protein functions.
Our findings suggest that the evolutionary stability of the Y is not limited to the more conserved single-copy region but also extends to ampliconic regions of the MSY. While human and chimpanzee MSYs differ drastically in both their gene repertoire and structure (Hughes et al. 2010), the gene content of felid Y Chromosomes appears to be highly conserved (and amplified) across the family, although copy number and chromosome structure appear to vary greatly between species. The stability in ampliconic gene content is especially impressive given that the length of time that these gene families have been expanded and retained on the Y is far greater than the estimated age of the felid MRCA (Supplemental Figs. S7–S13). The conservation of ampliconic loci in the face of frequent structural rearrangements, processes like Muller's ratchet, and hitchhiking of deleterious alleles suggests a high level of selective pressure to retain high copy numbers of these male-specific genes. This selective pressure enforces a type of chromosomal stability that differs from conventional ideas of genomic conservation (e.g., synteny), retaining certain functional Y-linked ampliconic genes while mitigating the erosive effects of replication without recombination.
In summary, our analysis of Y Chromosome evolution within the cat family illustrates the dynamic evolutionary forces that shape this chromosome. We provide, to our knowledge, the first evidence that genomic conservation is not limited to the shared (albeit depleted) X-degenerate regions but is extended to the ampliconic gene families and sequences that putatively play a large role in male reproductive success. We have shown these ampliconic gene families were amplified early (before the MRCA of this family) and have been maintained in high copy number throughout the Felidae radiation. This finding is in contrast to what has been shown in primate amplicons, which exhibit a high degree of gene content turnover. Within Felidae, ampliconic genes are retained, while the MSY is still serving as a dynamic substrate upon which novel genes and co-opted genomic elements undergo selection, most frequently in processes like male reproductive fitness, transmission distortion, and, perhaps, the establishment of nascent species boundaries.
Methods
Sequencing and assembly of the ampliconic Y
Clones containing Y Chromosome fragments were identified by screening the male cat RPCI 86 BAC library using an equimolar pool of overgo probes derived from known Y-linked ampliconic genes: CCDC71LY, CUL4BY, TETY1, CLDN34Y, and TSPY1 (Murphy et al. 2006). Overgo probe design and filter hybridization methods were performed as previously described (Pearks Wilkerson et al. 2008). Selected clones were cultured and DNA was extracted using standard protocols. Extracted DNA was then sequenced on the PacBio RS II system using the P6-P4 chemistry and/or on the Illumina MiSeq platform. Mate pair libraries from the same male cat were also utilized, with insert sizes of ∼3 and 8 kb (SRX2376195 and SRX2376203, respectively). The mate pair libraries were processed using NextClip v1.3.1 (Leggett et al. 2014) and mapped to the domestic cat 8.0 genome assembly (Montague et al. 2014) using default settings in BWA-MEM (Li and Durbin 2009). Mate pairs that did not align or that had a mapping quality greater than 30 were kept and represented those mate pairs most likely to be derived from the Y Chromosome.
Sequenced Illumina libraries were trimmed using TrimGalore!, a wrapper tool around Cutadapt (Martin 2011), and were assessed for quality using FastQC. Trimmed libraries were mapped to the Escherichia coli K12 MG1655 genome using default settings in BWA-MEM (Li and Durbin 2009). Accurately aligned reads (mapping quality greater than or equal to 30) were removed from the libraries. Filtered libraries were then split into subsets to target 100× coverage, and each subset was assembled using A5-miseq (Coil et al. 2015) with default settings. The target coverage for each clone was adjusted to optimize assembly length and contiguity. Contigs were filtered for quality requiring an average per base phrap score of 75 or greater. Assembled sequences were then aligned back to the E. coli K12 MG1655 genome using BLAST (Altschul et al. 1990) to further remove any bacterial contamination.
After the assembly for each clone was quality filtered and had the vector sequence removed, we aligned the 3- and 8-kb mate pair libraries using Newbler version 2.9, requiring 40-bp overlap and 99% pairwise identity. This program was used in order to produce the output required for Consed (Gordon and Green 2013), which allowed us to easily visualize the alignments, read depths, and mate pair connections for manual curation (Li et al. 2013). We then mapped the filtered paired-end libraries back to the complete scaffold using the program Bowtie 2 (Langmead et al. 2009) in Geneious (Kearse et al. 2012) with default settings for a final quality assessment. Mapping results were visually inspected for conspicuous anomalies in read depth coverage (e.g., the standard deviation of read depth was greater than the mean), and those without uniform coverage were not considered accurately assembled and were not used in downstream analyses.
Barcoded PacBio libraries were assembled using the PBcR pipeline with Celera Assembler version 8.3rc2 (Koren et al. 2012, 2013; Berlin et al. 2015). Assemblies were circularized using minimus2 in AMOS (Treangen et al. 2011). Custom scripts used in the assembly process are located in the Supplemental Material (Supplemental Scripts). All alignments generated in this study are provided in the Supplemental Material (Supplemental Alignments).
Sequence content of the ampliconic Y
We examined the sequence content of the assembled clones using a variety of tools in order to identify the origins, repeated units, and transcribed elements of the ampliconic Y. Repetitive elements were identified using the program RepeatMasker version open-4.0.5 (Smit et al. 2013–2015). We used GMAP (Wu and Watanabe 2005) to identify the locations of previously described Y-linked ampliconic genes. We iteratively aligned sections of assembled clones to the domestic cat genome assembly (Felis catus v8.0, which does not include the previous Y Chromosome assembly) using BLAT (Kent 2002) to delineate the boundaries of transposed segments.
Once the shared constitutive segments from each clone were identified, we aligned them with the corresponding autosomal and/or X-linked sequences from the domestic cat, tiger (Panthera tigris), cheetah (Acinonyx jubatus), domestic dog (Canis familiaris), and panda (Ailuropoda melanoleuca) using the program MAFFT (Katoh and Standley 2013). Columns in the alignment matrix for which >50% of the sites were missing were removed, and phylogenies and time trees were constructed using the programs RAxML (Stamatakis 2006) and MCMCTree (Yang 2007), respectively. Hard bound constraints for the base of Felidae were set at 10 and 24 Myr for divergence time estimation (Johnson et al. 2006). Some Y-linked segments were not included in order to maximize alignment lengths for this analysis. The transposition from Chr A2 at ∼77 Mb was the oldest transposition event to the ampliconic Y that we identified and, as a result, is highly fragmented. In order to increase the lengths of the alignments and number of sequences in each alignment, we built separate alignments and trees for different segments of this locus. Final alignment files are included in the Supplemental Materials (Supplemental Alignments).
We created an RNA-seq library from testis tissue taken from a mature domestic cat. The sequenced library was iteratively mapped to the repeat-masked domestic cat genome, each time containing a single repeat-masked assembled clone as well as the domestic cat Y Chromosome sequence published by Li et al. (2013), using the program STAR (Dobin et al. 2013). Transcripts were constructed and expression levels were estimated using Cufflinks (Trapnell et al. 2012). We also downloaded and mapped RNA-seq libraries from the SRA database (Supplemental Table S7) to characterize the tissue expression profiles of identified Y-linked transcripts.
After examining the tissue expression profile CCDC71L in humans and noting the highest levels of expression in the fallopian tubes, we created two RNA-seq libraries from the same domestic cat fallopian tube sample. Expression profiles for CCDC71L were quantified using the same protocol as above.
Estimating copy number of Y-linked ampliconic genes
We conducted qPCR on available felid species using the relative comparative threshold cycle (CT) method on a Roche 480 LightCycler. We used primers based on multispecies alignments for each ampliconic gene and for the single-copy Y-linked gene DDX3Y to serve as a control (Supplemental Tables S8, S9). Reactions were run in triplicate for each gene, and the copy numbers were calculated using the 2−ΔΔct method (Schmittgen and Livak 2008).
We also estimated ampliconic gene copy numbers in silico by analyzing changes in read depth coverage of WGS libraries (Supplemental Table S6) mapped to the domestic cat reference genome (felCat8) augmented with consensus sequences created from the assembled ampliconic regions of the domestic cat Y Chromosome, including a single representative sequence for each Y-linked gene. The resulting alignments were then analyzed using CNVnator (Abyzov et al. 2011), using a bin size of 150 for all genes except CLDN34Y, for which we used a bin size of 50 bp.
For the autosomal gene CCDC71L, we used calls from CNVnator for the regions immediately flanking this gene. This was necessary as the domestic cat genome assembly contains a gap in the sequence within the annotation of this gene, and thus, reads were unable to accurately map to this stretch of the genome. We also made estimates solely based on short-read libraries from female samples to avoid the possibility of overestimating copy number due to Y-derived paired-end reads incorrectly mapping to this autosomal progenitor.
FISH
FISH analyses were performed with biotin- and/or dioxigenin-labeled probes designed from cDNA sequences published by Murphy et al. (2006). These probes were hybridized to metaphase chromosomes from the Asian golden cat (C. temminckii), tigrina (L. tigrinus), bobcat (L. rufus), serval (L. serval), and the snow leopard (P. uncia) following established procedures (Raudsepp and Chowdhary 2008). A Zeiss Axioplan2 fluorescent microscope equipped with Cytovision/Genus V. 2.7 (Applied Imaging) was used to capture and analyze images from the metaphase spreads.
Data access
Y Chromosome sequence data generated in this study have been submitted to the NCBI Sequence Read Archive (SRA; https://www.ncbi.nlm.nih.gov/sra) under accession number SRP136722.
Supplementary Material
Acknowledgments
We thank Gang Li and Victor Mason for helpful comments and suggestions made throughout the duration of this study, Elaine Owens for help in chromosome preparation, and Mark Stickney for access to domestic cat fallopian tube tissue. Funding was provided by Morris Animal Foundation grants D12FE-019 and D16FE-011 to W.J.M., and by the Winn Feline Foundation under grant 07-034.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.237586.118.
References
- Abyzov A, Urban AE, Snyder M, Gerstein M. 2011. CNVnator: an approach to discover, genotype, and characterize typical and atypical CNVs from family and population genome sequencing. Genome Res 21: 974–984. 10.1101/gr.114876.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkan C, Sajjadian S, Eichler EE. 2011. Limitations of next-generation genome sequence assembly. Nat Methods 8: 61–65. 10.1038/nmeth.1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215: 403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Bachtrog D. 2008. The temporal dynamics of processes underlying Y chromosome degeneration. Genetics 179: 1513–1525. 10.1534/genetics.107.084012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D. 2013. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet 14: 113–124. 10.1038/nrg3366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, et al. 2014. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 508: 494–499. 10.1038/nature13206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlin K, Koren S, Chin CS, Drake JP, Landolin JM, Phillippy AM. 2015. Assembling large genomes with single-molecule sequencing and locality-sensitive hashing. Nat Biotechnol 33: 623–630. 10.1038/nbt.3238 [DOI] [PubMed] [Google Scholar]
- Campbell P, Nachman MW. 2014. X–Y interactions underlie sperm head abnormality in hybrid male house mice. Genetics 196: 1231–1240. 10.1534/genetics.114.161703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell P, Good JM, Dean MD, Tucker PK, Nachman MW. 2012. The contribution of the Y chromosome to hybrid male sterility in house mice. Genetics 191: 1271–1281. 10.1534/genetics.112.141804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carithers LJ, Ardlie K, Barcus M, Branton PA, Britton A, Buia SA, Compton CC, DeLuca DS, Peter-Demchok J, Gelfand ET, et al. 2015. A novel approach to high-quality postmortem tissue procurement: the GTEx project. Biopreserv Biobank 13: 311–319. 10.1089/bio.2015.0032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Clark AG. 2013. Efficient identification of Y chromosome sequences in the human and Drosophila genomes. Genome Res 23: 1894–1907. 10.1101/gr.156034.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaisson MJ, Wilson RK, Eichler EE. 2015. Genetic variation and the de novo assembly of human genomes. Nat Rev Genet 16: 627–640. 10.1038/nrg3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocquet J, Ellis PJ, Mahadevaiah SK, Affara NA, Vaiman D, Burgoyne PS. 2012. A genetic basis for a postmeiotic X versus Y chromosome intragenomic conflict in the mouse. PLoS Genet 8: e1002900 10.1371/journal.pgen.1002900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coil D, Jospin G, Darling AE. 2015. A5-miseq: an updated pipeline to assemble microbial genomes from Illumina MiSeq data. Bioinformatics 31: 587–589. 10.1093/bioinformatics/btu661 [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. 2014. Origins and functional evolution of Y chromosomes across mammals. Nature 508: 488–493. 10.1038/nature13151 [DOI] [PubMed] [Google Scholar]
- Davis BW, Seabury CM, Brashear WA, Li G, Roelke-Parker M, Murphy WJ. 2015. Mechanisms underlying mammalian hybrid sterility in two feline interspecies models. Mol Biol Evol 32: 2534–2546. 10.1093/molbev/msv124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MP, Carrieri C, Saini HK, van Dongen S, Leonardi T, Bussotti G, Monahan JM, Auchynnikava T, Bitetti A, Rappsilber J, et al. 2017. Transposon-driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Rep 18: 1231–1247. 10.15252/embr.201744059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca DS, Levin JZ, Sivachenko A, Fennell T, Nazaire MD, Williams C, Reich M, Winckler W, Getz G. 2012. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics 28: 1530–1532. 10.1093/bioinformatics/bts196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler EE, Clark RA, She X. 2004. An assessment of the sequence gaps: unfinished business in a finished human genome. Nat Rev Genet 5: 345–354. 10.1038/nrg1322 [DOI] [PubMed] [Google Scholar]
- Eizirik E, Murphy WJ, Koepfli KP, Johnson WE, Dragoo JW, Wayne RK, O'Brien SJ. 2010. Pattern and timing of diversification of the mammalian order Carnivora inferred from multiple nuclear gene sequences. Mol Phylogenet Evol 56: 49–63. 10.1016/j.ympev.2010.01.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershony LC, Penedo MC, Davis BW, Murphy WJ, Helps CR, Lyons LA. 2014. Who’s behind that mask and cape? The Asian leopard cat’s Agouti (ASIP) allele likely affects coat colour phenotype in the Bengal cat breed. Anim Genet 45: 893–897. 10.1111/age.12206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghenu AH, Bolker BM, Melnick DJ, Evans BJ. 2016. Multicopy gene family evolution on primate Y chromosomes. BMC Genomics 17: 157 10.1186/s12864-015-2187-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Good JM. 2012. The conflict within and the escalating war between the sex chromosomes. PLoS Genet 8: e1002955 10.1371/journal.pgen.1002955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D, Green P. 2013. Consed: a graphical editor for next-generation sequencing. Bioinformatics 29: 2936–2937. 10.1093/bioinformatics/btt515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA. 2016. Did sex chromosome turnover promote divergence of the major mammal groups? De novo sex chromosomes and drastic rearrangements may have posed reproductive barriers between monotremes, marsupials and placental mammals. Bioessays 38: 734–743. 10.1002/bies.201600019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray AP. 1972. Mammalian hybrids: a check-list with bibliography. Commonwealth Agricultural Bureaux, Farnham Royal, UK. [Google Scholar]
- The GTEx Consortium. 2013. The Genotype-Tissue Expression (GTEx) project. Nat Genet 45: 580–585. 10.1038/ng.2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallast P, Maisano Delser P, Batini C, Zadik D, Rocchi M, Schempp W, Tyler-Smith C, Jobling MA. 2016. Great-ape Y Chromosome and mitochondrial DNA phylogenies reflect subspecies structure and patterns of mating and dispersal. Genome Res 26: 427–439. 10.1101/gr.198754.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleu Q, Gérard PR, Montchamp-Moreau C. 2014. Sex chromosome drive. Cold Spring Harb Perspect Biol 7: a017616 10.1101/cshperspect.a017616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y, Wu Q, Ma S, Ma T, Shan L, Wang X, Nie Y, Ning Z, Yan L, Xiu Y, et al. 2017. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci 114: 1081–1086. 10.1073/pnas.1613870114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huddleston J, Ranade S, Malig M, Antonacci F, Chaisson M, Hon L, Sudmant PH, Graves TA, Alkan C, Dennis MY, et al. 2014. Reconstructing complex regions of genomes using long-read sequencing technology. Genome Res 24: 688–696. 10.1101/gr.168450.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Page DC. 2015. The biology and evolution of mammalian Y chromosomes. Annu Rev Genet 49: 507–527. 10.1146/annurev-genet-112414-055311 [DOI] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Pyntikova T, Graves TA, van Daalen SK, Minx PJ, Fulton RS, McGrath SD, Locke DP, Friedman C, et al. 2010. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature 463: 536–539. 10.1038/nature08700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, et al. 2012. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature 483: 82–86. 10.1038/nature10843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jobling MA, Tyler-Smith C. 2017. Human Y-chromosome variation in the genome-sequencing era. Nat Rev Genet 18: 485–497. 10.1038/nrg.2017.36 [DOI] [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ. 2006. The late Miocene radiation of modern Felidae: a genetic assessment. Science 311: 73–77. 10.1126/science.1122277 [DOI] [PubMed] [Google Scholar]
- Karmin M, Saag L, Vicente M, Wilson Sayres MA, Järve M, Talas UG, Rootsi S, Ilumäe AM, Mägi R, Mitt M, et al. 2015. A recent bottleneck of Y chromosome diversity coincides with a global change in culture. Genome Res 25: 459–466. 10.1101/gr.186684.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. 2013. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28: 1647–1649. 10.1093/bioinformatics/bts199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ. 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664. 10.1101/gr.229202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khost DE, Eickbush DG, Larracuente AM. 2017. Single-molecule sequencing resolves the detailed structure of complex satellite DNA loci in Drosophila melanogaster. Genome Res 27: 709–721. 10.1101/gr.213512.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitchener AC, Breitenmoser-Würsten C, Eizirik E, Gentry A, Werdelin L, Wilting A, Yamaguchi N, Abramov AV, Christiansen P, Driscoll C, et al. 2017. A revised taxonomy of the Felidae: the final report of the Cat Classification Task Force of the IUCN/SSC Cat Specialist Group. Cat News Special Issue 11: 1–80. [Google Scholar]
- Koren S, Schatz MC, Walenz BP, Martin J, Howard JT, Ganapathy G, Wang Z, Rasko DA, McCombie WR, Jarvis ED, et al. 2012. Hybrid error correction and de novo assembly of single-molecule sequencing reads. Nat Biotechnol 30: 693–700. 10.1038/nbt.2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, Mcvey SD, Radune D, Bergman NH, Phillippy AM. 2013. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol 14: R101 10.1186/gb-2013-14-9-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda-Kawaguchi T, Skaletsky H, Brown LG, Minx PJ, Cordum HS, Waterston RH, Wilson RK, Silber S, Oates R, Rozen S, et al. 2001. The AZFc region of the Y chromosome features massive palindromes and uniform recurrent deletions in infertile men. Nat Genet 29: 279–286. 10.1038/ng757 [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. 1997. Functional coherence of the human Y chromosome. Science 278: 675–680. 10.1126/science.278.5338.675 [DOI] [PubMed] [Google Scholar]
- Lamichhaney S, Berglund J, Almén MS, Maqbool K, Grabherr M, Martinez-Barrio A, Promerová M, Rubin CJ, Wang C, Zamani N, et al. 2015. Evolution of Darwin’s finches and their beaks revealed by genome sequencing. Nature 518: 371–375. 10.1038/nature14181 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leggett RM, Clavijo BJ, Clissold L, Clark MD, Caccamo M. 2014. NextClip: an analysis and read preparation tool for Nextera Long Mate Pair libraries. Bioinformatics 30: 566–568. 10.1093/bioinformatics/btt702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25: 1754–1760. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Davis BW, Raudsepp T, Pearks Wilkerson AJ, Mason VC, Ferguson-Smith M, O'Brien PC, Waters PD, Murphy WJ. 2013. Comparative analysis of mammalian Y chromosomes illuminates ancestral structure and lineage-specific evolution. Genome Res 23: 1486–1495. 10.1101/gr.154286.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Davis BW, Eizirik E, Murphy WJ. 2016. Phylogenomic evidence for ancient hybridization in the genomes of living cats (Felidae). Genome Res 26: 1–11. 10.1101/gr.186668.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Garber M, Zuk O, Lin MF, Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E et al. 2011. A high-resolution map of human evolutionary constraint using 29 mammals. Nature 478: 476–482. 10.1038/nature10530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17: 10–12. 10.14806/ej.17.1.200 [DOI] [Google Scholar]
- McLaughlin RN Jr, Malik HS. 2017. Genetic conflicts: the usual suspects and beyond. J Exp Biol 220: 6–17. 10.1242/jeb.148148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JRS, Lindblad-Toh K. 2017. Dissecting evolution and disease using comparative vertebrate genomics. Nat Rev Genet 18: 624–636. 10.1038/nrg.2017.51 [DOI] [PubMed] [Google Scholar]
- Meiklejohn CD, Tao Y. 2010. Genetic conflict and sex chromosome evolution. Trends Ecol Evol 25: 215–223. 10.1016/j.tree.2009.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melé M, Ferreira PG, Reverter F, DeLuca DS, Monlong J, Sammeth M, Young TR, Goldmann JM, Pervouchine DD, Sullivan TJ, et al. 2015. Human genomics: the human transcriptome across tissues and individuals. Science 348: 660–665. 10.1126/science.aaa0355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague MJ, Li G, Gandolfi B, Khan R, Aken BL, Searle SM, Minx P, Hillier LW, Koboldt DC, Davis BW, et al. 2014. Comparative analysis of the domestic cat genome reveals genetic signatures underlying feline biology and domestication. Proc Natl Acad Sci 111: 17230–17235. 10.1073/pnas.1410083111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy WJ, Wilkerson AJP, Raudsepp T, Agarwala R, Schäffer AA, Stanyon R, Chowdhary BP. 2006. Novel gene acquisition on carnivore Y chromosomes. PLoS Genet 2: e43 10.1371/journal.pgen.0020043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oetjens MT, Shen F, Emery SB, Zou Z, Kidd JM. 2016. Y-chromosome structural diversity in the bonobo and chimpanzee lineages. Genome Biol Evol 8: 2231–2240. 10.1093/gbe/evw150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paria N, Raudsepp T, Pearks Wilkerson AJ, O'Brien PC, Ferguson-Smith MA, Love CC, Arnold C, Rakestraw P, Murphy WJ, Chowdhary BP. 2011. A gene catalogue of the euchromatic male-specific region of the horse Y chromosome: comparison with human and other mammals. PLoS One 6: e21374 10.1371/journal.pone.0021374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearks Wilkerson AJ, Raudsepp T, Graves T, Albracht D, Warren W, Chowdhary BP, Skow LC, Murphy WJ. 2008. Gene discovery and comparative analysis of X-degenerate genes from the domestic cat Y chromosome. Genomics 92: 329–338. 10.1016/j.ygeno.2008.06.012 [DOI] [PubMed] [Google Scholar]
- Poznik GD, Xue Y, Mendez FL, Willems TF, Massaia A, Wilson Sayres MA, Ayub Q, McCarthy SA, Narechania A, Kashin S, et al. 2016. Punctuated bursts in human male demography inferred from 1,244 worldwide Y-chromosome sequences. Nat Genet 48: 593–599. 10.1038/ng.3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. 2008. FISH for mapping single copy genes. Methods Mol Biol 422: 31–49. 10.1007/978-1-59745-581-7_3 [DOI] [PubMed] [Google Scholar]
- Repping S, Skaletsky H, Lange J, Silber S, Van Der Veen F, Oates RD, Page DC, Rozen S. 2002. Recombination between palindromes P5 and P1 on the human Y chromosome causes massive deletions and spermatogenic failure. Am J Hum Genet 71: 906–922. 10.1086/342928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmittgen TD, Livak KJ. 2008. Analyzing real-time PCR data by the comparative CT method. Nat Protoc 3: 1101–1108. 10.1038/nprot.2008.73 [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. 2003. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 423: 825–837. 10.1038/nature01722 [DOI] [PubMed] [Google Scholar]
- Skinner BM, Sargent CA, Churcher C, Hunt T, Herrero J, Loveland JE, Dunn M, Louzada S, Fu B, Chow W, et al. 2016. The pig X and Y chromosomes: structure, sequence, and evolution. Genome Res 26: 130–139. 10.1101/gr.188839.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013–2015. RepeatMasker Open-4.0. http://www.repeatmasker.org
- Soh YQ, Alföldi J, Pyntikova T, Brown LG, Graves T, Minx PJ, Fulton RS, Kremitzki C, Koutseva N, Mueller JL, et al. 2014. Sequencing the mouse Y chromosome reveals convergent gene acquisition and amplification on both sex chromosomes. Cell 159: 800–813. 10.1016/j.cell.2014.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soumillon M, Necsulea A, Weier M, Brawand D, Zhang X, Gu H, Barthès P, Kokkinaki M, Nef S, Gnirke A et al. 2013. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep 3: 2179–2190. 10.1016/j.celrep.2013.05.031 [DOI] [PubMed] [Google Scholar]
- Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22: 2688–2690. 10.1093/bioinformatics/btl446 [DOI] [PubMed] [Google Scholar]
- Tao Y, Masly JP, Araripe L, Ke Y, Hartl DL. 2007. A sex-ratio meiotic drive system in Drosophila simulans. I: an autosomal suppressor. PLoS Biol 5: e292 10.1371/journal.pbio.0050292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaszkiewicz M, Rangavittal S, Cechova M, Campos Sanchez R, Fescemyer HW, Harris R, Ye D, O'Brien PC, Chikhi R, Ryder OA, et al. 2016. A time- and cost-effective strategy to sequence mammalian Y Chromosomes: an application to the de novo assembly of gorilla Y. Genome Res 26: 530–540. 10.1101/gr.199448.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Touré A, Grigoriev V, Mahadevaiah SK, Rattigan A, Ojarikre OA, Burgoyne PS. 2004. A protein encoded by a member of the multicopy Ssty gene family located on the long arm of the mouse Y chromosome is expressed during sperm development. Genomics 83: 140–147. 10.1016/S0888-7543(03)00216-7 [DOI] [PubMed] [Google Scholar]
- Touré A, Clemente EJ, Ellis PJ, Mahadevaiah SK, Ojarikre OA, Ball PA, Reynard L, Loveland KL, Burgoyne PS, Affara NA. 2005. Identification of novel Y chromosome encoded transcripts by testis transcriptome analysis of mice with deletions of the Y chromosome long arm. Genome Biol 6: R102 10.1186/gb-2005-6-12-r102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7: 562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Sommer DD, Angly FE, Koren S, Pop M. 2011. Next generation sequence assembly with AMOS. Curr Protoc Bioinformatics Chapter 11: Unit 11.8 10.1002/0471250953.bi1108s33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Cuomo CA, Zeng Q, Wortman J, Young SK, Earl AM. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9: e112963 10.1371/journal.pone.0112963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werren JH. 2011. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc Natl Acad Sci 108: 10863–10870. 10.1073/pnas.1102343108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, Watanabe CK. 2005. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21: 1859–1875. 10.1093/bioinformatics/bti310 [DOI] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. 10.1093/molbev/msm088 [DOI] [PubMed] [Google Scholar]
- Zhang G, Li C, Li Q, Li B, Larkin DM, Lee C, Storz JF, Antunes A, Greenwold MJ, Meredith RW, et al. 2014. Comparative genomics reveals insights into avian genome evolution and adaptation. Science 346: 1311–1320. 10.1126/science.1251385 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.