Abstract
Purpose
The purpose of this study was to explore differences in genotype, ocular surface microbiome, tear inflammatory markers, and environmental and behavioral exposures in soft contact lens (SCL) wearers with and without a history of corneal infiltrative events (CIEs).
Methods
Nine SCL wearers with a recent CIE and nine age-, sex-, and SCL material- and modality-matched controls were enrolled. The Contact Lens Risk Survey, slit-lamp examination data, basal tears, conjunctival microbial cultures, and peripheral blood samples were collected. Tear inflammatory mediator concentrations, genomic DNA from swabs, and whole exome sequencing of blood samples were quantified.
Results
There were no marked differences in SCL wear behaviors or exposures between case and control subjects. Predominant organisms detected among case and control subjects were Staphylococcus, Propionibacterium, Streptococcus, and Corynebacterium. Marginally higher levels of Neisseria were found in three of nine cases but zero of nine control samples (P = 0.056). A potentially deleterious missense single nucleotide polymorphism (SNP) variant in IL-6 Signal Transducer (IL6ST) was found in seven of eight cases and zero of nine controls (rs2228046; P = 0.03). The concentration of tear IL-6 was significantly higher in cases (4.5 [range, 2.1 to 6.2] pg/mL) versus controls (3.5 [range, 2.5 to 6.6] Pg/mL; = 0.02).
Conclusions
Tear IL-6 concentration was higher, and SNP variants were detected in subjects with a history of CIEs compared with healthy controls. The synthesis, signaling, and ocular surface cytokine concentration of IL-6 may be related to susceptibility to CIE. A larger study population is required to further explore relationships between genetic variations, the ocular surface microbiome, inflammatory mediators, and environmental exposures.
Keywords: tear cytokines, ocular microbiome, whole exome sequencing, soft contact lenses, corneal infiltrative events
Corneal infiltrative events (CIEs) occur in 7% to 44% of soft contact lens wearers per year and can be associated with significant morbidity and economic cost.1,2 Nearly 41 million adults in the United States wear contact lenses, and the annual rate of contact lens complications translates into approximately 1 million doctor visits at a cost of more than $175 million dollars per year.3,4 Contact lens wear can lead to CIEs due to mechanical, chemical, or hypoxic stress or via introduction of bacteria and associated toxins.5–8
Pioneering work by Keijser et al. and Carnt et al. demonstrated that genetic variations in single nucleotide polymorphism (SNP) variants of IL-1β, IL-6, IL-10, and IL-12 were associated with susceptibility to and severity of CIEs in soft contact lens wearers.9–11 Carriers of a SNP in the intron region of the TLR5 gene have been associated with a higher risk of developing keratitis, and an SNP in the exome of TLR4 has been associated with a lower risk of keratitis (Willcox M, et al. IOVS 2015;56:ARVO E-Abstract 4812). The human leukocyte antigen (HLA)-DQ genotype has been shown to influence staphylococcal colonization of the gut.12 To our knowledge, these are the only genetic variations that have been studied in patients with a history of CIEs.
Tear inflammatory mediators are proteins expressed by cells of the ocular surface during inflammation and are critical to understanding the eye's inflammatory/immune response to contact lens wear.13,14 Both routine contact lens wear and contact lens–related complications can regulate the levels of inflammatory mediators in tears.13,15–19
It is thought that the ocular surface may not elicit a full immune response against certain core microbes.20,21 However, routine soft contact lens wear may increase the number and diversity of bacteria on the ocular surface.22,23 Contact lens wearers with active CIEs harbor higher levels of bacteria on their ocular surface compared to healthy contact lens wearers.7,23 The presence of colonized bacteria on contact lenses increases the risk of CIE by three to eight times.23 It is known that many microbes cannot be grown in the laboratory, and therefore, genetic-based deep sequencing techniques (i.e., 16/18s rRNA) have recently been used to identify the presence of bacteria, fungi, or viruses on the ocular surface.24–26 Biome representational in silico karyotyping (BRiSK) detects a large number of DNA tags, including those of human, microbial, fungal, viral, and parasitic origin. This improves the accuracy of mapping of amplifications and deletions compared with 16s metagenomic techniques.27,28
This pilot study aimed to (1) demonstrate feasibility of study methodology and (2) explore differences in key genetic exome sequences, the ocular surface microbiome, and tear inflammatory proteins between contact lens wearers with and without a history of CIEs.
Methods
A single-visit cohort study was conducted at the State University of New York (SUNY), College of Optometry. Subjects were recruited from the SUNY College of Optometry and the surrounding New York City metropolitan area. The study followed the tenets of the Declaration of Helsinki. The Institutional Review Board at the SUNY College of Optometry approved the study before data collection began. Written informed consent was obtained from all participants prior to participation in the study.
Nine soft contact lens wearers with a medical record documented history of a CIE (contact lens peripheral ulcer [CLPU], contact lens–induced acute red eye [CLARE], infiltrative keratitis [IK], or microbial keratitis [MK]) were enrolled 3 to 15 months after treatment. As a control group, nine healthy established full-time soft contact lens wearers (≥1 year of full-time wear, >5 days/wk) were also enrolled, and matched by sex, contact lens material (silicone hydrogel or hydrogel), and modality (daily or reusable wear). All subjects were between 18 and 40 years of age. Exclusion criteria for all subjects included active eye infection or inflammatory disease (e.g., allergy, blepharitis, meibomian gland disease), a prior history of refractive/eye surgery, ocular trauma or systemic disease likely to affect the ocular surface (e.g., thyroid disease, diabetes, autoimmune disease), family history of systemic autoimmune or inflammatory disease, oral or ocular treatment with anti-inflammatory or antibiotic within 1 month prior to the study visit, and pregnancy or breastfeeding during the study period.
Measurements were carried out in the order of least to most invasive as described below. The Contact Lens Risk Survey (CLRS)29,30 was used to collect data on demographics, environmental exposures, and behaviors that may affect the ocular surface microbiome (i.e., water exposure and use of multipurpose solutions).4 A slit-lamp examination was conducted, and limbal and conjunctival redness and palpebral conjunctival roughness (papillae/follicles) were graded using the Cornea and Contact Lens Research Unit (CCLRU) scale (0 to 4 each eye); a total score from both eyes was recorded for analysis.31 Approximately 15 μL basal tears were collected from both eyes and prepared as previously described for batch analysis (see Tear Analysis section).32,33
Anesthetic (0.5% proparacaine; AKORN, Lake Forest, IL, USA) was instilled prior to culturing of the ocular surface as previously described.26 Briefly, two DNA free swabs (SK-2S-T swabs; Baca Scientific, Boca Raton, FL, USA) were used to collect samples from the upper and lower bulbar and palpebral conjunctiva of each eye. Negative controls were also prepared after every fourth subject by applying one drop of anesthetic to a new swab. All swabs were stored at −80°C until analysis (see Microbiome Analysis section). A second slit-lamp examination was conducted to assess corneal (0 to 5 per eye) and conjunctival staining (0 to 5 per region at temporal and nasal per eye) using sodium fluorescein, Lissamine green, and the modified Oxford grading scale.34 Approximately 10 mL peripheral blood was collected in EDTA tubes and stored at −80°C for batch analysis (see Whole Exome Sequencing section).
Tear Analysis
Tear inflammatory mediator concentrations (IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12(p70), IL-13, and TNFα) were determined using V-plex Multiplex assays (Meso Scale Discovery Japan, Tokyo, Japan) in 1:10 dilution. The concentration of secretory IgA (sIgA) was analyzed using standard ELISA (human secretory IgA ELISA; Alpco, Salem, NH, USA). Tear sIgA concentration has been used to discriminate basal tears from reflex tears.35 Therefore, sIgA concentration was compared between groups to ensure there was no difference in the type of tears collected, which may confound interpretation of the inflammatory mediators between controls and cases.
Microbiome Analysis
Genomic DNA collected from swabs were extracted using the DNeasy Blood and Tissue Kit (Qiagen, Inc., Venlo, the Netherlands), quantified using the NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE, USA) and analyzed using BRiSK and 16SrDNA metagenomic amplification and sequencing.27 Quantitative PCR (qPCR) for actin and 16SrDNA was performed to determine total bacterial load on individual swabs as previously described by comparing with a database in the GenBank data for known microbial species using the basic local alignment software tool.36 Phi29 amplification was performed for BRiSK analysis on genomic DNA and obtained microbial DNA.27 The 33-bp DNA sequence tags obtained from BRISK were compared with a database for known microbial species.27
Raw sequence data from 16SrDNA were processed with MOTHUR (https://www.mothur.org, provided by the University of Michigan, Ann Arbor, MI, USA).37 Taxonomic assignment and RDP II Classifier were used for the filtered sequences that were classified with the TUIT algorithm from the GenBank database. BRiSK analysis was performed using custom built scripts and databases. The reconstruction of ocular surface microbiomes allows the analyses of diversity, relative abundance per subject, and between-group comparisons to assess CIE-related differences. Visual comparison of the groups was performed with MEGAN4 (www-ab.informatik.uni-tuebingen.de/software/megan) (Shalabi NM, et al. IOVS 2014;55:ARVO E-Abstract 6288).
Whole Exome Sequencing Analysis
Blood samples were whole exome sequenced on the Illumina HiSeq 3000 (Illumina, San Diego, CA, USA) at the University of Miami John P. Hussman Institute for Human Genomics. Base calls were determined by the Illumina CASAVA pipeline. After filtering for base quality and adapter sequences, the sequencing reads were aligned to the human reference genome (hg19) using bowtie2 and formatted for input into the Genome Analysis Toolkit (GATK) from the Broad Institute (Cambridge, MA, USA; https://software.broadinstitute.org/gatk/). The pipeline is based on the Broad Institutes' Best Practices Guideline including local realignment, removal of PCR duplicates, and base quality recalibration. Single nucleotide variants and small insertion-deletion variants (indels) were called by GATK's HaplotypeCaller, and variants for each sample were consolidated with GenotypeGVCFs. The combined variant call format file (VCF) was then annotated with ANNOVAR (http://annovar.openbioinformatics.org/en/latest/, available in the public domain) and filtered for quality with VCFtools (http://vcftools.sourceforge.net/, available in the public domain). Variants were annotated with their frequency in the European population using the National Heart, Lung, and Blood Institute (NHLBI), Exome Sequencing Project's Exome Variant Server (ESP), Exome Aggregation Consortium (ExAC), and 1000 Genomes databases using ANNOVAR. Variants were annotated for region and exonic function by reference to refSeq and annotated for predicted impact by reference to PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/, available in the public domain) (benign, possibly damaging, probably damaging) and SIFT (http://sift.jcvi.org/, provided by the J. Craig Venter Institute, Rockville, MD, USA) (tolerated or damaging) using ANNOVAR. Variants with genotype quality (GQ) <30, depth (DP) <8, or Phred-scaled likelihood of reference genotypes (PL) <99 were excluded. Variants in IFN-γ, IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12(p70), IL-13, TNF-α, TLR-4, and HLA-DQ and related genes were prioritized for relevance based on previous studies. Additional hard filtering was applied as an alternative to VQSR using values suggested by GATK best practices.38 Filters included the following: quality by depth (QD) <2.0, Fisher strand (FS) >60.0, root mean square of the mapping quality (MQ) <40.0, Mapping quality rank sum test (MQRankSum) <−12.5, read position rank sum test (ReadPosRankSum) <−8.0, strand odds ratio (SOR) >3.0. Filters were applied manually in R version 3.4.2.39
Statistical Analysis
Descriptive data of the CLRS were generated to describe the demographic, environmental, and behavioral risk factors of CIEs of the study sample.30 A Mann-Whitney U-test was carried out to examine differences in tear concentration of inflammatory mediators and clinical indicators between case and control subjects. Relative abundance of pathogens detected in BRiSK were calculated, and significant differences between case and control subjects were determined after estimating the false discovery rate using a Monte Carlo analysis permutation test with 100,000 permutations.40 The Mann-Whitney U test with false discovery rate correction (Benjamini-Hochberg) was carried out to confirm the differences in the microbial compositions and diversity between two groups. For the whole exome sequencing, variants were analyzed with Fisher's exact test (PFET) according to recessive and dominant models. Significance was determined as P < 0.05.
Results
Sample Population and Clinical Results
Nine case and nine control subjects completed the study (four male in each group). Blood samples with adequate volume were only able to be acquired from eight of the nine case subjects. The case subjects all had a history of at least one serious or significant CIE (three subjects with CLPU and six subjects with CLARE with infiltrates). There were no reported cases of microbial keratitis. Each group was comprised of six daily disposable (three silicone hydrogel and three hydrogel) and three reusable silicone hydrogel contact lens wearers. All reusable contact lens wearers reported using multipurpose solutions. The mean age of case and control subjects were 25.9 ± 5.0 and 25.4 ± 2.3 years, respectively (P = 0.79). The mean age at which subjects began SCL wear was at 15.4 ± 5.5 years in the case group and 14.4 ± 2.5 years in the control group (P = 0.75). Table 1 shows other subject demographic information and some of the primary contact lens–related risk factors obtained from the CLRS. There were five (56%) African-American subjects in the case group compared with zero in the control group. Case subjects also reported sleeping in contact lenses more often after alcohol consumption than control subjects (Table 1). There were no other differences seen in any contact lens related risk factors between groups, and further statistical testing was not done (not all data shown). All subjects had normal ocular adnexa via slit-lamp examination, and bulbar and limbal redness and corneal and conjunctival staining were not significantly different between case and control groups (Table 2). Corneal scars were observed in seven of nine cases but zero controls.
Table 1.
Primary Demographic Information and Contact Lens Exposures for the Study Population
Table 2.
Summary of Group Mean and SD or Median and IQR, as Appropriate, for the Slit-Lamp Findings and Tear Proteins
Tear Cytokine Results
IFN-γ, IL-2, IL-4, IL-10, IL-12(p70), IL-13, and TNF-α were not detected in tear samples. There was no difference in IL-1β and IL-8 between groups, but there was a significantly higher concentration of IL-6 in the case group (Table 2). There were no significant differences in tear sIgA between groups (Table 2).
Ocular Microbiome Results
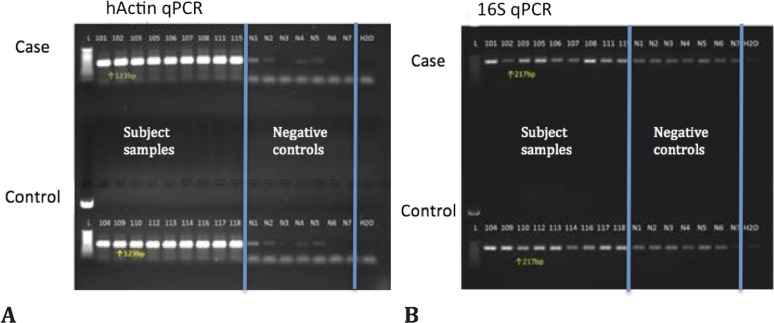
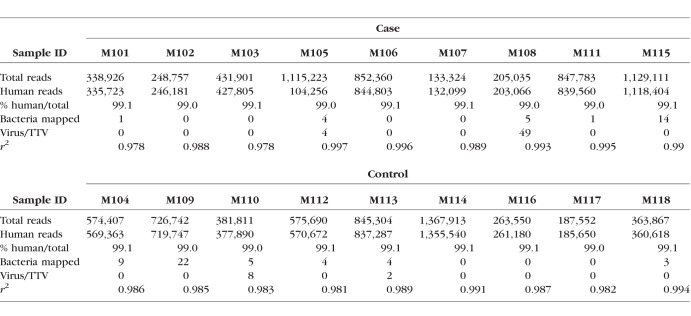
Actin and 16S bacterial ribosomal DNA were examined in all the conjunctival samples and negative controls using PCR (Fig. 1). All conjunctival samples showed strong actin results (Fig. 1A); some contamination was seen in the negative control samples but at much lower levels than the subject samples. Bacteria were recovered from all samples and very slight bands often seen from reagent contamination were observed in the negative controls (Fig. 1B).
Figure 1.
PCR results for actin (A) and 16S bacterial ribosomal DNA (B).
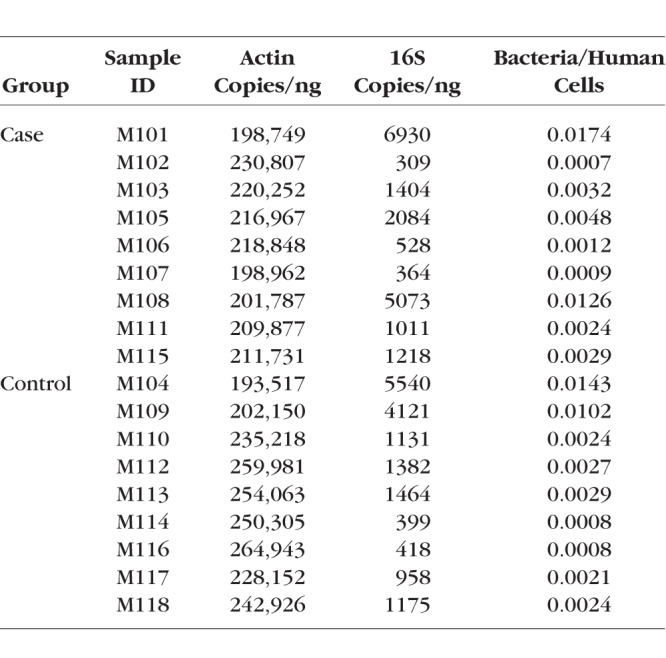
Table 3 shows the qPCR results in actin and 16S copies/ng, as well as the ratio of bacteria to human cells for each subject. There was no significant difference in the bacteria/human cell ratio between groups (Table 3; case average: 0.0052 ± 0.0056, control average: 0.0043 ± 0.0047; P = 0.74). Even though individual variance was observed in bacterial loads, the proportions of human DNA findings were consistent between subjects.
Table 3.
Quantitative PCR Actin and 16S Bacterial rDNA Copies/ng and the Ratio of Bacteria to Human Cells in All Subjects
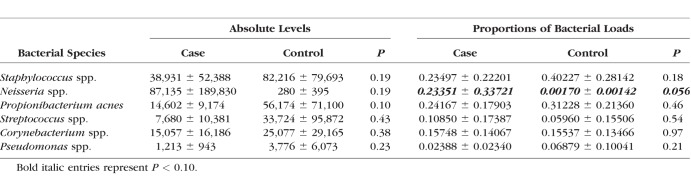
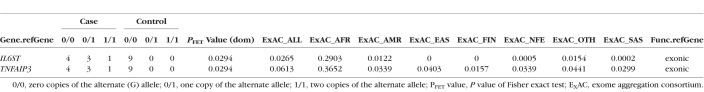
All samples yielded good sequences from BRiSK, and 263,550 tags were recovered, of which 99.1% mapped to human (mammalian) sequences. The distribution of the human sequences per chromosome had an r2 of 0.988 from expected, indicating high correlation. Tables 4 and 5 show the BRiSK and 16S results. Limited contamination was observed in the negative controls (see Supplementary Tables S1 and S2 for absolute levels of 16S bacterial rDNA results and the proportions of bacteria loads in all subjects). No significant differences were found in absolute levels or proportions of bacterial loads in the six major genera (Table 5) except for the proportion of Neisseria. The Neisseria genera was marginally higher in the case versus control group (P = 0.056); however, 16S bacterial rDNA results tend to be unreliable in extremely paucibacterial samples, therefore, the differences in Neisseria proportion between groups would need to be confirmed by directed qPCR.
Table 4.
BRiSK Reads for Human, Bacteria, and Virus for Control Subjects and Case
Table 5.
Group Mean and SD in Absolute Levels and Proportions of Bacterial Loads Using 16S Bacterial rDNA Analysis
Whole Exome Sequencing Results
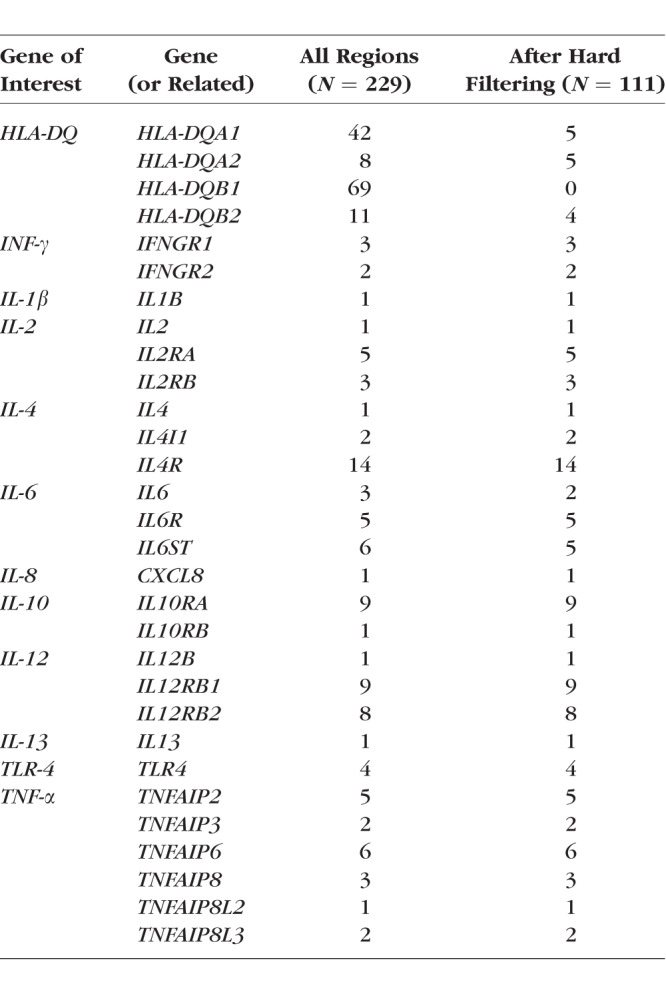
A total of 2,097,409 variants were available. After filtering on mapping quality (MQ) >30.0 and a maximum missing value of 0.5, 794,187 variants remained, and 767,401 had available annotation data. Variants were restricted to those with exonic function within the prespecified regions of interest (IL-1β, IL-2, IL-4, IL-6, IL-8, IL-10, IL-12, IL-13, HLA-DQ, TLR-4, TNF-α, and related genes) including a total of 229 exonic variants, 111 of which satisfied all filters (Table 6).
Table 6.
Examined Variants by Gene Region Before and After Hard Filtering
Two variants reached a nominal significance threshold (PFET < 0.05; Table 7). rs2228046 (PFET = 0.0294) is a missense single nucleotide variation (SNV) in IL6ST transcript variant 1 mRNA, located on chromosome 5q11.2 (position 55954899) that results in an Ile→Thr substitution at amino acid position 454 of exon 11. Three cases had one copy of the alternate allele, and one case had two copies of the alternate allele. Zero control subjects had copies of the alternate allele. rs2230926 (PFET = 0.0294) is a missense SNV in Homo sapiens TNFα-induced protein 3 (TNFAIP3), transcript variant 1, mRNA, located on chromosome 6q23.3 (position 137874929) that results in an Phe→Cys substitution at amino acid position 127 of exon 3. Three cases had one copy of the alternate allele, and one case had two copies of the alternate allele. Zero control subjects had copies of the alternate allele. Allele frequencies across varying populations included in the ExAC browser (http://exac.broadinstitute.org/; the Broad Institute), as well as predictions from various additional prediction sources for these variants, as annotated by ANNOVAR, are in Tables 7 and 8.
Table 7.
Single Nucleotide Variants in Genes of Interest With Suggestive PFET < 0.05
Table 8.
Single Nucleotide Variants in Genes of Interest With Suggestive PFET < 0.05
Correlations Between Exome Sequencing, Tear Cytokines, and Microbiome Results
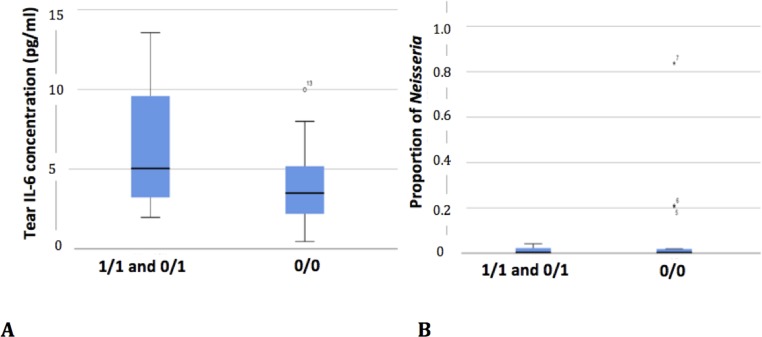
Subjects with a present (1/1 or 0/1) or absent (0/0) allele variation in IL-6 were plotted against the statistically significant tear cytokine and ocular microbiome data (Fig. 2). Given the pilot nature of the study, statistical tests were not performed; however, the direction of the tear cytokine concentration is in line with the hypothesis that a deleterious SNP in IL-6 receptor would lead to subsequent overproduction of cytokines at the ocular surface. There was no clear relationship between the genetic variation and Neisseria levels.
Figure 2.
The concentration of IL-6 in tears (A) and proportion (B) of Neisseria spp. isolated from ocular swabs in people homologous or heterologous for the IL-6ST variant. 1/1, those homologous for mutation; 1/0, those heterologous for the mutation; 0/0, subjects who possess the normal gene; box, interquartile range and median; whiskers, 1.5 times interquartile range. Outliers are indicated.
Discussion
The first aim of this pilot study was to demonstrate the ability to collect high-quality ocular and blood samples from subjects and send the samples for processing at remote sites. Nine subjects with a history of CIE and nine matched controls meeting all of the prespecified entry criteria were successfully enrolled. All enrolled subjects completed all study procedures; however, an adequate blood sample volume could not be obtained from one subject. The tears, ocular surface cultures, and blood samples obtained were of high quality and able to be processed by multiplex assay, BRISK/16srDNA, and whole exome sequencing.
The second aim of the study was to explore differences in key genetic exome sequences, the ocular surface microbiome, and tear inflammatory proteins between contact lens wearers with and without a history of CIEs. The sample population enrolled was matched by age, sex, and contact lens material and modality. Slit-lamp examination revealed that subjects had no active clinical signs of inflammation. The reported contact lens wear and care behaviors and environmental exposures associated with CIEs have been reported previously,41–43 and this pilot study was not powered to analyze these factors.
No significant difference in ocular surface microbiome was shown between groups in this sample of 18 soft contact lens wearers. A previous study using traditional culturing techniques has suggested an increased risk of harboring Gram-negative bacteria with contact lens wear.44 There have been only a few reports of the ocular surface microbiome by deep sequencing, which is known to show more diverse bacterial findings than traditional culturing methods.25,27,44 The ocular surface of contact lens wearer in this study harbored primarily Staphylococcus, Neisseria, Propionibacterium, Streptococcus, and Corynebacterium spp. This is in agreement with previous studies conducted in our laboratories that quantified the ocular surface microbiome of non–contact lens wearers using similar techniques.24,26 Shin et al. reported higher levels of Methylobacterium, Lactobacillus, Acinetobacter, and Pseudomonas spp. and lower levels of Haemophilus, Streptococcus, Staphylococcus, and Corynebacterium spp. among 9 soft contact lens wearers compared with 11 non–contact lens wearers.45 Zhang et al.46 reported no statistically significant differences in the ocular surface microbiome of 12 soft contact lens wearers compared with 12 non–contact lens wearers; however, they suggested levels of Delftia spp. may be decreased and Elizabethkingia spp. increased in contact lens wearers.
The presence of resident viruses on the ocular surface is even less well understood. Two case subjects and one healthy control harbored the torque teno virus (TTV). TTV was previously reported on the ocular surface of healthy adults in our study of healthy non–contact lens wearers.26 Given TTVs association with seasonal hyperacute panuveitis, culture-negative endophthalmitis and ability to induce an adaptive immune response, further research is needed to explore potential clinical implications of resident TTV on the ocular surface.
Differences in the ocular surface microbiome have been reported by region, sex, and age.24–26 There is some evidence of a minimal core microbiome.20,24 The potential effects of daily disposable or reusable contact lens wear, and use of hydrogen peroxide or multipurpose solution are yet to be fully explored; however, a study using similar techniques suggests that the ocular surface microbiome of contact lens wearers with or without a history of CIE may not be substantially different from each other or from non–lens wearers.46
The tear cytokine analysis showed higher levels of IL-6 in case subjects versus controls. Previous work has shown no significant day to day variation in IL-6 after discontinuing contact lens wear in healthy subjects, nor changes in IL-6 with one continuous day and night of hydrogel or silicone hydrogel contact lens wear among established contact lens wearers.32,47–49 Increases in IL-6 have been associated with the use of certain multipurpose cleaning solutions,50 and reuseable wearers have been shown to have higher levels of IL-6 than daily disposable soft contact lens wearers.51 Poyraz et al.52 examined changes in tear cytokines when neophytes were first fitted with either hydrogel or silicone hydrogel contact lenses and found early increases in IL-6. Taken together, these findings could indicate an initial upregulation of the ocular inflammatory status occurs on initiation of lens wear, which may become chronic in some wearers, especially those using multipurpose cleaners and reuseable lenses. A previous study also explored individual inflammatory cytokine responses of established wearers and showed a few subjects with much larger than average changes in IL-6 with overnight contact lens wear.49 An exaggerated or long-term inflammatory response to contact lens wear may be an early signal that some patients are predisposed to ocular inflammatory complications and are not good candidates for extended or continuous wear lenses.
Whole exome sequencing analysis revealed a significant variation in SNPs in IL-6 signal transducer for cases versus controls. We found that rs2228046 is predicted to be deleterious by the SIFT prediction tool and to be possibly damaging by the PolyPhen-2 prediction tool. The overall allele frequency in the ExAC is 0.2903 for this variant. rs2230926 is predicted to be tolerated by the SIFT prediction tool and to be benign by the PolyPhen-2 prediction tool. The overall allele frequency in the ExAC is 0.0613 for this variant. Carnt et al.10 found that variations in rs1800795 and rs1800797 in the IL-6 promoter region were associated with greater risk of and more severe microbial keratitis in a cohort of more than 2500 contact lens wearers. Importantly, these genetic variations were not present in cases with sterile (versus microbial) keratitis.10 In this study, all four cases with IL-6 variations in SNPs were African Americans. Coe et al.53 found that African Americans had higher serum IL-6 and lower soluble IL-6 receptor concentrations than Japanese Americans and whites. They suggested that higher body mass index (BMI) was a significant confounder related to higher levels of IL-6 in African Americans53 and that further research was needed in this area. No previous study has reported variations in IL-6 by race or ethnicity, and there is no reported difference in risk of CIE by race/ethnicity. Obesity has been identified as a risk factor for inflammatory disease. A larger, more diverse study sample with balanced racial groups and information on BMI is needed to explore the relationship between race/ethnicity/BMI and genetic variations in IL-6.
IL-6 is a pleotropic mediator with both pro- and anti-inflammatory properties.54,55 It has been shown to be directly involved in both the defense and homeostatic maintenance of the ocular surface.14 IL-6 helps regulate the initial acute phase of the inflammatory response by recruiting polymorphonuclear cells (PMNs) and also the later clearing of inflammation via lowering the production of IL-1 and TNF.56 Thakur et al.57 used a mouse model to demonstrate that concentrations of IL-6 varied significantly by type of Pseudomonas aeruginosa infecting the cornea. Cole et al.58 showed more severe Pseudomonas keratitis in the absence of IL-6. Hume et al.59 found that administration of exogenous IL-6 during infection with Staphylococcus aureus in mice decreased the numbers of Staphylococci and improved outcomes. Although not found in this pilot study, mutations in IL-6 could affect the diversity and density of bacterial loads on the ocular surface. CLPU is commonly associated with S. aureus colonization of lenses,6,23,60 whereas CLARE is commonly associated with Gram-negative, including P. aeruginosa bacterial colonization of contact lenses.23,61,62 However, as only three CLPU (presumed Gram positive) and six CLARE (presumed Gram negative) subjects were included in this study, subanalysis by bacterial type could not be conducted. Larger studies are needed to examine the potential effect of IL-6 SNPs on specific corneal infiltrative events and microbial colonization.
SNP variants in IL-6 have been associated with chronic inflammatory diseases including arthritis, diabetes, and cardiovascular disease.63–65 IL-6 and IL-6 receptors bind together with the signal-transducing subunit (also known as gp130).56 Impaired IL-6 trans-signaling (gp130) can lead to impairment of bacterial response including impaired influx of monocytes, excessive retention of neutrophils, and greater tissue damage.54,56 Thus, the SNP variants in IL-6ST found in case subjects in this study may support an increased inflammatory response and increased risk for the development of CIEs or other microbial infections. Longitudinal cohort studies would be needed to identify the direct causal relationship between possession of IL-6 SNPs, concentration of IL-6 in tears, and the risk of ocular surface infection or inflammation.
There are limitations to interpretation of the study results due to the small sample size. All 18 subjects were recruited from a single urban site and included a mix of contact lens wearers (i.e., material, modality, solution use) and races/ethnicities. In this pilot study, four of the nine cases were African American, whereas none of the controls were African American. Future studies with racially balanced samples are required to further explore potential associations between SNP variants and the ocular surface cytokine and bacterial levels. Variations in genes may lead to production of more or less proteins at the ocular surface, thus affecting the ocular inflammatory state and response. It is likely that changes in ocular surface proteins may also affect the ocular surface microbiome, especially because certain cytokines are known to have antimicrobial functions.
In summary, in this small pilot study, we demonstrated feasibility for measuring multiple internal and external risk factors for CIEs using a multisite collection and analysis protocol. Contact lens wear can serve as a useful “trigger” to elicit ocular surface inflammatory responses, as contact lenses are the most common cause of CIEs in the United States.4 Further research in large, diverse populations, enrolled from multiple geographic sites, is necessary to understand the complex relationship between variations in genetic profiles, ocular surface protein expression, and the ocular surface microbiome and how such changes may predispose patients to ocular surface infection and inflammation.
Supplementary Material
Acknowledgments
The authors thank Jonathan Haines, PhD, Tyler G. Kinzy, and Ernest R. Chan, PhD, at Case Western Reserve University for laboratory and technical support for the whole exome sequencing analysis.
Supported by a Johnson & Johnson Vision Care, Inc. investigator-initiated study grant (KR), SUNY Schnurmacher Institute for Vision Research pilot grant (KR, CC, MW), National Institutes of Health Grants R01EY022038 (RVG) and P30EY001730 (RVG), an unrestricted grant from Research to Prevent Blindness (RVG), and National Institutes of Health National Center for Advancing Translational Science (NCATS) Grant KL2TR000440 (JCB).
Disclosure: C. Chao, None; L. Akileswaran, None; J.N. Cooke Bailey, None; M. Willcox, None; R. Van Gelder, None; C. Lakkis, Johnson & Johnson Vision Care, Inc. (E); F. Stapleton, Alcon Ltd. (F), Allergan Ltd. (F), Coopervision (F), Menicon (F); K. Richdale, None
References
- 1.Stapleton F, Keay L, Jalbert I, Cole N. The epidemiology of contact lens related infiltrates. Optom Vis Sci. 2007;84:257–272. doi: 10.1097/OPX.0b013e3180485d5f. [DOI] [PubMed] [Google Scholar]
- 2.Sankaridurg PR, Sweeney DF, Sharma S, et al. Adverse events with extended wear of disposable hydrogels: results for the first 13 months of lens wear. Ophthalmology. 1999;106:1671–1680. doi: 10.1016/S0161-6420(99)90346-9. [DOI] [PubMed] [Google Scholar]
- 3.Collier SA, Gronostaj MP, MacGurn AK, et al. Estimated burden of keratitis: United States, 2010. Morb Mortal Wkly Rep. 2014;63:1027–1030. [PMC free article] [PubMed] [Google Scholar]
- 4.Cope JR, Collier SA, Rao MM, et al. Contact lens wearer demographics and risk behaviors for contact lens-related eye infections: United States, 2014. Morb Mortal Wkly Rep. 2015;64:865–870. doi: 10.15585/mmwr.mm6432a2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin MC, Yeh TN. Mechanical complications induced by silicone hydrogel contact lenses. Eye Contact Lens. 2013;39:115–124. doi: 10.1097/ICL.0b013e31827c77fd. [DOI] [PubMed] [Google Scholar]
- 6.Wu PZ, Thakur A, Stapleton F, Willcox MD. Staphylococcus aureus causes acute inflammatory episodes in the cornea during contact lens wear. Clin Exp Ophthalmol. 2000;28:194–196. doi: 10.1046/j.1442-9071.2000.00293.x. [DOI] [PubMed] [Google Scholar]
- 7.Szczotka-Flynn L, Lass JH, Sethi A, et al. Risk factors for corneal infiltrative events during continuous wear of silicone hydrogel contact lenses. Invest Ophthalmol Vis Sci. 2010;51:5421–5430. doi: 10.1167/iovs.10-5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robertson DM, Cavanagh HD. The clinical and cellular basis of contact lens-related corneal infections: a review. Clin Ophthalmol. 2008;2:907–917. doi: 10.2147/opth.s3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carnt NA, Willcox MD, Hau S, et al. Immune defense single nucleotide polymorphisms and recruitment strategies associated with contact lens keratitis. Ophthalmology. 2012;119:1997–2002. doi: 10.1016/j.ophtha.2012.05.020. [DOI] [PubMed] [Google Scholar]
- 10.Carnt NA, Willcox MD, Hau S, et al. Association of single nucleotide polymorphisms of interleukins-1beta, -6, and -12B with contact lens keratitis susceptibility and severity. Ophthalmology. 2012;119:1320–1327. doi: 10.1016/j.ophtha.2012.01.031. [DOI] [PubMed] [Google Scholar]
- 11.Keijser S, Kurreeman FA, de Keizer RJ, et al. IL-10 promotor haplotypes associated with susceptibility to and severity of bacterial corneal ulcers. Exp Eye Res. 2009;88:1124–1128. doi: 10.1016/j.exer.2009.01.018. [DOI] [PubMed] [Google Scholar]
- 12.Olivares M, Laparra JM, Sanz Y. Host genotype, intestinal microbiota and inflammatory disorders. Br J Nutr. 2013;109(suppl 2):S76–S80. doi: 10.1017/S0007114512005521. [DOI] [PubMed] [Google Scholar]
- 13.Thakur A, Willcox MD. Contact lens wear alters the production of certain inflammatory mediators in tears. Exp Eye Res. 2000;70:255–259. doi: 10.1006/exer.1999.0767. [DOI] [PubMed] [Google Scholar]
- 14.Chao C, Richdale K, Jalbert I, Doung K, Gokhale M. Non-invasive objective and contemporary methods for measuring ocular surface inflammation in soft contact lens wearers: a review. Cont Lens Anterior Eye. 2017;40:273–282. doi: 10.1016/j.clae.2017.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Poyraz C, Irkec M, Mocan MC. Elevated tear interleukin-6 and interleukin-8 levels associated with silicone hydrogel and conventional hydrogel contact lens wear. Eye Contact Lens. 2012;38:146–149. doi: 10.1097/ICL.0b013e3182482910. [DOI] [PubMed] [Google Scholar]
- 16.Thakur A, Willcox MD. Cytokine and lipid inflammatory mediator profile of human tears during contact lens associated inflammatory diseases. Exp Eye Res. 1998;67:9–19. doi: 10.1006/exer.1998.0480. [DOI] [PubMed] [Google Scholar]
- 17.Lema I, Duran JA, Ruiz C, Diez-Feijoo E, Acera A, Merayo J. Inflammatory response to contact lenses in patients with keratoconus compared with myopic subjects. Cornea. 2008;27:758–763. doi: 10.1097/ICO.0b013e31816a3591. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalez-Perez J, Villa-Collar C, Sobrino Moreiras T, et al. Tear film inflammatory mediators during continuous wear of contact lenses and corneal refractive therapy. Br J Ophthalmol. 2012;96:1092–1098. doi: 10.1136/bjophthalmol-2012-301527. [DOI] [PubMed] [Google Scholar]
- 19.Lakkis C, Ames S, Connellan PA, Banbury LK, Morris CA. 5th International Conference on the Tear Film & Ocular Surface: Basic Science and Clinical Relevance. Taormina, Italy: Tear Film & Ocular Surface Society;; 2007. Assessment of cytokine levels in the tears of contact lens wearers and non-contact lens wearers. [Google Scholar]
- 20.Ueta M, Kinoshita S. Ocular surface inflammation mediated by innate immunity. Eye Contact Lens. 2010;36:269–281. doi: 10.1097/ICL.0b013e3181ee8971. [DOI] [PubMed] [Google Scholar]
- 21.Dong Q, Brulc JM, Iovieno A, et al. Diversity of bacteria at healthy human conjunctiva. Invest Ophthalmol Vis Sci. 2011;52:5408–5413. doi: 10.1167/iovs.10-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Willcox MD. Characterization of the normal microbiota of the ocular surface. Exp Eye Res. 2013;117:99–105. doi: 10.1016/j.exer.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Willcox M, Sharma S, Naduvilath TJ, Sankaridurg PR, Gopinathan U, Holden BA. External ocular surface and lens microbiota in contact lens wearers with corneal infiltrates during extended wear of hydrogel lenses. Eye Contact Lens. 2011;37:90–95. doi: 10.1097/ICL.0b013e31820d12db. [DOI] [PubMed] [Google Scholar]
- 24.Ozkan J, Nielsen S, Diez-Vives C, Coroneo M, Thomas T, Willcox M. Temporal stability and composition of the ocular surface microbiome. Sci Rep. 2017;7:9880. doi: 10.1038/s41598-017-10494-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wen X, Miao L, Deng Y, et al. The influence of age and sex on ocular surface microbiota in healthy adults. Invest Ophthalmol Vis Sci. 2017;58:6030–6037. doi: 10.1167/iovs.17-22957. [DOI] [PubMed] [Google Scholar]
- 26.Doan T, Akileswaran L, Andersen D, et al. Paucibacterial microbiome and resident dna virome of the healthy conjunctiva. Invest Ophthalmol Vis Sci. 2016;57:5116–5126. doi: 10.1167/iovs.16-19803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Muthappan V, Lee AY, Lamprecht TL, et al. Biome representational in silico karyotyping. Genome Res. 2011;21:626–633. doi: 10.1101/gr.115758.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee AY, Akileswaran L, Tibbetts MD, Garg SJ, Van Gelder RN. Identification of torque teno virus in culture-negative endophthalmitis by representational deep DNA sequencing. Ophthalmology. 2015;122:524–530. doi: 10.1016/j.ophtha.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam D, Richdale K, Kinoshita B, et al. Repeatability of the contact lens risk survey (E-Abstract 135144) Optom Vis Sci. 2013;90 [Google Scholar]
- 30.Chalmers RL, Wagner H, Mitchell GL, et al. Age and other risk factors for corneal infiltrative and inflammatory events in young soft contact lens wearers from the Contact Lens Assessment in Youth (CLAY) study. Invest Ophthalmol Vis Sci. 2011;52:6690–6696. doi: 10.1167/iovs.10-7018. [DOI] [PubMed] [Google Scholar]
- 31.Terry RL, Schnider CM, Holden BA, et al. CCLRU standards for success of daily and extended wear contact lenses. Optom Vis Sci. 1993;70:234–243. doi: 10.1097/00006324-199303000-00011. [DOI] [PubMed] [Google Scholar]
- 32.Chao C, Golebiowski B, Stapleton F, Richdale K. Changes in tear cytokine concentrations following discontinuation of soft contact lenses-a pilot study. Eye Contact Lens. 2016;42:237–243. doi: 10.1097/ICL.0000000000000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chao C, Stapleton F, Zhou X, Chen S, Zhou S, Golebiowski B. Structural and functional changes in corneal innervation after laser in situ keratomileusis and their relationship with dry eye signs and symptoms. Graefe's Arch Clin Exp Ophthalmol. 2015;253:2029–2039. doi: 10.1007/s00417-015-3120-1. [DOI] [PubMed] [Google Scholar]
- 34.Bron AJ, Evans VE, Smith JA. Grading of corneal and conjunctival staining in the context of other dry eye tests. Cornea. 2003;22:640–650. doi: 10.1097/00003226-200310000-00008. [DOI] [PubMed] [Google Scholar]
- 35.Masoudi S, Zhao Z, Stapleton F, Willcox M. Contact lens–induced discomfort and inflammatory mediator changes in tears. Eye Contact Lens. 2017;43:40–45. doi: 10.1097/ICL.0000000000000237. [DOI] [PubMed] [Google Scholar]
- 36.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 37.Schloss PD, Westcott SL, Ryabin T, et al. Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van der Auwera GA, Carneiro MO, Hartl C, et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protocol Bioinformatics. 2013;43:11–33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.RCoreTeam. A Language and Environment for Statistical Computing. Vienna, Austria: RCoreTeam; 2017. [Google Scholar]
- 40.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Richdale K, Lam DY, Wagner H, et al. Case-control pilot study of soft contact lens wearers with corneal infiltrative events and healthy controls. Invest Ophthalmol Vis Sci. 2016;57:47–55. doi: 10.1167/iovs.15-18512. [DOI] [PubMed] [Google Scholar]
- 42.Wagner H, Richdale K, Mitchell GL, et al. Age, behavior, environment, and health factors in the soft contact lens risk survey. Optom Vis Sci. 2014;91:252–261. doi: 10.1097/OPX.0000000000000164. [DOI] [PubMed] [Google Scholar]
- 43.Sorbara L, Zimmerman AB, Mitchell GL, et al. Multicenter testing of a risk assessment survey for soft contact lens wearers with adverse events: a contact lens assessment in youth study. Eye Contact Lens. 2018;44:21–28. doi: 10.1097/ICL.0000000000000305. [DOI] [PubMed] [Google Scholar]
- 44.Boost M, Cho P, Wang Z. Disturbing the balance: effect of contact lens use on the ocular proteome and microbiome. Clin Exp Optom. 2017;100:459–472. doi: 10.1111/cxo.12582. [DOI] [PubMed] [Google Scholar]
- 45.Shin H, Price K, Albert L, Dodick J, Park L, Dominguez-Bello MG. Changes in the eye microbiota associated with contact lens wearing. mBio. 2016;7:e00198. doi: 10.1128/mBio.00198-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Zhao F, Hutchinson DS, et al. Conjunctival microbiome changes associated with soft contact lens and orthokeratology lens wearing. Invest Ophthalmol Vis Sci. 2017;58:128–136. doi: 10.1167/iovs.16-20231. [DOI] [PubMed] [Google Scholar]
- 47.Willcox MD, Zhao Z, Naduvilath T, Lazon de la Jara P. Cytokine changes in tears and relationship to contact lens discomfort. Mol Vis. 2015;21:293–305. [PMC free article] [PubMed] [Google Scholar]
- 48.Thakur A, Willcox MD. Contact lens wear alters the production of certain inflammatory mediators in tears. Exp Eye Res. 2000;70:255–259. doi: 10.1006/exer.1999.0767. [DOI] [PubMed] [Google Scholar]
- 49.Duong K, Chao C, Willcox M, Richdale K. Changes in tear cytokines following a short period of daily and overnight silicone hydrogel lens wear. J Contact Lens Res Sci. 2017;1:3–11. [Google Scholar]
- 50.Kalsow CM, Reindel WT, Merchea MM, Bateman KM, Barr JT. Tear cytokine response to multipurpose solutions for contact lenses. Clin Ophthalmol. 2013;7:1291–1302. doi: 10.2147/OPTH.S44642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chao C, Stapleton F, Willcox MDP, Golebiowski B, Richdale K. Preinflammatory signs in established reusable and disposable contact lens wearers. Optom Vis Sci. 2017;94:1003–1008. doi: 10.1097/OPX.0000000000001129. [DOI] [PubMed] [Google Scholar]
- 52.Poyraz C, Irkec M, Mocan MC. Elevated tear interleukin-6 and interleukin-8 levels associated with silicone hydrogel and conventional hydrogel contact lens wear. Eye Contact Lens. 2012;38:146–149. doi: 10.1097/ICL.0b013e3182482910. [DOI] [PubMed] [Google Scholar]
- 53.Coe CL, Love GD, Karasawa M, et al. Population differences in proinflammatory biology: Japanese have healthier profiles than Americans. Brain Behav Immun. 2011;25:494–502. doi: 10.1016/j.bbi.2010.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Scheller J, Chalaris A, Schmidt-Arras D, Rose-John S. The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim Biophys Acta. 2011;1813:878–888. doi: 10.1016/j.bbamcr.2011.01.034. [DOI] [PubMed] [Google Scholar]
- 55.Heinrich PC, Behrmann I, Haan S, Hermanns HM, Muller-Newen G, Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1–20. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rose-John S, Winthrop K, Calabrese L. The role of IL-6 in host defence against infections: immunobiology and clinical implications. Nat Rev Rheumatol. 2017;13:399–409. doi: 10.1038/nrrheum.2017.83. [DOI] [PubMed] [Google Scholar]
- 57.Thakur A, Xue M, Stapleton F, Lloyd AR, Wakefield D, Willcox MD. Balance of pro- and anti-inflammatory cytokines correlates with outcome of acute experimental Pseudomonas aeruginosa keratitis. Infect Immunity. 2002;70:2187–2197. doi: 10.1128/IAI.70.4.2187-2197.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cole N, Bao S, Stapleton F, et al. Pseudomonas aeruginosa keratitis in IL-6-deficient mice. Int Arch Allergy Immunol. 2003;130:165–172. doi: 10.1159/000069006. [DOI] [PubMed] [Google Scholar]
- 59.Hume EB, Cole N, Garthwaite LL, Khan S, Willcox MD. A protective role for IL-6 in staphylococcal microbial keratitis. Invest Ophthalmol Vis Sci. 2006;47:4926–4930. doi: 10.1167/iovs.06-0340. [DOI] [PubMed] [Google Scholar]
- 60.Jalbert I, Willcox MD, Sweeney DF. Isolation of Staphylococcus aureus from a contact lens at the time of a contact lens-induced peripheral ulcer: case report. Cornea. 2000;19:116–120. doi: 10.1097/00003226-200001000-00023. [DOI] [PubMed] [Google Scholar]
- 61.Holden BA, La Hood D, Grant T, et al. Gram-negative bacteria can induce contact lens related acute red eye (CLARE) responses. CLAO J. 1996;22:47–52. [PubMed] [Google Scholar]
- 62.Sankaridurg PR, Sharma S, Willcox M, et al. Bacterial colonization of disposable soft contact lenses is greater during corneal infiltrative events than during asymptomatic extended lens wear. J Clin Microbiol. 2000;38:4420–4424. doi: 10.1128/jcm.38.12.4420-4424.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu Y, Berthier-Schaad Y, Fallin MD, et al. IL-6 haplotypes, inflammation, and risk for cardiovascular disease in a multiethnic dialysis cohort. J Am Soc Nephrol. 2006;17:863–870. doi: 10.1681/ASN.2005050465. [DOI] [PubMed] [Google Scholar]
- 64.Valdes AM, Arden NK, Tamm A, et al. A meta-analysis of interleukin-6 promoter polymorphisms on risk of hip and knee osteoarthritis. Osteoarthritis Cartilage. 2010;18:699–704. doi: 10.1016/j.joca.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 65.Kristiansen OP, Mandrup-Poulsen T. Interleukin-6 and diabetes: the good, the bad, or the indifferent? Diabetes. 2005;54(suppl 2):S114–S124. doi: 10.2337/diabetes.54.suppl_2.s114. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.