Abstract
Spinal muscular atrophy (SMA) is an autosomal-recessive pediatric neurodegenerative disease characterized by selective loss of spinal motor neurons. It is caused by mutation in the survival of motor neuron 1 , SMN1 , gene and leads to loss of function of the full-length SMN protein. microRNAs (miRNAs) are small RNAs that are involved in post-transcriptional regulation of gene expression. Prior studies have implicated miRNAs in the pathogenesis of motor neuron disease. We hypothesized that motor neuron-specific miRNA expression changes are involved in their selective vulnerability in SMA. Therefore, we sought to determine the effect of SMN loss on miRNAs and their target mRNAs in spinal motor neurons. We used microarray and RNAseq to profile both miRNA and mRNA expression in primary spinal motor neuron cultures after acute SMN knockdown. By integrating the miRNA:mRNA profiles, a number of dysregulated miRNAs were identified with enrichment in differentially expressed putative mRNA targets. miR-431 expression was highly increased, and a number of its putative mRNA targets were significantly downregulated in motor neurons after SMN loss. Further, we found that miR-431 regulates motor neuron neurite length by targeting several molecules previously identified to play a role in motor neuron axon outgrowth, including chondrolectin. Together, our findings indicate that cell-type-specific dysregulation of miR-431 plays a role in the SMA motor neuron phenotype.
Introduction
Spinal muscular atrophy (SMA) is a severe neurodegenerative disease that results in degeneration and cell death of lower motor neurons in the spinal cord. It is the leading genetic cause of infant mortality with an incidence of 1:11 000 births ( 1 ). SMA is caused by homozygous deletions or mutations involving the survival of motor neuron 1 ( SMN1 ) gene that encodes the SMN protein ( 2 ). SMN protein is ubiquitously expressed in all cells of the body very early in development. In neurons, SMN has been found to play roles in snRNP biogenesis and alternative splicing as part of the SMN complex ( 3 ). Loss of SMN leads to aberrant splicing ( 4 , 5 ) and widespread transcriptional changes in several SMA models ( 6–10 ). In neurites, SMN is involved in trafficking of RNA binding proteins and local translation of target mRNAs ( 11–16 ). In vitro loss of SMN leads to significant deficits in motor neuron viability and neurite outgrowth ( 17–19 ). While SMN is ubiquitously expressed, the selective vulnerability of motor neurons to cell death in SMA remains an important and unanswered question. microRNAs (miRNAs) are a class of small non-coding RNAs, which are approximately 22 nucleotides in length and are endogenous mediators of RNA silencing. They function by translational repression and decay of target mRNAs via complementary binding to sequences in the 3′UTR. miRNAs are able to regulate a number of targets simultaneously and may work on whole networks of genes to modify protein expression ( 20 ). In spinal motor neurons, miRNAs are essential regulators of key facets of development ( 21 ), differentiation ( 22 , 23 ), synaptic morphology ( 24 ) and axon outgrowth ( 25 ). Interestingly, abolishing proper miRNA biogenesis through deletion of Dicer in post-mitotic spinal motor neurons leads to an SMA-like neurodegenerative phenotype causing motor neuron dysfunction and cell death ( 26 ). This indicates that miRNAs in mature spinal motor neurons are necessary for cell health and survival.
Dysregulation of RNA processing and miRNA expression has been identified as a common feature of motor neuron diseases such as SMA and amyotrophic lateral sclerosis (ALS) ( 27 ). Recent work in ALS has shown that aberrant miRNA expression contributes to disease pathophysiology and can be an important target for intervention ( 28–30 ). To date, studies of miRNA expression in SMA have identified differentially expressed miRNAs in primary cortical neurons ( 31 ) and increased expression of miR-9 in embryonic stem cell-derived motor neurons from an SMA mouse model ( 26 ). We hypothesized that cell-type-specific expression patterns of miRNAs in motor neurons, the cell type most affected by SMN loss, play an important role in cellular morphology and viability of motor neurons in SMA.
In this study, we report dysregulation of 17 miRNAs in primary motor neuron cultures following acute SMN loss, in particular miR-431. Together with transcriptome profiling of mRNA by RNA-seq, we determined the impact of miRNA expression changes on their putative target genes using Weighted Gene Coexpression Network Analysis (WGCNA). This analysis identified a set of miR-431 target genes that are significantly downregulated and are highly correlated with acute SMN knockdown in spinal motor neurons. Exogenous overexpression of miR-431 expression inhibits motor neuron neurite outgrowth in vitro . Inhibition of miR-431 in SMN-deficient neurons is sufficient to rescue the motor neuron neurite length phenotype. We show that chondrolectin (Chodl), a type 1 transmembrane protein and member of the c-type lectin domain-containing family ( 32 ), is a direct target of miR-431. Chodl has been shown to regulate motor neuron axon outgrowth. This therefore suggests that spinal motor neuron-specific dysregulation of Chodl expression by miR-431 overexpression is involved in the deficits of axonal outgrowth in SMA.
Results
miRNA expression profiling in an in vitro primary motor neuron model of SMA
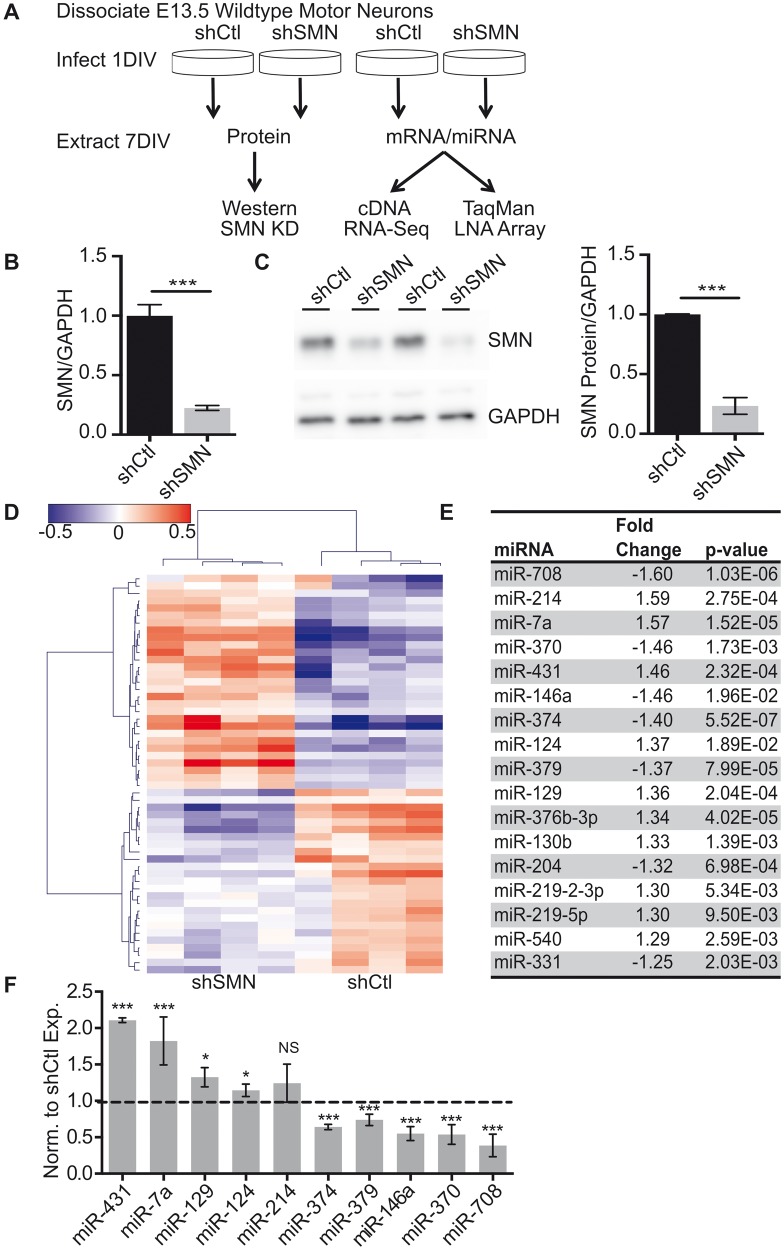
We utilized an in vitro primary spinal motor neuron system to evaluate miRNA expression. Wild-type embryonic rat spinal motor neurons were cultured as previously described ( 13 ), and motor neuron identity confirmed by immunostaining ( Supplementary Material, Fig S1A–C ). Cultures were infected with lentiviral-shRNA specific to SMN (shSMN) or a control (shCtl) hairpin sequence on day 1 in vitro (1DIV). SMN knockdown was assessed by both Western blot and quantitative reverse transcriptase polymerase chain reaction (qRT-PCR) 6 days after viral infection (7DIV) ( Fig. 1 A). These studies revealed an 80% and 75% reduction in SMN mRNA and protein levels, respectively ( Fig. 1 B and C). At 7DIV, there was no significant change in the motor neuron marker choline acetyltransferase or a marker of endoplasmic reticulum stress induction, C/EBP homologous protein ( Supplementary Material, Fig. S1D and E ) and we did not observe an upregulation of cleaved caspase 3, a marker of apoptosis, until 9DIV consistent with prior data ( 31 ). Therefore, we chose the 7DIV time point allowing us to focus on identification of early drivers of motor neuron phenotypes independent of cell death.
Figure 1.
miRNA expression changes in spinal motor neurons after acute SMN knockdown. ( A ) Schematic of experimental paradigm for SMN knockdown experiments. shRNA-mediated knockdown significantly decreases SMN mRNA (in B ) and protein (in C ) after 6 days of infection with shSMN or shCtl, normalized to Gapdh expression. N = 8 from four biological replicates, *** P < 0.001, unpaired two-tailed Student’s t -test. ( D ) LNA-based miRNA microarray identified 56 differentially expressed miRNAs ranked by hierarchical clustering following loss of SMN expression. ( E ) Table of top differentially expressed miRNA with P -value <0.02 after multiple corrections and > 1.25-fold change. ( F ) qRT-PCR validation of expression changes of the top 10 miRNAs identified by microarray normalized to miR-191 and to shCtl expression. N = 6 from four different experiments. * P < 0.05, *** P < 0.001 by unpaired two-tailed Student’s t -test. Data are represented as mean ± SEM.
Our previous work in cortical neurons showed significant dysregulation of miRNA expression after SMN loss ( 31 ). We hypothesized that motor neuron-specific changes in the miRNA expression may contribute to the SMA phenotype and in part underlie the specific vulnerability of this cell type to loss of SMN. We used LNA-based miRNA microarray analysis (Exiqon) to measure miRNA expression profiles in control versus SMN knockdown primary motor neurons. After normalization, we identified 54 miRNAs as differentially expressed using a two-sided t -test and Benjamini–Hochberg multiple hypothesis correction ( Fig. 1 D; Supplementary Material, Table S1 ). There was an even distribution between miRNAs that were upregulated and downregulated, suggesting that there was no alteration in overall miRNA biogenesis or stability in this model. We found 17 differentially expressed miRNAs that had a >1.25-fold change and a P- value <0.02 ( Fig. 1 E). qRT-PCR validation in SMN knockdown motor neurons revealed that 9 of the 10 most differentially expressed miRNAs identified by sequencing were altered in the expected direction with a magnitude comparable to the microarray results ( Fig. 1 F). miRNA expression was normalized to miR-191 due to its stability in the array samples (S1F-G).
miR-431 expression changes are motor neuron specific
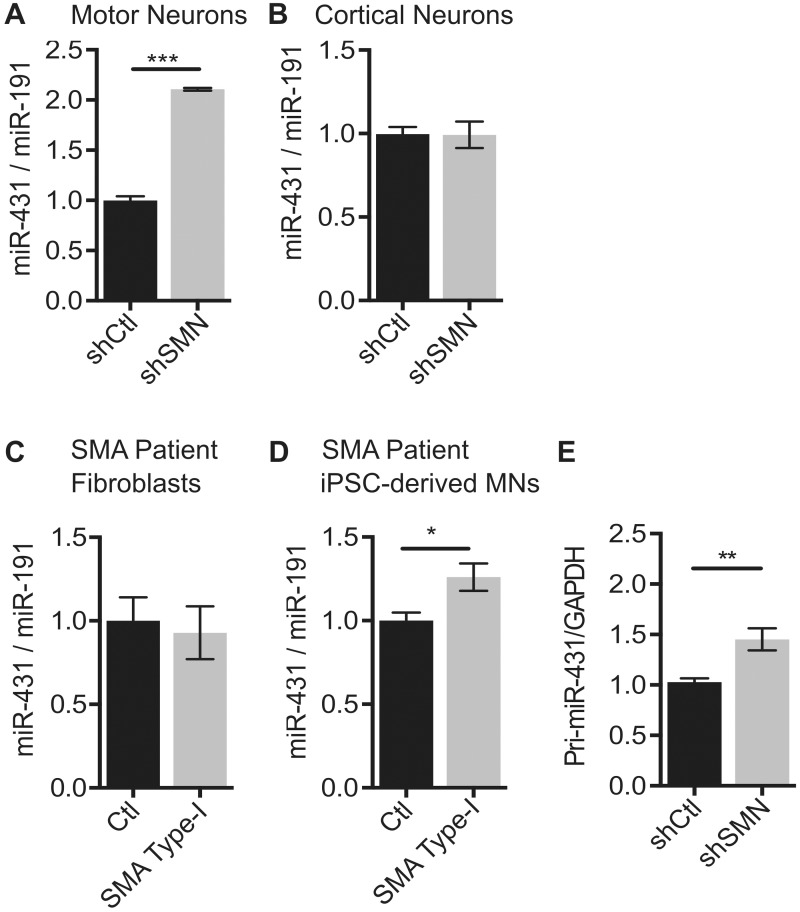
Among the 17 miRNAs identified in the array, we focused on miR-431 because it was the most highly dysregulated miRNA ( Figs 1 F and 2A ) and was previously reported to be expressed in the brain and spinal cord during embryonic development ( 33 ). To rule out off-target effects of our shRNA hairpin, we used two additional hairpins targeting separate regions of the Smn sequence to knockdown SMN protein in motor neurons and found that miR-431 is overexpressed in response to those hairpins as well ( Supplementary Material, Fig. S2A–C ). To further determine the extent of miR-431 dysregulation in SMA, we measured miR-431 expression in several different cell types. Cultured primary cortical neurons were infected with lentivirus expressing the same shRNA for SMN at 1DIV. RNA and protein were collected to confirm SMN knockdown ( Supplementary Material, Fig. S2D and E ). Unlike SMN knockdown in motor neurons, SMN knockdown in cortical neurons did not induce aberrant expression changes in miR-431 ( Fig. 2 B). While we detected an increased trend in miR-431 expression in P4 lumbar spinal cords and E13 whole spinal cords from the SMNΔ7 mouse model, it did not reach statistical significance ( Supplementary Material, Fig. S2F and G ). This may be because expression changes observed in miR-431 are motor neuron specific, and motor neurons only comprise approximately 5% of the spinal cord ( 34 ).
Figure 2.
Increased miR-431 expression is specific to SMN deficient motor neurons. ( A ) qRT-PCR of miR-431 expression in SMN knockdown motor neurons ( N = 8 from 4 experiments), ( B ) cortical neurons ( N = 6 from 3 experiments), ( C ) SMA patient fibroblasts ( N = 6 from 2 control and 2 SMA patient cell lines) and ( D ) SMA patient iPSC-derived motor neurons ( N = 4 from two separate differentiations). ( E ) Pri-miR-431 expression normalized to GAPDH control in SMN knockdown motor neurons ( N = 6 from 4 experiments). Significance was determined by unpaired Student’s test * P < 0.05, ** P < 0.01, and *** P < 0.001. Data are represented as mean ± SEM.
To determine the relevance of our findings to human disease, we assayed miR-431 expression in human SMA patient samples. Importantly, in unsorted severe type I SMA patient iPSC-derived motor neurons, there is a significant increase in miR-431 ( Fig. 2 D). These motor neurons only account for 20–30% of the total cells in culture after differentiation. Therefore, the smaller magnitude of change in the mixed culture could reflect a larger change in the human SMA motor neurons that is diluted by other neuronal subtypes and glia in the cultures. In contrast, miR-431 expression does not increase in human SMA type I patient fibroblasts when compared with control fibroblasts ( Fig. 2 C; Supplementary Material, Fig. S2H ). Together these findings suggest that the increase in miR-431 expression in response to SMN-deficiency is motor neuron specific.
To determine whether the increase in miR-431 was due to biogenesis or stability, we measured the primary miR-431 transcript (pri-miR-431). Pri-miR-431 was significantly increased ( Fig. 2 E), suggesting that the mechanism underlying the miR-431 increase may be partially dependent on increased primary transcription. It is not yet known whether alterations in RNA processing or stability contribute to the increased miR-431 levels.
mRNA expression profiling with RNA-Seq
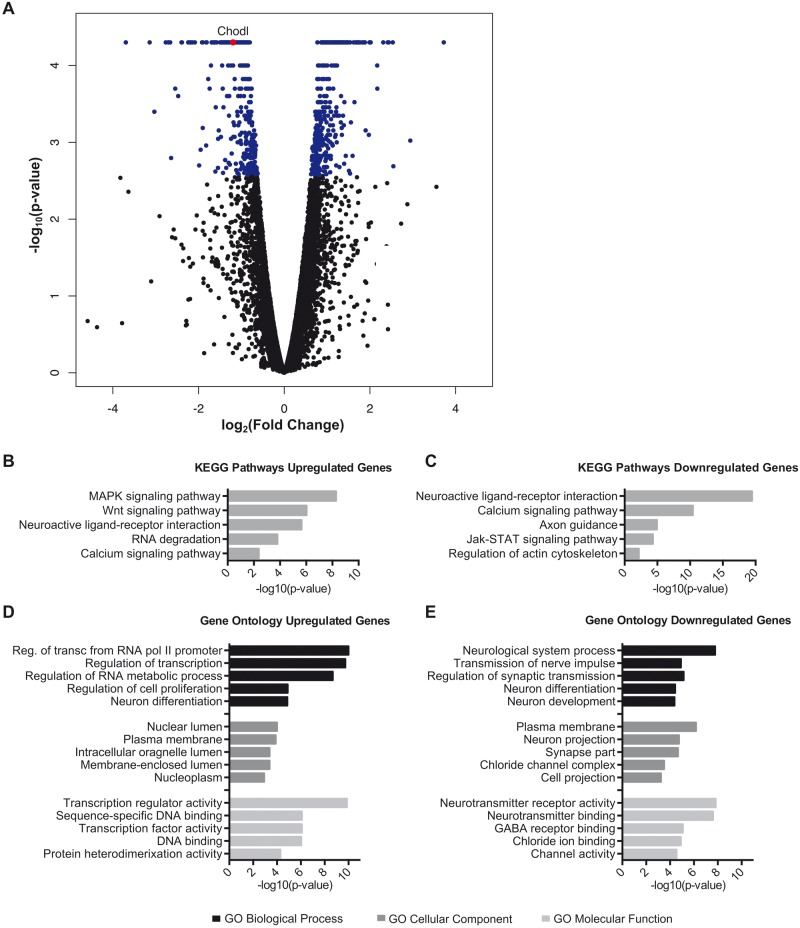
We hypothesized that miRNA changes that impact motor neuron phenotypes are reflected in target gene expression. While aberrant alternative splicing is a feature of SMA, miRNAs effect target expression by mRNA decay. Therefore, we focused on identifying transcriptional changes by RNA-Seq in our in vitro motor neuron model. RNA from four biological replicates each of shCtl- and shSMN-infected primary motor neurons were sequenced using single-end 50 bp reads. Mapping and differential analysis were performed using the Tuxedo suite of tools ( 35 ). The total raw read count was between 20 and 30 million reads per sample, and an average of 60% of reads per sample mapped to the Rn5 rat transcriptome ( Supplementary Material, Fig. S3C ). Overall mRNA expression and density of reads were not significantly altered between the shSMN and shCtl conditions ( Supplementary Material, Fig. S3A ). After filtering low expression transcripts with fewer than 10 reads, 13,943 genes were detected. Differential expression analysis revealed that 748 transcripts were differentially expressed in shSMN motor neurons versus controls with a corrected P -value <0.05. Of the differentially expressed transcripts in the SMN knockdown neurons, 391 were upregulated and 356 were downregulated when compared with controls ( Fig. 3 A; Supplementary Material, Fig. S3B and Table S2 ) indicating no overall direction of transcriptional change.
Figure 3.
mRNA transcriptome profiling in SMN knockdown motor neurons. ( A ) Volcano plot of log 2 (fold change) versus –log 10 ( P- value) of all 13,943 genes detected above threshold shows a subset of differentially expressed genes but no change in overall gene expression. Differentially expressed transcripts are indicated in blue, corrected P- value <0.05. The DAVID bioinformatics tool was used to complete Pathway and GO analysis of differentially expressed genes. KEGG pathway analysis shows the most significant biological pathways enriched in the ( B ) upregulated and ( C ) downregulated genes. ( D ) GO analysis of enriched terms in upregulated and ( E ) downregulated genes. Significance was determined using minimum number of hits = 2 and Bonferroni P < 0.05.
Next, by pathway analysis, we tested whether the differentially expressed transcripts are enriched in biological pathways that have potential implications in SMA. In particular, genes in the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathways of ‘Neuroactive Ligand-Receptor Interaction’, ‘Axon Guidance’ and ‘Calcium Signaling Pathway’ may relate to the altered excitability and neurite morphology found in SMA motor neurons ( 17 ) ( Fig. 3 B and C). Moreover, Gene Ontology (GO) analysis revealed upregulation of a number of genes related to RNA metabolism and transcription ( Fig. 3 D), which is consistent with the role of the SMN complex in RNA processing. Conversely, GO analysis of the downregulated genes identified a number of cellular components and molecular functions related to neurite development and synaptic transmission ( Fig. 3 E). Comparison of the differentially expressed transcripts shows overlap of several GO categories, as well as a number of specific genes found in transcriptome profiling of other SMA models ( 6–10 ). These data demonstrate that our model displays perturbations in known pathways and processes related to SMN loss of function.
Integration of miRNA: mRNA expression data
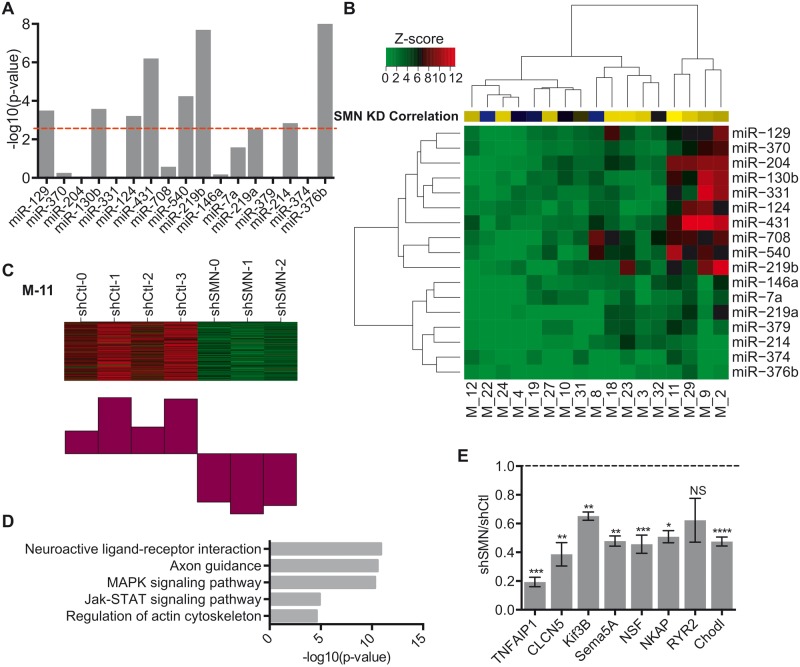
We next sought to integrate the gene and miRNA expression data to identify the potential effects of miRNA dysregulation on the transcriptome. We utilized the PITA algorithm ( 36 ) to identify putative miRNA binding sites within transcripts detected by RNA-seq. We then evaluated whether the putative target transcripts were enriched among genes whose expression changed in the opposite direction of the miRNA of interest using Gene Set Enrichment Algorithm (GSEA) ( 37 ). GSEA analysis showed that of the top 17 differentially expressed miRNAs, eight show significant enrichment in putative mRNA targets after correction for multiple comparisons (miR-376b, miR-431, miR-124, miR-219b, miR-129, miR-540, miR-214, miR-130b) with the top three miRNAs being miR-431, miR-376b and miR-219b ( Fig. 4 A). GSEA analysis of miRNAs with targets changing in the same direction did not show any enrichment for these eight miRNAs ( Supplementary Material, Fig. S4A ) supporting the specificity of the interaction.
Figure 4.
WGCNA of miRNA effect of co-expressed putative target genes. ( A ) Bar plot of P -values demonstrating the enrichment of PITA targets using GSEA. P -values were transformed using −log 10 and correction for multiple comparisons was performed using a Bonferroni correction P < 0.0025 (red line). ( B ) Heatmap of z -scores for miRNA target enrichment in WGCNA. Yellow–blue Heatmap indicates correlation with SMN knockdown for each module. Interactions with modules were significant at z -score > 3.6 corresponding to P < 0.00017. ( C ) Heatmap and corresponding chart of downregulated gene expression of shCtl versus shSMN RNA-Seq samples in Module-11. ( D ) Bar plots of GO analysis of Module 11 genes. Significance was determined by minimum number of hits = 2 and Bonferroni P < 0.05. ( E ) qRT-PCR of relative mRNA expression of nine putative miR-431 targets in shCtl vs. shSMN. N = 4 from four experiments, * P -value < 0.05, ** P < 0.01, *** P < 0.001, unpaired two-tailed Student’s t -test. Data are represented as mean ± SEM.
To evaluate gene expression from a network perspective and gain further insight into the mechanisms by which miRNA changes might be influencing gene expression, we performed WGCNA ( 38 ). WGCNA identified 34 modules of co-expressed genes. To assess the correlation of each module to the miRNA expression profile, the PITA algorithm identified predicted targets within each module for the top 17 differentially expressed miRNAs. The number of targets for each module:miRNA combination was compared with a null distribution and expressed as a z score ( Fig. 4 B). Interestingly, the targets of the most highly upregulated miRNA, miR-431, were also the most significantly enriched in modules associated with downregulation of expression, M-2, M-9, M-29 and M-11 ( Fig. 4 C; Supplementary Material, Fig. S4B–D ).
Module-11 was of particular interest because it contained a set of genes with decreased expression and was most highly correlated with SMN knockdown ( r = 0.97, Fig. 4 C). Disruptions in motor neuron axon and neurite outgrowth are a hallmark of a number of SMA models ( 17 , 18 , 39–41 ). Pathway and GO analysis of Module-11 revealed significant enrichment in a number of genes related to morphology including Actin Cytoskeleton Organization, Axon Guidance and Neuronal and Cell Projections ( Fig. 4 D; Supplementary Material, Fig. S4E ). Neuronal excitability is also perturbed in SMA in part due to abnormal expression of axonal calcium channels ( 17 ). GO analysis also revealed enrichment in molecular function categories related to aberrant excitability including ‘Ion Channel Activity’. These results indicate that the network of genes co-expressed in Module-11 is highly relevant to SMA phenotypes. Furthermore, the interaction of miR-431 with specific target genes in Module-11 suggests that miR-431-dependent regulation may underlie dysregulation of these pathways.
Expression of miR-431 target mRNAs is decreased by SMN loss in motor neurons
To further characterize the genes within Module-11, we used the module eigengene-based measure of connectivity ( kME )( 42 ). We identified 833 genes with significant connectivity within Module-11 ( kME > 0.75). Out of those 833 genes, we found that 450 were predicted to be the targets of miR-431. Using the results of the bioinformatic analysis of Module-11 ( Fig. 4 D; Supplementary Material, Fig. S4E ), we selected a number of genes related to neurite outgrowth and neuronal excitability including Tnfaip1 ( kME = 0.97), Clcn5 ( kME = 0.93), Kif3B ( kME = 0.96), Sema5A ( kME = 0.93), Nsf ( kME = 0.96), Nkap ( kME = 0.88), Ryr2 ( kME = 0.96) and Chodl ( kME = 0.78). We confirmed that seven of the eight selected genes were significantly downregulated in SMN knockdown motor neurons by qRT-PCR ( Fig. 4 E). Interestingly, chondrolectin (Chodl) mRNA was previously shown to have roles in axon development and is reduced in mouse models of SMA ( 43 , 44 ).
Aberrant Chodl expression is specific to motor neurons
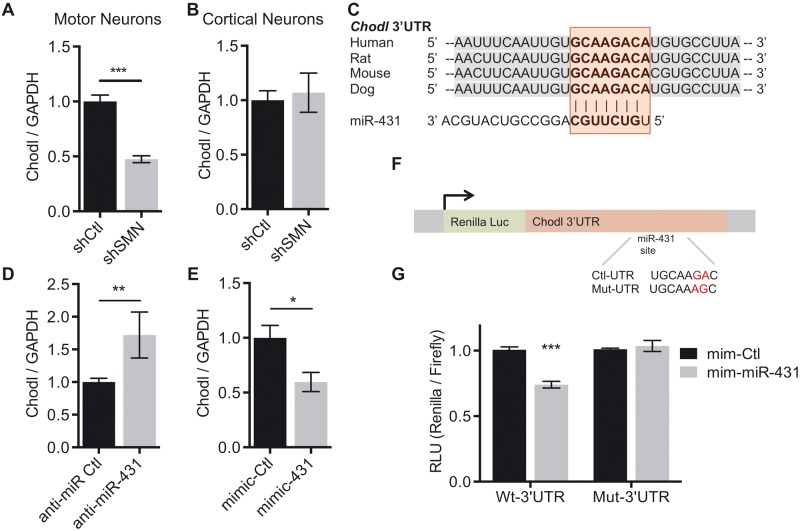
Given the high connectivity in Module-11, the predicted miR-431 interaction and the prior data suggesting a role in neurite growth, we focused on the interaction between miR-431 and chondrolectin. In the transcriptome profile (highlighted in red in Fig. 3 A), expression of Chodl was decreased by 2.29-fold with a P- value of 0.003. In qRT-PCR validation studies, Chodl expression decreased by 53% in SMN knockdown motor neurons ( Fig. 5 A). Chodl mRNA was previously shown to be alternatively spliced in a mouse model of SMA ( 4 ). Therefore, we used primers specific to Chodl variants 1 and 2 ( Chodl-001 and Chodl-002 ) and found that both Chodl variants were decreased significantly (S5A-B), indicating that splicing is not affected at this time point in our in vitro model.
Figure 5.
Chondrolectin is a direct target of miR-431 and is decreased in SMN deficient motor neurons. ( A ) qRT-PCR of Chodl confirms reduced expression in SMN knockdown motor neurons. N = 8 from 4 experiments, *** P < 0.001, unpaired two-tailed Student’s t -test. ( B ) qRT-PCR in cortical neurons shows Chodl is not reduced. N = 7 from 3 experiments. ( C ) Schematic depiction of the evolutionary conservation of the miR-431 binding site (highlighted in red) in the Chodl 3′UTR in mammals. The site is predicted by both the TargetScan −0.29 and PITA −10.17 algorithms. ( D ) Transfection of anti-miR-431 in motor neurons increases Chodl expression. N = 6 from 3 experiments, ** P < 0.01, **** P < 0.0001, unpaired two-tailed Student’s t -test. ( E ) Transfection of a miR-431 mimic in motor neurons decreases expression of Chodl . N = 8 from 4 experiments. * P < 0.05, unpaired two-tailed Student’s t -test. ( F ) Schematic of control and mutated (highlighted in red) luciferase constructs of the binding site in the Chodl 3′UTR. ( G ) Relative Renilla /firefly luciferase expression following miR-431 mimic expression in Chodl3′UTR and Chodl3′UTRmut psicheck-2 transfected HEK293 cells at 24 h. Wild-type Chodl3′UTR N = 7 from 3 experiments. Chodl3′UTRmut N = 5 from two experiments. Significance was determined by two way ANOVA *** P < 0.001. Data are represented as mean ± SEM.
To determine whether the reduction in Chodl was a general effect of SMN loss or whether it is motor neuron specific, Chodl expression after acute shSMN infection in primary cortical neurons was also measured ( Fig. 5 B). While we detected expression of both miR-431 and Chodl , neither total Chodl nor either transcript variant was decreased in cortical neurons ( Supplementary Material, Fig. S5B ). This finding indicates that the loss of Chodl expression is not a feature of SMN knockdown in all neuronal subtypes.
Chodl is a direct target of miR-431
We identified Chodl as a putative miR-431 target by both the TargetScan and PITA prediction algorithms. It has a canonical eight nucleotide miR-431 binding site in the 3′UTR ( Fig. 5 C). To confirm the functionality of this interaction, we transfected the wild-type motor neurons with an anti-miR to inhibit miR-431 expression. In the presence of anti-miR-431 inhibitor, Chodl showed increased expression when compared with control ( Fig. 5 D; Supplementary Material, Fig. S5C ). Further, we transfected wild-type primary motor neurons with a miR-431 mimic and found that Chodl mRNA is significantly decreased in response to miR-431 mimic when compared with control ( Fig. 5 E; Supplementary Material, Fig. S5D ). This modulation is not specific to the transcript variants as they share the 3′UTR binding site; therefore, both Chodl-001 and Chodl-002 are reduced by miR-431 expression ( Supplementary Material, Fig. S5E ). We were unable to resolve Chodl protein by western blot following miR-431 modulation due to low total expression of Chodl and lack of working antibodies; therefore, the impact of miR-431 on Chodl protein expression remains a question for future studies.
We then investigated direct targeting of miR-431 to Chodl via the predicted 3′UTR site using a luciferase reporter assay. The full-length Chodl 3′UTR sequence was ligated to a Renilla luciferase reporter to measure 3′UTR activity ( Fig. 5 F). We transfected either control or Chodl 3′UTR-containing vector into HEK293 cells with either a mimic-control or mimic-miR-431 for 24 h and measured the luciferase activity. Overexpression of the miR-431 mimic decreased the Chodl 3′UTR luciferase activity to 0.71 ± 0.058 RLU of the control. This reduction was abolished when two nucleotides in the seed region of the 3′UTR miR-431 binding site were mutated ( Fig. 5 G). Together, these data indicate that there is a direct interaction between miR-431 and the chodl 3′UTR at this site.
Phenotypic consequences of miR-431 expression changes in motor neurons
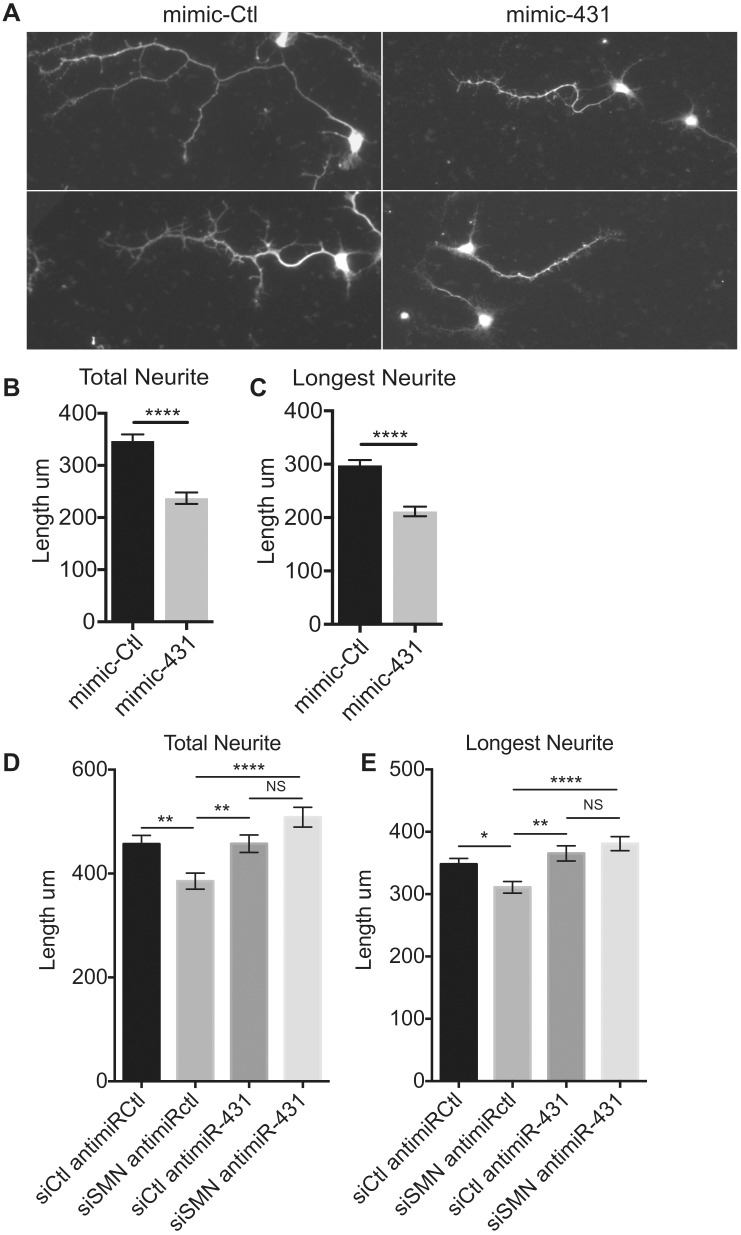
To observe the effect of aberrant expression, miR-431 mimic or control mimic was transfected into wild-type primary motor neuron cultures at 1DIV for 72 h. The neurons were fixed at 4DIV and stained with anti-tau antibody to observe neurite outgrowth and morphology ( Fig. 6 A). At this time point in culture, axons have not been fully specified; therefore, we measured primary neurites longer than three times the width of the cell body and any branches. Quantification of neurite length revealed a significant decrease in tau-positive neurite length after transfection of miR-431 mimic ( Fig. 6 B and C). In our previous work, we showed that the loss of SMN achieved by transfection of an siRNA against SMN (siSMN) inhibits the total neurite outgrowth by 40% and the length of the longest neurite by 30% ( 31 ). Here we see a 27% decrease in both total neurite length and in longest primary neurite length, when compared with the control neurites. These data support that exogenously increasing miR-431 expression inhibits neurite outgrowth. Conversely, inhibition of miR-431 expression by anti-miR-431 transfection did not have an effect on neurite length in wild-type motor neurons ( Supplementary Material, Fig. S6A and B ).
Figure 6.
Inhibition of miR-431 expression after acute SMN knockdown ameliorates the motor neuron neurite outgrowth phenotype. ( A ) Two representative images of Tau staining neurites in low density cultured wild-type spinal motor neurons transfected with 30 nM mimic-Ctl or mimic-431 for 72 h and fixed at 4DIV. Neurite tracing of ( B ) the total neurite and ( C ) longest Tau-stained neurite with a minimum cutoff of 3× the length of the cell body. N > 148 from 3 experiments. Mann–Whitney, **** P < 0.0001. ( D ) Antimir inhibition of miR-431 in SMN knockdown motor neurons rescues both total neurite length and ( E ) the longest neurite length. N > 160 from 3 experiments. Kruskal–Wallis with Dunns Multiple Comparisons post-test, * P < 0.05, ** P < 0.01, **** P < 0.0001. Data are represented as mean ± SEM.
To determine whether correcting miR-431 expression would ameliorate the SMA-like neurite outgrowth phenotype, we transfected the neurons with siRNA against SMN (siSMN) and control anti-miR or anti-miR-431. Knockdown of SMN by siSMN transfection ( Supplementary Material, Fig. S6C and D ) lead to a significant decrease in both the total neurite and the longest neurite length compared with controls ( Fig. 6 C and D). Inhibition of miR-431 by anti-miR in the siSMN-transfected motor neurons fully rescued both the total and the longest neurite length ( Fig. 6 D and E). This indicates that modulation of miR-431 expression is capable of rescuing the SMA motor neuron neurite length phenotype.
Discussion
A number of miRNAs highly expressed in the central nervous system have been implicated in motor neuron disease ( 30 ). Work in SMN mutant mouse embryonic stem cell-derived motor neurons ( 26 ) and SMN knockdown cortical neurons ( 31 ) showed that miRNA expression in these cells is affected by SMN loss. Our earlier work found that expression of a specific miRNA, miR-183, is increased in cortical neurons, motor neurons, fibroblasts and spinal cords in both cellular and animal models of SMA. However, follow-up analysis in our motor neuron model revealed cell-type-specific differences in miRNA expression following SMN knockdown. miRNAs are differentially expressed in distinct cell types, which leads to differential target regulation. Cellular context is crucial and can affect miRNA target interactions in several ways including differential availability of 3′UTR sites, altered expression of RNA binding proteins aiding or inhibiting miRNA repression and alternative polyadenylation ( 45 ). Transcriptome profiling in SMA motor neuron models has uncovered altered expression of a number of genes involved in ER stress ( 10 ), synapses ( 6 , 8 ), ribosomal RNA binding ( 9 ) and development ( 7 ). The aim of this study was to identify changes in miRNA expression in spinal motor neurons due to a loss of Smn and determine the functional impact of these changes on gene expression and motor neuron function.
We identified multiple miRNAs that were dysregulated by acute SMN knockdown in motor neurons and focused on miR-431. miR-431 has been reported to have high expression in the brain and spinal cord during embryonic development ( 33 ). In sensory neurons of the dorsal root ganglion, miR-431 was found to be upregulated after crush injury and to stimulate axon regeneration ( 46 ). Outside of the nervous system, it is involved in skeletal muscle myogenesis ( 47 , 48 ). Here, we demonstrate the specificity of miR-431 dysregulation in motor neurons following SMN loss. Further, miR-431 was differentially expressed in type I SMA patient iPSC-derived motor neurons but not fibroblasts. Therefore, our data indicate that aberrant miR-431 expression is a common feature of both rodent and human models of SMA. Consequently, miR-431 loss may be important for the progression of human disease in motor neurons.
The SMN protein forms a complex with Gemin3-7 and participates in the assembly of snRNPs, which are crucial for RNA processing ( 3 ). Distinct from the SMN complex, Gemin3 and Gemin4 can interact with argonaute-2 in the RNA induced silencing complex ( 49 , 50 ) and may control its turnover ( 51 ). This implies that SMN loss could affect miRNA processing by alteration of SMN complex components. We found a moderate increase in levels of pri-miR-431 RNA, suggesting that increased transcription or stabilization of the transcript could mediate the upregulation after SMN knockdown, but the precise mechanism is not yet known. Recent evidence in developing muscle shows miR-431 expression is mediated by myostatin signaling and found that Stat1 and interferon-stimulated response elements (ISRE) are potential transcriptional regulators ( 52 ). Previous work on dysregulation of miR-183 in our cortical neuron model did not reveal a change in its transcription ( 31 ). Therefore, we believe that dysregulation of miRNA expression in SMA may be influenced by both primary transcription and mature miRNA stability and is an intriguing question for future studies.
Integration of miRNA and mRNA profiles is a powerful technique to identify the impact of miRNA expression on individual targets, as well as larger gene networks. Several algorithms exist for integration of miRNA and mRNA profiles ( 53 ). In this study, we used a combination of the PITA algorithm to predict targets of specific miRNAs and WGCNA to organize gene expression into co-expression modules. We chose to focus on Module-11 because it was most highly correlated with SMN knockdown (0.967) and was significantly enriched for predicted targets of miR-431. There are several other co-expression modules that are significantly enriched for targets of dysregulated miRNAs, and these other miRNA:mRNA interactions may also contribute to the motor neuron SMA phenotype. Further investigation of these miRNAs and their targets may increase understanding of the dysregulation of these gene networks in SMA.
Several genes with high connectivity to Module-11 are involved in regulating axonal morphology, signaling and cell health including tumor necrosis factor alpha-induced protein 1 ( TNFAIP1 ) ( 54 , 55 ), Semaphorin 5a ( Sema5a ) ( 56 ) and Chodl . Chodl was of particular interest because it is a key regulator of motor axon outgrowth in vivo ( 44 ). miR-431 expression was not significantly increased in SMNΔ7 spinal cords at E13 or P4; however, decreased expression of Chodl has been reported in several SMA motor neuron models ( 6 , 7 , 9 ) but not in kidney or muscle ( 4 ). While it is expressed elsewhere in the body ( 57 ), in the spinal cord Chodl is a specific marker for a sub-population of large fast-spiking alpha motor neurons ( 58 ). In SMA, there is evidence of early denervation of fast alpha motor neurons in muscle biopsies ( 59 ). Notably, in ALS models, these neurons are particularly vulnerable to cell death ( 60 ), and Chodl expression is lost early in disease progression ( 61 ). Further, the rescue of Chodl expression has been shown to ameliorate the motor axon phenotype in a model of SMA ( 43 ). Here, we show that Chodl expression is directly regulated by miR-431 via interaction with the Chodl 3′UTR and that dysregulation of Chodl by miR-431 is specific to SMN-deficient motor neurons. These studies identify Chodl as an important regulator of motor neuron cell health and provide evidence of its misregulation in motor neuron disease.
In this study, we demonstrate that miR-431 is a regulator of motor neuron neurite outgrowth in vitro . Exogenous miR-431 overexpression significantly decreases motor neuron neurite length by a similar magnitude previously characterized in in vitro models of SMN loss ( 17 , 18 , 31 ). Importantly, inhibition of miR-431 overexpression ameliorates the SMN-induced neurite outgrowth deficit returning neurite length to wild-type levels. We hypothesize that miR-431 inhibits neurite outgrowth through downregulation of the target genes that we have identified, including Chodl . Inhibition of aberrant miR-431 expression impacts not only Chodl but also other dysregulated putative targets including those identified in our GSEA and WGCNA analyses. Therefore, manipulation of miR-431 function provides an attractive target in SMA because it can modulate several targets and have a profound impact on motor neuron axons and cell health that cannot be achieved by rescue of a single gene. Together, these studies provide evidence for the role of miR-431 in SMA pathogenesis through the downregulation of multiple target genes involved in axon outgrowth.
Materials and Methods
Primary neuron culture
All experimental procedures were performed in compliance with the Boston Children’s Hospital IACUC animal protocol no. 1207227R. Motor neurons were prepared as previously described in ( 13 ). Briefly, the ventral spinal cords of wild-type E13.5 Long Evans rat embryos (Charles River) were dissected, trypsinized at 37 °C and dissociated via mechanical trituration with DNAse and 0.4% BSA then passed through a 100 µm filter to remove debris. Cells were suspended in motor neuron media containing NB media supplemented with B27 (Life Technologies), l -glutamine and penicillin/streptomycin with additional growth factors CNTF (50 ng/µl), BDNF (50 ng/µl) and GDNF (25 ng/µl) (Peprotech). Primary cortical neurons were isolated as previously described ( 62 ). Briefly, cortices of E17 rats are isolated in HBSS with 10 m m Hepes, 10 m m MgCl 2 and 1 m m kynurenic acid. Cells are dissociated for 5 min at 37 °C in Papain (30 U/ml) followed by mechanical trituration. Neurons were plated in NB/B27. Neurons were then plated on 6 or 12-well plastic TC dishes coated with Poly- d -Lysine (PDL) or on glass coverslips in 24-well dishes coated with PDL and laminin. shCtl and shSMN virus ( 63 ) were produced by the Boston Children’s Hospital virus core and neurons were infected as previously described ( 31 ).
Exiqon LNA miRNA microarray data analysis
One microgram of total RNA isolated using the mirVANA kit was sent to Exiqon for analysis using the seventh generation miRCURY LNA miRNA Array no. 208500. Four shCtl and four shSMN knockdown samples were used for the miRNA microarray. RNA quality was assessed on a Bioanalyzer and the RIN numbers of all eight samples were >8.8. miRNAs were labeled with miRCURY LNA Hi-Power Labeling Kit (Hy3/Hy5) and hybridized to the array, scanned and the data returned after quality control and filtering based on spike in controls and background intensity levels.
Log2-transformed data were normalized across samples by a scaling factor and the addition of a constant on median-filtered data to remove outliers. One of the shCtl samples was found to have overall decreased hybridization quality and was thus removed for the rest of the analysis. miRNA passenger strands and all miRNAs with expression levels inferior to five were filtered from the probe list. This left a list of 250 miRNAs detected in the array above this threshold. Changes in miRNA expression were identified by two-sided t -test and Benjamini–Hochberg multiple hypothesis correction.
mRNA expression profiling by RNAseq
RNA profiling was completed at the Harvard Biopolymers Core. For library construction, 500 ng isolated mRNA was placed into the Wafergen Apollo, and the PrepX Complete ILMN DNA Library Kit (Wafergen Cat 400039) was used according to the manufacturer’s protocol. Samples were run on the PCR machine with indexed primers, and then cleaned using the ‘PCR clean’ on the Wafergen Apollo according to the manufacturer’s instructions. Quality control was performed on Agilent 2200 Tape Station on a D1000 High Sensitivity Tape and qPCR analysis using the SYBR green KAPA SYBR® FAST Universal 2X qPCR Master Mix (Kapa Biosystems, Cat#KK4602) and primers to the P5 and P7 regions of the Illumina adapters. Diluted PhiX was used to generate a standard curve to determine concentration. Samples were clustered and sequenced on a single lane of an Illumina HiSeq2500 for a 50-cycle 50-bp single end run using protocols HCS 1.5.15.1—RTA 1.13.48. The RNAseq reads were assessed using CASAVA 1.8.2 (Illumina) for demultiplexing indexed samples and quality control with FastQC. RNAseq alignment, assembly and differential analysis were performed on the Harvard Orchestra high performance-computing cluster. The data were analyzed with the Tuxedo suite of tools using the protocol described in ( 35 ). Rat reference sequence information and annotation was obtained from Illumina iGenomes using the Rattus norvegicus assembly, Rn5.
Identification of putative miRNA target mRNA interactions
The miRNA target prediction algorithm PITA was used to identify target genes ( http://genie.weizmann.ac.il/pubs/mir07/index.html , last accessed March 25, 2016) ( 36 ) on the entire mRNA sequence using a Δ−Δ G score of less than −10 as a threshold for putative targets. Transcriptome data were filtered with a threshold of average expression >15 across all samples and a coefficient of variation >13, resulting in the selection of 9163 genes. Exact transcript boundaries for these genes were determined, and we obtained corresponding sequences for all of these transcripts from the rn5 genome.
GSEA ( 37 ) was used to determine whether the predicted PITA targets of a specific miRNA were enriched in genes that were differentially expressed in the opposite direction to the miRNA of interest. The Pearson correlation between mRNA expression and a binary vector corresponding to knockdown was calculated, and genes were ordered based on their correlation with knockdown in the opposite direction to the miRNA of interest. Putative mRNA targets of the specific miRNA were identified within this list and assigned a positive value (step-up), while the other genes were assigned a negative value (step-down). The step-up and step-down values are determined so that the sum total of the vector is zero. A running sum of the vector was determined, and the maximum value of this vector was identified. Significance was determined by calculating a null distribution of maximum running sum values using 1000 randomly ordered vectors. This distribution was fit with a skewed normal distribution, and the P- value for the maximum value of the running sum for a specific miRNA was calculated based on this distribution.
Weighted gene coexpression network analysis
The co-expression analysis was performed using WGCNA methods as described previously ( 38 ). Correlation analyses demonstrated that the two sample types (shSMN and shCtl) were separated using hierarchical clustering, but there were no clear outlier samples. We then generated an adjacency matrix by calculating the Pearson correlation between all genes, and it was scaled using a beta value of 8 to approximate scale free topology. A topological overlap matrix was calculated using the adjacency matrix ( 64 ), and genes were clustered using the average hierarchical clustering algorithm. Modules were obtained from these clusters using the dynamic tree-cutting algorithm ( 65 ).
Transcript boundaries for all genes within all modules were determined and sequences for the corresponding transcripts were obtained. The PITA algorithm was used to predict whether these sequences were targets of the 17 differentially expressed miRNAs, as determined previously. The number of targets for a specific miRNA within a specific module was determined. A null distribution was generated by determining the number of targets within a random set of mRNAs that was the same size as the specific module. A z -score was calculated by comparing the number of targets within a module to the null distribution. This was repeated for all combinations of miRNAs and modules.
Pathway and GO analysis
The Database for Annotation Visualization and Integrated Discovery (DAVID) Functional Annotation tool ( http://david.abcc.ncifcrf.gov/home.jsp , last accessed March 25, 2016) ( 66 , 67 ) was used to identify biological pathways and processes that were significantly enriched in the gene sets. The background list was composed of all genes expressed over a threshold of 15 in the RNAseq analysis. Pathways were identified as significant by DAVID default settings.
Exogenous modulation of miRNA expression
mirVANA miRNA mimics (Life Technologies) and mirVANA miRNA-inhibitors (Life Technologies) were used for miR-431 modulation experiments; 30 nM mirVANA mimic or anti-miR inhibitors for miR-431 and the corresponding negative controls were transfected into primary spinal motor neuron cultures at 1 DIV with Lipofectamine 2000 as previously described ( 31 ). Complexes were incubated with the cells for 90 min for efficient transfection of small RNA complexes without induction of motor neuron cell death. Forty-eight hours after transfection, RNA or protein was extracted. Seventy-two hours after transfection, cells in 24-well dishes on glass coverslips were fixed with 4% PFA and stained. Transfection with siGlo (Dharmacon) was used to control for transfection efficiency in the primary motor neuron cultures ( Supplementary Material, Fig. S5F ).
Chondrolectin 3′UTR luciferase assays
The full-length rat chondrolectin 3′UTR was amplified from wild-type rat total RNA using the primers: Fwd 3′-GTCTCGAGGTTGTTATTCCAATTTACAGTG-5′ and Rev 3′-GTGCGGCCGCAGAAACAAGAACTCTTTATTTGG-5′. The vector and insert were cloned into the Psi-check2 vector (Promega) by digestion with Not1 and Xho1 (New England Biolabs) in NEB buffer 3 for 2 h at 37 °C. Cut vector and insert were run on a 1% agarose gel, purified by gel purification kit (Qiagen) and ligated at 4 °C overnight with T4 Ligase Buffer (NEB). Bacteria were transformed with the ligated plasmids and sent for sequencing at the Boston Children’s Hospital IDDRC Molecular Genetics core.
Site directed mutagenesis was performed using the Quickchange II site-directed mutagenesis kit (Agilent) according to the manufacture’s protocol. Briefly, primers were designed with two mismatches at the putative miR-431 3′UTR site 5′-TCTAACTTCAATTGTGCAA ag CATGTGCCTTACAATTA-3′ Rev 5′-TAATTGTAAGGCACATG ct TTGCACAATTGAAGTTAGA-3′. pfuTurbo DNA polymerase (2.5U/µl) amplification was performed at 98 °C for 30 s, 55 °C for 1.5 min and 68 °C for 2 min for 18 cycles followed by DPN1 digestion for 1 h at 37 °C.
The Promega Dual Luciferase assay (E1910) was used according to the manufacture’s protocol. HEK293T cells were plated in 24-well dishes at 80% confluency. At 1DIV, cells were transfected with the chodl3′UTR or chodl3′UTRmut construct as well as 30 nM miR-431 mimic or control mimic. 24 h after transfection, cells were lysed for 20 min at room temperature in passive lysis buffer. Ten microliters of cell lysate were added per well of a 96-well plate in triplicate, and the plate was read for Firefly and Renilla luciferase on the Victor2 plate reader. Relative Renilla expression was determined by normalization to firefly activity.
Neurite length measurement
To quantify morphological phenotypes, cells were stained with Tau and DAPI to identify neurites and healthy nuclei respectively. Slides were imaged on the Zeiss Imager.Z2 with the CoolSnap HQ2 camera and Axiovision 4.8.2 software in the Boston Children’s Hospital IDDRC Cellular Imaging Core. Image analysis was completed in ImageJ. The NeuronJ plugin was used for neurite tracing and quantification. Neurites greater than three times the cell body (100 µM) that initiated at the cell body were designated as ‘primary’. Branches originating from the other neurites >50 µM were measured and counted as secondary or tertiary neurites.
SMA patient samples
RNA was obtained from SMA patient iPSCs differentiated into mixed cultures containing approximately 30% human spinal motor neurons. The lines used were wild type—BG SEV, SMA type I—1-38G and SMA type II—1-51N, which were previously characterized by the Rubin lab ( 10 ). SMA patient fibroblasts were obtained from the Pediatric Neuromuscular Clinical Research Network. The human fibroblast cell lines used were from patients with SMA type I and carry two copies of the SMN2 genes 1–24 and 3–11.
Supplementary Material
Acknowledgements
We are grateful to members of the Sahin lab for critical reading of the manuscript. Human tissue was obtained from the Pediatric Neuromuscular Clinical Research Network (PNCRN). The role of the PNCR tissue repository is to distribute tissue and therefore cannot endorse the studies performed or the interpretation of the results.
Conflict of Interest statement . None declared.
Funding
The Slaney Family Fund, CureSMA and Children’s Hospital Boston Translational Research Program (to M.S.) and the Children’s Hospital Boston Intellectual and Developmental Disabilities Research Center (P30 HD18655).
References
- 1. Sugarman E.A., Nagan N., Zhu H., Akmaev V.R., Zhou Z., Rohlfs E.M., Flynn K., Hendrickson B.C., Scholl T., Sirko-Osadsa D.A, Allitto B. A. ( 2012. ) Pan-ethnic carrier screening and prenatal diagnosis for spinal muscular atrophy: clinical laboratory analysis of >72,400 specimens . Eur. J. Hum. Genet ., 20 , 27 – 32 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M . et al. . ( 1995. ) Identification and characterization of a spinal muscular atrophy-determining gene . Cell , 80 , 155 – 165 . [DOI] [PubMed] [Google Scholar]
- 3. Gubitz A.K., Feng W., Dreyfuss G. ( 2004. ) The SMN complex . Exp. Cell Res ., 296 , 51 – 56 . [DOI] [PubMed] [Google Scholar]
- 4. Baumer D., Lee S., Nicholson G., Davies J.L., Parkinson N.J., Murray L.M., Gillingwater T.H., Ansorge O., Davies K.E., Talbot K. ( 2009. ) Alternative splicing events are a late feature of pathology in a mouse model of spinal muscular atrophy . PLoS Genet ., 5 , e1000773.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Liu H., Shafey D., Moores J.N., Kothary R. ( 2010. ) Neurodevelopmental consequences of Smn depletion in a mouse model of spinal muscular atrophy . J. Neurosci. Res ., 88 , 111 – 122 . [DOI] [PubMed] [Google Scholar]
- 6. Zhang Z., Pinto A.M., Wan L., Wang W., Berg M.G., Oliva I., Singh L.N., Dengler C., Wei Z., Dreyfuss G. ( 2013. ) Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy . Proc. Natl. Acad. Sci. USA ., 110 , 19348 – 19353 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Maeda M., Harris A.W., Kingham B.F., Lumpkin C.J., Opdenaker L.M., McCahan S.M., Wang W., Butchbach M.E. ( 2014. ) Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells . PLoS One , 9 , e106818.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Saal L., Briese M., Kneitz S., Glinka M., Sendtner M. ( 2014. ) Subcellular transcriptome alterations in a cell culture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation . RNA , 20 , 1789 – 1802 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Murray L.M., Beauvais A., Gibeault S., Courtney N.L., Kothary R. ( 2015. ) Transcriptional profiling of differentially vulnerable motor neurons at pre-symptomatic stage in the Smn (2b/-) mouse model of spinal muscular atrophy . Acta Neuropathol. Commun ., 3 , 55.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ng S.Y., Soh B.S., Rodriguez-Muela N., Hendrickson D.G., Price F., Rinn J.L., Rubin L.L. ( 2015. ) Genome-wide RNA-Seq of Human Motor Neurons Implicates Selective ER Stress Activation in Spinal Muscular Atrophy . Cell Stem Cell , 17 , 569 – 584 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piazzon N., Rage F., Schlotter F., Moine H., Branlant C., Massenet S. ( 2008. ) In vitro and in cellulo evidences for association of the survival of motor neuron complex with the fragile X mental retardation protein . J. Biol. Chem ., 283 , 5598 – 5610 . [DOI] [PubMed] [Google Scholar]
- 12. Tadesse H., Deschenes-Furry J., Boisvenue S., Cote J. ( 2008. ) KH-type splicing regulatory protein interacts with survival motor neuron protein and is misregulated in spinal muscular atrophy . Hum. Mol. Genet ., 17 , 506 – 524 . [DOI] [PubMed] [Google Scholar]
- 13. Akten B., Kye M.J., Hao le T., Wertz M.H., Singh S., Nie D., Huang J., Merianda T.T., Twiss J.L., Beattie C.E, Steen J. A., Sahin M. ( 2011. ) Interaction of survival of motor neuron (SMN) and HuD proteins with mRNA cpg15 rescues motor neuron axonal deficits . Proc. Natl. Acad. Sci. USA ., 108 , 10337 – 10342 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fallini C., Zhang H., Su Y., Silani V., Singer R.H., Rossoll W., Bassell G.J. ( 2011. ) The survival of motor neuron (SMN) protein interacts with the mRNA-binding protein HuD and regulates localization of poly(A) mRNA in primary motor neuron axons . J. Neurosci ., 31 , 3914 – 3925 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hubers L., Valderrama-Carvajal H., Laframboise J., Timbers J., Sanchez G., Cote J. ( 2011. ) HuD interacts with survival motor neuron protein and can rescue spinal muscular atrophy-like neuronal defects . Hum. Mol. Genet ., 20 , 553 – 579 . [DOI] [PubMed] [Google Scholar]
- 16. Fallini C., Rouanet J.P., Donlin-Asp P.G., Guo P., Zhang H., Singer R.H., Rossoll W., Bassell G.J. ( 2014. ) Dynamics of survival of motor neuron (SMN) protein interaction with the mRNA-binding protein IMP1 facilitates its trafficking into motor neuron axons . Dev. Neurobiol ., 74 , 319 – 332 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jablonka S., Beck M., Lechner B.D., Mayer C., Sendtner M. ( 2007. ) Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy . J. Cell Biol ., 179 , 139 – 149 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. ( 2003. ) Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons . J. Cell Biol ., 163 , 801 – 812 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ning K., Drepper C., Valori C.F., Ahsan M., Wyles M., Higginbottom A., Herrmann T., Shaw P., Azzouz M., Sendtner M. ( 2010. ) PTEN depletion rescues axonal growth defect and improves survival in SMN-deficient motor neurons . Hum. Mol. Genet ., 19 , 3159 – 3168 . [DOI] [PubMed] [Google Scholar]
- 20. Bartel D.P., Chen C.Z. ( 2004. ) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs . Nat. Rev. Genet ., 5 , 396 – 400 . [DOI] [PubMed] [Google Scholar]
- 21. Asli N.S., Kessel M. ( 2010. ) Spatiotemporally restricted regulation of generic motor neuron programs by miR-196-mediated repression of Hoxb8 . Dev. Biol ., 344 , 857 – 868 . [DOI] [PubMed] [Google Scholar]
- 22. Luxenhofer G., Helmbrecht M.S., Langhoff J., Giusti S.A., Refojo D., Huber A.B. ( 2014. ) MicroRNA-9 promotes the switch from early-born to late-born motor neuron populations by regulating Onecut transcription factor expression . Dev. Biol ., 386 , 358 – 370 . [DOI] [PubMed] [Google Scholar]
- 23. Otaegi G., Pollock A., Hong J., Sun T. ( 2011. ) MicroRNA miR-9 modifies motor neuron columns by a tuning regulation of FoxP1 levels in developing spinal cords . J. Neurosci ., 31 , 809 – 818 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Loya C.M., McNeill E.M., Bao H., Zhang B., Van Vactor D. ( 2014. ) miR-8 controls synapse structure by repression of the actin regulator enabled . Development , 141 , 1864 – 1874 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nesler K.R., Sand R.I., Symmes B.A., Pradhan S.J., Boin N.G., Laun A.E., Barbee S.A. ( 2013. ) The miRNA pathway controls rapid changes in activity-dependent synaptic structure at the Drosophila melanogaster neuromuscular junction . PLoS One , 8 , e68385.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Haramati S., Chapnik E., Sztainberg Y., Eilam R., Zwang R., Gershoni N., McGlinn E., Heiser P.W., Wills A.M., Wirguin I . et al. . ( 2010. ) miRNA malfunction causes spinal motor neuron disease . Proc. Natl. Acad. Sci. USA ., 107 , 13111 – 13116 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yamazaki T., Chen S., Yu Y., Yan B., Haertlein T.C., Carrasco M.A., Tapia J.C., Zhai B., Das R., Lalancette-Hebert M . et al. . ( 2012. ) FUS-SMN protein interactions link the motor neuron diseases ALS and SMA . Cell Rep ., 2 , 799 – 806 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ishtiaq M., Campos-Melo D., Volkening K., Strong M.J. ( 2014. ) Analysis of novel NEFL mRNA targeting microRNAs in amyotrophic lateral sclerosis . PLoS One , 9 , e85653.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Parisi C., Arisi I., D'Ambrosi N., Storti A.E., Brandi R., D'Onofrio M., Volonte C. ( 2013. ) Dysregulated microRNAs in amyotrophic lateral sclerosis microglia modulate genes linked to neuroinflammation . Cell Death Dis ., 4 , e959.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kye M.J., Goncalves Ido C. ( 2014. ) The role of miRNA in motor neuron disease . Front Cell Neurosci ., 8 , 15.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kye M.J., Niederst E.D., Wertz M.H., Goncalves Ido C., Akten B., Dover K.Z., Peters M., Riessland M., Neveu P., Wirth B . et al. . ( 2014. ) SMN regulates axonal local translation via miR-183/mTOR pathway . Hum. Mol. Genet ., 23 , 6318 – 6331 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weng L., Smits P., Wauters J., Merregaert J. ( 2002. ) Molecular cloning and characterization of human chondrolectin, a novel type I transmembrane protein homologous to C-type lectins . Genomics , 80 , 62 – 70 . [DOI] [PubMed] [Google Scholar]
- 33. Wheeler G., Ntounia-Fousara S., Granda B., Rathjen T., Dalmay T. ( 2006. ) Identification of new central nervous system specific mouse microRNAs . FEBS Lett ., 580 , 2195 – 2200 . [DOI] [PubMed] [Google Scholar]
- 34. Arce V., Garces A., de Bovis B., Filippi P., Henderson C., Pettmann B., deLapeyriere O. ( 1999. ) Cardiotrophin-1 requires LIFRbeta to promote survival of mouse motoneurons purified by a novel technique . J. Neurosci. Res ., 55 , 119 – 126 . [DOI] [PubMed] [Google Scholar]
- 35. Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. ( 2012. ) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks . Nat. Protoc ., 7 , 562 – 578 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kertesz M., Iovino N., Unnerstall U., Gaul U., Segal E. ( 2007. ) The role of site accessibility in microRNA target recognition . Nat. Genet ., 39 , 1278 – 1284 . [DOI] [PubMed] [Google Scholar]
- 37. Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S . et al. . ( 2005. ) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles . Proc. Natl. Acad. Sci. USA ., 102 , 15545 – 15550 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhang B., Horvath S. ( 2005. ) A general framework for weighted gene co-expression network analysis . Stat. Appl. Genet. Mol. Biol ., 4 , Article17. [DOI] [PubMed] [Google Scholar]
- 39. Van Bergeijk J., Rydel-Konecke K., Grothe C., Claus P. ( 2007. ) The spinal muscular atrophy gene product regulates neurite outgrowth: importance of the C terminus . FASEB J ., 21 , 1492 – 1502 . [DOI] [PubMed] [Google Scholar]
- 40. Dimitriadi M., Sleigh J.N., Walker A., Chang H.C., Sen A., Kalloo G., Harris J., Barsby T., Walsh M.B., Satterlee J.S . et al. . ( 2010. ) Conserved genes act as modifiers of invertebrate SMN loss of function defects . PLoS Genet ., 6 , e1001172.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kwon J.E., Kim E.K., Choi E.J. ( 2011. ) Stabilization of the survival motor neuron protein by ASK1 . FEBS Lett ., 585 , 1287 – 1292 . [DOI] [PubMed] [Google Scholar]
- 42. Langfelder P., Horvath S. ( 2007. ) Eigengene networks for studying the relationships between co-expression modules . BMC Syst. Biol ., 1 , 54.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sleigh J.N., Barreiro-Iglesias A., Oliver P.L., Biba A., Becker T., Davies K.E., Becker C.G., Talbot K. ( 2014. ) Chondrolectin affects cell survival and neuronal outgrowth in in vitro and in vivo models of spinal muscular atrophy . Hum. Mol. Genet ., 23 , 855 – 869 . [DOI] [PubMed] [Google Scholar]
- 44. Zhong Z., Ohnmacht J., Reimer M.M., Bach I., Becker T., Becker C.G. ( 2012. ) Chondrolectin mediates growth cone interactions of motor axons with an intermediate target . J. Neurosci ., 32 , 4426 – 4439 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Nam J.W., Rissland O.S., Koppstein D., Abreu-Goodger C., Jan C.H., Agarwal V., Yildirim M.A., Rodriguez A., Bartel D.P. ( 2014. ) Global analyses of the effect of different cellular contexts on microRNA targeting . Mol. Cell , 53 , 1031 – 1043 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu D., Murashov A.K. ( 2013. ) MicroRNA-431 regulates axon regeneration in mature sensory neurons by targeting the Wnt antagonist Kremen1 . Front. Mol. Neurosci ., 6 , 35.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu R., Li H., Zhai L., Zou X., Meng J., Zhong R., Li C., Wang H., Zhang Y., Zhu D. ( 2015. ) MicroRNA-431 accelerates muscle regeneration and ameliorates muscular dystrophy by targeting Pax7 in mice . Nat. Commun ., 6 , 7713.. [DOI] [PubMed] [Google Scholar]
- 48. Lee K.P., Shin Y.J., Panda A.C., Abdelmohsen K., Kim J.Y., Lee S.M., Bahn Y.J., Choi J.Y., Kwon E.S., Baek S.J . et al. . ( 2015. ) miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4 . Genes Dev ., 29 , 1605 – 1617 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mourelatos Z., Dostie J., Paushkin S., Sharma A., Charroux B., Abel L., Rappsilber J., Mann M., Dreyfuss G. ( 2002. ) miRNPs: a novel class of ribonucleoproteins containing numerous microRNAs . Genes Dev ., 16 , 720 – 728 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Murashov A.K., Chintalgattu V., Islamov R.R., Lever T.E., Pak E.S., Sierpinski P.L., Katwa L.C., Van Scott M.R. ( 2007. ) RNAi pathway is functional in peripheral nerve axons . FASEB J ., 21 , 656 – 670 . [DOI] [PubMed] [Google Scholar]
- 51. Gibbings D., Mostowy S., Jay F., Schwab Y., Cossart P., Voinnet O. ( 2012. ) Selective autophagy degrades DICER and AGO2 and regulates miRNA activity . Nat. Cell Biol ., 14 , 1314 – 1321 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Wu R., Li H., Li T., Zhang Y., Zhu D. ( 2015. ) Myostatin regulates miR-431 expression via the Ras-Mek-Erk signaling pathway . Biochem. Biophys. Res. Commun ., 461 , 224 – 229 . [DOI] [PubMed] [Google Scholar]
- 53. Muniategui A., Pey J., Planes F.J., Rubio A. ( 2013. ) Joint analysis of miRNA and mRNA expression data . Brief Bioinform ., 14 , 263 – 278 . [DOI] [PubMed] [Google Scholar]
- 54. Sailland J., Tribollet V., Forcet C., Billon C., Barenton B., Carnesecchi J., Bachmann A., Gauthier K.C., Yu S., Giguere V . et al. . ( 2014. ) Estrogen-related receptor alpha decreases RHOA stability to induce orientated cell migration . Proc. Natl. Acad. Sci. USA ., 111 , 15108 – 15113 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang C.L., Wang C., Yan W.J., Gao R., Li Y.H., Zhou X.H. ( 2014. ) Knockdown of TNFAIP1 inhibits growth and induces apoptosis in osteosarcoma cells through inhibition of the nuclear factor-kappaB pathway . Oncol. Rep ., 32 , 1149 – 1155 . [DOI] [PubMed] [Google Scholar]
- 56. Hilario J.D., Rodino-Klapac L.R., Wang C., Beattie C.E. ( 2009. ) Semaphorin 5A is a bifunctional axon guidance cue for axial motoneurons in vivo . Dev. Biol ., 326 , 190 – 200 . [DOI] [PubMed] [Google Scholar]
- 57. Weng L., Hubner R., Claessens A., Smits P., Wauters J., Tylzanowski P., Van Marck E., Merregaert J. ( 2003. ) Isolation and characterization of chondrolectin (Chodl), a novel C-type lectin predominantly expressed in muscle cells . Gene , 308 , 21 – 29 . [DOI] [PubMed] [Google Scholar]
- 58. Enjin A., Rabe N., Nakanishi S.T., Vallstedt A., Gezelius H., Memic F., Lind M., Hjalt T., Tourtellotte W.G., Bruder C . et al. . ( 2010. ) Identification of novel spinal cholinergic genetic subtypes disclose Chodl and Pitx2 as markers for fast motor neurons and partition cells . J. Comput. Neurol ., 518 , 2284 – 2304 . [DOI] [PubMed] [Google Scholar]
- 59. Dubowitz V. ( 1978. ) Muscle disorders in childhood . Major Probl. Clin. Pediatr ., 16 , iii – xiii . 1–282. [PubMed] [Google Scholar]
- 60. Kanning K.C., Kaplan A., Henderson C.E. ( 2010. ) Motor neuron diversity in development and disease . Annu. Rev. Neurosci ., 33 , 409 – 440 . [DOI] [PubMed] [Google Scholar]
- 61. Wootz H., Fitzsimons-Kantamneni E., Larhammar M., Rotterman T.M., Enjin A., Patra K., Andre E., Van Zundert B., Kullander K., Alvarez F.J. ( 2013. ) Alterations in the motor neuron-renshaw cell circuit in the Sod1(G93A) mouse model . J. Comput. Neurol ., 521 , 1449 – 1469 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Nie D., Di Nardo A., Han J.M., Baharanyi H., Kramvis I., Huynh T., Dabora S., Codeluppi S., Pandolfi P.P., Pasquale E.B . et al. . ( 2010. ) Tsc2-Rheb signaling regulates EphA-mediated axon guidance . Nat. Neurosci ., 13 , 163 – 172 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Zou T., Yang X., Pan D., Huang J., Sahin M., Zhou J. ( 2011. ) SMN deficiency reduces cellular ability to form stress granules, sensitizing cells to stress . Cell Mol. Neurobiol ., 31 , 541 – 550 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ravasz E., Somera A.L., Mongru D.A., Oltvai Z.N., Barabasi A.L. ( 2002. ) Hierarchical organization of modularity in metabolic networks . Science , 297 , 1551 – 1555 . [DOI] [PubMed] [Google Scholar]
- 65. Langfelder P., Zhang B., Horvath S. ( 2008. ) Defining clusters from a hierarchical cluster tree: the Dynamic Tree Cut package for R . Bioinformatics , 24 , 719 – 720 . [DOI] [PubMed] [Google Scholar]
- 66. Huang da W., Sherman B.T., Lempicki R.A. ( 2009. ) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources . Nat. Protoc ., 4 , 44 – 57 . [DOI] [PubMed] [Google Scholar]
- 67. Huang da W., Sherman B.T., Lempicki R.A. ( 2009. ) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists . Nucleic Acids Res ., 37 , 1 – 13 . [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.