Abstract
Sex differences in pituitary growth hormone (GH) secretion (pulsatile in males vs near continuous/persistent in females) impart sex-dependent expression to hundreds of genes in adult mouse liver. Signal transducer and activator of transcription (STAT) 5, a GH-activated transcription factor that is essential for liver sexual dimorphism, is dynamically activated in direct response to each male plasma GH pulse. However, the impact of GH-induced STAT5 pulses on liver chromatin accessibility and downstream transcriptional events is unknown. In this study, we investigated the impact of a single pulse of GH given to hypophysectomized mice on local liver chromatin accessibility (DNase hypersensitive site analysis), transcription rates (heterogeneous nuclear RNA analysis), and gene expression (quantitative polymerase chain reaction and RNA sequencing) determined 30, 90, or 240 minutes later. The STAT5-dependent but sex-independent early GH response genes Igf1 and Cish showed rapid, GH pulse–induced increases in chromatin accessibility and gene transcription, reversing the effects of hypophysectomy. Rapid increases in liver chromatin accessibility and transcriptional activity were also induced in hypophysectomized male mice for some (Ces2b, Ugt2b38) but not for other liver STAT5-dependent male-biased genes (Cyp7b1). Moreover, in pituitary-intact male mice, Igf1, Cish, Ces2b, and Ugt2b38 all showed remarkable cycles of chromatin opening and closing, as well as associated cycles of induced gene transcription, which closely followed each endogenous pulse of liver STAT5 activity. Thus, the endogenous rhythms of male plasma GH pulsation dynamically open and then close liver chromatin at discrete, localized regulatory sites in temporal association with transcriptional activation of Igf1, Cish, and a subset of STAT5-dependent male-biased genes.
Igf1, Cish, and a subset of male-biased liver genes undergo rapid pulsatile increases in chromatin accessibility and gene transcription with each male plasma GH pulse–induced cycle of STAT5 activation.
Humans are exposed to thousands of lipophilic foreign chemicals, many of which are detoxified by hepatic drug-metabolizing enzymes and transporters (1, 2) but in some cases are converted to mutagenic and carcinogenic metabolites (3, 4). Many of these enzymes also metabolize cholesterol, steroid hormones, and other endogenous lipophiles, impacting liver physiology, endocrine homeostasis, and disease states (5–7). Cyp genes (8, 9) and other steroid-metabolizing gene families (Sult, Hsd, Ugt) (10–12) show sex-biased expression in both mouse and rat liver, enabling each sex to meet its own particular metabolic and hormonal requirements. Human orthologs of certain sex-biased rodent genes active in steroid and lipid metabolism show sex-biased liver expression (13–16) and may contribute to sex-differential cardiovascular disease risk (17). The adult pattern of sex-biased hepatic gene expression is determined by neonatal androgen exposure (18–20), which programs the hypothalamus and its orchestration of the sex-specific pituitary growth hormone (GH) secretion profiles that emerge at puberty (21, 22). These sex-dependent plasma GH profiles, in turn, activate two mutually exclusive patterns of liver gene expression, depending on whether the liver is stimulated by GH in a persistent manner (female liver) or intermittently (male liver), as best exemplified by studies of sex- and GH-regulated Cyp genes in both mouse and rat models (23–29).
Signal transducer and activator of transcription (STAT)5 is a GH-activated transcription factor (30, 31) that plays a key role in liver metabolism and pathophysiology (32) and is essential for the sex-dependent effects of GH on liver gene expression (8). STAT5 is activated in male liver in an intermittent manner (33) in direct response to each plasma GH pulse beginning at puberty (34), when >80% of male-specific genes are induced in mouse liver (35). In female liver, STAT5 activity persists over time due to the near continuous activation of STAT5 by circulating GH (36–38). Mouse knockout models show that the sex-dependent expression of >1000 liver-expressed genes (39, 40) requires STAT5. Thus, in male Stat5b knockout mice (41–43), 90% of male-biased genes are repressed and 60% of female-biased genes are induced (de-repressed) (39, 44). STAT5a, a closely related transcription factor, is unable to compensate for the loss of STAT5b (42, 43) but is essential for expression of a unique subset of female-biased genes in female mouse liver (45). Genome-wide mapping of liver binding sites for STAT5b and STAT5a (collectively, STAT5) identified ∼3500 sites with a strong sex bias in binding activity (38). Male-biased STAT5 binding is enriched near male-biased genes, and female-biased STAT5 binding is enriched near female-biased genes (38). Liver sex differences are enforced by two STAT5-regulated repressors, BCL6, which is expressed in a male-biased manner (38, 46), and CUX2, which shows strongly female-specific expression (47, 48). Robust sex differences can thus be achieved for hundreds of sex-biased genes by the complex interplay of STAT5 and other GH-regulated transcription factors.
Changes in chromatin structure and accessibility are a hallmark of epigenetic regulation and developmental plasticity (49) and can be probed by global analysis of open chromatin sites by DNase I digestion followed by high-throughput sequencing (DNase sequencing) (50, 51). Using this technique, we identified ∼71,000 open chromatin regions as DNase hypersensitive sites (DHSs) in male and female mouse liver (51). DHSs include promoters, enhancers, silencers, and insulators (52) and encompass up to 93% of genome-wide binding sites for several major liver transcription factors (51). Several thousand liver DHSs show sex differences in hypersensitivity (i.e., sex differences in chromatin accessibility) and are responsive to changes in circulating GH profiles (51). Sex-biased liver DHSs show strong enrichment for proximity to expressed genes of a corresponding sex bias, indicating they harbor positive regulatory elements (51). Indeed, the sex-differential binding of STAT5 to mouse liver chromatin seen at many promoters and enhancers linked to sex-specific genes shows strong enrichment for sex-biased DHSs (38). Sex differences in liver chromatin accessibility are thus a key feature of sex-differential hepatic gene expression. However, the mechanisms whereby sex-specific plasma GH profiles establish and maintain sex differences in liver chromatin accessibility are unknown and present a major challenge. One possibility is that liver chromatin accessibility at DHSs is directly responsive to the repeated on–off pulses of GH stimulation and STAT5 activation. In this study, we use the mouse liver model to investigate this hypothesis through studies of the dynamic effects of male plasma GH pulses on chromatin accessibility and gene transcription rates for both the classical GH/STAT5-regulated gene targets Igf1 (53–56) and Cish (55, 57, 58) and for several GH-responsive, STAT5-dependent male-biased genes.
Materials and Methods
Animal treatments
All mouse work was carried out in compliance with the requirements of the Boston University Institutional Animal Care and Use Committee. Male and female CD-1 mice (ICR strain) were obtained from Charles River Laboratories (Wilmington, MA). Mice were intact or were hypophysectomized by the supplier at 7 weeks of age. Mice were kept on a 12-hour light/dark cycle with food and water without restriction. Body weights were monitored for 4 to 5 weeks after surgery to verify the absence of weight gain above presurgery level, which was taken as an indicator of complete hypophysectomy. Urine collected from each mouse was analyzed on a 12% sodium dodecyl sulfate–polyacrylamide gel stained with Coomassie brilliant blue to detect the prominent low–molecular-mass Mup protein band, which was visually absent in hypophysectomized mouse urine. Mice showing weight gain above presurgery levels or detectable Mup protein in urine following hypophysectomy were excluded from the study. Hypophysectomized male mice were given a single intraperitoneal injection of vehicle (control) without or with recombinant rat GH at 125 ng of GH per gram of body weight (59). This dose approximates a physiological replacement dose and is 12-fold lower than the supraphysiological doses historically used in studies of GH-induced Igf1 gene transcription (54, 60–62). Mice were euthanized by cervical dislocation 30, 90, or 240 minutes after GH injection. In other experiments, fresh livers collected from individual untreated, intact mice (8 to 10 weeks of age) were used to prepare nuclei for DHS analysis and for nuclear RNA isolation, as described below. Portions of each liver were flash frozen in liquid N2, stored at −80°C, and used to isolate total liver RNA or to prepare a protein extract for analysis of liver STAT5 activity by electrophoretic mobility shift assay (EMSA) to identify STAT5-high activity and STAT5-low activity livers (see later).
Isolation of liver nuclei and purification of nuclear RNA
Nuclei were isolated from individual livers using a high sucrose–based homogenization buffer (51) to minimize disruption of chromatin structure. Each liver was minced, washed in ice cold diethyl pyrocarbonate–treated phosphate buffered saline, and homogenized in a Potter-Elvehjem homogenizer in 8 mL of cold homogenization buffer [10% (volume-to-volume ratio) glycerol, 2 M sucrose, 10 mM HEPES (pH 7.9), 25 mM KCl, 0.15 mM spermine, 0.5 mM spermidine trihydrochloride, 1 mM EDTA, 0.05 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM NaF, and one tablet of complete protease inhibitor cocktail (Roche) per 50 mL buffer]. The homogenate was layered over a 3-mL cushion of homogenization buffer in an ultracentrifuge tube and spun at 25,000 rpm for 30 minutes at 4°C in an SW-41Ti rotor. The supernatant was discarded by inverting the tube. The inner walls of the centrifuge tube were cleaned with a Kimwipe soaked in diethyl pyrocarbonate–treated phosphate buffered saline, and then cleaned with a dry Kimwipe. Nuclei were resuspended in nuclear storage buffer [20 mM Tris HCl (pH 8.0), 75 mM NaCl, 0.5 mM EDTA, 50% (volume-to-volume ratio) glycerol, 0.85 mM dithiothreitol, and 0.125 mM phenylmethylsulfonyl fluoride) using a Dounce homogenizer, counted using a hemocytometer, and stored in aliquots at −80°C. Nuclear RNA was purified from 25 to 50 million nuclei, corresponding to ∼25% of a liver, using TRIzol LS reagent. The final nuclear RNA preparation was validated by the presence of intact nuclear ribosomal RNA, which gave a characteristic pattern on agarose gel electrophoresis.
Reverse transcription–quantitative polymerase chain reaction analysis of heterogeneous nuclear RNA and messenger RNA levels of GH-responsive genes
Complementary DNA (cDNA) synthesized from TRIzol-extracted liver total RNA (intact mice and hypophysectomized mice, with or without exogenous GH treatment) or liver nuclear RNA (STAT5-high and STAT5-low activity livers) using the Applied Biosystems high-capacity cDNA reverse transcription (RT) kit (Thermo Fisher Scientific, catalog no. 43-688-14) was used to assay individual RNA transcripts by RT-quantitative polymerase chain reaction (RT-qPCR). Relative RNA levels were determined from the qPCR data by the ΔCt method after normalization to the 18S RNA content of each liver cDNA sample. qPCR primers were designed using Primer Express software (Thermo Fisher Scientific). Each primer was verified to be specific for its target sequence using the Blat function of the University of California Santa Cruz browser after extending the sequence to include 3, 5, or 10 nucleotides of flanking sequence on each end to compensate for the inability of Blat to reliably find closely related sequences when short sequences are analyzed. Primary RNA transcripts [heterogeneous nuclear RNA (hnRNA)] were assayed using a primer pair that spans an exon–intron (or an intron–exon) junction. Primers used to assay mature (spliced) messenger RNAs (mRNAs) were targeted to adjacent exons separated by an intron >1 kb in length to minimize the likelihood that contaminating genomic DNA contributes to the qPCR signal. All primer pairs except for those for Ces2b mRNA (intron length 0.65 kb) and Cish mRNA (intron length 0.22 kb) met this requirement. The melting curves of each qPCR amplicon were symmetric and free of secondary peaks, consistent with the amplicon largely being free of sequences derived from contaminating genomic DNA. qPCR primer sequences and their exonic and intronic locations are shown in Supplemental Table 1.
EMSA gels
STAT5 DNA-binding activity was assayed in total liver homogenate prepared from frozen individual mouse livers as described (36) using ∼50 mg of frozen liver tissue in hypotonic homogenization buffer [10 mM Tris buffer (pH 7.6) containing 1 mM EDTA, 250 mM sucrose, protease inhibitor, and phosphatase inhibitors]. DNA-binding activity was assayed on EMSA gels (36, 63) using an EMSA probe for the STAT5/MGF response element of the rat CSN2 (β-casein) gene and labeled with γP32–adenosine triphosphate. Gels were fixed in 20% methanol and 10% glacial acetic acid, dried, exposed overnight to an imaging screen, and scanned using a Typhoon Trio phosphorimager (GE Healthcare Life Sciences).
DHS analysis
DNase I digestion of mouse liver nuclei prepared from individual livers and purification of the released DNA fragments were carried out essentially as described (51) using 25 × 106 frozen mouse liver nuclei per sample. Buffer A was kept on ice; stop buffer and buffer D were kept at 37°C. Nuclei were washed twice in ice-cold buffer A, the supernatant was discarded, and warm buffer D was added. Enzyme buffer [buffer D containing 32 U of RQ1 RNase-free DNase I (1 U/μL; Promega) per 100 μL of buffer] was then prepared. Microcentrifuge tubes (2 mL) were placed in a 37°C water bath. Enzyme buffer (100 μL) was added to each tube, 850 μL of warmed nuclei in buffer D was added, and the mixture was incubated for 2 minutes at 37°C. Stop buffer (950 μL) was then added and the samples were incubated at 55°C for 15 minutes. Proteinase K (15 μL; 20 mg/mL; Bioline) was added followed by incubation overnight at 55°C. Samples were extracted with phenol/chloroform and then loaded on a sucrose gradient (11.4 mL of sample layered on top of a 24-mL 10% to 20% sucrose step gradient, composed of (top to bottom) 3 mL each of 10%, 12.5%, 15%, and 17.5% sucrose, and then 12 mL of 20% sucrose). Samples were centrifuged at 25,000 rpm for 24 hours at 25°C. Individual fractions (1.9 mL each) were collected and analyzed to determine the DNA fragment size distribution on a SYBR Green–stained 1.2% agarose gel after visualization on a Typhoon Trio phosphorimager. Fractions containing DNA fragments ∼100 bp to 1 kb in size were pooled in Qiagen Buffer PB, and DNA was purified on a QIAprep 2.0 DNA purification column (Qiagen). Final selection of fragments 125 to 400 bp in size was achieved using Agencourt AMPure XP beads. DNA concentrations were measured by a Quant-iT PicoGreen assay kit (Invitrogen), except that final DHS samples were quantified by Qbit analysis (Invitrogen) to equalize the input DNA for qPCR analysis of specific genomic DNA sequences in the released DHS fragments.
RNA sequencing and data analysis
RNA sequencing (RNA-seq) libraries were prepared from total liver RNA isolated from intact or hypophysectomized male and female mouse liver, and from livers of hypophysectomized male mice given a single injection of GH and euthanized 30, 90, or 240 minutes later. Two libraries (biological replicates) were prepared for each treatment group, with each library representing a pool of n = 4 independent liver RNA samples. Sex-biased liver genes were identified by RNA-seq analysis of three libraries per sex, with each library comprised of liver RNA pooled from n = 12 to 17 independent livers (biological replicates). Libraries were prepared from poly(A)-selected total liver RNA using NEBNext Ultra directional RNA library preparation kit for Illumina (New England Biolabs) or, in some cases, an Illumina TruSeq RNA library preparation kit. Where specified, pooled RNA samples were treated with DNase and then depleted of ribosomal RNA using the NEBNext ribosomal RNA (rRNA) depletion kit. Multiplexed sequencing (50-, 100-, or 125-nucleotide paired-end reads, as indicated in individual Gene Expression Omnibus submission files; see below) of each library was performed on an Illumina HiSeq 2000/2500 instrument.
RNA-seq data were analyzed using a custom pipeline that we developed to quantify both mature and primary (hnRNA) transcripts, as follows. The pipeline processes RNA-seq raw FASTQ files and outputs various quality control metrics, including FastQC reports (FASTX-Toolkit v0.0.13.2), confirmation of read length, absence of read strand bias, and alignment to functional genomic location classes (i.e., coding, intronic, untranslated region, intergenic, ribosomal) (Picard v1.123). Reads were mapped to the mouse genome (mm9) using TopHat (v2.1.1). FeatureCounts (64) was used to obtain read counts for RefSeq genes, and EdgeR (65) was used to calculate differential gene expression and significance values. Adjusted P values (i.e., false discovery rate) < 0.05 were considered significant. The pipeline generated data files (bigWig or BAM files) and gene-feature tracks (gene transfer format files) for visualization on the University of California Santa Cruz Genome Browser. For each gene symbol (University of California Santa Cruz Table Browser, February 2015), the RNA-seq pipeline counted RNA-seq reads mapping to three sets of genomic regions (features), which we termed collapsed exon counting, exonic-only counting, and intronic-only counting. Mature (spliced) mRNAs were quantified using either collapsed exon or exonic-only counting. The collapsed exon method counts sequence reads that map to a concatenated sequence comprised of the union of the exonic regions for all isoforms of a given gene, whereas exonic-only counting is limited to the intersection of exonic regions across all isoforms, and thus excludes exonic regions that overlap an intron in one or more isoforms of the gene. Intronic-only counting was used to quantify primary RNA transcripts (hnRNA) and is based on the intersection of all intronic sequences across all isoforms of a given gene; it thus excludes intronic regions that overlap an exon in one or more isoforms of the gene. Others have confirmed the suitability of this approach for quantifying primary RNA transcripts (66, 67). Exonic-only counting regions were identified for each gene symbol by subtracting the union of collapsed intronic sequences across all isoforms from the collapsed exonic regions, and intronic-only counting regions were identified for each gene symbol by subtracting the union of collapsed exonic sequences across all isoforms from the collapsed intronic regions (University of California Santa Cruz Genome Browser annotations). Collapsed exon, exonic-only, and intronic-only counting regions identified for each gene were collected in custom gene transfer format files (available upon request), which were used in relevant steps in the RNA-seq pipeline. We also identified a subset comprised of 2035 RefSeq gene symbols that have substantial overlap with other RefSeq genes, resulting in <130 nucleotides of nonoverlapping sequence; we determined empirically that this was too short a length for mapping sufficient numbers of RNA-seq reads for reliable quantification of gene expression. For these 2035 genes, we used the (-O) option of featureCounts, which allows sequence reads/fragments to be assigned to all overlapping gene features. Raw and processed RNA-seq data are available at Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/gds/) under accession number GSE93382.
Statistical analysis
Graphical and statistical analyses were performed using GraphPad Prism 7 software. RNA and DHS qPCR data are expressed as mean values ± standard error of the mean (SEM) for up to 18 individual mouse livers per group, as specified in each figure legend. A two-tailed Student t test was used when the means of two groups were compared (i.e., STAT5-high vs STAT5-low). One-way analysis of variance (ANOVA) with a Dunnett multiple comparisions test was used when comparing GH-treated hypophysectomized male mice to untreated hypophysectomized controls, and one-way ANOVA with a Sidak multiple comparisons test was used when comparing hypophysectomized mice to the same-sex intact controls. Significance values are indicated in each figure by † when comparing intact males to intact females, by # when comparing hypophysectomized mice to the same sex intact controls, and by * when comparing GH-treated hypophysectomized male mice to untreated hypophysectomized controls, or when comparing STAT5-high to STAT5-low male livers. Statistical significance reached by a t test but not ANOVA is indicated by ^. A single symbol (†, #, *, or ^) indicates significance at P < 0.05, a double symbol indicates P < 0.01, and a triple symbol indicates P < 0.001.
Results
GH-induced liver transcriptional responses in hypophysectomized mouse model
Hypophysectomized mice were used to investigate the GH pulse responsiveness of several STAT5-regulated genes. The completeness of hypophysectomy was validated by the cessation of body weight gain [Fig. 1(a)] and by the characteristic near total loss of urinary MUP proteins [Fig. 1(b)], which are produced in the liver under the control of GH and other pituitary-dependent hormones (68). RNA-seq analysis revealed that hypophysectomy led to strong and significant downregulation of all 19 Mup genes in male mouse liver, in almost all cases by >25-fold (Supplemental Table 2). Expression levels for individual Mup genes varied >5000-fold in intact male liver, and in all but two cases (Mup2, Mup8) showed significant male-biased expression (up to ∼70-fold male bias for the highly expressed Mup7 and Mup20). Fifteen of the 19 Mup genes were also significantly downregulated in female liver following hypophysectomy (Supplemental Table 2).
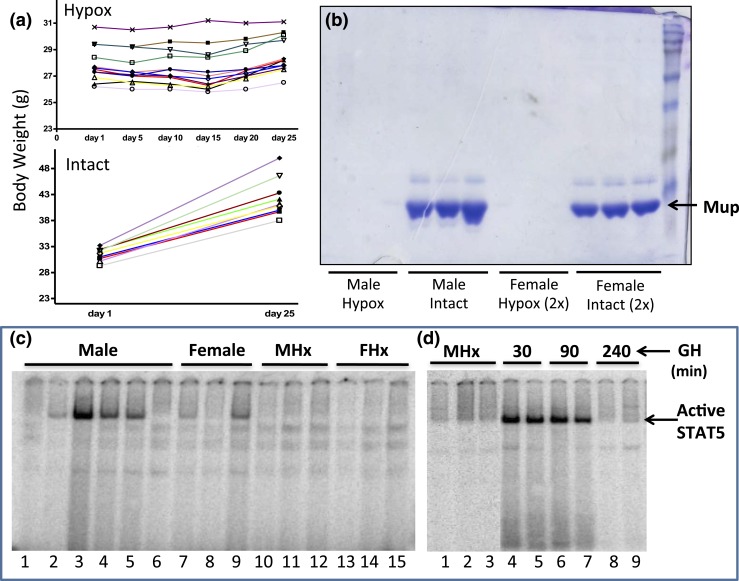
Figure 1.
Hypophysectomy abolishes GH-induced liver STAT5 activity. (a) Mice were hypophysectomized (Hypox) and body weights were recorded for ∼25 days after surgery. Body weight gain of pituitary-intact mice (Intact) is shown as a control. The cessation of weight gain is indicative of complete hypophysectomy. (b) Urine collected from individual mice (1 μL for males, 2 μL for females) was analyzed on an sodium dodecyl sulfate–polyacryamide gel stained with Coomassie blue. Major urinary proteins (Mup) were undetectable in the urine of hypophysectomized mice. (C and D) EMSA analysis of whole-liver extract prepared from individual mice using a STAT5 EMSA probe. STAT5 DNA-binding activity (arrow) varies markedly in individual livers, most notably in males [c, lanes 1 to 6; see text and Fig. 4(a)], and is abolished by hypophysectomy of males (MHx) or females (FHx) and restored by a single pulse of GH (d, lanes 4 to 7 vs lanes 1 to 3). A decline in STAT5 activity back to baseline is seen 240 minutes after GH injection (D, lanes 8-9).
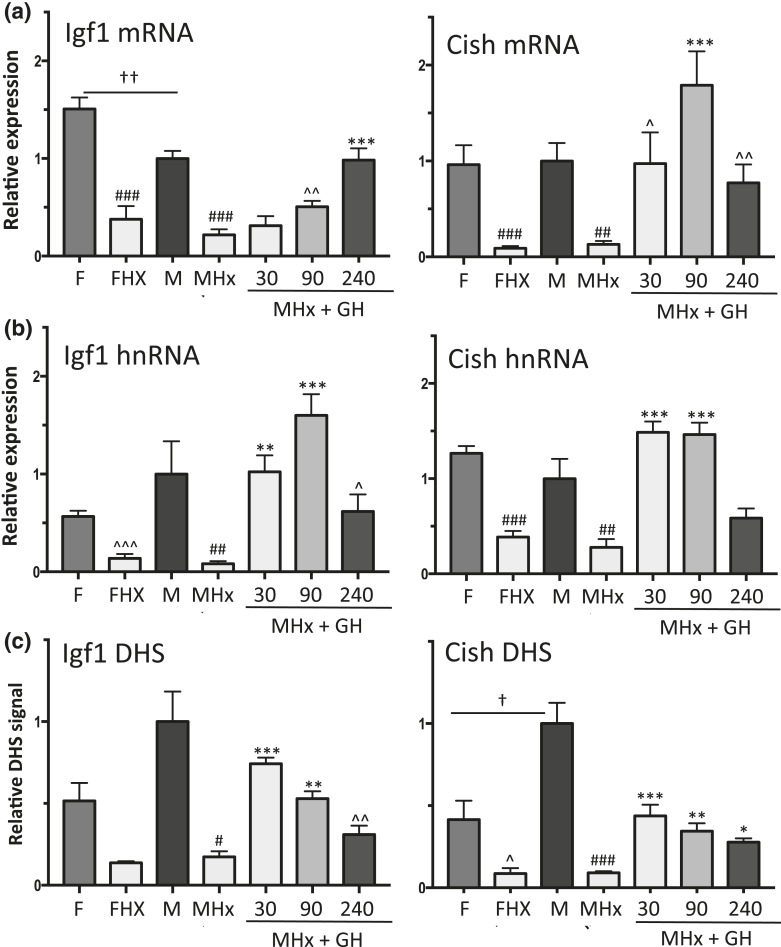
Liver STAT5 DNA-binding activity declined following hypophysectomy [Fig. 1(c), lanes 10 to 15 vs lanes 1 to 9], consistent with the loss of pituitary GH secretion and GH-induced liver STAT5 activation. Hypophysectomy also led to significant downregulation of two well-characterized GH- and STAT5-responsive genes, Igf1 and Cish, in both male and female liver, as shown by qPCR analysis of liver RNA using primers specific to the mature (spliced) mRNAs [Fig. 2(a)]. A time-dependent increase in expression back to at least intact mouse liver levels was seen for each gene when male hypophysectomized mice were given a single pulse of GH at a near physiological replacement dose [Fig. 2(a)]. Similar responses to hypophysectomy and GH pulse treatment were apparent when unspliced hnRNA was assayed using qPCR primers that span an exon–intron junction, except that maximal responses occurred earlier than the changes in mRNA levels [peak at 90 vs 240 minutes for Igf1; peak at 30 vs 90 minutes for Cish; Fig. 2(b) vs Fig. 2(a)]. Igf1 and Cish hnRNA levels, which are indicative of gene transcription rates (66, 67), declined by 240 minutes after GH injection [Fig. 2(b)], consistent with the decline of liver STAT5 DNA-binding activity back to basal level at this point in time [Fig. 1(d), lanes 8 to 9].
Figure 2.
Effects of a single GH pulse on chromatin accessibility and transcriptional response of sex-independent STAT5 target genes Igf1 and Cish. (a and b) RT-qPCR analysis of liver total RNA isolated from intact female (F) and male (M) mice, hypophysectomized female (FHx) and hypophysectomized male mice (MHx), and hypophysectomized male mice treated with a single pulse of GH and euthanized 30, 90, or 240 minutes later. qPCR was performed using primers that detect mature Igf1 and Cish RNA (a, mRNA) or primary unspliced RNA transcripts (b, hnRNA). RNA qPCR data were normalized to the 18S rRNA content of each liver RNA sample. (c) qPCR analysis of genomic DNA fragments released from DNase I–digested liver nuclei isolated from individual mice from the same treatment groups shown in (a) and (b). qPCR was carried out using primers that target open chromatin regions identified previously (51), which are marked DHS in Fig. 3(a) (Igf1) and Fig. 3(b) (Cish; also see Supplemental Table 1). DHS qPCR data were normalized to a DNase hypersensitive site at the Alb promoter [see Fig. 4(a)]. Data shown are mean ± SEM values based on n = 8 to 10 livers for each group (a and b) or n = 5 to 6 livers per group (c), except for the intact male group of (c), where n = 18 livers. Relative expression level and relative DHS activity of the intact male groups are set to 1. Significance values by ANOVA are indicated in each figure as follows: for intact male vs intact female, †P < 0.05, ††P < 0.01, †††P < 0.001; for hypophysectomized mice compared with intact mice of the same sex, #P < 0.05, ##P < 0.01, ###P < 0.001; and for GH-treated vs untreated hypophysectomized mice, *P < 0.05, **P < 0.01, ***P < 0.001. The symbol ^ indicates that statistical significance was reached only by t test and not ANOVA, at ^P < 0.05, ^^P < 0.01, and ^^^P < 0.001. Primers used for qPCR analysis are shown in Supplemental Table 1.
GH rapidly increases liver chromatin accessibility at early GH response genes
GH-activated STAT5 binds to mouse liver chromatin at multiple sites within or nearby Igf1 and Cish, as determined by global analysis of STAT5 binding by chromatin immunoprecipitation sequencing (ChIP-seq) (38). Many of these STAT5 binding sites contain a STAT5 motif, indicating direct STAT5 binding. STAT5 binding shows strong enrichment for regions of open (accessible) chromatin, which are identified by their sensitivity to cleavage by the enzyme DNase I (DHSs) (51). ChIP-seq analysis of a panel of livers collected from mice euthanized at either a peak or a trough of liver STAT5 activity indicated that STAT5 binds to these open chromatin regions in repeated cycles of binding and release in direct response to each plasma GH pulse (STAT5-high vs STAT5-low livers; Fig. 3). It is not known, however, whether these STAT5 binding sites are constitutively open or whether they dynamically open and then close in response to plasma GH pulse stimulation and liver STAT5 activation. To address this question, we isolated liver nuclei from intact and hypophysectomized male mice, and from hypophysectomized male mice given a single pulse of GH, and then euthanized 30, 90, or 240 minutes later. Nuclei were digested with low concentrations of DNase I, and the released genomic DNA fragments were purified and assayed by qPCR to quantify DNase I–accessible genomic regions at or near Igf1 and Cish (51) that contain STAT5 binding sites identified previously (38) (DHS regions are marked in Fig. 3 and detailed in Supplemental Table 1). Results show that chromatin accessibility at STAT5 binding sites at or near Igf1 and Cish decreased following hypophysectomy and was induced by GH pulse treatment [Fig. 2(c)]. The increased accessibility of these sites peaked 30 minutes after a single GH pulse, and thereafter began to decline, in parallel to the decline in liver STAT5 activity [Fig. 1(d)]. Moreover, chromatin accessibility (DHS activity) began to decline at 90 minutes, prior to the decline in Igf1 and Cish transcription rate (hnRNA analysis) [Fig. 2(c) vs Fig. 2(b)]. Thus, liver chromatin is dynamically responsive to GH pulse–induced signaling, which stimulates a cycle of chromatin opening associated with STAT5 binding to liver chromatin and transcriptional activation, followed by chromatin closing associated with STAT5 deactivation, loss of STAT5 binding, and a decline in gene transcription.
Figure 3.
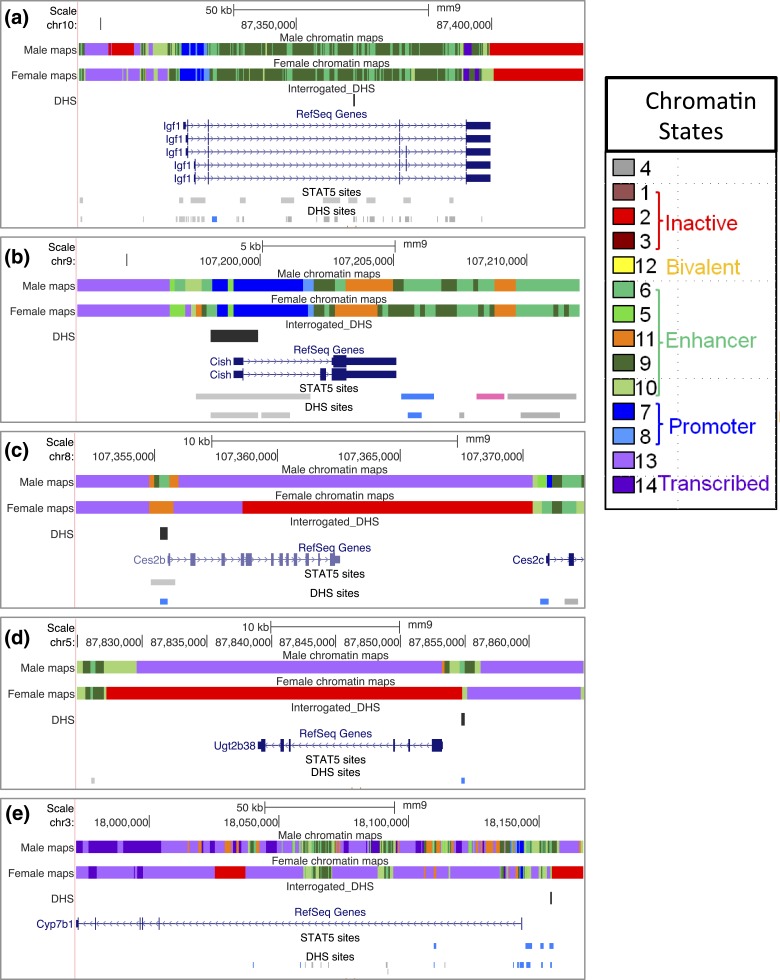
(a–e) Mouse genome browser screenshots showing for each of the five indicated genes (a–e) specific genomic regions assayed for hnRNA, mRNA, and DHS, as marked in the track “Sequence from Blat Search.” Primary RNA transcripts (hnRNA) were quantified using primer pairs that span an exon–intron (or an intron–exon) junction. Primers used to quantify mature (spliced) mRNAs were targeted to adjacent exons separated by an intron. Open chromatin regions were those previously identified as DHS (merged DHS sites, top track) (51) and are color coded to indicate the sex bias of chromatin accessibility: gray indicates no sex bias; blue indicates male bias, with darker shades of blue used to indicate greater/more significant male bias. The specific open chromatin regions interrogated by qPCR are marked DHS. These DHS all overlap regions of STAT5 binding in STAT5-high male mouse liver identified previously by ChIP-seq (38), as shown in the bottom track. STAT5 DNA binding is greatly reduced in mice euthanized when liver STAT5 activity is low [compare with Fig. 4(a)] (STAT5-low track).
Chromatin opening and STAT5-activated transcription are pulsatile in intact mouse liver
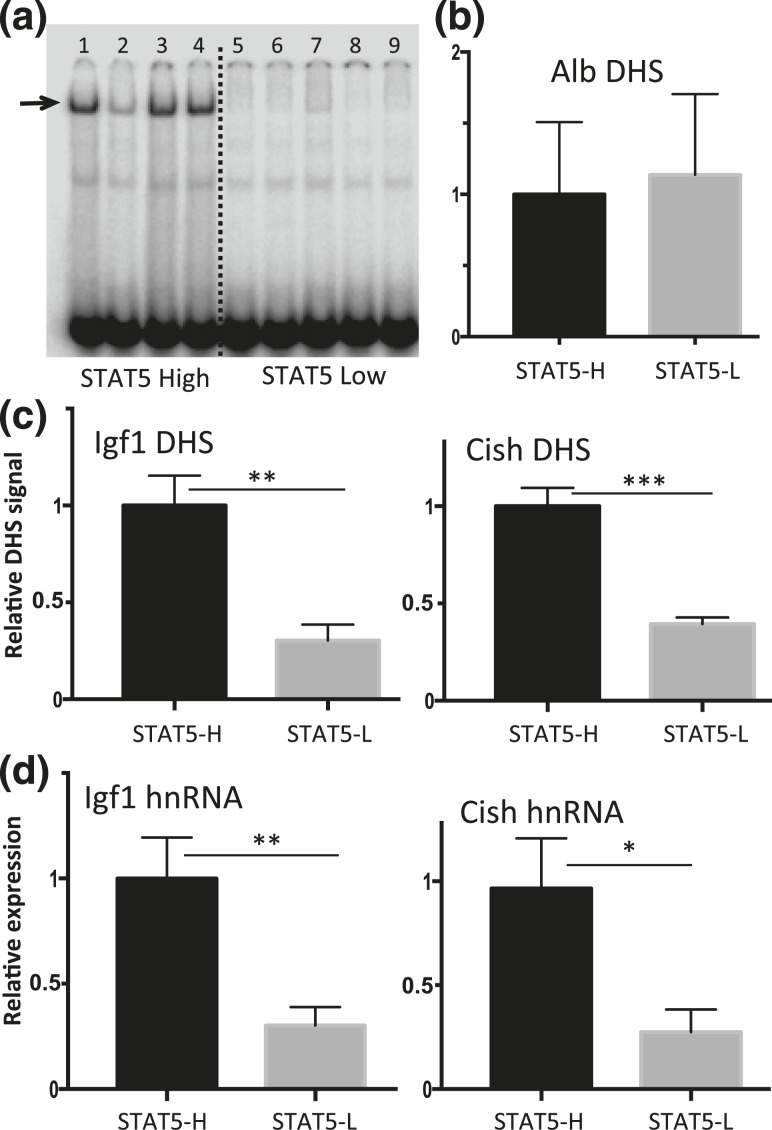
The above studies establish, in a hypophysectomized mouse model, a close association between exogenous GH pulse stimulation, liver chromatin opening, and GH-induced transcription of two primary STAT5 target genes. To ascertain whether chromatin opening and target gene transcription are dynamically responsive to endogenous pulses of pituitary GH release, we collected livers from a large set of individual male mice and then determined the STAT5 DNA-binding activity status of each liver by EMSA analysis. A binary pattern of STAT5 activity characterized most livers, which showed either high STAT5 activity or low STAT5 activity [Fig. 4(a), lanes 1, 3, and 4 vs lanes 5 to 9]. A few livers showed an intermediate level of STAT5 activity (lane 2). This same overall pattern was previously seen in livers of young adult male rats, where the STAT5 activity status of each liver was shown to directly reflect whether the liver was collected during a pulse of pituitary GH release (STAT5-high livers) or during the plasma GH-free period between GH pulses (STAT5-low livers) (34, 69). Chromatin accessibility analysis revealed that the STAT5-bound DHSs near Igf1 and Cish showed significantly higher chromatin accessibility in STAT5-high livers than in STAT5-low livers [Fig. 4(c)]. In contrast, no difference in DNase hypersensitivity was seen between STAT5-high and STAT5-low livers at a prominent DHS in the Albumin promoter [Fig. 4(b)], whose qPCR signal intensity was used to normalize the recovery of DNase fragments released from each liver. Furthermore, gene transcription rates, indicated by nuclear hnRNA analysis, were significantly higher for both Igf1 and Cish in STAT5-high livers compared with STAT5-low livers [Fig. 4(d)]. We conclude that endogenous pulses of GH and liver STAT5 activity dynamically induce liver chromatin opening, pulsatile STAT5 binding to DHS regulatory regions, and pulsatile transcription of Igf1 and Cish.
Figure 4.
Pulsatility of chromatin accessibility and STAT5-stimulated transcription in pituitary-intact male mouse liver. (a) EMSA analysis of whole liver extract using STAT5 EMSA probe. Individual intact male mouse livers show a binary pattern of liver STAT5-binding activity (STAT5-high, lanes 1, 3, and 4; STAT5-low, lanes 5 to 9; lane 2 shows a liver with STAT5-intermediate activity). Arrow indicates STAT5 activity band. (b and c) Isolated liver nuclei were digested with DNase I and the released DNA fragments were purified and assayed by qPCR for Alb, Igf1, and Cish DHS sites using primers shown in Supplemental Table 1. (b) Chromatin accessibility at the Alb promoter showed no significant difference between STAT5-high (STAT5-H) and STAT5-low (STAT5-L) livers and was used to normalize the DHS qPCR signal of the corresponding liver sample at all other DHS sites. (c) In contrast to the Alb DHS, chromatin accessibility was significantly greater in STAT5-high livers than in STAT5-low livers at the STAT5-bound DHS nearby Igf1 and Cish. (d) Transcription rates determined by RT-qPCR analysis of liver nuclear RNA using hnRNA primers specific to Igf1 and Cish were significantly higher in STAT5-high compared with STAT5-low livers. Data shown are mean values ± SEM for n = 9 livers per group (b and c) or n = 10 to 12 livers per group (d). The relative DHS activity and expression level of the STAT5-high groups are set to 1. *P < 0.05, **P < 0.01, ***P < 0.001 by two-tailed t test.
Four classes of pituitary hormone-dependent sex-biased genes
In contrast to the primary STAT5 target genes Igf1 and Cish, which are expressed at similar levels in male and female mouse liver (less than twofold sex difference), several hundred other STAT5-dependent genes show strong differential expression between the sexes. The STAT5 dependence of these sex-biased genes is evident in both global and liver-specific STAT5 knockout mouse models (39, 44). STAT5 regulates these genes directly, as indicated by the strong enrichment of male-biased STAT5 binding at DHS that map to genes showing male-biased expression in liver, and by the enrichment of female-biased STAT5 binding at DHS near female-biased genes (38). It is unknown, however, whether GH pulse–activated STAT5 stimulates male-biased gene transcription acutely, and in a pulsatile manner, as is shown above for the sex-independent STAT5 gene targets Igf1 and Cish.
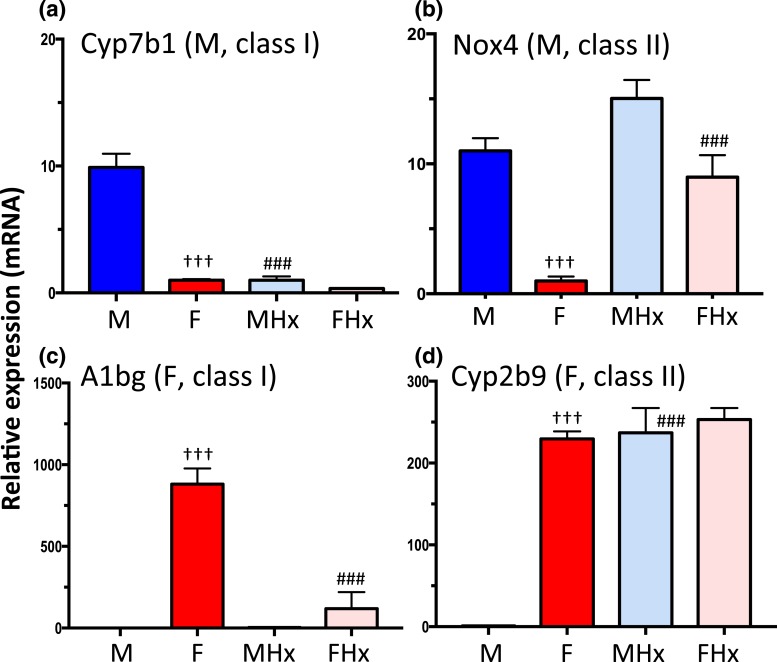
Two distinct classes of male-biased genes can be identified: (1) class I male-biased genes show decreased expression in male liver after hypophysectomy owing to their strong dependence on the male plasma GH pulse pattern for expression; additional male pituitary-dependent hormones are required for full expression of some class I male-biased genes [e.g., Mup genes (68)]; and (2) class II male-biased genes are repressed by the female pituitary profile and, consequently, they are strongly de-repressed in female liver following hypophysectomy. We carried out RNA-seq analysis of poly(A)-selected liver RNA, isolated from intact and hypophysectomized male and female mice, to identify male-biased genes belonging to each class on a genome-wide basis. We also identified corresponding sets of class I and class II female-biased genes; that is, genes whose expression in female liver decreases after hypophysectomy owing to a requirement for the near-continuous female GH pattern for expression (class I), and genes whose expression is repressed by the male pituitary profile and consequently show strong de-repression in hypophysectomized male liver (class II). Figure 5 shows RT-qPCR data for sex-biased genes representative of each gene class. Results based on global RNA-seq analysis of the same set of livers are summarized in Table 1 and detailed in Supplemental Table 3. These analyses complement an earlier microarray study (59) where the array probes cannot adequately distinguish closely related genes within large gene families and subfamilies, and whose members often show distinct patterns of sex-biased expression (e.g., genes in the Cyp2b, Cyp2c, Cyp3a, and Mup families).
Figure 5.
Classification of pituitary hormone-dependent sex-biased genes. RT-qPCR analysis of total liver RNA using gene-specific mRNA primers for the indicated class I and class II sex-biased genes, defined based on the effects of hypophysectomy on gene expression (see text). (a) Cyp7b1, a class I male-biased gene, is dependent on the male pituitary hormone profile, as indicated by its decreased expression in male liver after hypophysectomy. (b) Nox4, a class II male-biased gene, is repressed by the female pituitary profile, and is thus strongly de-repressed (i.e., upregulated) in female liver following hypophysectomy. (c) A1bg, a class I female-biased gene, shows decreased expression in female liver after hypophysectomy. (d) Cyp2b9, a class II female-biased gene, is repressed by the male pituitary profile, and thus is strongly de-repressed in male liver following hypophysectomy. Data shown are mean ± SEM values based on n = 9 livers per group. The relative expression level was set to 1 for the female liver samples (a and b) or for the male liver samples (c and d). Significance values (ANOVA) are marked as described in the Fig. 2 legend.
Table 1.
Classification of Pituitary Hormone–Dependent Sex-Biased Genes
| Class | Number of Genes (%) | Response in MHx | Response in FHx | Gene Subclass | Number of Genes | Examples |
|---|---|---|---|---|---|---|
| Male class I | 152 (78) | Down | — | IA | 83 | Ces2b, Pitx3, Mup20 |
| Down | Down | IB | 65 | Ugt2b38, Cyp7b1, Elovl3 | ||
| Down | Up | IC | 4 | Cyp2d9, Cyp4a12b | ||
| Male class II | 43 (22) | — | Up | IIA | 29 | Cyp4a12a, Nox4, Sstr2 |
| Up | Up | IIB | 14 | Serpina7, Susd4, Snhg11 | ||
| Female class I | 84 (38) | — | Down | IA | 48 | Cyp3a16, Fmo3, A1bg, Cux2 |
| Down | Down | IB | 24 | Cyp3a41a, Cyp2c69, Cyp2c40 | ||
| Up | Down | IC | 12 | Sult3a1, Sult2a1, Cyp2a4 | ||
| Female class II | 138 (62) | Up | — | IIA | 101 | Sult2a5, Cyp2b9, Hao2 |
| Up | Up | IIB | 37 | Sult2a6, Cyp2b13, Acot3 |
Of 530 sex-biased genes, 417 respond to hypophysectomy at |fold change| > 2 and an adjusted P value of <0.05 in either male or female mouse liver. Those genes are classified based on their response to hypophysectomy in male and female liver, as shown. Class I sex-biased genes are those that are downregulated by hypophysectomy in the sex where they show the higher expression in intact mice. Class II sex-biased genes are those that are upregulated by hypophysectomy in the sex where they show the lower expression in intact mice. Subclasses A, B, and C indicate the response to hypophysectomy in the dominant sex (class II genes) or in the opposite sex (class I genes), as indicated. See Supplemental Table 3 for a complete listing of genes.
Abbreviations: FHx, hypophysectomized female; MHx, hypophysectomized male; —, no significant change in expression.
Dynamic GH pulse responsiveness of class I male-biased genes Ces2b and Ugt2b38
Given the pituitary hormone/GH pulse dependence that characterizes class I male-biased genes, we investigated whether these genes show the dynamic regulation by GH pulse-activated STAT5 seen above for Igf1 and Cish. Intronic sequence reads were extracted from our RNA-seq datasets (see Materials and Methods) to quantify unspliced gene transcripts (i.e., hnRNA) that are downregulated by hypophysectomy and show a strong and significant increase in expression (more than fourfold increase in intronic reads at adjusted P value of <0.05) in hypophysectomized male mouse liver, either 30 minutes or 90 minutes after GH pulse treatment. Thirty-five genes were identified, including Igf1, Cish, and Socs2. The strongest response, a >500-fold increase, was seen for Ces2b at 90 minutes (adjusted P value = 1.14E-19) (Supplemental Table 4A). Six of the 35 genes showed consistent male-biased expression in intact mouse liver across multiple RNA-seq datasets (Supplemental Table 4C). Male-biased expression was strongest for Cyp4a12b (125-fold), Ugt2b38 (15-fold), and Ces2b (15-fold). Ces2b and Ugt2b38 are both class I male-biased genes (Table 1; Supplemental Table 3); they show strong decreases in male liver expression following hypophysectomy, as well as strong increases in transcription rate (566- and 30-fold, respectively) 90 minutes after GH pulse stimulation, based on our intronic RNA-seq analysis (Supplemental Table 4A). In contrast, Cyp4a12b is downregulated only twofold by hypophysectomy and showed a more modest 5.5-fold increase in transcription in the 90-minute GH pulse–stimulated livers. Similar results were obtained when we sequenced rRNA-depleted RNA rather than poly(A)-selected RNA (Supplemental Table 4B).
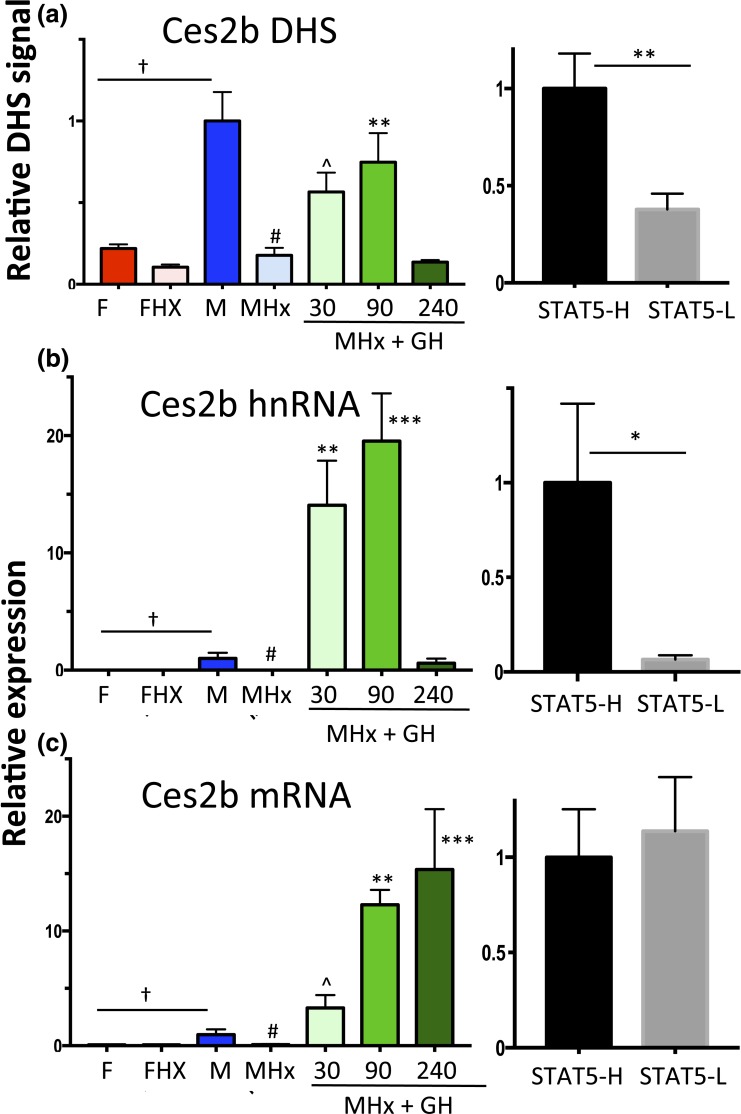
Ces2b has a male-biased DHS in its proximal promoter [Fig. 3(c); Supplemental Table 1] that overlaps a binding site for STAT5. In hypophysectomized male liver (where GH-activated STAT5 is deficient), chromatin accessibility at this DHS decreased to the level of intact female or hypophysectomized female liver [Fig. 6(a), left]. GH pulse treatment rapidly reversed this decline, as shown by the opening of this DHS within 30 minutes. DHS opening was sustained at 90 minutes, but then declined back to baseline at 240 minutes, coincident with the decline of liver STAT5 activity [Fig. 1(d)]. This close association between Ces2b proximal promoter–associated DHS opening and liver STAT5 activation/STAT5 binding was also seen in intact mouse liver, where STAT5-high livers showed significantly higher chromatin accessibility compared with STAT5-low livers [Fig. 6(a), right].
Figure 6.
Regulation of Ces2b, a class I male-biased gene, by GH pulse–activated STAT5. (a) DHS analysis at a male-biased STAT5 binding site in the Ces2b promoter [Fig. 3(c); Supplemental Table 1] carried out for intact and hypophysectomized male and female mouse livers, with a time course of GH treatment (left) as in Fig. 2. Chromatin accessibility at this DHS was significantly higher in STAT5-high than in STAT5-low livers (right). (b and c) RT-qPCR analysis of Ces2b hnRNA and Ces2b mRNA. Data shown in the left set of graphs for (a)–(c), that is, the hypophysectomy series, are mean ± SEM values for n = 5 to 6 livers per group, except for the intact male group, where n = 18. The right set of graphs (STAT5-high, STAT5-low) are based on n = 9 to 12 livers per group. Relative DHS activity and expression level of the intact male (left) and the STAT5-high groups (right) are set to 1. Significance values by ANOVA or two-tailed t test are marked as described in the Fig. 2 legend.
Next, we assayed Ces2b primary transcripts (hnRNA) to determine the functional consequences Ces2b DHS opening. Male-specific transcription of Ces2b was abolished by hypophysectomy [Fig. 6(b)], consistent with Ces2b being a class I male-biased gene (Table 1). GH pulse treatment induced Ces2b hnRNA to supraphysiological levels, as seen at 30 minutes and 90 minutes, followed by a dramatic decline back to intact male levels at 240 minutes. GH pulse-induced Ces2b transcription (i.e., hnRNA levels) showed an overall time dependence similar to that of DHS opening and closing [Fig. 6(b) vs Fig. 6(a)], although the magnitude of the transcriptional response was much greater, as noted. Mature (i.e., spliced) Ces2b mRNA showed a markedly delayed response, with a large, time-dependent accumulation evident at both 90 and 240 minutes [Fig. 6(c)]. The ability of endogenous GH pulse–activated STAT5 to stimulate pulsatile Ces2b transcription was established in intact male mouse liver by the significantly higher Ces2b hnRNA levels in STAT5-high liver nuclei compared with STAT5-low liver nuclei [Fig. 6(b), right]. There was no corresponding difference in mature Ces2b nuclear mRNA between STAT-high and STAT5-low livers [Fig. 6(c), right]. This reflects the half-life of Ces2b mRNA, which is apparently long compared with the 3- to 4-hour period between plasma GH pulses, as indicated by the rapid decline in Ces2b hnRNA from 90 to 240 minutes (indicating a short half-life) but not Ces2b mRNA (indicating a longer half-life) [Fig. 6(c) vs Fig. 6(b), left].
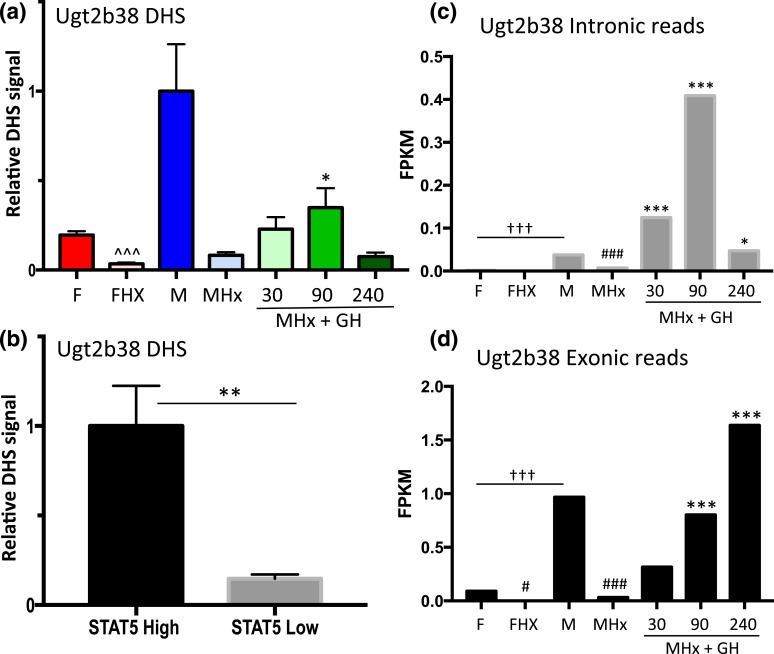
Similar findings were obtained for a second class I male-biased gene, Ugt2b38, where chromatin accessibility at a promoter-proximal male-biased DHS [Fig. 3(d); Supplemental Table 1] decreased significantly following hypophysectomy and increased following GH pulse treatment [Fig. 7(a)]. Furthermore, this DHS showed a marked increase in accessibility in STAT5-high compared with STAT5-low livers [Fig. 7(b)]. We were unable to design gene-specific RT-qPCR primers for Ugt2b38 owing to its close sequence similarity to Ugt2b37 and Ugt2b5. However, analysis of RNA-seq reads that map uniquely to Ugt2b38 introns verified the anticipated rapid (and supraphysiological) transcriptional response of this gene to GH pulse stimulation [Fig. 7(c)] followed by slower accumulation of Ugt2b38 exon-specific sequences [Fig. 7(d)]. As with Ces2b, Ugt2b38 hnRNA declined rapidly from 90 to 240 minutes [Fig. 7(c)], indicating a short half-life.
Figure 7.
Regulation of Ugt2b38, a class I male-biased gene, by GH pulse–activated STAT5. (a) DHS analysis at a male-biased STAT5 binding site in the Ugt2b38 promoter [Fig. 3(d); Supplemental Table 1] was carried out for intact and hypophysectomized male and female mouse livers, with a time course of GH treatment as in Fig. 2. (b) Chromatin accessibility at this DHS was significantly higher in STAT5-high than in STAT5-low livers. (c and d) RNA-seq analysis of rRNA-depleted RNA was performed, sequence reads mapping uniquely to introns and exons were counted, fragments per kilobase of transcript per million mapped reads (FPKM) expression values were calculated, and significance, marked here, was assessed by edgeR analysis (see Materials and Methods). Male-biased expression of Ugt2b38 was abolished by hypophysectomy and rapidly induced by a single GH pulse. Analysis of intronic-specific sequence reads as a measure of recent transcription showed that transcription declined 240 minutes after the GH pulse (c), whereas mature mRNA levels increased (d). Data shown in (a) are mean ± SEM values for n = 5 to 6 livers per group, except for the intact male group, where n = 18; data shown in (b) are mean ± SEM values for n = 9 livers per group. Relative activity of the intact male group (a) and the STAT5-high group (b) is set to 1. Significance values by ANOVA or two-tailed t test are marked as described in the Fig. 2 legend.
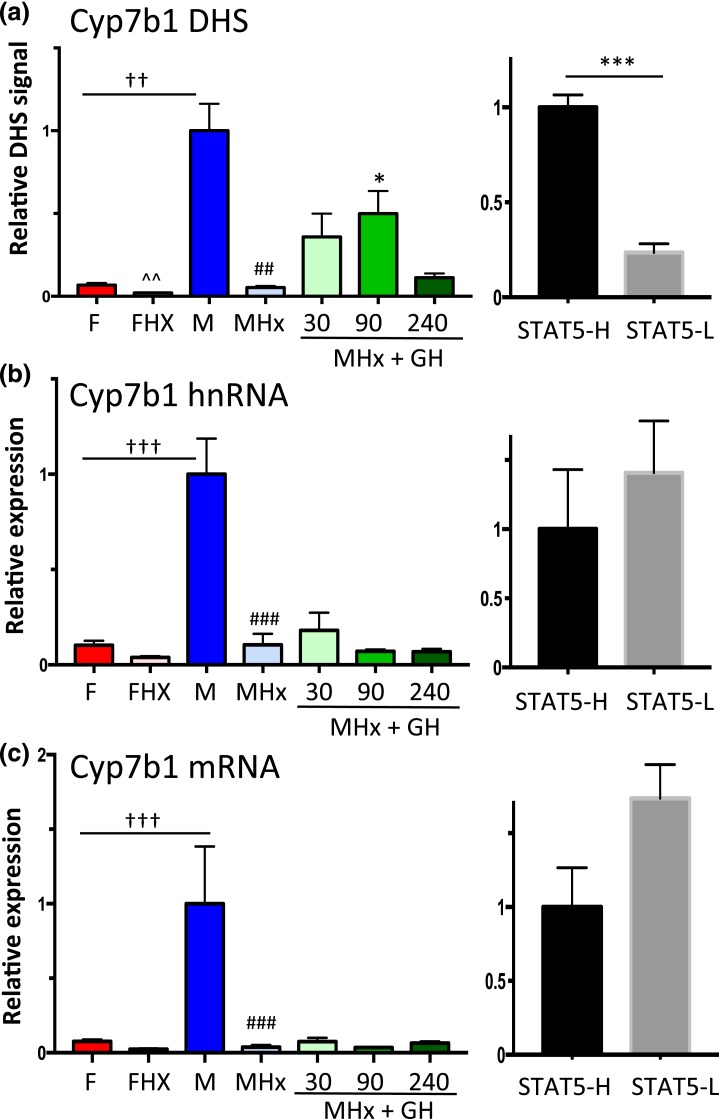
GH pulse responsiveness of Cyp7b1 DHS but not gene transcription
Cyp7b1 is a class I male-biased gene [Fig. 5(a); Table 1] with upstream DHS showing a strong male bias in accessibility [Fig. 3(e)]. Chromatin accessibility at the DHS chosen for analysis (Supplemental Table 1) was abolished in hypophysectomized male mouse liver, and it increased significantly following an exogenous GH pulse (albeit not to the full, intact male mouse liver level, similar to Ugt2b38) [Fig. 8(a)]. DHS analysis of individual STAT5-high and STAT5-low livers revealed that this site opens and closes in concert with the pulsatile activation and deactivation of STAT5 [Fig. 8(a), right], in a manner indistinguishable from the other two class I male-biased genes investigated [Figs. 6(a) and 7(b)]. However, in contrast to those two genes, Cyp7b1 transcription was not induced in livers of hypophysectomized male mice by GH pulse treatment, as indicated by hnRNA and mRNA analysis [Fig. 8(b) and 8(c)]. Furthermore, STAT5-high and STAT5-low livers were indistinguishable in terms of Cyp7b1 transcription rate and steady-state mRNA levels [Fig. 8(b) and 8(c), right panels]. Thus, although GH rapidly induces chromatin opening at the Cyp7b1 DHS, this does not lead to the pulsatile increase in gene transcription seen for the other two class I male-biased genes.
Figure 8.
GH-activated STAT5 induces Cyp7b1 DHS chromatin opening but not Cyp7b1 transcription. (a) qPCR analysis of Cyp7b1 male-biased DHS [Fig. 3(e)] for intact and hypophysectomized male and female mouse livers, with a time course of GH treatment as in Fig. 2. (b and c) RT-qPCR analysis of Cyp7b1 hnRNA and mRNA reveals that male-specific transcription is abolished by hypophysectomy, but fails to be induced by a GH pulse. Transcription rates and mRNA levels are not significantly different between STAT5-high and STAT5-low livers. Data shown in the left set of graphs (hypophysectomy series) are mean ± SEM values for n = 5 to 9 livers per group, except for DHS analysis of the intact male group, where n = 18. The right set of graphs (STAT5-high, STAT5-low) are based on n = 9 to 12 livers per group. Relative DHS activity and expression level of the intact male (left) and the STAT5-high groups (right) are set to 1. Significance values by ANOVA or two-tailed t test are indicated as described in the Fig. 2 legend.
Discussion
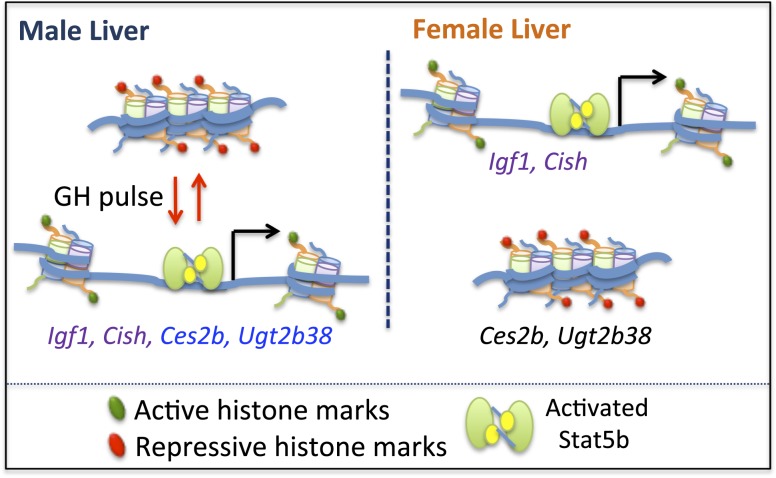
Igf1, Cish, and a small number of other GH-regulated genes have long been known to be activated in a STAT5-dependent manner when supraphysiological GH doses are given to hypophysectomized rats and mice (54, 60–62), but it was unclear whether, and by which mechanisms, these genes respond to naturally occurring, physiological GH pulses. In this study, we show that GH pulse–stimulated gene activation is associated with a rapid but short-lived increase in chromatin accessibility at STAT5-binding enhancers in livers of hypophysectomized mice given a single replacement dose of GH. GH-induced increases in chromatin accessibility were assayed by the increases in DNase hypersensitivity at specific genomic regions near each gene, that is, at chromosomal locations previously identified as DHS/open chromatin sites (51) and where STAT5 binds following GH pulse stimulation (38). We show that the opening and closing of these DHSs parallels GH pulse–stimulated increases, followed by decreases, in liver STAT5 DNA-binding activity and gene transcription rates, for both Igf1 and Cish. Importantly, these responses, seen in hypophysectomized mouse liver, were recapitulated in livers of pituitary-intact male mice, where the natural, endogenous plasma pulses of GH, and the pulses of liver STAT5 activity that they stimulate, were linked temporally to the GH pulse–induced increases in liver chromatin accessibility and Igf1 and Cish transcriptional activation. Dynamic, GH pulse–induced chromatin opening and closing linked to pulsatile increases in target gene transcription (Fig. 9) was also seen for some (Ces2b, Ugt2b38) but not for other (Cyp7b1) GH pulse–dependent male-biased genes. Thus, the natural, endogenous rhythm of male plasma GH pulsation induces dynamic opening and then closing of liver chromatin at discrete, localized regulatory sites in temporal association with transcriptional activation of Igf1, Cish, and a subset of STAT5-dependent male-biased genes.
Figure 9.
Model for GH pulse-induced chromatin opening at STAT5-responsive genes. In male liver, GH pulse activation of STAT5 is associated with chromatin remodeling linked to chromatin opening, which is proposed to involve acquisition of activating marks and loss of repressive marks, and enables STAT5 binding and transcriptional activation of Igf1, Cish, Ces2b, and Ugt2b38. Termination of a GH pulse is followed by cessation of nuclear STAT5 signaling with loss of STAT5 binding, local recompaction of chromatin associated with reversal of histone mark changes, and termination of gene transcription. In female liver, STAT5 is activated in a more continuous manner than in male liver, owing to its persistent stimulation by circulating plasma GH. STAT5 can thus bind to, and trans-activate, Igf1 and Cish via the same open chromatin sites (DHS) as in male liver, albeit with different kinetics: intermittent in male liver vs more persistent in female liver. GH-activated STAT5 is proposed to be unable to bind to and trans-activate the male-biased genes Ces2b and Ugt2b38 in female liver owing to the compaction of these genes in a repressive chromatin state [see Fig. 10(c) and 10(d)], which is resistant to decompaction and GH/STAT5-induced chromatin opening.
Targeted disruption of STAT5 (specifically, the Stat5b gene) has established that this GH-activated transcription factor is essential for the sex-biased expression of >1000 genes in mouse liver (39). Livers of young adult male STAT5b-deficient mice are substantially feminized, with 90% of male-biased genes downregulated to female liver-like levels, and 61% of female-biased genes upregulated to normal female liver levels (39). Global mapping of STAT5 binding sites in male and female mouse liver has shown that sex-biased STAT5 binding sites are strongly enriched at sex-biased DHS and in proximity to sex-biased genes (38). It was unclear, however, whether GH pulse–activated STAT5 directly activates transcription of the GH/STAT5-dependent male-biased genes or, alternatively, whether the STAT5 dependence of these genes involves other, more complex processes and an indirect dependence on STAT5. Our findings in the present study show that two GH pulse–dependent, male-biased genes, Ces2b and Ugt2b38, directly respond to GH pulse–activated STAT5 in a manner indistinguishable from that of Igf1 and Cish; namely, a single plasma GH pulse induces chromatin opening, which exposes sequences containing STAT5 motifs and enables STAT5 binding and induction of gene transcription (Fig. 9). Importantly, these responses to GH were seen in both a hypophysectomized mouse liver model and in intact male mice, where the chromatin and transcriptional responses closely followed the endogenous rhythm of plasma GH pulsation. Thus, the male-biased genes Ces2b and Ugt2b38 are direct STAT5 targets that are transcribed in a pulsatile manner in male mouse liver in direct response to each plasma GH pulse–stimulated cycle of STAT5 activation.
An important question raised by our findings is why male-biased gene expression is a characteristic of Ces2b and Ugt2b38, but not of Igf1 or Cish, given the very similar response profiles of all four genes to male plasma GH pulse stimulation. One possibility is that Ces2b and Ugt2b38, but not Igf1 and Cish, are in a repressed epigenetic state in female liver, which in turn renders them resistant to the GH/STAT5-induced chromatin opening that is a prerequisite for STAT5 binding and transcriptional activation (Fig. 9, right). Consistent with this proposal, the proximal promoter DHSs associated with Ces2b and Ugt2b38 show significant male bias in their accessibility [Fig. 3(c) and 3(d)], indicating that their STAT5 binding sites are comparatively inaccessible in female liver. Furthermore, chromatin state analysis, based on liver DHS profiles and a set of six histone marks (70), indicates that extensive portions of Ces2b and Ugt2b38 and nearby genomics sequences are in an inactive chromatin state in female, but not male, liver [Fig. 10(c) and 10(d)]. In contrast, neither Igf1 nor Cish is in an inactive chromatin state in female liver [Fig. 10(a) and 10(b)]. These findings strongly suggest that all four genes are able to respond rapidly and directly to male plasma GH pulse activation of STAT5 in male liver because they are in an active chromatin state, whereas Ces2b and Ugt2b38 are resistant to GH-activated STAT5 in female liver because they are in a repressed chromatin state (Fig. 9). Of note, STAT5 is efficiently activated in mouse liver by both the female and the male plasma GH profiles, albeit with different temporal kinetics, that is, pulsatile STAT5 activation in male liver vs persistent activation in female liver (38).
Figure 10.
Chromatin states of GH/STAT5-responsive genes. Shown are University of California Santa Cruz mouse genome browser screenshots displaying chromatin state maps for each of the five genes examined in this study, along with the locations of gene-proximal DHS interrogated by qPCR. Also marked are all of the STAT5 binding sites and DHS in the genomic region displayed (bottom two tracks of each panel). Blue indicates a male-biased STAT5 binding site or DHS site, pink indicates a female-biased site, and gray indicates a sex-independent site. Chromatin state maps were discovered using ChromHMM (71) based on genome-wide profiles for DHS and six chromatin marks determined in both male and female liver (70). Each consecutive 200-bp segment of the genome is assigned to one of 14 chromatin states in male liver, and separately in female liver, and is represented by a distinct color, as specified in the chromatin states legend on the right). The 14 states can be grouped into five superstates, three of which are active (enhancer, promoter, transcribed), one is inactive, and one is poised for activation (bivalent). These descriptions reflect the established functional annotations of the major chromatin marks associated with each state. (a) Igf1 is in an active chromatin state in both male and female liver, with large numbers of sex-independent DHS and STAT5 binding sites across the gene body and in the 5′-flank. (b) Cish shows very similar chromatin states in both sexes, and similar to Igf1, is devoid of inactive chromatin states. Cish contains one downstream STAT5-bound male-biased DHS and one downstream female-biased STAT5 binding site, both of unknown significance. (c and d) Ces2b and Ugt2b38 are in an active chromatin state in male liver but have extensive sequence in the inactive state (red) in female liver. These male-biased genes both have very few nearby DHS and STAT5 binding sites. (e) Cyp7b1 is primarily characterized by an active chromatin state in both male and female liver; however, a portion of intron 1 and an extended genomic segment that begins just upstream of the 5′-flank DHS interrogated in Fig. 8 are in an inactive chromatin state (red).
Little is known about the mechanisms by which a plasma GH pulse rapidly increases chromatin accessibility linked to activation of the four GH pulse-responsive genes characterized in this study. These changes in chromatin accessibility likely involve acquisition of activating histone marks and local chromatin remodeling linked to STAT5 DNA binding, as suggested by the increase in histone acetylation that accompanies STAT5 binding to a subset of regulatory sites associated with rat IGF1 (61). They could also involve other GH receptor–activated signaling events (55), as well as actions of STAT5-interacting partners, such as glucocorticoid receptor (72), NFIB (73), and other liver transcription factors whose binding (74) [or whose motifs (38)] are frequently found near STAT5 binding sites. GH-induced chromatin opening may be associated with linker histone H1 dissociation induced by the chromatin-modifying protein HMGN2, as was recently described for prolactin-activated STAT5 in breast cancer cells (75). Alternatively, binding by pioneer factors such as FOXA1 (76) may facilitate chromatin opening and enable the GH pulse–induced binding of STAT5 that we observed by ChIP-seq (38). The dynamic hormone responsiveness of GH/STAT5-induced chromatin opening and closing described in this study may be analogous to glucocorticoid receptor–induced epigenomic effects described in mouse mammary epithelial cells, where pulsatile corticosteroid treatment activates glucocorticoid receptor and induces transient chromatin opening at some sites but persistent chromatin opening at other sites (77). There are also similarities to the prolactin-responsive STAT5 binding events recently reported for mouse mammary gland, where termination of lactation and a decline in prolactin levels are associated with loss of STAT5 binding within 24 hours at many mammary gland–specific STAT5 binding sites, in particular sites that require high STAT5 concentrations for STAT5 binding (78).
Few details are known about the mechanisms whereby chromatin accessibility reverts back to the relatively closed basal state and gene transcription terminates at the conclusion of a GH pulse (Fig. 9). Presumably, this epigenetic deactivation involves a loss of activating chromatin marks and perhaps reacquisition of repressive marks associated with chromatin closing. Deactivation of STAT5 signaling is a consequence of the termination of GH-stimulated GH receptor signaling to STAT5 and the subsequent deactivation of nuclear STAT5 by phosphotyrosine dephosphorylation, a prerequisite for the recycling of inactive STAT5 protein back to the cytosol (79). Pulsatile activation of liver nuclear STAT5 in direct response to each plasma GH pulse and the termination of hepatic nuclear STAT5 activity at the conclusion of a GH pulse have been directly established in the rat model (69). STAT5 activity pulsation is also evident from our EMSA analysis of individual male mouse livers [STAT5-high vs STAT5-low livers; Fig. 4(a)]. Although deactivation of nuclear STAT5 is expected to be a key step in the reversal of GH/STAT5-induced chromatin opening that we observed, termination of other, STAT5-independent GH receptor–activated signaling pathways (55) could also contribute to the reversal of chromatin opening.
Cyp7b1 did not show a direct transcriptional response to GH pulse treatment, despite its having an upstream male-biased DHS that undergoes reversible opening and closing with GH/STAT5 pulsation in a manner indistinguishable from the DHS associated with Ces2b and Ugt2b38. The absence of a Cyp7b1 transcriptional response to GH was apparent, both in the hypophysectomized mouse model and from our analysis of Cyp7b1 transcriptional rates in livers of STAT5-high vs STAT5-low male mice. All three male-biased genes show male-biased expression dependent on STAT5b (39), are positively regulated by the male pituitary hormone profile (i.e., are class I male-biased genes; Table 1), and have proximal male-biased DHSs (Fig. 3; Supplemental Table 1) that contain binding sites for STAT5 (38). Cyp7b1 has an extended 5′-proximal region in a repressed chromatin state beginning just upstream of the male-biased DHS that we examined [Fig. 10(e)]. Transcriptional activation of Cyp7b1 in response to plasma GH pulse stimulation can be achieved in livers of hypophysectomized male mice, but it requires that GH pulse injections be given repeatedly during a 7-day period (80). This suggests that the transcriptional activation of Cyp7b1 by plasma GH pulses is more complex and involves slower, secondary events not required for activation of Ces2b and Ugt2b38; these events may include more extensive chromatin remodeling and activation of additional genomic regulatory regions, beyond the rapidly responding 5′-proximal DHS examined in this study. The multiple DHSs located upstream of Cyp7b1 and across an extended portion of Cyp7b1 intron 1 [Fig. 10(e)] contrasts with the much simpler array of regulatory elements surrounding Ces2b and Ugt2b38 [Fig. 10(c) and 10(d)] and is consistent with Cyp7b1 being under a more complex regulatory regimen.
In conclusion, endogenous GH pulses induce rapid increases in hepatic gene transcription, as shown by the rapid induction of unspliced hnRNA transcripts, both in livers of hypophysectomized mice and in response to the natural on–off rhythm of GH release in pituitary-intact adult male mice. These pulsatile increases in gene transcription characterize the well-established STAT5 target genes Igf1 and Cish as well as a subset of STAT5-dependent male-biased genes, but were not mirrored by a corresponding pulsatility for the mature RNA transcripts owing to their apparently long RNA half-lives. Whereas sex-biased epigenetic and transcriptional responses to GH pulse–activated STAT5, including the pulsatile activation of Ces2b and Ugt2b38 transcription shown in this study, are an essential part of the overall mechanism whereby GH and STAT5 regulate sex-biased gene expression in the liver, the physiological significance of pulsatile transcription of non–sex-biased genes, such as Igf1, is unclear. Further studies are needed to address this question and to elucidate the underlying molecular mechanisms for GH pulse–induced opening and closing of liver chromatin at highly localized regulatory sites and the role of STAT5 and other cooperating factors in these processes.
Supplementary Material
Acknowledgments
The authors thank Tisha Melia of this laboratory for contributions to the custom pipeline for RNA-seq analysis used in this study and Gracia Bonilla for assistance with the extended Blat analysis script used to validate the specificity of qPCR primers and in chromatin state analysis.
This work was supported in part by National Institutes of Health Grant DK33765 (to D.J.W.).
Author contributions: J.C. carried out the animal studies and laboratory experiments, including EMSA, nuclei isolation, DHS preparation and RNA purification, and together with D.L.C. carried out qPCR analysis; D.L.C. prepared sequencing libraries, analyzed the RNA-seq and qPCR data, and prepared figures and tables for publication; A.R. developed and automated the pipeline for RNA-seq analysis, including intronic analysis; D.J.W. and J.C. conceived and designed the study; and D.J.W. wrote the manuscript, which was reviewed by all the authors.
Disclosure Summary: The authors have nothing to disclose.
Abbreviations:
- ANOVA
analysis of variance
- cDNA
complementary DNA
- ChIP-seq
chromatin immunoprecipitation sequencing
- DHS
DNase hypersensitive site
- EMSA
electrophoretic mobility shift assay
- GH
growth hormone
- hnRNA
heterogeneous nuclear RNA
- mRNA
messenger RNA
- qPCR
quantitative polymerase chain reaction
- RNA-seq
RNA sequencing
- rRNA
ribosomal RNA
- RT
reverse transcription
- RT
reverse transcription
- SEM
standard error of the mean
- STAT
signal transducer and activator of transcription.
References
- 1. Zanger UM, Schwab M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. 2013;138(1):103–141. [DOI] [PubMed] [Google Scholar]
- 2. Klaassen CD, Aleksunes LM. Xenobiotic, bile acid, and cholesterol transporters: function and regulation. Pharmacol Rev. 2010;62(1):1–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gonzalez FJ. Role of cytochromes P450 in chemical toxicity and oxidative stress: studies with CYP2E1. Mutat Res. 2005;569(1-2):101–110. [DOI] [PubMed] [Google Scholar]
- 4. Guengerich FP. Cytochrome P450s and other enzymes in drug metabolism and toxicity. AAPS J. 2006;8(1):E101–E111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tessari P, Coracina A, Cosma A, Tiengo A. Hepatic lipid metabolism and non-alcoholic fatty liver disease. Nutr Metab Cardiovasc Dis. 2009;19(4):291–302. [DOI] [PubMed] [Google Scholar]
- 6. Luoma PV. Cytochrome P450—physiological key factor against cholesterol accumulation and the atherosclerotic vascular process. Ann Med. 2007;39(5):359–370. [DOI] [PubMed] [Google Scholar]
- 7. Caro AA, Cederbaum AI. Role of cytochrome P450 in phospholipase A2- and arachidonic acid-mediated cytotoxicity. Free Radic Biol Med. 2006;40(3):364–375. [DOI] [PubMed] [Google Scholar]
- 8. Waxman DJ, Holloway MG. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol Pharmacol. 2009;76(2):215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Renaud HJ, Cui JY, Khan M, Klaassen CD. Tissue distribution and gender-divergent expression of 78 cytochrome P450 mRNAs in mice. Toxicol Sci. 2011;124(2):261–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Alnouti Y, Klaassen CD. Mechanisms of gender-specific regulation of mouse sulfotransferases (Sults). Xenobiotica. 2011;41(3):187–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albiston AL, Smith RE, Krozowski ZS. Sex- and tissue-specific regulation of 11β-hydroxysteroid dehydrogenase mRNA. Mol Cell Endocrinol. 1995;109(2):183–188. [DOI] [PubMed] [Google Scholar]
- 12. Buckley DB, Klaassen CD. Mechanism of gender-divergent UDP-glucuronosyltransferase mRNA expression in mouse liver and kidney. Drug Metab Dispos. 2009;37(4):834–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wolbold R, Klein K, Burk O, Nüssler AK, Neuhaus P, Eichelbaum M, Schwab M, Zanger UM. Sex is a major determinant of CYP3A4 expression in human liver. Hepatology. 2003;38(4):978–988. [DOI] [PubMed] [Google Scholar]
- 14. Parkinson A, Mudra DR, Johnson C, Dwyer A, Carroll KM. The effects of gender, age, ethnicity, and liver cirrhosis on cytochrome P450 enzyme activity in human liver microsomes and inducibility in cultured human hepatocytes. Toxicol Appl Pharmacol. 2004;199(3):193–209. [DOI] [PubMed] [Google Scholar]
- 15. Thangavel C, Boopathi E, Shapiro BH. Intrinsic sexually dimorphic expression of the principal human CYP3A4 correlated with suboptimal activation of GH/glucocorticoid-dependent transcriptional pathways in men. Endocrinology. 2011;152(12):4813–4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Court MH. Interindividual variability in hepatic drug glucuronidation: studies into the role of age, sex, enzyme inducers, and genetic polymorphism using the human liver bank as a model system. Drug Metab Rev. 2010;42(1):209–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang Y, Klein K, Sugathan A, Nassery N, Dombkowski A, Zanger UM, Waxman DJ. Transcriptional profiling of human liver identifies sex-biased genes associated with polygenic dyslipidemia and coronary artery disease. PLoS One. 2011;6(8):e23506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Waxman DJ, Dannan GA, Guengerich FP. Regulation of rat hepatic cytochrome P-450: age-dependent expression, hormonal imprinting, and xenobiotic inducibility of sex-specific isoenzymes. Biochemistry. 1985;24(16):4409–4417. [DOI] [PubMed] [Google Scholar]
- 19. Cadario BJ, Bellward GD, Bandiera S, Chang TK, Ko WW, Lemieux E, Pak RC. Imprinting of hepatic microsomal cytochrome P-450 enzyme activities and cytochrome P-450IIC11 by peripubertal administration of testosterone in female rats. Mol Pharmacol. 1992;41(5):981–988. [PubMed] [Google Scholar]
- 20. Ramirez MC, Luque GM, Ornstein AM, Becu-Villalobos D. Differential neonatal testosterone imprinting of GH-dependent liver proteins and genes in female mice. J Endocrinol. 2010;207(3):301–308. [DOI] [PubMed] [Google Scholar]
- 21. Jansson JO, Edén S, Isaksson O. Sexual dimorphism in the control of growth hormone secretion. Endocr Rev. 1985;6(2):128–150. [DOI] [PubMed] [Google Scholar]
- 22. Shapiro BH, Agrawal AK, Pampori NA. Gender differences in drug metabolism regulated by growth hormone. Int J Biochem Cell Biol. 1995;27(1):9–20. [DOI] [PubMed] [Google Scholar]
- 23. Waxman DJ, O’Connor C. Growth hormone regulation of sex-dependent liver gene expression. Mol Endocrinol. 2006;20(11):2613–2629. [DOI] [PubMed] [Google Scholar]
- 24. Li J, Wan Y, Na S, Liu X, Dong G, Yang Z, Yang J, Yue J. Sex-dependent regulation of hepatic CYP3A by growth hormone: roles of HNF6, C/EBPα, and RXRα. Biochem Pharmacol. 2015;93(1):92–103. [DOI] [PubMed] [Google Scholar]
- 25. Banerjee S, Das RK, Shapiro BH. Growth hormone-independent suppression of growth hormone-dependent female isoforms of cytochrome P450 by the somatostatin analog octreotide. Eur J Pharmacol. 2013;715(1-3):256–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheung C, Yu AM, Chen CS, Krausz KW, Byrd LG, Feigenbaum L, Edwards RJ, Waxman DJ, Gonzalez FJ. Growth hormone determines sexual dimorphism of hepatic cytochrome P450 3A4 expression in transgenic mice. J Pharmacol Exp Ther. 2006;316(3):1328–1334. [DOI] [PubMed] [Google Scholar]
- 27. Endo M, Takahashi Y, Sasaki Y, Saito T, Kamataki T. Novel gender-related regulation of CYP2C12 gene expression in rats. Mol Endocrinol. 2005;19(5):1181–1190. [DOI] [PubMed] [Google Scholar]
- 28. Sueyoshi T, Yokomori N, Korach KS, Negishi M. Developmental action of estrogen receptor-α feminizes the growth hormone-Stat5b pathway and expression of Cyp2a4 and Cyp2d9 genes in mouse liver. Mol Pharmacol. 1999;56(3):473–477. [DOI] [PubMed] [Google Scholar]
- 29. Ström A, Eguchi H, Mode A, Legraverend C, Tollet P, Strömstedt PE, Gustafsson JA. Characterization of the proximal promoter and two silencer elements in the CYP2C11 gene expressed in rat liver. DNA Cell Biol. 1994;13(8):805–819. [DOI] [PubMed] [Google Scholar]
- 30. Rotwein P. Mapping the growth hormone–Stat5b–IGF-I transcriptional circuit. Trends Endocrinol Metab. 2012;23(4):186–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waters MJ, Brooks AJ. Growth hormone and cell growth. Endocr Dev. 2012;23:86–95. [DOI] [PubMed] [Google Scholar]
- 32. Baik M, Yu JH, Hennighausen L. Growth hormone-STAT5 regulation of growth, hepatocellular carcinoma, and liver metabolism. Ann N Y Acad Sci. 2011;1229:29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Waxman DJ, Ram PA, Park SH, Choi HK. Intermittent plasma growth hormone triggers tyrosine phosphorylation and nuclear translocation of a liver-expressed, Stat 5-related DNA binding protein. Proposed role as an intracellular regulator of male-specific liver gene transcription. J Biol Chem. 1995;270(22):13262–13270. [DOI] [PubMed] [Google Scholar]
- 34. Choi HK, Waxman DJ. Plasma growth hormone pulse activation of hepatic JAK-STAT5 signaling: developmental regulation and role in male-specific liver gene expression. Endocrinology. 2000;141(9):3245–3255. [DOI] [PubMed] [Google Scholar]
- 35. Conforto TL, Waxman DJ. Sex-specific mouse liver gene expression: genome-wide analysis of developmental changes from pre-pubertal period to young adulthood. Biol Sex Differ. 2012;3:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Choi HK, Waxman DJ. Growth hormone, but not prolactin, maintains, low-level activation of STAT5a and STAT5b in female rat liver. Endocrinology. 1999;140(11):5126–5135. [DOI] [PubMed] [Google Scholar]
- 37. Ji S, Frank SJ, Messina JL. Growth hormone-induced differential desensitization of STAT5, ERK, and Akt phosphorylation. J Biol Chem. 2002;277(32):28384–28393. [DOI] [PubMed] [Google Scholar]
- 38. Zhang Y, Laz EV, Waxman DJ. Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol Cell Biol. 2012;32(4):880–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Clodfelter KH, Holloway MG, Hodor P, Park SH, Ray WJ, Waxman DJ. Sex-dependent liver gene expression is extensive and largely dependent upon signal transducer and activator of transcription 5b (STAT5b): STAT5b-dependent activation of male genes and repression of female genes revealed by microarray analysis. Mol Endocrinol. 2006;20(6):1333–1351. [DOI] [PubMed] [Google Scholar]
- 40. Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16(8):995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davey HW, Park SH, Grattan DR, McLachlan MJ, Waxman DJ. STAT5b-deficient mice are growth hormone pulse-resistant. Role of STAT5b in sex-specific liver P450 expression. J Biol Chem. 1999;274(50):35331–35336. [DOI] [PubMed] [Google Scholar]
- 42. Park SH, Liu X, Hennighausen L, Davey HW, Waxman DJ. Distinctive roles of STAT5a and STAT5b in sexual dimorphism of hepatic P450 gene expression. Impact of Stat5a gene disruption. J Biol Chem. 1999;274(11):7421–7430. [DOI] [PubMed] [Google Scholar]
- 43. Udy GB, Towers RP, Snell RG, Wilkins RJ, Park SH, Ram PA, Waxman DJ, Davey HW. Requirement of STAT5b for sexual dimorphism of body growth rates and liver gene expression. Proc Natl Acad Sci USA. 1997;94(14):7239–7244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Holloway MG, Cui Y, Laz EV, Hosui A, Hennighausen L, Waxman DJ. Loss of sexually dimorphic liver gene expression upon hepatocyte-specific deletion of Stat5a-Stat5b locus. Endocrinology. 2007;148(5):1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clodfelter KH, Miles GD, Wauthier V, Holloway MG, Zhang X, Hodor P, Ray WJ, Waxman DJ. Role of STAT5a in regulation of sex-specific gene expression in female but not male mouse liver revealed by microarray analysis. Physiol Genomics. 2007;31(1):63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Meyer RD, Laz EV, Su T, Waxman DJ. Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol Endocrinol. 2009;23(11):1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Laz EV, Holloway MG, Chen CS, Waxman DJ. Characterization of three growth hormone-responsive transcription factors preferentially expressed in adult female liver. Endocrinology. 2007;148(7):3327–3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Conforto TL, Zhang Y, Sherman J, Waxman DJ. Impact of CUX2 on the female mouse liver transcriptome: activation of female-biased genes and repression of male-biased genes. Mol Cell Biol. 2012;32(22):4611–4627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500. [DOI] [PubMed] [Google Scholar]
- 50. Boyle AP, Davis S, Shulha HP, Meltzer P, Margulies EH, Weng Z, Furey TS, Crawford GE. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132(2):311–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Ling G, Sugathan A, Mazor T, Fraenkel E, Waxman DJ. Unbiased, genome-wide in vivo mapping of transcriptional regulatory elements reveals sex differences in chromatin structure associated with sex-specific liver gene expression. Mol Cell Biol. 2010;30(23):5531–5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Thurman RE, Rynes E, Humbert R, Vierstra J, Maurano MT, Haugen E, Sheffield NC, Stergachis AB, Wang H, Vernot B, Garg K, John S, Sandstrom R, Bates D, Boatman L, Canfield TK, Diegel M, Dunn D, Ebersol AK, Frum T, Giste E, Johnson AK, Johnson EM, Kutyavin T, Lajoie B, Lee BK, Lee K, London D, Lotakis D, Neph S, Neri F, Nguyen ED, Qu H, Reynolds AP, Roach V, Safi A, Sanchez ME, Sanyal A, Shafer A, Simon JM, Song L, Vong S, Weaver M, Yan Y, Zhang Z, Zhang Z, Lenhard B, Tewari M, Dorschner MO, Hansen RS, Navas PA, Stamatoyannopoulos G, Iyer VR, Lieb JD, Sunyaev SR, Akey JM, Sabo PJ, Kaul R, Furey TS, Dekker J, Crawford GE, Stamatoyannopoulos JA. The accessible chromatin landscape of the human genome. Nature. 2012;489(7414):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alzhanov D, Mukherjee A, Rotwein P. Identifying growth hormone-regulated enhancers in the Igf1 locus. Physiol Genomics. 2015;47(11):559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Santhanam M, Chia DJ. Hepatic-specific accessibility of Igf1 gene enhancers is independent of growth hormone signaling. Mol Endocrinol. 2013;27(12):2080–2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chia DJ. Minireview: mechanisms of growth hormone-mediated gene regulation. Mol Endocrinol. 2014;28(7):1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Laz EV, Sugathan A, Waxman DJ. Dynamic in vivo binding of STAT5 to growth hormone-regulated genes in intact rat liver. Sex-specific binding at low- but not high-affinity STAT5 sites. Mol Endocrinol. 2009;23(8):1242–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Matsumoto A, Masuhara M, Mitsui K, Yokouchi M, Ohtsubo M, Misawa H, Miyajima A, Yoshimura A. CIS, a cytokine inducible SH2 protein, is a target of the JAK-STAT5 pathway and modulates STAT5 activation. Blood. 1997;89(9):3148–3154. [PubMed] [Google Scholar]
- 58. Lin G, LaPensee CR, Qin ZS, Schwartz J. Reciprocal occupancy of BCL6 and STAT5 on growth hormone target genes: contrasting transcriptional outcomes and promoter-specific roles of p300 and HDAC3. Mol Cell Endocrinol. 2014;395(1–2):19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wauthier V, Sugathan A, Meyer RD, Dombkowski AA, Waxman DJ. Intrinsic sex differences in the early growth hormone responsiveness of sex-specific genes in mouse liver. Mol Endocrinol. 2010;24(3):667–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chia DJ, Rotwein P. Defining the epigenetic actions of growth hormone: acute chromatin changes accompany GH-activated gene transcription. Mol Endocrinol. 2010;24(10):2038–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chia DJ, Varco-Merth B, Rotwein P. Dispersed chromosomal Stat5b-binding elements mediate growth hormone-activated insulin-like growth factor-I gene transcription. J Biol Chem. 2010;285(23):17636–17647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Woelfle J, Billiard J, Rotwein P. Acute control of insulin-like growth factor-I gene transcription by growth hormone through Stat5b. J Biol Chem. 2003;278(25):22696–22702. [DOI] [PubMed] [Google Scholar]
- 63. Gebert CA, Park SH, Waxman DJ. Regulation of signal transducer and activator of transcription (STAT) 5b activation by the temporal pattern of growth hormone stimulation. Mol Endocrinol. 1997;11(4):400–414. [DOI] [PubMed] [Google Scholar]
- 64. Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. [DOI] [PubMed] [Google Scholar]
- 65. Lun AT, Chen Y, Smyth GK. It’s DE-licious: a recipe for differential expression analyses of RNA-seq experiments using quasi-likelihood methods in edgeR. Methods Mol Biol. 2016;1418:391–416. [DOI] [PubMed] [Google Scholar]
- 66. Madsen JG, Schmidt SF, Larsen BD, Loft A, Nielsen R, Mandrup S. iRNA-seq: computational method for genome-wide assessment of acute transcriptional regulation from total RNA-seq data. Nucleic Acids Res. 2015;43(6):e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gaidatzis D, Burger L, Florescu M, Stadler MB. Analysis of intronic and exonic reads in RNA-seq data characterizes transcriptional and post-transcriptional regulation. Nat Biotechnol. 2015;33(7):722–729. [DOI] [PubMed] [Google Scholar]
- 68. Knopf JL, Gallagher JF, Held WA. Differential, multihormonal regulation of the mouse major urinary protein gene family in the liver. Mol Cell Biol. 1983;3(12):2232–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Tannenbaum GS, Choi HK, Gurd W, Waxman DJ. Temporal relationship between the sexually dimorphic spontaneous GH secretory profiles and hepatic STAT5 activity. Endocrinology. 2001;142(11):4599–4606. [DOI] [PubMed] [Google Scholar]
- 70. Sugathan A, Waxman DJ. Genome-wide analysis of chromatin states reveals distinct mechanisms of sex-dependent gene regulation in male and female mouse liver. Mol Cell Biol. 2013;33(18):3594–3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Ernst J, Kellis M. ChromHMM: automating chromatin-state discovery and characterization. Nat Methods. 2012;9(3):215–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Engblom D, Kornfeld JW, Schwake L, Tronche F, Reimann A, Beug H, Hennighausen L, Moriggl R, Schütz G. Direct glucocorticoid receptor–Stat5 interaction in hepatocytes controls body size and maturation-related gene expression. Genes Dev. 2007;21(10):1157–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Robinson GW, Kang K, Yoo KH, Tang Y, Zhu BM, Yamaji D, Colditz V, Jang SJ, Gronostajski RM, Hennighausen L. Coregulation of genetic programs by the transcription factors NFIB and STAT5. Mol Endocrinol. 2014;28(5):758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Conforto TL, Steinhardt GF IV, Waxman DJ. Cross talk between GH-regulated transcription factors HNF6 and CUX2 in adult mouse liver. Mol Endocrinol. 2015;29(9):1286–1302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. doi: 10.1074/jbc.M116.764233. Schauwecker SM, Kim JJ, Licht JD, Clevenger CV. Histone H1 and chromosomal protein HMGN2 regulate prolactin-induced STAT5 transcription factor recruitment and function in breast cancer cells. J Biol Chem. 2017;292(6):2237–2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Iwafuchi-Doi M, Donahue G, Kakumanu A, Watts JA, Mahony S, Pugh BF, Lee D, Kaestner KH, Zaret KS. The pioneer transcription factor FoxA maintains an accessible nucleosome configuration at enhancers for tissue-specific gene activation. Mol Cell. 2016;62(1):79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Stavreva DA, Coulon A, Baek S, Sung MH, John S, Stixova L, Tesikova M, Hakim O, Miranda T, Hawkins M, Stamatoyannopoulos JA, Chow CC, Hager GL. Dynamics of chromatin accessibility and long-range interactions in response to glucocorticoid pulsing. Genome Res. 2015;25(6):845–857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Willi M, Yoo KH, Wang C, Trajanoski Z, Hennighausen L. Differential cytokine sensitivities of STAT5-dependent enhancers rely on Stat5 autoregulation. Nucleic Acids Res. 2016;44(21):10277–10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Gebert CA, Park SH, Waxman DJ. Termination of growth hormone pulse-induced STAT5b signaling. Mol Endocrinol. 1999;13(1):38–56. [DOI] [PubMed] [Google Scholar]
- 80. Holloway MG, Laz EV, Waxman DJ. Codependence of growth hormone-responsive, sexually dimorphic hepatic gene expression on signal transducer and activator of transcription 5b and hepatic nuclear factor 4α. Mol Endocrinol. 2006;20(3):647–660. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.